Abstract
The genus Methylocystis belongs to the class Alphaproteobacteria, the family Methylocystaceae, and encompasses aerobic methanotrophic bacteria with the serine pathway of carbon assimilation. All Methylocystis species are able to fix dinitrogen and several members of this genus are also capable of using acetate or ethanol in the absence of methane, which explains their wide distribution in various habitats. One additional trait that enables their survival in the environment is possession of two methane-oxidizing isozymes, the conventional particulate methane monooxygenase (pMMO) with low-affinity to substrate (pMMO1) and the high-affinity enzyme (pMMO2). Here, we report the finished genome sequence of Methylocystis bryophila S285, a pMMO2-possessing methanotroph from a Sphagnum-dominated wetland, and compare it to the genome of Methylocystis sp. strain SC2, which is the first methanotroph with confirmed high-affinity methane oxidation potential. The complete genome of Methylocystis bryophila S285 consists of a 4.53 Mb chromosome and one plasmid, 175 kb in size. The genome encodes two types of particulate MMO (pMMO1 and pMMO2), soluble MMO and, in addition, contains a pxmABC-like gene cluster similar to that present in some gammaproteobacterial methanotrophs. The full set of genes related to the serine pathway, the tricarboxylic acid cycle as well as the ethylmalonyl-CoA pathway is present. In contrast to most described methanotrophs including Methylocystis sp. strain SC2, two different types of nitrogenases, that is, molybdenum–iron and vanadium–iron types, are encoded in the genome of strain S285. This unique combination of genome-based traits makes Methylocystis bryophila well adapted to the fluctuation of carbon and nitrogen sources in wetlands.
Keywords: methanotrophs, Methylocystis, finished genome, comparative genomics, methane monooxygenase, nitrogenase
Introduction
Aerobic methanotrophs (methane-oxidizing bacteria, MOB) are a unique subset of methylotrophic bacteria that can utilize methane (CH4) as their sole source of energy. They use methane monooxygenase (MMO) enzymes to oxidize methane to methanol (Hanson and Hanson 1996; Trotsenko and Murrell 2008). Methanotrophic capabilities relying on MMO activity are currently recognized in members of the bacterial phyla Proteobacteria, Verrucomicrobia, and the candidate division NC10 (Stein et al. 2012). Of these, methanotrophic Proteobacteria are represented by the greatest number of characterized isolates. Belonging to the classes Gamma- and Alphaproteobacteria, they are classified as type I and type II MOB, respectively. Members of these two groups differ in their cellular ultrastructure, C1-utilization pathway, fatty acid composition, and other physiological and biochemical characteristics.
The genus Methylocystis is one of the first described and historically recognized genera of aerobic methanotrophic bacteria (Whittenbury et al. 1970). It belongs to the class Alphaproteobacteria, the family Methylocystaceae, and encompasses obligate and restricted facultative methanotrophic bacteria with the serine pathway of carbon assimilation (Belova et al. 2013). All members of this genus possess a membrane-bound or particulate methane monooxygenase (pMMO), whereas some also contain a soluble form of this enzyme (sMMO). Representatives of this genus inhabit a wide variety of terrestrial and aquatic ecosystems and display a number of environmental adaptations. Thus, in the absence of methane, some species of this genus are capable of slow growth on acetate and ethanol (Belova et al. 2011; Im et al. 2011; Belova et al. 2013). Another ecologically important adaptation of these methanotrophs is their ability to produce two pMMO isozymes, the conventional form with low affinity to methane (pMMO1), and the high-affinity enzyme (pMMO2) (Baani and Liesack 2008). The isozymes are encoded by pmoCAB1 and pmoCAB2, respectively. To date, the complete genome sequence has been reported for only a single pMMO2-possessing member of the genus Methylocystis, Methylocystis sp. strain SC2 (Dam et al. 2012).
In this study, we obtained the complete genome sequence of another pMMO2-possessing methanotroph, Methylocystis bryophila S285. The species Methylocystis bryophila accommodates facultative methanotrophs, which were isolated from acidic Sphagnum-dominated wetlands and are capable of slow growth on acetate in the absence of methane (Belova et al. 2011). Members of this species account for 20–50% of all the methanotroph cells detectable in acidic peat by fluorescence in situ hybridization (Belova et al. 2011). The taxonomic description of Methylocystis bryophila was based on the characterization of two isolates, strains H2sT and S284 (Belova et al. 2013). One additional representative of this species, strain S285, was later obtained from the same peat sample as strain S284. These two isolates share identical 16S rRNA gene sequences, which also match the 16S rRNA gene sequence of strain H2sT, the type strain of Methylocystis bryophila (GenBank accession number FN422003). Our further analysis of pMMO-encoding genes revealed that, in contrast to strain S284, only the pmoA2 gene fragments could be PCR-amplified from DNA of strain S285 by using the primer combination A189f–A682b (Holmes et al. 1995). These primers are routinely employed for pmoA1 detection in methanotrophs. To conclusively verify the absence or presence of pMMO1 and to get an insight into genome-encoded features of a pMMO2-possessing methanotroph from wetlands, we have determined and analyzed the complete genome sequence of Methylocystis bryophila S285.
Materials and Methods
Bacterial Strain Isolation and Characterization
Strain S285 was isolated from a methanotrophic enrichment culture that was established from an acidic peat soil (pH 3.8) sampled at a depth of 10 cm from Sphagnum peat bog Staroselsky moss (56°34′ N, 32°46′ E), Tver region, Russia, in August 2008. The enrichment culture was obtained from this peat sample using liquid mineral medium M2 of pH 5.0 (Belova et al. 2013) and 30% (vol/vol) methane in the headspace. Strain S285 was isolated from this enrichment culture by means of surface plating onto the agar medium M2. For genome analysis, cells of strain S285 were grown in liquid medium M2 with 20% methane and harvested after 2 weeks of incubation at 24°C.
Genome Sequencing and Assembly
Genome sequencing of Methylocystis bryophila S285 was performed at the Max Planck Genome Centre Cologne (MP-GCC), using the PacBio RSII platform (Pacific Biosciences, Menlo Park, California). De novo assembly was done using the hierarchical genome-assembly process (HGAP2) via the SMRT Portal v.2.0 offered by Pacific Biosciences.
Genome Annotation
With default parameters, the genome was annotated using RAST v. 2.0 (Rapid Annotation using Subsystem Technology) (Overbeek et al. 2014) and BASys (Bacterial Annotation System) (Van Domselaar et al. 2005). Furtherly, functional roles of protein-encoding genes were analyzed using SEED Subsystems (Overbeek et al. 2014).
Genome Comparison
We thoroughly compared the genome of Methylocystis bryophila S285 with that of the obligate methanotroph Methylocystis sp. SC2. To date, these two strains are the only members of the Methylocystaceae for which complete genome sequences are available. For reference, we included in our comparison the genome of Methylocella silvestris BL2. This facultative sMMO-containing methanotroph is a member of the family Beijerinckiaceae. The map of all three finished genomes was constructed using BRIG (BLAST Ring Image Generator), a prokaryote genome comparison software (Alikhan et al. 2011). To determine the overall genome similarities among methanotrophs and the distribution of particular genomic traits, we expanded this comparison to 15 methanotrophs encompassing the genus Methylocystis (seven genomes), other type II MOB (one genome each for the genera Methylosinus, Methylocella, Methylocapsa, and Methyloferula), type I MOB (one genome each for the genera Methylococcus, Methylomonas, and Methylomicrobium), and one genome representing Verrucomicrobia-like methanotrophs (“Methylacidiphilum”). Their genomic similarities were estimated using the average nucleotide identity calculator JSpeciesWS (Richter et al. 2016).
Phylogenetic Analysis
The relationships between the deduced amino acid sequences of pmoA1, pmoA2, and pxmA of methanotrophic bacteria as well as those of their symporter genes were calculated using MEGA7 (Molecular Evolutionary Genetics Analysis version 7.0) (Kumar et al. 2016).
Results and Discussion
Finished Genome of Methylocystis bryophila S285
A total of 50,623 PacBio reads were obtained with a mean length of 3,228 bp. These reads enabled the assembly of the complete genome sequence of Methylocystis bryophila S285. The finished genome consists of one circular chromosome of 4,532,950 bp and a single plasmid of 175,021 bp. The chromosome contains two identical rrn operon copies (16S-23S-5S rRNA), a full complement of 47 tRNA genes and 4,387 CDS (table 1). The distribution of protein-coding genes into SEED subsystem is shown in supplementary table S1, Supplementary Material online.
Table 1.
General Genomic Features of Methylocystis bryophila S285, Methylocystis sp. SC2, and Methylocella silvestris BL2
| Features | S285 | SC2 | BL2 |
|---|---|---|---|
| Accession number | CP019948 | HE956757 | CP001280 |
| Size (Mb) | 4.53 | 3.77 | 4.3 |
| G+C (%) | 63 | 63 | 63 |
| Genes (total) | 4,444 | 3,677 | 4,014 |
| CDS (total) | 4,387 | 3,623 | 3,956 |
| Genes (coding) | 4,285 | 3,583 | 3,875 |
| Pseudogenes | 102 | 40 | 81 |
| Genes (RNA) | 57 | 54 | 58 |
| rRNAs (5S, 16S, 23S) | 2, 2, 2 | 1, 1, 1 | 2, 2, 2 |
| tRNAs | 47 | 47 | 48 |
| ncRNAs | 4 | 4 | 4 |
| pmoCAB1 operon | 2 | 2 | Absent |
| pmoCAB2 operon | 1 | 1 | Absent |
| Monocistronic pmoC | 2 | 3 (1 in plasmid) | Absent |
| pxmABC operon | 1 | Absent | Absent |
| sMMO operon | 1 | Absent | 1 |
| Serine pathway genes | Present | Present | Present |
| RuMP pathway genes | Absent | Absent | Absent |
| Plasmid(s) | 1 | 2 | NR |
ncRNAs, noncoding RNAs; NR, not reported.
Genome Alignment
The complete genome map (fig. 1) of the three alphaproteobacterial methanotrophs suggests that the genomes of Methylocystis bryophila S285 and Methylocystis sp. SC2 display high synteny. In fact, the comparison of average nucleotide identities (ANIs) between the seven Methylocystis genomes and the eight genomes from other methanotrophic taxa revealed that Methylocystis bryophila S285 shares highest ANI values with the genomes of the other six Methylocystis spp. and Methylosinus trichosporium OB3b (72–74%) (supplementary table S2, Supplementary Material online). The further comparison of particular gene traits between Methylocystis bryophila S285 and the other six Methylocystis genomes provided additional evidence for high genomic similarity between Methylocystis bryophila S285 and Methylocystis sp. SC2. These two type II MOB share the largest number of common features (88.2%) and differ in the least number of unique traits (5.2%) among the seven Methylocystis genomes (supplementary table S3, Supplementary Material online). By contrast, Methylocella silvestris BL2 shows low similarity to Methylocystis bryophila S285 and Methylocystis sp. SC2, with regard to both complete genome (fig. 1) and particular gene traits (table 1 and supplementary tables S1 and S4, Supplementary Material online).
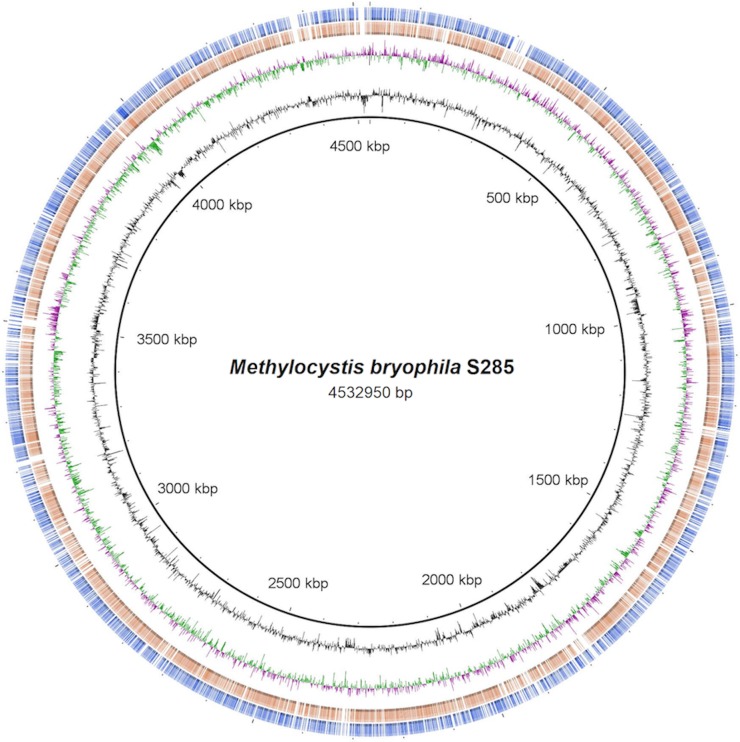
Fig. 1.
—Genomic map of Methylocystis bryophila S285 in comparison to those of Methylocystis sp. SC2 and Methylocella silvestris BL2. The concentric circles denote the following features (from inside to outside): genome of Methylocystis bryophila S285 with coordinates (chromosome region, black circle), the GC content, GC skew (green and red indicate that the nucleotides Guanine and Cytosine are over-respectively underrepresented) and the genome alignment of Methylocystis sp. SC2 and Methylocella silvestris BL2 against Methylocystis bryophila S285, where color indicates a BLAST match of nucleotide sequence identity of 70–100% (based on BLASTn) between central genome (Methylocystis bryophila S285) and comparative genomes, Methylocystis sp. SC2 (brown) and Methylocella silvestris BL2 (blue).
Diverse Methane Monooxygenase Genes
Despite the inability to detect the pMMO1-encoding genes in Methylocystis bryophila S285 by PCR, our genomic analysis revealed that this strain possesses both types of pmoCAB: two copies of pmoCAB1 and one copy of pmoCAB2 (table 1 and supplementary fig. S1, Supplementary Material online). The reason for the failure to detect pmoA1 is not fully clear but may be due to the fact that, if using primer set A189f–A682b, pmoA2 of strain S285 is more efficiently PCR-amplified than its pmoA1 counterpart. Although primer A189f is perfectly matching to its target site in either pmoA1 or pmoA2, primer A682b has one mismatch in its target site of both pmoA1 and pmoA2 (supplementary table S5, Supplementary Material online). However, this mismatch position differs between pmoA1 (G–G mismatch in primer position 14 from 3′-end) and pmoA2 (C-T mismatch in position 13 from 3′-end). In addition, adjacent primer positions are defined by N (position 12 from 3′-end) and S (position 15 from 3′-end). Thus, our experimental results indicate that the G–G mismatch (pmoA1) in primer position 14 is more detrimental for efficient pmoA amplification than the C–T mismatch (pmoA2) in position 13.
Notably, Radajewski et al. (2002) also detected only pmoA2 but not pmoA1 in their study on metabolically active methanotrophs in an acidic forest soil. This group of researchers also used the primer set A189f–A682b for their analysis. The inferred peptide sequences of pmoA clones retrieved by means of stable isotope probing (SIP) technique (accession numbers AY080950, AY080958, AY080959) showed high similarity (97.7–98.3%) to PmoA2 from Methylocystis bryophila S285, whereas no pmoA1 fragments were obtained from Methylocystis bryophila-like methanotrophs (Radajewski et al. 2002).
In addition to pMMO, strain S285 is able to produce the soluble form of MMO. Although pMMO is encoded by three genes (pmoCAB), the sMMO operon encompasses five consecutive genes (mmoYZXBC). Two monocistronic pmoC genes were also identified (table 1). Based on our survey of currently available genomes (including complete and draft genomes), Methylocystis bryophila S285 and Methylocystis sp. strain LW5 are the only type II MOB which harbor all three types of MMO: pMMO1, pMMO2, and sMMO. Strain SC2 does not produce sMMO (table 1 and supplementary table S4, Supplementary Material online).
A particular trait of strain S285 is the presence of a pxmABC-like gene cluster. The pxmABC operon is predicted to encode a member of the copper-containing membrane-bound monooxygenase (Cu-MMO) protein family, pXMO. Its function and substrate are not yet known (Tavormina et al. 2011). Recent evidence for pxmABC expression in response to hypoxia suggests that pXMO is important for survival of methanotrophs under O2 limitation (Kits et al. 2015). The pxmABC-like gene clusters are widely distributed among gammaproteobacterial type I MOB, including strains of the genera Methylomonas, Methylobacter, and Methylomicrobium (Tavormina et al. 2011). Among type II MOB, the pxmABC-like gene cluster has previously been detected only in Methylocystis rosea SV97 and Methylocystis sp. strain SB2 (Knief 2015) (supplementary table S4, Supplementary Material online). Thus, Methylocystis bryophila S285 is the only third type II MOB shown to possess pxmABC genes. Notably, its PxmA fragment clusters together with a large group of environmental pxmA transcripts obtained from a subarctic peatland (AFY11631.1 and AFY11641.1 in supplementary fig. S1, Supplementary Material online; Liebner and Svenning 2013) as well as with PxmA fragments from two type I MOB, Methylomonas sp. strain M5 and Methylococcaceae bacterium M200. Like Methylocystis bryophila S285, these two strains were isolated from a Sphagnum wetland (Kip et al. 2011). This wide array of different MMO types, including MMO-like enzymes, may ensure survival of strain S285-like methanotrophs in environments with fluctuating methane concentrations as well as under copper or O2 limitation.
Multiple Carbon Assimilation Pathways
All the genes required for the serine pathway are present in the genome of Methylocystis bryophila S285 and those of two key enzymes of the ribulose monophosphate cycle (RuMP) are absent. The serine pathway is the main carbon assimilation pathway of type II MOB. In addition to the complete tricarboxylic acid (TCA) cycle, the genomes of Methylocystis bryophila S285 and Methylocystis sp. strain SC2 encode the complete ethylmalonyl-CoA (EMC) pathway. This pathway is also present in Methylocystis sp. strain SB2, which has proven ability to grow on acetate or ethanol (Im et al. 2011; Vorobev et al. 2014). We also identified the genes encoding alcohol dehydrogenase and aldehyde dehydrogenase in the S285 genome. These two enzymes convert ethanol to acetate. Thus, presence of the ethanol-converting enzymes coupled with EMC pathway provides the possibility for growth of Methylocystis bryophila S285 on ethanol or acetate. Strain S285 may convert ethanol or acetate via acetyl-coenzyme A synthetase (gene locus: B1812_19345) to acetyl-CoA, which is then funneled into the TCA cycle for energy generation or incorporated into biomass via the EMC pathway. Utilization of these two-carbon compounds was confirmed for strains H2sT and S284, the two taxonomically characterized—but not genome-sequenced—representatives of Methylocystis bryophila (Belova et al. 2013). Thus, facultative methanotrophy represents an important alternative strategy for life in peatlands, allowing survival if no methane is available (Belova et al. 2011).
Even to date, the exact metabolic basis for facultative methanotrophy is not known, but it has been suggested that a key determinant of obligate methanotrophy is the restricted ability to transport potential substrates across the membrane (Tamas et al. 2014). Correspondingly, the total number of membrane transporters encoded in the genome of strain S285 exceeds that in strain SC2. Notably, the genome of Methylocella silvestris BL2 even encodes a greater repertoire of membrane transporters than the two Methylocystis spp., strains S285 and SC2 (supplementary table S1, Supplementary Material online). This finding corresponds well to the fact that acetate is the preferred substrate for growth of Methylocella spp. In addition, M. silvestris BL2 also grows on ethanol, pyruvate, succinate, malate, propanol, propanediol, acetone, methyl acetate, acetol, glycerol, propionate, tetrahydrofuran, gluconate, ethane, and propane (Dedysh and Dunfield 2016).
The repertoire of membrane transporters encoded by the S285 genome also includes acetate permease ActP (cation: acetate symporter; gene locus: B1812_15125). ActP has proved function for acetate transportation in E. coli (Gimenez et al. 2003). However, the mere genomic presence of actP is not a valid indicator for facultative methanotrophy. Genes encoding putative ActP are also present in Methylocystis spp. hitherto characterized as obligate methanotrophs such as, for example, Methylocystis sp. strain SC2, Methylocystis rosea SV97, and Methylocystis sp. strain Rockwell (supplementary fig. S2 and table S4, Supplementary Material online). Alternatively, it may be that these methanotrophs can uptake and utilize acetate for growth but that appropriate growth conditions have yet to be identified. The genome of M. silvestris BL2 also encodes ActP, but its sequence clusters on a branch separate from those of the Methylocystaceae spp. (supplementary fig. S2, Supplementary Material online).
Nitrogen Fixation
Nitrogenase is a metalloprotein complex that comprises two components, a nitrogenase iron protein and a dinitrogenase reductase (McGlynn et al. 2012). At least three genetically distinct but homologous nitrogenase systems have been identified until now (Hu and Ribbe 2015). These O2-sensitive nitrogenases are primarily distinguished by the metal composition of their active-site metallocluster: conventional molybdenum–iron nitrogenase and the alternative vanadium–iron type, and iron-only nitrogenase (Eady 1996; Hu and Ribbe 2015). Genes encoding the molybdenum–iron (Mo) and vanadium–iron (V) types of nitrogenase were identified in the S285 genome (supplementary table S4, Supplementary Material online). The Mo-nitrogenase is the most universally distributed nitrogenase in nature. The V-nitrogenase is present only in a limited number of microorganisms such as, for example, Azotobacteriaceae. The V-nitrogenase, encoded along with the Mo-nitrogenase, is expressed in the case of Mo-deficiency and has therefore been considered an alternative or “back-up” system (Rehder 2000; Zhao et al. 2006). However, despite the high structural similarity between the two nitrogenases, V-nitrogenase can also reduce CO (Hu et al. 2012). The ability of V-nitrogenase to catalyze the reduction of both CO and N2 suggests a potential link between the evolution of carbon and nitrogen cycles (Lee et al. 2010). We conducted a survey (August 10, 2017) on the distribution of V-nitrogenase-coding gene clusters among all 49 public methanotroph genomes that are available in NCBI GenBank. The survey showed that the genes encoding V-nitrogenase are present in only two type II MOB: Methylocystis bryophila S285 and Methylocystis parvus OBBP, but not in any type I MOB. The V-nitrogenase is presumably a primitive form that in ancient microbes, functioned in both nitrogen and carbon fixation (Lee et al. 2010). Thus, the rare inherited trait of V-nitrogenase in methanotrophs, but identified in Methylocystis bryophila S285, may indicate that in peatlands, the fluctuation of both carbon and nitrogen sources might retard the evolution of carbon fixation system.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgment
This study was supported by the Deutsche Forschungsgemeinschaft (DFG) through Collaborative Research Center SFB 987.
Literature Cited
- Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA.. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baani M, Liesack W.. 2008. Two isozymes of particulate methane monooxygenase with different methane oxidation kinetics are found in Methylocystis sp. strain SC2. Proc Natl Acad Sci U S A. 105(29):10203–10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belova SE. et al. 2011. Acetate utilization as a survival strategy of peat-inhabiting Methylocystis spp. Environ Microbiol Rep. 3(1):36–46.http://dx.doi.org/10.1111/j.1758-2229.2010.00180.x [DOI] [PubMed] [Google Scholar]
- Belova SE, Kulichevskaya IS, Bodelier PL, Dedysh SN.. 2013. Methylocystis bryophila sp. nov., a facultatively methanotrophic bacterium from acidic Sphagnum peat, and emended description of the genus Methylocystis (ex Whittenbury et al. 1970) Bowman et al. 1993. Int J Syst Evol Microbiol. 63(Pt 3):1096–1104. [DOI] [PubMed] [Google Scholar]
- Dam B, Dam S, Kube M, Reinhardt R, Liesack W.. 2012. Complete genome sequence of Methylocystis sp. strain SC2, an aerobic methanotroph with high-affinity methane oxidation potential. J Bacteriol. 194(21):6008–6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedysh SN, Dunfield PF.. 2016. Methylocella. Bergey’s Manual of Systematics of Archaea and Bacteria. Online © 2015 Bergey’s Manual Trust. Published by John Wiley & Sons, Inc., in association with Bergey’s Manual Trust. doi: 10.1002/9781118960608.gbm00797.pub2. [Google Scholar]
- Eady RR. 1996. Structure-function relationships of alternative nitrogenases. Chem Rev. 96(7):3013–3030.http://dx.doi.org/10.1021/cr950057h [DOI] [PubMed] [Google Scholar]
- Gimenez R, Nuñez MF, Badia J, Aguilar J, Baldoma L.. 2003. The gene yjcG, cotranscribed with the gene acs, encodes an acetate permease in Escherichia coli. J Bacteriol. 185(21):6448–6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson RS, Hanson TE.. 1996. Methanotrophic bacteria. Microbiol Rev. 60(2):439–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes AJ, Costello A, Lidstrom ME, Murrell JC.. 1995. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol Lett. 132(3):203–208. [DOI] [PubMed] [Google Scholar]
- Hu Y, Lee CC, Ribbe MW.. 2012. Vanadium nitrogenase: a two-hit wonder? Dalton Trans. 41(4):1118–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Ribbe MW.. 2015. Nitrogenase and homologs. J Biol Inorg Chem. 20(2):435–445.http://dx.doi.org/10.1007/s00775-014-1225-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im J, Lee S-W, Yoon S, DiSpirito AA, Semrau JD.. 2011. Characterization of a novel facultative Methylocystis species capable of growth on methane, acetate and ethanol. Environ Microbiol Rep. 3(2):174–181.http://dx.doi.org/10.1111/j.1758-2229.2010.00204.x [DOI] [PubMed] [Google Scholar]
- Kip N, et al. 2011. Detection, isolation, and characterization of acidophilic methanotrophs from Sphagnum mosses. Appl Environ Microbiol. 77(16):5643–5654.http://dx.doi.org/10.1128/AEM.05017-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kits KD, Klotz MG, Stein LY.. 2015. Methane oxidation coupled to nitrate reduction under hypoxia by the Gammaproteobacterium Methylomonas denitrificans, sp. nov. type strain FJG1. Environ Microbiol. 17(9):3219–3232.http://dx.doi.org/10.1111/1462-2920.12772 [DOI] [PubMed] [Google Scholar]
- Knief C. 2015. Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Front Microbiol. 6:1346.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.http://dx.doi.org/10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CC, Hu Y, Ribbe MW.. 2010. Vanadium nitrogenase reduces CO. Science 329(5992):642..http://dx.doi.org/10.1126/science.1191455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebner S, Svenning MM.. 2013. Environmental transcription of mmoX by methane-oxidizing Proteobacteria in a subarctic Palsa Peatland. Appl Environ Microbiol. 79(2):701–706.http://dx.doi.org/10.1128/AEM.02292-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlynn SE, Boyd ES, Peters JW, Orphan VJ.. 2012. Classifying the metal dependence of uncharacterized nitrogenases. Front Microbiol. 3:419.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R, et al. 2014. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 42(Database issue):D206–D214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radajewski S, et al. 2002. Identification of active methylotroph populations in an acidic forest soil by stable-isotope probing. Microbiology 148(8):2331–2342.http://dx.doi.org/10.1099/00221287-148-8-2331 [DOI] [PubMed] [Google Scholar]
- Rehder D. 2000. Vanadium nitrogenase. J Inorg Biochem. 80(1–2):133–136.http://dx.doi.org/10.1016/S0162-0134(00)00049-0 [DOI] [PubMed] [Google Scholar]
- Richter M, Rossello-Mora R, Oliver Glöckner F, Peplies J.. 2016. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32(6):929–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LY, Roy R, Dunfield PF.. 2012. Aerobic methanotrophy and nitrification: processes and connections In: Battista J, editor. Encyclopedia of life sciences. Chichester (UK: ): John Wiley & Sons. [Google Scholar]
- Tamas I, Smirnova AV, He Z, Dunfield PF.. 2014. The (d)evolution of methanotrophy in the Beijerinckiaceae–a comparative genomics analysis. ISME J. 8(2):369–382.http://dx.doi.org/10.1038/ismej.2013.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavormina PL, Orphan VJ, Kalyuzhnaya MG, Jetten MS, Klotz MG.. 2011. A novel family of functional operons encoding methane/ammonia monooxygenase-related proteins in gammaproteobacterial methanotrophs. Environ Microbiol Rep. 3(1):91–100. [DOI] [PubMed] [Google Scholar]
- Trotsenko YA, Murrell JC.. 2008. Metabolic aspects of aerobic obligate methanotrophy. Adv Appl Microbiol. 63:183–229. [DOI] [PubMed] [Google Scholar]
- Van Domselaar GH, et al. 2005. BASys: a web server for automated bacterial genome annotation. Nucleic Acids Res. 33(Web Server issue):W455–W459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobev A, et al. 2014. Genomic and transcriptomic analyses of the facultative methanotroph Methylocystis sp. strain SB2 grown on methane or ethanol. Appl Environ Microbiol. 80(10):3044–3052.http://dx.doi.org/10.1128/AEM.00218-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittenbury R, Phillips KC, Wilkinson JF.. 1970. Enrichment, isolation and some properties of methane-utilizing bacteria. J Gen Microbiol. 61(2):205–218.http://dx.doi.org/10.1099/00221287-61-2-205 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Bian SM, Zhou HN, Huang JF.. 2006. Diversity of nitrogenase systems in diazotrophs. J Integrat Plant Biol. 48(7):745–755.http://dx.doi.org/10.1111/j.1744-7909.2006.00271.x [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.