Abstract
Malignant glioblastoma multiforme is one of the most aggressive human cancers, with very low survival rates. Recent studies have reported that glioma stem-like cells transdifferentiate into endothelial cells, indicating a new mechanism for tumor angiogenesis and potentially providing new therapeutic options for glioblastoma treatment. Glioma malignancy is strongly associated with altered expression of N-linked oligosaccharide structures on the cell surface. We have previously reported that β1,4-galactosyltransferase V (β1,4GalTV), which galactosylates the GlcNAcβ1–6Man arm of the branched N-glycans, is highly expressed in glioma and promotes glioma cell growth in vitro and in vivo. However, the mechanism by which β1,4GalTV stimulates glioma growth is unknown. Here we demonstrate that short hairpin RNA–mediated β1,4GalTV knockdown inhibits the tumorigenesis of glioma stem-like cells and reduces their transdifferentiation into endothelial cells. We also found that β1,4GalTV overexpression increased glioma stem-like cell transdifferentiation into endothelial cells and that this effect required β1,4GalTV galactosylation activity. Moreover, β1,4GalTV promoted β1,4-galactosylation of Notch1 and increased Notch1 protein levels. Of note, ectopic expression of activated Notch1 rescued the inhibitory effect of β1,4GalTV depletion on glioma stem-like cell transdifferentiation. In summary, our findings indicate that β1,4GalTV stimulates transdifferentiation of glioma stem-like cells into endothelial cells by activating Notch1 signaling. These detailed insights shed important light on the mechanisms regulating glioma angiogenesis.
Keywords: glioblastoma, endothelial cell, glycosyltransferase, cancer stem cells, differentiation, β4GalTV, Notch1, glioma stem-like cell, transdifferentiation
Introduction
Malignant glioblastoma multiforme (GBM)5 is one of the most aggressive human cancers, with a median survival of about 1 year (1). A number of anti-angiogenic treatment regimens are currently under clinical investigation (2). However, some studies have shown that tumor cells become more aggressive after anti-angiogenic therapy (3, 4), implying the existence of VEGF-independent angiogenesis. Supporting this notion, a series of recent papers has shown that glioma stem-like cells could transdifferentiate into tumor vascular endothelial cells (5–7), which might be one of the resistance mechanisms against anti-VEGF therapy. Thus, clarifying the mechanisms of transdifferentiation of glioma stem-like cell into endothelial cell helps to develop new anti-angiogenic therapeutic strategies.
Increased expression of highly branched N-glycans of cell surface glycoproteins is one of the most malignant prominent transformation-associated changes in the sugar chains of glycoproteins (8, 9). Recent experimental evidence from our laboratory and others has suggested that changes in the N-glycan structure of glycoproteins in glioma cells affect cell migration and tumor malignancy (10–13). For example, β1,4-galactosyltransferase V (β1,4GalTV), which galactosylates the GlcNAcβ1-6 branch arm of the highly branched N-glycan, is highly expressed in glioma (10, 14–16). Furthermore, β1,4GalTV promotes glioma cell growth and invasiveness (11, 17, 18). Recently, we have confirmed that β1,4GalTV could regulate the self-renewal of glioma-initiating cells (19). N-acetylglucosaminyltransferase V promotes the self-renewal and tumorigenicity of colon cancer stem cells (20). The N-glycan inhibitor tunicamycin inhibits the self-renewal ability and tumorigenesis of glioma stem-like cells (21). Thus, N-glycans perform critical roles in cancer stem cells.
N-glycosylation frequently regulates cell behavior by regulating membrane protein signaling (22). Notch1 signaling is widely known to regulate cell differentiation, proliferation, and apoptosis (23, 24). Inactivation of Notch signaling with γ-secretase inhibition or Notch1 knockdown inhibits transdifferentiation of glioma stem-like cells into endothelial cells (5, 25). The glycans, including N-glycan, O-fucose glycan, and O-glycan, regulate the activation of Notch signaling by manipulating Notch receptor–ligand interaction (26–28). Therefore, studying the role of N-glycans in the transdifferentiation of glioma stem-like cells into endothelial cells contributes to elucidation of the mechanisms of glioma vasculogenesis.
Here we evaluate the role and mechanism of β1,4GalTV in the transdifferentiation of glioma stem-like cells into endothelial cells. Our findings will provides a framework for a better understanding of the role of glycosylation in tumor development and might offer a new strategy to manipulate Notch signaling for therapeutic purposes.
Results
β1,4GalTV depletion inhibits transdifferentiation of glioma stem-like cells into endothelial cells in vitro
To determine whether β1,4GalTV regulates the transdifferentiation of glioma stem-like cells into endothelial cells, T698968 glioma stem-like cells were isolated from glioblastoma surgical biopsy specimens as described previously (29, 30). Under serum-free culture conditions, T698968 cells formed typical neurosphere structures (Fig. S1A). These neurospheres expressed the neural stem cell marker Nestin (Fig. S1B). Upon exposure to differentiation conditions, a mixture of cells expressed the markers of neurons and astrocytes (Fig. S1B). Thus, T698968 cells have self-renewal capability and multilineage differentiation capacity in vitro. Similarly, we isolated T109002 glioma stem-like cells from glioblastoma surgical biopsy specimens using the same method.
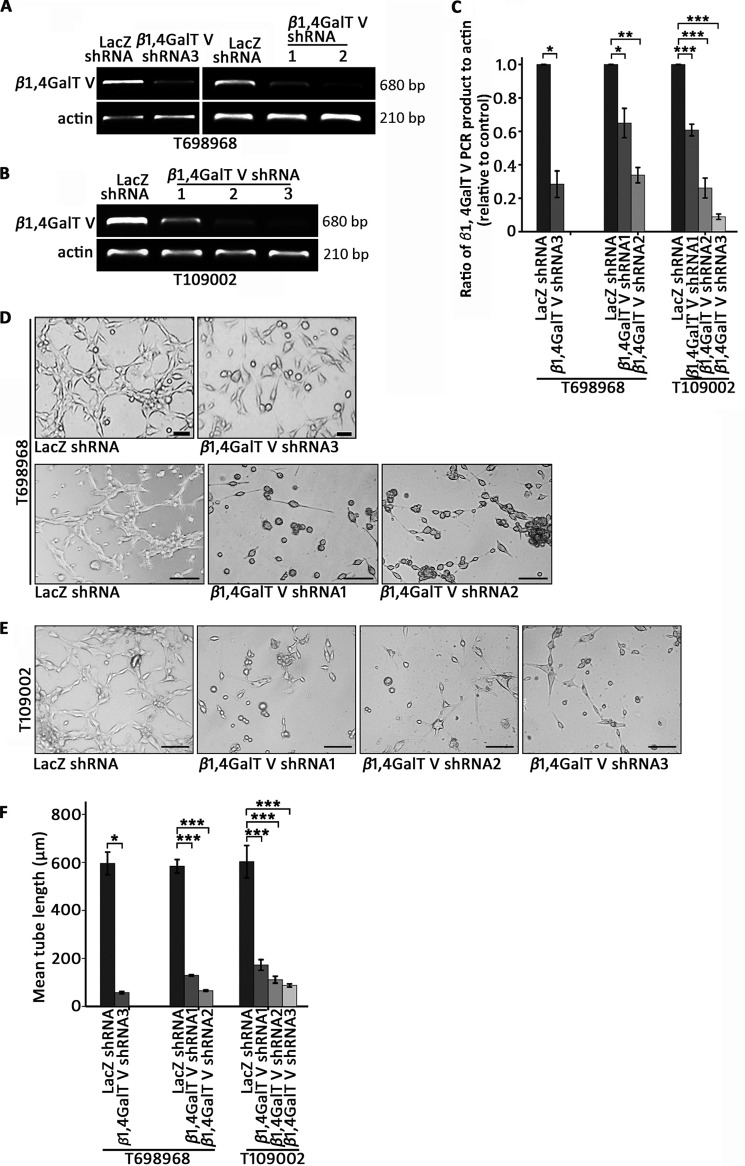
Next we constructed lentiviral shRNA vectors targeting human β1,4GalTV. RT-PCR results showed that β1,4GalTV shRNA reduced β1,4GalTV mRNA expression in T698968 cells and T109002 cells (Fig. 1, A–C). β1,4GalTV galactosylates the GlcNAcβ1–6Man arm of the highly branched N-glycan (31). Using a lectin blot with RCA-1 lectin, which interacts with Galβ1,4GlcNAc, a significant decrease in the binding of total glycoprotein with RCA-1 was observed in cells expressing β1,4GalTV shRNA compared with control cells (Fig. S1C).
Figure 1.
β1,4GalTV depletion inhibits the transdifferentiation of glioma stem-like cells in vitro. A, RT-PCR analysis of β1,4GalTV mRNA expression in T698968 cells expressing control or β1,4GalTV shRNA. Actin mRNA expression served as a loading control. B, RT-PCR analysis of β1,4GalTV mRNA expression in T109002 cells expressing control or β1,4GalTV shRNA. Actin mRNA expression served as a loading control. C, the relative densities of β1,4GalTV PCR product levels in A and B were quantified using densitometry. Values are normalized to that of cells expressing control shRNA. Results are expressed as mean ± S.D. (n = 3; *, p < 0.05; **, p < 0.01; ***, p < 0.001). D and E, representative images of tube formation derived from T698968 (D) or T109002 (E) cells expressing control or β1,4GalTV shRNA. Scale bars = 20 μm. F, quantification of mean tube length in D and E. The results are expressed as mean ± S.E. (n = 3; *, p < 0.05; ***, p < 0.001).
To investigate the role of β1,4GalTV in the transdifferentiation process of glioma stem-like cells into endothelial cells in vitro, a tube formation assay was performed. β1,4GalTV knockdown inhibited tube formation and decreased the length of tubes in T698968 or T109002 cells (Fig. 1, D–F). CD31 is generally considered a specific marker for endothelial cells (5, 6). The RT-PCR assay showed that β1,4GalTV knockdown significantly decreased CD31 expression (Fig. S1, D and E). Consistent with this, immunofluorescent staining showed that β1,4GalTV depletion decreased CD31 expression in the tube formation assay (Fig. S1, F and G). Thus, β1,4GalT V could regulate the endothelial cell transdifferentiation of glioma stem-like cell in vitro.
Reduction of β1,4GalTV expression inhibits transdifferentiation of glioma stem-like cells into endothelial cells in vivo
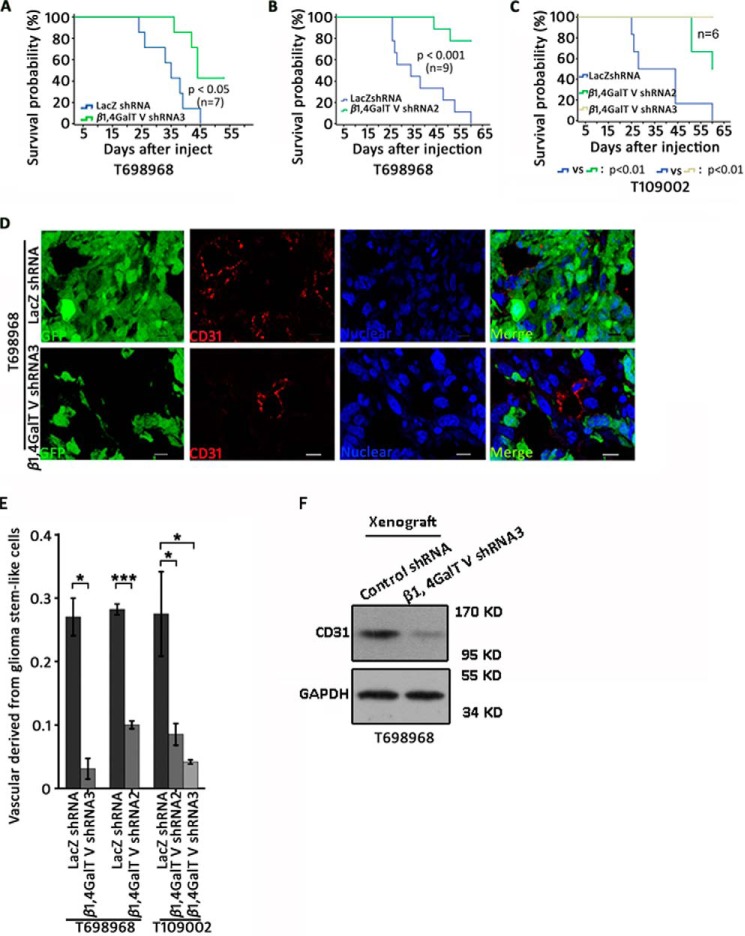
We used an intracranial glioma model to evaluate the contribution of β1,4GalTV in the transdifferentiation of glioma stem-like cells into endothelial cells in vivo. Glioma stem-like cells marked with GFP were implanted into the frontal lobe of nude mice. β1,4GalTV knockdown reduced tumor burdens in nude mice (Fig. S2, A and B). Depletion of β1,4GalTV in glioma stem-like cells significantly increased the overall survival of tumor-bearing nude mice (Fig. 2, A–C). Thus, β1,4GalTV depletion inhibits gliomagenesis in vivo.
Figure 2.
Reduction of β1,4GalTV expression inhibits gliomagenesis and transdifferentiation of glioma stem-like cells into the endothelium in vivo. A–C, Kaplan–Meier survival curves of nude mice injected with T698968 (A and B) or T109002 (C) cells expressing control or β1,4GalTV shRNA. D, confocal immunofluorescence analysis shows co-localization of the human endothelial cell marker CD31 and the tumor cell marker GFP in tumor xenografts formed by T698968 cells expressing GFP and control or β1,4GalTV shRNA. Scale bars = 10 μm. E, the relative vascular density derived from glioma stem-like cells was quantified. Results are expressed as mean ± S.E. (n = 3; *, p < 0.05; ***, p < 0.001). F, Western blot analysis of the endothelial cell marker CD31 in xenografts formed by T698968 cells expressing control or β1,4GalTV shRNA. GAPDH protein expression served as a loading control.
To examine whether β1,4GalTV regulates endothelial cell transdifferentiation of glioma stem-like cell in vivo, co-immunofluorescence analysis of GFP and the endothelial cell marker CD31 was performed in tumor xenografts formed by T698968 cells or T109002 cells marked with GFP. β1,4GalTV depletion significantly reduced the number of CD31+ endothelial cells co-expressing GFP in xenografts formed by both T698968 cells and T109002 cells (Fig. 2, D and E, and Fig. S2, C and D). Consistent with this, β1,4GalTV depletion significantly reduced the protein level of CD31 (Fig. 2F). β1,4GalTV knockdown inhibits transdifferentiation of glioma stem-like cells into endothelial cells in vivo.
β1,4GalT V regulates the transdifferentiation of glioma stem-like cells into endothelial cells depending on its galactosylation activity
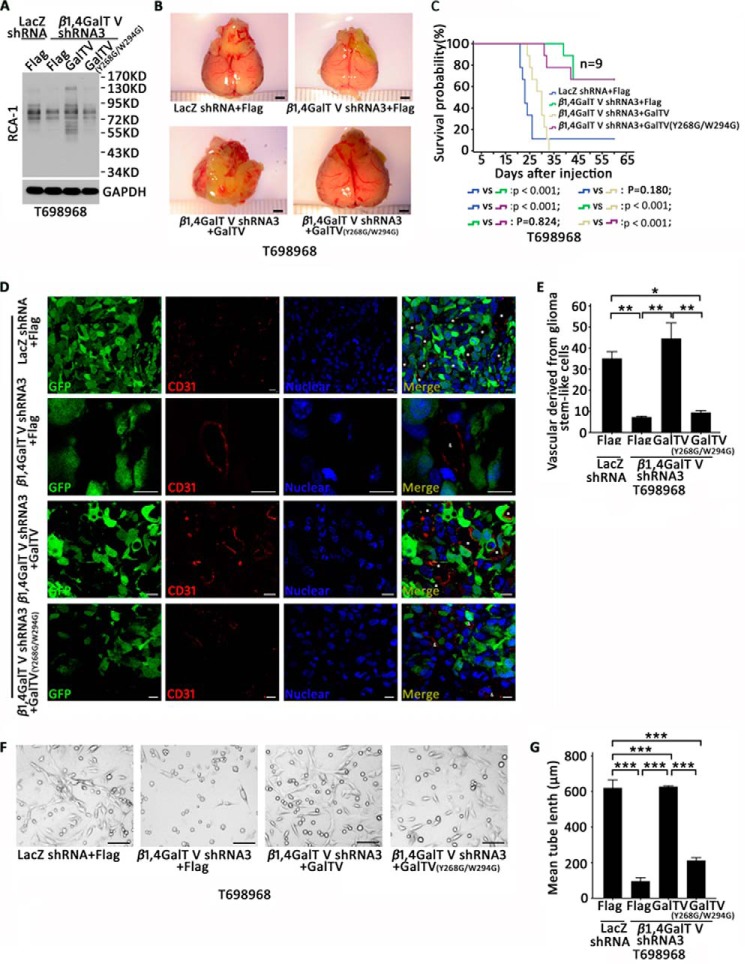
β1,4GalTV protein consists of a short NH2-terminal cytoplasmic domain, a stem region, and a catalytic domain that contains two conserved residues (Tyr268/Trp294), which are important for its galactosylation activity (32–34). To investigate the contribution of β1,4GalTV galactosylation activity in the transdifferentiation of glioma stem-like cells into endothelial cells, we constructed shRNA-resistant, hemagglutinin-tagged point mutant of β1,4GalTV (Y268G/W294G). Using a lectin blot assay, overexpression of wildtype β1,4GalTV, but not the Y268G/W294G mutant, could rescue the decreased Galβ1,4GlcNAc-glycosylated protein (43- to 130-kDa protein bands) by β1,4GalTV shRNA (Fig. 3A). The expression of wildtype β1,4GalTV, but not the Y268G/W294G mutant, could rescue gliomagenesis and mouse survival in the β1,4GalTV knockdown group (Fig. 3, B and C). Thus, β1,4GalTV regulates gliomagenesis depending on its galactosylation activity.
Figure 3.
β1,4GalTV regulates gliomagenesis and the transdifferentiation of glioma stem-like cells into endothelial cells depending on its galactosylation activity. A, total lysates of T698968 cells expressing LacZ shRNA + FLAG, β1,4GalTV shRNA3 + FLAG, β1,4GalTV shRNA3 + GalTV, or β1,4GalTV shRNA3 + GalTV (Y268G/W294G) were analyzed with an RCA-1 lectin blot. The protein expression of GAPDH served as a loading control. B, nude mice were injected with T698968 cells expressing LacZ shRNA + FLAG, β1,4GalTV shRNA3 + FLAG, β1,4GalTV shRNA3 + GalTV, or β1,4GalTV shRNA3 + GalTV (Y268G/W294G). Four weeks later, photos were taken for the xenograft. Scale bars = 2 mm. C, Kaplan–Meier survival curves of nude mice injected with T698968 cells expressing LacZ shRNA + FLAG, β1,4GalTV shRNA3 + FLAG, β1,4GalTV shRNA3 + GalTV, or β1,4GalTV shRNA3 + GalTV (Y268G/W294G). D, immunofluorescence analysis shows co-localization of the human endothelial cell marker CD31 and the tumor cell marker GFP in tumor xenografts formed by T698968 cells expressing GFP and LacZ shRNA + FLAG, β1,4GalTV shRNA3 + FLAG, β1,4GalTV shRNA3 + GalTV, or β1,4GalTV shRNA3 + GalTV (Y268G/W294G). Scale bars = 10 μm. E, the relative vascular density derived from glioma stem-like cells was quantified. Results are expressed as mean ± S.E. (n = 3; *, p < 0.05; **, p < 0.01). F, representative images of tube formation derived from T698968 cells expressing LacZ shRNA + FLAG, β1,4GalTV shRNA3 + FLAG, β1,4GalTV shRNA3 + GalTV, or β1,4GalTV shRNA3 + GalTV (Y268G/W294G). Scale bars = 20 μm. G, quantification of mean tube length in F. The results are expressed as mean ± S.E. (n = 3; ***, p < 0.001).
Co-immunofluorescence staining in tumor xenografts also showed that overexpression of wildtype β1,4GalTV, but not the β1,4GalTV mutant (Y268G/W294G), increased the number of CD31+ endothelial cells co-expressing GFP in the β1,4GalTV knockdown group (Fig. 3, D and E). Furthermore, a tube formation assay also showed that overexpression of wildtype β1,4GalTV reduced the inhibitory effect of β1,4GalTV depletion on tube formation (Fig. 3, F and G). Together, β1,4GalTV regulates the transdifferentiation of glioma stem-like cells into endothelial cells depending on its galactosylation activity.
β1,4GalTV regulates Notch signaling depending on its galactosylation activity
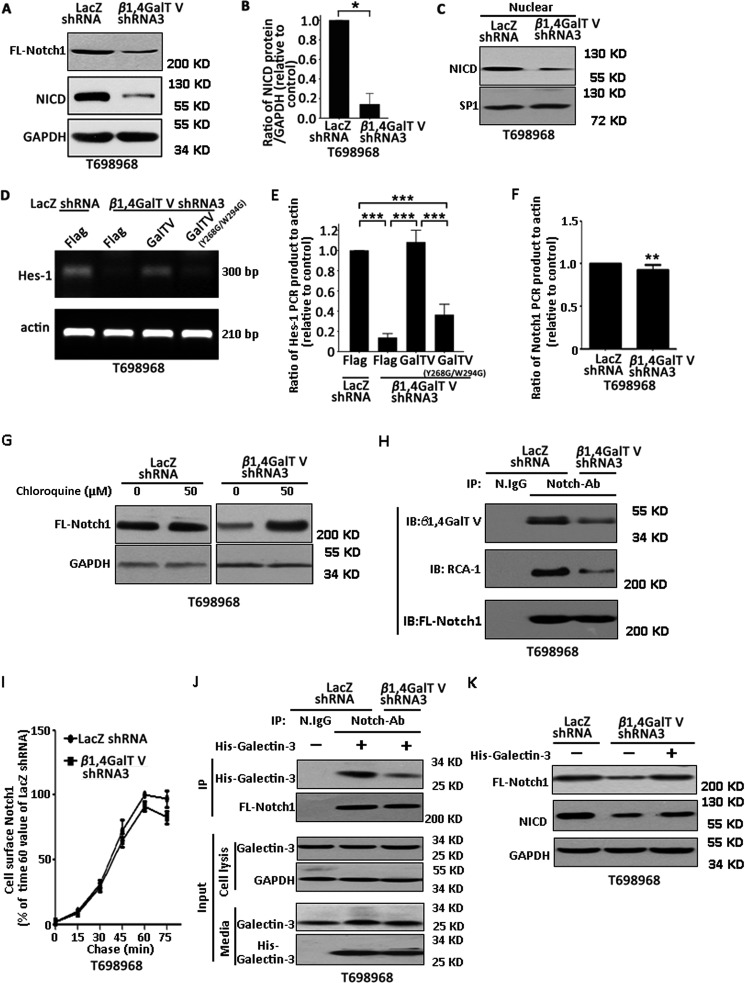
We next examined the mechanisms by which β1,4GalTV regulates the transdifferentiation of glioma stem-like cells into endothelial cells. It has been reported that the Notch signaling pathway regulates endothelial transdifferentiation of glioma stem-like cells (5, 25). Thus, we primarily examined whether β1,4GalTV regulates Notch signaling. Each activated Notch1 molecule is consumed to produce a Notch1 intracellular domain (NICD) (35). Western blotting using an antibody against the Notch1 intracellular domain showed that β1, 4GalTV depletion significantly decreased the expression of full-length Notch1 and NICD protein (Fig. 4, A and B). β1, 4GalTV depletion also significantly decreased the expression of NICD in the nucleus (Fig. 4C). Consistent with this, β1,4GalTV depletion inhibited the mRNA expression of a canonical Notch target gene, Hes-1, which could be rescued by wildtype β1,4GalTV other than the β1,4GalTV mutant (Y268G/W294G) (Fig. 4, D and E). Thus, β1,4GalTV regulates Notch signaling.
Figure 4.
β1,4GalTV regulates the activity of Notch1 signaling. A, total lysates of T698968 cells expressing control or β1,4GalTV shRNA3 were analyzed with Notch1 antibody by Western blotting. The protein expression of GAPDH served as a loading control. B, the relative densities of NICD protein levels in A were quantified using densitometry. Values are normalized to that of T698968 cells expressing LacZ shRNA. Results are expressed as mean ± S.D. (n = 3; *, p < 0.05). C, Western blot analysis of nuclear NICD expression in T698968 cells expressing control or β1,4GalTV shRNA3. Sp1 served as a nuclear marker. D, RT-PCR analysis of Hes-1 expression in T698968 cells expressing LacZ shRNA + FLAG, β1,4GalTV shRNA3 + FLAG, β1,4GalTV shRNA3 + GalTV, or β1,4GalTV shRNA3 + GalTV (Y268G/W294G). E, the relative densities of Hes-1 PCR product levels were quantified using densitometry. Values are normalized to that of cells expressing LacZ shRNA + FLAG. Results are expressed as mean ± S.D. (n = 3; ***, p < 0.001). F, RT-PCR analysis of Notch1 expression in T698968 cells expressing control or β1,4GalTV shRNA. Actin expression served as a loading control. The relative densities of Notch1 PCR product levels were quantified using densitometry. Values are normalized to that of cells expressing control shRNA. **, p < 0.01. G, Western blot analysis of Notch1 from lysates of T698968 cells expressing control or β1,4GalTV shRNA in the absence or presence of the lysosome inhibitor chloroquine overnight. H, Notch1 proteins immunoprecipitated from T698968 cells expressing control or β1,4GalTV shRNA were resolved by SDS-PAGE and transferred to a polyvinylidene difluoride membrane. The membrane was overlaid with Notch1 antibody to verify the immunoprecipitation efficiency, with lectin RCA-1 to confirm β1,4-linked galactosylation, and with β1,4GalTV antibody to examine the interaction between β1,4GalTV and Notch1. IB, immunoblot. I, trafficking of newly synthesized Notch1 to the cell surface in T698968 cells expressing LacZ shRNA or β1,4GalTV shRNA3. Pulse labeling was performed with 1 mm AHA (a structural analog of methionine). The AHA-labeled Notch1 trafficked to the cell surface was examined using Western blotting. Results are expressed as percent of AHA-labeled Notch1 in cells expressing LacZ shRNA at 60 min. Data are represented as the means ± S.D. from three separate experiments. J, Notch1 proteins immunoprecipitated from T698968 cells expressing control or β1,4GalTV shRNA treated with His-tagged galectin-3 for 30 min were resolved by SDS-PAGE and transferred to a polyvinylidene difluoride membrane. The membrane was overlaid with Notch1 antibody to verify the IP efficiency and with His antibody to examine the interaction between His-tagged galectin-3 and Notch1. K, total lysates of T698968 cells expressing control or β1,4GalTV shRNA3 treated with or without His-tagged galectin-3 protein for 24 h were analyzed with Notch1 antibody by Western blotting. The protein expression of GAPDH served as a loading control. FL-Notch1, full-length Notch1.
Next, we examined the mechanism of β1,4GalTV regulating Notch1 expression. β1,4GalTV knockdown slightly reduced the level of Notch1 mRNA (Fig. 4F). Previous studies have shown that Notch1 could be degraded by the lysosome (36). Thus, we examined the effect of β1,4GalTV knockdown on Notch1 lysosome-mediated degradation. We found no obvious effect of the lysosomal inhibitor chloroquine on Notch 1 levels in control cells, whereas chloroquine significantly increased the level of Notch 1 protein in cells expressing β1,4GalTV shRNA (Fig. 4G). Thus, β1,4GalTV depletion might promote Notch1 lysosome-mediated degradation.
A previous report has shown that Notch1 is modified with N-glycan and that N-glycan regulates Notch1 stability (37). This finding motivated us to examine whether Notch could be β1,4-galactosylated using RCA-1 lectin. Notch1 protein immunoprecipitated from T698968 cells reacted with the lectin RCA-I. Furthermore, β1,4GalTV depletion inhibited the β1,4-galactosylation of Notch1. In addition, β1,4GalTV could be immunoprecipitated with Notch1 (Fig. 4H). Thus, Notch might be a substrate of β1,4GalTV.
Glycosylation is required for protein processing and transport to the plasma membrane (38). Thus, we next examined the effect of β1,4GalTV knockdown on the delivery of Notch1 to the cell surface. The kinetics of Notch1 synthesis and cell surface appearance were measured using metabolic labeling. Notch1 synthesis and trafficking to the cell surface took 50–60 min in cells. This was only slightly decreased by β1,4GalTV knockdown (Fig. 4I). β1,4-galactosylation could provide galactose for β1,4-galactosylated protein to interact with galectin (39). The galactose-dependent interaction between galectin and membrane protein frequently increases the membrane protein level by inhibiting its degradation (40). It has been reported that galectin-3 binds to Notch1 in a galactose-dependent manner (41). Thus, we examined the effect of β1,4GalTV knockdown on the interaction between galectin-3 and Nocth1. β1,4GalTV knockdown obviously inhibited the interaction between Notch1 and galectin-3 (Fig. 4J). Furthermore, galectin-3 treatment could recue the inhibitory effect of β1,4GalTV knockdown on the expression of full-length Notch 1 and the cleavage of Notch 1 (Fig. 4K). β1,4GalTV regulates Notch signaling depending on its galactosylation activity.
The inhibitory effect of β1,4GalTV depletion on the transdifferentiation of glioma stem-like cells into endothelial cells could be rescued by ectopic expression of NICD
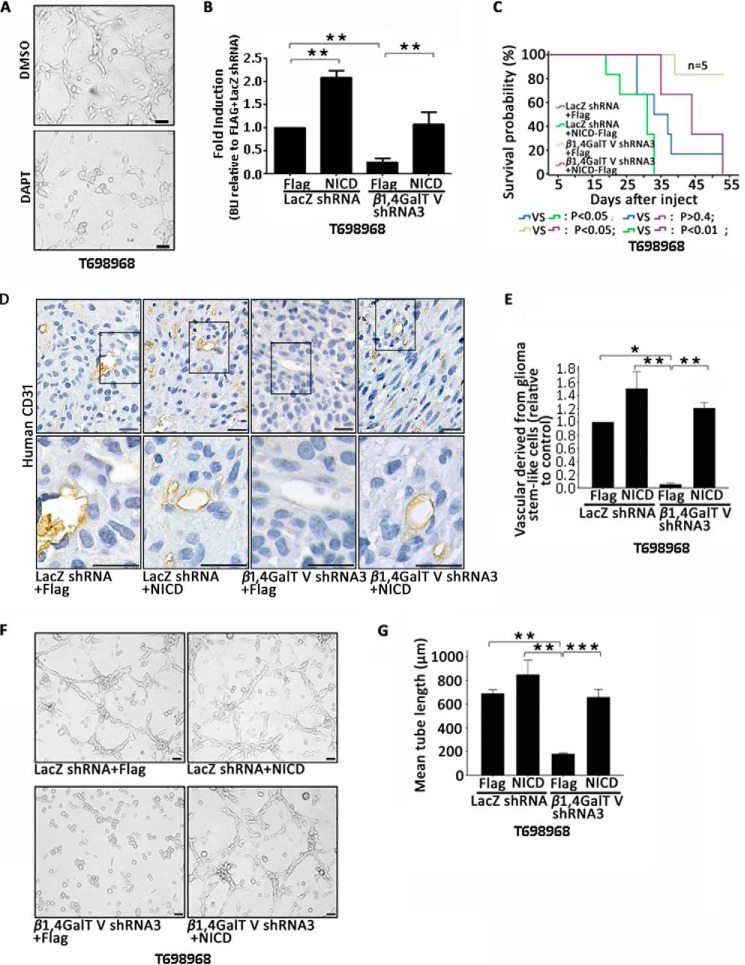
Next, we explored the role of Notch1 signaling in β1,4GalTV promotion of endothelial cell transdifferentiation of glioma stem-like cells. First we tested the effect of DAPT on the transdifferentiation of glioma stem-like cells. DAPT is a γ-secretase inhibitor that effectively inhibits Notch signaling (42). A tube formation assay showed that DAPT treatment decreased tube formation in vitro (Fig. 5A).
Figure 5.
The effect of β1,4GalTV depletion on transdifferentiation of glioma stem-like cells could be rescued by NICD. A, representative images of tube formation derived from T698968 cells treated with the γ-secretase inhibition DAPT. Scale bar = 20 μm. B–E, T698968 cells expressing LacZ shRNA + FLAG, LacZ shRNA + NICD, β1,4GalTV shRNA + FLAG, or β1,4GalTV shRNA + NICD were injected into the frontal lobe of nude mice. Four weeks later, the size of tumor xenografts was quantified using bioluminescent imaging (BLI). Results are expressed as mean ± S.D. (n = 6; *, p < 0.05; **, p < 0.01). The ratio of BLI was standardized to that of cells expressing LacZ shRNA + FLAG (B). C, Kaplan–Meier survival curves of nude mice injected with T698968 cells expressing LacZ shRNA + FLAG, LacZ shRNA + NICD, β1,4GalTV shRNA3 + FLAG, or β1,4GalTV shRNA3 + NICD. D, immunohistochemistry analysis of human CD31+ vascular cells in tumor xenografts initiated by T698968 cells expressing LacZ shRNA + FLAG, LacZ shRNA + NICD, β1,4GalTV shRNA3 + FLAG, or β1,4GalTV shRNA3 + NICD using anti-human-CD31 antibody. Scale bars = 20 μm. E, the relative vascular density derived from glioma stem-like cells in D was quantified. Values are normalized to that of T698968 cells expressing LacZ shRNA. Results are expressed as mean ± S.E. (n = 3; *, p < 0.05; **, p < 0.01). F, representative images of tube formation derived from T698968 cells expressing LacZ shRNA + FLAG, LacZ shRNA + NICD, β1,4GalTV shRNA3 + FLAG, or β1,4GalTV shRNA3 + NICD. Scale bar = 20 μm. G, quantification of mean tube length in F, shown as mean ± S.E. (n = 3; **, p < 0.01; ***, p < 0.001).
Next we used an intracranial glioma model to evaluate the contribution of Notch1 signaling during β1,4GalTV regulation of the transdifferentiation process from glioma stem-like cells in vivo. T698968 cells expressing β1,4GalTV shRNA3 and NICD were implanted into the frontal lobe of nude mice. Bioluminescent imaging (BLI) targeting GFP showed that NICD protein overexpression rescued the inhibitory effects of β1,4GalTV knockdown on gliomagenesis (Fig. 5B). Additionally, NICD protein overexpression reversed the effects of β1,4GalTV knockdown on tumor-bearing mouse survival (Fig. 5C).
Next we performed immunohistochemistry staining with tumor xenografts formed by β1,4GalTV knockdown T698968 cells overexpressed with NICD protein with the human CD31 antibody. Overexpression of NICD protein could rescue the inhibitory effect of β1,4GalTV knockdown on endothelial cell transdifferentiation of glioma stem-like cells (Fig. 5, D and E). Overexpression of NICD protein could reverse the inhibitory effect of β1,4GalTV knockdown on tube formation (Fig. 5, F and G). Taken together, these data demonstrate that β1, 4GalTV regulates the transdifferentiation of glioma stem-like cells into endothelial cells through Notch1 signaling.
Nucleus NICD expression in human GBM correlates with β1,4GalTV protein level
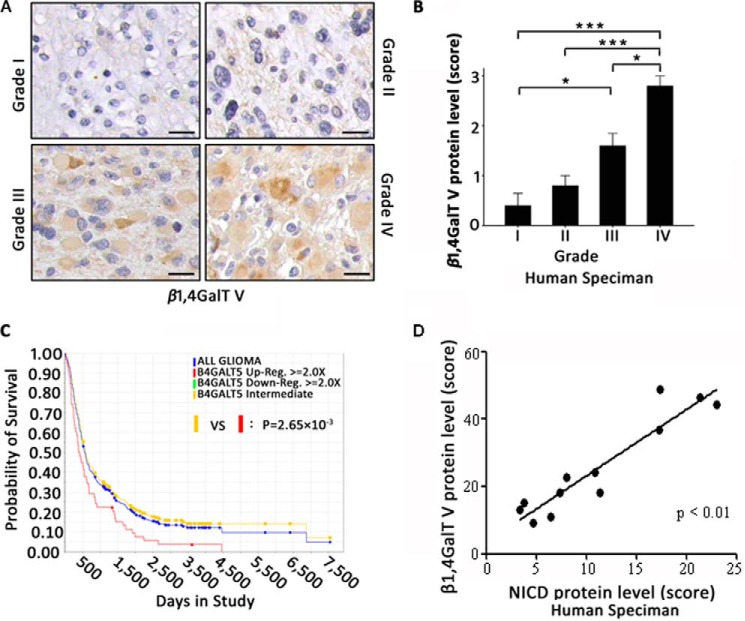
To further investigate the critical role of β1,4GalTV in glioma malignancy, we performed immunohistochemistry with human brain tumor specimens classified according to the World Health Organization. The expression of β1,4GalTV protein increased, accompanied by tumor malignancy (Fig. 6, A and B). This result was consistent with the Repository of Molecular Brain Neoplasia Data (REMBRANDT) database of the NCI, National Institutes of Health, which showed a significant decrease in the probability of survival with elevated β1,4GalTV expression (Fig. 6C). To further confirm the correlation between Notch1 signaling activation and β1,4GalTV expression, immunohistochemistry was performed on human GBM cells. The immunohistochemistry assay showed a strong linear relationship between β1,4GalTV expression and nucleus NICD expression (Fig. 6D). These data strongly support that β1,4GalTV regulates Notch1 signaling.
Figure 6.
Nucleus NICD expression in human GBMs correlates with β1,4GalTV protein level. A, expression of β1,4GalTV protein was examined via immunohistochemistry staining in 20 brain tumor sections. Representative β1,4GalTV protein expression levels is shown in four human brain tumor sections. Scale bars = 20 μm. B, quantitative result of β1,4GalTV protein expression in A is shown as mean ± S.E. (*, p < 0.05; ***, p < 0.001). The staining index of β1,4GalTV protein was scored as 0 to 4. C, Kaplan–Meier curve showing the overall survival of patients with glioma from the NCI, National Institutes of Health The Cancer Genome Atlas (TCGA) database (the REMBRANDT database of the NCI, National Institutes of Health) in which clinical outcome data were available (n = 343). Patients with high expression (red line) have significantly shorter survival than those with an intermediate (yellow line) or low (green line) level of β1,4GalTV (log-rank test, p = 2.65 × 10−3). D, correlation between nucleus NICD protein expression and β1,4GalTV protein expression in human GBM specimens. The scores for quantitative staining of the tissue sections were determined. The correlation between nucleus NICD protein expression and β1,4GalTV protein expression was significant as determined by Kendall correlation test (r = 0.815, p < 0.01).
Discussion
Here we report for the first time that β1,4GalTV can regulate the transdifferentiation of glioma stem-like cells into endothelial cells in vitro and in vivo. β1,4GalTV depletion could significantly improve the survival of tumor-bearing mice. Additionally, the REMBRANDT database indicated a significant decrease in the probability of survival with high β1,4GalTV expression. The expression of β1,4GalTV was correlated with glioma grade. Thus, β1,4GalTV might be a therapeutic target for GBM.
Tumor angiogenesis is fundamental to glioma development (43, 44). The regulation of glioma angiogenesis is a complex and intricately orchestrated process involving multiple interrelated signaling pathways, such as VEGF/VEGFR2 and HIF-1 (45–48). Increasing evidence has revealed that N-glycan plays a critical role in tumor angiogenesis (49, 50). Recent studies have also reported that tunicamycin, a widely known N-glycosylation inhibitor, inhibited angiogenesis in breast tumors (51). Therefore, studying the role of N-glycan in tumor angiogenesis might help to elucidate the mechanism of angiogenesis. β1,4GalTV regulates the transdifferentiation of glioma stem-like cells into endothelial cells depending on its galactosylation activity. To our knowledge, our study reports for the first time that glycosyltransferase regulates the transdifferentiation process from glioma stem-like cells into endothelial cells. Our findings might help to understand the role of N-glycan chains in tumor development.
Another interesting finding was that β1,4GalTV regulated Notch signaling. Results from previous studies have demonstrated that the Notch1 signaling pathway is critical for the transdifferentiation process from glioma stem-like cells into vascular endothelial cells (5, 25). Our results demonstrate that decreasing the expression of β1,4GalTV reduces Notch1 cleavage. Ectopic expression of the Notch1 intracellular domain could rescue the inhibitory effect of β1,4GalTV knockdown on transdifferentiation from glioma stem-like cells. Thus, we speculate that β1,4GalTV regulates the transdifferentiation process from glioma stem-like cells into endothelial cells through the Notch1 signaling pathway.
The extracellular domains of Notch receptors contain numerous potential sites for N-linked and O-linked glycosylation (28, 52, 53). Fringe is a β 3N-acetylglucosaminyltransferase (β3GlcNAcT) that transfers GlcNAc to O-fucose in epidermal growth factor-like repeats of Notch. Fringe regulates the response of Notch to ligand (54, 55). The glycosyltransferase GnT-III activates Notch signaling through inhibition of Notch degradation (37). Our data suggest that Notch1 has β1,4-galactosylation. In addition, β1,4GalTV can be immunoprecipitated with Notch. Thus, these data suggest that Notch might be a substrate of β1,4GalTV. Furthermore, β1,4GalTV knockdown obviously inhibited the interaction between Notch1 and galectin-3. Galectin-3 treatment could recue the inhibitory effect of β1,4GalTV knockdown on Notch1 expression. However, the mechanisms by which β1,4GalTV regulates Notch1 lysosome-mediated degradation need further exploration.
Gliomas develop as a result of stepwise accumulations of multiple genetic alterations that result in the activation of oncogenes such as Ras/Raf/extracellular signal-regulated kinase and phosphatidylinositol 3-kinase/Akt signaling pathways (56). Our previous studies have shown that β1,4GalTV expression is activated by Ras/Raf/extracellular signal-regulated kinase and Akt signaling pathways (33). Thus, our finding that β1,4GalTV regulates the transdifferentiation of glioma stem-like cells into endothelial cells might provide cues for understanding the mechanisms of the Akt and Ras signaling pathways promoting glioma development.
Our findings expand the understanding of the mechanism involved in tumor angiogenesis and provide a novel strategy for glioma therapy. The molecular mechanism of β1,4GalTV regulating Notch signaling should be further explored.
Experimental procedures
Cell culture
The tumor sample studies were approved by the Research Ethics Committee of Fudan University, and informed consent was obtained from all brain tumor patients contributing tumor specimens. T698968 cells and T109002 cells were obtained from primary human brain tumor patient specimens. Briefly, tumors were digested with collagenase (type V, Sigma) and filtered with a 70-μm cell strainer to remove tissue pieces. Cells were then cultured under non-adherent conditions in neural stem cell culture medium composed of Dulbecco's modified Eagle's medium (Gibco) and Ham's F-12 medium supplemented with serum-free B27 (Gibco), 50 mm Hepes, 2 μg/ml heparin (Sigma), 20 ng/ml epidermal growth factor (Chemicon), and 20 ng/ml FGF-2 (Chemicon) in a humidified CO2 incubator (5% CO2, 95% air).
Lentivirus production and cell infection
The pLL3.7 vector containing a GFP expression cassette was a gift from Prof. V. Parijs (Massachusetts Institute of Technology, Cambridge, MA). Hairpin sequences used for the β1,4GalTV shRNA were as follows: β1,4GalTV (human) shRNA1, forward 5′-TGATGACGACCTCTGGAACATTCAAGAGATGTTCCAGAGGTCGTCATCTTTTTTC-3′ and reverse 5′-TCGAGAAAAAAGATGACGACCTCTGGAACATCTCTTGAATGTTCCAGAGGTCGTCATCA-3′; β1,4GaTV (human) shRNA2, forward 5′-TGACATCACATACGACGCCTTTTCAAGAGAAAGGCGTCGTATGTGATGTTTTTTTC-3′ and reverse 5′-TCGAGAAAAAAACATCACATACGACGCCTTTCTCTTGAAAAGGCGTCGTATGTGATGTCA-3′; β1,4GalTV (human) shRNA3, forward 5′-TCGGAGTGAGTGGCTTAACATTCAAGAGATGTTAAGCCACTCACTCCGTTTTTTC-3′ and reverse 5′-TCGAGAAAAAACGGAGTGAGTGGCTTAACATCTCTTGAATGTTAAGCCACTCACTCCGA-3′. The β1,4GalTV shRNA sequenceswere cloned into the pLL3.7 vector to generate a lentiviral expression vector. β1,4GalTV (human) was cloned into the LV-FLAG vector using BamHI and AgeI restriction enzymes. The β1,4GalTV (human) primers were as follows: forward 5′-GGGTGGATCCATGTACCCATACGATGTTCCAGATTACGCTCGCGCCCGCCG-3′ and reverse 5′-CGGACCGGTCCGTACTCGTTCACCT-3′. NICD was subcloned into the LV-FLAG vector using BamHI and AgeI restriction enzymes. The NICD primers were as follows: forward 5′-CGCATGGATCCATGCGGCGGCAGCATGGCCAG-3′ and reverse 5′-AGGCACCGGTGCCTTGAAGGCCTCCGGAAT-3′. Lentivirus production was done by transfection of 293T cells using calcium phosphate transfection. Supernatants were collected 72 h after transfection and filtered; the viral titers were then determined by FACS 48 h post-transduction. Subconfluent cells were infected with the lentivirus at a multiplicity of infection of 5 in the presence of 8 μg/ml Polybrene (Sigma-Aldrich).
Tube formation assay
The in vitro tube formation assay was performed as described previously (57). In brief, 12 μl of tail collagen was dropped onto glass coverslips on 12-well plates and allowed to polymerize for 1 h at 37 °C. Cells (1 × 104) were then suspended in 2 ml of endothelial basal medium (Gibco) containing 2% fetal bovine serum and incubated in a humidified CO2 incubator (5% CO2, 95% air) for 7 days.
Data were photographically recorded every day. Images were acquired using Motic Microscopy connected to a computer with the online image acquisition software WinFast PVR2. For quantification of tube lengths, images were exported to Image-Pro Plus software.
Immunoblot analysis
The Western blot assay was performed as described previously (33). The following primary antibodies were used: mouse monoclonal anti-Notch1 (BD Pharmingen, catalog no. 552466), rabbit polyclonal anti-FLAG (Sigma, catalog no. F7425), rabbit polyclonal anti-galectin-3 (Abcam, catalog no. 31707), and rabbit polyclonal anti-β1,4GalTV (Santa Cruz Biotechnology, catalog no. sc-22289). Horseradish peroxidase (HRP)–conjugated secondary antibodies were as follows: goat anti-mouse (Santa Cruz Biotechnology, catalog no. sc-2005) and goat anti-rabbit (Santa Cruz Biotechnology, catalog no. sc-2004). Relative protein levels were quantified by scanning densitometry. The gray value of the protein level was measured with National Institutes of Health ImageJ Software.
Lectin blots were also performed as described previously (33). The primary antibody was biotinylated lectin Ricinus communis agglutinin I (RCA-1) (Vector, catalog no. B-1085). The secondary antibody was HRP conjugated with streptavidin (Southern Biotech, catalog no. 7100-05).
Immunofluorescence
Immunofluorescence assays were performed on cells and frozen sections following protocols described previously (58). The following primary antibodies were used: mouse monoclonal anti-Nestin (Millipore, catalog no. MAB5326), rabbit polyclonal anti-GFAP (glial fibrillary acidic protein) (Millipore, catalog no. AB5804), rabbit polyclonal anti-β-tubulin III (Sigma, catalog no. T2200), goat polyclonal anti-CD31 (Santa Cruz Biotechnology, catalog no. sc-1506), and goat polyclonal anti-Notch1 (Santa Cruz Biotechnology, catalog no. sc6014). The secondary antibodies used were as follows: Alexa Fluor 594 anti-mouse IgG, Alexa Fluor 594 anti-rabbit IgG, and Alexa Fluor 594 anti-goat IgG. Nuclei were stained with Hoechst (Sigma, catalog no. 33258). The images were obtained by confocal laser-scanning microscopy (Leica TCS SP5), and the obtained images were processed with LAS-AF-Lite software.
Tumor formation assay
For intracranial xenografts, 4-week-old male nude mice were intracranially injected with 1.0 × 105 T698968 cells or T109002 cells into the right frontal lobes under the Fudan University Animal Care Committee protocol. Four weeks later, bioluminescence imaging was performed on nude mice to measure tumor size. All mice were maintained until development of neurologic signs, sacrificed, and perfused with 4% paraformaldehyde.
Immunohistochemistry
Human brain tumor specimens of different grades were provided by Zhongshan Hospital of Fudan University and Provincial Hospital affiliated with Shandong University in accordance with the appropriate institutional review boards. Immunohistochemistry studies were performed on paraffin-embedded primary human glioblastoma sections and mouse intracranial glioblastoma xenograft sections following a protocol described previously (59). The following primary antibodies were used: mouse monoclonal anti-human-CD31 (BD Pharmingen, catalog no. 550389) and rabbit polyclonal anti-β1,4GalTV (Santa Cruz Biotechnology, catalog no. sc-22289). Secondary antibodies conjugated with HRP were as follows: goat anti-mouse (Santa Cruz Biotechnology, catalog no. sc-2005) and goat anti-rabbit (Santa Cruz Biotechnology, catalog no. sc-2004). Tris-EDTA buffer (pH 9.0) was used for antigen retrieval in all cases. Images were acquired using Motic Microscopy connected to a computer with the online image acquisition software WinFast PVR2. For quantification, images were exported to Image-Pro Plus software.
RT-PCR
Total RNA (1 μg) extracted was used as a template for complementary DNA synthesis with a TaKaRa RNA PCR kit and specific primers as follows: β1,4GalTV (human), forward 5′-GCTGCTGTACTTCGTCTATGTGGCGC-3′ and reverse 5′-GCCTCGGCATCTGTCCACATCC-3′; CD31 (human), forward 5′-ATTGCAGTGGTTATCATCGGAGTG-3′ and reverse 5′-CTCGTTGTTGGAGTTCAGAAGTGG-3′. Amplification was carried out for 22–27 cycles under saturation (each at 94 °C, 45 s; 60 °C, 45 s; and 72 °C, 1 min) in a 50-μl reaction mixture containing 2 μl of each complementary DNA, 0.2 μm each primer, 0.2 mm dNTP, and 2.5 units of DNA polymerase (Sino Develop). After amplification, 10 μl of each reaction mixture was analyzed by 1%∼2.5% agarose gel electrophoresis, and the bands were then visualized by ethidium bromide staining. Images were acquired by Gel Imaging (Xia Ri Technology). System levels of mRNA were quantified with National Institutes of Health ImageJ software.
Pulse–chase analysis
Cells were incubated with Dulbecco's modified Eagle's medium/F12 medium depleted of methionine for 1 h at 37 °C. Pulse labeling was performed with 1 mm AHA (l-azidohomoalanine, a structural analog of methionine) for 60 min as described previously (60). Cells were returned to the above culture conditions containing methionine for 0, 15, 30, 45, 60, or 75 min. Then cell surface protein was eluted using a cell surface protein isolation kit (Pierce, 89881). AHA-labeled protein on the cell surface was purified by immunoprecipitation. Immunoprecipitates were subjected to SDS-PAGE, and AHA-labeled Notch1 was examined.
Statistical analysis
All in vitro experiments were repeated at least three times. All numerical data are expressed as mean ± S.D. or mean ± S.E. as indicated. Data were analyzed using Student's t test or one-way analysis of variance. Survival curves were generated using Kaplan–Meier methods and compared using a log-rank test. Kendall correlation analysis was used to analyze the correlation between nucleus NICD protein expression and β1,4GalTV expression.
Author contributions
C. C., X. C., Y. Liu, B. C., Y. X., C.L., F.Y., Y. Li, T.Y., and J.J. data curation; C.C., X.C., Y. Liu, B.C., and Y.X. validation; X.C. formal analysis; L.H. and M.T. resources; Y.W., Y.G., and J.J. supervision; Y.W. and J.J. writing-review and editing; J.J. writing-original draft.
Supplementary Material
Acknowledgments
We thank Prof. Yalin Huang (Institute of Stem Cell and Regenerative Medicine, Institute of Biomedical Sciences, Fudan University) for providing confocal laser-scanning microscopy technology.
This work was supported by the Program for National Natural Scientific Foundation of China (31370807, 81472724, 81773164, and 31770856) and the National Key R&D Program of China (2016YFA0501303). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and S2.
- GBM
- glioblastoma multiforme
- VEGF
- vascular endothelial growth factor
- β1,4GalTV
- β1,4-galactosyltransferase V
- shRNA
- short hairpin RNA
- NICD
- Notch1 intracellular domain
- BLI
- bioluminescent imaging
- HRP
- horseradish peroxidase
- AHA
- l-azidohomoalanine
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- DAPT
- N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester
- LV
- lentiviral.
References
- 1. Maher E. A., Furnari F. B., Bachoo R. M., Rowitch D. H., Louis D. N., Cavenee W. K., and DePinho R. A. (2001) Malignant glioma: genetics and biology of a grave matter. Genes Dev. 15, 1311–1333 10.1101/gad.891601 [DOI] [PubMed] [Google Scholar]
- 2. Vredenburgh J. J., Desjardins A., Herndon J. E. 2nd, Dowell J. M., Reardon D. A., Quinn J. A., Rich J. N., Sathornsumetee S., Gururangan S., Wagner M., Bigner D. D., Friedman A. H., and Friedman H. S. (2007) Phase II trial of bevacizumab and irinotecan in recurrent malignant glioma. Clin. Cancer Res. 13, 1253–1259 10.1158/1078-0432.CCR-06-2309 [DOI] [PubMed] [Google Scholar]
- 3. Bergers G., and Hanahan D. (2008) Modes of resistance to anti-angiogenic therapy. Nat. Rev. Cancer 8, 592–603 10.1038/nrc2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shojaei F., and Ferrara N. (2008) Refractoriness to antivascular endothelial growth factor treatment: role of myeloid cells. Cancer Res. 68, 5501–5504 10.1158/0008-5472.CAN-08-0925 [DOI] [PubMed] [Google Scholar]
- 5. Wang R., Chadalavada K., Wilshire J., Kowalik U., Hovinga K. E., Geber A., Fligelman B., Leversha M., Brennan C., and Tabar V. (2010) Glioblastoma stem-like cells give rise to tumour endothelium. Nature 468, 829–833 10.1038/nature09624 [DOI] [PubMed] [Google Scholar]
- 6. Ricci-Vitiani L., Pallini R., Biffoni M., Todaro M., Invernici G., Cenci T., Maira G., Parati E. A., Stassi G., Larocca L. M., and De Maria R. (2010) Tumour vascularization via endothelial differentiation of glioblastoma stem-like cells. Nature 468, 824–828 10.1038/nature09557 [DOI] [PubMed] [Google Scholar]
- 7. Soda Y., Marumoto T., Friedmann-Morvinski D., Soda M., Liu F., Michiue H., Pastorino S., Yang M., Hoffman R. M., Kesari S., and Verma I. M. (2011) Transdifferentiation of glioblastoma cells into vascular endothelial cells. Proc. Natl. Acad. Sci. U.S.A. 108, 4274–4280 10.1073/pnas.1016030108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Couldrey C., and Green J. E. (2000) Metastases: the glycan connection. Breast Cancer Res. 2, 321–323 10.1186/bcr75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rebbaa A., Yamamoto H., Saito T., Meuillet E., Kim P., Kersey D. S., Bremer E. G., Taniguchi N., and Moskal J. R. (1997) Gene transfection-mediated overexpression of β1,4-N-acetylglucosamine bisecting oligosaccharides in the glioma cell line U373 MG inhibits epidermal growth factor receptor function. J. Biol. Chem. 272, 9275–9279 10.1074/jbc.272.14.9275 [DOI] [PubMed] [Google Scholar]
- 10. Yamamoto H., Swoger J., Greene S., Saito T., Hurh J., Sweeley C., Leestma J., Mkrdichian E., Cerullo L., Nishikawa A., Ihara Y., Taniguchi N., and Moskal J. R. (2000) 1,6-N-acetylglucosamine-bearing N-glycans in human gliomas: implications for a role in regulating invasivity. Cancer Res. 60, 134–142 [PubMed] [Google Scholar]
- 11. Xu S., Zhang S., Chen C., Yan J., Cai M., Zhu X., and Gu J. (2002) Over-expression of β-1,4-galactosyltransferase V increases the growth of astrocytoma cell line. J. Exp. Clin. Cancer Res. 21, 409–414 [PubMed] [Google Scholar]
- 12. Hakomori S. (1996) Tumor malignancy defined by aberrant glycosylation and sphingo(glyco)lipid metabolism. Cancer Res. 56, 5309–5318 [PubMed] [Google Scholar]
- 13. Dawson G., Moskal J. R., and Dawson S. A. (2004) Transfection of 2,6 and 2,3-sialyltransferase genes and GlcNAc-transferase genes into human glioma cell line U-373 MG affects glycoconjugate expression and enhances cell death. J. Neurochem. 89, 1436–1444 10.1111/j.1471-4159.2004.02435.x [DOI] [PubMed] [Google Scholar]
- 14. Sato T., and Furukawa K. (2007) Sequential action of Ets-1 and Sp1 in the activation of the human β-1,4-galactosyltransferase V gene involved in abnormal glycosylation characteristic of cancer cells. J. Biol. Chem. 282, 27702–27712 10.1074/jbc.M611862200 [DOI] [PubMed] [Google Scholar]
- 15. Sato T., Guo S., and Furukawa K. (2001) Occurrence of poly-N-acetyllactosamine synthesis in Sf-9 cells upon transfection of individual human β-1,4-galactosyltransferase I, II, III, IV, V and VI cDNAs. Biochimie 83, 719–725 10.1016/S0300-9084(01)01304-9 [DOI] [PubMed] [Google Scholar]
- 16. Xu S., Zhu X., Zhang S., Yin S., Zhou L., Chen C., and Gu J. (2001) Over-expression of β-1,4-galactosyltransferase I, II, and V in human astrocytoma. J. Cancer Res. Clin. Oncol. 127, 502–506 10.1007/s004320100246 [DOI] [PubMed] [Google Scholar]
- 17. Jiang J., Wei Y., Shen J., Liu D., Chen X., Zhou J., Zong H., Yun X., Kong X., Zhang S., Yang Y., and Gu J. (2007) Functional interaction of E1AF and Sp1 in glioma invasion. Mol. Cell. Biol. 27, 8770–8782 10.1128/MCB.02302-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wei Y., Liu D., Ge Y., Zhou F., Xu J., Chen H., Yun X., Gu J., and Jiang J. (2008) Down-regulation of β1,4GalT V at protein level contributes to arsenic trioxide-induced glioma cell apoptosis. Cancer Lett. 267, 96–105 10.1016/j.canlet.2008.03.019 [DOI] [PubMed] [Google Scholar]
- 19. Wei Y., Zhou F., Ge Y., Chen H., Cui C., Li Q., Liu D., Yang Z., Wu G., Sun S., Gu J., and Jiang J. (2010) β1,4-galactosyltransferase V regulates self-renewal of glioma-initiating cell. Biochem. Biophys. Res. Commun. 396, 602–607 10.1016/j.bbrc.2010.04.110 [DOI] [PubMed] [Google Scholar]
- 20. Guo H., Nagy T., and Pierce M. (2014) Post-translational glycoprotein modifications regulate colon cancer stem cells and colon adenoma progression in Apc(−/+) mice through altered Wnt receptor signaling. J. Biol. Chem. 289, 31534–31549 10.1074/jbc.M114.602680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xing Y., Ge Y., Liu C., Zhang X., Jiang J., and Wei Y. (2016) ER stress inducer tunicamycin suppresses the self-renewal of glioma-initiating cell partly through inhibiting Sox2 translation. Oncotarget 7, 36395–36406 10.18632/oncotarget.8954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dennis J. W., Lau K. S., Demetriou M., and Nabi I. R. (2009) Adaptive regulation at the cell surface by N-glycosylation. Traffic 10, 1569–1578 10.1111/j.1600-0854.2009.00981.x [DOI] [PubMed] [Google Scholar]
- 23. Ehebauer M., Hayward P., and Arias A. M. (2006) Notch, a universal arbiter of cell fate decisions. Science 314, 1414–1415 10.1126/science.1134042 [DOI] [PubMed] [Google Scholar]
- 24. Fernandez-Valdivia R., Takeuchi H., Samarghandi A., Lopez M., Leonardi J., Haltiwanger R. S., and Jafar-Nejad H. (2011) Regulation of mammalian Notch signaling and embryonic development by the protein O-glucosyltransferase Rumi. Development 138, 1925–1934 10.1242/dev.060020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hovinga K. E., Shimizu F., Wang R., Panagiotakos G., Van Der Heijden M., Moayedpardazi H., Correia A. S., Soulet D., Major T., Menon J., and Tabar V. (2010) Inhibition of notch signaling in glioblastoma targets cancer stem cells via an endothelial cell intermediate. Stem Cells 28, 1019–1029 10.1002/stem.429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jafar-Nejad H., Leonardi J., and Fernandez-Valdivia R. (2010) Role of glycans and glycosyltransferases in the regulation of Notch signaling. Glycobiology 20, 931–949 10.1093/glycob/cwq053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Batista F., Lu L., Williams S. A., and Stanley P. (2012) Complex N-glycans are essential, but core 1 and 2 mucin O-glycans, O-fucose glycans, and NOTCH1 are dispensable, for mammalian spermatogenesis. Biol. Reprod. 86, 179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haines N., and Irvine K. D. (2003) Glycosylation regulates Notch signalling. Nat. Rev. Mol. Cell Biol. 4, 786–797 10.1038/nrm1228 [DOI] [PubMed] [Google Scholar]
- 29. Galli R., Binda E., Orfanelli U., Cipelletti B., Gritti A., De Vitis S., Fiocco R., Foroni C., Dimeco F., and Vescovi A. (2004) Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 64, 7011–7021 10.1158/0008-5472.CAN-04-1364 [DOI] [PubMed] [Google Scholar]
- 30. Ge Y., Zhou F., Chen H., Cui C., Liu D., Li Q., Yang Z., Wu G., Sun S., Gu J., Wei Y., and Jiang J. (2010) Sox2 is translationally activated by eukaryotic initiation factor 4E in human glioma-initiating cells. Biochem. Biophys. Res. Commun. 397, 711–717 10.1016/j.bbrc.2010.06.015 [DOI] [PubMed] [Google Scholar]
- 31. Ishida H., Togayachi A., Sakai T., Iwai T., Hiruma T., Sato T., Okubo R., Inaba N., Kudo T., Gotoh M., Shoda J., Tanaka N., and Narimatsu H. (2005) A novel β1,3-N-acetylglucosaminyltransferase (β3Gn-T8), which synthesizes poly-N-acetyllactosamine, is dramatically upregulated in colon cancer. FEBS Lett. 579, 71–78 10.1016/j.febslet.2004.11.037 [DOI] [PubMed] [Google Scholar]
- 32. Lo N. W., Shaper J. H., Pevsner J., and Shaper N. L. (1998) The expanding β 4-galactosyltransferase gene family: messages from the databanks. Glycobiology 8, 517–526 10.1093/glycob/8.5.517 [DOI] [PubMed] [Google Scholar]
- 33. Fang J., Meng Q., Vogt P. K., Zhang R., and Jiang B. H. (2006) A downstream kinase of the mammalian target of rapamycin, p70S6K1, regulates human double minute 2 protein phosphorylation and stability. J. Cell. Physiol. 209, 261–265 10.1002/jcp.20749 [DOI] [PubMed] [Google Scholar]
- 34. Sato T., and Furukawa K. (2004) Transcriptional regulation of the human β-1,4-galactosyltransferase V gene in cancer cells: essential role of transcription factor Sp1. J. Biol. Chem. 279, 39574–39583 10.1074/jbc.M405805200 [DOI] [PubMed] [Google Scholar]
- 35. Andersson E. R., Sandberg R., and Lendahl U. (2011) Notch signaling: simplicity in design, versatility in function. Development 138, 3593–3612 10.1242/dev.063610 [DOI] [PubMed] [Google Scholar]
- 36. Platonova N., Manzo T., Mirandola L., Colombo M., Calzavara E., Vigolo E., Cermisoni G. C., De Simone D., Garavelli S., Cecchinato V., Lazzari E., Neri A., and Chiaramonte R. (2015) PI3K/AKT signaling inhibits NOTCH1 lysosome-mediated degradation. Genes Chromosomes Cancer 54, 516–526 10.1002/gcc.22264 [DOI] [PubMed] [Google Scholar]
- 37. Harvey B. M., Rana N. A., Moss H., Leonardi J., Jafar-Nejad H., and Haltiwanger R. S. (2016) Mapping sites of O-glycosylation and fringe elongation on Drosophila Notch. J. Biol. Chem. 291, 16348–16360 10.1074/jbc.M116.732537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vagin O., Kraut J. A., and Sachs G. (2009) Role of N-glycosylation in trafficking of apical membrane proteins in epithelia. Am. J. Physiol. Renal Physiol. 296, F459–F469 10.1152/ajprenal.90340.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hughes R. C. (1997) The galectin family of mammalian carbohydrate-binding molecules. Biochem. Soc. Trans. 25, 1194–1198 10.1042/bst0251194 [DOI] [PubMed] [Google Scholar]
- 40. Funasaka T., Raz A., and Nangia-Makker P. (2014) Galectin-3 in angiogenesis and metastasis. Glycobiology 24, 886–891 10.1093/glycob/cwu086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nakajima K., Kho D. H., Yanagawa T., Harazono Y., Gao X., Hogan V., and Raz A. (2014) Galectin-3 inhibits osteoblast differentiation through notch signaling. Neoplasia 16, 939–949 10.1016/j.neo.2014.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schuster-Gossler K., Harris B., Johnson K. R., Serth J., and Gossler A. (2009) Notch signalling in the paraxial mesoderm is most sensitive to reduced Pofut1 levels during early mouse development. BMC Dev. Biol. 9, 6 10.1186/1471-213X-9-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kaur B., Khwaja F. W., Severson E. A., Matheny S. L., Brat D. J., and Van Meir E. G. (2005) Hypoxia and the hypoxia-inducible-factor pathway in glioma growth and angiogenesis. Neuro-oncology 7, 134–153 10.1215/S1152851704001115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Folkman J. (2002) Role of angiogenesis in tumor growth and metastasis. Semin. Oncol. 29, 15–18 10.1053/sonc.2002.37263 [DOI] [PubMed] [Google Scholar]
- 45. De Falco S., Gigante B., and Persico M. G. (2002) Structure and function of placental growth factor. Trends Cardiovasc. Med. 12, 241–246 10.1016/S1050-1738(02)00168-8 [DOI] [PubMed] [Google Scholar]
- 46. Plate K. H., Breier G., Weich H. A., and Risau W. (1992) Vascular endothelial growth factor is a potential tumour angiogenesis factor in human gliomas in vivo. Nature 359, 845–848 10.1038/359845a0 [DOI] [PubMed] [Google Scholar]
- 47. Jackson M. W., Roberts J. S., Heckford S. E., Ricciardelli C., Stahl J., Choong C., Horsfall D. J., and Tilley W. D. (2002) A potential autocrine role for vascular endothelial growth factor in prostate cancer. Cancer Res. 62, 854–859 [PubMed] [Google Scholar]
- 48. Maxwell P. H., Dachs G. U., Gleadle J. M., Nicholls L. G., Harris A. L., Stratford I. J., Hankinson O., Pugh C. W., and Ratcliffe P. J. (1997) Hypoxia-inducible factor-1 modulates gene expression in solid tumors and influences both angiogenesis and tumor growth. Proc. Natl. Acad. Sci. U.S.A. 94, 8104–8109 10.1073/pnas.94.15.8104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Banerjee D. K., and Vendrell-Ramos M. (1993) Is asparagine-linked protein glycosylation an obligatory requirement for angiogenesis? Indian J. Biochem. Biophys. 30, 389–394 [PubMed] [Google Scholar]
- 50. Nguyen M., Strubel N. A., and Bischoff J. (1993) A role for sialyl Lewis-X/A glycoconjugates in capillary morphogenesis. Nature 365, 267–269 10.1038/365267a0 [DOI] [PubMed] [Google Scholar]
- 51. Banerjee A., Lang J. Y., Hung M. C., Sengupta K., Banerjee S. K., Baksi K., and Banerjee D. K. (2011) Unfolded protein response is required in nu/nu mice microvasculature for treating breast tumor with tunicamycin. J. Biol. Chem. 286, 29127–29138 10.1074/jbc.M110.169771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bush G., diSibio G., Miyamoto A., Denault J. B., Leduc R., and Weinmaster G. (2001) Ligand-induced signaling in the absence of furin processing of Notch1. Dev. Biol. 229, 494–502 10.1006/dbio.2000.9992 [DOI] [PubMed] [Google Scholar]
- 53. Moloney D. J., Shair L. H., Lu F. M., Xia J., Locke R., Matta K. L., and Haltiwanger R. S. (2000) Mammalian Notch1 is modified with two unusual forms of O-linked glycosylation found on epidermal growth factor-like modules. J. Biol. Chem. 275, 9604–9611 10.1074/jbc.275.13.9604 [DOI] [PubMed] [Google Scholar]
- 54. Brückner K., Perez L., Clausen H., and Cohen S. (2000) Glycosyltransferase activity of Fringe modulates Notch-Delta interactions. Nature 406, 411–415 10.1038/35019075 [DOI] [PubMed] [Google Scholar]
- 55. Moloney D. J., Panin V. M., Johnston S. H., Chen J., Shao L., Wilson R., Wang Y., Stanley P., Irvine K. D., Haltiwanger R. S., and Vogt T. F. (2000) Fringe is a glycosyltransferase that modifies Notch. Nature 406, 369–375 10.1038/35019000 [DOI] [PubMed] [Google Scholar]
- 56. Cavenee W. K. (1992) Accumulation of genetic defects during astrocytoma progression. Cancer 70, 1788–1793 10.1002/1097-0142(19920915)70:4+%3C1788::AID-CNCR2820701621%3E3.0.CO%3B2-L [DOI] [PubMed] [Google Scholar]
- 57. Kolmakova A., Rajesh M., Zang D., Pili R., and Chatterjee S. (2009) VEGF recruits lactosylceramide to induce endothelial cell adhesion molecule expression and angiogenesis in vitro and in vivo. Glycoconj. J. 26, 547–558 10.1007/s10719-008-9206-9 [DOI] [PubMed] [Google Scholar]
- 58. Fang D., Hawke D., Zheng Y., Xia Y., Meisenhelder J., Nika H., Mills G. B., Kobayashi R., Hunter T., and Lu Z. (2007) Phosphorylation of β-catenin by AKT promotes beta-catenin transcriptional activity. J. Biol. Chem. 282, 11221–11229 10.1074/jbc.M611871200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Beck H., Acker T., Wiessner C., Allegrini P. R., and Plate K. H. (2000) Expression of angiopoietin-1, angiopoietin-2, and tie receptors after middle cerebral artery occlusion in the rat. Am. J. Pathol. 157, 1473–1483 10.1016/S0002-9440(10)64786-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang J., Zhang J., Lee Y. M., Ng S., Shi Y., Hua Z. C., Lin Q., and Shen H. M. (2017) Nonradioactive quantification of autophagic protein degradation with L-azidohomoalanine labeling. Nat. Protoc. 12, 279–288 10.1038/nprot.2016.160 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.