Abstract
Background and Aims
Roots facilitate acquisition of macro- and micronutrients, which are crucial for plant productivity and anchorage in the soil. Phosphorus (P) is rapidly immobilized in the soil and hardly available for plants. Adaptation to P scarcity relies on changes in root morphology towards rooting systems well suited for topsoil foraging. Root-system architecture (RSA) defines the spatial organization of the network comprising primary, lateral and stem-derived roots and is important for adaptation to stress conditions. RSA phenotyping is a challenging task and essential for understanding root development.
Methods
In this study, 19 traits describing RSA were analysed in a diversity panel comprising 194 sorghum genotypes, fingerprinted with a 90-k single-nucleotide polymorphism (SNP) array and grown under low and high P availability.
Key Results
Multivariate analysis was conducted and revealed three different RSA types: (1) a small root system; (2) a compact and bushy rooting type; and (3) an exploratory root system, which might benefit plant growth and development if water, nitrogen (N) or P availability is limited. While several genotypes displayed similar rooting types in different environments, others responded to P scarcity positively by developing more exploratory root systems, or negatively with root growth suppression. Genome-wide association studies revealed significant quantitative trait loci (P < 2.9 × 10−6) on chromosomes SBI-02, SBI-03, SBI-05 and SBI-09. Co-localization of significant and suggestive (P < 5.7 × 10−5) associations for several traits indicated hotspots controlling root-system development on chromosomes SBI-02 and SBI-03.
Conclusions
Sorghum genotypes with a compact, bushy and shallow root system provide potential adaptation to P scarcity in the field by allowing thorough topsoil foraging, while genotypes with an exploratory root system may be advantageous if N or water is the limiting factor, although such genotypes showed highest P uptake levels under the artificial conditions of the present study.
Keywords: Root-system architecture, root-system classification, sorghum, phosphorus scarcity, genome-wide association studies
INTRODUCTION
Sorghum is the fifth most important cereal grown worldwide as staple food and fodder. Recently, sorghum has emerged as an alternative to maize as a bioenergy crop in Central Europe due to its high biomass potential and tolerant to many biotic and abiotic stresses (Mace and Jordan, 2011; Zheng et al., 2011; Fiedler et al., 2012, 2014; Theuretzbacher et al., 2013). As a C4 crop, sorghum produces high amounts of fermentable stem sugars and biomass even when grown under adverse conditions, including drought stress (Trouche et al., 2014). However, crop growth and productivity are limited by the availability of nutrients in the soil and the availability of phosphorus (P), a key component of many essential macromolecules (Hawkesford et al., 2012; Zhang et al., 2014), is declining in agricultural soils worldwide. Intensive P fertilization is uncommon in developing countries, while high fertilizer inputs are frequent in developed countries in order to increase plant-available P at early growth stages, which may be detrimental as a long-term strategy since losses from agricultural soils lead to surface-water eutrophication. Phosphorus is highly immobile in the soil, since it is fixed in organic compounds or as Ca, Fe or Al phosphates (Shen et al., 2011). Therefore, a plant-based strategy to improve P uptake would be a seismic shift towards crops using soil P more efficiently and leading to reduced fertilizer inputs, which is crucial for sustainable cropping systems (Horst et al., 2001; Ramaekers et al., 2010; Simpson et al., 2011; Shen et al., 2013). Root mechanisms to enhance P acquisition are focused around two strategies: increasing soil exploration and mobilization of phosphate from poorly available P pools (Lynch, 2011; Richardson et al., 2011). Identification of genotypes with root traits enhancing P acquisition was suggested as a suitable strategy to achieve genetic progress instead of genotype selection based on yield in contrasting P environments (Lynch, 2011).
Root-system architecture (RSA) defines the spatial organization of primary roots and root- and stem-derived branches (lateral roots), which play a fundamental role in plant adaptation to adverse soil conditions by improving plant growth and productivity (Smith and Smet, 2012). The cereal root system is fibrous and composed of seminal roots, which appear at germination, and nodal, crown or adventitious roots, which emerge later from the shoot (Coudert et al., 2010; Singh et al., 2010). Development of the seminal root-system is largely determined by the genetic background, while post-embryonic roots, crown and brace roots produce the major portion of root biomass at the adult stage and react strongly to environmental conditions (Hochholdinger et al., 2004; Singh et al., 2010). Lateral roots have a large influence on root architecture and their function in water and nutrient uptake is essential (Lynch, 2013). Since root growth and development are highly plastic processes, guided by environmental factors and genetic components, root ecotype characterization will enable breeders to select plants with root systems for improved P uptake efficiency.
In general, an RSA with increased P uptake capacity under stress conditions enhances topsoil foraging, expands the effective depletion zones around roots and reduces the metabolic cost of soil exploration (Lynch, 2011; Richardson et al., 2011). Shallower growth angles of axial roots, enhanced adventitious rooting and greater dispersion of lateral roots improve topsoil foraging, while longer root hair and higher root hair densities increase root depletion zones (Richardson et al., 2011). Rocha et al. (2010) found that total root length and root surface area apparently were more important for adaptation to low P levels than root volume or diameter, and sorghum productivity under P scarcity was related to specific root length and tissue density, i.e. to root mass per unit root volume.
The main objectives of the present study were to characterize the RSA of a sorghum diversity set, grown in contrasting P environments and harvested at different growth stages, and to elucidate the genetic background of root-system development via marker–trait associations. Sorghum RSA was phenotyped after growth in germination paper rolls and mini-rhizotrons and 19 traits describing the rooting network were analysed. Root traits that may serve as selection criteria for breeders were identified and the 194 sorghum genotypes were classified into different rooting types. Selected genome regions might be useful for the breeding of new sorghum cultivars with improved P acquisition.
MATERIALS AND METHODS
Experiments to assess traits related to RSA
The diversity set used in the present study included 194 Sorghum bicolor and S. bicolor sudanenese genotypes. Plants were grown from April to August 2014 in a greenhouse at an air temperature of 22/18 °C (day/night) at the Faculty of Biology, University of Rostock (54°5′32″ N, 12°5′56″ E). In order to assay the complete plant root system, two different growing methods were used: (1) germination paper to assess root morphology at growth stage 12, the two-leaf stage according to the extended BBCH scale (Lancashire et al., 1991); and (2) mini-rhizotrons for RSA analysis at growth stage 16, after emergence of nodal roots.
For method (1), a sheet of Whatman® grade 1 filter paper (29 × 29 cm) was rolled to make a cigar roll. One seed per genotype was placed 2 cm below the top of each cigar roll after moistening with ddH2O. The 194 genotypes were randomly arranged in plastic boxes filled with 6 L of ddH2O. Each genotype was replicated five times using five plastic boxes. The germination paper was maintained moist by wrapping all the rolls with vinyl paper and watering the boxes daily with ddH2O, while nutrients were not applied. For germination, boxes were covered with black plastic bags for 2 d. Twelve days after sowing (DAS), plants were harvested and roots were carefully collected for further analysis.
For method (2), mini-rhizotrons were built using polystyrene square bioassay plates (24 × 24 cm, Thermo Scientific™ Nunc™) and nylon meshes with 20 µm pore size (Klein & Wieler, Königswinter, Germany) following the construction principles described by Hylander (2002). The top of each Petri plate was cut with a bandsaw and a sheet of nylon was fitted between the plate’s bottom and lid after the bottom of each mini-rhizotron had been filled with 1.6 kg of sand and fertilized with macro- and micronutrients. Rubber bands were used to keep the parts in position (Supplementary Data Fig. S1).
The sand was watered to water-holding capacity and three seeds per genotype were sown per mini-rhizotron. Seeds were placed between the lid and nylon mesh on top of the sand-filled Petri plate. The nylon mesh prevented root penetration into the sand compartment, while water, nutrients and root exudates could flow through. Mini-rhizotrons were stacked at an angle of ~70° into plastic boxes. The boxes were covered with black plastic bags during the first 2 DAS for germination. At 3 DAS, two of the three seedlings were removed from each mini-rhizotron. The experiment was conducted with two replications per treatment and genotype. Roots and shoots were harvested 27 DAS, when most plants were in the sixth-leaf stage (BBCH 16). Roots were protected from light by covering the mini-rhizotrons with black plastic.
In the mini-rhizotron experiment, plants were fertilized with a nutrient solution at the beginning of the experiment. Afterwards, plants were irrigated with ddH2O. The following initial nutrient concentrations were attained by fertilization: 140 mg N as NH3NO3, 63 mg K as KH2PO4, 120 mg Ca as CaCl2, 42 mg Mg as MgSO4, 0.5 mg B as H3BO3, 2 mg Fe as FeSO4, 0.3 mg Mn as MnSO4, 0.4 mg Zn as ZnSO4, 0.01 mg Cu as CuSO4, 0.05 mg Mo as Na2MoO4 and 58 mg S per kg dry sand. Phosphorus concentration was set to 50 or 1 mg P per kg dry sand for high and low P treatments, respectively, using KH2PO4. To equilibrate the K concentration in the low P treatment and keep the amount of N similar in both treatments, 155 mg KNO3 was added per kg of sand while the concentration of NH3NO3 was reduced to 350 mg (instead of 400 mg). With these changes the N concentration of the low P treatment was 144 mg N per kg dry sand.
Root image acquisition and analysis
Images of the collected fresh roots at BBCH stages 12 and 16 were acquired using a scanner (CanoScan LiDE 210, Canon, Krefeld, Germany). Image resolution was set to 300 dpi. A black background maximized the contrast. Image processing and phenotyping were carried out with GiA Roots (Galkovskyi et al., 2012), a free software package for RSA phenotyping. Images were scaled and cropped, a grey-scale image was created, and a double adaptive image threshold was applied using preset parameters. The thresholded images were segmented into foreground (root) and background. Pixels were counted and results were converted into centimetre values. Nineteen parameters describing the size, extent, shape and distribution of the root network were analysed (Table 1) for high and low P but only 16 traits at BBCH 12 (ERD), since maximum and median number of roots (MNR and MeNR) and network bushiness (NB) were not estimated at ERD.
Table 1.
Abbreviations, units, root trait categories and descriptions of root traits evaluated with GiA Roots (Galkovskyi et al., 2012, adapted from Topp et al., 2013)
| Trait | Abbreviation | Category | Description |
|---|---|---|---|
| Network depth | ND (cm) | Extent | Number of pixels in the vertical direction from the uppermost network pixel to the lowermost network pixel |
| Network width | NW (cm) | Extent | Number of pixels in the horizontal direction from the left-most network pixel to the right-most network pixel |
| Network convex area | NCA (cm2) | Extent | Area of the convex hull that encompasses the image |
| Major ellipse axis | MaEA (cm) | Extent | Length of the major axis of the ellipse best fitting the network |
| Minor ellipse axis | MiEA (cm) | Extent | Length of the minor axis of the ellipse best fitting the network |
| Total root length | NL (cm) | Size | Total number of pixels in the network skeleton |
| Network area | NAR (cm2) | Size | Number of network pixels in the image |
| Network perimeter | NP (cm) | Size | Total number of pixels connected to a background pixel |
| Network surface area | NSA (cm2) | Size | Sum of the local surface area at each pixel of the network skeleton, as approximated by a tubular shape whose radius is estimated from the image |
| Network volume | NV (cm3) | Size | Sum of the local volume at each pixel of the network skeleton, as approximated by a tubular shape whose radius is estimated from the image |
| Average root width | ARW (cm) | Size | Mean value of the root width estimation computed for all pixels of the medial axis of the entire root system. Corresponds to the diameter of the root |
| Network bushiness | NB | Distribution | Ratio of maximum to median number of roots |
| Network length distribution | NLD | Distribution | Fraction of the network pixel found in the lower two-thirds of the network |
| Network solidity | NS (cm2 cm−2) | Distribution | Total network area divide by the convex area |
| Maximum number of roots | MNR | Distribution | After sorting the number of roots crossing a horizontal line from smallest to largest, the maximum is considered to be the 84th percentile value |
| Median number of roots | MeNR | Distribution | Result of a vertical line sweep in which the number of roots that crossed a horizontal line was estimated, and then the median of all values for the extent of the horizontal network was calculated |
| Specific root length | SRL (cm cm−3) | Distribution | Total root length divide by network volume |
| Ellipse axis ratio | EAR (cm cm−1) | Shape | Ratio of the minor to the major axis of best-fitting ellipse |
| Network width to depth ratio | NWDR (cm cm−1) | Shape | Value of the network width divided by the value of the network depth |
Plant dry matter and P content
After image acquisition, shoots and roots were pre-dried and ground, dried at 105 °C, weighed and burnt in a muffle furnace at 550 °C. Phosphorus was extracted using 25 % HCl and measured following the methodology described by Page et al. (1982) using a Perkin Elmer Optima 8300 DV ICP-OES spectrometer (Perkin Elmer, Waltham, MA, USA). Uptake of P (PUP) was estimated as the total amount of P in the plant material and P concentration (PCONC) as mg P per g plant dry matter.
Data analyses
Analysis of variance (ANOVA), Pearson’s correlation coefficients, principal components analysis (PCA), cluster analysis and genome-wide association studies (GWAS) were carried out using R (R Core Team, 2014). RSA classification of sorghum genotypes was done following the procedure described by Bodner et al. (2013) based on principal components and computed with FactoMineR (Lê et al., 2008). Cluster analysis was carried out using the hierarchical agglomerative method implemented in the R package hclust (Murtagh and Legendre, 2014). Before PCA and cluster analysis, root trait means obtained for each genotype in each growing condition were standardized using z-scores (Kreyszig, 1979). Root phenotypes were grouped in each experiment and environment according to clusters obtained from PCA at high P.
ANOVA was carried out on root traits with the factor genotype at early development (ERD) and the factors environment and genotype at high and low P. Statistical significance was assumed at P < 0.05. Broad-sense heritability (h2) of each trait was estimated according to Hill et al. (1998) using data obtained in high and low P conditions:
where σ2G is the genotypic variance, σ2G×E is the genotype × environment interaction variance, σ2 is the error variance, n is the number of environments and r is the number of replications.
Significant marker–trait associations were identified using a two-step mixed linear model (Stich, 2008) and the R package GenABEL (Aulchenko et al., 2007). Previously, the diversity panel was fingerprinted with a single-nucleotide polymorphism (SNP) chip composed of 90 000 markers, which were used to conduct association mapping. Prior to association mapping the number of markers was reduced to 44 515 after removing those with a minor allele frequency <5 % and a missing call rate >10 % in the population. To account for population structure, a kinship matrix (identity by state) and PCA were conducted using the 44 515 SNPs. The first three principal components and genetic relatedness were used as cofactors in the model. The association study was conducted for all trait means at each growing condition. The P-value thresholds for significant and suggestive associations were adjusted using the effective number of independent markers (N) (Gao et al., 2010; Yang et al., 2014). Marker–trait associations were considered to be suggestive (1/N) at P < 5.7 × 10−5 and significant after Bonferroni adjustment (0.05/N) at P < 2.9 × 10−6. Local linkage disequilibrium (LD) was calculated in regions around significant marker–trait associations.
An interval of 100 ± 50 kb around significant marker–trait associations was used to look for candidate genes. Such an interval was considered based on global sorghum LD analysis conducted by Morris et al. (2013), who estimated a 50 % LD decay within 75–150 kb based on a diversity panel comprising 971 genotypes and 265 487 SNPs. Global LD decay to 50 % of the initial value was estimated at 108 kb in the present population. The LD decay was estimated according to Marroni et al. (2011). Candidate genes were identified using physical SNP positions in the S. bicolor reference genome (http://plants.ensembl.org/Sorghum_bicolor/Info/Index) and their putative functions were assigned using the R package biomaRt (Durinck et al., 2009).
RESULTS
P scarcity induced increased root growth in relation to largely decreased plant dry matter
Differences among genotypes were statistically significant for six out of 16 traits describing the extent, size and distribution of the sorghum root system at BBCH 12 (Table 2). ANOVA of data from BBCH 16 revealed 12 traits to be significantly different between high and low P environments and 16 traits among genotypes. Genotype–treatment interaction was significant only for the network volume (NV) (Table 2). The extent of the root system was larger at low P due to increased width (NW) and depth (ND) of the network with higher MeNR, which is an indicator for more lateral roots (Table 2). Since roots were on average thicker (ARW) at high P, NV and network surface area (NSA) were higher under high P than under low P. In consequence, specific root lengths (SRL) increased due to thinner roots at low P, making it possible to explore the soil more thoroughly. Higher root network solidity (NS) and NB indicate a more compact RSA of plants grown in high P conditions in comparison with those grown at low P. Network length distribution (NLD) was significantly different between treatments: at high P, 13.3 % of total root lengths were located in the lower two-thirds of the network (NLD), while at low P only 9.8 % were located in the lower two-thirds (Table 2). Heritability was highest for network convex area (NCA), ARW and NLD. Network bushiness and NS had lowest heritability estimates among all root traits (Table 2). Differences in PUP and PCONC were statistically not significant between genotypes; the heritability of PUP and PCONC was estimated at 0.05 and 0.03, respectively, although huge variation was found among genotypes (Supplementary Data Fig. S2).
Table 2.
Means, standard deviations and heritabilities (h2) for root system architecture at early development at BBCH 12 (ERD) and BBCH 16 under high (HP) and low (LP) P availability, total dry matter (TDM), P uptake (PUP) and P concentration (PCONC)
| Traita | Category | ERD | HP | LP | ANOVAb | h 2 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | s.d. | Gb | Mean | s.d. | Mean | s.d. | E | G | G×E | |||
| ND (cm) | Extent | 25.44 | 5.56 | ns | 25.13 | 3.07 | 25.56 | 2.27 | * | ns | ns | 0.14 |
| NW (cm) | Extent | 3.15 | 3.09 | ns | 12.35 | 4.27 | 12.98 | 3.49 | * | *** | ns | 0.28 |
| NCA (cm2) | Extent | 68.43 | 59.14 | *** | 188.95 | 84.03 | 192.90 | 67.64 | ns | *** | ns | 0.45 |
| MaEA (cm) | Extent | 27.93 | 7.50 | ** | 18.29 | 3.64 | 17.57 | 3.15 | ** | *** | ns | 0.37 |
| MiEA (cm) | Extent | 2.97 | 3.12 | ns | 10.66 | 3.63 | 10.31 | 2.93 | ns | *** | ns | 0.34 |
| NL (cm) | Size | 94.77 | 59.21 | ** | 880.68 | 471.99 | 934.64 | 341.49 | * | *** | ns | 0.30 |
| NAR (cm2) | Size | 3.34 | 1.64 | *** | 24.83 | 13.24 | 24.22 | 8.79 | ns | *** | ns | 0.35 |
| NP (cm) | Size | 189.08 | 116.07 | ** | 1542.28 | 795.03 | 1715.32 | 607.84 | *** | *** | ns | 0.33 |
| NSA (cm2) | Size | 12.40 | 6.47 | *** | 98.14 | 53.94 | 93.82 | 34.17 | ns | *** | ns | 0.34 |
| NV (cm3) | Size | 0.171 | 0.178 | ns | 1.183 | 0.743 | 0.973 | 0.411 | *** | *** | ** | 0.37 |
| ARW (cm) | Size | 0.046 | 0.012 | ns | 0.036 | 0.004 | 0.032 | 0.004 | *** | *** | ns | 0.39 |
| NB | Distribution | – | – | – | 2.456 | 1.190 | 1.896 | 0.516 | *** | ns | ns | 0.06 |
| NLD | Distribution | 1.006 | 0.594 | ** | 0.133 | 0.200 | 0.098 | 0.132 | ** | *** | ns | 0.39 |
| NS (cm2 cm−2) | Distribution | 0.073 | 0.044 | ns | 0.132 | 0.036 | 0.127 | 0.027 | * | ns | ns | 0.05 |
| MNR | Distribution | – | – | – | 37.469 | 16.352 | 38.899 | 13.647 | ns | *** | ns | 0.22 |
| MeNR | Distribution | – | – | – | 17.187 | 9.309 | 21.466 | 7.990 | *** | ** | ns | 0.14 |
| SRL (cm cm−3) | Distribution | 604.750 | 240.157 | ns | 786.470 | 179.665 | 997.648 | 242.306 | *** | *** | ns | 0.33 |
| EAR (cm cm−1) | Shape | 0.106 | 0.105 | ns | 0.592 | 0.192 | 0.597 | 0.169 | ns | *** | ns | 0.17 |
| NWDR (cm cm−1) | Shape | 0.123 | 0.132 | ns | 0.492 | 0.166 | 0.509 | 0.138 | ns | *** | ns | 0.19 |
| TDM (g) | – | – | – | – | 254.18 | 135.74 | 143.39 | 48.89 | *** | * | * | 0.32 |
| PCONC (mg g−1) | – | – | – | – | 2.65 | 2.88 | 0.62 | 0.43 | *** | ns | ns | 0.05 |
| PUP (mg) | – | – | – | – | 0.59 | 0.59 | 0.08 | 0.05 | *** | ns | ns | 0.03 |
aTrait abbreviations are explained in Table 1.
bANOVA results for genotypes (G) at ERD and environments (E), genotypes, and genotype × environment interactions (G×E) at HP and LP. *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant.
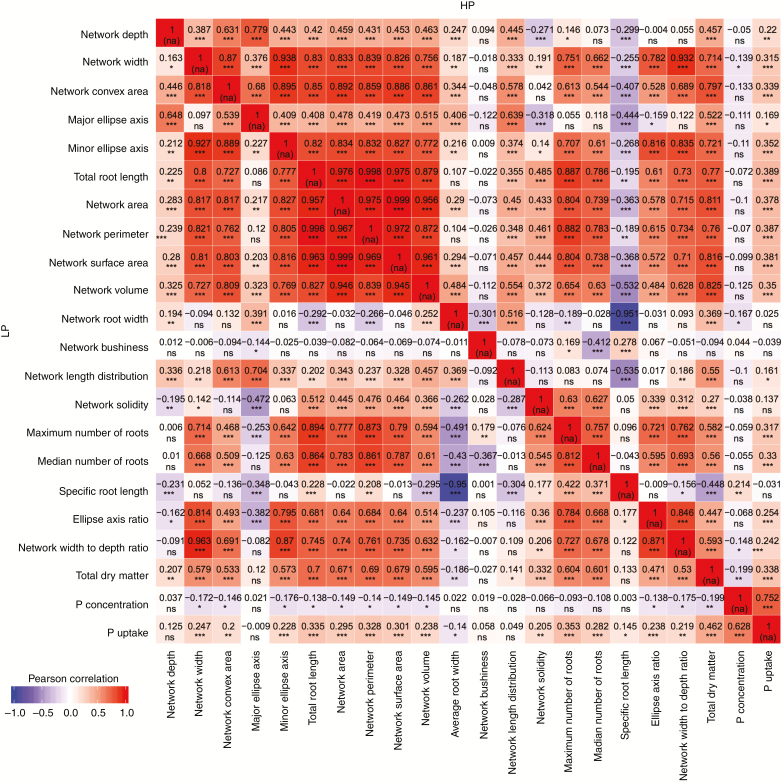
High correlations were observed for RSA data at BBCH 16 if traits belonging to the same category were compared (Fig. 1). However, root traits assessed in contrasting P environments at BBCH 16 also showed highly significant correlations for traits describing the extent [major ellipse axis (MaEA), minor ellipse axis (MiEA) and NCA] and size [NL, network area (NAR), network perimeter (NP), NSA, NV and ARW] of RSA. Pearson’s correlation coefficients were particularly low if ND was compared with other traits. Plant dry matter and PUP had slightly higher correlations with traits describing root size than with traits describing extent and distribution. PCONC was negatively correlated with nearly all RSA traits. Most of the traits describing RSA were significantly correlated among growth stages and P treatments (Supplementary Data Fig. S2). Huge variation in RSA parameters was observed across genotypes, with mostly a greater difference between growth stages than between contrasting P environments (Supplementary Data Fig. S3). Comparing RSA at BBCH 12 and under low P conditions at BBCH 16 revealed high correlation coefficients for intrinsic root size traits (NL, NAR, NSA and ARW). However, NLD was negatively correlated when early root development was compared with the later BBCH stage, but the correlation was positive when high and low P treatments at BBCH 16 were compared.
Fig. 1.
Heat map showing Person’s correlation coefficients among z-score standardized root traits assessed at high (HP) and low P (LP). *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant.
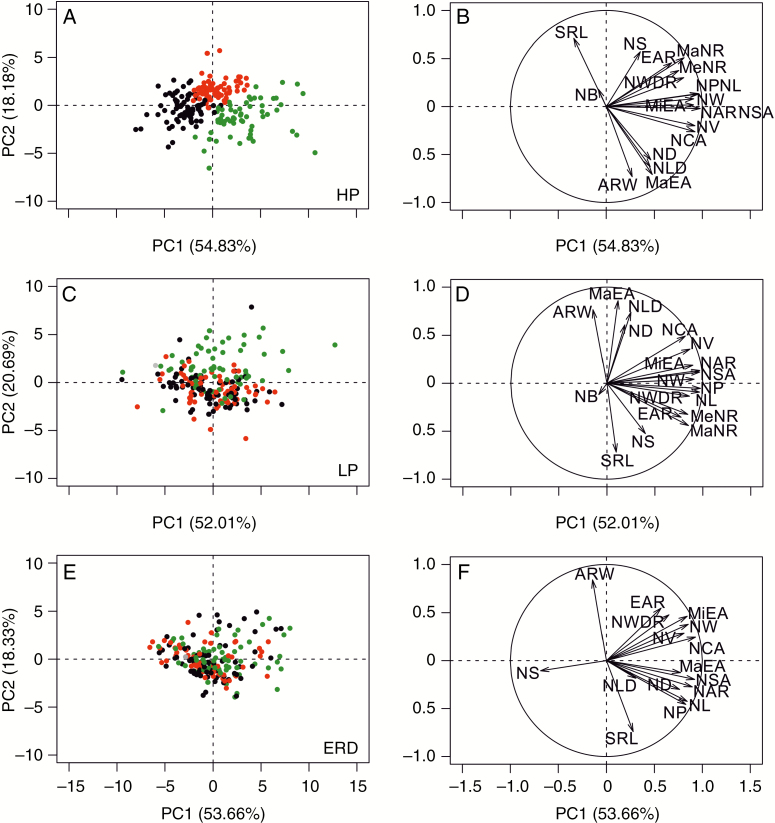
RSA classification by cluster analysis and PCA showed different rooting types at high P
After data standardization by means of z-scores, cluster analysis was carried out and root traits were merged into independent composite variables (principal components). Cluster analysis revealed three distinct rooting types (indicated by the three colours green, black and red) (Fig. 2) when plants were grown in an optimal environment (Figs 2 and 3). Under all growing conditions, two principal components (PC1 and PC2) accounted for the majority of RSA variation (>70 %, Fig. 4A, C, E), while no other component explained >10 % of RSA variation (Supplementary Data Table S1) and >50 % was explained by PC1. Network convex area showed a higher vector loading to PC1 (0.93) and ARW to PC2 (0.84, Supplementary Data Table S1). Under high P conditions, rooting type clusters were located along PC1 and differed mainly in extent and size-related traits (Fig. 4B, D, F). However, genotypes of the green cluster showed considerable variation along PC2 as well. At low P, genotypes displayed slightly higher variation along PC2 and differed mainly in ARW, SRL, MaEA, NLD and NS. Vectors describing the intrinsic root network size were in the positive PC1 direction, indicating high variation within the diversity set for NL, NAR, NV, NP and NSA. Traits describing root distribution were in the direction of PC2. Genotypes grouped in the green cluster had the largest root networks in high P but only some of them held the position in low P, while root-system size, especially for some of the black cluster genotypes, increased. Root systems of the red cluster were compact and bushy at high P; however, red cluster genotypes did not consistently form such a cluster at low P. Although several of the green cluster genotypes had the largest root systems at ERD as well, there was no clear clustering at all, nor was clustering consistent with clusters at high P.
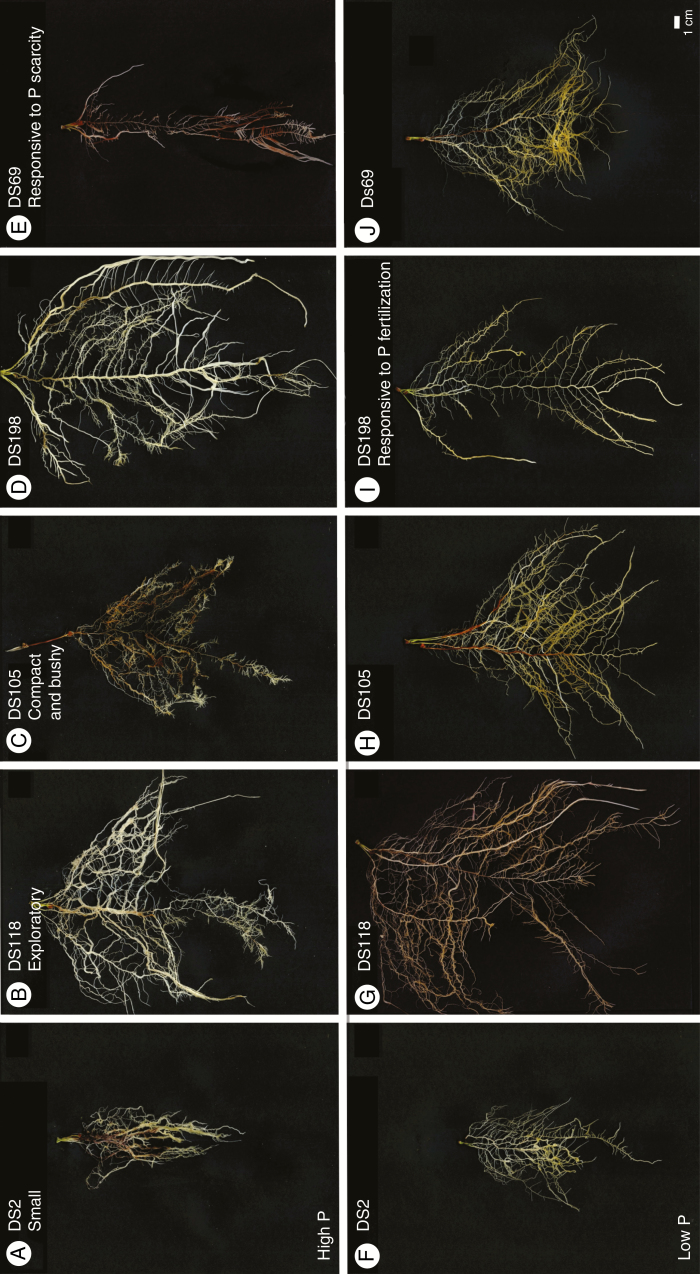
Fig. 2.
Unweighted pair group method with arithmetic means dendrogram displaying clusters with small (black), compact and bushy (red) and exploratory (green) rooting types at high P.
Fig. 3.
Typical root systems belonging to the clusters of small (A, F), exploratory (B, G) and compact and bushy (C, H) systems according to cluster analysis in high P conditions and genotypes with contrasting root system development in both environments, i.e. responsive to P fertilization (D, I) and responsive to P scarcity (E, J).
Fig. 4.
Biplots generated from PCA with the percentage of variance explained by the first two components for root traits assessed at early development (A, B), in high P (C, D) and in low P (E, F). Directional vectors (B, D, F) represent traits and points (A, C, E) represent sorghum genotypes. Colours of dots display the rooting types small (black), compact and bushy (red) and exploratory (green) as defined by cluster analysis at high P.
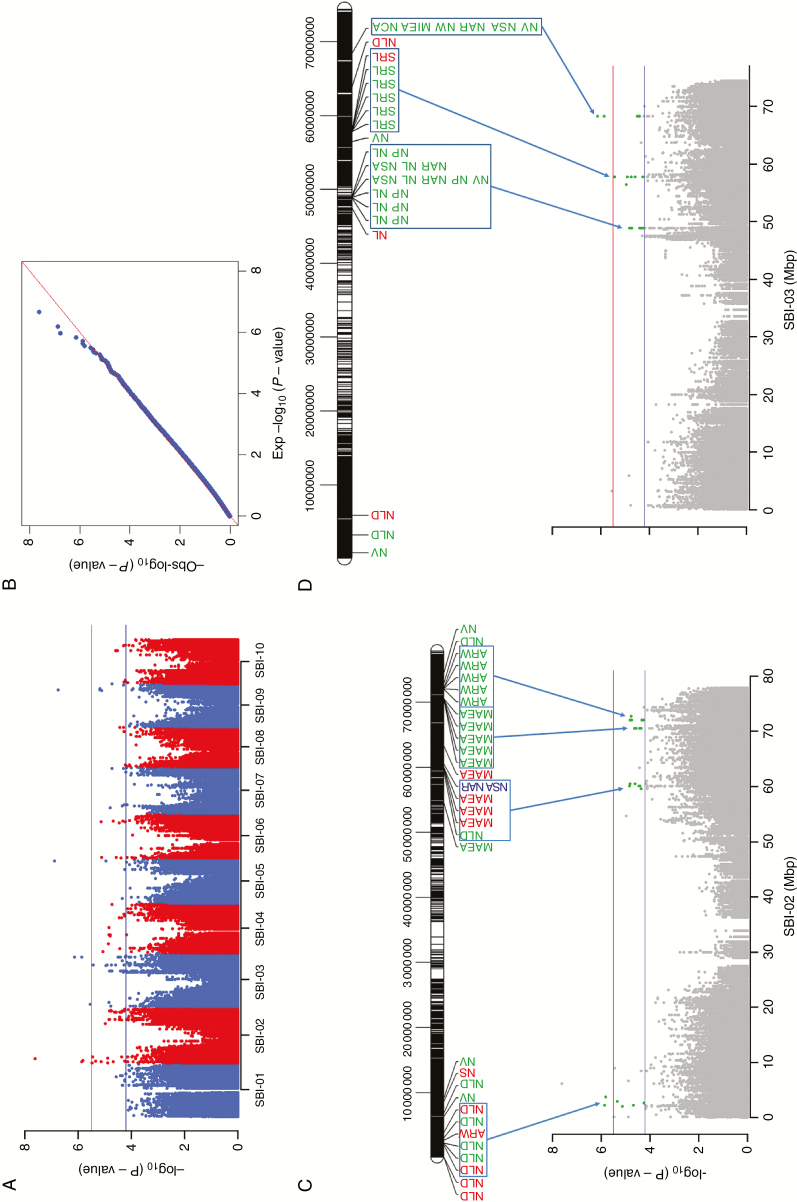
GWAS revealed quantitative trait loci for root architectural traits on four chromosomes
Genome-wide association studies were carried out separately for development stages and P environments on all 19 root traits. Eight significant associations (P < 2.9 × 10−6), seven at high and one at low P, were observed for NV, MiEA, NCA and NLD on chromosomes SBI-02, -03, -05 and -09 (Table 3, Fig. 5A). In addition, 108 suggestive marker–trait associations (P < 5.7 × 10−5) were detected on the ten sorghum chromosomes (Supplementary Data Table S2). At high P, low P and ERD, 72, 33 and 3 suggestive marker–trait associations were identified, respectively. Twelve markers showed co-localizations with more than one root trait on chromosomes SBI-01, -02, -03, -04, -07 and -08. Hotspots with significant and suggestive marker–trait associations were observed on SBI-02 and -03 (Fig. 5C, D). Network length distribution, MaEA and ARW were associated with SNPs on SBI-02, while NP, NL, NV and SRL were associated with SNPs on SBI-03. A co-localization of suggestive and significant quantitative trait loci (QTL) for the six traits NAR, NP, NSA, NCA, NV and MiEA was found on SBI-03 at 68.3 Mbp. Local LD investigated for significant marker–trait associations revealed that the marker associated with NLD at 2.2 Mbp on SBI-02 is in LD with two SNPs associated with MaEA and NLD (Supplementary Data Fig. S4). Two markers associated with NLD are in LD at 51.1 Mbp on SBI-09.
Table 3.
Significant marker–trait associations for the traits network lengths distribution (NLD), network volume (NV), network convex area (NCA) and minor eclipse axis (MiEA)
| Marker–trait association | Nearest gene | ||||||
|---|---|---|---|---|---|---|---|
| Trait | Treatment | Chromosome | Position (bp) | P-value | ID | Start position (bp) | Description |
| NLD | HP | SBI-02 | 2 209 409 | 1.35E-06 | Sb02g002180 | 2 205 025 | Uncharacterized |
| NLD | LP | SBI-02 | 3 725 908 | 1.56E-06 | Sb02g003260 | 3 724 427 | Armadillo-type fold, Fanconi anaemia protein FANCD2 |
| NV | HP | SBI-02 | 6 171 759 | 2.46E-08 | Sb02g005290 | 6 174 218 | Uncharacterized |
| NLD | HP | SBI-03 | 3 201 076 | 2.86E-06 | Sb03g003110 | 3 207 828 | Zinc finger, C3H-type transcription factor |
| NCA | HP | SBI-03 | 68 303 195 | 7.40E-07 | Sb03g040800 | 68 297 624 | Serine/threonine-protein kinase, active site |
| MiEA | HP | SBI-03 | 68 303 195 | 1.36E-06 | Sb03g040800 | 68 297 624 | Serine/threonine-protein kinase, active site |
| NLD | HP | SBI-05 | 58 660 907 | 1.34E-07 | Sb05g025360 | 58 660 524 | Protein of unknown function DUF594 |
| NLD | HP | SBI-09 | 51 109 156 | 1.76E-07 | Sb09g021620 | 51 107 666 | Fructose-1,6-bisphosphatase class 1/ Sedoheputulose-1,7-bisphosphatase |
Fig. 5.
(A) Manhattan plot showing P-values and suggestive and significant marker–trait associations for all traits. Horizontal red and blue lines indicate thresholds for significant marker–trait associations after Bonferroni correction (P < 2.9 × 10−6) and suggestive associations (P < 5.7 × 10−5). (B) QQ plot. (C, D) QTL hotspots for the traits network length distribution (NLD), average root width (ARW), network volume (NV), network solidity (NS), major eclipse axis (MaEA), network surface area (NSA), network area (NAR), total root length (NL), network perimeter (NP), specific root length (SRL), minor eclipse axis (MiEA) and network convex area (NCA) on SBI-02 (C) and SBI-03 (D) at early development (blue), high (green) and low P (red).
Annotation analysis was conducted for significant marker–trait associations ±50 kbp. In total, 77 different transcripts near the seven QTL positions were found, and hint that candidate genes may putatively function at different biological and molecular processes (Supplementary Data Table S3). Markers at 3.7 Mbp on SBI-02, 58.6 Mbp on SBI-05 and 51.1 Mbp on SBI-09 were located in the coding sequences of three different genes (Table 3). Another five transcripts were in LD with the SNP at 51.1 Mbp on SBI-09 (Table S3). Annotation analysis showed that genes may function in DNA repair, sugar metabolism and transport and as inorganic pyrophosphatase (Table 3 and Supplementary Data Table S3). Particular attention was given to genes encoding transcription factors (TFs), transcriptional regulators (TRs) and protein kinases (PKs) found in the vicinity (±50 kbp) of significant marker–trait associations. Seven putative TFs and three putative PKs were identified by the annotation analysis on chromosomes SBI-03 and -05. The TFs belong to the C3H, MYB and PHD classes while the PKs belong to the Aurora and NEK domain classes. Based on the gene ontology output, two genes, Sb03g040800 and Sb05g025390, are potentially related to root development. The first corresponds to the nearest gene to the marker associated with NCA and MiEA on SBI-03, while the latter is 14.33 kbp from the SNP associated with NLD on SBI-05. Additionally, Sb03g040830, a gene that may encode a glucosyl transferase involved in cell wall biosynthesis, was identified in the vicinity of a QTL for NCA and MiEA on SBI-03.
DISCUSSION
High variability in RSA may aid in improving adaptability of sorghum to P scarcity
In order to ameliorate P efficiency in cropping systems and reduce environmental side effects of intensive fertilization practices, research has focused on root traits that may improve P acquisition (Gahoonia and Nielsen, 2004; Yan et al., 2004; Lambers et al., 2006; Ramaekers et al., 2010; De Smet et al., 2012). In this study, a huge variability in RSA, illustrating the high adaptation capability of sorghum to certain stress conditions, was observed within the diversity set. Similarly to the present study, Chen et al. (2011) found evidence for large root-system variability in wild lupins, which possibly benefits the breeding of cultivars adapted to abiotic stress. Adu et al. (2014) found that variation coefficients for root traits in oilseed rape (Brassica rapa) seedlings ranged from 5.8 to 83.2 % when high-throughput phenotyping experiments were conducted. A root-system classification, as performed in the present study by assigning genotypes to distinct RSA phenotype clusters, might be helpful in identifying potentially efficient genotypes. Subsequently, these can be selected and introduced to breeding programmes aiming at the development of cultivars with improved P efficiency. Multivariate approaches were shown to be useful tools for capturing the development of the whole root system (Bodner et al., 2013) in order to clarify root network behaviour in, and its interaction with, the environment and to define groups of contrasting phenotypes potentially adapted to specific environmental scenarios.
One of the main mechanisms by which sorghum plants cope with P scarcity was to stabilize the number and length of lateral roots despite reduced overall growth, which led to slightly increased network width and depth. In favour to develop root systems that occupy a large space, bushiness and solidity of root networks were reduced under low P conditions. Under field conditions, deep-rooting networks may not always be beneficial since P is relatively immobile in the soil, with higher amounts in the topsoil resulting from fertilization practices. It was shown that crops such as maize (Zea mays), rice (Oryza sativa), common bean (Phaseolus vulgaris), white lupin (Lupinus albus), tomato (Solanum lycopersicum) and also arabidopsis reduce lengths of primary roots and increase amounts and lengths of lateral roots if grown at low P, while root growth might be more directed towards increasing network width when P is mainly available from topsoil layers in the field (Borch et al., 1999; Mollier and Pellerin, 1999; Shimizu et al., 2004; Kim et al., 2008; Lambers et al., 2011; Jin et al., 2012; Niu et al., 2012).
Root-system classification reveals distinct rooting types
Apart from general rooting network characteristics, three rooting types were identified in the present study at high P: firstly, a small, shallow and narrow root system with few and short lateral roots, occupying only a small fraction of the available space; secondly, an exploratory root system, which shows the deepest and widest network but less bushiness and solidity and comprises long lateral roots; and thirdly, a compact and bushy root system composed of relatively short lateral roots, which thoroughly explores the upper available space.
The architecture of the compact and bushy root system perfectly matches ideotype descriptions to maximize P uptake if plants are grown on P-poor soils (Lynch, 2011; Richardson et al., 2011). However, plants with compact root systems may have limited access to water and N in deeper soil layers (Zurek et al., 2015). In contrast to P, N mobility in the soil is very high. Since nitrate is diluted in soil water, deep percolation of water, e.g. as a consequence of erratic rainfall events, requires deep rooting to improve N uptake efficiency. Exploratory root systems were suggested to be potentially better adapted to drought as well as to N scarcity, since such root networks may reach deep soil layers better and earlier during crop development (Lynch, 2013; Trachsel et al., 2013; Borrell et al., 2014). Since bushiness was significantly reduced under low P conditions in the present study, the main strategy to cope with P scarcity may have been to explore as much space as possible. However, the situation in mini-rhizotrons is different from that in the field since P can be assumed to be evenly distributed without elevated concentrations in topsoil layers, as is typical for field conditions.
Multivariate analysis may allow identification of stress-adapted phenotypes
Higher genotypic variation was observed at low P in the present study, which held true especially for traits describing root-system distribution. Among these traits, network bushiness and network solidity had low heritabilities, indicating that P availability had a large effect on the traits. However, comparatively low heritabilities were observed for many of the described traits, which may result in part from strongly contrasting phenotyping environments. Root system characteristics of genotypes DS198 and DS69 (Fig. 3) represent extremes with different adaptation processes to P scarcity: while DS198 reduced root system size at low P, DS69 adapted by developing a shallow and bushy rooting type, which was considered to be advantageous at limited P availability soils (Lynch, 2011; Richardson et al., 2011). We suggest that univariate analysis of root systems, e.g. by measuring a few simple traits like total root lengths and root dry matter, is less straightforward as a means of selecting ideotypes for specific environments if P or other environmental constraints are the limiting factors of cultivation. Moreover, a complex analysis of rooting types may help to analyse the plasticity of root networks in order to identify genotypes ideally adapted to different environmental conditions. An ideotype with high plasticity may develop bushy networks in P-scarce environments for optimized topsoil foraging, while explorative networks with increased rooting depths are developed if N or soil water are the limiting factors and not too much energy is spent in root-system development if soil water and nutrient status are optimal. Contrasting root phenotypes adapted only to e.g. N or P scarcity may develop identical root lengths in different situations, which makes breeding strategies difficult if total root lengths, root dry weight and specific root lengths are the only selection criteria. However, correlations between network bushiness and P uptake were statistically not significant in the present study. Highest correlations were observed between P uptake in the low P conditions and total root lengths and maximum number of roots, which enabled a thorough exploration of the available space and thus may have maximized P uptake in the artificial rhizotron environment. Similar results were found for rice genotypes, showing higher P uptake when plants developed larger root systems (Wissuwa and Ae, 2001).
Exploratory RSAs are evident only in part at early growth stages
Since root phenotyping is a time-consuming and complex task at the adult stage, early assessment of root traits might enable the identification of genotypes with root systems matching desired traits for improving P uptake from P-poor soils. Traits explaining the intrinsic size and extension of the root system such as NL, NAR and NCA showed stronger positive correlations under different growing conditions, suggesting that these seedling root traits may be used to predict root characteristics at later developmental stages. In concordance with the present study, a positive correlation in root dry weight was observed between maize seedlings and adult plants grown in high and low N conditions (Abdel-Ghani et al., 2013). In a recent study, it was found that oilseed rape root traits at the seedling stage positively correlated with crop vigour and seed yield (Thomas et al., 2016). However, although several studies suggest that seedling root traits might be used to predict adult root characteristics (cf. Pace et al., 2015), in the present study only a few genotypes showed similar RSAs during early development and later in the high and low P environments (Fig. 4A, C, E). In particular, genotypes with large root-system sizes under any environmental condition might be interesting for breeders if nutritional or drought stress are random but frequently occurring factors since plants are well prepared if a stressor becomes evident.
For crop breeding programmes, other attractive targets are root hair densities and lengths, which have large variation between different species and cultivars (Datta et al., 2011). Root hairs play a crucial role in P acquisition by increasing the absorption area of the root network and it was suggested that they are under simple genetic control (Jungk, 2001; Lynch, 2011; Richardson et al., 2011). Experiments conducted with soybean showed that P uptake efficiency was largely related to root hair length and density (Vandamme et al., 2013). Similar results were obtained in beans, when P uptake was analysed in the field and was shown to be positively correlated with basal root-hair length and density (Yan et al., 2004). Therefore, an additional characterization of root hairs in our sorghum diversity panel might be useful to identify genotypes with improved P uptake from P-scarce soils.
Genes involved in signal transduction pathways are close to QTL for RSA in sorghum
Previous studies on sorghum root traits showed a suggestive QTL for root dry matter on SBI-02, identified in a biparental cross (Mace et al., 2012). Two significant associations close to the cited QTL were detected for network length distribution in the present study. Bekele et al. (2013) found several QTL for total root length and root fresh and dry weights in a recombinant inbred population when genotypes were grown at chilling temperatures, and suggested hotspots with QTL co-localizations observed under different growing conditions on SBI-01, -04 and -06. After annotation analysis, they identified numerous genes within the hotspots encoding TFs and PKs.
Annotation analysis showed that the transcripts Sb09g021620, Sb09g021630 and Sb09g021645, which were in LD with a significant marker–trait association on SBI-09, encode proteins related to sugar metabolism and transport. In plants, sucrose may function as a short- and long-distance signalling molecule and plays, together with auxins and cytokinins, a crucial role in growth and development of roots and shoots (Ljung et al., 2015). The transcript Sb09g021610, located in the same chromosomal region, encodes a pyrophosphatase. In arabidopsis, rice and tomato, overexpression of the proton-pyrophosphatase AVP1DOX resulted in increased root mass, root branching and P uptake (Yang et al., 2007).
In the present study, annotation analysis was focused on TFs, TRs and PKs since these proteins regulate signal transduction pathways and the expression of target genes and alter protein activity (Liu et al., 2013; Jin et al., 2014; Nakashima et al., 2014; Stelpflug et al., 2016; Zheng et al., 2016). Observed TFs belong to the classes MYB, C3H and PHD (Jin et al., 2014, 2015). Sb03g003100, a gene putatively encoding an MYB TF, is located close to the QTL for NLD at 3.2 Mbp on SBI-03. Expression of MYB-class TFs was observed during root development from primary root until lateral root formation and they were shown to be part of a complex regulatory network involved in the formation of root hairs in arabidopsis (Montiel et al., 2004). It was observed that expression of MYB TFs is induced under P deficiency in seedling roots of both maize and arabidopsis (Osmont et al., 2007; Lin et al., 2013).
The putative PKs Sb03g003130, Sb03g003140 and Sb03g04800 were identified near SNPs significantly associated with root traits at 3.20 and 68.3 Mbp on SBI-03. In rice, the PK PSTOL1, a serine/threonine kinase, confers tolerance of P deficiency. Transgenic rice seedlings overexpressing PSTOL1 showed higher total root length and surface area when grown in both high and low P conditions (Gumayao et al., 2012). Our annotation analysis of the PKs Sb03g003130 and Sb03g04800 shows that they may contain serine/threonine domains.
Alteration of the expression of genes involved in cell wall remodelling is an early transcriptional response associated with root development and sensing of inorganic P (Zhang et al., 2014). The putative glucosyl transferase Sb003g040830 involved in the polysaccharide xyloglucan biogenesis of the cell wall (Pauly and Keegstra, 2016) was located near an SNP significantly associated with NCA and MiEA on SBI-03.
Further investigations focusing on transcriptome analysis using sorghum genotypes with contrasting RSAs under high and low P treatments could provide complementary data to elucidate the genetic basis of root development and the signal transduction network regulating adaptation mechanisms. Sequencing of candidate genes to analyse allele diversity and carrying out regional association studies may enable the development of stable markers for marker-assisted selection.
Different types of root networks potentially adapted to environmental constraints were observed in the present study. Of particular interest were genotypes with a compact, bushy and shallow root system, which provides potential adaptation to P scarcity in the field by allowing thorough topsoil foraging. Genotypes with an exploratory root system at both high and limited P availability, which may be advantageous if N or water is the limiting factor, showed the highest P uptake levels under the artificial conditions of the present study. Traits such as total root length, network area and network surface area can be assessed in seedlings in order to predict performance of the root system at later development stages.
The most interesting genome regions influencing root system development were found on SBI-02 and SBI-03. Among the genes located near significant QTL on SBI-03 were MYB TFs, PKs and a glucosyl transferase, which were suggested by earlier studies to be involved in the regulation of root development.
SUPPLEMENTARY DATA
Supplementary data are available at https://academic.oup.com/aob and consist of the following. Table S1: percentage of variance explained and vector loadings for the first three principal components. Table S2: list of all significant (P < 2.9 × 10−6) and suggestive (P < 5.7 × 10−5) marker–trait associations. Table S3: list of all genes located near significant QTL positions (±50 kbp). Figure S1: (A) sand-filled mini-rhizotrons with sorghum seedlings, sown between the front of the rhizotron and a nylon mesh to prevent roots growing into the sand; (B) filter paper rolls used for root phenotyping at early development. Figure S2: heat map showing Pearson’s colour-coded correlation coefficients between the growing conditions early development (ERD), high P (HP, 50 mg P per kg soil) and low P (LP, 1 mg P per kg soil). *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant. Figure S3: frequency distributions of the evaluated root traits, P uptake, P concentration and total dry matter. Vertical dotted lines indicate the mean of the trait in each growing condition. Figure S4: local LD for significant and suggestive marker–trait associations on chromosomes SBI-02 (A, B, C), SBI-03 (D, E), SBI-05 (F) and SBI-09 (G). Horizontal lines show P-value thresholds for suggestive marker–trait associations (red, P < 5.7 × 10−5), marker–trait associations after Bonferroni correction (blue, P < 2.9 × 10−6) and P < 0.001 (black).
ACKNOWLEDGEMENTS
The authors gratefully acknowledge financial support from ERA-net Bioenergy and the Fachagentur Nachwachsende Rohstoffe e.V. (FKZ 22001213). Further support was given by the Leibniz Science Campus Phosphorus Research Rostock.
LITERATURE CITED
- Abdel-Ghani AH, Kumar B, Reyes-Matamoros J et al. 2013. Genotypic variation and relationships between seedling and adult plant traits in maize (Zea mays L.) inbred lines grown under contrasting nitrogen levels. Euphytica 189: 123–133. [Google Scholar]
- Adu MO, Chatot A, Wiesel L et al. 2014. A scanner system for high-resolution quantification of variation in root growth dynamics of Brassica rapa genotypes. Journal of Experimental Botany 65: 2039–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. 2007. GenABEL: an R library for genome-wide association analysis. Bioinformatics 23: 1294–1296. [DOI] [PubMed] [Google Scholar]
- Bekele WA, Wieckhorst S, Friedt W, Snowdon RJ. 2013. High-throughput genomics in sorghum: from whole-genome resequencing to a SNP screening array. Plant Biotechnology Journal 11: 1112–1125. [DOI] [PubMed] [Google Scholar]
- Borch K, Bouma TJ, Lynch JP, Brown KM. 1999. Ethylene: a regulator of root architectural responses to soil phosphorus availability. Plant, Cell & Environment 22: 425–431. [Google Scholar]
- Bodner G, Leitner D, Nakhforoosh A, Sobotik M, Moder K, Kaul HP. 2013. A statistical approach to root system classification. Frontiers in Plant Science 4: 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrell AK, Mullet JE, George-Jaeggli B et al. 2014. Drought adaptation of stay-green sorghum is associated with canopy development, leaf anatomy, root growth and water uptake. Journal of Experimental Botany 62: 6251–6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YL, Dunbabin V, Diggle AJ, Siddique KHM, Rengel Z. 2011. Assessing variability in root traits of wild Lupinus angustifolius germplasm: basis for modelling root system structure. Plant and Soil 354: 141–155. [Google Scholar]
- Coudert Y, Périn C, Courtois B, Khong NG, Gantet P. 2010. Genetic control of root development in rice, the model cereal. Trends in Plant Science 15: 219–226. [DOI] [PubMed] [Google Scholar]
- Datta S, Kim CM, Pernas M et al. 2011. Root hairs: development, growth and evolution at the plant-soil interface. Plant and Soil 346: 1–14. [Google Scholar]
- Durinck S, Spellman P, Birney E, Huber W. 2009. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nature Protocols 4: 1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler K, Bekele WA, Friedt W et al. 2012. Genetic dissection of the temperature dependent emergence processes in sorghum using a cumulative emergence model and stability parameters. Theoretical and Applied Genetics 125: 1647–1661. [DOI] [PubMed] [Google Scholar]
- Fiedler K, Bekele WA, Duensing R et al. 2014. Genetic dissection of temperature-dependent sorghum growth during juvenile development. Theoretical and Applied Genetics 127: 1935–1948. [DOI] [PubMed] [Google Scholar]
- Gahoonia TS, Nielsen NE. 2004. Root traits as tools for creating phosphorus efficient crop varieties. Plant and Soil 260: 47–57. [Google Scholar]
- Galkovskyi T, Mileyko Y, Bucksch A et al. 2012. GiA Roots: software for the high-throughput analysis of plant root system architecture. BMC Plant Biology 12: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Becker LC, Becker DM, Starmer JD, Province MA. 2010. Avoiding the high Bonferroni penalty in genome-wide association studies. Genetic Epidemiology 34: 100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumayao R, Chin JH, Pariasca-Tanaka J et al. 2012. The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature 488: 535–539. [DOI] [PubMed] [Google Scholar]
- Hawkesford M, Horst W, Kichey T et al. 2012. Functions of macronutrients. In: Marschner P. ed. Marschner’s mineral nutrition of higher plants, 3rd edn London: Academic Press, 135–189. [Google Scholar]
- Hill J, Becker HC, Tigerstedt PMA. 1998. Quantitative and ecological aspects of plant breeding. London: Chapman and Hall. [Google Scholar]
- Hochholdinger F, Woll K, Sauer M, Dembinsky D. 2004. Genetic dissection of root formation in maize (Zea mays) reveals root-type specific developmental programmes. Annals of Botany 93: 359–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst WJ, Kamh M, Jibirin JM, Chude VO. 2001. Agronomic measures for increasing P availability to crops. Plant and Soil 237: 211–223. [Google Scholar]
- Hylander LD. 2002Improvements of rhizoboxes used for studies of soil-root interactions. Communications in Soil Science and Plant Analysis 33: 155–161. [Google Scholar]
- Jin J, Tang CX, Amstrong R, Sale P. 2012. Phosphorus supply enhances the response of legumes to elevated CO2 (FACE) in phosphorus-deficient vertisol. Plant and Soil 358: 86–99. [Google Scholar]
- Jin JP, Zhang H, Kong L, Gao G, Luo JC. 2014. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Research 42: D1182–D1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin JP, He K, Tang X et al. 2015. An Arabidopsis transcriptional regulatory map reveals distinct functional and evolutionary features of novel transcription factors. Molecular Biology and Evolution 32: 1767–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungk A. 2001. Root hairs and the acquisition of plants nutrients from soil. Journal of Plant Nutrition and Soil Science 164: 121–129. [Google Scholar]
- Kim HJ, Lynch JP, Brown KM. 2008. Ethylene insensitivity impedes a subset of responses to phosphorus deficiency in tomato and petunia. Plant, Cell & Environment 31: 1744–1755. [DOI] [PubMed] [Google Scholar]
- Kreyszig E. 1979. Advanced engineering mathematics, 4th edn Rostock: John Wiley & Sons. [Google Scholar]
- Lambers H, Shane MW, Cramer MD, Pearse SJ, Veneklaas EJ. 2006. Root structure and functioning for efficient acquisition of phosphorus: matching morphological and physiological traits. Annals of Botany 98: 693–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambers H, Finnegan PM, Laliberté E et al. 2011. Phosphorus nutrition of Proteaceae in severely phosphorus-impoverished soils: are there lessons to be learned for future crops?Plant Physiology 156: 1058–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancashire P, Bleiholder H, Boom T et al. 1991. A uniform decimal code for growth stages of crops and weeds. Annals of Applied Biology 119: 561–601. [Google Scholar]
- Lê S, Josse J, Husson F. 2008. FactoMineR: an R package for multivariate analysis. Journal of Statistical Software 25: 1–18. [Google Scholar]
- Lin HJ, Gao J, Zhang ZM et al. 2013. Transcriptional response of maize seedling root to phosphorus starvation. Molecular Biology Reports 40: 5359–5379. [DOI] [PubMed] [Google Scholar]
- Liu S, Wang X, Wang H et al. 2013. Genome-wide analysis of ZmDREB genes and their association with natural variation in drought tolerance at seedling stage of Zea mays L. PLoS Genetics 9: e1003790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung K, Nemhauser JL, Perata P. 2015. New mechanistic links between sugar and hormone signalling networks. Current Opinion in Plant Biology 25: 130–137. [DOI] [PubMed] [Google Scholar]
- Lynch JP. 2011. Root phenes for enhanced soil exploration and phosphorus acquisition: tools for future crops. Plant Physiology 156: 1041–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JP. 2013. Steep, cheap and deep: an ideotype to optimize water and N acquisition by maize root systems. Annals of Botany 112: 347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace ES, Jordan DR. 2011. Integrating sorghum whole genome sequence information with a compendium of sorghum QTL studies reveals uneven distribution of QTL and of gene-rich regions with significant implications for crop improvement. Theoretical and Applied Genetics 123: 169–191. [DOI] [PubMed] [Google Scholar]
- Mace ES, Singh V, van Oosterom EJ, Hammer GL, Hunt CH, Jordan DR. 2012. QTL for nodal root angle in sorghum (Sorghum bicolor L. Moench) co-locate with QTL for traits associated with drought adaptation. Theoretical and Applied Genetics 124: 97–109. [DOI] [PubMed] [Google Scholar]
- Marroni F, Pinosio S, Zaina G et al. 2011. Nucleotide diversity and linkage disequilibrium in Populus nigra cinnamyl alcohol dehydrogenase (CAD4) gene. Tree Genetics & Genomes 7: 1011–1023. [Google Scholar]
- Mollier A, Pellerin S. 1999. Maize root system growth and development as influenced by phosphorus deficiency. Journal of Experimental Botany 50: 487–497. [Google Scholar]
- Montiel G, Gantet P, Jay-Allemand C, Breton C. 2004. Transcription factor networks. Pathways to the knowledge of root development. Plant Physiology 136: 3478–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GP, Ramu P, Deshpande SP et al. 2013. Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proceedings of the National Academy of Sciences of the USA 110: 453–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtagh F, Legendre P. 2014. Ward’s hierarchical agglomerative clustering method: which algorithms implement Ward’s criterion?Journal of Classification 31: 274–195. [Google Scholar]
- Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K. 2014. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold and heat. Frontiers in Plant Science 5: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu YF, Chai RS, Jin GL, Wang H, Tang CX, Zhang YS. 2012. Responses of root architecture development to low phosphorus availability: a review. Annals of Botany 112: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmont KS, Sibout R, Hardtke CS. 2007. Hidden branches: developments in root system architecture. Annual Review of Plant Biology 58: 93–113. [DOI] [PubMed] [Google Scholar]
- Pace J, Gardener C, Romay C, Ganapathysubramanian B, Lübberstedt T. 2015. Genome-wide association analysis of seedling root development in maize (Zea mays L.). BMC Genomics 16: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page AL, Miller RH, Kenney DR. 1982. Methods of soil analysis. Part 2. Chemical and microbial properties.Madison WI: American Society of Agronomy, Soil Science Society of America. [Google Scholar]
- Pauly M, Keegstra K. 2016. Biosynthesis of the plant cell wall matrix polysaccharide xyloglucan. Annual Review of Plant Biology 67: 235–259. [DOI] [PubMed] [Google Scholar]
- R Core Team 2014. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; http://www.R-project.org. [Google Scholar]
- Ramaekers L, Remans R, Rao IM, Blair MW, Vanderleyden J. 2010. Strategies for improving phosphorus acquisition efficiency of crop plants. Field Crops Research 117: 169–176. [Google Scholar]
- Richardson AE, Lynch JP, Ryan PR et al. 2011. Plant and microbial strategies to improve the phosphorus efficiency of agriculture. Plant and Soil 349: 121–156. [Google Scholar]
- Rocha MC, Miranda GV, Vasconcelos MJV et al. 2010. Characterization of root morphology in contrasting genotypes of sorghum at low and high phosphorus level. Revista Brasileira de Milho e Sorgo 9: 65–78. [Google Scholar]
- Shen J, Yuan L, Zhang J et al. 2011. Phosphorus dynamics: from soil to plant. Plant Physiology 156: 997–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Li C, Mi G et al. 2013. Maximizing root/rhizosphere efficiency to improve crop productivity and nutrient use efficiency in intensive agriculture of China. Journal of Experimental Botany 64: 1181–1192. [DOI] [PubMed] [Google Scholar]
- Singh V, Van Oosterom EJ, Jordan DR, Messina CD, Cooper M, Hammer GL. 2010. Morphological and architectural development of root systems in sorghum and maize. Plant and Soil 333: 287–299. [Google Scholar]
- Simpson RJ, Oberson A, Culvenor RA et al. 2011. Strategies and agronomic interventions to improve the phosphorus-use efficiency of farming systems. Plant and Soil 349: 89–120. [Google Scholar]
- Shimizu A, Yanagihara S, Kawasaki S, Ikehashi H. 2004. Phosphorus deficiency-induced root elongation and its QTL in rice (Oryza sativa L.). Theoretical and Applied Genetics 109: 1361–1368. [DOI] [PubMed] [Google Scholar]
- De Smet I, White PJ, Bengough AG et al. 2012. Analyzing lateral root development: how to move forward. Plant and Cell 24: 15–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, De Smet I. 2012. Root system architecture: insights from Arabidopsis and cereal crops. Philosophical Transactions of the Royal Society of London, Series B 367: 1441–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelpflug SC, Sekhon RS, Vaillancourt B et al. 2016. An expanded maize gene expression atlas based on RNA sequencing and its use to explore root development. Plant Genome 9: 1–16. [DOI] [PubMed] [Google Scholar]
- Stich B, Möhring J, Piepho HP, Heckenberger M, Buckler ES, Melchinger AE. 2008. Comparison of mixed-model approaches for association mapping. Genetics 178: 1745–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theuretzbacher F, Bauer A, Lizasoain J et al. 2013. Potential of different Sorghum bicolor (L. moench) varieties for combined ethanol and biogas production in the Pannonian climate of Austria. Energy 55: 107–113. [Google Scholar]
- Topp CN, Iyer-Pascuzzi AS, Anderson JT et al. 2013. 3D phenotyping and quantitative trait locus mapping identify core regions of the rice genome controlling root architecture. Proceedings of the National Academy of Sciences of the USA 110: E1695–E1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CL, Graham NS, Hayden R et al. 2016. High-throughput phenotyping (HTP) identifies seedling root trait linked to variation in seed yield and nutrient capture in field-grown oilseed rape (Brassica napus L.). Annals of Botany 118: 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachsel S, Kaeppler SM, Brown KM, Lynch JP. 2013. Maize root growth angles become steeper under low N conditions. Field Crops Research 140: 18–31. [Google Scholar]
- Trouche G, Bastianelli D, Hamadou TVC, Chantereau J, Rami JF, Pot D. 2014. Exploring the variability of photoperiod-insensitive sorghum genetic panel for stem composition and related traits in temperate environments. Field Crops Research 166: 72–81. [Google Scholar]
- Vandamme E, Renkens M, Pypers P, Smolders E, Vanluawe B, Merckx R. 2013. Root hairs explain P uptake efficiency of soybean genotypes grown in a P-deficient Ferralsol. Plant and Soil 369: 269–282. [Google Scholar]
- Wissuwa M, Ae N. 2001. Genotypic variation for tolerance to phosphorus deficiency in rice and the potential for its exploitation in rice improvement. Plant Breeding 120: 43–48. [Google Scholar]
- Yan X, Liao H, Beebe SE, Blair MW, Lynch JP. 2004. QTL mapping of root hair and acid exudation traits and their relationship to phosphorus uptake in common bean. Plant and Soil 265: 17–29. [Google Scholar]
- Yang H, Knapp J, Koirala P et al. 2007. Enhanced phosphorus nutrition in monocots and dicots over-expressing a phosphorus responsive type I H+-pyrophosphatase. Plant Biotechnology Journal 5: 735–745. [DOI] [PubMed] [Google Scholar]
- Yang W, Guo Z, Huang C et al. 2014. Combining high-throughput phenotyping and genome-wide association studies to reveal natural genetic variation in rice. Nature Communications 5: 5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Liao H, Lucas WJ. 2014. Molecular mechanism underlying phosphate sensing, signaling, and adaptation in plants. Journal of Integrative Plant Biology 56: 192–220. [DOI] [PubMed] [Google Scholar]
- Zheng LY, Guo XS, He B et al. 2011. Genome-wide patterns of genetic variation in sweet and grain sorghum (Sorghum bicolor). Genome Biology 12: R114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Jiao C, Sun H et al. 2016. iTAK: a program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators and protein kinases. Molecular Plant 9: 1667–1670. [DOI] [PubMed] [Google Scholar]
- Zurek PR, Topp CN, Benfey PN. 2015. Quantitative trait locus mapping reveals regions of the maize genome controlling root system architecture. Plant Physiology 167: 1487–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.