Abstract
Neisseria gonorrhoeae is one of the most prevalent sexually transmitted infections worldwide. This obligate human pathogen has been extensively studied in vitro, where bacterial factors that are known to contribute to gonococcal disease and their regulation are relatively well defined. However, these in vitro experimental conditions only loosely replicate the host specific environment encountered by the bacteria in vivo. We recently reported on the complete gonococcal transcriptome expressed during natural human mucosal infection using RNA-seq analysis. Gene transcripts expressed in vivo (in vivo expressed factors) included genes encoding antibiotic resistance determinants, and a large number of hypothetical genes. A comparison of the gonococcal transcriptome expressed in vivo with the corresponding strain grown in vitro identified sets of genes regulated by infection, including those regulated by iron and the transcriptional regulatory protein Fur. We highlight here the role of Fur and gonococcal-specific regulatory processes important for infection and pathogenicity. We have determined that the genes controlled by Fur follow the same expression pattern in vivo as described previously in vitro, confirming Fur's regulatory role during infection. Collectively, these studies provide new insights into how bacterial fitness and pathogenicity are modulated during human mucosal infection.
Keywords: Neisseria gonorrhoeae, gene regulation, iron, transcriptome, mucosal infection
This article reviews how combining transcriptomics on pathogens from naturally infected hosts and other mechanistic studies can improve our understanding the genes involved in bacterial pathogenesis and how they are regulated.
INTRODUCTION
The Gram-negative obligate human pathogen Neisseria gonorrhoeae is the causative agent of the sexually transmitted infection gonorrhea, the second most common reportable disease in the USA. According to the Centers for Disease Control and Prevention (CDC) and the World Health Organization (WHO), there are approximately 800 000 new cases of gonorrhea in the USA and 106 million cases worldwide each year (World Health Organization 2012, 2016; Newman et al.2015; CDC 2016). Neisseria gonorrhoeae infects the genitourinary tract of men and women; however, the disease sequelae differ between genders (Edwards and Apicella 2004). In men, gonorrhea is defined by a marked infiltration of polymorphonuclear neutrophils (PMNs), resulting in symptomatic urethritis and epididymitis (World Health Organization 2012, 2016; Yu and Genco 2012a; Goire et al.2014). In women, the disease is more insidious, remaining asymptomatic and often resulting in spread of the infection to the ascending genitourinary tract (Walker and Sweet 2011; Islam et al.2015; Newman et al.2015). If left untreated in women, gonococcal infection can result in pelvic inflammatory disease, endometriosis, ectopic pregnancy and ultimately, infertility (Fichorova et al.2001; World Health Organization 2012, 2016; Islam et al.2015). Although the genitourinary tract is the primary site of infection in humans, N. gonorrhoeae can also infect extragenital sites such as the oropharynx and the rectum (Chan et al.2016).
A rising trend in incidence of N. gonorrhoeae has recently been reported, and this is expected to continue to rise, due in part to increased antibiotic resistance of the organism and the lack of a preventative vaccine (Unemo et al.2012; Unemo and Shafer 2014; Jerse, Bash and Russell 2015; Gottlieb et al.2016). A better understanding of N. gonorrhoeae pathogenic mechanisms during human mucosal disease would aide in development of novel therapeutics. There are a few in vivo models of gonococcal genital infection, but these present with several limitations. The human male urethral N. gonorrhoeae challenge model uses experimental infection of male volunteers as a model of urethritis (Cornelissen et al.1998; Biswas et al.1999). While studies in this model have expanded our understanding of acute infection, they do not address the consequences of long-term infection or infection specifically in women.
A mouse model of female gonococcal infection was developed in the late 1990s based on estradiol treatment of female mice, which allows for N. gonorrhoeae genitourinary tract infection in this otherwise resistant species (Jerse 1999; Jerse et al.2011). Although successful, this model represents a surrogate of female infection in humans in an artificial hormone condition. A more recent model based on generation of ‘humanized’ mice expressing the human carcinoembryonic antigen cell adhesion molecule (CEACAM) 1 and 5, required for N. gonorrhoeae opacity (Opa) proteins interactions with host cells overcomes some of these limitations (Schmitter et al.2007; Sadarangani, Pollard and Gray-Owen 2011). While this model has expanded the study of N. gonorrhoeae infection in both male and female mice without the use of estradiol (Gu et al.2010; Sintsova et al.2015), it does not mimic the unique environment of the human genital tract.
GONOCOCCAL GENE EXPRESSION DURING INFECTION
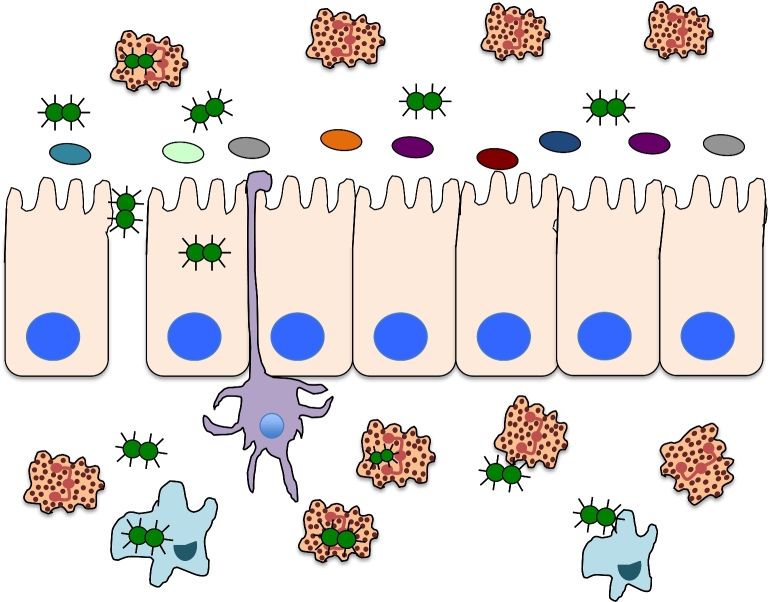
Like other human pathogens, Neisseria gonorrhoeae has evolved mechanisms to adapt to the specific environments encountered during infection. However, the majority of studies aimed at characterizing this response of the gonococcus to different environmental stimuli have examined N. gonorrhoeae cultured in in vitro conditions that do not completely replicate the environment encountered in the human host (Biswas et al.1999; Grifantini et al.2003; Agarwal et al.2008). In the female genital tract, N. gonorrhoeae is exposed to an environment characterized by low pH, varying oxygen and iron levels, and the presence of additional microbes and host cells (Fig. 1) (O’Hanlon, Moench and Cone 2013; McClure et al.2015; Zozaya et al.2016). Free iron is scarce in the female host and is complexed to host iron-binding proteins including lactoferrin or transferrin, making the female genital tract iron deplete (Agarwal et al.2008). During menses, iron levels can rise and N. gonorrhoeae responds to these changing levels through regulation of gene expression (Anderson et al.2001; Jerse et al.2002; McClure et al.2015). During infection, the gonococcus encounters several different host cells such as epithelial cells and PMNs, the latter of which function to engulf and degrade the organism (Fig. 1). However, N. gonorrhoeae has been demonstrated to survive and replicate within epithelial cells and PMNs and to evade the antibacterial actions of PMNs (Criss and Seifert 2012). The gonococcus also encounters other microbes during genital tract infection in men and women (Weis and Nelson 2006; O’Hanlon, Moench and Cone 2013). Thus, the complexity of environmental signals during human mucosal infection is difficult to recapitulate by in vitro studies.
Figure 1.
Mucosal environment during gonococcal infection. During infection, the gonococcus encounters several different host cells such as epithelial cells and PMNs, the latter of which function to engulf and degrade the organism. Gonococcal infection in men typically results in a robust immune infiltration by PMNs, whereas in women asymptomatic infection is common and N. gonorrhoeae exists as a biofilm. In both men and women, the genitourinary tract mucosa is an iron-deplete environment with most bioavailable iron being bound host proteins such as transferrin. The gonococcus also encounters other microbes during genital tract infection in men and women, although the microbiota in men is typically less robust and diverse than that of females. Gonococci have also been demonstrated to transverse the epithelial barrier.
GONOCOCCAL GENE EXPRESSION DURING NATURAL MUCOSAL INFECTION IN HUMANS
We previously reported on the expression of a subset of iron-regulated genes during human mucosal infection in men and women. These studies revealed that the gene encoding the transcriptional regulatory protein Fur (fur) and the Fur-regulated tbpA/B and fbpA genes were expressed in mucosal samples obtained from men and women with uncomplicated gonorrhea (Agarwal et al.2005). However, these studies were carried out using microarray and qRT-PCR analysis and were limited in their sensitivity and detection (Agarwal et al.2005, 2008). To define gonococcal global gene responses during human mucosal infection, we recently utilized RNA-seq analysis to define the complete gonococcal transcriptome in cervico-vaginal lavage samples from naturally infected female subjects. These studies revealed that 65% of the gonococcal genome was expressed during natural mucosal infection of the lower genitourinary tract in women. We detected expression of 1700 gonococcal genes which represented 22 functional categories and included large groups of hypotheticals, rRNA, sRNA, phage-associated and translation-related genes all (Fig. 2). Within these categories, we observed high expression of genes encoding antimicrobial efflux pumps, iron transport, phage, pilin, outer membrane and hypothetical proteins (McClure et al.2015).
Figure 2.
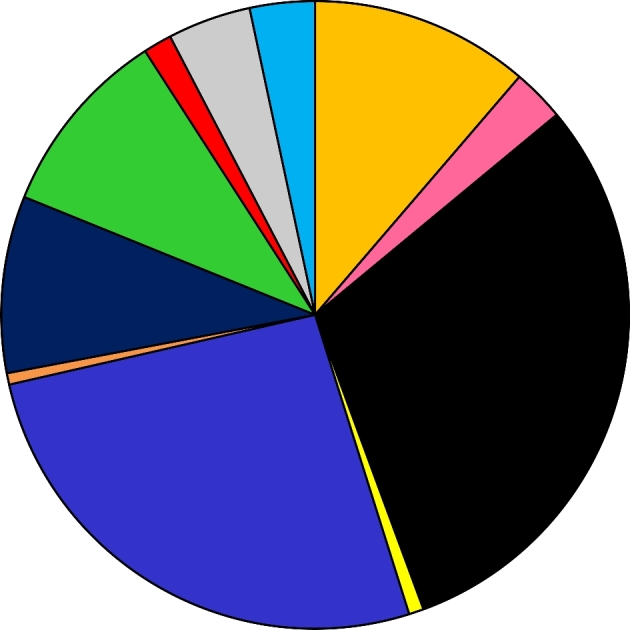
Gonococcal gene expression during mucosal infection in women. Approximately 1700 gonococcal genes are expressed during natural mucosal infection in women. Genes with RPKM values above 10 based on analysis with Rockhopper are grouped in 11 representative categories. The largest categories are hypothetical protein genes (517), indicated in black, and metabolism associated genes (447), which include general metabolism, energy, DNA and amino acid metabolism (blue). Expression-associated genes (192), including transcription, translation, rRNA synthesis and transcription factors, are indicated in goldenrod; sRNA genes (166) are in green; phage-associated genes (155), which include dsDNA, filamentous phages and transposase genes, are in dark blue; transport-associated genes (73) are in gray; tRNA genes (57) are in light blue. Host interaction-associated genes (46) include generally membrane-associated proteins such as adhesins, pilin biosynthesis and functions, lipoproteins, etc. and are indicated in pink; stress-associated genes (25) are in red; iron-associated genes (13) are in yellow and genes categorized as other (10) are in orange.
The strains isolated from cervico-vaginal lavage specimens were also grown in vitro and resulting transcriptomes compared to those expressed during infection to define infection-specific expression profiles. This analysis established that a large portion of the gonococcal genome was regulated during mucosal infection relative to in vitro growth. Genes involved in DNA/RNA processing, genetic regulation, sugar uptake, amino acid processing and phage-associated proteins all displayed large differences in expression among these two datasets. Furthermore, expression levels of a group of gonococcal hypothetical genes were observed to vary between in vivo and in vitro conditions, leading to speculations that new metabolic or regulatory pathways may be discovered, as well as bacterial factors involved in virulence, evasion of host defense mechanisms (i.e. antibiotic resistance) and novel proteins that could represent new therapeutic targets (McClure et al.2015). Our analysis also revealed increased expression of Fur and iron-regulated genes during infection in women as compared to growth in vitro, suggesting that during infection of the genital tract in women, the gonococcus is exposed to an iron-deplete environment.
TRANSCRIPTIONAL CONTROL BY FUR
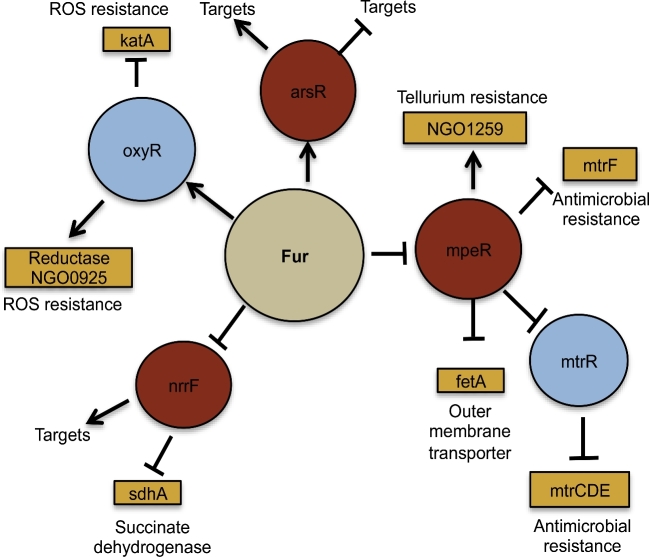
The gonococcal Fur protein controls expression of iron homeostasis genes in response to intracellular iron levels (Fig. 3). This ensures a crucial balance between the requirement for iron, as an essential element for growth, and the avoidance of iron toxicity, which occurs via production of hydroxyl or peroxide radicals (Cornelissen et al.1998; Touati 2000; Seib et al.2006; Bartnikas 2012; Troxell and Hassan 2013). Classically, Fur binds directly to DNA sequences to inhibit transcription of downstream genes (Mellin et al.2007; Carpenter, Whitmire and Merrell 2009; Yu and Genco 2012a,b). One of the best-known examples involves transcriptional control of genes that scavenge iron from the host, including the transferrin binding proteins (tbpAB) and the ferric binding protein (fbp) (Gray-Owen and Schryvers 1996; Cornelissen et al.1998; Agarwal et al.2005; McClure et al.2015). In the absence of iron, Fur exists as an inactive monomer that becomes active when intracellular levels of iron are high allowing Fur to bind to promoter regions bearing the Fur box (Fig. 3) (Bagg and Neilands 1987; Troxell and Hassan 2013). While this interaction typically acts to block subsequent binding by RNA polymerase, Fur can also function as a transcriptional activator (Yu and Genco 2012a). Fur can also regulate genes indirectly by repressing a series of trans elements that regulate downstream targets. For example, sRNAs can act as repressors as is the case for the Fur repressed sRNA NrrF, which controls transcription of the sdhC/A genes (Fig. 3). Fur-mediated repression of NrrF results in increased translation of sdhC/A transcripts; thus, expression of functional SdhC/A proteins (succinate dehydrogenases involved in the TCA cycle) is indirectly activated by Fur (Ducey et al.2005; Agarwal et al.2008; Jackson et al.2010). Fur can also control additional regulatory proteins including ArsR, MpeR and OxyR (Fig. 3). OxyR is known to regulate resistance of the gonococcus to ROS (Seib et al.2007). MpeR targets genes involved in antimicrobial resistance including MtrR (Lee et al.2003; Warner et al.2007; Jackson et al.2010; Mercante et al.2012; Yu et al.2016). ArsR is a regulator of the norB gene involved in nitrous oxide reduction, as well as predicted to regulate NGO1411 and NGO1646 (encoding a hypothetical and phage-associated gene respectively) and has been shown to be important for intracellular survival in endocervical cells (Isabella et al.2008; Yu et al.2016).
Figure 3.
Fur interaction with regulatory proteins and sRNAs. Fur has been shown or hypothesized to interact with other regulatory proteins and sRNAs. Interactions between Fur (tan circle) and other regulators (light blue circles) are shown. Downstream targets of these regulators are shown and defined as indirect targets of Fur. Rust colored circles indicate regulators identified in our studies. Expression under iron-replete conditions is depicted.
Fur regulation of regulatory and stress proteins in vitro
Superoxide and other oxide-containing effectors are stresses that Neisseria gonorrhoeae encounters within the human genital tract. In response to infection, epithelial cells and resident macrophages produce nitric oxide (NO) via AKT kinase activation and iNOS in vitro (Householder et al.2000; Seib et al.2006, 2007; Isabella et al.2008; Edwards 2010). To combat the effects of NO, N. gonorrhoeae expresses NorB, a NO reductase whose expression is repressed by the Fur-regulated ArsR regulatory protein (Isabella et al.2008). In iron-replete conditions in vitro, Fur functions as a transcriptional activator of arsR, a gene that is essential for survival within endocervical cells in vitro (Yu et al.2016).
Intrinsically, high levels of intracellular iron can react with the reduction and oxidation of NADH resulting in the formation of damaging reactive oxygen species (ROS) (Touati 2000; Seib et al., 2006, 2007). PMNs also express hydrogen peroxide (H2O2) and other oxidative species as antimicrobial compounds (Criss and Seifert 2012). The transcriptional regulator OxyR that is, in turn, regulated by active Fur regulates the ROS protection regulon; when Fur is active, it upregulates oxyR expression (Fig. 3). Thus, genes under the control of OxyR are also upregulated in iron-deplete environments (Seib et al.2006, 2007; Criss and Seifert 2012).
During growth under iron-deplete conditions, Fur is inactive and cannot repress the sRNA NrrF in vitro, which regulates target genes via post-transcriptional regulation (Mellin et al.2007; Jackson et al.2013). Thus, NrrF indirectly through Fur can regulate a series of genes, including the stress protein TdfF. TdfF is an essential gene for gonococcal intracellular survival in vitro in endocervical cells, and is downregulated in response to increased levels of NrrF (Jackson et al.2013). Fur-regulated TdfH is important for N. gonorrhoeae extracellular survival in the presence of PMNs. This is due to the zinc sequestration function exhibited by TdfH that can inactivate calprotectin in neutrophil extracellular traps (Jean et al.2016). In the female genitourinary tract mucosa, N. gonorrhoeae can form a biofilm, further advancing its transition toward anaerobic respiration (Steichen et al.2008, 2011; Phillips et al.2012). NrrF has also been shown to inactivate the sdh operon, which encodes an iron-containing enzyme involved in the TCA cycle aspect of aerobic respiration. In the anaerobic and iron-limiting conditions observed in vivo, downregulation of this operon is predicted to enhance N. gonorrhoeae survival (Jackson et al.2013; Jean et al.2016).
Epithelial cells and PMNs expose N. gonorrhoeae to host-derived antimicrobials that serve as primary immune defenses such as fatty acids, defensins and hydrogen peroxide (Quayle 2002; Lee et al.2003; Johnson et al.2015). Most charged and non-charged antimicrobial compounds are exported from gonococci via efflux pumps such as FarAB and MtrCDE (Rouquette, Harmon and Shafer 1999; Lee et al.2003). Expression of the genes encoding these efflux pumps is tightly controlled due to their important role in the development of resistance to antimicrobial and antibiotic stress. Fur plays an integral role in regulation of the antimicrobial resistance efflux pump system encoded by the mtr locus via upstream regulation of the MtrR transcriptional regulatory protein (Lee et al.2003; Yu et al.2016). During growth under iron-deplete conditions, such as those observed during human mucosal infection, Fur is inactive and as such expression of MpeR, the transcriptional regulator of the mtrR, is increased resulting in depression of mtrR expression (Fig. 3). The inhibition of mtrR expression results in expression of the mtrCDE operon that encodes the multidrug efflux pump proteins (Mercante et al.2012). Interestingly, our analysis of gonococcal gene expression profiles in cervico-vaginal lavage specimens revealed that both mtrR and mtrCDE expression levels were similar to the levels expressed during in vitro culture in the presence of iron (McClure et al.2015). These observations lead us to speculate that loss in regulation of these genes during growth in vivo could results from promoter allele changes altering the interaction between the repressor, MtrR, and the cis elements in the promoter. Indeed, allele changes in these operons have been reported to be involved in increases in antimicrobial resistance as well as intracellular survival in the gonococcus (Warner et al.2007; Kirkcaldy, Kidd and Weinstock 2013; Unemo and Shafer 2014; Grad et al.2016).
Fur control of gonococcal factors involved in host–pathogen interactions
During initial mucosal infection, the gonococcus interacts with epithelial cells via the Opa proteins and their cognate receptors, CEACAM 1, 3, 5 and 6 on the epithelial cell surface. This results in downstream signaling that favors subsequent gonococcal invasion of epithelial cells (Schmitter et al.2007; Sadarangani, Pollard and Gray-Owen 2011; Tchoupa, Schuhmacher and Hauck 2014; Sintsova et al.2015). Fur has been demonstrated to bind to and repress the gonococcal opa promoter during growth of the gonococcus under iron-replete conditions in vitro (Sebastian et al.2002). In concert with these observations, Opa-negative strains are predominantly recovered from infected female subjects during menses, when iron is abundant in the lower genitourinary tract (Sebastian et al.2002; Folster et al.2009; Jackson et al.2010; Sadarangani, Pollard and Gray-Owen 2011; Yu and Genco 2012b).
Intracellular survival of N. gonorrhoeae within endocervical cells in vitro as well as colonization within the female mouse model is partially mediated by the expression of a Fur-controlled phage repressor npr (Neisseria phage repressor) (Daou et al.2013). Despite the ability of Fur to bind the common promoter between npr and the four genes immediately downstream, we demonstrated that expression of npr is not regulated by iron or Fur in contradiction to previous studies (Ducey et al.2005; Jackson et al.2010; Daou et al.2013).
Opa proteins also mediate interaction of the gonococcus with PMNs (Johnson et al.2015). PMNs are the primary immune cells observed during N. gonorrhoeae infection in symptomatic males and females (albeit to a lesser extent in females compared to males) and the gonococcus has adapted mechanisms that enable the organism to survive within PMNs (Edwards and Apicella 2004). In women, N. gonorrhoeae infection is also associated with biofilm formation, protecting the organisms from immune cells such PMNs (Fig. 1). Evidence suggests that iron is more replete within a biofilm based on expression of iron-responsive gene products that are upregulated during growth in iron-deplete conditions, such as FetA and TbpA/B (Phillips et al.2012). Eventually, however, some bacteria escape the biofilm and enter into the iron-deplete luminal space where they make contact with PMNs (Steichen et al.2011; Phillips et al.2012). Survival after internalization of N. gonorrhoeae into PMNs in vitro depends on Opa expression. Expression of Opa proteins results in efficient clearance mediated by serine protease activity (Ball and Criss 2013; Johnson and Criss 2013; Johnson et al.2015).
OUTLOOK/CONCLUSIONS
Neisseria gonorrhoeae is one of the most common bacterial sexually transmitted infections worldwide (World Health Organization 2016). The incidence of infections has increased due in part to the continuing evolution of bacterial mechanisms of antimicrobial resistance and the asymptomatic nature of the disease which results in increased transmission. The lack of a good experimental model that can replicate the various microenvironmental conditions N. gonorrhoeae is exposed to within the human host has led to a significant gap in our understanding of its pathogenic strategies during natural mucosal infection. Transcriptome studies of gonococci during natural human mucosal infection reported by our group in combination with in vitro-based regulatory studies have shown that several regulatory circuits control N. gonorrhoeae genes important for colonization and survival. We demonstrated that the Fur transcriptional regulatory protein extends not only to iron-regulated genes, but also to genes involved in a number of other regulatory networks. All of these studies show that the general trend of activation or repression of genes by Fur observed in vitro is mimicked in vivo, further showing the importance of Fur as a central regulator during infection. A comparison of the gonococcal transcriptome expressed in vivo to the corresponding strain grown in vitro revealed increased expression of genes involved in iron transport including tbpAB and fbpABC in vivo (Agarwal et al.2005, 2008; McClure et al.2015). As TbpA and TbpB have been considered potential gonococcal vaccine candidates, defining the details of their regulation in vivo compared to in vitro, may be beneficial for future characterization of the therapeutic potential of these proteins. In addition, it is possible that other regulated factors, among both the known and hypothetical gene products revealed by our transcriptome studies, may represent novel targets for therapeutic and preventive strategies, i.e. new antimicrobial factors or vaccine candidates.
Current studies are focused on gonococcal transcriptional profiles expressed during infection in men. These studies have revealed a divergence in both the disease presentation in men and women, and gonococcal transcriptional programming. Initial results suggest that there is significant overlap in genes expressed by the gonococcus during mucosal infection in both men and women, including genes related to general metabolism and transport. Distinct gene sets expressed during infection in men were enriched for genes encoding host interaction and membrane-associated proteins. In contrast, distinct gene sets expressed during infection in women were enriched for iron and DNA metabolism genes (Nudel et al., unpublished). Further analysis of these unique data sets obtained from infected male and female subjects will define gonococcal adaptions during natural mucosal infection, a long-term goal of our studies. Taken together, these studies offer powerful new insights into the pathobiology of N. gonorrhoeae and will not only lead to a better understanding of the mechanisms of gene regulation employed by N. gonorrhoeae during infection, but ultimately will allow for the identification of novel virulence factors and consequently expand the potential for preventive and therapeutic strategies against N. gonorrhoeae infection.
Conflict of interest. None declared.
REFERENCES
- Agarwal S, King CA, Klein EK et al. The gonococcal Fur-regulated tbpA and tbpB genes are expressed during natural mucosal gonococcal infection. Infect Immun 2005;73:4281–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal S, Sebastian S, Szmigielski B et al. Expression of the gonococcal global regulatory protein Fur and genes encompassing the Fur and iron regulon during in vitro and in vivo infection in women. J Bacteriol 2008;190:3129–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JE, Leone PA, Miller WC et al. Selection for expression of the gonococcal hemoglobin receptor during menses. J Infect Dis 2001;184:1621–3. [DOI] [PubMed] [Google Scholar]
- Bagg A, Neilands JB. Ferric uptake regulation protein acts as a repressor, employing iron (II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 1987;26:5471–7. [DOI] [PubMed] [Google Scholar]
- Ball LM, Criss AK. Constitutively opa-expressing and opa-deficient neisseria gonorrhoeae strains differentially stimulate and survive exposure to human neutrophils. J Bacteriol 2013;195:2982–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartnikas TB. Known and potential roles of transferrin in iron biology. BioMetals 2012;25:677–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas GD, Anderson JE, Chen CJ et al. Identification and functional characterization of the Neisseria gonorrhoeae lbpB gene product. Infect Immun 1999;67:455–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter BM, Whitmire JM, Merrell DS. This is not your mother's repressor: the complex role of fur in pathogenesis. Infect Immun 2009;77:2590–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC. Addressing the Threat of Drug-Resistant Gonorrhea. Atlanta, GA: Centers for Disease Control and Prevention, 2016:2015–7. [Google Scholar]
- Chan PA, Robinette A, Montgomery M et al. Extragenital infections caused by Chlamydia trachomatis and Neisseria gonorrhoeae: a review of the literature. Infect Dis Obstet Gynecol 2016;2016, DOI: 10.1155/2016/5758387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelissen CN, Kelley M, Hobbs MM et al. The transferrin receptor expressed by gonococcal strain FA1090 is required for the experimental infection of human male volunteers. Mol Microbiol 1998;27:611–6. [DOI] [PubMed] [Google Scholar]
- Criss AK, Seifert HS. A bacterial siren song: intimate interactions between Neisseria and neutrophils. Nat Rev Microbiol 2012;10:178–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daou N, Yu C, Mcclure R et al. Neisseria prophage repressor implicated in gonococcal pathogenesis. Infect Immun 2013;81:3652–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducey TF, Carson MB, Orvis J et al. Identification of the iron-responsive genes of Neisseria gonorrhoeae by microarray analysis in defined medium. J Bacteriol 2005;187:4865–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JL. Neisseria gonorrhoeae survival during primary human cervical epithelial cell infection requires nitric oxide and is augmented by progesterone. Infect Immun 2010;78:1202–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JL, Apicella MA. The molecular mechanisms used by Neisseria gonorrhoeae to initiate infection differ between men and women. Clin Microbiol Rev 2004;17:965–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichorova RN, Desai PJ, Iii CG et al. Distinct proinflammatory host responses to Neisseria gonorrhoeae infection in immortalized human cervical and vaginal epithelial cells. Infect Immun 2001;69:5840–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folster JP, Johnson PJT, Jackson L et al. MtrR modulates rpoH expression and levels of antimicrobial resistance in Neisseria gonorrhoeae. J Bacteriol 2009;91:287–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goire N, Lahra MM, Chen M et al. Molecular approaches to enhance surveillance of gonococcal antimicrobial resistance. Nat Rev Microbiol 2014;12:223–9. [DOI] [PubMed] [Google Scholar]
- Gottlieb SL, Deal CD, Giersing B et al. The global roadmap for advancing development of vaccines against sexually transmitted infections: update and next steps. Vaccine 2016;34:2939–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grad YH, Harris SR, Kirkcaldy RD et al. Genomic epidemiology of gonococcal resistance to extended-spectrum cephalosporins, macrolides, and fluoroquinolones in the United States, 2000–2013. J Infect Dis 2016;214:1579–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray-Owen SD, Schryvers AB. Bacterial transferrin and lactoferrin receptors. Trends Microbiol 1996;4:185–91. [DOI] [PubMed] [Google Scholar]
- Grifantini R, Sebastian S, Frigimelica E et al. Identification of iron-activated and -repressed Fur-dependent genes by transcriptome analysis of Neisseria meningitidis group B. P Natl Acad Sci USA 2003;100:9542–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu A, Zhang Z, Zhang N et al. Generation of human CEACAM1 transgenic mice and binding of Neisseria Opa protein to their neutrophils. PLoS One 2010;5, DOI: 10.1371/journal.pone.0010067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Householder TC, Fozo EM, Jean A et al. Gonococcal nitric oxide reductase is encoded by a single gene, norB, which is required for anaerobic growth and is induced by nitric oxide. Appl Environ Microb 2000;68:5241–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabella V, Wright LF, Barth K et al. cis- and trans-acting elements involved in regulation of norB (norZ), the gene encoding nitric oxide reductase in Neisseria gonorrhoeae. Microbiology 2008;154:226–39. [DOI] [PubMed] [Google Scholar]
- Islam EA, Shaik-Dasthagirisaheb Y, Kaushic C et al. The reproductive cycle is a pathogenic determinant during gonococcal pelvic inflammatory disease in mice. Mucosal Immunol 2015;9:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson LA, Ducey TF, Day MW et al. Transcriptional and functional analysis of the Neisseria gonorrhoeae fur regulon. J Bacteriol 2010;192:77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson LA, Pan JC, Day MW et al. Control of RNA stability by NrrF, an iron-regulated small RNA in Neisseria gonorrhoeae. J Bacteriol 2013;195:5166–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean S, Juneau RA, Criss AK et al. Neisseria gonorrhoeae evades calprotectin-mediated nutritional immunity and survives neutrophil extracellular traps by production of TdfH. Infect Immun 2016;84:2982–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerse AE. Experimental gonococcal genital tract infection and opacity protein expression in estradiol-treated mice. Infect Immun 1999;67:5699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerse AE, Bash MC, Russell MW. Vaccines against gonorrhea: current status and future challenges. Vaccine 2015;33:395–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerse AE, Crow ET, Bordner AN et al. Growth of Neisseria gonorrhoeae in the female mouse genital tract does not require the gonococcal transferrin or hemoglobin receptors and may be enhanced by commensal lactobacilli. Infect Immun 2002;70:2549–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerse AE, Wu H, Packiam M et al. Estradiol-treated female mice as surrogate hosts for neisseria gonorrhoeae genital tract infections. Front Microbiol 2011;2:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MB, Ball LM, Daily KP et al. Opa+ Neisseria gonorrhoeae exhibits reduced survival in human neutrophils via Src family kinase-mediated bacterial trafficking into mature phagolysosomes. Cell Microbiol 2015;17:648–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MB, Criss AK. Neisseria gonorrhoeae phagosomes delay fusion with primary granules to enhance bacterial survival inside human neutrophils. Cell Microbiol 2013;15:1323–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkcaldy R, Kidd S, Weinstock H. Trends in antimicrobial resistance in Neisseria gonorrhoeae in the USA: the Gonococcal Isolate Surveillance Project (GISP), January 2006–June 2012. Sex Transm 2013;89;5–10. [DOI] [PubMed] [Google Scholar]
- Lee EH, Rouquette-Loughlin C, Folster JP et al. FarR regulates the farAB-encoded efflux pump of Neisseria gonorrhoeae via an MtrR regulatory mechanism. J Bacteriol 2003;185:7145–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure R, Nudel K, Massari P et al. The gonococcal transcriptome during infection of the lower genital tract in women. PLoS One 2015;10:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellin JR, Goswami S, Grogan S et al. A novel fur- and iron-regulated small RNA, NrrF, is required for indirect fur-mediated regulation of the sdhA and sdhC genes in Neisseria meningitidis. J Bacteriol 2007;189:3686–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercante AD, Jackson L, Johnson PJT et al. MpeR regulates the mtr efflux locus in Neisseria gonorrhoeae and modulates antimicrobial resistance by an iron-responsive mechanism. Antimicrob Agents Ch 2012;56:1491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman L, Rowley J, Hoorn SV et al. Global estimates of the prevalence and incidence of four curable sexually transmitted infections in 2012 based on systematic review and global reporting. PLoS One 2015;10:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hanlon DE, Moench TR, Cone RA. Vaginal pH and microbicidal lactic acid when lactobacilli dominate the microbiota. PLoS One 2013;8:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips NJ, Steichen CT, Schilling B et al. Proteomic analysis of Neisseria gonorrhoeae biofilms shows shift to anaerobic respiration and changes in nutrient transport and outermembrane proteins. PLoS One 2012;7, DOI: 10.1371/journal.pone.0038303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quayle AJ. The innate and early immune response to pathogen challenge in the female genital tract and the pivotal role of epithelial cells. J Reprod Immunol 2002;57:61–79. [DOI] [PubMed] [Google Scholar]
- Rouquette C, Harmon JB, Shafer WM. Induction of the mtrCDE-encoded efflux pump system of Neisseria gonorrhoeae requires MtrA, an AraC-like protein. Mol Microbiol 1999;33:651–8. [DOI] [PubMed] [Google Scholar]
- Sadarangani M, Pollard AJ, Gray-Owen SD. Opa proteins and CEACAMs: pathways of immune engagement for pathogenic Neisseria. FEMS Microbiol Rev 2011;35:498–514. [DOI] [PubMed] [Google Scholar]
- Schmitter T, Pils S, Weibel S et al. Opa proteins of pathogenic neisseriae initiate src kinase-dependent or lipid raft-mediated uptake via distinct human carcinoembryonic antigen-related cell adhesion molecule isoforms. Infect Immun 2007;75:4116–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian S, Agarwal S, Murphy JR et al. The gonococcal Fur regulon: identification of additional genes involved in major catabolic, recombination, and secretory pathways. J Bacteriol 2002;184:3965–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seib KL, Wu HJ, Kidd SP et al. Defenses against oxidative stress in Neisseria gonorrhoeae: a system tailored for a challenging environment. Microbiol Mol Biol R 2006;70:344–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seib KL, Wu HJ, Srikhanta YN et al. Characterization of the OxyR regulon of Neisseria gonorrhoeae. Mol Microbiol 2007;63:54–68. [DOI] [PubMed] [Google Scholar]
- Sintsova A, Wong H, MacDonald KS et al. Selection for a CEACAM receptor-specific binding phenotype during Neisseria gonorrhoeae infection of the human genital tract. Infect Immun 2015;83:1372–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steichen CT, Cho C, Shao JQ et al. The Neisseria gonorrhoeae biofilm matrix contains DNA, and an endogenous nuclease controls its incorporation. Infect Immun 2011;79:1504–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steichen CT, Shao JQ, Ketterer MR et al. Gonococcal cervicitis: a role for biofilm in pathogenesis. J Infect Dis 2008;198:1856–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchoupa AK, Schuhmacher T, Hauck CR. Signaling by epithelial members of the CEACAM family - mucosal docking sites for pathogenic bacteria. Cell Commun Signal 2014;12:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touati D. Iron and oxidative stress in bacteria. Arch Biochem Biophys 2000;373:1–6. [DOI] [PubMed] [Google Scholar]
- Troxell B, Hassan HM. Transcriptional regulation by ferric uptake regulator (Fur) in pathogenic bacteria. Front Cell Infect Microbiol 2013;3:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unemo M, Golparian D, Nicholas R et al. High-level cefixime- and ceftriaxone-resistant Neisseria gonorrhoeae in France: novel penA mosaic allele in a successful international clone causes treatment failure. Antimicrob Agents Ch 2012;56:1273–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unemo M, Shafer WM. Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: past, evolution, and future. Clin Microbiol Rev 2014;27:587–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CK, Sweet RL. Gonorrhea infection in women: prevalence, effects, screening, and management. Int J Womens Health 2011;3:197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner DM, Folster JP, Shafer WM et al. Regulation of the MtrC‐MtrD‐MtrE efflux‐pump system modulates the in vivo fitness of Neisseria gonorrhoeae. J Infect Dis 2007;196:1804–12. [DOI] [PubMed] [Google Scholar]
- Weis WI, Nelson WJ. Re-solving the cadherin-catenin-actin conundrum. J Biol Chem 2006;281:35593–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Global Incidence and Prevalence of Selected Curable Sexually Transmitted Infections-2008. 2012:1–28. [Google Scholar]
- World Health Organization. WHO Guidelines for the Treatment of Neisseria gonorrhoeae. 2016. [PubMed] [Google Scholar]
- Yu C, Genco CA. Fur-mediated global regulatory circuits in pathogenic Neisseria species. J Bacteriol 2012a;194:6372–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Genco CA. Fur-mediated activation of gene transcription in the human pathogen Neisseria gonorrhoeae. J Bacteriol 2012b;194:1730–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, McClure R, Nudel K et al. Characterization of the Neisseria gonorrhoeae iron and Fur regulatory network. J Bacteriol 2016;198:JB.00166–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zozaya M, Ferris MJ, Siren JD et al. Bacterial communities in penile skin, male urethra, and vaginas of heterosexual couples with and without bacterial vaginosis. Microbiome 2016;4:16. [DOI] [PMC free article] [PubMed] [Google Scholar]