Abstract
Integrins are vital transmembrane receptors that mediate cell-cell and cell extra-cellular matrix (ECM) interactions and signaling. Talin is a 270 kDa protein and is considered a key regulator of integrin activity. The interaction between talin and integrin is commonly regarded as the final step of inside-out activation. In the cytosol, talin adopts an auto-inhibited conformation, in which the C-terminal rod domain binds the N-terminal head domain, preventing the interactions of the head domain with the membrane surface and the integrin cytoplasmic domain. It has long been suggested that the presence of Phosphatidylinositol-4, 5-bisphosphate (PIP2) at focal adhesions plays a role in activating talin. However, a detailed picture and mechanism of PIP2 activation of auto-inhibited talin remains elusive. Here, we use a FRET based binding assay to measure affinity of talin and lipid bilayers harboring anionic lipids. Results show that the R9 and R12R13 segments of the talin rod domain inhibit the binding of the talin head domain (THD) to anionic lipid bilayers. In contrast, we show that the binding of THD to bilayers containing PIP2 is insensitive to the presence of the inhibitor domains thereby directly implicating PIP2 as an effective activator of talin. Furthermore, we have mapped the activation to the interaction of PIP2 with the F2F3 domain of the talin head, showing that PIP2 plays a critical role in the regulation of the auto-inhibited form of talin and stimulates recruitment of talin to the membrane, which is essential for integrin inside-out signaling.
Keywords: talin, auto-inhibition, lipid bilayer, phosphatidylinositol, phosphatidylserine
Graphical Abstract
INTRODUCTION
Talin plays a pivotal role in regulating the activities of integrins, which are a family of heterodimeric membrane receptors composed of one α and one β subunit. In the integrin resting state, the extracellular domains of the subunits exist in a closed and bent conformation giving rise to low affinity for extracellular ligands.1 Intracellular events stimulate large conformational changes (inside-out signaling) that allow integrin to connect the Extra Cellular Matrix (ECM) to the actin cytoskeleton. A critical step in the mechanism of inside-out signaling is the binding of talin to the cytoplasmic tail of integrin which promotes the destabilization of essential interactions required for maintaining the integrin in the inactive state.2,3 A recent study reports that talin also alters the tilt angle of the integrin transmembrane domain within the bilayer.4 The topology change of the transmembrane helix promotes the disruption of the interactions that stabilize the low affinity state of the integrin receptors and transduce the signal across membrane to extracellular domain.5 Conversely, the extracellular regions can transduce signals to the intracellular space upon interaction with ECM ligands (outside-in signaling).6 Talin also plays an undisputed role as an adapter molecule through the interaction of the FERM (band 4.1/ezrin/radixin/moesin) domain with integrin tails and the membrane surface and the rod domain with the actin cytoskeleton.7
Talin is a large cytosolic adapter protein that consists of a 45-kDa N-terminal head domain and a large 200 kDa rod domain.8 Compared to canonical FERM domains, which consists of F1, F2, and F3, THD contains an additional 85 amino acid ubiquitin-like subunit, F0, preceding F1.9,10 The four subdomains are assembled into an extended linear conformation rather than the traditional trefoil arrangement seen in other FERM domains.11,12 Previous Studies show that the charge on the THD is asymmetrically distributed. Several positively charged amino acids, known as the membrane orientation patch (MOP), on the F2 domain are crucial for recruitment of talin to the membrane surface.13–15 Charge reversal mutations in the MOP strongly diminish talin binding to anionic lipid vesicles. The F3 domain is known to interact with the Phospho-Tyrosine Binding (PTB) NPxY motif on the β-integrin cytoplasmic tail thereby disrupting a key salt bridge that stabilizes the integrin α/β heterodimer.16
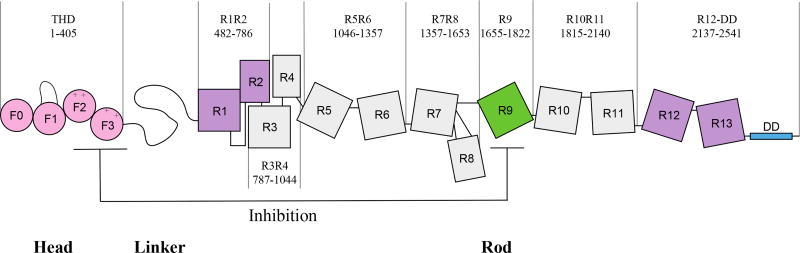
The talin rod domain is composed of 13 helical bundles and an N-terminal dimerization sequence (figure 1).17 In the inactive state, the talin rod packs against the THD and inhibits interactions with the phospholipid bilayer.17–20 Previous structural and NMR studies suggest that segments R1R2, and R9 make contact with the PTB binding site within the F2F3 subdomains.17,21 Goult et al. suggest that full length talin exists as a donut shaped dimer structure with the rod domains encircling the head domain in the auto-inhibited state.17 Multiple pathways have been shown to disrupt the interactions between the head domain and the road domain; these include interactions with complexes of Ras-related protein 1 (Rap1) and Rap1-interacting adapter molecule (RIAM),22–24 talin linker specific digestion by calpain,25–27 and binding of phosphatidylinositol-4-phosphate 5-kinase-γ (PIP5KI-γ),28–30 which subsequently generates Phosphatidylinositol 4,5-bisphosphate (PIP2) from phoshpaidylinositol-4-phosphate (PI4P).31,32 It is interesting to note that PIP2 has also been implicated in the activations of auto-inhibited talin.20,21
Figure 1.
Schematic representation of full-length talin. The previous identified inhibitory rod subdomain R9 is labeled in green. Two suggested inhibitory rod segments are R1R2 and R12R13 labeled in purple. The dimerization domain is presented in cyan.
PIP2 only comprises a small fraction (1~2%) of the plasma membrane phospholipid and is relatively evenly distributed throughout the membrane.33 PIP2 has been recognized as an important messenger molecule and plays a significant role in focal adhesion assembly.34,35 Many allosteric regulatory mechanisms rely on the presence of PIP2. For example, vinculin is expressed in an inactive, closed conformation in the cytosol. PIP2 binding to the regulatory site of the vinculin tail-domain opens up the closed form.36 Also PIP2 is critical for Focal Adhesion Kinase (FAK) function through interactions with the basic patch on the FERM domain resulting in activation of the auto-inhibited conformation.37 Although it has been known that the affinity of THD for lipid bilayers is mainly controlled by overall membrane charge,38 the presence of PIP2 facilitates talin membrane localization.39 Early studies have shown that talin binds to various phosphatidylinositides resulting in the inhibition of the head to rod domain interactions that are important in maintaining talin in the auto-inhibited form.39 In addition, the presence of PIP2 has been implicated in the talin mediated integrin activation and clustering.40 The local concentration of PIP2 is enriched at focal adhesion due the recruitment of PIP5KI-γ which is responsible for converting PI4P into PIP2.31 Specific deletion of PIP5KI-γ in focal adhesions results in remarkably reduced talin recruitment to the membrane and adhesion sites.41 Surface Plasmon Resonance (SPR) and NMR studies using soluble C4- or C8-PIP2 demonstrate that talin binds to PIP2 through several residues in F2F3 domain, and disrupts the THD/talin R9 interaction. A “push-pull” mechanism of talin activation has recently been proposed where PIP2 establishes a charge mismatch to simultaneously attract THD and release the inhibitory effects by talin R9.21
Here, we examine the affinities of the talin head domain toward various phospholipid bilayers by utilizing Nanodisc technology to provide a native membrane environment. Nanodiscs are phospholipid bilayers stabilized by membrane scaffold proteins (MSP) derived from apolipoprotein A1.42,43 Nanodiscs allow the study of protein-membrane interactions in an aqueous environment with controlled lipid composition. To investigate the role of PIP2 in the activation of auto-inhibited talin, we developed a Fluorescence Resonance Energy Transfer (FRET) based assay to measure the interaction of THD-membrane association in the presence of purified rod domain segments and β3 integrin transmembrane and cytoplasmic domains. We report that talin rod subdomains R9 and R12R13 strongly inhibited THD binding to negatively charged lipid bilayers with the exception of PIP2. PIP2 “activates” the auto-inhibited THD and can support tight binding of THD in the presence of talin rod inhibitory subdomains. We also show that PIP2 specifically targets the talin F2F3 domain to activate auto-inhibited THD.
EXPERIMENTAL PROCEEDURES
Materials
Phospholipids 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), 1,2-dimyristoyl-sn-glycero-3-phospho-L-serine (DMPS), 1,2-dimyristoyl-sn-glycero-3-phosphoglycerol (DMPG), 1,2-dimyristoyl-sn-glycero-3-phosphate (DMPA), brain PIP2 and brain PI4P were purchased from Avanti Polar Lipids. Membrane scaffold proteins were expressed in E. coli BL21 DE3 and purified as previously described.42 Tetra-methyl rhodamine (5 and 6) maleimide (TAMRA) was purchased from Anaspec Inc. Uniblue A (UA) was obtained from Sigma-Aldrich. iProof polymerase was obtained from BioRad Inc. A DNA G-block sequence encoding the integrin β3 transmembrane-cytoplasmic domain was purchased from Integrated DNA Technologies Inc. pET30a-THD for expression of talin-1 head domain was a kind gift from Dr. Mark Ginsberg.
Cloning, expression and purification of talin-1 Head domains, talin R9, talin R12R13, talin R1R2, and integrin β3 transmembrane-cytoplasmic domain
Cysteine mutations were engineered at position F123 or I398 of talin-1 to provide an accessible thiol-reactive labeling sites using pET30a-THD as template for PCR mutagenesis. iProof polymerase was used for all PCR reactions. Primers were purchased from Integrated DNA Technologies. All DNA constructs were inserted between the Xho1 and Nde1 restriction sites of the vector pET30a with a C-terminal 6×His tag for purification. The talin head domain contains the entire talin FERM domain (F0–F3) from the N terminus to residue 433. F0F1 subdomains extend from residues 1–195, and F2F3 subdomains extend from residues 196 to 433. Talin R9 was cloned between talin-1 residues 1654 to 1848, talin R12R13 extends from 2225 through 2344, and talin R1R2 spans from residues 482 to 786. Sequencing was performed by ACGT Inc.
Cloned DNA constructs in the vector pET30A were transformed into E. coli BL21 (DE3) gold (Novagen Inc.) Cell cultures were incubated on a platform shaker at 37 °C and 250 RPM until optical density at 600 nm reached 0.8. Protein expression was induced by adding 1mM IPTG followed by a 4-hour incubation at 37°C. Cell pellets were collected by centrifugation and resuspended in 20mM Tris-HCL buffer pH 7.4, 500mM NaCl, 20mM imidazole. After sonication on ice, cell lysates were cleared by centrifugation. Supernatants were loaded on to a Ni+ Nitrilotriacetic affinity column. The column was washed with 20 mM imidazole followed by elution with buffer containing 500mM imidazole. Elution products were dialyzed into 50 mM Tris, 150 mM KCl, and pH 7.0.
To generate the MBP-β3 TM/tail expression vector, the DNA sequence of integrin β3 TM/tail (residues 685–762) was engineered to have an AvrII site on 5’-terminus and an XbaI site on 3’-terminus for subsequent digestion and ligation. In addition, a poly-histidine sequence and a TEV digestion site was inserted upstream to integrin coding sequence to facilitate subsequent purification and Nanodisc assembly. Blunt-ended DNA fragments were synthesized by Integrated DNA Technologies Inc. and cut with XbaI and AvrII to create sticky ends. The linearized pMAL-c2e vector which possesses marching restricted digestion sites was used for expression vector. The reconstructed plasmid was enriched by transforming into competent NEB turbo E. Coli cell follow by plasmid extraction using Miniprep spin column kit. DNA sequence confirmation was performed by ACGT using M13 promoter primers.
The expression vector pMal c2e harboring the integrin β3 TM/tail construct was transformed into E. coli BL21 (DE3) gold (Novagen Inc.). Cell cultures were grown at 37°C in rich broth containing glucose and ampicillin (per liter broth: 10 g tryptone, 5 g yeast extract, 5 g NaCl, 2 g glucose, 100 mg ampicillin). Cells were allowed to grow to an OD of 0.5. Protein expression was induced by the addition of IPTG to a final concentration of 0.3 mM. Cell pellets were harvested by centrifugation 2 hours after induction. Cell pellets were resuspended in 5 volumes of column buffer (20 mM Tris-HCl pH 7.4, 200 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, 1 mM DTT). Protease inhibitor cocktail tablets (1 tablet per 10 g of cell pellet) and DNase (1 mg per 5 g of cell pellet) were used to prevent target degradation and facilitate lysis. The completed suspension was placed on ice and incubated with 0.25 mg/mL lysozyme over night with gentle stirring. The cell lysate was centrifuged at 30,000 RPM for 30 minutes to remove cell debris. The supernatant was passed over an amylose affinity column equilibrated with column buffer. The column was then washed with 10 column volumes of column buffer. The fusion protein was eluted with column buffer containing 10 mM maltose. The final product was dialyzed into 50 mM Tris, 150 mM KCl, pH 7.0.
Protein labeling
Purified talin head domains (THD) I398C, THD F0F1 F123C, THD F2F3 I398C were incubated with 4-fold molar excess tris(2-carboxyethyl)phosphine (TCEP) in 50 mM Tris, 150 mM KCl, pH 7.0 at room temperature for 10 min. A 2-fold molar excess of Uniblue A dissolved in dry DMSO was added to the reduced THD. The mixture was incubated at room temperature for 1 hour. Labeled protein was then purified by Sephadex G-25 size exclusion column. MSP1D1 D73C was diluted to 100 µM in 20 mM Tris HCL pH 7.4, 100 mM NaCl, 0.1 mM NaN3. A 4-fold molar excess of TCEP was added to the reaction and incubated at room temperature for 10min under an argon atmosphere. A 10-fold molar excess of TAMRA was mixed with the reduced THD and incubated at room temperature for 4 hours then placed at 4°C overnight. The labeling mixture was incubated with an equal volume of Amberlite XAD-2 to remove excess TAMRA. Labeled MSP was then purified by Sephadex G-25 size exclusion column.
Preparation of Nanodiscs
The protocol for Nanodisc preparation is adapted from a previous publication.43 In short, phospholipids, solubilized in chloroform, were dried under steady stream of nitrogen gas and further dried under vacuum for 4 hours. The dried lipid powder was then re-suspended and solubilized in 200 mM sodium cholate by shaking vigorously and sonication. TAMRA labeled MSP was added to the solubilized lipid to reach final ratio of 125:1 lipid/MSP. Detergents were removed by incubating with equal volume of Amberlite XAD-2 beads for 4 hours at room temperature. Nanodiscs were then purified by Superdex 200 Increase size exclusion column (GE Healthcare) equilibrated in 10 mM HEPES, 125 mM KCl, 4 mM KH2PO4, 14 mM NaCl, 1 mM MgCl2, 0.02 mM EGTA, 0.01% NaN3, PH 7.2 (binding buffer) at flow rate 0.75 mL/min.
Integrin TM/tail Nanodisc assembly
To favor the insertion of a single integrin TM/tail domain into the Nanodisc bilayer a large excess Nanodiscs were used for assembly. The typical stoichiometry of lipid:TAMRA MSP: integrin TM/tail is 1050:10:1. The purified MBP-integrin β3 TM/tail protein was concentrated in 10,000 Da cutoff centrifugal filters to 50 µM. The protein stock was incubated with 6 M urea for 1 hour at room temperature. The lipid stocks were dried as described above and then resuspended in 100 mM sodium cholate to a final concentration of 50 µ. Lipids, TAMRA labeled MSP, and denatured integrin β3 TM/tail were mixed incubated at room temperature for 45 minutes. The self-assembly process was initiated through dialysis into detergent and urea free buffer (20 mM Tris-HCl pH 7.4, 150 mM NaCl) at room temperature. The dialyzed Nanodisc sample was further purified using an amylose affinity resin in batch mode. The integrin containing Nanodiscs were eluted with 10 mM maltose. The Nanodisc fractions were pooled and digested with TEV protease (100:1 protein to protease molar ratio) overnight at 4 degrees to remove the maltose binding protein. The sample was passed over a second amylose affinity column to remove cleaved MBP. The column flow through and wash fractions were combined and further purified using a Superdex 200 increase size exclusion column equilibrated in 10 mM HEPES, 125 mM KCl, 4 mM KH2PO4, 14 mM NaCl, 1 mM MgCl2, 0.02 mM EGTA, 0.01% NaN3, PH 7.2 (binding buffer) at flow rate 0.75 mL/min. The fraction containing Nanodiscs were pooled and stored at 4°C until use.
Fluorescence Resonance Energy Transfer (FRET) THD-Nanodisc binding assay
The FRET based THD-Nanodisc was performed by titrating THD stepwise into 100 nM Nanodisc solutions in the presence of talin rod subdomains. Fluorescence was monitored using a Hitachi 3010 fluorometer equipped with a circulating water bath for temperature control. The samples were excited at 557 nm and the emission was monitored at 578 nm. Experiments were done in binding buffer to mimic cytosolic conditions. The FRET efficiencies were calculated as (F0-F)/F0 after correction for dilution. Typical binding isotherms are shown in figure 2. Fitting was performed by using the Matlab software suite (MathWorks) to obtain the dissociation constants and maximum FRET efficiencies using the equation below:
| (1) |
in which, F is the measured FRET efficiency, Fmax is the FRET efficiency of the bound complex,×is the total THD concentration, ND is twice the Nanodisc concentration (two leaflets per Nanodisc). Fitting the data to equation 1 gives rise to Fmax and the dissociation constant.
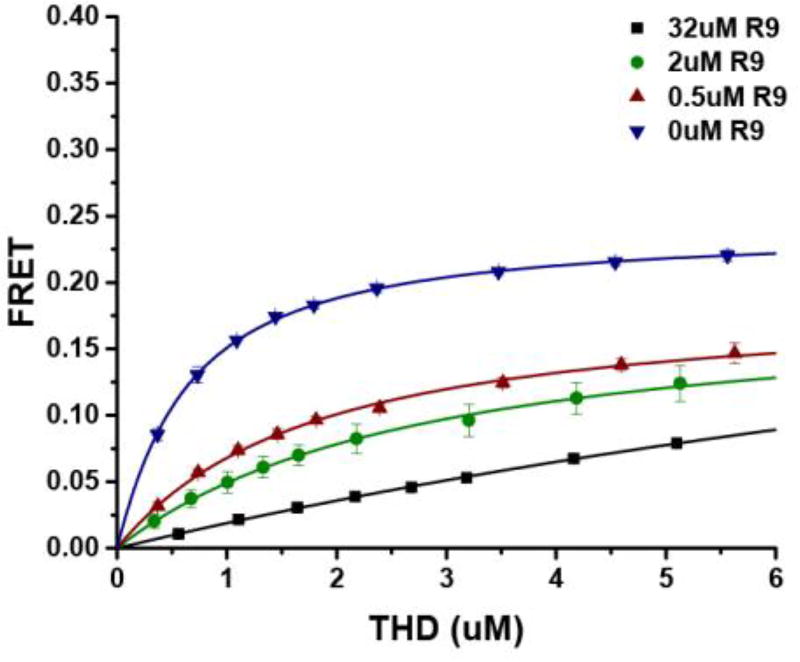
Figure 2.
Binding isotherms of THD on DMPS with talin R9. Data for 32 (µM), 2 (µM), 0.5 (µM), and 0 (µM) of isolated talin R9 domains, are shown in blue, green, red, and blue, respectively. Higher anionic lipid ratios promote tighter binding and increased maximum FRET efficiency.
RESULTS
Segments of talin Rod domain inhibit THD binding to the membrane
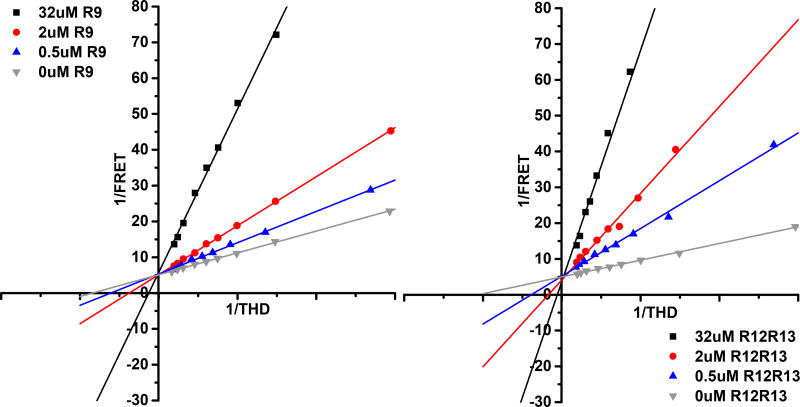
Previous studies showed that talin R9 (1655–1822) binds to F3 of the talin head domain and competes with the interaction of THD with the β3-integrin cytoplasmic tail. Talin R12R13 (1984–2344) is also known to interact with talin F3 domain albeit with a weaker affinity compared to R9.17,20 To understand the interplay of protein-protein and protein-membrane interactions, we used Nanodiscs of a defined lipid composition. We first tested the R9 and R12R13 regions of the talin rod. Using our FRET based assay, we directly assessed the ability of the talin rod domains to inhibit the interaction of THD with phospholipid bilayers. Uniblue A labeled THD was titrated into TAMRA labeled Nanodiscs containing 50% DMPS and the results are summarized in Table 1. THD binds to 50% DMPS Nanodiscs with a dissociation constant of 0.58 µM, which agrees with values previously reported.38 In the presence of 0.5 µM talin R9 the dissociation constant increases 3-fold and increases further with increasing R9 concentrations. On the other hand, the maximum FRET efficiency obtained from isotherm fitting is not affected by the presence of R9. In our experiments since there is no catalytic activity for the THD Nanodisc complex (kcat = 0, therefore Kd = Km) we can make use of a modified Michaelis-Menton equiation to analyze our data and provide insight into the mechanism of inhibition. Figure 3 shows Lineweaver-Burk plots of the titrations at increasing concentrations of R9 (panel left) and R12R13 (panel right). The shared Y intercept in the titrations indicate that both talin R9 and R12R13 are classic competitive inhibitors of the interaction between talin FERM domains and DMPS membranes. In addition since our assay is sensitive the dye separation distances, an identical FRETmax suggests that the bound complex is in a similar conformation in the presence of inhibitor, which is also consistent with a competitive mechanism. Previous NMR studies have shown that R1R2 interacts with the FERM domain suggesting that R1R2 may also be involved in the regulation of talin.17 We see that R1R2 only has modest effect on talin FERM membrane binding displaying only a 4-fold increase in dissociation constant (Table 1).
Table 1.
THD binding to 50% DMPS Nanodiscs in the presence of talin rod subdomains
| Rod Domain | Kd (µM) | Fold Increase |
|---|---|---|
|
| ||
| None | 0.58 ± 0.04 | NA |
| 0.5 µM R9 | 1.6 ± 0.1 | 2.8 |
| 2 µM R9 | 2.6 ± 0.5 | 4.6 |
| 32 µM R9 | 18 ± 10 | 30 |
|
| ||
| 0.5 µM R12-13 | 2.2 ± 0.2 | 3.8 |
| 2 µM R12-13 | 4.4 ± 0.5 | 7.5 |
| 32 µM R12-13 | 180 ± 40 | 300 |
|
| ||
| 0.5 µM R1R2 | 0.43 ± 0.01 | 0.7 |
| 2 µM R1R2 | 0.65 ± 0.06 | 1.1 |
| 32 µM R1R2 | 2.4 ± 0.2 | 4.2 |
Figure 3.
Linweaver-Burk Plot of fluorescence titrations in the presence of talin rod subdomains. Subdomain concentrations 32 (µM), 2 (µM), 0.5 (µM), and 0 (µM). Panel left – R9 subdomain, Panel right - R1213 subdomain. The shared Y intercepts suggest that both talin rod inhibitory subdomains are competitive inhibitor for talin membrane interactions.
PIP2 activates inhibited THD
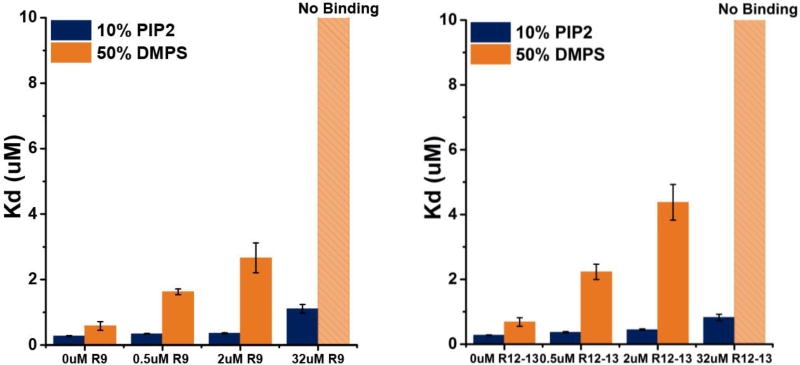
At sites of focal adhesion, recruitment of PIP5KI-γ greatly elevates the local concentration of PIP2.31 In order to quantify the effects of PIP2 on auto-inhibited talin, binding of THD to 10% PIP2 Nanodiscs was measured in the presence of talin rod subdomains. Since the charge on PIP2 at pH 7.2 is between -4 and -5, depending on the counter ions and the local environment,33 we use 50% DMPS Nanodiscs as a convenient comparison to 10% PIP2 owing to a similar overall Nanodisc charge. Figure 4 compares the affinity of PIP2 and DMPS Nanodiscs in the presence of the rod subdomains R9 and R12R13. Both R9 and R12R13 have very little effect on THD-PIP2 interactions, with only a 4-fold increase in the dissociation constants, which is in stark contrast to the inhibition seen on DMPS Nanodiscs (Figure 4).
Figure 4.
Affinities of THD on DMPS and PIP2 in the presence of talin rod inhibitory subdomains R9 (left), R12R13 (right). THD interaction with a PIP2 membrane surface is insensitive to the inhibitory effect by talin rod subdomains.
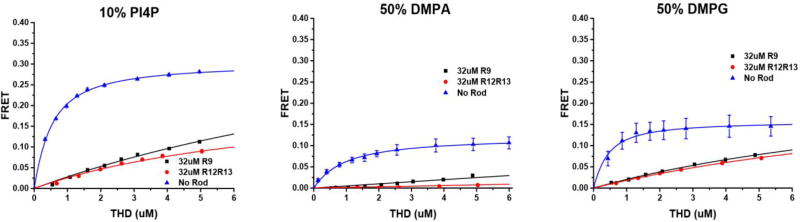
To further inspect the activation effects of the lipid headgroup charge, we prepared Nanodiscs with 50% DMPG, 50% DMPA, and 10% phosphatidylinositol-4-phosphate (PI4P). Here again we use Nanodiscs with similar overall charge for direct comparison. Previously, we have shown that THD binds to DMPG, DMPA, and PI4P with sub-micromolar dissociation constants similar to that of DMPS and PIP244. However, neither DMPA nor DMPG can support high affinity binding of THD in the presence of talin rod subdomains R9 and R12R13 (Figure 5). Interestingly, PI4P also did not support THD binding in the presence of rod subdomains despite the fact that the molecular structure of PI4P differ only by a single phosphate group.
Figure 5.
Binding isotherms of THD on DMPA, DMPG, and PI4P in the presence of talin rod domains. PI4P, DMPA, and DMPG cannot support tight binding of THD in the presence of talin R9 or R12R13.
PIP2 targets F2F3 but not F0F1
In order to further understand the role of the THD subdomains in the interaction with the phospholipid bilayer, we measured the affinity of isolated F0F1 and F2F3 to DMPS and PIP2 Nanodiscs (Table 2). We found that F2F3 bound to 10% PIP2 with a dissociation constant of 0.18 µM, whereas the dissociation constant for 50% DMPS was 1.2 µM, an increase of 6-fold. F0F1 bound to 10% PIP2 and 50% DMPS with a dissociation constant of 0.86 µM and 0.45 µM respectively. These results suggest that F0F1 does not discriminate between anionic lipids, while F2F3 has a strong preference for PIP2 over DMPS. When binding experiments of the isolated subdomains are performed in the presence of the inhibitor domains, both talin R9 and R12R13 domains impede the binding F0F1 to 10% PIP2 by a factor of 10 and completely inhibits binding to 50% DMPS. In marked contrast, adding rod subdomains led to 10-fold higher dissociation constant on F2F3 binding to 50% DMPS but fail to impede F2F3 binding to 10% PIP2 with a tight dissociation constant of ~0.35 µM.
Table 2.
THD F0F1 and F2F3 binding to Nanodiscs in the presence of talin rod subdomains
| F0F1 Kd (µM) |
F2F3 Kd (µM) |
|||
|---|---|---|---|---|
|
| ||||
| Rod Domain | 50% DMPS | 10% PIP2 | 50%DMPS | 10% PIP2 |
|
| ||||
| None | 0.45 ± 0.09 | 0.86 ± 0.25 | 1.2 ± 0.3 | 0.18 ± 0.01 |
| 32 µM R9 | No Binding | 14 ± 1 | 11 ± 2 | 0.36 ± 0.03 |
| 32 µM R12R13 | No Binding | 9 ± 4 | 10 ± 3 | 0.36 ± 0.03 |
Binding of talin to membrane embedded integrin β3 tail
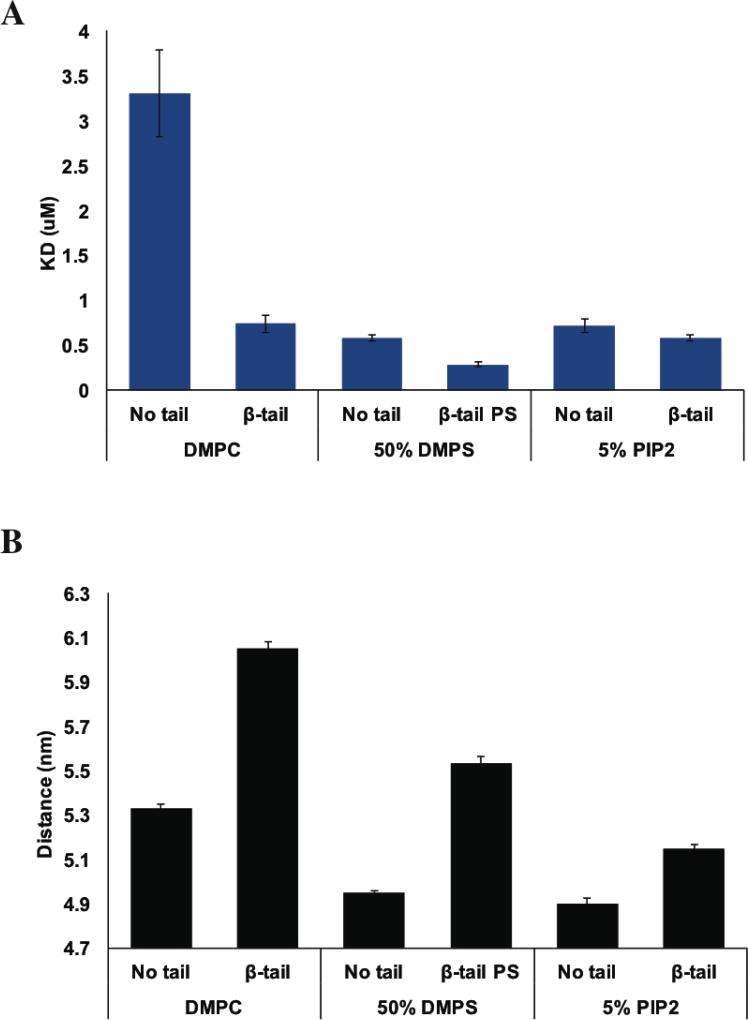
To investigate the mechanism of talin membrane recruitment in the presence of integrin β3 TM/tail, FRET based binding assay was performed with TAMRA labeled Nanodiscs with an embedded integrin tail. Figure 6A summaries the fitting results of Uniblue A labeled THD to Nanodiscs containing integrin β3 TM/tail. THD binds to β3 tail DMPC Nanodisc with a dissociation constant of 0.74 µM. Compared to the plain DMPC membrane surface, the mere presence of integrin tail promotes the affinity of THD by a factor of 5. On 50% DMPS membranes there is a two-fold increase in affinity in the presence of integrin tails. Intriguingly, on a 5% PIP2 membrane surface, adding integrin β3 tail only results in a slightly tighter binding, from 0.72 µM to 0.58 µM with integrin tail.
Figure 6.
Results of FRET based binding assay with Nanodiscs containing integrin β3 tails. (A) THD binding to Nanodiscs with or without integrin β3 tail. (B) Comparison of the fluorophore separation distances calculated from maximum FRET efficiencies.
To further understand the function of lipid composition in the talin mediated integrin activation, the dye separation distances were calculated and summarized in Figure 6B. The maximum FRET efficiencies on DMPC and DMPS surface were significantly altered by the presence of integrin β3 tail indicating an increase of 0.6 nm between the donor and acceptor dyes. Yet on PIP2 membrane, the dye separation distances are modestly affected with only 0.2 nm increase in the presence of integrin β3 tail (Figure 6B).
Talin auto-inhibition and activation in the presence of integrin β tail
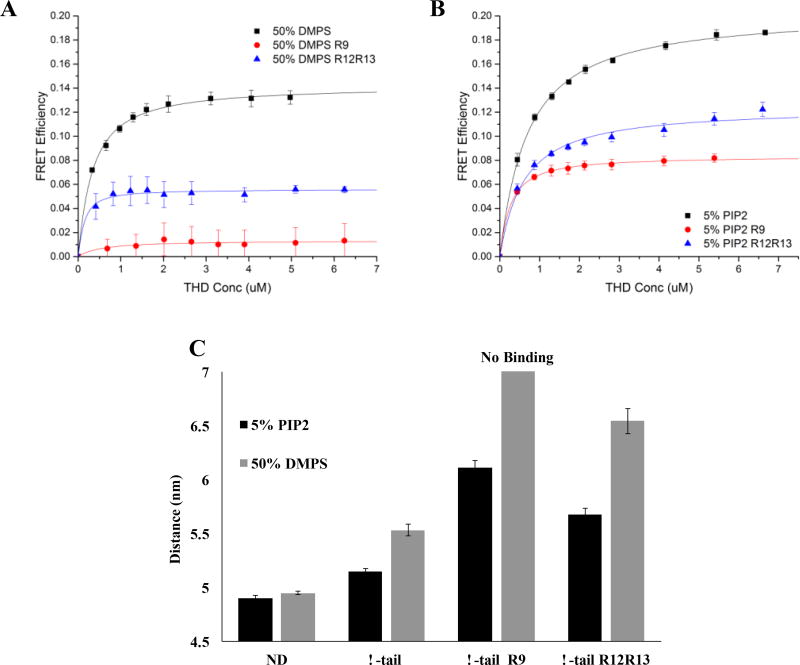
To further investigate the talin auto inhibition, binding assays were performed with integrin tail inserted Nanodiscs in the presence of talin rod inhibitory domains. We chose 50% DMPS and 5% PIP2 Nanodiscs for comparison since they have similar affinities toward THD and since PIP2 is insensitive to R9 or R12R13 inhibition of THD membrane interactions.
Figure 7A shows the binding isotherms of THD binding to integrin inserted Nanodiscs. 32 µM talin R9 subdomain completely abrogates the interactions of THD with 50% DMPS membrane and integrin tail. The dissociation constant is not noticeably affected by adding same concentration of the talin R12R13 segments and the maximum FRET efficiency is reduced to 5% from 13%. On 5% PIP2 Nanodisc (Figure 7B), the presence of both talin inhibitory subdomains leads to comparable dissociation affinities and 10% decrease in maximum FRET efficiencies. Figure 7C summarizes distance calculations from the measured maximum FRET efficiencies. 5% PIP2 Nanodiscs promote a closer binding geometry than found on 50% DMPS. Particularly in the presence of talin R9 inhibitory subdomains, adding THD yields undetectable change on 50% DMPS membrane while 5% PIP2 Nanodiscs still give rise to a dye separation distance of 6 nm. The inhibitory effect of R12R13 on THD binding topology is weaker on both PS and PIP2 membranes inserted with integrin tail with 0.5~0.9 nm increased FRET pair distance.
Figure 7.
Interaction of THD Nanodiscs containing integrin β3 tails in the presence of talin rod inhibitory subdomains. (A and B) The binding isotherms of THD / 50% DMPS and 5% PIP2 Nanodiscs. (C) Fluorophore distance of THD bound to integrin inserted Nanodiscs.
DISCUSSION
Integrin activation and its downstream events, such as actin coupling or cell matrix recognition, are pivotal for cell-ECM dependent functions.1 Significant efforts have been spent investigating the protein-protein interactions involved in many of these processes, yet the role of the membrane in mediating these interactions at the cytoplasmic face of the cell has not been extensively studied. In this investigation we seek to understand the molecular basis of talin auto-inhibition and how the membrane environment modulates talin activation. We combined Nanodisc technology, which provides a precisely controlled lipid bilayer environment, with a FRET based binding assay, to yield information on protein-membrane affinity and protein-protein interactions.
Previous studies have focused on dissecting the mechanism of talin auto-inhibition and inter-domain interactions. A middle segment of the talin rod, residues 1654–2344, covers the integrin tail binding site in the talin FERM domain.20 A more specific region, residues 1655–1822, strongly interacts with the F3 domain and it was shown that vesicles consisting of a mixture of PC and PIP2 disruopt this interaction.19 Figure 2 shows representative binding isotherms of THD binding to 50% DMPS Nanodisc membranes in the presence of the R9 sub-domain. Table 1 shows that the dissociation constant for the interaction increases dramatically as the R9 and R12R13 concentration rises. The local concentration of the rod domains is expected to be much higher than what we tested here. The volume of the full-length talin dimer calculated from a previously reported structure is approximately 1300 nm3, which represents a ~2 mM inter-domain concentration considering the dimer contains two rod domains.17 Thus, both R9 and R12R13 segments of the talin rod domain are strong inhibitors that modulate the membrane recruitment of the THD under physiological conditions. The shared Y-axis intercept in the Lineweaver-Burk plots (Figure 3) indicates that the talin rod subdomains are competitive inhibitors of THD binding to DMPS Nanodiscs. The maximum FRET efficiency between the donor and acceptor is unchanged and suggests that the structure of the complex that is formed is similar in the presence and absence of the inhibitor domains. These results suggest that the rod subdomains are directly interacting with the membrane orientation patch (MOP), consistent with previous NMR studies, yet does not perturb the overall topology of THD/membrane complex.19–21 Thus, the association of THD and membrane is tightly regulated through competitive inhibition by R9 and R12R13 via masking the membrane and/or integrin binding sites on THD. This is in close agreement with previous structural studies that suggest the talin F2F3/R9 interface sterically blocks the portion of membrane proximal region in integrin β1 cytoplasmic tail.21,45 Another important function of the talin rod domain is the ability to bind the actin cytoskeleton and its role in mechanical force transduction and signaling. One of two recognized actin binding sites within the talin rod is located in the R13 subdomain.46 Several vinculin binding sites have also been identified in talin R1-R3, R6-R8, R10-R11 subdomains.47 Thus, the interaction of R9 and R12R13 with the THD may play a dual role regulating integrin activation and talin functions, such as integrin clustering, actin binding, and vinculin engagement.
A more recent structural study proposed a novel compact auto-inhibited dimer conformation, suggesting an additional inhibitory region at R1R2 rod domain.17 However, we did not detect a significant change in talin FERM binding to Nanodiscs in the presence of R1R2, which is consistent with other NMR studies that report talin R1 does not promote strong chemical shifts. 18,20 Thus, binding of talin R1 to F2F3 is weak and non-specific.18 Talin R1R2 has a closely packed arrangement with R3R4 and does not play a role in talin inhibition but may be implicated in occluding vinculin binding sites of the rod domain in the compact form of talin in the absence of force.48
A key unknown in talin mediated integrin signaling is the mechanism of activating the auto-inhibited form of full-length talin, in particular the role of the membrane surface. Several previous studies indicate that membranes containing PIP2 are potential activators of talin. At sites of focal adhesions, synthesis of PIP2 from PI4P is promoted by the recruitment of PIP5KI-γ and in turn the presence of PIP2 enhances the membrane localization of talin.39 NMR and SPR studies demonstrated a unique bivalent mode of PIP2 binding with THD, revealing that the binding interfaces are located in F2 membrane orientation patch and at residues K322-K324 of the F3 domain.21 Our results here show that 10% PIP2 supports tight binding of the THD in the presence of the inhibitor subdomains R9 and R12R13, while other anionic phospholipids are significantly inhibited (figure 4). The affinity of THD for PIP2 membranes remains at or below 1 µM regardless of the concentration of rod subdomains present. This shows that the mere presence of PIP2 at the membrane surface is sufficient to relieve the auto inhibitory interactions and activate talin.
An atypical feature of talin FERM domains is the large unstructured insert (~30 amino acids) within F1 domain. A “Flying cast” model has been proposed where the F1 loop has a propensity to form a α-helix when in contact with anionic lipids and is thought to be involved in talin-membrane interactions and talin activation. Deletion of this F1 loop has been shown to substantially impede talin mediated integrin activation, but not alter the affinity of the talin FERM domain for integrin tail.9 In this work, both talin F0F1 and F2F3 subdomains readily bound to anionic membranes at sub-micromolar dissociation constants (Table 2). F2F3 has a strong binding preference of PIP2 over PS, with the dissociation constant increasing by a factor of six. F0F1, on the other hand, shows no preference for PIP2 over PS. The similar affinities of talin FERM subdomains and the intact THD with PS membrane surface suggest that there is little cooperativity between F0F1 and F2F3. In comparison, the clear preferential binding of PIP2 to talin F2F3 over F0F1 is consistent with the previous NMR study that the binding interface between PIP2 and talin resides in the F2F3 subdomain.19,38 Both talin R9 and R12R13 are able to inhibit both F0F1 and F2F3 domain interactions with PS membranes and it is not clear how F0F1 interacts with talin rod subdomains. One likely explanation could be the electrostatic interaction between the F0F1 positively charged loop and the extensively negatively charged surface on talin R9.21 Surprisingly, the presence of R9 or R12R13 has little effect on F2F3 binding to PIP2 membranes yet significantly inhibits F0F1 interaction. Our results are not consistent with the previously suggested “Fly casting” model.10,49 We propose that the rod domains inhibit F2F3 by a tight specific interaction that is efficiently disrupted by the presence of PIP2, while the talin rod segments may mask membrane-binding sites of F0F1 through general electrostatic interaction by competing with negatively charged bilayers.
Interestingly, THD binds to PI4P with high affinity yet the absence of the phosphate group on the five position renders this interaction sensitive to the inhibition by the rod subdomains (figure 5). Furthermore, both subdomains of talin rod also strongly inhibit talin head binding to PG and PA Nanodiscs. This finding demonstrates that the presence of PIP2 at the protein membrane interface is a key player in dislodging talin rod inhibitory domains and activating talin. At focal adhesion sites, the presence of PIP5K-γ, which is responsible for transforming PI4P into PIP2, may be essential for talin activation.
Early studies of talin interactions with integrin often incorporate integrin tails into lipid vessiles39 or tether integrin peptides on lipid coated surfaces38. However, it is difficult to resolve the insertion orientation and the oligomeric state while providing a native like membrane environment. Here, Nanodisc technology was employed to assemble Nanodiscs with a controlled lipid composition and an unclustered integrin β3 TM/tail. The results of FRET binding assays reveal that THD readily binds to integrin tails with high affinity. The presence of anionic lipids does not significantly increase the affinities of the THD – Nanodcisc interaction, suggesting that the integrin cytoplasmic tail provides most of the binding free energy..
It has been shown that the binding geometry of talin F3 subdomain is sensitive to the lipid headgroup identity. Higher charge density of the lipid headgroup, like phosphatidylinositols, gives rise to a conformation in which F3 is closely associated with the bilayer 44. Here, we analyzed the maximum FRET efficiencies of titration to further understand the effects of the lipid and integrin on talin binding geometry. Surprisingly, the presence of integrin tail results in an ~0.6 nm increased FRET pair distances on DMPC and DMPS surface. This observation may be explained by looking at the known talin – integrin interactions. Integrin tails contain two NPxY motifs targeted by the PTB domain of talin in addition to the inner membrane clasp. Although talin preferably binds to the membrane proximal NPxY motifs, it is possible that talin also interacts with the distal NPxY motif and results in a longer FRET pair distance. Similarly to Nanodiscs without embedded integrin tails, PIP2 Nanodiscs promotes the closest binding topology at 5 nm dye-dye separation, which is only 0.2 nm longer than that on plain PIP2 surface. This close approach of the F3 domain may be responsible for efficient disruption of the outer membrane clasp between the α and β integrin subunits.
The presence of integrin β3 tail provides insight into the inhibitory effects of talin R9 and R12R13 subdomains. On 50% DMPS membrane (Figure 7A), talin R9 completely inhibited the binding of THD while talin R12R13 strongly alters the conformation of THD – nanodisc complex yet does not alter the affinity. On 10% PIP2 membranes (Figure 7B), both inhibitory rod subdomains exhibit similar effects resulting larger FRET pair distance with unchanged affinities. The results indicate that the interfaces of THD, integrin tail and PS membrane, namely the F3 domain, may be sterically blocked by talin R9 but not R12R13. This is distinctive from what was previously observed where both talin inhibitory subdomains are classic competitive inhibitors on DMPS Nanodiscs to THD. The R12R13 might inhibit the talin membrane recruitment by covering the F2 MOP region to impede THD membrane binding, yet the NPxY motif on integrin tail is still accessible for THD. Thus integrin tail could still interact with THD but in an altered biding topology due to the lack of MOP interactions. The result also confirm the role of PIP2 as an activator during talin membrane engagement due to the fact that the presence of 5% PIP2 on lipid bilayer partially promotes high affinities and closer binding geometry in the presence of the rod subdomains compared to PS membrane. It should be noted that the formal charge of 5% PIP2 roughly equals to only 20–30% DMPS, thus the high charge density on the PIP2 lipid headgroup may be key player in the disruptions of THD - talin rod domain interactions. However, the PIP2 alone does not fully rescue talin activation, other protein partners, such as RIAM-RAP1, PIPKα, and calpain, need to work in concert with PIP2 for complete talin activation and optimal integrin activation.
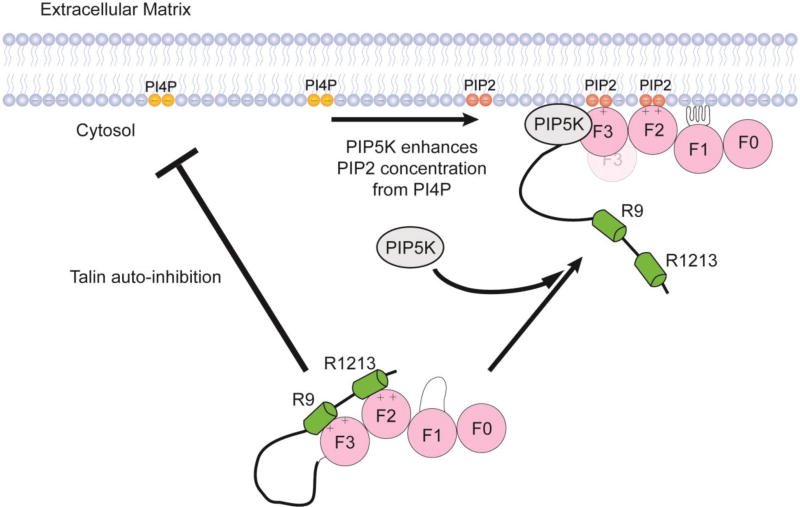
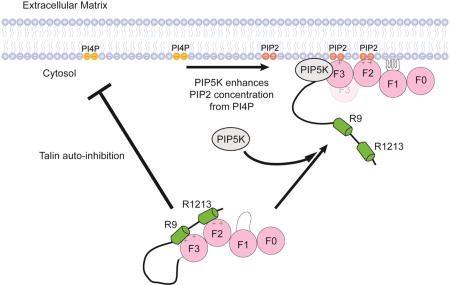
Figure 8 shows a schematic description of the lipid dependent mechanism of talin activation through a complex interplay of protein-protein and protein-membrane interactions.
Figure 8.
Regulation of talin activation by PIP2. The presence of PIP2 in the bilayer promotes the dissociation of talin R9 and R12R13.
CONCLUSION
In summary, we have described a unique role of the membrane in talin activation and recruitment. Our studies show that PIP2 can promote the disruption of the head to rod domain interactions through a process that is extremely sensitive to the headgroup identity. Simply converting PI4P to PIP2 at the membrane promotes the unraveling of the auto-inhibited form of talin. This results in the unmasking talin N-terminal FERM domain from C-terminal Rod subdomains, allowing effective communication with integrin cytoplasmic tails. Our results point to the importance of the interplay between talin, the membrane surface, and PIP5KI-γ during the talin mediated integrin activation. Our proposed mechanism suggests that the presence of PIP5KI-γ at the site of focal adhesion results in the amplification of talin activation through the production of PIP2 from PI4P. Clearly, among common phospholipid head groups, PIP2 seems to be a key player in an orchestra involving many other adaptor protein partners, such as Rap1-RIAM, calpain, and the aforementioned PIP5KI-γ, that promotes efficient talin mediated integrin activation.
Acknowledgments
We would like to thank Dr. Mark H. Ginsberg and Dr. Feng Ye for the gift of the THD expression vector.
Funding Sources
This work was funded by NIH grant GM101048 and GM33775 to SGS.
ABBREVIATIONS
- ECM
extracellular matrix
- THD
talin head domain
- FERM
band 4.1/ezrin/radixin/moesin
- MOP
membrane orientation patch
- PTB
phospho-tyrosine binding
- Rap1
Ras-related protein 1
- RIAM
Rap1-interacting adapter protein
- PIP5K- γ
phosphatidylinositol-4-phosphate 5-kinase-γ
- PIP2
phoshpaidylinositol-4, 5-bisphosphate
- PI4P
phoshpaidylinositol-4-phosphate
- SPR
surface Plasmon resonance
- MSP
Membrane Scaffold Protein
- FRET
fluorescence resonance energy transfer
- DMPC
1,2-dimyristoyl-sn-glycero-3-phosphocholine
- DMPS
1,2-dimyristoyl-sn-glycero-3-phospho-L-serine
- DMPG
1,2-dimyristoyl-sn-glycero-3-phosphoglycerol
- DMPA
1,2-dimyristoyl-sn-glycero-3-phosphate
- TAMRA
Tetramethylrhodamine-5-(and -6) C2 maleimide
- UA
Uniblue A
Footnotes
Author Contributions
XY performed the titration experiments, analyzed the data, and contributed to the writing of the paper, MAM conceived the idea for the project, analyzed the data, and wrote and edited the paper, SGS contributed to the project design and edited this communication.
References
- 1.Hynes RO. Integrins: Bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 2.Lau TL, Kim C, Ginsberg MH, Ulmer TS. The structure of the integrin alphaIIbbeta3 transmembrane complex explains integrin transmembrane signalling. EMBO J. 2009;28:1351–1361. doi: 10.1038/emboj.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calderwood Da, Zent R, Grant R, Rees DJG, Hynes RO, Ginsberg MH. The Talin head domain binds to integrin beta subunit cytoplasmic tails and regulates integrin activation. J. Biol. Chem. 1999;274:28071–28074. doi: 10.1074/jbc.274.40.28071. [DOI] [PubMed] [Google Scholar]
- 4.Kim C, Ye F, Hu X, Ginsberg MH. Talin activates integrins by altering the topology of the beta transmembrane domain. J. Cell Biol. 2012;197:605–611. doi: 10.1083/jcb.201112141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diaz-Gonzalez F. Breaking the Integrin Hinge. J. Biol. Chem. 1996;271:6571–6574. doi: 10.1074/jbc.271.12.6571. [DOI] [PubMed] [Google Scholar]
- 6.Shattil SJ, Kim C, Ginsberg MH. The final steps of integrin activation: the end game. Nat. Rev. Mol. Cell Biol. 2010;11:288–300. doi: 10.1038/nrm2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tadokoro S, Shattil SJ, Eto K, Tai V, Liddington RC, de Pereda JM, Ginsberg MH, Calderwood DA. Talin binding to integrin beta tails: a final common step in integrin activation. Science. 2003;302:103–106. doi: 10.1126/science.1086652. [DOI] [PubMed] [Google Scholar]
- 8.Roberts GCK, Critchley DR. Structural and biophysical properties of the integrin-associated cytoskeletal protein talin. Biophys. Rev. 2009;1:61–69. doi: 10.1007/s12551-009-0009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goult BT, Bouaouina M, Elliott PR, Bate N, Patel B, Gingras AR, Grossmann JG, Roberts GCK, Calderwood DA, Critchley DR, Barsukov IL. Structure of a double ubiquitin-like domain in the talin head: a role in integrin activation. EMBO J. 2010;29:1069–1080. doi: 10.1038/emboj.2010.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Domadia PN, Li Y-F, Bhunia A, Mohanram H, Tan S-M, Bhattacharjya S. Functional and structural characterization of the talin F0F1 domain. Biochem. Biophys. Res. Commun. 2010;391:159–65. doi: 10.1016/j.bbrc.2009.11.024. [DOI] [PubMed] [Google Scholar]
- 11.Campbell ID. The talin FERM domain is not so FERM. Structure. 2010;18:1222–1223. doi: 10.1016/j.str.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Elliott PR, Goult BT, Kopp PM, Bate N, Grossmann JG, Roberts GCK, Critchley DR, Barsukov IL. The Structure of the Talin Head Reveals a Novel Extended Conformation of the FERM Domain. Structure. 2010;18:1289–1299. doi: 10.1016/j.str.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anthis NJ, Wegener KL, Ye F, Kim C, Goult BT, Lowe ED, Vakonakis I, Bate N, Critchley DR, Ginsberg MH, Campbell ID. The structure of an integrin/talin complex reveals the basis of inside-out signal transduction. EMBO J. 2009;28:3623–3632. doi: 10.1038/emboj.2009.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalli AC, Wegener KL, Goult BT, Anthis NJ, Campbell ID, Sansom MSP. The structure of the talin/integrin complex at a lipid bilayer: an NMR and MD simulation study. Structure. 2010;18:1280–1288. doi: 10.1016/j.str.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arcario MJJ, Tajkhorshid E. Membrane-Induced Structural Rearrangement and Identification of a Novel Membrane Anchor in Talin F2F3. Biophys. J. 2014;107:2059–2069. doi: 10.1016/j.bpj.2014.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim C, Lau T-LL, Ulmer TS, Ginsberg MH. Interactions of platelet integrin alphaIIb and beta3 transmembrane domains in mammalian cell membranes and their role in integrin activation. Blood. 2009;113:4747–4753. doi: 10.1182/blood-2008-10-186551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goult BT, Xu XP, Gingras AR, Swift M, Patel B, Bate N, Kopp PM, Barsukov IL, Critchley DR, Volkmann N, Hanein D. Structural studies on full-length talin1 reveal a compact auto-inhibited dimer: Implications for talin activation. J. Struct. Biol. 2013;184:21–32. doi: 10.1016/j.jsb.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banno A, Goult BT, Lee H, Bate N, Critchley DR, Ginsberg MH. Subcellular Localization of Talin Is Regulated by Inter-domain Interactions. J. Biol. Chem. 2012;287:13799–13812. doi: 10.1074/jbc.M112.341214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goult BT, Bate N, Anthis NJ, Wegener KL, Gingras AR, Patel B, Barsukov IL, Campbell ID, Roberts GCK, Critchley DR. The structure of an interdomain complex that regulates Talin activity. J. Biol. Chem. 2009;284:15097–15106. doi: 10.1074/jbc.M900078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goksoy E, Ma Y-Q, Wang X, Kong X, Perera D, Plow EF, Qin J. Structural Basis for the Autoinhibition of Talin in Regulating Integrin Activation. Mol. Cell. 2008;31:124–133. doi: 10.1016/j.molcel.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song X, Yang J, Hirbawi J, Ye S, Perera HD, Goksoy E, Dwivedi P, Plow EF, Zhang R, Qin J. A novel membrane-dependent on/off switch mechanism of talin FERM domain at sites of cell adhesion. Cell Res. 2012;22:1533–1545. doi: 10.1038/cr.2012.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee HS, Lim CJ, Puzon-McLaughlin W, Shattil SJ, Ginsberg MH. RIAM activates integrins by linking talin to Ras GTPase membrane-targeting sequences. J. Biol. Chem. 2009;284:5119–5122. doi: 10.1074/jbc.M807117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang J, Zhu L, Zhang H, Hirbawi J, Fukuda K, Dwivedi P, Liu J, Byzova T, Plow EF, Wu J, Qin J. Conformational activation of talin by RIAM triggers integrin-mediated cell adhesion. Nat. Commun. 2014;5:5880. doi: 10.1038/ncomms6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goult BT, Zacharchenko T, Bate N, Tsang R, Hey F, Gingras AR, Elliott PR, Roberts GCK, Ballestrem C, Critchley DR, Barsukov IL. RIAM and vinculin binding to talin are mutually exclusive and regulate adhesion assembly and turnover. J. Biol. Chem. 2013;288:8238–8249. doi: 10.1074/jbc.M112.438119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan B, Calderwood Da, Yaspan B, Ginsberg MH. Calpain cleavage promotes talin binding to the beta 3 integrin cytoplasmic domain. J. Biol. Chem. 2001;276:28164–28170. doi: 10.1074/jbc.M104161200. [DOI] [PubMed] [Google Scholar]
- 26.Franco SJ, Rodgers MA, Perrin BJ, Han J, Bennin DA, Critchley DR, Huttenlocher A. Calpain-mediated proteolysis of talin regulates adhesion dynamics. Nat. Cell Biol. 2004;6:977–983. doi: 10.1038/ncb1175. [DOI] [PubMed] [Google Scholar]
- 27.Mattheij NJa, Gilio K, Van Kruchten R, Jobe SM, Wieschhaus AJ, Chishti AH, Collins P, Heemskerk JWM, Cosemans JMEM. Dual mechanism of integrin αIIbβ3 closure in procoagulant platelets. J. Biol. Chem. 2013;288:13325–13336. doi: 10.1074/jbc.M112.428359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Zhao L, Suzuki A, Lian L, Min SH, Wang Z, Litvinov RI, Stalker TJ, Yago T, Klopocki AG, Schmidtke DW, Yin H, Choi JK, McEver RP, Weisel JW, Hartwig JH, Abrams CS. Platelets lacking PIP5KIγ have normal integrin activation but impaired cytoskeletal-membrane integrity and adhesion. Blood. 2013;121:2743–2752. doi: 10.1182/blood-2012-07-445205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Pereda JM, Wegener KL, Santelli E, Bate N, Ginsberg MH, Critchley DR, Campbell ID, Liddington RC. Structural basis for phosphatidylinositol phosphate kinase type Igamma binding to talin at focal adhesions. J. Biol. Chem. 2005;280:8381–8386. doi: 10.1074/jbc.M413180200. [DOI] [PubMed] [Google Scholar]
- 30.Barsukov IL, Prescot A, Bate N, Patel B, Floyd DN, Bhanji N, Bagshaw C, Letinic KR, Di Paolo G, De Camilli P, Roberts GCK, Critchley DR. Phosphatidylinositol phosphate kinase type 1γ and β1-integrin cytoplasmic domain bind to the same region in the talin FERM domain. J. Biol. Chem. 2003;278:31202–9. doi: 10.1074/jbc.M303850200. [DOI] [PubMed] [Google Scholar]
- 31.van den Bout I, Divecha N. PIP5K-driven PtdIns(4,5)P2 synthesis: regulation and cellular functions. J. Cell Sci. 2009;122:3837–3850. doi: 10.1242/jcs.056127. [DOI] [PubMed] [Google Scholar]
- 32.Hammond GRV, Fischer MJ, Anderson KE, Holdich J, Koteci A, Balla T, Irvine RF. PI4P and PI(4,5)P2 Are Essential But Independent Lipid Determinants of Membrane Identity. Science. 2012;337:727–730. doi: 10.1126/science.1222483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McLaughlin S, Wang J, Gambhir A, Murray D. PIP2 and Proteins: Interactions, Organization, and Information Flow. Annu. Rev. Biophys. Biomol. Struct. 2002;31:151–175. doi: 10.1146/annurev.biophys.31.082901.134259. [DOI] [PubMed] [Google Scholar]
- 34.Zhou J, Bronowska A, Le Coq J, Lietha D, Gräter F. Allosteric Regulation of Focal Adhesion Kinase by PIP2 and ATP. Biophys. J. 2015;108:698–705. doi: 10.1016/j.bpj.2014.11.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raucher D, Stauffer T, Chen W, Shen K, Guo S, York JD, Sheetz MP, Meyer T. Phosphatidylinositol 4,5-bisphosphate functions as a second messenger that regulates cytoskeleton-plasma membrane adhesion. Cell. 2000;100:221–228. doi: 10.1016/s0092-8674(00)81560-3. [DOI] [PubMed] [Google Scholar]
- 36.Gilmore AP, Burridge K. Regulation of vinculin binding to talin and actin by phosphatidyl-inositol-4-5-bisphosphate. Nature. 1996;381:531–535. doi: 10.1038/381531a0. [DOI] [PubMed] [Google Scholar]
- 37.Goni GM, Epifano C, Boskovic J, Camacho-Artacho M, Zhou J, Bronowska A, Martin MT, Eck MJ, Kremer L, Grater F, Gervasio FL, Perez-Moreno M, Lietha D. Phosphatidylinositol 4,5-bisphosphate triggers activation of focal adhesion kinase by inducing clustering and conformational changes. Proc. Natl. Acad. Sci. 2014;111:E3177–E3186. doi: 10.1073/pnas.1317022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore DT, Nygren P, Jo H, Boesze-Battaglia K, Bennett JS, DeGrado WF. Affinity of talin-1 for the beta3-integrin cytosolic domain is modulated by its phospholipid bilayer environment. Proc. Natl. Acad. Sci. U. S. A. 2012;109:793–798. doi: 10.1073/pnas.1117220108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martel V, Racaud-Sultan C, Dupe S, Marie C, Paulhe F, Galmiche A, Block MR, Albiges-Rizo C. Conformation, localization, and integrin binding of talin depend on its interaction with phosphoinositides. J. Biol. Chem. 2001;276:21217–21227. doi: 10.1074/jbc.M102373200. [DOI] [PubMed] [Google Scholar]
- 40.Saltel F, Mortier E, Hytonen VP, Jacquier M-C, Zimmermann P, Vogel V, Liu W, Wehrle-Haller B. New PI(4,5)P2- and membrane proximal integrin-binding motifs in the talin head control beta3-integrin clustering. J. Cell Biol. 2009;187:715–731. doi: 10.1083/jcb.200908134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Legate KR, Takahashi S, Bonakdar N, Fabry B, Boettiger D, Zent R, Fässler R. Integrin adhesion and force coupling are independently regulated by localized PtdIns(4,5)2 synthesis. EMBO J. 2011;30:4539–4553. doi: 10.1038/emboj.2011.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bayburt TH, Grinkova YV, Sligar SG. Self-Assembly of Discoidal Phospholipid Bilayer Nanoparticles with Membrane Scaffold Proteins. Nano Lett. 2002;2:853–856. [Google Scholar]
- 43.Denisov IG, Grinkova YV, Lazarides aa, Sligar SG. Directed Self-Assembly of Monodisperse Phospholipid Bilayer Nanodiscs with Controlled Size. J. Am. Chem. Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 44.Ye X, McLean MA, Sligar SG. Conformational equilibrium of talin is regulated by anionic lipids. Biochim. Biophys. Acta - Biomembr. 2016;1858:1833–1840. doi: 10.1016/j.bbamem.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang H, Chang Y, Huang Q, Brennan ML, Wu J. Structural and Functional Analysis of a Talin Triple-Domain Module Suggests an Alternative Talin Autoinhibitory Configuration. Structure. 2016;24:721–729. doi: 10.1016/j.str.2016.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gingras AR, Ziegler WH, Bobkov AA, Joyce MG, Fasci D, Himmel M, Rothemund S, Ritter A, Grossmann JG, Patel B, Bate N, Goult BT, Emsley J, Barsukov IL, Roberts GCK, Liddington RC, Ginsberg MH, Critchley DR. Structural determinants of integrin binding to the talin rod. J. Biol. Chem. 2009;284:8866–76. doi: 10.1074/jbc.M805937200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gingras AR, Ziegler WH, Frank R, Barsukov IL, Roberts GC, Critchley DR, Emsley J. Mapping and consensus sequence identification for multiple vinculin binding sites within the talin rod. J. Biol. Chem. 2005;280:37217–37224. doi: 10.1074/jbc.M508060200. [DOI] [PubMed] [Google Scholar]
- 48.Rio A, Perez-jimenez R, Liu R, Roca-cusachs P, Fernandez JM, Sheetz MP. Stretching Single Talin Rod. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bouaouina M, Goult BT, Huet-Calderwood C, Bate N, Brahme NN, Barsukov IL, Critchley DR, Calderwood DA. A conserved lipid-binding loop in the kindlin FERM F1 domain is required for kindlin-mediated αIIbβ3 integrin coactivation. J. Biol. Chem. 2012;287:6979–90. doi: 10.1074/jbc.M111.330845. [DOI] [PMC free article] [PubMed] [Google Scholar]