Synopsis
Objectives
To improve the prediction of the impact of HIV-1 protease mutations in different viral subtypes on virologic response to darunavir.
Methods
Darunavir-containing treatment change episodes (TCE) in patients previously failing protease inhibitors, were selected from large European databases. HIV-1 subtype B-infected patients were used as derivation dataset, cases with non-B subtypes as validation dataset. The adjusted association of each mutation with week-8 HIV-RNA change from baseline was analyzed by linear regression. A prediction model was derived based on best subset least squares estimation with mutational weights corresponding to regression coefficients. Virological outcome prediction accuracy was compared with that from existing genotypic resistance interpretation systems (GIS: ANRS 2013, Rega 9.1.0, HIVdb 7.0).
Results
TCE were selected from 681 subtype B and 199 non-B infected adults. Accompanying drugs were NRTIs in 87%, NNRTIs in 27%, and raltegravir or maraviroc or enfuvirtide in 53%. The prediction model included weighted protease mutations, HIV RNA, CD4 and activity of accompanying drugs. The model’s association with week-8 HIV-RNA change in the subtype B (derivation) set was R2=0.47 (average square error, ASE=0.67, p<10-6); in the non-B (validation) set, ASE was 0.91. Accuracy investigated by means of Area Under the Receiver Operating Characteristic curves with a binary response (> threshold value of HIV-RNA reduction) shows that our final model outperformed models with existing interpretation systems in both training and validation sets.
Conclusion
A model with a new darunavir weighted mutation score outperformed existing GIS in both B and non-B subtypes in predicting virological response to darunavir.
Keywords: HIV-1 drug resistance, interpretation, virologic response
Introduction
HIV-1 drug resistance is a major limitation to the efficacy of combination antiretroviral therapy. 1,2 With several available drug options, the aim of combination antiretroviral therapy (cART) is to achieve full virological suppression even in treatment-experienced patients with multi-class drug resistance.3–5 Darunavir is a protease inhibitor (PI) requiring pharmacokinetic boosting with ritonavir, with preserved antiviral activity against several PI-resistant HIV-1 isolates,6–9 and has a documented activity against non-B HIV-1 subtypes.10 The tight binding between darunavir and the active viral protease site as well as its ability to inhibit protease dimerization form the basis for this preserved activity and high genetic barrier to resistance.6–15
The higher efficacy of darunavir/ritonavir as compared to other PI has been demonstrated in highly treatment-experienced patients with extensive drug resistance.12 Nonetheless, in the presence of a high number of resistance mutations in the viral protease, combination regimens with darunavir/ritonavir will ultimately fail. It is therefore important to identify the combination of protease substitutions which result in a loss of darunavir activity. Several scores and algorithms for the interpretation of darunavir genotypic resistance (genotypic interpretation systems, GIS) have been constructed.16–25 However, different GIS vary with respect to the mutations that are taken into account and the weights assigned to the individual mutations. This variation may be explained by different source studies included and different analytical approaches. Moreover, although most knowledge on darunavir resistance is based on subtype B viruses, other subtypes are becoming increasingly relevant due to the extension of cART coverage in African and Asian countries where non-B subtypes prevail, and to the spread of non-B viruses in Europe and North America.26–28 Therefore, the precise impact of HIV-1 protease mutations and their combination on virologic response to darunavir remains to be fully elucidated, particularly with different subtypes.
We aimed to derive a refined genotypic interpretation score for darunavir based on virologic response in patients harboring subtype B HIV-1, and at validating its performance for non-B HIV-1 subtypes.
Materials and Methods
Case selection and definition of treatment change episodes (TCE)
We selected darunavir-containing TCE from European observational cohort studies sharing their data through the CHAIN project,29 namely Euresist (gathering data from Italy, Germany, Sweden, Belgium and Portugal),30 the Swiss HIV cohort study,31 the Aquitaine cohort,32 cohorts from Paris and Rome, and through the Treatment Change Episodes Database of COHERE in EuroCoord, a collaboration of European cohorts (www.cohere.org). COHERE’s Treatment Change Episodes Database was pooled within the EuroCoord network in 2012 (www.EuroCoord.net).
Cases were required to be HIV-infected adults (>= 18 years old). A TCE was defined as the start of a darunavir-containing regimen including at least two additional antiretroviral agents in patients with a previous history of virological failure to at least one PI (HIV RNA >1,000 copies/mL after at least 3 months of treatment). Not all participating cohorts were able to provide information on darunavir dose; for those that did, only cases of 600 mg twice daily were included. Cases were required to have an available drug resistance genotype (protease and reverse transcriptase sequences obtained through standard population sequencing) and an HIV-1 RNA quantification and CD4 count performed within the period 6 months before to 1 week after baseline (the start date of the darunavir regimen). Additional variables necessary for inclusion were: prior treatment history, drugs accompanying darunavir and HIV-1 RNA at week 8 (window +/- 4 weeks) after baseline; gender and age were not required for inclusion but were collected. No regimen change was allowed between the date of sample collection for pre-baseline viral load and resistance genotyping and the baseline, and the darunavir-containing regimen had to be prescribed for at least 12 weeks without changes or interruptions. All cases gave written informed consent within the respective cohorts. Viral subtype was determined using the Rega 2.0 subtyping system. Unassigned subtypes were classified as non-B.
Statistical analysis
Definition of main outcome variable
The main outcome was change in log10 HIV-RNA from baseline to week 8. An undetectable viral load at week 8 was imputed with a value of 50 copies/mL (main analysis) or 25 copies/mL (sensitivity analysis).
Definition of PI mutations
Any protease substitution at any codon (compared with consensus B reference sequence) with a frequency >2% was considered as a candidate PI mutation.
Prediction model derivation strategy
We used a standard linear regression model applied to the derivation dataset to identify predictive mutations from the candidate protease mutations. Regression models included candidate protease mutations, adjusting for the following variables: baseline log10 viral load, baseline CD4 cell count and Genotypic Susceptibility Score (GSS) of drugs prescribed with darunavir. GSS of the drugs accompanying darunavir was calculated summing the number of active drugs using the interpretation of the ANRS 2013 algorithm (HIVdb 7.0 with three resistance categories21 was used in sensitivity analyses): individual drug activity was scored as 1 if no evidence of resistance, 0.5 if intermediate resistance, and 0 if resistance. New and recycled use of raltegravir, enfuvirtide and maraviroc were scored 1 and 0, respectively.
The derivation set was comprised of all TCE cases with subtype B virus, and the validation dataset was comprised of all non-B subtype cases.
Least squared estimation (LSE) was used to select the “best” subset of mutations. The LSE was chosen to minimize the average squared error (ASE) based on a five-fold cross-validation (CV). Briefly, 5-fold CV works by dividing the derivation dataset randomly into 5 equal parts. The method fits the model to four-fifths of the data and then computes the prediction error on the remaining one-fifth. This procedure was applied to each fifth of the dataset and the 5 prediction error estimates were averaged. From this procedure we obtained an estimated prediction of the 5-fold CV error (CV PRESS) curve, which was used to establish a threshold for stopping the inclusion of the covariates. In sensitivity analyses, we also investigated allowing for 2-way interactions between mutation terms and the use of ten-fold cross validation. Cross-validation was applied to the derivation dataset to select the best model providing the final set of mutations defining the score. The final prediction model included the individual selected mutations, along with baseline CD4 and HIV RNA and GSS of the accompanying drugs, which were forced into the model.
Validation of the score model
The performance of the final model was evaluated in different ways. First, the ASE on both derivation and validation sets were compared. For completeness the R-square on the derivation set was also shown. Second, we computed prediction accuracy of the final model and compared it with a model including only the adjustment variables (the “base model” with baseline log10 viral load, baseline CD4 cell count and GSS of the accompanying drugs) and with models including adjustment variables and the darunavir level of resistance from three main existing GIS (ANRS 2013, Rega 9.1.0 and HIVdb 7.0) using Area Under the Receiver Operating Characteristic (AUROC) curves. This was applied to both the derivation and the validation set. For each exiting GIS a dummy variable indicating resistance was created: for ANRS and REGA “no resistance” and “intermediate resistance” were considered as “no resistance”; for HIVdb, “susceptible”, “potential low-level resistance” and “low-level resistance” were considered as “no resistance”. That required also the transformation of the viral load reduction at week 8 into a binary outcome (response) using three different levels of HIV-1 RNA reduction from baseline ( >1 log10, >1.5 log10 and >2 log10 copies/mL). Statistical analyses were performed using procedures GLMSELECT and LOGISTIC in SAS 9.3 (SAS institute, Cary, NC, USA, 2013).
Results
Baseline patients characteristics
Eight-hundred eighty cases fulfilled the selection criteria: 681 (77.4%) were infected with a subtype B virus (derivation dataset) and 199 (22.6%) with a non-B subtype (validation set). Non-B subtypes consisted mainly of subtype A (19.6%), CRF02_AG (18.6%), C (14.1%), D (7%), F (6.5%), unique recombinant forms (5.5%), G (4.5%), BF recombinants (2.5%), others (12.5%), and unassigned (9%). The main baseline characteristics of study patients overall and divided by derivation and validation datasets are summarized in table 1. The numbers contributed by each cohort are illustrated in online table S1. The patients were heavily pre-treated with a previous experience of a median of 5 different nucleos(t)ide reverse transcriptase inhibitors [N(t)RTI] and 3 different PI, including tipranavir in one quarter. The majority had also experienced non-NRTI (NNRTI) and almost one half other drug classes. Patients infected harboring non-B viruses differed from those infected with subtype B, with higher proportion of females, a lower median HIV-1 RNA, slightly lower CD4 counts and a smaller number of previously experienced antiretroviral drugs.
Table 1.
Patients characteristics at baseline
| All (n=880) |
Subtype B (n=681) |
Non-B subtypes (n=199) |
|
|---|---|---|---|
| Age, years, median (IQR) | 42.7 (41.6-44.6) | 42.6 (41.5-44.6) | 43.4 (42.1-44.7) |
| Male gender, n (%) | 688 (78.2) | 583 (85.6) | 105 (52.8) |
| HIV RNA log10 copies/mL, median (IQR) | 4.3 (3.3-4.9) | 4.4 (3.5-4.9) | 3.9 (3.1-4.8) |
| CD4 counts cells/mm3, median (IQR) | 215 (96-363) | 220 (98-370) | 197 (90-330) |
| Median nr of previously used ARV drugs (IQR) | 9 (4-13) | 10 (4-13) | 7 (3-11) |
| Median nr of previously used NRTI (IQR) | 5 (2-6) | 5 (3-6) | 4 (2-6) |
| Median nr of previously used NNRTI (IQR) | 1 (0-2) | 1 (0-2) | 1 (0-1) |
| Median nr of previously used PI (IQR) | 3 (1-5) | 3 (1-5) | 2 (1-4) |
| Previous lopinavir use, % | 70.1 | 70.2 | 69.8 |
| Previous tipranavir use, % | 25.3 | 26.7 | 20.6 |
| Previous etravirine, % | 8.8 | 9.1 | 7.5 |
| Previous enfuvirtide use, % | 27.3 | 29.4 | 20.1 |
| Previous raltegravir use, % | 10.0 | 10.0 | 10.1 |
| Previous maraviroc use, % | 8.1 | 7.9 | 8.5 |
Abbreviations: ARV, antiretroviral; IQR, interquartile range; Nr, number; NRTI: nucleos(t)ide reverse transcriptase inhibitor; non-NRTI, NNRTI; PI, protease inhibitor.
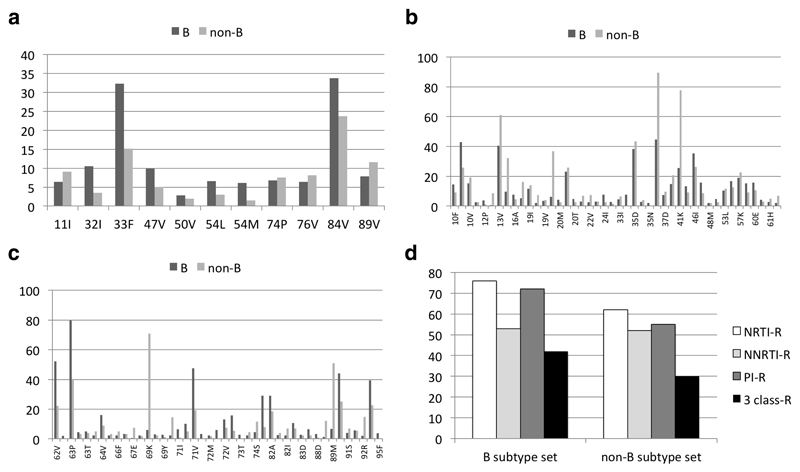
The distribution of protease substitutions from consensus B virus by viral subtype category is illustrated in figure 1. Among the mutations included in the IAS-USA list of major and minor darunavir resistance mutations16 (figure 1a), V32I, L33F, I47V and I84V were more frequent in the subtype B subset as compared to the non-B, while L89V was more frequent in non-B subtypes. For the other mutations (Figure 1b, c) there were differences in frequency at several polymorphic positions, while among substitutions associated with resistance to PI there was a higher frequency for L10I, M46I/L, A71V, G73S, V77I, V82A and L90M in subtype B, whereas K20I, M36I, K70R, T74S, L89I/M prevailed among the non-B subtypes. Figure 1d illustrates the proportion of cases with full resistance to at least one drug in each class and the proportion with resistance to at least one drug to all three historical classes, in subtype B and non-B subtype TCE, according to HIVdb interpretation. Subtype B cases had a higher proportion with resistance to NRTI, PI and a higher proportion of 3-class resistance, compared to non-B subtypes.
Figure 1.
Resistance mutations and interpretations in baseline genotypes of TCE with subtype B virus (derivation set) and non-B subtypes (validation set): proportion of genotypes with IAS-USA 2014 darunavir mutations (a), proportions with any protease mutation from consensus B with a frequency of >2% (b,c); proportion with full resistance to at least one drug in each class and to the 3 historical classes, based on HIVdb 9.0 interpretation.
Antiretroviral drugs accompanying darunavir
The antiretrovirals used in combination with darunavir/ritonavir and their GSS are summarized in table 2. About one of four patients used NNRTI and around one half were prescribed an integrase inhibitor or entry inhibitor. The single drugs more frequently used in combination with darunavir/ritonavir were tenofovir, raltegravir, enfuvirtide and etravirine, most of these reflecting the treatments employed in heavily experienced patients with multi-class resistant viruses. Individuals with non-B viruses were treated less frequently with raltegravir, enfuvirtide and etravirine, while showing a slightly higher GSS of the accompanying drugs.
Table 2.
Antiretrovirals accompanying darunavir/ritonavir
| All (n=880) |
Subtype B (n=681) |
Non-B subtypes (n=199) |
|
|---|---|---|---|
| Any NRTI (%) | 87.2 | 86.9 | 87.9 |
| Any NNRTI (%) | 27.5 | 28.3 | 24.6 |
| Any raltegravir, enfuvirtide or maravicoc (%) | 52.8 | 54.6 | 46.7 |
| Tenofovir (%) | 62.5 | 62.3 | 63.3 |
| Nevirapine (%) | 1.8 | 1.5 | 3.0 |
| Efavirenz (%) | 3.0 | 3.2 | 2.0 |
| Etravirine (%) | 23.1 | 24.1 | 19.6 |
| Enfuvirtide (%) | 24.2 | 25.6 | 19.6 |
| Enfuvirtide first time use (%) | 13.4 | 13.7 | 12.6 |
| Raltegravir (%) | 28.8 | 29.2 | 27.1 |
| Raltegravir first time use (%) | 24.2 | 24.8 | 23.1 |
| Maraviroc (%) | 5.9 | 5.9 | 6.0 |
| Maraviroc first time use (%) | 4.5 | 5.0 | 3.0 |
| GSS of the accompanying drugs | |||
| 0 to 1 | 41.3 | 43.4 | 34.2 |
| >1 to 2 | 41.3 | 39.4 | 48.2 |
| >2 | 17.3 | 17.3 | 17.6 |
Abbreviations: ARV, antiretroviral; GSS, Genotypic Susceptibility Score; IQR, interquartile range; NRTI, nucleos(t)ide reverse transcriptase inhibitor; non-NRTI: NNRTI; PI, protease inhibitor.
Association of protease mutations with 8-week viral load changes in the derivation set (subtype B) and model derivation
In the derivation dataset, median week 8 HIV-1 RNA change from baseline was -2.01 (IQR -2.68, -1.03) log10copies/mL, with 41.4% achieving less than 50 copies/mL at this time point. In the validation set the week 8 HIV-1 RNA change was -1.57 (IQR -2.43, -0.52) log10copies/mL, with 41.1% achieving less than 50 copies/mL. Five mutations (L10F, V11L, I54M, T74P and V82I) were associated with reduced response while 6 substitutions (K20T, E34D, I64L, V82A, I85V and I93L) correlated with improved response (see table 3). The final model included the selected protease mutations, baseline CD4, HIV RNA and GSS of the accompanying drugs. The parameters of the model are reported, with the regression coefficient of each individual component indicating its predicted contribution to the 8-week change from baseline HIV RNA (in log10 copies/mL): intercept +1.499; baseline HIV RNA (per 1 log higher) -0.728; baseline CD4 count (per 100 cells/mm3 higher) -0.048; GSS (by ANRS 2013, per 1 point higher) -0.083; L10F +0.319; V11L +0.405; I54M +0.747; T74P +0.401; V82I +0.537; K20T -0.335; E34D -0.752; I64L -0.471; V82A -0.194; I85V -0.276; I93L -0.161. For each individual case, the predicted 8-week HIV RNA change (in log10 copies/mL) is the result of the algebraic sum of the products of the individual components value times the respective regression coefficient.
Table 3.
Linear regression model of the association of protease mutations with 8-week viral load changes. Subtype B=derivation set (n=681)
| Protease substitution from consensus B | Frequency | Adjusted mean difference in VL change at week 8 (log10 copies/mL)* | SE* | P-value |
|---|---|---|---|---|
| L10F | 14.5% | +0.32 | 0.09 | 0.0006 |
| V11L | 2.6% | +0.40 | 0.20 | 0.045 |
| I54M | 6.1% | +0.75 | 0.14 | <0.0001 |
| T74P | 6.9% | +0.40 | 0.13 | 0.0022 |
| V82I | 2.5% | +0.54 | 0.21 | 0.011 |
| K20T | 4.9% | -0.34 | 0.15 | 0.026 |
| E34D | 2.1% | -0.75 | 0.23 | 0.001 |
| I64L | 2.5% | -0.47 | 0.21 | 0.029 |
| V82A | 29.2% | -0.19 | 0.07 | 0.007 |
| I85V | 6.6% | -0.28 | 0.13 | 0.036 |
| I93L | 39.5% | -0.16 | 0.07 | 0.017 |
results represent mean+/- standard error (SE) of 5-fold cross-validation. Positive values indicate less virologic response (resistance), negative values indicate better virologic response (hypersusceptibility). Associations are adjusted for baseline HIV RNA and CD4 and GSS of the companion drugs. Abbreviations: VL, viral load.
Derived model performance and comparison with available GIS
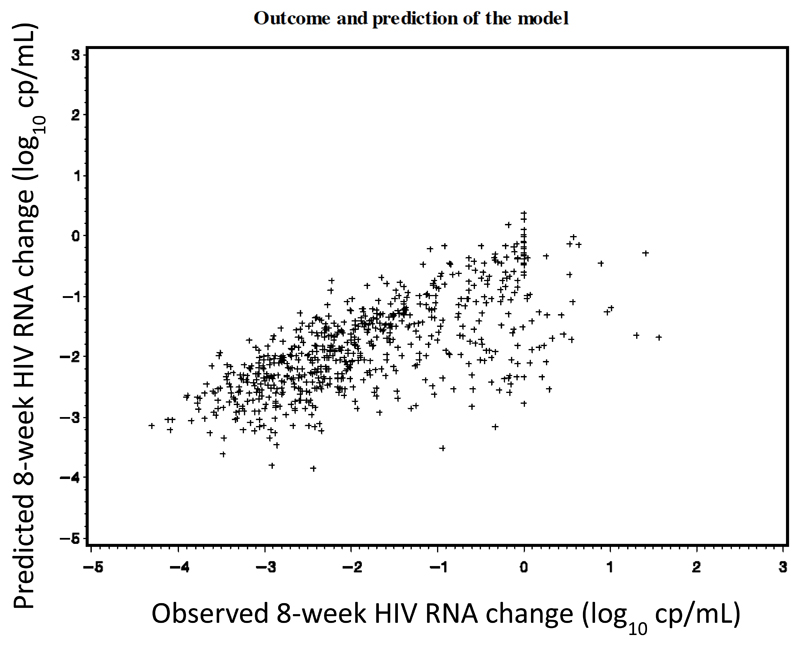
The correlation between the 8-week viral load changes predicted by the derived score and those observed in the derivation set is illustrated in figure 2. Performance of the final regression model and the different models from sensitivity analyses are summarized in Table 4. As expected, the derived score performed better in training than in validation sets. In the training set, our final model performed much better than the model without any resistance variables (base model) and better than models with existing GIS. There was no such difference in the validation set. In sensitivity analyses, the performance of these different models was broadly similar, except when undetectable viral load was fixed at 25 copies/mL, and most of the mutations kept in the final set were similar.
Figure 2.
Eight-week HIV RNA changes from baseline: observed versus predicted outcomes in the derivation set (subtype B TCE).
Table 4.
Average squared error and R-square for the final model and models investigated in sensitivity analyses on the derivation and validation sets.
| Derivation set | Validation set | ||
|---|---|---|---|
| Models | ASE | R-square | ASE |
| Final model* | 0.67 | 0.47 | 0.91 |
| Base model# | 0.76 | 0.40 | 0.92 |
| ANRS 2013§ | 0.72 | 0.44 | 0.94 |
| HIVdb 7.0§ | 0.73 | 0.43 | 0.95 |
| REGA 9.1.0§ | 0.74 | 0.42 | 0.91 |
| Sensitivity analyses on the derived model | |||
| 10-fold | 0.67 | 0.47 | 0.91 |
| 25 copies/mL | 0.77 | 0.41 | 1.04 |
| 2 ways interactions | 0.68 | 0.48 | 0.91 |
| GSS HIVdb | 0.68 | 0.46 | 0.93 |
Least squared estimation (LSE) using the model including the derived weighted darunavir mutations, baseline CD4 and HIV-1 RNA and GSS of the accompanying drugs (according to ANRS 2013 interpretation)
LSE using the final model without the derived weighted darunavir mutations (i.e. using only baseline CD4 and HIV-1 RNA and GSS of the accompanying drugs)
LSE models including the available darunavir genotypic interpretations, baseline CD4, HIV-1 RNA and GSS of the accompanying drugs
10-fold = 10-fold cross validation was used instead of 5-fold
25 copies/mL =undetectable value replaced with 25 copies/mL
2-way interactions = significant 2-way interactions between mutations terms retained in the final model
GSS HIVdb = the final model is adjusted for the GSS of the drugs accompanying darunavir computed according to HIVdb 7.0 instead of ANRS 2013.
Accuracy = percentage of cases correctly classified
Table 5 summarizes the accuracy of the prediction of viral load “response” at 8 weeks (using a threshold of 1.5 log10 copies/mL HIV-1 RNA reduction) for the model with the newly derived darunavir score and three existing GIS (see online Table S2 for results with threshold values of 1 and 2 log10 copies/mL) using AUROC curves. Among the 681 patients infected with a subtype B virus, 444 (65%) patients had a >1.5 log10 copies/mL reduction (response) at 8 weeks and 101 (51%) among the 199 patients infected with a non-B subtypes. Two sets of comparisons were performed. First we compared the accuracy of the response prediction by the different models including darunavir genotypic resistance interpretation (“final model” in the table and three available GIS models) with the “base model”, which incorporated only CD4, HIV-1 RNA and GSS of the accompanying drugs. Of note, all darunavir resistance interpretation systems showed a better prediction over the “base model”, demonstrating the added value of the different darunavir resistance interpretations. Second, we compared the existing darunavir GIS with the “final model”. In the derivation set, the new score of the “final model” consistently out-performed the existing GIS in terms of accuracy, regardless of the definition of viral load response.
Table 5.
Comparative Area Under the Receiver Operating Characteristic (AUROC) curves of different models using a binary outcome at week 8 (HIV-1 RNA reduction < or ≥ 1.5 log10 copies/mL). All models include baseline CD4 and HIV-1 RNA and GSS of the accompanying drugs. The base model includes only baseline CD4 and HIV-1 RNA and GSS of the accompanying drugs. The final model includes the weighted mutations as listed in Table 3, the other models include a dummy variable indicating susceptibility or resistance according to the three existing interpretations systems (see Methods)
| Models | AUROC | Difference in AUROC | P-value* | Difference in AUROC | P-value§ |
|---|---|---|---|---|---|
| Subtype B = derivation set | |||||
| Base model | 0.804 | ref | |||
| Final model | 0.844 | 0.040 | <0.001 | ref | |
| ANRS 2013 | 0.830 | 0.026 | 0.003 | -0.014 | 0.108 |
| HIVdb 7.0 | 0.824 | 0.020 | 0.002 | -0.020 | 0.025 |
| REGA 9.1.0 | 0.817 | 0.013 | 0.009 | -0.027 | 0.002 |
| subtype non-B = validation set | |||||
| Base model | 0.838 | ref | |||
| Final model | 0.872 | 0.034 | 0.015 | ref | |
| ANRS 2013 | 0.840 | 0.002 | 0.390 | -0.032 | 0.016 |
| HIVdb 7.0 | 0.840 | 0.001 | 0.690 | -0.032 | 0.015 |
| REGA 9.1.0 | 0.853 | 0.015 | 0.140 | -0.019 | 0.190 |
comparison with the base model
comparison with the final model
In the validation set, consisting of non-B subtypes, the greater accuracy of the new score was evident in several analyses. Indeed, the “final model” was the only showing consistently higher AUROC values as compared to the base model in predicting the 8-weeks virologic response, using all viral load reduction thresholds (table 5 and online supplementary table S2). Moreover, the “final model” showed higher AUROC as compared to the models of the existing darunavir GIS with significant differences over ANRS and HIVdb at the 1.5 log copies/mL threshold (table 5), over ANRS at the 1.0 log copies/mL and over all three GIS at the 2.0 log copies/mL threshold (supplementary table S2).
Discussion
Darunavir with ritonavir is one of the most widely used drugs in patients with virological failure, and is an essential component of salvage therapy for those who have previously failed other PI therapy.33 Accurate information of the predicted activity of darunavir based on the results of resistance genotyping enables decisions on whether it should be prescribed in the new regimen, as well as the requirement for active companion drugs. Several previous studies have focused on the interpretation of darunavir resistance.17–20,24,34,35 However analyses were either based on genotypic analysis of clinical trials, which used specific selection criteria, or were performed using observational data with a limited size24 or without external validation. Moreover, the performance of predictions was mostly based on sets of cases infected with subtype B virus, yet non-B variants predominate in resource-limited settings and are spreading in Europe. Given the polymorphic variability in the viral protease across different subtypes, it is essential to test how darunavir resistance interpretation performs in non-B subtypes.
In this study, we used the largest dataset of darunavir-based TCE from PI-experienced HIV-1 infected patients collected to date. This allowed us to use two independent sets of data: one, consisting of subtype B TCE, to derive a new weighted score, and a separate second set, consisting of non-B subtypes TCE, for validation.
The main finding of this study was that a model including the new weighted score was more accurate in predicting the virological response at 8 weeks compared to three popular existing GIS for darunavir. While this was expected for B subtypes, due to the training on the same dataset, the model including the new darunavir score also outperformed GIS prediction of virological outcome in an independent validation set of non-B subtypes.
Previously existing GIS were consistently less accurate in predicting virological response in non-B subtypes. In particular, none of the existing GIS significantly improved prediction of response to darunavir over a prediction made computing CD4, viral load and GSS of the accompanying drugs (the “base model”). This indicates that renewed efforts are necessary to improve the understanding of protease resistance interpretation, in non-B subtypes. Notably, the weighted darunavir resistance score developed in the present study included in the final prediction model resulted in an improved prediction of 8-week viral load outcome in non-B subtypes, both over the “base model” and over other existing GIS, using different virologic response thresholds, indicating that the weighted mutation score included in the models is of relevance in this context.
The derived score includes five protease mutations that were associated, with different weights, with reduced response to the darunavir regimens. Two, I54M and T74P, are included in the IAS-USA 2014 mutation list as major and minor darunavir resistance mutation, respectively.16 Consistent with this, our score gives the highest resistance weight to I54M and a somewhat lower weight to T74P. On the other hand, V11I but not V11L, which was found in our score, is considered a minor resistance mutation in that list. However, V11L and L10F, are considered accessory PI-resistance mutations that are associated with minimal reductions in darunavir susceptibility at the same level as V11I.21 Finally, V82I is a highly polymorphic mutation that is not selected by PIs and is the consensus amino acid in subtype G viruses.21 We hypothesize that it may represent a marker or be in linkage disequilibrium with other mutational haplotypes, either in or outside the viral protease, translating into reduced in vivo susceptibility to darunavir. Six additional protease mutations correlated with improved response to darunavir and received negative weights in the derived score. With the exception of V82A, that has already been associated with improved response to darunavir,19,36 no other of these mutations has been associated with decreased or increased susceptibility to darunavir. Three, E34D, I64L, and I93L are considered natural polymorphisms, K20T is associated with variable resistance to other PI, and I85V is selected by PI but has no impact on susceptibility to any of them.21 It should be underscored that the developed prediction model includes baseline viral load, CD4 counts and GSS of the antiretroviral drugs co-prescribed with darunavir and that each contribute to improve the prediction, as previously shown by others,36 but it is noteworthy that the inclusion of the darunavir mutational score improved the prediction over the simple use of these background features.
The findings of this paper are subject to several limitations. First, we were not able to develop a subtype-specific tool predicting virological response to darunavir. Indeed, despite this study included the largest standardized darunavir TCE set ever analysed, the number of cases with individual non-B subtypes was limited and did not allow for specific predictions. In order to overcome this limitation, future studies will need to address the role of specific protease mutations in the context of the most common non-B subtypes, such as C, A, D, and recombinants AE and AG, either in vitro using site-directed mutagenesis experiments or in vivo using well defined TCE from large cohorts in specific African or Asian settings, as darunavir becomes to be more widely employed as rescue therapy in these countries. Second, some of the established darunavir resistance mutations did not appear in the derived score. As a possible explanation we hypothesize that since patients from this study were selected from clinical practice, most were prescribed darunavir based on results of a genotypic resistance test, as indicated by the manufacturer, and thus darunavir was included in the salvage treatment mostly in the absence of known darunavir resistance mutations. Since a selection bias against these mutations has been applied in this dataset, their absence in this newly derived score should be interpreted with caution. Indeed, a prudent approach would be not to ignore mutations that were identified to be predictive of reduced response based on previous clinical trial analyses16,17,19,35 and to use the newly derived score as an additional, refined darunavir genotypic resistance interpretation tool. Moreover, rare mutations, which may play a relevant role in predicting the virologic outcome, may have been missed due to their low prevalence in this study dataset. Finally, this study allowed us to only predict the short-term virologic outcome, which may differ from more long-term outcomes. However, short-term resistance genotype-guided virologic responses are significantly associated with long-term virologic outcomes.37 Moreover, short-term responses are more strongly influenced by the antiviral activity of the regimen and by its relationship with baseline resistance mutations, and less by other confounders such as medication adherence and toxicity, which are more influential on the longer-term.
The strong points of this study are the large size of darunavir TCE utilized, the uniform standardization of the data used to define eligible TCE and the significant proportion of non-B subtypes analyzed.
In conclusion, using a large, standardized dataset of genotype-response cases including darunavir-based regimens we derived and validated a new weighted score that outperformed state-of-the-art darunavir GIS in predicting virologic response both in B and non-B subtypes. This score may be used for predicting response to darunavir in individuals failing previous PI with different subtypes and thus be of help in designing the most appropriate salvage regimens. However, mutations found in previous large clinical trials as predictive of reduced virologic response and not included in this score should also be considered in treatment decisions. After predicting treatment outcome of a darunavir-based regimen using the new score, a prudent solution for a user could be to check for the presence of additional major darunavir resistance mutations on the IAS-USA list.16
Supplementary Material
Acknowledgements
Conjunct CHAIN and COHERE in EuroCoord Darunavir Resistance Working Group
Andrea De Luca, Philippe Flandre, Diane Descamps project leads
COHERE representatives: Antonella Castagna (San Raffaele), Alessandro Cozzi-Lepri (EuroSIDA), Duncan Churchill, David Dunn (UK CHIC and UK HIV Drug Resistance Database) Stéphane De Wit (St Pierre), Wolfgang Fuchs (KOMPNET), Federico Garcia (Co-RIS), Arkaitz Imaz (PISCIS), Theodoros Kordossis (AMACS), Cristina Mussini (MODENA cohort), Niels Obel (Danish HIV cohort), Bernardino Roca (VACH), Carlo Torti(MASTER), Ard van Sighem, Annemarie Wensing (ATHENA), Robert Zangerle (AHIVCOS), Linda Wittkop (Bordeaux RCC), Peter Reiss (Copenhagen – CHIP, RCC)
CHAIN representatives: Francesca Ceccherini-Silberstein, Maria Mercedes Santoro, Carlo Federico Perno (Rome cohort), Huldrych Günthard (Swiss HIV Cohort Study), Eugen Schülter, Maurizio Zazzi (EURESIST), Linda Wittkop(Bordeaux), Diane Descamps (ANRS).
COHERE in EuroCoord
Steering Committee - Contributing Cohorts: Robert Zangerle (AHIVCOS),Giota Touloumi (AMACS), Josiane Warszawski (ANRS CO1 EPF/ANRS CO11 OBSERVATOIRE EPF), Laurence Meyer (ANRS CO2 SEROCO), François Dabis (ANRS CO3 AQUITAINE), Murielle Mary Krause (ANRS CO4 FHDH), Jade Ghosn (ANRS CO6 PRIMO), Catherine Leport (ANRS CO8 COPILOTE), Linda Wittkop (ANRS CO13 HEPAVIH), Peter Reiss (ATHENA), Ferdinand Wit (ATHENA), Maria Prins (CASCADE), Heiner Bucher (CASCADE), Diana Gibb (CHIPS), Gerd Fätkenheuer (Cologne-Bonn), Julia Del Amo (CoRIS), Niels Obel (Danish HIV Cohort), Claire Thorne (ECS), Amanda Mocroft (EuroSIDA), Ole Kirk (EuroSIDA), Christoph Stephan (Frankfurt), Santiago Pérez-Hoyos (GEMES-Haemo), Osamah Hamouda (German ClinSurv), Barbara Bartmeyer (German ClinSurv), Nikoloz Chkhartishvili (Georgian National HIV/AIDS), Antoni Noguera-Julian (CORISPE-cat), Andrea Antinori (ICC), Antonella d’Arminio Monforte (ICONA), Norbert Brockmeyer (KOMPNET), Luis Prieto (Madrid PMTCT Cohort), Pablo Rojo Conejo (CORISPES-Madrid), Antoni Soriano-Arandes (NENEXP), Manuel Battegay (SHCS), Roger Kouyos (SHCS), Cristina Mussini (Modena Cohort), Pat Tookey (NSHPC), Jordi Casabona (PISCIS), Jose M. Miró (PISCIS), Antonella Castagna (San Raffaele), Deborah_Konopnick (St. Pierre Cohort), Tessa Goetghebuer (St Pierre Paediatric Cohort), Anders Sönnerborg (Swedish InfCare), Carlo Torti (The Italian Master Cohort), Caroline Sabin (UK CHIC), Ramon Teira (VACH), Myriam Garrido (VACH). David Haerry (European AIDS Treatment Group)
Executive Committee: Stéphane de Wit (Chair, St. Pierre University Hospital), Jose Mª Miró (PISCIS), Dominique Costagliola (FHDH), Antonella d’Arminio-Monforte (ICONA), Antonella Castagna (San Raffaele), Julia del Amo (CoRIS), Amanda Mocroft (EuroSida), Dorthe Raben (Head, Copenhagen Regional Coordinating Centre), Geneviève Chêne (Head, Bordeaux Regional Coordinating Centre). Paediatric Cohort Representatives: Ali Judd, Pablo Rojo Conejo.
Regional Coordinating Centres: Bordeaux RCC: Diana Barger, Christine Schwimmer, Monique Termote, Linda Wittkop; Copenhagen RCC: Maria Campbell, Casper M. Frederiksen, Nina Friis-Møller, Jesper Kjaer, Dorthe Raben, Rikke Salbøl Brandt.
Project Leads and Statisticians: Juan Berenguer, Julia Bohlius, Vincent Bouteloup, Heiner Bucher, Alessandro Cozzi-Lepri, François Dabis, Antonella d’Arminio Monforte, Mary-Anne Davies, Julia del Amo, Maria Dorrucci, David Dunn, Matthias Egger, Hansjakob Furrer, Marguerite Guiguet, Sophie Grabar, Ali Judd, Ole Kirk, Olivier Lambotte, Valériane Leroy, Sara Lodi, Sophie Matheron, Laurence Meyer, Jose Mª Miró, Amanda Mocroft, Susana Monge, Fumiyo Nakagawa, Roger Paredes, Andrew Phillips, Massimo Puoti, Michael Schomaker, Colette Smit, Jonathan Sterne, Rodolphe Thiebaut, Claire Thorne, Carlo Torti, , Marc van der Valk, Linda Wittkop, Natasha Wyss.
The members of the Swiss HIV Cohort Study are: Aubert V, Battegay M, Bernasconi E, Böni J, Bucher HC, Burton-Jeangros C, Calmy A, Cavassini M, Dollenmaier G, Egger M, Elzi L, Fehr J, Fellay J, Furrer H (Chairman of the Clinical and Laboratory Committee), Fux CA, Gorgievski M, Günthard H (President of the SHCS), Haerry D (deputy of "Positive Council"), Hasse B, Hirsch HH, Hoffmann M, Hösli I, Kahlert C, Kaiser L, Keiser O, Klimkait T, Kouyos R, Kovari H, Ledergerber B, Martinetti G, Martinez de Tejada B, Metzner K, Müller N, Nadal D, Nicca D, Pantaleo G, Rauch A (Chairman of the Scientific Board), Regenass S, Rickenbach M (Head of Data Center), Rudin C (Chairman of the Mother & Child Substudy), Schöni-Affolter F, Schmid P, Schüpbach J, Speck R, Tarr P, Telenti A, Trkola A, Vernazza P, Weber R, Yerly S.
Contributors to the UK HIV Drug Resistance Database are listed at http://www.hivrdb.org/.
Funding: The COHERE study group has received unrestricted funding from: Agence Nationale de Recherches sur le SIDA et les Hépatites Virales (ANRS), France; HIV Monitoring Foundation, The Netherlands; and the Augustinus Foundation, Denmark. COHERE receives funding from the European Union Seventh Framework Programme (FP7/2007-2013) under EuroCoord grant agreement n° 260694. A list of the funders of the participating cohorts can be found at www.COHERE.org. CHAIN has received funding from the EU 7th Framework Programme (FP7/2007-2013) grant nr 223131. This Swiss HIV Cohort Study is supported by the Swiss National Science Foundation (grant # 148522) and by the SHCS research foundation, the Yvonne-Jacob Foundation. HFG was supported by the university of Zurich’s clinical research priority program: viral infectious diseases.
This work was presented in part at the RW2014 - International Workshop on Antiviral Drug Resistance, June 3-7, 2014, Berlin, Germany, abstract 99.
Footnotes
Transparency declaration: Dr. De Luca reports grants and personal fees from ViiV Healthcare, personal fees from Gilead, grants and personal fees from Merck, personal fees from Janssen, personal fees from Abbvie, personal fees from Roche, personal fees from Novartis, personal fees from Bristol-Myers Squibb, outside the submitted work. Dr. Wittkop reports personal fees from BMS, outside the submitted work; Dr. DESCAMPS reports personal fees and non-financial support from ViiV heathcare, personal fees and non-financial support from Gilead Sciences, personal fees and non-financial support from MSD, personal fees and non-financial support from Janssen-Cilag , personal fees and non-financial support from BMS, from null, outside the submitted work; Dr. FLANDRE reports personal fees and non-financial support from ViiV Healthcare, personal fees and non-financial support from Janssen Cilag, personal fees and non-financial support from Bristol Myers Squibb, outside the submitted work; Dr. van Sighem reports grants from Dutch Ministry of Health, Welfare and Sport, grants from Gilead Sciences, during the conduct of the study; Dr. García reports grants from Abbvie, grants from Roche, grants from Gilead, outside the submitted work; H.F.G. has been an adviser and/or consultant for the following companies: GlaxoSmithKline, Abbott, Gilead, Novartis, Boehringer Ingelheim, Roche, Tibotec, Pfizer and Bristol-Myers Squibb, and has received unrestricted research and educational grants from Roche, Abbott, Bristol-Myers Squibb, Gilead, Astra-Zeneca, GlaxoSmithKline, and Merck Sharp & Dohme. Dr. Wensing reports travel grants from MSD, Viiv Healthcare, Janssen, Virology Education, BMS and Gilead; Consultancy fee from Janssen, BMS, Gilead, Viiv Healthcare; Investigator initiated grants from Janssen, Viiv Healthcare; Fee for accredited course from Virology Education, all outside the submitted work, all paid to her institution; Dr. Mussini reports personal fees from Gilead, personal fees from ViiV, personal fees from MSD, personal fees from Bristol-Myers Squibb, personal fees from Abbvie, grants from Johnson and Johnson, grants from Bristol-Myers Squibb, outside the submitted work. Dr. Carlo Federico Perno CF has received funds for attending symposia; speaking, organizing educational activities, grant research support, consultancy from Abbott, Bristol Myers Squibb, Gilead, Merck Sharp & Dohme, Janssen Cilag, Pfizer, Roche, ViiV Healthcare. Dr. Reiss reports grants from Gilead Sciences, grants from ViiV Healthcare, grants from Janssen Pharmaceutica, grants from Bristol Myers Squibb, grants from Merck & Co, other from Gilead Sciences, other from Janssen Pharmaceutica, other from ViiV Healthcare, outside the submitted work; Dr. Santoro has received funds for attending symposia, speaking and organizing educational activities from Abbott, Bristol Myers Squibb, Merck Sharp & Dohme, ViiV Healthcare and Janssen Cilag. CT received a grant for research from Gilead Sciences as a result of a open competition (Fellowship Program); Dr. Zazzi reports grants and personal fees from ViiV Healthcare, personal fees from Janssen, personal fees from Abbott Molecular, personal fees from Merck Sharp and Dohme, outside the submitted work. All other authors: none to declare
References
- 1.Zaccarelli M, Tozzi V, Lorenzini P, et al. Multiple drug class-wide resistance associated with poorer survival after treatment failure in a cohort of HIV-infected patients. AIDS. 2005;19:1081–9. doi: 10.1097/01.aids.0000174455.01369.ad. [DOI] [PubMed] [Google Scholar]
- 2.Di Giambenedetto S, Colafigli M, Pinnetti C, et al. Genotypic resistance profile and clinical progression of treatment-experienced HIV type 1-infected patients with virological failure. AIDS Res Hum Retroviruses. 2008;24:149–54. doi: 10.1089/aid.2007.0070. [DOI] [PubMed] [Google Scholar]
- 3.Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. Department of Health and Human Services; [Section accessed November 3, 2015]. Available at http://aidsinfo.nih.gov/ContentFiles/AdultandAdolescentGL.pdf. [Google Scholar]
- 4.European Aids Clinical Society (EACS) Guidelines version 8.0. [last access November 3, 2015];2015 Oct; http://www.eacsociety.org/guidelines/eacs-guidelines/eacs-guidelines.html.
- 5.British HIV Association guidelines for the treatment of HIV-1-positive adults with antiretroviral therapy 2015. [last access November 3, 2015]; doi: 10.1111/hiv.12426. Available at http://www.bhiva.org/HIV-1-treatment-guidelines.aspx. [DOI] [PubMed]
- 6.Kovalevsky AY, Liu F, Leshchenko S, et al. Ultra-high resolution crystal structure of HIV-1 protease mutant reveals two binding sites for clinical inhibitor TMC114. J Mol Biol. 2006;363:161–73. doi: 10.1016/j.jmb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kovalevsky AY, Tie Y, Liu F, et al. Effectiveness of nonpeptide clinical inhibitor TMC-114 on HIV-1 protease with highly drug resistant mutations D30N, I50V, and L90M. J Med Chem. 2006;49:1379–87. doi: 10.1021/jm050943c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Meyer S, Azijn H, Surleraux D, et al. TMC114, a novel human immunodeficiency virus type 1 protease inhibitor active against protease inhibitor-resistant viruses, including a broad range of clinical isolates. Antimicrob Agents Chemother. 2005;49:2314–21. doi: 10.1128/AAC.49.6.2314-2321.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sayer JM, Liu F, Ishima R, et al. Effect of the active site D25N mutation on the structure, stability, and ligand binding of the mature HIV-1 protease. J Biol Chem. 2008;283:13459–70. doi: 10.1074/jbc.M708506200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dierynck I, De Meyer S, Lathouwers E, et al. In vitro susceptibility and virological outcome to darunavir and lopinavir are independent of HIV type-1 subtype in treatment-naive patients. Antivir Ther. 2010;15:1161–9. doi: 10.3851/IMP1697. [DOI] [PubMed] [Google Scholar]
- 11.Koh Y, Matsumi S, Das D, et al. Potent inhibition of HIV-1 replication by novel non-peptidyl small molecule inhibitors of protease dimerization. J Biol Chem. 2007;282:28709–20. doi: 10.1074/jbc.M703938200. [DOI] [PubMed] [Google Scholar]
- 12.Clotet B, Bellos N, Molina JM, et al. Efficacy and safety of DRV-rtv at week 48 in treatment-experienced patients with HIV-1 infection in POWER 1 and 2: a pooled subgroup analysis of data from two randomised trials. Lancet. 2007;369:1169–78. doi: 10.1016/S0140-6736(07)60497-8. [DOI] [PubMed] [Google Scholar]
- 13.Madruga JV, Berger D, McMurchie M, et al. Efficacy and safety of DRV-rtv compared with that of lopinavir-rtv at 48 weeks in treatment-experienced, HIV-infected patients in TITAN: a randomised controlled phase III trial. Lancet. 2007;370:49–58. doi: 10.1016/S0140-6736(07)61049-6. [DOI] [PubMed] [Google Scholar]
- 14.Ortiz R, Dejesus E, Khanlou H, et al. Efficacy and safety of once-daily DRV/rtv versus lopinavir/rtv in treatment-naive HIV-1-infected patients at week 48. AIDS. 2008;22:1389–97. doi: 10.1097/QAD.0b013e32830285fb. [DOI] [PubMed] [Google Scholar]
- 15.Mills AM, Nelson M, Jayaweera D, et al. Once-daily DRV/rtv vs. lopinavir/rtv in treatment-naive, HIV-1-infected patients: 96-week analysis. AIDS. 2009;23:1679–88. doi: 10.1097/QAD.0b013e32832d7350. [DOI] [PubMed] [Google Scholar]
- 16.Wensing AM, Calvez V, Günthard H, et al. 2014 Update of the Drug Resistance Mutations in HIV-1. Topics in Antivir Med. 2014;22:642–650. [PMC free article] [PubMed] [Google Scholar]
- 17.De Meyer S, Vangeneugden T, van Baelen B, et al. Resistance profile of DRV: combined 24-week results from the POWER trials. AIDS Res Hum Retroviruses. 2008;24:379–88. doi: 10.1089/aid.2007.0173. [DOI] [PubMed] [Google Scholar]
- 18.Pellegrin I, Wittkop L, Joubert LM, et al. Virological response to DRV/rtv-based regimens in antiretroviral-experienced patients (PREDIZISTA study) Antivir Ther. 2008;13:271–9. [PubMed] [Google Scholar]
- 19.Descamps D, Lambert-Niclot S, Marcelin AG, et al. Mutations associated with virological response to DRV/rtv in HIV-1-infected protease inhibitor-experienced patients. J Antimicrob Chemother. 2009;63:585–92. doi: 10.1093/jac/dkn544. [DOI] [PubMed] [Google Scholar]
- 20.Delaugerre C, Pavie J, Palmer P, et al. Pattern and impact of emerging resistance mutations in treatment experienced patients failing DRV-containing regimen. AIDS. 2008;22:1809–13. doi: 10.1097/QAD.0b013e328307f24a. [DOI] [PubMed] [Google Scholar]
- 21.Stanford University HIV Drug Resistance Database. [Accessed November 3, 2015]; Available at: http://hivdb.stanford.edu.
- 22.Laboratory for Clinical and Evolutionary Virology, Rega Institute for Medical Research, Katholieke Universiteit Leuven. Rega algorithm. [Accessed November 3, 2015]; Available at: https://rega.kuleuven.be/cev/avd/software/rega-algorithm.
- 23.HIV-1 genotypic drug resistance interpretation’s algorithms. [Accessed November 3, 2015]; available at http://www.hivfrenchresistance.org/table.html.
- 24.De Luca A, Di Giambenedetto S, Maserati R, et al. Interpretation of genotypic HIV-1 resistance to darunavir and virological response: validation of available systems and of a new score. Antivir Ther. 2011;16:489–97. doi: 10.3851/IMP1799. [DOI] [PubMed] [Google Scholar]
- 25.Zazzi M, Romano L, Venturi G, et al. Comparative evaluation of three computerized algorithms for prediction of antiretroviral susceptibility from human immunodeficiency virus type 1 genotype. J Antimicrob Chemother. 2004;53:356–60. doi: 10.1093/jac/dkh021. [DOI] [PubMed] [Google Scholar]
- 26.von Wyl V, Kouyos RD, Yerly S, et al. Swiss HIV Cohort Study The role of migration and domestic transmission in the spread of HIV-1 non-B subtypes in Switzerland. J Infect Dis. 2011;204:1095–103. doi: 10.1093/infdis/jir491. [DOI] [PubMed] [Google Scholar]
- 27.Chaix ML, Seng R, Frange P, et al. ANRS PRIMO Cohort Study Group Increasing HIV-1 non-B subtype primary infections in patients in France and effect of HIV subtypes on virological and immunological responses to combined antiretroviral therapy. Clin Infect Dis. 2013;56:880–7. doi: 10.1093/cid/cis999. [DOI] [PubMed] [Google Scholar]
- 28.Pyne MT, Hackett J, Jr, Holzmayer V, et al. Large-scale analysis of the prevalence and geographic distribution of HIV-1 non-B variants in the United States. J Clin Microbiol. 2013;51:2662–9. doi: 10.1128/JCM.00880-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wittkop L, Günthard HF, de Wolf F, et al. EuroCoord-CHAIN study group Effect of transmitted drug resistance on virological and immunological response to initial combination antiretroviral therapy for HIV (EuroCoord-CHAIN joint project): a European multicohort study. Lancet Infect Dis. 2011;11:363–71. doi: 10.1016/S1473-3099(11)70032-9. [DOI] [PubMed] [Google Scholar]
- 30.Zazzi M, Incardona F, Rosen-Zvi M, et al. Predicting response to antiretroviral treatment by machine learning: the EuResist project. Intervirology. 2012;55:123–7. doi: 10.1159/000332008. [DOI] [PubMed] [Google Scholar]
- 31.Swiss HIV Cohort Study. Schoeni-Affolter F, Ledergerber B, et al. Cohort profile: the Swiss HIV Cohort study. Int J Epidemiol. 2010;39:1179–89. doi: 10.1093/ije/dyp321. [DOI] [PubMed] [Google Scholar]
- 32.Wittkop L, Breilh D, Da Silva D, et al. ANRS CO3 Aquitaine Cohort Virological and immunological response in HIV-1-infected patients with multiple treatment failures receiving raltegravir and optimized background therapy, ANRS CO3 Aquitaine Cohort. J Antimicrob Chemother. 2009;63:1251–5. doi: 10.1093/jac/dkp114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Günthard HF, Aberg JA, Eron JJ, et al. International Antiviral Society-USA Panel Antiretroviral treatment of adult HIV infection: 2014 recommendations of the International Antiviral Society-USA Panel. JAMA. 2014;312:410–25. doi: 10.1001/jama.2014.8722. [DOI] [PubMed] [Google Scholar]
- 34.Young J, Scherrer AU, Günthard HF, et al. Efficacy, tolerability and risk factors for virological failure of darunavir-based therapy for treatment-experienced HIV-infected patients: the Swiss HIV Cohort Study. HIV Med. 2011;12:299–307. doi: 10.1111/j.1468-1293.2010.00885.x. [DOI] [PubMed] [Google Scholar]
- 35.De Meyer S, Descamps D, Van Baelen B, et al. Confirmation of the negative impact of protease mutations I47V, I54M, T74P and I84V and the positive impact of protease mutation V82A in virological response to darunavir/ritonavir. XVIII International HIV Drug Resistance Workshop, Fort Myers, Florida June 9-13, 2009, abstract 126. Antivir Ther. 2009;14(Suppl 1):A147. [Google Scholar]
- 36.Revell AD, Wang D, Boyd MA, et al. The development of an expert system to predict virological response to HIV therapy as part of an online treatment support tool. AIDS. 2011;25:1855–63. doi: 10.1097/QAD.0b013e328349a9c2. [DOI] [PubMed] [Google Scholar]
- 37.De Luca A, Di Giambenedetto S, Cingolani A, et al. Three-year clinical outcomes of resistance genotyping and expert advice: extended follow-up of the Argenta trial. Antivir Ther. 2006;11:321–7. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.