Abstract
Background
2,3-butanediol (2,3-BD) is a bulk platform chemical with various potential applications such as aviation fuel. 2,3-BD has three optical isomers: (2R, 3R)-, (2S, 3S)- and meso-2,3-BD. Optically pure 2,3-BD is a crucial precursor for the chiral synthesis and it can also be used as anti-freeze agent due to its low freezing point. 2,3-BD has been produced in both native and non-native hosts. Several pathogenic bacteria were reported to produce 2,3-BD in mixture of its optical isomers including Klebsiella pneumoniae and Klebsiella oxytoca. Engineered hosts based on episomal plasmid expression such as Escherichia coli, Saccharomyces cerevisiae and Bacillus subtilis are not ideal for industrial fermentation due to plasmid instability.
Results
Pichia pastoris is generally regarded as safe and a well-established host for high-level heterologous protein production. To produce pure (2R, 3R)-2,3-BD enantiomer, we developed a P. pastoris strain by introducing a synthetic pathway. The alsS and alsD genes from B. subtilis were codon-optimized and synthesized. The BDH1 gene from S. cerevisiae was cloned. These three pathway genes were integrated into the genome of P. pastoris and expressed under the control of GAP promoter. Production of (2R, 3R)-2,3-BD was achieved using glucose as feedstock. The optical purity of (2R, 3R)-2,3-BD was more than 99%. The titer of (2R, 3R)-2,3-BD reached 12 g/L with 40 g/L glucose as carbon source in shake flask fermentation. The fermentation conditions including pH, agitation speeds and aeration rates were optimized in batch cultivations. The highest titer of (2R, 3R)-2,3-BD achieved in fed-batch fermentation using YPD media was 45 g/L. The titer of 2,3-BD was enhanced to 74.5 g/L through statistical medium optimization.
Conclusions
The potential of engineering P. pastoris into a microbial cell factory for biofuel production was evaluated in this work using (2R, 3R)-2,3-BD as an example. Engineered P. pastoris could be a promising workhorse for the production of optically pure (2R, 3R)-2,3-BD.
Electronic supplementary material
The online version of this article (10.1186/s13068-018-1031-1) contains supplementary material, which is available to authorized users.
Keywords: Pichia pastoris; 2,3-Butanediol; Metabolic engineering; Medium optimization
Background
Driven by depleting fossil fuels and growing environmental concern, production of bulk chemicals from renewable sources via biosynthesis is becoming increasingly desirable [1]. 2,3-Butanediol (2,3-BD) is a crucial platform chemical with enormous applications. It can be converted to other platform chemicals such as methyl ethyl ketone, gamma-butyrolactone and 1,3-butadiene [2]. 2,3-BD has three stereoisomers: (2R, 3R)-, (2S, 3S)- and meso-2,3-BD [3]. Enantiopure isomers of 2,3-BD could serve as the precursor for asymmetric synthesis and synthetic rubber. It is also used as an anti-freeze agent due to its low freezing point. 2,3-BD is also a good alternative liquid fuel owing to its high energy capacity. Biosynthesis of 2,3-BD has been reported in many microorganisms. Native producers belonging to Enterobacteriaceae family such as Klebsiella pneumoniae, Klebsiella oxytoca and Enterobacter aerogenes were able to produce high titer of 2,3-BD in mixture of its optical isomers [1]. Despite the high titer achieved in those hosts, chiral purity of 2,3-BD was not satisfactory. Moreover, the use of risk group 2 microorganisms projects a safety concern for industrial fermentation. Production of 2,3-BD via fermentation of GRAS (generally regarded as safe) bacteria has also been reported. Several Bacillus species including B. subtilis [4], B. amyloliquefaciens [5] and B. licheniformis [6] are effective producers of 2,3-BD. Jurchescu and coworkers reported 144.7 g/L 2,3-BD production using B. licheniformis DSM 8785, a risk group 1 bacterium [7]. Paenibacillus polymyxa ZJ9 was reported to produce 36.92 g/L (2R, 3R)-2,3-BD with 98% purity from Jerusalem artichoke tubers under optimized conditions [8]. Engineered hosts based on plasmid expression of heterologous pathway genes from the aforementioned microorganisms were reported. The use of non-native hosts such as Escherichia coli [9, 10] and Saccharomyces cerevisiae [11–13] offers several advantages due to their well-established genetics, higher metabolic rate, simpler process control and lack of competing pathways. Lian and coworkers reported the production of 100 g/L 2,3-BD in S. cerevisiae using glucose and galactose as carbon source [14].
The methylotrophic yeast Pichia pastoris has been recognized as an excellent host for heterologous protein production. Its success as a workhorse for recombinant protein production was attributed to the ability to perform post-translational modifications, high secretion efficiency of proteins and simple nutrition requirements [15]. It is also ideal for industrial fermentation due to the GRAS status. P. pastoris can be grown to high cell density on defined medium using crude glycerol, a by-product of biodiesel production, as carbon source [16]. P. pastoris has been engineered into microbial cell factory for the production of biofuel in recent years. P. pastoris was used as whole cell catalysts to produce biodiesel through surface display of lipases [17, 18]. Other bio-products produced by engineered P. pastoris included lycopene [19], β-carotene [20], xanthophylls [21], (+)-nootkatone [22], dammarenediol-II [23] and lactic acid [24]. In this work, we aim to produce chiral pure (2R, 3R)-2,3-BD in P. pastoris using metabolic and process engineering strategies. The potential of P. pastoris as a promising host for 2,3-BD production was demonstrated for the first time.
Methods
Strains, plasmids and reagents
Escherichia coli XL1-Blue was used for plasmid cloning and propagation. P. pastoris X33 was used as host for metabolic engineering. E. coli was grown in low-salt LB broth (1% peptone, 0.5% yeast extract and 0.5% sodium chloride) supplemented with 25 µg/mL of Zeocin where appropriate. P. pastoris was cultivated in YPD media (1% yeast extract, 2% peptone and 2% dextrose) with 100 µg/mL of Zeocin. YPDS plates consisting of 1% yeast extract, 2% peptone, 2% dextrose, 18% sorbitol and 2% agar were used for yeast transformant screening. Plasmid pGAPZαA (Invitrogen, USA) was used for gene cloning. Chemicals of analytical grade and restriction enzymes used in this work were purchased from Sigma-Aldrich (USA) or Fisher scientific (Canada). DNA primers were ordered from Invitrogen (USA) and nucleotide sequences of primers are listed in Table 1. E. coli or yeast genomic DNA was purified using Gentra Puregene Yeast/Bac. Kit (Qiagen, Germany). Polymerase chain reaction (PCR) was performed using KAPA HiFi Hotstart Readymix PCR Kit (Kapa Biosystems, USA).
Table 1.
Primers used in this study
| Primer name | Sequence (5′–3′) |
|---|---|
| AlsS-f | CGGGGTACCATGTTGACTAAGGCTACTAAGGAACAAA |
| AlsS-r | TCCCCGCGGTTACAAAGCTTTAGTCTTCATCAAC |
| AlsD-f | CGGAATTCAAAATGAAGAGAGAGTCCAACATCCAAG |
| AlsD-r | CGGGGTACCTTATTCTGGTGATCCCTCGGTTGTT |
| BDH1-f | CGGAATTCAAAATGAGAGCTTTGGCATATTTCAAGA |
| BDH1-r | CGGGGTACCTTACTTCATTTCACCGTGATTGTTA |
| udhA-f | CGGGGTACCAAAATGCCACATTCCTACGATTACGATG |
| udhA-r | TGCTCTAGATTAAAACAGGCGGTTTAAACCGTTT |
| HIS4-f | GAAGATCTATGACATTTCCCTTGCTACCTGC |
| HIS4-r | CGGGATCCTTAAATAAGTCCCAGTTTCTCCATACG |
Restriction sites are underlined
DNA manipulation
To clone the 2,3-BD biosynthesis genes under the control of GAP promoter, pGAPZαA was digested with Bsp119I and EcoRI to remove the α-signal sequence. The large fragment was gel-purified and blunted by DNA blunting enzyme (CloneJET PCR cloning kit, Thermo scientific, USA). The treated fragment was self-ligated to result in a plasmid pGAPZ which is used for gene expression in this work. DNA sequences of biosynthesis pathway genes B. subtilis 168 alsS and alsD were retrieved from National Center for Biotechnology Information (NCBI). Sequences of alsS and alsD were codon-optimized according to the P. pastoris codon usage to achieve better expression. The optimized genes of alsS and alsD were synthesized by Genscript (USA) and inserted into vector pUC57, resulting in pUC57-AlsS and pUC57-AlsD, respectively. alsS gene was amplified from pUC57-AlsS using AlsS-f and AlsS-r. The PCR product was digested with KpnI and SacII and ligated into pGAPZ digested with the same enzymes to obtain pGAPZ-AlsS. alsD was cloned into pGAPZ to result in pGAPZ-AlsD with the same manner using EcoRI and KpnI. 2,3-Butanediol dehydrogenase gene BDH1 was amplified from S. cerevisiae genomic DNA using BDH1-f and BDH1-r and cloned into pGAPZ between EcoRI and KpnI to obtain pGAPZ-BDH1. pGAPZ-AlsD was digested with BamHI and BglII to obtain a fragment containing PGAP-AlsD-tAOX1. pGAPZ-AlsS was linearized with BamHI and ligated with PGAP-AlsD-tAOX1 to result in plasmid pGAPZ-SD. BDH1 was cloned into pGAPZ-SD in the same manner to result in pGAPZ-SDB. To integrate the pathway genes into HIS4 locus of the P. pastoris genome, HIS4 gene was PCR amplified from P. pastoris X33 genomic DNA using primers HIS4-f and HIS4-r. HIS4 was cloned into pGAPZ-SD and pGAPZ-SDB to result in pGAPZ-SDH and pGAPZ-SDBH, respectively. E. coli udhA gene was amplified from E. coli XL1-blue genomic DNA using primers udhA-f and udhA-r. udhA was first cloned into pGAPZ to result in pGAPZ-udhA and then inserted into pGAPZ-SD to obtain pGAPZ-SDU. HIS4 was inserted into pGAPZ-SDU, resulting in pGAPZ-SDUH. All plasmid constructs were confirmed by PCR and sequencing.
Yeast transformation and screening
Pichia pastoris X33 was made competent and transformed with various plasmids (Table 2) using an electroporator (Eppendorf, Canada) according to the manufacturer’s instructions. Briefly, 5 µg of pGAPZ-SDH, pGAPZ-SDBH, pGAPZ-SDUH and pGAPZ-SDBUH were linearized with NheI and transformed into competent P. pastoris X33 cells to obtain strains X33-SD, X33-SDB and X33-SDU, respectively. Yeast transformants were screened on YPDS plates by incubating at 30 °C for 2–3 days. Yeast colonies were picked up and grown in YPD media. Genomic DNA was extracted and used as PCR templates for the confirmation of pathway gene integration.
Table 2.
Plasmids and strains used in this study
| Strains or plasmids | Description | Source |
|---|---|---|
| E. coli XL1-blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F´ proAB lacIq Z∆M15 Tn10 (Tetr)] | Lab stock |
| P. pastoris strains | ||
| X33 | Wilde type | Lab stock |
| X33-SD | P. pastoris X33 harbouring codon-optimized AlsS and AlsD gene | This study |
| X33-SDB | P. pastoris X33 harbouring codon-optimized AlsS and AlsD and S. cerevisiae BDH1 gene | This study |
| X33-SDU | P. pastoris X33 harbouring codon-optimized AlsS and AlsD and E. coli udhA gene | This study |
| Plasmids | ||
| pGAPZαA | GAP promoter, α-signal, Zeocin resistance | Invitrogen |
| pGAPZ | GAP promoter, Zeocin resistance | This study |
| pGAPZ-AlsS | pGAPZ harbouring AlsS gene | This study |
| pGAPZ-AlsD | pGAPZ harbouring AlsD gene | This study |
| pGAPZ-BDH1 | pGAPZ harbouring BDH1 gene | This study |
| pGAPZ-udhA | pGAPZ harbouring udhA gene | This study |
| pGAPZ-SD | pGAPZ harbouring AlsS and AlsD genes | This study |
| pGAPZ-SDB | pGAPZ harbouring AlsS, AlsD and BDH1 genes | This study |
| pGAPZ-SDU | pGAPZ harbouring AlsS, AlsD and udhA genes | This study |
| pGAPZ-SDH | pGAPZ harbouring AlsS, AlsD and HIS4 genes | This study |
| pGAPZ-SDBH | pGAPZ harbouring AlsS, AlsD, BDH1 and HIS4 genes | This study |
| pGAPZ-SDUH | pGAPZ harbouring AlsS, AlsD, udhA and HIS4 genes | This study |
Shake flask cultivation
Single yeast colony was inoculated into 50-mL Falcon tube containing 10 mL YPD media and grown overnight. Shake flask cultivation was performed by inoculating 1 mL of overnight culture into 100 mL YP media (1% yeast extract, 2% peptone) containing various concentrations of glucose in a 500-mL shake flask. Aliquots were taken every 4–6 h. Samples were centrifuged at 13,000 rpm for 5 min. Supernatant was filtered through 0.22-µm filter and used for further analysis.
Bioreactor setup
Batch and fed-batch fermentations were performed in 5-L bioreactors (Bioflo320, Eppendorf, Canada) equipped with two Rushton impellers. Overnight yeast culture (5% v/v) was inoculated into the fermentation media. Bioreactors were kept at 30 °C. The cultivation was performed at the optimal condition with pH 5.0 (± 0.02), agitation speed of 300 rpm and aeration rate of 0.5 vvm unless otherwise specified. The pH was maintained constant by adding 30% ammonia hydroxide through on-line pH monitoring. Feeding solution was fed to the bioreactor once the glucose was depleted. Glucose feed rate was adjusted between 0.2 and 0.8 mL/min to maintain a low concentration of glucose to avoid substrate inhibition.
Effect of cultivation conditions on titer of 2,3-BD in YPD
Batch fermentation of P. pastoris strain was performed in 5-L bioreactors (Bioflo320, Eppendorf, Canada) containing 3 L YPD media. Bioreactor inoculum was prepared by streaking a single colony from the plate and growing in a 500-mL shake flask containing 150 mL YPD media overnight (30 °C and 250 rpm). Effect of fermentation conditions was investigated by varying one parameter (pH, agitation speed and aeration rate) at a time. Other cultivation conditions were kept unchanged when one parameter was varied.
Statistical medium optimization
Basal salt medium (BSM) consisting of per liter: 42.9 g KH2PO4, 14.33 g K2SO4, 5.17 g (NH4)2SO4, 5.7 g MgSO4·7H2O, 0.6 g CaSO4·2H2O and 4 mL PTM1 was used for medium optimization. Trace metal solution PTM1 consists of per liter: 6 g CuSO4·5H2O, 0.08 g NaI, 3 g MnSO4·H2O, 0.2 g Na2MoO4·2H2O, 0.02 g HBO3, 0.5 g CoCl2, 20 g ZnCl2, 65 g FeSO4·7H2O, 0.2 g biotin and 5 mL H2SO4. Minitab 15 was used to generate Plackett–Burman and Box–Behenken design matrix and perform statistical analysis. Shake flask cultivation was performed to screen the significant factors affecting the titer of 2,3-BD. The responses (2,3-BD titer) were used to fit a first-order model. Box–Behnken design was employed to determine the optimal level of the three significant factors. The 2,3-BD titers obtained in shake flask cultivation were used to fit a quadratic model. Fed-batch cultivation using the optimized medium recipe was performed at 30 °C, pH 5.0, 300 rpm and 0.5 vvm.
Analytical methods
Biomass was monitored by measuring optical density at 600 nm (OD600) with Ultraspec 60 (UK). Glucose concentration was measured using YSI2900 Bio-analyzer (Mandel, Canada) installed with a glucose membrane. 2,3-BD isomers were differentiated using gas chromatography (GC, Agilent 6850 series, Santa Clara, CA, USA) equipped with flame ionized detector (FID). For GC-FID analysis, yeast cultures were centrifuged at 13,000 rpm for 5 min. The supernatant was extracted with equal volume of ethyl acetate. Organic phase was dehydrated with sodium sulphate before GC analysis. The capillary GC column (Supelco Astec CHIRALDEX™ B-PM, 35 m × 0.25 mm × 0.12 µm) was used in this study. Helium was used as the carrier gas with a flow rate of 2 mL/min. The injector and detector were kept at 250 °C. Injection volume was 1 µL. Oven temperature program [25] was as follows: 1.5 min at 50 °C, programmed to increase up to 160 °C at a rate of 8.8 °C/min and hold for 5 min at 160 °C. Extracellular metabolites were determined by high-performance liquid chromatography (HPLC, Agilent 1200 series, Santa Clara, CA, USA) equipped with a Shodex Sugar SH1011 column (8 mm ID × 300 mm, 6 µm; Showa Denko, Tokyo, Japan). The column was maintained at 60 °C and eluted with 5 mM sulphuric acid at a flow rate of 0.6 mL/min. Acetoin was detected with variable wavelength detector at 214 nm. Other metabolites were detected with refractive index detector.
Statistical analysis
Student test was used to compare mean values using Origin 8.0. For medium optimization, statistical analysis was carried out using Minitab 15. Difference with confidence level of 95% (P < 0.05) was considered statistically significant.
Results
Construction of P. pastoris strains for the production of (2R, 3R)-2,3-BD
Pichia pastoris is an industrially relevant host for recombinant protein production and has not been reported to produce 2,3-BD from glucose up to date. Previous study described the conversion of acetoin to 2,3-BD using P. pastoris through NADH (nicotinamide adenine dinucleotide) regeneration [26]. 2,3-BD is produced from pyruvate via three enzymatic steps (Fig. 1). To introduce a synthetic route for 2,3-BD production in P. pastoris, B. subtilis α-acetolactate synthase AlsS, B. subtilis α-acetolactate decarboxylase AlsD and S. cerevisiae (2R, 3R)-2,3-BD dehydrogenase BDH1 were selected to direct the carbon flux towards 2,3-BD synthesis. Constitutive promoter PGAP is commonly used for protein expression in P. pastoris due to its high-level constitutive expression and growth-associated product formation [27]. The three pathway genes were cloned under the control of PGAP for constitutive expression. Codon optimization of alsS and alsD was performed to achieve better expression in P. pastoris [28]. The pathway genes were assembled into a single plasmid for chromosome integration at the HIS4 locus of Pichia genome. As shown in Fig. 2, alsS, alsD, udhA and BDH1 were cloned into vector pGAPZ and successfully integrated into P. pastoris genome via homologous recombination. The constructed strains were grown in YPD media to verify 2,3-BD production. The optical purity of 2,3-BD is determined by the stereo-specificity of 2,3-BD dehydrogenase (Fig. 1). As shown in Fig. 3, recombinant P. pastoris strains X33-SD, X33-SDB and X33-SDU were able to produce enantiopure (2R, 3R)-2,3-BD using glucose as feedstock. Optical purity was determined to be over 99%. Trace amount of meso-2,3-BD was produced. (2S, 3S)-2,3-BD was not detected, indicating the stereo-specificity of endogenous 2,3-butanediol dehydrogenase (2,3-BDH) is specific for (2R, 3R)-2,3-BD synthesis.
Fig. 1.
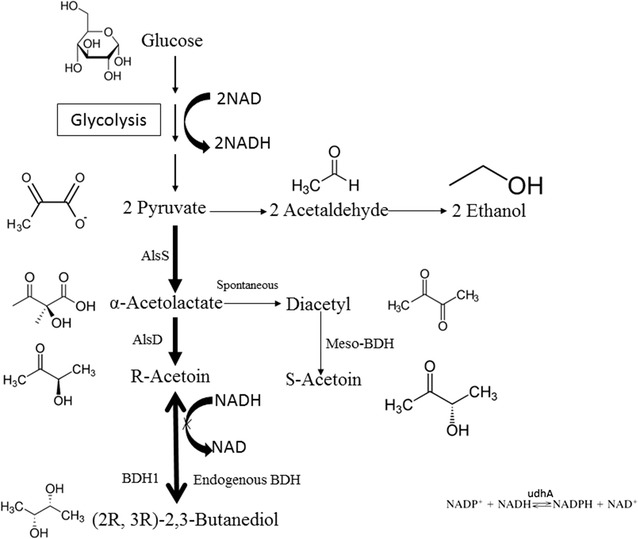
Engineered 2,3-BD synthesis pathway in this work. The 2,3-BD synthesis pathway was indicated by bold arrows
Fig. 2.
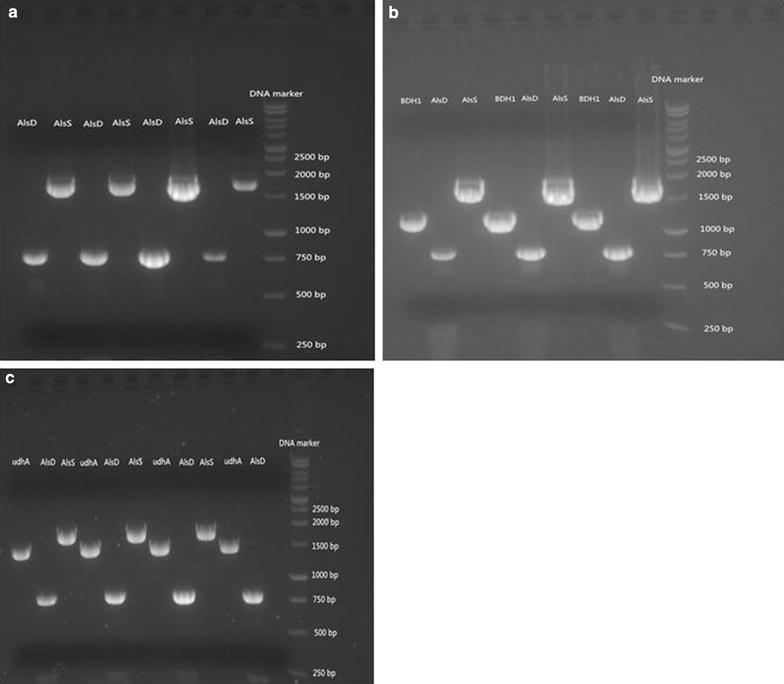
PCR confirmation of constructed strains. a PCR verification of X33-SD. Genomic DNA of four yeast colonies was extracted and used as templates for PCR. b PCR verification of X33-SDB. Genomic DNA of three yeast colonies was extracted and used as templates for PCR. c PCR verification of X33-SDU. Genomic DNA of four yeast colonies was extracted and used as templates for PCR. DNA bands and DNA markers are indicated in the pictures
Fig. 3.
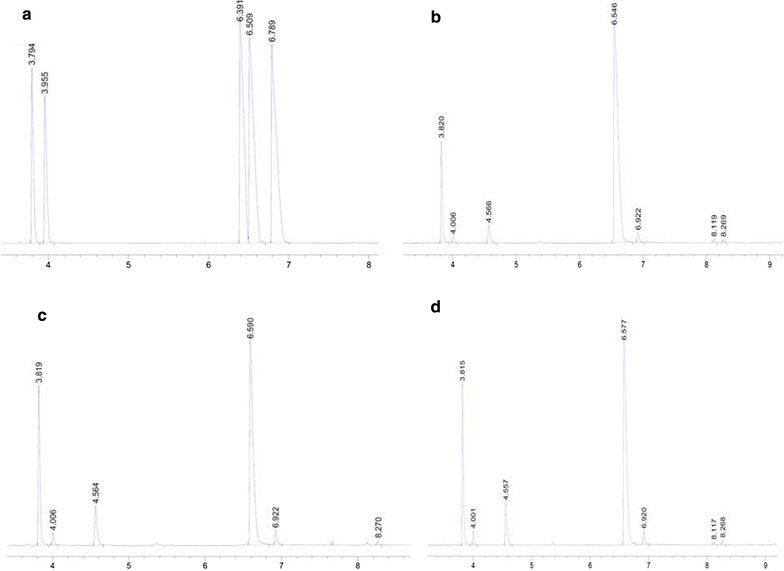
GC profiles of metabolites in different strains. a GC profile of standards. Retention times: 3S-acetoin, 3.8 min; 3R-actoin, 4.0 min; (2S, 3S)-2,3-BD, 6.4 min; (2R, 3R)-2,3-BD, 6.5 min; meso-2,3-BD, 6.8 min. b GC profile of extract of culture of X33-SD. c GC profile of extract of culture of X33-SDB. d GC profile of extract of culture of X33-SDU
Screening of different strains for high 2,3-BD production
Three strains constructed in this work were cultivated to evaluate their potential for 2,3-BD production in shake flasks containing 40 g/L glucose (Fig. 4). Yeast cultures were grown for 36 h and aliquots were analysed. As shown in Fig. 4, glucose was depleted at 24 h. Major by-products detected in the broth were glycerol, acetoin and ethanol. No acetic acid was detected in all three strains. Cell growth was not significantly affected by the 2,3-BD synthetic pathway (Table 3). Strain X33-SD, X33-SDB and X33-SDU produced 2,3-BD with a titer of 12.2 ± 0.84, 8.04 ± 0.95 and 7.44 ± 0.19 g/L, respectively. Surprisingly, overexpression of S. cerevisiae BDH1 was not beneficial to boost 2,3-BD production. This result is contradictory with previous study [26]. The E. coli udhA gene for NADH regeneration from NADPH (nicotinamide adenine dinucleotide phosphate) did not improve 2,3-BD titer as well. The highest acetoin titer was observed in strain X33-SD, reaching 1.84 ± 0.76 g/L. Acetoin consists mainly of S-acetoin with minor R-acetoin detected based on the GC analysis (Fig. 3). Despite P. pastoris is generally recognized as Crabtree effect-negative, ethanol is produced as a by-product under anaerobic condition. Wild-type P. pastoris X33 produced 15 g/L ethanol after 36 h cultivation (data not shown). The highest ethanol titer of X33-SD, X33-SDB and X33-SDU was detected at 24 h, with 2.28 ± 0.36 g/L, 6.43 ± 0.07 g/L and 4.96 ± 0.07 g/L, respectively. Strain X33-SD produced 0.21 ± 0.19 g/L ethanol after 36 h, in contrast with 4.3 ± 0.52 g/L and 1.99 ± 0.01 g/L obtained with X33-SDB and X33-SDU, respectively. Ethanol titer decreased at the end of cultivation because it could be consumed after glucose depletion. Compared with wild-type X33, the heterologous 2,3-BD synthetic pathway effectively redirected the carbon flux from ethanol formation towards 2,3-BD synthesis.
Fig. 4.
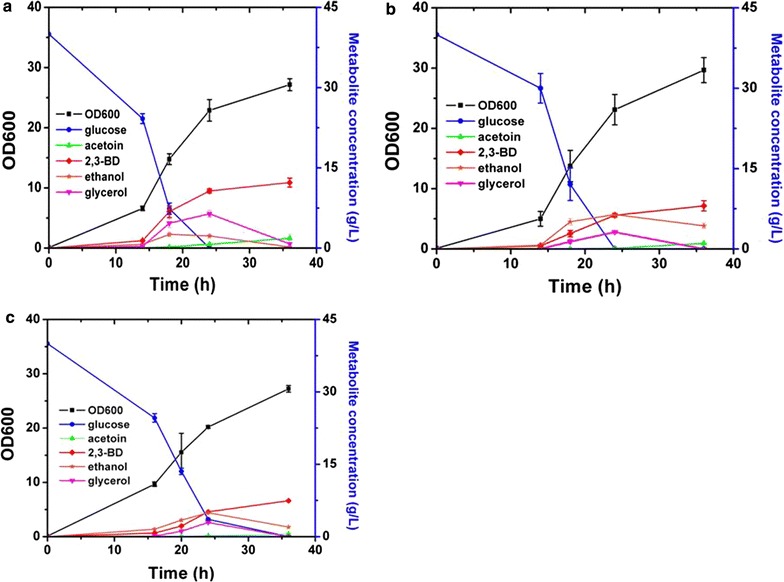
Screen of recombinant strains in shake flasks. a Time course of X33-SD in 500-mL shake flask containing 100 mL YP media with 40 g/L glucose. b Time course of X33-SDB in 500-mL shake flask containing 100 mL YP media with 40 g/L glucose. c Time course of X33-SDU in 500-mL shake flask containing 100 mL YP media with 40 g/L glucose. Cultivation last for 36 h. Error bars indicate the standard deviation of three replicate experiments
Table 3.
Comparison of metabolite production of three P. pastoris strains
| Strains | X33-SD | X33-SDB | X33-SDU |
|---|---|---|---|
| OD600 | 27.15 ± 1α | 29.68 ± 2.08α | 27.23 ± 0.62α |
| 2,3-BD (g/L) | 12.2 ± 0.84α | 8.04 ± 0.95β | 7.44 ± 0.19β |
| Acetoin (g/L) | 1.84 ± 0.76α | 1.03 ± 0.09α | 0.54 ± 0.07β |
| Glycerol (g/L) | 6.42 ± 0.62α | 3.14 ± 0.28β | 2.94 ± 0.23β |
| Ethanol (g/L) | 2.28 ± 0.36α | 6.43 ± 0.08β | 4.96 ± 0.07γ |
Cultivation of three strains was carried out in 500-mL shake flasks containing 100 mL YP media with 40 g/L glucose. Cultivation last for 36 h. Data indicates average values of triplicate experiments with standard deviation. Values with different Greek symbols between columns were significantly different (P < 0.05). ND indicates not detected
Glycerol was produced as a major by-product as a channel for the regeneration of NAD+ in engineered S. cerevisiae for the production of 2,3-BD [29]. Notably, 6.42 ± 0.62, 3.14 ± 0.28 and 2.94 ± 0.23 g/L glycerol was accumulated in strain X33-SD, X33-SDB and X33-SDU, respectively. Production of glycerol in P. pastoris has never been reported in the literature. Analysis of the P. pastoris genome sequence could offer a possible mechanism for glycerol formation. Two putative glycerol-3-phosphate dehydrogenases (GPDs) were present in P. pastoris genome. Putative GPD (PAS_ch2_2_0111) shares 57% amino acid similarity with GPD2 in S. cerevisiae. Further investigation is imperative to verify the enzyme activity of the putative GPDs in P. pastoris. Strain X33-SD is the most promising strain in terms of highest 2,3-BD production and least ethanol accumulation and is thus used for further study (Table 3).
Effect of glucose concentration
Yeast strain X33-SD was cultivated in shake flasks with YPD medium containing different initial concentrations of glucose. As shown in Fig. 5, higher titer of 2,3-BD was obtained at higher glucose concentration, reaching 5.64 ± 0.16, 9.22 ± 0.12 and 12.24 ± 0.15 g/L with glucose concentration of 20, 30 and 40 g/L, respectively. Higher concentrations of ethanol and glycerol were also observed with higher glucose concentrations. Acetoin was detected below 1 g/L for all three glucose concentrations. The 2,3-BD yield on glucose remained about 0.3 g/g for the three glucose concentrations tested, which is 60% of the theoretical yield (0.5 g/g).
Fig. 5.
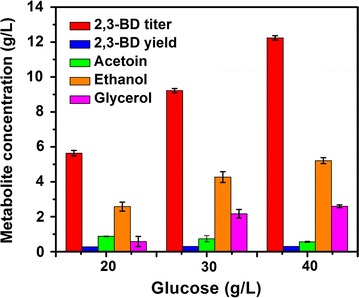
Effect of glucose concentration. Strain X33-SD was cultured in 500-mL shake flask containing 100 mL YP media containing 20, 30 and 40 g/L glucose. Error bars indicate standard deviation of three replicate experiments
Effect of fermentation conditions on 2,3-BD production
Effect of agitation
Bioreactor cultivations of strain X33-SD were performed to investigate its potential for large-scale production of (2R, 3R)-2,3-BD. Fermentation conditions such as agitation speeds, aeration rates and pH were optimized under controlled conditions. Effect of agitation is illustrated in Fig. 6a. Three agitation speeds were examined: 300, 400 and 500 rpm with an aeration rate of 1.5 vvm. It was found that metabolite profile was closely related to agitation speed. Production of 2,3-BD was detected under 300 and 400 rpm but not detected under 500 rpm. The highest titer of 2,3-BD was achieved at 300 rpm, reaching 8.34 ± 0.62 g/L. Acetoin was the major product at 500 rpm with a titer of 9.27 ± 0.81 g/L. Glycerol and ethanol were accumulated only at 300 rpm. The highest biomass concentration was obtained at 500 rpm with an OD600 of 42. 35 ± 2.61, compared with 23.65 ± 1.12 and 34.8 ± 1.89 at 300 rpm and 400 rpm, respectively. It was reported that lower dissolved oxygen (DO) level favours the reduction of acetoin to 2,3-BD [1]. DO is normally controlled by cascading agitation and aeration where agitation is more prominent to DO control. Optimization of agitation speed for improved production of 2,3-BD has been reported in previous studies. Xu and coworkers optimized the agitation speed within the range of 200 to 500 rpm using engineered E. coli and found that 400 rpm was optimal in terms of high biomass accumulation and 2,3-BD production [30]. Two-stage agitation control strategy has been implemented to cultivate K. oxytoca to achieve a balance between biomass production and 2,3-BD synthesis. Agitation was maintained at 300 rpm in the first 15 h for biomass growth and lowered to 200 rpm for the production of 2,3-BD [31]. Two-stage agitation improved the titer by 6.2% compared with constant speed of 200 rpm. The agitation-associated metabolite profile suggests that strain X33-SD could be used to produce acetoin under higher agitation speed.
Fig. 6.
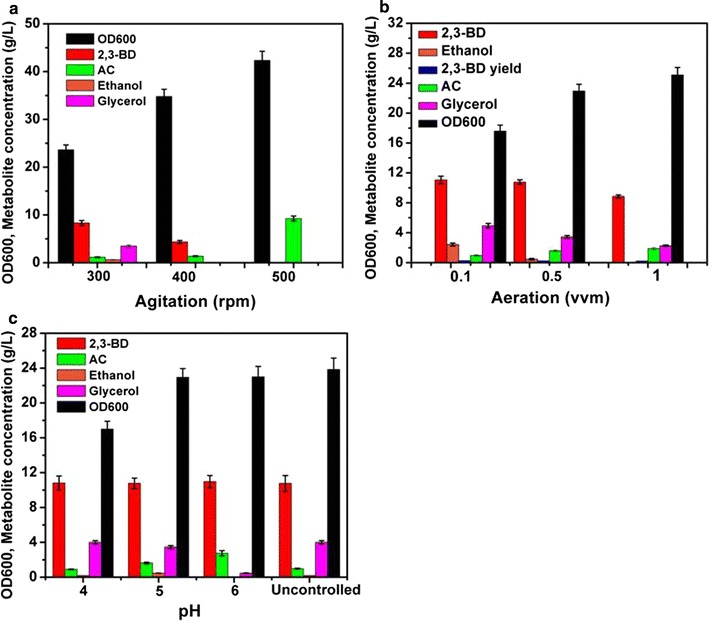
Effect of cultivation conditions on 2,3-BD titer. Strain X33-SD was cultivated in 3 L YPD media in 5-L Bioflo320 reactors. Glucose concentration was 40 g/L. Error bars indicate standard deviation of duplicate experiments. a Effect of agitation. b Effect of aeration. c Effect of pH
Effect of aeration
Aeration is another major factor to determine DO level. We tested three aeration rates: 0.1, 0.5 and 1 vvm with agitation speed of 300 rpm. As was shown in Fig. 6b, higher aeration resulted in higher biomass production. OD600 reached 25.1 ± 1.2 at 1 vvm compared with 17.6 ± 0.9 at 0.1 vvm. The titer of 2,3-BD obtained at 0.1 vvm was slightly higher than that at 0.5 vvm, reaching 11.07 ± 0.81 and 10.78 ± 0.65 g/L, respectively. Only 8.87 ± 0.43 g/L 2,3-BD was detected at 1 vvm. Acetoin was produced at 0.97 ± 0.03, 1.6 ± 0.07 and 1.89 ± 0.15 g/L at 0.1, 0.5 and 1 vvm, respectively. The highest glycerol concentration of 4.95 ± 0.56 g/L was observed at 0.1 vvm. Ethanol was not detected at 1 vvm whilst 0.46 and 2.43 ± 0.23 g/L were produced at 0.5 vvm and 1 vvm, respectively. Aeration rate of 0.5 vvm was used further study for high production of biomass and 2,3-BD.
Effect of pH
The acidity of fermentation is critical to the cell growth and metabolite production. In the case of 2,3-BD production, it was reported that the formation of neutral 2,3-BD could be a countermeasure against acidification [1]. It was found that the production of 2,3-BD was triggered by pH drop in K. pneumoniae G31 [32]. Forced pH fluctuation was implemented to increase the titer of 2,3-BD to 70 g/L in comparison with 52.5 g/L without pH control. In this work, we examined the effect of pH through cultivation ranging from pH 4 to pH 6 and without pH control. As illustrated in Fig. 6c, the production of 2,3-BD in X33-SD was not affected by the pH, reaching a final titer of about 10.8 g/L. The highest acetoin titer was observed at pH 6. The lowest glycerol concentration was detected at pH 6, reaching 0.46 g/L. At pH 4, cell growth was significantly retarded (data not shown). When pH was not controlled, pH value dropped from initial pH 6.8 to pH 5.2 at the end of the batch. In previous studies, pH was reported to have an impact on the activity of acetoin reductase. Through a two-stage pH control strategy, cell growth of K. oxytoca was favoured in the first stage where pH was uncontrolled and maintained at a set pH for 2,3-BD production [33]. The results obtained in this study indicate that P. pastoris could be a robust host for the production of 2,3-BD within a wide range of pH. We selected pH 5 for fed-batch cultivation, as optimal cell growth was achieved under this pH value.
Results of effect of cultivation conditions are summarized in Table 4. Comparison of results was performed using statistical analysis. The following conditions were selected for fed-batch cultivation: pH 5, 300 rpm and 0.5 vvm.
Table 4.
Effect of cultivation conditions
| Agitation (rpm) | Aeration (vvm) | pH | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 300 | 400 | 500 | 0.1 | 0.5 | 1.0 | 4 | 5 | 6 | Uncontrolled | |
| OD600 | 23.65 ± 1.5α | 34.8 ± 1.9β | 42.35 ± 2.61γ | 17.6 ± 0.8δ | 22.95 ± 0.85α | 25.1 ± 1α | 17 ± 0.9δ | 22.95 ± 1α | 23 ± 1.2α | 23.85 ± 1.3α |
| 2,3-BD (g/L) | 8.34 ± 0.62α | 4.36 ± 32β | ND | 11.07 ± 0.5γ | 10.78 ± 0.3γ | 8.87 ± 0.2α | 10.82 ± 0.2γ | 10.78 ± 0.6γ | 10.97 ± 0.7γ | 10.77 ± 0.9γ |
| Acetoin (g/L) | 1.17 ± 0.02α | 1.37 ± 0.02β | 9.27 ± 0.81γ | 0.97 ± 0.08δ | 1.6 ± 0.12ε | 1.89 ± 0.15ε | 0.91 ± 0.04α | 1.63 ± 0.1γ | 2.75 ± 0.3ζ | 0.99 ± 0.05α |
| Glycerol (g/L) | 3.49 ± 0.1α | ND | ND | 4.95 ± 0.3β | 3.45 ± 0.2α | 2.26 ± 0.2γ | 4 ± 0.2δ | 3.46 ± 0.18α | 0.46 ± 0.03ε | 4 ± 0.3δ |
| Ethanol (g/L) | 0.66 ± 0.01α | ND | ND | 2.43 ± 0.2β | 0.46 ± 0.1α | ND | 0.2 ± 0.01γ | 0.47 ± 0.05α | 0.01 ± 0δ | 0.2 ± 0.01γ |
Data indicate average values of duplicate experiments with standard deviation. Values with different Greek symbols between columns were significantly different (P < 0.05). ND indicates not detected
Fed-batch cultivation in YPD
To scale up the production of 2,3-BD using strain X33-SD, fed-batch cultivation was performed. Batch cultivation was started in 2 L YPD media with initial glucose concentration of 40 g/L. The time course of fed-batch fermentation is shown in Fig. 7. Glucose was depleted at 19 h and feeding solution consisting of 500 g/L glucose was fed to the bioreactor. Fermentation lasted for 135 h with a final volume of approximately 2.5 L. Biomass and the titer of 2,3-BD steadily increased. The final OD600 reached 42.5 at the end of fed-batch. The production of 2,3-BD stagnated after 100 h and a final titer of 45.8 g/L was achieved. Ethanol was detected below 1 g/L. Final acetoin concentration was found to be 15.9 g/L. Glycerol production was 17.5 g/L at 66 h and remained unchanged at the end of cultivation. The yield of 2,3-BD on glucose of the whole fed-batch cultivation was 0.197 g/g, which is 39.4% of the theoretical yield. The productivity achieved in fed-batch fermentation was 0.34 g/L/h. Compared with shake flask cultivation, 2,3-BD yield obtained in fed-batch cultivation was relatively low. This could be due to the inhibitory effect of much higher concentration of glycerol and 2,3-BD in fed-batch cultivation. Moreover, YPD might not be a suitable medium for long-time fermentation. Medium optimization could be performed to boost the titer and yield of 2,3-BD.
Fig. 7.
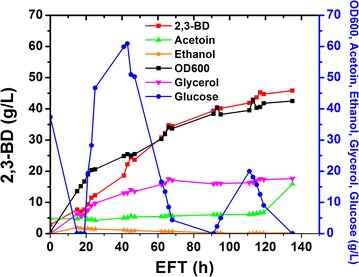
Time course of fed-batch cultivation. Fermentation was performed in 5-L Bioflo320 bioreactor with 2 L initial YPD media. 1 L of 500 g/L glucose was fed to bioreactor after glucose depletion
Statistical medium optimization
Effect of yeast extract
BSM is a defined medium commonly used for high cell density cultivation of P. pastoris. BSM consists of mineral salts without complex nitrogen source. To develop an appropriate medium for 2,3-BD production, we first investigated the effect of complex nitrogen source on the titer of 2,3-BD. Media containing 40 g/L glucose were used for the production of 2,3-BD. It was shown that only 0.5 ± 0.05 g/L 2,3-BD was detected in BSM, whilst 4.5 ± 0.7 g/L was obtained in BSM supplemented with 10 g/L yeast extract (Additional file 1: Figure S1). It has been previously reported that the addition of complex nitrogen source such as yeast extract and corn steep liquor was beneficial to boost the titer of 2,3-BD [33, 34]. Therefore, it is reasonable to factor in yeast extract for optimization of BSM in this work.
Plackett–Burman and Box–Behnken design
Plackett–Burman design was used to screen significant factors affecting the production of 2,3-BD. Twelve-run shake flask cultivation was carried out based on the matrix (Additional file 4: Tables S1–S2) generated by Minitab 15. The responses showed good fit to the first-order model (Additional file 4: Equation S1). The F value for the fitted equation is 6.93 with P = 0.04, indicating the good fitness of the model. Statistical analysis (Additional file 4: Table S3 and Additional file 2: Figure S2) revealed that significance of medium components impacting the titer of 2,3-BD in the following order: KH2PO4 > yeast extract > MgSO4 > PTM1 > K2SO4 > (NH4)2SO4 > CaSO4. Apparently, KH2PO4, yeast extract and MgSO4 were the key variables and were chosen to determine their optimal levels using Box–Behnken design (Additional file 4: Tables S4–S7). The responses showed good fit to the quadratic model. It was found that the highest titer of 2,3-BD was achieved when the concentration of yeast extract, KH2PO4 and MgSO4 was 15, 21.5 and 2.85 g/L, respectively.
Fed-batch cultivation using optimized BSMY media
The optimized BSMY media consist of 15 g/L yeast extract, 21.5 g/L KH2PO4 and 2.85 g/L MgSO4. Other medium components were kept the same as the standard BSM. Three fed-batch cultivations were performed to evaluate the performance of the optimized medium. Upon the depletion of initial glucose in the batch within about 18 h, feeding solution containing 250 or 500 g/L glucose was fed to the bioreactor. Glucose feed rate was adjusted to maintain a low glucose concentration. In fed-batch cultivation with 40 g/L initial glucose concentration and 1 L of 250 g/L glucose feeding solution, the fermentation was completed within 49 h (Additional file 3: Figure S3). The final titer of 2,3-BD was 41 g/L with a yield on glucose of 0.31 g/g. To boost the titer, higher concentrations of initial glucose and feeding glucose were studied. As can be seen from Figs. 8 and 9, fed-batch cultivation was completed within 90 h, which was significantly shortened in comparison with YPD (135 h). The highest titer of 2,3-BD was achieved when 60 g/L of initial glucose and 1 L of 500 g/L glucose feeding solution were used, reaching 74.5 g/L. In contrast, the highest titer obtained in YPD was 45 g/L in fed-batch fermentation with a much longer cultivation time. Comparison of the fed-batch cultivation results are summarized in Additional file 4: Table S8. Compared with YPD media, optimized BSMY could support faster glucose consumption without accumulating excessive glucose in the culture. Glycerol was the major by-product, reaching about 35 g/L in the final culture. Ethanol and acetoin were detected at low titer.
Fig. 8.
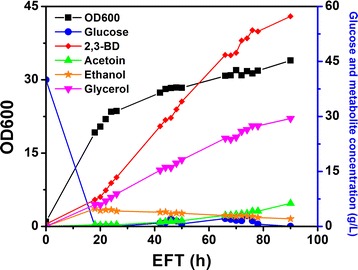
Time course of fed-batch fermentation using optimized BSMY medium. Bioreactor contained 2 L of initial media. 40 g/L glucose was used as initial substrate and 1 L of 500 g/L glucose solution was used as feeding solution in fed-batch
Fig. 9.
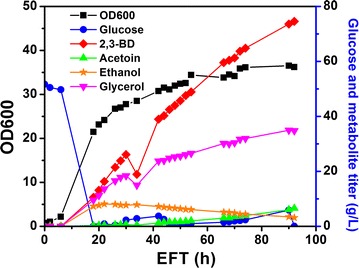
Time course of fed-batch fermentation using optimized BSMY medium. Bioreactor contained 2 L of initial media. 60 g/L glucose was used as initial substrate and 1 L of 500 g/L glucose solution was used as feeding solution in fed-batch
Discussion
Biological production of bulk chemicals has garnered increasing attention in recent years. Production of optically pure 2,3-BD has been extensively investigated due to the wide applications of this platform chemical. Selection of a suitable host for metabolic engineering is crucial to the economic feasibility of the process. Engineering of native producers such as K. pneumoniae [47], S. marcescens [48] and B. licheniformis [49] requires the deletion of 2,3-BDH. An alternative strategy to produce pure optical isomer of 2,3-BD was to engineer heterologous hosts such as E. coli [10], S. cerevisiae [14] and L. lactis [25]. To boost the titer of 2,3-BD, metabolic engineering and process engineering strategies could be employed. Metabolic engineering strategies include overexpression of the 2,3-BD synthesis pathway genes, inactivation of the competing pathway genes and co-factor engineering. Overexpression of budA and budB in a K. pneumoniae strain resulted in 60% increase of 2,3-BD compared with parental strain [50]. Minimization of by-products was necessary to increase the yield of 2,3-BD. Lactic acid was formed as by-product in K. pneumoniae, K. oxytoca [51] and E. cloacae [37], and deletion of ldhA (encoding lactate dehydrogenase) enhanced the production of 2,3-BD in those strains.
Co-factor imbalance is a major bottleneck for the production of 2,3-BD from glucose. Two moles of NADH were generated during the glycolysis of 1 mol of glucose. Only 1 mol of NADH was consumed to reduce acetoin to 2,3-BD, causing a surplus of NADH. NAD+ can be recycled through NADH consumption processes such as the formation of lactic acid, ethanol and glycerol. As was mentioned previously, glycerol was produced as the major by-product in engineered P. pastoris strains, similar to that observed in S. cerevisiae [11, 12]. In S. cerevisiae, NADH reoxidation could be achieved through several mechanisms such as alcoholic fermentation, glycerol formation and respiration [52]. Under anaerobic conditions where respiration cannot occur, glycerol formation was employed by P. pastoris as a major redox sink to recycle NAD+, as P. pastoris is a Crabtree effect-negative yeast and produces much less ethanol than S. cerevisiae. Similar glycerol formation mechanism may also exist in engineered Z. mobilis [46]. More glycerol was produced when more 2,3-BD was formed. In fed-batch cultivation, 35 g/L glycerol was observed with 74.5 g/L 2,3-BD (Fig. 9). In contrast, no glycerol was detected when acetoin was the major product and no 2,3-BD was formed at agitation speed of 500 rpm (Fig. 6). In S. cerevisiae, glycerol is formed through the two GPDs. Disruption of the two GPDs in S. cerevisiae almost abolished glycerol formation and enhanced 2,3-BD production [29]. Similar strategy could be implemented in P. pastoris to minimize glycerol synthesis and improve 2,3-BD titer. Co-factor engineering is an efficient strategy to balance the NAD+/NADH ratio. Expression of NADH oxidase gene L. lactis noxE in engineered S. cerevisiae strains lacking pyruvate decarboxylase resulted in increased 2,3-BD yield and productivity [29, 35, 53]. Overexpression of E. coli udhA gene (encoding a transhydrogenase) also led to enhanced production of 2,3-BD in engineered B. subtilis [39]. However, expression of E. coli udhA was found not beneficial to boost the titer of 2,3-BD in our study.
Cultivation conditions such as pH and DO should be optimized to achieve optimal production of 2,3-BD. Cultivation pH can affect the production of 2,3-BD as well as cell growth and thus should be investigated. It was found that metabolite profile was highly associated with the cultivation pH in Klebsiella sp. Zmd30 and optimal existed for high production of 2,3-BD [54]. Biomass accumulation and 2,3-BD production may favour different pH. Two-stage pH control scheme by controlling pH at 7.5 for cell growth and then reduced to 6.5 for 2,3-BD synthesis has been successfully implemented in E. cloacae [55]. In this work, variation of pH between 4 and 6 did not show a significant difference of 2,3-BD titer, albeit cell growth was retarded at pH 4. DO is critical for achieving high titer of 2,3-BD. Complicated DO control schemes such as volumetric oxygen transfer coefficient KLa [34] and respiratory quotient [56] have been reported. Since DO is governed by agitation speed and aeration rate, it would be easier to control DO through optimizing those two parameters. Chan et al. [57] implemented a “one variable a time” strategy to optimize pH, agitation speed, aeration rate and maltodextrin concentration for the production of 2,3-BD in K. oxytoca. Three-stage agitation speed increased the titer of 2,3-BD by 9.8% in B. amyloliquefaciens [58]. In our work, product profile was related to agitation speed and it could be manipulated to achieve production of acetoin or 2,3-BD.
YPD is a commonly used rich medium for yeast cultivation. However, it is not viable to use such expensive medium for industrial-scale production due to the costly yeast extract and peptone. Moreover, glucose accumulation was observed during long-time fermentation (Fig. 9). The slow utilization of glucose was also observed in the fermentation of engineered S. cerevisiae strains using YPD where cultivation time was over 300 h [12, 14]. Statistical medium optimization is an efficient approach to develop a suitable medium for enhanced production of 2,3-BD. It has been successfully used to develop industrial medium for other 2,3-BD producers [59, 60]. Despite minimal medium should be employed for industrial fermentation to reduce cost, the addition of complex nitrogen source such as yeast extract, casamino acids and corn steep liquor has been reported to increase the titer and yield of 2,3-BD in K. oxytoca [33], P. polymyxa [34], Raoultella ornithinolytica B6 [61] and B. subtilis [62]. Cultivation of P. pastoris strains in minimal salt medium led to very low titer of 2,3-BD. Therefore, yeast extract was used for the optimization of BSM to avoid compromising the titer of 2,3-BD. The three significant variables were identified through statistical analysis. KH2PO4 was found to have a negative impact on the titer of 2,3-BD and should be reduced. This is consistent with a previous report optimizing BSM for enhanced phytase production in P. pastoris [16]. The optimized medium showed more robust performance than YPD in terms of higher yield, shorter fermentation time, faster glucose consumption and higher 2,3-BD titer.
Yeast extract was still used in the optimized medium due to its role in enhancing 2,3-BD titer. Feasibility of using other inexpensive nitrogen source such as corn steep liquor [63, 64] to partially substitute yeast extract could be explored with similar medium optimization strategy described in this work.
A comparison of the production of 2,3-BD using various strains is summarized in Table 5. Efficient producers of 2,3-BD such as K. pneumoniae and K. oxytoca are predominantly native hosts. The highest titer of 2,3-BD in bacteria was reported in K. pneumoniae SDM, reaching 150 g/L with a productivity of 4.21 g/L/h [43]. The highest production yield of 2,3-BD on glucose was 0.49 g/g achieved in K. oxytoca [42], close to the theoretical yield of 0.5 g/g. The yield also reached 0.487 g/g in an engineered B. subtilis [39]. The best yield in a heterologous host was reported in an engineered E. coli, reaching 0.49 g/g in shake flask cultivation with a titer of 54 g/L [65]. Engineered S. cerevisiae has achieved success in the production of 2,3-BD with the best reported titer being 154 g/L [35]. However, the yield obtained in S. cerevisiae was relatively low, mostly between 0.2 and 0.4 g/g. In this work, engineered P. pastoris strain was able to achieve 0.3 g/g production yield, comparable to that of S. cerevisiae. Further metabolic engineering should be performed to increase the yield and productivity thereby enhancing the economic viability.
Table 5.
Comparison of 2,3-BD production in various microorganisms
| Species | Titer (g/L) | Operation strategy | Enantiopurity (%) | Yield (g/g) | Feed stock | 2,3-BD productivity (g/L/h) | References |
|---|---|---|---|---|---|---|---|
| S. cerevisiae | 43.6 | Fed-batch | 97 | 0.27 | Xylose | 0.15 | [12] |
| S. cerevisiae | 96.2 | Fed-batch | NA | 0.28 | Glucose | 0.39 | [11] |
| S. cerevisiae | 154.3 | Fed-batch | NA | 0.404 | Glucose | 1.97 | [35] |
| S. cerevisiae | 100 | Fed-batch | 98 | 0.35 | Glucose and galactose | 0.33 | [14] |
| E. coli | 6.9 | Shake flask | 99 | 0.31 | Glucose | 0.14 | [36] |
| E. coli | 73.8 | Fed-batch | NA | 0.41 | Glucose | 1.19 | [30] |
| E. coli | 115 | Fed-batch | 99 | 0.42 | Glucose | 1.44 | [9] |
| E. aerogenes | 152 | Fed-batch | 97.5 | 0.489 | Glucose and xylose | 3.5 | [37] |
| E. aerogenes | 140 | Fed-batch | NA | NA | Sugarcane molasses | 2.59 | [38] |
| B. subtilis | 49.29 | Shake flask | 99 | 0.47 | Glucose | 0.224 | [4] |
| B. subtilis | 130.7 | Fed-batch | 99 | 0.487 | Glucose | 0.459 | [39] |
| B. licheniformis | 123.7 | Fed-batch | 99 | NA | Glucose | 2.95 | [6] |
| B. amyloliquefaciens | 132.9 | Fed-batch | NA | 0.45 | Glucose | 2.95 | [40] |
| K. oxytoca | 131.5 | Fed-batch | NA | 0.44 | Crude glycerol | 0.84 | [41] |
| K. oxytoca | 117.4 | Fed-batch | NA | 0.49 | Glucose | 1.2 | [42] |
| K. pneumoniae | 150 | Fed-batch | NA | NA | Glucose | 4.21 | [43] |
| S. marcescens | 152 | Fed-batch | NA | 0.463 | Sucrose | 2.67 | [44] |
| P. polymyxa | 36.92 | Batch | 98 | Raw inulin extract | 0.88 | [8] | |
| C. glutamicum | 6.3 | Batch | NA | 0.33 | Glucose | 0.2 | [45] |
| Z. mobilis | 15 | Batch | NA | 0.16 | Glucose, xylose | 0.3 | [46] |
| P. pastoris | 75 | Fed-batch | 99 | 0.3 | Glucose | 0.81 | This study |
Construction of engineered hosts generally requires antibiotic resistance as selection marker. The use of antibiotic-resistant microorganisms may not be ideal for industrial applications due to possible environmental impact. This concern could be addressed using nutritional auxotrophy as selection marker such as histidine-deficiency (his4) in P. pastoris. The antibiotic resistance gene could also be removed through proper molecular techniques such as flippase-mediated homologous recombination [66].
Compared with other engineered heterologous systems depending on plasmid-based expression of 2,3-BD synthesis genes, gene expression in P. pastoris was based on genome-targeting cassette and eliminated the need of episomal plasmids. Combined with the high growth rate, high alcohol tolerance and GRAS status, engineered P. pastoris could become a robust host for the production of 2,3-BD.
Conclusions
Pichia pastoris was engineered into a microbial cell factory to produce bulk chemical (2R, 3R)-2,3-BD using glucose as feedstock for the first time. The endogenous 2,3-BDH could support efficient conversion of acetoin to 2,3-BD. Statistical medium optimization was a useful tool to boost the titer of 2,3-BD. The highest titer of (2R, 3R)-2,3-BD reached 74.5 g/L in a fed-batch cultivation using optimized medium. P. pastoris was proved a versatile platform for biofuel production other than heterologous protein production. The engineered P. pastoris strain could be a good starting point for further metabolic and process engineering to achieve a novel host for cost-effective production of (2R, 3R)-2,3-BD.
Additional files
Additional file 1: Figure S1. Effect of yeast extract on 2,3-BD titer. 100 mL media containing 40 g/L glucose were used to cultivate strain X33-SD. Error bar indicate standard deviation of three replicate experiments.
Additional file 2: Figure S2. Pareto graph of the seven variables.
Additional file 3: Figure S3. Time course of fed-batch fermentation using optimized BSMY medium. Bioreactor contained 2 L of initial media. 40 g/L glucose as initial substrate and 1 L of 250 g/L glucose solution as feeding solution.
Additional file 4. Supplementing tables in this work. Table S1. Level code for variables based on Plackett-Burman design. Table S2. Cultivation results of Plackett-Burman design. Table S3. Analysis results of Plackett-Burman design. Table S4. Design of Box-Behnken experiments. Table S5. Concentration level for Box-Behnken design. Table S6. Cultivation results of Box-Behnke design. Table S7. Analysis results of Box-Behnken design. Table S8. Comparison of fed-batch cultivations.
Authors’ contributions
ZY designed the experiments. ZY performed the experiments and data analysis. ZY wrote the manuscript. ZZ coordinated the manuscript draft and revision. ZZ supervised the work. Both authors read and approved the final manuscript.
Acknowledgements
The authors are grateful to the financial support from the Natural Sciences and Engineering Research Council (NSERC) of Canada. Zhiliang Yang is the recipient of a doctoral scholarship from the China Scholarship Council for the duration of this work.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Not applicable.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This work was supported by a Discovery grant from the National Sciences and Engineering Council (NSERC) of Canada.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13068-018-1031-1) contains supplementary material, which is available to authorized users.
Contributor Information
Zhiliang Yang, Email: zyang079@uottawa.ca.
Zisheng Zhang, Phone: +1 613 562 5800 (6110), Email: zzhang@uottawa.ca.
References
- 1.Ji XJ, Huang H, Ouyang PK. Microbial 2,3-butanediol production: a state-of-the-art review. Biotechnol Adv. 2011;3:351–364. doi: 10.1016/j.biotechadv.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 2.Bialkowska AM. Strategies for efficient and economical 2,3-butanediol production: new trends in this field. World J Microbiol Biotechnol. 2016;12:200. doi: 10.1007/s11274-016-2161-x. [DOI] [PubMed] [Google Scholar]
- 3.Celinska E, Grajek W. Biotechnological production of 2,3-butanediol—current state and prospects. Biotechnol Adv. 2009;6:715–725. doi: 10.1016/j.biotechadv.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Fu J, Wang Z, Chen T, Liu W, Shi T, Wang G, Tang YJ, Zhao X. NADH plays the vital role for chiral pure D-(−)-2,3-butanediol production in Bacillus subtilis under limited oxygen conditions. Biotechnol Bioeng. 2014;10:2126–2131. doi: 10.1002/bit.25265. [DOI] [PubMed] [Google Scholar]
- 5.Yang T, Rao Z, Zhang X, Lin Q, Xia H, Xu Z, Yang S. Production of 2,3-butanediol from glucose by GRAS microorganism Bacillus amyloliquefaciens. J Basic Microbiol. 2011;6:650–658. doi: 10.1002/jobm.201100033. [DOI] [PubMed] [Google Scholar]
- 6.Ge Y, Li K, Li L, Gao C, Zhang L, Ma C, Xu P. Contracted but effective: production of enantiopure 2,3-butanediol by thermophilic and GRAS Bacillus licheniformis. Green Chem. 2016;17:4693–4703. doi: 10.1039/C6GC01023G. [DOI] [Google Scholar]
- 7.Jurchescu IM, Hamann J, Zhou X, Ortmann T, Kuenz A, Prusse U, Lang S. Enhanced 2,3-butanediol production in fed-batch cultures of free and immobilized Bacillus licheniformis DSM 8785. Appl Microbiol Biotechnol. 2013;15:6715–6723. doi: 10.1007/s00253-013-4981-z. [DOI] [PubMed] [Google Scholar]
- 8.Gao J, Xu H, Li QJ, Feng XH, Li S. Optimization of medium for one-step fermentation of inulin extract from Jerusalem artichoke tubers using Paenibacillus polymyxa ZJ-9 to produce R, R-2,3-butanediol. Bioresour Technol. 2010;18:7076–7082. doi: 10.1016/j.biortech.2010.03.143. [DOI] [PubMed] [Google Scholar]
- 9.Ji XJ, Liu LG, Shen MQ, Nie ZK, Tong YJ, Huang H. Constructing a synthetic metabolic pathway in Escherichia coli to produce the enantiomerically pure (R, R)-2,3-butanediol. Biotechnol Bioeng. 2015;5:1056–1059. doi: 10.1002/bit.25512. [DOI] [PubMed] [Google Scholar]
- 10.Tong YJ, Ji XJ, Shen MQ, Liu LG, Nie ZK, Huang H. Constructing a synthetic constitutive metabolic pathway in Escherichia coli for (R, R)-2,3-butanediol production. Appl Microbiol Biotechnol. 2016;2:637–647. doi: 10.1007/s00253-015-7013-3. [DOI] [PubMed] [Google Scholar]
- 11.Kim SJ, Seo SO, Jin YS, Seo JH. Production of 2,3-butanediol by engineered Saccharomyces cerevisiae. Bioresour Technol. 2013;146:274–281. doi: 10.1016/j.biortech.2013.07.081. [DOI] [PubMed] [Google Scholar]
- 12.Kim SJ, Seo SO, Park YC, Jin YS, Seo JH. Production of 2,3-butanediol from xylose by engineered Saccharomyces cerevisiae. J Biotechnol. 2014;192:376–382. doi: 10.1016/j.jbiotec.2013.12.017. [DOI] [PubMed] [Google Scholar]
- 13.Yamada R, Wakita K, Mitsui R, Nishikawa R, Ogino H. Efficient production of 2,3-butanediol by recombinant Saccharomyces cerevisiae through modulation of gene expression by cocktail delta-integration. Bioresour Technol. 2017;245:1558–1566. doi: 10.1016/j.biortech.2017.05.034. [DOI] [PubMed] [Google Scholar]
- 14.Lian J, Chao R, Zhao H. Metabolic engineering of a Saccharomyces cerevisiae strain capable of simultaneously utilizing glucose and galactose to produce enantiopure (2R,3R)-butanediol. Metab Eng. 2014;23:92–99. doi: 10.1016/j.ymben.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 15.Potvin G, Ahmad A, Zhang Z. Bioprocess engineering aspects of heterologous protein production in Pichia pastoris: a review. Biochem Eng J. 2012;64:91–105. doi: 10.1016/j.bej.2010.07.017. [DOI] [Google Scholar]
- 16.Tang S, Boehme L, Lam H, Zhang Z. Pichia pastoris fermentation for phytase production using crude glycerol from biodiesel production as the sole carbon source. Biochem Eng J. 2009;2:157–162. doi: 10.1016/j.bej.2008.09.020. [DOI] [Google Scholar]
- 17.Liu Y, Zhang R, Lian Z, Wang S, Wright AT. Yeast cell surface display for lipase whole cell catalyst and its applications. J Mol Catal B Enzym. 2014;106:17–25. doi: 10.1016/j.molcatb.2014.04.011. [DOI] [Google Scholar]
- 18.Liu Z, Ho SH, Hasunuma T, Chang JS, Ren NQ, Kondo A. Recent advances in yeast cell-surface display technologies for waste biorefineries. Bioresour Technol. 2016;215:324–333. doi: 10.1016/j.biortech.2016.03.132. [DOI] [PubMed] [Google Scholar]
- 19.Bhataya A, Schmidt-Dannert C, Lee PC. Metabolic engineering of Pichia pastoris X-33 for lycopene production. Process Biochem. 2009;10:1095–1102. doi: 10.1016/j.procbio.2009.05.012. [DOI] [Google Scholar]
- 20.Araya-Garay JM, Feijoo-Siota L, Rosa-dos-Santos F, Veiga-Crespo P, Villa TG. Construction of new Pichia pastoris X-33 strains for production of lycopene and beta-carotene. Appl Microbiol Biotechnol. 2012;6:2483–2492. doi: 10.1007/s00253-011-3764-7. [DOI] [PubMed] [Google Scholar]
- 21.Araya-Garay JM, Ageitos JM, Vallejo JA, Veiga-Crespo P, Sánchez-Pérez A, Villa TG. Construction of a novel Pichia pastoris strain for production of xanthophylls. AMB Express. 2012;1:24. doi: 10.1186/2191-0855-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wriessnegger T, Augustin P, Engleder M, Leitner E, Muller M, Kaluzna I, Schurmann M, Mink D, Zellnig G, Schwab H, et al. Production of the sesquiterpenoid (+)-nootkatone by metabolic engineering of Pichia pastoris. Metab Eng. 2014;24:18–29. doi: 10.1016/j.ymben.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 23.Liu XB, Liu M, Tao XY, Zhang ZX, Wang FQ, Wei DZ. Metabolic engineering of Pichia pastoris for the production of dammarenediol-II. J Biotechnol. 2015;216:47–55. doi: 10.1016/j.jbiotec.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 24.de Lima PB, Mulder KC, Melo NT, Carvalho LS, Menino GS, Mulinari E, de Castro VH, Dos Reis TF, Goldman GH, Magalhaes BS, et al. Novel homologous lactate transporter improves l-lactic acid production from glycerol in recombinant strains of Pichia pastoris. Microb Cell Fact. 2016;1:158. doi: 10.1186/s12934-016-0557-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J, Chan SH, Brock-Nannestad T, Chen J, Lee SY, Solem C, Jensen PR. Combining metabolic engineering and biocompatible chemistry for high-yield production of homo-diacetyl and homo-(S, S)-2,3-butanediol. Metab Eng. 2016;36:57–67. doi: 10.1016/j.ymben.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 26.Schroer K, Peter Luef K, Stefan Hartner F, Glieder A, Pscheidt B. Engineering the Pichia pastoris methanol oxidation pathway for improved NADH regeneration during whole-cell biotransformation. Metab Eng. 2010;1:8–17. doi: 10.1016/j.ymben.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 27.Çalık P, Ata Ö, Güneş H, Massahi A, Boy E, Keskin A, Öztürk S, Zerze GH, Özdamar TH. Recombinant protein production in Pichia pastoris under glyceraldehyde-3-phosphate dehydrogenase promoter: from carbon source metabolism to bioreactor operation parameters. Biochem Eng J. 2015;95:20–36. doi: 10.1016/j.bej.2014.12.003. [DOI] [Google Scholar]
- 28.Yang Z, Zhang Z. Engineering strategies for enhanced production of protein and bio-products in Pichia pastoris: a review. Biotechnol Adv. 2017 doi: 10.1016/j.biotechadv.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 29.Kim S, Hahn JS. Efficient production of 2,3-butanediol in Saccharomyces cerevisiae by eliminating ethanol and glycerol production and redox rebalancing. Metab Eng. 2015;31:94–101. doi: 10.1016/j.ymben.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 30.Xu Y, Chu H, Gao C, Tao F, Zhou Z, Li K, Li L, Ma C, Xu P. Systematic metabolic engineering of Escherichia coli for high-yield production of fuel bio-chemical 2,3-butanediol. Metab Eng. 2014;23:22–33. doi: 10.1016/j.ymben.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 31.Ji X-J, Huang H, Du J, Zhu J-G, Ren L-J, Hu N, Li S. Enhanced 2,3-butanediol production by Klebsiella oxytoca using a two-stage agitation speed control strategy. Bioresour Technol. 2009;13:3410–3414. doi: 10.1016/j.biortech.2009.02.031. [DOI] [PubMed] [Google Scholar]
- 32.Petrov K, Petrova P. Enhanced production of 2,3-butanediol from glycerol by forced pH fluctuations. Appl Microbiol Biotechnol. 2010;3:943–949. doi: 10.1007/s00253-010-2545-z. [DOI] [PubMed] [Google Scholar]
- 33.Cho S, Kim T, Woo HM, Lee J, Kim Y, Um Y. Enhanced 2,3-butanediol production by optimizing fermentation conditions and engineering Klebsiella oxytoca M1 through overexpression of acetoin reductase. PLoS ONE. 2015;9:e0138109. doi: 10.1371/journal.pone.0138109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hassler T, Schieder D, Pfaller R, Faulstich M, Sieber V. Enhanced fed-batch fermentation of 2,3-butanediol by Paenibacillus polymyxa DSM 365. Bioresour Technol. 2012;124:237–244. doi: 10.1016/j.biortech.2012.08.047. [DOI] [PubMed] [Google Scholar]
- 35.Kim JW, Kim J, Seo SO, Kim KH, Jin YS, Seo JH. Enhanced production of 2,3-butanediol by engineered Saccharomyces cerevisiae through fine-tuning of pyruvate decarboxylase and NADH oxidase activities. Biotechnol Biofuels. 2016;9:265. doi: 10.1186/s13068-016-0677-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan Y, Lee CC, Liao JC. Enantioselective synthesis of pure (R, R)-2,3-butanediol in Escherichia coli with stereospecific secondary alcohol dehydrogenases. Org Biomol Chem. 2009;19:3914–3917. doi: 10.1039/b913501d. [DOI] [PubMed] [Google Scholar]
- 37.Li L, Li K, Wang Y, Chen C, Xu Y, Zhang L, Han B, Gao C, Tao F, Ma C, et al. Metabolic engineering of Enterobacter cloacae for high-yield production of enantiopure (2R,3R)-2,3-butanediol from lignocellulose-derived sugars. Metab Eng. 2015;28:19–27. doi: 10.1016/j.ymben.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 38.Jung MY, Jung HM, Lee J, Oh MK. Alleviation of carbon catabolite repression in Enterobacter aerogenes for efficient utilization of sugarcane molasses for 2,3-butanediol production. Biotechnol Biofuels. 2015;8:106. doi: 10.1186/s13068-015-0290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu J, Huo G, Feng L, Mao Y, Wang Z, Ma H, Chen T, Zhao X. Metabolic engineering of Bacillus subtilis for chiral pure meso-2,3-butanediol production. Biotechnol Biofuels. 2016;9:90. doi: 10.1186/s13068-016-0502-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang T, Rao Z, Zhang X, Xu M, Xu Z, Yang ST. Improved production of 2,3-butanediol in Bacillus amyloliquefaciens by over-expression of glyceraldehyde-3-phosphate dehydrogenase and 2,3-butanediol dehydrogenase. PLoS ONE. 2013;10:e76149. doi: 10.1371/journal.pone.0076149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cho S, Kim T, Woo HM, Kim Y, Lee J, Um Y. High production of 2,3-butanediol from biodiesel-derived crude glycerol by metabolically engineered Klebsiella oxytoca M1. Biotechnol Biofuels. 2015;8:146. doi: 10.1186/s13068-015-0336-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jantama K, Polyiam P, Khunnonkwao P, Chan S, Sangproo M, Khor K, Jantama SS, Kanchanatawee S. Efficient reduction of the formation of by-products and improvement of production yield of 2,3-butanediol by a combined deletion of alcohol dehydrogenase, acetate kinase-phosphotransacetylase, and lactate dehydrogenase genes in metabolically engineered Klebsiella oxytoca in mineral salts medium. Metab Eng. 2015;30:16–26. doi: 10.1016/j.ymben.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 43.Ma C, Wang A, Qin J, Li L, Ai X, Jiang T, Tang H, Xu P. Enhanced 2,3-butanediol production by Klebsiella pneumoniae SDM. Appl Microbiol Biotechnol. 2009;1:49–57. doi: 10.1007/s00253-008-1732-7. [DOI] [PubMed] [Google Scholar]
- 44.Zhang L, Sun JA, Hao Y, Zhu J, Chu J, Wei D, Shen Y. Microbial production of 2,3-butanediol by a surfactant (serrawettin)-deficient mutant of Serratia marcescens H30. J Ind Microbiol Biotechnol. 2010;8:857–862. doi: 10.1007/s10295-010-0733-6. [DOI] [PubMed] [Google Scholar]
- 45.Rados D, Carvalho AL, Wieschalka S, Neves AR, Blombach B, Eikmanns BJ, Santos H. Engineering Corynebacterium glutamicum for the production of 2,3-butanediol. Microb Cell Fact. 2015;14:171. doi: 10.1186/s12934-015-0362-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang S, Mohagheghi A, Franden MA, Chou YC, Chen X, Dowe N, Himmel ME, Zhang M. Metabolic engineering of Zymomonas mobilis for 2,3-butanediol production from lignocellulosic biomass sugars. Biotechnol Biofuels. 2016;1:189. doi: 10.1186/s13068-016-0606-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen C, Wei D, Shi J, Wang M, Hao J. Mechanism of 2,3-butanediol stereoisomer formation in Klebsiella pneumoniae. Appl Microbiol Biotechnol. 2014;10:4603–4613. doi: 10.1007/s00253-014-5526-9. [DOI] [PubMed] [Google Scholar]
- 48.Bai F, Dai L, Fan J, Truong N, Rao B, Zhang L, Shen Y. Engineered Serratia marcescens for efficient (3R)-acetoin and (2R,3R)-2,3-butanediol production. J Ind Microbiol Biotechnol. 2015;5:779–786. doi: 10.1007/s10295-015-1598-5. [DOI] [PubMed] [Google Scholar]
- 49.Qiu Y, Zhang J, Li L, Wen Z, Nomura CT, Wu S, Chen S. Engineering Bacillus licheniformis for the production of meso-2,3-butanediol. Biotechnol Biofuels. 2016;9:117. doi: 10.1186/s13068-016-0522-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim B, Lee S, Park J, Lu M, Oh M, Kim Y, Lee J. Enhanced 2,3-butanediol production in recombinant Klebsiella pneumoniae via overexpression of synthesis-related genes. J Microbiol Biotechnol. 2012;9:1258–1263. doi: 10.4014/jmb.1201.01044. [DOI] [PubMed] [Google Scholar]
- 51.Park JM, Song H, Lee HJ, Seung D. In silico aided metabolic engineering of Klebsiella oxytoca and fermentation optimization for enhanced 2,3-butanediol production. J Ind Microbiol Biotechnol. 2013;9:1057–1066. doi: 10.1007/s10295-013-1298-y. [DOI] [PubMed] [Google Scholar]
- 52.Bakker BM, Overkamp KM, van Maris AJ, Kötter P, Luttik MA, van Dijken JP, Pronk JT. Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS Microbiol Rev. 2001;1:15–37. doi: 10.1111/j.1574-6976.2001.tb00570.x. [DOI] [PubMed] [Google Scholar]
- 53.Kim JW, Seo SO, Zhang GC, Jin YS, Seo JH. Expression of Lactococcus lactis NADH oxidase increases 2,3-butanediol production in Pdc-deficient Saccharomyces cerevisiae. Bioresour Technol. 2015;191:512–519. doi: 10.1016/j.biortech.2015.02.077. [DOI] [PubMed] [Google Scholar]
- 54.Wong CL, Yen HW, Lin CL, Chang JS. Effects of pH and fermentation strategies on 2,3-butanediol production with an isolated Klebsiella sp. Zmd30 strain. Bioresour Technol. 2014;152:169–176. doi: 10.1016/j.biortech.2013.10.101. [DOI] [PubMed] [Google Scholar]
- 55.Priya A, Dureja P, Talukdar P, Rathi R, Lal B, Sarma PM. Microbial production of 2,3-butanediol through a two-stage pH and agitation strategy in 150 l bioreactor. Biochem Eng J. 2016;105:159–167. doi: 10.1016/j.bej.2015.09.016. [DOI] [Google Scholar]
- 56.Zhang L, Yang Y, Sun JA, Shen Y, Wei D, Zhu J, Chu J. Microbial production of 2,3-butanediol by a mutagenized strain of Serratia marcescens H30. Bioresour Technol. 2010;6:1961–1967. doi: 10.1016/j.biortech.2009.10.052. [DOI] [PubMed] [Google Scholar]
- 57.Chan S, Jantama SS, Kanchanatawee S, Jantama K. Process optimization on micro-aeration supply for high production yield of 2,3-butanediol from maltodextrin by metabolically-engineered Klebsiella oxytoca. PLoS ONE. 2016;9:e0161503. doi: 10.1371/journal.pone.0161503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang T, Rao Z, Zhang X, Xu M, Xu Z, Yang ST. Enhanced 2,3-butanediol production from biodiesel-derived glycerol by engineering of cofactor regeneration and manipulating carbon flux in Bacillus amyloliquefaciens. Microb Cell Fact. 2015;14:122. doi: 10.1186/s12934-015-0317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ji XJ, Huang H, Du J, Zhu JG, Ren LJ, Li S, Nie ZK. Development of an industrial medium for economical 2,3-butanediol production through co-fermentation of glucose and xylose by Klebsiella oxytoca. Bioresour Technol. 2009;21:5214–5218. doi: 10.1016/j.biortech.2009.05.036. [DOI] [PubMed] [Google Scholar]
- 60.Adlakha N, Yazdani SS. Efficient production of (R, R)-2,3-butanediol from cellulosic hydrolysate using Paenibacillus polymyxa ICGEB2008. J Ind Microbiol Biotechnol. 2015;1:21–28. doi: 10.1007/s10295-014-1542-0. [DOI] [PubMed] [Google Scholar]
- 61.Kim T, Cho S, Woo HM, Lee SM, Lee J, Um Y, Seo JH. High production of 2,3-butanediol from glycerol without 1,3-propanediol formation by Raoultella ornithinolytica B6. Appl Microbiol Biotechnol. 2017;7:2821–2830. doi: 10.1007/s00253-017-8094-y. [DOI] [PubMed] [Google Scholar]
- 62.Yang TW, Rao ZM, Zhang X, Xu MJ, Xu ZH, Yang ST. Effects of corn steep liquor on production of 2,3-butanediol and acetoin by Bacillus subtilis. Process Biochem. 2013;11:1610–1617. doi: 10.1016/j.procbio.2013.07.027. [DOI] [Google Scholar]
- 63.Li L, Zhang L, Li K, Wang Y, Gao C, Han B, Ma C, Xu P. A newly isolated Bacillus licheniformis strain thermophilically produces 2,3-butanediol, a platform and fuel bio-chemical. Biotechnol Biofuels. 2013;1:123. doi: 10.1186/1754-6834-6-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vellanki RN, Potumarthi R, Doddapaneni KK, Anubrolu N, Mangamoori LN. Constitutive optimized production of streptokinase in Saccharomyces cerevisiae utilizing glyceraldehyde 3-phosphate dehydrogenase promoter of Pichia pastoris. BioMed Res Int. 2013 doi: 10.1155/2013/268249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nakashima N, Akita H, Hoshino T. Establishment of a novel gene expression method, BICES (biomass-inducible chromosome-based expression system), and its application to the production of 2,3-butanediol and acetoin. Metab Eng. 2014;25:204–214. doi: 10.1016/j.ymben.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 66.Zhu XD, Sadowski PD. Cleavage-dependent ligation by the flp recombinase characterization of a mutant flp protein with an alteration in a catalytic amino acid. J Biol Chem. 1995;39:23044–23054. doi: 10.1074/jbc.270.39.23044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Effect of yeast extract on 2,3-BD titer. 100 mL media containing 40 g/L glucose were used to cultivate strain X33-SD. Error bar indicate standard deviation of three replicate experiments.
Additional file 2: Figure S2. Pareto graph of the seven variables.
Additional file 3: Figure S3. Time course of fed-batch fermentation using optimized BSMY medium. Bioreactor contained 2 L of initial media. 40 g/L glucose as initial substrate and 1 L of 250 g/L glucose solution as feeding solution.
Additional file 4. Supplementing tables in this work. Table S1. Level code for variables based on Plackett-Burman design. Table S2. Cultivation results of Plackett-Burman design. Table S3. Analysis results of Plackett-Burman design. Table S4. Design of Box-Behnken experiments. Table S5. Concentration level for Box-Behnken design. Table S6. Cultivation results of Box-Behnke design. Table S7. Analysis results of Box-Behnken design. Table S8. Comparison of fed-batch cultivations.
Data Availability Statement
Not applicable.


