Abstract
Feedback control is a key mechanism in signal transduction, intimately involved in regulating the outcome of the cellular response. Here, we report a novel mechanism by which PHLDA1, Pleckstrin homology-like domain, family A, member 1, negatively regulates ErbB receptor signaling by inhibition of receptor oligomerization. We have found that the ErbB3 ligand, heregulin, induces PHILDA1 expression in MCF-7 cells. Transcriptionally-induced PHLDA1 protein directly binds to ErbB3, whereas knockdown of PHLDA1 increases complex formation between ErbB3 and ErbB2. To provide insight into the mechanism for our time-course and single-cell experimental observations, we performed a systematic computational search of network topologies of the mathematical models based on receptor dimer-tetramer formation in the ErbB activation processes. Our results indicate that only a model in which PHLDA1 inhibits formation of both dimers and tetramer can explain the experimental data. Predictions made from this model were further validated by single-molecule imaging experiments. Our studies suggest a unique regulatory feature of PHLDA1 to inhibit the ErbB receptor oligomerization process and thereby control the activity of receptor signaling network.
Keywords: breast cancer, mathematical modeling, oligomerization, receptor tyrosine kinase, signal transduction
Introduction
The ErbB receptor signaling pathway plays important roles in a variety of physiological processes in mammalian cells, and its dysregulation is frequently associated with development of human cancers (1). Therefore, a system level understanding of the ErbB signaling network is important for uncovering the regulatory mechanisms of disease progression. The ErbB receptors, EGFR5 (ErbB1), ErbB2, ErbB3, and ErbB4, are activated by ligand binding and trans-phosphorylated through their homo- and heterodimerization. Ligand-stimulated, tyrosine-phosphorylated receptors recruit adaptor proteins and effector kinases. This signal transduction cascade subsequently activates extracellular signal-regulated kinase (ERK) and Akt, which turn on the transcriptional program (2–6). At present, there are 13 known ErbB ligands, including epidermal growth factor (EGF) and heregulin (HRG) (7). The combination of those ErbB ligands and receptors enables this signaling pathway to evoke a wide range of quantitatively different responses. The potency and duration of ErbB signaling responses are also controlled by feedback mechanisms. EGF-activated EGFR is rapidly internalized from the cell surface and decreased in abundance by ubiquitination (8, 9). The activity of EGF-activated ERK is decreased by Raf-1 negative feedback (10). Negative feedback regulation mediated by post-translational modifications rapidly attenuates the input signal and thus induces transient responses. There is an additional class of transcriptionally-inducible negative feedback regulators in ErbB signaling pathways. Such examples include Mig6 and dual specificity MAPK phosphatase, which are induced upon receptor activation to suppress EGFR and ERK activities, respectively (11, 12). In general, in contrast to the rapid feedback regulation mediated by post-translational modification of signaling proteins, transcriptionally-induced negative regulators modulate signaling activity on a longer time scale, intimately crucial consequences for the time-dependent process of cellular transitions.
Pleckstrin homology-like domain, family A, member 1 (PHLDA1), has been implicated in the regulation of cell death (13) and suppression of metastasis (14), and its mRNA expression is often reduced in human cancers (15). PHLDA2 and PHLDA3, other PHLDA family proteins, were known to attenuate oncogenic PI3K-Akt activity (16, 17). PHLDA1 is one of the early response genes in growth factor-stimulated cells (18–20). Although PHLDA1 has been reported to be a negative regulator of ErbB-signaling pathways and significantly enhances the sensitivity of ErbB2-positive breast cancer cells to lapatinib (21), it has not been demonstrated how PHLDA1 regulates ErbB signaling at a network level. In this study, we have found using liquid chromatography-mass spectrometry (LC/MS) that PHLDA1 targets ErbB3 and thereby inhibits phosphorylation of ErbB receptors in HRG-stimulated MCF-7 cells. Although these experimental results suggest a role for PHLDA1 in negative regulation of the receptors, single-cell data have shown that the expression of PHLDA1 and phospho-ErbB2 are positively correlated, even at the time when phosphorylation of ErbB2 is attenuated and PHLDA1 expression is increased. These results suggested a complex inhibitory mode of PHLDA1 in ErbB receptor activation. Mathematical models, including ErbB receptor activation processes such as dimerization, phosphorylation, and tetramer formation with different inhibitory modes of PHLDA1, demonstrated that only a model containing inhibition of both dimer and tetramer formation could explain the experimental data. Live cell single-molecule imaging analysis demonstrated that ligand–receptor interactions closely mimicked the computational predictions. Our study suggests that PHLDA1 inhibits higher-order oligomerization of the ErbB receptor via a transcriptionally-induced feedback mechanism.
Results
PHLDA1 induced by HRG stimulation modulates the ErbB receptor signaling pathway
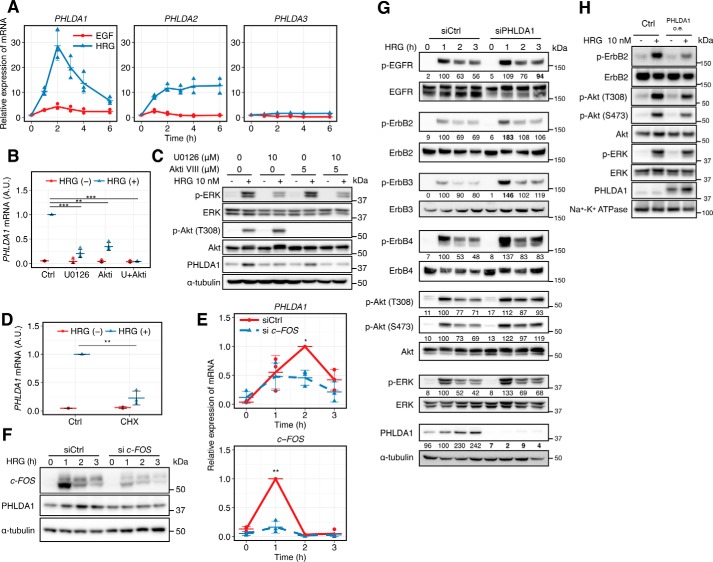
We first used qRT-PCR to examine time-course mRNA expression of the PHLDA family genes, PHLDA1, PHLDA2, and PHLDA3, in HRG-stimulated MCF-7 cells (Fig. 1A). Expression of PHLDA1 mRNA increased about 30-fold after HRG ligand stimulation, with a peak maximum at 120 min. PHLDA2 mRNA showed a sustained increase, but the amount of PHLDA3 mRNA was not increased by HRG. Expression levels of PHLDA1 and PHLDA2 were more increased by HRG compared with EGF. We tested several kinase inhibitors, U0126 (a mitogen-activated protein kinase/extracellular signal-regulated kinase kinase inhibitor), wortmannin (a PI3K inhibitor), and trastuzumab (an ErbB2 inhibitor), to identify the induction pathways using a microarray platform (Fig. S1). As a result, expression of PHLDA1 was suppressed by all three inhibitors. As shown in Fig. 1B, U0126 and the Akt inhibitor VIII, a specific inhibitor targeting Akt1 and -2, decreased the induction of PHLDA1 mRNA at 2 h after HRG stimulation. These results suggest that PHLDA1 mRNA induction is dependent on both Ras-ERK and PI3K-Akt pathways. These pathways also affected PHLDA1 protein levels at 3 h after HRG stimulation (Fig. 1C, quantification values are shown in Fig. S2). PHLDA1 mRNA expression induced by HRG is suppressed by the protein synthesis inhibitor cycloheximide (CHX) (Fig. 1D) and siRNA targeting c-FOS (Fig. 1E) as well, suggesting that de novo synthesis of the c-FOS transcription factor is necessary prior to PHLDA1 mRNA expression. We confirmed that c-FOS knockdown decreased the induction of PHLDA1 proteins (Figs. 1F and Fig. S3). In contrast, PHLDA1 siRNA moderately increased phosphorylation of ErbB receptors, Akt (Thr-308 and Ser-473) and ERK (Fig. 1G). Among the molecules we analyzed for phosphorylation, ErbB2 was most affected (1.8 times higher than the control), and the phosphorylation of EGFR, ErbB2, and ErbB3 was significantly up-regulated by PHLDA1 knockdown (p < 0.05, Welch's statistical test, Fig. S4). Consistent with the above findings, PHLDA1 overexpression inhibited phosphorylation of ErbB2, Akt, and ERK in the plasma membrane fraction with statistical significance (Fig. 1H and Fig. S5), implying that PHLDA1 is responsible for negative regulation of the ErbB signaling pathway.
Figure 1.
PHLDA1 inhibits the ErbB receptor pathway. A, time course of relative amounts of PHLDA gene family transcripts in ligand-stimulated MCF-7 cells. The blue line shows the cells stimulated with HRG, and the red line shows stimulation with EGF. Data were normalized so that the non-stimulated condition is designated as 1. B, effect of U0126 (10 μm) and Akt inhibitor VIII (5 μm) on PHLDA1 induction at 2 h after HRG stimulation. Data were normalized so that the HRG-stimulated condition is designated as 1. C, effect of U0126 (10 μm) and Akt inhibitor VIII (5 μm) on PHLDA1 protein induction at 3 h after HRG stimulation. The blotting-determined PHLDA1 levels are shown in Fig. S2. D, effect of cycloheximide (CHX) (10 μg/ml) on PHLDA1 mRNA induction at 2 h after HRG stimulation. Data normalization was done the same way as in B. Ctrl, control. E and F, effect of c-FOS siRNA on PHLDA1 mRNA (E) and protein (F) expression levels. E, data were normalized so that the highest value in all conditions is designated as 1. G, effect of PHLDA1 knockdown on ErbB receptor signaling. After transfection of PHLDA1 or control siRNA, MCF-7 cells were stimulated with 10 nm HRG for the indicated time periods and subjected to Western blotting. The digital values were annotated under each lane. The band intensities of phosphorylated proteins were quantified by dividing the total protein, and the band intensities of PHLDA1 were quantified by dividing α-tubulin. Then the values were normalized so that the value of the siCtrl sample with HRG treatment for 1 h is designated as 100. The values that have statistical significance are presented in boldface. H, effect of PHLDA1 overexpression on the plasma membrane fraction. After vector transfection, MCF-7 cells were stimulated with 10 nm HRG for 5 min. A, B, D, and E, each point represents the results of an independent experiment; colored bars indicate the average value of all experiments, and error bars denote standard deviation (S.D.) calculated from biological independent experiments (n = 3). The digital values of the band intensities in F, G, and H are shown in Fig. S3–S5, respectively. Data in B, D, and E, two-tailed Welch's test: *, p < 0.05; **, p < 0.01; ***, p < 0.001. A.U. indicates relative expression value of PHLDA1 mRNA in B and D.
HRG titration experiments under conditions where PHLDA1 was overexpressed showed that its inhibitory effect on ErbB2 phosphorylation was only significant at higher HRG concentrations (Fig. S6A). Overexpression of PHLDA1 suppressed ErbB2 phosphorylation at higher ligand doses but did not affect the EC50 (4.1 nm in control and 6.0 nm in PHLDA1 overexpression conditions) (Fig. S6A). This non-competitive inhibitory profile indicates that PHLDA1 may indirectly inhibit ErbB2 phosphorylation by modulating unknown regulatory molecules or by inducing conformational changes but not by competing with ErbB2 kinase activity. A similar phenomenon was also observed with cells that were first treated with 1 nm HRG for 180 min followed by a second treatment with different amounts of HRG (Fig. S6B). These results suggest that the ErbB signaling network is negatively regulated by mechanisms that at least in part include PHLDA1. From the current experimental results (Fig. 1, G and H, and S6A) and our previous study (18), we concluded that 10 nm HRG is sufficient to induce phosphorylation of ErbB2 and expression of PHLDA1 for the following experiments.
PHLDA1 negatively regulates ErbB2 through interaction with ErbB3
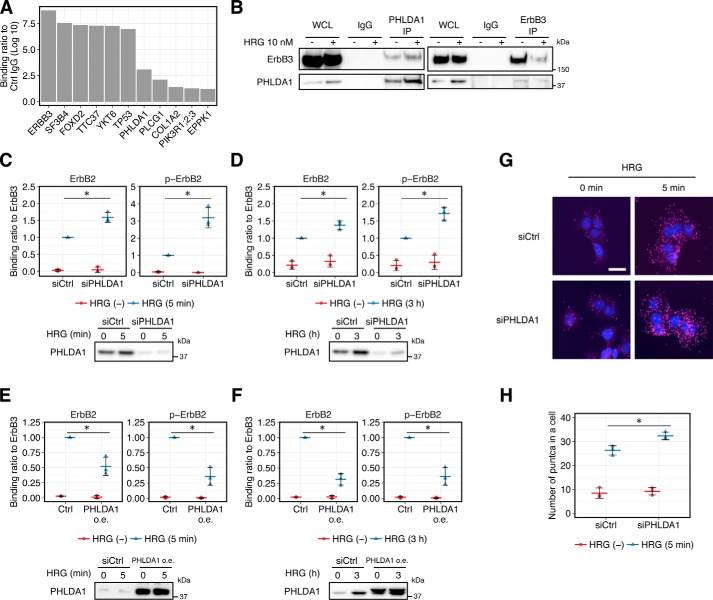
Next, to further clarify the inhibitory mechanism of PHLDA1 on ErbB activation, we investigated PHLDA1-binding partners using LC/MS. Immunoprecipitates of HRG-stimulated MCF-7 samples using an anti-PHLDA1 antibody contained proteins such as ErbB3, TP53, PLCG1, and PIK3R1, -2, or -3 (PIK3R1/2/3) with ErbB3 having the highest score (Fig. 2A). The ErbB3–PHLDA1 interaction was further confirmed by co-immunoprecipitation (co-IP) and immunoblot analysis (Fig. 2B). In this experiment, less ErbB3 is immunoprecipitated under HRG-stimulated conditions, which may be due to modification of the antibody recognition site on ErbB3. Association between PHLDA1 and ErbB3 was also observed in an earlier study (22). Therefore, we hypothesized that PHLDA1 might interrupt phosphorylation of ErbB receptors by binding to ErbB3.
Figure 2.
Effect of PHLDA1 on ErbB receptor activation. A, binding score of the proteins that were co-immunoprecipitated with PHLDA1 antibody in HRG-stimulated MCF-7 cells. Proteins co-IP with the anti-PHLDA1 antibody were identified by LC/MS analysis. The indicated values are the log10-transformed ratio of the LFQ intensities of PHDLA1-IP over the negative control mouse IgG. The proteins with less than 1 (log10 ratio) are not shown. The graph is a representative of two experiments. B, co-IP experiment with PHLDA1 antibody or ErbB3 antibody (sc-7390). WCL, whole-cell lysate. C and D, top, effect of PHLDA1 knockdown on the physical interaction between ErbB3 and phosphorylated ErbB2 or ErbB2 at 5 min (C) or 3 h (D) after 10 nm HRG stimulation. Bottom, blot confirming the knockdown of PHLDA1. E and F, top, effect of PHLDA1 overexpression on the physical interaction between ErbB3 and phosphorylated ErbB2 or ErbB2 at 5 min (E) or 3 h (F) after 10 nm HRG stimulation. Bottom, blot confirming the overexpression of PHLDA1. oe, overexpression. C, D, E, and F, graphs show the relative intensities of the phospho-ErbB2 or ErbB2 bands divided by that of total ErbB3. Data were normalized so that the value of the HRG-stimulated control (Ctrl) condition is designated as 1, n = 3. Each point represents the result of an independent experiment, and colored bars indicate the average value of all experiments and error bars denote S.D. Two-tailed Welch's test: *, p < 0.05. Representative raw blotting data are shown in Fig. S7. G, effect of PHLDA1 knockdown on hetero-oligomerization between phospho-ErbB2 and ErbB3 by PLA. DAPI staining is in blue and magenta puncta represent individual oligomers. Scale bar, 30 μm. H, quantification of magenta puncta per cell in the PLA. Each point represents the result of an independent experiment, and colored bars indicate average values of all experiments, and error bars denote S.D. Two-tailed Welch's test: *, p < 0.05.
HRG is a growth factor that preferentially binds to ErbB3 and ErbB4 receptors and induces strong phosphorylation of the ErbB2 receptor through receptor heterodimerization (23). In MCF-7 cells, it is thought that the main partner of ErbB3 in the heterodimer is ErbB2, because ErbB4 is only weakly expressed (24, 25). We therefore examined whether the amount of PHLDA1 expression affects the interaction between ErbB3 and phosphorylated ErbB2 at 5 and 180 min after HRG stimulation, when phosphorylation of the ErbB receptor reaches its peak maximum and when the cells show a sufficient amount of PHLDA1 expression, respectively. As a result, knockdown of PHLDA1 increased the interaction between ErbB3 and phospho-ErbB2 as well as the interaction ErbB3 and ErbB2 after HRG stimulation at both 5 and 180 min (Fig. 2, C and D), whereas overexpression of PHLDA1 decreased this interaction (Fig. 2, E and F). These results suggest that PHLDA1 affects not only the amount of phosphorylated ErbB receptor but also the amount of ErbB receptor oligomers containing both ErbB2–ErbB3 association. We also confirmed the effect of PHLDA1 knockdown on receptor dimerization by using proximity ligation assay (PLA). PLA is a technology that enables detection of protein–protein interaction, similar to a colocalization analysis in immunostaining, and it can be applied for detection of ErbB receptor complex formation (26, 27). In this assay, bright fluorescent puncta can be detected only when two antibodies recognizing ErbB3 and phospho-ErbB2 are in proximal regions (i.e. these proteins form a complex). We found that knockdown of PHLDA1 increases complex formation between phosphorylated ErbB2 and total ErbB3 proteins after HRG stimulation (Fig. 2, G and H). Overall, these data support the hypothesis that PHLDA1 negatively regulates the transactivation of ErbB2 receptor through interaction with ErbB3.
Despite its negative regulatory role, PHLDA1 expression positively correlates with ErbB2 phosphorylation at a single-cell level
ErbB receptor signal response in MCF-7 cells is heterogeneous across cell populations (28, 29), and the amount of PHLDA1 expression is moderate. Therefore, it is necessary to quantitatively assess the activation status of the ErbB-signaling pathway and PHLDA1 expression at a single-cell level to confirm the inhibitory function of PHLDA1.
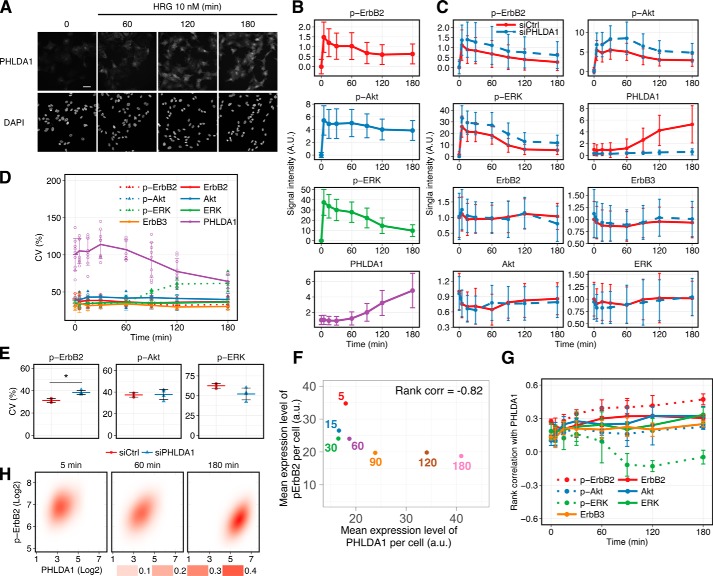
We obtained the averaged single-cell time course of PHLDA1 expression and phosphorylation of ErbB2, ERK, and Akt using immunofluorescence-based imaging cytometry (Fig. 3, A and B). These data are consistent with mRNA expression and Western blotting data obtained from bulk cell experiments (Fig. S8). Cell population average behaviors of the same molecules in the PHLDA1 knockdown condition are also consistent with the Western blotting data (Figs. 1G and 3C and Fig. S4). Regardless of the large standard deviation (S.D.) of signal intensities in each cell population (because these values are dependent on sample size; in our experiments >1,500 cells in each condition), statistical analysis supported the hypothesis that the amount of phospho-ErbB2, phospho-ERK, and phospho-Akt in control and PHLDA1 knockdown conditions are statistically different (p value <1.0 × 10−20, Welch's statistical test). The coefficient of variation (CV) is one of the indexes for evaluating cell-to-cell variability in a population. The CV of PHLDA1 decreased over time while its expression is increased (Fig. 3D). Knockdown of PHLDA1 increased the CV of phospho-ErbB2, although it did not significantly affect those of phospho-Akt and phospho-ERK at 180 min after HRG stimulation (Fig. 3E). Thus, elimination of PHLDA1 from the ErbB network resulted in increased cell-to-cell variation in phospho-ErbB2.
Figure 3.
HRG-induced PHLDA1 expression and ErbB phosphorylation were determined by using imaging cytometry. A, immunostaining of PHLDA1 in MCF-7 cells. Cells were treated with/without 10 nm HRG. Top, PHLDA1; bottom, DAPI. White scale bar, 50 μm. B, time-course pattern of PHLDA1 expression and phosphorylation of ErbB2, ERK, and Akt. The graphs represent the average dynamics of single-cell measurements. Error bars denote S.D. of signal intensities in a single cell. Similar results obtained by Western blotting are shown in Fig. S8. C, PHLDA1 knockdown experiments using an imaging cytometer. The graphs represent the average dynamics of single-cell measurements in control and knockdown conditions. Error bars denote S.D. of signal intensities in a single cell. D, coefficient of variation at each time point was calculated from the results of single-cell imaging. Error bars denote S.D. from at least three independent experimental values. E, effect of PHLDA1 knockdown on the CV of ErbB signaling at 180 min after HRG stimulation. Each point indicates the result of an independent experiment, and colored bars indicate the average value of all experiments. Error bars denote S.D., n = 4. Two-tailed Welch's test: *, p = 1.3 × 10−3. F, relationship between PHLDA1 and phospho-ErbB2 in single-cell measurement experiments. Mean expression levels of both proteins were calculated from experimental data (details are described under “Mean expression level per cell” under “Experimental procedures”). The numbers represent time points. Spearman's rank correlation coefficient was −0.82. G, rank correlation coefficient at each time point was calculated from the results of single-cell imaging. Error bars indicate S.D. of at least three independent experiments. H, 2D probabilistic density of phospho-ErbB2 and PHLDA1 in a cell population stimulated with 10 nm HRG. Each panel contains at least 1500 cells. B and C, method for data normalization is described in detail under “Experimental procedures.” A.U. indicates relative signal intensity.
Mean expression levels of PHLDA1 and phospho-ErbB2 per cell at each time point (see “Mean expression level per cell” under “Experimental procedures” for details) (Fig. 3F) indicated that the mean expression level of PHLDA1 increased along with decreased phospho-ErbB2 levels (rank correlation = −0.82) indicating that PHLDA1 negatively affects the phosphorylation of ErbB2 after HRG stimulation. However, despite these data, in individual cells the PHLDA1 expression level positively correlated with the phospho-ErbB2 expression level at each time point (Fig. 3, G and H). We confirmed that this positive correlation was not due to an artificial effect of the phosphorylated ErbB2-specific antibody (Fig. S9). To explain this discrepancy, we therefore hypothesized that PHLDA1 might not directly inhibit receptor phosphorylation but instead might inhibit other steps in the ErbB receptor activation processes, for example, formation of receptor dimers and oligomers. Indeed, several studies have demonstrated the existence of higher-order ErbB receptor oligomers (30–33). Moreover, an earlier study suggested that tetramer formation between ErbB2 and ErbB3 is functionally important for potent signal transduction (34). Therefore, we further examined the effect of PHLDA1 on activation of ErbB receptors, including higher-order oligomer formation.
Prediction of the PHLDA1 inhibition mode using simple mathematical models
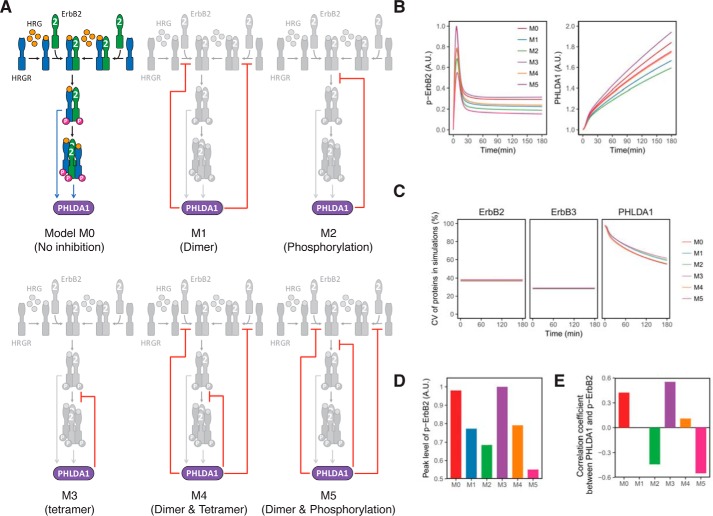
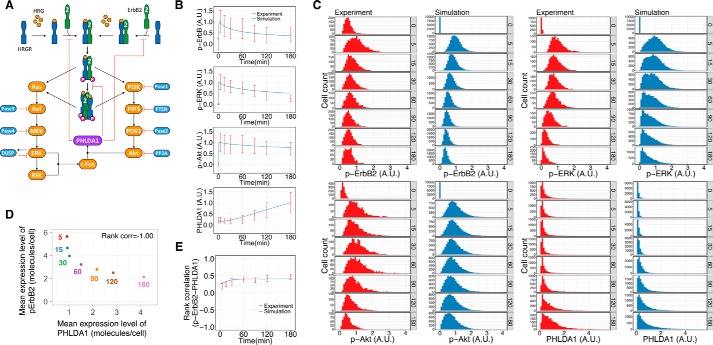
To identify the inhibitory mode of PHLDA1 in HRG-induced ErbB receptor activation processes, we constructed six simple mathematical models to explore network topology that can explain our experimental data. For simplification purposes, the models are described in a way such that phosphorylated ErbB heterodimers and tetramers directly induce PHLDA1 expression. In the model, we considered that the main population of HRG-binding ErbB receptors in MCF-7 cells, termed HRGR, is ErbB3, because its amount is about 70 times higher than ErbB4 (25). Based on an earlier study (34), the models include the formation of tetramers composed of the orphan receptor ErbB2 and HRGR complexes (Fig. 4A). The ErbB receptor activation scheme is described as follows: 1) formation of inactive heterodimers between ErbB2 and HRGR prior to HRG stimulus; 2) binding of HRG to HRGR, both monomers and heterodimers; 3) formation of heterodimers between ErbB2 and HRG-bound HRGR (ErbB2/HRGR); 4) phosphorylation of ErbB2/HRGR; and 5) formation of tetramers consisting of two phosphorylated ErbB2/HRGR. For the PHLDA1-mediated regulation, six types of inhibitory modes are considered as follows: model M0, no inhibition from PHLDA1 to HRGR activation; model M1, inhibition of the 1st and 3rd reaction steps; model M2, inhibition of the 4th reaction step; model M3, inhibition of the 5th reaction step; model M4, inhibition of the 1st, 3rd, and 5th reaction steps; and model M5, inhibition of the 1st, 3rd, and 4th reaction steps (Fig. 4A, see Tables S1 to S5, and supporting Methods for details of the models). We performed stochastic simulation of each model using experimentally obtained CVs of ErbB2, ErbB3, and PHLDA1 expression (Fig. 3D). The averaged dynamics of phospho-ErbB2 and PHLDA1 (Figs. 3B and 4B), in addition to time courses of CVs of ErbB2, ErbB3, and PHLDA1 proteins (Figs. 3D and 4C) in all models and the experimental data, were consistent with each other. However, model M3 was excluded from the network candidates because the peak intensities of phosphorylated ErbB2 were down-regulated in models M1, M2, M4, and M5 relative to PHLDA1, which is consistent with the data, but not in model M3 (Figs. 1, G and H, and 4D).
Figure 4.
Simple mathematical models of the activation of ErbB receptors. A, six models describing the inhibitory function of PHLDA1 on ErbB receptor activation. B, computational simulation of phospho-ErbB2 and PHLDA1 in each model. The graphs represent the average dynamics of 10,000 simulations. The colored lines correspond to the six models shown in A. A.U. indicates relative signal intensity of p-ErbB2 (left) and relative abundance of PHLDA1. C, time-course pattern of CVs of total ErbB2, total ErbB3, and PHLDA1 in each simulation model. D, peak intensities of phospho-ErbB2 in each model. A.U. indicates relative peak intensity of p-ErbB2. E, rank correlation between phospho-ErbB2 and PHLDA1 in each model at 180 min after HRG stimulation.
Next, we calculated rank correlation coefficients between PHLDA1 and phosphorylated ErbB2 in the models to evaluate the single-cell experimental data. The analysis revealed that the models that contain PHLDA1 inhibition of dimer formation or phosphorylation tend to show negative correlation coefficients (such as model M2 where PHLDA1 inhibits dimer phosphorylation) (Fig. 4E). In contrast, the models containing inhibition of tetramer formation show positive correlation coefficients (model M3) (Fig. 4E). Those inhibitory effects seemed to be additive because model M4 (in which PHLDA1 inhibits dimer and tetramer formation) showed a positive correlation, and model M5 (in which PHLDA1 inhibits both dimer formation and phosphorylation) showed a negative correlation. Thus, we hypothesized that the correlation coefficients between PHLDA1 expression and phospho-ErbB2 could be modulated by the PHLDA1-mediated inhibition of receptor oligomerization. Thus, only model M4 could satisfy our experimental observations (Figs. 3G and 4E) and the apparently contradictory experimental data (Figs. 1G and 3H).
Model with inhibition of receptor oligomerization could account for single-cell signal response
To confirm that the simple topological model of M4, in which PHLDA1 inhibits the higher-order oligomerization of ErbB receptors, quantitatively reflects the pathway response, we constructed a detailed mathematical model of the entire ErbB signaling pathway, including downstream Ras-ERK and PI3K-Akt modules and c-FOS-mediated PHLDA1 induction (Fig. 5A). A detailed scheme of our model is described in the supporting Methods and Tables S6–S9. The kinetic parameters in the model were fitted to account for the average time course of phospho-ErbB, phospho-ERK, phospho-Akt, and PHLDA1 obtained from single-cell experiments (Fig. 5B). We performed stochastic simulations with cell-to-cell variability using experimentally obtained CVs of ErbB2, ErbB3, ERK, Akt, and PHLDA1 (Fig. 3D). The resulting simulations reproduced the heterogeneous responses of those molecules at a single-cell level (Fig. 5C). As shown in Fig. 5D, the mean expression level of PHLDA1 per cell increased with decreasing phosphorylated ErbB2 as well as in the experimental results (Fig. 3F). In addition, the time-course pattern of rank correlation between phosphorylated ErbB2 and PHLDA1 calculated from simulation results reasonably fitted that observed experimentally (Fig. 5E). Thus, our simulation results suggested that a mechanism in which PHLDA1 inhibits ErbB2-ErbB3 oligomer formation can explain the experimentally observed time-course profiles of the receptor, Akt and ERK, activities suppressed by transcriptionally-induced PHLDA1 and their single-cell positive correlation.
Figure 5.
Mathematical simulation considering PHLDA1 and experiments of HRG-induced ErbB receptor signaling including PHLDA1. A, mathematical model of the ErbB-PHLDA1 network. Details of the model construction are described in the supporting information. B, time-course kinetics of phospho-ErbB2, phospho-Akt, phospho-ERK, and PHLDA1 expression after treatment of MCF-7 cells with 10 nm HRG. Red plots represent average signal intensity detected experimentally by imaging cytometry (shown in Fig. 3B). Blue lines represent averaged dynamics of each species in the simulation results. Each time-course plot is normalized so that the maximum value is designated as 1. Top, A.U. indicates relative signal intensity of p-ErbB2; 2nd from top, A.U. indicates relative signal intensity of p-ERK; 3rd from top, A.U. indicates relative signal intensity of p-Akt; bottom, A.U. indicates relative abundance of PHLDA1. C, time-series histogram of phospho-ErbB2, phospho-Akt, phospho-ERK, and PHLDA1 in a cell population stimulated with 10 nm HRG (red, single-cell experiment by imaging cytometry; blue, 10,000 times of simulation). Each plot is normalized so that the maximum of average signal intensity of a cell population in the time-course is designated as 1. Upper panel from left to right: A.U. indicates relative signal intensity of p-ErbB2 in 1st and 2nd panels and indicates relative signal intensity of p-ERK in 3rd and 4th panels. Lower panel from left to right: A.U. indicates relative signal intensity of p-Akt in 1st and 2nd panels and indicates relative abundance of PHLDA1 in 3rd and 4th panels. D, relationship between PHLDA1 and pErbB2 in the simulation. Mean expression levels of both proteins were calculated from simulation results (details are described under “Mean expression level per cell under the “Experimental procedures”). The numbers represent time points. Spearman's rank correlation coefficient was −1.00. E, time-course patterns of rank correlation between phospho-ErbB2 and PHLDA1 (red, experiment; blue, simulation). Error bars denote S.D., n = 3.
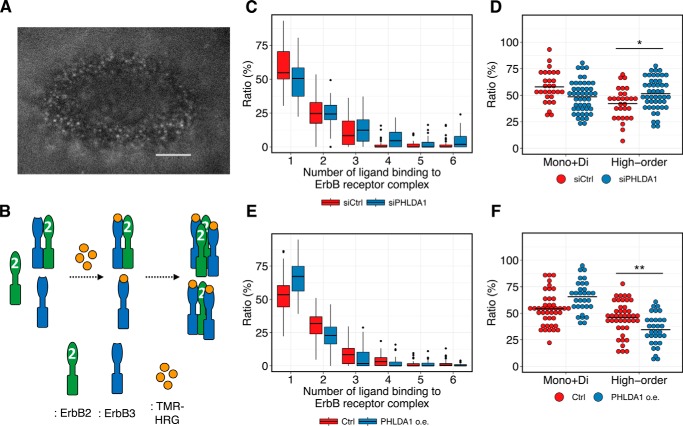
Single-molecule imaging of HRG–HRGR complexes confirmed that PHLDA1 modulates the amount of higher-order ErbB receptor oligomers
To experimentally test the model-driven hypothesis that PHLDA1 inhibits oligomerization of ErbB receptors, we examined the association of fluorescent-labeled HRG (carboxytetramethylrhodamine (TMR)-HRG) with ErbB receptors on the apical surface of living MCF-7 cells using oblique illumination fluorescence microscopy (Fig. 6A) (35). A fluorescent spot emitted by a single TMR-HRG molecule detected in this experiment indicates the presence of either an HRG-bound ErbB3 monomer or a heterodimer between HRG-bound ErbB3 and an unliganded partner such as ErbB2 (Fig. 6B, middle complexes). Otherwise, a fluorescent spot whose intensity indicates more than one TMR-HRG molecule suggests the existence of an ErbB heterooligomer containing at least two HRG-bound ErbB3 receptors and a heterooligomeric partner such as ErbB2 (Fig. 6B, right complexes). Although it is thought that HRG-bound ErbB3 cannot form a homodimer (36), a few studies have suggested that this is a possibility (37, 38). However, it is still unclear whether the direct interaction of HRG-bound ErbB3 homodimers exists and functions as a signal initiator in MCF-7 cells; therefore, we did not take the ErbB3 homodimer into consideration in our model. In our experiments, we could quantify the amount of higher-order ErbB receptor oligomers that contain at least two ErbB3 molecules, which may possibly include an HRG-bound ErbB3 monomer and a heterodimer between HRG-bound ErbB3 and ErbB2. Using this approach, we could predict the degree of ErbB receptor association by measuring the fluorescent intensity of each spot and then calculate the ratio of ErbB higher-order oligomers to the total number of HRG-bound ErbB receptors. Knockdown of PHLDA1 increased this ratio (Fig. 6C, the ratio of 2–6 HRG-bound ErbB receptors per all HRG-bound ErbB receptors) and decreased the fraction of both liganded monomer and heterodimer (Fig. 6C, the ratio of one HRG-bound ErbB receptor per all HRG-bound ErbB receptors; summarized in Fig. 6D). Consistent with the above, overexpression of PHLDA1 decreased the ratio of ErbB higher-order oligomers and increased the ratio of both liganded monomers and heterodimers (Fig. 6, E and F). These experimental data confirm our modeling studies and indicate that PHLDA1 indeed modulates ErbB receptor oligomer formation in MCF-7 cells.
Figure 6.
Single-molecule imaging of TMR-HRG on the cell surface of MCF-7 cells. A, representative image of single-molecule imaging. Scale bar, 5 μm. B, illustration of interpretation of the results of single-molecule imaging. C–F, box plots of the ratio of ErbB higher-order oligomers affected by PHLDA1 knockdown (C) and overexpression (E). Summarized plots generated from the same data are shown in D and F, respectively. Each point indicates a result in a single cell. Black horizontal lines indicate the mean value of each condition. Two-tailed Welch's test was performed: *, p = 9.7 × 10−3; **, p = 2.2 × 10−3.
Knockdown of PHLDA1 accelerates differentiation of MCF-7 cells
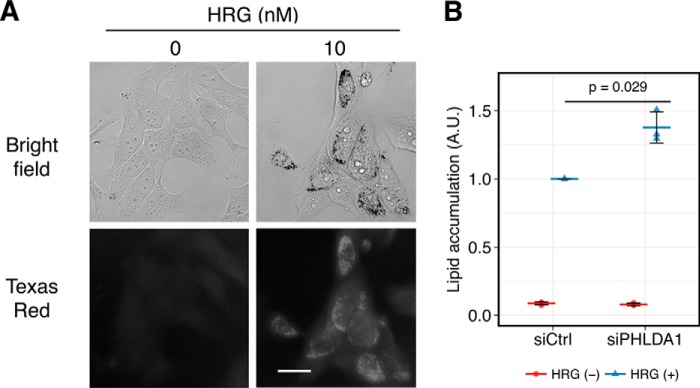
Finally, we examined the biological function of PHLDA1 in the MCF-7 system. In a previous study, it was shown that HRG-stimulated MCF-7 cells undergo cellular differentiation as indicated by lipid accumulation (18, 39). We confirmed that HRG treatment induced lipid accumulation (Fig. 7A), and this process was accelerated by knockdown of PHLDA1 (Fig. 7B). Thus, our data indicate that PHLDA1 negatively controls cell differentiation through inhibition of ligand-dependent ErbB receptor activation.
Figure 7.
Effect of PHLDA1 knockdown on differentiation of MCF-7 cells. A, Oil Red O staining of serum-starved MCF-7 cells treated with/without 10 nm HRG. Top, bright field; bottom, Texas Red fluorescence. Scale bar, 30 μm. B, total intensities of Oil Red-positive puncta in a single cell were measured by imaging cytometry. The values were normalized so that the value of HRG-treated siCtrl samples was 1. Each point is an independent result of an experiment, and colored bars indicate the average value of all experiments. Error bars denote S.D., n = 3. A.U., arbitrary units.
Discussion
Our study revealed that PHLDA1 is transcriptionally induced by HRG-mediated ErbB receptor activation via the Ras-ERK and PI3K-Akt pathways, and it inhibits oligomerization of ErbB2-ErbB3 receptors, suppressing their downstream signaling. Using a proteomics approach, we detected several proteins, including TP53, PLCG1, and PIK3R1, -2, and -3, in addition to ErbB3, as PHLDA1-binding proteins (Fig. 2A). PIK3R1, also called p85, is known as a PI3K regulatory subunit, and it binds to ErbB3 when the ErbB receptor complex is activated. Therefore, the binding of p85 to ErbB3 can be detected via PHLDA1-ErbB3 binding. To date, a number of reports have demonstrated that PHLDA1 has both pro- and anti-tumorigenic function, depending on the cellular context. PHLDA1 was first identified as a modulator of T cell apoptosis (13). Later, it was found that PHLDA1 is responsible for regulation of apoptosis, autophagy, and chemotaxis in normal tissues as well as several types of cancer (14, 15, 40–42). In contrast, PHLDA1 is overexpressed in human tumors and contributes to cell migration and tumorigenesis (43, 44). In our analysis, PHLDA1 knockdown accelerated HRG-mediated differentiation of MCF-7 cells, as manifested by accumulation of lipid droplets (Fig. 7), in a manner similar to that previously suggested for 3T3-L1 cells (45). Therefore, the role of PHLDA1 in cell differentiation seemed to be inhibitory.
From a systems biology point of view, as a transcriptionally-inducible negative feedback regulator, PHLDA1 has functions in common with other inducible feedback inhibitors, such as MIG6, SOCS4, and SOCS5, in EGFR signaling. For example, MIG6 is transcriptionally induced by EGFR activation and directly and specifically binds to the active form of the EGFR kinase domain (46). However, because PHLDA1 inhibits ErbB receptor oligomer formation, the inhibitory mechanisms of MIG6 and PHLDA1 to attenuate the pathway are distinct from each other. These studies indicate that multilayered negative feedback mechanisms cooperate to ensure the suppression of ErbB receptor activity. In general, a negative feedback mechanism can increase the signal-to-noise ratio in system output by decreasing cell-to-cell variation (47). Our results showed that PHLDA1 also functions to suppress cell-to-cell variability of phospho-ErbB2 (Fig. 3E). In this study, we demonstrate that whereas ErbB2 phosphorylation is a crucial step in pathway activation, measuring its average value in a population of cells is not sufficient for predicting regulatory mechanisms of pathways. Our mathematical analysis together with quantitative single-cell analysis proved to be a useful combination for identifying the novel function of this novel signal regulator.
Experimental procedures
Cell culture, treatment, and fractionation
Cultivation of the MCF-7 cell line and stimulation with growth factors were performed as described previously (48). For inhibitor assays, U0126, Akt inhibitor VIII (Merck Millipore, Billerica, MA), and cycloheximide (Nacalai Tesque, Kyoto, Japan) were added 20 min prior to HRG stimulation. For preparation of total cell lysate, cells were lysed with Bio-Plex lysis buffer (Bio-Rad) after cell treatment and were centrifuged at 12,000 × g for 15 min. The supernatant was used as the total cell lysate fraction. For preparation of the plasma membrane fraction and the corresponding cytosol fraction, a protocol earlier described by Dunn and Connor (49) was used.
Gene silencing with siRNA
Reverse transfection was performed by using Hiperfect reagent (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Trypsinized MCF-7 cells were resuspended in antibiotic-free medium and then mixed with a suspension of Opti-MEM (Thermo Fisher Scientific, Grand Island, NY) containing 10 nm siRNA and Hiperfect reagent in 100-mm dishes (for membrane fractionation and co-IP), 12-well plates (for Western blotting), or 96-well plates (for immunostaining). SMARTpool ON-TARGETplus siRNA targeting PHLDA1 (L-012389-00) and c-FOS (L-003265-00), and ON-TARGETplus Non-targeting Pool (D-001810-10) were purchased from Dharmacon (GE Healthcare).
Gene overexpression
MCF-7 cells were seeded at 3 × 106 cells per 100-mm dish. After overnight incubation, cells were transfected with 5 μg of expression vector using Lipofectamine LTX and Plus Reagent (Thermo Fisher Scientific) in Opti-MEM according to the manufacturer's protocol. After 48 h, cells were starved for 16 h in serum-free Dulbecco's modified Eagle's medium, then stimulated with 10 nm HRG for the designated periods, harvested, and then lysed for assays.
Western blot analysis
Protein phosphorylation and total proteins were analyzed as described previously (48). Antibodies specific for the following proteins were purchased: ErbB3 (sc-285), ErbB4 (sc-283), and PHLDA1 (sc-23866) from Santa Cruz Biotechnology (Santa Cruz, CA); phospho-Akt (Ser-473, no. 9271), phospho-Akt (Thr-308, no. 2965), pan-Akt (40D4, no. 2920), phospho-EGFR (Tyr-1068, no. 2234), phospho-ErbB2 (Tyr-1221/1222, no. 2249), phospho-ErbB3 (Tyr-1289, no. 4791), phospho-ErbB4 (Tyr-1284, no. 4757), phospho-ERK1/2 (Thr-202/Tyr-204, no. 4370), ErbB2 (no. 2165), and ERK1/2, (no. 9102) from Cell Signaling Technology (Beverly, MA); α1 sodium potassium ATPase (ab7671) and α-tubulin (ab15246) from Abcam (Cambridge, MA); and EGFR (20-ES04) from Fitzgerald (North Acton, MA). Blots show representative results from one of at least three independent experiments. After Western blotting, protein band intensities were quantified using ImageJ software.
qRT-PCR
cDNA synthesis was done by using ReverTra Ace® (TOYOBO, Osaka, Japan). Equivalent volumes of cDNA were used for all PCRs, which were performed using KOD SYBR® qPCR Mix (TOYOBO) in the Thermal Cycler Dice Real Time System TP800 (Takara Bio, Otsu, Japan). The standard curve method was used to determine relative quantification of mRNA abundance with technical triplicates. For normalization of the qRT-PCR data, GAPDH expression was used as a control. Primers designed for PHLDA1 (PPH10228B) and PHLDA3 (PPH15380B) were purchased from Qiagen. The other primers designed for quantitative real-time RT-PCR analysis were as follows: PHLDA2, 5′-aatcacttggccagtttgct-3′ and 5′-gactggatgagggtgtcctg-3′; c-FOS, 5′-ctaccactcacccgcagact-3′ and 5′-aggtccgtgcagaagtcct-3′; and GAPDH, 5′-caaagttgtcatggatgacc-3′ and 5′-ccatggagaaggctgggg-3′.
Co-immunoprecipitation and LC/MS analysis
MCF-7 cells were washed on ice with ice-cold PBS twice and then collected in a lysis buffer containing 150 mm NaCl, 50 mm Tris-HCl (pH 7.5), 2 mm EDTA, 1% Nonidet P-40, supplemented with Complete protease inhibitor mixture and PhosSTOP phosphatase inhibitor mixture (Roche Diagnostics, Basel, Switzerland). Lysates were incubated for 15 min on ice and then centrifuged at 12,000 × g for 15 min at 4 °C. Supernatants containing the proteins were transferred into new micro tubes, and then 10 μl of beads and antibody were added to each tube. Protein G-agarose (Thermo Fisher Scientific) and PHLDA1 antibody (sc-23866, Santa Cruz Biotechnology) were used for LC/MS analysis to detect PHLDA1-binding partners, and ErbB3 antibody (sc-73964, Santa Cruz Biotechnology) was used for detecting interaction between ErbB3 and ErbB2. The supernatant was incubated for 1 h (for LC/MS) at 4 °C. After incubation, the beads were washed three times with a detergent-free lysis buffer and then subjected to further experimental analysis. LC/MS analysis was performed as described previously (50).
Proximity ligation assay
MCF-7 cells were seeded in 96-well plates, and the following day, cells were exposed to serum-free medium for 16 h. Then the cells were stimulated with HRG for 5 min, fixed with ice-cold MeOH for 5 min, and blocked blocking buffer (10% fetal bovine serum in Blocking ONE (Nacalai Tesque)). After blocking, cells were incubated with a combination of primary antibodies (against phospho-ErbB2 (06-229, Millipore) and ErbB3 (sc-81455, Santa Cruz Biotechnology)). Subsequent hybridization and ligation of PLA probes, amplification, and detection were performed using the manufacturer's instruction (Sigma). Fluorescence images were obtained using InCell Analyzer 2000 (GE Healthcare), and quantification of puncta was done using Developer toolbox software (GE Healthcare).
Immunostaining and imaging cytometry
MCF-7 cells were seeded at a density of 1 × 104 cells/well in 96-well plates for fluorescent imaging. The following day, culture medium was replaced with serum-free medium. After 16 h, cells were stimulated with HRG for the indicated period, fixed with 4% paraformaldehyde in PBS, and permeabilized with 0.1% Triton X-100 in PBS for 5 min. After washing with PBS, the cells were incubated in blocking buffer for 1 h and then stained with primary antibody at 4 °C. The next day, the cells were stained with fluorescent-labeled secondary antibodies (Dylight488-anti-mouse IgG and Dylight550-anti-rabbit-IgG, Thermo Fisher Scientific), and then stained with DAPI for detecting nuclei. Fluorescence images were obtained using InCell Analyzer 2000 (GE Healthcare), and image analysis was done using Developer tool software. The signal intensity of the protein expression at each time point was normalized to the average intensity of the value at time 0 (the average intensity at time 0 was set as 1). The signal intensity of each phosphorylated protein was normalized in the same way, and then the normalized intensity at time 0 was subtracted from that at each time point (the average intensity at time 0 was set as 0). Error bars denote the S.D. of signal intensities in a cell population.
Mean expression level per cell
For each time point (t), the mean expression level per cell (M) of phospho-ErbB2 and PHLDA1 is calculated from the normalized signal intensity of the protein of interest using Equation 1.
| (1) |
Here, ni is the number of cells with the protein expression level ei within i-th bin. We used 50 bins to perform the calculations, using the corresponding histograms of the numbers of cells with the expression intensity within each bin. The supporting information shows that the influence of the bin size on the calculated values vanishes if the number of bins is over 20 (Fig. S10A). Using M(t), Spearman's rank correlation coefficient between phospho-ErbB2 and PHLDA1 was calculated. Note that to compare our experimental data with simulation results, the data at time 0 were removed. This is because basal expression of phospho-ErbB2 is not considered in our mathematical model.
Mathematical modeling
We developed two types of mathematical models, the simple and the expanded models. The simple model was developed to simulate the regulation between ErbB and PHLDA1, and the expanded model was developed to simulate the entire ErbB-signaling pathway. The biochemical reactions in both models were described by ordinary differential equations (Tables S1 and S6), and the simulations were conducted using XPPAUT (51). The kinetic parameters in the simple model were constrained to satisfy detailed valance. In addition, the kinetic parameters in the expanded model, which reproduce the experimental data (Fig. 3B), were obtained by the evolutionary algorithm AGLSDC (52). In this study, cell-to-cell variability was defined as the difference in the signaling protein abundance between the individual cells, which was represented by sampling from log-normal distributed protein concentrations with various CV. Detailed descriptions and the simulation method are described in the supporting Methods.
Single-molecule imaging
The protocol for single-molecule imaging using TMR-labeled HRG has been described previously (35). In brief, MCF-7 cells were seeded onto glass coverslips. Overnight before the experiments, the culture medium was replaced with Dulbecco's modified Eagle's medium without fetal bovine serum and phenol red. Before the experimental observations, the culture medium was replaced with HBSS, and the coverslip was mounted on a metal culture chamber (Thermo Fisher Scientific), and the cells were observed with an oblique illumination microscope based on a Nikon TE2000 inverted fluorescence microscope. On the microscope, HBSS in the chamber was discarded, and then 600 μl of a 6 nm TMR-HRG solution was added. These operations were done at room temperature. Images of single TMR-HRG molecules on the cell surfaces were acquired using an EM-CCD camera (ImageEM; Hamamatsu Photonics, Hamamatsu, Japan) and were analyzed using in-house software.
Oil Red O staining
We slightly modified the previously published method (39) as follows: 0.4 × 105 cells/well were seeded in standard 24-well plates. Culture medium was replaced with serum-free medium 24 h prior to stimulation, and cells were stimulated with 10 nm HRG. HRG-containing medium was changed after 2 days. Cells were grown in the constant presence of stimuli for 5 days and then fixed with 4% paraformaldehyde for 1 h. Then cells were washed once with PBS, once with 60% isopropyl alcohol for 5 min, dried completely, and then stained with Oil Red O solution (Sigma) for 10 min. Stained cells were washed with water three times and then stained with a DAPI solution. Fluorescence images were obtained using InCell Analyzer 2000 (GE Healthcare), and image analysis was done to calculate total signal intensities of lipid particles per cell using Developer tool software (GE Healthcare).
Author contributions
M. O.-H. designed the study. S. M., A. G.-M., A. K., and N. Y. performed experiments and analyzed data. B. N. K. and N. V. performed analysis of proteome data. S. M. and T. N. analyzed microarray data. K. I., K. T., and B. N. K. constructed mathematical models and performed simulation. K. I. and S. K. performed parameter optimization. S. M., M. H., and Y. S. performed single-molecule imaging of TMR-HRG and analyzed the data. R. O. provided materials. S. M., K. I., and M. O.-H. wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Kaoru Takahashi, Sewon Ki, Akihiro Yamamoto, and Hiromi Sato for technical assistance and Dr. Masaki Nomura for fruitful discussions about modeling.
This work supported in part by JSPS KAKENHI Grant 15KT0084, RIKEN Epigenome and Single Cell Project Grants, the Cooperative Research Program of Institute for Protein Research, Osaka University Grant CRa-17-01, Nagase Science Technology Foundation, and Astellas Foundation for Research on Metabolic Disorders (to M. O.-H.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S10, Tables S1–S9, supporting Methods, and supporting Refs. 1–10.
- EGFR
- epidermal growth factor receptor
- EGF
- epidermal growth factor
- HRG
- heregulin
- HRGR
- HRG receptor
- ERK
- extracellular signal-regulated kinase
- DUSP
- dual-specificity MAPK phosphatase
- PHLDA1
- Pleckstrin homology-like domain, family A, member 1
- co-IP
- co-immunoprecipitation
- CV
- coefficient of variation
- MAPK
- mitogen-activated protein kinase
- qRT-PCR
- quantitative RT-PCR
- PI3K
- phosphatidylinositol 3-kinase
- TMR
- carboxytetramethylrhodamine
- PLA
- proximity ligation assay
- HBSS
- Hanks' buffered saline solution.
References
- 1. Citri A., and Yarden Y. (2006) EGF-ERBB signalling: towards the systems level. Nat. Rev. Mol. Cell Biol. 7, 505–516 10.1038/nrm1962 [DOI] [PubMed] [Google Scholar]
- 2. Nakaoka Y., Nishida K., Narimatsu M., Kamiya A., Minami T., Sawa H., Okawa K., Fujio Y., Koyama T., Maeda M., Sone M., Yamasaki S., Arai Y., Koh G. Y., Kodama T., et al. (2007) Gab family proteins are essential for postnatal maintenance of cardiac function via neuregulin-1/ErbB signaling. J. Clin. Invest. 117, 1771–1781 10.1172/JCI30651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shi Z. Q., Yu D. H., Park M., Marshall M., and Feng G. S. (2000) Molecular mechanism for the Shp-2 tyrosine phosphatase function in promoting growth factor stimulation of Erk activity. Mol. Cell. Biol. 20, 1526–1536 10.1128/MCB.20.5.1526-1536.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Turke A. B., Song Y., Costa C., Cook R., Arteaga C. L., Asara J. M., and Engelman J. A. (2012) MEK inhibition leads to PI3K/AKT activation by relieving a negative feedback on ERBB receptors. Cancer Res. 72, 3228–3237 10.1158/0008-5472.CAN-11-3747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Grossmann K. S., Wende H., Paul F. E., Cheret C., Garratt A. N., Zurborg S., Feinberg K., Besser D., Schulz H., Peles E., Selbach M., Birchmeier W., and Birchmeier C. (2009) The tyrosine phosphatase Shp2 (PTPN11) directs Neuregulin-1/ErbB signaling throughout Schwann cell development. Proc. Natl. Acad. Sci. U.S.A. 106, 16704–16709 10.1073/pnas.0904336106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sithanandam G., Smith G. T., Fields J. R., Fornwald L. W., and Anderson L. M. (2005) Alternate paths from epidermal growth factor receptor to Akt in malignant versus nontransformed lung epithelial cells: ErbB3 versus Gab1. Am. J. Respir. Cell Mol. Biol. 33, 490–499 10.1165/rcmb.2005-0049OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Holbro T., Civenni G., and Hynes N. E. (2003) The ErbB receptors and their role in cancer progression. Exp. Cell Res. 284, 99–110 10.1016/S0014-4827(02)00099-X [DOI] [PubMed] [Google Scholar]
- 8. Haj F. G., Verveer P. J., Squire A., Neel B. G., and Bastiaens P. I. (2002) Imaging sites of receptor dephosphorylation by PTP1B on the surface of the endoplasmic reticulum. Science 295, 1708–1711 10.1126/science.1067566 [DOI] [PubMed] [Google Scholar]
- 9. Dikic I. (2003) Mechanisms controlling EGF receptor endocytosis and degradation. Biochem. Soc. Trans. 31, 1178–1181 10.1042/bst0311178 [DOI] [PubMed] [Google Scholar]
- 10. Dougherty M. K., Müller J., Ritt D. A., Zhou M., Zhou X. Z., Copeland T. D., Conrads T. P., Veenstra T. D., Lu K. P., and Morrison D. K. (2005) Regulation of Raf-1 by direct feedback phosphorylation. Mol. Cell 17, 215–224 10.1016/j.molcel.2004.11.055 [DOI] [PubMed] [Google Scholar]
- 11. Amit I., Citri A., Shay T., Lu Y., Katz M., Zhang F., Tarcic G., Siwak D., Lahad J., Jacob-Hirsch J., Amariglio N., Vaisman N., Segal E., Rechavi G., Alon U., et al. (2007) A module of negative feedback regulators defines growth factor signaling. Nat. Genet. 39, 503–512 10.1038/ng1987 [DOI] [PubMed] [Google Scholar]
- 12. Descot A., Hoffmann R., Shaposhnikov D., Reschke M., Ullrich A., and Posern G. (2009) Negative regulation of the EGFR-MAPK cascade by actin-MAL-mediated Mig6/Errfi-1 induction. Mol. Cell 35, 291–304 10.1016/j.molcel.2009.07.015 [DOI] [PubMed] [Google Scholar]
- 13. Park C. G., Lee S. Y., Kandala G., Lee S. Y., and Choi Y. (1996) A novel gene product that couples TCR signaling to Fas(CD95) expression in activation-induced cell death. Immunity 4, 583–591 10.1016/S1074-7613(00)80484-7 [DOI] [PubMed] [Google Scholar]
- 14. Neef R., Kuske M. A., Pröls E., and Johnson J. P. (2002) Identification of the human PHLDA1/TDAG51 gene: down-regulation in metastatic melanoma contributes to apoptosis resistance and growth deregulation. Cancer Res. 62, 5920–5929 [PubMed] [Google Scholar]
- 15. Nagai M. A., Fregnani J. H., Netto M. M., Brentani M. M., and Soares F. A. (2007) Down-regulation of PHLDA1 gene expression is associated with breast cancer progression. Breast Cancer Res. Treat. 106, 49–56 10.1007/s10549-006-9475-6 [DOI] [PubMed] [Google Scholar]
- 16. Kawase T., Ohki R., Shibata T., Tsutsumi S., Kamimura N., Inazawa J., Ohta T., Ichikawa H., Aburatani H., Tashiro F., and Taya Y. (2009) PH domain-only protein PHLDA3 is a p53-regulated repressor of Akt. Cell 136, 535–550 10.1016/j.cell.2008.12.002 [DOI] [PubMed] [Google Scholar]
- 17. Wang X., Li G., Koul S., Ohki R., Maurer M., Borczuk A., and Halmos B. (2015) PHLDA2 is a key oncogene-induced negative feedback inhibitor of EGFR/ErbB2 signaling via interference with AKT signaling. Oncotarget 10.18632/oncotarget.3674 10.18632/oncotarget.3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nagashima T., Shimodaira H., Ide K., Nakakuki T., Tani Y., Takahashi K., Yumoto N., and Hatakeyama M. (2007) Quantitative transcriptional control of ErbB receptor signaling undergoes graded to biphasic response for cell differentiation. J. Biol. Chem. 282, 4045–4056 10.1074/jbc.M608653200 [DOI] [PubMed] [Google Scholar]
- 19. Mullenbrock S., Shah J., and Cooper G. M. (2011) Global expression analysis identified a preferentially nerve growth factor-induced transcriptional program regulated by sustained mitogen-activated protein kinase/extracellular signal-regulated kinase (ERK) and AP-1 protein activation during PC12 cell differentiation. J. Biol. Chem. 286, 45131–45145 10.1074/jbc.M111.274076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Toriseva M., Ala-aho R., Peltonen S., Peltonen J., Grénman R., and Kähäri V.-M. (2012) Keratinocyte growth factor induces gene expression signature associated with suppression of malignant phenotype of cutaneous squamous carcinoma cells. PLoS ONE 7, e33041 10.1371/journal.pone.0033041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li G., Wang X., Hibshoosh H., Jin C., and Halmos B. (2014) Modulation of ErbB2 blockade in ErbB2-positive cancers: the role of ErbB2 mutations and PHLDA1. PLoS ONE 9, e106349 10.1371/journal.pone.0106349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li J., Bennett K., Stukalov A., Fang B., Zhang G., Yoshida T., Okamoto I., Kim J.-Y., Song L., Bai Y., Qian X., Rawal B., Schell M., Grebien F., Winter G., et al. (2013) Perturbation of the mutated EGFR interactome identifies vulnerabilities and resistance mechanisms. Mol. Syst. Biol. 9, 705 10.1038/msb.2013.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Holbro T., Beerli R. R., Maurer F., Koziczak M., Barbas C. F. 3rd., and Hynes N. E. (2003) The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc. Natl. Acad. Sci. U.S.A. 100, 8933–8938 10.1073/pnas.1537685100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Karunagaran D., Tzahar E., Beerli R. R., Chen X., Graus-Porta D., Ratzkin B. J., Seger R., Hynes N. E., and Yarden Y. (1996) ErbB-2 is a common auxiliary subunit of NDF and EGF receptors: implications for breast cancer. EMBO J. 15, 254–264 [PMC free article] [PubMed] [Google Scholar]
- 25. Aguilar Z., Akita R. W., Finn R. S., Ramos B. L., Pegram M. D., Kabbinavar F. F., Pietras R. J., Pisacane P., Sliwkowski M. X., and Slamon D. J. (1999) Biologic effects of heregulin/neu differentiation factor on normal and malignant human breast and ovarian epithelial cells. Oncogene 18, 6050–6062 10.1038/sj.onc.1202993 [DOI] [PubMed] [Google Scholar]
- 26. Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K.-J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L.-G., and Landegren U. (2006) Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 10.1038/nmeth947 [DOI] [PubMed] [Google Scholar]
- 27. Karamouzis M. V., Dalagiorgou G., Georgopoulou U., Nonni A., Kontos M., and Papavassiliou A. G. (2016) HER-3 targeting alters the dimerization pattern of ErbB protein family members in breast carcinomas. Oncotarget 7, 5576–5597 10.18632/oncotarget.6762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nagashima T., Inoue N., Yumoto N., Saeki Y., Magi S., Volinsky N., Sorkin A., Kholodenko B. N., and Okada-Hatakeyama M. (2015) Feedforward regulation of mRNA stability by prolonged extracellular signal-regulated kinase activity. FEBS J. 282, 613–629 10.1111/febs.13172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mina M., Magi S., Jurman G., Itoh M., Kawaji H., Lassmann T., Arner E., Forrest A. R., Carninci P., Hayashizaki Y., Daub C. O., FANTOM Consortium, Okada-Hatakeyama M., and Furlanello C. (2015) Promoter-level expression clustering identifies time development of transcriptional regulatory cascades initiated by ErbB receptors in breast cancer cells. Sci. Rep. 5, 11999 10.1038/srep11999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang G. C., Ouyang X., and Epstein R. J. (1998) Proxy activation of protein ErbB2 by heterologous ligands implies a heterotetrameric mode of receptor tyrosine kinase interaction. Biochem. J. 331, 113–119 10.1042/bj3310113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Furuuchi K., Berezov A., Kumagai T., and Greene M. I. (2007) Targeted antireceptor therapy with monoclonal antibodies leads to the formation of inactivated tetrameric forms of ErbB receptors. J. Immunol. 178, 1021–1029 10.4049/jimmunol.178.2.1021 [DOI] [PubMed] [Google Scholar]
- 32. Webb S. E., Roberts S. K., Needham S. R., Tynan C. J., Rolfe D. J., Winn M. D., Clarke D. T., Barraclough R., and Martin-Fernandez M. L. (2008) Single-molecule imaging and fluorescence lifetime imaging microscopy show different structures for high- and low-affinity epidermal growth factor receptors in A431 cells. Biophys. J. 94, 803–819 10.1529/biophysj.107.112623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Clayton A. H., Walker F., Orchard S. G., Henderson C., Fuchs D., Rothacker J., Nice E. C., and Burgess A. W. (2005) Ligand-induced dimer-tetramer transition during the activation of the cell surface epidermal growth factor receptor-A multidimensional microscopy analysis. J. Biol. Chem. 280, 30392–30399 10.1074/jbc.M504770200 [DOI] [PubMed] [Google Scholar]
- 34. Zhang X., Meng J., and Wang Z.-Y. (2012) A switch role of Src in the biphasic EGF signaling of ER-negative breast cancer cells. PLoS ONE 7, e41613 10.1371/journal.pone.0041613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hiroshima M., Saeki Y., Okada-Hatakeyama M., and Sako Y. (2012) Dynamically varying interactions between heregulin and ErbB proteins detected by single-molecule analysis in living cells. Proc. Natl. Acad. Sci. U.S.A. 109, 13984–13989 10.1073/pnas.1200464109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Berger M. B., Mendrola J. M., and Lemmon M. A. (2004) ErbB3/HER3 does not homodimerize upon neuregulin binding at the cell surface. FEBS Lett. 569, 332–336 10.1016/j.febslet.2004.06.014 [DOI] [PubMed] [Google Scholar]
- 37. Steinkamp M. P., Low-Nam S. T., Yang S., Lidke K. A., Lidke D. S., and Wilson B. S. (2014) erbB3 is an active tyrosine kinase capable of homo- and heterointeractions. Mol. Cell. Biol. 34, 965–977 10.1128/MCB.01605-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McCabe Pryor M., Steinkamp M. P., Halasz A. M., Chen Y., Yang S., Smith M. S., Zahoransky-Kohalmi G., Swift M., Xu X.-P., Hanein D., Volkmann N., Lidke D. S., Edwards J. S., and Wilson B. S. (2015) Orchestration of ErbB3 signaling through hetero and homo-interactions. Mol. Biol. Cell 26, 4109–4123 10.1091/mbc.E14-06-1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Volinsky N., McCarthy C. J., von Kriegsheim A., Saban N., Okada-Hatakeyama M., Kolch W., and Kholodenko B. N. (2015) Signalling mechanisms regulating phenotypic changes in breast cancer cells. Biosci. Rep. 35, e00178 10.1042/BSR20140172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moad A. I., Muhammad T. S., Oon C. E., and Tan M. L. (2013) Rapamycin induces apoptosis when autophagy is inhibited in T-47D mammary cells and both processes are regulated by Phlda1. Cell Biochem. Biophys. 66, 567–587 10.1007/s12013-012-9504-5 [DOI] [PubMed] [Google Scholar]
- 41. Johnson E. O., Chang K.-H., de Pablo Y., Ghosh S., Mehta R., Badve S., and Shah K. (2011) PHLDA1 is a crucial negative regulator and effector of Aurora A kinase in breast cancer. J. Cell Sci. 124, 2711–2722 10.1242/jcs.084970 [DOI] [PubMed] [Google Scholar]
- 42. Park E.-S., Kim J., Ha T.-U., Choi J.-S., Soo Hong K., and Rho J. (2013) TDAG51 deficiency promotes oxidative stress-induced apoptosis through the generation of reactive oxygen species in mouse embryonic fibroblasts. Exp. Mol. Med. 45, e35 10.1038/emm.2013.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sakthianandeswaren A., Christie M., D'Andreti C., Tsui C., Jorissen R. N., Li S., Fleming N. I., Gibbs P., Lipton L., Malaterre J., Ramsay R. G., Phesse T. J., Ernst M., Jeffery R. E., Poulsom R., et al. (2011) PHLDA1 expression marks the putative epithelial stem cells and contributes to intestinal tumorigenesis. Cancer Res. 71, 3709–3719 10.1158/1538-7445.AM2011-3709,10.1158/0008-5472.CAN-10-2342 [DOI] [PubMed] [Google Scholar]
- 44. Kastrati I., Canestrari E., and Frasor J. (2015) PHLDA1 expression is controlled by an estrogen receptor-NFκB-miR-181 regulatory loop and is essential for formation of ER+ mammospheres. Oncogene 34, 2309–2316 10.1038/onc.2014.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Basseri S., Lhoták S., Fullerton M. D., Palanivel R., Jiang H., Lynn E. G., Ford R. J., Maclean K. N., Steinberg G. R., and Austin R. C. (2013) Loss of TDAG51 results in mature-onset obesity, hepatic steatosis, and insulin resistance by regulating lipogenesis. Diabetes 62, 158–169 10.2337/db12-0256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Segatto O., Anastasi S., and Alemà S. (2011) Regulation of epidermal growth factor receptor signalling by inducible feedback inhibitors. J. Cell Sci. 124, 1785–1793 10.1242/jcs.083303 [DOI] [PubMed] [Google Scholar]
- 47. Yu R. C., Pesce C. G., Colman-Lerner A., Lok L., Pincus D., Serra E., Holl M., Benjamin K., Gordon A., and Brent R. (2008) Negative feedback that improves information transmission in yeast signalling. Nature 456, 755–761 10.1038/nature07513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Birtwistle M. R., Hatakeyama M., Yumoto N., Ogunnaike B. A., Hoek J. B., and Kholodenko B. N. (2007) Ligand-dependent responses of the ErbB signaling network: experimental and modeling analyses. Mol. Syst. Biol. 3, 144 10.1038/msb4100188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dunn E. F., and Connor J. H. (2011) Dominant inhibition of Akt/protein kinase B signaling by the matrix protein of a negative-strand RNA virus. J. Virol. 85, 422–431 10.1128/JVI.01671-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Turriziani B., Garcia-Munoz A., Pilkington R., Raso C., Kolch W., and von Kriegsheim A. (2014) On-beads digestion in conjunction with data-dependent mass spectrometry: a shortcut to quantitative and dynamic interaction proteomics. Biology 3, 320–332 10.3390/biology3020320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ermentrout B. (2002) Simulating, Analyzing, and Animating Dynamical Systems: A Guide to XPPAUT for Researchers and Students, the Society for Industrial and Applied Mathematics, Philadelphia, PA [Google Scholar]
- 52. Kimura S., Nakakuki T., Kirita S., and Okada M. (2011) AGLSDC: a genetic local search suitable for parallel computation. SICE JCMSI 4, 105–113 10.9746/jcmsi.4.105-4889 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.