Abstract
Cellular metabolites act as important signaling cues, but are subject to complex unknown chemistry. Kynurenine is a tryptophan metabolite that plays a crucial role in cancer and the immune system. Despite its atypical, non-ligand-like, highly polar structure, kynurenine activates the aryl hydrocarbon receptor (AHR), a PER, ARNT, SIM (PAS) family transcription factor that responds to diverse environmental and cellular ligands. The activity of kynurenine is increased 100–1000-fold by incubation or long-term storage and relies on the hydrophobic ligand-binding pocket of AHR, with identical structural signatures for AHR induction before and after activation. We purified trace-active derivatives of kynurenine and identified two novel, closely related condensation products, named trace-extended aromatic condensation products (TEACOPs), which are active at low picomolar levels. The synthesized compound for one of the predicted structures matched the purified compound in both chemical structure and AHR pharmacology. Our study provides evidence that kynurenine acts as an AHR pro-ligand, which requires novel chemical conversions to act as a receptor agonist.
Keywords: aryl hydrocarbon receptor (AhR) (AHR), structure-function, dioxin, ligand-binding protein, chemical biology, Ah receptor, dioxin, kynurenine, pro-ligand, structural determinants
Introduction
Kynurenine is a tryptophan metabolite generated by the enzymes indoleamine 2,3-dioxygenase and tryptophan 2,3,-dioxygenase. The cellular levels of kynurenine and its downstream metabolites play crucial roles in regulating the immune system, vascular biology, and neurological function (1–4). Disorders of kynurenine metabolism are associated with a variety of human health issues, including cancer, hypertension, chronic inflammation, and neurodegenerative disorders (1, 5, 6). A number of recent studies have suggested a link between the physiological effects of kynurenine and the aryl hydrocarbon receptor (AHR)3 (7–9). The AHR is a PAS (PER, ARNT, SIM) family transcription factor that is essential for development and normal function of vascular and immune systems (10–13). In support of this relationship are the numerous observations that kynurenine levels influence a variety of immune responses in an AHR-dependent manner (7, 14, 15). The underlying mechanism of the AHR in kynurenine action is currently uncertain. Although it has been shown that kynurenine is a receptor activator, its structure does not conform to many of the rules that correlate with high-affinity binding to the AHR (Fig. 1A) (16–19).
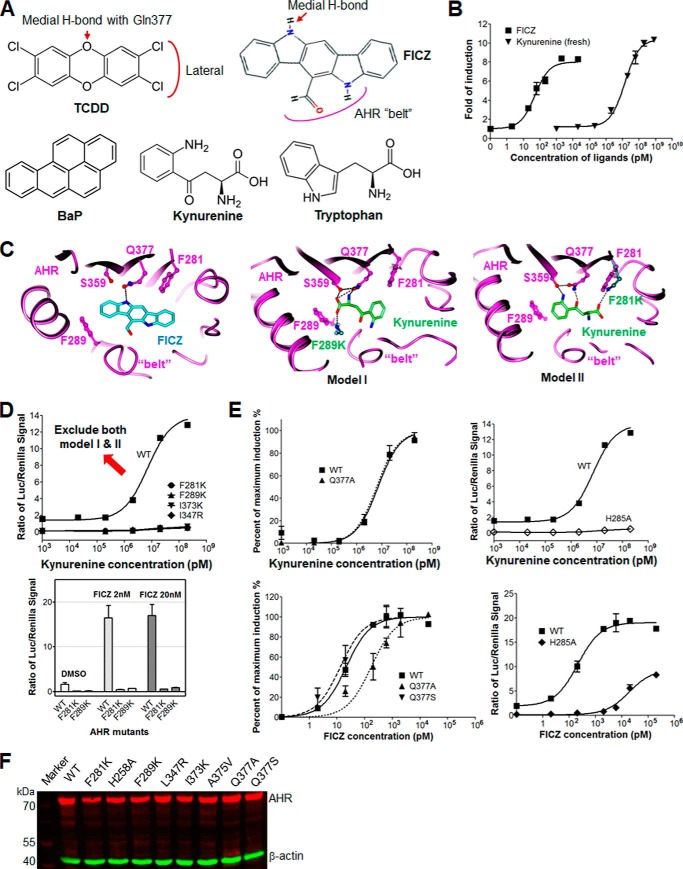
Figure 1.
Kynurenine has low in vitro efficacy, and models of AHR binding contradict predicted structural signatures. A, chemical structures of well-known AHR ligands, TCDD, FICZ, BaP, and kynurenine, compared with tryptophan. The chemical signatures of TCDD and FICZ with medial H-bond potential, facing lateral extension, and the belt of the AHR ligand-binding pocket are illustrated. B, dose-dependent response curves of AHR ligands, FICZ and kynurenine, in induction of the transcriptional activity of mAHR. The induction level of the AHR activity was measured by reporter luciferase activity and normalized to the signal of Renilla luciferase. C, two hypothetical structural models of kynurenine bound to the AHR ligand-binding pocket, compared with the structural model of FICZ bound to AHR. Key residues of AHR are shown in a ball-and-stick representation and colored by atom type. Intermolecular H-bonds are shown by black dashed lines. Kynurenine and FICZ are shown by sticks and colored green and cyan, respectively, and by atom type. D, dose-dependent response curves of WT and mutant AHR, F289K, F281K, I373K, and L347R to kynurenine, determined as in B (top). Induction of the transcriptional activity of WT and mutant mAHR, F289K and F281K, by FICZ (2 and 20 nm) is shown for comparison (bottom). E, dose-dependent response curves of kynurenine in activation of WT and mutant AHR bearing mutation to the residue with medial H-bond potential, Q377A (top left) or the residue that forms part of the H-bond network lining the ligand-binding pocket, H285A. The level of AHR activity was normalized to the maximum induction or to the signal of Renilla luciferase (top right). The results for FICZ are shown at the bottom for comparison. The loss of medial H-bond potential by Q377A reduced the activity of FICZ by 5-fold but barely affected the activity of kynurenine. F, expression level of WT and mutant mAHR in COS-1 cells was examined by Western blotting. β-Actin was detected as a loading control. Error bars, S.E.
Like kynurenine, many cellular metabolites that activate the AHR are derived from tryptophan. For example, exposure to UV radiation in the skin converts tryptophan to 6-formylindolo[3,2-b]carbazole (FICZ) (20–22), stomach acid converts dietary indole-3-carbinol to indolo[3,3-b]carbazole (ICZ), the enzyme d-amino acid oxidase converts tryptophan to indole 3-pyruvic acid, and gut microbiota generate tryptophan-derived AHR activators that are crucial for curtailing inflammatory bowl disease and central nervous system inflammation (23–26). In addition to endogenous ligands, the AHR also responds to numerous xenobiotic ligands to influence a wide variety of toxicological, immunological, and cardiovascular end points (27). Knowledge of AHR pharmacology has arisen from studying xenobiotic agonists like the halogenated dibenzo-p-dioxins (e.g. 2,3,7,8-tetrachlorodibenzo-p-dioxin; TCDD) and polycyclic aromatic hydrocarbons (e.g. benzo[a]pyrene; BaP) (16–19). These studies show that AHR prefers elongated planar compounds with large lateral extension and small medial extension with specific medial H-bond potential (Fig. 1A). Thousands of xenobiotic compounds and cellular metabolites with diverse shape and chemical properties have been reported to bind AHR (28, 29). Whereas a majority of AHR ligands have an overall elongated planar shape, some ligands barely have any AHR ligand structural signatures. Kynurenine is one such ligand that is much smaller, polar, and irregular in shape (Fig. 1A). Using homology models of the ligand-binding domain of AHR (AHR-LBD) bound to TCDD and BaP, we previously accurately identified key structural signatures for AHR binding that differentially affect the efficacy of these two prototype AHR ligands and flexible structural elements that are essential for tolerating diverse ligands (19). A flexible extended loop of AHR, named the “belt,” is longer and more flexible than other PAS family transcription factors, underlying the unique ability of AHR to respond to diverse ligands.
Based on our previous models of AHR-LBD bound to TCDD and BaP (19), we generated a model of AHR-LBD bound to FICZ that is consistent with AHR structural signatures controlling FICZ binding but could not build a model for AHR bound to a single kynurenine molecule. This observation, coupled with our previous speculation that kynurenine “breakdown products” or metabolites were the actual AHR ligands (7), prompted us to test the idea that kynurenine is a pro-ligand of AHR, which spontaneously converts in solution to yield trace-extended aromatic condensation products (TEACOPs) that act as high-affinity AHR ligands. By extensive fractionation and characterization, we identified two closely related novel kynurenine derivatives with picomolar EC50 values. The predicted structures were confirmed through synthesis of a compound that matches one of the purified compounds in both chemical structure and AHR induction.
Results
Low in vitro potency and structural signatures governing AHR binding oppose models of single kynurenine bound to the AHR-LBD
We compared the ability of kynurenine and FICZ, another tryptophan derivative that acts as a potent AHR ligand (20, 21), to activate the mouse AHR (mAHR B1-allele, simplified as AHR or mAHR if not specified) using a luciferase reporter gene assay (30). COS-1 cells expressing mAHR were treated with increasing doses of FICZ and kynurenine for 4 h. Consistent with previous observations, FICZ exhibits a very high AHR activation activity with a measured EC50 of 36 pm (Fig. 1B). This value is similar to the measured KD value of 70 pm (20) and the EC50 of 34 pm for MH1C1 rat hepatoma cells exposed for 3 h (31–33). As observed previously (14), kynurenine induces a maximum AHR activation that is ∼40% higher than that induced by FICZ (Fig. 1B). The EC50 of kynurenine was measured to be ∼13 μm (Fig. 1B). The low biological potency of kynurenine that we observed is consistent with its predicted “non-ligand-like” structure but could not account for its in vivo function via AHR, suggesting that a single kynurenine might not directly act as an AHR ligand.
The high potency of FICZ in AHR activation can be readily predicted from its chemical structure and its predicted fit to our structural model of AHR-LBD (19). As an aromatic heterocyclic multi-ring compound, FICZ has an overall planar and elongated shape similar to TCDD (Fig. 1A), one of the most active AHR ligands ever identified (34). The relatively even top edge of FICZ gives minimal medial extension similar to TCDD and harbors an -NH group facing the medial residue of the AHR-LBD, Gln-377, which defines the H-bond potential and explains the preferences of AHR for small extensions at the medial position (19). In this orientation, the larger aldehyde extension at the opposite side would be placed near, and be well accommodated by, the flexible belt (Fig. 1A). To confirm this prediction, we performed molecular docking of FICZ to our model of AHR-LBD. The binding mode of FICZ to the AHR-LBD (Fig. 1C) is exactly as predicted above.
In contrast, kynurenine fits poorly to the AHR ligand-binding pocket in two potential orientations (Fig. 1B), and the calculated docking energies are at least 3 kcal weaker than that for FICZ. In addition, the terminal carboxylate group of kynurenine is placed near Phe-289 or Phe-281 within highly hydrophobic portions of the pocket. Nonetheless, the predicted extensive H-bond interactions between kynurenine terminal carboxylate and amine groups and the AHR medial residues, Gln-377 and Ser-359, might alleviate these unfavorable acidic-hydrophobic contacts. These models suggest that mutation of Phe-289 or Phe-281 in AHR to the positively charged lysine would enhance the interaction of kynurenine with AHR and thus increase its AHR induction activity. Contrary to our reasoning, neither F289K nor F281K could enhance the activity of kynurenine; instead, both completely abolished the responsiveness of AHR to kynurenine (Fig. 1D). Similar effects of these AHR mutations were observed for FICZ (Fig. 1D). Mutation of other hydrophobic residues to the positively charged lysine or arginine also completely abolished the activity of kynurenine (Fig. 1D), suggesting that, despite its high polarity, kynurenine prefers the hydrophobic AHR ligand-binding pocket for AHR activation. Furthermore, mutation of Gln-377 to alanine barely affected kynurenine activity (Fig. 1E), whereas both models predicted that this mutation should abolish medial H-bond interactions with kynurenine (Fig. 1C). Instead, Q377A reduced the efficacy of FICZ by 5-fold (Fig. 1E), consistent with the medial H-bond interaction between FICZ and Gln-377 (Fig. 1C). The AHR mutation H285A, which abolishes an H-bond network crucial to defining the overall shape of the ligand-binding pocket (19), completely disrupts kynurenine activity but remains partially receptive to FICZ binding (Fig. 1E). This is counterintuitive, given the much smaller size of kynurenine compared with FICZ. For all mutational analysis, mutant mAHRs were expressed at a level similar to that of the wildtype (Fig. 1F). Collectively, these data support our model of FICZ bound to AHR but are not consistent with either model of AHR bound to kynurenine (Fig. 1C). This conclusion is consistent with the non-ligand-like structure of kynurenine (Fig. 1A).
Activated and fresh kynurenine share identical structural signatures in AHR binding and activate both the B- and D-alleles of AHR
The low in vitro potency of kynurenine in AHR activation (Fig. 1B) is difficult to reconcile with its known action through AHR in vivo. In initial examinations of the idea that kynurenine is converted to a more potent structure in vitro or in vivo, we discovered that kynurenine solution stored at −20 °C for 2 years exhibited >1000-fold higher activity than a freshly prepared solution of kynurenine (EC50 = 5 nm) (Fig. 2A). We refer to this “aged” kynurenine as “activated kynurenine.” Like fresh kynurenine (Fig. 1B), activated kynurenine exhibited a higher level of maximum activity in AHR activation compared with other ligands, such as FICZ and BaP (Fig. 2A). Furthermore, the structural signatures, as defined by mutational analysis, governing AHR binding are identical for fresh and activated kynurenine (Figs. 1 (D and E) and 2 (B and C)). Similar to fresh kynurenine, activated kynurenine prefers the overall hydrophobic AHR ligand-binding pocket and fails to activate mutant AHR with alterations to its hydrophobic residues, such as F281K, F289K, I373K, and I347R (Fig. 2B). AHR induction by activated kynurenine was barely affected by AHR mutation Q377A and was completely abolished by H285A (Fig. 2C), just like fresh kynurenine (Fig. 1E). The identical structural signatures governing AHR binding let us predict that the same chemical compounds function as AHR ligands in both fresh and activated kynurenine. It seems plausible that these compounds are spontaneous kynurenine derivatives, and their abundance increases >1000-fold after 2-year storage.
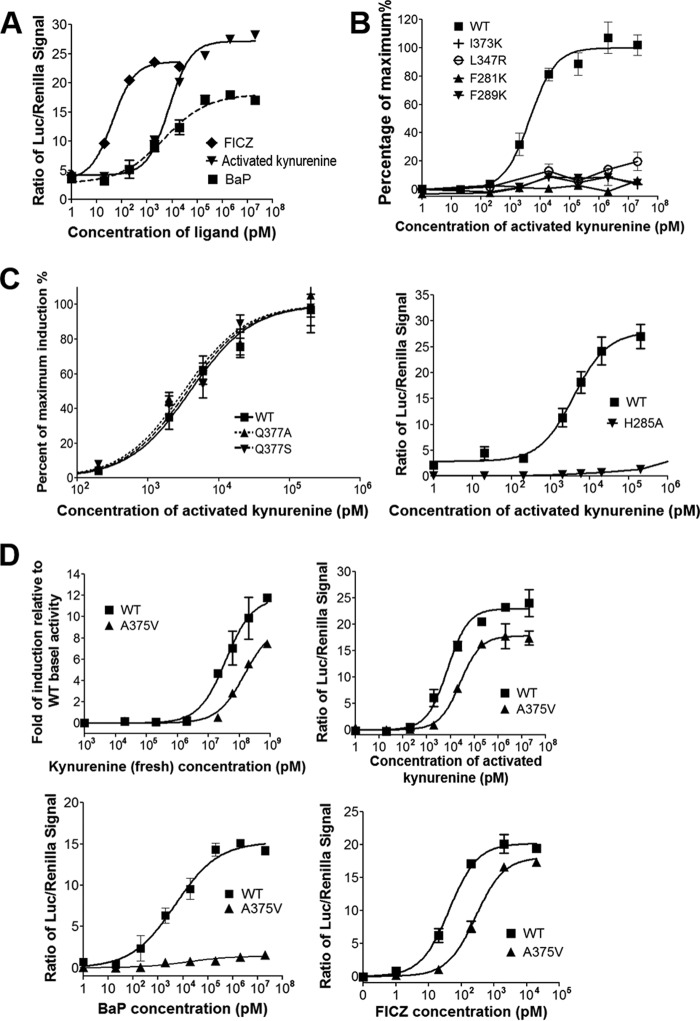
Figure 2.
Activated and fresh kynurenine share identical structural signatures in AHR induction and activate both B- and D-alleles of AHR. A, dose-dependent response curves of AHR ligands in induction of the transcriptional activity of mAHR, with activated kynurenine compared with FICZ and BaP, similar to Fig. 1B. B, dose-dependent response curves of WT and mutant AHR, F289K, F281K, I373K, and L347R, to the activated kynurenine, determined as in Fig. 1B. The level of AHR activity was normalized to the maximum induction. C, dose-dependent response curves of activated kynurenine in activation of WT and mutant AHR bearing mutation to the residue with medial H-bond potential, Q377A (left), or the residue that forms part of the H-bond network lining the ligand-binding pocket, H285A. The level of AHR activity was determined as in Fig. 1E. D, dose-dependent response curves of WT mAHR (B-allele) and its genetic variant A375V (D-allele) to fresh (top left) and activated (top right) kynurenine, BaP (bottom left), and FICZ (bottom right). The level of AHR activity was normalized to the -fold induction relative to WT basal activity or to the signal of Renilla luciferase, followed by normalization of basal activity to zero. Error bars, S.E.
The murine Ahr B1 and D alleles are two well-known genetic variants that harbor a key sequence variation at residue 375, with alanine in the B-allele and valine in the D-allele (A375V). Residue 375 is located near the medial position of the AHR-LBD. The A375V variant has weakened binding to TCDD and other xenobiotic AHR ligands (35–37). This replacement increases the steric hindrance to the medial positions of ligands (19), particularly environmental ligands that are bulkier in the medial direction, such as BaP, which could barely stimulate the activity of the A375V D-allele (Fig. 2D) (19). In sharp contrast, both fresh and activated kynurenine can readily activate the B-allele and the A375V D-allele, and their AHR induction activities were affected similarly by this genetic variant (Fig. 2D). A similar observation was made using FICZ ligand (Fig. 2D). Thus, unlike BaP or other environmental compounds, kynurenine and FICZ are capable of activating both B- and D-alleles of AHR. Collectively, our study showed that fresh and activated kynurenine share the same modes of interaction with AHR, which partially resembles the binding mode of FICZ. We thus predicted that spontaneous chemical conversion of kynurenine to trace-extended aromatic condensation products (TEACOPs) might result in heterocyclic multi-ring aromatic compounds, with ligand binding affinities similar to FICZ.
Spontaneous chemical conversion of kynurenine
Based on the above ideas, we examined whether chemical conversion of kynurenine to activated kynurenine can be sped up by increasing incubation temperature. Incubation of kynurenine solution at room temperature or 37 °C continuously increased AHR induction activity by kynurenine. Induction increased ∼100-fold by incubation after 3 days at 37 °C or after 18 days at room temperature (Fig. 3A). Consistent with this observation, AHR activation by kynurenine in COS-1 cells for 8 h gave a higher biological response compared with a 4-h induction (Fig. S1A). In contrast, FICZ activity was slightly reduced in the 8-h versus 4-h induction and further reduced in the 20-h induction (Fig. S1B). These results suggest that whereas active derivatives of kynurenine accumulated continuously during AHR induction, longer incubation times with FICZ have lower efficacy due to cellular turnover or FICZ metabolism.
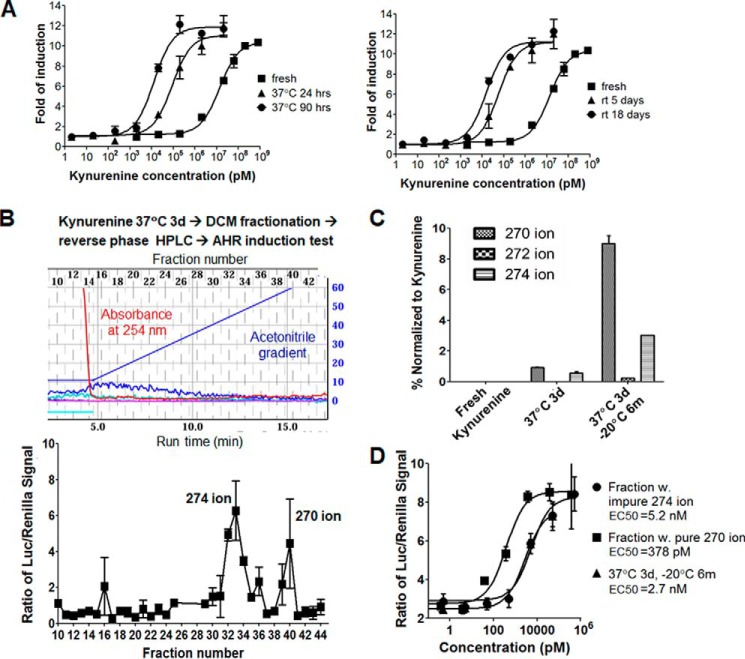
Figure 3.
Temperature-dependent spontaneous chemical conversion of kynurenine and fractionation and identification of novel derivatives with potent AHR induction activity. A, dose-dependent response curves of mAHR to kynurenine samples that were freshly prepared or incubated for the indicated amounts of time at 37 °C (left) or room temperature (right). B, fractionation of kynurenine derivatives and monitoring of their AHR induction activity. Experimental scheme is shown at the top, the profile of reverse phase HPLC is in the middle, and the transcriptional activity of mAHR induced by HPLC fractions is at the bottom. Key ions associated with each active peak are indicated. C, quantitative LC/MS analysis of ions of kynurenine derivatives for three indicated samples, fresh kynurenine, kynurenine incubated at 37 °C for 3 days (3d), and kynurenine incubated for at 37 °C 3 days followed by storage at −20 °C for 6 months (3d, 6m). The average peak areas of 270, 272, and 274 ions were normalized to that of kynurenine mother ions. D, dose-dependent response curves of mAHR to kynurenine derivative fractions associated with the 274 and 270 ions, respectively, compared with kynurenine (3d, 6m) used in C. The EC50 of each sample is shown. Error bars, S.E.
Kynurenine is metabolized to different compounds by diverse enzymes in the kynurenine pathway (1), and many of these metabolic intermediates (other than kynurenine) are not active in AHR activation assays (7). As kynurenine is also known to be chemically converted to several other well-characterized derivatives (38–40) (Fig. S2A), we next determined whether any known chemical derivatives of kynurenine could activate AHR. In this regard, kynurenic acid exhibited a low activity in AHR induction, and 4(1H)-quinolinone (4-HQ) did not show any activity. Given that the responses to these known kynurenine derivatives are much lower than those to fresh kynurenine (Fig. S2B), they are unlikely to account for the activity of activated kynurenine. Other compounds in the chemical conversion scheme shown in Fig. S2A are similar to either kynurenic acid, 4-HQ, or kynurenine itself. This led us to conclude that the AHR ligands derived from kynurenine are most likely unknown trace condensation products that have not yet been identified.
Identification of two potent and closely related trace derivatives of kynurenine
To identify the putative active kynurenine products, we incubated 200 mg of kynurenine at 37 °C for 3 days, followed by phase separation between dichloromethane (DCM) and water, with the DCM phase retaining nearly 50% of the total activity. Based on our prediction of the extended multi-ring aromatic structure of the active derivatives, we expected that the DCM phase would enrich these compounds over the abundant kynurenine and its polar derivatives. Consistently, reverse-phase preparative HPLC fractionation of compounds from the DCM phase gave a spectrum with barely any absorbance peaks at 254 nm during acetonitrile gradient, but AHR induction tests detected two prominent peaks that could activate AHR (Fig. 3B). These observations support our earlier supposition that the active derivatives of kynurenine are highly potent and are only present in trace amounts. Mass spectrometry coupled to HPLC detected a 274 ion and 270 ion in the two peaks, respectively. The peak of the 270 ion appeared at a higher acetonitrile gradient, suggesting a higher hydrophobicity.
Analysis of fresh and 3-day incubated kynurenine by high-resolution mass spectrometry detected known kynurenine derivatives at a high intensity after 3-day incubation at 37 °C, such as 4-HQ and kynurenic acid. Intriguingly, the intensity of the 274 ion was most increased among all ions detected (Fig. S3). Close examination of the mass spectra also identified the closely related 272 and 270 ions in the 3-day incubation sample (Fig. S3), which hinted to us that the 270 ion might be a two-round dehydrogenation product of the 274 ion. To accurately quantify the level of 270, 272, and 274 ions in kynurenine samples, LC-MS was performed, and the peak area for each ion was normalized to the kynurenine mother ion. Whereas the 270, 272, and 274 ions were barely detected in fresh kynurenine, the 270 and 274 ions were detected in <1% compared with the kynurenine mother ion in the 3-day incubation sample (Fig. 3C). After 3-day incubation at 37 °C, the sample that had been stored at −20 °C for 6 months yielded almost 10% 270 ion, 3% 274 ion, and readily detectable 272 ion (Fig. 3C). Consistently, this sample gave an EC50 of 2.7 nm for AHR activation (Fig. 3D), more active than the activated kynurenine tested earlier (Fig. 2A).
To facilitate chemical and functional characterization of the 274 and 270 ions, we scaled up the purification of active AHR ligands from 2 g of kynurenine after 3-day 37 °C incubation following procedures shown in Fig. 3A. Additional rounds of HPLC purification were performed to gain purity. The 270 ion fraction was relatively pure, but the 274 ion fraction contained a few other more dominant ions. The 270 ion gave an EC50 of 378 pm for induction of cellular AHR (Fig. 3D), making it a novel, highly potent AHR ligand. The 274 ion fraction gave an EC50 of ∼5 nm, which is expected to give a higher activity if pure material were obtained.
Deciphering the structures and mode of interactions of potent kynurenine derivatives
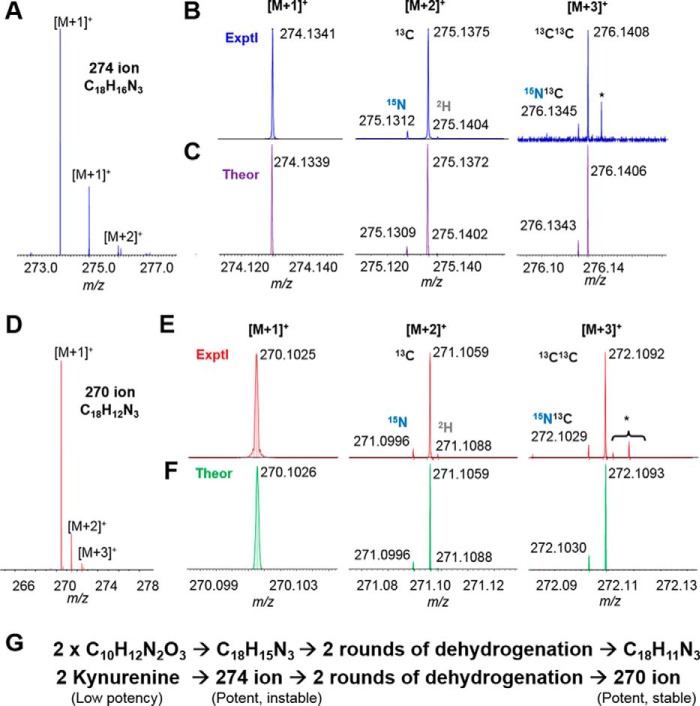
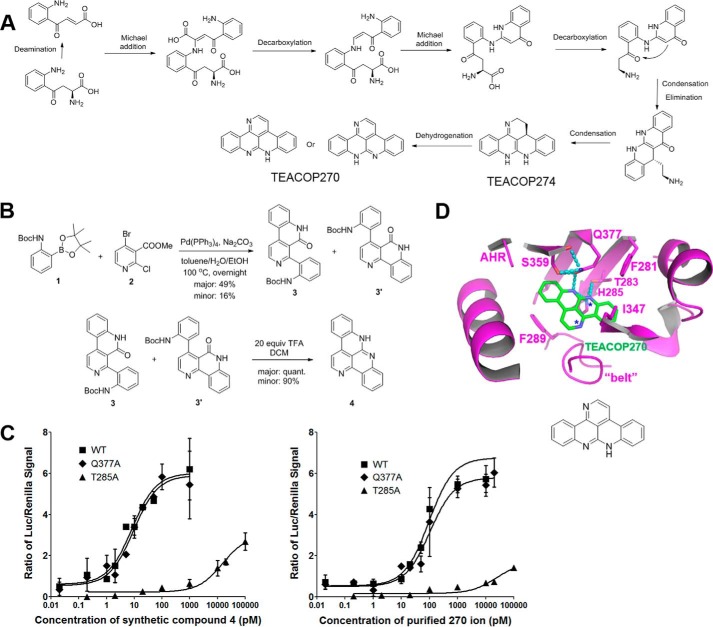
To characterize the chemical structures of active kynurenine derivatives, the peaks with the 270 and 274 ions from the large scale purification described above were analyzed with ultrahigh-resolution Fourier transform mass spectrometry (UH-FTMS). The chemical formulas for the 270 and 274 ions were determined: C18H16N3 for the 274 ion and C18H12N3 for the 270 ion (Fig. 4, A–F). Consistent with our earlier prediction that the 270 ion is derived from the 274 ion, the two compounds differed by exactly four hydrogen atoms, most likely by two rounds of dehydrogenation (Fig. 4G).
Figure 4.
UH-FTMS determined the chemical formula of the trace 274 and 270 ions of kynurenine derivatives. Shown is C18H16N3+ isotopic fine structure determined by FTMS for the 274 ion: isotopic clusters (A) with a zoom-in of individual clusters from experimental (B) and theoretical (C) results. *, belonging to another ion. Likewise, C18H12N3+ isotopic fine structure was determined by FTMS for the 270 ion: isotopic clusters (D) with zoom-in of individual clusters from experimental (E) and theoretical (F) results. *, belonging to other ions. G, initial prediction of chemical conversions of two kynurenine molecules to the 274 ion and then to the 270 ion based on the chemical formula determined in A–F.
The peak containing the 270 ion was further analyzed with NMR experiments (Supporting Method). 1H NMR showed 10 aromatic protons (Fig. S4A), which were grouped in three sets based on COSY (homonuclear correlation spectroscopy) data (Fig. S4B), although lipid contaminations were also found present in the isolated material. The quality of the 13C NMR spectrum is poor due to the limited amount of the isolated material and its low solubility in commonly used solvents (CDCl3, CD3OD, DMSO-d6, etc.). Based on the expected planar structures, formulas determined by mass spectrometry, and NMR results, we were able to predict the chemical structure of the 270 ion as well as a possible reaction pathway leading to it from kynurenine (Fig. 5A). The predicted structures for the 270, 272, and 274 ions are extended molecules with polyaromatic rings. Due to the trace amount of these compounds in kynurenine derivatives, we term them TEACOPs.
Figure 5.
Predicted chemical conversion schemes of kynurenine to potent ligands, synthesis of a potent kynurenine derivative, and its mode of interaction with AHR. A, chemical reaction schemes that give rise to the active 274 and 270 ions from kynurenine. The predicted structures of the two active ions are consistent with the NMR spectra provided in Fig. S4 and confirmed by the synthesized compound. B, synthesis of the predicted compound (compound 4) for the 270 ion. NMR spectra and assignment of all protons and carbons are provided in Fig. S5 (A–G). C, dose-dependent response curves of the synthesized compound 4 in the free base form and the purified 270 ion in activation of WT AHR and AHR mutants, Q377A and H285A. The level of AHR activity was determined as in Fig. 1A. D, structural model of the 270 ion bound to the AHR-LBD. Key residues of AHR are shown in stick representations and colored by atom type. Intermolecular H-bonds are shown as cyan dashed lines. The 270 ion are shown in stick representations and colored green and by atom type. Error bars, S.E.
To confirm the predicted structure of TEACOP270 (Fig. 5A), a chemical synthesis was performed (compound 4; Fig. 5B). The TFA salt of the synthesized compound 4 showed good solubility in CDCl3 and thus was fully characterized (Fig. S5). Similar to that of the purified TEACOP270, we were not able to fully characterize the free base form of compound 4 due to the poor solubility, but compound 4 clearly showed a 1H NMR spectrum almost identical to that of the purified TEACOP270 except for the N–H proton (Figs. S4A and S6). The difference in the N–H proton is probably due to lipid contamination in the purified TEACOP270 sample (Fig. S4) or different levels of water molecules in the two samples. Furthermore, the purified TEACOP270, the synthesized free base compound 4, and its TFA salt were compared by liquid chromatography–mass spectrometry analysis, and the observed single peak from mixtures of each two as well as a mixture of the three apparently showed that the synthesized compound 4 was identical to the purified TEACOP270 (Fig. S7).
We further showed that compound 4 and the purified TEACOP270 exhibited the same structural signatures in AHR induction except that the measured activity of the purified TEACOP270 is >10-fold lower than that of compound 4 (Fig. 5C). The lower activity of the purified TEACOP270 is consistent with the fact that the sample was largely contaminated by lipids (Fig. S4B) and that the purified TEACOP270 has 10-fold lower absorbance at the UV and visible light wavelengths than the synthesized TEACOP270 (Fig. S9). Similar to the fresh and activated kynurenine (Figs. 1 and 2), the activity of both compound 4 and the purified TEACOP270 in AHR induction was barely affected by the AHR Q377A mutation, but drastically affected by T285A mutation (Fig. 5C).
Discussion
Although kynurenine-mediated AHR signaling has been increasingly implicated in many aspects of biological function, how kynurenine activates AHR has remained highly puzzling. Whereas both FICZ and kynurenine are potent AHR activators, the structure of FICZ resembles prototype AHR ligands, but not kynurenine, a much smaller, polar, and charged compound (Fig. 1A). The ligand-binding pocket of AHR is highly hydrophobic, and many of its hydrophobic residues are important for ligand binding (16–19). It is unlikely that a highly polar/charged molecule such as kynurenine is compatible with the hydrophobic ligand-binding pocket of AHR. By harnessing a novel and powerful combination of research approaches, including structural modeling, structural signatures of AHR for ligand binding, cell biology, small-molecule chemistry, advanced MS technologies, and NMR spectroscopy, we identified two novel TEACOPs of kynurenine that are potent at low picomolar concentrations and elucidated their structural and chemical basis in AHR activation. Given that these active derivatives are present in only trace amounts in kynurenine mixtures (Fig. 3, A and B), it is not surprising that the two novel AHR ligands had not been previously identified among either cellular kynurenine metabolites (1, 41–43) or in vitro chemical derivatives (38–40). Whereas TEACOPs identified here might account for the AHR induction activity of kynurenine in mammalian cells, the chemical reaction schemes of kynurenine that lead to active AHR ligands (Fig. 5A) might follow modified reaction schemes in complex cellular environments. For example, kynuramine and 5-hydroxykynuramine, other cellular metabolites of tryptophan (1), might provide the active amine group in place of kynurenine for intermolecular Michael addition to the deaminated kynurenine (Fig. 5A).
Besides the hydrophobic nature of the AHR ligand-binding cavity, our previous study identified key structural signatures for several important aspects of AHR-ligand interactions (19). For example, Gln-377 defines medial H-bond interactions and preferences of AHR toward small medial extension of AHR ligands; His-285 defines an H-bond network in controlling the overall shape of the ligand-binding cavity (19). Furthermore, the belt and other flexible structural elements allow AHR to recognize diverse ligands with distinctly different shapes and chemical properties (19). These structural signatures also helped us to understand AHR interaction with kynurenine derivatives and FICZ. We docked the chemical structure of TEACOP270 to our previous model of AHR-LBD (Fig. 5D). The predicted binding energy of the structure is much higher than that for single kynurenine (Fig. 1B) and close to that for FICZ (Fig. S2A). Examination of the docked model supports the structural signatures in AHR-ligand interactions revealed by mutagenesis analysis. Whereas TEACOP270, which is highly similar to other TEACOPs identified here, forms more medial H-bond interactions with AHR than FICZ does (Fig. 5D and Fig. S2A), the Q377A mutation reduced FICZ activity by 5-fold (Fig. S2C) but barely affected the activity of TEACOPs in AHR induction (Figs. 2C and 5C). This is probably due to the fact that the smaller size and polarity of Q377A allows residue 377 to accommodate the larger, less polar medial edge of TEACOP270 so that its polar edge forms H-bond interactions with the belt backbone carboxylate group of AHR (Fig. S8). This mode of ligand flipping would not happen for FICZ because Q377A would not allow residue 377 to accommodate the larger, polar edge of FICZ that faces the AHR belt for wildtype AHR (Fig. S2A).
The fact that kynurenine activates both the B- and D-alleles of mAHR in vitro (Fig. 2D) indicates that these trace derivatives of kynurenine might mediate the normal physiological functions of these two alleles. Human AHR is similar to the D-allele of mAHR, which is much less active or inert to many environmental ligands, such as BaP (Fig. 2D). Our recent advance in understanding the structural basis of AHR signaling revealed a versatile allosteric structural pathway from the AHR-LBD to the N-terminal nuclear localization signal and DNA-reading head (44). Due to directionality of H-bond interactions, the extensive H-bond interactions of TEACOPs to the AHR-LBD (Fig. 5D) are expected to induce a more defined conformation in AHR-LBD than BaP. The latter interacts with AHR by only hydrophobic contacts, which are expected to be more malleable than H-bond interactions (19). This probably explains why the maximum activity of kynurenine is 60% higher than that of BaP (Fig. 2A). Reduced structural dynamics of TEACOPs-bound AHR-LBD might also account for the distinct role of kynurenine in broad physiological functions. Our study here provides an important structural and chemical basis for further understanding of kynurenine in broad human physiology.
Experimental procedures
Modeling AHR-LBD bound to FICZ, kynurenine, and kynurenine derivatives
Built on our previous structural model of AHR-LBD (19), single FICZ, kynurenine, and kynurenine derivatives (TEACOP270 and TEACOP274) were docked to the AHR ligand-binding cavity using the Autodock program (45). This was followed by energy minimization and optimization of backbone conformation and rotamer usage. Iterative model building, ligand docking, energy minimization, and optimization of backbone conformation and rotamer usage were performed until satisfying results were obtained.
Cloning and expression of recombinant mAHR
The wildtype and mutant mAHR were cloned into the XhoI/SalI cloning sites of pTARGET (Promega, Madison, WI) using routine PCR and molecular cloning procedures using the pSport-mAHR plasmid (PL65) as template (46). For recombinant expression of mAHR, COS-1 cells were cultured in 6-cm dishes and transfected with 2 μg of wildtype or mutant mAHR expression vector. Twenty-four hours after transfection, cells were collected, and the whole-cell extracts were prepared by celLytic M reagent (Sigma-Aldrich). To normalize the total protein, the protein concentrations of the whole-cell lysates were measured by Bradford assay, and 50 μg of total proteins was analyzed by Western blotting using antibody that specifically recognizes the bacterially expressed mAHR (BEAR-4) (47). β-Actin was used as a loading control (Sigma-Aldrich). Note that several AHR mutants were previously proved to not affect the cellular expression level of AHR (19).
Luciferase reporter gene assay
COS-1 cells were cultured in 96-well plates and transiently transfected with pTarget vector containing the expression cassette of wildtype or mutant mAHR or empty vector (3 ng), together with pGudLu6.1 DREs-driven luciferase reporter vector (14 ng) (30) and TK-Renilla luciferase vector (3 ng) (Invitrogen). Six hours after transfection, cells were treated with titrated or fixed concentrations of FICZ, kynurenine, BaP, or vehicle alone (0.1% DMSO) for 4 h (or 20 h when specifically indicated) and assayed with the Dual-Luciferase reporter assay system (Promega, Madison, WI). The expressed luciferase activity was measured by an ENSPIRE plate reader (PerkinElmer Life Sciences). Data analysis and simulation of dose-response curves were performed using GraphPad Prism version 5 (GraphPad Software Inc., La Jolla, CA). The experiments were performed in triplicate and repeated at least three times. Representative results of one repeat are shown as mean ± S.E.
Kynurenine activation and phase separation
Fresh crystalline l-kynurenine (Sigma-Aldrich; product number K8625) was dissolved in DMSO at a concentration of 10 mg/ml and incubated at 37 °C for 3 days, followed by temporary or long-term storage at −20 °C. To separate derivatives of kynurenine, kynurenine with a 3-day incubation was diluted with water to a concentration of 1 mg/ml and mixed thoroughly with an equal volume of DCM by shaking in a separator funnel. The two phases (aqueous upper layer/organic bottom layer) were clearly separated after 1 h of standing still. The bottom layer was collected for further fractionation by HPLC.
HPLC
The combined DCM phases from phase separation for 200 mg or 2 g of activated kynurenine were concentrated under reduced pressure with a Buchi Rotavapor R-300 system to generate a crude mixture with residual DMSO residue. The crude mixture of DCM phases of activated kynurenine was purified using the Teledyne Isco CombiFlash EZprep system with a RediSep Prep C18 column (particle size 5 μm, size 20 × 150 mm) via mass-directed fractionation (phase A: 0.1% formic acid, 5% CH3CN in H2O; phase B: 0.1% formic acid in CH3CN; flow rate 18 ml/min; gradient (B%): 0–1 min 10%, 1–21 min 10–100%, 21–23 min 100%). For the 200-mg pilot test, the AHR induction activity of each fraction was tested, and an initial prediction of the compounds associated with activity was made based on MS ions in the active fractions. For large-scale fractionation, the 274 and 270 ion fractions were collected, combined, and dried in a Genevac EZ-2 Elite centrifugal evaporator at 30 °C. The resulting product (∼30 mg) was further purified (five repeated injections) on Agilent 1200 series HPLC with an Eclipse XDB-C18 column (particle size 5 μm; size 9.4 × 250 mm; phase A: 0.1% formic acid, 5% CH3CN in H2O; phase B: 0.1% formic acid in CH3CN; flow rate 2 ml/min; gradient (B%): 0–4 min 5%, 4–42 min 10–50%, 42–44 min 50–100%, 44–50 min 100%). Fractions were collected based on UV absorption at 254 nm and dried in the Genevac EZ-2 Elite at 30 °C. Mass analysis of the resulting dry materials was performed on a Waters Autopure system with a QDa electrospray ionization mass spectrometer.
High-resolution mass spectrometry and LC-MS
For the initial investigation of kynurenine conversion to active products, we used mass spectrometry to analyze fresh kynurenine and kynurenine after a 3-day incubation at 37 °C. Samples were diluted 1:1000 and infused into a Bruker MaXis 4G ultrahigh-resolution time-of-flight mass spectrometer (Bruker Daltonics, Billerica, MA). Samples were infused at 3 μl/min, and spectra were collected in positive mode for 2 min. For data analysis, we averaged spectra for >1 min and manually compared peaks in each sample.
For subsequent relative quantitation, we compared the area under the curve of peaks of interest from samples of fresh kynurenine, kynurenine after a 3-day incubation at 37 °C, and kynurenine with 3-day incubation at 37 °C followed by storage at −20 °C for 6 months following LC-MS analysis. Samples were diluted 1:3000 in solvent containing an internal standard (100 ng/ml D3-naproxen) before separation on a Waters Acquity UPLC system (Waters Corp., Milford, MA) and then analyzed on the MaXis. Samples (5 μl) were injected on a 2.1 × 100-mm Kinetex XB-C18 column (Phenomenex, Torrance, CA) with 2.6-μm particles equipped with a guard column. Analytes were separated using an increasing gradient of acetonitrile in LC separation created by solvent A (water with 0.1% formic acid (v/v)) and solvent B (acetonitrile with 0.1% formic acid) at a flow rate of 0.3 ml/min. The gradient was started with 2% B held for 2 min, followed by a gradient to 75% B in 15 min and ramp to 95% B in 1 min. Spectra were collected over a mass range of 50–1750 m/z. All data were analyzed using Bruker Data Analysis software. Briefly, extracted ion chromatograms for masses of interest were created with a window of 0.005 m/z, and areas for peaks of interest were determined using the software. Peak areas were determined from two replicate reactions, and each was injected twice (technical replicates). Areas of kynurenine mother ions and ions of interest were normalized to the area under the curve for the internal standard. The relative area of each of the two ions of interest was then normalized to the relative kynurenine area in the same run to determine the percent abundance of the new compounds.
UH-FTMS
Samples containing the 274 and 270 ions were diluted to 10–20 μg/ml in acetonitrile, and diluted samples were directly infused to a 12T solariX FTMS (Bruker Daltonics) by a TriVersa NanoMate (Advion) with spray voltage of 1300 V and gas pressure of 0.25 p.s.i. The FT data size was set to 8,000,000 (3.5-s transient length, 1,390,000 resolving power at m/z 274 and 270, respectively). The isotopic clusters were isolated in a 5–8 m/z window. The experimental spectra were averaged by 50–200 scans. The MS results were analyzed by SmartFormula manually integrated into the DataAnalysis software (Bruker Daltonics). The mass tolerance was set to 2 ppm. Only one candidate was found for each sample, and the theoretical isotopic distribution was compared with the experimental data.
Author contributions
Y. X. and C. A. B. conceived the study; Y. X. and S.-H. S. performed structural and functional characterization, assisted by K. A. S., Hui Chen., Hongbo Chen., C. R. J., and M. C. .; Z.-X. M. performed chemical characterization and synthesis, guided by J. B. F., W. T., and Y. X. and assisted by S.-H. S.; C. S., Z. L., S.-H. S., and Y. X. performed MS analysis, guided by Y. G.; Y. X. and Z.-X. M. predicted the reaction schemes; Y. X. wrote the manuscript.
Supplementary Material
This work was supported by National Institutes of Health Grants R01 GM096060 (to Y. X.), R35028377 (to C. A. B.), P30CA014520, and UL1TR000427, as well as the McArdle New Direction Fund. All authors have intellectual property associated with this work. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S9 and Methods.
- AHR
- aryl hydrocarbon receptor
- mAHR
- mouse AHR
- PAS
- PER, ARNT, SIM
- FICZ
- 6-formylindolo[3,2-b]carbazole
- ICZ
- indolo[3,3-b]carbazole
- TCDD
- 2,3,7,8-tetrachlorodibenzo-p-dioxin
- BaP
- benzo[a]pyrene
- H-bond
- hydrogen bond
- LBD
- ligand-binding domain
- TEACOP
- trace-extended aromatic condensation products
- 4-HQ
- 4(1H)-quinolinone
- DCM
- dichloromethane
- UH-FTMS
- ultrahigh-resolution Fourier transform mass spectrometry.
References
- 1. Stone T. W., and Darlington L. G. (2002) Endogenous kynurenines as targets for drug discovery and development. Nat. Rev. Drug. Discov. 1, 609–620 10.1038/nrd870 [DOI] [PubMed] [Google Scholar]
- 2. Jasiewicz M., Moniuszko M., Pawlak D., Knapp M., Rusak M., Kazimierczyk R., Musial W. J., Dabrowska M., and Kaminski K. A. (2016) Activity of the kynurenine pathway and its interplay with immunity in patients with pulmonary arterial hypertension. Heart 102, 230–237 10.1136/heartjnl-2015-308581 [DOI] [PubMed] [Google Scholar]
- 3. Polyzos K. A., and Ketelhuth D. F. (2015) The role of the kynurenine pathway of tryptophan metabolism in cardiovascular disease: an emerging field. Hamostaseologie 35, 128–136 10.5482/HAMO-14-10-0052 [DOI] [PubMed] [Google Scholar]
- 4. Rudzite V., Sileniece G., Liepina D., Dalmane A., and Zirne R. (1991) Impairment of kynurenine metabolism in cardiovascular disease. Adv. Exp. Med. Biol. 294, 663–667 10.1007/978-1-4684-5952-4_89 [DOI] [PubMed] [Google Scholar]
- 5. Changsirivathanathamrong D., Wang Y., Rajbhandari D., Maghzal G. J., Mak W. M., Woolfe C., Duflou J., Gebski V., dos Remedios C. G., Celermajer D. S., and Stocker R. (2011) Tryptophan metabolism to kynurenine is a potential novel contributor to hypotension in human sepsis. Crit. Care. Med. 39, 2678–2683 10.1097/CCM.0b013e31822827f2 [DOI] [PubMed] [Google Scholar]
- 6. Oxenkrug G. F. (2010) Metabolic syndrome, age-associated neuroendocrine disorders, and dysregulation of tryptophan-kynurenine metabolism. Ann. N.Y. Acad. Sci. 1199, 1–14 10.1111/j.1749-6632.2009.05356.x [DOI] [PubMed] [Google Scholar]
- 7. Mezrich J. D., Fechner J. H., Zhang X., Johnson B. P., Burlingham W. J., and Bradfield C. A. (2010) An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J. Immunol. 185, 3190–3198 10.4049/jimmunol.0903670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bessede A., Gargaro M., Pallotta M. T., Matino D., Servillo G., Brunacci C., Bicciato S., Mazza E. M., Macchiarulo A., Vacca C., Iannitti R., Tissi L., Volpi C., Belladonna M. L., Orabona C., et al. (2014) Aryl hydrocarbon receptor control of a disease tolerance defence pathway. Nature 511, 184–190 10.1038/nature13323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lanis J. M., Alexeev E. E., Curtis V. F., Kitzenberg D. A., Kao D. J., Battista K. D., Gerich M. E., Glover L. E., Kominsky D. J., and Colgan S. P. (2017) Tryptophan metabolite activation of the aryl hydrocarbon receptor regulates IL-10 receptor expression on intestinal epithelia. Mucosal. Immunol. 10, 1133–1144 10.1038/mi.2016.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Savouret J. F., Berdeaux A., and Casper R. F. (2003) The aryl hydrocarbon receptor and its xenobiotic ligands: a fundamental trigger for cardiovascular diseases. Nutr. Metab. Cardiovasc. Dis. 13, 104–113 10.1016/S0939-4753(03)80026-1 [DOI] [PubMed] [Google Scholar]
- 11. Korashy H. M., and El-Kadi A. O. (2006) The role of aryl hydrocarbon receptor in the pathogenesis of cardiovascular diseases. Drug. Metab. Rev. 38, 411–450 10.1080/03602530600632063 [DOI] [PubMed] [Google Scholar]
- 12. Stevens E. A., Mezrich J. D., and Bradfield C. A. (2009) The aryl hydrocarbon receptor: a perspective on potential roles in the immune system. Immunology 127, 299–311 10.1111/j.1365-2567.2009.03054.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Esser C., Rannug A., and Stockinger B. (2009) The aryl hydrocarbon receptor in immunity. Trends Immunol. 30, 447–454 10.1016/j.it.2009.06.005 [DOI] [PubMed] [Google Scholar]
- 14. Opitz C. A., Litzenburger U. M., Sahm F., Ott M., Tritschler I., Trump S., Schumacher T., Jestaedt L., Schrenk D., Weller M., Jugold M., Guillemin G. J., Miller C. L., Lutz C., Radlwimmer B., et al. (2011) An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 478, 197–203 10.1038/nature10491 [DOI] [PubMed] [Google Scholar]
- 15. Nguyen N. T., Kimura A., Nakahama T., Chinen I., Masuda K., Nohara K., Fujii-Kuriyama Y., and Kishimoto T. (2010) Aryl hydrocarbon receptor negatively regulates dendritic cell immunogenicity via a kynurenine-dependent mechanism. Proc. Natl. Acad. Sci. U.S.A. 107, 19961–19966 10.1073/pnas.1014465107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bisson W. H., Koch D. C., O'Donnell E. F., Khalil S. M., Kerkvliet N. I., Tanguay R. L., Abagyan R., and Kolluri S. K. (2009) Modeling of the aryl hydrocarbon receptor (AhR) ligand binding domain and its utility in virtual ligand screening to predict new AhR ligands. J. Med. Chem. 52, 5635–5641 10.1021/jm900199u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pandini A., Soshilov A. A., Song Y., Zhao J., Bonati L., and Denison M. S. (2009) Detection of the TCDD binding-fingerprint within the Ah receptor ligand binding domain by structurally driven mutagenesis and functional analysis. Biochemistry 48, 5972–5983 10.1021/bi900259z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Procopio M., Lahm A., Tramontano A., Bonati L., and Pitea D. (2002) A model for recognition of polychlorinated dibenzo-p-dioxins by the aryl hydrocarbon receptor. Eur. J. Biochem. 269, 13–18 10.1046/j.0014-2956.2002.02619.x [DOI] [PubMed] [Google Scholar]
- 19. Xing Y., Nukaya M., Satyshur K. A., Jiang L., Stanevich V., Korkmaz E. N., Burdette L., Kennedy G. D., Cui Q., and Bradfield C. A. (2012) Identification of the Ah-receptor structural determinants for ligand preferences. Toxicol. Sci. 129, 86–97 10.1093/toxsci/kfs194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rannug A., Rannug U., Rosenkranz H. S., Winqvist L., Westerholm R., Agurell E., and Grafström A. K. (1987) Certain photooxidized derivatives of tryptophan bind with very high affinity to the Ah receptor and are likely to be endogenous signal substances. J. Biol. Chem. 262, 15422–15427 [PubMed] [Google Scholar]
- 21. Helferich W. G., and Denison M. S. (1991) Ultraviolet photoproducts of tryptophan can act as dioxin agonists. Mol. Pharmacol. 40, 674–678 [PubMed] [Google Scholar]
- 22. Rannug U., Rannug A., Sjöberg U., Li H., Westerholm R., and Bergman J. (1995) Structure elucidation of two tryptophan-derived, high affinity Ah receptor ligands. Chem. Biol. 2, 841–845 10.1016/1074-5521(95)90090-X [DOI] [PubMed] [Google Scholar]
- 23. Rothhammer V., Mascanfroni I. D., Bunse L., Takenaka M. C., Kenison J. E., Mayo L., Chao C. C., Patel B., Yan R., Blain M., Alvarez J. I., Kébir H., Anandasabapathy N., Izquierdo G., Jung S., et al. (2016) Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 22, 586–597 10.1038/nm.4106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hubbard T. D., Murray I. A., Bisson W. H., Lahoti T. S., Gowda K., Amin S. G., Patterson A. D., and Perdew G. H. (2015) Adaptation of the human aryl hydrocarbon receptor to sense microbiota-derived indoles. Sci. Rep. 5, 12689 10.1038/srep12689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zelante T., Iannitti R. G., Cunha C., De Luca A., Giovannini G., Pieraccini G., Zecchi R., D'Angelo C., Massi-Benedetti C., Fallarino F., Carvalho A., Puccetti P., and Romani L. (2013) Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 39, 372–385 10.1016/j.immuni.2013.08.003 [DOI] [PubMed] [Google Scholar]
- 26. Lamas B., Richard M. L., Leducq V., Pham H. P., Michel M. L., Da Costa G., Bridonneau C., Jegou S., Hoffmann T. W., Natividad J. M., Brot L., Taleb S., Couturier-Maillard A., Nion-Larmurier I., Merabtene F., et al. (2016) CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 22, 598–605 10.1038/nm.4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McIntosh B. E., Hogenesch J. B., and Bradfield C. A. (2010) Mammalian Per-Arnt-Sim proteins in environmental adaptation. Annu. Rev. Physiol. 72, 625–645 10.1146/annurev-physiol-021909-135922 [DOI] [PubMed] [Google Scholar]
- 28. Schmidt J. V., and Bradfield C. A. (1996) Ah receptor signaling pathways. Annu. Rev. Cell Dev. Biol. 12, 55–89 10.1146/annurev.cellbio.12.1.55 [DOI] [PubMed] [Google Scholar]
- 29. Nguyen L. P., and Bradfield C. A. (2008) The search for endogenous activators of the aryl hydrocarbon receptor. Chem. Res. Toxicol. 21, 102–116 10.1021/tx7001965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Han D., Nagy S. R., and Denison M. S. (2004) Comparison of recombinant cell bioassays for the detection of Ah receptor agonists. BioFactors 20, 11–22 10.1002/biof.5520200102 [DOI] [PubMed] [Google Scholar]
- 31. Wei Y. D., Helleberg H., Rannug U., and Rannug A. (1998) Rapid and transient induction of CYP1A1 gene expression in human cells by the tryptophan photoproduct 6-formylindolo[3,2-b]carbazole. Chem. Biol. Interact. 110, 39–55 10.1016/S0009-2797(97)00111-7 [DOI] [PubMed] [Google Scholar]
- 32. Wei Y. D., Bergander L., Rannug U., and Rannug A. (2000) Regulation of CYP1A1 transcription via the metabolism of the tryptophan-derived 6-formylindolo[3,2-b]carbazole. Arch. Biochem. Biophys. 383, 99–107 10.1006/abbi.2000.2037 [DOI] [PubMed] [Google Scholar]
- 33. Bergander L., Wincent E., Rannug A., Foroozesh M., Alworth W., and Rannug U. (2004) Metabolic fate of the Ah receptor ligand 6-formylindolo[3,2-b]carbazole. Chem. Biol. Interact. 149, 151–164 10.1016/j.cbi.2004.08.005 [DOI] [PubMed] [Google Scholar]
- 34. Bjeldanes L. F., Kim J. Y., Grose K. R., Bartholomew J. C., and Bradfield C. A. (1991) Aromatic hydrocarbon responsiveness-receptor agonists generated from indole-3-carbinol in vitro and in vivo: comparisons with 2,3,7,8-tetrachlorodibenzo-p-dioxin. Proc. Natl. Acad. Sci. U.S.A. 88, 9543–9547 10.1073/pnas.88.21.9543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Poland A., Palen D., and Glover E. (1994) Analysis of the four alleles of the murine aryl hydrocarbon receptor. Mol. Pharmacol. 46, 915–921 [PubMed] [Google Scholar]
- 36. Ema M., Ohe N., Suzuki M., Mimura J., Sogawa K., Ikawa S., and Fujii-Kuriyama Y. (1994) Dioxin binding activities of polymorphic forms of mouse and human arylhydrocarbon receptors. J. Biol. Chem. 269, 27337–27343 [PubMed] [Google Scholar]
- 37. Chang C., Smith D. R., Prasad V. S., Sidman C. L., Nebert D. W., and Puga A. (1993) Ten nucleotide differences, five of which cause amino acid changes, are associated with the Ah receptor locus polymorphism of C57BL/6 and DBA/2 mice. Pharmacogenetics 3, 312–321 10.1097/00008571-199312000-00005 [DOI] [PubMed] [Google Scholar]
- 38. Tokuyama T., Senoh S., Sakan T., Brown K. S. Jr, and Witkop B. (1967) The photoreduction of kynurenic acid to kynurenine yellow and the occurrence of 3-hydroxy-l-kynurenine in butterflies. J. Am. Chem. Soc. 89, 1017–1021 10.1021/ja00980a046 [DOI] [PubMed] [Google Scholar]
- 39. Zelentsova E. A., Sherin P. S., Snytnikova O. A., Kaptein R., Vauthey E., and Tsentalovich Y. P. (2013) Photochemistry of aqueous solutions of kynurenic acid and kynurenine yellow. Photochem. Photobiol. Sci. 12, 546–558 10.1039/C2PP25357G [DOI] [PubMed] [Google Scholar]
- 40. Brown K. S., and Becher D. (1967) The mass spectra of the kynurenines. Tetrahedron Lett. 18, 1721–1726 10.1016/S0040-4039(00)90709-3 [DOI] [Google Scholar]
- 41. Hubbard T. D., Murray I. A., and Perdew G. H. (2015) Indole and tryptophan metabolism: endogenous and dietary routes to Ah receptor activation. Drug. Metab. Dispos. 43, 1522–1535 10.1124/dmd.115.064246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bohár Z., Toldi J., Fülöp F., and Vécsei L. (2015) Changing the face of kynurenines and neurotoxicity: therapeutic considerations. Int. J. Mol. Sci. 16, 9772–9793 10.3390/ijms16059772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Leklem J. E. (1971) Quantitative aspects of tryptophan metabolism in humans and other species: a review. Am. J. Clin. Nutr. 24, 659–672 [DOI] [PubMed] [Google Scholar]
- 44. Seok S. H., Lee W., Jiang L., Molugu K., Zheng A., Li Y., Park S., Bradfield C. A., and Xing Y. (2017) Structural hierarchy controlling dimerization and target DNA recognition in the AHR transcriptional complex. Proc. Natl. Acad. Sci. U.S.A. 114, 5431–5436 10.1073/pnas.1617035114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Goodsell D. S., Morris G. M., and Olson A. J. (1996) Automated docking of flexible ligands: applications of AutoDock. J. Mol. Recognit. 9, 1–5 10.1002/(SICI)1099-1352(199601)9:1%3C1::AID-JMR241%3E3.0.CO%3B2-6 [DOI] [PubMed] [Google Scholar]
- 46. Dolwick K. M., Swanson H. I., and Bradfield C. A. (1993) In vitro analysis of Ah receptor domains involved in ligand-activated DNA recognition. Proc. Natl. Acad. Sci. U.S.A. 90, 8566–8570 10.1073/pnas.90.18.8566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Poland A., Glover E., and Bradfield C. A. (1991) Characterization of polyclonal antibodies to the Ah receptor prepared by immunization with a synthetic peptide hapten. Mol. Pharmacol. 39, 20–26 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.