Abstract
Introduction: Current prognostic models for acute myeloid leukemia (AML) are inconsistent at predicting clinical outcomes for individual patients. Variability in the quality of specimens utilized for biomarker discovery and validation may contribute to this prognostic inconsistency.
Methods: We evaluated the impact of sample heterogeneity on prognostic biomarkers and methods to mitigate any adverse effects of this heterogeneity in 240 cryopreserved bone marrow and peripheral blood specimens from AML patients enrolled on SWOG (Southwest Oncology Group) trials.
Results: Cryopreserved samples displayed a broad range in viability (37% with viabilities ≤60%) and nonleukemic cell contamination (13% with lymphocyte percentages >20%). Specimen viability was impacted by transport time, AML immunophenotype, and, potentially, patients' age. The viability and cellular heterogeneity in unsorted samples significantly altered biomarker results. Enriching for viable AML blasts improved the RNA quality from specimens with poor viability and refined results for both DNA and RNA biomarkers. For example, FLT3-ITD allelic ratio, which is currently utilized to risk-stratify AML patients, was on average 1.49-fold higher in the viable AML blasts than in the unsorted specimens.
Conclusion: To our knowledge, this is the first study to provide evidence that using cryopreserved specimens can introduce uncontrollable variables that may impact biomarker results and enrichment for viable AML blasts may mitigate this impact.
Keywords: : AML, biomarkers, biorepository, cryopreservation, flow cytometry, immunophenotype
Introduction
Acute myeloid leukemia (AML) is one of the most common and devastating hematopoietic malignancies.1–3 Similar to most malignancies, AML is primarily a disease of older adults (median age at diagnosis 65 years). Although progress has been made in treating this disease, clinical outcomes remain poor for most patients and especially for older adults who frequently die of relapse or treatment-related complications.1 Many of the advances in the care for patients with AML have hinged on better risk stratification at the time of initial therapy and shepherding high-risk patients to allogeneic transplant or novel therapies.2–4 Current risk-stratification models incorporate clinical factors, cytogenetics, and molecular biomarkers, yet remain relatively imprecise for risk-stratifying patients for clinical outcome.2,3 As such, many patients continue to receive ineffective conventional therapies and forgo the opportunity of potential novel experimental approaches or upfront allogeneic transplant.4
Several potential explanations may account for the imprecision of risk-stratification models. Current risk-assessment models may lack biomarkers that are capable of estimating long-term responses. Armed with a more comprehensive molecular landscape of AML blasts, researchers may be able to determine the most informative biomarkers for individual patients and more precisely risk-stratify them. Certainly, the movement to molecularly profile patients has garnered much attention, being described as the first step to personalized medicine for cancer patients.
Another potential barrier to improving the precision of risk-stratification models may be the heterogeneous nature of the tested samples. Out of necessity, the majority of biomarker discovery and validation studies in AML have examined cryopreserved samples, which comprised heterogeneous mononuclear cells (MNCs).5–22 These MNCs include both AML blasts of varying differentiation stages and nonleukemic cells (e.g., lymphocytes and monocytes). Repository samples certainly provide an invaluable resource for such studies, but factors such as specimen source, handling, processing, and cryopreservation may impact the composition and quality of the cells. The National Cancer Institute (NCI), the International Society for Biological and Environmental Repositories (ISBER), and others have provided recommendations to ensure a more standardized approach for the collection, storage, and distribution of repository samples.23–39 Many of these recommendations are based on studies examining the impact of specimen handling and processing on the overall quality of normal or solid tumor samples.32,39–45 Fewer studies have examined the impact of cryopreservation on the morphology, function, and molecular profiles of AML blasts,46–48 although our group recently reported that delayed processing may adversely alter the leukemic transcriptome.49,50 Therefore, we systematically evaluated the quality of cryopreserved AML blasts from a large cooperative group, examining the impact of viability and the cellular composition on the quality of nucleic material and biomarker results. Further, we determined whether the potential adverse impact of dying and nonmalignant cells on the prognostic biomarkers is mitigated by evaluating viable AML blasts.
Materials and Methods
Patient materials
The SWOG (Southwest Oncology Group) Myeloid Repository collects and curates diagnostic specimens from patients with AML for correlative research from several treatment trials. For our study, we had access to diagnostic specimens with ≥3 cryopreserved vials from 383 out of 1042 previously untreated (i.e., de novo) AML patients who received cytarabine (Ara-C) and daunorubicin (DNR)-based induction chemotherapy on treatment protocols SWOG-9031, SWOG-9333, S0106, and S0112.51–54 Details on chemotherapy regimens for each of the protocols have been previously described.51–54 For these treatment protocols, the patient's AML diagnosis was confirmed by using cellular morphology and blast percentage by using established guidelines at the time of the diagnosis.51–54 Diagnostic bone marrow (BM, N = 124) and peripheral blood (PB, N = 116) from 190 out of 383 patients were randomly selected, that is, no specific algorithm, such as morphology, flow cytometry, and chromosomal abnormalities were employed when selecting the specimens for this study. All participants provided written informed consent to participate in research studies in compliance with the Declaration of Helsinki, and all studies were conducted with approval of the Fred Hutch Institution Review Board. Cytogenetics were available for 72% of evaluated patients.6,55,56 The specimen handling and cell processing of samples for correlative studies on these SWOG treatment protocols was consistent through the years of registration on the trials (SWOG-9031: 1992–1994, SWOG-9333: 1995–1998, S0106: 2004–2009, S0112: 2001–2003). Briefly, BM and PB samples were collected in SWOG-provided vacuum tubes containing RPMI 1640, 10% fetal calf serum, and EDTA (20 mg/m) and shipped overnight at room temperature to the processing laboratories. On receipt, MNCs were isolated via Ficoll-Hypaque density gradient under sterile conditions and cryopreserved in aliquots (1 mL volume in 2 mL cryovial) in 90% fetal calf serum and 10% dimethyl sulfoxide (DMSO). The cryopreserved aliquots of specimens were kept at −135°C until retrieval for studies. The time elapsed between specimen collection and processing was not recorded.57
Thawing of samples, FACS preparation and analyses, and nucleic acid extraction
Cryopreserved samples were thawed as previously described.57 Briefly, the vials with cryopreserved cells were thawed in a 37°C water bath and cells were quickly transferred into 15 mL falcon tubes. Prewarmed thawing media (20% fetal bovine serum in RPMI, both from Thermofisher Scientific, Waltham, MA) were slowly added to the thawed cells to gradually wash out the DMSO, all while continuously agitating the tube. Resuspended thawed cells were pelleted at 1200 rpm for 5 minutes, after which the supernatant was removed and the pellet was resuspended in FACS Buffer (Thermofisher Scientific). An aliquot of cells was diluted with Trypan Blue (Thermofisher Scientific) to assess the viability, and a portion of unsorted MNCs was lysed in RLT-Plus buffer (Qiagen, Valencia, CA) supplemented with beta-mercaptoethanol, a reducing agent that deactivates intracellular RNases (Sigma-Aldrich, St. Louis, MO). The rest of the MNCs were stained with CD45-APC-H7 (to identify lymphoid and myeloid populations, Beckman Coulter, Pasadena, CA), CD34-APC, and CD117-PE (to identify the immunophenotype of leukemic blasts, BioLegend, San Diego, CA) at recommended concentrations, incubated for 30 minutes, washed, resuspended in FACS buffer with 4,6-diamidino-2-phenylindole (DAPI) (to identify viable cells, Life Technologies), and sorted on BD FACSAria II instruments for viable AML blasts as described in Supplementary Methods (Supplementary Data are available online at www.liebertpub.com/bio). RNA and DNA from unsorted MNCs and enriched viable leukemic blast populations were extracted by using the AllPrep DNA/RNA Mini kit (Qiagen) and quantified by using the Nanodrop spectrophotometer (Thermofisher Scientific). RNA integrity number (RIN) was determined on Agilent 2200 TapeStation (Agilent Technologies, Santa Clara, CA).
Genomic and transcriptional biomarker assessment
Mutations in FLT3 and NPM1 genes are currently employed in prognosticating patients with AML. The presence of internal tandem duplication in FLT3 (FLT3-ITD) and insertion mutations in NPM1 were assayed as previously described.6,19 Allelic ratios (ARs) were computed as the ratio of the mutated product to the wild-type products.58 In the absence of the wild-type FLT3 allele, a value of 20, which represents approximately the highest AR previously described in such cases, was assigned.59 The expression of transcriptional biomarkers was evaluated by quantitative reverse transcription polymerase chain reaction (qRT-PCR) as previously described.60 Taqman gene expression assays for BAALC, CEBPA, CCNA1, CD34, ERG1, EVI1, FLT3, GATA2, IL3RA, JAG1, KIT, MN1, RUNX1, and WT1 were utilized to quantify the fold change (FC) by using housekeeping gene beta-glucuronidase (GUSB) to normalize expression and pooled nonmalignant PB MNCs as a calibrator, as previously described61 except for WT1 expression, which was calibrated to the WT1-expressing LAMA-84 cell line (Supplementary Methods and Supplementary Table S1).60,62 The FC was censored at maximal cycle threshold, Ct, of 45 for those without evidence of expression by qRT/PCR. Each gene was assayed in experimental duplicates, and the geometric mean of the replicates was used for downstream analyses. If either of the replicates was censored, their mean was also marked as censored. If only one replicate was available, we used the expression and censoring for that replicate.
Statistical analyses
Since specimen abundance was an eligibility criterion for this study, the 190 included patients were compared with the 852 who were excluded by random selection or by absence of a sufficient specimen. Patient and disease characteristics and treatment outcomes were compared between the included and excluded groups by using chi-square, Fisher's exact, or Wilcoxon rank-sum tests as appropriate. Overall survival (OS) and relapse-free survival (RFS) of included versus excluded patients were compared by using Cox regression models. Quantitative variables across two or more groups (e.g., viability by sex) were compared by using Wilcoxon rank-sum or Kruskal–Wallis tests, as appropriate. Spearman's correlation coefficient (rs) was used to evaluate associations between two quantitative variables. Paired t-tests were used to assess differences in mean expression in matched pairs of unsorted cells and enriched blasts from the same patient. Wilcoxon signed-rank tests were used to compare FLT3-ITD AR and NPM1 AR in matched pairs of enriched blasts and MNCs and in matched pairs of BM and PB samples. McNemar's test was used to compare the distribution of dichotomous FLT3-ITD AR (<0.5 vs. ≥0.5) in matched pairs of BM and PB samples.
The association of sorting and tissue type with log-transformed FLT3-ITD AR and NPM1 AR was evaluated by using linear mixed models, adjusting for age, year of patient registration to the clinical trial, BM cellularity, WBC count, BM and peripheral blasts (determined at diagnosis), hemoglobin, and platelet count. These models included a random effect for patients to account for correlation of AR values between specimen source (BM vs. PB) and subpopulations (unsorted MNCs vs. AML blasts) from the same patient. Gene expression was analyzed by using nonlinear mixed models with gene expression left-censored at the limit of detection and included a random effect for patients. Two-sided p-values were reported.
Results
Comparisons of included and excluded patient populations
Evaluated patients (N = 190) had significantly higher WBC counts and blast percentages than the not analyzed patients (p < 0.001 for both, Supplementary Table S2), which is likely an artifact that arose from the use of repository specimens, some of which have been heavily drawn on for previous studies; thus, the remaining specimens available for this study tended to be from patients with higher counts. There were more specimens from the S0106 study than other studies, given the larger number of available specimens from patients enrolled on this protocol (p < 0.001; Supplementary Table S2). Despite these differences, the analyzed and not analyzed patients did not significantly differ in clinical outcomes (complete remission [CR] rates, 63% vs. 61%, p = 0.62; OS, HR 1.06, p = 0.56; RFS, hazard ratio [HR] 1.14, p = 0.29; Supplementary Fig. S1).
Biologic and molecular heterogeneity of cryopreserved repository samples
Differences in immunophenotypes
Immunophenotype (IP) was evaluable for 187 out of 190 patients, with three samples displaying such poor viability that an accurate assessment of IP was not possible. Approximately half of the specimens (101/187, 54%) were CD34-expressing, with >10% of cells exceeding 1 × 104 mean fluorescent intensity (MFI), and with the remaining samples either expressing CD117 (35/187, 19%) at a predetermined cutoff of >4 × 103 MFI or not expressing either (51/187, 27%; Supplementary Fig. S2). The IPs were identical between the 50 paired BM and PB samples.
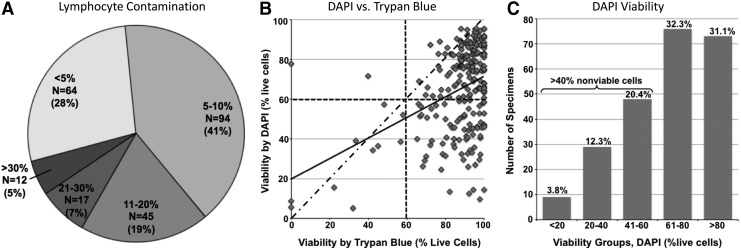
Marked variations in lymphocyte percentages
Given that Ficoll density gradient processing and subsequent cryopreservation and thawing eliminate polynucleated cells, we chose a defined population of MNCs that are not derived from the malignant leukemic stem cells as a marker of nonmalignant cell contamination. Monocytes, another component of cryopreserved MNCs of myeloid lineage, can be normal in origin or may represent a more differentiated population of malignant cells. Therefore, the percentage of lymphocytes in thawed MNCs was evaluated as a marker of nonmalignant cell contamination. Lymphocyte percentage varied substantially between samples (Fig. 1A) and was inversely correlated with the reported clinical blast percentage (BM, rs = −0.26, p = 0.004 and PB, rs = −0.37, p < 0.001). The lymphocyte percentage in the BM samples was significantly lower than in the PB (mean 9.0% [0.4%–36.8%] and 12.7% [1.6%–75.0%], respectively, p = 0.005). In the paired samples from the 50 patients, this trend was less significant, yet persisted, with the average BM lymphocyte percentage of 7.4% (1.3%–30.8%) and 11.9% (1.6%–75.0%) in PB (p = 0.03). Lymphocyte percentage did not vary significantly among the three IP groups.
FIG. 1.
Variation in cell composition and viability among the evaluated samples. (A) Figure shows the abundance of lymphocytes in cryopreserved MNCs. Thirty-two percent of specimens have >10% lymphocytes in unsorted MNCs. (B) Figure shows the relationship between viability determined by trypan blue (x-axis) and DAPI (y-axis) staining. However, most (∼70%) of the samples have similar viabilities via both methods; although trypan blue does overestimate viability relative to DAPI, >30% of samples have overestimated viabilities by trypan blue (bottom right quadrant). Solid line—actual trendline; dash-dotted line—expected trendline if two methods fully correlated; dashed lines—60% marks. (C) Figure shows the viability as defined by DAPI staining (% of DAPI-negative, i.e., viable cells, x-axis) relative to the number of cryopreserved AML SWOG samples (y-axis). More than one-third of samples (>37%) contain >40% dying or dead cells. AML, acute myeloid leukemia; DAPI, 4,6-diamidino-2-phenylindole; MNCs, mononuclear cells; SWOG, Southwest Oncology Group.
Decreased viability in large percentage of samples
The viability of thawed cells by Trypan Blue (manual count) and DAPI (FACS) was significantly correlated in the BM (rs = 0.40, p < 0.001), but it was not significant in the PB (rs = 0.16, p = 0.09). DAPI was more sensitive at detecting dead and dying cells, with 96% of BM and 89% of PB specimens having overestimation of viable cells by Trypan Blue (Fig. 1B), and DAPI was used in the remaining viability analyses. The cellular viability of BM and PB samples varied substantially (p < 0.001, mean 60.8% [5.2%–95.5%] and 69.4% [14.3%–95.8%], respectively). Further analyses in paired BM and PB from the same patients confirmed a decreased DAPI viability in BM versus PB (mean 64.3 and 70.4, respectively, p = 0.02). More than a third of the specimens were composed of ≥40% nonviable cells in unsorted MNCs (Fig. 1C).
The majority of the specimens reached the processing facility within 2 days of collection (i.e., the next day after collection, 91% of BM specimens, 85% of PB specimens), with the remaining samples taking 2 or more days. An analysis evaluating the impact of the delayed processing demonstrated a nonsignificant reduction in mean viability for samples processed <2 versus ≥2 days as measured by DAPI (64.4% vs. 57.7%, respectively, p = 0.14; Supplementary Table S3A). Viability was significantly correlated with the patient's age (p < 0.001 for BM, p < 0.001 for PB) and year of entry into clinical trial (p < 0.001 for BM, p < 0.001 for PB), and it varied significantly among the four trials (p < 0.001 for BM, p < 0.001 for PB; Supplementary Table S4). However, these three factors are very strongly associated with each other: Patients from studies SWOG-9031, SWOG-9333, and S0112, for age >55, were enrolled during 1992–2003; those from S0106, for age ≤60, were enrolled during 2004–2009. Thus, the older patients' specimens were collected during the earliest years, and the younger patients' specimens were collected more recently. Any changes in specimen handling at sites and the repository, as well as duration of cryopreservation, are, therefore, correlated with patient age. It is difficult to determine which one or more of these factors may explain the effect on viability. The viability also varied significantly among the three IP groups (p = 0.002 for BM, p < 0.001 for PB), being the highest in patient samples with CD34-expressing blasts and the poorest in CD34-CD117 blasts (Supplementary Table S5). The association of viability with FAB (French-American-British Classification system) was significant for PB (p = 0.04) but not BM (p = 0.26). The viability was not significantly associated with gender, race, performance status, or cytogenetic groups. Among the 50 patients with paired BM and PB specimens, viability was significantly correlated between the two sample sources (rs = 0.68, p < 0.001; Supplementary Fig. S3).
Impact of viability on RNA quality in unsorted MNCs and AML blasts
Increasing GUSB Ct, corresponding to degrading RNA, was correlated with decreasing specimen viability in both BM and PB specimens (rs = −0.42, p < 0.001, and rs = −0.47, p < 0.001, respectively). In addition, higher GUSB Ct was correlated with higher lymphocyte percentages in BM and PB specimens (rs = 0.35, p < 0.001, and rs = 0.28, p = 0.005, respectively). There were no significant associations between GUSB expression and IP groups, although most of the specimens with higher GUSB Ct were from patients with CD34-CD117- IP. Since the majority of specimens with GUSB Ct over 30 cycles had lower viability (<50%), downstream gene expression analyses were limited to specimens with GUSB Ct <30.
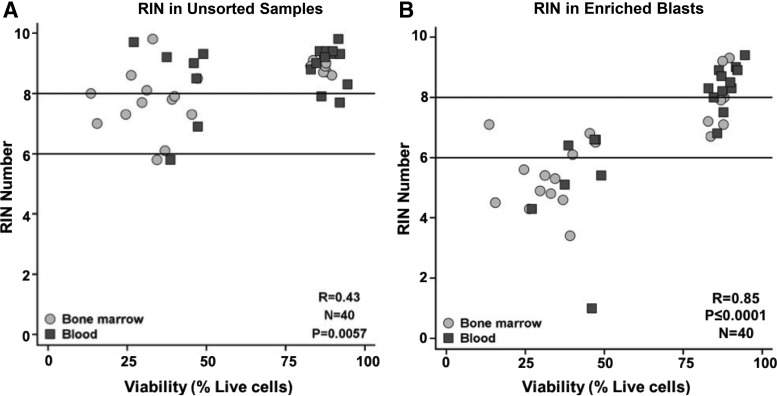
We then examined the global nucleic acid quality via RIN, with a higher RIN number associated with improved quality of RNA. For these analyses, we randomly selected 40 patients with RNA from both MNCs and enriched blasts, half from the lower tertile of DAPI viability and half from the upper tertile of DAPI viability. The RIN number for the MNCs was significantly correlated with viability in both BM and PB specimens (rs = 0.72, p < 0.001 and rs = 0.89, p < 0.001, respectively) and remained highly significant when BM and PB data from MNCs were combined for analyses (rs = 0.85, p < 0.001). RIN was not significantly associated with lymphocyte percentage in BM (p = 0.27), but it was modestly associated with lymphocyte percentage in PB (p = 0.02). The latter finding was primarily driven by two PB specimens with the highest lymphocytes and very low RINs. There were no significant associations between IP groups and RINs, although the RIN tended to be the lowest for patients within the CD34-CD117- group. Not surprisingly, there was an inverse relationship between RIN and GUSB Ct for MNCs from the BM (rs = −0.37, p = 0.11) and PB (rs = −0.59, p = 0.005). The positive impact of enrichment for viable blasts was readily demonstrated at the global RNA level—the RIN was on average 1.9-fold and 1.5-fold higher, respectively, in enriched blasts than unsorted cells (BM: 95% CI 1.2–2.6, p = 0.009, PB: 95% CI 0.4–2.6, p < 0.001; Fig. 2) and was not correlated with time in transit (Supplementary Table S3B).
FIG. 2.
Impact of enrichment for viable leukemic blasts on RNA quality. Figure shows the positive impact of enrichment on the global quality of RNA. Specimens in the lowest and highest tertiles of viability by DAPI are included. Figure shows the correlation between viability (DAPI, x-axis) and RIN (y-axis) in unsorted MNCs (A) and enriched blasts (B) from bone marrow (light gray circles) and peripheral blood (dark gray squares). The majority of specimens with poor viability and low RIN numbers (i.e., suboptimal RNA quality) in unsorted MNCs demonstrated an improvement in RIN numbers after enrichment for viable AML blasts (RIN <6, suboptimal RNA quality, lower black line; RIN >8, higher quality RNA, upper black line). RIN, RNA integrity number.
Impact of enrichment for AML blasts on mutation ARs
To investigate the impact of factors contributing to mutation ARs, initial analyses focused on the correlation of ARs derived from the 50 paired BM and PB samples. FLT3-ITD and NPM1 ARs, measured as continuous variables, were not significantly different between BM and PB in either unsorted MNCs (p = 0.67 for FLT3-ITD and p = 0.10 for NPM1-AR) or enriched AML blasts (p > 0.99 for FLT3-ITD and p = 0.55 for NPM1-AR). FLT3-ITD AR, when treated as a dichotomous variable (FLT3-ITD AR <0.5 vs. ≥0.5), was also not significantly different between BM and PB, whether examining MNCs or AML blasts (p = 0.32 and p = 0.56, respectively). Therefore, the mutation data from all available samples were combined for further analyses, using the BM data for the 50 patients with paired samples. The overall frequencies of mutations for all patients were 28.9% (54/187) for FLT3-ITDs and 32.6% (61/187) for NPM1 mutations. One patient demonstrated a complete loss of the wild-type FLT3 in the enriched blasts from BM and PB. FLT3-ITDs and NPM1 mutations were associated with normal cytogenetics, as well as with higher WBC and blast percentages (Supplementary Tables S6 and S7).
An increasing FLT3-ITD AR in unsorted MNCs was associated with higher WBC and BM blast% (p = 0.09 and p = 0.08, respectively; Supplementary Table S8). The mean FLT3-ITD AR was also significantly higher in AML blasts than in the MNCs from PB samples (p < 0.001), but not from the BM samples (p = 0.27). Similarly, increasing NPM1 AR in MNCs was significantly correlated with higher WBC and PB blast% (p = 0.003 and p = 0.01, respectively; Supplementary Table S9). The NPM1 ARs were also significantly higher in AML blasts than in unsorted MNCs from both the PB and BM (p < 0.001 for both). Higher lymphocyte percentage (i.e., inverse marker of blast percentage) was significantly associated with lower NPM1 AR in the MNCs (p < 0.001) and AML blasts (p < 0.001). There were no significant associations of ARs with any of the measures of specimen quality (i.e., viability) or IP subtype.
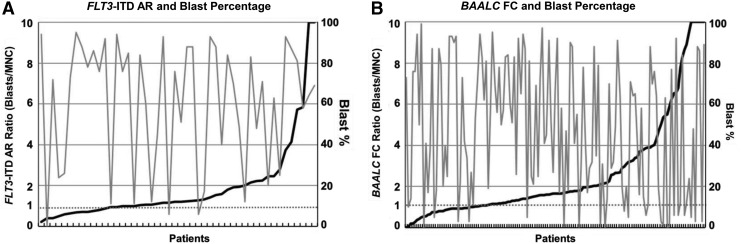
In linear mixed models, FLT3-ITD ARs in AML blasts were on average 1.49 times higher than ARs in unsorted MNCs (p < 0.001), but there was no significant difference between sources of sample (p = 0.57). Similarly, NPM1 AR was 1.14 times higher in AML blasts than in unsorted MNCs (p = 0.02) and there was no significant difference between BM and PB (p = 0.08). The increased AR in blasts relative to MNCs did not appear to be correlated with the clinically reported blast percentages (Fig. 3A).
FIG. 3.
Reported blast percentage does not explain differences in biomarker results in viable blasts and MNCs. (A) Figure shows the ratio of FLT3-ITD AR in enriched blasts relative to AR in unsorted MNCs (left y-axis, black solid line) and the reported blast percentage (right y-axis, gray solid line) in patients with FLT3-ITD mutations (N = 48, x-axis). The dashed line at 1 represents the ratio that one would expect if the AR was the same in both enriched viable blasts and unsorted MNCs. (B) Figure shows the ratio of FC of BAALC expression in viable blasts relative to unsorted MNCs (left y-axis, black solid line) and the reported blast percentage (right y-axis, gray solid line) in patients with a known blast percentage (N = 139, x-axis). The dashed line at 1 represents the ratio one that would expect if the FC was the same in both enriched viable blasts and unsorted MNCs. FC, fold change.
Impact of enrichment for AML blasts on expression of transcript biomarkers
To examine the impact of enrichment on expression, we selected a finite list of transcriptional biomarkers that had previously been associated with clinical outcomes.21,60,63 Variance component analyses of the qRT/PCR expression data assessed the intra- and inter-patient differences between the replicates for each sample source (BM vs. PB), cellular subpopulation (MNCs vs. AML blasts). The majority of the genes displayed intra-patient variance <25% of the total variance in MNCs and AML blasts, indicating a high correlation between the replicates (Supplementary Table S10).
Univariate analyses demonstrated that biomarker expression for all genes examined except EVI1 was significantly associated with FAB in both unsorted MNCs and AML blasts. CEBPA, CD34, ERG1, EVI1, KIT, and MN1 were significantly associated with cytogenetics in unsorted MNCs and AML blasts. Half of the biomarkers were significantly associated with normal cytogenetics in both unsorted MNCs and AML blasts, with RUNX1 only being significantly associated with normal cytogenetics in MNCs. BAALC, CCNA, ERG1, GATA2, IL3RA, JAG1, RUNX1, and WT1 were positively correlated with PB blast percentages in unsorted MNCs and AML blasts. EVI1, FLT3, and KIT in unsorted MNCs and CEBPA in enriched blasts were significantly correlated with PB blast percentage. Similarly, CCNA1, CD34, ERG1, IL3RA, JAG1, KIT, and MN1 were significantly correlated with WBC in MNCs and blasts, whereas EVI1 was significantly correlated with WBC in unsorted MNCs only (Supplementary Table S11).
Mixed-model analyses of gene expression were performed with cellular subpopulation as a fixed effect, allowing for censoring and intra-patient correlations and without adjustment for any other factors. In the BM specimens, BAALC and FLT3 were expressed at higher levels in AML blasts than MNCs (p < 0.001 and 0.004, respectively), whereas the expression of JAG1 was lower in the AML blasts (p < 0.001). In the PB samples, the expression of BAALC was significantly higher in AML blasts than MNCs (p = 0.01), whereas the expression of CEBPA and JAG1 was lower in AML blasts (p = 0.04 and 0.004) (Supplementary Table S12 and Supplementary Fig. S4). The increase or decrease in transcript expression in blasts relative to MNCs did not appear to correlate with the clinically reported blast percentages (Fig. 3B). The correlation of biomarker expression between BM and PB was quite high in unsorted MNCs (rs ≥0.79) and AML blasts (rs ≥0.69) from the same patient (Supplementary Table S13); thus, we combined expression data from BM and PB samples for further analyses.
Multivariate mixed-model analyses were fit for each gene, allowing the mean log expression levels to vary with the following factors: sample source (PB vs. BM), cellular subpopulation (AML Blasts vs. Total MNCs), cytogenetics (normal, other, rejected, not submitted), IP group (CD34+, CD34−CD117+, and CD34−CD117−), FLT3-ITD, and NPM1 mutation status (positive vs. negative) while allowing for correlation of expression between tissues and cellular subpopulations from the same patient (Table 1). Expression levels of CCNA1, FLT3, and JAG1 were significantly higher in BM than PB samples. BAALC and FLT3 expression levels were higher in AML blasts than in the unsorted MNCs, whereas the expression of CEBPA and JAG1 was lower in AML blasts than in the MNCs. The majority (10/14) of the genes' expression levels correlated with the IP of the leukemic blasts, such that the expression levels were higher in CD34+ cells than non-CD34-expressing samples. Expression levels of CCNA1, ERG1, FLT3, GATA2, IL3RA, and WT1 were significantly higher in samples with FLT3 mutations. The expression of BAALC, CD34, and MN1 was significantly lower in samples with NPM1 mutations, whereas the expression of CCNA1, CEBPA, FLT3, GATA2, JAG1, RUNX1, and WT1 was significantly higher in samples from patients with NPM1 mutations. The expression of WT1 was significantly lower in patients with normal cytogenetics.
Table 1.
Multivariable Mixed Model of Gene Expression Fold Changes
| Sample source | Subpopulation | Immunophenotype | FLT3 mutation | NPM1 mutation | Cytogenetics | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PB vs. BM | Blast vs. MNC | CD117 and other vs. CD34 | Pos vs. neg | Pos vs. neg | Oth, NS, and Rej vs. NN | ||||||||||
| Gene | Coef. | p | Coef. | p | 1 vs. 0 Coef. | 2 vs. 0 Coef. | p | Coef. | p | Coef. | p | Oth vs. NN Coef. | NS vs. NN Coef. | Rej vs. NN Corr. | p |
| BAALC | 1.10 | 0.48 | 1.57 | <0.001 | 0.25 | 0.07 | <0.001 | 1.37 | 0.27 | 0.21 | <0.001 | 1.39 | 0.98 | 0.98 | 0.67 |
| CCNA1 | 0.72 | 0.003 | 1.14 | 0.0742 | 0.75 | 0.37 | 0.06 | 2.11 | 0.03 | 5.00 | <0.001 | 0.48 | 1.21 | 0.99 | 0.14 |
| CD34 | 1.07 | 0.71 | 1.16 | 0.2864 | 0.02 | 0.01 | <0.001 | 1.49 | 0.27 | 0.23 | 0.001 | 1.09 | 1.43 | 2.52 | 0.24 |
| CEBPA | 0.93 | 0.29 | 0.89 | 0.0190 | 0.84 | 0.63 | 0.14 | 1.12 | 0.54 | 1.91 | 0.003 | 1.01 | 1.43 | 1.49 | 0.25 |
| ERG1 | 0.87 | 0.41 | 1.10 | 0.4464 | 0.33 | 0.09 | <0.001 | 2.01 | 0.01 | 0.92 | 0.80 | 1.36 | 1.05 | 1.39 | 0.65 |
| EVI1 | 0.27 | 0.05 | 1.61 | 0.2452 | 0.88 | 0.53 | 0.94 | 2.02 | 0.63 | 0.12 | 0.21 | 5.8 | 0.52 | 0.10 | 0.23 |
| FLT3 | 0.78 | 0.001 | 1.11 | 0.0347 | 0.77 | 0.59 | 0.05 | 1.47 | 0.03 | 1.62 | 0.02 | 1.25 | 1.21 | 1.32 | 0.53 |
| GATA2 | 1.22 | 0.13 | 0.90 | 0.2585 | 0.48 | 0.18 | <0.001 | 2.23 | 0.003 | 2.85 | 0.001 | 1.3 | 1.98 | 2.07 | 0.11 |
| IL3RA | 1.22 | 0.09 | 0.91 | 0.2902 | 0.83 | 0.41 | 0.001 | 1.83 | 0.002 | 1.47 | 0.09 | 1.04 | 1.15 | 1.06 | 0.97 |
| JAG1 | 0.60 | 0.01 | 0.46 | <0.001 | 1.84 | 1.71 | 0.55 | 1.53 | 0.36 | 7.88 | <0.001 | 2.77 | 2.46 | 3.07 | 0.13 |
| KIT | 0.80 | 0.07 | 1.01 | 0.8941 | 0.51 | 0.11 | <0.001 | 1.34 | 0.21 | 1.02 | 0.95 | 1.68 | 1.48 | 2.12 | 0.05 |
| MN1 | 1.22 | 0.30 | 1.04 | 0.7599 | 0.17 | 0.04 | <0.001 | 1.24 | 0.55 | 0.34 | 0.01 | 1.2 | 1.58 | 1.76 | 0.60 |
| RUNX1 | 0.90 | 0.37 | 0.99 | 0.9097 | 0.47 | 0.41 | <0.001 | 1.35 | 0.08 | 1.62 | 0.02 | 1.27 | 1.45 | 1.60 | 0.14 |
| WT1 | 0.95 | 0.82 | 1.15 | 0.3540 | 0.11 | 0.03 | <0.001 | 6.20 | 0.001 | 13.49 | <0.001 | 2.23 | 8.90 | 3.89 | 0.04 |
Results of multivariate mixed-model analyses fit for each gene, allowing the mean log expression levels to vary with the following factors: sample source (PB vs. BM), cellular subpopulation (Blasts vs. MNCs), cytogenetics (normal [NN], other [Oth, any abnormal clone], rejected [Rej], not submitted [NS]), IP group (coded in this table as 0 = CD34+, 1 = CD34−CD117+, and 2 = Other, CD34−CD117−), FLT3-ITD, and NPM1 mutation status (positive [Pos] vs. negative [Neg]) while allowing for correlation of expression between sample sources and cellular subpopulations from the same patient. Entries in the table are average fold changes between categories of each factor (coef. = coefficient).
BM, bone marrow; MNCs, mononuclear cells; PB, peripheral blood.
Discussion
This is the first article that systematically examined the quality of AML specimens from a large cooperative group repository. Our studies demonstrated that the cryopreserved SWOG AML samples display wide heterogeneity in viability, nonleukemic cell percentage, and overall differentiation status of the leukemic blasts. Time to processing (in hours) could not be used as a continuous variable because it was not measured when these specimens were collected, but our analyses suggest that samples that reached processing facilities within a day after collection (i.e., within 2 days of draw) tended to have greater percentages of viable cells. Further, our results suggest that other factors may impact the quality of specimens, such as the age of patients, AML blast immunophenotype, and duration of storage. Our results suggested that MNCs from PB demonstrated higher viability after thawing than MNCs from the BM. There are multiple factors that can contribute to this finding. There may have been differences between the time of collection and processing for BM and PB, even though they were shipped at the same time. Another possibility is that the differences in volumes collected for BM and PB specimens may have skewed concentrations of vacutainer contents, thus impacting the viability. Lastly, the viability of the MNCs was assessed on the total MNCs, and PB contained higher percentages of lymphocytes that are more resilient to freeze/thaw,64 thus masking the signature of leukemic biomarkers. In addition, we demonstrated that the quality of the specimens and the heterogeneity of the examined cells significantly impacted biomarker results. Thus, investigators need to recognize these potential limitations when using cryopreserved samples for biomarker studies.
Despite the heterogeneity of cryopreserved AML specimens and its impact on biomarkers, paired BM and PB specimens provided similar results. The two specimen sources correlated with respect to cell viability, immunophenotype, and quality of RNA. Further, the performance of genomic and transcriptional biomarkers was comparable between BM and PB. ARs for both FLT3-ITD and NPM1 in paired BM and PB samples were highly correlated, whether examining the unsorted MNCs or enriched viable AML blasts. Similarly, most of the transcript biomarkers demonstrated a high degree of correlation between the two sample sources. This has practical implications in that PB can be reliably utilized in the absence of diagnostic BM, the more coveted sample source. These findings are consistent with previous observations of genomic mutations in MDS and methylation markers in AML.65,66
We acknowledge that researchers will continue to rely on specimens collected through cooperative group repositories to obtain statistical power and patient heterogeneity for many types of studies, including biomarker discovery and validation. In some cases, these samples have been collected over a large period from multiple trials. For example, the earliest SWOG leukemia samples were collected in 1983 and are still available for use. Our results and data from other studies examining different tissue types provide solid support for the efforts to standardize and harmonize the preanalytical variables across biorepositories.32,50 Biorepositories and Biospecimen Research Branch (BBRB), which was founded in 2005 by NCI, is leading a national initiative to provide guidance to biorepositories on collection, handling, and annotation of specimens for broad investigational use.
As demonstrated in our study, preanalytical variables are not the only factors that may impact biomarker results. There were associations between viability and other factors, including the immunophenotype of AML blasts and the percentage of nonleukemic cells within the specimen. For example, the specimens from patients with more differentiated blasts (i.e., CD34−CD117−) tended to have a low viability post-thaw, as has been previously described.46 In contrast, the specimens from patients with less differentiated AML blasts (i.e., CD34+) were not as impacted by cryopreservation and thawing. Unfortunately, the immunophenotype at diagnosis was not available for the majority of patient samples, and thus, we were unable to determine how sampling, processing, and thawing may impact immunophenotype.
The results suggest that contaminating nonleukemic cells can alter the signal of genomic and transcript biomarkers. Although these findings may be expected, the implications of the findings are highly relevant to current biomarker tests, especially given the emerging clinical importance of quantitative genomic (e.g., FLT3-ITD AR) and expression biomarkers.67–69 Unfortunately, the reported blast percentage showed no association with AML blasts/MNCs ratios for the examined biomarkers (Fig. 3A, B), limiting the ability to accurately correct biomarker signals through the use of reported blast percentage. Potential explanations for the lack of relationship between reported blast percentage and biomarker results from cryopreserved samples may be due to multiple factors: hemodilution secondary to multiple aspirations, variability in defining the blast population, and the heterogeneity of the blast survival during processing. Given the clinical influence of quantitative biomarkers on the risk-stratification of AML patients, it is imperative to determine whether biomarkers are more informative in viable AML blasts rather than the MNCs that are presently being tested. In addition, it will be imperative to evaluate similar studies using freshly collected and noncryopreserved cells, given that the vast majority of clinical assays utilize fresh samples containing granulocytes and, thus, harboring even larger numbers of nonleukemic cells.
The results, however, show that the enrichment of specimens for viable AML blasts allows one to salvage specimens with poor viability for downstream analyses such as RNA sequencing. In our study, 92% of MNC specimens with low viability and poor RNA quality (RIN <6) displayed marked improvements in RNA quality after selecting for viable blasts (Fig. 2).70 These findings suggest that one can improve the quality of RNA from samples with moderately poor viability by selecting for viable blasts, and some of these poor viability samples can then readily meet quality controls criteria for assays such as RNA sequencing. The implications of these findings are that investigators may be able to increase the number of potentially useful samples through such preprocessing methods, which would improve the power to detect prognostic factors and reduce potential transcriptional noise caused by dying cells.
Although the study provides strong support for examining more restricted viable AML blasts, there are some natural limitations that require further investigations. Our study focused on a patient population from a single cooperative group and multiple clinical trials, which spanned over two decades of collecting, processing, storing, and curating samples. Changes in methods and technologies for repositories are evolving, such that the demonstrated heterogeneity in specimen characteristics in our report may not be generalizable to all repositories, and certainly, the results may not be applicable to research specimens collected for ongoing and future trials.
We restricted randomly selected samples from patients with three or more vials. The rationale behind this restriction and the number of examined samples was practical in nature, given the resources required and the potential number of needed vials for the studies. If there were no major differences in biomarkers between MNCs and AML blasts in this set of samples, it may be difficult to justify pursuing larger studies looking at enriching for viable AML blasts. Despite the random selection, there was a bias for patients with higher WBC and blast percentage in the examined samples. It is conceivable that the biological variables examined could differ between the included and excluded samples. However, the clinical outcomes were similar between the two groups. Most discovery and validation studies for biomarkers in AML still utilize cryopreserved material with an understanding that “fresh” materials may provide different results.47,71 Such prospective analyses are extremely difficult to perform in large numbers of samples in the cooperative group setting. Unfortunately, fresh material was not available for the samples in this study.
The genomic mutations for this analysis were limited to NPM1 and FLT3-ITD ARs, displaying significant differences in the MNCs and AML blasts—especially for FLT3-ITD. It is conceivable that other mutations that occur earlier in the course of leukemogenesis (e.g., DNMT3A and TET2) may actually be present in more differentiated leukemic cells as well as in the leukemic blast population. In this case, enriching for AML blasts may not have as great of an impact for the identification and quantification of these driver mutations. Nevertheless, these results highlight overlooked and potentially uncontrollable variables that may impact biomarker discovery and validation, especially given that the vast majority of previous studies relied on cryopreserved specimens. Hence, there is a need to systematically evaluate the heterogeneity of cryopreserved specimens and its impact on biomarker performance.
Supplementary Material
Acknowledgments
The authors wish to gratefully acknowledge the important contributions of the late Dr. Stephen H. Petersdorf to SWOG and to the study S0106. The development of flow sorting assays, blast enrichment, and biomarker assay development was accomplished with the use of specimens from the Fred Hutch and University of Washington Leukemia Repository. Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Numbers CA160872, CA114563, CA180861, CA180819, CA180828, and CA180888. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Authorship Contributions
Concept and design: E.L.P.A., A.M., M.O., J.P.R., S.M., and D.L.S.; Financial support: S.M. and D.L.S.; Provision of study materials or patients: F.R.A., T.C., I.M.L.C., H.P.E., J.E.G., A.F.L., J.P.R., C.L.W., and D.L.S.; Collection and assembly of the data: E.L.P.A., A.M., M.O., I.M.L.C., M.F., K.J.K., G.L.P., C.L.W., B.L.W., S.M., and D.L.S.; Data analysis and interpretation: E.L.P.A., A.M., M.O., F.R.A., T.C., I.M.L.C., H.P.E., J.E.G., M.F., K.J.K., A.F.L., G.L.P., J.P.R., C.L.W., B.L.W., S.M., and D.L.S.; Manuscript writing: E.L.P.A., A.M., M.O., F.R.A., T.C., I.M.L.C., H.P.E., J.E.G., M.F., K.J.K., A.F.L., G.L.P., J.P.R., C.L.W., B.L.W., S.M., and D.L.S.; Final approval of the article: E.L.P.A., A.M., M.O., F.R.A., T.C., I.M.L.C., H.P.E., J.E.G., M.F., K.J.K., A.F.L., G.L.P., J.P.R., C.L.W., B.L.W., S.M., and D.L.S.
Author Disclosure Statement
No conflicting financial interests exist.
References
- 1.Stein EM, Tallman MS. Emerging therapeutic drugs for AML. Blood 2016;127:71–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Donnell MR, Abboud CN, Altman J, et al. Acute myeloid leukemia. J Natl Compr Canc Netw 2012;10:984–1021 [DOI] [PubMed] [Google Scholar]
- 3.Mrozek K, Marcucci G, Nicolet D, et al. Prognostic significance of the European LeukemiaNet standardized system for reporting cytogenetic and molecular alterations in adults with acute myeloid leukemia. J Clin Oncol 2012;30:4515–4523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornelissen JJ, Blaise D. Hematopoietic stem cell transplantation for patients with AML in first complete remission. Blood 2016;127:62–70 [DOI] [PubMed] [Google Scholar]
- 5.Meshinchi S, Woods WG, Stirewalt DL, et al. Prevalence and prognostic significance of Flt3 internal tandem duplication in pediatric acute myeloid leukemia. Blood 2001;97:89–94 [DOI] [PubMed] [Google Scholar]
- 6.Stirewalt DL, Kopecky KJ, Meshinchi S, et al. FLT3, RAS, and TP53 mutations in elderly patients with acute myeloid leukemia. Blood 2001;97:3589–3595 [DOI] [PubMed] [Google Scholar]
- 7.Preudhomme C, Sagot C, Boissel N, et al. Favorable prognostic significance of CEBPA mutations in patients with de novo acute myeloid leukemia: A study from the Acute Leukemia French Association (ALFA). Blood 2002;100:2717–2723 [DOI] [PubMed] [Google Scholar]
- 8.Schnittger S, Schoch C, Dugas M, et al. Analysis of FLT3 length mutations in 1003 patients with acute myeloid leukemia: Correlation to cytogenetics, FAB subtype, and prognosis in the AMLCG study and usefulness as a marker for the detection of minimal residual disease. Blood 2002;100:59–66 [DOI] [PubMed] [Google Scholar]
- 9.Thiede C, Koch S, Creutzig E, et al. Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML). Blood 2006;107:4011–4020 [DOI] [PubMed] [Google Scholar]
- 10.Bullinger L, Rucker FG, Kurz S, et al. Gene-expression profiling identifies distinct subclasses of core binding factor acute myeloid leukemia. Blood 2007;110:1291–1300 [DOI] [PubMed] [Google Scholar]
- 11.Marcucci G, Maharry K, Whitman SP, et al. High expression levels of the ETS-related gene, ERG, predict adverse outcome and improve molecular risk-based classification of cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B Study. J Clin Oncol 2007;25:3337–3343 [DOI] [PubMed] [Google Scholar]
- 12.Gale RE, Green C, Allen C, et al. The impact of FLT3 internal tandem duplication mutant level, number, size, and interaction with NPM1 mutations in a large cohort of young adult patients with acute myeloid leukemia. Blood 2008;111:2776–2784 [DOI] [PubMed] [Google Scholar]
- 13.Langer C, Marcucci G, Holland KB, et al. Prognostic importance of MN1 transcript levels, and biologic insights from MN1-associated gene and microRNA expression signatures in cytogenetically normal acute myeloid leukemia: A cancer and leukemia group B study. J Clin Oncol 2009;27:3198–3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Damm F, Oberacker T, Thol F, et al. Prognostic importance of histone methyltransferase MLL5 expression in acute myeloid leukemia. J Clin Oncol 2011;29:682–689 [DOI] [PubMed] [Google Scholar]
- 15.Staffas A, Kanduri M, Hovland R, et al. Presence of FLT3-ITD and high BAALC expression are independent prognostic markers in childhood acute myeloid leukemia. Blood 2011;118:5905–5913 [DOI] [PubMed] [Google Scholar]
- 16.Patel JP, Gonen M, Figueroa ME, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med 2012;366:1079–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaidzik VI, Schlenk RF, Paschka P, et al. Clinical impact of DNMT3A mutations in younger adult patients with acute myeloid leukemia: Results of the AML Study Group (AMLSG). Blood 2013;121:4769–4777 [DOI] [PubMed] [Google Scholar]
- 18.Damm F, Markus B, Thol F, et al. TET2 mutations in cytogenetically normal acute myeloid leukemia: Clinical implications and evolutionary patterns. Genes Chromosomes Cancer 2014;53:824–832 [DOI] [PubMed] [Google Scholar]
- 19.Ostronoff F, Othus M, Lazenby M, et al. Prognostic significance of NPM1 mutations in the absence of FLT3-internal tandem duplication in older patients with acute myeloid leukemia: A SWOG and UK National Cancer Research Institute/Medical Research Council report. J Clin Oncol 2015;33:1157–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson CS, Davidson GS, Martin SB, et al. Gene expression profiling of adult acute myeloid leukemia identifies novel biologic clusters for risk classification and outcome prediction. Blood 2006;108:685–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rockova V, Abbas S, Wouters BJ, et al. Risk stratification of intermediate-risk acute myeloid leukemia: Integrative analysis of a multitude of gene mutation and gene expression markers. Blood 2011;118:1069–1076 [DOI] [PubMed] [Google Scholar]
- 22.Marcucci G, Yan P, Maharry K, et al. Epigenetics meets genetics in acute myeloid leukemia: Clinical impact of a novel seven-gene score. J Clin Oncol 2014;32:548–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engel KB, Vaught J, Moore HM. National Cancer Institute Biospecimen Evidence-Based Practices: A novel approach to pre-analytical standardization. Biopreserv Biobank 2014;12:148–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeMartino JK. NCCN Work Group Report: Emerging issues in tissue allocation. J Natl Compr Canc Netw 2016;14:265–271 [DOI] [PubMed] [Google Scholar]
- 25.Institute NC. Biorepositories and Biospecimen Research Branch. Vol. 2016 Rockville, MD: National Cancer Institute, NIH; 2016 [Google Scholar]
- 26.Lehmann S, Guadagni F, Moore H, et al. Standard preanalytical coding for biospecimens: Review and implementation of the Sample PREanalytical Code (SPREC). Biopreserv Biobank 2012;10:366–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Betsou F, Gunter E, Clements J, et al. Identification of evidence-based biospecimen quality-control tools: A report of the International Society for Biological and Environmental Repositories (ISBER) Biospecimen Science Working Group. J Mol Diagn 2013;15:3–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matzke EA, O'Donoghue S, Barnes RO, et al. Certification for biobanks: The program developed by the Canadian Tumour Repository Network (CTRNet). Biopreserv Biobank 2012;10:426–432 [DOI] [PubMed] [Google Scholar]
- 29.Barnes R, Albert M, Damaraju S, et al. Generating a comprehensive set of standard operating procedures for a biorepository network-The CTRNet experience. Biopreserv Biobank 2013;11:387–396 [DOI] [PubMed] [Google Scholar]
- 30.Langseth H, Luostarinen T, Bray F, Dillner J. Ensuring quality in studies linking cancer registries and biobanks. Acta Oncol 2010;49:368–377 [DOI] [PubMed] [Google Scholar]
- 31.Yong WH, Dry SM, Shabihkhani M. A practical approach to clinical and research biobanking. Methods Mol Biol 2014;1180:137–162 [DOI] [PubMed] [Google Scholar]
- 32.Caixeiro NJ, Lai K, Lee CS. Quality assessment and preservation of RNA from biobank tissue specimens: A systematic review. J Clin Pathol 2016;69:260–265 [DOI] [PubMed] [Google Scholar]
- 33.Betsou F, Bulla A, Cho SY, et al. Assays for qualification and quality stratification of clinical biospecimens used in research: A technical report from the ISBER Biospecimen Science Working Group. Biopreserv Biobank 2016;14:398–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miranda LB, Wyatt K, Johnston I, Milljanic M, Chaffey J. “Proof of concept” pilot study: Bioprocess chain of custody and bioresource sample management temperature observations. Sample level temperature trends and stability data obtained via utilization of bluechiip((R)) temperature tracking technology. Biopreserv Biobank 2013;11:115–121 [DOI] [PubMed] [Google Scholar]
- 35.Sun J, Kil C, Stankewich MC, Yao Z, Li J, Vortmeyer AO. A 10-minute prototype assay for tissue degradation monitoring in clinical specimens. Exp Mol Pathol 2015;99:86–94 [DOI] [PubMed] [Google Scholar]
- 36.Kofanova OA, Mathieson W, Thomas GA, Betsou F. DNA fingerprinting: A quality control case study for human biospecimen authentication. Biopreserv Biobank 2014;12:151–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore HM, Kelly A, Jewell SD, et al. Biospecimen reporting for improved study quality. Biopreserv Biobank 2011;9:57–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alsayed H, Owaidah T, Al Rawas F. Validation of a modified cryopreservation method for leukemic blasts for flow cytometry assessment. Hematol Oncol Stem Cell Ther 2008;1:94–97 [DOI] [PubMed] [Google Scholar]
- 39.Ellervik C, Vaught J. Preanalytical variables affecting the integrity of human biospecimens in biobanking. Clin Chem 2015;61:914–934 [DOI] [PubMed] [Google Scholar]
- 40.Hubel A, Spindler R, Skubitz AP. Storage of human biospecimens: Selection of the optimal storage temperature. Biopreserv Biobank 2014;12:165–175 [DOI] [PubMed] [Google Scholar]
- 41.Sewart S, Barraclough R, Rudland PS, West CR, Barraclough DL. Molecular analysis of a collection of clinical specimens stored at 4 degrees C as an alternative to snap-freezing. Int J Oncol 2009;35:381–386 [PubMed] [Google Scholar]
- 42.Kofanova OA, Davis K, Glazer B, et al. Viable mononuclear cell stability study for implementation in a proficiency testing program: Impact of shipment conditions. Biopreserv Biobank 2014;12:206–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olson WC, Smolkin ME, Farris EM, et al. Shipping blood to a central laboratory in multicenter clinical trials: Effect of ambient temperature on specimen temperature, and effects of temperature on mononuclear cell yield, viability and immunologic function. J Transl Med 2011;9:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greytak SR, Engel KB, Bass BP, Moore HM. Accuracy of molecular data generated with FFPE biospecimens: Lessons from the literature. Cancer Res 2015;75:1541–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bass BP, Engel KB, Greytak SR, Moore HM. A review of preanalytical factors affecting molecular, protein, and morphological analysis of formalin-fixed, paraffin-embedded (FFPE) tissue: How well do you know your FFPE specimen? Arch Pathol Lab Med 2014;138:1520–1530 [DOI] [PubMed] [Google Scholar]
- 46.Lowenthal RM, Park DS, Goldman JM, Hill RS, Whyte G, Th'ng KH. The cryopreservation of leukaemia cells: Morphological and functional changes. Br J Haematol 1976;34:105–117 [DOI] [PubMed] [Google Scholar]
- 47.Lanza F, Moretti S, Castagnari B, et al. Assessment of distribution of CD34 epitope classes in fresh and cryopreserved peripheral blood progenitor cells and acute myeloid leukemic blasts. Haematologica 1999;84:969–977 [PubMed] [Google Scholar]
- 48.Campos L, Guyotat D, Larese A, et al. Expression of immunological markers on leukemic cells before and after cryopreservation and thawing. Cryobiology 1988;25:18–22 [DOI] [PubMed] [Google Scholar]
- 49.Radich JP, Mao M, Stepaniants S, et al. Individual-specific variation of gene expression in peripheral blood leukocytes. Genomics 2004;83:980–988 [DOI] [PubMed] [Google Scholar]
- 50.Dvinge H, Ries RE, Ilagan JO, Stirewalt DL, Meshinchi S, Bradley RK. Sample processing obscures cancer-specific alterations in leukemic transcriptomes. Proc Natl Acad Sci U S A 2014;111:16802–16807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anderson JE, Kopecky KJ, Willman CL, et al. Outcome after induction chemotherapy for older patients with acute myeloid leukemia is not improved with mitoxantrone and etoposide compared to cytarabine and daunorubicin: A Southwest Oncology Group study. Blood 2002;100:3869–3876 [DOI] [PubMed] [Google Scholar]
- 52.Petersdorf SH, Rankin C, Head DR, et al. Phase II evaluation of an intensified induction therapy with standard daunomycin and cytarabine followed by high dose cytarabine for adults with previously untreated acute myeloid leukemia: A Southwest Oncology Group study (SWOG-9500). Am J Hematol 2007;82:1056–1062 [DOI] [PubMed] [Google Scholar]
- 53.Godwin JE, Kopecky KJ, Head DR, et al. A double-blind placebo-controlled trial of granulocyte colony-stimulating factor in elderly patients with previously untreated acute myeloid leukemia: A Southwest oncology group study (9031). Blood 1998;91:3607–3615 [PubMed] [Google Scholar]
- 54.List AF, Kopecky KJ, Willman CL, et al. Benefit of cyclosporine modulation of drug resistance in patients with poor-risk acute myeloid leukemia: A Southwest Oncology Group study. Blood 2001;98:3212–3220 [DOI] [PubMed] [Google Scholar]
- 55.Stirewalt DL, Kopecky KJ, Meshinchi S, et al. Size of FLT3 internal tandem duplication has prognostic significance in patients with acute myeloid leukemia. Blood 2006;107:3724–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ostronoff F, Othus M, Ho PA, et al. Mutations in the DNMT3A exon 23 independently predict poor outcome in older patients with acute myeloid leukemia: A SWOG report. Leukemia 2013;27:238–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leith CP, Chen IM, Kopecky KJ, et al. Correlation of multidrug resistance (MDR1) protein expression with functional dye/drug efflux in acute myeloid leukemia by multiparameter flow cytometry: Identification of discordant MDR-/efflux+ and MDR1+/efflux- cases. Blood 1995;86:2329–2342 [PubMed] [Google Scholar]
- 58.Meshinchi S, Alonzo TA, Stirewalt DL, et al. Clinical implications of FLT3 mutations in pediatric AML. Blood 2006;108:3654–3661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thiede C, Steudel C, Mohr B, et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: Association with FAB subtypes and identification of subgroups with poor prognosis. Blood 2002;99:4326–4335 [DOI] [PubMed] [Google Scholar]
- 60.Stirewalt DL, Meshinchi S, Kopecky KJ, et al. Identification of genes with abnormal expression changes in acute myeloid leukemia. Genes Chromosomes Cancer 2008;47:8–20 [DOI] [PubMed] [Google Scholar]
- 61.Pogosova-Agadjanyan EL, Kopecky KJ, Ostronoff F, et al. The prognostic significance of IRF8 transcripts in adult patients with acute myeloid leukemia. PLoS One 2013;8:e70812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402–408 [DOI] [PubMed] [Google Scholar]
- 63.Bruserud O, Reikvam H, Fredly H, et al. Expression of the potential therapeutic target CXXC5 in primary acute myeloid leukemia cells - high expression is associated with adverse prognosis as well as altered intracellular signaling and transcriptional regulation. Oncotarget 2015;6:2794–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Verschoor CP, Kohli V, Balion C. A comprehensive assessment of immunophenotyping performed in cryopreserved peripheral whole blood. Cytometry B Clin Cytom 2017. [Epub ahead of print]; http://onlinelibrary.wiley.com/doi/10.1002/cyto.b.21526/epdf (accessed November7, 2017) [DOI] [PubMed]
- 65.Mohamedali AM, Alkhatabi H, Kulasekararaj A, et al. Utility of peripheral blood for cytogenetic and mutation analysis in myelodysplastic syndrome. Blood 2013;122:567–570 [DOI] [PubMed] [Google Scholar]
- 66.Qu X, Othus M, Davison J, et al. Prognostic methylation markers for overall survival in cytogenetically normal patients with acute myeloid leukemia treated on SWOG trials. Cancer 2017;123:2472–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dohner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017;129:424–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ho PA, Alonzo TA, Gerbing RB, et al. High EVI1 expression is associated with MLL rearrangements and predicts decreased survival in paediatric acute myeloid leukaemia: A report from the children's oncology group. Br J Haematol 2013;162:670–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kuhnl A, Valk PJ, Sanders MA, et al. Downregulation of the Wnt inhibitor CXXC5 predicts a better prognosis in acute myeloid leukemia. Blood 2015;125:2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gallego Romero I, Pai AA, Tung J, Gilad Y. RNA-seq: Impact of RNA degradation on transcript quantification. BMC Biol 2014;12:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aasebo E, Mjaavatten O, Vaudel M, et al. Freezing effects on the acute myeloid leukemia cell proteome and phosphoproteome revealed using optimal quantitative workflows. J Proteomics 2016;145:214–225 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.