Abstract
Background and Aims
Extensive clonal (vegetative) reproduction in lianas is a common and important life history strategy for regeneration and colonization success. However, few studies have evaluated the contribution of clonal reproduction to stand-level distribution of lianas in their natural habitat using genetic tools. The objectives of the present study were to investigate (1) the contribution of clonal reproduction to the distribution of Wisteria floribunda, (2) the size of clonal patches and (3) how the distribution patterns of W. floribunda clones are affected by micro-topography.
Methods
The contribution of clonal reproduction to the distribution of the deciduous liana species W. floribunda was evaluated using genetic analysis across a 6-ha plot of an old-growth temperate forest in Japan and preference in landform between clonal ramets and non-clonal ramets was assessed.
Key Results
Of the 391 ramets sampled, clonal reproduction contributed to 71 and 62 % of the total abundance and basal area, respectively, or 57 and 31 % when the largest ramet within a genet was excluded. The large contribution of clonal reproduction to the density and basal area of W. floribunda was consistent with previous observational studies. The largest genet included a patch size of 0.47 ha and ranged over 180 m. Preferred landforms of clonal and non-clonal ramets were significantly different when evaluated by both abundance and basal area. Non-clonal ramets distributed more on lower part of the slope than other landforms in comparison with clonal ramets and trees, possibly reflecting the limitation of clonal growth by stolons.
Conclusions
Using genetic analysis, the present study found evidence of a large contribution of clonal reproduction on the distribution of W. floribunda in its natural habitat. The results indicate that clonal reproduction plays an important role not only in the formation of populations but also in determining the distribution patterns of liana species.
Keywords: Clone patch size, clonal structure, genetic analysis, liana, Ogawa Forest Reserve, stolon, Wisteria floribunda, vegetative growth
INTRODUCTION
Lianas are woody vine species that require mechanical support (i.e. host tree) to grow up to the forest canopy. Lianas are not only able to establish from seeds and suppressed saplings but also via clonal (vegetative) reproduction, whereas most tree species reproduce in only the two former ways (Schnitzer and Bongers, 2011). Extensive clonal reproduction in lianas is thought to be a major driver of their regeneration and colonization success (Schnitzer et al., 2012; Yorke et al., 2013; Ledo and Schnitzer, 2014), with a recent increase in their abundance and basal area reported in temperate and tropical forests around the world (Phillips et al., 2002; Allen et al., 2007; Yorke et al., 2013). It is also important to note that clonal reproduction is known to be one of the major potential drivers of spatial distribution patterns of liana species in tropical forests (Ledo and Schnitzer, 2014). However, studies of liana distribution patterns are often conducted in tropical forests while studies in temperate forests are lacking. Therefore, studies of liana clonal reproduction are essential to understanding their distribution and growth patterns in temperate forests.
Previous studies of clonal reproduction in lianas have provided important insight into their regeneration processes at the stand level in both temperate and tropical forests; however, most of these studies are still limited in their ability to accurately identify genetically distinct individuals (i.e. genets, clones). These studies utilized above-ground (i.e. liana inventory; Schnitzer et al., 2012; Yorke et al., 2013; Ledo and Schnitzer, 2014; Mori et al., 2016a) and below-ground censuses (i.e. excavation; Peñalosa, 1984; Putz, 1984; Sakai et al., 2002). An above-ground census detects rooted stems that have above-ground physical connections with other rooted stems (i.e. rooted ramets), which is different from branches in terms of connectivity to the ground (Gerwing et al., 2006; Schnitzer et al., 2008, 2012); however, these methods are unable to detect clonally reproduced ramets below the forest floor or ramets that were previously connected to each other (i.e. clonal ramets) (Parks and Werth, 1993; Suyama et al., 2000; Schnitzer et al., 2012). While a below-ground census detects the presence of below-ground connections, it is often difficult to conduct a below-ground census in a large area as it requires the destruction of the surrounding vegetation, thus making it difficult to apply this methodology widely to study plots. Furthermore, a below-ground census can only detect the current connectivity between ramets.
Clonal analysis using genetic tools such as microsatellite markers is an effective approach for the precise evaluation of clonal reproduction in lianas. Although some previous studies have applied genetic analysis to some liana species (Foster and Sork, 1997; Grashof-Bokdam et al., 1998; Arnold and Schnitzler, 2010; Kartzinel et al., 2015), these studies were either conducted in rural ecosystems and disturbed forests, and/or were based on non-continuous sampling within small plots. To the best of our knowledge, no studies have conducted genetic analyses on lianas with a focus on the contribution of clonal reproduction to regeneration processes and distribution patterns under natural conditions. In addition, genetic analysis for the evaluation of the liana clonal structure and distribution patterns of clones should be conducted in large plots (i.e. >1 ha). Sakai et al. (2002) showed that individual ramets of the deciduous liana species Wisteria floribunda can be up to 310.6 m in total length. It is also important to test the clonal ability of liana species in various topographies to assess any potential impact of the spatial heterogeneity of the forest floor environment on clonal reproduction, as clonal growth below the ground and on the forest floor via rhizomes and stolons – horizontal connections between plant organisms – is often sensitive to microsite characteristics (Parks and Werth, 1993; Suyama et al., 2000; Waters and Watson, 2015). In contrast, micro-topography often has a relatively small impact on liana distribution patterns in both tropical and temperate forests (Dalling et al., 2012; Mori et al., 2016a). Thus, it is necessary to assess the contribution of clonal reproduction on the distribution patterns of liana species over various micro-topographies.
To evaluate the contribution of clonal reproduction on the distribution of a liana species in its natural habitat, we conducted a genetic analysis using ten microsatellite markers in the deciduous liana species W. floribunda, with continuous sampling of a 6-ha plot in an old-growth cool temperate forest of Japan. Specifically, the following questions were addressed: (1) Does clonal reproduction significantly contribute to the distribution of W. floribunda? (2) How large and long is the clone (i.e. genet) patch and what is the distance between ramets within one genet? I3) Are distribution patterns of W. floribunda clones affected by micro-topography? Answering these questions is important not only to further elucidate the clonal structure of this species, but also to evaluate the contribution clonal ability makes to liana distribution and survival strategies.
MATERIALS AND METHODS
Study site
This study was conducted in the Ogawa Forest Reserve (OFR) in a 6-ha plot located in Ibaraki Prefecture, Japan (Nakashizuka and Matsumoto, 2002). The OFR is an old-growth cool temperate forest. The dominant canopy tree species ordered by basal area are Quercus serrata Murray, Fagus japonica Maxim. and Fagus crenata Blume (Masaki et al., 1992). The forest canopy in the OFR is mostly continuous without conspicuously large gaps (Nakashizuka et al., 1995). In this study plot, tree inventories for all trees larger than 5 cm in diameter at breast height (dbh) have been conducted since 1987. Data from a tree inventory conducted in 2013 was used for this analysis. Diameter at breast height, species name, host tree and location of all lianas on trees (dbh ≥ 5 cm) higher than 1.3 m above the ground were measured in 2013 (Mori et al., 2016a).
This study plot includes a variety of landforms including a crest slope (CS), head hollow (HH), upper side slope (US), lower side slope (LS), flood terrace (FT) and river bed (RB) (Fig. S1; Yoshinaga et al., 2002; see illustration in Nagamatsu and Miura, 1997), which are landform classifications proposed by Tamura (1981). These landform types were classified based on discontinuities in slope angles and the forms of adjacent units (i.e. convexity and concavity; for details, see Tamura, 1981). CS and US are stable landforms that are characterized by decreased soil disturbance, whereas the other landforms often experience more frequent soil disturbances (Nagamatsu and Miura, 1997). A small stream runs through the centre of the plot, and thus the variety of its topographic conditions is suitable for the evaluation of clonal reproduction in lianas on the forest floor.
Study species
Wisteria floribunda (Willd.) DC. (Fabaceae) is a deciduous, stem twining liana species that is widely distributed throughout Japan (Ohashi, 1989). This species produces stolons on the ground surface via clonal reproduction (Fig. S2; Sakai et al., 2002). Wisteria floribunda often predominates temperate forests, representing 57 % of the abundance and 85 % of the basal area of the liana community in this study plot (Mori et al., 2016a). Other liana species in this study plot are mostly root climbers (e.g. Euonymus fortunei, Schizophragma hydrangeoides, Hydrangea petiolaris, Rhus ambigua). Wisteria floribunda and other root-climbing species accounted for 98 and 97 % of the total abundance and basal area in this liana community, respectively. Wisteria floribunda often reaches the forest canopy and covers the host tree canopy, while root climbers are suppressed under the host tree canopy (Ichihashi and Tateno, 2011; Mori et al., 2016a). There are no significantly exclusive distribution patterns between the liana species in this study plot (Mori et al. 2016a).
Sampling, DNA extraction and genotyping
Following the liana census conducted in 2013 (Mori et al. 2016a), we collected either fresh leaf or inner bark samples from all W. floribunda individuals that were higher than 1.3 m above-ground in 2015. We defined individuals as being independently rooted ramets that had no apparent above-ground connection to other rooted ramets (‘apparent genet’; Gerwing et al., 2006). Within the study plot, we did not observe any rooted ramets with obvious connections to other rooted ramets. Therefore, the term ‘ramets’ used in this study refers to rooted above-ground stems that sprouted from stolons or emerged from seeds. Inner bark samples were collected in cases where leaf samples could not be collected owing to the difficulty in distinguishing individuals, which often occurred when no leaves were available under the forest canopy and/or multiple individuals were tangled on the same host tree. The collection of leaf and inner bark samples (N = 391; 326 leaves and 65 bark samples) were stored at –30 °C prior to DNA extraction. DNA extraction was conducted using the DNeasy kit (Qiagen, Valencia, CA, USA). The polymerase chain reaction (PCR) was performed using the ten microsatellite markers designed for W. floribunda as described by H. Mori et al. (2016b). Genotyping data were checked and binned with Geneious R9.0 (Kearse et al., 2012).
Data analysis
Clones were identified with standardized methods (Arnaud-Haond et al., 2007). In brief, the ability of ten microsatellite markers to distinguish multilocus genotypes (MLGs) was examined by calculating the distinct number of MLGs for all combinations of a given locus. The results were confirmed from the plateau of the genotype accumulation curve (Fig. S3). To ascertain whether ramets of the same MLG belonged to the same clone, the probability of a given MLG occurring in a population under Hardy–Weinberg equilibrium was calculated (Pgen) (Parks and Werth, 1993):
where fi is the frequency of each allele at the ith locus estimated with a round-robin method, and h is the number of heterozygous loci. The probability of obtaining n repeated MLGs from a population more than once by chance in N samples (Psex) was calculated (Parks and Werth, 1993):
To ascertain each distinct MLG that belonged to a distinct clone, multilocus lineages (MLLs) were defined based on pairwise genetic distances. This procedure was necessary to prevent the false detection of clones owing to slightly different MLGs resulting from somatic mutation and genotyping errors. The threshold of pairwise genetic distance was determined as 1 by changing the threshold from 0 to 5 following the recommendations of Meirmans and Van Tienderen (2004) (Table S1). The MLLs are equivalent to the term ‘genet’ in ecological studies and thus we will use this term hereafter for MLLs to avoid further confusion.
Genets were mapped to show the clonal structure across the study plot. The patch size for all genets with three or more ramets was measured by the area of a convex hull polygon for each genet calculated in QGIS version 2.16.2 (QGIS Development Team, 2009). The robustness of habitat composition based on the landforms of both clonal and non-clonal ramets was tested using the jack-knife procedure: mean and 95 % confidence intervals for habitat composition of landforms were obtained from datasets that were generated excluding one genet. This was done for all combinations of genets. If the contribution of a single genet on the habitat composition of landforms was large, then a 95 % confidence interval would be a high value compared to that of the mean value. To evaluate the degree of aggregation and intermingling between genets, the aggregation index (Ac) was defined as Ac = (Psg – Psp)/Psg, where Psg is the average probability of clonal identity for all pairs and Psp is the average probability for clonal identity among the nearest neighbours. Ac is 0 when all genets are intermingled and 1 when all genets are distinctly distributed. Finally, to evaluate the clonal diversity the following indices were obtained: clonal richness (R), Simpson’s evenness index (V) and the Pareto index (β). The equation for clonal richness (genotypic richness) was R = (G –1)/(N –1), where G is the number of genets in N samples. Simpson’s evenness index was then estimated as V = (D – Dmin)/(Dmax – Dmin) where D is the Simpson index , , , G is the number of unique MLGs, and N is the number of samples. The Pareto index is the negative value of the slope of the power law (Pareto) distribution of clonal membership. The equation for the Pareto index is , where is the number of genets containing X or more ramets. The Pareto index (β) is higher when genets have a higher density and the distribution of ramets per genet has higher evenness. Pgen and Psex were calculated in R version 3.3.2 (R Development Core Team, 2016) with the ‘poppr’ package version 2.2.0 (Kamvar et al., 2014, 2015). Determination of genets (i.e. MLLs) was conducted in GenoDive version 2.0 (Meirmans and Van Tienderen, 2004). The calculation of clonal indices was done in R version 3.3.2 (R Development Core Team, 2016) with the ‘RClone’ package version 1.0 (Bailleul et al., 2016).
RESULTS
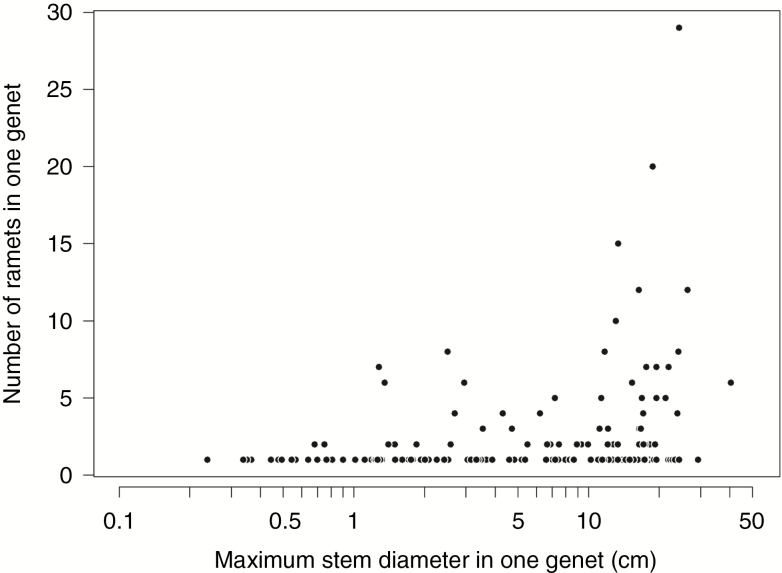
All ramets (N = 391; 326 leaf and 65 bark samples) sampled from the study plot were genotyped, and a total of 168 genets were detected. There was a low probability of obtaining a given genotype (Pgen < 0.001) and obtaining repeated genotypes that originated from distinct sexual reproductive events by chance (Psex < 0.001); thus, it was assumed that errors were unlikely to occur in the identification of clones. The number of ramets in one genet ranged from one to 29 (Supplementary Data Figs S4a, b). Clones contributed 71 and 62 % to abundance and total basal area, respectively, or 57 and 31 % when the largest ramet within a genet (presumed parent ramet) was excluded. The number of unique genets (i.e. a single-ramet genet) was 29 % of all ramets sampled in the study plot, while multiple-ramet genets with 2–4, 5–9 or 10 or more ramets comprised 21, 24 and 26 % of all individuals, respectively. Similarly, the basal area of unique genets was 38 %, while multiple-ramet genets with 2–4, 5–9 or 10 or more ramets were found to make up 20, 26 and 16 % of the sampled individuals, respectively. The clonal richness (R), Shannon evenness index (V), Pareto index (β) and aggregation index (Ac) were 0.43, 0.96, 0.76 (P < 0.001) and 0.38 (P < 0.001), respectively. The number of clonally reproduced ramets and the maximum stem diameter within one genet were positively correlated (Fig. 1).
Fig. 1.
Relationship between the maximum stem diameter and number of ramets in one genet. Note that the x-axis is log transformed.
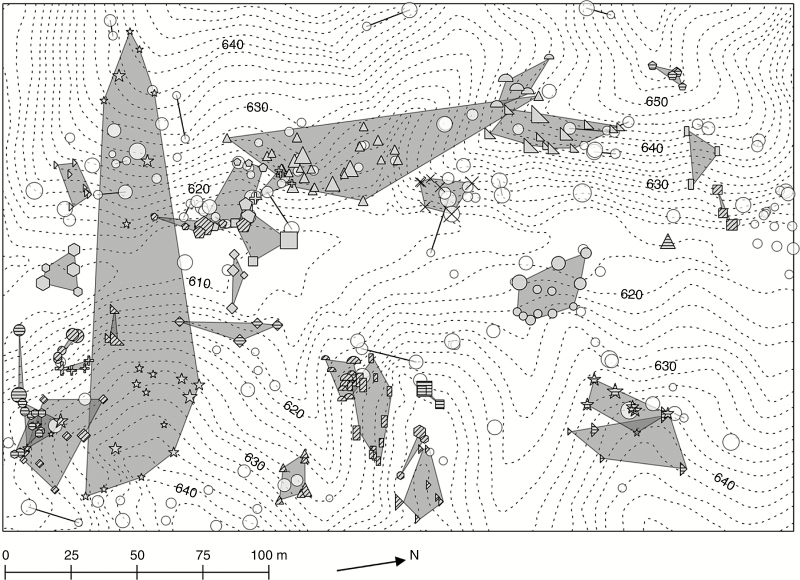
Spatial distribution maps of W. floribunda clones are shown in Fig. 2. Clones ranged over a distance of 180 m in the study plot. With the exception of the two largest genets, most of the clones did not spread over both sides of the slope, which was divided by a stream or valley landform (Fig. 2). The largest genet had a patch size of 0.47 ha (Figs S4 and 2). The total area of a clone patch size for all W. floribunda clones detected in the study plot was approximately 1.1 ha or 18 % of the plot.
Fig. 2.
Clonal patch size of Wisteria floribunda genets. Symbol size represents the dbh of each ramet. Polygons indicate the clone patches that have three or more ramets in one genet. Solid lines indicate clones that have two ramets in one genet. The different genets with three or more ramets are represented by closed symbols using combinations of different patterns and shapes, while those with one or two ramets are represented as open circles with a grey outline. Contour is presented in two meters. Numerals on contour lines indicate the altitude (m).
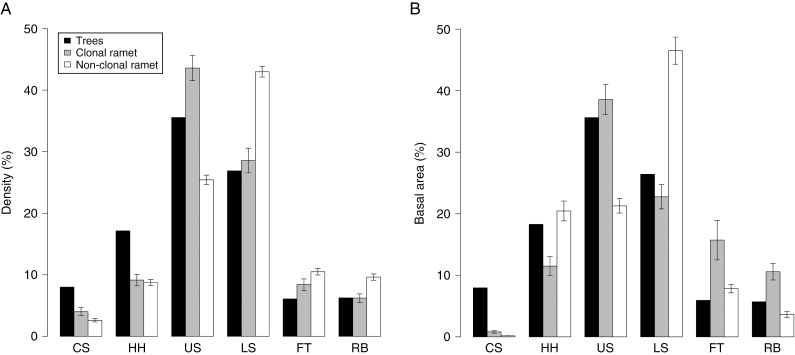
The composition of microscale landforms was significantly different between clonal and non-clonal ramets in terms of abundance (chi-square test; d.f. = 5; P < 0.05) and basal area (chi-square test; d.f. = 5; P < 0.001; Fig. 3). The non-clonal ramets were distributed to a greater extent on LS in both abundance and basal area than were trees and clonal-ramets in comparison to other landforms. These results indicate that the clonal reproduction of W. floribunda led to different distribution patterns over the micro-topography.
Fig. 3.
Habitat composition of microscale landforms of the (A) abundance and (B) basal area of trees, clonal and non-clonal ramets. Clonal and non-clonal ramets represent genets with two or more ramets and genets with one ramet, respectively. The robustness of habitat composition for clonal and non-clonal ramets was tested by generating datasets that excluded one genet from all combinations. Error bars represent a 95 % confidence interval for the generated datasets. Abbreviations of landforms are as follows: CS, crest slope; HH, head hollow; US, upper side slope; LS, lower side slope; FT, flood terrace; RB, river bed.
DISCUSSION
Genetic analysis of the W. floribunda population revealed that clonal reproduction makes a large contribution to abundance (71 %) and basal area (62 %) in this study plot. This contribution was 57 and 31 %, respectively, when the largest ramet within a genet was excluded, still indicating a significant contribution. To our knowledge, this is the first study to provide direct evidence for the clonal reproduction of a liana species in natural forest conditions using genetic tools with fine-scale sampling. Yorke et al. (2013) found that in old-growth tropical forests, long-distance clonal reproduction contributed 19 and 60 % to abundance and basal area, respectively. Schnitzer et al. (2012) reported that clonal rooted stems contributed 43 % to the total density and 23 % to total basal area in a 50-ha plot on Barro Colorado Island in Panama. They also found a strong relationship between stem diameter and the number of rooted ramets that had apparent connections to other rooted ramets. A similar correlation was found in the present study: a positive correlation was observed between maximum dbh and the number of ramets within one genet (Fig. 1). This indicates that larger lianas could produce a large amount of clonal ramets above (i.e. apparent rooted clones) or below (i.e. stolons) ground. Overall, the large contribution of clonal reproduction to W. floribunda abundance and basal area is consistent with previous observational studies of lianas in tropical forests.
We did not observe rooted clones of W. floribunda with apparent connections to each other in this study plot (H. Mori, personal observation); however, we did observe a substantial contribution of clonal reproduction to the abundance and basal area of W. floribunda in the present study. Our results indicate that genetic tools are useful in evaluating the clonal structure of liana populations in cases where physical connections between ramets are not entirely apparent (e.g. when they are connected below ground), and where the connections have been lost in the past. In fact, underground clonal reproduction could be fairly common among liana species. For example, Putz (1984) excavated small individuals (<50 cm tall) of five common liana species on Barro Colorado Island and found that 15–90 % of these plants were not true seedlings but clonal ramets connected by stolons or rhizomes. Thus, while previous studies have observed the considerable contributions of clonal reproduction to the dynamics of tropical liana communities (Schnitzer et al., 2012; Ledo and Schnitzer, 2014), the magnitude of the contribution may be even greater if unapparent (below-ground) connections between individuals are included. This possibility may be addressed in the future by means of genetic analyses.
There was a slight intermingling of genets, as indicated by the aggregation index of 0.38. The aggregation index of W. floribunda was comparable with those of other non-liana clonal woody plant species. For example, the Gondwanan conifer species Athrotaxis cupressoides had a mean aggregation index of 0.43 (Worth et al., 2016), while the aggregation index is reported to vary widely, both within species (0.1–0.79: A. curpressoides, Worth et al., 2016) and between species (0.03: Nothofagus pumilio, Mathiasen and Premoli, 2013; 0.62: Populus nigra, Chenault et al., 2011). A slightly intermingled distribution pattern of genets was observed in the present study, while clonal patches of W. floribunda seemed to exhibit a lower degree of overlapping distribution (Fig. 2). This could be because many non-clonal ramets (i.e. single-ramet genet) were distributed within patches of large genets. These non-clonal ramets could have originated from sexual reproduction (i.e. germinated from seed) that was observed as one unique genet in the study plot by genetic analysis. Further observations on seed production and seedling distribution using both ecological and genetic approaches would provide important insight into the contribution of seed reproduction on liana distribution.
It is also important to note that the wide variation in patch size and range of W. floribunda clones (Figs 1 and 2) was consistent with other non-liana clonal plants. For example, clones of the deciduous tree species Padus ssiori formed a large 0.4-ha genet, but it also had many small and unique genets distributed within the same population (Y. Mori et al., 2009). Wide variation in clone patch size was also reported in Fagus grandifolia (Kitamura et al., 2001) and in dwarf bamboo (Suyama et al., 2000). The heterogeneous clonal structure was also indicated by clonal diversity indices. The clonal richness (R = 0.43) and Pareto index (β = 0.76) were comparable with the average value of other clonal plants [R = 0.44; terrestrial and marine plant species: β = 0.60–1.49 as reviewed by Honnay and Jacquemyn (2008) and Ohsako (2010); kudzu (Pueraria lobata): β = 0.03–1.47; Kartzinel et al. (2015)], whereas the evenness (V = 0.96) was relatively higher in the liana we measured in comparison to that of other clonal plants (clonal plant average: V = 0.74; Honnay and Jacquemyn, 2008). Kartzinel et al. (2015) showed that the clonal structure of kudzu (Pueraria lobata), which is a deciduous vine species that grows along roadsides and in abandoned farmlands, forms both small and large patches of clones. Although clonal indices are an effective way to compare clonal abilities between species, information of the clonal indices of lianas is currently lacking. Further studies are needed to accumulate clonal indices for a general understanding of the relative importance of the clonal reproduction of liana species and how they relate to non-liana clonal plant species.
Clonal ramets were distributed more on the upper side of slopes (US) than the lower side of slopes (LS), whereas non-clonal ramets were distributed more on LS than on US (Fig. 3). LS is the steepest slope of the landforms in our study plot and has the most intense amount of soil surface disturbance (Nagamatsu and Miura, 1997). Consequently, non-liana trees in the study plot are more abundant on US than on LS (Fig. 3) areas, in agreement with a previous study that vegetation is often more developed on US but scarcer on LS (Nagamatsu and Miura, 1997). The contrasting distribution pattern of these ramets was more evident in basal area than in density (Fig. 3). This indicates that non-clonal ramets on LS are relatively mature, as indicated by their large stem diameter, number of ramets larger than 5 cm dbh for clonal ramets (US: 39, LS: 21) and number of non-clonal ramets (US: 16, LS: 20). Although larger ramets are able to produce more clonal ramets, clonal ramets were found more on the US than on the LS. Thus, the differences in preferred landforms between clonal and non-clonal ramets may reflect limitations of clonal growth on the forest floor by stolons. In summary, lianas derived from seeds do not particularly prefer growing in LS; however, those established in LS rarely succeed in clonal reproduction, leading to an accumulation of large single-ramet plants in this area.
The two largest genets (0.47 and 0.19 ha) were found to range over both sides of the slopes, which were divided by a stream and valley landform (Fig. 2). These results indicate that these clones were somehow able to override these topographical barriers. One possible explanation could be ‘laddering’. By laddering host tree crowns to one another, lianas can expand their distribution despite the topographical limitations. When a tree carrying stem twiners falls, lianas will also be pulled down to the forest floor and re-root from the stem and/or produce stolons (Schnitzer et al., 2000, 2004; Gerwing, 2004). In the present study site, we found stolons that had been produced from the fallen stems of W. floribunda (H. Mori, personal observation), while we did not observe any rooted ramet higher than 1.3 m above ground that had apparent connections to another rooted ramet, as previously mentioned. This could be because stolons can be easily buried in the ground, making the observation of ramets produced from fallen stems more difficult. An alternative explanation could be the expansion of stolons over the streams and valleys; however, no stolons were observed to have crossed over or started to cross over the stream, despite the presence of stolons derived from mother ramets along the streams at the study site (H. Mori, personal observation). Even if the stolons succeeded at crossing over the streams, ramets need to re-root to become independent from the mother ramet, making the probability that stolons can overcome topographical barriers minimal. The fact that the same clones were detected across streams and valleys implies that W. floribunda expanded its distribution clonally in two different layers of this forest, which were most likely the canopy and understorey.
We found a significant contribution of clonal reproduction to the distribution of a liana species (W. floribunda) that had no apparent rooted ramets to other rooted plant bodies. Clonal reproduction played an important role, not only in forming a population but also in determining the distribution patterns of a liana species over different micro-topographies, where clonal reproduction was reduced in steeper slopes, probably owing to the limitation of clonal growth via stolons in habitat with intense disturbances to the soil surface. As this study was conducted for one dominant liana species in one location, further studies are required to compare the contribution of clonal reproduction to the distribution of different species and/or in multiple forest stands, which is essential to provide a deeper understanding of clonal reproduction in liana populations. More importantly, defining clonal reproduction as a potential driver of liana distribution patterns will contribute to a comprehensive understanding of the distribution patterns of liana species in temperate forests.
SUPPLEMENTARY INFORMATION
Supplementary data are available at https://academic.oup.com/aob and consist of the following. Figure S1: Spatial distribution map of Wisteria floribunda genets and landforms. Figure S2: Photograph of Wisteria floribunda ramets and stolons in the study plot. Figure S3: Genotype accumulation curve. Figure S4: Histogram of (a) the number of ramets per genet and (b) clonal patch size. Table S1: Number of multilocus lineages (MLLs) and clonal diversity indices with changing thresholds of genetic distance.
Supplementary Material
ACKNOWLEDGMENTS
We thank Takahito Nishihira, Kana Furuki, Singgih Utomo and Yoshihiro Yamazaki for their help with liana sampling. We also thank Dr Kentaro Uchiyama and other members of the Department of Forest Genetics in the Forestry and Forest Products Research Institute for their technical help with genetic experiments, Dr Shoji Naoe and Dr Kaoru Niiyama for valuable advice during the revision, and Dr James Worth for revising the manuscript. Finally, we would like to thank Hajime Yamagata, who was the owner of the research station near the study plot. Sadly, he passed away before this study was published. This study was supported by a Grant-in-Aid for JSPS Research Fellow (Grant No. 16J00768) from the Japan Society for the Promotion of Science. This study was partly funded by the Ministry of Environment, Japan (Monitoring Sites 1000), and JSPS KAKENHI (Grant No. 15H04517).
LITERATURE CITED
- Allen BP, Sharitz RR, Goebel PC. 2007. Are lianas increasing in importance in temperate floodplain forests in the southeastern United States?Forest Ecology and Management 242: 17–23. [Google Scholar]
- Arnaud-Haond S, Duarte CM, Alberto F, Serrão EA. 2007. Standardizing methods to address clonality in population studies. Molecular Ecology 16: 5115–5139. [DOI] [PubMed] [Google Scholar]
- Arnold C, Schnitzler A. 2010. Historical reconstruction of a relictual population of wild grapevines (Vitis vinifera subsp. sylvestris, Gmelin, Hegi) in a floodplain forest of the upper Seine valley, France. River Research and Applications 26: 904–914. [Google Scholar]
- Bailleul D, Stoeckel S, Arnaud-Haond S. 2016. RClone: a package to identify MultiLocus Clonal Lineages and handle clonal data sets in r. Methods in Ecology and Evolution 7: 966–970. [Google Scholar]
- Chenault N, Arnaud-Haond S, Juteau M et al. 2011. SSR-based analysis of clonality, spatial genetic structure and introgression from the Lombardy poplar into a natural population of Populus nigra L. along the Loire River. Tree Genetics and Genomes 7: 1249–1262. [Google Scholar]
- Dalling JW, Schnitzer SA, Baldeck C et al. 2012. Resource-based habitat associations in a neotropical liana community. Journal of Ecology 100: 1174–1182. [Google Scholar]
- Foster PF, Sork VL. 1997. Population and genetic structure of the West African rain forest liana Ancistrocladus korupensis (Ancistrocladaceae). American Journal of Botany 84: 1078–1091. [PubMed] [Google Scholar]
- Gerwing JJ. 2004. Life history diversity among six species of canopy lianas in an old-growth forest of the eastern Brazilian Amazon. Forest Ecology and Management 190: 57–72. [Google Scholar]
- Gerwing JJ, Schnitzer SA, Burnham RJ et al. 2006. A standard protocol for liana censuses. Biotropica 38: 256–261. [Google Scholar]
- Grashof-Bokdam CJ, Jansen J, Smulders MJM. 1998. Dispersal patterns of Lonicera periclymenum determined by genetic analysis. Molecular Ecology 7: 165–174. [Google Scholar]
- Honnay O, Jacquemyn H. 2008. A meta-analysis of the relation between mating system, growth form and genotypic diversity in clonal plant species. Evolutionary Ecology 22: 299–312. [Google Scholar]
- Ichihashi R, Tateno M. 2011. Strategies to balance between light acquisition and the risk of falls of four temperate liana species: to overtop host canopies or not?Journal of Ecology 99: 1071–1080. [Google Scholar]
- Kamvar ZN, Tabima JF, Grunwald NJ. 2014. Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2: e281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamvar ZN, Brooks JC, Grunwald NJ. 2015. Novel R tools for analysis of genome-wide population genetic data with emphasis on clonality. Frontiers in Genetics 6: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartzinel TR, Hamrick JL, Wang C, Bowsher AW, Quigley BGP. 2015. Heterogeneity of clonal patterns among patches of kudzu, Pueraria Montana var. lobata, an invasive plant. Annals of Botany 116: 739–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. Retrieved from http://www.geneious.com [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura K, Homma K, Takasu H et al. 2001. Demographic genetics of the American beech, Fagus grandifolia. Ii. Genet substructure of populations for the blue ridge, piedmont and the Great Smoky Mountains. Plant Species Biology 16: 219–230. [Google Scholar]
- Ledo A, Schnitzer SA. 2014. Disturbance and clonal reproduction determine liana distribution and maintain liana diversity in a tropical forest. Journal of Ecology 95: 2169–2178. [DOI] [PubMed] [Google Scholar]
- Masaki T, Suzuki W, Niiyama K, Iida S, Tanaka H, Nakashizuka T. 1992. Community structure of a species-rich temperate forest, Ogawa Forest Reserve, central Japan. Vegetatio 98: 97–111. [Google Scholar]
- Mathiasen P, Premoli AC. 2013. Fine-scale genetic structure of Nothofagus pumilio (lenga) at contrasting elevations of the altitudinal gradient. Genetica 141: 95–105. [DOI] [PubMed] [Google Scholar]
- Meirmans PG, Van Tienderen PH. 2004. GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Molecular Ecology Notes 4: 792–794. [Google Scholar]
- Mori H, Kamijo T, Masaki T. 2016a. Liana distribution and community structure in an old-growth temperate forest: the relative importance of past disturbances, host trees, and microsite characteristics. Plant Ecology 217: 1–12. [Google Scholar]
- Mori H, Ueno S, Matsumoto A et al. 2016b. Development and characterization of 10 microsatellite markers from Wisteria floribunda (Fabaceae). Silvae Genetica 65: 55–58. [Google Scholar]
- Mori Y, Nagamitsu T, Kubo T. 2009. Clonal growth and its effects on male and female reproductive success in Prunus ssiori (Rosaceae). Population Ecology 51: 175–186. [Google Scholar]
- Nagamatsu D, Miura O. 1997. Soil disturbance regime in relation to micro-scale landforms and its effects on vegetation structure in a hilly area in Japan. Plant Ecology 133: 191–200. [Google Scholar]
- Nakashizuka T, Matsumoto Y. 2002. Diversity and interaction in a temperate forest community: Ogawa Forest Reserve of Japan. Springer Japan; Retrieved from https://books.google.co.jp/books?id=Vbd9CAAAQBAJ [Google Scholar]
- Nakashizuka T, Katsuki T, Tanaka H. 1995. Forest canopy structure analyzed by using aerial photographs. Ecological Research 10: 13–18. [Google Scholar]
- Ohashi H. 1989. Wisteria, p. 247–248. In: Satake Y, Hara H, Watari S, Tominari T, eds. Wild flowers of Japan: woody plants I.Tokyo: Heibonsha, (in Japanese). [Google Scholar]
- Ohsako T. 2010. Clonal and spatial genetic structure within populations of a coastal plant, Carex kobomugi (Cyperaceae). American Journal of Botany 97: 458–470. [DOI] [PubMed] [Google Scholar]
- Parks JC, Werth CR. 1993. A study of spatial features of clones in a population of bracken fern, Pteridium aquilinum (Dennstaedtiaceae). American Journal of Botany 80: 537–544. [DOI] [PubMed] [Google Scholar]
- Peñalosa J. 1984. Basal branching and vegetative spread in two tropical rain forest lianas. Biotropica 16: 1–9. [Google Scholar]
- Phillips OL, Vásquez Martínez R, Arroyo L et al. 2002. Increasing dominance of large lianas in Amazonian forests. Nature 418: 770–774. [DOI] [PubMed] [Google Scholar]
- Putz FE. 1984. The natural history of lianas on Barro Colorado Island, Panama. Journal of Ecology 65: 1713–1724. [Google Scholar]
- QGIS Development Team 2009. QGIS Geographic Information System. Retrieved from http://qgis.osgeo.org [Google Scholar]
- R Development Core Team 2016. R: A Language and Environment for Statistical Computing. Vienna, Austria: Retrieved from https://www.r-project.org/ [Google Scholar]
- Sakai A, Nomiya H, Suzuki W. 2002. Horizontal distribution of stolons of a temperate liana Wisteria floribunda DC. and its ecological significance. Journal of Forest Research 7: 125–130. [Google Scholar]
- Schnitzer SA, Bongers F. 2011. Increasing liana abundance and biomass in tropical forests: Emerging patterns and putative mechanisms. Ecology Letters 14: 397–406. [DOI] [PubMed] [Google Scholar]
- Schnitzer SA, Dalling JW, Carson WP. 2000. The impact of lianas on tree regeneration in tropical forest canopy gaps: evidence for an alternative pathway of gap-phase regeneration. Journal of Ecology 88: 655–666. [Google Scholar]
- Schnitzer SA, Parren MPE, Bongers F. 2004. Recruitment of lianas into logging gaps and the effects of pre-harvest climber cutting in a lowland forest in Cameroon. Forest Ecology and Management 190: 87–98. [Google Scholar]
- Schnitzer SA, Rutishauser S, Aguilar S. 2008. Supplemental protocol for liana censuses. Forest Ecology and Management 255: 1044–1049. [Google Scholar]
- Schnitzer SA, Mangan SA, Dalling JW et al. 2012. Liana abundance, diversity, and distribution on Barro Colorado Island, Panama. PLoS ONE 7: e52114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama Y, Obayashi K, Hayashi I. 2000. Clonal structure in a dwarf bamboo (Sasa senanensis) population inferred from amplified fragment length polymorphism (AFLP) fingerprints. Molecular Ecology 9: 901–906. [DOI] [PubMed] [Google Scholar]
- Tamura T. 1981. Multiscale landform classification study in the hills of Japan: Part Ⅱ Application of the multiscale landform classification system to pure geomorphological studies of the hills of Japan. The Science Reports of Tohoku University 31: 85–184. [Google Scholar]
- Waters EM, Watson MA. 2015. Live substrate positively affects root growth and stolon direction in the woodland strawberry, Fragaria vesca. Frontiers in Plant Science 6: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worth JRP, Sakaguchi S, Rann KD et al. 2016. Gondwanan conifer clones imperilled by bushfire. Scientific Reports 6: 33930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorke SR, Schnitzer SA, Mascaro J, Letcher SG, Carson WP. 2013. Increasing liana abundance and basal area in a tropical forest: the contribution of long-distance clonal colonization. Biotropica 45: 317–324. [Google Scholar]
- Yoshinaga S, Takahashi M, Aizawa S. 2002. Landforms and soil characteristics in Ogawa Forest Reserve. In: Nakashizuka T, Matsumoto Y, eds. Diversity and interaction in a temperate forest community: Ogawa Forest Reserve of Japan Toyko: Springer Japan, 19–26. Retrieved from https://books.google.co.jp/books?id=Vbd9CAAAQBAJ [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.