Abstract
Recent studies on brain imaging analysis witnessed the core roles of machine learning techniques in computer-assisted intervention for brain disease diagnosis. Of various machine-learning techniques, sparse regression models have proved their effectiveness in handling high-dimensional data but with a small number of training samples, especially in medical problems. In the meantime, deep learning methods have been making great successes by outperforming the state-of-the-art performances in various applications. In this paper, we propose a novel framework that combines the two conceptually different methods of sparse regression and deep learning for Alzheimer’s disease/mild cognitive impairment diagnosis and prognosis. Specifically, we first train multiple sparse regression models, each of which is trained with different values of a regularization control parameter. Thus, our multiple sparse regression models potentially select different feature subsets from the original feature set; thereby they have different powers to predict the response values, i.e., clinical label and clinical scores in our work. By regarding the response values from our sparse regression models as target-level representations, we then build a deep convolutional neural network for clinical decision making, which thus we call ‘ Deep Ensemble Sparse Regression Network.’ To our best knowledge, this is the first work that combines sparse regression models with deep neural network. In our experiments with the ADNI cohort, we validated the effectiveness of the proposed method by achieving the highest diagnostic accuracies in three classification tasks. We also rigorously analyzed our results and compared with the previous studies on the ADNI cohort in the literature.
Keywords: Alzheimer’s disease, Convolutional neural network, Deep ensemble learning, Sparse regression model
1. Introduction
With the advent and advance of brain imaging techniques such as Magnetic Resonance Imaging (MRI), Positron Emission Tomography (PET), Diffusion Tensor Imaging (DTI), functional MRI (fMRI), imaging-based brain disorder diagnosis or prognosis has always been of great interest in computer-assisted interventions (Davatzikos et al., 2008; Fan et al., 2008; Cuingnet et al., 2011). However, it was limited to analyze the high dimensional neuroimaging data until the application of machine learning techniques, which are now playing core roles in the field (Davatzikos et al., 2008).
Given a brain image, to identify whether a subject has a certain brain disorder can be regarded as a classification task. From a machine learning standpoint, the prevalent framework for brain imaging data analysis for diagnosis can be summarized as preprocessing, feature extraction/selection, and classifier learning. Although machine learning techniques can be basically involved in all of these steps, in this paper, we mainly focus on the steps of feature extraction/selection and classifier learning. For applications of machine learning in brain imaging analysis, e.g., tissue segmentation, registration, atlas construction, etc., please refer to Powell et al. (2008), Liao et al. (2012) and Wang and Summers (2014).
One of the main challenges in brain imaging analysis is the high dimensionality of data, but a small number of samples are available. While various methods have been proposed for dimensionality reduction in the field of machine learning, due to interpretational requirement, it is limited for the applicable methods. Further, motivated by the principle of parsimony in many areas of science, i.e., the simplest explanation of a given observation should be preferred over more complicated ones, sparsity-inducing penalization is considered as one of the key techniques in machine learning. In light of these, sparse regression methods with different forms of regularization terms (Tibshirani, 1996; Zou and Hastie, 2005; Yuan and Lin, 2006), and their variants (Liu et al., 2009; Wang et al., 2011; Zhang and Shen, 2012; Zhou et al., 2012; Zhu et al., 2014; Suk et al., 2015b) have been proposed and demonstrated their validity for feature selection in medical problems.
Meanwhile, deep representation learning has recently been showing state-of-the-art performances in various fields of computer vision, speech recognition, natural language processing, and medical image analysis. To this end, it has also been considered as one of the major tools in brain imaging data analysis (Li et al., 2014b; Plis et al., 2014; Suk et al., 2015a; 2016b; Pereira et al., 2016; Brosch et al., 2016; Dou et al., 2016) by achieving promising performances. In deep learning, a Convolutional Neural Network (CNN) is of main stream for image analysis thanks to its modeling characteristic that helps discover local structural or configural relations in observations. While it is desirable to apply CNNs to learn feature representations from a whole-brain MRI for brain disease diagnosis, it is still limited because of its huge number of network parameters, e.g., millions number of parameters, that should be learned from a small number data, e.g., less than 1000 samples. In this regard, Brosch and Tam (2013) downsampled images before training a convolutional restricted Boltzmann machine for manifold learning of brain MRIs. However, image downsampling inevitably causes information loss of subtle structural changes especially in Mild Cognitive Impairment (MCI). Unlike the conventional methods that were mostly used for feature representation learning, in this work, we utilize a CNN for ensemble modeling by finding a non-linear mapping function from matrix-formed predictions, whose dimension is considerably lower than the original dimension of imaging features, to make a clinical decision.
One prevalent step of the sparsity-inducing regularization methods is to choose the optimal value of a regularization control parameter, mostly via cross-validation with a grid search strategy. It is, however, possible that for different validation sets, it can be different for the optimal value of a regularization control parameter. In this work, instead of choosing a single value for the regularization control parameter, we propose to build a set of sparse regression models, each of which is trained with different values of the regularization control parameter. We regard outputs of the sparse regression models, e.g., clinical label and clinical scores in our work, as target-level representations. This is comparable to the previous work that mostly used the sparse regression method for feature selection and then trained a classifier such as support vector machine based on the selected features only. In our work, the sparse regression model plays the role of predicting target outputs, which we consider as abstract feature representation, along with informative features selection. It is noteworthy that the predicted values from multiple sparse regression models are stacked in an ascending or descending 1 order based on the regularization control parameter values. In this way, we can exploit the prediction values of “neighboring” sparse regression models, which are trained with relatively similar regularization control parameter values, as additional information. We then build a CNN that hierarchically combines the prediction values from sparse regression models, i.e., target-level representations, for MRI-based brain disease diagnosis, especially, Alzheimer’s Disease (AD) and its prodromal stage, MCI.
Our deep network discovers the optimal weights to ensemble multiple sparse regression models in a hierarchical and non-linear way, which thus we call ‘Deep Ensemble Sparse Regression Network’ (DeepESRNet). The proposed method can be understood as ensemble of expert systems. That is, sparse regression models with different regularization control values can be considered as experts that output their own prediction values. The prediction values of different experts are then combined by CNN that finds non-linear weighting coefficients among them.
To our knowledge, one of the beauties of CNN is to hierarchically integrate information distributed in the input by means of convolution and pooling operations. We utilize this feature to integrate the outputs from multiple sparse regression models, thus to build a strong classifier. In a convolution layer, the learnable kernels find relations among outputs, i.e., predicted clinical label and clinical scores, from different sparse regression models. In a pooling layer, the max operation plays the role of drawing a mid-level decision among neighboring regression models. By repeating these operations over layers, our CNN finally make a decision by integrating all the information from multiple sparse regression models.
The rationale of using a CNN is two aspects: (1) In machine learning, it is generally known that combining multiple classifiers is helpful for the improvement of the performance of individual classifiers (Kittler et al., 1998). In this work, we use a CNN with a target-level representation as input to combine the outputs of multiple sparse regression models. Due to the use of non-linear activation functions, our CNN finds non-linear weight coefficients in combining the outputs from multiple sparse regression models. (2) In our target-level representation, element values (i.e., predicted clinical label and clinical scores) in the same row are obtained from the same sparse regression model, thus they are naturally related to each other. In the meantime, element values of neighboring rows are from different sparse regression models trained with different values of the regularization control parameter. Note that our target-level representation is obtained by stacking the outputs of multiple sparse regression models in an ordered way based on their regularization control parameter values. Sparse regression models trained with slightly different regularization control parameter values are likely to produce similar outputs. Thus, it is expected that values of the neighboring rows are more related to each other than those of the rows apart in the target-level representation. A CNN is suitable to well discover such local configural features and to combine the overall information hierarchically.
The main contributions of our work can be two-fold: 1) The different values of the regularization control parameter are used to select different feature subsets with different weight coefficients. Hence, different regression models can predict both clinical status and clinical scores in different feature spaces. Also, the use of target-level representations from multiple sparse regression models has the effect of reducing dimensionality of an observation, allowing us to use CNN with less concern of data insufficiency. 2) Our DeepESRNet built on the outputs of the sparse regression models finds a (sub)optimal way of combining the regression models in a non-linear manner with the following rationales. First, the target-level representation values (i.e., response values of clinical label and clinical scores in our work) of a sparse regression model are highly related to each other (intra-model relation). Second, sparse regression models trained with similar regularization control parameter values tend to find similar weight coefficients, thus similar target-representations (inter-model relation). In these regards, we couple the target-level representation of the same sparse response model(s) and of the neighboring sparse response models via local learnable kernels. The local combination allows us to exploit complementary information and their hierarchical and non-linear integration over the whole target-level representation helps make a robust clinical decision. It is also noteworthy that since we treat the outputs from individual sparse regression models as target-level representation, we justify to re-use the training data for learning network parameters after training sparse regression models.
Methodologically, our method can be considered as a cascading classifier, which is one case of ensemble learning. Our sparse regression models take a vector of features and output clinical label and clinical scores, which are usually used for making a decision. As a cascading classifier, our method further uses such outputs as inputs to another model, i.e., CNN.
The rest of the paper is organized as follows: In Section 2, we review the previous work on brain imaging-based AD disease diagnosis in the literature. We then describe the dataset used in this work and explain the steps involved in preprocessing for feature extraction in Section 3. We elaborate upon the basics in sparse linear/logistic regression and propose a novel deep ensemble sparse regression network in Section 4. The experimental results on the public ADNI dataset are presented by comparing with competing methods in Section 5 and the discussion on our experimental results and comparison with state-of-the-art results in the literature are detailed in Section 6. We conclude this paper by summarizing the proposed method and suggesting future research directions in Section 7.
2. Related work
The small number of training samples compared to high-dimension of imaging data has been one of the main challenges in brain imaging-based disease diagnosis. To circumvent the problem, various machine learning techniques were proposed in the literature. Depending on their strategies, techniques can be categorized into 1) feature embedding (Roweis and Saul, 2000; He et al., 2006; Liu et al., 2013) and 2) feature selection (Wang et al., 2011; Zhu et al., 2014; Tohka et al., 2016). Basically, the common goal of the methods of both categories is to learn dimension-reduced features or representations from a small number of training samples, while still pertaining useful information for target tasks. Specifically, the feature embedding methods find a latent low-dimensional space, where the original features can be efficiently represented without losing much information. Meanwhile, the feature selection methods try to find associations among features, based on which they select features informative to identify the incidence of a brain disease. From a clinical perspective, the interpretability of the features involved in diagnosis is of great importance. In this regard, the feature selection methods are generally preferred to the feature embedding methods, and thus we consider the feature selection methods in this paper.
Among various feature selection methods, sparse regression models have shown good promises in the small sample size problem with different forms of regularization. The Least Absolute Shrinkage and Selection Operator (LASSO) regression model (Tibshirani, 1996) uses an ℓ1-norm to induce sparsity in the regression coefficients for each target response variable independently. To circumvent the limitation of LASSO, i.e., independence in sparsity of coefficients, Zou and Hastie (2005) proposed the elastic net penalty which is capable of retaining the sparse property of LASSO and utilizing correlation among predictors simultaneously. Yuan and Lin (2006) proposed the group LASSO penalty that leads a group sparse property by imposing penalties in a group-wise manner via an ℓ2, 1-norm, and thus select features whose weights are non-zero to all predictors within the selected groups. This group LASSO has been successfully applied in brain imaging-based AD/MCI diagnosis (Wang et al., 2011; Zhang and Shen, 2012; Wee et al., 2012; Suk et al., 2015b). For example, Zhang and Shen (2012) designed a multi-task sparse regression model with an ℓ2, 1-norm for AD diagnosis. Wang et al. (2011) proposed sparse joint classification and regression with logistic regression function for classification and linear regression function for regression. Zhou et al., introduced a smoothness constraint along with ℓ1- and ℓ2, 1-norm penalties for predicting disease progression. Suk et al. (2015b) devised a discriminative group sparse representation method by penalizing a large within-class variance and a small between-class variance in estimating functional connectivities.
In the meantime, inspired by great successes in the fields of computer vision and speech recognition, deep learning methods have been applied to discover hierarchical features in brain images, too. For example, Suk et al. (2015a) used a stacked auto-encoder to find non-linear relations among gray matter volumes of brain regions. Suk et al. (2014) also proposed a generative deep learning to integrate structural and functional patterns inherent in MRI and PET, respectively, by learning shared representations. Similarly, Ithapu et al. (2015) also devised a deep learning algorithm called a randomized denoising auto-encoder marker to integrate multimodal data of PET and MRI. However, due to complex composition of non-linear patterns in deep learning, it still remains challenging to interpret the learned representations. Hence, unlike other fields such as computer vision and speech recognition, it is still popular to use hand-crafted features for neuroscientific interpretations in the field of brain disease diagnosis. In this paper, we propose a novel brain disease diagnosis system based on sparse regression models with interpretable volumetric features, and further use a deep learning method to combine the regression models to make a clinical decision.
3. Materials and image processing
3.1. Dataset
We analyzed a baseline MRI dataset from the ADNI database (http://www.loni.ucla.edu/ADNI). The dataset included 805 subjects of 186 (AD), 393 (MCI), and 226 (Normal Control, NC). For the MCI subjects, they were clinically further subdivided into 167 progressive MCI (pMCI), who progressed to AD in 18 months, and 226 stable MCI (sMCI), who did not progress to AD in 18 months. Each subject had both Mini-Mental State Examination (MMSE) and Alzheimer’s Disease Assessment Scale - Cognition (ADAS-Cog) scores recorded. The subjects were in the age between 55 and 90, with a study partner, who provided an independent evaluation of functioning. All of 805 subjects met the following general inclusion criteria: (a) NC subjects: an MMSE between 25 and 30 (inclusive), a clinical dementia rating (CDR) of 0, non-depressed, non-MCI, and non-demented; (b) mild AD subjects: an MMSE score between 18 and 27 (inclusive), a CDR of 0.5 or 1.0, and met the National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer’s Disease and Related Disorders Association (NINCDS/ADRDA) criteria for probable AD. Demographic and clinical information of subjects is provided in Table 1.
Table 1.
Demographic and clinical information of the subjects. (pMCI: progressive MCI, sMCI: stable MCI, SD: Standard Deviation).
| AD | pMCI | sMCI | NC | |
|---|---|---|---|---|
| Number of subjects | 186 | 167 | 226 | 226 |
| Female/male | 87/99 | 65/102 | 75/151 | 108/118 |
| Age (Mean ± SD) | 75.37 ± 7.55 | 74.89 ± 6.83 | 75.00 ± 7.63 | 75.96 ± 5.04 |
| Education years (Mean ± SD) | 14.70 ± 3.13 | 15.69 ± 2.87 | 15.62 ± 3.18 | 16.03 ± 2.88 |
| MMSE (Mean ± SD) | 23.28 ± 2.02 | 26.59 ± 1.71 | 27.28 ± 1.77 | 29.11 ± 1.00 |
| ADAS-Cog (Mean ± SD) | 18.44 ± 6.71 | 13.30 ± 4.05 | 10.33 ± 4.31 | 6.21 ± 2.93 |
| CDR (Mean ± SD) | 0.75 ± 0.25 | 0.50 ± 0.00 | 0.50 ± 0.03 | 0.00 ± 0.00 |
3.2. Preprocessing
All structural MRI data in this study were acquired using 1.5T scanners. The baseline MRI data were downloaded from ADNI in the Neuroimaging Informatics Technology Initiative (NIfTI) format, which had already been processed for spatial distortion correction caused by gradient nonlinearity and B1 field inhomogeneity. We further processed the MR images by applying the typical procedures of Anterior Commissure (AC)-Posterior Commissure (PC) correction, skull-stripping, and cerebellum removal. Specifically, we used MIPAV software 2 for AC-PC correction, resampled images to 256 × 256 × 256, and applied N3 algorithm (Sled et al., 1998) to correct intensity inhomogeneity. An accurate and robust skull stripping (Wang et al., 2014) was performed along with cerebellum removal. To ensure the clean and dura removal, we manually reviewed the skull-stripped images. Then, FAST in FSL package 3 was used for structural MRI image segmentation into three tissue types of Gray Matter (GM), White Matter (WM), and CerebroSpinal Fluid (CSF). 4 We finally parcellated them into 93 Regions Of Interest (ROIs), whose list is provided in Table B1, by warping the atlas of Kabani et al. (1998), which has been widely used for AD/MCI diagnosis studies (Davatzikos et al., 2011; Negash et al., 2013; Suk et al., 2015a) to each subject’s space via HAMMER (Shen and Davatzikos, 2002), although many other advanced registration methods could be also used (Avants and Gee, 2004; Zacharaki et al., 2009; Xue et al., 2006; Shen et al., 1999). Next, we generated the regional volumetric maps, called RAVENS maps, using a tissue preserving image warping method (Davatzikos et al., 2001). In this work, we considered only the spatially normalized GM densities, due to its relatively high relevance to AD compared to WM and CSF (Liu et al., 2012). For each of the 93 ROIs, we computed the GM tissue volumes, which is widely used in the field for AD/MCI diagnosis (Davatzikos et al., 2011; Hinrichs et al., 2011; Zhang and Shen, 2012; Suk et al., 2015a), as features, i.e., 93-dimensional features from an MR image. 5
4. Deep ensemble sparse regression network
4.1. Notations
Throughout this paper, we denote matrices as boldface uppercase letters, vectors as boldface lowercase letters, and scalars as normal italic letters, respectively. For a matrix X = [xi j], its i-th row and j-th column are denoted as xi and xj, respectively. Also, we denote the Frobenius norm and ℓ2, 1-norm of a matrix X as and , respectively. We use a superscript ⊤ for transpose of a vector or a matrix.
4.2. Sparse linear/logistic regression
Assume that we are given N samples of a training set 𝒟 = {X, T}, where X ∈ ℝ D ×N and T ∈ ℝ L ×N denote, respectively, D-dimensional brain imaging features and the corresponding L-dimensional target values. Sparse regression models are formulated in the form of an optimization problem as follows:
| (1) |
where ℒ (𝒟; Θ) denotes a loss function and Ω(Θ) denotes a regularization term over a parameter set Θ, and λ is a regularization control parameter.
In terms of brain disease diagnosis, xi can include brain imaging features, e.g., GM tissue volumes of 93 ROIs xi ∈ ℝ93 in our work, and T can be defined with clinical outputs, e.g., clinical label6 and two clinical scores of MMSE and ADAS-Cog ti ∈ ℝ4 in our work. Earlier, Zhang and Shen (2012) and Wang et al. (2011) independently designed a sparse multi-task learning model with the rationale that since the prediction of clinical label and clinical scores are inter-related it is reasonable to learn the models jointly rather than each prediction task, separately. The main difference between these two methods lies in the way of defining a loss function ℒ. Specifically, Zhang and Shen used a single least square error function for both clinical label prediction (classification task) and clinical scores prediction (regression task) as follows
| (2) |
where Θ = W ∈ ℝ D ×L. In the meantime, Wang et al. used different error functions for classification and regression tasks, namely, a cross-entropy function for classification and a least square error function for regression as follows
| (3) |
| (4) |
| (5) |
where C denotes the number of classes, Θce = Z ∈ ℝ D ×C, ti j = 1 iff xi belongs to the class j, T̃ = [t1; …; tR], R (= L − C) denotes the number of clinical scores, and Θls = P ∈ ℝ D ×R.
However, these loss functions themselves do not enforce sparsity. The regularization term Ω(Θ) plays the role of selecting informative features for target tasks and different forms of the regularization term produce different sets of selected features. Among different forms of regularization in the literature, in this paper, we consider an ℓ2, 1-norm regularizer that induces sparsity in a group-wise manner by following Wang et al. (2011) and Zhang and Shen (2012). Let Θ denote a weight coefficient matrix, i.e., Θ = W for Eq. (2) and Θ = [Z P] for Eq. (3). Note that each element in a column θj of Θ assigns a weight to each of the observed features in predicting the j-th response t̂i j for xi. The regularizer of Ω(Θ) = ||Θ||2, 1 penalizes all coefficients in the same row of Θ and thus guides to select features that are informative to predict all the response variables jointly (Liu et al., 2009).
After finding the optimal weight coefficients Θ̂ with respect to a certain value of the control parameter λ in Eq. (1), we can then predict the response values for an input vector x* as follows:
| (6) |
We consider the predicted vector t̂ as target-level representations obtained from a sparse regression model and utilize it as input to our CNN as described below for clinical decision making.
4.3. Deep ensemble sparse regression network
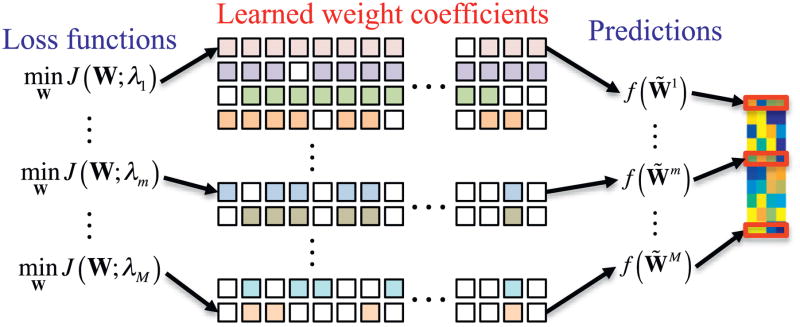
Unlike the conventional approach that finds the optimal control parameter λ in Eq. (1) via cross-validation from a predefined parameter space, in this work, we build multiple sparse regression models, as many as the size M of the predefined parameter space Λ. 7 It is noteworthy that we use the predicted vector obtained by Eq. (6) as a target-level representation of the original input vector. Specifically, we exploit the M learned sparse regression models with different values of the control parameter λ as ‘target-level representation learners’ that transform an input vector xi into a matrix form , where denotes the predicted vector by a regression model trained with λ equal to the m-th value in the parameter set Λ. Fig. 1 illustrates the construction of a target-level representation matrix from M sparse regression models, each of which was trained with different value of a regularization control parameter. The target-level representation matrix is then fed into our CNN for clinical decision making as described below.
Fig. 1.
Multiple sparse regression models with different values of a sparse control parameter, where λ1 < ··· < λm < ··· < λM. The prediction function f (·) is defined by Eq. (6).
Note that different values of a regularization control parameter λ cause to select different feature subsets in learning sparse regression models. Therefore, our M sparse regression models produce different target-level representations, estimated from possibly different feature subsets. However, it is likely that sparse regression models trained with similar regularization control parameter values tend to have similar weight coefficients and thus, to select similar feature subsets. There can be high relations among neighboring rows of a target-level representation in Fig. 1. In the meantime, since the target-level representations are basically related to clinical status and clinical scores jointly estimated from the original brain imaging features, the values are highly related to each other. Thus, there can be high relations among elements of the same row in a target-level representation. In these regards, we couple elements of the same row (intra-model relation) and elements of neighboring rows (inter-model relation) in a target-level representation Ti together via learnable kernels that find configural relations locally. For hierarchical integration of the input representation Ti, we employ a CNN that can discover configural relations inherent in inputs. It should be emphasized that to our best knowledge, this is the first work that systematically combines multiple regression models via CNN for classification.
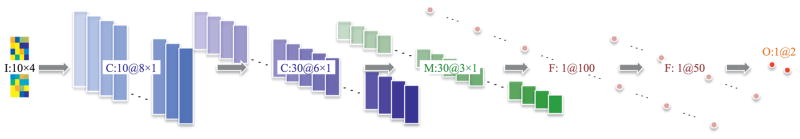
Fig. 2 illustrates our CNN that takes target-level representations obtained from multiple sparse regression models as input and discovers non-linear relations among them in a hierarchical manner for brain disease diagnosis. From a pattern classification standpoint, our CNN is an ensemble classifier that systematically finds the relations of different sparse regression models. Thus, we call our network as ‘Deep Ensemble Sparse Regression Network’ (DeepESRNet).
Fig. 2.
Proposed convolutional neural network of modeling deep ensemble sparse regressions for brain disorder diagnosis. (I: input, C: convolution, M: max-pooling, F: fully-connect, O: output). The online color version provides a clearer view. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
In our DeepESRNet, we have three types of layers, namely, convolution layer, pooling layer, and fully connected layer. At a convolution layer, the previous layer’s outputs (called feature maps) are convolved with learnable kernels and go through a non-linear activation function 8 to form the feature maps of the current layer as follows:
| (7) |
where * is a convolution operator, a superscript (ℓ) denotes a layer index, and F (l − 1) are, respectively, the j-th feature map and the index set of feature maps in the layer (l − 1), is a trainable kernel between the i-th feature map in the layer (l − 1) and j-th feature map in the layer l, is a bias term for a feature map j, and f (·) is a non-linear activation function. A pooling layer is interspersed with the convolution layer for reducing the resolution of feature maps. In our DeepESRNet, we assign a max-pooling layer that partitions an input feature map into a set of non-overlapping regions and outputs the maximum value for each region. Lastly, the fully connected layer is the same as the conventional multilayer neural network such that the inter-layer units are fully connected but with no units within the same layer connected. With this fully connected layer, we finally integrate all the information from the outputs from multiple sparse regression models, i.e., predicted clinical status and clinical scores, for making a clinical decision at the top output layer of our DeepESRNet.
Fig. 2 shows an example of applying our DeepESRNet to target-level representations obtained from 10 sparse regression models. Given a target-level representation , the first convolution layer with 10 feature maps couples target-level representation values in the same row and target-level representation values of neighboring rows in via a kernel in size according to Eq. (7), resulting in . A second convolution layer with 30 feature maps follows to find associations among the values within the same feature maps and also across different feature maps simultaneously via a kernel followed by a non-linear transformation, producing . We then apply a max-pooling operation to each feature map in the second layer, which downsamples the features maps into with a non-overlapping sliding window of 2 × 1 in size. Beyond the max-pooling layer, the network corresponds to a conventional multi-layer neural network with two hidden layers (100 units and 50 units, respectively) and one output layer by taking vectorized values of the 30 feature maps in the max-pooling layer as input. For the output layer, we use a softmax function for classification.
In our DeepESRNet, we have network parameters, i.e., kernels and biases in convolutional layers and also connection weights and biases in the top three multi-layer neural network, that should be learned from data. To train our DeepESRNet, we use a backpropagation algorithm (Rumelhart et al., 1986) with a mini-batch gradient descent method (Cotter et al., 2011).
5. Experimental results
In this section, we validate the effectiveness of the proposed method for AD/MCI diagnosis or prognosis with MRI by comparing with competing methods. We consider three binary classification problems: AD vs. NC, MCI vs. NC, pMCI vs. sMCI. In MCI vs. NC classification, the samples of both pMCI and sMCI subjects were used for MCI class. Due to the limited number of samples, we applied a 10-fold cross validation technique. Specifically, we randomly partitioned the dataset into 10 subsets, each of which included 10% of the samples per class. We repeated experiments for each classification problem 10 times, by using 9 out of 10 subsets for training and the remaining one for testing at each time.
5.1. Experimental settings
With regard to the structure of deep neural networks, i.e., the number of layers and the number of feature maps in each layer, there is no golden rule for those. In this work, we empirically designed our DeepESRNet with two convolutional layers followed by one max-pooling layer, two fully connected layers, and one output layer as shown in Fig. 2. The kernels for two convolution layers were 3 × 4 and 3 × 1 in size, respectively, with a stride of 1. In the max-pooling layer, a kernel of 2 × 1 in size was used with a stride of 2. For two fully connected layers, we set 100 hidden units and 50 hidden units sequentially. We also applied a batch normalization (Ioffe and Szegedy, 2015) to convolution and fully-connected layers except for the last output layer for fast training. No dropout (Srivastava et al., 2014) was involved based on the work of Ioffe and Szegedy (2015), where they empirically presented that dropout could be removed in a batch-normalized network. The network parameters were trained with a stochastic gradient descent approach (Li et al., 2014a) with a mini-batch size of 50, a learning rate of 0.001, a weight decay of 0.005, and a momentum factor of 0.9. We used a MatConvNet toolbox 9 (Vedaldi and Lenc, 2015) to train our DeepESRNet. In regard to computational time, for training our DeepESRNet, it took less than 1 minute in a computer with 3.4 GHz Intel(R) Core i7 CPU and 16GB 1333 MHz DDR3 RAM. All computations were conducted with CPU only without involving GPU computation. The short training time of our CNN was resulted from the greatly reduced input size after applying sparse regression models.
We considered two sparse regression models, namely, 1) Multi-Output Linear Regression with ℓ2, 1-norm regularization (MOLR) (Yuan and Lin, 2006; Zhang and Shen, 2012) and 2) Joint Linear and Logistic Regression with ℓ2, 1-norm regularization (JLLR) (Wang et al., 2011) with the loss function defined by Eqs. (2) and (3), respectively. We set the space of the sparsity control parameter λ with M = 10 values equally spaced between 0.01 and 0.3. 10 By taking the outputs of 10 regression models for each of MOLR and JLLR, we trained MOLR+DeepESRNet and JLLR+DeepESRNet, separately.
To validate the effectiveness of the proposed method, we compared our method with the two baseline sparse regression models and their variants of the following methods
MOLR+SVM: This method first finds the optimal weight coefficients Ŵ in Eq. (2) and then selects the informative features based on the learned weight coefficients. Specifically, after optimizing Eq. (2), we had some zero row vectors in Ŵ and discarded the corresponding features. With the selected features only, we then trained a linear Support Vector Machine (SVM) for classification.
JLLR+SVM: This method jointly finds the optimal weight coefficients P̂ and Ẑ in Eq. (3) and then allows to select the informative features based on the learned weight coefficients. Specifically, after optimizing Eq. (3), we had some zero row vectors in [Ẑ P̂] and discarded the corresponding features. With the selected features only, we then trained a linear SVM for classification.
For a linear SVM in the competing methods of MOLR+SVM and JLLR+SVM, we determined the model parameter C via 5-fold nested cross-validation with the space of C defined as {10−5, 10−4, …, 105} and trained by using the LIBSVM toolbox.11
5.2. Performance evaluation and comparison
Let TP, TN, FP, and FN denote, respectively, True Positive, True Negative, False Positive, and False Negative. For quantitative evaluation and comparison among the competing methods, we considered the metrics of accuracy = (TP + TN)/(TP + TN + FP + FN), sensitivity = TP/(TP + FN), specificity = TN/(TN + FP), Balanced Accuracy (BA) = (sensitivity+specificity)/2, Positive Predictive Value (PPV) = TP/(TP + FP), and Negative Predictive Value (NPV) = TN/(TN + FN). We also considered Area Under the receiver operating characteristics Curve (AUC), which is widely used to evaluate the performance of diagnostic tests in brain disease diagnosis as well as other medical areas.
5.2.1. MOLR as a baseline regression model
The performance comparison among the competing methods with MOLR involved in different ways is presented in Table 2. The proposed method of MOLR+DeepESRNet achieved mean classification accuracies of 90.28% (AD vs. NC), 74.20% (MCI vs. NC) and 73.28% (pMCI vs. sMCI). Our MORL+DeepESRNet improved the mean accuracies by 5.35% (AD vs. NC), 9.54% (MCI vs. NC), and 9.93% (pMCI vs. sMCI) in comparison with MOLR and by 3.41% (AD vs. NC), 7.58% (MCI vs. NC), and 6.62% (pMCI vs. sMCI) in comparison with MOLR+SVM.
Table 2.
Performance comparison to the competing methods with MOLR as a baseline model. (BA: Balanced Accuracy; PPV: Positive Predictive Value; NPV: Negative Predictive Value; AUC: Area Under receiver operating characteristics Curve).
| Tasks | Accuracy (%) | Sensitivity (%) | Specificity (%) | BA (%) | PPV (%) | NPV (%) | AUC | |
|---|---|---|---|---|---|---|---|---|
| MOLR | AD vs. NC | 84.93 ± 6.30 | 83.87 | 87.05 | 85.46 | 84.39 | 85.42 | 0.9165 |
| MCI vs. NC | 64.66 ± 8.16 | 79.40 | 51.67 | 65.53 | 59.87 | 73.08 | 0.7242 | |
| pMCI vs. sMCI | 63.35 ± 7.88 | 56.10 | 71.44 | 63.77 | 64.63 | 62.35 | 0.6824 | |
| MOLR + SVM (Zhang and Shen, 2012) | AD vs. NC | 86.87 ± 4.80 | 88.42 | 86.35 | 87.38 | 82.22 | 90.67 | 0.9230 |
| MCI vs. NC | 66.62 ± 7.35 | 72.80 | 55.26 | 64.03 | 75.65 | 50.97 | 0.7190 | |
| pMCI vs. sMCI | 66.66 ± 6.58 | 62.06 | 70.02 | 66.04 | 56.32 | 74.31 | 0.7124 | |
| MOLR + DeepESRNet | AD vs. NC | 90.28 ± 5.26 | 92.65 | 89.05 | 90.85 | 85.50 | 94.25 | 0.9260 |
| MCI vs. NC | 74.20 ± 6.16 | 78.74 | 66.30 | 72.52 | 81.92 | 60.69 | 0.7662 | |
| pMCI vs. sMCI | 73.28 ± 6.35 | 70.61 | 75.29 | 72.95 | 64.12 | 80.12 | 0.7192 |
For the metrics of sensitivity and specificity, our MOLR+DeepESRNet also outperformed the competing methods for the classification tasks of AD vs. NC and pMCI vs. sMCI. It is remarkable that our method enhanced the sensitivity by 14.51% (vs. MOLR) and 8.55% (vs. MOLR+SVM) for the most challenging task of pMCI vs. sMCI. For MCI vs. NC classification, MOLR achieved the highest sensitivity of 79.40%. However, in terms of the balanced accuracy that avoids inflated performance estimates on imbalanced dataset, e.g., MCI vs. NC classification in our case, compared to the competing methods (MOLR/MOLR+SVM), our MOLR+DeepESRNet improved by 5.39%/3.47% (AD vs. NC), 6.99%/8.49% (MCI vs. NC), and 9.18%/6.91% (pMCI vs. sMCI).
Regarding PPV and NPV, our MOLR+DeepESRNet achieved the highest PPVs of 85.50% in AD vs. NC and 81.92% in MCI vs. NC, and the highest NPVs of 94.25% in AD vs. NC and 80.12% in pMCI vs. sMCI. In the meantime, MOLR showed the highest PPV of 64.63% in pMCI vs. sMCI and the highest NPV of 73.08% in MCI vs. NC.
It is noteworthy that in terms of the AUC, which can be thought as a measure of the overall performance of a diagnostic test, the MOLR+DeepESRNet showed the best AUCs of 0.9260 in AD vs. NC, 0.7662 in MCI vs. NC, and 0.7192 in pMCI vs. sMCI. Compared to MOLR/MOLR+SVM, our method increased the AUCs by 0.0095/0.0030 (AD vs. NC), 0.0420/0.0472 (MCI vs. NC), and 0.0368/0.0068 (pMCI vs. sMCI).
5.2.2. JLLR as a baseline regression model
Table 3 shows the performance of our JLLR+DeepESRNet as well as the performance of the competing methods. Our JLLR+DeepESRNet achieved the mean accuracies of 91.02% (AD vs. NC), 73.02% (MCI vs. NC), and 74.82% (pMCI vs. sMCI). Compared to the competing methods, i.e., JLLR/JLLR+SVM, our method improved the mean accuracies by 6.33%/4.63% (AD vs. NC), 4.47%/6.24% (MCI vs. NC), and 7.14%/8.43% (pMCI vs. sMCI).
Table 3.
Performance comparison to the competing methods with JLLR as a baseline model. (BA: Balanced Accuracy; PPV: Positive Predictive Value; NPV: Negative Predictive Value; AUC: Area Under receiver operating characteristics Curve).
| Tasks | Accuracy (%) | Sensitivity (%) | Specificity (%) | BA (%) | PPV (%) | NPV (%) | AUC | |
|---|---|---|---|---|---|---|---|---|
| JLLR (Wang et al., 2011) | AD vs. NC | 84.69 ± 4.03 | 84.95 | 84.95 | 84.95 | 80.58 | 88.04 | 0.9213 |
| MCI vs. NC | 68.55 ± 7.10 | 73.24 | 59.42 | 66.33 | 79.46 | 49.58 | 0.7330 | |
| pMCI vs. sMCI | 67.68 ± 6.31 | 64.13 | 70.24 | 67.18 | 55.66 | 76.52 | 0.7301 | |
| JLLR + SVM | AD vs. NC | 86.39 ± 5.54 | 87.62 | 86.18 | 86.90 | 82.25 | 89.80 | 0.9220 |
| MCI vs. NC | 66.78 ± 7.36 | 72.87 | 55.69 | 64.28 | 75.90 | 50.97 | 0.7206 | |
| pMCI vs. sMCI | 66.39 ± 6.56 | 61.94 | 69.39 | 65.66 | 55.07 | 74.76 | 0.7124 | |
| JLLR + DeepESRNet | AD vs. NC | 91.02 ± 4.29 | 92.72 | 89.94 | 91.33 | 87.08 | 94.23 | 0.9272 |
| MCI vs. NC | 73.02 ± 6.44 | 77.60 | 68.22 | 72.91 | 82.96 | 55.77 | 0.7361 | |
| pMCI vs. sMCI | 74.82 ± 6.80 | 70.93 | 78.82 | 74.87 | 71.43 | 77.47 | 0.7539 |
In regard to the fact that the higher the sensitivity, the lower the chance of mis-diagnosing patients, which is of great importance in the clinic, it is promising that our JLLR+DeepESRNet overwhelmed the competing methods in sensitivity. Specifically, our method improved the sensitivity, compared to JLLR/JLLR+SVM, by 7.77%/5.1% (AD vs. NC), 4.36%/4.73% (MCI vs. NC), and 6.8%/8.99% (pMCI vs. sMCI). It was also observed for high improvements in specificity across the three classification tasks, i.e., 4.99%/3.76% (AD vs. NC), 8.8%/12.53% (MCI vs. NC), and 8.58%/9.43% (pMCI vs. sMCI) in comparison with JLLR/JLLR+SVM.
In PPV and NPV, our JLLR+DeepESRNet showed the highest PPVs of 87.08% (AD vs. NC), 82.96% (MCI vs. NC), and 71.43% (pMCI vs. sMCI) and the highest NPVs of 94.23% (AD vs. NC), 55.77% (MCI vs. NC), and 77.47% (pMCI vs. sMCI). It is remarkable that in the task of pMCI vs. sMCI, the improvements for PPV by our JLLR+DeepESRNet were 15.77% (vs. JLLR) and 16.36% (vs. JLLR+SVM).
Regarding the AUC metric, our JLLR+DeepESRNet showed the best AUCs of 0.9272 in AD vs. NC, 0.7361 in MCI vs. NC, and 0.7539 in pMCI vs. sMCI. Compared to JLLR+SVM, our method increased the AUCs by 0.0052 (AD vs. NC), 0.0155 (MCI vs. NC), and 0.0415 (pMCI vs. sMCI).
6. Discussion
6.1. Visual inspection of target-level representation
To validate our rationale of using CNN, which is designed to extract local relationship in our target-level representation and to hierarchically integrate information, we first computed correlation coefficients among rows or columns of the target-level representations. Fig. 3 presents samples of the target-level representations and the correlation matrix estimated from our dataset, for which a sliding window of 4 × 4 in size, i.e., 4 consecutive sparse regression models, was considered with a stride of 1. Specifically, for the number Q of target-level representations of 10 × 4 in size, we shaped them into a 3-dimensional tensor of 4 × 4 × (7 × Q), where 7 comes from the use of a sliding window of 4 × 4 in size. The large tensor was then used to compute the 4 × 4 correlation matrix. Clearly, there exists the correlations among target-level representations predicted from sparse regression models with similar control parameter values.
Fig. 3.
Samples of target-level representations, which were Gaussian normalized by first subtracting with means and then dividing with standard deviations, and the correlation matrix that represents relations among four sparse regression models.
6.2. Performance comparison
To better justify the effectiveness of the proposed method, we further conducted statistical significance testing. The null hypothesis was that the combination of sparse regression models with the proposed DeepESRNet, i.e., MOLR+DeepESRNet and JLLR+DeepESRNet, produces the same mean accuracies with the competing methods, i.e., MOLR, MOLR+SVM, JLLR, and JLLR+SVM. We used the Wilcoxon signed rank test (Wilcoxon, 1945) to assess whether the differences in classification accuracies between two methods are at a significant level. The resulting p-values are presented in Table 4, where it is noticeable that all the null hypothesis can be rejected beyond the 95 percent confidence level (i.e., p-value < 0.05). Hence, we can say that the proposed method outperformed the competing methods in terms of statistical significance by rejecting the null hypothesis beyond the 95 percent confidence level.
Table 4.
Statistical significance test.
| Task | MOLR + DeepESRNet
|
JLLR + DeepESRNet
|
||
|---|---|---|---|---|
| MOLR | MOLR + SVM | JLLR | JLLR + SVM | |
| AD vs. NC | 0.0078 | 0.0312 | 0.0020 | 0.0059 |
| MCI vs. NC | 0.0020 | 0.0039 | 0.0234 | 0.0078 |
| pMCI vs. sMCI | 0.0020 | 0.0039 | 0.0117 | 0.0020 |
Although it is not clear why some people with MCI progress to AD and some do not, MCI is considered as an early stage of dementia in the particular form and it is estimated that approximately 10–15% of individuals with MCI progress to AD in one year (Alzheimer’s Association, 2012). In this perspective, it is momentous to correctly discriminate pMCI from sMCI so that pMCI subjects can take benefit from a proper treatment for possible delay of progressing to AD. In our experiments, our MOLR+DeepESRNet and JLLR+DeepESRNet improved, respectively, by 6.62% (vs. MOLR+SVM) and 7.14% (vs. JLLR) compared to the maximal accuracies of their counterpart methods. It is also noteworthy that our JLLR+DeepESRNet achieved the best performance across seven metrics considered in our work.
From a clinical standpoint, the PPV is also of great importance, which measures the proportion of correctly diagnosed subjects belonging to AD, MCI, or pMCI in the tasks of AD vs. NC, MCI vs. NC, and pMCI vs. sMCI, respectively. Based on a recent report by Alzheimer’s Association (2012), the AD prevalence is projected to be 11 million to 16 million by 2050. For MCI and pMCI, although there is high variation among reports depending on definitions, the median of the prevalence estimates of MCI or pMCI in the literature is 26.4% (MCI) and 4.9% (amnestic MCI) (Ward et al., 2012). Regarding the AD prevalence by 2050, our JLLR+DeepESRNet, which maximally achieved the PPV of 87.08% in the classification of AD and NC, can correctly identify 9.5788 million to 13.9328 million of subjects with AD, while JLLR+SVM, whose respective PPV was 82.25%, can identify 9.0475 million to 13.16 million of subjects with AD. Accordingly, our method can correctly identify as many as 0.5313 million to 0.7728 million of subjects more than JLLR+SVM.
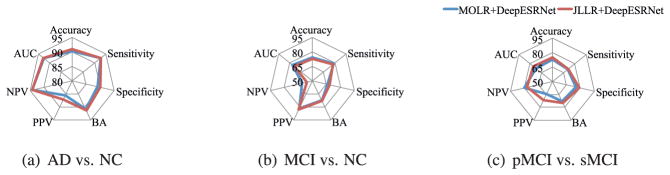
We also compared our two methods, each of which involved different baseline regression models but with the same Deep-ESRNet architecture, and summarized results in Fig. 4. From the figure, there is no significant difference between them across all three tasks, i.e., AD vs. NC, MCI vs. NC, and pMCI vs. sMCI. In our statistical significance testing with the Wilcoxon signed rank test (Wilcoxon, 1945), the p-values were 0.25 (AD vs. NC), 0.4570 (MCI vs. NC), and 0.2109 (pMCI vs. sMCI), for which we cannot reject the null hypothesis that their mean accuracies are the same in the different tasks.
Fig. 4.
Performance comparison between MOLR+DeepESRNet and JLLR+DeepESRNet.
6.3. Comparison with (deep) neural networks
It may be possible to think of mapping the ensemble of sparse regression models into a conventional multi-layer perceptron or a deep neural network by regarding a subset of hidden units being one sparse-model and other subsets of hidden units as other sparse-models, etc. In this regard, we conducted experiments with (deep) neural networks, into which the 93 MR features were fed, by varying the number of hidden layers and their respective units. For all the networks, we initialized the parameters, i.e., connection weights and biases, via greedy layer-wise pretraining (Hinton and Salakhutdinov, 2006) by using a Stacked Auto-Encoder (SAE) (Bengio et al., 2007),12 and trained with a stochastic gradient descent approach (Li et al., 2014a) with a learning rate of 0.01, a weight decay of 0.0001, and a momentum factor of 0.9. We summarized the results in Table 5. Overall, for the classification of AD vs. NC, there were no significant differences in performance among the neural networks with different architectures considered in our experiments. However, for the task of pMCI vs. sMCI, the 3-layer neural network, i.e., 93(input)-50(hidden)-30(hidden)-2(output), showed the superiority to the other networks. When comparing with the performances of our method in Tables 2 and 3, the proposed method outperformed the (deep) neural network-based methods.
Table 5.
Performance of (deep) neural networks, pretrained with Stacked Auto-Encoder (SAE), with 93 regional features. The boldface denotes the best performance for each metric in each classification task.
| # of hidden units | Tasks | Accuracy (%) | Sensitivity (%) | Specificity (%) | BA (%) | PPV (%) | NPV (%) | AUC | |
|---|---|---|---|---|---|---|---|---|---|
| 2-layer SAE + NN | 50 | AD vs. NC | 86.16 ± 4.28 | 87.04 | 86.59 | 86.82 | 82.22 | 89.41 | 0.9321 |
| MCI vs. NC | 67.39 ± 6.84 | 73.50 | 56.36 | 64.93 | 76.11 | 52.31 | 0.7141 | ||
| pMCI vs. sMCI | 64.39 ± 8.64 | 57.81 | 69.67 | 63.74 | 57.50 | 69.49 | 0.6954 | ||
| 100 | AD vs. NC | 86.64 ± 4.92 | 87.15 | 87.12 | 87.14 | 83.30 | 89.41 | 0.9306 | |
| MCI vs. NC | 69.19 ± 8.19 | 74.46 | 58.96 | 66.71 | 78.42 | 53.16 | 0.7233 | ||
| pMCI vs. sMCI | 66.39 ± 7.48 | 60.97 | 70.46 | 65.71 | 57.39 | 73.00 | 0.7179 | ||
| 3-layer SAE + NN | 50-30 | AD vs. NC | 86.87 ± 5.14 | 86.24 | 88.19 | 87.22 | 84.91 | 88.50 | 0.9249 |
| MCI vs. NC | 69.02 ± 6.00 | 74.67 | 58.97 | 66.82 | 77.40 | 54.49 | 0.7387 | ||
| pMCI vs. sMCI | 69.18 ± 6.65 | 64.52 | 72.74 | 68.63 | 61.07 | 75.14 | 0.7401 | ||
| 100-30 | AD vs. NC | 87.85 ± 5.33 | 88.99 | 88.12 | 88.56 | 84.36 | 90.73 | 0.9272 | |
| MCI vs. NC | 67.58 ± 8.65 | 72.97 | 57.02 | 65.00 | 77.65 | 50.14 | 0.7103 | ||
| pMCI vs. sMCI | 68.93 ± 7.17 | 64.30 | 72.37 | 68.33 | 60.99 | 74.80 | 0.7347 | ||
| 4-layer SAE + NN | 50-50-30 | AD vs. NC | 85.43 ± 4.99 | 85.37 | 86.61 | 85.99 | 82.75 | 87.61 | 0.9201 |
| MCI vs. NC | 67.40 ± 6.87 | 74.10 | 55.60 | 64.85 | 74.83 | 54.49 | 0.7194 | ||
| pMCI vs. sMCI | 66.65 ± 8.21 | 60.33 | 72.27 | 66.30 | 62.17 | 69.92 | 0.7059 | ||
| 100-50-30 | AD vs. NC | 87.11 ± 5.78 | 86.92 | 88.25 | 87.58 | 84.91 | 88.91 | 0.9212 | |
| MCI vs. NC | 68.69 ± 7.51 | 74.72 | 58.29 | 66.51 | 76.62 | 54.94 | 0.7182 | ||
| pMCI vs. sMCI | 67.37 ± 7.09 | 62.04 | 71.85 | 66.94 | 62.21 | 71.19 | 0.7224 |
In the meantime, in order to validate the use of a CNN in our method, we also performed experiments with (deep) neural networks by taking the vectorized target-level representations as input. We considered the same network architectures in Table 5 and trained networks with the same parameter settings. The results are summarized in Table 6. In comparison of the results in Table 6 with the performances in Tables 2 and 3, the proposed method again outperformed (deep) neural networks that took the vectorized target-level representation as input.
Table 6.
Performance of (deep) neural networks, pretrained with Stacked Auto-Encoder (SAE), taking vectorized target-level representations as input. The boldface denotes the best performance for each metric in each classification task.
| # of hidden units | Tasks | Accuracy (%) | Sensitivity (%) | Specificity (%) | BA (%) | PPV (%) | NPV (%) | AUC | |
|---|---|---|---|---|---|---|---|---|---|
| 2-layer SAE + NN | 50 | AD vs. NC | 86.86 ± 4.97 | 81.70 | 91.11 | 86.40 | 88.88 | 86.29 | 0.9300 |
| MCI vs. NC | 68.38 ± 7.60 | 78.18 | 51.38 | 64.78 | 73.63 | 58.23 | 0.7233 | ||
| pMCI vs. sMCI | 66.93 ± 7.01 | 55.66 | 75.22 | 65.44 | 62.69 | 70.06 | 0.7183 | ||
| 100 | AD vs. NC | 86.38 ± 3.39 | 82.19 | 89.80 | 86.00 | 87.67 | 86.48 | 0.9219 | |
| MCI vs. NC | 68.38 ± 6.90 | 78.43 | 50.95 | 64.69 | 73.50 | 58.27 | 0.7212 | ||
| pMCI vs. sMCI | 67.94 ± 8.68 | 59.19 | 74.35 | 66.77 | 63.28 | 71.55 | 0.7232 | ||
| 3-layer SAE + NN | 50–30 | AD vs. NC | 86.36 ± 5.01 | 81.14 | 90.65 | 85.90 | 88.47 | 85.99 | 0.9274 |
| MCI vs. NC | 69.18 ± 6.24 | 77.41 | 54.90 | 66.16 | 74.94 | 59.14 | 0.7236 | ||
| pMCI vs. sMCI | 67.70 ± 7.00 | 57.43 | 75.24 | 66.33 | 63.56 | 70.72 | 0.7453 | ||
| 100–30 | AD vs. NC | 86.73 ± 5.37 | 81.14 | 90.67 | 85.91 | 88.25 | 85.93 | 0.9341 | |
| MCI vs. NC | 68.22 ± 7.45 | 75.37 | 55.83 | 65.60 | 74.86 | 56.99 | 0.7303 | ||
| pMCI vs. sMCI | 67.44 ± 7.99 | 58.01 | 74.33 | 66.17 | 62.87 | 70.84 | 0.7277 | ||
| 4-layer SAE + NN | 50–50–30 | AD vs. NC | 86.60 ± 5.01 | 81.67 | 90.65 | 86.16 | 88.63 | 86.24 | 0.9155 |
| MCI vs. NC | 68.69 ± 6.19 | 76.15 | 55.77 | 65.96 | 75.12 | 57.96 | 0.7268 | ||
| pMCI vs. sMCI | 66.93 ± 7.76 | 58.01 | 73.48 | 65.75 | 62.05 | 70.55 | 0.7326 | ||
| 100–50–30 | AD vs. NC | 86.36 ± 5.01 | 81.67 | 90.22 | 85.94 | 88.05 | 86.18 | 0.9026 | |
| MCI vs. NC | 67.39 ± 6.01 | 74.85 | 54.47 | 64.66 | 74.12 | 56.07 | 0.7231 | ||
| pMCI vs. sMCI | 67.70 ± 8.20 | 59.82 | 73.44 | 66.63 | 63.24 | 71.49 | 0.7374 |
6.4. Comparison with previous studies on ADNI dataset
We compared the maximal accuracies achieved by our JLLR+DeepESRNet with the accuracies of the previous studies of MRI-based AD/MCI diagnosis on the ADNI cohort in the literature. Note that, due to the difference in dataset size and different approaches for extracting features (they all belong to the volumetric methods, though), it is not fair to directly compare the performances among the methods. However, since those performances were obtained with the same ADNI cohort, it still deserves to compare their performances. Since previous work mostly focused on classification tasks of AD vs. NC and pMCI vs. sMCI, we summarize here only for these two tasks in Tables 7 and 8, respectively. First, in the AD vs. NC task, our method of JLLR+DeepESRNet achieved the highest accuracy and sensitivity. When comparing with Liu et al.’s results, the results are competitive. However, we would like to emphasize the improvements of 6.4% in sensitivity, which is clinically regarded more important than other metrics. In the meantime, for the task of pMCI vs. sMCI, Moradi et al.’s (2015) method that combined MRI features with age and cognitive measures for classification showed the highest accuracy and sensitivity. However, they considered a smaller-sized dataset than ours and their very low sensitivity raises doubts about an overfitting problem in their classifier, possibly due to a much smaller number of sMCI samples than the number of pMCI samples. Except for Moradi et al.’s method, our JLLR+DeepESRNet achieved the best accuracy, sensitivity, and specificity on the largest dataset, i.e., 167 pMCI subjects and 226 sMCI subjects.
Table 7.
Comparison with the previous studies of AD vs. NC classification on ADNI dataset. The boldface denotes the best performance for each metric. (GM: Gray Matter; SVM: Support Vector Machine; CT: Cortical Thickness; PCA: Principal Component Analysis; LDA: Linear Discriminant Analysis; ROI: Region Of Interest; QDA: Quadratic Discriminant Analysis; RLR: Regularized Linear Regression; SAE: Stacked Auto-Encoder).
| Method | Feature Type | Classifier | Subjects (AD/NC) | Accuracy (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|---|
| Zhang and Shen (2012) | GM volumes | SVM | 45/50 | 84.80 | – | – |
| Cho et al. (2012) | CT | PCA+LDA | 128/160 | – | 82.00 | 93.00 |
| Coupé et al. (2012) | HP/EC volumes | QDA | 60/60 | 90.00 | 88.00 | 92.00 |
| Liu et al. (2012) | GM voxels | Ensemble SRC | 198/229 | 90.80 | 86.32 | 94.76 |
| Casanova et al. (2013) | GM voxels | RLR | 171/188 | 87.10 | 84.30 | 88.90 |
| Eskildsen et al. (2013) | ROI CT | LDA | 194/226 | 84.50 | 79.40 | 88.90 |
| Schmitter et al. (2015) | 10 Volumes | SVM | 221/276 | – | 86.00 | 91.00 |
| Suk et al. (2015a) | GM volumes+SAE | SVM | 51/52 | 88.20 | – | – |
| Proposed | GM volumes | JLLR+DeepESM | 186/226 | 91.02 | 92.72 | 89.94 |
Table 8.
Comparison with the previous studies of pMCI vs. sMCI classification on ADNI dataset. The boldface denotes the best performance for each metric. (GM: Gray Matter; SVM: Support Vector Machine; CT: Cortical Thickness; PCA: Principal Component Analysis; LDA: Linear Discriminant Analysis; ROI: Region Of Interest; RLR: Regularized Linear Regression; LDS: Low Density Separation; SAE: Stacked Auto-Encoder).
| Method | Feature Type | Classifier | Subjects (pMCI/sMCI) | Accuracy (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|---|
| Zhang and Shen (2012) | GM volumes | SVM | 43/48 | 62.00 | 56.60 | 60.20 |
| Cho et al. (2012) | CT | PCA+LDA | 72/131 | – | 63.00 | 76.00 |
| Eskildsen et al. (2013) | ROI CT | LDA | 61/134 | 66.70 | 59.00 | 70.20 |
| Casanova et al. (2013) | GM voxels | RLR | 153/182 | 61.50 | 45.80 | 75.50 |
| Moradi et al. (2015) | GM voxels+age +clinical scores | LDS | 164/100 | 76.61 | 88.85 | 51.59 |
| Schmitter et al. (2015) | 10 volumes | SVM | 147/254 | – | 67.00 | 71.00 |
| Suk et al. (2015a) | GM volumes+SAE | SVM | 43/56 | 69.30 | – | – |
| Proposed | GM volumes | JLLR+DeepESM | 167/226 | 74.82 | 70.93 | 78.82 |
6.5. Potential imaging biomarkers
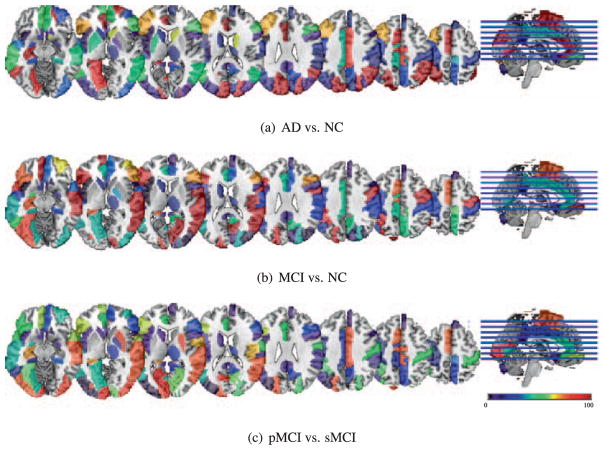
It is worthy to understand and visualize ROIs selected in sparse regression models that provide useful information to extract target-representations, which are finally used by our deep model. Note that it doesn’t mean those selected ROIs used by deep learning directly. Fig. 5 shows the frequency distribution of the selected ROIs over 10-fold cross validation for three different classification tasks by our multiple JLLR. To identify brain regions that can be regarded as potential imaging biomarkers, we first identified the most selected features. We defined the most selected features based on their frequency, which should be higher than 95,13 and then found the commonly selected ones across three tasks. The brain regions corresponding to the commonly selected features are as follows: insula right, precentral gyrus right, lateral front-orbital gyrus right, frontal lobe WM left, cingulate region left, hippocampal formation right, superior parietal lobule left, middle temporal gyrus left, temporal lobe WM left, superior parietal lobule right, lateral front-orbital gyrus left, inferior temporal gyrus left, lateral occipitotemporal gyrus right, hippocampal formation left, medial occipitotemporal gyrus left, middle temporal gyrus right, and lateral occipitotemporal gyrus left.
Fig. 5.
Distribution of the selected ROIs by JLLR for different classification tasks. The color denotes the frequency of being selected in 10-fold cross-validation. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
7. Conclusions
In this paper, we proposed a novel method that combines two conceptually different models of sparse regression and deep CNN. Specifically, we proposed to build multiple sparse regression models with different values of a regularization control parameter. Next, we devised a CNN by taking the predictions from the multiple regression models as input for final clinical decision making. With an MRI dataset of 805 subjects from the ADNI cohort, our methods outperformed the competing methods in terms of statistical significance, rejecting the null hypothesis beyond the 95 percent confidence level. One of the limitations in our current work is related to the predefined number of regularization control parameter values. From a machine learning perspective, it is necessary to find the optimal number of regularization control parameter values from training data solely, which should be our future research issue. In the meantime, as end-to-end learning has verified its effectiveness, especially in deep learning (Long et al., 2015; Yang et al., 2016), we believe that there is a way to further improve the proposed method in that direction. That is, it is desirable to train parameters of both sparse regression models and a CNN jointly in a systematic manner.
As deep learning has achieved the state-of-the-art performance over different artificial intelligence applications, its use for performance enhancement is also one of the major steps in medical imaging. However, performance improvement in brain disease diagnosis is still minor. In order to foster the use of deep learning for imaging-based brain disease diagnosis, we suggest some directions. First, as witnessed in computer vision, where breakthrough improvements could be achieved by exploiting large amounts of training data, e.g., more than 1 million annotated images in ImageNet (Russakovsky et al., 2015), it would be one direction to build such big publicly available brain imaging datasets. These will help deep models to find more generalized features for brain disease diagnosis, thus allowing making a leap in performance. Second, it is necessary to develop algorithmic techniques to efficiently handle images acquired with different scanning protocols, by which there is no need to train scanning protocol-specific deep models. Third, it is desirable to develop a systematic framework for constructing an optimal network architecture, instead of empirical design. The performances are generally sensitive to the varying number of layers or units per layer (e.g., Tables 5 and 6). Last but not least, it is important to develop a method for identifying or interpreting deep learned features that mostly devoted to the predicted outputs, thus allowing for practical use in clinic.
Acknowledgments
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (http://www.loni.ucla.edu/ADNI). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report.
This work was partly supported by Institute for Information & communications Technology Promotion (IITP) grant funded by the Korea government (MSIP) (No. B0101-15-0307, Basic Software Research in Human-level Lifelong Machine Learning (Machine Learning Center)), by the Brain Research Program through the National Research Foundation of Korea funded by the Ministry of Science, ICT & Future Planning (NRF-2014M3C7A1046050), and by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (NRF-2015R1C1A1A01052216). This work was also supported in part by NIH grants (EB006733, EB008374, MH100217, MH108914, AG041721, AG049371, AG042599, AG053867, EB022880).
Appendix A. Performance by DBN+DNN
For a comparison purpose, we have also conducted an experiment with deep neural networks, whose parameter values were pretrained with Deep Belief Network (DBN) (Hinton and Salakhutdinov, 2006) by stacking multiple Restricted Boltzmann Machines (RBMs). Specifically, we exploited a Gaussian RBM for the bottom input-hidden RBM and binary RBMs for the upper hidden layers. For a Gaussian RBM, we fixed the standard deviations to 1 to reduce the number of parameters, thus avoiding overfitting and lessening training time (Nair and Hinton, 2008). In order for that, the training samples were first Gaussian normalized by subtracting with mean and then dividing with standard deviation. Regarding the DBN training, we used a contrastive-divergence algorithm (Hinton et al., 2006) with 100 epochs, a learning rate of 0.01, and a batch size of 10. The pretained DBN was then transformed into a deep neural network for which we attached a top output layer for classification. The performance is summarized in Table A1.
Table A1.
Performance of (deep) neural networks, pretrained with Deep Belief Network (DBN), with 93 regional volume features. The boldface denotes the best performance for each metric in each classification task.
| # of hidden units | Tasks | Accuracy (%) | Sensitivity (%) | Specificity (%) | BA (%) | PPV (%) | NPV (%) | AUC | |
|---|---|---|---|---|---|---|---|---|---|
| 2-layer DBN + NN | 50 | AD vs. NC | 85.41 ± 4.80 | 85.15 | 86.58 | 85.87 | 82.75 | 87.57 | 0.9197 |
| MCI vs. NC | 65.46 ± 5.42 | 69.66 | 56.17 | 62.91 | 80.73 | 38.99 | 0.6963 | ||
| pMCI vs. sMCI | 62.85 ± 7.80 | 57.72 | 66.81 | 62.26 | 53.31 | 69.90 | 0.6786 | ||
| 100 | AD vs. NC | 83.97 ± 4.22 | 85.09 | 83.87 | 84.48 | 78.51 | 88.48 | 0.9174 | |
| MCI vs. NC | 66.43 ± 6.33 | 70.34 | 56.95 | 63.64 | 81.24 | 40.69 | 0.6966 | ||
| pMCI vs. sMCI | 63.33 ± 7.42 | 57.73 | 67.03 | 62.38 | 51.47 | 72.08 | 0.6707 | ||
| 3-layer DBN + NN | 50-30 | AD vs. NC | 85.91 ± 5.32 | 85.55 | 86.91 | 86.23 | 83.30 | 88.04 | 0.9082 |
| MCI vs. NC | 65.78 ± 6.53 | 69.44 | 56.97 | 63.21 | 82.24 | 37.13 | 0.6980 | ||
| pMCI vs. sMCI | 59.48 ± 7.75 | 52.53 | 63.64 | 58.09 | 45.26 | 69.84 | 0.6194 | ||
| 100-30 | AD vs. NC | 84.68 ± 6.08 | 86.59 | 84.25 | 85.42 | 78.51 | 89.80 | 0.9102 | |
| MCI vs. NC | 64.32 ± 4.76 | 68.86 | 53.89 | 61.37 | 79.97 | 37.15 | 0.6848 | ||
| pMCI vs. sMCI | 65.35 ± 7.60 | 60.11 | 69.21 | 64.66 | 56.25 | 72.13 | 0.6651 | ||
| 4-layer DBN + NN | 50-50-30 | AD vs. NC | 85.40 ± 6.32 | 85.68 | 85.81 | 85.84 | 81.17 | 88.87 | 0.9062 |
| MCI vs. NC | 62.71 ± 4.79 | 68.55 | 51.05 | 59.80 | 77.46 | 37.09 | 0.6438 | ||
| pMCI vs. sMCI | 59.01 ± 5.42 | 52.36 | 61.68 | 57.02 | 34.49 | 77.00 | 0.6146 | ||
| 100-50-30 | AD vs. NC | 82.77 ± 5.77 | 82.89 | 83.48 | 83.19 | 78.01 | 86.72 | 0.8867 | |
| MCI vs. NC | 65.93 ± 3.05 | 69.16 | 57.36 | 63.26 | 84.02 | 34.31 | 0.6667 | ||
| pMCI vs. sMCI | 57.24 ± 7.27 | 50.98 | 61.42 | 56.20 | 40.74 | 69.41 | 0.5822 |
Appendix B. List of 93 ROIs
See Table B1.
Table B1.
A list of 93 ROIs considered in this work.
| medial front-orbital gyrus right | middle frontal gyrus right | lateral ventricle left |
| insula right | precentral gyrus right | lateral front-orbital gyrus right |
| cingulate region right | lateral ventricle right | medial frontal gyrus left |
| superior frontal gyrus right | globus palladus right | globus palladus left |
| putamen left | inferior frontal gyrus left | putamen right |
| frontal lobe WM right | parahippocampal gyrus left | angular gyrus right |
| temporal pole right | subthalamic nucleus right | nucleus accumbens right |
| uncus right | cingulate region left | fornix left |
| frontal lobe WM left | precuneus right | subthalamic nucleus left |
| posterior limb of internal capsule inc. cerebral peduncle left | posterior limb of internal capsule inc. cerebral peduncle right | hippocampal formation right |
| inferior occipital gyrus left | superior occipital gyrus right | caudate nucleus left |
| supramarginal gyrus left | anterior limb of internal capsule left | occipital lobe WM right |
| middle frontal gyrus left | superior parietal lobule left | caudate nucleus right |
| cuneus left | precuneus left | parietal lobe WM left |
| temporal lobe WM right | supramarginal gyrus right | superior temporal gyrus left |
| uncus left | middle occipital gyrus right | middle temporal gyrus left |
| lingual gyrus left | superior frontal gyrus left | nucleus accumbens left |
| occipital lobe WM left | postcentral gyrus left | inferior frontal gyrus right |
| precentral gyrus left | temporal lobe WM left | medial front-orbital gyrus left |
| perirhinal cortex right | superior parietal lobule right | lateral front-orbital gyrus left |
| perirhinal cortex left | inferior temporal gyrus left | temporal pole left |
| entorhinal cortex left | inferior occipital gyrus right | superior occipital gyrus left |
| lateral occipitotemporal gyrus right | entorhinal cortex right | hippocampal formation left |
| thalamus left | parietal lobe WM right | insula left |
| postcentral gyrus right | lingual gyrus right | medial frontal gyrus right |
| amygdala left | medial occipitotemporal gyrus left | parahippocampal gyrus right |
| anterior limb of internal capsule right | middle temporal gyrus right | occipital pole right |
| corpus callosum | amygdala right | inferior temporal gyrus right |
| superior temporal gyrus right | middle occipital gyrus left | angular gyrus left |
| medial occipitotemporal gyrus right | cuneus right | lateral occipitotemporal gyrus left |
| thalamus right | occipital pole left | fornix right |
Footnotes
In our work, we stack in an ascending order. However, there is no difference in the framework.
Available at ‘ http://mipav.cit.nih.gov/clickwrap.php’
Available at ‘ http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/’
Note that when normalization is conducted with the whole-brain images, it can introduce unexpected structural differences for tissues. That is, there is no any difference but it can be caused for the difference by normalization due to the effect of the neighboring tissue that does show structural difference. One common way of minimizing such unexpected error is to normalize with the tissue-segmented images, instead of the whole-brain images (Mechelli et al., 2005).
In this paper, we exploit regional gray matter tissue volumes based on the fact that brain atrophy is one of the main neuropathological changes with AD. However, visual features with Scale-Invariant Feature Transform (SIFT) (Lowe, 1999) or Histogram of Oriented Gradients (HOG) (Dalal and Triggs, 2005) can also be good alternatives to the ROI-based regional features.
We use a class indicator vector ti ∈ {0, 1}2 with a zero-one encoding, i.e., ti j = 1 iff xi belongs to the class j; otherwise ti j = 0.
In our implementation, we used a SLEP toolbox for sparse model learning, which requires a regularization control parameter, i.e., λ, to be in the range of [0, 1], and rescales the value internally. Based on our earlier work (Suk et al., 2015b; 2016a), where we observed that parameter values of higher than 0.3 were not useful and never chosen in cross-validation, we thus defined the parameter space with 10 values equally spaced between 0.01 and 0.3. As for the size of parameter space (M = 10), it is determined empirically.
As for the activation function, while the logistic sigmoid function or hyperbolic tangent function has been commonly used, thanks to its great success in recent applications, we used a Rectified Linear Unit (ReLU) (Nair and Hinton, 2010), defined as f (a) = max (0, a).
Available at ‘ http://www.vlfeat.org/matconvnet/’
For sparse model training, we used a SLEP toolbox available at’ http://www.public.asu.edu/~jye02/Software/SLEP/index.htm,’ where the control parameter is required to be set between 0 and 1 because its value is internally rescaled (Liu et al., 2010).
Available at ‘ http://www.csie.ntu.edu.tw/~cjlin/libsvm/’.
We also conducted experiments by pretraining the network with Deep Belief Network (DBN) (Hinton and Salakhutdinov, 2006) and reported the results in Appendix A.
The maximal frequency in our work is 100.
References
- Alzheimer’s Association. 2012 Alzheimer’S disease facts and figures. Alzheimer Dement. 2012;8:131–168. doi: 10.1016/j.jalz.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Avants B, Gee JC. Geodesic estimation for large deformation anatomical shape averaging and interpolation. Neuroimage. 2004;23:S139–150. doi: 10.1016/j.neuroimage.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Bengio Y, Lamblin P, Popovici D, Larochelle H. Greedy layer-wise training of deep networks. In: Schölkopf B, Platt J, Hoffman T, editors. Advances in Neural Information Processing Systems. Vol. 19. MIT Press; Cambridge, MA: 2007. pp. 153–160. [Google Scholar]
- Brosch T, Tam R. Manifold learning of brain mris by deep learning. Proceedings of International Conference on Medical Image Computing and Computer-Assisted Intervention; 2013. pp. 633–640. [DOI] [PubMed] [Google Scholar]
- Brosch T, Tang LYW, Yoo Y, Li DKB, Traboulsee A, Tam R. Deep 3D convolutional encoder networks with shortcuts for multiscale feature integration applied to multiple sclerosis lesion segmentation. IEEE Trans Med Imaging. 2016;35:1229–1239. doi: 10.1109/TMI.2016.2528821. [DOI] [PubMed] [Google Scholar]
- Casanova R, Hsu FC, Sink KM, Rapp SR, Williamson JD, Resnick SM, Espeland MA for the Alzheimer’s Disease Neuroimaging Initiative. Alzheimer’s disease risk assessment using large-scale machine learning methods. PLoS One. 2013;8:1–13. doi: 10.1371/journal.pone.0077949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y, Seong JK, Jeong Y, Shin SY. Individual subject classification for Alzheimer’s disease based on incremental learning using a spatial frequency representation of cortical thickness data. Neuroimage. 2012;59:2217–2230. doi: 10.1016/j.neuroimage.2011.09.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter A, Shamir O, Srebro N, Sridharan K. Better mini-batch algorithms via accelerated gradient methods. In: Shawe-Taylor J, Zemel R, Bartlett P, Pereira F, Weinberger K, editors. Advances in Neural Information Processing Systems. Vol. 24. 2011. pp. 1647–1655. [Google Scholar]
- Coupé P, Eskildsen SF, Manjn JV, Fonov VS, Collins DL. Simultaneous segmentation and grading of anatomical structures for patient’s classification: application to Alzheimer’s disease. Neuroimage. 2012;59:3736–3747. doi: 10.1016/j.neuroimage.2011.10.080. [DOI] [PubMed] [Google Scholar]
- Cuingnet R, Gerardin E, Tessieras J, Auzias G, Lehéricy S, Habert MO, Chupin M, Benali H, Colliot O The Alzheimer’s Disease Neuroimaging Initiative. Automatic classification of patients with Alzheimer’s disease from structural MRI: a comparison of ten methods using ADNI database. Neuroimage. 2011;56:766–781. doi: 10.1016/j.neuroimage.2010.06.013. [DOI] [PubMed] [Google Scholar]
- Dalal N, Triggs B. Histograms of oriented gradients for human detection. Proceedings of the 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’05); 2005. pp. 886–893. [Google Scholar]
- Davatzikos C, Bhatt P, Shaw LM, Batmanghelich KN, Trojanowski JQ. Prediction of MCI to AD conversion, via MRI, CSF biomarkers, and pattern classification. Neurobiol Aging. 2011;32:2322e19–2322.e27. doi: 10.1016/j.neurobiolaging.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davatzikos C, Fan Y, Wu X, Shen D, Resnick SM. Detection of prodromal Alzheimer’s disease via pattern classification of magnetic resonance imaging. Neurobiol Aging. 2008;29:514–523. doi: 10.1016/j.neurobiolaging.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davatzikos C, Genc A, Xu D, Resnick SM. Voxel-based morphometry using the RAVENS maps: methods and validation using simulated longitudinal atrophy. Neuroimage. 2001;14:1361–1369. doi: 10.1006/nimg.2001.0937. [DOI] [PubMed] [Google Scholar]
- Dou Q, Chen H, Yu L, Zhao L, Qin J, Wang D, Mok VC, Shi L, Heng PA. Automatic detection of cerebral microbleeds from MR images via 3D convolutional neural networks. IEEE Trans Med Imaging. 2016;35:1182–1195. doi: 10.1109/TMI.2016.2528129. [DOI] [PubMed] [Google Scholar]
- Eskildsen SF, Coup P, Garca-Lorenzo D, Fonov V, Pruessner JC, Collins DL. Prediction of Alzheimer’s disease in subjects with mild cognitive impairment from the ADNI cohort using patterns of cortical thinning. Neuroimage. 2013;65:511–521. doi: 10.1016/j.neuroimage.2012.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Batmanghelich N, Clark CM, Davatzikos C the Alzheimer’s Disease Neuroimaging Initiative. Spatial patterns of brain atrophy in MCI patients, identified via high-dimensional pattern classification, predict subsequent cognitive decline. Neuroimage. 2008;39:1731–1743. doi: 10.1016/j.neuroimage.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Cai D, Niyogi P. Laplacian score for feature selection. In: Weiss Y, Schölkopf B, Platt JC, editors. Advances in Neural Information Processing Systems. Vol. 18. 2006. pp. 507–514. [Google Scholar]
- Hinrichs C, Singh V, Xu G, Johnson SC. Predictive markers for AD in a multi-modality framework: an analysis of MCI progression in the ADNI population. Neuroimage. 2011;55:574–589. doi: 10.1016/j.neuroimage.2010.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton GE, Osindero S, Teh Y-W. A fast learning algorithm for deep belief nets. Neural Comput. 2006;18:1527–1554. doi: 10.1162/neco.2006.18.7.1527. [DOI] [PubMed] [Google Scholar]
- Hinton GE, Salakhutdinov RR. Reducing the dimensionality of data with neural networks. Science. 2006;313:504–507. doi: 10.1126/science.1127647. [DOI] [PubMed] [Google Scholar]
- Ioffe S, Szegedy C. Batch normalization: accelerating deep network training by reducing internal covariate shift. Proceedings of the 32nd International Conference on Machine Learning; 2015. pp. 448–456. [Google Scholar]
- Ithapu VK, Singh V, Okonkwo OC, Chappell RJ, Dowling NM, Johnson SC the Alzheimer’s Disease Neuroimaging Initiative. Imaging-based enrichment criteria using deep learning algorithms for efficient clinical trials in mild cognitive impairment. Alzheimer Dement. 2015;11:1489–1499. doi: 10.1016/j.jalz.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani N, MacDonald D, Holmes C, Evans A. A 3D atlas of the human brain. Neuroimage. 1998;7:S717. [Google Scholar]
- Kittler J, Hatef M, Duin RPW, Matas J. On combining classifiers. IEEE Trans Pattern Anal Mach Intell. 1998;20:226–239. [Google Scholar]
- Li M, Zhang T, Chen Y, Smola AJ. Efficient mini-batch training for stochastic optimization. Proceedings of the 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; 2014a. pp. 661–670. [Google Scholar]
- Li R, Zhang W, Suk H-I, Wang L, Li J, Shen D, Ji S. Deep learning based imaging data completion for improved brain disease diagnosis. Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention; 2014b. pp. 305–312. Vol. 8675 of Lecture Notes in Computer Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S, Jia H, Wu G, Shen D for the Alzheimer’s Disease Neuroimaging Initiative. A novel framework for longitudinal atlas construction with group-wise registration of subject image sequences. Neuroimage. 2012;59:1275–1289. doi: 10.1016/j.neuroimage.2011.07.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Suk H-I, Wee C-Y, Chen H, Shen D. High-order graph matching based feature selection for Alzheimer’s disease identification. In: Mori K, Sakuma I, Sato Y, Barillot C, Navab N, editors. Proceedings of International Conference on Medical Image Computing and Computer-Assisted Intervention. 2013. pp. 311–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Ji S, Ye J. Multi-task feature learning via efficient ℓ2, 1-norm minimization. Proceedings of the Twenty-Fifth Conference on Uncertainty in Artificial Intelligence; 2009. pp. 339–348. [Google Scholar]
- Liu J, Ji S, Ye J. SLEP: Sparse Learning with Efficient Projections. Arizona State University; 2010. [Google Scholar]
- Liu M, Zhang D, Shen D. Ensemble sparse classification of Alzheimer’s disease. Neuroimage. 2012;60:1106–1116. doi: 10.1016/j.neuroimage.2012.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2015. pp. 3431–3440. [DOI] [PubMed] [Google Scholar]
- Lowe DG. Object recognition from local scale-invariant features. Proceedings of the Seventh IEEE International Conference on Computer Vision; 1999. pp. 1150–1157. [Google Scholar]
- Mechelli A, Price CJ, Friston KJ, Ashburner J. Voxel-based morphometry of the human brain: methods and applications. Curr Med Imaging Rev. 2005;1:105–113. [Google Scholar]
- Moradi E, Pepe A, Gaser C, Huttunen H, Tohka J. Machine learning framework for early MRI-based Alzheimer’s conversion prediction in MCI subjects. Neuroimage. 2015;104:398–412. doi: 10.1016/j.neuroimage.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair V, Hinton GE. Implicit mixtures of restricted Boltzmann machines. In: Koller D, Schuurmans D, Bengio Y, Bottou L, editors. Advances in Neural Information Processing Systems. 2008. pp. 1145–1152. [Google Scholar]
- Nair V, Hinton GE. Rectified linear units improve restricted Boltzmann machines. Proceedings of the 27th International Conference on Machine Learning; 2010. pp. 807–814. [Google Scholar]
- Negash S, Xie S, Davatzikos C, Clark CM, Trojanowski JQ, Shaw LM, Wolk DA, Arnold SE. Cognitive and functional resilience despite molecular evidence of Alzheimer’s disease pathology. Alzheimer Dement. 2013;9:e89–e95. doi: 10.1016/j.jalz.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira S, Pinto A, Alves V, Silva CA. Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans Med Imaging. 2016;35:1240–1251. doi: 10.1109/TMI.2016.2538465. [DOI] [PubMed] [Google Scholar]
- Plis SM, Hjelm D, Salakhutdinov R, Allen EA, Bockholt HJ, Long JD, Johnson HJ, Paulsen J, Turner JA, Calhoun VD. Deep learning for neuroimaging: a validation study. Front Neurosci. 2014;8:1–11. doi: 10.3389/fnins.2014.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell S, Magnotta VA, Johnson H, Jammalamadaka VK, Pierson R, Andreasen NC. Registration and machine learning-based automated segmentation of subcortical and cerebellar brain structures. Neuroimage. 2008;39:238–247. doi: 10.1016/j.neuroimage.2007.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roweis ST, Saul LK. Nonlinear dimensionality reduction by locally linear embedding. Science. 2000;290:2323–2326. doi: 10.1126/science.290.5500.2323. [DOI] [PubMed] [Google Scholar]
- Rumelhart DE, Hinton GE, Williams RJ. Learning representations by back--propagating errors. Nature. 1986;323:533–536. [Google Scholar]
- Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, Huang Z, Karpathy A, Khosla A, Bernstein M, Berg AC, Fei-Fei L. Imagenet large scale visual recognition challenge. Int J Comput Vis. 2015;115:211–252. [Google Scholar]
- Schmitter D, Roche A, Marchal B, Ribes D, Abdulkadir A, Bach-Cuadra M, Daducci A, Granziera C, Klõppel S, Maeder P, Meuli R, Krueger G. An evaluation of volume-based morphometry for prediction of mild cognitive impairment and Alzheimer’s disease. NeuroImage: Clinical. 2015;7:7–17. doi: 10.1016/j.nicl.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D, Davatzikos C. HAMMER: Hierarchical attribute matching mechanism for elastic registration. IEEE Trans Med Imaging. 2002;21:1421–1439. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- Shen D, Wong W, Ip HHS. Affine-invariant image retrieval by correspondence matching of shapes. Image and Vision Computing. 1999;17:489–499. [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res. 2014;15:1929–1958. [Google Scholar]
- Suk H-I, Lee S-W, Shen D. Latent feature representation with stacked auto-encoder for AD/MCI diagnosis. Brain Struct Funct. 2015a;220:841–859. doi: 10.1007/s00429-013-0687-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suk H-I, Lee S-W, Shen D. Deep sparse multi-task learning for feature selection in Alzheimer’s disease diagnosis. Brain Struct Funct. 2016a;221:2569–2587. doi: 10.1007/s00429-015-1059-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suk HI, Lee SW, Shen D for the Alzheimer’s Disease Neuroimaging Initiative. Hierarchical feature representation and multimodal fusion with deep learning for AD/MCI diagnosis. Neuroimage. 2014;101:569–582. doi: 10.1016/j.neuroimage.2014.06.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suk HI, Wee CY, Lee SW, Shen D. Supervised discriminative group sparse representation for mild cognitive impairment diagnosis. Neuroinformatics. 2015b;13:277–295. doi: 10.1007/s12021-014-9241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suk HI, Wee CY, Lee SW, Shen D. State-space model with deep learning for functional dynamics estimation in resting-state fmri. Neuroimage. 2016b;129:292–307. doi: 10.1016/j.neuroimage.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the LASSO. J R Stat Soc. 1996;58:267–288. [Google Scholar]
- Tohka J, Moradi E, Huttunen H. Comparison of feature selection techniques in machine learning for anatomical brain MRI in dementia. Neuroinformatics. 2016;14:279–296. doi: 10.1007/s12021-015-9292-3. [DOI] [PubMed] [Google Scholar]
- Vedaldi A, Lenc K. MatConvNet: convolutional neural networks for MATLAB. Proceedings of ACM International Conference on Multimedia.2015. [Google Scholar]
- Wang H, Nie F, Huang H, Risacher S, Saykin AJ, Shen L. Identifying AD-sensitive and cognition-relevant imaging biomarkers via joint classification and regression. In: Fichtinger G, Martel A, Peters T, editors. International Conference on Medical Image Computing and Computer-Assisted Intervention. 2011. pp. 115–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Summers RM. Machine learning and radiology. Med Image Anal. 2014;16:933–951. doi: 10.1016/j.media.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Nie J, Yap PT, Li G, Shi F, Geng X, Guo L, Shen D. Knowledge-guided robust MRI brain extraction for diverse large-scale neuroimaging studies on humans and non-human primates. PLoS One. 2014;9:e77810. doi: 10.1371/journal.pone.0077810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward A, Arrighi HM, Michels S, Cedarbaum JM. Mild cognitive impairment: disparity of incidence and prevalence estimates. Alzheimer Dement. 2012;8:14–21. doi: 10.1016/j.jalz.2011.01.002. [DOI] [PubMed] [Google Scholar]
- Wee C-Y, Yap P-T, Zhang D, Wang L, Shen D. Constrained sparse functional connectivity networks for MCI classification. In: Ayache N, Delingette H, Golland P, Mori K, editors. Proceedings of International Conference on Medical Image Computing and Computer-Assisted Intervention. 2012. pp. 212–219. Vol. 7511 of Lecture Notes in Computer Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcoxon F. Individual comparisons by ranking methods. Biom Bull. 1945;1:80–83. [Google Scholar]
- Xue Z, Shen D, Karacali B, Stern J, Rottenberg D, Davatzikos C. Simulating deformations of MR brain images for validation of atlas-based segmentation and registration algorithms. Neuroimage. 2006;33:855–866. doi: 10.1016/j.neuroimage.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Ouyang W, Li H, Wang X. End-to-end learning of deformable mixture of parts and deep convolutional neural networks for human pose estimation. Proceedings of 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016. pp. 3073–3082. [Google Scholar]
- Yuan M, Lin Y. Model selection and estimation in regression with grouped variables. J R Stat Soc Ser B. 2006;68:49–67. [Google Scholar]
- Zacharaki EI, Hogea CS, Shen D, Biros G, Davatzikos C. Non-diffeomorphic registration of brain tumor images by simulating tissue loss and tumor growth. Neuroimage. 2009;46:762–774. doi: 10.1016/j.neuroimage.2009.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Shen D. Multi-modal multi-task learning for joint prediction of multiple regression and classification variables in Alzheimer’s disease. Neuroimage. 2012;59:895–907. doi: 10.1016/j.neuroimage.2011.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Liu J, Narayan VA, Ye J. Modeling disease progression via fused sparse group lasso. Proceedings of ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; 2012. pp. 1095–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Suk HI, Shen D. A novel matrix-similarity based loss function for joint regression and classification in AD diagnosis. Neuroimage. 2014;100:91–105. doi: 10.1016/j.neuroimage.2014.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H, Hastie T. Regularization and variable selection via the elastic net. J R Stat Soc Ser B. 2005;67:301–320. [Google Scholar]