Abstract
Background and Aims
Disentangling the relative roles of past fragmentation (vicariance), colonization (dispersal) and post-divergence gene flow in the genetic divergence of continental island organisms remains a formidable challenge. Amplified fragment length polymorphisms (AFLPs) were used to (1) gain further insights into the biogeographical processes underlying the Pleistocene diversification of the Aegean Nigella arvensis complex; (2) evaluate the role of potential key factors driving patterns of population genetic variability (mating system, geographical isolation and historical contingencies); and (3) test the robustness of conclusions previously drawn from chloroplast (cp) DNA.
Methods
Genetic diversity was analysed for 235 AFLP markers from 48 populations (497 individuals) representing 11 taxa of the complex using population genetic methods and Bayesian assignment tests.
Key Results
Most designated taxa are identifiable as genetically distinct units. Both fragmentation and dispersal-driven diversification processes occurred at different geological time scales, from Early to Late Pleistocene, specifically (1) sea barrier-induced vicariant speciation in the Cyclades, the Western Cretan Strait and Ikaria; and (2) bi-regional colonizations of the ‘Southern Aegean Island Arc’ from the Western vs. Eastern Aegean mainland, followed by allopatric divergences in Crete vs. Rhodos and Karpathos/Kasos. Outcrossing island taxa experienced drift-related demographic processes that are magnified in the two insular selfing species. Population genetic differentiation on the mainland seems largely driven by dispersal limitation, while in the Central Aegean it may still be influenced by historical events (island fragmentation and sporadic long-distance colonization).
Conclusions
The biogeographical history of Aegean Nigella is more complex than expected for a strictly allopatric vicariant model of divergence. Nonetheless, the major phylogeographical boundaries of this radiation are largely congruent with the geography and history of islands, with little evidence for ongoing gene exchange between divergent taxa. The present results emphasize the need to investigate further biological and landscape features and contemporary vs. historical processes in driving population divergence and taxon diversification in Aegean plant radiations.
Keywords: Aegean archipelago, AFLP, colonization, dispersal, gene flow, Nigella, palaeogeography, phylogeography, population genetic structure, vicariance
INTRODUCTION
‘Recent continental shelf or land-bridge islands’ (sensuWhittaker and Fernández-Palacios, 2007; Ali et al., 2017) form part of the same continental shelf and have become separated from each other and/or adjacent landmasses through rising sea levels, most recently during Pleistocene inter- and/or post-glacial periods. This island type has been instrumental in elucidating the effects of past range fragmentation and geographical isolation on (incipient) allopatric vicariant speciation (e.g. Wallace, 1890; Wright, 1940; Mayr, 1954). In general, recent molecular phylogenetic and phylogeographical studies have clarified that most evolutionary diversification in islands occurred via allopatric speciation (e.g. Comes et al., 2008; Esselstyn et al., 2009; Li et al., 2010; Salvo et al., 2010; Mayol et al., 2012; Zhai et al., 2012; Poulakakis et al., 2015; Sfentourakis and Triantis, 2017; for a review, see also Warren et al., 2015). However, some researchers have also challenged this paradigm as too simplistic, especially for recent continental shelf or land-bridge islands, given the increased opportunities for (re-)colonization/dispersal events and/or the genetic exchange between divergent populations afforded by the recurrent formation of land-bridges (and/or the narrowing of sea straits) during low sea level periods of the last glacial(s) (e.g. Li et al., 2010; Zhai et al., 2012). Hence, disentangling the relative roles of past fragmentation (vicariance), colonization (dispersal) and post-divergence gene flow on the genetic divergence of continental island organisms remains a formidable challenge, especially in island-rich systems with great geographical heterogeneity and complex palaeogeographical history.
These latter properties certainly apply to the Aegean archipelago (Fig. 1A), which comprises some 7500 islands and islets at a variety of isolation levels between the Greek peninsula in the west and the Turkish coast of Asia Minor in the east (Sfenthourakis and Triantis, 2017). This mostly continental island system has always attracted the interests of plant biologists, given its high species diversity and endemism (e.g. Strid, 1996; Perissoratis and Conispoliatis, 2003; Georghiou and Delipetrou, 2010), but has hitherto received far less attention from plant molecular phylogeographers (but see below). Nonetheless, phytogeographers have long been aware of the Aegean featuring strong boundaries, despite relatively homogenous climatic and habitat conditions (Rechinger, 1950; Strid, 1997; see Fig. 1A). The most well-known floristic division is ‘Rechinger’s line’ (Strid, 1996) between the Central and East Aegean islands, which coincides with the north-central part of the ‘mid-Aegean Trench’ (MAT), an ancient sea transgression [approx. 12–9 million years ago (Ma); Dermitzakis and Papanikolaou, 1981] that quickly (re-)established after the Messinian Salinity Crisis (6.0–5.3 Ma; Duggen et al., 2003) and has remained a sea barrier ever since. Another example is the ‘Cycladic Window’ (Rechinger, 1950), which broadly delimitates an ancient Central Aegean peninsula (‘Cycladia’) that persisted until the end of the Pliocene/Early Pleistocene (approx. 2 Ma; Chatzimanolis et al., 2003) but then underwent cyclical fragmentation induced by sea level cycles (Lambeck, 1996). Recent floristic analyses at more regional scales likewise support the long-standing view that distribution patterns of the Aegean flora tend to reflect the area’s palaeogeographical history (e.g. Trigas et al., 2013; Kougioumoutzis et al., 2014).
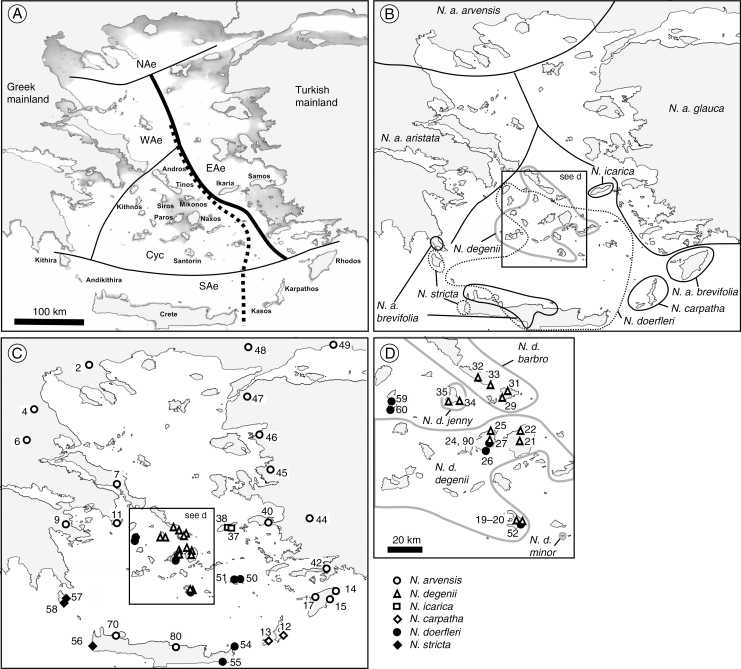
Fig. 1.
(A) Floristic sub-division of the Aegean region (after Rechinger, 1950) into five major zones, namely the Western (WAe), Northern (NAe) and Eastern Aegean (EAe), the Cyclades (Cyc) and the ‘Southern Aegean Island Arc’ (SAe; Crete, Karpathos/Kasos and Rhodos), with ‘Rechinger’s line’ (Strid, 1996) highlighted in bold. The stippled line shows the position of the mid-Aegean trench (MAT; after Poulakakis et al., 2015). The shadowed areas indicate tentative coastal configurations at the sea level lowstand (sea level at –120 m) during the Last Glacial Maximum (approx. 21 500 years ago; modified after Perissoratis and Conispoliatis, 2003). (B) The approximate distribution ranges of the six species (12 taxa) of the Aegean Nigella arvensis complex (after Strid, 2002). Solid vs. dashed lines delineate outcrossing vs. selfing taxa. Note that the range of N. arvensis ssp. brevifolia on Crete comprises only four known populations. (C) Geographical locations of the 48 populations representing 11 taxa of the complex surveyed for AFLPs (except N. degenii ssp. minor). (D) Inset showing an expanded view of the Cyclades. Taxon abbreviations (in B and D): N. a., N. arvensis; N. d., N. degenii. Population numbers correspond to those in Table 1.
The Aegean region has also served as a popular study area to investigate the patterns and underlying causes of local differentiation and speciation in a range of ecologically largely unspecialized plant groups (e.g. Snogerup, 1967; Strid, 1970; Stork, 1972; Bentzer, 1973; von Bothmer, 1987). These biosystematic studies underscored the importance of palaeogeographical events in the evolution of the Aegean flora but also offered a clear hypothesis for non-adaptively driven allopatric differentiation largely caused by random genetic drift (Wright, 1940; Runemark, 1969) in small, isolated populations (‘non-adaptive radiation’; Rundell and Price, 2009). The idea that organismal distributions in the Aegean often mirror palaeogeographical patterns and allopatric processes of divergence has received considerable support over the last years, particularly from phylogenetic, phylogeographical and population genetic studies on animals (reviewed in Poulakakis et al., 2015; Sfenthourakis and Triantis, 2017). Yet, to date, plants have remained grossly understudied in this regard (but see Brassica cretica, Edh et al., 2007; Nigella arvensis alliance, Bittkau and Comes, 2005, 2009; Comes et al., 2008; N. degenii, Jorgensen et al., 2006; South Aegean Campanula, Cellinese et al., 2009; Campanula subg. Roucela, Crowl et al., 2015). Hence, our understanding of how the geographical ranges of closely related Aegean plant species evolved in response to palaeogeographical events is still far from complete.
Here we revisit the Nigella arvensis L. complex (Ranunculaceae), which represents one of the most intriguing radiations of the Aegean region (reviewed in Comes et al., 2008). It comprises four predominantly outcrossing and two selfing species (12 taxa in total) that are mostly allo-/or parapatrically distributed on numerous islands and surrounding mainland areas of Greece and Turkey (Strid, 1970; Fig. 1B). Previous phylogeographical work focused on chloroplast (cp) restriction fragment length polymorphisms (RFLPs) in the outcrossing species (termed ‘N. arvensis alliance’; Bittkau and Comes, 2005). This analysis pointed at sea barrier-induced vicariance, restricted gene flow and genetic drift as major processes shaping the evolutionary history of this group, and identified a major genetic break along Rechinger’s line. A phylogenetic study of Nigella L. using time-calibrated sequences of nuclear ribosomal DNA (Bittkau and Comes, 2009) inferred an Early/Mid-Pleistocene crown age for the entire complex [approx. 0.78 (± 0.39) – 0.16 (± 0.08) Ma], while its stem age could only be dated very roughly (approx. 6.2–1.3 Ma). Only a preliminary survey has been done to date on the complex using amplified fragment length polymorphisms (AFLPs) (U Jaros, unpubl. res.; but see Comes et al., 2008). This analysis of 110 individuals (42 populations) indicated that these fast-evolving nuclear markers have a high potential to delimitate taxonomic boundaries, as known from other studies where traditional sequence-based methods failed to resolve relationships among rapidly evolving lineages (e.g. Després et al., 2002; Vitales et al., 2014).
In the present study, we used AFLPs to investigate the evolutionary history of the entire N. arvensis complex and the role of potential key factors driving intraspecific patterns of population genetic variability (i.e. mating system, geographical isolation and historical contingencies). In sum, we have surveyed 11 of the 12 taxa (48 populations, 497 individuals), including the majority of populations studied for cpDNA (Bittkau and Comes, 2005; see Fig. 1C, D; Table 1). Specifically, we aimed to (1) evaluate the genetic and biogeographical boundaries of designated (sub)species; (2) generate the first comprehensive hypothesis of evolutionary relationships between them; and (3) infer the prevalent geographical modes (vicariance vs. dispersal/colonization) and relative timings of taxon diversification in the context of Aegean palaeogeography. In addition, we (4) tested for potential effects of selfing and/or the occurrence on islands per se on patterns of population genetic diversity and structure; and (5) asked to what extent present-day population genetic differentiation is driven by contemporary processes (e.g. gene flow and drift) or past range dynamics (e.g. island fragmentation and colonization).
Table 1.
Locations of 11 taxa (48 populations, 497 individuals) of the Nigella arvensis complex sampled in the Aegean region together with estimates of genetic (AFLP) diversity for each population with sample sizes of n ≥7 (42 populations, 478 individuals)
| Taxon/population no.* | Region/island, locality† | Latitude | Longitude | n | PPF | H E | SI |
|---|---|---|---|---|---|---|---|
| N. arvensis L. | |||||||
| ssp. arvensis | |||||||
| 2 (1) | Macedonia (GR), Megali Volvi | 40°41′32″ | 23°30’16″ | 9 | 58.72 | 0.2241 | 0.2511 |
| ssp. aristata (Sibth. & Sm.) Nym. | |||||||
| 4 (2) | Thessalia (GR), SW of Goni | 39°50′54″ | 22°27’10″ | 14 | 61.27 | 0.1992 | 0.2326 |
| 6 (4) | Thessalia (GR), W of Farsala | 39°16′40″ | 22°19’80″ | 11 | 60.42 | 0.2219 | 0.2538 |
| 7 (5) | Euboea (GR), Aliveri | 38°24′31″ | 24°10’34″ | 9 | 60.00 | 0.2102 | 0.2396 |
| 9 (8) | Peloponnese (GR), E of Ligourio | 37°38′23″ | 23°40’50″ | 11 | 68.51 | 0.2516 | 0.2882 |
| 11 (9) | Attiki (GR), Sounio, Acropolis | 37°39′19″ | 24°10’49″ | 9 | 60.85 | 0.2232 | 0.2556 |
| ssp. brevifolia Strid | |||||||
| 70 (26) | Crete (GR), W of Chania | 35°30′36″ | 24°00’58″ | 4 | NC | NC | NC |
| 80 (27) | Crete (GR), near Knossos | 35°18′00″ | 25°10’12″ | 2 | NC | NC | NC |
| 17 (21) | Rhodos (GR), near Kallithea | 36°22′29″ | 28°13’41″ | 15 | 31.91 | 0.1050 | 0.1333 |
| 14 (22) | Rhodos (GR), W of Archangelos | 36°13′80″ | 28°50’53″ | 10 | 45.53 | 0.1802 | 0.2162 |
| 15 (24) | Rhodos (GR), Kamiros to Kritina | 36°15′44″ | 27°48’50″ | 11 | 51.06 | 0.1795 | 0.2006 |
| ssp. glauca (Boiss.) Terracc. | |||||||
| 40 (20) | Samos (GR), NW of Pithagorio | 37°40′53″ | 26°56′80″ | 10 | 52.76 | 0.1970 | 0.2259 |
| 42 (17) | Marmaris (TR), Bencik to Emecik | 36°47′50″ | 28°20′20″ | 16 | 65.95 | 0.2009 | 0.2249 |
| 44 (15) | Aydin (TR), near Kocarli | 37°45′58″ | 27°43′15″ | 9 | 55.74 | 0.2140 | 0.2334 |
| 45 (14) | Menemen (TR), Menemen to Aliaga | 38°41′00″ | 26°57′59″ | 13 | 63.83 | 0.2022 | 0.2304 |
| 46 (13) | Ayvalik (TR), Ayvalik to Keremköy | 39°20′26″ | 26°45′55″ | 18 | 69.78 | 0.2282 | 0.2588 |
| 47 (12) | Canakkale (TR), W of Orada | 40°40′45″ | 26°32′39″ | 14 | 76.17 | 0.2488 | 0.2773 |
| 48 (11) | Thrakia (TR), Ibriktepe to Türkobasi | 41°10′44″ | 26°32′50″ | 13 | 62.12 | 0.2025 | 0.2312 |
| 49 (10) | Thrakia (TR), W of Silivri | 41°40′21″ | 28°90′40″ | 12 | 42.12 | 0.1365 | 0.1671 |
| N. degenii Vierh. | |||||||
| ssp. degenii | |||||||
| 19 (43) | Santorini (GR), S of Fira | 36°24′40″ | 25°26′15″ | 8 | 40.42 | 0.1723 | 0.2080 |
| 20 (42) | Santorini (GR), N of Kamari | 36°23′52″ | 25°28′57″ | 7 | 40.42 | 0.1670 | 0.2013 |
| 21 (40) | Naxos (GR), W of Himaros | 37°30′36″ | 25°27′39″ | 8 | 51.48 | 0.2041 | 0.2220 |
| 22 (39) | Naxos (GR), near Moni Faneromenis | 37°80′45″ | 25°27′59″ | 12 | 54.89 | 0.1885 | 0.2271 |
| 25 (38) | Paros (GR), S of Lefkes | 37°30′80″ | 25°12′45″ | 7 | 40.42 | 0.1609 | 0.1892 |
| 27 (36) | Paros (GR), S of Cape Korakas | 37°80′52″ | 25°13′20″ | 8 | 40.00 | 0.1588 | 0.1895 |
| ssp. barbro Strid | |||||||
| 29 (32) | Mikonos (GR), W of Ornos | 37°25′13″ | 25°19′50″ | 15 | 51.91 | 0.1693 | 0.2092 |
| 31 (30) | Mikonos (GR), Panormos beach | 37°28′27″ | 25°21′42″ | 11 | 40.42 | 0.1428 | 0.1654 |
| 32 (29) | Tinos (GR), E of Agios Fokas beach | 37°31′54″ | 25°13′11″ | 15 | 43.83 | 0.1409 | 0.1758 |
| 33 (28) | Tinos (GR), SE of Kardiani | 37°35′28″ | 25°60′58″ | 12 | 34.04 | 0.1091 | 0.1347 |
| ssp. jenny Strid | |||||||
| 34 (35) | Siros (GR), E of Vari | 37°23′51″ | 24°57′46″ | 8 | 51.48 | 0.2012 | 0.2379 |
| 35 (34) | Siros (GR), Kokkina beach (Finikas) | 37°23′44″ | 24°52′16″ | 14 | 48.93 | 0.1691 | 0.2114 |
| N. icarica Strid | |||||||
| 37 (45) | Ikaria (GR), SW of Chrisostomos | 37°34′30″ | 26°12′46″ | 16 | 50.21 | 0.1619 | 0.2042 |
| 38 (44) | Ikaria (GR), S of Kosikia | 37°34′41″ | 26°90′51″ | 16 | 40.43 | 0.1251 | 0.1670 |
| N. carpatha Strid | |||||||
| 12 (46) | Karpathos (GR), E of Pigadia | 35°30′22″ | 27°13′18″ | 16 | 59.57 | 0.1856 | 0.2327 |
| 13 (47) | Kasos (GR), Phry | 35°24′51″ | 26°55′44″ | 11 | 36.95 | 0.1356 | 0.1625 |
| N. doerfleri Vierh. | |||||||
| 50 | Astipaleia (GR), Maltezana | 36°34′34″ | 26°23′38″ | 7 | 20.85 | 0.0908 | 0.1757 |
| 51 | Astipaleia (GR), Mesaria to Panormoy | 36°34′55″ | 26°17′44″ | 8 | 23.83 | 0.0977 | 0.1281 |
| 54 | Crete (GR), S of Akra Sideros | 35°17′20″ | 26°17′36″ | 12 | 22.55 | 0.0752 | 0.1067 |
| 55 | Crete (GR), Moni Kapsa, Perivolakia | 35°10′27″ | 26°30′30″ | 5 | NC | NC | NC |
| 59 | Kithnos (GR), Episkopi | 37°23′53″ | 24°23′45″ | 12 | 20.85 | 0.0687 | 0.0974 |
| 60 | Kithnos (GR), S of the island | 37°19′80″ | 24°22′54″ | 8 | 16.60 | 0.0635 | 0.0847 |
| 26 | Paros (GR), S of the island | 36°58′58″ | 25°10′38″ | 2 | NC | NC | NC |
| 90 | Paros (GR), near Lefkes (1) | 37°30′50″ | 25°12′41″ | 11 | 21.70 | 0.0786 | 0.1088 |
| 24 | Paros (GR), near Lefkes (2) | 37°30′50″ | 25°12′41″ | 2 | NC | NC | NC |
| 52 | Santorini (GR), Kamari to Perisa | 36°22′40″ | 25°28′23″ | 4 | NC | NC | NC |
| N. stricta Strid | |||||||
| 56 | Crete (GR), Moni Chrisoskalitisas | 35°20′52″ | 23°32′18″ | 13 | 31.06 | 0.1036 | 0.1482 |
| 57 | Kithira (GR), beach of Paleopoleos | 36°13′33″ | 23°30′59″ | 8 | 25.53 | 0.0955 | 0.1099 |
| 58 | Kithira (GR), Halkos, E of Kapsali | 36°80′60″ | 23°20′80″ | 11 | 16.60 | 0.0568 | 0.0794 |
Standard deviations are given in parentheses.
See Table 3 for taxon-wide levels of diversity.
n, number of individuals assayed; PPF, percentage of polymorphic fragments; HE, Nei’s (1987) unbiased gene diversity; SI, Shannon’s index of phenotypic diversity (Lewontin, 1972); NC, not calculated due to small sample sizes (n ≤ 5).
*Numbers in parentheses refer to population codes in Bittkau and Comes (2005).
†Country codes: GR, Greece; TR, Turkey.
MATERIALS AND METHODS
Study system
Following Strid (1970), and based on molecular evidence (Bittkau and Comes, 2009), the Aegean N. arvensis complex is monophyletic with six annual, diploid (2n = 12) and self-compatible species. The four members of the ‘alliance’ (N. arvensis, N. degenii, N. icarica and N. carpatha) are summer-flowering, cross-compatible species with relatively large, nectariferous and mostly bee-pollinated flowers (approx. 1.2–3.4 cm wide). In contrast, N. doerfleri and N. stricta are reproductively isolated inter se and towards the alliance; they are spring-flowering, smaller sized species, in which selfing regularly takes place within flowers (approx. 0.9–1.5 cm wide) since the styles twist around the dehiscing anthers. The fruit (capsule) releases numerous, small seeds (<4 mm) without adaptations for attaching and transport. Populations generally occur in disturbed habitats (e.g. phrygana, stony sea shores, roadsides or abandoned/cultivated fields), whereby selfers tend to occupy more arid sites (Strid 1970).
Most taxa are allo-/or parapatrically distributed (Strid, 2002; Fig. 1B). Within the alliance, N. arvensis displays more or less continuous morphological variation from southern Greece (ssp. aristata) via northern Greece (ssp. arvensis) to western Turkey (ssp. glauca), while ssp. brevifolia occurs in Elafonisos (south-east of the Peloponnese), Crete and Rhodos; the four subspecies of N. degenii are endemic to particular islands of the Cyclades; and N. icarica and N. carpatha are respective endemics of Ikaria and Karpathos/Kasos. Also the two selfers have non-overlapping ranges: N. doerfleri is widespread in the Cyclades (and peripheral arid islets) but also found in Andikithira (south-east of the Peloponnese) and central/eastern Crete, whereas N. stricta is restricted to Kithira and south-west Crete. The exceptional range overlap between N. doerfleri and N. degenii in the Cyclades suggests that only species differing in mating system (and associated traits) are able to live in broad sympatry (Comes et al., 2008).
Plant material and sampling
The present study includes collections from 48 populations, representing all six species and 11 of the 12 taxa of the N. arvensis complex, with the exception of N. degenii Vierh. ssp. minor Strid, endemic to a southern Cycladic islet (Pakhia, south of Anafi) (Table 1; Fig. 1C, D). These samples were obtained almost exclusively from silica-dried leaf material collected in the field by Christiane Bittkau (May–July 2002, except for two populations of N. a. ssp. brevifolia from Crete, collected by Zacharias Kypriotakis, October 2002). In a few cases, fresh leaf material was obtained from seedlings raised in the glasshouse from field-collected seed. This sampling represents 35 of the 47 populations previously surveyed for cpDNA variation (Bittkau and Comes, 2005). On average, 4–5 populations were sampled per taxon (mean 4.4 ± 2.8 s.d.); taxa with only two or three populations involve several island endemics (N. degenii ssp. jenny, N. icarica, N. carpatha and N. stricta), while a single population represents N. a. ssp. arvensis, given its ‘rarity’ at the time of collection (in 2002). With few exceptions, between seven and 18 individuals were genotyped per population (mean 10.3 ± 3.8 s.d.), with 497 individuals in total. Populations with fewer sample sizes (n ≤5) involved N. a. ssp. brevifolia from Crete (population nos 70 and 80) as well as N. doerfleri from Paros (24 and 26), Santorini (52) and Crete (55) (see Table 1).
AFLP genotyping
Total genomic DNA was extracted and purified from silica-dried (or more rarely fresh) leaf tissue as detailed in Bittkau and Comes (2005). The AFLP analysis was performed according to Vos et al. (1995), with minor modifications as described by Jaros et al. (2016) for the use of fluorescent dye-labelled primers. For a preliminary AFLP screen, we selected 12 individuals representing four species (i.e. N. arvensis, N. degenii, N. doerfleri and N. stricta), using 30 selective primer pair combinations with three or four selective nucleotides (data not shown). Based on this, the following primer combinations that produced the best results with respect to polymorphism and clarity of AFLP profiles were chosen for the complete survey (fluorescent dye in parentheses): EcoRI-ACA/MseI-CAGC (6-FAM), EcoRI-AGA/MseI-CAGA (VIC) and EcoRI-ACC/MseI-CTCA (NED). The selective PCR products were purified using Sephadex G-50 Superfine Resin (GE Healthcare BioSciences, Uppsala, Sweden) following the manufacturer’s instructions. AFLP fragments were separated on a MegaBACE™ 1000 (GE Healthcare Biosciences, Pittsburgh, PA, USA), using the ET550-R-Rox-MegaBACE™ sizing standard. Using DAX ver. 8.0 (Van Mierlo Software Consultancy, Eindhoven, The Netherlands), polymorphisms were manually scored as presence (1) or absence (0), whereby a fragment was considered polymorphic if at least one individual showed a variant pattern. The reproducibility of markers was checked by repeating the analysis from the restriction/ligation step onward on 11 samples (representing all 11 taxa) for each pair of primers (Bonin et al., 2007). The genotyping error rate obtained (6.7 %) was only slightly higher than reported for most plant species (approx. 2–5 %; Bonin et al., 2007).
Genetic distance and structure analyses
Genetic relationships between populations of the entire data set were investigated using two approaches. First, we calculated Nei’s (1972) standard genetic distance (D) among all possible pairs of populations from allele frequencies estimated in POPGENE ver. 1.32 (Yeh and Boyle, 1997) and assuming Hardy–Weinberg equilibrium for outcrossing (FIS = 0) and full inbreeding for selfing (FIS = 1) taxa (only N. doerfleri and N. stricta). Secondly, we calculated pairwise FST values from fragment frequencies in AFLP-SURV ver. 1.0 (Vekemans et al., 2002). For either approach, 10 000 genetic distance matrices were constructed in AFLP-SURV by bootstrapping over loci, i.e. fragments. The procedures NEIGHBOR and CONSENSE in PHYLIP ver. 3.63 (Felsenstein, 2005) were then used to generate unrooted Neighbor–Joining (NJ) networks and to infer bootstrap values on their internal edges.
We employed Bayesian clustering in STRUCTURE ver. 2.2 (Pritchard et al., 2000) to assign individuals to genotypically distinct clusters (K) and infer their potentially ordered splitting in consecutive runs (see below). As four of our study species are interfertile, we assumed the ‘admixture’ model, as well as independent allele frequencies across populations, as proposed for multispecies AFLP studies (e.g. Pachschwöll et al., 2015). The number of K was set to vary from 1 to 12. For each value of K, we performed eight runs with a burn-in length of 200 000 and a run length of 500 000 Markov chain Monte Carlo (MCMC) replications. To identify the most likely number of clusters, we computed for each K the posterior probability of each run [lnp(D)] and the similarity coefficient between runs according to Pritchard et al. (2000) and Nordborg et al. (2005), respectively, using the R-scripts STRUCTURE-SUM ver. 2.2 and 2009 (Ehrich et al., 2007). The optimal number of clusters was identified as the value of K where the likelihood started to plateau, the results of replicate runs were consistent, and the mean similarity was at the maximum. However, we also considered several ‘sub-optimal’ solutions to infer hierarchical patterns in clustering as proxy of the relative temporal order of splitting events (Rosenberg et al., 2002; Nordborg et al., 2005). Bar plots with cluster membership coefficients for all individuals were generated in Microsoft Excel after choosing the replicate run with the highest level of probability for each selected K. In addition, we conducted principal co-ordinates analyses (PCoAs) in PAST ver. 1.40 (Hammer et al., 2001) on either the entire N. arvensis complex or a sub-set of taxa to visualize genetic patterns among individuals based on Dice’ similarity coefficient.
Population genetic analyses of diversity and differentiation
The degree of genetic variability within each population and taxon was assessed by the percentage of polymorphic fragments (PPF), Nei’s (1987) gene diversity (HE) and Shannon’s index (SI) of phenotypic diversity (Lewontin, 1972). The AFLPDAT R package (Ehrich, 2006) was used to calculate the PPF and HE, and POPGENE was used to estimate the SI. Differences in within-population diversities (PPF, HE and SI) between outcrossing and selfing taxa, or between mainland and island taxa (outcrossers only), were tested by means of non-parametric Mann–Whitney U-tests (two-tailed) using the ‘Social Science Statistics’ online calculators (http://www.socscistatistics.com/). The six populations with insufficient sample sizes (n ≤5) were excluded from these diversity analyses.
Measures of population differentiation (ΦST) within taxa and/or lineages were calculated by non-hierarchical analyses of molecular variance (AMOVAs) in ARLEQUIN ver. 3.11 (Excoffier et al., 2007). Hierarchical AMOVAs were further performed at various levels to test the data support of a priori population structure hypotheses. Thus, for both the entire complex and the alliance, populations were grouped according to taxonomic (subspecies) designation (with the single population of N. a. ssp. arvensis excluded but material from Rhodos treated separately; see the Results). For the alliance, populations were further assigned to five phytogeographical zones defined by Rechinger (1950), namely the Western, Northern and Eastern Aegean, the Cyclades and the ‘Southern Aegean Island Arc’ (Crete, Karpathos/Kasos, Rhodos; see also Fig. 1A). At the species level, hierarchical AMOVAs were restricted to those taxa with sufficient population sampling, namely N. arvensis (sspp. aristata/glauca), N. degenii and N. doerfleri. These four latter taxa were also used separately to test for isolation-by-distance (IBD) effects by regressing ΦST /(1 – ΦST) against the natural logarithm of geographical distance for all pairs of populations (Rousset, 1997). The significance of regression slopes was evaluated by Mantel tests in ARLEQUIN (1023 permutations).
RESULTS
AFLP marker characteristics
The three primer combinations employed with samples from 497 individuals (48 populations) of the N. arvensis complex (six species, 11 taxa) generated a total of 235 fragments, ranging from 74 to 434 bp, of which 233 AFLP markers were polymorphic (PPF = 99 %; see Supplementary Data Table S1). Because of this high degree of polymorphism, all individuals were distinguishable as separate AFLP phenotypes, with an overall mean of 50.0 (± 11.2 s.d.) fragments per individual (Nfind) across primer pairs. Values of Nfind were not significantly different for all six species (see overlapping standard deviations in Table S1). However, the total number of fragments (Nftot) and values of PPF per taxon tended to be higher in the four outcrossing species (Nftot = 143–233; PPF = 95–99) when compared with the selfing species N. doerfleri/N. stricta (Nftot = 134/115; PPF = 95/84), and this trend was unaffected by sample size (n) per species (Spearman’s r = 0.551 and 0.605; P = 0.257 and 0.205, respectively).
Major AFLP lineages and relationships within the N. arvensis complex
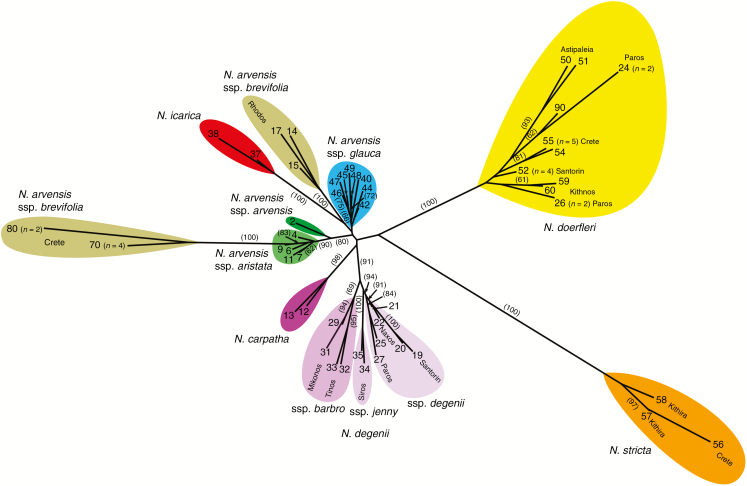
The population-level NJ network using Nei’s genetic distances (Fig. 2) resolved most designated taxa as distinct units, including N. degenii (bootstrap support, BS = 91 %) and its subspecies (degenii, barbro and jenny; 69–100 %), N. icarica (100 %), N. carpatha (98 %), N. doerfleri (100 %) and N. stricta (100 %). The four subspecies of N. arvensis grouped next to each other, but did not form a well-supported lineage. Instead, populations of ssp. brevifolia from Crete (100 %) nested within a cluster of sspp. arvensis/aristata (80 %), while those from Rhodos (100 %) grouped next to ssp. glauca (<60 %) and N. icarica, but without support. The NJ network based on FST distances (Supplementary Data Fig. S1) largely retrieved the same clusters, but recovered N. a. ssp. glauca as a distinct lineage (90 %). However, both network approaches largely failed to resolve interspecific relationships due to weakly supported internal edges (e.g. N. doerfleri/N. stricta; BS = 63 % in Fig. S1).
Fig. 2.
Unrooted Neighbor–Joining (NJ) network depicting AFLP-derived Nei’s (1972) standard genetic distances (D) between 48 populations (11 taxa) of the Aegean Nigella arvensis complex calculated from allele frequencies under the assumptions of Hardy–Weinberg equilibrium for outcrossing taxa and full inbreeding for selfing taxa (see text). Bootstrap values (≥60 %) based on 10 000 replicates are shown in parentheses along internal edges. Population numbers on terminal branches are identified in Table 1. For five populations (nos 24, 26, 52, 70 and 80), comprising only a few (≤5) individuals, their respective sample sizes are indicated.
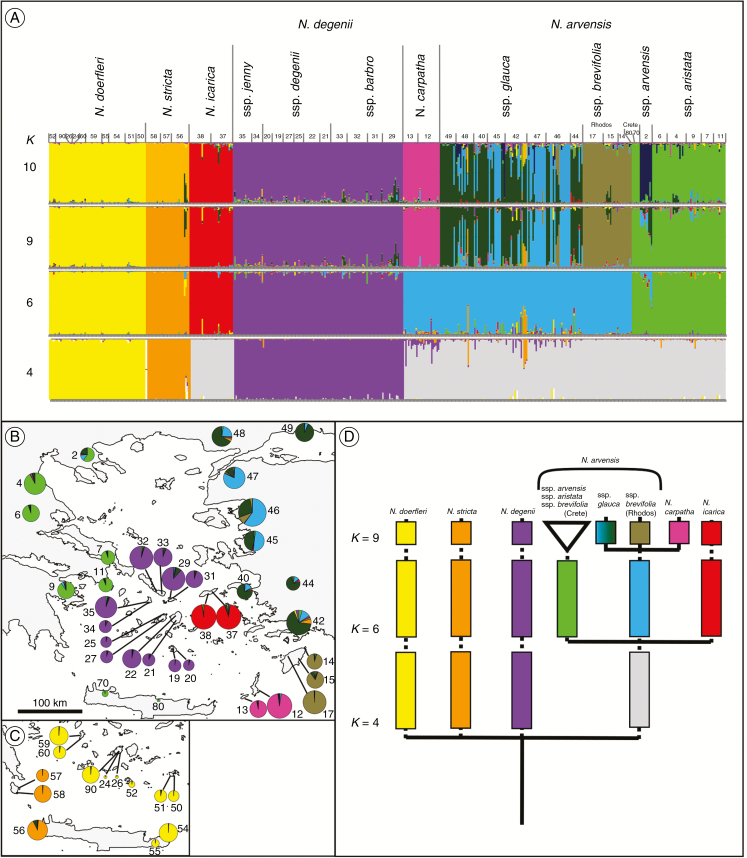
For the entire complex, the STRUCTURE analysis inferred K = 9 as the optimal number of clusters based on two lines of evidence (see Supplementary Data Fig. S2): first, the posterior probabilities of the data [lnp(D)] progressively increased up to K = 9, where they started to flatten out; and, secondly, the similarity coefficient between replicated runs reached a maximum (1.0) at this value, even though K = 4, 6 and 10 also gave moderate to high mean values of similarity (approx. 0.6–0.8). Bar plots of all four solutions are displayed in Fig. 3A, while the geographical distributions of clusters at K = 9 are shown in Fig. 3B and C. These nine clusters or ‘gene pools’ were almost entirely congruent with the species or lineages identified in the network approach, including regional groups of N. arvensis from the Greek mainland/Crete (green), Rhodos (brown) and Turkey/Samos (mostly blue or blue-green), as well as N. degenii (purple), N. icarica (red), N. carpatha (pink), N. doerfleri (yellow) and N. stricta (orange). However, a remarkable hierarchical structure resulted from the ordered splitting of clustering groups with successively increasing numbers of clusters (K = 4, 6 and 9; see Fig. 3D). Accordingly, the first four clusters emerging at K = 4 separated N. degenii, N. doerfleri and N. stricta from each other and the remainder (grey cluster). Within this latter, ‘circum-Aegean’ cluster, three additional groups appeared at K = 6, including: (1) N. icarica (red); (2) an unresolved ‘western Aegean’ lineage (green) comprising N. a. sspp. arvensis/aristata/brevifolia (Crete); and (3) an ‘eastern Aegean’ lineage (blue). At K = 9, the latter split into ssp. glauca (Turkey/Samos), ssp. brevifolia (Rhodos) and N. carpatha. The single population of N. a. ssp. arvensis appeared to be admixed with the eastern (ssp. glauca) gene pool at K = 6 and 9 but resolved at K = 10 (dark green; see Fig. 3A).
Fig. 3.
(A) Bar plots of the STRUCTURE assignment tests for all 497 AFLP phenotypes of the Aegean Nigella arvensis complex for K = 4, 6, 9 and 10, assuming the admixture model, with the smallest vertical bar representing one individual. The y-axis of each bar plot presents the estimated membership coefficient (Q) for each individual in the respective coloured-coded clusters. The x-axis corresponds to population numbers as identified in Table 1. (B, C) Geographical distribution of STRUCTURE clusters at K = 9 (i.e. optimal solution) within and among populations of (B) the outcrossing and interfertile N. arvensis alliance (nine taxa, 35 populations) and (C) the reproductively isolated selfers (N. doerfleri and N. stricta; 13 populations in total). Sizes of pie charts are proportional to sample size (n), with the smallest and largest circles representing n = 2 and 18, respectively (see Table 1). (D) Diagram showing the hierarchical structure of taxon relationships resulting from the ordered splitting into clustering groups with successively increasing numbers of clusters (K = 4, 6 and 9).
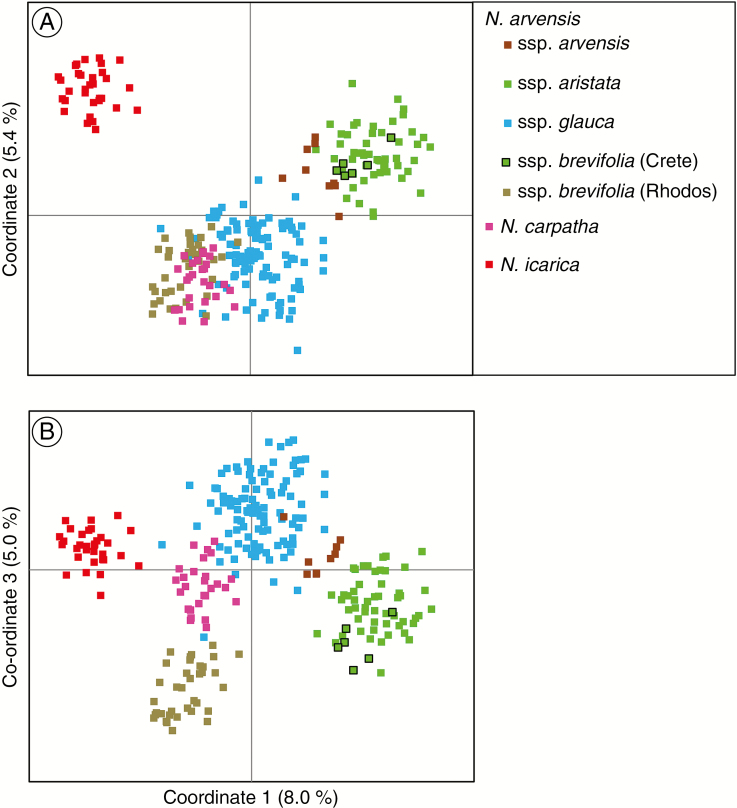
The individual-based PCoA plot of the entire complex for the first two axes displayed patterns largely consistent with the STRUCTURE clustering at K = 4, while the first and third axes only separated N. doerfleri and N. stricta from the remainder, including N. degenii (see Supplementary Data Fig. S3). A subsequent PCoA without these latter three species was conducted to gain additional insights into relationships among the remaining taxa. In this analysis, the first two axes (Fig. 4A) displayed the K = 6 pattern by resolving the western and eastern Aegean lineages as well as N. icarica; however, the first and third axes (Fig. 4B) clustered most individuals according to taxonomy and/or geography (consistent with K = 9), whereby N. a. ssp. arvensis grouped between sspp. aristata/brevifolia (Crete) and ssp. glauca, and N. carpatha between the latter and ssp. brevifolia (Rhodos).
Fig. 4.
Principal co-ordinates analysis (PCoA) for 268 AFLP phenotypes (six taxa, 23 populations) of the Aegean Nigella arvensis complex, with N. doerfleri, N. stricta and N. degenii excluded. Plots of (A) the first and second, and (B) the first and third axes, explaining 8.0, 5.4 and 5.0 % of the total variance, respectively. See Supplementary Data Fig. S3 for the PCoA of the entire complex.
Hierarchical components of genetic variation
For the complex as a whole, hierarchical AMOVA (Table 2) indicated a strong and significant relationship between subspecies/lineage designations and AFLP variation. With N. a. ssp. arvensis excluded and ssp. brevifolia/Rhodos treated separately, 31 % of the total variance was explained between the 11 taxa/lineages (P < 0.001), while 13 % resided among populations within each group, and 56 % was within populations. When the alliance was analysed separately, the strongest relationships were obtained when populations were grouped according to taxonomy, i.e. genetic cluster affinity (24 %), while 12 % were attributed to the among-population component. Designating the same populations to five phytogeographical zones (Rechinger, 1950) resulted in only 14 % of the total variance at the group level, whereas the among-population component increased to 22 % (Table 2).
Table 2.
Hierarchical analyses of molecular variance (AMOVAs) for AFLP variation based on several groupings of populations from the Nigella arvensis complex in the Aegean region
| Grouping/source of variation | d.f. | SS | VC | Variation (%) | Φ-statistic |
|---|---|---|---|---|---|
| Nigella arvensis complex | |||||
| Among 11 taxa* | 10 | 5119.027 | 10.341 | 31.04 | Φ CT = 0.301 |
| Among populations within taxa | 36 | 2246.787 | 4.323 | 12.98 | Φ SC = 0.188 |
| Within populations | 442 | 8245.437 | 18.654 | 55.99 | Φ ST = 0.440 |
| Nigella arvensis alliance | |||||
| Among 9 taxa* | 8 | 3124.250 | 7.946 | 24.08 | Φ CT = 0.241 |
| Among populations within taxa | 25 | 1636.739 | 3.994 | 12.10 | Φ SC = 0.169 |
| Within populations | 351 | 7391.193 | 21.057 | 68.82 | Φ ST = 0.361 |
| Among five phytogeographical zones† | 4 | 1798.828 | 4.533 | 13.73 | Φ CT = 0.137 |
| Among populations within zones | 30 | 3070.659 | 7.309 | 22.14 | Φ SC = 0.257 |
| Within populations | 395 | 7601.860 | 21.175 | 64.13 | Φ ST = 0.359 |
| N. arvensis (sspp. aristata/glauca) | |||||
| Among two mainland regions/taxa‡ | 1 | 393.284 | 4.180 | 12.94 | Φ CT = 0.129 |
| Among populations within regions/taxa | 12 | 770.895 | 3.295 | 10.20 | Φ SC = 0.117 |
| Within populations | 154 | 3824.940 | 24.837 | 76.86 | Φ ST = 0.231 |
| N. degenii (sspp. degenii, barbro, jenny) | |||||
| Among six islands§ | 5 | 889.307 | 6.634 | 23.86 | Φ CT = 0.239 |
| Among populations within islands | 6 | 240.622 | 2.075 | 7.46 | Φ SC = 0.098 |
| Within populations | 113 | 2157.847 | 19.096 | 68.68 | Φ ST = 0.313 |
| N. doerfleri | |||||
| Among four islands¶ | 3 | 294.664 | 3.779 | 13.73 | Φ CT = 0.239 |
| Among populations within islands | 5 | 133.422 | 2.877 | 22.14 | Φ SC = 0.240 |
| Within populations | 58 | 528.989 | 9.120 | 64.13 | Φ ST = 0.422 |
All Φ values were significant (P < 0.001) based on 1023 permutations in ARLEQUIN.
d.f., degrees of freedom; SS, sum of squares; VC, variance components.
*With N. arvensis ssp. arvensis excluded and N. a. ssp. brevifolia/Rhodos treated as a separate unit.
†Western, Eastern, Northern Aegean; Cyclades; ‘Southern Aegean Island Arc’ (after Rechinger, 1950; Fig. 1A).
‡Mainland Greece (ssp. aristata) vs. Anatolia and the off-shore island Samos (ssp. glauca).
§Paros/Naxos/Santorini (ssp. degenii), Mikonos/Tinos (ssp. barbro) and Siros (ssp. jenny).
¶Kithnos, Paros, Crete and Astipaleia.
Population genetic diversity, differentiation and isolation-by-distance
Estimates of AFLP diversity (in terms of PPF, HE or SI) for each population are shown in Table 1, and their averages for each taxon are summarized in Table 3. In general, values of PPF, HE and SI were positively correlated with each other (all P < 0.001), but hardly with the number of individuals sampled per population (except for PPF; P = 0.024) and, when averaged, not at all with the number of individuals or populations per taxon (Spearman’s |r|: 0.2–0.11; all P ≥ 0.56). These diversity parameters most strongly differed between the outcrossing and selfing taxa (Table 3). For example, gene diversity (HE) ranged from 0.221 (± 0.018 s.d.) in N. a. ssp. aristata to 0.144 (± 0.026) in N. icarica, compared with 0.080 (± 0.013) in N. doerfleri and 0.085 (± 0.025) in N. stricta. In fact, for all three parameters, within-population diversities were significantly higher in the outcrossing taxa by Mann–Whitney U-tests (mean PPF, 51.9 vs. 22.2; HE, 0.18 vs. 0.08; SI, 0.21 vs. 0.11; all P < 0.001). The same, however, was also true for the outcrossing taxa from the mainland (N. a. sspp. aristata/glauca) when compared with their counterparts from the islands (N. a. ssp. brevifolia/Rhodos, N. degenii, N. icarica, N. carpatha; mean PPF, 61.3 vs. 44.9; HE, 0.21 vs. 0.16; SI, 0.24 vs. 0.20; all P < 0.001).
Table 3.
Summary of mean estimates of within-population AFLP diversity for 11 (outcrossing or selfing) taxa of the Nigella arvensis complex from the Aegean region based on 478 individuals representing 42 populations with sample sizes of n ≥7 (see Table 1)
| Mating system/taxon | N pop* | N ind | PPF | H E | SI |
|---|---|---|---|---|---|
| Outcrossing | |||||
| N. arvensis | |||||
| ssp. arvensis | 1 | 9 | 58.72 | 0.2241 | 0.2511 |
| ssp. aristata | 5 | 54 | 62.21 (3.55) | 0.2212 (0.0176) | 0.2540 (0.0214) |
| ssp. brevifolia (Rhodos) | 3 | 36 | 42.83 (9.86) | 0.1549 (0.0043) | 0.1834 (0.0441) |
| ssp. glauca | 8 | 105 | 61.06 (10.64) | 0.2038 (0.0323) | 0.2311 (0.0318) |
| N. degenii | |||||
| ssp. degenii | 6 | 50 | 44.61 (6.73) | 0.1753 (0.0177) | 0.2062 (0.0160) |
| ssp. barbro | 4 | 53 | 42.55 (7.44) | 0.1405 (0.0246) | 0.1713 (0.0307) |
| ssp. jenny | 2 | 22 | 51.32 (1.80) | 0.1797 (0.0227) | 0.2119 (0.0187) |
| N. icarica | 2 | 32 | 45.32 (6.92) | 0.1435 (0.0260) | 0.1856 (0.0263) |
| N. carpatha | 2 | 27 | 48.08 (16.25) | 0.1606 (0.0353) | 0.1976 (0.0496) |
| Selfing | |||||
| N. doerfleri | 6 | 58 | 20.58 (2.46) | 0.0799 (0.0130) | 0.1020 (0.0149) |
| N. stricta | 3 | 32 | 24.40 (7.30) | 0.0853 (0.0250) | 0.1125 (0.0344) |
| Overall total | 42 | 478 | 99.14 | 0.2737 | 0.3094 |
| Overall mean | – | 11.38 (2.98) | 45.61 (13.49) | 0.1608 (0.0481) | 0.1915 (0.0495) |
Standard deviations are given in parentheses.
N pop, number of populations; Nind, number of individuals assayed; PPF, percentage of polymorphic fragments; HE, Nei’s (1987) unbiased gene diversity; SI, Shannon’s index of phenotypic diversity (Lewontin, 1972).
*Six populations were excluded due to small (n ≤ 5) sample sizes (see text and Table 1).
The above trends of genetic diversity were reversed when taxon-wide levels of population differentiation (ΦST) were evaluated by non-hierarchical AMOVAs (Supplementary Data Table S2). Thus, generally lower but still markedly different values of ΦST were observed in the outcrossing taxa, ranging from 0.08 in N. a. ssp. aristata to 0.29 in N. degenii, while ΦST levels were highest in the selfers N. stricta (0.34) and N. doerfleri (0.41). Hierarchical AMOVAs of the three selected species (Table 2) further revealed lower levels of differentiation between N. a. sspp. aristata and glauca (ΦCT = 0.129) compared with the various island population groups of N. degenii and N. doerfleri (both ΦCT = 0.239); however, by taking this geographical sub-structure into account, population differentiation within regions/islands now proved to be of comparable magnitude in N. arvensis (ΦSC = 0.117) and N. degenii (ΦSC = 0.098), while still remaining high in N. doerfleri (ΦSC = 0.240) (Table 2). Overall, the higher ΦCT value of N. degenii compared with N. arvensis illustrates how geographical (island) isolation increases spatial genetic sub-structure (unaffected by mating type), while the higher ΦSC value of N. doerfleri relative to N. degenii highlights how selfing increases among-population differentiation (unaffected by the occurrence on islands per se).
By taking these four taxa separately, Mantel tests revealed significant IBD effects in N. a. ssp. aristata (rM = 0.644, P = 0.004), ssp. glauca (rM = 0.489, P = 0.011) and N. degenii (rM = 0.618, P < 0.001). Although there was only a weak and marginally significant effect in N. doerfleri (rM = 0.265, P = 0.06), this nonetheless proved significant after excluding two populations (nos 24 and 26) with exceptionally small sample sizes (rM = 0.467, P = 0.016).
DISCUSSION
The Aegean archipelago has become a focal area for evolutionary and biogeographical research at a global level, especially for continental shelf (land-bridge) islands. Based on many phylogeographical investigations recently carried out in this area, in particular on animals (reviewed in Poulakakis et al., 2015; Sfenthourakis and Triantis, 2017), the general picture emerging is one that identifies Late Tertiary/Pleistocene alterations in sea level as an important vicariant factor driving lineage diversification. However, our AFLP study indicates that the current distributions of Aegean Nigella mainly result from an interplay between island fragmentation and colonization events at different temporal and spatial scales, but have also been affected by the biological properties (e.g. mating system) of particular taxa. Hence, our data suggest a more complex biogeographical history than expected for a strictly allopatric vicariant model of divergence. Nonetheless, the major phylogeographical patterns observed are largely congruent with the geography and history of islands, and there is little evidence to suggest that the radiation of this classic Aegean plant system is constrained by ongoing gene exchange between divergent (sub)species (see below).
Major phylogeographical patterns within the Aegean N. arvensis complex
Our comprehensive analysis of six species (11 taxa, 497 individuals) of the Aegean N. arvensis complex with 235 AFLP markers revealed three major findings from a taxonomic–phylogeographical perspective. First, most designated taxa of this complex (Strid, 1970), except for N. a. ssp. brevifolia, are identifiable as genetically distinct units by means of genetic distance, STRUCTURE and PCoA (Figs 2–4). Secondly, this AFLP study revealed high levels of phylogeographical diversity and structure across the entire complex (Fig. 3B, C). Finally, consecutive STRUCTURE runs with different numbers of K (Fig. 3A) indicated an unresolved polytomy in the complex (Fig. 3D), consisting of N. doerfleri, N. stricta, N. degenii and a ‘circum-Aegean’ cluster, with the latter comprised of N. icarica as well as western and eastern Aegean lineages, both with affinities to the South Aegean. As we currently lack a resolved and time-calibrated phylogeny of the complex, any genealogical or temporal interpretations of such a sequential order of AFLP clustering has to be treated with caution. Nonetheless, as discussed below, this hierarchical structure suggests a sequence of divergences that are fairly reasonable when considered together with previous inferences from cpDNA (Bittkau and Comes, 2005) and on the backdrop of Aegean palaeogeography.
Evolutionary and biogeographical history of the Aegean N. arvensis complex
In the Central Aegean, the phylogeographical distinctiveness of N. degenii (Fig. 3B) is consistent with the floristic concepts of a ‘Cycladic’ window (Rechinger, 1950) and its eastern border, ‘Rechinger’s line’ (Strid, 1996), which separates the Cyclades from the eastern Aegean along the northern MAT. At the cpDNA level, however, the western borderline of this ‘window’ was not clearly indicated due to the presence of a ‘Cycladic’ haplotype in mainland Greece (Bittkau and Comes, 2005). Otherwise, both AFLPs and cpDNA concur that N. degenii represents a relatively ancient species, which probably originated after the separation of the Cycladic landmass (peninsula) from the Greek mainland/Euboea approx. 2.0 Ma (Chatzimanolis et al., 2003).
As regards N. doerfleri and N. stricta, inferences about their evolutionary history have so far been impeded by their near fixation for an ancestral cpDNA haplotype (A), which is also particularly common in N. degenii (Bittkau and Comes, 2005; C Bittkau, unpubl. res.). In our AFLP networks (Fig. 2; Supplementary Data Fig. S1), these two selfing species are only weakly clustered (BS ≤63 %), suggesting the hierarchical STRUCTURE analysis (Fig. 3D) more reliably identifies them as relatively ancient, distinct lineages that largely evolved separately from each other. Moreover, N. doerfleri and N. degenii are broadly co-distributed, and often sympatric, in the Cyclades, and thus may have been similarly affected by palaeo-events having shaped this region. We therefore propose that N. doerfleri likewise evolved via sea barrier-induced vicariant speciation in the Cyclades during the Early Pleistocene (see above) and was only secondarily dispersed to central-eastern Crete and numerous peripheral islets to attain its presently wider range (Fig. 1B). This dispersal scenario gains support from the network (Fig. 2), where populations from Crete (54 and 55) and Astipaleia (50 and 51) hold different, but nested, and thus potentially derived positions relative to those from the Cyclades (Kithnos, Paros and Santorini). Unfortunately, samples of N. doerfleri from Andikithira (south-east of the Peloponnese/West Cretan Strait) were not available to test the genetic position of this disjunct population, also in relation to N. stricta. Nonetheless, the latter species is only known from nearby Kithira and a few locations in South-west Crete (Strid, 1970). In this region, rises and falls in sea level during the Mid-/Late Pleistocene have repeatedly disconnected and connected Kithira/Andikithira with the southern Peloponnese (Schüle, 1996). For example, at low sea level stages, the sea strait separating these islands from Crete may have been as narrow as 2–3 nautical miles (Sakellariou and Galanidou, 2016). As the Cretan population studied (56) is nested within those from Kithira (57 and 58; Fig. 2), N. stricta might have originated in Kithira and then dispersed to Crete, with both events facilitated by sea level oscillations.
In the Eastern Aegean (EAe), the genetic distinctiveness of N. icarica is stronger when inferred from AFLPs (Fig. 3B) than from cpDNA, given the fixation of a derived ‘Anatolian’ haplotype in this island endemic (Bittkau and Comes, 2005). Nonetheless, this latter finding agrees with the hierarchical AFLP clustering (Fig. 3D) in suggesting a more recent origin of N. icarica by comparison with N. degenii, N. doerfleri and N. stricta. Hence, the present data support our earlier hypothesis (Bittkau and Comes, 2005) that N. icarica evolved from an EAe stock due to rising sea levels that separated Ikaria from Asia Minor during the Last Interglacial (approx. 125 000–75 000 years ago; Beerli et al., 1996) or even more recently (<21 500 years ago; Perissoratis and Conispoliatis, 2003; Tourloukis, 2010).
The AFLPs also identified strong genetic breaks around Rhodos (N. a. ssp. brevifolia) and Karpathos/Kasos (N. carpatha) (Fig. 3C). These patterns agree with floristic divisions across the Strait of Marmara (Rechinger, 1950) and between Rhodos and Karpathos (Strid, 1997) but conflict with more inclusive concepts such as the ‘Southern Aegean Island Arc’ (SAe) (Rechinger, 1950) or the ‘Kriti–Karpathos’ region (Strid, 1997). A similar ambiguity was seen in the cpDNA data, which assigned N. carpatha from Karpathos to a derived ‘Anatolian’ clade but conspecifics from Kasos and N. a. ssp. brevifolia from Crete/Rhodos to an ancestral, central/western Aegean clade (Bittkau and Comes, 2005). In contrast, the AFLPs clearly indicate that N. a. ssp. brevifolia from Crete belongs to a Western Aegean (WAe) lineage together with sspp. aristata/arvensis, while ssp. brevifolia from Rhodos and N. carpatha are members of the Eastern Aegean (EAe) lineage together with N. a. ssp. glauca (Fig. 3D). All these SAe taxa, including Cretan ssp. brevifolia, form well-supported clusters in the network (Fig. 2). Hence, their genetic affinities for extant mainland taxa of N. arvensis most probably reflect historical associations rather than ongoing gene flow. In turn, this would imply two independent colonizations of the SAe from the Greek and Turkish mainland, respectively. Since the WAe and EAe lineages share the same hierarchical cluster level with N. icarica (Fig. 3D), such colonizations might have occurred relatively recently, e.g. via island hopping during low sea level periods of the last glacial(s), when exposed shelf areas projected from the southern Peloponnese to the West Cretan Strait (Sakellariou and Galanidou, 2016; see above) and from the south-western edge of the Turkish mainland to the East Aegean Sea (possibly connecting Rhodos; Perissoratis and Conispoliatis, 2003; Tourloukis, 2010). This ‘bi-regional colonization’ scenario also accords with recent diversity studies on the South Aegean flora (Trigas et al., 2013), suggesting that Crete functions as a dispersal filter between the continents. Notably, too, Strid (1970) had pointed out the occurrence of ‘arvensis-like’ forms of N. a. ssp. brevifolia in Crete (sometimes accorded specific status, N. cretensis Stevens) and ‘glauca-like’ variants in Rhodos. Our AFLP data agree with this phenotypic variability within ssp. brevifolia and stress the need for a thorough taxonomic revision of this subspecies.
When combined, the above scenarios suggest that the Pleistocene radiation of the Aegean N. arvensis complex built up over time, and not all at once, involving two major diversification processes: (1) relatively ancient sea barrier-induced vicariant speciation in the Cyclades (N. degenii and N. doerfleri) and the West Cretan Strait (N. stricta), and more recently in the EAe (N. icarica); and (2) likewise recent bi-regional colonizations of the SAe by differentiated mainland WAe vs. EAe lineages (similar to extant N. a. aristata/arvensis vs. glauca) followed by allopatric (sub-)speciation in situ within islands (i.e. ssp. brevifolia in Crete vs. Rhodos, and N. carpatha in Karpathos/Kasos).
Effects of mating system and geographical isolation on patterns of genetic variability
For Aegean Nigella, the present AFLP data showed significantly lower within-population diversity (in terms of PPF, HE and SI; all U-tests: P < 0.001) and generally higher levels of population differentiation (ΦST) in selfing than outcrossing taxa (see Table 3; Supplementary Data Table S2). Moreover, by taking island sub-structure into account, we observed a >2-fold higher level of population differentiation (ΦSC) in N. doerfleri relative to its outcrossing island counterpart N. degenii (Table 2). These differences agree with expectations from theory (Holsinger, 2000; Charlesworth, 2003) and empirical data (e.g. Nybom, 2004), and can be generally understood as a consequence of selfing per se by decreasing effective population size owing to reduced recombination and increased linkage disequilibrium (Charlesworth and Pannell, 2001).
However, mating type differences are not the only factors affecting genetic variability and, especially in the present study system, might be confounded with others, in particular geographical isolation on islands and/or population history (e.g. Barrett, 1996; Stuessy et al., 2014). Nonetheless, by controlling for mating type, our comparison of mainland and island taxa within the outcrossing alliance revealed significantly lower diversity (P < 0.001) and increased population differentiation in the island group (Table 3; Supplementary Data Table S2), and a near 2-fold higher geographical range substructure (ΦCT) in N. degenii relative to N. arvensis (Table 2). This strongly suggests that, due to their smaller population sizes and geographical isolation, island taxa experience a greater impact of genetic drift and decreased gene flow than their mainland counterparts, as often reported for other plant and animal groups (e.g. Barrett, 1996; Frankham, 1997; Lecocq et al., 2013; but see García-Verdugo et al., 2015). Evidently, such island and drift effects are magnified in N. doerfleri and N. stricta (see above), which is not unexpected because selfing accentuates population sub-division, isolation and bottlenecks (Charlesworth and Pannell, 2001).
Contrasting inferences from isolation-by-distance patterns in mainland vs. island taxa
All four of the selected taxa from either the mainland (N. a. sspp. arvensis and glauca) or the islands (N. degenii and N. doerfleri) showed a significant IBD pattern. Taken at face value, this would imply restricted, but still ongoing nearest-site gene exchange, i.e. ‘isolation by dispersal limitation’ (IBDL; sensuOrsini et al., 2013). This explanation seems plausible for the mainland taxa, given the limited seed dispersal potential of Nigella, but, for the very same reason, appears to be unrealistic for N. degenii and N. doerfleri populations currently separated by sea barriers. This paradox, however, can be solved if we consider that an IBD pattern generated by IBDL can be confounded with the one generated by a population history of step-wise (‘serial’) colonization (Orsini et al., 2013). Although difficult to disentangle, this latter model predicts relatively high among-population differentiation, as actually observed for N. degenii/N. doerfleri (ΦST = 0.29/0.41) but not for N. a. sspp. arvensis/glauca (ΦST = 0.08/0.12). Furthermore, the intraspecific relationships of N. degenii (Fig. 2) are broadly congruent with the temporal sequence of Cycladic island formation towards the end of the Pleistocene (approx. 14 000–10 000 years ago; Lambeck, 1996), with Tinos/Mikonos (ssp. barbro) separating first, followed by Siros (ssp. jenny), and Paros and Naxos last (ssp. degenii) (for details, see Comes et al., 2008). It is feasible, therefore, that the IBD pattern of N. degenii largely reflects sequential temporal population splitting, resulting from the latest ‘sinking of Cycladia’. A similar hypothesis could apply to N. doerfleri, albeit modified to accommodate sporadic long-distance colonization of peripheral islands (see above). However, our population sampling of this species is too limited to support this claim further.
Conclusions
This follow-up study employing AFLP markers has provided important new insights into the taxonomic boundaries, phylogeographical structure and evolutionary history of the Aegean N. arvensis complex. Although in need of further time-calibrated studies, our results indicate that this Pleistocene radiation comprises both fragmentation and dispersal-driven diversification processes at different geological time scales, from Early to Late Pleistocene, specifically (1) sea barrier-induced vicariant speciation in the Cyclades, the Western Cretan Strait and Ikaria; and (2) bi-regional colonizations of the ‘Southern Aegean Island Arc’ by differentiated West vs. East Aegean mainland lineages, followed by allopatric divergences in Crete vs. Rhodos and Karpathos/Kasos. As a result, major phylogeographical boundaries are not only located between the Central and Eastern Aegean (i.e. along ‘Rechinger’s line’), as previously inferred from cpDNA (Bittkau and Comes, 2005), but also between Crete and Karpathos–Kasos, thus coinciding with the entire MAT (see Fig. 1A). However, despite the importance of the MAT as a barrier to dispersal and gene flow in Aegean biota (e.g. Runemark, 1980; Fattorini, 2002; Jesse et al., 2011; Crowl et al., 2015; Poulakakis et al., 2015; Sfenthourakis and Triantis, 2017), this ancient (Upper Miocene) sea strait does not seem to have played a causal (vicariant) role in shaping diversification within Nigella. Hence, a priori assumptions about this sea strait as a vicariant factor and its usage for calibrating molecular clocks may not be warranted unless its role in vicariance is verified (Papadopoulou et al., 2010; De Baets et al., 2016).
In addition, this study has demonstrated the profound influences of differences in mating system and geographical isolation in shaping intra(sub)specific patterns of genetic variability. Compared with the outcrossing mainland taxa, isolation of their insular counterparts has led to drift-related demographic processes (loss of genetic diversity, increased population genetic differentiation and spatial sub-structure), which are magnified in the insular selfers. Moreover, population genetic differentiation on the mainland appears to be largely driven by dispersal limitation, while in the Central Aegean it may still be influenced by historical events (island fragmentation or long-distance colonization). In sum, the present results emphasize the need to investigate further the complex interactions between biological and landscape features and contemporary vs. historical processes in driving population divergence and taxon diversification in Aegean plant radiations.
DATA ACCESSIBILITY
The AFLP data matrix generated for this study is deposited in Dryad: https://doi.org/10.5061/dryad.961t3.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Table S1: AFLP marker characteristics per species. Table S2: non-hierarchical AMOVAs for eight taxa. Figure S1: population-level NJ network using FST. Figure S2: STRUCTURE results for identifying the optimal number of K. Figure S3: individual-based PCoA for the entire complex.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Zacharias Kypriotakis (Technicon, Irákleion, Crete) for collecting samples of N. arvensis ssp. brevifolia from Crete; Marion Kever, Petra Siegert (Mainz University), Michaela Eder and Matthias Affenzeller (Salzburg University) for laboratory assistance; Albin Blaschka (Agricultural Research and Education Centre Raumberg-Gumpenstein, Austria) for computational support; and two anonymous reviewers and the handling editor (Professor Elena Conti) for valuable comments on earlier versions of this paper. This work was supported by the Deutsche Forschungsgemeinschaft (DFG grant Co254/3-1 to H.P.C.), the Stiftungs- und Förderungsgesellschaft der Paris–Lodron Universität Salzburg (PLUS), and a PLUS MSc grant to U.J. This paper is dedicated to the memory of Dr Christiane Bittkau, who laid the foundations of this work.
LITERATURE CITED
- Ali JR. 2017. Islands as biological substrates: classification of the biological assemblage components and the physical island types. Journal of Biogeography 44: 984–994. [Google Scholar]
- Barrett SCH. 1996. The reproductive biology and genetics of island plants. Philosophical Transactions of the Royal Society B: Biological Sciences 351: 725–733. [Google Scholar]
- Beerli P, Hotz H, Uzzell T. 1996. Geologically dated sea barriers calibrate a protein clock for Aegean water frogs. Evolution 50: 1676–1687. [DOI] [PubMed] [Google Scholar]
- Bentzer B. 1973. Taxonomy, variation and evolution in representatives of Leopoldia Parl. (Liliaceae) in the southern and central Aegean. Botaniska Notiser 126: 69–132. [Google Scholar]
- Bittkau C, Comes HP. 2005. Evolutionary processes in a continental island system: molecular phylogeography of the Aegean Nigella arvensis alliance (Ranunculaceae) inferred from chloroplast DNA. Molecular Ecology 14: 4065–4083. [DOI] [PubMed] [Google Scholar]
- Bittkau C, Comes HP. 2009. Molecular inference of a Late Pleistocene diversification shift in Nigella s. lat. (Ranunculaceae) resulting from increased speciation in the Aegean archipelago. Journal of Biogeography 36: 1346–1360. [Google Scholar]
- Bonin A, Ehrich D, Manel S. 2007. Statistical analysis of amplified fragment length polymorphism data: a toolbox for molecular ecologists and evolutionists. Molecular Ecology 16: 3737–3758. [DOI] [PubMed] [Google Scholar]
- Cellinese N, Smith SA, Edwards EJ et al. 2009. Historical biogeography of the endemic Campanulaceae of Crete. Journal of Biogeography 36: 1253–1269. [Google Scholar]
- Charlesworth D. 2003. Effects of inbreeding on the genetic diversity of populations. Philosophical Transactions of the Royal Society B: Biological Sciences 358: 1051–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Pannell JR. 2001. Mating systems and population genetic structure in the light of coalescent theory. In: Silvertown J, Antonovics J, eds. Integrating ecology and evolution in a spatial context. Oxford: Blackwell Science, 73–95. [Google Scholar]
- Chatzimanolis S, Trichas A, Mylonas M. 2003. Phylogenetic analysis and biogeography of Aegean taxa of the genus Dendarus (Coleoptera: Tenebrionidae). Insect Systematics and Evolution 34: 295–312. [Google Scholar]
- Comes HP, Tribsch A, Bittkau C. 2008. Plant speciation in continental island floras as exemplified by Nigella in the Aegean Archipelago. Philosophical Transactions of the Royal Society B: Biological Sciences 363: 3083–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowl AA, Visger CJ, Mansion G et al. 2015. Evolution and biogeography of the endemic Roucela complex (Campanulaceae: Campanula) in the Eastern Mediterranean. Ecology and Evolution 5: 5329–5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Baets K, Antonelli A, Donoghue PCJ. 2016. Tectonic blocks and molecular clocks. Philosophical Transactions of the Royal Society B: Biological Sciences 371: 20160098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermitzakis DM, Papanikolaou DJ. 1981. Paleogeography and geodynamics of the Aegean region during the Neogene. Annales Géologiques des Pays Helléniques 30: 245–289. [Google Scholar]
- Després L, Loriot S, Gaudeul M. 2002. Geographic pattern of genetic variation in the European globeflower Trollius europaeus L. (Ranunculaceae) inferred from amplified fragment length polymorphism markers. Molecular Ecology 11: 2337–2347. [DOI] [PubMed] [Google Scholar]
- Duggen S, Hoernle K, Van den Bogaard P, Rüpke L, Morgan JP. 2003. Deep roots of the Messinian salinity crisis. Nature 422: 602–606. [DOI] [PubMed] [Google Scholar]
- Edh K, Widén B, Ceplitis A. 2007. Nuclear and chloroplast microsatellites reveal extreme population differentiation and limited gene flow in the Aegean endemic Brassica cretica (Brassicaceae). Molecular Ecology 16: 4972–4983. [DOI] [PubMed] [Google Scholar]
- Ehrich D. 2006. Aflpdat: a collection of R functions for convenient handling of AFLP data. Molecular Ecology Notes 6: 603–604. [Google Scholar]
- Ehrich D, Gaudeul M, Assefa A et al. 2007. Genetic consequences of Pleistocene range shifts: contrast between the Arctic, the Alps and the East African mountains. Molecular Ecology 16: 2542–2559. [DOI] [PubMed] [Google Scholar]
- Esselstyn JA, Timm RM, Brown RM. 2009. Do geological or climatic processes drive speciation in dynamic archipelagos? The tempo and mode of diversification in Southeast Asian shrews. Evolution 63: 2595–2610. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. 2007. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online 1: 47–50. [PMC free article] [PubMed] [Google Scholar]
- Fattorini S. 2002. Biogeography of the tenebrionid beetles (Coleoptera, Tenebrionidae) on the Aegean Islands (Greece). Journal of Biogeography 29: 49–67. [Google Scholar]
- Felsenstein J. 2005. PHYLIP (Phylogeny Inference Package) Version 3.6. Distributed by the author. Seattle: Department of Genome Sciences, University of Washington. [Google Scholar]
- Frankham R. 1997. Do island populations have lower genetic variation than mainland populations?Heredity 78: 311–327. [DOI] [PubMed] [Google Scholar]
- García-Verdugo C, Sajeva M, La Mantia T, Harrouni C, Msanda F, Caujapé-Castells J. 2015. Do island plant populations really have lower genetic variation than mainland populations? Effects of selection and distribution range on genetic diversity estimates. Molecular Ecology 24: 726–741. [DOI] [PubMed] [Google Scholar]
- Georghiou K, Delipetrou P. 2010. Patterns and traits of the endemic plants of Greece. Botanical Journal of the Linnean Society 162: 130–422. [Google Scholar]
- Hammer Ø, Harper DAT, Ryan PD. 2001. PAST: paleontological statistics software package for education and data analysis. Palaeontologia Electronica, 4 Website: http: //palaeo-electronica.org/2001_1/past/issue1_01.htm (last accessed 14 April 2017). [Google Scholar]
- Holsinger KE. 2000. Reproductive systems and evolution in vascular plants. Proceedings of the National Academy of Sciences, USA 97: 7037–7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaros U, Fischer GA, Pallier T, Comes HP. 2016. Spatial patterns of AFLP diversity in Bulbophyllum occultum (Orchidaceae) indicate long-term refugial isolation in Madagascar and long-distance colonization effects in La Réunion. Heredity 116: 434–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesse R, Grudinski M, Klaus S, Streit B, Pfenninger M. 2011. Evolution of freshwater crab diversity in the Aegean region (Crustacea: Brachyura: Potamidae). Molecular Phylogenetics and Evolution 59: 23–33. [DOI] [PubMed] [Google Scholar]
- Jorgensen TH, Richardson DS, Andersson S. 2006. Comparative analysis of population structure in two subspecies of Nigella degenii: evidence for diversifying selection on pollen-color dimorphisms. Evolution 60: 518–528. [PubMed] [Google Scholar]
- Kougioumoutzis K, Simaiakis SM, Tiniakou A. 2014. Network biogeographical analysis of the central Aegean archipelago. Journal of Biogeography 41: 1848–1858. [Google Scholar]
- Lambeck K. 1996. Sea-level change and shore-line evolution in Aegean Greece since Upper Palaeolithic time. Antiquity 70: 588–611. [Google Scholar]
- Lecocq T, Vereecken NJ, Michez D et al. 2013. Patterns of genetic and reproductive traits differentiation in mainland vs. Corsican populations of bumblebees. PLoS One 8: e65642 doi.org/10.1371/journal.pone.0065642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin RC. 1972. The apportionment of human diversity. Evolutionary Biology 6: 381–398. [Google Scholar]
- Li JW, Yeung CKL, Tsai PI et al. 2010. Rejecting strictly allopatric speciation on a continental island: prolonged postdivergence gene flow between Taiwan (Leucodioptron taewanus, Passeriformes Timaliidae) and Chinese (L. canorum canorum) hwameis. Molecular Ecology 19: 494–507. [DOI] [PubMed] [Google Scholar]
- Mayol M, Palau C, Rosselló JA, González-Martínez SC, Molins A, Riba M. 2012. Patterns of genetic variability and habitat occupancy in Crepis triasii (Asteraceae) at different spatial scales: insights on evolutionary processes leading to diversification in continental islands. Annals of Botany 109: 429–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr E. 1954. Change of genetic environment and evolution. In: Huxley J, Hardy AC, Ford EB, eds. Evolution as a process. London: Allen and Unwin, 157–180. [Google Scholar]
- Nei M. 1972. Genetic distance between populations. American Naturalist 106: 283–292. [Google Scholar]
- Nei M. 1987. Molecular evolutionary genetics. New York: Columbia University Press. [Google Scholar]
- Nordborg M, Hu TT, Ishino Y et al. 2005. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biology 3: e196 doi.org/10.1371/journal.pbio.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybom H. 2004. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Molecular Ecology 13: 1143–1155. [DOI] [PubMed] [Google Scholar]
- Orsini L, Vanoverbeke J, Swillen I, Mergeay J, De Meester L. 2013. Drivers of population genetic differentiation in the wild: isolation by dispersal limitation, isolation by adaptation and isolation by colonization. Molecular Ecology 22: 5983–5999. [DOI] [PubMed] [Google Scholar]
- Pachschwöll C, Escobar García P, Winkler M, Schneeweiss GM, Weiss-Schneeweiss H, Schönswetter P. 2015. Polyploidisation and Pleistocene range shifts drive diversification in the European high mountain Doronicum clusii aggregate (Asteraceae). PLoS One 10: e0118197 doi.org/10.1371/journal.pone.0118197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulou A, Anastasiou I, Vogler AP. 2010. Revisiting the insect mitochondrial molecular clock: the mid-Aegean trench calibration. Molecular Biology and Evolution 27: 1659–1672. [DOI] [PubMed] [Google Scholar]
- Perissoratis C, Conispoliatis N. 2003. The impacts of sea-level changes during latest Pleistocene and Holocene times on the morphology of the Ionian and Aegean seas (SE Alpine Europe). Marine Geology 196: 145–156. [Google Scholar]
- Poulakakis N, Kapli P, Lymberakis P et al. 2015. A review of phylogeographic analyses of animal taxa from the Aegean and surrounding regions. Journal of Zoological Systematics and Evolutionary Research 53: 18–32. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechinger KH. 1950. Grundzüge der Pflanzenverbreitung in der Aegäis I–III. Vegetatio 2: 55–119, 239–308, 365–386. [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL et al. 2002. The genetic structure of human populations. Science 298: 2381–2385. [DOI] [PubMed] [Google Scholar]
- Rousset F. 1997. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145: 1219–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundell RJ, Price TD. 2009. Adaptive radiation, nonadaptive radiation, ecological speciation and nonecological speciation. Trends in Ecology and Evolution 24: 394–399. [DOI] [PubMed] [Google Scholar]
- Runemark H. 1969. Reproductive drift, a neglected principle in reproductive biology. Botaniska Notiser 122: 90–129. [Google Scholar]
- Runemark H. 1980. Studies in the Aegean Flora XXIII. The Dianthus fruticosus complex (Caryophyllaceae). Botaniska Notiser 133: 475–490. [Google Scholar]
- Sakellariou D, Galanidou N. 2016. Pleistocene submerged landscapes and Palaeolithic archaeology in the tectonically active Aegean region. In: Harff J, Bailey G, Lüth F, eds. Geology and archaeology: submerged landscapes of the continental shelf. London: Geological Society, Special Publications, 411, 145–178. [Google Scholar]
- Salvo G, Ho SYW, Rosenbaum G, Ree R, Conti E. 2010. Tracing the temporal and spatial origins of island endemics in the Mediterranean region: a case study from the Citrus family (Ruta L., Rutaceae). Systematic Biology 59: 705–222. [DOI] [PubMed] [Google Scholar]
- Schüle W. 1993. Mammals, vegetation and the initial human settlement of the Mediterranean Islands – a paleoecological approach. Journal of Biogeography 20: 399–412. [Google Scholar]
- Sfenthourakis S, Triantis KA. 2017. The Aegean archipelago: a natural laboratory of evolution, ecology and civilisations. Journal of Biological Research-Thessaloniki 24: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snogerup S. 1967. Studies in the Aegean flora. IX. Erysimum sect. Cheiranthus. B. Variation and evolution in the small-population system. Opera Botanica 14: 1–86. [Google Scholar]
- Stork AL. 1972. Studies in the Aegean flora. XX. Biosystematics of the Malcolmia maritima complex. Opera Botanica 33: 1–118. [Google Scholar]
- Strid A. 1970. Studies in the Aegean flora. XVI. Biosystematics of the Nigella arvensis complex. Opera Botanica 28: 1–169. [Google Scholar]
- Strid A. 1996. Phytogeographia Aegaea and the Flora Hellenica database. Annalen des Naturhistorischen Museums in Wien 96 B (Suppl.): 279–289. [Google Scholar]
- Strid A. 1997. Introduction. In: Strid A, Tan K, eds. Flora Hellenica, Vol. 1 Koenigstein: Koeltz Scientific Books, ix–xxxv. [Google Scholar]
- Strid A. 2002. Nigella L. In: Strid A, Tan K, eds. Flora Hellenica, Vol. 2 Koenigstein: Koeltz Scientific Books, 5–13. [Google Scholar]
- Stuessy TF, Takayama K, López-Sepúlveda P, Crawford DJ. 2014. Interpretation of patterns of genetic variation in endemic plant species of oceanic islands. Botanical Journal of the Linnean Society 174: 276–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourloukis V. 2010. The Early and Middle Pleistocene archaeological record of Greece: current status and future prospects. Leiden: Leiden University Press. [Google Scholar]
- Trigas P, Panitsa M, Tsiftsis S. 2013. Elevational gradient of vascular plant species richness and endemism in Crete—the effect of post-isolation mountain uplift on a continental island system. PLoS One 8: e59425 doi.org/10.1371/journal.pone.0059425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vekemans X, Beauwens M, Lemaire M, Roldán-Ruiz I. 2002. Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size. Molecular Ecology 11: 139–151. [DOI] [PubMed] [Google Scholar]
- Vitales D, García-Fernández A, Pellicer J et al. 2014. Key processes for Cheirolophus (Asteraceae) diversification on oceanic islands inferred from AFLP data. PLoS One 9: e113207 doi.org/10.1371/journal.pone.0113207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Bothmer R. 1987. Differentiation patterns in the E. Mediterranean Scutellaria rubicunda group (Lamiaceae). Plant Systematics and Evolution 155: 219–249. [Google Scholar]
- Vos P, Hogers R, Bleeker M et al. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research 23: 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace AR. 1880. Island life. London: Macmillan & Co. [Google Scholar]
- Warren BH, Simberloff D, Ricklefs RE et al. 2015. Islands as model systems in ecology and evolution: progress and prospects fifty years after MacArthur–Wilson. Ecology Letters 18: 200–217. [DOI] [PubMed] [Google Scholar]
- Whittaker RJ, Fernández-Palacios JM. 2007. Island biogeography: ecology, evolution, and conservation, 2nd edn Oxford: Oxford University Press. [Google Scholar]
- Wright S. 1940. The statistical consequences of Mendelian heredity in relation to speciation. In: Huxley JS, ed. The new systematics. Oxford: Clarendon Press, 161–183. [Google Scholar]
- Yeh FC, Boyle TJB. 1997. Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belgian Journal of Botany 129: 157–163. [Google Scholar]
- Zhai SN, Comes HP, Nakamura K, Yan HF, Qiu YX. 2012. Late Pleistocene lineage divergence among populations of Neolitsea sericea (Lauraceae) across a deep sea-barrier in the Ryukyu Islands. Journal of Biogeography 39: 1347–1360. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The AFLP data matrix generated for this study is deposited in Dryad: https://doi.org/10.5061/dryad.961t3.