Abstract
Background and Aims
Plant growth-promoting rhizobacteria (PGPR) strains can influence plant–insect interactions. However, little is known about the effect of changes in the soil bacterial community in general and especially the loss of rare soil microbes on these interactions. Here, the influence of rare soil microbe reduction on induced systemic resistance (ISR) in a wild ecotype of Arabidopsis thaliana against the aphid Myzus persicae was investigated.
Methods
To create a gradient of microbial abundances, soil was inoculated with a serial dilution of a microbial community and responses of Arabidopsis plants that originated from the same site as the soil microbes were tested. Plant biomass, transcription of genes involved in plant defences, and insect performance were measured. In addition, the effects of the PGPR strain Pseudomonas fluorescens SS101 on plant and insect performance were tested under the influence of the various soil dilution treatments.
Key Results
Plant biomass showed a hump-shaped relationship with soil microbial community dilution, independent of aphid or Pseudomonas treatments. Both aphid infestation and inoculation with Pseudomonas reduced plant biomass, and led to downregulation of PR1 (salicylic acid-responsive gene) and CYP79B3 (involved in synthesis of glucosinolates). Aphid performance and gene transcription were unaffected by soil dilution.
Conclusions
Neither the loss of rare microbial species, as caused by soil dilution, nor Pseudomonas affect the resistance of A. thaliana against M. persicae. However, both Pseudomonas survival and plant biomass respond to rare species loss. Thus, loss of rare soil microbial species can have a significant impact on both above- and below-ground organisms.
Keywords: Arabidopsis thaliana, induced systemic resistance, low-abundance soil microbes, Myzus persicae, PGPR
INTRODUCTION
There is increasing awareness that some soil bacterial strains may have beneficial effects on plants (Compant et al., 2010). Most intensively studied and applied are the so-called plant growth-promoting rhizobacteria (PGPR). These bacteria can directly promote plant growth, and also systemically enhance plant defences against above- and below-ground pathogens and herbivores (Lugtenberg and Kamilova, 2009; Pieterse et al., 2014). A widely known mechanism through which beneficial soil microorganisms can affect plant defence is designated induced systemic resistance (ISR) (Van der Ent et al., 2009). This type of resistance is dependent on jasmonic acid (JA) and ethylene (ET), but can also be mediated via salicylic acid (SA) signalling and is triggered by interaction with non-pathogenic microbes. Colonization of plant roots by certain PGPR may prime plant defences, resulting in a faster and stronger defence response upon pathogen or insect attack (Van Wees et al., 2008). Induced systemic resistance is most effective against necrotrophic pathogens and insect herbivores (Pieterse and Dicke, 2007). However, little is known about how the induction of resistance by single PGPR strains compares with the effect of a natural soil microbial community on plant resistance.
Many PGPR have been recognized to be able to induce systemic resistance in the plant. Especially fluorescent pseudomonads have high effectiveness in the protection of plant health. Different bacterial species or strains have been found to induce resistance via different pathways. Some interact with only particular plant hosts (Pieterse et al., 2002, 2003). For example, Pseudomonas fluorescens WCS417 protects Arabidopsis thaliana against a variety of pathogens via priming of the JA signalling pathway, whereas P. fluorescens SS101 has been found to increase plant resistance by inducing SA-dependent defences (Pieterse et al., 2002; Van de Mortel et al., 2012; Pangesti et al., 2017). Other bacterial species, such as Bacillus amyloliquefaciens, Bacillus subtilis and Serratia marcescens, have also shown protective ability (Mauch-Mani and Métraux, 1998; Van Loon and Bakker, 2006). In order to acquire effective induced resistance in plants, high densities of ≥105 colony-forming units (CFU) per gram of root of a previously cultured PGPR strain in soil are required (Raaijmakers et al., 1995). However, the bacterial numbers often rapidly decline once inoculated into soil, as a result of competition with the microbial community that already exists in the soil (Mallon et al., 2015; Adam et al., 2016). Reduced species richness could be expected to reduce competition and lead to increased survival of PGPR.
Besides well-studied PGPR strains, soil is also inhabited by numerous other bacteria and other microbial species carrying out many different functions (Bent and Forney, 2008). Whereas soil bacterial diversity may relate positively to specific ecosystem functions (Griffiths et al., 2004; Cook et al., 2006), there is increasing evidence that the presence and absence of certain species, as well as their relative abundances, might be even more important than species diversity (Strickland et al., 2009). Especially low-abundant bacterial species, also termed the ‘rare biosphere’ (Sogin et al., 2006), can have a greater impact on certain ecosystem processes than predicted based on their abundance (Jousset et al., 2017). Rare taxa may contribute disproportionally to microbial community dynamics and activity (Jones and Lennon, 2010; Shade et al., 2014; Wilhelm et al., 2014). Rare microbes that are specialized on recalcitrant substrates may play a crucial role in decomposition of soil organic matter. In addition, the presence of a high diversity of rare species may increase the community resistance to pathogen invasion, thereby enhancing soil disease suppressiveness (Van Elsas et al., 2012; Baumann et al., 2013). However, high diversity might also reduce the survival of introduced beneficial microbes. Rare soil microbes have also been shown to influence plant defences (Hol et al., 2010). In that study, the loss of rare microbes from soil was correlated with a decrease in plant defensive compounds and an increase in aphid performance. However, the molecular mechanisms behind this decrease still remain unresolved.
The effect of specific plant growth-promoting bacterial strains on ISR in plants has been intensively studied. In contrast, the effect of soil microbial community composition, especially the low-abundant members of the community, are poorly studied. Recent reports on the possible roles of the rare biosphere in ecosystem functioning raise questions about their contribution to plant defence induction relative to introduced PGPR (Van Elsas et al., 2012; Delgado-Baquerizo et al., 2016). Therefore, the aim of the present study was to test how (loss of) rare soil microbial species influences plant performance and induction of plant defences against above-ground insect herbivores. Furthermore, we investigated how loss of rare soil microbes affects defence induction by a known PGPR strain.
Our first hypothesis was that plants growing in soils containing a complete assemblage of low-abundant microbes will be more resistant to insect herbivores than plants growing in soils that lack a large fraction of these low-abundant microbes, based on our previous research (Hol et al., 2010). We expected that this resistance would be based on induced systemic resistance, which is known to be effective against aphid herbivores (De Vos et al., 2007). We also expected that soil with a full microbial community compared with soil with a community reduced in rare microbes would lead to (1) a less negative effect of above-ground aphids on the biomass of Arabidopsis thaliana, (2) priming of enhanced expression of defence-marker genes in A. thaliana via the ISR pathway and consequently higher expression of JA- and ET-dependent genes in the presence of herbivores, and (3) decreased insect fecundity. Our second hypothesis was that the addition of the PGPR strain Pseudomonas fluorescens SS101 would prime plant defences against insect herbivory, and that this effect would be reduced in microbial communities containing low-abundance species (Mallon et al., 2015).
MATERIALS AND METHODS
To test these hypotheses, we performed a soil dilution experiment [using a similar approach to that described by Hol et al. (2010)], in which plants of a wild ecotype of Arabidopsis thaliana originating from a grassland were grown in soil with six subsequent dilutions of a microbial community from the same grassland. Pseudomonas bacteria and the aphid herbivore Myzus persicae were added in a full factorial design. Myzus persicae is a common herbivore of Arabidopsis in the field and Pseudomonas has been shown to induce systemic resistance in this plant species (Harvey et al., 2007; Van de Mortel et al., 2012). After 24 h of aphid feeding the expression of several genes involved in induced plant defences was measured. After 2 weeks of aphid feeding, plant biomass and aphid reproductive success were assessed.
Soil collection and treatment
Ten soil samples (referred to as soil origins in the following) were collected from a biodiversity field experiment near Ede (The Netherlands). (For more details see Supplementary Data, Soil collection.) Soil inocula were made by suspending 30 g of each sample in 100 mL of phosphate buffer (1 g L−1 KH2PO4; pH 6.5). The suspensions were shaken for 1.5 h at 120 rpm on a flatbed shaker, sonicated twice for 1 min, followed by another 0.5 h of shaking. Subsequently they were sieved through a 45-µm sieve. Thus, the mesofauna was excluded but microorganisms, such as bacteria, archaea, fungi and protozoa, were kept. The resulting suspension represents the undiluted inoculum (or 100 dilution). Four subsequent 1:100 dilutions were prepared from all ten suspensions and used as inocula. Consequently, the five dilutions were 100, 102, 104, 106 and 108.
Three kilograms of sterilized bulk soil in autoclavable polypropylene plastic bags was inoculated with 100 mL of each dilution from each of the ten soil origins. Ten bags, serving as the sterile control, received 100 mL of sterile phosphate buffer similar to the dilution treatments, resulting in 60 bags of 3 kg each (five dilution treatments + control × ten soil origins). The bags were closed with a cotton wool plug to allow gas exchange. Then, the bags were stored at room temperature for a period of 25 weeks, during which the soil was homogenized every week by turning over the bags several times. This incubation was done in order to enable the microbial communities to reach similar cell densities independent of the dilution treatment (Griffiths et al., 2004; Hol et al., 2010). It was expected that in all soil origins the dilution treatment would reduce the rare species, as has been found in numerous previous dilution studies (Van Elsas et al., 2012; Yan et al., 2015; Roger et al., 2016).
Pseudomonas fluorescens SS101 culture
A rifampicin-resistant natural mutant of Pseudomonas fluorescens SS101 was cultured in liquid lysogeny broth (LB) medium for 24 h at 25 °C with continuous shaking, washed three times in sterile MgSO4 buffer (2.64 g L−1 MgSO4) and adjusted to a final concentration of 109 CFU per mL (OD600 1.0). One day prior to planting, all bags with 3 kg of soil from all dilution treatments, including the sterile control, were split into two bags of 1.5 kg each. One bag of 1.5 kg for each treatment was inoculated with 15 ml of Pseudomonas suspension, resulting in a final concentration of 107 CFU g−1 soil (+ Pseudomonas). Soil receiving no Pseudomonas inoculum was mock-treated with 15 mL of sterile MgSO4 buffer (− Pseudomonas).
Plant and insect material
The ecotype of A. thaliana (Msl) used in this study was obtained from the Laboratory of Molecular Biology of Wageningen University (Wageningen, The Netherlands) and was originally obtained from the site where the soil had been collected. All seeds used in this study originated from an inbred line and represented the third generation since collection from the field. The seeds were surface vapour-sterilized (van de Mortel et al., 2012). Five seeds per plate were sown on half-strength Murashige–Skoog agar (4.3 g L−1 Murashige–Skoog salts with vitamins, 103 g L−1 sucrose, 83 g L−1 plant agar) and germinated for 7 d in a climate chamber with a day/night cycle of 8/16 h, 21/21 °C, 200 µmol m−2 s−1 light intensity at plant level and 70 % relative humidity.
The generalist aphid species M. persicae was reared in a growth cabinet under a day/night cycle of 16/8 h, 22/22 °C and 50–70 % relative humidity. Aphids were reared on A. thaliana plants of the same ecotype as used in the experiment. One day prior to plant infestation adult aphids were isolated in a separate net cage to enable the collection of neonates.
Experimental setup and maintenance
Each bag of soil was used to fill three pots of 500 g soil each, resulting in 360 pots [ten soil origins × six dilution treatments × two Pseudomonas treatments (+ Pseudomonas/− Pseudomonas) × three pots]. The soil was saturated with autoclaved demineralized water (Demi) and one 7-d-old Arabidopsis seedling was planted in each pot. The plants were grown in a growth chamber in a randomized block design under the same conditions as during germination. Each block consisted of three trays each with 12 pots of a single soil origin with every dilution treatment present in duplicate (with and without Pseudomonas) and distributed randomly across the tray. This resulted in 27 trays with nine soil origins (Supplementary Data Fig. S2). Plants in soil from origin CA5 were randomly distributed over the 27 trays. Once a week, trays moved position in the growth chamber while keeping the trays for soil origins adjacent to each other. The plants were watered three times a week with a total volume of 50 mL of demineralized water per pot.
Five weeks after transfer of the seedlings to the soil, plants on two trays of each soil origin (two out of three pots with the same treatment) were infested with aphids, resulting in a full factorial experiment with all ten soil origins, all five dilution treatments and the sterile control, the Pseudomonas treatment (+ Pseudomonas/− Pseudomonas) and the aphid treatment (+ aphids/− aphids). This resulted in a pot number of n = 10 for treatments without aphids, whereas treatments with aphids had a pot number of n = 20 (for all treatments see overview in Supplementary Data Fig. S1).
One-half of the plants that were chosen for the aphid treatment were infested with six aphids of mixed ages from the general rearing. The other half of these plants were infested with five aphids from the mixed rearing and one neonate aphid (<24 h), which was placed in a clipcage so that time until first reproduction and number of offspring could be measured (Supplementary Data Fig. S3). Clipcages are round cages of 2.3 cm diameter that are attached to a leaf and allow an insect to feed on this leave, but prevent it from moving freely on the plant. This setup resulted in six aphids for each plant in total. After aphid infestation, all trays were placed individually in insect cages to prevent movement of aphids between infested and uninfested plants. One fully expanded leaf was harvested from each plant 24 h after aphid infestation to analyse gene expression. For aphid-treated plants, leaves containing at least one aphid were selected, and harvested after removal of the insect from the leaf. Leaves were shock-frozen in liquid nitrogen after clipping and stored at −80 °C until RNA isolation. The expression of five genes was assessed by real time quantitative PCR (RT-qPCR): CYB79B3, HEL, PDF1.2 and VSP2, involved in JA- and ET-dependent plant defence pathways, and PR1, involved in SA-dependent defences (for more details see Supplementary Data: Supporting methods, RNA extraction and quantitative RT-PCR analysis).
The reproduction of the aphids in the clipcages was observed continuously and aphids reproduced latest at an age of 10 d. Two weeks after aphid infestation the aphids in the clipcages were counted and all aphids were removed from the plants by careful brushing. On the same day plant shoots and roots were harvested. Subsequently they were oven-dried for assessment of dry weight. Prior to root washing, rhizosphere soil was collected for + Pseudomonas treatments and several − Pseudomonas soils to control for contamination. The roots were carefully removed from the bulk soil and shaken gently. The remaining attached soil was designated as rhizosphere soil, transferred into a 1.5-mL Eppendorf tube and stored at 4 °C for a maximum of 2 weeks for Pseudomonas quantification (Supplementary Data, Supporting methods, Pseudomonas quantification).
Statistical analysis
All statistical analyses were carried out in R (R Core Team, 2016).
The effects of dilution treatment (specified as a discrete factor), Pseudomonas inoculation and aphid infestation on the response variables shoot and root biomass, the number of Pseudomonas, the number of aphid offspring, the time until first aphid reproduction and the expression of all five genes were analysed using linear mixed effect models. The fixed factors were nested in the random factor soil origin. For plant shoot biomass, time until first aphid reproduction and the expression of the genes CYB79B3 and PR1, the function lmer() from the lme4 package was used together with the step function from the lmerTest package (Bates et al., 2015). Due to non-normal distribution of residuals, the glmmPQL() function from the package MASS was used to fit mixed models for plant root biomass, Pseudomonas numbers and the expression of the genes HEL, PDF1.2 and VSP2 (Venables and Ripley, 2002). For the number of aphid offspring the glmer() function from the lme4 package was used. For the number of Pseudomonas a quasipoisson error distribution was specified, for the gene expression and root biomass a Gaussian distribution with a log link, and for the number of aphid offspring a gamma distribution with a log link.
In addition, the relationship between the number of Pseudomonas and gene expression, as well as plant biomass and the relationship between the number of aphid offspring and plant biomass, were analysed with the lmer() function with soil origin as a random factor.
RESULTS
Plant biomass response to dilution, Pseudomonas inoculation and aphid infestation
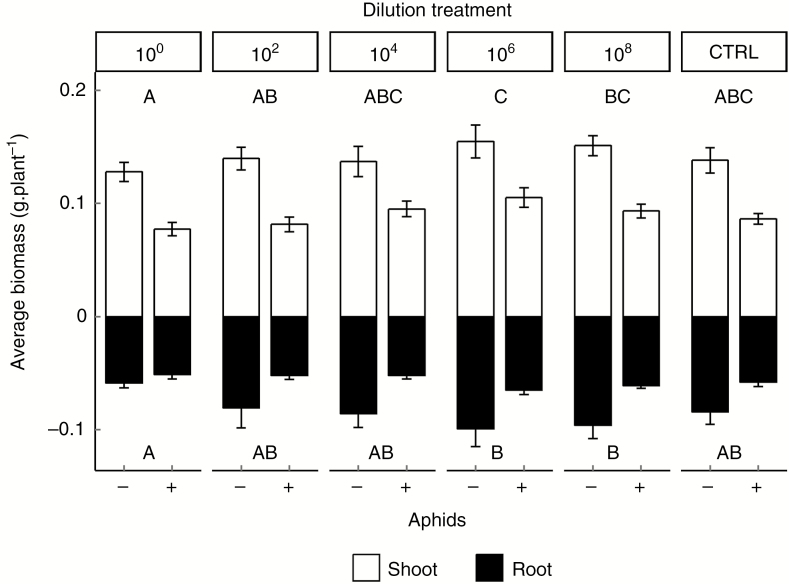
Shoot and root biomass were significantly affected by dilution treatment (for statistical results see Table 1). Both shoot and root biomass showed a hump-shaped relationship with dilution treatment, increasing from the 100 to the 106 dilution by 23 and 18 %, respectively (Fig. 1). Biomass then decreased from the 106 dilution to the sterilized control by 15 % in shoots and 8 % in roots; however, this decrease was not significant. Aphids overall decreased shoot and root biomass by 46 and 25 %, respectively, but this reduction was independent of the dilution treatment (Fig. 1). There was considerable variation in biomass response to dilution and aphid treatment due to large differences among the soil origins (e.g. shoot biomass; Supplementary Data Fig. S4). The largest plant had on average twice as much biomass as the smallest plant (from soil origin LD2 and NC2, respectively).
Table 1.
Statistical results of linear mixed models in this study assessed with 1 the lmer function, 2 the glmmPQL function with a Gaussian error distribution, 3 the glmmPQL function with a quasipoisson error distribution, 4 the glmmPQL function with a gamma error distribution and log link. Besides the main effects, the table shows the standard deviation of soil origin (a random factor), and the standard deviation of the residuals.
| Shoot biomass1 | Root biomass2 | CYB79B3 1 | HEL 2 | PDF1.2 2 | PR1 1 | VSP2 2 | Pseudomonas count3 | Aphid reproduction time1 | No. of aphid offspring4 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Dilution | ||||||||||
| d.f. | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| Stat | 2.5 | 19.2 | 0.6 | 4.1 | 5.1 | 0.4 | 6.9 | 34.7 | 2.5 | 3.6 |
| P | 0.04 | <0.01 | 0.74 | 0.53 | 0.4 | 0.81 | 0.23 | <0.01 | 0.78 | 0.61 |
| Aphid | ||||||||||
| d.f. | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||
| Stat | 176.3 | 58.4 | 8.1 | 0.9 | 0.1 | 5.3 | 2.3 | 0.7 | ||
| P | <0.01 | <0.01 | <0.01 | 0.34 | 0.75 | 0.02 | 0.25 | 0.4 | ||
| Pseudomonas | ||||||||||
| d.f. | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Stat | 59.1 | 1.3 | 12.3 | 0.9 | 1 | 5.7 | 1 | 0.002 | 0.4 | |
| P | 0.05 | 0.025 | <0.01 | 0.34 | 0.31 | 0.02 | 0.31 | 0.96 | 0.52 | |
| Dilution:aphid | ||||||||||
| d.f. | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | ||
| Stat | 0.4 | 6.8 | 0.7 | 5.2 | 7.8 | 1.5 | 7.4 | 19.9 | ||
| P | 0.82 | 0.23 | 0.64 | 0.39 | 0.17 | 0.2 | 0.19 | <0.01* | ||
| Dilution:Pseudomonas | ||||||||||
| d.f. | 5 | 5 | 5 | 5 | 5 | 1 | 5 | 5 | 5 | |
| Stat | 0.2 | 2.6 | 1.1 | 9.9 | 4 | 1.5 | 5.1 | 1.7 | 2.77 | |
| P | 0.97 | 0.77 | 0.36 | 0.08 | 0.54 | 0.21 | 0.4 | 0.88 | 0.74 | |
| Aphid:Pseudomonas | ||||||||||
| d.f. | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||
| Stat | 0 | 0.6 | 0.4 | 0.2 | 0.5 | 1.1 | 0.9 | |||
| P | 0.97 | 0.45 | 0.51 | 0.68 | 0.48 | 0.29 | 0.34 | |||
| Dilution:aphid:Pseudomonas | ||||||||||
| d.f. | 5 | 5 | 5 | 5 | 5 | 5 | 5 | |||
| Stat | 2 | 3.4 | 0.7 | 3.7 | 8.8 | 0.8 | 6.1 | |||
| P | 0.07 | 0.64 | 0.64 | 0.59 | 0.12 | 0.54 | 0.29 | |||
| Random effects s.d. | ||||||||||
| Soil origin | 0.02 | 0.02 | 4.0·10–9 | 0.13 | 8.6·10–5 | 2.9·10–7 | 8.15 | 0.36 | 0.14 | 0.12 |
| Residual | 0.03 | 0.03 | 0.59 | 1.01 | 3.22 | 1.59 | 664.92 | 105.12 | 0.62 | 0.29 |
*Stat represents the F or Χ2 value for analyses carried out using the lmer or the glmmPQL function.
Fig. 1.
Shoot and root biomass (g plant−1) of plants grown in soil with different dilution treatments with and without aphid infestation averaged over Pseudomonas treatments because there were no significant interactions between the treatments (100, 102,104, 106 and 108 represent the different dilutions; CTRL, control soil). White bars represent shoot biomass; black bars root biomass and error bars the standard error (n = 20 for − aphid treatment, n = 40 for + aphid treatment); different letters represent significant differences (P < 0.05).
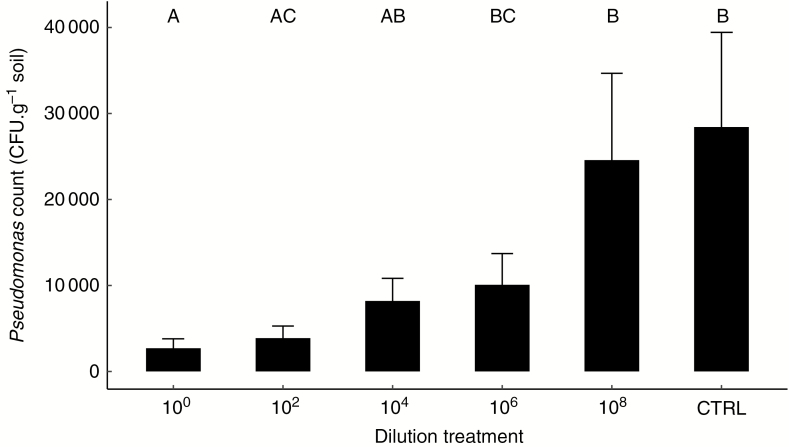
At the end of the experiment, 7 weeks after Pseudomonas inoculation, this strain could still be recovered from soil. The number of Pseudomonas CFU had decreased in all soils compared with the density that was inoculated (107 CFU g−1 soil). The number of recovered Pseudomonas was positively associated with increasing soil dilution (Fig. 2). Pseudomonas decreased least in the sterilized control soil, to 37 000 CFU g−1 soil (0.37 % of the original inoculum) and most in the 100 dilution, to 2700 CFU g−1 soil (0.027 % of the original inoculum). In spite of the substantial effect of dilution treatment on CFU g−1 soil, the inoculation effect on shoot biomass did not depend on dilution treatment: Pseudomonas reduced shoot biomass by an average of 7.8 % regardless of soil community dilution. Root biomass was unaffected by Pseudomonas inoculation (Supplementary Data Fig. S5). Pseudomonas numbers also differed between soil origins, with an ~19-fold higher number in LD5 soil than in CA4 soil.
Fig. 2.
Pseudomonas CFU g−1 in soils subjected to different dilution treatments. Values represent averages of two technical replicates and aphid treatments and error bars indicate the standard error (n = 10); different letters represent significant differences (P < 0.05). CTRL, control soil.
Plant gene expression in response to dilution, Pseudomonas inoculation and aphid infestation
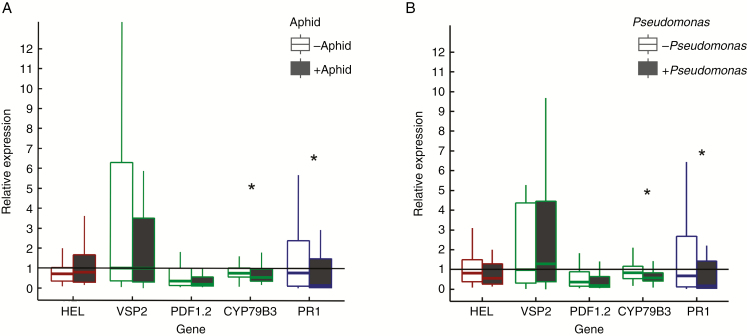
Infestation by aphids led to significant downregulation of the expression of CYP79B3 by a factor of 0.6. The expression of PR1 was significantly but slightly repressed 24 h after infestation (Fig. 3A). Similarly, the addition of Pseudomonas significantly repressed the genes CYP79B3 and PR1 (Fig. 3B). HEL, VSP2 and PDF1.2 expressions were not significantly affected by aphid or Pseudomonas treatment. Although both aphid infestation and Pseudomonas inoculation showed the same effect on gene expression, there was no interaction between the two treatments (for statistical results see Table 1). The number of Pseudomonas CFU recovered from soil was not correlated to gene expression. There was no effect of dilution treatment on plant gene expression (Supplementary Data Fig. S6). There was no interaction between dilution and Pseudomonas or aphid infestation effects.
Fig. 3.
(A) Relative gene expression in plant leaves without or with aphid treatments 24 h after infestation, averaged over soil community dilution and Pseudomonas treatments since there was no significant interaction between treatments. *Significant difference between treatments without and with aphids (P < 0.05). (B) Relative gene expression in plant leaves without or with Pseudomonas. Gene expression was averaged over soil community dilution and aphid treatments since there was no significant interaction with the Pseudomonas treatments. Relative expression of 1 indicates no change in gene expression. Genes regulated by the ET, JA and SA pathways are shown by different colours (ET, red; JA, green; SA, blue); whiskers represent the maximum and minimum, the top and bottom of the boxes represent the 25 and 75 % quartiles and the middle line represents the median. *Significant difference between treatments without or with Pseudomonas (P < 0.05).
Aphid reproductive success
Although plant biomass varied between dilutions and Pseudomonas treatments, aphid reproduction (time until first reproduction and total number of offspring) did not differ between dilution treatments (Supplementary Data Fig. S5) or Pseudomonas treatments (Table 1). The pre-reproductive period was on average 7.1 d (minimum 6, maximum 10). The number of offspring per neonate aphid after 2 weeks varied between 1 and 26, with an average of 15 newborn aphids. The number of aphid offspring was significantly positively associated with plant shoot biomass (t1,56 = 3.1, P < 0.01) (Supplementary Data Fig. S7), but showed no relation with root biomass or with the number of Pseudomonas CFU recovered from soil (root biomass t1,76 = 1.3, P = 0.19; Pseudomonas CFU, t1,33 = 1.2, P = 0.24). Furthermore, the number of neonate aphids showed a maximum difference of 63 % between the soil origins (LD2 having the highest average number of offspring and CA5 the lowest number).
DISCUSSION
In spite of the many studies on the effects of single PGPR strains on plant growth and resistance to pathogens and insect herbivores, little is known about such effects when considering their effects in the presence of the entire soil microbiome, or subsets of it. Low-abundance soil microbes are known to contain species with a high potential impact on various ecosystem functions (Delgado-Baquerizo et al., 2016). Therefore, we examined whether the absence of some of the rare microbes may play a role in priming plant defences against insect herbivores. We examined the effects of soil dilution on plant defence against aphids and show that there was no consistent effect of loss of rare microbes, indicating that species that are rare in the soil microbiome had no effect on plant resistance against herbivory by aphids. Interestingly, random removal of low-abundance soil microbial species by dilution generally led to an initial increase in plant biomass. This was unexpected given that bacterial diversity and the presence of rare species have been found to be positively associated with several functions, such as decomposition and the suppression of plant pathogens (Salonius, 1981; Van Elsas et al., 2012). However, a previous study also showed that high microbial diversity negatively influenced plant biomass compared with a community with fewer rare species (Hol et al., 2010). The authors of that study suggested that more inter-bacterial interactions at a higher diversity might have led to increased production of phytotoxic compounds.
We used an Arabidopsis ecotype originating from the same site as the microbial community. Many other studies on Arabidopsis have confronted plant ecotypes with non-co-evolved soil microbes, which may help to understand interaction mechanisms, but not their ecological roles. We observed substantial differences between the dilutions that were prepared from soil samples collected from ten different plots in the field. This contributed to the high variation in biomass in the dilution treatments. Although we expected that the loss of rare species would have similar effects on plant growth and defence irrespective of the site of collection, our results indicate that the effect of dilution will depend on the initial soil community composition. Thus, this composition might have differed between the plots. This conclusion is supported by Hol et al. (2015), who found the effect of changes in community composition by dilution on plant biomass to be dependent on soil origin and consequently initial microbiome community composition. Our results suggest that the absence or presence of certain species in the soil microbiome will be of importance for both plant performance and resistance.
Effects of our dilution treatment might not solely be due to the loss of rare species. While dilution has been shown to reliably reduce the number of low-abundance species, it can also lead to an overall change in composition of the remaining species, for example because dominant species are released from (interference) competition. In addition, random sampling processes during dilution and cell regrowth could have led to differences among the dilutions of the different soil origins (Yan et al., 2015). Moreover, this study focused on the effect of soil bacteria since low-abundance bacterial species have frequently been demonstrated to affect other community members and ecosystem functioning. However, our experimental design did include other soil microorganisms, such as fungi, archaea and protozoa. Little is known about rare microbial species other than bacteria. Therefore, we cannot speculate on how they might have affected the present results.
In accordance with the overall lack of effect on plant biomass loss by aphid feeding, dilution treatments had no effect on plant defence-related gene expression. However, aphid infestation did affect the expression of several defence-related genes. In contrast to our expectation that aphid feeding would induce JA-dependent defence, they downregulated the expression of CYP79B3 involved in indole glucosinolate synthesis (Mikkelsen et al., 2003) and had no effect on the two other JA-responsive genes, VSP2 and PDF1.2. In addition, aphids repressed PR1, a marker gene of the SA pathway. Several studies have reported highly variable effects of aphids and other phloem feeders on different plant defence pathways (Thompson and Goggin, 2006). It has been suggested that phloem-feeding insects may use specific saliva components, such as glucose oxidase, to repress or redirect defence signalling in the plant (Giordanengo et al., 2010). Especially JA-dependent defences, which are supposed to be most effective against aphids, may be suppressed by phloem-feeding insects through the induction of SA-dependent defences (Zhu-Salzman et al., 2004; Walling, 2008; Kant et al., 2015). Kim and Jander (2007) also reported that M. persicae feeding decreased total indole glucosinolates, which are dependent on the enzyme encoded by CYP79B3. Nevertheless, our findings strongly indicate a reduction in plant defences following aphid infestation independent of microbial interactions.
We did not detect an effect of soil dilution treatment on induction of systemic resistance by aphids. Nevertheless, we cannot exclude that rare microbial species loss had an effect on resistance induction. On the one hand, induction of defences might have been restricted to the site of aphid feeding (De Vos et al., 2005). Variation in the number of aphids feeding on the leaves that were sampled for RNA expression might have led to high variation in gene expression. On the other hand, the pooling of leaves might have obscured potential differences in the measured gene expression.
Upon inoculation of Pseudomonas, we found repression of genes involved in SA- dependent defences and in the glucosinolate biosynthesis gene CYP79B3, and no effect on aphid performance. These results suggest that Pseudomonas did not induce systemic resistance in A. thaliana against aphid herbivores. Moreover, the presence of Pseudomonas decreased shoot biomass in all soil dilution and aphid treatments. Our results are unexpected for several reasons. First, the PGPR strain of Pseudomonas is known to act via both SA- dependent and SA-independent defences in plants and to lead to an increase in both aliphatic and indolyl glucosinolate levels (Tran et al., 2007; Van de Mortel et al., 2012). Second, Pseudomonas was previously found to induce resistance against leaf-chewing herbivores and to promote plant growth (Van de Mortel et al., 2012; Park et al., 2015; Pangesti et al., 2017). Third, survival of the strain decreased dramatically with increasing dilution treatment, which is in accordance with previous studies finding decreasing success of invading strains in more diverse communities (Mallon et al., 2015), but this has never been shown for plant growth-promoting rhizobacteria. Although there are several possible explanations, our results demonstrate that low-abundance soil microbes may be a key component in the establishment of inoculated biocontrol strains in the soil. Whether the growth-reducing effects of Pseudomonas are caused by the strain itself or by its interaction with other members of the microbiome cannot be determined without more detailed studies on microbiome composition. Interactions of Pseudomonas with other microbial species might explain the negative effect of the PGPR strain even if it is reduced to low cell densities.
Plant interactions with PGPR may vary between species and even between bacterial strains and plant genotypes (Liu et al., 1995; Smith et al., 1999; Van Loon, 2007; Wintermans et al., 2016). It has also been shown that different plant ecotypes select for specific rhizosphere microbial communities by secreting a particular blend of root exudates (Hartmann et al., 2009). This mechanism enables plants to recruit specific beneficial bacterial taxa upon attack by herbivores, which in turn induce plant resistance against insects (Yi et al., 2011). Species-specific interactions might also have played a role in the lack of growth promotion of Pseudomonas as observed here. Previous studies on this bacterial strain report largely positive effects on plant biomass and defence. However, these studies used the A. thaliana Col-0 accession, whereas in our study an accession (Msl) was used that was growing at the same site where the soil for the dilution experiment had been collected. The discrepancy between our findings and the results of studies using the Col-0 accession indicate that the plant response to PGPR strains may depend on the plant genotype (Wintermans et al., 2016). Hence, matching plant accessions, bacterial communities and insect species should be used to create an ecologically relevant experimental system.
We conclude that the loss of rare microbial species from soil may affect plant performance, as well as the abundance of a single microbial strain of PGPR, but not the resistance of A. thaliana to leaf-sucking herbivores. However, different types of herbivores (e.g. phloem sucking versus chewing, generalists versus specialists) have been found to differentially trigger plant defence responses (Ali and Agrawal, 2012; Erb et al., 2012). It is very possible that generalist chewers will cause stronger responses. Moreover, the high variation in biomass between plants grown in soil with inocula from the different soil origins indicates that differences in microbial communities, for example as a result of different plant community compositions, could be more important for plant performance than previously thought. Therefore, we suggest that variation in microbial community composition should be taken into account in studies on plant–insect interactions.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Figure S1: overview of applied treatments. Figure S2: scheme of tray positions on the growth chamber. Figure S3: picture of a clipcage. Figure S4: plant biomass reduction in response to dilution treatments of the different soil replicates. Figure S5: effect of Pseudomonas treatment on Arabidopsis shoot and root biomass. Figure S6: relative gene expression with different dilution treatments. Figure S7: aphid fecundity on plants grown in soil with different dilution treatments. Table S1: description of sites of soil origin. Table S2: primer sequences for RT-qPCR.
ACKNOWLEDGEMENTS
We thank Sebastian Yanore and employees of the NIOO-KNAW for their help with the set-up, maintenance and harvest of the plant experiment. In addition, we are grateful to Julie Ferreira de Carvalho and Koen Verhoeven for help with the optimization and interpretation of the RT-qPCR assay. We also thank Léon Westerd for providing us with aphids and Xu Cheng and Ton Bisseling from Wageningen University for the provision of Arabidopsis Ms1 seeds and their kind advice. This is publication 6377 of NIOO-KNAW.
LITERATURE CITED
- Adam E, Groenenboom AE, Kurm V et al. 2016. Controlling the microbiome: microhabitat adjustments for successful biocontrol strategies in soil and human gut. Frontiers in Microbiology 7: 1079 http://journal.frontiersin.org/article/10.3389/fmicb.2016.01079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali JG, Agrawal AA. 2012. Specialist versus generalist insect herbivores and plant defense. Trends in Plant Science 17: 293–302. [DOI] [PubMed] [Google Scholar]
- Bates D, Maechler M, Bolker B, Walker S. 2015. Fitting linear mixed-effects models using lme4. Journal of Statistical Software 67: 1–48. [Google Scholar]
- Baumann K, Dignac M-F, Rumpel C et al. 2013. Soil microbial diversity affects soil organic matter decomposition in a silty grassland soil. Biogeochemistry 114: 201–212. [Google Scholar]
- Bent SJ, Forney LJ. 2008. The tragedy of the uncommon: understanding limitations in the analysis of microbial diversity. ISME Journal 2: 689–695. [DOI] [PubMed] [Google Scholar]
- Compant S, Clément C, Sessitsch A. 2010. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biology and Biochemistry 42: 669–678. [Google Scholar]
- Cook KL, Garland JL, Layton AC, Dionisi HM, Levine LH, Sayler GS. 2006. Effect of microbial species richness on community stability and community function in a model plant-based wastewater system processing system. Microbial Ecology 52: 725–737. [DOI] [PubMed] [Google Scholar]
- Delgado-Baquerizo M, Giaramida L, Reich PB et al. 2016. Lack of functional redundancy in the relationship between microbial diversity and ecosystem functioning. Journal of Ecology, 104: 936–946. [Google Scholar]
- Van Elsas JD, Chiurazzi M, Mallon CA, Elhottová D, Kristufek V, Falcao Salles J. 2012. Microbial diversity determines the invasion of soil by a bacterial pathogen. Proceedings of the National Academy of Sciences of the USA 109: 1159–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Ent S, Van Wees SCM, Pieterse CMJ. 2009. Jasmonate signaling in plant interactions with resistance-inducing beneficial microbes. Phytochemistry 70: 1581–1588. [DOI] [PubMed] [Google Scholar]
- Erb M, Meldau S, Howe GA. 2012. Role of phytohormones in insect-specific plant reactions. Trends in Plant Science 17: 250–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordanengo P, Brunissen L, Rusterucci C et al. 2010. Compatible plant-aphid interactions: How aphids manipulate plant responses. Comptes Rendus Biologies 333: 516–523. [DOI] [PubMed] [Google Scholar]
- Griffiths BS, Kuan HL, Ritz K, Glover LA, McCaig AE, Fenwick C. 2004. The relationship between microbial community structure and functional stability, tested experimentally in an upland pasture soil. Microbial Ecology 47: 104–113. [DOI] [PubMed] [Google Scholar]
- Hartmann A, Schmid M, Tuinen Dv, Berg G. 2009. Plant-driven selection of microbes. Plant and Soil 321: 235–257. [Google Scholar]
- Harvey JA, Witjes LMA, Benkirane M, Duyts H, Wagenaar R. 2007. Nutritional suitability and ecological relevance of Arabidopsis thaliana and Brassica oleracea as foodplants for the cabbage butterfly, Pieris rapae. Plant Ecology 189: 117–126. [Google Scholar]
- Hol WHG, De Boer W, Termorshuizen A et al. 2010. Reduction of rare soil microbes modifies plant-herbivores interactions. Ecology Letters 13: 292–301. [DOI] [PubMed] [Google Scholar]
- Hol WHG, De Boer W, De Hollander M, Kuramae EE, Meisner A, Van der Putten WH. 2015. Context dependency and saturating effects of loss of rare soil microbes on plant productivity. Frontiers in Plant Science 6: 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SE, Lennon JT. 2010. Dormancy contributes to the maintenance of microbial diversity. Proceedings of the National Academy of Sciences of the USA 107: 5881–5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousset A, Bienhold C, Chatzinotas A et al. 2017. Where less may be more: how the rare biosphere pulls ecosystems strings. ISME Journal 11: 853–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant MR, Jonckheere W, Knegt B et al. 2015. Mechanisms and ecological consequences of plant defence induction and suppression in herbivore communities. Annals of Botany 115: 1015–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Jander G. 2007. Myzus persicae (green peach aphid) feeding on Arabidopsis induces the formation of a deterrent indole glucosinolate. Plant Journal 49: 1008–1019. [DOI] [PubMed] [Google Scholar]
- Van Loon LC. 2007. Plant responses to plant growth-promoting rhizobacteria. European Journal of Plant Pathology 119: 243–254. [Google Scholar]
- Van Loon L, Bakker P. 2006. Root-associated bacteria inducing systemic resistance. In Gnanamanickam SS. ed. Plant-associated bacteria. Dordrecht: Springer. [Google Scholar]
- Liu L, Kloepper JW, Tuzun S. 1995. Induction of systemic resistance in cucumber by plant growth-promoting rhizobacteria – duration of protection and effect of host-resistance on protection and root colonization. Phytopathology 85: 1064–1068. [Google Scholar]
- Lugtenberg B, Kamilova F. 2009. Plant-growth-promoting rhizobacteria. Annual Review of Microbiology 63: 541–556. [DOI] [PubMed] [Google Scholar]
- Mallon CA, Poly F, Le Roux X, Marring I, Van Elsas JD, Salles JF. 2015. Resource pulses can alleviate the biodiversity–invasion relationship in soil microbial communities. Ecology 96: 915–926. [DOI] [PubMed] [Google Scholar]
- Mauch-Mani B, Métraux J-P. 1998. Salicylic acid and systemic acquired resistance to pathogen attack. Annals of Botany 82: 535–540. [Google Scholar]
- Mikkelsen MD, Petersen BL, Glawischnig E, Jensen AB, Andreasson E, Halkier BA. 2003. Modulation of CYP79 genes and glucosinolate profiles in Arabidopsis by defense signaling pathways. Plant Physiology 131: 298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Mortel JE, De Vos RCH, Dekkers E et al. 2012. Metabolic and transcriptomic changes induced in Arabidopsis by the rhizobacterium Pseudomonas fluorescens SS101. Plant Physiology 160: 2173–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pangesti N, Vandenbrande S, Pineda A, Dicke M, Raaijmakers JM, Van Loon JJA. 2017. Antagonism between two root-associated beneficial Pseudomonas strains does not affect plant growth promotion and induced resistance against a leaf-chewing herbivore. FEMS Microbiology Ecology 93: doi: 10.1093/femsec/fix038. [DOI] [PubMed] [Google Scholar]
- Park Y-S, Dutta S, Ann M, Raaijmakers JM, Park K. 2015. Promotion of plant growth by Pseudomonas fluorescens strain SS101 via novel volatile organic compounds. Biochemical and Biophysical Research Communications 461: 361–365. [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Dicke M. 2007. Plant interactions with microbes and insects: from molecular mechanisms to ecology. Trends in Plant Science 12: 564–569. [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Van Wees SCM, Ton J, Van Pelt JA, Van Loon LC. 2002. Signalling in rhizobacteria-induced systemic resistance in Arabidopsis thaliana. Plant Biology 4: 535–544. [Google Scholar]
- Pieterse CM, Van Pelt JA, Verhagen BW et al. 2003. Induced systemic resistance by plant growth-promoting rhizobacteria. Symbiosis 35: 39–54. [Google Scholar]
- Pieterse CMJ, Zamioudis C, Berendsen RL, Weller DM, Van Wees SCM, Bakker PAHM. 2014. Induced systemic resistance by beneficial microbes. Annual Review of Phytopathology 52: 347–375. [DOI] [PubMed] [Google Scholar]
- R Core Team 2016. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Raaijmakers JM, Leeman M, Vanoorschot MMP, Vandersluis I, Schippers B, Bakker PAHM. 1995. Dose-response relationships in biological-control of fusarium wilt of radish by Pseudomonas spp. Phytopathology 85: 1075–1081. [Google Scholar]
- Roger F, Bertilsson S, Langenheder S, Osman OA, Gamfeldt L. 2016. Effects of multiple dimensions of bacterial diversity on functioning, stability and multifunctionality. Ecology 97: 2716–2728. [DOI] [PubMed] [Google Scholar]
- Salonius PO. 1981. Metabolic capabilities of forest soil microbial-populations with reduced species-diversity. Soil Biology & Biochemistry 13: 1–10. [Google Scholar]
- Shade A, Jones SE, Caporaso JG et al. 2014. Conditionally rare taxa disproportionately contribute to temporal changes in microbial diversity. mBio 5: e01371–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KP, Handelsman J, Goodman RM. 1999. Genetic basis in plants for interactions with disease-suppressive bacteria. Proceedings of the National Academy of Sciences of the USA 96: 4786–4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin ML, Morrison HG, Huber JA et al. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proceedings of the National Academy of Sciences of the USA 103: 12115–12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland MS, Lauber C, Fierer N, Bradford MA. 2009. Testing the functional significance of microbial community composition. Ecology 90: 441–451. [DOI] [PubMed] [Google Scholar]
- Thompson GA, Goggin FL. 2006. Transcriptomics and functional genomics of plant defence induction by phloem-feeding insects. Journal of Experimental Botany 57: 755–766. [DOI] [PubMed] [Google Scholar]
- Tran H, Ficke A, Asiimwe T, Höfte M, Raaijmakers JM. 2007. Role of the cyclic lipopeptide massetolide A in biological control of Phytophthora infestans and in colonization of tomato plants by Pseudomonas fluorescens. New Phytologist 175: 731–742. [DOI] [PubMed] [Google Scholar]
- Venables WN, Ripley BD. 2002. Modern applied statistics with S, 4th edn. New York: Springer. [Google Scholar]
- Walling LL. 2008. Avoiding effective defenses: strategies employed by phloem-feeding insects. Plant Physiology 146: 859–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wees SCM, Van der Ent S, Pieterse CMJ. 2008. Plant immune responses triggered by beneficial microbes. Current Opinion in Plant Biology 11: 443–448. [DOI] [PubMed] [Google Scholar]
- De Vos M, Van Oosten VR, Van Poecke RMP et al. 2005. Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Molecular Plant-Microbe Interactions 18: 923–937. [DOI] [PubMed] [Google Scholar]
- De Vos M, Kim JH, Jander G. 2007. Biochemistry and molecular biology of Arabidopsis–aphid interactions. BioEssays 29: 871–883. [DOI] [PubMed] [Google Scholar]
- Wilhelm L, Besemer K, Fasching C et al. 2014. Rare but active taxa contribute to community dynamics of benthic biofilms in glacier-fed streams. Environmental Microbiology 16: 2514–2524. [DOI] [PubMed] [Google Scholar]
- Wintermans PCA, Bakker PAHM, Pieterse CMJ. 2016. Natural genetic variation in Arabidopsis for responsiveness to plant growth-promoting rhizobacteria. Plant Molecular Biology 90: 623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Kuramae EE, Klinkhamer PGL, Van Veen JA. 2015. Revisiting the dilution procedure used to manipulate microbial biodiversity in terrestrial systems. Applied and Environmental Microbiology 81: 4246–4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H-S, Yang JW, Ghim S-Y, Ryu C-M. 2011. A cry for help from leaf to root. Plant Signaling & Behavior 6: 1192–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu-Salzman K, Salzman RA, Ahn J-E, Koiwa H. 2004. Transcriptional regulation of sorghum defense determinants against a phloem-feeding aphid. Plant Physiology 134: 420–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.