The ancient Greeks attributed life to Mother Nature, complaining that she was fond of hiding the workings of her creations. Although we no longer attribute the mysteries of life to Mother Nature, multiple facets of living systems still evade us. The complexity of the human proteome and proteoform families are examples. Until recently we believed that each of the ~20 000 protein-coding genes in humans produced a single protein,1 assuming this small number of proteins was sufficient to sustain life. Wrong! We were off by 1–2 orders of magnitude. The human proteome is more likely to be made up of a million protein species.2–4 Then there is the question of how these proteins arise and what they do. Mass spectrometry (MS) has been enormously helpful in proteomics, but gas phase ions do not reveal in vivo biosynthesis and function. Isolating proteins will still be necessary to establish their 3D structure and function.
The discussion below is directed toward analytical and separation strategies involved in the discovery and identification of previously unidentified proteins, necessitating the focus on genes and histones found below. Monitoring changes in the structure and concentration of known proteins as part of cellular regulation and diagnostics is an equally important part of proteomics but not addressed in this review.
Genomic Component of Protein Synthesis
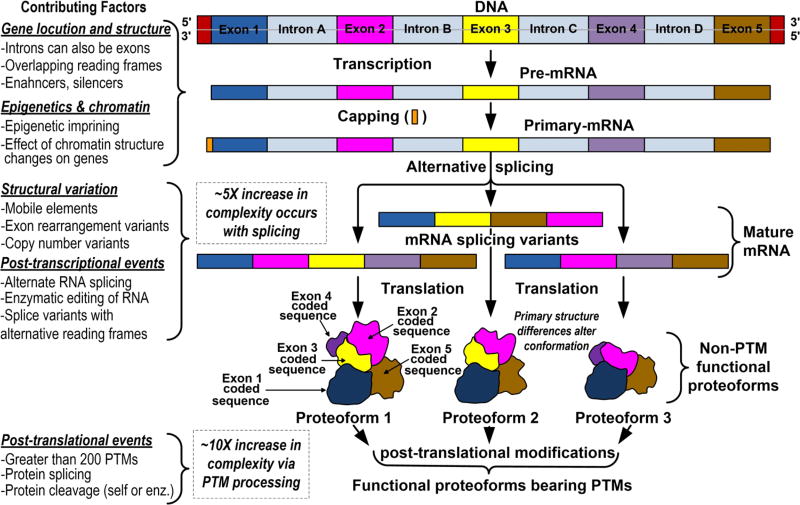
The question of how individual proteins arise and even the definition of a gene are still in flux. A gene is being defined herein as the “union of genomic sequences encoding a coherent set of potentially overlapping functional products5”, those gene sequences being exons in protein-coding genes. An exon from a single gene can supply sequence code for multiple proteins6 (Figure 1). Proteins in these genetically related families of structural isoforms are referred to in general as “proteoforms”.7
Figure 1.
Origin of proteoform complexity. Illustrations of a gene, RNA species, and proteoforms above are intended to show the origin and positioning of structural elements; not actual structures. The “contributing factors” column shows that individual steps in the illustration can occur in multiple ways.
Our current understanding of the origin of proteoform families is that their synthesis starts with DNA transcription via the formation of pre- and primary-mRNA species (Figure 1). The process involves a combination of intron excisions,8 exon rearrangements9 and/or shuffling,10 exon fusion,11 RNA copy number regulation,12 and epigenetic imprinting,13 enabled by a series of enhancers14 and silencers.15 How these various components collaborate in proteoform synthesis varies between proteins, organelles, tissues, and organisms.
Post-transcriptional processing subsequently leads to the production of mature mRNA (Figure 1), the process being accompanied by variations in splicing,16 enzymatic editing,17 and reading frame shifts.18 Five or more mRNA species on average can arise from a single protein-coding gene during the course of these processing steps, each of which produce different proteins. Alternative splicing of mRNA, single amino acid polymorphisms (SAPs), and post-translational modifications (PTMs)19 play an additional role in proteoform formation. Clearly the potential for variation and regulatory control within these processes is large.
The objective of the illustration in Figure 1 is not to provide a detailed mechanism by which protein isoforms are synthesized. Well known steps involved in mRNA translation and post-translational modifications along with how cotranslational translocation20,21 occurences are omitted. The figure is meant to reveal the multiple routes by which proteoform complexity can arise at the DNA and mRNA levels along with more than 200 types of PTMs.22 Even higher levels of complexity arise from sequential modification of a PTM, as with glycoproteins.23 Serial addition of different monosaccharides along with glycan branching as glycoproteins pass through the Golgi can lead to the production of an additional 10–50 glycoforms of a protein. PTMs collectively increase the complexity of proteoform families ~10× beyond the 5× increase provided by transcriptional processing. At the extreme, a proteoform family can be composed of 50 or more members.24 This is the secret to how so many proteins are fabricated from such a small number of protein-coding genes. It reveals the magnitude of the separation problems that will be encountered in isolating these closely related species.
An important ramification of Figure 1 is that proteoforms derived from the same protein-coding gene will have multiple structural features that are identical, although not necessarily at the same location in the 3D structure of all proteoform family members. It is important that structure and function are likely to be proteoform specific; an issue best examined with pure proteins. Validated analytical methods that qualitatively and quantitatively differentiate between proteoforms are also best established with pure proteins. Confirmation that an analytical method detects a single proteoform is critical.
Epigenetics and the Nucleosome
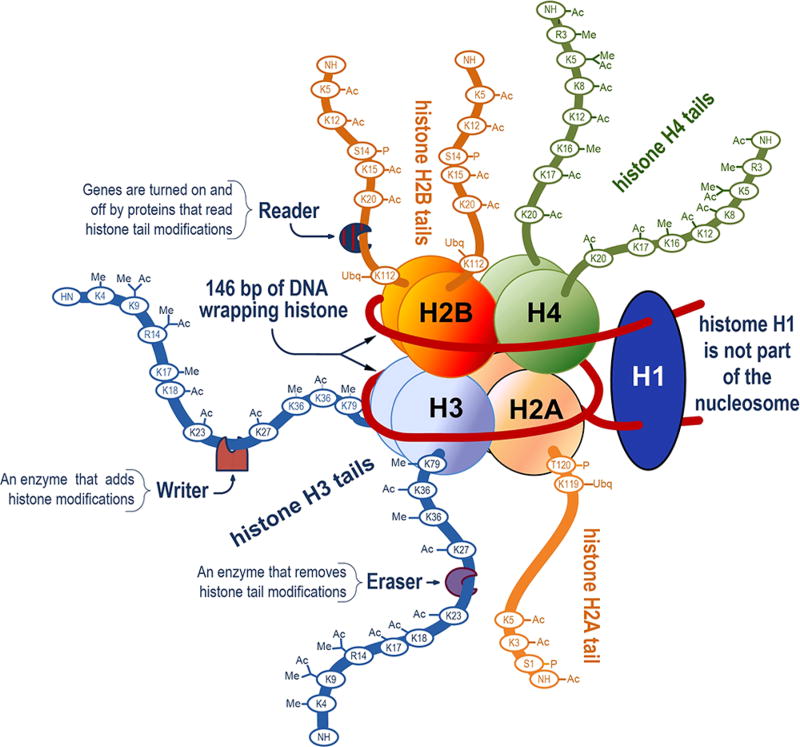
It appears from Figure 1 that the flow of information in living systems is exclusively one way, from genes to proteins. What this figure does not show is the additional process known as epigenetics in which a series of PTMs in histones contained within a nucleosome (Figure 2) enable information flow back to the gene level. Significantly, the sites of PTM modification on histone tails are conserved but not necessarily the patterns of modification.25 This enables a combinatorial set of PTM based modifications among histone proteoforms (Figure 2), probably numbering into thousands that direct the alteration of DNA bases in genes without altering their sequence. Features common to all nucleosomes are the 146 bp of genomic DNA encircling the protein octamer complex formed by two copies of histones H2A, H2B, H3, and H4.26 Moreover, the sequence of these histones is highly conserved.25 Histone H1 is not part of the nucleosome. Its function is to maintain the number of DNA bases enclosing histones at 146 bp and control the number of bases linking nucleosomes. This allows the close packing of nucleosomes into chromatin structures that are tightly linked to transcription regulation.27 Lysine acetylation is generally associated with decreasing nucleosome packing density, enhancing transcription.28
Figure 2.
Illustration of a hypothetical nucleosome, showing extensive posttranslational modification in the histone tails at conserved sites which enable epigenetics. Nucleosomes package DNA into a compact, dense structure that prevents DNA damage and allows the formation of chromatin structures. Enzymes that modify histone tails are indicated in the figure as Writers and Erasers while Reader proteins ultimately determine the functional outcome of specific modifications.
DNA modifications triggered by epigenetics are often heritable, at least for a few generations. Epigenetically driven alterations in protein-coding genes ultimately loop back to the protein level through adaptations in protein expression illustrated in Figure 1. Environmental stimuli cause a flow of information back to DNA through nucleosomes (Figure 2). The objective of epigenetic modifications in DNA bases is to enable different genes to be switched on and off as a means to restructure proteome composition and adjust particular cellular activities in response to environmental stimuli. This means that in addition to the evolution of long-term gene codes for directing protein synthesis, nature has made provisions for short-term environmentally driven adaptations in genes as well.
Epigenetics is easily demonstrated in mice and C. elegans. Periodic application of a physical stress to mice for short periods during pregnancy can result in (i) passage of a stimulus aversion to progeny, (ii) inheritance of the aversion for several successive generations, and (iii) no response to the stimulus in nonstressed controls.29 Similarly, with C. elegans30 adaptations to a temperature stress were inherited for at least 14 generations.31 Inheritance in this case was associated with altered trimethylation of histone H3 lysine 9 (H3K9me3) (Figure 2). In contrast, epigenetic changes in the life span of C. elegans are associated with H3K4me332 (Figure 2). The fact that the H3K4me3 mark and H3K9me3 mark are associated with completely different changes in gene expression in the same organism attests to the subtleties involved in epigenomics. (Altered sites and structural types of epigentically driven modifications on histones and DNA are often referred to as a “mark”.) Cancer and other diseases are known to epigenetically enhance the prospects of their own survival via histone marks.33 It is also highly significant that cocaine34 and drugs35 have recently been found to alter the human epigenome. This suggest that drugs might be developed that can block disease progression epigenetically.
A major question is how large numbers of environmental stimuli can be coded at the histone level and transmitted to the genome. The “histone code” hypothesis36 suggests this is possible through combinatorial modifications in the tails of histones driven by nonhistone “reader”, “writer”, and “eraser” proteins.37 These proteins are involved in modifying each histone at 6–10 sites (Figure 2) in multiple ways.38 Lysine and arginine modifications each accommodate four types of marks; the native form along with mono-, di-, or trimethylation. Acetylation, propionylation, an malonylation, in addition to ubiquitination are other ways lysine can be modified. Histone marks are conveyed by serine (pS), threonine (pT), and tyrosine (pY) phosphorylation as well. Considering the aggregate of possible PTM combinations, this theoretically allows the production of a huge number of histone proteoforms.39
A critical element of histone coding is the need for nonhistone reader proteins40 that bind to specific histone sequences as a component of initiating signaling.41 Bromodomain (BRD) reader proteins are an example. They bind to acetylated lysine residues on the H3 and H4 histone tails as a means to direct signaling,42 often favoring lysine methylation, acetylated lysine,43 or a ubiquitin modification.44 Within lysine acetylated BRD readers, there are eight subfamilies representing 61 diverse BRDs from 46 different proteins.39 The fact that reader domains preferring the acetyllysine mark have been shown to be “druggable”45 suggests there is therapeutic potential in manipulating reader proteins.
Nonhistone “writer” and “eraser” enzymes are responsible for the highly dynamic addition and removal PTM marks.46 Acetyltransferases and histone methyltransferases are typical writers, attaching covalent marks at specific sites on histones. Histone deacetylases and histone demethylases are eraser enzymes.47
Collectively it is probable that millions of nucleosomes, each with a specific histone code are required to enclose the 3 billion base pairs in the human genome. Again, the number of PTM based proteoforms involved is likely to be enormous. How stimuli are transduced into histone codes and the actual coding mechanism are unknown.
GENERAL ANALYTICAL ISSUES
Clearly, proteomics is enabled by mass spectrometry, the critical element being that the mass of amino acids and proteins can be readily tied to nucleotide sequences by the genetic code and genomic databases. This led to the initial hypothesis it would be easier to determine the structure of proteins through DNA sequencing and gene sequence data than by sequencing proteins directly. It was reasoned that LC–MS/MS systems would provide a platform for the global analysis of proteomes based on linking the mass of parent ions and their fragments to a protein-coding gene in a database. A constant problem with this hypothesis from the beginning has been sample diversity. With samples of 105 to 106 components peaks from chromatographic and electrophoretic separations bear hundreds to thousands of proteins or peptides. Eluting fractions this complex into a mass spectrometer has a series of ramifications. Among the more serious are that (i) abundant species mask those of low abundance, (ii) ions in the first or second dimension of mass analysis can be of similar mass or isobaric, (iii) enrichment of low-abundance species is difficult, (iv) no structural information is being derived from chromatographic retention times or electrophoretic mobility, and (v) there is no correlation a priori between a protein or peptide in any particular chromatographic peak and the protein-coding gene from which it was derived. Second, it was not recognized at the inception of proteomics that protein-coding genes produce multiple proteoforms as noted above. The fact that multiple proteins in a mixture will be of very similar structure complicates the interpretation of mass spectral data, particularly in the case of bottom-up proteomics. Third, as seen Figure 1 there may be no direct relationship between protein-coding gene sequence and protein structure. Some proteoforms are not yet in genomic databases.48,49 This complicates (i) the identification of signature peptides unique to a single protein species and (ii) their use in multiple reaction monitoring (MRM)50 or SILAC based quantification.51
FRACTIONATION OF STRUCTURALLY SIMILAR PROTEINS
The HUPO Human Proteome Project (HPP) was launched in September 2011, a major aim being to identify and characterize at least one protein from each of the ~20 000 predicted protein-coding genes in the human genome along with other family members when possible.52 A challenge is how to find and know that the protein being identified is a single protein. As noted above, there is the issue that a protein-coding gene can produce 10–50 proteoforms. A huge number of combinations are possible as was seen above with histones. Second, lower abundance proteoforms compound the problem. It is difficult to obtain enough protein for structure analyses.
The discussion below suggests strategies that deal with these issues. Step one would be to structure specifically select and enrich targeted proteins accompanied by the elimination of nonanalytes. Chromatographic separations would target specific structures and greater than 99% of irrelevant protein mass would be eliminated from samples in most cases. Part two would be to extract a single proteoform from the remaining sample based on some combination of chromatography and mass spectrometry.
Proteomics is in the early stages of using exon coded structural features to enrich and purify proteoform families. Use of solid phase media with immobilized antibodies, affimers, or natural binding proteins would be the most obvious selector medium. Moreover, there would be a high level of certainty that most of the polypeptides selected are of some degree of interest. This would be more efficient than searching through millions of mass spectra from hundreds of IEC, RPC, HILIC, HIC, or IMAC column fractions. The discussion below focuses on these strategies.
Polyclonal Antibody Selectors
Obviously structure specific selection of a proteoform family depends on having an affinity sorbent capable of selectively binding proteins on the basis of amino acid sequence and conformation. Antibodies do this well; but they have been developed to select single analytes, not families of proteins. Antibody cross-reactivity is generally considered to be undesirable. In contrast, cross-reactivity would be desirable in proteoform selection. The discussion below examines antibody production in terms of proteoform cross-reactivity.
Mammalian B cells produce 108 cells daily with large numbers of randomly specific surface receptors to which foreign proteins bind.53 Binding accelerates B cell production along with triggering a large increase in coding sequence mutations responsible for producing antigen-binding antibodies.54 Through iterations of antigen binding, B cell mutations, and antibody synthesis, the association constant for a protein target increases, eventually reaching or exceeding 106 M−1 within a few weeks.
Immunogens frequently bear multiple antigenic determinants on their surface,55 each of which leads to a different B cell clone and production of an antibody specific for the epitope matching the antigenic determinant. The resulting mixture of polyclonal antibodies is ideal for proteoform selection. However, immunization has some variability. Different animals, even of the same species and gender do not respond exactly the same way to an immunogen. This means that polyclonal antibodies from different suppliers against the same substance can vary in epitope binding affinity and ratio of antibodies against specific epitopes. Antibodies from the same source must be used to achieve comparable results within and between laboratories.
As seen in Figure 3, one or all the proteoforms in a family bear exon derived antigenic determinants that trigger antibody production. When a single proteoform is used as the immunogen, antibody selectivity of two specific types will be produced. One group will recognize epitopes unique to that single proteoform alone. The second will select family members bearing the same antigenic determinants. Epitopes from different proteins that react with the same antibody are often referred to as “cross-reacting epitopes” (CREs). Proteoform family members are likely to have CREs, but they may not be identical in structure and sequence. Their association constant only has to be sufficiently large to cause Ab binding. The attractive feature of polyclonal antibodies is the high probably that a whole proteoform family is likely to be recognized based on the presence of CREs, irrespective of the proteoform used as the immunogen.
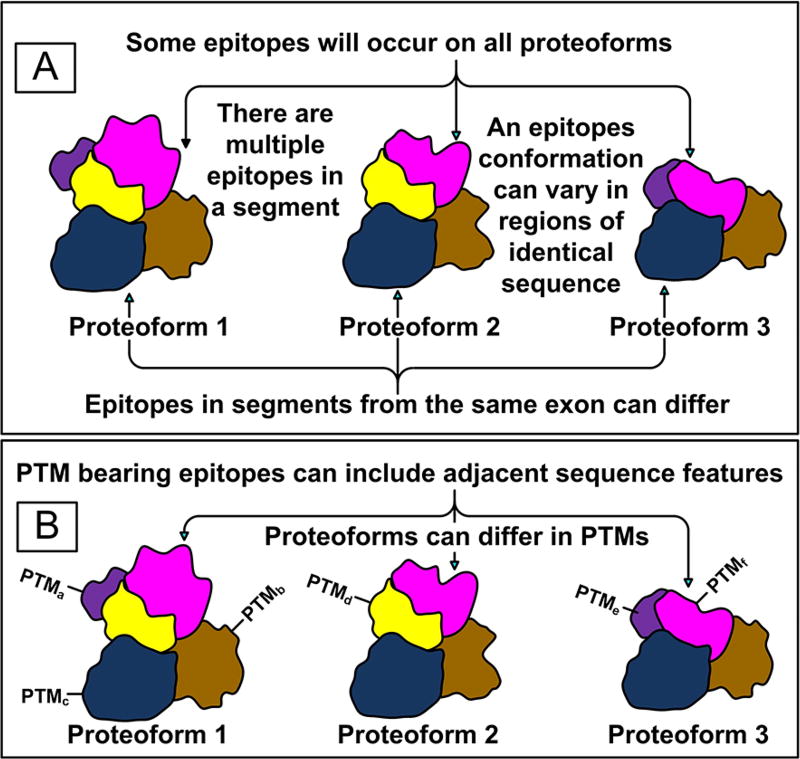
Figure 3.
Contributing factors in the production of polyclonal antibodies that target a proteoform family. Panel A illustrates the fact that the conformation of a protein domain depends to some extent on other portions of the structure. Panel B suggests that antibodies targeting a specific PTM can have bias for the surrounding amino acid sequence.
An important feature of immunogens is that their antigenic determinants and thus epitopes ultimately targeted can be linear sequences of a few amino acids or discontinuous sections of amino acid sequence that are conformationally juxtapositioned. The presence of conformational (discontinuous) epitopes allows antibodies to sense protein conformation. Even so, regions of identical sequence in proteoforms will not necessarily present the same antigenic determinants. This would be true when adjacent regions in proteoforms cause conformational differences (Figure 3).
A PTM, the sequence around the PTM, and the impact of that PTM on conformation play a role in forming antigenic determinants56 (Figure 3). It is easily seen how one of the antibodies in a pAb mixture could be sensitive to the conformation of a PTM bearing epitope in a single proteoform while others bind to CREs common to the whole family. Polyclonal antibodies that target PTMs are likely to be of more limited value in the selection of proteoform families for this reason. Monoclonal antibodies that target PTMs will in contrast be very useful as has been found with antihistone H3 (dimethyl-K4),57 antihistone H3 (phospho-S10),58 and antihistone H3 (acetyl-K4)58 antibodies.
A concern is whether proteoforms can be missed with the antibody selection strategy described above. Theoretically there are several scenarios in which this could happen. One would be the case in which sites of structural complimentarity are nonimmunogenic.59,60 There would be no antibodies to target these regions of complimentarity. A second would be the case in which amino acid substitutions, splice variations, or PTMs have changed the association constants of epitopes in a specific proteoform to the point there is no binding. This could easily occur where modifications cause large changes in conformation.
Still another issue is the structure of antigen(s) during immunization versus analysis. Partial denaturation during immunization is common. This destroys some conformational epitopes while exposing or creating new epitopes.61 When this occurs, native proteoforms will present a different set of epitopes than were present during immunization. This means some epitopes common to a proteoform family can be missed if they were absent during immunization.
Finally there is the issue of antibody selectivity and reproducibility. It will be seen throughout this review that polyclonal and monoclonal antibodies vary substantially between suppliers. Some will provide wider proteoform family selection than others. Widespread use of immunological assays in clinical diagnostics means the problem can be managed in high volume, routine analyses. Whether that is true at the research level remains to be determined.
Monoclonal Antibodies
Production of a monoclonal antibody (mAb) is achieved in a different way,62 but the selectivity rules are the same. As the name implies, mAbs are derived from a single clone and are directed against a single epitope. Whether that epitope is common to the family, present in a group within the family, or unique to a single proteoform determines how the mAb can be used. There must be a method to determine which members of the family are being selected in mAb production.
Signature Peptide Capture
The terms signature and surrogate peptide have been used interchangeably,63 designating peptides unique to a single proteoform. The degree to which signature peptides reported in the proteomics literature belong to multiple proteins in a proteoform family is not clear. As noted above, it is the practice in bottom-up proteomics to identify proteins based on peptides derived from trypsin digestion of samples. Subsequent to trypsin digestion, peptides from all proteoforms will be mixed. On the basis of the fact that not all proteoforms are in a database,48,49 it cannot be assumed that a peptide derived from a tryptic digest is unique to a single protein without independent confirmation based on something other than databases.
A trypsin digest of a complex sample can contain millions of peptides. Antibody selection of a peptide common to a proteoform family or a single proteoform would be an obvious asset. This has been accomplished in two ways. One is via antibody capture of a parent protein and identification of its peptides by LC–MS following trypsin digestion.64 Depending on the selectivity of the antibody, the peptides in the digest may have come from multiple proteoforms or a single proteoform. An advantage of this approach is that the antibody has specifically selected proteoform(s) having common epitopes. This eliminates the concern that what is being treated as a signature peptide of a proteoform family might also be present in other proteins not in the family. The rare case in which this would not be true is when a nonspecifically bound protein bears some of the same peptides. A second advantage is that a single antibody is required. The disadvantage is that the proteoform selectivity of the antibody should be validated.
The second strategy is to antibody select signature peptides from a trypsin digest directly. The SISCAPA approach65 to quantification is the most well-known of these methods. Three signature peptides from the same protein are generally used to ensure accuracy in quantification. Heavy isotope labeled signature peptides from the targeted protein are added to a sample as internal standards after which the sample is trypsin digested and the signature peptides purified by immunosorbent selection on magnetic particles using a series of washes.65 Signature peptides thus purified are then desorbed from the immunosorbent, identified, and quantified by RPC-MS/MS. The rationale in using three signature peptides is to (i) identify and/or circumvent problems arising from under-digestion at specific sites and (ii) diminish the possibility of interfering isobaric fragments from other peptides interfering with quantification. The great advantage of this approach is that it is self-validating. The isotope ratio of all signature/standard peptides should theoretically be the same. When true there is a high probability that all three peptides came from the same proteoform. If there is a low-abundance proteoform bearing the same “signature” peptide, it will not be seen. Differences in signature peptide ratios can also result from incomplete proteolysis. Disadvantages of SISCAPA are that (i) incomplete trypsin digestion or formation of miscleavage fragments is relatively common, (ii) three antibodies are required per target protein, and (iii) some signature peptides might be of limited immunogenicity.
Validation of proteoform specificity in bottom-up methods requires either (i) purification to homogeneity prior to proteolysis or (ii) that top-down sequencing methods have been used to eliminate the possibility of coexisting isobaric proteoforms.
Peptidomics
There are hundreds of hormones, cytokines, toxins, neuropeptides, and in vivo protein degradation fragments of less than 10 kDa. Polypeptides of this size are generally categorized as being part of the peptidome. As with proteins, they bear structure specific signatures that assist in MS based sequencing. The fact that polypeptides this small breach the blood–brain barrier, tumors, the vascular system, and interstitial space make them of interest as both therapeutic agents and biomarkers.66 Their size and abundance enable a series of purification methods that differentiate them from proteins. Among the methods most widely used are (i) size fraction methods, (ii) organic solvent extraction,67 and (iii) immunoaffinity based abundant protein removal.68 These methods exploit the removal of abundant nonanalytes as a prelude to finding and identifying lower abundance peptides of unknown structure and properties.
Affinity based purification methods used in peptidomics are of two types. One uses monoclonal69 or polyclonal antibodies70 in a manner similar to that in SISCAPA,65 targeting polypeptides of known structure. Phage display selectors can be used in this approach as well, an advantage being that they are easier to produce than monoclonal antibodies.71 Subsequent to trypsin digestion of the peptidome and fractionation, peptides are identified by LC–MS/MS.
Affinity purification of unknown in vivo peptides must take a different route. They are not available for use as immunogens. Instead, they can be targeted indirectly as with tumors that produce protein antigens which are degraded to peptides in vivo.72 These peptide fragments associate with soluble class I human leukocyte antigen (HLA), appearing on the surface of nucleated cells. Antigen derived peptide fragments in these complexes are then presented in vivo to T cells in the immune system. Melanoma cells, for example, present HLA bound peptides from tumor rejection antigens that appear in patient plasma. The HLA class I pan-reactive antibody W6/32S has been used to select and purify HLA:peptide complexes in vitro as a group.73 Following low pH dissociation of captured HLA:peptide complexes, 972 peptides were identified from 5 mL of serum, including peptides from known tumor-associated antigens. At present, 27 862 unique HLA bound peptides from five human cancer cell lines have been identified in this way, most of which are 8–11 amino acids in length. Clearly this HLA capture approach could be used in many other cases seeking to recognize disease specific antigens.
PTM Targeting
Affinity methods have been used in the isolation of a variety of proteins and peptides carrying PTMs, phosphorylation, acetylation, methylation, and ubiquitination being most frequently targeted. The degree to which anti-PTM antibodies are pan-specific is variable and antibody specific. Use of PTM targeting antibodies to select all proteins that carry a particular PTM is referred to herein as the PTM-1 selection mode. This method has been used with immobilized ant-Lewis × Ab to select glycoproteins bearing conjugated Lewis × antigens in cancer patients.74,75 One third of the proteins identified via the bottom-up method were elevated in cancer patients. Histones76 and proteins vulnerable to oxidative stress77 have been identified in similar fashion by targeting other forms of PTM. Although a number of PTM modification sites can be identified with bottom-up proteomics, it is difficult to identify the proteoforms involved. Top-down identification helps,78 but locating PTM modification sites in the primary structure of a protein requires gas phase sequencing.
With the PTM-1 mode of analysis the proteoform family is selected first followed by identification of member(s) bearing PTMs as with hemoglobin A1c.79 Following capture with hemoglobin specific immunosorbent, the glycan portion is identified in either of two ways. One is by addition of a second primary antibody specific for the glycated terminus. Detection is achieved by ELISA or an electrochemical method.80 The second means of glyan detection is via the glycan associated mass shift in top-down MALDI-MS spectra.81
FTICR MS/MS takes top-down proteomics to a higher level. Using a 21 T LC–FTICR instrument to examine intact proteins derived from human colorectal cancer cell lysate it has been possible to identify a combined total of 684 unique proteins from 3238 proteoforms at a 1% false discovery rate, including 372 over 30 kDa in size.82
The PTM-2 approach is to select PTM bearing peptides from proteolytic digests. Separation of peptide fragments can be achieved directly or following an affinity chromatographic purification using an immunosorbent,83 immobilized lectin,84 a natural binding protein,85,86 or by IMAC.87 A heterozygous lysine-to-methionine point mutation (H3K27M) in histone H3 occurs in more than 80% of pediatric tumors in diffuse intrinsic pontine glioma patients.88 The epigenome of H3K27 M mutant cells was profiled after histone isolation and trypsin digestion, showing increased H3K27 acetylation (H3K27ac). The majority of the heterotypic H3K27M-K27ac nucleosomes colocalize with bromodomain proteins at the loci of actively transcribed genes. This is in effect a PTM-2 analysis with a nonantibody affinity selector. It was demonstrated that blocking the recruitment of bromodomain proteins by heterotypic H3K27M-K27ac nucleosomes inhibited tumor progression.
Phosphorylation
The most widely studied forms of protein phosphorylation are on serine (pS), threonine (pT), and tyrosine (pY). Less abundant, more labile forms of phosphorylation occur as phosphoramidates of histidine, arginine, and lysine along with the phosphorothioate of cysteine.89 Although phosophorylation at a single site is common, hundreds of proteins are phosphorylated at 2–6 sites.90,91 Multiple site phosphorylation is especially important in signaling proteins,92 transcription factors,93 and transcriptional coactivators.94 As noted above, phosphorylation at (n) sites could theoretically produce n2 proteoforms. The degree to which this is seen in vivo remains to be determined. Through the presence of opposing kinases and phosphatases phosphorylation is highly dynamic.95 This is an important issue in sampling. These enzymes can continue to alter the proteome after sample collection.96 Enzyme activity must be arrested immediately.
Multiple solid phase capture schemes are being used in the isolation of phosphopeptides for MS analysis.97 Among the antibody based methods, most are being used to purify pY bearing phosphoryled peptides.98–100 Phosphopeptide specific selection based on structural features surrounding the tyrosine phosphate residue is possible101,102 but not common. The ideal pY specific antibody (anti-pY Ab) would be sequence neutral, allowing universal selection of pY peptides without sequence bias. Recent studies have found that unfortunately the anti-pY antibodies 4G10, pY20, and p-TYR-100 in common use are not totally sequence neutral.103,104 Using peptide microarray technology to examine the specificity of these anti-pY antibodies, it has been demonstrated that they show amino acid biases within 1–3 amino acid residues of pY. Moreover, these antibodies vary from each other. The 4G10 and p-TYR-100 antibodies showed the least bias. Using these two antibodies together in an antibody cocktail would perhaps provide a solution to this specificity problem. The significance of these studies is that they show how affinity selector specificity can impact relative and absolute quantification accuracy within and between laboratories.
Purification of pS and pT peptides is most frequently achieved by IMAC105 prior to MS analysis. Although phosphorylated peptides106 and proteins107 bind to Fe3+, Al3+, Ga3+, and Zn2+, Ga3+-IMAC has the highest selectivity in minimizing the isolation of nonphosphorylated, acidic peptides. Phosphate versus carboxyl discrimination was enhanced even more with Fe(III)-nitrilotriacetic acid (NTA) IMAC resin by using acetonitrile in the mobile phase.108 With this method 512 phosphorylation sites were identified in 162 cytosolic phosphoproteins. A total of 97% of these phosphorylation sites were located outside of structural domains, mostly in regions of intrinsic sequence disorder. These regions were at least 40 amino acids in length.
Phosphorylated protein109 and more commonly phosphorylated peptide110 enrichment and purification has also been achieved with aluminum hydroxide (Al(OH)3), titanium dioxide (TiO2), and zirconium dioxide (ZrO2) in the nanofiber mode.111 Metal oxide/hydroxide affinity chromatographic (MOAC) enrichment of both proteins and peptides has been described.112
Although not an affinity selection method, nearly pure phosphopeptides can be obtained by multidimensional IEC followed by RPC.113 A highly enriched mixture of 4 045 MS identified phosphopeptides was obtained by batch selection with a strong cation exchange (SCX) chromatography sorbent. When further split into 14 fractions with weak cation exchange (WCX) chromatography and analyzed by RPC-MS, a total of 7 251 unique phosphopeptides were identified. Still higher resolution was achieved by increasing the RPC gradient time, leading to the detection of over 11 000 unique phosphopeptides.
Lysine Methylation
Lysine methylation plays a major role in locus-specific modulation of protein histones114 and in multiple, nonhistone signaling networks.115 Methylation of proteins occurs through the addition of one to three methyl groups on a specific lysine residue (Figure 4A). In histones, lysine methyltransferases along with demethylases modulate the activity of transcription factors and transcription coregulators. The degree of methylation matches the methyl-specific reader domain of a protein partner to which the transferase binds.116 H3 lysine-4 methyltransferase Set9 monomethylates lysine-630 on a Drosophila androgen receptor (AR) is an example.117 The association between endogenous AR and Set9 is sufficiently strong that they can be coimmunoprecipitated. The lysine methylation mechanism is variable, occurring with a single enzyme on some regulatory proteins and multiple enzymes on others.118 The same is true with demethylases. Deciphering lysine methylation mechanisms will require new analytical methods that are faster and more precise.
Figure 4.
Methylated forms of lysine and arginine along with a supramolecular affinity selector was designed to select methylated amine species via a cation-exchange process. Lysine can also be acetylated as seen in panel A.
Purification of proteins and peptides bearing methylated lysine residues has been achieved in multiple ways. One is by immunosorbent selection. Lysine 51 in the RNA-binding region of Tat is a substrate for the lysine methyltransferases KMT1E (SETDB1) and KMT7 (Set7/9).119 A cocktail of pAbs for the selection of mono-, di-, and trimethylated lysine 51 in the HIV transactivator (Tat) was used to affinity select a pAb subclass of narrower selectivity through monomethyllysine affinity chromatography. If the selectivity of these two antibody mixtures was sufficiently different, it was possible to establish that cellular Tat is predominantly monomethylated at lysine 51.
Synthesis and use of a solid phase supramolecular p-sulfonocalix(4)arene affinity selector is another approach to methylated lysine selection119 (Figure 4C). When used in an affinity chromatography format, this anionic matrix is capable of forming host–guest complexes with methylated lysine peptides. Selectivity of the p-sulfonocalix(4)arene for methyllysine residues in peptides is reported to be superior to that of a cation-exchange chromatography resin. Attaching this supramolecular affinity selector to agarose120 (Figure 4C) allowed the enrichment of peptides bearing methylated lysine residues from a trypsin digested nuclear extract prior to a bottom-up LC–MS/MS analysis. The advantages of this method are the enhanced selectivity and enrichment of methylated lysine carrying species. A probable limitation would be poor selectivity and adsorption bias between the cationic isoforms of methyllysine.
A soluble version of the p-sulfonocalix(4)arene affinity selector has also been recently reported in which detection is achieved via fluorescent labeling.121
Arginine methylation generates three isoforms, a monomethylated and two dimethylated forms; the dimethylated arginines have either asymmetric or symmetric derivatization (Figure 4B). Dimethyl arginine-specific antibodies have been used in the identification of ~200 putatively argininemethylated proteins.122 Major protein complexes identified in this way include those required for pre-mRNA splicing, polyadenylation, transcription, signal transduction, and DNA repair.
Lysine Acetylation
There are multiple reports of lysine acetylation and deacetylation via histone acetyltransferases and deacetylases being involved in the regulation of gene expression and epigenetics (Figure 2). Site modification selectivity with these enzymes suggests that in addition to the presence of lysine or acetyllysine, the surrounding sequence biases binding. As noted above, bromodomain readers bind to acetylated lysine residues on H3 and H4 histone tails pan-specifically.35 The resulting bromodomain:histone complexes are part of the system altering nucleosome structure in epigenetic modifications.
Lysine acetylation is also important for p53 functioning and microtubule stabilization, being found in bacteria, yeast, insects, and human cell lines.123–126 Moreover, it plays a role in the regulation of protein synthesis, the citric acid (TCA) cycle, fatty acid metabolism, glycolysis/gluconeogenesis, and secondary metabolism.
Microarrays displaying acetyllysine-containing peptides have been used to establish that pan-specific antiacetyllysine antibodies have a nonlysine amino acid bias within 1–3 amino acids of the lysine acetylation site.127 Alternatively a random library of acetylated peptide antigens does the same.128 The problem can be ameliorated by using a cocktail of monoclonal antibodies.129 In this case, the majority of acetylated lysine residues identified with the cocktail were distinct from those enriched by the polyclonal antibody alone. Taken together these studies suggest that immunoaffinity enrichment of acetylated peptides will be limited to some extent by antibody specificity. Lysine acetylated peptides of the same concentration will not be equally enriched. Even with the selectivity limitations of antiacetyllysine antibodies, affinity enrichment with this approach allowed a total of 5 775 proteins bearing 1 124 lysine acetylation sites to be identified in AML HL60 cells.99 A similar study with Mycobacterium tuberculosis antiacetyllysine antibody enrichment enabled the identification of 1 128 lysine acetylation sites on 658 proteins.130 Among the lysine-acetylated proteins identified, 20 showed homology with acetylated proteins from Escherichia coli, Salmonella enterica, Bacillus subtilis, and Streptomyces roseosporus, suggesting that these are conserved proteins.
Antiacetyllysine pAb selection of lysine-acetylated peptides from Saccharopolyspora erythraea isolated 664 unique lysine-acetylated peptides from 363 proteins.131 Favored motif sequences in the acetylation sites were KAcH, KAcY, KAcXXXXR, and KAcXXXXK. With human cell lines,132 3600 lysine acetylation sites on 1750 proteins have been identified. Additionally, 15 474 acetylation sites have been identified in 16 different rat tissues.133,134
Glycosylation
Antibody selection of glycoproteins is similar. Selection of Lewis X or sialyl-Lewis X antigen-bearing glycoproteins from cancer patient plasma samples with a single antibody generally yields mixtures of 50–100 proteins, depending on the type of cancer.135,136 Lectins have also been widely used in glycoprotein and glycopeptide selection as well.137 There are more than 50 lectins, each of differing selectivity.138 Lectin affinity chromatography in proteomics is discussed more extensively in a recent review.139
FRACTIONATION BY CONVENTIONAL LC
Affinity chromatography and conventional chromatography vary substantially in their selection mechanisms. Selectivity in immunoaffinity chromatography revolves around recognition of a spatially defined set of structural features contained within a small area on the analyte surface that is complementary to affinity selector structure. All molecular species bearing this specific epitope or very similar structural forms of the epitope will be captured with high affinity while nonantigens are ignored. A limitation of this approach is the inability to differentiate between proteoforms thus selected. Association of cross-reacting epitopes with the affinity selector will generally be so strong that elution requires antibody denaturation. This precludes differential elution of proteoforms. Family members will be eluted together.
It is critical to have methods that separate proteoforms. Studies that determined the degree to which this is possible with conventional modes of chromatography such as ion exchange, reversed phase, hydrophobic interaction, hydrophilic interaction, and immobilized affinity chromatography are in their infancy. A general characteristic of conventional separation modes of chromatography is that they lack analyte structure complimentarity and bind substances having little structural similarity. Reversed phase chromatography (RPC) columns bind most of the substances in a sample for example. Even weakly hydrophobic substances adsorb to an octadecyl stationary phase.
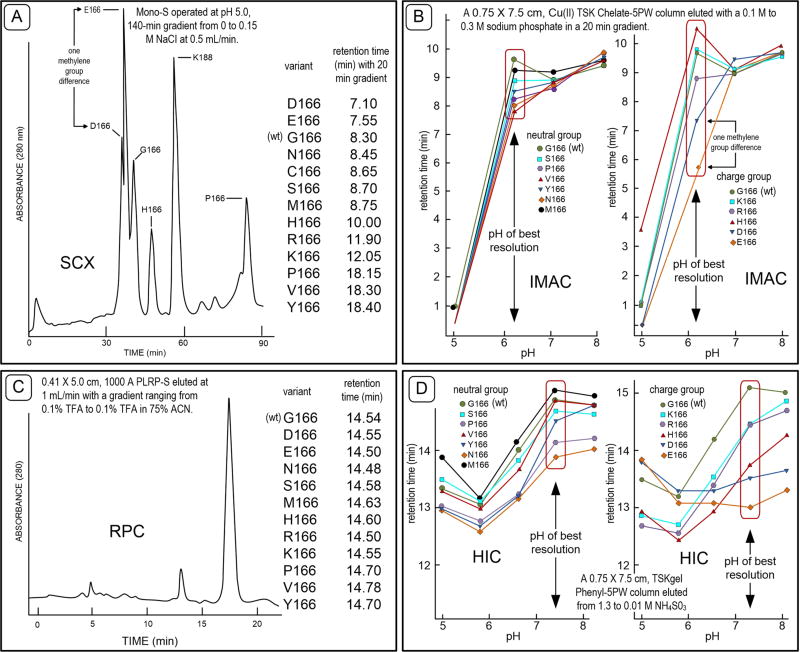
A priori these facts would seem to diminish the probability that conventional modes of chromatography can differentiate between proteoforms differing slightly in structure. Investigations of this have been undertaken with synthetic and natural proteoforms. Synthetic proteoforms of Bacillus amyloliquefaciens subtilisin were created by site-directed mutagenesis in which 15 different amino acid substitutions were made at residue 166 in the hydrophobic pocket of the catalytic site.140 At 1.8 Å resolution X-ray crystallography showed these synthetic variants to be conformationally identical.141 Any difference in chromatographic behavior among these 27.5 kDa variants would have to be from single amino acid contributions.
Strong cation-exchange (SCX) chromatography resolved six of the subtilisin variants by 30 s or more142 (Figure 5A), showing that both neutral and charged amino acid substitutions impacted SCX behavior. Apparently alterations in pKa values and hydrogen bonded water in the interfacial microenvironment between the protein and stationary phase played a key role in selectivity. Glutamate and aspartate variants differing by a single methylene group were partially resolved. This is remarkable. It is equally surprising that the G166 → V166 substitution increased retention on a hydrophilic SCX column by 10 min while the G166 → S166 substitution had little impact on retention.
Figure 5.
Chromatographic behavior of subtilisin variants, the wild type (wt) being G166. Panel A shows variant elution times from a strong cation-exchange column operated at pH 5.0. Panel B is an examination of the same variants in the IMAC mode. Neutral and charged variant behavior is plotted separately and compared to the wild type (wt) control. Panel C demonstrates variant separations on a macroporous poly(styrenedivinylbenzene) type reversed phase column. Panel D illustrates the differences in chromatographic behavior of the subtilisin variants in hydrophobic interaction chromatography using a 0.75 cm × 7.5 cm Cu(II) TSK Chelate-5PW column.
IMAC, in contrast to ion exchange is based on interactions between electron donors such as histidine, tryptophan, and cysteine on polypeptide surfaces and immobilized metal atoms such as (Cu(II), Ni(II), Ca(II), Zn(II), Fe(II), and Fe(III)) that act as electron acceptors. Protein adsorption occurs best in IMAC with an alkaline mobile-phase where electron donor groups will be at least partially unprotonated (Figure 5B). Highest resolution of the variants occurred at pH 6.2. Among the chelated metals, Cu(II) provided the best resolution of the subtilisin variants. Variants bearing a charged amino acid substitution at G166 showed the largest alteration in chromatographic behavior.143 Again there was the dramatic difference in elution behavior between the aspartate and glutamate variants. Lysine and arginine variants were resolved as well but not as dramatically. Resolution of neutral amino acid variants was substantial. This study demonstrates again that amino acid substitutions adjacent to groups actively involved in binding to a stationary phase secondarily influence chromatographic retention of a protein. The X-ray crystallographic structure of subtilisin suggests that six histidines at three locations could interact with an IMAC column.130 Two are on the side away from G166, while the third bearing His 64 and His 226 are in the active site cleft near G166. With the Gly-to-His substitution at position 166, three histidine resides are together at the residue 166 site. Surprisingly, addition of the third histidine in the chromatographic contact region had relatively little effect on IMAC retention.
Hyrophobic interaction chromatography was also quite effective in resolving the subtilisin variants (Figure 5D).144 Descending ammonium sulfate gradient elution from 1.3 to 0.01 M over a wide pH range showed pH 7.4 to be optimal. Separation of neutral amino acid variants was best with HIC, as expected. The fact that HIC was more effective in separating charged amino acid variants than SCX was unexpected. Having charged groups in the hydrophobic pocket of the enzyme obviously diminished surface hydrophobicity. This had a large impact on chromatographic behavior. The difference in HIC retention time seen with the G166 → H166 substitution was larger than in the IMAC mode.
Reversed phase chromatography (RPC) showed little resolution of the site-166 subtilisin variants (Figure 5C),134 probably due to denaturation during elution. Proteins are generally gradient eluted in RPC with a mobile phase ranging from an acidic aqueous phase to an acidic phase rich in organic solvent. Acidic mobile phases suppress silanol effects while the organic solvent disrupts hydrophobic association of proteins with the stationary phase. The impact of organic solvent driven denaturation on resolution is case specific as will be shown below.
Increasing the number of theoretical plates in columns helps resolution in all cases. Using a 0.47 µm particle diameter RPC sorbent in a 4 cm length column operated in the slip flow mode led to a protein peak capacity of 195 in a 10 min elution gradient with an ACN/TFA mobile phase.145 The peak capacity of this system increased to 750 in a 60 min gradient. When used in histone proteoform separations with TFA, a C18 slip flow column was superior to RPC columns packed with fully porous 5 µm particles or superficially porous 3 µm particles.146 It was also noted that TFA caused less band spreading than difluoroacetic acid. Proteoforms of ribonuclease A, carbonic anhydrase, superoxide dismutase, and trypsin inhibitor were resolved with the slip flow RPC column. The degree to which complete proteoform resolution was achieved in these cases is unknown.
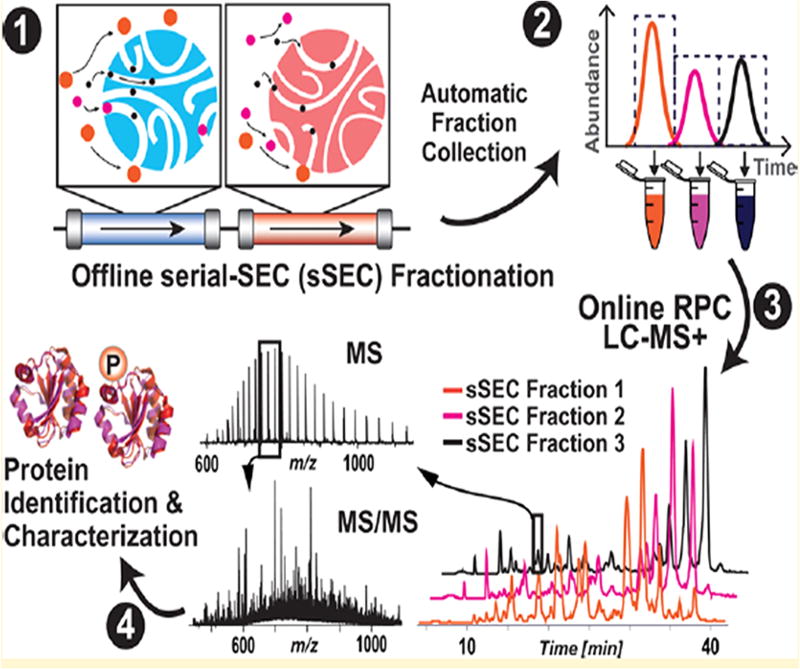
Directly coupling of serial size exclusion chromatography (sSEC) columns to RPC is still another approach (Figure 6).147 Using this strategy intact proteins of 10–223 kDa were initially separated into fractions of differing molecular weight before transfer to the RPC column where they were further resolved and transported to a high-resolution ESI-MS/MS instrument. Multidimensional 2D sSEC-RPC to 1D RPC enabled the identification of 4044 proteoforms with a 15-fold increase in sensitivity above 60 kDa (Figure 6). An advantage of this approach is that proteins eluting from the SEC column adsorb and refocus at the inlet of the RPC column. Proteoforms up to 223 kDa in size were enriched in this way.
Figure 6.
Illustration of serial size exclusion chromatography followed by RPC-MS/MS. Reproduced from Cai, W.; Tucholski, T.; Chen, B.; Alpert, A.J.; McIlwain, S.; Kohmoto, T.; Jin, S.; Ge, Y. Anal. Chem. 2017, 89, 5467–5475 (ref 147). Copyright 2017 American Chemical Society.
Hydrophilic interaction liquid chromatography (HILIC) also uses high concentrations of organic solvents in the mobile phase but for a different reason. Organic solvent is used in loading HILIC columns to drive hydrophilic portions of proteins to interact with a hydrophilic stationary phase. Gradient elution from organic solvent to an aqueous buffer allows desorption and elution of proteins from the column. The potential of this separation mechanism in the resolution of the five RNase B glycoforms was demonstrated using a TSKgel Amide-80 column eluted in a 20 min mobile phase gradient ranging from 75% to 65% acetonitrile followed by isocratic elution at 65% acetonitrile for 10 min;148 all solvents contained 10 mM HClO4. RNase B glycoforms varying in chitobiose core mannose content (GlcNAc2Man5 → GlcNAc2Man9) were resolved into four peaks, generally by a min or more with increasing numbers of mannose residues eluting last. It is remarkable that proteoforms of ~14.7 kDa varying in single mannose residues could be separated this widely. Clearly the polar glycan portion of the protein dominates the interaction with the polar amide stationary phase. It also shows that organic solvent drives the requisite polar–polar interaction between the glycan portion of the protein and stationary phase.
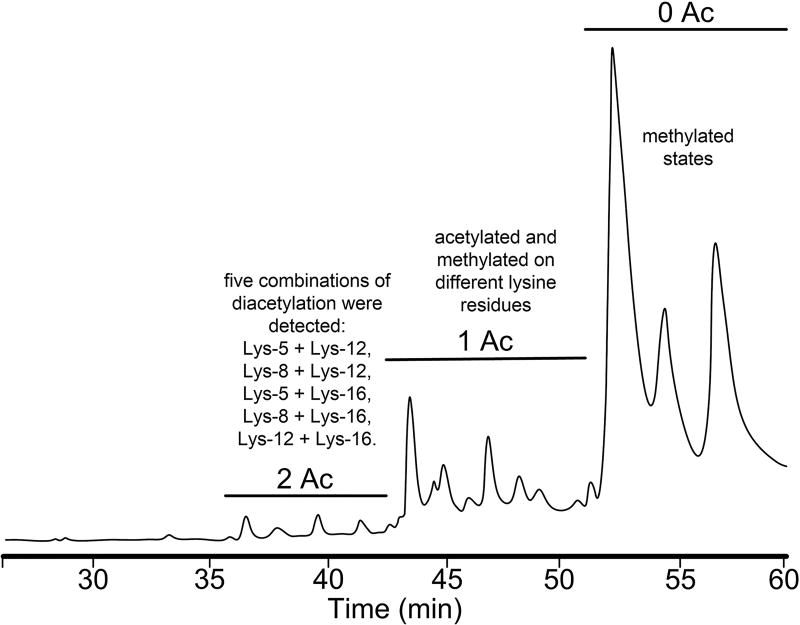
Efforts directed at elucidating the “histone code”149 in histone H3150 and H4151 have focused on the use of RPC and HILIC.152,153 Separations of histone proteoforms via HILIC were executed in this case through a mixed mode retention mechanism.154 Although HILIC is normally carried out with a neutral, hydrophilic stationary phase, HILIC separations can also be achieved with weak ion exchange columns, especially when the ion exchanging groups are attached to a polar organic matrix. This separation mode has been referred to as WCX-HILIC.155 Using salt-free mobile phases allows direct MS analysis. Mixtures from histone H3.2 and H4 were Glu-C or Asp-N digested, respectively, and applied to a polyaspartic acid stationary phase (WCX) column, and here they were gradient eluted from solvent A (75% ACN, 20 mM propionic acid adjusted to pH 6.0) to B (25% ACN, 500 mM ammonium acetate adjusted to pH 6.0 using ammonium hydroxide). Peptides in WCX-HILIC are resolved by HILIC partitioning alone or a combination of ionic displacement and HILIC partitioning. The advantage of this approach is that two modes of selectivity are being applied simultaneously. Obviously WCX-HILIC can also be used in a 2D mode where histones are fractionated by RPC and following proteolysis the fragments can be separated by HILIC.156 A total of 708 histone proteoforms (H4, 105; H2B, 110; H2A, 77; H3, 416) were identified in this approach. The tremendous potential of HILIC is seen in Figure 7 where histone proteoforms are being resolved based on their degree of lysine acetylation and methylation.157 This work has been summarized in an excellent review.158
Figure 7.
HILIC separation of H4 histones prefractionated by RPC. The HILIC separation was achieved with a PolyCAT A column (4.6 mm × 150 mm packed 3 µm, 1000 Å pore diameter sorbent) eluted with a gradient ranging from buffer A (70% CH3CN, 20 mM triethylamine (TEA), pH 4.0 with H3PO4) to buffer B (20% CH3CN, 20 mM TEA, 500 mM NaClO4, pH 4.0, with H3PO4) in 60 min. Effluent was transported directly into an LC–MS. H4 proteoforms eluted in order of increasing hydrophilicity with relative retention inversely related to their degree of ε-N-acetylation and Lys-20 methylation. Data in this figure was derived from reference Pesavento et al.157
CONCLUSIONS
Among the several “take-away” messages from this review, the first would be that affinity targeting protein sequence and conformation is likely to increase in the design of chromatographic methods. “Exon”, “gene”, and “protein-coding” will appear more frequently in the key word list of separations papers. This will be especially true as it relates to using affinity methods to rapidly remove nonanalytes from complex biological samples, enrichment of low abundance species, and chromatographically target expression products.
Second, when most of the structural features of a protein family are very similar, conventional modes of chromatography are capable of recognizing small structural differences between the species involved. This is good news in proteoform separations. The caveat is that nonanalytes must be removed to preclude interference in proteoform detection. Yet to be determined is whether proteoform separations depend on structural differences being at specific sites or they can be anywhere in a protein or peptide.
Third, it is clear that proteoform purification will be best achieved by combining affinity and conventional modes of chromatography. Affinity methods select species with cross-reacting epitopes while convention columns differentiate between them.
And finally, there is the signature peptide conundrum. Many of the identification and quantification methods used in proteomics are based on the use of signature peptides. The fact that some proteoforms are not in databases suggests that signature peptide identification has been invalid in some cases. Signature peptide validation should perhaps be a prerequisite in bottom-up proteomics.
Acknowledgments
The authors gratefully acknowledge support of their work by SBIR Grant R43-GM116663-01 from the National Institutes of Health.
Biographies
Fred E. Regnier is currently the Chief Executive Officer at Novilytic and J.H. Law Distinguished Professor of Chemistry, Emeritus at Purdue University. He has coauthored more than 300 peer reviewed publications and 50 patents in the fields of chromatography, proteomics, metabolomics, and microfabricated devices. The current focus of his work is on rapid, miniaturized sample preparation methods in proteomics and diagnostics, mobile affinity sorbent chromatography, and miniature membrane laboratories being examples.
JinHee Kim is the Director of Analytics at Novilytic. She received her Ph.D. in chemistry from Purdue University in 2009. Following work on cancer proteomics as a postdoctoral research fellow with the “Proteomics Assessment for Cancer Center” at Purdue University and at the Bindley Bioscience Center, she joined Novilytic. Her research focuses throughout her career have been on developing analytical methods for proteins, peptides, and metabolites identification and quantification by mass spectrometry.
Footnotes
The authors declare the following competing financial interest(s): Fred Regnier and JinHee Kim are employed by Novilytic.
References
- 1.International Human Genome Sequencing Consortium. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 2.Ponomarenko EA, Kopylov AT, Lisitsa AV. J. Proteome Res. 2014;13:183–190. doi: 10.1021/pr400883x. [DOI] [PubMed] [Google Scholar]
- 3.Archakov A, Lisitsa A, Ponomarenko E, Zgoda V. Expert Rev. Proteomics. 2015;12:111–113. doi: 10.1586/14789450.2015.1018895. [DOI] [PubMed] [Google Scholar]
- 4.Muñoz J, Heck AJR. Angew. Chem., Int. Ed. 2014;53:10864–10866. doi: 10.1002/anie.201406545. [DOI] [PubMed] [Google Scholar]
- 5.Gerstein MB, Bruce C, Rozowsky JS, Zheng D, Du J, Korbel JO, Emanuelsson O, Zhang Z, Weissman D, Snyder M. Genome Res. 2007;17:669–681. doi: 10.1101/gr.6339607. [DOI] [PubMed] [Google Scholar]
- 6.Mouilleron H, Delcourt V, Roucou X. Nucleic Acids Res. 2016;44:14–23. doi: 10.1093/nar/gkv1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith LM, Kelleher NL. Nat. Methods. 2013;10:186–187. doi: 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Semlow DR, Blanco MR, Walter NG, Staley JP. Cell. 2016;164:985–998. doi: 10.1016/j.cell.2016.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang F, Khajavi M, Connolly AM, Towne CF, Batish SD, Lupski JR. Nat. Genet. 2009;41:849–853. doi: 10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dietschi Q, Tuberosa J, Rosingh L, Loichot G, Ruedi M, Carleton A, Rodriguez I. Proc. Natl. Acad. Sci. U. S. A. 2017;114:7397–7402. doi: 10.1073/pnas.1704009114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan J, Ma Z, Xu X, Guo A-Y. Mol. Genet. Genomics. 2014;289:25–36. doi: 10.1007/s00438-013-0786-0. [DOI] [PubMed] [Google Scholar]
- 12.Inniss MC, Bandara K, Zhang L, Jusiak B, Lu TK, Weiss R, Wroblewska L. Biotechnol. Bioeng. 2017;114:1837–1846. doi: 10.1002/bit.26268. [DOI] [PubMed] [Google Scholar]
- 13.van Otterdijk SD, Michels KB. FASEB J. 2016;30:2457–2465. doi: 10.1096/fj.201500083. [DOI] [PubMed] [Google Scholar]
- 14.Sen R. J. Immunol. 2017;199:3–4. doi: 10.4049/jimmunol.1700680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu X, McCarthy PJ, Wang Z, Gorlen DA, Mertz JE. J. Virology. 2012;86:8086–8096. doi: 10.1128/JVI.00234-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonner TI. Br. J. Pharmacol. 2014;171:1231–1240. doi: 10.1111/bph.12526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Q, Li X, Qi R, Billiar T. Genes. 2017;8:41. doi: 10.3390/genes8010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daniels G. Hum. Genet. 2009;126:729–742. doi: 10.1007/s00439-009-0738-2. [DOI] [PubMed] [Google Scholar]
- 19.Ponomarenko EA, Poverennaya EV, Ilgisonis EV, Pyatnitskiy MA, Kopylov AT, Zgoda VG, Lisitsa AV, Archakov AI. Int. J. Anal. Chem. 2016;2016:7436849. doi: 10.1155/2016/7436849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graves PR, Haystead TAJ. Microbiol Mol. Biol. Rev. 2002;66:39–63. doi: 10.1128/MMBR.66.1.39-63.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfeffer S, Dudek J, Gogala M, Becker T, Beckmann R, Schorr St, Linxweiler J, Lang S, Zimmermann R, Forster F. Nat. Commun. 2014;5:3072. doi: 10.1038/ncomms4072. [DOI] [PubMed] [Google Scholar]
- 22.Funk A, van der Donk WA. Acc. Chem. Res. 2017;50:1577–1586. doi: 10.1021/acs.accounts.7b00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Y, Barendregt A, Kamerling JP, Heck AJR. Anal. Chem. 2013;85:12037–12045. doi: 10.1021/ac403057y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Archakov A, Ivanov Y, Lisitsa A, Zgoda V. Proteomics. 2009;9:1326–1343. doi: 10.1002/pmic.200800598. [DOI] [PubMed] [Google Scholar]
- 25.Mujahid H, Meng X, Xing S, Peng X, Wang C, Peng Z. J. Proteomics. 2018;170:88–98. doi: 10.1016/j.jprot.2017.08.021. [DOI] [PubMed] [Google Scholar]
- 26.Perez-Montero S, Carbonell A, Azorin F. Chromosoma. 2016;125:1–13. doi: 10.1007/s00412-015-0517-x. [DOI] [PubMed] [Google Scholar]
- 27.Huang P, Keller CA, Giardine B, Grevet JD, Davies JOJ, Hughes JR, Kurita R, Nakamura Y, Hardison RC, Blobel GA. Genes Dev. 2017;31:1704–1713. doi: 10.1101/gad.303461.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allfrey VG, Faulkner R, Mirsky AE. Proc. Natl. Acad. Sci. U. S. A. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matrisciano F, Tueting P, Dalal I, Kadriu B, Grayson DR, Davis JM, Nicoletti F, Guidotti A. Neuropharmacology. 2013;68:184–194. doi: 10.1016/j.neuropharm.2012.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong E, Dzitoyeva SG, Matrisciano F, Tueting P, Grayson DR, Guidotti A. Biol. Psychiatry. 2015;77:589–596. doi: 10.1016/j.biopsych.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klosin A, Casas E, Hidalgo-Carcedo C, Vavouri T, Lehner B. Science. 2017;356:320–323. doi: 10.1126/science.aah6412. [DOI] [PubMed] [Google Scholar]
- 32.Greer EL, Maures TJ, Ucar D HAG, Mancini E, Lim JP. Nature. 2011;479:365–371. doi: 10.1038/nature10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramadoss M, Mahadevan V. Drug Discovery Today. 2017 doi: 10.1016/j.drudis.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 34.Anier K, Zharkovsky A, Kalda A. Int. J. Neuropsychopharmacol. 2013;16:2053–2066. doi: 10.1017/S1461145713000394. [DOI] [PubMed] [Google Scholar]
- 35.Chistiakov DA, Myasoedova VA, Orekhov AN, Bobryshev YV. Curr. Pharm. Des. 2017;23:1167–1174. doi: 10.2174/1381612822666161021110827. [DOI] [PubMed] [Google Scholar]
- 36.Jenuwein T, Allis C. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 37.Jain AK, Barton MC. J. Mol. Biol. 2017;429:2003–2010. doi: 10.1016/j.jmb.2016.11.020. [DOI] [PubMed] [Google Scholar]
- 38.Fulton MD, Zhang J, He M, Ho M-C, Zheng YG. Biochemistry. 2017;56:3539–3548. doi: 10.1021/acs.biochem.7b00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh T-Y, Peng W, Zhang MQ, Zhao K. Nat. Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wierer M, Mann M. Hum. Mol. Genet. 2016;25:R106–R114. doi: 10.1093/hmg/ddw208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Nat. Rev. Drug Discovery. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 42.Padmanabhan B, Mathur S, Manjula R, Tripathi SJ. J. Biosci. 2016;41:295–311. doi: 10.1007/s12038-016-9600-6. [DOI] [PubMed] [Google Scholar]
- 43.Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, Felletar I, Volkmer R, Müller S, Pawson T, Gingras AC, Arrowsmith CH, Knapp S. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elliott PR. Biochem. Soc. Trans. 2016;44:1581–1602. doi: 10.1042/BST20160227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patel DJ. Cold Spring Harbor Perspect. Biol. 2016;8:a018754. doi: 10.1101/cshperspect.a018754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garay PM, Wallner MA, Iwase S. Epigenomics. 2016;8:1689–1708. doi: 10.2217/epi-2016-0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Micelli C, Rastelli G. Drug Discovery Today. 2015;20:718–735. doi: 10.1016/j.drudis.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 48.Nilsson CL, Mostovenko E, Lichti CF, Ruggles K, Fenyo D, Rosenbloom KR, Hancock WS, Paik Y-K, Omenn GS, LaBaer J, Kroes RA, Uhlen M, Hober S, Vegvari A, Andren PE, Sulman EP, Lang FF, Fuentes M, Carlsohn E, Emmett MR, Moskal JR, Berven FS, Fehniger TE, Marko-Varga G. J. Prot. Res. 2015;14:603–608. doi: 10.1021/pr500564q. [DOI] [PubMed] [Google Scholar]
- 49.Kumar D, Yadav AK, Jia X, Mulvenna J, Dash D. Mol. Cell. Proteomics. 2016;15:329–339. doi: 10.1074/mcp.M114.047126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anderson L, Hunter CL. Mol. Cell. Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 51.Andersen JS, Lam YW, Leung AKL, Ong SE, Lyon CE, Lamond AI, Mann M. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 52.Paik Y-Ki. J. Prot. Res. 2015;14:3409–3414. doi: 10.1021/acs.jproteome.5b00785. [DOI] [PubMed] [Google Scholar]
- 53.Jostock T, Knopf H-P. Therapeutic Proteins. Vol. 899. Methods in Molecular Biology; Humana Press; Totowa, NJ: 2012. pp. 227–238. [Google Scholar]
- 54.Marchalonis JJ, Kaveri S, Lacroix-Desmazes SB, Kazatchkine MD. FASEB J. 2002;16:842–848. doi: 10.1096/fj.01-0953hyp. [DOI] [PubMed] [Google Scholar]
- 55.Wong JL, Liu DZ, Zheng YT. J. Pept. Res. 2004;63:171–174. doi: 10.1111/j.1399-3011.2003.00119.x. [DOI] [PubMed] [Google Scholar]
- 56.Gianazza E, Parravicini C, Primi R, Miller I, Eberini I. J. Proteomics. 2016;134:65–75. doi: 10.1016/j.jprot.2015.09.026. [DOI] [PubMed] [Google Scholar]
- 57.Adkins MW, Carson JJ, English CM, Ramey CJ, Tyler JK. J. Biol. Chem. 2007;282:1334–1340. doi: 10.1074/jbc.M608025200. [DOI] [PubMed] [Google Scholar]
- 58.Tochigi Y, Iwasaki Y, Sano M, Yasuda H, Katayama K, Suzuki H. Biochem. Biophys. Res. Commun. 2017;486:958–964. doi: 10.1016/j.bbrc.2017.03.137. [DOI] [PubMed] [Google Scholar]
- 59.Guillet JG, Hoebeke J, Lengagne R, Tate K, Borras-Herrera F, Strosberg AD, Borras-Cuesta F. J. Mol. Recognit. 1991;4:17–25. doi: 10.1002/jmr.300040104. [DOI] [PubMed] [Google Scholar]
- 60.Zhong W, Reche PA, Lai C-C, Reinhold B, Reinherz EL. J. Biol. Chem. 2003;278:45135–45144. doi: 10.1074/jbc.M307417200. [DOI] [PubMed] [Google Scholar]
- 61.Cierniewski CS, Budzynski AZ. J. Biol. Chem. 1987;262:13896–13901. [PubMed] [Google Scholar]
- 62.Bolton GR, Mehta KK. Biotechnol. Prog. 2016;32:1193–1202. doi: 10.1002/btpr.2324. [DOI] [PubMed] [Google Scholar]
- 63.Chen Q, Pan X-D, Huang B-F. RSC Adv. 2017;7:32903–32908. [Google Scholar]
- 64.Hsieh F, Wang H, Elicone C, Mark J, Martin SA, Regnier F. Anal. Chem. 1996;68:455–62. doi: 10.1021/ac950421c. [DOI] [PubMed] [Google Scholar]
- 65.Anderson NL, Anderson NG, Haines LR, Hardie DB, Olafson RW, Pearson TW. J. Prot. Res. 2004;3:235–244. doi: 10.1021/pr034086h. [DOI] [PubMed] [Google Scholar]
- 66.Mahboob S, Mohamedali A, Ahn SB, Schulz-Knappe P, Nice E, Baker MS. J. Proteomics. 2015;127:300–309. doi: 10.1016/j.jprot.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 67.Chertov O, Biragyn A, Kwak LW, Simpson JT, Boronina T, Hoang VM, Prieto DA, Conrads TP, Veenstra TD, Fisher RJ. Proteomics. 2004;4:1195–203. doi: 10.1002/pmic.200300677. [DOI] [PubMed] [Google Scholar]
- 68.Baracat-Pereira MC, Barbosa MdO, Magalhaes MJ, Jr, Carrijo LC, Games PD, Almeida HO, Sena Netto JF, Pereira MR, Goncalves de Barros E. Genet. Mol. Biol. 2012;35:283–291. doi: 10.1590/S1415-47572012000200009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fiedler GM, Leichtle AB, Kase J, Baumann S, Ceglarek U, Felix K, Conrad T, Witzigmann H, Weimann A, Schütte C, Hauss J, Büchler M, Thiery J. Clin. Cancer Res. 2009;15:3812–3819. doi: 10.1158/1078-0432.CCR-08-2701. [DOI] [PubMed] [Google Scholar]
- 70.Page NM, Weston-Bell NJ. Methods Mol. Biol. 2010;615:293–312. doi: 10.1007/978-1-60761-535-4_22. [DOI] [PubMed] [Google Scholar]
- 71.Zhang F, Dulneva A, Bailes J, Soloviev M. Methods Mol. Biol. 2010;615:313–344. doi: 10.1007/978-1-60761-535-4_23. [DOI] [PubMed] [Google Scholar]
- 72.Pisano MR, Ford M. BioTechniques. 2017;63:86–87. [Google Scholar]
- 73.Ritz D, Gloger A, Weide B, Garbe C, Neri D, Fugmann T. Proteomics. 2016;16:1570–1580. doi: 10.1002/pmic.201500445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cho W, Jung K, Regnier FE. Anal. Chem. 2008;80:5286–5292. doi: 10.1021/ac8008675. [DOI] [PubMed] [Google Scholar]
- 75.Teresa DB, Santos RA, Takahashi CS, Carrara HH, Moreira HW, Mattos LC, Lia-Neto N, Cunha LA, Bassi CL, Soares EG. Tumor Biol. 2010;31:401–409. doi: 10.1007/s13277-010-0048-2. [DOI] [PubMed] [Google Scholar]
- 76.Castro LSEPW, Kviecinski MR, Ourique F, Parisotto EB, Grinevicius VMAS, Correia JFG, Wilhelmm FD, Pedrosa RC. Redox Biol. 2016;10:90–99. doi: 10.1016/j.redox.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Furukawa A, Kawamoto Y, Chiba Y, Takei S, Hasegawa-Ishii S, Kawamura N, Yoshikawa K, Hosokawa M, Oikawa S, Kato M, Oikawa S. Neurobiol. Dis. 2011;43:706–714. doi: 10.1016/j.nbd.2011.05.024. [DOI] [PubMed] [Google Scholar]
- 78.Zheng Y, Huang X, Kelleher NL. Curr. Opin. Chem. Biol. 2016;33:142–150. doi: 10.1016/j.cbpa.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boonyasit Y, Laiwattanapaisal W, Chailapakul O, Emneus J, Heiskanen AR. Anal. Chem. 2016;88:9582–9589. doi: 10.1021/acs.analchem.6b02234. [DOI] [PubMed] [Google Scholar]
- 80.Chen H-H, Wu C-H, Tsai M-L, Huang Y-J, Chen S-H. Anal. Chem. 2012;84:8635–8641. doi: 10.1021/ac301756d. [DOI] [PubMed] [Google Scholar]
- 81.Li L, Zang W, Zhang X. Anal. Bioanal. Chem. 2016;408:1507–1513. doi: 10.1007/s00216-015-9258-1. [DOI] [PubMed] [Google Scholar]
- 82.Anderson LC, DeHart CJ, Kaiser NK, Fellers RT, Smith DF, Greer JB, LeDuc RD, Blakney GT, Thomas PM, Kelleher NL, Hendrickson CL. J. Proteome Res. 2017;16:1087–1096. doi: 10.1021/acs.jproteome.6b00696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhu D, Hou L, Hu B, Zhao H, Sun J, Wang J, Meng X. Sci. Rep. 2016;6:37478. doi: 10.1038/srep37478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Carpentieri A, Giangrande C, Pucci P, Amoresano A. Eur. Mass Spectrom. 2010;16:123–149. doi: 10.1255/ejms.1035. [DOI] [PubMed] [Google Scholar]
- 85.Valero ML, Sendra R, Pamblanco M. J. Proteomics. 2016;136:183–192. doi: 10.1016/j.jprot.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 86.Bryson BD, Del Rosario AM, Gootenberg JS, Yaffe MB, White FM. Proteomics. 2015;15:1470–1475. doi: 10.1002/pmic.201400401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yu Q, Shi X, Feng Y, Kent KC, Li L. Anal. Chim. Acta. 2017;968:40–49. doi: 10.1016/j.aca.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Piunti A, Hashizume R, Morgan MA, Bartom ET, Horbinski CM, Marshall SA, Rendleman EJ, Ma Q, Takahashi Y–h, Woodfin AR, Kelleher N. Nat. Med. 2017;23:493–500. doi: 10.1038/nm.4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hauser A, Penkert M, Hackenberger CPR. Acc. Chem. Res. 2017;50:1883–1893. doi: 10.1021/acs.accounts.7b00170. [DOI] [PubMed] [Google Scholar]
- 90.Nishi H, Demir E, Panchenko AR. J. Mol. Biol. 2015;427:511–520. doi: 10.1016/j.jmb.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Thomson M, Gunawardena J. Nature. 2009;460:274–277. doi: 10.1038/nature08102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cha H, Shapiro P. J. Cell Biol. 2001;153:1355–1367. doi: 10.1083/jcb.153.7.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pufall MA, Lee GM, Nelson ML, Kang HS, Velyvis A, Kay LE, McIntosh LP, Graves BJ. Science. 2005;309:142–145. doi: 10.1126/science.1111915. [DOI] [PubMed] [Google Scholar]
- 94.Wu RC, Qin J, Yi P, Wong J, Tsai SY, Tsai M-J, O’Malley BW. Mol. Cell. 2004;15:937–949. doi: 10.1016/j.molcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 95.Prabakaran S, Everley RA, Landrieu I, Wieruszeski J-M, Lippens G, Steen H, Gunawardena J. Mol. Syst. Biol. 2011;7:482–497. doi: 10.1038/msb.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Raffick RAR, Remaley ATR. Biochemia Medica. 2014;24:31–44. doi: 10.11613/BM.2014.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dunn JD, Reid GE, Bruening ML. Mass Spectrom. Rev. 2010;29:29–54. doi: 10.1002/mas.20219. [DOI] [PubMed] [Google Scholar]
- 98.Bergstroem LS, Artemenko KA, Elfineh L, Mayrhofer C, Zubarev RA, Bergquist J, Pettersson U. Cell. Signalling. 2011;23:1387–1395. doi: 10.1016/j.cellsig.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 99.Artemenko KA, Lind SB, Elfineh L, Mayrhofer C, Zubarev RA, Bergquist J, Pettersson U. Analyst. 2011;136:1971–1978. doi: 10.1039/c0an00649a. [DOI] [PubMed] [Google Scholar]
- 100.Nagano K, Shinkawa T, Yabuki N, Mutoh H, Inomata N, Watanabe Y, Ashihara M, Nagahashi S, Ishii N, Aoki Y, Haramura M. J. Proteomics. 2011;74:319–326. doi: 10.1016/j.jprot.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 101.Zhang H, Zha X, Tan Y, Hornbeck PV, Mastrangelo AJ, Alessi DR, Polakiewicz RD, Comb M. J. Biol. Chem. 2002;277:39379–39387. doi: 10.1074/jbc.M206399200. [DOI] [PubMed] [Google Scholar]
- 102.Astoul E, Laurence AD, Totty N, Beer S, Alexander DR, Cantrell DA. J. Biol. Chem. 2003;278:9267–9275. doi: 10.1074/jbc.M211252200. [DOI] [PubMed] [Google Scholar]
- 103.Tinti M, Nardozza AP, Ferrari E, Sacco F, Corallino S, Castagnoli L, Cesareni G. New Biotechnol. 2012;29:571–577. doi: 10.1016/j.nbt.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 104.Zerweck J, Masch A, Schutkowski M. Methods Mol. Biol. 2009;524:169–180. doi: 10.1007/978-1-59745-450-6_12. [DOI] [PubMed] [Google Scholar]
- 105.Matheron L, van den Toorn H, Heck AJR, Mohammed S. Anal. Chem. 2014;86:8312–8320. doi: 10.1021/ac501803z. [DOI] [PubMed] [Google Scholar]
- 106.Collins MO, Yu L, Campuzano I. Mol. Cell. Proteomics. 2008;7:1331–1348. doi: 10.1074/mcp.M700564-MCP200. [DOI] [PubMed] [Google Scholar]
- 107.Cantin GT, Yi W, Lu B. J. Proteome Res. 2008;7:1346–1351. doi: 10.1021/pr0705441. [DOI] [PubMed] [Google Scholar]
- 108.Ye J, Zhang X, Young C, Zhao X, Hao Q, Cheng L, Jensen O. J. Prot. Res. 2010;9:3561–3573. doi: 10.1021/pr100075x. [DOI] [PubMed] [Google Scholar]
- 109.Fíla J, Honys D. Amino Acids. 2012;43:1025–1047. doi: 10.1007/s00726-011-1111-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Machida M, Kosako H, Shirakabe K. FEBS J. 2007;274:1576–1587. doi: 10.1111/j.1742-4658.2007.05705.x. [DOI] [PubMed] [Google Scholar]
- 111.Wang H, Duan Y, Zhong W. ACS Appl. Mater. Interfaces. 2015;7:26414–26420. doi: 10.1021/acsami.5b09348. [DOI] [PubMed] [Google Scholar]
- 112.Wolschin F, Wienkoop S, Weckwerth W. Proteomics. 2005;5:4389–4397. doi: 10.1002/pmic.200402049. [DOI] [PubMed] [Google Scholar]
- 113.Hennrich ML, Groenewold V, Kops GJPL, Heck AJR, Mohammed S. Anal. Chem. 2011;83:7137–7143. doi: 10.1021/ac2015068. [DOI] [PubMed] [Google Scholar]
- 114.Hiroi H, Christenson LK, Chang L, Sammel MD, Berger SL, Strauss JF. Mol. Endocrinol. 2004;18:791–806. doi: 10.1210/me.2003-0305. [DOI] [PubMed] [Google Scholar]
- 115.Wu Z, Connolly J, Biggar KK. FEBS J. 2017;284:2732–2744. doi: 10.1111/febs.14056. [DOI] [PubMed] [Google Scholar]
- 116.Carlson SM, Gozani O. J. Mol. Biol. 2014;426:3350–3362. doi: 10.1016/j.jmb.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ko S, Ahn J, Song CS, Kim S, Knapczyk-Stwora K, Chatterjee B. Mol. Endocrinol. 2011;25:433–444. doi: 10.1210/me.2010-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dobson THW, Hatcher RJ, Swaminathan J, Das CM, Shaik S, Tao R-H, Milite C, Castellano S, Taylor PH, Sbardella G, Gopalakrishnan V. Mol. Cancer Res. 2017;15:1073–1084. doi: 10.1158/1541-7786.MCR-16-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pagans S, Sakane N, Schnolzer M, Ott M. Methods. 2011;53:91–96. doi: 10.1016/j.ymeth.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Garnett GAE, Starke MJ, Shaurya A, Li J, Hof F. Anal. Chem. 2016;88:3697–3703. doi: 10.1021/acs.analchem.5b04508. [DOI] [PubMed] [Google Scholar]
- 121.Gober IN, Waters MLJ. J. Am. Chem. Soc. 2016;138:9452–9459. doi: 10.1021/jacs.6b02836. [DOI] [PubMed] [Google Scholar]
- 122.Ong S-E, Mittler G, Mann M. Nat. Methods. 2004;1:119–126. doi: 10.1038/nmeth715. [DOI] [PubMed] [Google Scholar]
- 123.Xie L, Zeng J, Zhou M, Duan X, Li Q, Zhang Z, Luo H, Pang L, Li W, Liao G. Int. J. Biochem. Cell Biol. 2015;59:193–202. doi: 10.1016/j.biocel.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 124.Huang D, Li Z–H, You D, Zhou Y, Ye B-C. Appl. Microbiol. Biotechnol. 2015;99:1399–1413. doi: 10.1007/s00253-014-6144-2. [DOI] [PubMed] [Google Scholar]
- 125.Kwon OK, Sim J, Kim SJ, Oh HR, Nam DH, Lee S. Biochimie. 2016;121:219–227. doi: 10.1016/j.biochi.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 126.Zhu X, Liu X, Cheng Z, Zhu J, Xu L, Wang F, Qi W, Yan J, Liu N, Sun Z. Sci. Rep. 2016;6:19926. doi: 10.1038/srep19926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Komatsu Y, Iwabata H, Yoshida M. Anti-Acetyllysine Antibody: A Useful Tool for Listening to Posttranslational Language. In: Simmons MA, editor. Trends Monoclonal Antibody Research. Nova Science Publishers, Inc.; Hauppauge, NY: 2005. pp. 37–57. [Google Scholar]
- 128.Kim S-Y, Sim CK, Zhang Q, Tang H, Brunmeir R, Pan H, Karnani N, Han W, Zhang K, Xu F. PLoS One. 2016;11:e0162528. doi: 10.1371/journal.pone.0162528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shaw PG, Chaerkady R, Zhang Z, Davidson NE, Pandey A. Anal. Chem. 2011;83:3623–3626. doi: 10.1021/ac1026176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Xie L, Zeng J, Zhou M, Duan X, Li Q, Zhang Z, Luo H, Pang L, Li W, Liao G. Int. J. Biochem. Cell Biol. 2015;59:193–202. doi: 10.1016/j.biocel.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 131.Huang D, Li Z–H, You D, Zhou Y, Ye B-C. Appl. Microbiol. Biotechnol. 2015;99:1399–1413. doi: 10.1007/s00253-014-6144-2. [DOI] [PubMed] [Google Scholar]
- 132.Kim SY, Zhang Q, Brunmeir R, Han W, Xu F. PLoS One. 2015;10:e0133448. doi: 10.1371/journal.pone.0133448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kim SY, Sim CK, Tang H, Han W, Zhang K, Xu F. PLoS One. 2015;10:e0140619. doi: 10.1371/journal.pone.0140619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Colaert N, Helsens K, Martens L, Vandekerckhove J, Gevaert K. Nat. Methods. 2009;6:786–787. doi: 10.1038/nmeth1109-786. [DOI] [PubMed] [Google Scholar]
- 135.Shu H, Zhang S, Kang X, Li S, Qin X, Sun C, Lu H, Liu Y. Acta Biochim. Biophys. Sin. 2011;43:528–534. doi: 10.1093/abbs/gmr038. [DOI] [PubMed] [Google Scholar]
- 136.Cho W, Jung K, Regnier FE. J. Sep. Sci. 2010;33:1438–1447. doi: 10.1002/jssc.200900860. [DOI] [PubMed] [Google Scholar]
- 137.Liu Y, He J, Lubman DM. Methods Mol. Biol. 2013;951:69–77. doi: 10.1007/978-1-62703-146-2_6. [DOI] [PubMed] [Google Scholar]
- 138.Xin A-J, Cheng L, Diao H, Wang P, Gu Y-H, Wu B, Wu Y-C, Chen G-W, Zhou S–M, Guo S-J. Clin. Proteomics. 2014;11:10. doi: 10.1186/1559-0275-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.O’Connor BF, Monaghan D, Cawley J. Methods Mol. Biol. 2017;1485:411–420. doi: 10.1007/978-1-4939-6412-3_23. [DOI] [PubMed] [Google Scholar]
- 140.Bonneau PR, Graycar TP, Estell DA, Jones JB. J. Am. Chem. Soc. 1991;113:1026–1030. [Google Scholar]
- 141.Bott R, Ultsch M, Wells J, Powers D, Burdick D, Struble M, Burnier J, Estell D, Miller J, Graycar T, Adams R, Power S. Importance of Conformational Variability in Protein Engineering of Subtilisin. Biotechnology in Agricultural Chemistry; ACS Symposium Series, Vol. 334; American Chemical Society; Washington, DC. 1987. pp. 139–147. [DOI] [Google Scholar]
- 142.Chicz RM, Regnier FE. Anal. Chem. 1989;61:2059–2066. doi: 10.1021/ac00193a012. [DOI] [PubMed] [Google Scholar]
- 143.Chicz RM, Regnier FE. Anal. Chem. 1989;61:1742–1749. doi: 10.1021/ac00190a030. [DOI] [PubMed] [Google Scholar]
- 144.Chicz RM, Regnier FE. J. Chromatogr. 1990;500:503–518. doi: 10.1016/s0021-9673(00)96088-6. [DOI] [PubMed] [Google Scholar]
- 145.Wu Z, Wei B, Zhang X, Wirth MJ. Anal. Chem. 2014;86:1592–1598. doi: 10.1021/ac403233d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhou Y, Zhang X, Fornelli L, Compton PD, Kelleher N, Wirth MJ. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2017;1044–1045:47–53. doi: 10.1016/j.jchromb.2016.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cai W, Tucholski T, Chen B, Alpert AJ, McIlwain S, Kohmoto T, Jin S, Ge Y. Anal. Chem. 2017;89:5467–5475. doi: 10.1021/acs.analchem.7b00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Pedrali A, Tengattini S, Marrubini G. Molecules. 2014;19:9070–9088. doi: 10.3390/molecules19079070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pesavento JJ, Bullock CR, LeDuc RD, Mizzen CA, Kelleher NL. J. Biol. Chem. 2008;283:14927–14937. doi: 10.1074/jbc.M709796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Garcia BA, Pesavento JJ, Mizzen CA, Kelleher NL. Nat. Methods. 2007;4:487–489. doi: 10.1038/nmeth1052. [DOI] [PubMed] [Google Scholar]
- 151.Pesavento JJ, Mizzen CA, Kelleher NL. Anal. Chem. 2006;78:4271–4280. doi: 10.1021/ac0600050. [DOI] [PubMed] [Google Scholar]
- 152.Pesavento JJ, Yang H, Kelleher NL, Mizzen CA. Mol. Cell. Biol. 2008;28:468–486. doi: 10.1128/MCB.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Toby TK, Abecassis M, Kim K, Thomas PM, Fellers RT, Le Duc RD, Kelleher NL, Demetris J, Levitsky J. Am. J. Transplant. 2017;17:2458–2467. doi: 10.1111/ajt.14359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hancock WS, Sparrow JT. J. Chromatogr. 1981;206:71–82. doi: 10.1016/s0021-9673(00)82606-0. [DOI] [PubMed] [Google Scholar]
- 155.Young NL, DiMaggio PA, Plazas-Mayorca MD, Baliban RC, Floudas CA, Garcia BA. Mol. Cell. Proteomics. 2009;8:2266–2284. doi: 10.1074/mcp.M900238-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tian Z, Tolic N, Zhao R, Moore RJ, Hengel SM, Robinson EW, Stenoien DL, Wu S, Smith RD, Pasa-Tolic L. Gen. Biol. 2012;13:R86. doi: 10.1186/gb-2012-13-10-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Pesavento JJ, Bullock CR, LeDuc RD, Mizzen CA, Kelleher NL. J. Biol. Chem. 2008;283:14927–14937. doi: 10.1074/jbc.M709796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Toby TK, Fornelli L, Kelleher NL. Annu. Rev. Anal. Chem. 2016;9:499–519. doi: 10.1146/annurev-anchem-071015-041550. [DOI] [PMC free article] [PubMed] [Google Scholar]