Abstract
Phytochelatins (PCs) catalyzed by phytochelatin synthases (PCS) are important for the detoxification of metals in plants and other living organisms. In this study, we isolated a PCS gene (VsPCS1) from Vicia sativa and investigated its role in regulating cadmium (Cd) tolerance. Expression of VsPCS1 was induced in roots of V. sativa under Cd stress. Analysis of subcellular localization showed that VsPCS1 was localized in the cytoplasm of mesophyll protoplasts of V. sativa. Overexpression of VsPCS1 (35S::VsPCS1, in wild-type background) in Arabidopsis thaliana could complement the defects of Cd tolerance of AtPCS1-deficent mutant (atpcs1). Compared with atpcs1 mutants, 35S::VsPCS1/atpcs1 (in AtPCS1-deficent mutant background) transgenic plants significantly lowered Cd-fluorescence intensity in mesophyll cytoplasm, accompanied with enhanced Cd-fluorescence intensity in the vacuoles, demonstrating that the increased Cd tolerance may be attributed to the increased PC-based sequestration of Cd into the vacuole. Furthermore, overexpressing VsPCS1 could enhance the Cd tolerance in 35S::VsPCS1, but have no effect on Cd accumulation and distribution, showing the same level of Cd-fluorescence intensity between 35S::VsPCS1 and wild-type (WT) plants. Further analysis indicated this increased tolerance in 35S::VsPCS1 was possibly due to the increased PCs-chelated Cd in cytosol. Taken together, a functional PCS1 homolog from V. sativa was identified, which hold a strong catalyzed property for the synthesis of high-order PCs that retained Cd in the cytosol rather the vacuole. These findings enrich the original model of Cd detoxification mediated by PCS in higher plants.
Keywords: VsPCS1, phytochelatins, Cd, tolerance, Arabidopsis
Introduction
Heavy metal contamination is a predominant environmental issue in the world. Cadmium (Cd) is one of highly toxic metals for all organisms and is of particular concern to human health since Cd can readily uptake by plant roots from polluted soils and transported to shoots (Wagner, 1993). This element enters the environment mainly through mining operations, smelting of metals, electroplating, municipal wastes, and phosphate fertilizers. Excess Cd can inhibit numerous biochemical and physiological processes in plants including photosynthesis and pigment synthesis, respiration, nitrogen and protein metabolism, nutrient uptake, transpiration, and plant–water relationships (Clemens, 2006).
To cope with an exposure to toxic levels of heavy metals, several mechanisms have been developed for metal detoxification, including exclusion, compartmentalization, chelating, and binding to organic ligands such as organic acids, amino acids, phytochelatins (PCs), and metallothioneins (MTs). Numerous studies have demonstrated the critical roles of PCs in metal tolerance and translocation in plants exposed to Cd, As, Hg, Pb, and Zn (Tennstedt et al., 2009; Mendoza-Cozatl et al., 2011; Franchi et al., 2014; Song et al., 2014; Shi et al., 2017; Xia et al., 2018). PC is a family of peptides with the general structure (Glu-Cys)n-Gly, where n is in the range of 2–11 (Cobbett and Goldsbrough, 2002). They form stable metal complexes and are subsequently sequestrate from the cytosol into vacuoles. PCs are synthesized directly from reduced glutathione (GSH) by the enzyme phytochelatin synthase (PCS). PCS is constitutively expressed, but is activated by metal ions, especially Cd2+ (Degola et al., 2014). PCS genes have been identified in some plants including Arabidopsis (Ha et al., 1999), Oryza sativa (Das et al., 2017), Triticum aestivum (Wang et al., 2012), Nelumbo nucifera (Liu et al., 2012), Lotus japonicas (Ramos et al., 2007), and Ceratophyllum demersum (Shukla et al., 2012). PC-deficient mutants of Arabidopsis and yeast are hypersensitive to Cd (Cobbett, 2000). In Arabidopsis, γ-glutamylcysteine synthetase (γ-ECS) and glutathione synthetase (GS) mutant plants were hypersensitive to Cd owing to its lower content of GSH and PC (Jobe et al., 2012). atpcs1 was firstly identified as a knockout mutant of PCS1 in Arabidopsis, and sensitive to Cd and As (Nahar et al., 2014). Heteroexpression of PCS coming from various species increased PC levels, Cd tolerance and Cd accumulation in Arabidopsis (Guo et al., 2008; Liu et al., 2012) and Nicotiana tabacum (Pomponi et al., 2006; Liu et al., 2011). In contrast, some studies showed that increased PCs production is not the primary tolerance mechanism to Cd (De Knecht et al., 1992; Wójcik et al., 2015), indicating a complicated mechanism underlying the PCs involved Cd tolerance.
The legume Vicia sativa is ordinarily farmed as food for humans and fodder for livestock. It also acts as green manure to improve soil fertility, especially in dry-land farming operations. Our previous studies have investigated the mechanisms of V. sativa responses to Cd stress, demonstrating that Cd toxicity most likely induced hydrogen peroxide (H2O2) and lignification in the roots of V. sativa by increasing apoplastic POD activity (Zhang et al., 2009, 2011; Rui et al., 2016). However, the molecular mechanisms underlying the Cd tolerance of V. sativa are unclear. In this study, we cloned a V. sativa PCS gene and determined its tissue expression patterns. Moreover, we ectopically overproduced VsPCS1 in Arabidopsis and generated 35S::VsPCS1 (in wild-type background) and 35S::VsPCS1/atpcs1 (in AtPCS1-deficent mutant background) and examined their growth under Cd stress. The metal accumulation and distribution were also investigated in these transgenic plants. Our study aimed to unravel the physiological roles of VsPCS1 and the Cd-tolerance mechanisms in higher plants.
Materials and Methods
Plant Materials, Growth Conditions, and Treatments
The ZM variety of V. sativa was used in this study, the culture process of plants was described as previously (Rui et al., 2016). For VsPCS1 expression analysis, 7-day-old plants were treated with 0, 5, and 50 μM Cd for different days.
Sterile seeds of Arabidopsis wild-type (Columbia, WT), mutant atpcs1, transgenic plants were surface sterilized and germinated on 1/2 Murashige & Skoog (MS) agar medium (pH 5.8) containing the different concentrations of Cd. After 3 days at 4°C in the dark, seeds were germinated in a growth chamber (22/18°C day/night temperatures, 16/8 h day/night photoperiod) for 8 days, the length of plants roots, fresh weight, and dry weight were measured.
To analysis GHS and PCs content and Cd transport, the seeds of WT, atpcs1, 35S::VsPCS1, and 35S::VsPCS1/atpcs1 were first grown on half-strength MS agar plates for 2 weeks, then the seedlings were grown hydroponically in 1/4 strength Hoagland’s nutrient solution (Liu et al., 2017) for 2 weeks, after, the plants was placed in the solution with or without 10 μM CdCl2 (Remans et al., 2008; Cuypers et al., 2011) for 24 h to detect the expression level of AtABCC1, 3 [C-type ATP-binding cassette (ABC) transporter], AtCAX2 (Calcium exchanger 2), AtNRAMP3 (Natural resistance-associated macrophage proteins 3), and for 5 days to analyze NPT content and intracellular Cd localization.
Isolation of Full-Length VsPCS1 cDNA
The nucleic acid sequences of other plant phytochelatin synthases were searched from NCBI database1. The sequences were analyzed using DNAMAN software (Version 6.0.3.99 LynnonBioSoft, Foster City, CA, United States). Primers were designed according to the highly conserved region of PCS1 for VsPCS1 isolation. Total RNA was isolated by total RNA extraction Kit (TakaRa) from V. sativa. The first strand cDNA was synthesized by RT-PCR using PrimeScript (TaKaRa) and oligo (dT) primers, PCR amplifications were performed with VsPCS1 primers. The PCR products were sequenced, then the sequences of VsPCS1 were tested by BLAST2.
Plants Transformation and Selection
Wild-type Arabidopsis (Col-0) and its mutant, atpcs1 (SAIL_650_C12), with T-DNA insertion in an exon of AtPCS1 gene was available from public repositories3 for this study. Homozygous atpcs1 lines were identified by the method of Yi and An (2013).
pCAMBIA1304 was used as the plant expression vectors. CDS of VsPCS1 was cloned into pCAMBIA1304 using primers of PCS1-F2 and PCS1-R2. The confirmed plasmid were transformed into Arabidopsis WT Col-0 and the mutant atpcs1 plants via standard floral dip transformation using Agrobacterium tumefaciens (Clough and Bent, 1998), which were named 35S::VsPCS1 and 35S::VsPCS1/atpcs1, respectively. Homozygotic transgenic lines of T3 progeny were used for subsequent study.
Quantitative RT-PCR Analysis
RNA isolation and cDNA synthesis followed the procedure above mentioned. The quantitative RT-PCR was performed with SYBR pre-mix EX Taq (TaKaRa) by a Real-time PCR system (Eppendorf, Mastercycler ep realplex, Germany) with VsPCS1 primers presented in Supplementary Table 2. V. sativa gene Actin11 was used as an internal control and relative expression levels of genes which were calculated by 2-∆∆CT method (Gao et al., 2016). The primers for AtABCC1, 3, AtCAX2, AtNRAMP3 genes presented in Supplementary Table 1, and AtActin2 gene was used as an internal control.
Protoplast Preparation
The mesophyll protoplasts of WT, atpcs1, 35S::VsPCS1, and 35S::VsPCS1/atpcs1 were isolated by enzyme solution containing 0.25% w/v macerozyme R-10 (Yakult), 1% w/v cellulase R-10 (Yakult), 0.4 M D-mannitol, 20 mM MES (pH 5.7), 10 mM CaCl2, 20 mM KCl, and 0.1% w/v bovine serum albumin (Yoo et al., 2007). The isolated cells were purified using 21% sucrose and counted on a hemacytometer.
V. sativa protoplasts were prepared from 14-days-old leaves using the same modifications as described (Wu et al., 2009). The isolated cells were purified and concentrated using 30% sucrose.
Subcellular Localization of VsPCS1
35S (CaMV35S) promoter-driven expression clones were generated in the pSGFP vectors, resulting in N-terminal green fluorescent protein (GFP)-protein fusions. The CDS of V. sativa PCS1 gene amplified using the primers of PCS1F1 and PCS1R1. PCR products were cloned into pSGFP vector and confirmed by sequencing. 35S::VsPCS1-GFP plasmid were transiently expressed in V. sativa mesophyll protoplasts via polyethylene glycol (4000)-calcium transfection (Yoo et al., 2007). Incubating at room temperature for 16 h, transformed cells were observed with a uitraviewvox confocal microscope (PerkinElmer, United States).
Determination of Cd Content
The plant materials were washed with 5 mM CaCl2 for half an hour, and then dried at 105°C for 15 min and 48 h at 80°C in an oven. The digestion of dried samples according to the method of Rui et al. (2016). Cd concentrations were determined using atomic absorption spectrophotometer (novAA® 400; Analytik Jena, Jena, Germany).
GSH and PCs Analysis
For Cys, GSH and PCs measurement, they were extracted by 1 ml TFA (0.1%) and 6.3 mM diethylene triaminepentaacetic acid, put the homogenate on ice for 15 min. Following centrifugation (13200 g, 30 min, 4°C), the supernatant were derivatized based on the methods of Kühnlenz et al. (2015). UPLC system (Agilent 1290 Infinity, Germany) was equipped with a ZORBAX Eclipse Plus C18 column (2.1 mm × 100 mm) used for the separation of the mBBr-labeled thiols. Thiols were detected using the fluorescence detector set at excitation and emission wavelengths of 380 and 470 nm. Quantification was performed via authentic Cys, GSH, PC2, PC3, and PC4 standards.
Intracellular Cd Localization through Cd-Sensing Fluorescent Dyes
The leaf protoplasts with or without Cd treatment from WT, atpcs1, 35S::VsPCS1, and 35S::VsPCS1/atpcs1 were loaded with 0.04% v/v Leadmium Green AM dye (Molecular Probes, Invitrogen, Carlsbad, CA, United States). Intracellular Cd localization was observed according to the method of Park et al. (2012) using a uitraviewvox confocal microscope (PerkinElmer, United States). The intensity of green fluorescence signal was quantified by ImageJ software.
Statistical Analysis
The data were analyzed by one-way analysis of variance, followed by multiple comparisons with the least significant difference (LSD) test (P < 0.05), using SPSS software (ver. 17.0; SPSS, Inc., Chicago, IL, United States), different letters indicate significant differences among treatments.
Results
Isolation of Full-Length VsPCS1 Gene
The complete full-length phytochelatin synthase cDNA (complementary DNA) was amplified from V. sativa cDNA using a pair of degenerate primers. VsPCS1 coding DNA sequence (CDS) contains 1500 base pairs that encodes 500 amino acids (Supplementary Data 1, 2). Sequences analysis of amino acids showed that PCS1 is highly conserved between V. sativa (VsPCS1) and Medicago truncatula (MtPCS1) with 89.4% identity and 94% similarity. Twenty Cys residues are present in VsPCS1. Seven Cys residues are conserved in plant kingdom, and one Cys specific residue is in VsPCS1 (Supplementary Figure 1). In the aspect of catalytic activity, VsPCS1 holds the same catalytic active sites as other plant PCS1. Phylogenetic analysis for PCS1 demonstrated that V. sativa was grouped with leguminous plants. These data suggested that the VsPCS1 obtained is a V. sativa PCS1 homolog (Figure 1).
FIGURE 1.
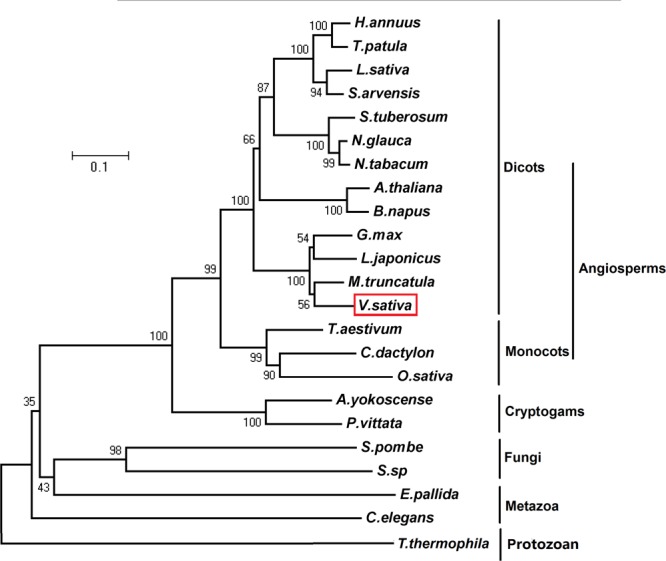
Phylogenetic analysis of PCS1. Phylogenetic relationship of PCS1 from Vicia sativa, Athyrium yokoscense (BAB64932.1), Arabidopsis thaliana (AAD50593.1), Brassica napus (AOV94290.1), Cynodon dactylon (AAO13810.2), Caenorhabditis elegans (NP_001122615.1), Exaiptasia pallida (KXJ16980.1), Glycine max (NP_001235576.1), Helianthus annuus (OTG28437.1), Lactuca sativa (AAU93349.1), Lotus japonicus (AAQ01752.1), Medicago truncatula (XP_013449920.1), Nicotiana glauca (ABX10958.1), Nicotiana tabacum (AAO74500.1), Oryza sativa (AAO13349.2), Pteris vittata (ADR51707.1), Sonchus arvensis (ACU44656.1), Schizosaccharomyces pombe (NP_593552.1), Solanum tuberosum (NP_001275308.1), Sporobolomyces sp. (AIS24729.1), Tagetes patula (AQT18915.1), Tetrahymena thermophila (AAY68362.2), and Triticum aestivum (AAD50592.1). The phylogenetic tree was constructed by neighbour-joining (NJ) method of MEGA5.01 with 1000 bootstraps replicates.
Expression Patterns of VsPCS1 in V. sativa
PCS homologs have been characterized as crucial factors for the detoxification of metals in organisms (Pomponi et al., 2006; Liu et al., 2011; Shukla et al., 2012; Kühnlenz et al., 2014, 2015). To test whether VsPCS1 hold the function to detoxify the heavy metals in V. sativa, we firstly determined the expression of VsPCS1 in response to excess Cd among different tissues, including leaves, stems, and roots. Through employing reverse transcript-polymerase chain reaction (RT-PCR) and quantitative RT-PCR, no significant difference on the VsPCS1 transcripts was detected in all the detected tissues under Cd-free and 5 μM Cd treatments. However, 50 μM Cd treatment dramatically induced the increase of VsPCS1 transcripts in roots, being about 4.2 times of Cd-free treatment (Figures 2A,B). Such Cd induced increase was not observed in leaves and stems under 50 μM Cd treatment for 24 h. Time-course analysis demonstrated that VsPCS1 in the roots rapidly and consistently responded to 50 μM Cd, showing a significant increase of transcript from 12 to 96 h (Figures 2C,D).
FIGURE 2.
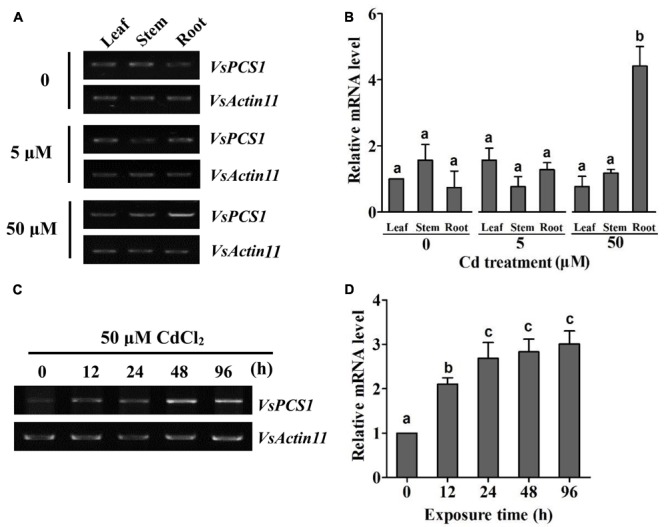
Expression analysis of VsPCS1 in V. sativa. RT-PCR (A) and quantitative RT-PCR (B) analysis the expression level of VsPCS1 in different organs (roots, stems, and leaves) of V. sativa under Cd treatment. RT-PCR (C) and quantitative RT-PCR (D) analysis the expression level of VsPCS1 under 50 μM Cd treatment for different time in roots of V. sativa. VsActin11 was used as an internal control. Values are means ± SD of three biological replicates. Columns labeled with distinct lowercase letters indicate statistically significant differences among treatments (P ≤ 0.05).
Subcellular Localization of VsPCS1 in V. sativa Mesophyll Protoplasts
To investigate the subcellular localization of VsPCS1, a fused protein of VsPCS1 with GFP was transiently expressed in mesophyll protoplasts of V. sativa. GFP signal mainly accumulated in the cytoplasm of VsPCS1-GFP transformed cells, whereas GFP signal is diffused everywhere in GFP transformed cells (Figure 3).
FIGURE 3.
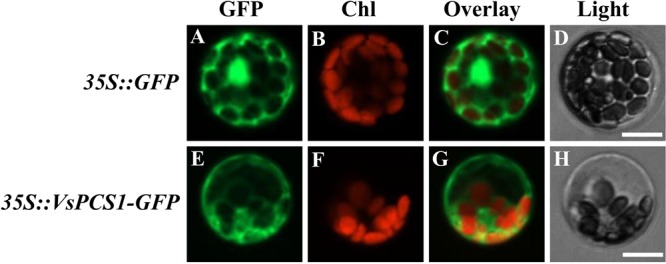
Subcellular localization of VsPCS1. V. sativa protoplasts transformed with 35S::GFP (A–D) or 35S::VsPCS1-GFP (E–H) plasmids. Confocal cross sections show green fluorescent protein (GFP) fluorescence (A,E), chlorophyll auto-fluorescence (B,F) and the overlay of GFP and chlorophyll auto-fluorescence (C,G). In (D,H) depicts the corresponding bright-field images. Bars = 14 μm.
Ectopic Expression of VsPCS1 Could Rescue the Cd Tolerance of Arabidopsis PCS1 Deficient Mutant
To test whether VsPCS1 is a functional homolog of Arabidopsis PCS1, we conducted a complementation experiment by ectopic expression of VsPCS1 in Arabidopsis PCS1 deficient mutant atpcs1 (designated as 35S::VsPCS1/atpcs1). Consistent with previous report (Nahar et al., 2014), atpcs1 was hypersensitive to Cd. Such Cd hypersensitivity of atpcs1 was restored when constitutively expressed VsPCS1 (Figure 4).
FIGURE 4.
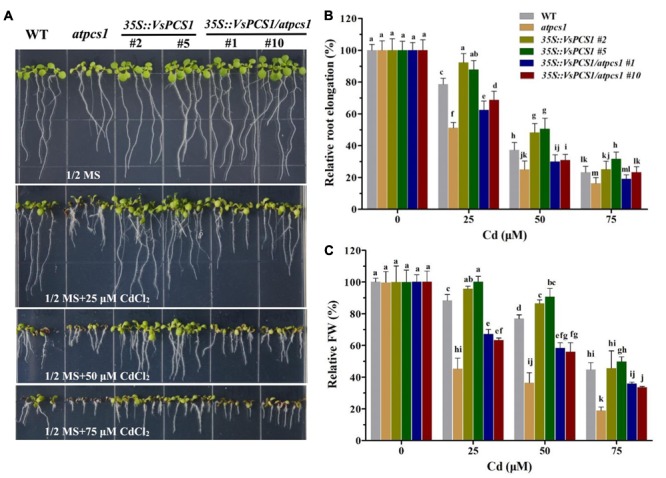
The VsPCS1 transgenic Arabidopsis shows tolerance to Cd toxicity. Seedlings grown in half-strength MS medium treated with or without of CdCl2 for 8 days (A). Relative root length (B) and fresh weight (C) of each genotype on plate treated with or without CdCl2. Values are means ± SD (n = 30–50). Columns labeled with distinct lowercase letters indicate statistically significant differences among treatments (P ≤ 0.05).
Cd Tolerance in VsPCS1-Overexpressing Arabidopsis
To further determine the function of VsPCS1 in detoxifying Cd, VsPCS1 was also ectopically expressed in wild-type Arabidopsis (Col-0). Fifteen independent 35-VsPCS1 lines were obtained by using hygromycin screen; six out of seven independent transgenic lines showed VsPCS1 overexpression; and two transgenic lines (#2 and #5) were selected for further analysis (Supplementary Figure 2). Ectopic expression of VsPCS1 has no effect on the levels of AtPCS1 in Arabidopsis (Supplementary Figure 3). To investigate Cd tolerance of these VsPCS1-expressing lines, Arabidopsis seeds of WT and homozygotic transgenic lines were germinated and grown on solid 1/2 MS medium contained 0, 25, 50, and 75 μM Cd for 8 days. No significant difference could be observed in the growth between WT and all of transgenic lines under Cd-free treatment (Figure 4A). When treated with 25 and 50 μM Cd, 35S::VsPCS1 (#2 and #5) had a longer root length and more fresh weight than WT (Figures 4B,C). These results demonstrate that overexpression of VsPCS1 increase Cd tolerance in 35S::VsPCS1.
Cd Accumulation in VsPCS1-Overexpressing Arabidopsis
Cd tolerance is tightly related to the Cd uptake from growth medium (Lin and Aarts, 2012). Since Cd tolerance of both 35S::VsPCS1 and 35S::VsPCS1/atpcs1 significantly increased, we investigated the effects of VsPCS1 overexpression on Cd concentration. Under control condition, Cd concentration was undetectable in all Arabidopsis lines (data not shown). After 8-day treatment with 25 and 50 μM Cd, a comparable Cd concentration was detected in WT and 35S::VsPCS1. Similarly, no difference in Cd concentration was observed between atpcs1 and 35S::VsPCS1/atpcs1 (Figure 5).
FIGURE 5.
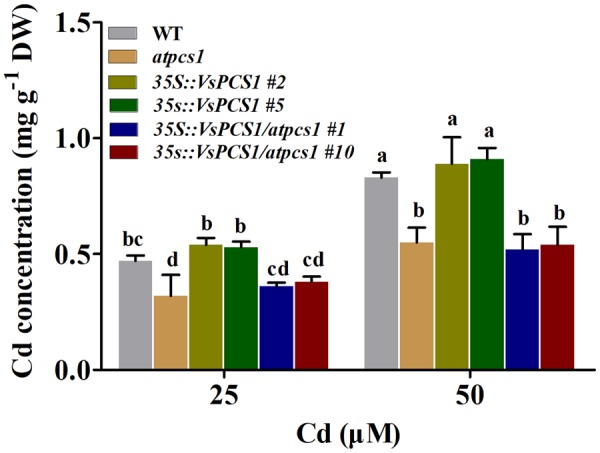
Effects of overexpression VsPCS1 on cadmium accumulation in Arabidopsis. Seedlings grown in half-strength MS medium in the presence or absence of CdCl2 for 8 days. Values are means ± SD of three biological replicates, columns labeled with distinct lowercase letters indicate statistically significant differences among treatments (P ≤ 0.05).
Cellular Distribution of Cd in VsPCS1-Overexpressing Arabidopsis
Apart from Cd uptake, the tolerance is also affected by cellular distribution of Cd. To examine whether the enhanced Cd tolerance conferred by overexpression of VsPCS1 resulted from the increased vacuolar sequestration, the cellular distribution of Cd was analyzed in the leaves of WT, atpcs1, 35S::VsPCS1 (#2 and #5), and 35S::VsPCS1/atpcs1 (#1 and #10) using a Cd-indicator dye, by which Cd concentration could be indicated by green fluorescence. In the absence of Cd, leaf cells from WT showed a negligible LeadmiumTM Green fluorescence signal (Figures 6A–C). After Cd treatment, observed green fluorescence was emitted in all of the detected lines. As expected, the green fluorescence signal mainly emitted from the vacuole of cells in wild-type (Figures 6D–F) and transgenic lines (35S::VsPCS1 #2, 35S::VsPCS1#5, 35S::VsPCS1/atpcs1#1, and 35S::VsPCS1/atpcs1#10) (Figures 6J–U). However, the green fluorescence signal in atpcs1 was detected mainly in cytoplasm and chloroplasts, being an orange-green signal when green LeadmiumTM Green and red chlorophyll auto-fluorescence were merged (Figures 6G–I). Quantitative analysis showed that average intensity of Cd-fluorescence was significantly lower in the vacuole and higher in the cytosol of atpcs1 than 35S::VsPCS1/atpcs1, demonstrating that VsPCS1 could rescue the defects of PC-based sequestration of Cd into the vacuole in atpcs1. Surprisingly, no difference in average intensity of Cd-fluorescence was observed between WT and 35S::VsPCS1 (Figure 6V).
FIGURE 6.

Analyses of cytosolic and vacuolar Cd in Arabidopsis leaf protoplasts. (A–C) WT protoplast treated without Cd. WT (D–F), atpcs1 (G–I), 35S::VsPCS1 (J–O) and 35S::VsPCS1/atpcs1 (P–U), protoplasts isolated from plants grown in10 μM CdCl2 for 5 days. LeadmiumTM Green signal was detected in protoplasts of the WT (D), 35S::VsPCS1 (J,M), 35S::VsPCS1/atpcs1 (P,S), and atpcs1 mutant (G). Merged images of green LeadmiumTM Green and red chlorophyll auto-fluorescence (B,E,H,K,N,Q,T) and bright-field images (C,F,I,L,O,R,U) are shown. Scale bars = 14 μm. (V) Fluorescence signal intensity in the cytosol and in the vacuole, values are means ± SD (n = 15–20), and columns labeled with distinct lowercase letters or uppercase indicate statistically significant differences in cytosol or in vacuole among treatments, respectively.
The PC-Cd complexes sequestrating to vacuole were mainly mediated by two ABCC proteins, AtABCC1 and AtABCC3 in Arabidopsis (Park et al., 2012; Brunetti et al., 2015). To exclude the possibility that the rescued Cd vacuole transport by overproducing VsPCS1 is owing to the increased ABCC proteins in Arabidopsis, we measured their expression in different Arabidopsis lines. As shown in Figure 7, the expression of AtABCC1 and AtABCC3 had the same transcription level in all lines under control condition. Although the transcript level of AtABCC3 increased in all lines after 10 μM Cd treatment, no significant increase were observed between VsPCS1 overproducing lines and WT. Unexpectedly, a higher AtABCC3 transcript was detected in the atpcs1 than other lines. Cd treatment did not affect the expression of AtABCC1.
FIGURE 7.
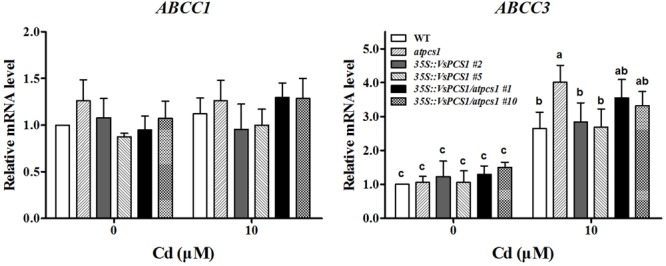
Effects of Cd treatment on gene expression of AtABCC1 and AtABCC3 in different Arabidopsis. Seedlings were exposed to 0 and 10 μM CdCl2 for 24 h. AtActin2 was used as an internal control. Values are means ± SD of three biological replicates. Columns labeled with distinct lowercase letters indicate statistically significant differences among treatments (P ≤ 0.05).
Content of PC in VsPCS1-Overexpressing Arabidopsis
Phytochelatin synthases are well-known to synthesize PCs from reduced glutathione (Noctor et al., 2011). To test whether PCS1 hold the catalytic activity for PCs synthesis, we measured the GSH and its high-order products PCs in the lines we used. As expected, atpcs1 mutant had the highest contents of GSH among all the detected lines. Consistently, a relative reduction of PCs was observed in atpcs1 compared with WT. Such change could be bounced back by introducing VsPCS1 into atpcs1 (Figure 8). Total PCs content in 35S::VsPCS1/atpcs1 was about twofold of that in the mutant atpcs1. Surprisingly, overexpressing VsPCS1 in Arabidopsis dramatically increase high-order products, especially PC4 in 35S::VsPCS1.
FIGURE 8.

The content of Cys, GSH, PC2, PC3, PC4, and total PCs under Cd treatment in Arabidopsis. Seedlings were treated with 10 μM CdCl2 for 5 days. Values are means ± SD of three biological replicates, and columns labeled with distinct lowercase letters indicate statistically significant differences among treatments (P ≤ 0.05).
Discussion
Phytochelatin synthases encoded by PCS genes. It have been identified that PCS has a role in the detoxification of diverse metals and metalloids (Grill et al., 1985; Ha et al., 1999). In this study, a phytochelatin synthase cDNA (VsPCS1) was isolated from V. sativa and the alignment of the amino acid sequences of VsPCS1 and other plant species was analyzed. As shown in Supplementary Figure 1, N-terminal amino acid residues of the plant PCS1 shows high conservation for approximately the first 221 amino acid residues, whereas the C-terminal, which contains rich cysteine residues, exhibits large variability, indicating that N-terminal is essential part for PCS1 function in plants. In support, three key amino acids, Cys56, His162, and Asp180 were located at the N-terminal of VsPCS1, the corresponding amino acids of which in AtPCS1 have been identified to be essential for AtPCS1 catalytic activity and Cd tolerance (Cobbett and Goldsbrough, 2002; Vivares et al., 2005). Additionally, compared with PCS1 in other plant species, a Cys16 residue only in VsPCS1 possibly hold a specific role in V. sativa.
It has been reported that AtPCS1 and AtPCS2 were observed to be constitutively expressed rather transcriptionally regulated by Cd (Ha et al., 1999; Cazalé and Clemens, 2001). In contrast, some studies reported that the transcriptional expression of PCS is regulated when exposed to heavy metals or metalloids (Ramos et al., 2007; Das et al., 2017). In this study, the VsPCS1 are expressed in leaves, stems and roots of V. sativa. When V. sativa was treated with 50 μM Cd, a significant induction of VsPCS1 expression was observed in root but not in stems and leaves (Figure 2). Similar expression pattern was reported for a rice PCS1 homolog, OsPCS1 under Cd stress (Das et al., 2017). This Cd-induced expression of PCS1 only in root may be the fact that roots are the major site of metal(loid)s accumulation in non-hyperaccumulator plants (Hernández et al., 2015).
Cytoplasm is a major formation site for the low molecular weight complexes of Cd and PC that are in turn transported into the vacuole to generating high molecular weight Cd and PC complexes (Cobbett, 2000). Consistently, VsPCS1 proteins were located in the cytoplasm. Similar cellular localization of AtPCS1 was observed in Arabidopsis (Blum et al., 2010). Besides cytoplasm localization of PCS1, other kind of cellular localization was also reported for PCS1 homolog. For an instance, a recent study indicates that SpPCS1 protein from Schizosaccharomyces pombe was localized to the mitochondria (Shine et al., 2015). This different cellular localization of PCS1 homolog among different species suggested that PCS1 might have the diversified roles in metal transport.
A decreased tolerance has been reported in mutant atpcs1 when exposed on Cd and As (Nahar et al., 2014). Similarly, a compromised Cd tolerance in atpcs1 mutant was observed in this study when compared with WT (Figure 4). Overexpressing VsPCS1 in the AtPCS1-deficent mutant atpcs1 can increase Cd tolerance. A similar increase of Cd tolerance in 35S::VsPCS1 (Figure 4). This increased Cd tolerance might be due to the elevated content of PCs under Cd stress because PCs contents is positively related Cd tolerances among different lines we tested (Figure 8). Furthermore, VsPCS1 homologs have been identified to catalyze the PCs synthesis and execute important role for the detoxification of metals in plants and other living organisms (Kühnlenz et al., 2015; Zanella et al., 2016). This increased Arabidopsis Cd tolerance was also reported by heteroexpression of PCS1 from Allium sativum (Guo et al., 2008) and Nelumbo nucifera (Liu et al., 2012). Additionally, tobacco overexpressing AtPCS1 and TcPCS1 from Thlaspi caerulescens also displayed increased Cd tolerance (Pomponi et al., 2006; Liu et al., 2011). By contrast, overproducing AtPCS1 in Arabidopsis reduced Cd tolerance despite enhanced PC production (Lee et al., 2003). This contradictory results on Cd tolerance among various transgenic plants might be due to the sequence variation of PCS genes from various sources or the difference in experimental conditions.
Apart from PCs contents, the increased Cd tolerance might be related to the availability of GSH. Pomponi et al. (2006) reported a direct correlation between the availability of GSH and the increase in Cd tolerance and accumulation in AtPCS1 overexpressing tobacco plants. Exogenous GSH increased the Cd tolerance of overexpressing AtPCS tobacco, but did not increase the Cd tolerance of overexpressing AtPCS Arabidopsis (Brunetti et al., 2011). Brunetti et al. (2011) observed that the endogenous content of PCs and GSH could affect Cd tolerance in AtPCS1 overexpressing Arabidopsis and tobacco. GSH functions as an antioxidant by scavenging free radicals and protects cells from the oxidative stress induced by heavy metals. GSH also serves as a direct precursor for PC synthesis. Some studies showed that Cd stress resulted in depletion of GSH, subsequently caused oxidative damage (Semane et al., 2007; Bankaji et al., 2015). In our study, the increase in PC synthesis did not cause a depletion of GSH in VsPCS1-expresing lines. No difference in GSH concentration was observed among all lines exposed to Cd except for atpcs1 (Figure 8), in which GSH was significantly accumulated due to inhibition of PC synthesis. The unchanged level of GSH, combined with the increased PC, showed that these transgenic plants had ability to restore the GSH pool used for PC synthesis.
Compared with that in atpcs1, average intensity of Cd-fluorescence in mesophyll cytoplasm was significantly lower in 35S::VsPCS1/atpcs1 lines, accompanied with higher intensity of Cd-fluorescence in vacuole, indicating that a role of VsPCS1 is involved in PC-based sequestration of Cd into the vacuole. However, this increased Cd sequestration into the vacuole was not observed in 35S::VsPCS1 lines although higher PCs contents were detected. Such difference might be explained by a higher PC4 in 35S::VsPCS1 than 35S::VsPCS1/atpcs1 under Cd stress. PCs with higher degree of polymerization were more efficient in the complexation of Cd (Gupta and Goldsbrough, 1991) and the small chain length peptide-Cd complexes can be more easily transported across the tonoplast than the longer peptide-Cd complexes (Shukla et al., 2013). VsPCS1 induced production of PC4 formed longer peptide-Cd complexes that could not transported across the tonoplast, finally resulting in no increased Cd sequestration into the vacuole in 35S::VsPCS1. Additionally, a high PC4 content and a proportion in the total PCs in 35S::VsPCS1 indicates that VsPCS1 hold a strong catalyzed property for the synthesis of high-order PCs, especially PC4. Surprisingly, no or less PC4 was detected in 35S::VsPCS1/atpcs1. Possibly, the loss of AtPCS1 in 35S::VsPCS1/atpcs1 lead to the deficiency of the precursors for high-order PCs, PC4. Up to date, only AtPCS1 was reported to promote the synthesis of PC4 under Cd stress (Brunetti et al., 2011).
Another possible explanation is that the subsequent sequestration of PC-Cd complexes from the cytosol into vacuoles might be limited by other factors such as the activity of vacuolar transporters. It has been reported that tonoplast transporters play a crucial role in transport of PCs-Cd complexes (Adamis et al., 2007; Park et al., 2012; Brunetti et al., 2015). Guo et al. (2012) reported that simultaneous expression of AsPCS1 and YCF1 (yeast cadmium factor 1, a member of vacuolar ATP-binding cassette transporter family) in Arabidopsis increased the tolerance and accumulation of Cd and As. Cd can be pumped directly into vacuoles by members of CAX (cation exchange transporters) family and HMA3 (Heavy metal ATPase 3) (Gravot et al., 2004; Mendoza-Cozatl et al., 2011). Cd can also be released from the vacuole by NRAMP-type transporters, AtNRAMP3 and AtNRAMP4 (Oomen et al., 2009). In this study, the transcription levels of AtCAX2 and AtNRAMP3 was not affected by Cd treatment in all lines (Supplementary Figure 4), which is in agreement with observations of Hirschi et al. (2000) and Oomen et al. (2009). Furthermore, although a lower Cd distribution is in vacuoles of atpcs1 than those of WT and 35S::VsPCS1/atpcs1 lines, atpcs1 mutant had higher transcript of ABCC3 than WT and a comparable level of 35S::VsPCS1/atpcs1 lines, suggesting the expression difference in ABCC transporters might not explain different patterns of Cd distribution in 35S::VsPCS1 and 35S::VsPCS1/atpcs1 lines.
Conclusion
We isolated a functional PCS1 homolog from V. sativa that located in the cytoplasm. Ectopic expressing of VsPCS1 in Arabidopsis increase Cd tolerance in 35S::VsPCS1 and 35S::VsPCS1/atpcs1, which is positively correlated with PC contents in plants. Surprisingly, VsPCS1 exhibited a strong catalyzed property for the synthesis of high-order PCs. Such property might be explained that a promoting effects on Cd transport into vacuole by overexpressing VsPCS1 in the 35S::VsPCS1/atpcs1, but not in the 35S::VsPCS1.
Author Contributions
XZ, ZS, and YX conceived, designed the experiments. ZS, XZ, YX, and ZH analyzed the data and revised the manuscript. FZ provided plants materials. XZ and HR carried out the experiments. All the authors read and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by the National Natural Science Foundation of China (31471938, 31672224, and 31160053).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00107/full#supplementary-material
References
- Adamis P. D. B., Panek A. D., Eleutherio E. C. A. (2007). Vacuolar compartmentation of the cadmium-glutathione complex protects Saccharomyces cerevisiae from mutagenesis. Toxicol. Lett. 173 1–7. 10.1016/j.toxlet.2007.06.002 [DOI] [PubMed] [Google Scholar]
- Bankaji I., Caçador I., Sleimi N. (2015). Physiological and biochemical responses of Suaeda fruticosa to cadmium and copper stresses: growth, nutrient uptake, antioxidant enzymes, phytochelatin, and glutathione levels. Environ. Sci. Pollut. Res. 22 13058–13069. 10.1007/s11356-015-4414-x [DOI] [PubMed] [Google Scholar]
- Blum R., Meyer K. C., Wünschmann J., Lendzian K. J., Grill E. (2010). Cytosolic action of phytochelatin synthase. Plant Physiol. 153 159–169. 10.1104/pp.109.149922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetti P., Zanella L., De Paolis A., Di Litta D., Cecchetti V., Falasca G., et al. (2015). Cadmium-inducible expression of the ABC-type transporter AtABCC3 increases phytochelatin-mediated cadmium tolerance in Arabidopsis. J. Exp. Bot. 66 3815–3829. 10.1093/jxb/erv185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetti P., Zanella L., Proia A., De Paolis A., Falasca G., Altamura M. M., et al. (2011). Cadmium tolerance and phytochelatin content of Arabidopsis seedlings over-expressing the phytochelatin synthase gene AtPCS1. J. Exp. Bot. 62 5509–5519. 10.1093/jxb/err228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalé A. C., Clemens S. (2001). Arabidopsis thaliana expresses a second functional phytochelatin synthase. FEBS Lett. 507 215–219. 10.1016/S0014-5793(01)02976-3 [DOI] [PubMed] [Google Scholar]
- Clemens S. (2006). Toxic metal accumulation, responses to exposure and mechanisms of tolerance in plants. Biochimie 88 1707–1719. 10.1016/j.biochi.2006.07.003 [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Cobbett C., Goldsbrough P. (2002). Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Annu. Rev. Plant Biol. 53 159–182. 10.1146/annurev.arplant.53.100301.135154 [DOI] [PubMed] [Google Scholar]
- Cobbett C. S. (2000). Phytochelatins and their roles in heavy metal detoxification. Plant Physiol. 123 825–832. 10.1104/pp.123.3.825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuypers A., Smeets K., Ruytinx J., Opdenakker K., Keunen E., Remans T., et al. (2011). The cellular redox state as a modulator in cadmium and copper responses in Arabidopsis thaliana seedlings. J. Plant Physiol. 168 309–316. 10.1016/j.jplph.2010.07.010 [DOI] [PubMed] [Google Scholar]
- Das N., Bhattacharya S., Bhattacharyya S., Maiti M. K. (2017). Identification of alternatively spliced transcripts of rice phytochelatin synthase 2 gene OsPCS2 involved in mitigation of cadmium and arsenic stresses. Plant Mol. Biol. 94 167–183. 10.1007/s11103-017-0600-1 [DOI] [PubMed] [Google Scholar]
- De Knecht J. A., Koevoets P. L. M., Verkleij J. A. C., Ernst W. H. O. (1992). Evidence against a role for phytochelatins in naturally selected increased cadmium tolerance in Silene vulgaris (Moench) Garcke. New Phytol. 122 681–688. 10.1111/j.1469-8137.1992.tb00097.x [DOI] [Google Scholar]
- Degola F., De Benedictis M., Petraglia A., Massimi A., Fattorini L., Sorbo S., et al. (2014). A Cd/Fe/Zn-responsive phytochelatin synthase is constitutively present in the ancient liverwort Lunularia cruciata (L.) dumort. Plant Cell Physiol. 55 1884–1891. 10.1093/pcp/pcu117 [DOI] [PubMed] [Google Scholar]
- Franchi N., Piccinni E., Ferro D., Basso G., Spolaore B., Santovito G., et al. (2014). Characterization and transcription studies of a phytochelatin synthase gene from the solitary tunicate Ciona intestinalis exposed to cadmium. Aquat. Toxicol. 152 47–56. 10.1016/j.aquatox.2014.03.019 [DOI] [PubMed] [Google Scholar]
- Gao S., Yang L., Zeng H. Q., Zhou Z. S., Yang Z. M., Li H., et al. (2016). A cotton miRNA is involved in regulation of plant response to salt stress. Sci. Rep. 6:19736. 10.1038/srep19736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravot A., Lieutaud A., Verret F., Auroy P., Vavasseur A., Richaud P. (2004). AtHMA3, a plant P1B-ATPase, functions as a Cd/Pb transporter in yeast. FEBS Lett. 561 22–28. 10.1016/s0014-5793(04)00072-9 [DOI] [PubMed] [Google Scholar]
- Grill E., Winnacker E. L., Zenk M. H. (1985). Phytochelatins: the principal heavy-metal complexing peptides of higher plants. Science 230 674–677. 10.1126/science.230.4726.674 [DOI] [PubMed] [Google Scholar]
- Guo J., Dai X., Xu W., Ma M. (2008). Overexpressing GSH1 and AsPCS1 simultaneously increases the tolerance and accumulation of cadmium and arsenic in Arabidopsis thaliana. Chemosphere 72 1020–1026. 10.1016/j.chemosphere.2008.04.018 [DOI] [PubMed] [Google Scholar]
- Guo J., Xu W., Ma M. (2012). The assembly of metals chelation by thiols and vacuolar compartmentalization conferred increased tolerance to and accumulation of cadmium and arsenic in transgenic Arabidopsis thaliana. J. Hazard. Mater. 199 309–313. 10.1016/j.jhazmat.2011.11.008 [DOI] [PubMed] [Google Scholar]
- Gupta S. C., Goldsbrough P. B. (1991). Phytochelatin accumulation and cadmium tolerance in selected tomato cell Lines. Plant Physiol. 97 306–312. 10.1104/pp.97.1.306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha S. B., Smith A. P., Howden R., Dietrich W. M., Bugg S., O’Connell M. J., et al. (1999). Phytochelatin synthase genes from Arabidopsis and the yeast Schizosaccharomyces pombe. Plant Cell 11 1153–1163. 10.1105/tpc.11.6.1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández L. E., Sobrino-Plata J., Montero-Palmero M. B., Carrasco-Gil S., Flores-Cáceres M. L., Ortega-Villasante C., et al. (2015). Contribution of glutathione to the control of cellular redox homeostasis under toxic metal and metalloid stress. J. Exp. Bot. 66 2901–2911. 10.1093/jxb/erv063 [DOI] [PubMed] [Google Scholar]
- Hirschi K. D., Korenkov V. D., Wilganowski N. L., Wagner G. J. (2000). Expression of Arabidopsis CAX2 in tobacco. Altered metal accumulation and increased manganese tolerance. Plant Physiol. 124 125–134. 10.1104/pp.124.1.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobe T. O., Sung D. Y., Akmakjian G., Pham A., Komives E. A., Mendoza-Cózatl D. G., et al. (2012). Feedback inhibition by thiols outranks glutathione depletion: a luciferase-based screen reveals glutathione-deficient γ-ECS and glutathione synthetase mutants impaired in cadmium-induced sulfate assimilation. Plant J. 70 783–795. 10.1111/j.1365-313X.2012.04924.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühnlenz T., Schmidt H., Uraguchi S., Clemens S. (2014). Arabidopsis thaliana phytochelatin synthase 2 is constitutively active in vivo and can rescue the growth defect of the PCS1-deficient cad1-3 mutant on Cd-contaminated soil. J. Exp. Bot. 65 4241–4253. 10.1093/jxb/eru195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühnlenz T., Westphal L., Schmidt H., Scheel D., Clemens S. (2015). Expression of Caenorhabditis elegans PCS in the AtPCS1-deficient Arabidopsis thaliana cad1-3 mutant separates the metal tolerance and non-host resistance functions of phytochelatin synthases. Plant Cell Environ. 38 2239–2247. 10.1111/pce.12534 [DOI] [PubMed] [Google Scholar]
- Lee S., Moon J. S., Ko T. S., Petros D., Goldsbrough P. B., Korban S. S. (2003). Overexpression of Arabidopsis phytochelatin synthase paradoxically leads to hypersensitivity to cadmium stress. Plant Physiol. 131 656–663. 10.1104/pp.014118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. F., Aarts M. G. (2012). The molecular mechanism of zinc and cadmium stress response in plants. Cell. Mol. Life Sci. 69 3187–3206. 10.1007/s00018-012-1089-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G. Y., Zhang Y. X., Chai T. Y. (2011). Phytochelatin synthase of Thlaspi caerulescens enhanced tolerance and accumulation of heavy metals when expressed in yeast and tobacco. Plant Cell Rep. 30 1067–1076. 10.1007/s00299-011-1013-2 [DOI] [PubMed] [Google Scholar]
- Liu Q., Luo L., Wang X., Shen Z., Zheng L. (2017). Comprehensive analysis of rice Laccase Gene (OsLAC) family and ectopic expression of OsLAC10 enhances tolerance to copper stress in Arabidopsis. Int. J. Mol. Sci. 18:209. 10.3390/ijms18020209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Gu C., Chen F., Yang D., Wu K., Chen S., et al. (2012). Heterologous expression of a Nelumbo nucifera phytochelatin synthase gene enhances cadmium tolerance in Arabidopsis thaliana. Appl. Biochem. Biotechnol. 166 722–734. 10.1007/s12010-011-9461-2 [DOI] [PubMed] [Google Scholar]
- Mendoza-Cozatl D. G., Jobe T. O., Hauser F., Schroeder J. I. (2011). Long-distance transport, vacuolar sequestration, tolerance, and transcriptional responses induced by cadmium and arsenic. Curr. Opin. Plant Biol. 14 554–562. 10.1016/j.pbi.2011.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahar N., Rahman A., Moś M., Warzecha T., Ghosh S., Hossain K., et al. (2014). In silico and in vivo studies of molecular structures and mechanisms of AtPCS1 protein involved in binding arsenite and/or cadmium in plant cells. J. Mol. Model. 20:2104. 10.1007/s00894-014-2104-0 [DOI] [PubMed] [Google Scholar]
- Noctor G., Queval G., Mhamdi A., Chaouch S., Foyer C. H. (2011). Glutathione. Arabidopsis Book 9 1–32. 10.1199/tab.0142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oomen R. J., Wu J., Lelièvre F., Blanchet S., Richaud P., Barbier-Brygoo H., et al. (2009). Functional characterization of NRAMP3 and NRAMP4 from the metal hyperaccumulator Thlaspi caerulescens. New Phytol. 181 637–650. 10.1111/j.1469-8137.2008.02694.x [DOI] [PubMed] [Google Scholar]
- Park J., Song W. Y., Ko D., Eom Y., Hansen T. H., Schiller M., et al. (2012). The phytochelatin transporters AtABCC1 and AtABCC2 mediate tolerance to cadmium and mercury. Plant J. 69 278–288. 10.1111/j.1365-313X.2011.04789.x [DOI] [PubMed] [Google Scholar]
- Pomponi M., Censi V., Di Girolamo V., De Paolis A., di Toppi L. S., Aromolo R., et al. (2006). Overexpression of Arabidopsis phytochelatin synthase in tobacco plants enhances Cd2+ tolerance and accumulation but not translocation to the shoot. Planta 223 180–190. 10.1007/s00425-005-0073-3 [DOI] [PubMed] [Google Scholar]
- Ramos J., Clemente M. R., Naya L., Loscos J., Pérez-Rontomé C., Sato S., et al. (2007). Phytochelatin synthases of the model legume Lotus japonicus. A small multigene family with differential response to cadmium and alternatively spliced variants. Plant Physiol. 143 1110–1118. 10.1104/pp.106.090894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remans T., Smeets K., Opdenakker K., Mathijsen D., Vangronsveld J., Cuypers A. (2008). Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta 227 1343–1349. 10.1007/s00425-008-0706-4 [DOI] [PubMed] [Google Scholar]
- Rui H., Chen C., Zhang X., Shen Z., Zhang F. (2016). Cd-induced oxidative stress and lignification in the roots of two Vicia sativa L. varieties with different Cd tolerances. J. Hazard. Mater. 301 304–313. 10.1016/j.jhazmat.2015.08.052 [DOI] [PubMed] [Google Scholar]
- Semane B., Cuypers A., Smeets K., Van Belleghem F., Horemans N., Schat H., et al. (2007). Cadmium responses in Arabidopsis thaliana: glutathione metabolism and antioxidative defence system. Physiol. Plant 129 519–528. 10.1111/j.1399-3054.2006.00822.x [DOI] [Google Scholar]
- Shi D., Zhuang K., Xia Y., Zhu C., Chen C., Hu Z., et al. (2017). Hydrilla verticillata employs two different ways to affect DNA methylation under excess copper stress. Aquat. Toxicol. 193 97–104. 10.1016/j.aquatox.2017.10.007 [DOI] [PubMed] [Google Scholar]
- Shine A. M., Shakya V. P., Idnurm A. (2015). Phytochelatin synthase is required for tolerating metal toxicity in a basidiomycete yeast and is a conserved factor involved in metal homeostasis in fungi. Fungal Biol Biotechnol. 2 1–13. 10.1186/s40694-015-0013-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla D., Kesari R., Mishra S., Dwivedi S., Tripathi R. D., Nath P., et al. (2012). Expression of phytochelatin synthase from aquatic macrophyte Ceratophyllum demersum L. enhances cadmium and arsenic accumulation in tobacco. Plant Cell Rep. 31 1687–1699. 10.1007/s00299-012-1283-3 [DOI] [PubMed] [Google Scholar]
- Shukla D., Tiwari M., Tripathi R. D., Nath P., Trivedi P. K. (2013). Synthetic phytochelatins complement a phytochelatin-deficient Arabidopsis mutant and enhance the accumulation of heavy metal (loid) s. Biochem. Biophy. Res. Commun. 434 664–669. 10.1016/j.bbrc.2013.03.138 [DOI] [PubMed] [Google Scholar]
- Song W. Y., Mendoza-Cózatl D. G., Lee Y., Schroeder J. I., Ahn S. N., Lee H. S., et al. (2014). Phytochelatin-metal(loid) transport into vacuoles shows different substrate preferences in barley and Arabidopsis. Plant Cell Environ. 37 1192–1201. 10.1111/pce.12227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennstedt P., Peisker D., Böttcher C., Trampczynska A., Clemens S. (2009). Phytochelatin synthesis is essential for the detoxification of excess zinc and contributes significantly to the accumulation of zinc. Plant Physiol. 149 938–948. 10.1104/pp.108.127472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivares D., Arnoux P., Pignol D. (2005). A papain-like enzyme at work: native and acyl-enzyme intermediate structures in phytochelatin synthesis. Proc. Natl. Acad. Sci. U.S.A. 102 18848–18853. 10.1073/pnas.0505833102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner G. J. (1993). Accumulation of cadmium in crop plants and its consequences to human health. Adv. Agron. 51 173–212. 10.1016/S0065-2113(08)60593-3 [DOI] [Google Scholar]
- Wang F., Wang Z., Zhu C. (2012). Heteroexpression of the wheat phytochelatin synthase gene (TaPCS1) in rice enhances cadmium sensitivity. Acta Biochim. Biophys. Sin. 44 886–893. 10.1093/abbs/gms073 [DOI] [PubMed] [Google Scholar]
- Wójcik M., Dresler S., Plak A., Tukiendorf A. (2015). Naturally evolved enhanced Cd tolerance of Dianthus carthusianorum L. is not related to accumulation of thiol peptides and organic acids. Environ. Sci. Pollut. Res. 22 7906–7917. 10.1007/s11356-014-3963-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F. H., Shen S. C., Lee L. Y., Lee S. H., Chan M. T., Lin C. S. (2009). Tape-Arabidopsis Sandwich-a simpler Arabidopsis protoplast isolation method. Plant Methods 5:16. 10.1186/1746-4811-5-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y., Liu J., Wang Y., Zhang X. X., Shen Z. G., Hu Z., et al. (2018). Ectopic expression of Vicia sativa Caffeoyl-CoA O-methyltransferase(VsCCoAOMT) increases the uptake and tolerance of cadmium in Arabidopsis. Environ. Exp. Bot. 145 47–53. 10.1016/j.envexpbot.2017.10.019 [DOI] [Google Scholar]
- Yi J., An G. (2013). Utilization of T-DNA tagging lines in rice. J. Plant Physiol. 56 85–90. 10.1007/s12374-013-0905-9 22228180 [DOI] [Google Scholar]
- Yoo S. D., Cho Y. H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2 1565–1572. 10.1038/nprot.2007.199 [DOI] [PubMed] [Google Scholar]
- Zanella L., Fattorini L., Brunetti P., Roccotiello E., Cornara L., D’Angeli S., et al. (2016). Overexpression of AtPCS1 in tobacco increases arsenic and arsenic plus cadmium accumulation and detoxification. Planta 243 605–622. 10.1007/s00425-015-2428-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Zhang H., Wang G., Xu L., Shen Z. (2009). Cadmium-induced accumulation of hydrogen peroxide in the leaf apoplast of Phaseolus aureus and Vicia sativa and the roles of different antioxidant enzymes. J. Hazard. Mater. 168 76–84. 10.1016/j.jhazmat.2009.02.002 [DOI] [PubMed] [Google Scholar]
- Zhang F., Zhang H., Xia Y., Wang G., Xu L., Shen Z. (2011). Exogenous application of salicylic acid alleviates cadmium toxicity and reduces hydrogen peroxide accumulation in root apoplasts of Phaseolus aureus and Vicia sativa. Plant Cell Rep. 30 1475–1483. 10.1007/s00299-011-1056-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


