Abstract
Background and Aims
Bread wheat (Triticum aestivum) has been through a severe genetic bottleneck as a result of its evolution and domestication. It is therefore essential that new sources of genetic variation are generated and utilized. This study aimed to generate genome-wide introgressed segments from Aegilops speltoides. Introgressions generated from this research will be made available for phenotypic analysis.
Methods
Aegilops speltoides was crossed as the male parent to T. aestivum ‘Paragon’. The interspecific hybrids were then backcrossed to Paragon. Introgressions were detected and characterized using the Affymetrix Axiom Array and genomic in situ hybridization (GISH).
Key Results
Recombination in the gametes of the F1 hybrids was at a level where it was possible to generate a genetic linkage map of Ae. speltoides. This was used to identify 294 wheat/Ae. speltoides introgressions. Introgressions from all seven linkage groups of Ae. speltoides were found, including both large and small segments. Comparative analysis showed that overall macro-synteny is conserved between Ae. speltoides and T. aestivum, but that Ae. speltoides does not contain the 4A/5A/7B translocations present in wheat. Aegilops speltoides has been reported to carry gametocidal genes, i.e. genes that ensure their transmission through the gametes to the next generation. Transmission rates of the seven Ae. speltoides linkage groups introgressed into wheat varied. A 100 % transmission rate of linkage group 2 demonstrates the presence of the gametocidal genes on this chromosome.
Conclusions
A high level of recombination occurs between the chromosomes of wheat and Ae. speltoides, leading to the generation of large numbers of introgressions with the potential for exploitation in breeding programmes. Due to the gametocidal genes, all germplasm developed will always contain a segment from Ae. speltoides linkage group 2S, in addition to an introgression from any other linkage group.
Keywords: Aegilops speltoides, Triticum aestivum, introgression, recombination, comparative synteny, gametocidal, genetic linkage mapping, GISH, chromosome transmission
INTRODUCTION
Bread wheat (Triticum aestivum), one of the world’s leading sources of food, is an allopolyploid (6x = AABBDD = 42) composed of the genomes of three different species. The A genome is derived from Triticum urartu (Dvorak et al., 1993), the B from Aegilops speltoides or a closely related species (Sarkar and Stebbins, 1956; Riley et al., 1958; Dvorak and Zhang, 1990; Maestra and Naranjo, 1998; Marcussen et al., 2014) and the D genome from Aegilops tauschii (McFadden and Sears, 1946). The final hybridization event between tetraploid wheat (AABB) and Ae. tauschii (DD) is thought to have occurred only once or twice, ~8000–10 000 years ago. As a result, wheat went through a significant genetic bottleneck. Thus, the significant yield gains achieved by wheat breeders to date have been via the exploitation of genetic variation that has arisen via gene mutation over the last 8000–10 000 years and rare outcrossing events with tetraploid wheat. In many parts of the world wheat yields are now starting to plateau and this is thought to be a direct result of a lack of genetic variation compounded by the changing environment at a time of increasing demand for food due to the increasing global population (Charmet, 2011; Grassini et al., 2013; Ray et al., 2013). It is therefore essential that new sources of genetic variation be found that will enable breeders to generate the next generation of high-yielding environmentally adapted wheat varieties (Tanksley and McCouch, 1997; Haussmann et al., 2004; Tester and Langridge, 2010; McCouch et al., 2013; Warschefsky et al., 2014; Brozynska et al., 2015; Zhang et al., 2017)
Unlike wheat, its progenitor species and wild relatives provide a vast and largely untapped source of genetic variation for most, if not all, traits of agronomic importance. Considerable efforts have been made to exploit the genetic variation within the progenitors and wild relatives, with some noticeable successes, for example the transfer of a segment of Aegilops umbellulata to wheat conferring resistance to leaf rust (Sears, 1955), the transfer of a segment from Aegilops ventricosa carrying resistance to eyespot (Maia, 1967; Doussinault et al., 1983) and its subsequent release in the variety Rendevouz, and the registration of ‘MACE’, a hard red winter wheat carrying a segment from Thinopyrum intermedium conferring resistance to wheat streak mosaic virus (Graybosch et al., 2009). Progress has been severely hindered, however, due to an inability to quickly and accurately identify and characterize interspecific transfers to wheat. However, with the advent of new molecular technologies coupled with specific crossing strategies, we can now systematically exploit the genetic variation available in the progenitors of wheat and its wild relatives. (e.g. Tiwari et al., 2014, 2015; King et al., 2016, 2017).
The transfer of genetic variation to wheat from its progenitors/wild relatives occurs via recombination between the chromosomes of wheat and those of its wild relative at meiosis in F1 hybrids or in their derivatives, e.g. addition and substitution lines (King et al., 2016, 2017). This results in the generation of interspecific recombinant chromosomes. Once identified, interspecific recombinant chromosomes are then recurrently backcrossed with wheat. The ultimate objective is to generate lines of wheat carrying a small chromosome segment from a progenitor/wild relative with a target gene but few, if any, deleterious genes.
One of the wild relatives of wheat that is of particular interest to researchers is Ae. speltoides, a rich source of genetic variation for resistance to a range of diseases of importance in wheat (Jia et al., 1996; Mago et al., 2009; Klindworth et al., 2012; Deek and Distelfeld, 2014). Aegilops speltoides, or an extinct close relative, has been proposed as the donor of the B genome of wheat (Sarkar and Stebbins, 1956; Dvorak and Zhang, 1990; Friebe and Gill, 1996; Tsunewaki, 1996; Dvorak and Akhunov, 2005; Marcussen et al., 2014). Aegilops speltoides is a facultative outbreeder, but can be readily inbred in the glasshouse.
Unlike most of the wild relatives of wheat, Ae. speltoides possesses the ability to suppress the wheat Ph1 locus (Dvorak, 1972; Dvorak et al., 2006). Recombination is normally restricted to homologous chromosomes from the same genome, i.e. although the A, B and D genomes of wheat and chromosomes from wild relatives are related, recombination can only occur within genomes in the presence of Ph1. However, if Ph1 is removed, related chromosomes (homoeologues) from different genomes can recombine at meiosis. Aegilops speltoides possesses genes on chromosomes 3S (Su1-Ph1) and 7S (Su2-Ph1) (Dvorak et al., 2006), with the ability to suppress Ph1. Thus, in F1 hybrids between wheat and Ae. speltoides Su1-Ph1 or Su2-Ph1 enables intergenomic homoeologous recombination to occur during meiosis.
Aegilops speltoides also carries gametocidal genes (Gc) (Tsujimoto and Tsunewaki, 1984, 1988; Ogihara et al., 1994), which are preferentially transmitted to the next generation. Individuals heterozygous for Gc gene(s) produce two types of gametes. Both male and female gametes that lack the Gc genes are generally not viable due to chromosome breakage (Finch et al., 1984). Gametes that carry the Gc genes, however, behave normally and thus it is only these gametes that are transmitted to the next generation. Aegilops speltoides carries two alleles of the gametocidal genes. Gc1a and Gc1b, both located on chromosome 2S, are transmitted at a very high frequency to the next generation and give rise to different morphological aberrations. Gc1a causes endosperm degeneration and chromosome aberrations while Gc1b results in the production of seeds that lack shoot primordia (Tsujimoto and Tsunewaki, 1988). The limitations of utilizing species with gametocidal genes for wild relative introgression are: (1) the fertility of hybrid/crossed grain is reduced by at least 50 %; and (2) all introgressions from wild relatives with Gc genes will always carry them and any other genes closely linked to them. Hence it is very difficult, if not impossible, to transfer genes from other regions of the genome in the absence of the Gc genes.
In this paper we investigate (1) the ability of the Ae. speltoides suppressors of the Ph1 locus located on chromosomes 3S and 7S to induce recombination between the chromosomes of wheat and those of Ae. speltoides, and (2) the frequency of transmission of the Ae. speltoides gametocidal chromosomes through the gametes in F1 hybrids and their derivatives.
MATERIALS AND METHODS
Generation of introgressions
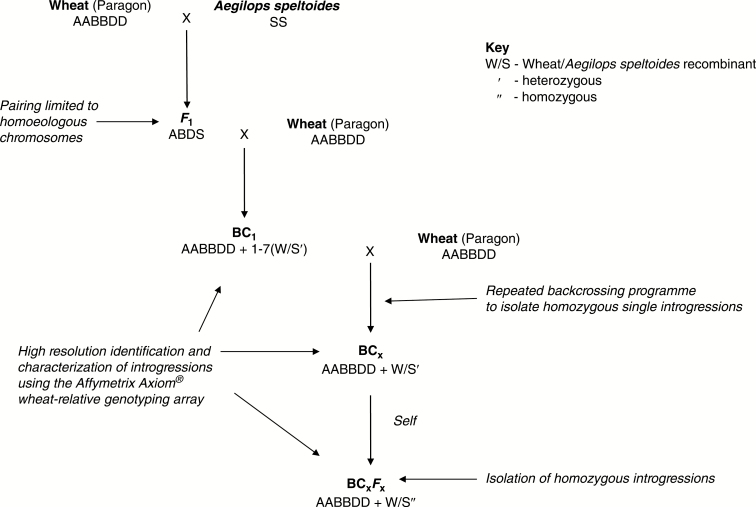
The method used to induce introgressions between wheat and Ae. speltoides is the same as that described by King et al. (2017). In summary, hexaploid wheat (‘Paragon’, obtained from the Germplasm Resources Unit at the JIC; code W10074) was pollinated with Ae. speltoides (accession 2140066 obtained from the Germplasm Resources Unit at the JIC) to produce F1 interspecific hybrids. These hybrids were then grown to maturity and backcrossed as the female with the wheat parent to generate BC1 populations. The BC1 individuals and their resulting progenies were then recurrently pollinated to produce BC2, BC3 etc. populations(Fig. 1).
Fig. 1.
Wheat/wild relative introgression strategy.
As a result of the presence of the suppressors of the Ph1 locus, homoeologous recombination was expected to occur at meiosis in the F1 wheat/Ae. speltoides hybrids, leading to the production of interspecific recombinant chromosomes/introgressions. Subsequent recurrent backcrossing of any BC1 progeny to the wheat parent was undertaken to transfer any interspecific recombinant chromosomes generated into a wheat (Paragon) background.
Detection of putative wheat/Ae. speltoides introgressions
A 35K Axiom® Wheat-Relative Genotyping Array (Affymetrix, Santa Clara, CA) was used to detect the presence of putative wheat/Ae. speltoides introgressions in each of the backcross generations (King et al., 2017). In summary, the array is composed of single-nucleotide polymorphisms (SNPs), each showing polymorphism for the ten wild relatives (under study at the Nottingham/BBSRC Wheat Research Centre), including Ae. speltoides (King et al., 2017). All the SNPs incorporated in this array formed part of the Axiom® 820K SNP array (Winfield et al., 2016). The dataset for the Axiom® 820K SNP array is available from www.cerealsdb.uk.net (Wilkinson et al., 2012, 2016). Table 1 shows the number of putative SNPs between Ae. speltoides and each of the wheat genotypes included on the array. The array has been constructed in such a way that up to 384 lines could be screened at one time. Thus, the array facilitated the high-throughput, high-resolution screening of wheat/Ae. speltoides introgressions. Genotyping was performed as described by King et al. (2017) with no modifications (see below).
Table 1.
Number of polymorphic SNPs between Ae. speltoides and hexaploid wheat based on the Affymetrix 35K array. A comparison is made between all calls (all markers showing polymorphism between wheat and Ae. speltoides) and Poly High Resolution (PHR) calls, which are codominant and polymorphic between wheat and Ae. speltoides with at least two examples of the minor allele
| Linkage group | Total | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | ||
| All calls (% of total) | 2770 (12.4) | 3992 (17.9) | 3358 (15.1) | 2798 (12.6) | 3646 (16.4) | 2530 (11.4) | 3164 (14.2) | 22 258 |
| PHR calls (% of total) | 69 (12.7) | 78 (14.3) | 82 (15.1) | 85 (15.6) | 90 (16.5) | 46 (8.5) | 94 (17.3) | 544 |
Genetic mapping of Ae. speltoides
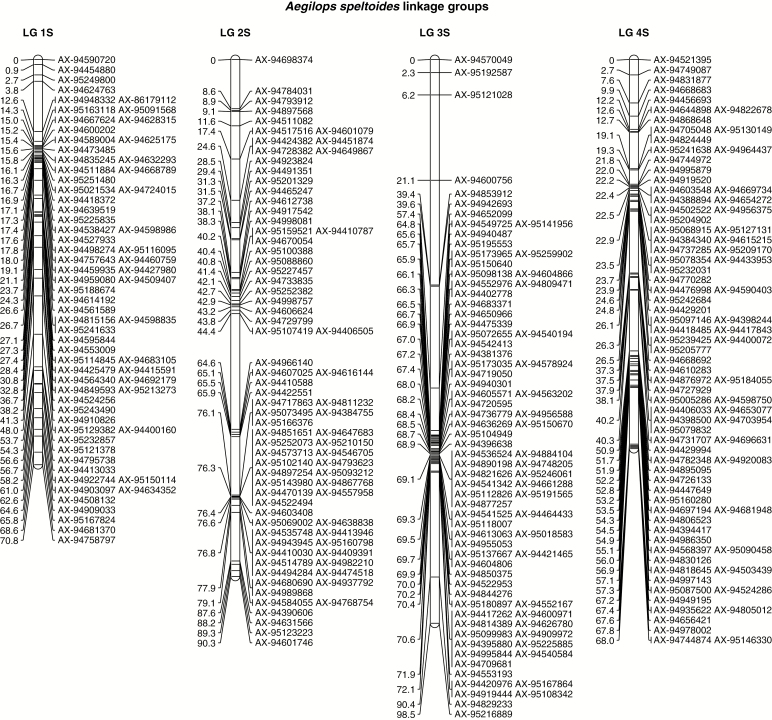
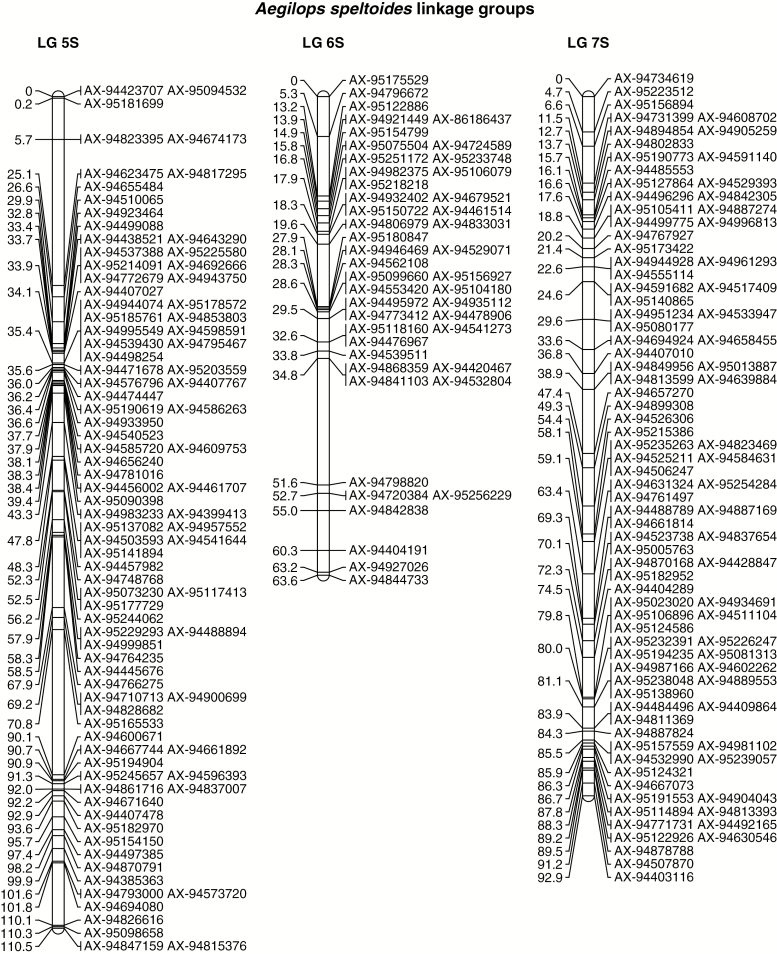
Genetic mapping was as described by King et al. (2017); in summary, DNA was extracted using a CTAB method (Zhang et al., 2013) from individuals of the back-crossed populations, BC1, BC2, BC3, BC4 and BC5 derived from the wheat/Ae. speltoides F1 hybrids. These populations were genotyped with the Axiom® Wheat-Relative Genotyping Array. Only Poly High Resolution (PHR) SNP markers, which were codominant and polymorphic, with at least two examples of the minor allele were used for genetic mapping (King et al., 2017). The SNP markers that showed (1) heterozygous calls for either parent(s), (2) no polymorphism between the wheat parents and Ae. speltoides, and/or (3) no calls for either parent(s) were removed using Flapjack™ (Milne et al., 2010; v.1.14.09.24). The resulting markers were sorted into linkage groups (Fig. 2) in JoinMap® 4.1 (van Ooijen, 2011) with a LOD score of 20 and a recombination frequency threshold of 0.1 using the Haldane mapping function (Haldane, 1919). All markers that did not show any heterozygous call or were unlinked were ignored and only the highest-ranking linkage groups with >30 markers were selected for map construction. These were exported and assigned to chromosomes using information from the Axiom® Wheat HD Genotyping Array (Winfield et al., 2016).
Fig. 2.
Genetic linkage map of Ae. speltoides.
Comparative analysis
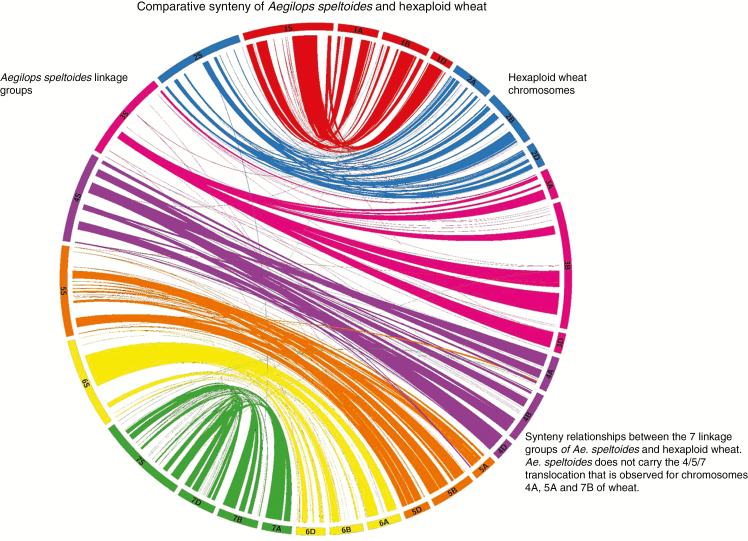
Synteny analysis was carried out using sequence information of the markers located on the present map of Ae. speltoides. The sequences of the mapped markers were compared using BLAST (e-value cut-off of 1e-05) against the wheat genome (http://plants.ensembl.org/Triticum_aestivum) to obtain the orthologous map positions of the top hits in the A, B and D genomes of wheat. To generate the figures, centimorgan (cM) distances on the linkage groups of the present map of Ae. speltoides were scaled up by a factor of 100 000 to match similar base-pair lengths of the chromosomes of the wheat genome. Figure 3 was visualized using Circos (v. 0.67; Krzywinski et al., 2009) in order to observe synteny between Ae. speltoides (genetic position in cM) and the wheat genome (physical position in Mb).
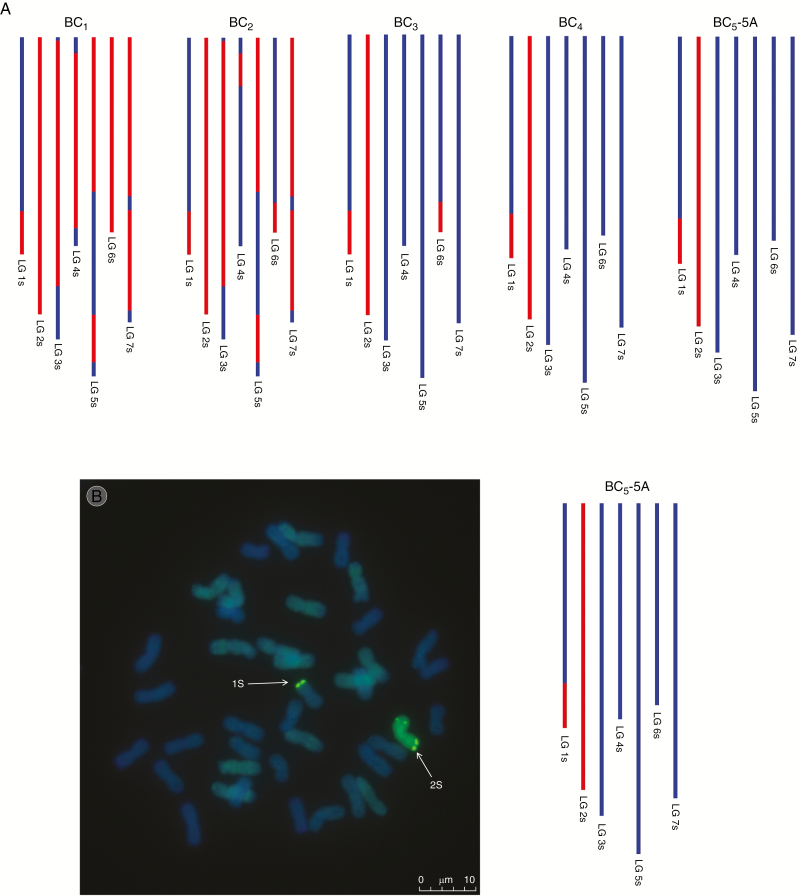
Fig. 3.
(A) SNP characterization of Ae. speltoides introgressions in five consecutive generations, i.e. BC1, BC2, BC3, BC4 and BC5. (B) Genomic in situ hybridization image of the BC5 genotype (aneuploid 41-chromosome plant). In the SNP characterization red colour is used to represent the presence of an Ae. speltoides introgression and blue colour represents wheat. It should be noted that these diagrams cannot be used to assess which wheat chromosomes the Ae. speltoides segments have recombined with. The GISH image shows a metaphase spread of BC5-5A probed with labelled genomic DNA of Ae. speltoides. Arrows show Ae. speltoides introgressions (green).
Cytogenetic analysis
The protocol for genomic in situ hybridization (GISH) was as described in Zhang et al. (2013), Kato et al. (2004) and King et al. (2017). In summary, genomic DNAs from young leaves of the three putative diploid progenitors of bread wheat, i.e. T. urartu (A genome), Ae. speltoides (B genome) and Ae. tauschii (D genome), were isolated using extraction buffer (0.1 m Tris–HCl pH 7.5, 0.05 m EDTA pH 8.0, 1.25% SDS). Samples were incubated at 65 °C for 1 h before being placed on ice and mixed with ice-cold 6 m NH4C2H3O for 15 min. The samples were then spun down, the supernatant was mixed with isopropanol to pellet the DNA and the isolated DNA was further purified with phenol/chloroform. The genomic DNAs of Ae. speltoides and T. urartu were labelled by nick translation with Chroma Tide Alexa Fluor 488-5-dUTP (Invitrogen, Carlsbad, CA; C11397). Genomic DNA of Ae. tauschii was labelled with Alexa Fluor 594-5-dUTP (Invitrogen; C11400). Genomic DNAs of Ae. speltoides and T. aestivum ‘Chinese Spring’ were fragmented to 300–500 bp in a heat block at 100 °C.
Preparation of chromosome spreads was as described in Kato et al. (2004) and King et al. (2017). Briefly, roots were excised from germinated seeds, treated with nitrous oxide gas at 10 bar for 2 h, fixed in 90 % acetic acid for 10 min and then washed three times in water on ice. Root tips were dissected and digested in 20 µL of 1 % pectolyase Y23 and 2 % cellulase Onozuka R-10 (Yakult Pharmaceutical, Tokyo) solution for 50 min at 37 °C and then washed three times in 70 % ethanol. Root tips were crushed in 70 % ethanol, cells collected by centrifugation at 2.5 g for 1 min, briefly dried and then re-suspended in 30–40 µL of 100 % acetic acid prior to being placed on ice. The cell suspension was dropped onto glass slides (6–7 µL per slide) in a moist box and dried slowly under cover.
Slides were initially probed using labelled genomic DNA of Ae. speltoides (100 ng) and fragmented genomic DNA of Chinese Spring (3000 ng) as blocker (in a ratio of 1: 30 per slide) to detect the Ae. speltoides introgressions. The slides were bleached (dipped in 2 × saline–sodium citrate (SSC) to remove the coverslip, transferred to 4 × SSC for 5 min and air-dried in the light) and re-probed with labelled DNAs of T. urartu (100 ng) and Ae. tauschii (200 ng) and fragmented DNA of Ae. speltoides (3000 ng) as blocker in a ratio of 1: 2:30 per slide to detect the AABBDD genomes of wheat. In both cases, the hybridization mix was made up to 10 µL with 2 × SSC in 1 × Tris/EDTA buffer (TE). Slides were incubated initially at 75 °C for 5 min and then overnight at 55 °C in a closed box before counterstaining with Vectashield mounting medium with 4',6-diamidino-2-phenylindole,dihydrochloride (DAPI), and analysed using a high-throughput, fully automated Zeiss Axio Imager Z2 upright epifluorescence microscope (Carl Zeiss, Oberkochen, Germany) with filters for DAPI (blue), Alexa Fluor 488 (green) and Alexa Fluor 594 (red). Photographs were taken using a MetaSystems Coolcube 1m CCD camera. Further slide analysis was carried out using Meta Systems ISIS and Metafer software (Metasystems, Altlussheim, Germany). This system enabled the fully automated capture of high- and low-power fluorescent images of root tip metaphase spreads. Slides with root tip preparations were automatically scanned and the images downloaded for analysis.
RESULTS
Generation of wheat/Ae. speltoides introgressions
A total of 3890 crosses were made between wheat and Ae. speltoides and their derivatives, leading to the generation of 9953 crossed seeds and 2029 self-seeds (Fig. 1). The number of seeds germinated, plants crossed and seed set are shown in Table 2. Every ear produced by the F1 was crossed with wheat. The F1 hybrids generated between wheat and Ae. speltoides showed the lowest frequency of germination. Only 15 % germinated compared with 100, 73, 70 and 78 % in the BC1, BC2, BC3 and BC4 generations. The F1 hybrids also showed the lowest fertility, with only 29 % of crossed ears producing any seeds compared with crossed ears from the BC1, BC2, BC3 and BC5 generations, which showed 58, 72, 83 and 84 % fertility, respectively.
Table 2.
Number of seed produced and germinated in relation to the number of crosses carried out for each generation of the introgression programme for Ae. speltoides into wheat
| Wheat × Ae. speltoides | F 1 | BC1 | BC2 | BC3 | BC4 | Total | |
|---|---|---|---|---|---|---|---|
| Number of seeds sown | NA | 20 | 22 | 400 | 187 | 173 | 802 |
| Number of seeds that germinated (%) | NA | 3 (15) | 22 (100) | 292 (73) | 130 (70) | 135 (78) | 582 |
| Number of plants that set seed (%) | NA | 3 (100) | 22 (100) | 261 (89) | 124 (95) | 117 (87) | 527 |
| Number of seed/total number of crosses (average number of seed set per crossed ear) | 127/26 (4.9) | 22/35 (0.6) | 509/357 (1.4) | 5743/2180 (2.6) | 3204/876 (3.7) | 348/416 (0.8) | 9963/3890 |
| Number of crosses producing seed (%) | 18 (69) | 10 (29) | 207 (58) | 1579 (72) | 730 (83) | 350 (84) | 2894 |
| Number of self seed produced | 0 | 0 | 21 | 520 | 753 | 735 | 2029 |
Of the 20 F1 seeds germinated, only three reached maturity and set seeds. Thus all of the wheat/Ae. speltoides introgressions generated in this programme were limited to these three viable F1 hybrids. Table 2 summarizes the number of seeds germinated and seed set for each of the backcross generations. In each generation, the average seed set per crossed ear was very low, i.e. the lowest average seed set per ear was 0.6 (observed in the F1 hybrid), while the highest average seed set per crossed ear was only 3.7 (observed in the BC3). The fertility of plants that had previously been self-fertilized at least once was considerably higher than that of plants in the backcross programme, although again variability was seen, e.g. BC3F1-16A was male-sterile and therefore produced no self-fertilized seed, while seed numbers from crossed ears ranged from 8 to 19. Some plants were also found to be asynchronous for anther and stigma maturity. For these plants with manual pollination, self-fertilized seeds were produced.
Detection of introgressions
Of the SNPs on the Axiom array, 22 258 were polymorphic between Ae. speltoides and wheat (Table 1). The Axiom array was used to screen genomic DNA prepared from 536 samples of BC1 to BC5 lines between wheat and Ae. speltoides. Genotype calls were generated, and the sample call rate (markers working in a particular genotype) ranged from 86.5 to 99.8 %, with an average of 98.8 % for the 536 samples. Affymetrix software classified the scores for each SNP into one of six cluster patterns. However, only those classified as Poly High Resolution (PHR) were used for genetic mapping as these are considered to be optimum quality. Linkage group 7 had the highest number of SNPs (17.3 %), while linkage group 6 had the lowest number (8.5 %).
JoinMap® (van Ooijen, 2011) was used to analyse the PHR SNPs and this led to the establishment of seven linkage groups. The genetic map was composed of 544 SNPs and represented the seven chromosomes of Ae. speltoides. Linkage groups 1–7 had 69, 78, 82, 85, 90, 46 and 94 SNPs respectively (Fig. 2). The lengths of linkage groups 1–7 were 70.8, 90.3, 98.5, 68, 110.5, 63.6 and 92.9 cM, respectively, with a total length of 594.6 cM and an average chromosome length of 84.9 cM.
Genomic in situ hybridization
In most cases the number of wheat/Ae. speltoides introgressions detected by genomic in situ hybridization (GISH) in BC4 and BC5 individuals corresponded exactly with the number detected via SNP analysis (Table 3 and Fig. 3). One exception was detected in the GISH analysis of BC5-4A. The SNP analysis of this plant showed the only segment present to be a complete 2S chromosome. However, the GISH image showed the presence of a large Ae. speltoides 2S segment (one telomere was missing from the chromosome) and a very small Ae. speltoides segment recombined with a different wheat chromosome.
Table 3.
Number of introgressed segments from Ae. speltoides present in BC4 and BC5 plants as detected by SNP genotyping and GISH. The Ae. speltoides chromosomes have been assigned to linkage groups via the comparative analysis of the SNPs with wheat
| Plant accession numbers | Number of segments | Ae. speltoides linkage group of segments | ||
|---|---|---|---|---|
| Backcross | Number | Genotyping | GISH | |
| BC4 BC5 |
45A | 3 | 3 | 2, 5, 6 |
| 45C | 1 | 1 | 2 | |
| 45D | 3 | 3 | 2, 5, 6 | |
| 1A | 3 | 3 | 2, 4, 5 | |
| 1B | 1 | 1 | 2 | |
| 1C | 1 | 1 | 2 | |
| 1D | 2 | 2 | 2, 5 | |
| 2 | 2 | 2 | 2, 5 | |
| 3 | 1 | 1 | 1 | |
| 4A | 1 | 1 + small end | 2,? | |
| 5A | 2 | 2 | 1, 2 | |
| 6A | 1 | 1 | 2 | |
| 6B | 2 | 2 | 1, 2 | |
| 6C | 1 | 1 | 2 | |
| 7A | 2 | 2 | 2, 5 | |
| 7C | 1 | 1 | 2 | |
| 8A | 1 | 1 | 2 | |
| 8B | 1 | 1 | 2 | |
| 8C | 1 | 1 | 2 | |
| 8D | 1 | 1 | 2 | |
Syntenic relationship
Figure 4 shows the syntenic relationship between the seven linkage groups of Ae. speltoides and the three genomes of wheat, with large ribbons showing significant synteny. Some gene rearrangements are indicated where single markers cross-map to non-collinear positions on wheat chromosomes. The only major disruption in synteny between the two species is that Ae. speltoides does not carry the 4/5/7 translocation observed for chromosomes 4A, 5A and 7B of wheat (Liu et al., 1992; Naranjo et al., 1987). These data demonstrate that there is a close syntenic relationship between the A, B and D genomes of wheat and Ae. speltoides.
Fig. 4.
Synteny of Ae. speltoides (genetic position in cM) with hexaploid wheat (physical position in Mb) [visualized using Circos v. 0.67 (Krzywinski et al., 2009)].
Preferential transmission
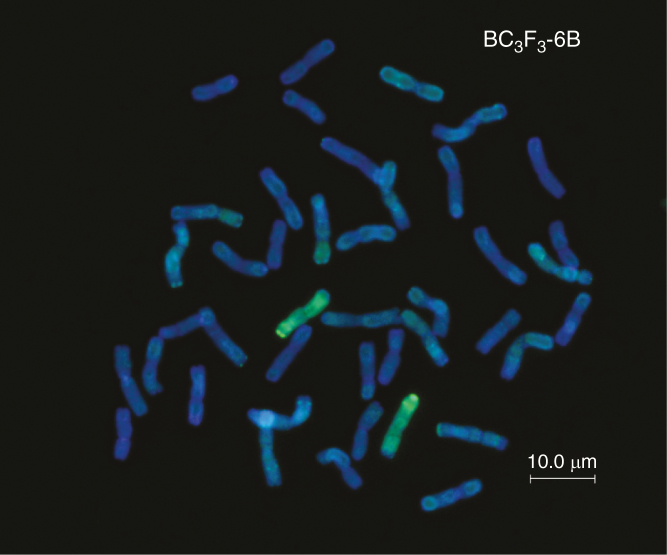
The data obtained from SNP analysis was used to determine the transmission frequency of chromosome segments from each of the seven Ae. speltoides linkage groups through the female gametes to the BC1, BC2, BC3, BC4 and BC5 generations. Chromosome segments from linkage group 2 were observed in all 536 backcross progeny observed (Table 4). The smallest segment carried only three SNP markers, AX94631566, AX95123223 and AX94601746, located near one of the terminal ends of the 2S linkage group (Fig. 2). These three SNP markers were found to be present in all the backcross individuals analysed. In order to determine whether the 2S chromosome was also preferentially transmitted through the male gametes, 41 BC3 and BC4 plants were allowed to self-fertilize and their progenies were analysed using GISH. The BC3/BC4 plants selected each carried a single copy of a 2S chromosome segment. Thus, these plants would produce two types of male and female gametes during gametogenesis: those that carried 2S and those that did not. If the 2S chromosome segment was preferentially transmitted through both the male and female gametes, only those that carried the 2S chromosome would be viable and as a result all the progeny observed in the BC3/BC4 self-populations would be homozygous for the 2S chromosome segment. Alternatively, if the 2S chromosome segment was not preferentially transmitted through the male or the female gametes, then the progeny would segregate for the presence of either one or two 2S chromosome segments. All progeny observed carried two copies of the 2S chromosome segment (Fig. 5), thus demonstrating that the 2S chromosome segment was preferentially transmitted through both the male and female gametes.
Table 4.
Transmission frequencies of the seven linkage groups of Ae. speltoides in the backcross populations (BC1 to BC5) analysed by SNP genotyping
| BC1 | BC2 | BC3 | BC4 | BC5 | Total | |
|---|---|---|---|---|---|---|
| Number of plants genotyped | 22 | 252 | 123 | 117 | 22 | 536 |
| Linkage group 1 (%) | 18 (82) | 124 (49) | 36 (29) | 13 (11) | 3 (14) | 194 (36) |
| Linkage group 2 (%) | 22 (100) | 252 (100) | 123 (100) | 117 (100) | 22 (100) | 536 (100) |
| Linkage group 3 (%) | 18 (81) | 135 (54) | 27 (22) | 20 (17) | 0 (0) | 200 (37) |
| Linkage group 4 (%) | 15 (68) | 89 (35) | 23 (19) | 7 (6) | 0 (0) | 134 (25) |
| Linkage group 5 (%) | 19 (86) | 162 (64) | 54 (44) | 32 (27) | 6 (27) | 273 (51) |
| Linkage group 6 (%) | 17 (77) | 115 (46) | 31 (25) | 25 (21) | 0 (0) | 188 (35) |
| Linkage group 7 (%) | 13 (59) | 80 (32) | 15 (12) | 9 (8) | 0 (0) | 117 (22) |
Fig. 5.
GISH of a complete one-cell metaphase spread of a genotype produced by self-fertilization (BC3F3-6B) with labelled genomic Ae. speltoides DNA as probe, showing a homozygous pair of Ae. speltoides linkage group 2 introgressed chromosomes (green).
The frequency of transmission of chromosome segments from other linkage groups in each of the backcross progenies was much lower than that observed for linkage group 2. Of the remaining linkage groups, segments from linkage group 5 of Ae. speltoides had the highest frequency of transmission in all the backcross progenies observed (Table 4)
DISCUSSION
Aegilops speltoides is a potentially important source of genetic variation for a range of agronomically important traits (Jia et al., 1996; Mago et al., 2009; Klindworth et al., 2012; Deek and Distelfeld, 2014). In the past, it has been difficult to detect and characterize wheat/wild relative introgressions due to the absence of high-throughput, genome-wide marker technologies. In this paper we have utilized an Affymetrix SNP array (King et al., 2017) complemented by a specific crossing strategy and the use of an automated, high-throughput microscope system for GISH image capture.
In this work we generated F1 interspecific hybrids between wheat and Aegilops speltoides and backcrossed these to wheat (Fig. 1). Hybrids were haploid for the A, B and D genomes of wheat and the S genome of Ae. speltoides. Thus, in the absence of homologous chromosomes the only recombination occurring at meiosis was between homoeologous chromosomes. This would normally be prevented by the presence of Ph1. However, the presence of the pairing promoters Su1-Ph1 and Su2-Ph1 located on chromosomes 3S and 7S of Ae. speltoides (Chen and Dvorak, 1984; Dvorak et al., 2006) enabled homoeologous recombination to occur.
Since the interspecific F1 hybrids generated were haploid for the A, B, D and S genomes, their fertility was predicted to be low and this was found to be the case (Table 1). However, their fertility of 29 % was higher than that previously observed in those between wheat and Amblyopyrum muticum, which was 16.2 % (King et al., 2017). The low fertility of the F1 hybrids resulted in the generation of only 22 BC1 seeds that grew to maturity and set seed. Hence, if recombination did not occur in later generations, i.e. in the gametes of the BC1, BC2, BC3 and BC4 generations, then the total number of introgressions that could be generated was limited to the 22 female F1 gametes giving rise to these 22 BC1 plants. However, the level of interspecific recombination detected by the genetic mapping was such that it was possible to assemble the seven linkage groups of Ae. speltoides. Using the genetic linkage map of Ae. speltoides, we estimated that 294 wheat/Ae. speltoides introgressions were generated spanning the entire Ae. speltoides genome. We were able to characterize these introgressions and track them through the backcross generations (Fig. 3). However, while using the genetic map to characterize the introgressions, it was important to treat the cM distances with caution, as the maps were not produced using proper mapping families. Tracking the introgressions through the different backcross generations via the SNP data showed that the majority of introgressions generated had occurred due to recombination in the F1 gametes and were therefore present in the BC1 plants.
Validation of introgressions identified by SNP analysis was carried out by GISH analysis. In most cases the number of wheat/Ae. speltoides introgressions detected by the SNP analysis corresponded to the number of introgressions detected by the GISH analysis (Table 3 and Fig. 3). The one exception (BC5-4A) can be explained in two possible ways. The first possibility is that linkage group 2S of Ae. speltoides has recombined with a wheat chromosome. Although the telomere is on a separate chromosome, the SNP markers would have detected the two introgressions as a complete chromosome. The second possibility is that the genetic linkage map of Ae. speltoides is not complete and therefore the smaller introgression has not been detected at all and the large segment has appeared complete in the SNP analysis.
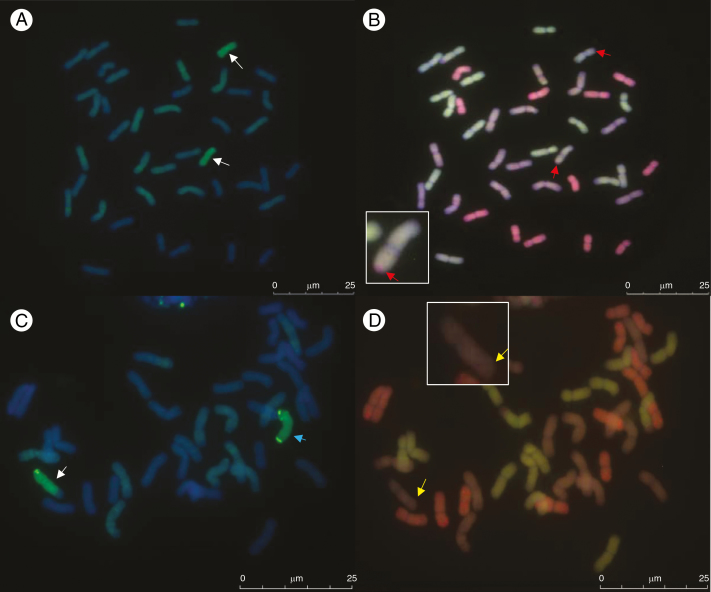
Multicolour GISH analysis was carried out to verify which of the wheat genomes were involved in recombination with Ae. speltoides (Fig. 6). The majority of recombination events were shown to have occurred between Ae. speltoides and the B genome of wheat (91 %). Recombination had also occurred between Ae. speltoides and the D genome (18 %) but we found no evidence of recombination between Ae. speltoides and the wheat A genome. As the B genome progenitor of wheat is thought to be Ae. speltoides or a close relative (Sarkar and Stebbins, 1956; Riley et al., 1958; Dvorak and Zhang, 1990; Maestra and Naranjo, 1998), the higher level of recombination between Ae. speltoides and the B genome was expected. The low level of recombination observed with the D genome of wheat would also suggest that Ae. speltoides is more closely related to the D genome than to the A genome. This situation closely mirrors that observed between wheat and Amblyopyrum muticum, where most of the recombination occurred between the chromosomes of the wild relative and the B and D genomes of wheat (King et al., 2017). In addition, although the genetic location of the Ph1 pairing suppressors located on Am. muticum are not currently known, the fact that both Ae. speltoides and Am. muticum carry suppressors indicates that the two species may have had a common ancestry.
Fig. 6.
Genomic in situ hybridization showing recombination between Ae. speltoides and the B and D genomes of wheat. (A) GISH analysis of a 41-chromosome BC3F1 individual. Metaphase spread with labelled genomic Ae. speltoides DNA as probe showing Ae. speltoides (green) introgressions (white arrows). (B) Same metaphase spread as (A) with three-colour GISH showing Ae. speltoides recombined with the D genome (red arrows). Enlarged inset shows one of the recombined chromosomes. This individual carries 14 A chromosomes (green), 13 B chromosomes (purple), 12 D chromosomes (red) and two wheat/Ae. speltoides 2S recombinant chromosomes (the segment of the D genome is red and marked with the red arrow). (C) GISH analysis of a 42-chromosome BC5 individual. Metaphase spread with labelled genomic Ae. speltoides (green) DNA as probe showing one complete Ae. speltoides chromosome (blue arrow) from linkage group 2S and one wheat/Ae. speltoides recombined chromosome (white arrow) from Ae. speltoides linkage group 5S. (D) Same metaphase spread as (C) with three-colour GISH showing Ae. speltoides recombined with the B genome (yellow arrow). Enlarged insert shows the recombined chromosome. This individual carries 14 A chromosomes (green), 12 B chromosomes (purple), 14 D chromosomes (red), one complete Ae. speltoides chromosome (purple) and one wheat/Ae. speltoides recombinant chromosome (purple).
We are currently unable to use the SNP markers to identify which of the chromosomes of wheat have recombined with the Ae. speltoides chromosomes. We are currently developing wheat chromosome-specific Kompetitive Allele Specific PCR (KASP) markers from the SNP markers on the 35K array. These markers will allow the analysis of large numbers of individuals, firstly to tag individual introgressions and track them through the generations (both via backcrossing and selfing) and secondly to identify which wheat chromosome(s) is (are) involved in each introgression once homozygous introgressions have been generated. Although considerably more labour-intensive and technically demanding, the latter could also be achieved using fluorescence in situ hybridization (FISH) with repetitive DNA sequences combined with GISH. This has been successfully demonstrated to identify the recipient wheat chromosomes with introgressions from Thinopyrum bessarabicum (Patokar et al., 2016), Th. intermedium and Secale cereale (Ali et al., 2016) and Aegilops markgrafii, Aegilops triuncialis and Aegilops cylindrica (Molnár et al., 2015).
In the Ae. speltoides accession used in this study, chromosome 2S was found to be preferentially transmitted at a frequency of 100 % through both the male and female gametes. This demonstrates that the gametocidal genes on chromosome 2S are extremely potent and resemble those in species such as Aegilops sharonensis (King et al., 1991a, b; King and Laurie, 1993). The transmission of the other six linkage groups of Ae. speltoides was also observed in the female gametes, and although linkage group 5 showed a potentially elevated rate of transmission, there is no evidence at this time to suggest that it carries a gene for preferential transmission.
It is important to note that the work undertaken was based on a single accession of Ae. speltoides and thus it is not possible to determine whether all other accessions carry gametocidal genes on chromosome 2S or other linkage groups. However, if they do, then it will be necessary to remove them if introgression lines without them are to be developed. In order to develop lines that lack the 2S gametocidal genes, but still carry introgressions from other linkage groups, we are currently undertaking research to mirror the work of Friebe et al. (2003), who successfully deleted the genes responsible for preferential transmission in Ae. sharonensis.
CONCLUSION
Researchers have been working to transfer genetic variation from its wild relatives into wheat for many years. However, although there have been some notable successes, we have barely scratched the surface of the genetic variation available for exploitation. This has been as a direct result of: (1) the difficulty of inducing wheat/wild relative introgressions and (2) an inability to detect the introgressions. The data obtained here clearly demonstrate that Ae. speltoides chromosomes recombine with the chromosomes of wheat at a very high frequency, giving rise to large numbers of introgressions in derivatives of the F1 hybrid. Considering the importance of the genetic variation identified in Ae. speltoides for a range of agronomic traits (Jia et al., 1996; Mago et al., 2009; Klindworth et al., 2012; Deek and Distelfeld, 2014), wheat/Ae. speltoides introgressions have the potential to play a critical role in the development of superior wheat varieties in the future.
ACKNOWLEDGEMENTS
This work was supported by the Biotechnology and Biological Sciences Research Council as part of the Wheat Improvement Strategic Programme [grant number BB/J004596/1]. The funding body played no role in the design of the study, collection, analysis and interpretation of data and in writing the manuscript.
LITERATURE CITED
- Ali N, Heslop-Harrison JS, Ahmad H, Graybosch RA, Hein GL, Schwarzacher T. 2016. Introgression of chromosome segments from multiple alien species in wheat breeding lines with wheat streak mosaic virus resistance. Heredity 117: 114–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brozynska M, Furtado A, Henry RJ. 2015. Genomics of crop wild relatives: expanding the gene pool for crop improvement. Plant Biotechnology Journal 14: 1070–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charmet G. 2011. Wheat domestication: lessons for the future. Comptes Rendus Biologies 334: 212–220. [DOI] [PubMed] [Google Scholar]
- Chen KC, Dvorak J. 1984. The inheritance of genetic variation in Triticum speltoides affecting heterogenetic chromosome pairing in hybrids with Triticum aestivum. Canadian Journal of Genetics and Cytology 26: 279–287. [Google Scholar]
- Deek J, Distelfeld A. 2014. Characterisation of Aegilops speltoides introgression lines conferring resistance to wheat leaf rust. Phytoparasitica 42: 651–672. [Google Scholar]
- Doussinault G, Delibes A, Sanchezmonge R, Garcia-Olmedo F. 1983. Transfer of a dominant gene for resistance to eyespot disease from a wild grass to hexaploid wheat. Nature 303: 698–700. [Google Scholar]
- Dvorak J. 1972. Genetic variability in Aegilops speltoides affecting homoeologous pairing in wheat. Canadian Journal of Genetics and Cytology 14: 371–380. [Google Scholar]
- Dvorak J, Akhunov E. 2005. Tempos of gene locus deletions and duplications and their relationship to recombination rate during diploid and polyploid evolution in the Aegilops-Triticum alliance. Genetics 171: 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak J, Zhang HB. 1990. Variation in repeated nucleotide sequences sheds light on the phylogeny of the wheat B and G genomes. Proceedings of the National Academy of Sciences of the USA 87: 9640–9644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak J, di Terlizzi P, Zhang HB, Resta P. 1993. The evolution of polyploid wheats – identification of the A-genome donor species. Genome 36: 21–31. [DOI] [PubMed] [Google Scholar]
- Dvorak J, Deal KR, Luo M-C. 2006. Discovery and mapping of wheat Ph1 suppressors. Genetics 174: 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch RA, Miller TE, Bennett MD. 1984. “Cuckoo” Aegilops addition chromosome in wheat ensures its transmission by causing chromosome breaks in meiospores lacking it. Chromosoma 90: 84–88. [Google Scholar]
- Friebe B, Gill BS. 1996. Chromosome banding and genome analysis in diploid and cultivated polyploid wheats. In: Jauhar PP. ed. Methods of genome analysis in plants. New York: CRC Press, 39–59. [Google Scholar]
- Friebe B, Zhang P, Nasuda S, Gill BS. 2003. Characterization of a knock-out mutation at the Gc2 locus in wheat. Chromosoma 111: 509–517. [DOI] [PubMed] [Google Scholar]
- Grassini P, Eskridge KM, Cassman KG. 2013. Distinguishing between yield advances and yield plateaus in historical crop production trends. Nature Communications 3: 2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybosch RA, Peterson CJ, Baenziger PS et al. . 2009. Registration of ‘MACE’ hard red winter wheat. Journal of Plant Registration 3: 51–56. [Google Scholar]
- Haldane JBS. 1919. The probable errors of calculated linkage values, and the most accurate method of determining gametic from certain zygotic series. Journal of Heredity 8: 291–297. [Google Scholar]
- Haussmann BIG, Parzies HK, Presterl T, Susic Z, Miedaner T. 2004. Plant genetic resources in crop improvement. Plant Genetic Resources 2: 3–21. [Google Scholar]
- Jia J, Devos KM, Chao S, Miller TE, Reader SM, Gale MD. 1996. RFLP-based maps of the homoeologous group 6 chromosomes of wheat and their application in the tagging of Pm12, a powdery mildew resistance gene transferred from Aegilops speltoides to wheat. Theoretical and Applied Genetics 92: 559–565. [DOI] [PubMed] [Google Scholar]
- Kato A, Lamb JC, Birchler JA. 2004. Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proceedings of the National Academy of Sciences of the USA 101: 13554–13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King IP, Laurie DA. 1993. Chromosome damage in early embryo and endosperm development in crosses involving the preferentially transmitted 4Sl chromosome of Aegilops sharonensis. Heredity 70: 52–59. [Google Scholar]
- King IP, Koebner RMD, Schlegel R, Reader SM, Miller TE, Law CN. 1991a. Exploitation of a preferentially transmitted chromosome from Aegilops sharonensis for the elimination of segregation for height in semidwarf bread wheat varieties. Genome 34: 944–949. [Google Scholar]
- King IP, Miller TE, Koebner RMD. 1991b. Determination of the transmission frequency of chromosome 4Sl of Aegilops sharonensis in a range of wheat genetic backgrounds. Theoretical and Applied Genetics 81: 519–523. [DOI] [PubMed] [Google Scholar]
- King J, Gustafson P, Allen A, King I. 2016. Exploitation of interspecific diversity in wheat. In: Bonjean AP, Angus WJ, van Ginkel M eds. The world wheat book: a history of wheat breeding, Vol. 3 Paris: Lavoisier, 1125–1139. [Google Scholar]
- King J, Grewal S, Yang C et al. . 2017. A step change in the transfer of interspecific variation into wheat from Amblyopyrum muticum. Plant Biotechnology Journal 15: 217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klindworth DL, Niu Z, Chao S et al. . 2012. Introgressions and characterisation of a goatgrass gene for a high level of resistance to Ug99 stem rust in tetraploid wheat. Gene, Genomes and Genetics 2: 665–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I et al. . 2009. Circos: an information aesthetic for comparative genomics. Genome Research 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CJ, Atkinson MD, Chinoy CN, Devos KM, Gale MD. 1992. Nonhomoeologous translocations between group 4, 5 and 7 chromosomes within wheat and rye. Theoretical and Applied Genetics 83: 305–312. [DOI] [PubMed] [Google Scholar]
- Maestra B, Naranjo T. 1998. Homoeologous relationships of Aegilops speltoides chromosomes to bread wheat. Theoretical and Applied Genetics 97: 181–186. [Google Scholar]
- Mago R, Zhang P, Banana HS et al. . 2009. Development of wheat lines carrying stem rust resistance gene Sr39 with reduced Aegilops speltoides chromatin and simple PCR markers for marker-assisted selection. Theoretical and Applied Genetics 119: 1441–1450. [DOI] [PubMed] [Google Scholar]
- Maia N. 1967. Obtention de blés tendres résistants au piétin-verse (Cercosporella herpotrichoides) par crôisements interspécifiques blés x Aegilops. Comptes Rendus de l’Académie d’Agriculture de France 53: 149–155. [Google Scholar]
- Marcussen T, Sandve SR, Heier L et al. . 2014. Ancient hybridizations among the ancestral genomes of bread wheat. Science 345: 1250092. [DOI] [PubMed] [Google Scholar]
- McCouch S, Baute GJ, Bradeen J et al. . 2013. Agriculture: feeding the future. Nature 499: 23–24. [DOI] [PubMed] [Google Scholar]
- McFadden E, Sears ER. 1946. The origin of Triticum spelta and its free threshing hexaploid relatives. Journal of Heredity 37: 81–107. [DOI] [PubMed] [Google Scholar]
- Milne I, Shaw P, Stephen G et al. . 2010. Flapjack – graphical genotype visualisation. Bioinformatics 26: 3133–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnár I, Vrána J, Farkas A et al. . 2015. Flow sorting of C-genome chromosomes from wild relatives of wheat Aegilops markgrafii, Ae. triuncialis and Ae. cylindrica and their molecular organization. Annals of Botany 116: 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naranjo T, Roca A, Goicoechea PG, Giraldez R. 1987. Arm homoeology of wheat and rye chromosomes. Genome 29: 873–882. [Google Scholar]
- Ogihara Y, Hasegawa K, Tsujimoto K. 1994. High-resolution cytological mapping of the long arm of chromosome 5A in common wheat using a series of deletion lines induced by gametocidal (Gc) genes of Aegilops speltoides. Molecular and General Genetics 244: 253–259. [DOI] [PubMed] [Google Scholar]
- van Ooijen JW. 2011. Multipoint maximum likelihood mapping in a full-sib family of outbreeding species. Genetics Research 93: 343–349. [DOI] [PubMed] [Google Scholar]
- Patokar C, Sepsi A, Schwarzacher T, Kishii M, Heslop-Harrison JS. 2016. Molecular cytogenetic characterization of novel wheat-Thinopyrum bessarabicum recombinant lines carrying intercalary translocations. Chromosoma 125: 163–172. [DOI] [PubMed] [Google Scholar]
- Ray DK, Mueller ND, West PC, Foley JA. 2013. Yield trends are insufficient to double global crop production by 2050. PLoS ONE 8: e66428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley R, Unrau J, Chapman V. 1958. Evidence on the origin of the B genome of wheat. Journal of Heredity 49: 91–98 [Google Scholar]
- Sarkar P, Stebbins GL. 1956. Morphological evidence concerning the origin of the B genome in wheat. American Journal of Botany 43: 297–304. [Google Scholar]
- Sears ER. 1955. An induced gene transfer from Aegilops to Triticum. Genetics 40: 595. [Google Scholar]
- Tanksley SD, McCouch SR. 1997. Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277: 1063–1066. [DOI] [PubMed] [Google Scholar]
- Tester M, Langridge P. 2010. Breeding technologies to increase crop production in a changing world. Science 327: 818–822. [DOI] [PubMed] [Google Scholar]
- Tiwari VK, Wang S, Sehgal S et al. . 2014. SNP discovery for mapping alien introgressions in wheat. BMC Genomics 15: 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari VK, Wang S, Danilova T et al. . 2015. Exploring the tertiary gene pool of bread wheat: sequence assembly and analysis of chromosome 5M9g0 of Aegilops geniculata. Plant Journal 84: 733–746. [DOI] [PubMed] [Google Scholar]
- Tsujimoto H, Tsunewaki K. 1984. Gc genes in wheat and its relatives. I. Genetic analyses in common heat of a Gc gene derived from Aegilops speltoides. Canadian Journal of Genetics and Cytology 26: 78–84. [Google Scholar]
- Tsujimoto H, Tsunewaki K. 1988. Gametocidal genes in wheat and its relatives. III. Chromosome location and effects of two Aegilops speltoides-derived Gc genes in common wheat. Genome 30: 239–244. [Google Scholar]
- Tsunewaki K. 1996. Plasmon analysis as the counterpart of genome analysis. In: Jauhar PP. ed. Methods of genome analysis in plants. New York: CRC Press, 271–299. [Google Scholar]
- Warschefsky E, Penmetsa RV, Cook DR, von Wettberg EJB. 2014. Back to the wilds: tapping evolutionary adaptations for resilient crops through systematic hybridization with crop wild relatives. American Journal of Botany 101: 1791–1800. [DOI] [PubMed] [Google Scholar]
- Wilkinson PA, Winfield MO, Barker GLA et al. . 2012. CerealsDB 2.0: an integrated resource for plant breeders and scientists. BMC Bioinformatics 13: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson PA, Winfield MO, Barker GLA et al. . 2016. CerealsDB 3.0: expansion of resources and data integration. BMC Bioinformatics 17: 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winfield MO, Allen AM, Burridge AJ et al. . 2016. High-density SNP genotyping array for hexaploid wheat and its secondary and tertiary gene pool. Plant Biotechnology Journal 14: 1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Bian Y, Gou X et al. . 2013. Persistent whole-chromosome aneuploidy is generally associated with nascent allohexaploid wheat. Proceedings of the National Academy of Sciences of the USA 110: 3447–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Mittal N, Leamy LJ, Barazani O, Song B-H. 2017. Back into the wild – apply untapped genetic diversity of wild relatives for crop improvement. Evolutionary Applications 10: 5–24. [DOI] [PMC free article] [PubMed] [Google Scholar]