Abstract
Introns are removed from eukaryotic mRNA precursors by the spliceosome in two transesterification reactions – branching and exon ligation. The mechanism of 3'-splice site recognition during exon ligation has remained unclear. Here we present the 3.7Å cryo-EM structure of the yeast P complex spliceosome immediately after exon ligation. The 3'-splice site AG dinucleotide is recognized through non-Watson-Crick pairing with the 5'-splice site and the branch point adenosine. After the branching reaction protein factors work together to remodel the spliceosome and stabilize a conformation competent for 3'-splice site docking, thereby promoting exon ligation. The structure accounts for the strict conservation of the GU and AG dinucleotides at the 5' and 3' ends of introns and provides insight into the catalytic mechanism of exon ligation.
Pre-mRNA splicing is catalyzed by a dynamic molecular machine called the spliceosome (1), which employs a single RNA-based active site (2, 3) to catalyze two sequential transesterification reactions that excise non-coding introns from pre-mRNA and ligate the coding exons to form mature mRNA. Introns are marked by GU and AG dinucleotides at the 5'- and 3'-splice sites (SS) respectively and a branch point (BP) adenosine upstream of the 3'SS; these nucleotides are invariant except in introns removed by the metazoa-specific minor spliceosome (4). The spliceosome assembles de novo on each pre-mRNA by the ordered joining of five small nuclear ribonucleoprotein particles (snRNPs) and the NineTeen and NineTeen-Related (NTC and NTR) protein complexes, along with additional protein factors (1). First the U1 and U2 snRNPs recognize the 5'SS and BP sequences of the pre-mRNA respectively, forming A complex. Next the pre-assembled U4/U6.U5 tri-snRNP joins to form pre-B complex, followed by spliceosome activation via B complex to form Bact complex. Subsequent Prp2-mediated remodeling yields B* complex, which is competent to carry out the first transesterification reaction, called branching, when the 2'-hydroxyl of the conserved BP adenosine in the intron performs a nucleophilic attack on the 5'SS, producing the cleaved 5'-exon and a branched lariat-intron intermediate. The resulting C complex is then remodeled to C* complex upon dissociation of step 1 (branching) factors by the DEAH-box ATPase Prp16 (5, 6). In C* complex, step 2 (exon ligation) factors promote docking of the 3'SS into the active site (7) and exon ligation, via nucleophilic attack of the 3'-hydroxyl of the 5'-exon at the 3'SS. The resulting P complex contains ligated exons (mRNA) and the excised lariat-intron. The newly formed mRNA is then released from P complex by the DEAH-box ATPase Prp22 (8), forming the intron-lariat spliceosome (ILS) which is disassembled by the DEAH-box ATPase Prp43 (1) to recycle the snRNPs for further rounds of splicing.
Recent cryo-electron microscopy (cryo-EM) studies of yeast (6, 9–15) and human spliceosomes (16, 17) have elucidated the configuration of the RNA-based active site and many mechanistic details of splice site recognition and catalysis, as well as the role of specific protein factors (18). Structures of C complex (6, 12) showed how the spliceosome recognizes and positions the 5'SS and BP sequences in the active site through RNA-RNA interactions with U6 snRNA and U2 snRNA, while the branching factors Cwc25, Yju2, and Isy1 lock these sequences in a conformation competent for catalysis. Structures of C and C* complex (6, 14–17) revealed how Prp16 remodels the spliceosome into the exon ligation conformation, which is stabilized by the exon ligation factors Prp18 and Slu7 (7, 19, 20). In both C and C* complexes the 5'SS and 5'-exon remain paired with U6 snRNA (21, 22) and loop 1 of the U5 snRNA (23, 24), respectively. However in the C* complex structures the 3’-exon and 3’SS are not yet docked into the active site. Thus it has remained unclear how the spliceosome selects and docks the 3'SS while aligning the 3'-exon for step 2 catalysis. Additionally, it was not known how Slu7 and Prp18 interact with the 3'SS to promote docking.
Here, we report the cryo-EM structure of the Saccharomyces cerevisiae post-catalytic P complex at near-atomic resolution, showing the catalytic step 2 configuration of the active site. The structure reveals how the 3'SS and 3'-exon are recognized by the spliceosome and shows the critical role of the branched lariat-intron in promoting the chemistry of exon ligation.
Overall architecture of P complex
We produced P complex by an in vitro splicing reaction supplemented with dominant-negative mutant Prp22 protein to prevent mRNA release from the spliceosome (25). Complexes with a docked 3'-exon were selectively purified via MS2-MBP fusion protein. The resulting spliceosomes contained spliced mRNA and excised intron and were enriched in Prp22, as expected (Fig. S1). We obtained a cryo-EM reconstruction at 3.7Å overall resolution and modeled 45 components (Figs. S2-3, Tables S1-2)
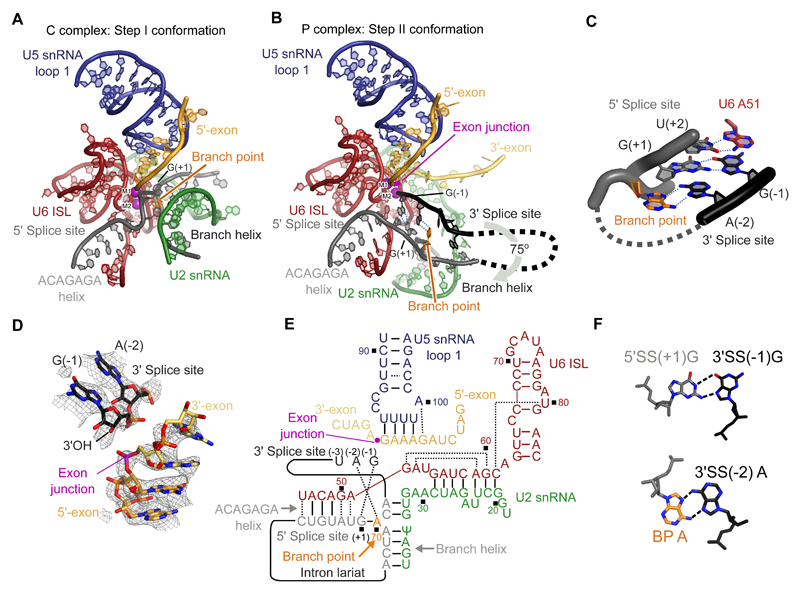
P complex has the same overall exon ligation conformation as C* complex (Fig. 1A). Relative to the branching conformation of C complex (6, 12), the branch helix between the intron and U2 snRNA has rotated 75 degrees out of the active site, extracting the BP adenosine and creating space for the incoming 3'-exon in the active center (Fig. 2A,B). Branch helix rotation is accompanied by movement of the U2 snRNP and its attached NTC protein Syf1, and Prp16-mediated release of the branching factors Cwc25, Isy1 and the N-terminal domain of Yju2 (14, 15) (Fig. S4A). The undocked branch helix is stabilized in its new position by the WD40 domain of Prp17, and by the Prp8 RNaseH domain, which has rotated to insert its β-hairpin near the BP (14, 15). The exon ligation factors Prp18 and Slu7 occupy the same locations as they do in C* complex, with the α-helical domain (26) of Prp18 binding the outer surface of the Prp8 RNaseH domain (Fig. 1C). While in C* complex we were only able to assign the C-terminal globular domain of Slu7, the higher quality density in P complex allowed us to essentially complete this model, revealing a sprawling architecture that spans 120Å of the spliceosome (Fig. 1A). Prp22, which replaces Prp16 in both C* and P complex, is stabilized onto the Prp8 N-terminal domain through interactions with the C-terminus of Yju2.
Figure 1. P complex structure.
A, Overview of the P complex spliceosome. NTC, NineTeen Complex. B, The same view of P complex with the path of the substrate intron and exons shown. Dotted lines indicate the path of nucleotides not visible in the density. C, Binding of substrate at the core of P complex. U6 snRNA and NTC/NTR proteins are omitted for clarity. 3'SS, 3' splice site; RT, Prp8 reverse transcriptase domain; CR, Prp18 conserved region.
Figure 2. Structure of the RNA catalytic core.
A, Key RNA elements at the active site of the C complex spliceosome. ISL, internal stem-loop; M1 and M2, catalytic metal ions one and two. B, Equivalent view to A of the active site of the P complex spliceosome. M1 was not visible in the density and its position is inferred from C and C* complexes. C, Non-Watson-Crick RNA-RNA interactions mediate recognition of the 3' splice site. Putative hydrogen bonds are shown with dotted blue lines. Branch point adenosine and U6 snRNA A51 are highlighted. D, CryoEM density around the exon junction for the 5'-exon, 3'-exon and 3' splice site. E, Base-pairing scheme of the P complex active site. Watson-Crick pairing is indicated with lines, other base pairs with dotted lines. Ψ, pseudo-uridine. F, Details of the pairing that mediates 3' splice site (3'SS) recognition. 5'SS, 5' splice site; BP A, branch point adenosine.
RNA active site
The RNA catalytic core of the P complex spliceosome remains essentially unchanged compared to C* complex (14–17) (Fig. 2A,B) , except that the 3'-exon is ligated to the 5'-exon and the 3'SS is docked into the space occupied by the branch helix in C complex (Figs. 1B,C, 2B, S3). The catalytic triplex formed by U2 and U6 snRNAs, harboring the catalytic metal ions, is unaltered (2, 3, 27), and the 5'-exon remains base-paired to loop 1 of U5 snRNA (23, 24). The position of the 3'SS relative to the 3'-exon suggests that prior to exon ligation, the pre-mRNA undergoes an almost 180° turn to expose the 3'SS scissile phosphate for nucleophilic attack by the 3'-hydroxyl of the 5'-exon. This deformation is similar to that seen during branching, when the 5'SS is highly bent to expose the scissile phosphate to the branch-point adenosine nucleophile (6, 12) (Fig. S5).
The first two bases of the 3'-exon are well ordered and extend the 5'-exon with A-form helical geometry and regular base stacking (Fig. 2D) whereas ten nucleotides of the 5'-exon are well ordered in the channel between the N-terminal and Large domains of Prp8. Density for the 3'-exon downstream of G(+2) becomes weaker and follows the surface of the Prp8 reverse transcriptase (RT) domain up towards Prp22 (Fig. S3F), consistent with crosslinking experiments (25). This arrangement is consistent with the role of Prp22 in pulling the ligated exon from the 3' direction to release mRNA (25). Our C complex structure revealed a similar mechanism for remodeling by Prp16 (6, 18). It was previously shown that U5 snRNA loop 1 aligns the 5'-exon and 3'-exon for ligation (23, 24). Indeed, U5 snRNA U96, which points away from the active site in Bact, C, and C* complexes, can pair with G(-1) of the 5'-exon in P complex (Fig. 2E), explaining previous genetic and crosslinking data (23, 24).
In yeast the intron sequence of the 3'SS immediately preceding the 3'-exon is stringently conserved as Y(-3)A(-2)G(-1), where Y is any pyrimidine (28) (Fig. S6A). In our P complex structure the phosphodiester bond at the 3'SS is cleaved, but the 3'-hydroxyl of the 3'SS nucleotide G(-1) remains close to the phosphate of the newly-formed exon junction, consistent with observations that exon ligation is reversible (29) (Fig. 2D). This suggests that our structure represents the state of the spliceosome immediately after exon ligation, allowing us to infer the mechanism of 3'SS recognition (Fig. 2C,F). Remarkably, the Hoogsteen edge of the 3'SS G(-1) forms a base pair with the Watson-Crick edge of the 5'SS G(+1), while stacking on U6 snRNA A51, which remains paired to U(+2) of the 5'SS as in C* complex (14). This arrangement allows the 3'-hydroxyl of 3'SS G(-1) to project towards the active site (Fig. 2D). The Hoogsteen edge of the 3'SS A(-2) interacts with the Hoogsteen edge of the branch point adenosine, which is still linked via its 2'-hydroxyl to the 5'SS G(+1). Thus, 3'SS recognition is achieved through RNA base-pairing with the 5'SS and the BP adenosine. This mechanism is consistent with the genetic interactions between the first and last bases of the intron (30, 31) and exon ligation defects observed in branch point mutants (32, 33) (Fig. S6). Importantly, mutations at 3'SS G(-1) that are proofread and undocked by Prp22 (7) would destabilize the interaction between the 5'SS and the 3'SS (Fig. S6), consistent with a proofreading mechanism in which Prp22 senses the stability of the docked 3'SS (7), although the structural basis for such sensing remains unknown. We additionally observed ordered density for 3'SS nucleotides (-3) to (-5), while the 20 nucleotides connecting the 3'SS to the branch helix were not visible and are likely disordered as they would loop out of the spliceosome from the branch helix to just upstream of the 3'SS.
Additional density adjacent to the 3'-hydroxyl of 3'SS G(-1) was putatively interpreted as the catalytic Mg2+ M2 because it is coordinated by phosphate oxygen ligands from U6 snRNA bases U80, A59, and G60, which were previously shown biochemically to bind M2 (2). In contrast, no density consistent with M1 was observed, as expected for P complex being in a post-catalytic state, whereas M1 was predicted to coordinate the nucleophile in the pre-catalytic state and density consistent with M1 was indeed visible in C* complex (14) (Figs. S3,S5).
Proteins around the active site
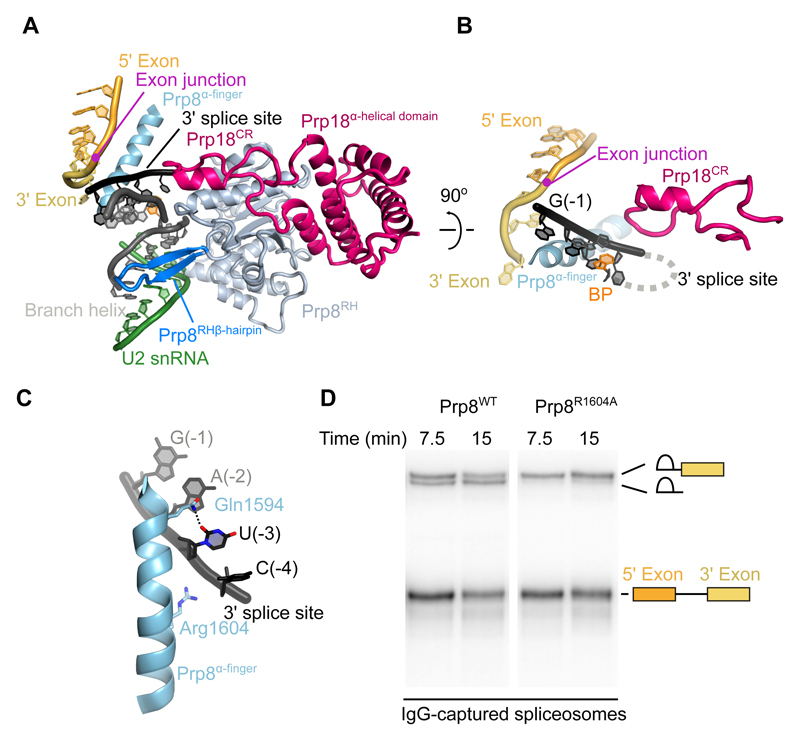
Compared to C* complex, which lacked the docked 3'-exon and 3'SS, new protein density is visible around the P complex active site. Prp8 and Prp18 cooperate to stabilize the docked 3'-splice site as well as the binding of the 3'-exon (Fig. 3A). The 3'-exon is sandwiched between the α-finger and the reverse transcriptase domains of Prp8 (Fig. 1C) (34). The α-finger (residues 1565-1610) of Prp8 forms an extended helix that wedges between the 3'-exon and 3'SS (Figs. 3B, C, S4). Before exon ligation, the contiguous 3'-exon and 3'SS would wrap around this helix, exposing the 3'SS phosphate for attack by the 5'-exon (Figs. 3B, S5). Alpha-finger residue Arg1604, which is essential for exon ligation (Figs. 3D, S7), contacts the phosphate backbone of the 3'SS residues -3 and -4. The highly conserved Gln1594 forms a hydrogen bond with the O2 carbonyl group of U(-3), and cytosine at the (-3) position could form an equivalent hydrogen bond, explaining the preference for pyrimidines at (-3) of the 3'SS (Figs. 3C, S6). The non-Watson-Crick RNA pairing between the BP, the 5'SS, and the 3'SS is reinforced by these sequences being clamped between the Prp8 α-finger and the β-hairpin of the Prp8 RNaseH domain (Fig. 3A). Consistent with the structure, mutations in the β-hairpin suppress the exon ligation defects of mutations at 3'SS A(-2) and the BP (35), further underscoring the essential structural role of Prp8 in 3'SS docking. The so-called conserved region of Prp18 (26) forms a loop that penetrates from outside the spliceosome through a channel formed by Prp8 into the active site, where it buttresses against nucleotides U(-3) and C(-4) of the 3'SS (Figs. 1C, 3A, B). Deletion of this conserved loop in Prp18 affects 3'SS selection (36), consistent with its direct involvement in exon ligation.
Figure 3. Proteins at the active site.
A, The Prp8 α-finger and β-hairpin clamp around the active site, with Prp18 bound on the outer face of the Prp8 RNaseH domain. CR, Prp18 conserved region; RH, Prp8 RNaseH domain. B, The Prp8 α-finger contacts both 3'-exon and 3' splice site; Prp18 CR loop contacts the 3'SS from the opposite side. C, Residues of the Prp8 α-finger that contact the 3' splice site. D, In vitro splicing reaction with wild-type (WT) and Arg1604 to Ala (R1604A) mutant Prp8. RNA species found in Prp8-immunoprecipitated spliceosomes are labelled schematically. R1604A causes a second-step defect, evidenced by accumulation of lariat intron-3'-exon intermediate.
Step 2 factors promote exon ligation
In C complex the branching factors Cwc25, Isy1, and Yju2 make extensive contacts with the branch and ACAGAGA helices and together directly stabilize the docking of the distorted branch helix into the active site to allow efficient branching (6, 12). In P complex, the exon ligation factors Prp18 and Slu7 make only one direct contact with the active site RNA, via the Prp18 conserved loop (Fig. 3A, B), while the α-helical domain of Prp18 binds the outer face of the Prp8 RNaseH domain (Fig. 4A). The N-terminus of Slu7 binds to the Cwc22 C-terminal and Prp8 linker and endonuclease domains. Slu7 then extends to the Prp8 N-terminal domain, where it is anchored by its zinc-knuckle domain before passing through the interface between the Prp8 N-terminal and endonuclease domains. After a helical interaction with Prp18 the globular C-terminal region of Slu7 binds the inner surface of the Prp8 RNaseH domain (Fig. 4A). This mostly peripheral binding of exon ligation factors suggests they promote splicing by a less direct mechanism than branching factors, which lock the branch helix in the active site.
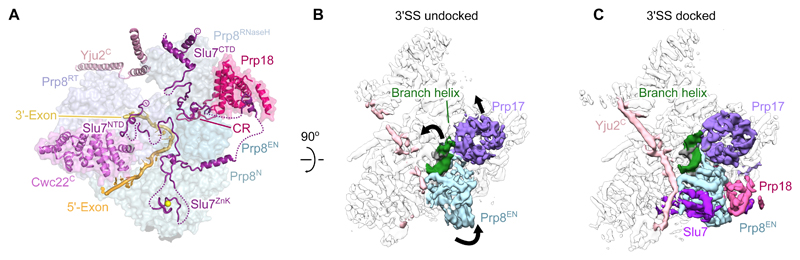
Figure 4. Docking of the 3' splice site is associated with binding of exon ligation factors.
A, The binding of exon ligation factors Slu7 and Prp18 to the surface of P complex. Disordered segments of Slu7 are shown with dotted lines. Domains of Prp8 are colored as indicated. N, Prp8 N-terminal domain; RT, Prp8 reverse transcriptase domain; EN, Prp8 endonuclease domain; ZnK, Slu7 zinc knuckle domain; CR, Prp18 conserved region. B, C, Cryo-EM density maps for P complex with the 3'SS docked and undocked. Maps were filtered to 5 Å resolution to aid visualization. Movements of the branch helix, Prp17, and the Prp8 endonuclease domain when changing into the docked conformation are indicated.
Docked and undocked conformations of P complex
We performed global classification of our cryo-EM dataset to assess the conformational dynamics of P complex spliceosomes. A subset comprising approximately half of the purified P complex particles lacks density for the 3'SS and Prp8 α-finger (Fig. S8). In this “undocked” conformation the junction between the 5'-exon and 3'-exon in ligated mRNA is still visible, confirming the undocked conformation represents a P complex state. These particles also lack density for Prp18 and Slu7, indicating that the presence of exon ligation factors correlates with stable docking of the 3'SS (Fig. 4B, C). This is consistent with previous biochemistry and genetics which suggest that Slu7 and Prp18 act after Prp16-mediated remodeling (7, 37) and promote juxtaposition of the splice sites in the exon ligation conformation (7). The branch helix, which is locked in place by exon ligation factors in the docked conformation, undergoes slight movement together with Prp17 in the undocked conformation and has weaker density, suggesting that it is more flexible in the absence of exon ligation factors (Figs. 4B, C, S8). Because the BP adenosine in the branch helix and the attached 5'SS G(+1) form the recognition platform for the 3'SS AG sequence, it is likely that the 3'SS can only stably dock when the branch helix is held rigid in the docked conformation.
Interestingly, the docked conformation competent for exon ligation is also associated with stronger density for two long collinear alpha helices spanning the width of the spliceosome from the C-terminus of Syf1 to the Prp8 RNaseH domain and contacting Cef1 (Fig. 4C). This density was previously seen weakly in C* complex (14) but limited local resolution precluded its assignment. The higher quality of the P complex density allowed assignment of these helices as the C-terminus of Yju2. In contrast, the N-terminus of Yju2 binds onto the branch helix and acts as a branching factor in C complex. However, the Yju2 N-terminus is no longer visible in C* or P complexes. Previous experiments showed that the N-terminal and C-terminal domains of Yju2 can act in trans; the N-terminal domain is essential for branching but impedes exon ligation, while the C-terminal domain promotes exon ligation, despite not being essential for viability (38). Our structure explains these apparently opposing roles of Yju2 and suggests the C-terminal domain stabilizes binding of Prp22 and Slu7 and acts as a brace, further restricting flexibility in the docked conformation. Thus Yju2 is both a branching and exon ligation factor and Prp16-dependent remodeling of C complex effects an exchange of its stable binding to the spliceosome from the N-terminal to the C-terminal domain (Fig. 5).
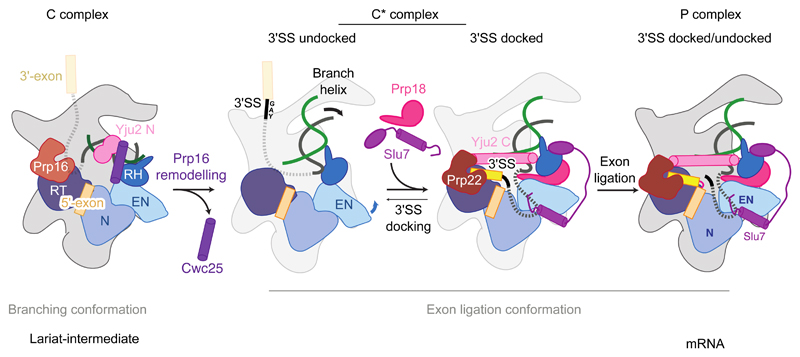
Figure 5. Model for the action of exon ligation factors.
Following Prp16-mediated remodeling of C complex the branching factors Cwc25, Yju2 N-domain and Isy1 (not depicted) are removed. The undocked branch helix is then locked in a conformation competent for second step catalysis by the binding of exon ligation factors Prp18 and Slu7 and the C-domain of Yju2. The 3'SS docked and undocked conformations may be in equilibrium due to flexibility of the branch helix and Prp8 Endonuclease domain in the C* state.
The two alternative forms of P complex suggest that exon ligation factors aid exon ligation in part by stabilizing the docked conformation: Prp18 and Slu7 bind together to the Prp8 RNaseH domain in its rotated conformation induced by Prp16 remodeling of C complex, while Slu7 anchors the RNaseH domain in place via multi-pronged interactions with the other domains of Prp8 (Fig. 5). Slu7 may also promote exon ligation by additional mechanisms. Slu7 is dispensable for exon ligation when the distance between the BP and 3'SS is less than nine nucleotides (19, 39), consistent with the seven ordered nucleotides we see in P complex, which could be joined by two additional nucleotides. The intron region between the BP and the docked 3'SS would likely protrude from the spliceosome through an opening between the Prp8 RNaseH, Linker, and RT domains. Intriguingly, the N-terminus of Slu7 binds at the base of this opening (Figs. 4A, 1B) and truncation of the N-terminus abolishes the ability of Slu7 to promote exon ligation for pre-mRNAs with long BP-3'SS distances (37). Thus the N-terminus of Slu7 could reduce the entropic cost of 3'SS docking. In conjunction with Prp18, Slu7 could also guide the 3'SS into the active site and promote the correct topology for stable 3'SS docking.
Conclusions
The molecular mechanism of 3'SS recognition during the catalytic phase of pre-mRNA splicing had been elusive despite decades of functional studies. Our P complex structure now shows that the 3'SS is recognized by pairing with the 5'SS and the branch point adenosine. This interaction involves all invariant nucleotides (GU and AG) at the 5' and 3' ends of the intron and the invariant branch point adenosine, making the exon ligation step a final quality check of the splicing reaction. Indeed, mutation of any of these invariant nucleotides impairs mRNA formation (40, 41). Thus, like the 5'SS GU dinucleotide and the branch point A nucleotide, recognition of the 3'SS AG dinucleotide is achieved through non-Watson-Crick RNA-RNA interactions stabilized by protein factors, explaining the conservation of these splice site sequences throughout eukaryotic evolution. An AU at the 5'SS and a AC at the 3'SS – a combination observed in the human minor spliceosome (4) and as a suppressor of 5'SS mutations in yeast (30) – could also be tolerated (Fig. S6), consistent with the major and minor spliceosome utilizing a similar mechanism for 3'SS selection.
Interestingly, although the structure of the RNA-based active site is remarkably conserved between the spliceosome and group II self-splicing introns (Fig. S5) (18, 42, 43), our P complex structure reveals that specific recognition of the 3'SS differs between the two splicing systems. Whereas in the group II intron the J2/3 junction interacts with the 3'SS (44), in the spliceosome the equivalent region in U6 snRNA (nucleotide A51) interacts instead with the 5'SS to stabilize the C* and P complex configuration of the active site (Fig. 2B) (14). Nonetheless, in both systems the 5'SS and the branch adenosine appear critical for both steps of splicing (Fig. 2) (43, 45), potentially explaining conservation of the 2'-5' linkage during evolution of both splicing systems.
Overall our P complex structure elucidates the mechanism of 3'-splice site selection, showing the crucial role of the excised lariat-intron in organizing the active site and splice sites for exon ligation. The structure now completes our basic understanding of the two-step splicing reaction, showing how the dynamic protein scaffold of the spliceosome cradles and modulates a fundamentally RNA-based mechanism for splice site recognition during catalysis (18).
Supplementary Material
Summary.
A CryoEM structure of the spliceosome shows how the 3' ends of introns are recognized and how exons are joined to form mature mRNA.
Acknowledgements
M.E.W. prepared the sample, made EM grids, collected and processed EM data, carried out model building, and refined the structure. S.M.F. suggested the RNaseH method, prepared Prp22, and assisted data acquisition. W.P.G. was involved in the early stage of the project. M.E.W., S.M.F., and K.N. analyzed the structure and drafted a manuscript. Mutagenesis and splicing assays were carried out by A.J.N. and C.M.N. The manuscript was finalized by M.E.W., S.M.F., and K.N. with input from all authors. K.N. initiated and coordinated the spliceosome project. We thank C. Savva, S. Chen, G. McMullan, G. Cannone, J. Grimmett, and T. Darling for smooth running of the EM and computing facilities; the mass spectrometry facility for help with protein identification; and the members of the spliceosome group for help and advice throughout the project. We thank J. Löwe, V. Ramakrishnan, D. Barford, and R. Henderson for their continuing support and C. Plaschka, P. C. Lin, C. Charenton, C. J. Oubridge, and L. Strittmatter for critical reading of the manuscript. The project was supported by the Medical Research Council (MC_U105184330) and European Research Council Advanced Grant (AdG-693087-SPLICE3D). M.E.W was supported by a Cambridge-Rutherford Memorial PhD Scholarship, S.M.F. was supported by a Marie Skłodowska-Curie fellowship. Cryo-EM maps are deposited in the Electron Microscopy Data Bank under accession numbers EMD-3979 (3'SS docked) and EMD-3980 (3'SS undocked); atomic models are deposited in the Protein Data Bank under accession numbers 6EXN.
References
- 1.Wahl MC, Will CL, Lührmann R. The Spliceosome: Design Principles of a Dynamic RNP Machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Fica SM, et al. RNA catalyses nuclear pre-mRNA splicing. Nature. 2013;503:229–234. doi: 10.1038/nature12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci USA. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel AA, Steitz JA. Splicing double: insights from the second spliceosome. Nat Rev Mol Cell Biol. 2003;4:960–970. doi: 10.1038/nrm1259. [DOI] [PubMed] [Google Scholar]
- 5.Schwer B, Guthrie C. PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature. 1991;349:494–499. doi: 10.1038/349494a0. [DOI] [PubMed] [Google Scholar]
- 6.Galej WP, et al. Cryo-EM structure of the spliceosome immediately after branching. Nature. 2016;537:197–201. doi: 10.1038/nature19316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Semlow DR, Blanco MR, Walter NG, Staley JP. Spliceosomal DEAH-Box ATPases Remodel Pre-mRNA to Activate Alternative Splice Sites. Cell. 2016;164:985–998. doi: 10.1016/j.cell.2016.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Company M, Arenas J, Abelson J. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature. 1991;349:487–493. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- 9.Yan C, et al. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science. 2015;349:1182–1191. doi: 10.1126/science.aac7629. [DOI] [PubMed] [Google Scholar]
- 10.Yan C, Wan R, Bai R, Huang G, Shi Y. Structure of a yeast activated spliceosome at 3.5 Å resolution. Science. 2016;353:904–911. doi: 10.1126/science.aag0291. [DOI] [PubMed] [Google Scholar]
- 11.Rauhut R, et al. Molecular architecture of the Saccharomyces cerevisiae activated spliceosome. Science. 2016;353:1399–1405. doi: 10.1126/science.aag1906. [DOI] [PubMed] [Google Scholar]
- 12.Wan R, Yan C, Bai R, Huang G, Shi Y. Structure of a yeast catalytic step I spliceosome at 3.4 Å resolution. Science. 2016;353:895–904. doi: 10.1126/science.aag2235. [DOI] [PubMed] [Google Scholar]
- 13.Plaschka C, Lin P-C, Nagai K. Structure of a pre-catalytic spliceosome. Nature. 2017;546:617–621. doi: 10.1038/nature22799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fica SM, et al. Structure of a spliceosome remodelled for exon ligation. Nature. 2017;542:377–380. doi: 10.1038/nature21078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan C, Wan R, Bai R, Huang G, Shi Y. Structure of a yeast step II catalytically activated spliceosome. Science. 2017;355:149–155. doi: 10.1126/science.aak9979. [DOI] [PubMed] [Google Scholar]
- 16.Bertram K, et al. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature. 2017;542:318–323. doi: 10.1038/nature21079. [DOI] [PubMed] [Google Scholar]
- 17.Zhang X, et al. An Atomic Structure of the Human Spliceosome. Cell. 2017;169:918–929.e14. doi: 10.1016/j.cell.2017.04.033. [DOI] [PubMed] [Google Scholar]
- 18.Fica SM, Nagai K. Cryo-electron microscopy snapshots of the spliceosome: structural insights into a dynamic ribonucleoprotein machine. Nat Struct Mol Biol. 2017;24:791–799. doi: 10.1038/nsmb.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohrt T, et al. Molecular dissection of step 2 catalysis of yeast pre-mRNA splicing investigated in a purified system. RNA. 2013;19:902–915. doi: 10.1261/rna.039024.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tseng CK, Liu HL, Cheng SC. DEAH-box ATPase Prp16 has dual roles in remodeling of the spliceosome in catalytic steps. RNA. 2010;17:145–154. doi: 10.1261/rna.2459611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lesser CF, Guthrie C. Mutations in U6 snRNA that alter splice site specificity: implications for the active site. Science. 1993;262:1982–1988. doi: 10.1126/science.8266093. [DOI] [PubMed] [Google Scholar]
- 22.Kandels-Lewis S, Séraphin B. Role of U6 snRNA in 5′ Splice Site Selection. Science, New Series. 1993;262:2035–2039. doi: 10.1126/science.8266100. [DOI] [PubMed] [Google Scholar]
- 23.Newman AJ, Norman C. U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell. 1992;68:743–754. doi: 10.1016/0092-8674(92)90149-7. [DOI] [PubMed] [Google Scholar]
- 24.Sontheimer EJ, Steitz JA. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science. 1993;262:1989–1996. doi: 10.1126/science.8266094. [DOI] [PubMed] [Google Scholar]
- 25.Schwer B. A conformational rearrangement in the spliceosome sets the stage for Prp22-dependent mRNA release. Mol Cell. 2008;30:743–754. doi: 10.1016/j.molcel.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang J, Horowitz DS, Xu RM. Crystal structure of the functional domain of the splicing factor Prp18. Proc Natl Acad Sci USA. 2000;97:3022–3027. doi: 10.1073/pnas.97.7.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fica SM, Mefford MA, Piccirilli JA, Staley JP. Evidence for a group II intron-like catalytic triplex in the spliceosome. Nat Struct Mol Biol. 2014;21:464–471. doi: 10.1038/nsmb.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spingola M, Grate L, Haussler D, Ares M. Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA. 1999;5:221–234. doi: 10.1017/s1355838299981682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tseng CK, Cheng SC. Both Catalytic Steps of Nuclear Pre-mRNA Splicing Are Reversible. Science. 2008;320:1782–1784. doi: 10.1126/science.1158993. [DOI] [PubMed] [Google Scholar]
- 30.Parker R, Siliciano PG. Evidence for an essential non-Watson–Crick interaction between the first and last nucleotides of a nuclear pre-mRNA intron. Nature. 1993;361:660–662. doi: 10.1038/361660a0. [DOI] [PubMed] [Google Scholar]
- 31.Luukkonen BG, Séraphin B. The role of branchpoint-3′ splice site spacing and interaction between intron terminal nucleotides in 3′ splice site selection in Saccharomyces cerevisiae. EMBO J. 1997;16:779–792. doi: 10.1093/emboj/16.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hornig H, Aebi M, Weissmann C. Effect of mutations at the lariat branch acceptor site on beta-globin pre-mRNA splicing in vitro. Nature. 1986;324:589–591. doi: 10.1038/324589a0. [DOI] [PubMed] [Google Scholar]
- 33.McPheeters DS. Interactions of the yeast U6 RNA with the pre-mRNA branch site. RNA. 1996;2:1110–1123. [PMC free article] [PubMed] [Google Scholar]
- 34.Galej WP, Oubridge C, Newman AJ, Nagai K. Crystal structure of Prp8 reveals active site cavity of the spliceosome. Nature. 2013;493:638–643. doi: 10.1038/nature11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu L, Query CC, Konarska MM. Opposing classes of prp8 alleles modulate the transition between the catalytic steps of pre-mRNA splicing. Nat Struct Mol Biol. 2007;14:519–526. doi: 10.1038/nsmb1240. [DOI] [PubMed] [Google Scholar]
- 36.Crotti LB, Bacíková D, Horowitz DS. The Prp18 protein stabilizes the interaction of both exons with the U5 snRNA during the second step of pre-mRNA splicing. Genes Dev. 2007;21:1204–1216. doi: 10.1101/gad.1538207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.James S-A, Turner W, Schwer B. How Slu7 and Prp18 cooperate in the second step of yeast pre-mRNA splicing. RNA. 2002;8:1068–1077. doi: 10.1017/s1355838202022033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiang T-W, Cheng S-C. A weak spliceosome-binding domain of Yju2 functions in the first step and bypasses Prp16 in the second step of splicing. Mol Cell Biol. 2013;33:1746–1755. doi: 10.1128/MCB.00035-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brys A, Schwer B. Requirement for SLU7 in yeast pre-mRNA splicing is dictated by the distance between the branchpoint and the 3′ splice site. RNA. 1996;2:707–717. [PMC free article] [PubMed] [Google Scholar]
- 40.Aebi M, Hornig H, Padgett RA, Reiser J, Weissmann C. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell. 1986;47:555–565. doi: 10.1016/0092-8674(86)90620-3. [DOI] [PubMed] [Google Scholar]
- 41.Vijayraghavan U, et al. Mutations in conserved intron sequences affect multiple steps in the yeast splicing pathway, particularly assembly of the spliceosome. EMBO J. 1986;5:1683–1695. doi: 10.1002/j.1460-2075.1986.tb04412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marcia M, Pyle AM. Visualizing Group II Intron Catalysis through the Stages of Splicing. Cell. 2012;151:497–507. doi: 10.1016/j.cell.2012.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robart AR, Chan RT, Peters JK, Rajashankar KR, Toor N. Crystal structure of a eukaryotic group II intron lariat. Nature. 2014;514:193–197. doi: 10.1038/nature13790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jacquier A, Michel F. Base-pairing interactions involving the 5′ and 3′ terminal nucleotides of group II self-splicing introns. J Mol Biol. 1990;213:437–447. doi: 10.1016/S0022-2836(05)80206-2. [DOI] [PubMed] [Google Scholar]
- 45.Costa M, Walbott H, Monachello D, Westhof E, Michel F. Crystal structures of a group II intron lariat primed for reverse splicing. Science. 2016;354 doi: 10.1126/science.aaf9258. aaf9258. [DOI] [PubMed] [Google Scholar]
- 46.Umen JG, Guthrie C. Prp16p, Slu7p, and Prp8p interact with the 3' splice site in two distinct stages during the second catalytic step of pre-mRNA splicing. RNA. 1995;1:584–597. [PMC free article] [PubMed] [Google Scholar]
- 47.Lin RJ, Newman AJ, Cheng SC, Abelson J. Yeast mRNA splicing in vitro. J Biol Chem. 1985;260:14780–14792. [PubMed] [Google Scholar]
- 48.Abelson J, Hadjivassiliou H, Guthrie C. Methods in Enzymology. Vol. 472. Elsevier; 2010. pp. 31–40. Methods in Enzymology. [DOI] [PubMed] [Google Scholar]
- 49.Wu TP, Ruan KC, Liu WY. A fluorescence-labeling method for sequencing small RNA on polyacrylamide gel. Nucleic Acids Res. 1996;24:3472–3473. doi: 10.1093/nar/24.17.3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 51.Schwer B, Meszaros T. RNA helicase dynamics in pre-mRNA splicing. EMBO J. 2000;19:6582–6591. doi: 10.1093/emboj/19.23.6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turner IA, Norman CM, Churcher MJ, Newman AJ. Dissection of Prp8 protein defines multiple interactions with crucial RNA sequences in the catalytic core of the spliceosome. RNA. 2006;12:375–386. doi: 10.1261/rna.2229706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scheres SHW. A Bayesian view on cryo-EM structure determination. J Mol Biol. 2012;415:406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kimanius D, Forsberg BO, Scheres SH, Lindahl E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife. 2016;5:e18722. doi: 10.7554/eLife.18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen S, et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang J, et al. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tauchert MJ, Fourmann J-B, Lührmann R, Ficner R. Structural insights into the mechanism of the DEAH-box RNA helicase Prp43. Elife. 2017;6:e21510. doi: 10.7554/eLife.21510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Popenda M, et al. Automated 3D structure composition for large RNAs. Nucleic Acids Res. 2012;40:e112. doi: 10.1093/nar/gks339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Afonine PV, et al. Joint X-ray and neutron refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2010;66:1153–1163. doi: 10.1107/S0907444910026582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 65.Leontis NB, Stombaugh J, Westhof E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.