Abstract
Chemical modifications of 2′-O-methyl (2′-OMe) and locked nucleic acid (LNA) of the nucleotides in the seed region (positions 2–8) of the small interfering RNA (siRNA) guide strand significantly reduced seed-matched (SM) off-target effects. The siRNA with 2′-OMe modifications inhibited the expression of a completely-matched (CM) target gene, whereas that with LNA modifications did not inhibit the expression of the CM target. By computational predictions of conformational changes of siRNA by these modifications, we revealed that both modifications in the siRNA seed region reduce SM off-target effects by steric hindrance to base-pairing with target transcripts but LNA modifications also disturb the association of the siRNA guide strand with the Argonaute (AGO) protein by altering RNA conformation. Thus, chemical modifications of the siRNA guide strand, which alter steric conformation to disturb base-pairing with target transcripts but do not disturb the association with the AGO protein, may successfully suppress off-target effects without substantial loss of RNA silencing activity.
Introduction
RNA interference (RNAi) is a prominent tool for functional genomics because small interfering RNAs (siRNAs) can effectively knock down the target genes with perfect sequence complementarity to the siRNA. siRNAs are double-stranded RNAs, 21 nucleotides (nts) in length with 2 nt 3′ overhangs (Figure 1). When an siRNA is transfected into cells, it is incorporated into the RNA-induced silencing complex (RISC), which includes the core protein Argonaute (AGO), and this process is referred to as RISC loading. Subsequently, the siRNA is unwound into single-stranded RNAs. Then, one RNA strand (guide strand) remains associated with AGO to form active RISC, whereas the other strand (passenger strand) is degraded.1 The guide strand forms complete base-pairing with a target transcript, and AGO cleaves the target to suppress its function.2
Figure 1.
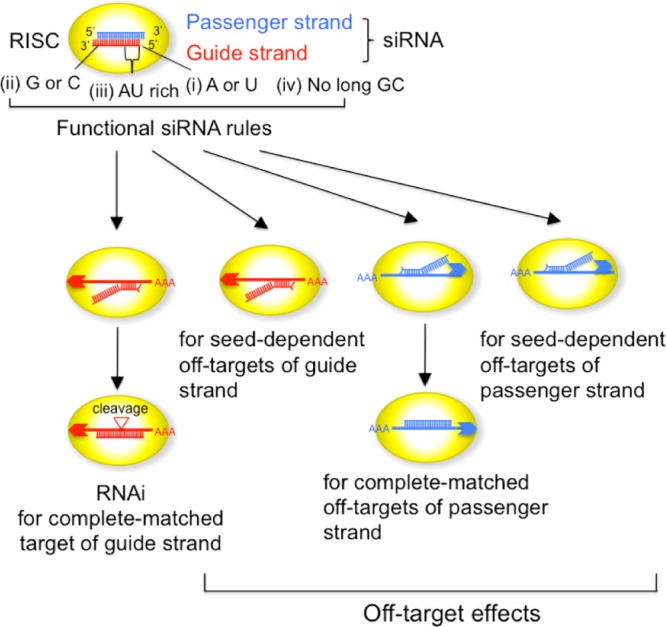
Schematic presentation of the siRNA-mediated RNAi pathway and off-target pathways. RNAi is induced by the completely complementary siRNA guide strand, and seed-dependent off-target effects are induced by seed-matched (SM) targets of the guide strand. The completely-matched (CM) or SM off-target effect of the passenger strand is also observed. The siRNA satisfying four functional siRNA rules was used.
Only a limited fraction of siRNA sequences are functional in mammalian cells. We revealed the guidelines for functional siRNA sequences in mammalian cells using various types of experiments.3 Functional siRNAs satisfied the following four conditions simultaneously: (i) A/U at the 5′-terminus of the siRNA guide strand, (ii) G/C at the 5′-terminus of the passenger strand, (iii) AU richness at the 5′ one-third of the guide strand, and (iv) absence of a long GC stretch (>9 nts) (Figure 1). Results from a number of studies suggest that asymmetry in the stability of the siRNA termini is essential for determining the unwinding direction of the siRNA duplex into single-stranded RNAs.3−5 An siRNA duplex is easily unwound from the more unstable terminus, and the unwound 5′-terminal nucleotide is anchored in the binding pocket in the mid domain of the AGO protein, in which A or U is preferentially anchored at an affinity up to 30-fold higher than that with either G or C.6 Thus, an RNA strand with the unstable 5′-terminus acts as a functional guide RNA. If the passenger strand of siRNA remains in the RISC, it may induce unintended off-target effects. However, as the siRNA selected according to the above criteria possesses a G or C residue at the 5′-terminus of the passenger strand, which has a lower affinity with the AGO pocket, the passenger-strand-dependent off-target effects are effectively eliminated even when the passenger strand remains in the RISC. A total of 56 375 087 (100%) 23-mer (19 nt double-stranded region with 2 nt overhangs at both termini) subsequences were obtained from human mRNAs registered in the RefSeq database (release 30). Bioinformatics analysis revealed that 14.7% of the human siRNA sequences satisfied all four criteria, (i)–(iv), for functional siRNAs; 98% of more than 100 different siRNAs satisfying these sequence rules were experimentally confirmed to be functional. Furthermore, at least one unique functional siRNA was selected for 92 and 99% of human and mouse genes, respectively.3
Partial complementarity of the siRNA guide strand with nontargeted mRNAs induces unintended off-target effects7−10 (Figure 1). Every backbone phosphate of the seed nucleotides at positions 2–8 from the anchored 5′-terminal nucleotide preordered on the AGO protein to make stable base-pairing between the siRNA seed region and target mRNA in an A-form helix.11,12 The efficiency of the off-target effect is positively correlated with the thermodynamic stability of the base pair between the guide strand seed region and SM transcripts.10 Thus, siRNAs with low seed–target stability may be a promising tool for target-specific RNAi with fewer off-target effects. However, the addition of a fifth condition of low thermodynamic stability in the seed region to the siRNA selection criteria (i)–(iv) decreased the percentage of selectable siRNA candidates from 14.7 to 2.1%.13 This means that at least one functional siRNA is selected for just 77.2% of human genes. To overcome such sequence limitations, we evaluated the impact of chemical modifications in this study. Many chemical modifications of various sites in the siRNA were examined for improvements in siRNA specificity, stability, and immunogenicity. Because the off-target effects of the guide strand are induced by seed–target complementarity,7−10 the effects of a few types of modifications in the seed region were examined.14−16 Among them, 2′-O-methyl (2′-OMe) modifications of the nucleotide at position 2 of the guide strand reduced the SM off-target effects without a severe reduction in RNAi activity.14 However, because systematic studies on the effects of chemical modifications in the seed region have not been performed, the mechanism of the reduced off-target effects is not well-elucidated. In this study, five different modifications, shown in Figure 2a, were introduced into the seed region of authentic siRNA, which has 19 nt double-stranded region with 2 nt overhangs in both termini (Figure 2b).
Figure 2.
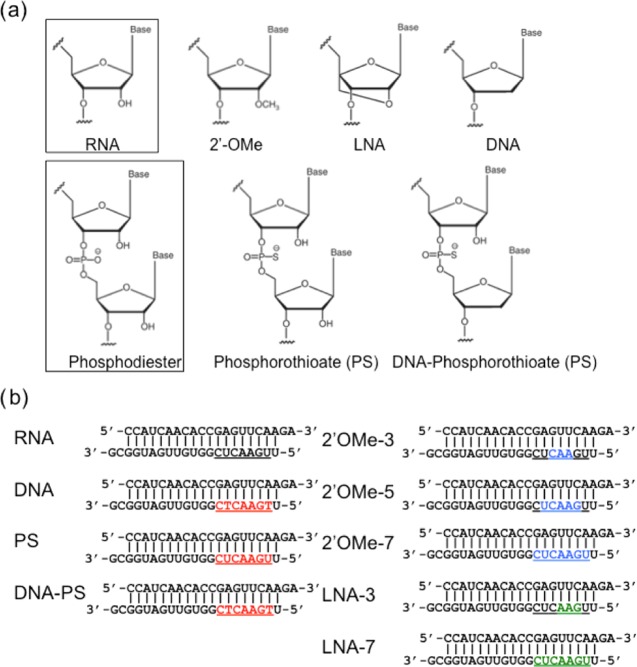
Chemical modifications used in this study. (a) Chemical structures of authentic RNA, 2′-OMe, locked nucleic acid (LNA), DNA, phosphorothioate (PS) linkage, DNA–PS. (b) siRNA sequences and the position of modified nucleotides. Red indicates DNA-, PS-, and DNA–PS-modified nucleotides. Blue indicates 2′-OMe-modified nucleotides. Green indicates LNA-modified nucleotides. Underlines indicate the nucleotide positions with each modification in the siRNA. The upper strand is the passenger strand. The lower strand is the guide strand.
Results and Discussion
To perform dual-luciferase reporter assays, four types of siRNA reporter plasmids were constructed (Figure 3a). psiCHECK-gCM with a CM sequence of the entire siRNA guide strand in the 3′UTR region of the Renilla luciferase gene was used to investigate RNAi activity of the guide strand. psiCHECK-gSM containing three tandem-repeated SM sequences of the guide strand was used to examine seed-dependent off-target effects (Figure 3a). psiCHECK-pCM with three tandem repeats of CM sequences on the passenger strand and psiCHECK-pSM with three tandem repeats of SM sequences were used to examine passenger-strand-dependent off-target effects (Figure S1a). Each of these plasmids was transfected into human HeLa cells with pGL3-Control, expressing the control firefly luciferase gene, and siRNA with or without chemical modifications in the seed region. The chemical modifications of deoxyribonucleic acid (DNA), PS, and DNA–PS were introduced into all of the seven nucleotides of the seed region of the siRNA guide strand (Figure 2b). In addition, 3 (positions 4–6), 5 (positions 3–7), and 7 (seed positions 2–8) nts in the guide strand were modified with 2′-OMe (named 2′-OMe-3, 2′-OMe-5, 2′-OMe-7); and 3 (positions 3–5) and 7 (seed positions 2–8) nts were modified with LNA (LNA-3, LNA-7) (Figure 2b). siRNA against an enhanced green fluorescent protein (siGY441) was used as a negative control. One day after transfection, firefly and Renilla luciferase activities were measured and relative luciferase activity was calculated as an indicator of the CM RNAi activity and SM off-target effects of the guide strand and CM and SM off-target effects of the passenger strand.
Figure 3.
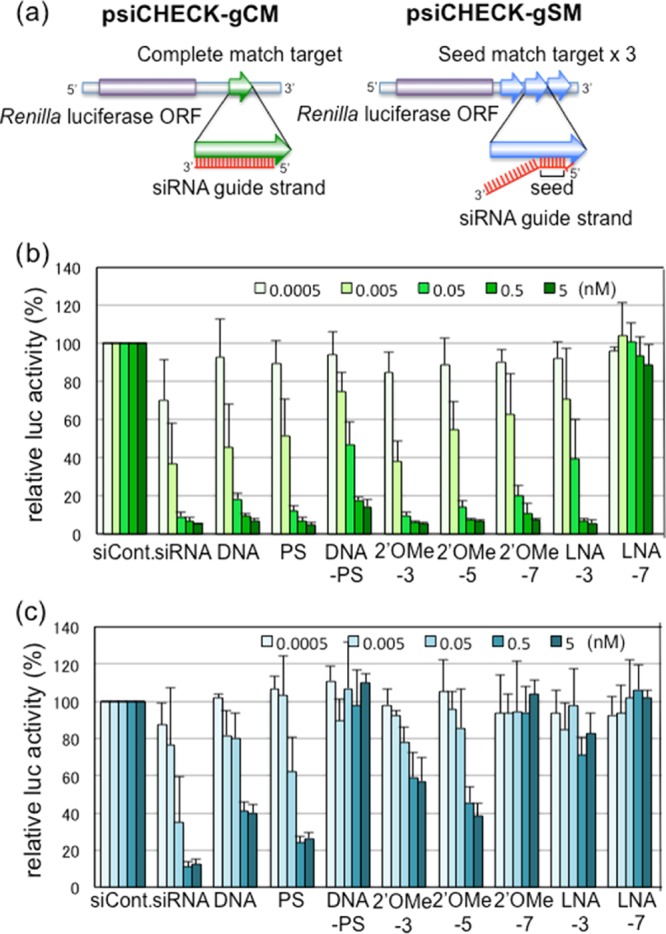
Results of luciferase reporter assays. (a) Structures of reporter constructs for luciferase reporter assays. psiCHECK-gCM contains a CM sequence of the siRNA guide strand. psiCHEK-gSM contains three tandem repeats of the SM sequences of the guide strand. (b) Results of RNAi activities on the CM target using psiCHECK-gCM with unmodified or modified siRNAs. (c) Results of seed-only silencing on the SM target using psiCHECK-gSM with unmodified or modified siRNAs. The data were averaged from three to four independent experiments, and the bar indicates the standard deviation.
Unmodified siRNA downregulated the Renilla luciferase activity via the guide strand CM target in a dose-dependent manner (Figure 3b). siRNA modified with DNA, PS, 2′-OMe (2′-OMe-3, -5, -7), or LNA (LNA-3) reduced the expression of the CM target of the guide strand at the equivalent level to that of unmodified siRNA, particularly at high concentrations (0.5 or 5 nM). PS is a modification in which one of the nonbridging oxygens is replaced by sulfur,17 which blocks the exonuclease action and increases its permeability through the lipid bilayer, but PS has little effect on base pair formation.18,19 In fact, PS-modified siRNA did not inhibit RNAi activity against the CM target. The various modifications at the 2′-position of the pentose sugar of siRNA are not required for RNAi20,21 and target recognition.15 DNA is modified with hydrogen at C2′, and the 5′ one-third of each RNA strand is capable of replacement with DNA without substantial loss of RNAi activity.15 Consistent with our previous report,15 DNA replacement at the seed region of the guide strand alone retained efficient RNAi activity on the CM target. 2′-OMe is a well-known C2′-modification that increases serum stability and specificity in base-pairing and abrogates immunogenicity.20,22−24 2′-OMe is tolerated at multiple positions in the siRNA guide strand because of its small size, comparable to the 2′-OH of natural RNA.25 Our results also showed that three 2′-OMe nucleotide modifications resulted in similar suppression levels on the CM target compared with unmodified RNA, although the efficiency was slightly reduced with increasing 2′-OMe modifications in the seed region (Figure 3b). LNAs contain a bridge connecting the 2′-oxygen with the 4′-carbon of the ribose ring, resulting in a greater stability compared to that of 2′-OMe.26,27 LNA-3 suppressed the expression of the CM target, but LNA-7 showed little to no activity.
The off-target effects on the SM target of the guide strand were observed for unmodified siRNAs in a reporter assay using psiCHECK-gSM (Figure 3c). siRNAs with PS modifications exhibited similar, but slightly weaker inhibitory effects compared to those from unmodified siRNAs, probably because PS modification has little effect on base pair formation.18,19 The inhibitory effect on the SM target of DNA-modified siRNAs was decreased, consistent with our previous results.15 DNA–PS-modified siRNA exhibited almost no effects. This may be because the base-pair stability of the DNA–RNA duplex is weaker than that of the RNA duplex.15 In contrast to DNA-modified siRNAs, the inhibitory effects on the SM target of the 2′-OMe and LNA modifications were strikingly decreased, even though both result in strong base pairs28 (Figure 3c).
Because we used siRNAs that satisfy the four functional siRNA sequence criteria,3 the guide strand is easily unwound from the 5′ terminus, but the passenger strand is not. In fact, almost no or little inhibitory effects on the CM or SM target of the passenger strand were observed in the reporter assays using psiCHECK-pCM or psciCHECK-pSM, respectively (Figure S1a).
Although DNA duplexes form B-form structures, structural superposition of the guide DNA–target RNA heteroduplex in the AGO complex fits better with the A-form helix of RNA duplexes than with the B-form.29 This implies that the duplex formed between a target transcript and an AGO-preloaded guide strand is modified to an A-form structure.
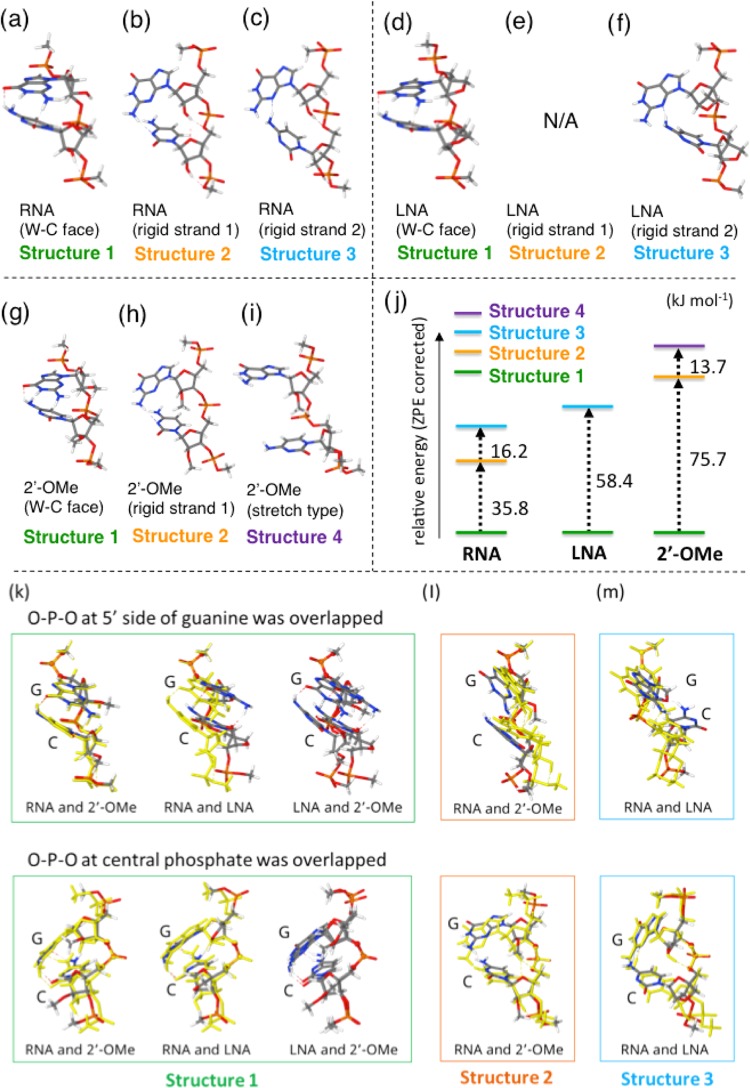
Computationally, we investigated how each chemical modification affects the single-stranded RNA structure that is not associated with protein. The single-stranded RNA structures of 5′-GC-3′ modified by 2′-OMe or LNA were calculated using density functional theory at the ωB97-XD/6-31G(d) level. The results of geometry optimization revealed that unmodified RNA can form at least three different stable conformation types: (i) the “Watson (W)–Crick (C) face form” in which two neighboring nucleotides are stabilized by two stacked hydrogen bonds, primed to form W–C base pairs when the complementary RNA strand is recognized (Figure 4a); (ii) “rigid strand 1” in which the OH residue at the C2′ position forms bifurcated hydrogen bonds with the sugar and downstream phosphate (Figure 4b); and (iii) “rigid strand 2” with different hydrogen bonds compared to those in structure 1 (Figure 4c). LNA-modified RNA had structures corresponding to structures 1 and 3 of unmodified RNA (Figure 4d–f), but we could not detect the corresponding structure 2. 2′-OMe-modified RNA had structures corresponding to structures 1 and 2 of unmodified RNA (Figure 4g,h). Additionally, structure 4, containing a stretched backbone, was also observed (Figure 4i). A comparison of the relative energies of these structures in each modification is shown in Figure 4j. In the case of 2′-OMe modifications, the W–C face form (structure 1) was the most stable and the relative energies of the other structures (structures 2 and 4) were rather high, which indicated that 2′-OMe modifications destabilized largely the other structures. In the case of unmodified RNA, even though structure 1 (W–C face form) is the most stable, other structures can be taken rather easily because of the hydrogen bonding ability of 2′-OH with the phosphate region of the backbone (Figure 4j). This suggests that 2′-OMe and LNA modifications inhibit the conformational variation of the RNA backbone and tend to keep the RNA structure suitable for base-pairing with target RNA; thus, the 2′-OMe modification results in the most stable W–C base-pairing and the LNA modification results in the second-most-stable pairing, with unmodified RNA having the lowest stability. Our previous results demonstrated that SM off-target effects are high when the stability of the seed–target duplex is high.10 However, both 2′-OMe and LNA modifications exhibited strong base-pairing stability with weaker off-target effects, in contrast to our previous results.
Figure 4.
Computational prediction of unmodified, LNA-modified, and 2′-OMe-modified RNA structures. Structures of unmodified RNA (a–c) and LNA-modified (d–f) and 2′-OMe-modified (g–i) GC RNAs with formations of W–C face (c, f, i), rigid strand 1 (b, e, h), rigid strand 2 (a, d), and stretch type (g). (j) The relative calibrated energies. Superposition of RNA and modified RNAs (k–m). The superposition patterns of O–P–O at the 5′ side of guanine (upper panels), and O–P–O at central phosphate (lower panels) was overlapped.
The phosphates of the siRNA guide strand interact with the amino acid side chains of the AGO protein.12 Therefore, it was speculated that 2′-OMe and LNA sugar modifications did not inhibit the association between the guide RNA and AGO. Our computational prediction revealed that structure 1 of the 2′-OMe-modified RNA was similar to that of the unmodified RNA (Figure 4k), consistent with a previous report.30 However, the structure of LNA-modified RNA differed from that of 2′-OMe-modified RNA (Figure 4k), probably, because the sugar puckering may affect the backbone conformation. Structure 2 of unmodified RNA and 2′-OMe-modified RNA exhibited only slight differences (Figure 4l), whereas structure 3 of unmodified RNA and LNA-modified RNA strongly differed in their overall structures, probably, because the sugar puckering is affected by the linkage between C2′ and C4′ (Figure 4m). Thus, the LNA modification affects the RNA structure more severely compared to the 2′-OMe modification.
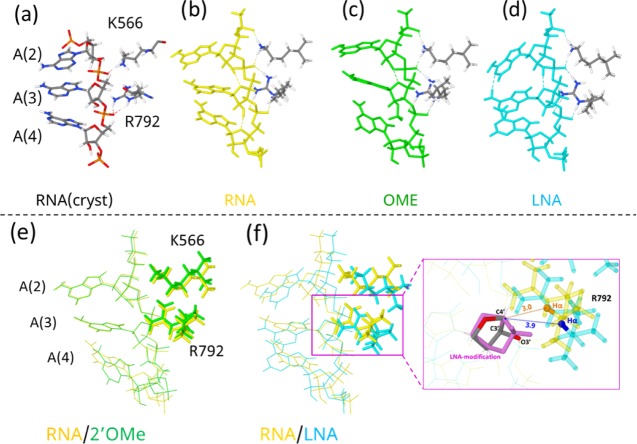
We further examined 2′-OMe- and LNA-modified single-stranded RNA structures on the AGO protein. The unmodified 5′-AAA-3′ RNA structure (nucleotides 2–4 (A(2), A(3), and A(4)) of the guide RNA) on the human AGO protein reported by Schirle et al.12 (Figure 5a) was used after optimization (Figure 5b). The side chains of the Arg (R792) and Lys (K566) residues in AGO form hydrogen bonds with the phosphates of the guide RNA (Figure 5a). Therefore, the full geometry optimization was achieved including R792 and K566, with capping each of the amino acid main chains with a methyl group (Figure 5b). The RNA structure in which the sugar of A(3) was modified with 2′-OMe (Figure 5c) or LNA (Figure 5d) was also optimized including R792 and K566. Overlapping of unmodified RNA and 2′-OMe-modified RNA and unmodified RNA and LNA-modified RNA on the AGO protein is shown in Figure 5e,f, respectively. As shown in Figure 5e, the 2′-OMe modification of A(3) led to little conformational change in A(2), A(3), R792, and K566. However, the 2′-OMe modification of the sugar in A(3) caused the repulsion of the methyl group with the nucleotide (A(4)) on the 3′ side and the conformation of the third adenine residues (A(4)) of 2′-OMe-modified RNA became different from that of unmodified RNA (Figure 5e). On the other hand, the LNA modification induced a different effect: the methylene group of the LNA caused steric hindrance to Hα of R792 and pushed R792 away, which would result in a large structural change of the AGO binding site, even though a stacked structure of LNA-modified RNA was almost kept (Figure 5f).
Figure 5.
Computational prediction of the unmodified, 2′-OMe-modified, and LNA-modified structures loaded on the AGO protein binding pocket. Unmodified AAA RNA with the R792 and K566 crystal structures from Schirle et al.12 (a), optimized structure of unmodified (b), 2′-OMe-modified (c), and LNA-modified RNA and (d) RNA with R792 and K566. Superposition at C4′-C3′-O3′ of A(3) of unmodified RNA with 2′-OMe-modified RNA (e) and unmodified RNA with LNA-modified RNA (f). The numbers in (f) are the distances (in Å) between C4′ (of A(3)) and Hα (of R792) atoms in unmodified RNA (in brown) and in LNA-modified RNA (in blue).
Thus, although both 2′-OMe and LNA are well-known modifications that enhance the base-pairing stability under protein-free conditions, our results strongly suggest that these modifications induce different steric hindrance, which would result in different effects on RNA silencing efficiency on CM and SM targets in terms of steric hindrance in association with the AGO protein or base-pairing with the target RNA. The 2′-OMe modification in the seed region of the siRNA guide strand was essentially stable on the AGO protein, as shown in the crystal structure by Schirle et al.,31 but it was revealed that the 2′-OMe modification may disturb base-pairing with the target RNA because of the conformational change in the 3′-side nucleotide (Figure 5e). Schirle et al. also showed that 2′-OMe of a specific nucleotide in 2′-OMe-modified guide RNA leads to a major positional shift of ∼6 Å (31). Such a positional shift of 2′-OMe-modified nucleotides propagates further and disorders the modified siRNA structure. Certainly, the suppression activity on the SM target was greatly reduced by the guide strand with 2′-OMe modifications in all of the seven nucleotides compared to that from those in three or five nucleotides, although the effect of siRNA with three 2′-OMe modifications was slightly weaker than that with five (Figure 3c). However, all of the 2′-OMe-modified siRNAs showed strong gene repression effects on the CM target (Figure 3b), although the inhibitory effects were slightly reduced according to the increase of the 2′-OMe modifications. The 5′-half of the siRNA guide strand is more sensitive sterically to chemical modifications than is the 3′-half with regard to RNAi activity against the CM target.32 Then, the nonseed region may play important roles in RNAi against the CM target. Recent single-molecule imaging revealed that the AGO protein reshapes the binding properties of the microRNA (miRNA) guide strand and serves as a specificity determinant with thermodynamic and kinetic properties typical of RNA-binding proteins.33,34 They showed that when RISC binds a target transcript through both miRNA seed and its 3′ supplementary nonseed region it dissociates nearly as rapidly as for seed-only binding and the rates of association and dissociation are very similar for these two binding modes. Our results clearly suggested that the seed–target base pair regulates the SM target expression. However, even when the expression of the SM target was not inhibited by siRNA with 2′-OMe or LNA modifications in the seed region, the expression of its CM target was significantly inhibited (Figure 3b,c). Thus, our result suggests that the nonseed region may compensate the incomplete seed–target base pair for inducing RNAi on the CM target.
The guide RNA structure with LNA modifications on the AGO protein was kinked by the sugar puckering by the linkage between C2′ and C4′ (Figure 4k–m), indicating that the base-pairing with target RNA is disturbed to some extent. Furthermore, the LNA-modified guide strand repulsed within the AGO binding site (Figure 5f). Thus, it was considered that siRNA modified with LNA in 7 nts at the seed region, LNA-7, did not associate with the AGO protein. Then, the expression of both the CM and SM targets may not be inhibited (Figure 3b,c). However, LNA-3 can associate with the AGO protein. Then, the SM off-target effects were reduced by the disturbance of base-pairing with the target RNA (Figure 3c) without substantial effects on the CM target (Figure 3b).
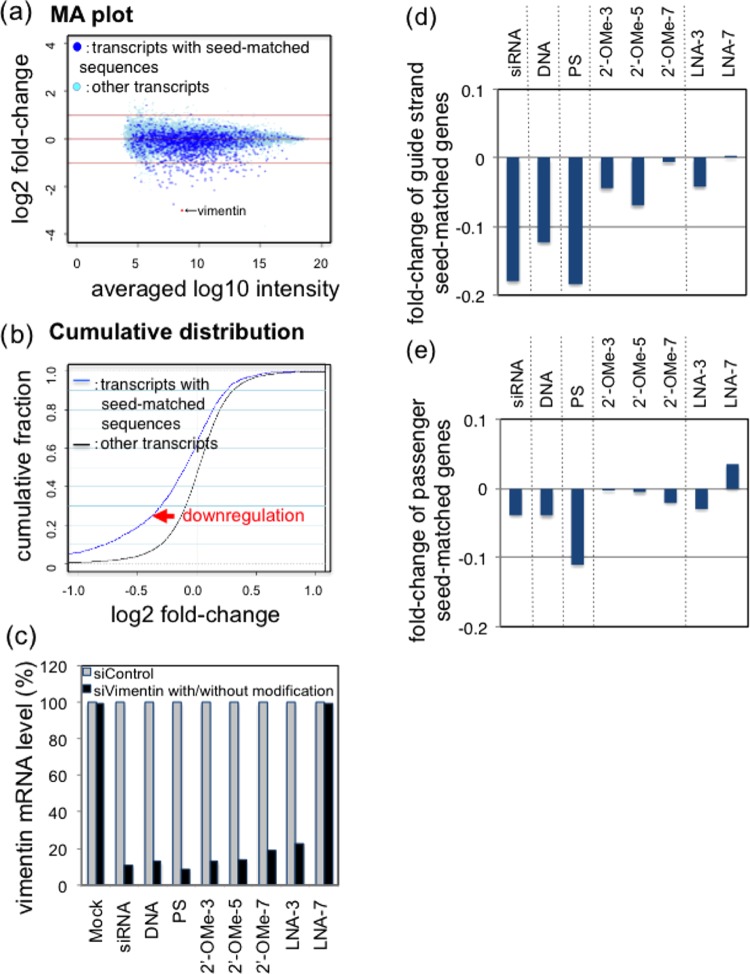
Microarray analysis was performed to examine the genome-wide off-target effects of siRNAs modified with DNA, PS, 2′-OMe, and LNA (Figure 2b). Unmodified and modified siRNAs were each transfected into HeLa cells. Total RNA was purified from cells 1 day later and subjected to microarray analyses. The MA plots (M = intensity ratio, A = average intensity) of the microarray data indicated changes in the expression levels of the annotated transcripts, and the cumulative distribution indicated the averaged fold-changes of the SM targets and control non-SM transcripts (Figures 6a,b, S2, and S3). The expression level of a CM target of vimentin gene was unambiguously downregulated by the unmodified siRNA to 11%, and the siRNAs modified with DNA, PS, 2′-OMe-3, 2′-OMe-5, 2′-OMe-7, and LNA-3 exhibited similar levels of RNAi activity as that of the unmodified siRNA, but LNA-7 modifications showed little to no activity (Figure 6c). The difference in the mean log 2 fold-changes of SM or non-SM transcripts was calculated as an indicator of the degree of off-target effects (Figure 6d,e). PS modifications had no effect on off-target effects, similar to the reporter assay results, compared to those of unmodified siRNA (Figure 3b), and DNA modifications reduced the off-target effects as reported previously.15 In the microarray experiments, off-target effects caused by the passenger strand were also observed. Unmodified and DNA-modified siRNAs showed similar levels of off-target effects as those of the passenger strand, but PS-modified siRNA exhibited stronger passenger-strand-dependent off-target effects (Figure 6e). Three 2′-OMe-modified siRNAs (2′-OMe-3, -5, and -7) and two LNA-modified siRNAs (LNA-3 and -7) exhibited reduced SM off-target effects in both strands compared to those of unmodified siRNA. These results also support the importance of steric hindrance in association with the AGO protein and base pair with target RNA. If asymmetry in siRNA terminal stability is the exclusive determinant of the siRNA unwinding efficiency, simultaneous reductions in the off-target effects of both strands cannot be explained. In this study, it was suggested that siRNAs modified by LNA in the seed region did not successfully associate with AGO. When the number of nucleotides with LNA modifications is small, a target gene could be successfully repressed, but when all of the seed nucleotides were modified, a CM target as well as SM target(s) should not be repressed. However, it was suggested that siRNAs modified by 2′-OMe in the seed region did not affect the association with the AGO protein but formed defective base pairs with target transcripts. This means that the 2′-OMe modification may exhibit the similar effect of siRNA with seed sequences composed of nucleotides with low thermodynamic stabilities, as shown in our previous report,15 because both commonly disturb the stable base pair between the siRNA seed region and its SM targets.
Figure 6.
Microarray analysis of the expression levels of the target vimentin gene and SM off-target genes. (a) MA plot. The vertical bar indicates the mean log 2 of signal intensities relative to those of mock transfection (M value), and the horizontal bar indicates the averaged log 10 signal intensities of mock and siRNA transfection (A value). The dark blue dots indicate the transcripts with SM sequences, and the light blue dots indicate the other transcripts. (b) Cumulative distribution. The horizontal axis indicates the M value of (a), and the vertical axis indicates the cumulative fraction of transcripts. The blue line indicates the cumulative curve of SM transcripts, and the black line indicates the cumulative curve of the other non-SM transcripts. The downregulation of SM transcripts is shown by the fold-change in the expression of SM transcripts compared to that of the other non-SM transcripts. (c) Expression levels of the target vimentin gene by the transfection of authentic siRNA or modified siRNAs. Seed-dependent off-target effects of the siRNA guide strand (d) and passenger strand (e). The vertical axis indicates the fold-change of off-target transcripts in the cells transfected with unmodified siRNA and modified siRNAs.
The off-target effects of the guide strand of 2′-OMe-modified siRNAs were greatest for the 2′-OMe-5 modification compared to those for the 2′-OMe-3 and 2′-OMe-7 modifications (Figure 6d). A similar pattern was also observed in the reporter assay (Figure 3c). These results may be explained by the counterbalance between two opposing factors regulating the seed-dependent off-target effects: base-pair stability in the seed–target duplex and steric hindrance in the base pair with the SM targets on the AGO protein. The 2′-OMe-7 modification may result in a more stable base pair in the seed–target duplex compared to that from the 2′-OMe-3 modification under protein-free conditions. However, the base pair with the SM target RNAs on AGO should be more sterically distorted with 2′-OMe-7 compared to that from 2′-OMe-3. With regard to 2′-OMe-5, the balance between seed–target base-pairing stability and steric hindrance may induce greatest SM off-target effects among the three 2′-OMe-modified siRNAs.
However, unlike those of the guide strand, the off-target effects of the passenger strand gradually increased with the number of 2′-OMe modifications in the guide strand seed region (Figure 6e). The results suggest that only the unwinding efficiency of each terminus may affect the off-target effect of the passenger strand. The 5′-terminal base-pairing stability in siRNA duplexes was highest for the 2′-OMe-7 modification and lowest for the 2′-OMe-3 modification, indicating that unwinding from the 5′-terminus of the guide strand is most favorable for 2′-OMe-3 but unfavorable for 2′-OMe-7. Thus, unwinding from the 5′-terminus of the passenger strand is easy for 2′-OMe-7 but difficult for 2′-OMe-3, leading to a stronger induction of off-target effects by the former compared to that by the latter.
Conclusions
In conclusion, the chemical modifications of DNA, PS, DNA–PS, 2′-OMe, and LNA in the seed region of the siRNA guide strand reduced the off-target effects of the guide strand SM targets and may also reduce those of the passenger strand CM and SM targets. Such off-target effects were greatly reduced without affecting the RNAi effects on the CM target using siRNA containing 2′-OMe modifications in the guide strand seed region. Our results suggest that appropriate introduction of 2′-OMe into siRNA can reduce off-target effects by inducing steric hindrance in duplex formation on the AGO protein. Such a chemical modification may function in a similar manner to the nucleotide sequences in the siRNA seed region with low thermodynamic stability. Because the off-target effects are serious problems that need to be addressed, the modifications like 2′-OMe in the seed region may overcome the limitations of siRNA sequences by reducing the off-target effects and may be useful as a potential therapeutic in the future.
Experimental Section
Preparation of Chemically Synthesized siRNA Duplexes
In this study, we used an siVIM-270,3 which satisfies four functional sequence conditions, to examine the seed-dependent off-target effects. The guide strand sequence of siRNA against human vimentin (siVIM-270) is 5′-UUGAACUCGGUGUUGAUGGCG-3′ and the passenger strand sequence is 5′-CCAUCAACACCGAGUUCAAGA-3′. RNA oligonucleotides of siRNA duplexes with and without modifications, shown in Figure 2a, were chemically synthesized (Genepharma).
Construction of Luciferase Reporters with CM and SM Sequences
All of the reporter plasmids were constructed from psiCHECK-1 (Promega). Oligonucleotides with the target sequence that is CM to the siRNA guide strand (5′-tcgaCGCCATCAACACCGAGTTCAAGA-3′ and 5′-aattTCTTGAACTCGGTGTTGATGGCG-3′) and three tandem repeats of the passenger strand CM target sequences (5′- tcgaTCTTGAACTCGGTGTTGATGGCGAATCTTGAACTCGGTGTTGATGGCGAATCTTGAACTCGGTGTTGATGGCGAA-3′ and 5′-aattTTCGCCATCAACACCGAGTTCAAGATTCGCCATCAACACCGAGTTCAAGATTCGCCATCAACACCGAGTTCAAGA-3′) were chemically synthesized with cohesive XhoI/EcoRI ends. They were annealed and inserted into psiCHECK-1 at the corresponding restriction enzyme sites and named psiCHECK-gCM and psiCHECK-pCM, respectively. Similarly, psciCHECK with three tandem repeats of SM sequences to the siRNA guide strand (5′-tcga AATGATGCACCAGGAGAGTTCAAAATGATGCACCAGGAGAGTTCAAAATGATGCACCAGGAGAGTTCAA-3′ and 5′-aattTTGAACTCTCCTGGTGCATCATTTTGAACTCTCCTGGTGCATCATTTTGAACTCTCCTGGTGCATCATT-3′) and the passenger strand (5′-tcga AATGATGCACCAGGAGTTGATGGAATGATGCACCAGGAGTTGATGGAATGATGCACCAGGAGTTGATGG-3′ and 5′-aattCCATCAACTCCTGGTGCATCATTCCATCAACTCCTGGTGCATCATTCCATCAACTCCTGGTGCATCATT-3′), each of which has complementarity with the 8 nt long seed-containing sequence but not with the nonseed region, were also generated and named psiCHECK-gSM and psiCHECK-pSM, respectively. Each of the inserted targets was expressed as part of the 3′-UTR region of Renilla luciferase mRNA in the transfected cells.
Cell Culture and the RNA Silencing Activity Assay Using the Firefly Luciferase Reporter System
Human HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco BRL) supplemented with 10% heat-inactivated fetal bovine serum (Mitsubishi Kagaku) at 37 °C. The cells inoculated in each well of 24-well plates at 1 × 105 cells/mL were transfected simultaneously with psiCHECK-gCM, -gSM, -pCM, and -pSM target constructs (100 ng), pGL3-Control (100 ng; Promega), and siRNA duplex (0.0005, 0.005, 0.05, 0.5, and 5 nM) using 2 μL of lipofectamine 2000 per well (Life Technologies). siGY441 was used as the control. The cells were harvested 24 h post-transfection, and the relative luciferase activity (Renilla luciferase activity/firefly luciferase activity) was measured using a dual-luciferase reporter assay system (Promega). The pGL3-Control encoding firefly luciferase served as a control for the calculation of the relative luciferase activity.
Computational Prediction of the Structure of Modified RNA
All of the geometries of single-stranded 5′-GC-3′ RNA structures [unmodified and modified (2′-OMe or LNA for both G and C)] were optimized at the theoretical level of ωB97-XD/6-31G(d). We confirmed that all of the optimized geometries are local minima with all positive harmonic frequencies. Zero-point energy (ZPE) was calculated using the harmonic frequencies for each of the local minima. The optimized geometries are shown in Figure 4, and the energies are listed in Table S1, together with all of the Cartesian coordinates.
The Cartesian coordinates of the crystal structure12 (PDB 4OLA) were used as an initial geometry for 5′-AAA-3′ RNA loaded on the AGO protein. Arg792 and Lys566 from the AGO protein were taken into the calculations, where the main chains of these amino acids were replaced by methyl groups (Figure S4). For 5′-AAA-3′ RNA, we used three types: unmodified and modified (2′-OMe or LNA for the central A). The full geometry optimization was performed for each of the models of the three types (5′-AAA-3′ RNA with Arg792 and Lys566) at the theoretical level of ωB97-XD/6-31G(d). The optimized geometries are shown in Figure 5, and the energies are listed in Table S2, together with all of the Cartesian coordinates. The Gaussian 0935 program package was used for all of the calculations. The calculations were carried out at the Center for Quantum Life Sciences (QuLiS) and at the Research Center for Computational Science, Okazaki National Research Institutes.
Microarray Analysis
The cells inoculated in each of 2 wells of 24-well plates at 1 × 105 cells/mL were transfected with 50 nM of siRNA duplex using 2 μL of Lipofectamine 2000. At 24 h post-transfection, total RNA was purified with an RNeasy kit (Qiagen) and RNA quality was assessed using a NanoDrop 2000 spectrophotometer (Thermo Scientific) and a Bioanalyzer (Agilent). cDNA was synthesized from each total RNA sample using an Agilent one-color spike mix kit (Agilent) and used for hybridization to an Agilent SurePrint G3 human GE microarray (8 × 60 K, ver. 2.0, 3.0) according to the manufacturer’s protocol. RNA from mock-transfected cells treated with the transfection reagent in the absence of siRNA was used as a control, and the distributions of the signal intensities of transcripts were normalized across all samples by quantile normalization.36 They were shown in MA plots and cumulative accumulations.
Acknowledgments
Theoretical calculations were partly performed using the Research Center for Computational Science, Okazaki, Japan. The English in this document has been checked by at least two professional editors, both native speakers of English.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.7b00291.
Results of luciferase reporter assays of the passenger strand; additional information on microarray analyses, MA plots and cumulative distributions of the SM off-target transcripts of the siRNA guide strand; additional information on microarray analyses, MA plots and cumulative distributions of the SM off-target transcripts of the siRNA passenger strand; supplementary computational data, total energy and ZPE-corrected energy of 5′-GC-3′ RNA; total energy of 5′-AAA-3′ RNA with the AGO protein (PDF)
Author Contributions
H.I., T.T., Y.K., and J.L. performed the reporter assays. Microarray experiments were carried out by T.T., and H.I. and T.T. analyzed the data. Computational predictions of nucleotide structures were performed by K.M. and M.A. M.A. and K.U.-T. discussed the experimental and computational data. The manuscript was written by K.U.-T. and reviewed by all authors. K.U.-T. designed the study and is responsible for scientific correspondence. All authors have given approval to the final version of the manuscript.
This work was financially supported by the Grants-in-Aid for Scientific Research (B) (No. 15H04319) and on Innovative Areas (No. 26102713) from the Ministry of Education, Culture, Sports, Science and Technology and Japan Society for the Promotion of Science, and by the grant from the Suzuken Memorial Foundation to K.U.-T.
The authors declare no competing financial interest.
Supplementary Material
References
- Hutvagner G.; Simard M. J. Nat. Rev. Mol. Cell Biol. 2008, 9, 22–32. 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- Elbashir S. M.; Martinez J.; Patkanlowska A.; Landeckeland W.; Tuschl T. EMBO J. 2001, 20, 6877–6888. 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ui-Tei K.; Naito Y.; Takahashi F.; Haraguchi H.; Ohki-Hamazaki H.; Juni A.; Ueda R.; Saigo K. Nucleic Acids Res. 2004, 32, 936–948. 10.1093/nar/gkh247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A.; Leake D.; Boese O.; Scaringe S.; Marsball W. S.; Khvorova A. Nat. Biotechnol. 2004, 22, 326–330. 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- Amarzguioui M.; Prydz H. Biochem. Biophys. Res. Commun. 2004, 316, 1050–1058. 10.1016/j.bbrc.2004.02.157. [DOI] [PubMed] [Google Scholar]
- Frank F.; Sonenberg N.; Nagar B. Nature 2010, 465, 818–822. 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- Jackson A. L.; Bartz S. R.; Schelter J.; Kobayashi S. V.; Burchard J.; Mao M.; Li B.; Cavet G.; Linsley P. S. Nat. Biotechnol. 2003, 21, 635–637. 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Lin X.; Ruan X.; Anderson M. G.; McDowell I. A.; Kroeger P. E.; Fesik S. W.; Shen Y. Nucleic Acids Res. 2005, 33, 4527–4535. 10.1093/nar/gki762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birmingham A.; Anderson E. M.; Reynolds A.; Ilsley-Tyree D.; Leake D.; Fedorow Y.; Baskerville S.; Maksinova E.; Robinson K.; Karpilow I.; Marshall W. S.; Khvorova A. Nat. Methods 2006, 3, 199–204. 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- Ui-Tei K.; Naito Y.; Nishi K.; Juni A.; Saigo K. Nucleic Acids Res. 2008, 36, 7100–7109. 10.1093/nar/gkn902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkayam E.; Kuhn C.-D.; Tocilj A.; Haase A. D.; Greene E. M.; Hannon G. J.; Joshua-Tor L. Cell 2012, 150, 100–110. 10.1016/j.cell.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirle N. T.; MacRae I. J. Science 2012, 336, 1037–1040. 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito Y.; Yoshimura J.; Morishita S.; Ui-Tei K. BMC Bioinf. 2009, 10, 392. 10.1186/1471-2105-10-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A. L.; Burchard J.; Leake D.; Reynolds A.; Schelter J.; Guo J.; Johnson J. M.; Lim L.; Karpilow J.; Nichols K.; Marshall W.; Khvorova A.; Linsley P. S. RNA 2006, 12, 1197–1205. 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ui-Tei K.; Naito Y.; Zenno S.; Nishi K.; Yamato K.; Takahash F.; Juni A.; Saigo K. Nucleic Acids Res. 2008, 36, 2136–2151. 10.1093/nar/gkn042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramsen J. B.; Pakula M. M.; Hansen T. B.; Bus C.; Langkjar N.; Okakzic D.; Smicius R.; Wengel S. L.; Chattopadhyaya J.; Engels J. W.; Herdewijn P.; Wengel J.; Kjems J. Nucleic Acids Res. 2010, 38, 5761–5773. 10.1093/nar/gkq341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckstein F.; Gish G. Trends Biochem. Sci. 1989, 14, 97–100. 10.1016/0968-0004(89)90130-8. [DOI] [PubMed] [Google Scholar]
- Crooke S. T.Handbook of Experimental Pharmacology; Springer Verlag: Berlin, 1998, p 1–50. [Google Scholar]
- Bennett C. F.; Swayze E. E. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 259–293. 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- Chiu Y. L.; Rana T. M. RNA 2003, 9, 1034–1048. 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czauderna F.; Fechtner M.; Dames S.; Aygun H.; Klippel A.; Pronk G. J.; Giese K.; Kaufmann J. Nucleic Acids Res. 2003, 31, 2705–2716. 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choung S.; Kim Y. J.; Kim S.; Park H. O.; Choi Y. C. Biochem. Biophys. Res. Commun. 2006, 342, 919–927. 10.1016/j.bbrc.2006.02.049. [DOI] [PubMed] [Google Scholar]
- Ge Q.; Dalla A.; Ilves H.; Shorenstein J.; Behlke M. A.; Johnston B. H. RNA 2010, 16, 118–130. 10.1261/rna.1901810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H.; Pardi A. Nucleic Acids Res. 2007, 35, 2965–2974. 10.1093/nar/gkm184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller E.; Propp S.; Murray H.; Lima W.; Bhat B.; Prakash T. P.; Allerson C. R.; Swayze E. E.; Marcusson E. G.; Dean N. M. Nucleic Acids Res. 2006, 34, 4467–4476. 10.1093/nar/gkl589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vester B.; Wengel J. Biochemistry 2004, 43, 13233–13241. 10.1021/bi0485732. [DOI] [PubMed] [Google Scholar]
- Shen L.; Johnson T. L.; Clugston S.; Huang H.; Butenhof K. J.; Stanton R. V. J. Chem. Inf. Model. 2011, 51, 1957–1965. 10.1021/ci200141j. [DOI] [PubMed] [Google Scholar]
- Elmén J.; Thonberg H.; Ljungberg K.; Frieden M.; Westergaard M.; Xu Y.; Wahren B.; Liang Z.; Ørum H.; Koch T.; Wahlestedt C. Nucleic Acids Res. 2005, 33, 439–447. 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Juranek S.; Li H.; Sheng G.; Tuschl T.; Patel D. J. Nature 2008, 456, 921–926. 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim I.; Kierzek E.; Kierzek R.; Schatz G. C. J. Phys. Chem. B. 2014, 118, 14177–14187. 10.1021/jp506703g. [DOI] [PubMed] [Google Scholar]
- Schirle N. T.; Kinberger G. A.; Murray H. F.; Lima W. F.; Prakash T. P.; MacRae I. J. J. Am. Chem. Soc. 2016, 138, 8694–8697. 10.1021/jacs.6b04454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrazas M.; Kool E. T. Nucleic Acids Res. 2009, 37, 346–353. 10.1093/nar/gkn958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee L. M.; Flores-Jasso C. F.; Salomon W. E.; Zamore P. D. Cell 2012, 151, 1055–1067. 10.1016/j.cell.2012.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon W. E.; Jolly S. M.; Moore M. J.; Zamore P. D.; Serebrov V. Cell 2015, 162, 84–95. 10.1016/j.cell.2015.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch M. J.; Trucks G. W.; Schlegel H. B.; Scuseria G. E.; Robb M. A.; Cheeseman J. R.; Scalmani G.; Barone V.; Mennucci B.; Petersson G. A.. et al. Gaussian 09, revision D.01; Gaussian, Inc.: Wallingford, CT, 2009.
- Bolstad B. M.; Irizarry R. A.; Gautier L.; Wu Z.. Bioinformatics and Computational Biology Solutions Using R and Bioconductor; Gentleman R., Carey V., Huber W., Irizarry R., Dudoit S., Eds.; Springer: New York, 2005, pp 13–32. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.