Abstract
Background:
Glioblastoma multiforme (GBM) is the most aggressive and deadly primary brain cancer that arises from astrocytes and classified as grade IV. Recently, exosomes have been reported as an essential mediator in diverse cancer carcinogenesis and metastasis. However, their role in GBM is still unclear. In this study, we aimed to investigate whether blood exosomes can be potential clinical diagnostic markers for GBM.
Methods:
We used a xenograft orthotopic mouse model to detect the differentially expressed genes in the brain and blood exosomes of original/recurrent GBM.
Results:
We found that recurrent GBM had stronger growth capacity and lethality than original GBM in the mouse model. A gene microarray of original tumors and blood exosomes from GBM orthotopic xenografts results showed that DNM3, p65 and CD117 expressions increased, whereas PTEN and p53 expressions decreased in both original tumors and blood exosomes. In the recurrent GBM tumor model, DNM3 and p65 showed increased expressions, whereas ST14 and p53 showed decreased expressions in tumor and blood exosomes of the recurrent GBM mouse model.
Conclusion:
In summary, we found that DNM3, p65 and p53 had a similar trend in brain and blood exosomes both for original and recurrent GBM, and could serve as potential clinical diagnostic markers for GBM.
Keywords: exosomes, DNM3, glioblastoma multiforme (GBM), p53, p65
Introduction
Glioblastoma multiforme (GBM) is the most aggressive and deadly primary brain cancer that arise from astrocytes and is classified as grade IV, with a 5-year survival rate of only 4–5%.1 The current standard treatment for GBM is surgery followed by the radiation and chemotherapy.2 Although treatments for GBM have been greatly improved, the recurrence rate of GBM remains high. Thus, there is an urgent need for early detection of local recurrence in GBM treatments.3
GBM is often initially diagnosed by computed tomography (CT) or magnetic resonance imaging (MRI). In most cases, GBM appears as ring-enhancing lesions under the view of MRI, which is not specific because other lesions may have a similar appearance.4 The suspected GBM requires definitive diagnosis such as craniotomy with tumor resection, stereotactic biopsy or pathologic confirmation. However, biopsy and resection often lead to under-grading of the lesion, as the tumor grade is determined by the most malignant portion of the tumor. Moreover, distinguishing the primary GBM from secondary GBM is essential, as primary GBM represents a worse prognosis and tumors with different biology may have different responses to therapy.5 Thus, it is important to find out an efficient method for GBM diagnosis.
Exosomes are cell-derived vesicles that are found in many fluids within the mammalian organism such as blood, urine and ascites, as well as in the medium of cell cultures.6 Exosomes were initially considered as cellular waste disposal.7 However, recent data support the idea that exosomes were cargo containers for eukaryotic cells to exchange biomolecules, such as proteins, mRNA and miRNA, which participate in cell–cell communication, cell migration, angiogenesis and tumor growth.8 The recipient cells could capture exosomes by targeting the surface proteins of exosomes, and the contents of exosomes could then alter the biological state of the recipient cells. For example, some full-length mRNAs found in exosomes might influence protein expressions in the recipient cells, even though most exosome mRNAs are degraded into short fragments that are <200 nucleotides.9
Studies have shown that aberrant exosomes or their altered pathological cargos facilitated tumor growth and metastasis.10,11 Tumor progression, metastasis and immune escaping could be modulated by the interaction between tumor cell-derived exosomes and recipient cells in the tumor microenvironment. Thus, researchers targeted tumor cell-derived exosomes to investigate their potential therapeutic roles in cancer.12 Evidence supports that the RNAs contained in exosomes are similar to the original cancer cells.13,14 The growing interests of exosomes focus on their potential applications as biomarkers in clinical diagnosis. Because of their excellent stability, exosomes exert higher specificity and sensitivity, compared with the conventional diagnostic materials such as urine or serum. Moreover, the contents of exosomes, such as proteins, mRNAs and microRNAs (miRNAs), provide them with potent properties to be clinical diagnosis biomarkers.9,15
As blood is the optimal specimen to detect the altered body exosomes, we aimed to investigate whether the GBM-associated exosomes could be detected in blood, and whether these exosomes could accurately represent the developing stages of GBM. Specifically, we established connections between blood exosomes and GBM by comparing their respective gene expressions. Moreover, we found that GBM-derived exosomes could be the biomarker for early stage clinical diagnosis of GBM.
Methods
Orthotopic xenograft model
The animal experiments were approved by The Second Hospital of Hebei Medical University, China and performed in accordance with standard policies. All efforts were made to minimize the number and the suffering of the animals. All procedures performed in studies involving human participants were in accordance with the ethics standards of the institutional and national research committee and with the 1964 Helsinki Declaration. Written consent was collected from all the participants. The study was approved by Ethics Committee of Second Hospital of Hebei Medical University. GBM tumor tissues were collected following previously established procedures.16 Briefly, the GBM tumor tissues were obtained from patients diagnosed with GBM and undergoing surgery. Excised patient’s tumor tissues were washed with Dulbecco’s modified Eagle’s medium (DMEM) and minced to small pieces. Then the Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) was added to the tumor tissue and planted subcutaneously to the nude mice. The primary tumors were allowed to grow up to 1 cm in diameter. The patient-derived orthotopic xenograft model was established as adapted from previously established protocols.17 Briefly, nude mice aged 6–8 weeks were anesthetized with sodium pentobarbital (50 mg/kg, intraperitoneal injection). Tumor cells (1 × 104) from patients suspended in 2 μl of culture medium (DMEM with 10% fetal bovine serum) were injected into the right cerebral hemisphere (1 mm to right of midline, 1.5 mm anterior to lambdoidal suture, 3 mm deep) via a 10 μl 26-gauge Gastight 1701 syringe needle (Hamilton Company, Allston, MA, USA). After the injection, mice were observed till they reached a moribund state.
To compare the proliferation rate between the primary and recurrent tumor, the mice were sacrificed 2 months after the injection. The tumor tissues were taken and the volumes were measured. A total of 4 patient-derived samples and 10 mice in each experimental group were used for each assay.
Microarray
The quantitative reverse transcription polymerase chain reaction (RT-PCR) arrays were used to compare the gene expression between the primary, recurrent GBM and the normal brain tissues, as well as the exosomes containing mRNA level between the xenograft and the normal mice. Gene expression was analyzed as ΔΔCt value normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) level.
Isolation of tissue exosome mRNA and quantitative polymerase chain reaction (qPCR)
Normal brain/tumor tissue punches with 0.5 mm in diameter were collected from brain regions and Tissuelyser II (Qiagen Valencia, CA, USA) was added to the tissues. The frozen tissues were homogenized and extraction buffer was directly added to the tissue powder. QiaZol and Qiagen RNeasy Mini Kit were used to extract the total RNA of tissues according to the manufacturer’s instruction. The exosome mRNA was isolated by ultracentrifugation as described previously.18 In brief, 0.3–0.5 ml whole blood was collected from xenograft mice (n = 10 each group) into an anticoagulation tube and then centrifuged at 300 × g for 10 min to pellet the cells. The supernatants were further centrifuged at 16,500 × g for 20 min and filtered through a 0.2-μm filter to remove cell debris. The filtered supernatant was ultracentrifuged at 120,000 × g for 60 min. After this step, the circulating cell-free nucleic acid was in the supernatant and removed, and the exosome content in the pellets were verified.
Exosome RNA was isolated using RNeasy mini kit (Qiagen,) following the manufactures protocol. The mRNAs were determined by qPCR with the following premiers: DNM3 (F, 5’-ACC CCA CAC CTG CAG AAG GT-3’; R 5’-TGG AGA GCA ACT GTC CCT GTA-3’), p65 (F, 5’-CTG CAG TTT GAT GAT GAA GA-3’; R, 5’-TAG GCG AGT TAT AGC CTC AG-3’), LLGL2 (F, 5’-ATG AGG CGG TTC CTG AGG CCA-3’; R, 5’-GAG GAA TGT GGC AGG ACC ACG-3’), ST14 (F, 5’-CAC GAA TGA TGT GTG TGG GTT-3’; R, 5’-CCT GGA ACA TTC GCC CAT CT-3’), PTEN (F, 5’-CAA GAT GAT GTT TGA AAC TAT-3’; R, 5’-CCT TTA GCT GGC AGA CCA CAA-3’) and GAPDH (F, 5’-CAC ATC AAG AAG GTG GTG-3’; R, 5’-TGT CAT ACC AGG AAA TGA-3’).
Western blots
Western blots (WBs) from the exosomes were performed according to the standard WB protocol. Briefly, the exosome pellets were dissolved with RIPA lysis buffer and the protein amount was determined using a BCA kit. A total of 20 μg protein was separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred to polyvinylidene fluoride (PVDF) membranes (Bio-Rad, Hercules, CA, USA). The membranes were blocked in 5% non-fat milk for 30 min, and then incubated with primary antibodies [anti-DNM3 (1:1000), anti-p65 (1:1000), anti-p53 (1:1000) or anti-beta-actin (1:10,000)] at 4°C overnight. All antibodies were purchased from Santa Cruz Biotechnology. Membranes were then incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies for another hour at room temperature and developed with an chemiluminescence kit (Bio-Rad). Signals were visualized using Chemidoc XRS+ (Bio-Rad). The ImageJ 1.50i software (Wayne Rasband, National Institute of Health, USA) was used to quantify the WB results.
Statistical analyses
All data were expressed as the mean ± standard deviation (SD). Student’s t tests (two-tailed) and one-way analysis of variance analysis were used to compare the differences between groups or among three groups. A p value <0.05 was considered as significant.
Results
Recurrent GBM cells are more lethal than primary GBM cells in mouse model
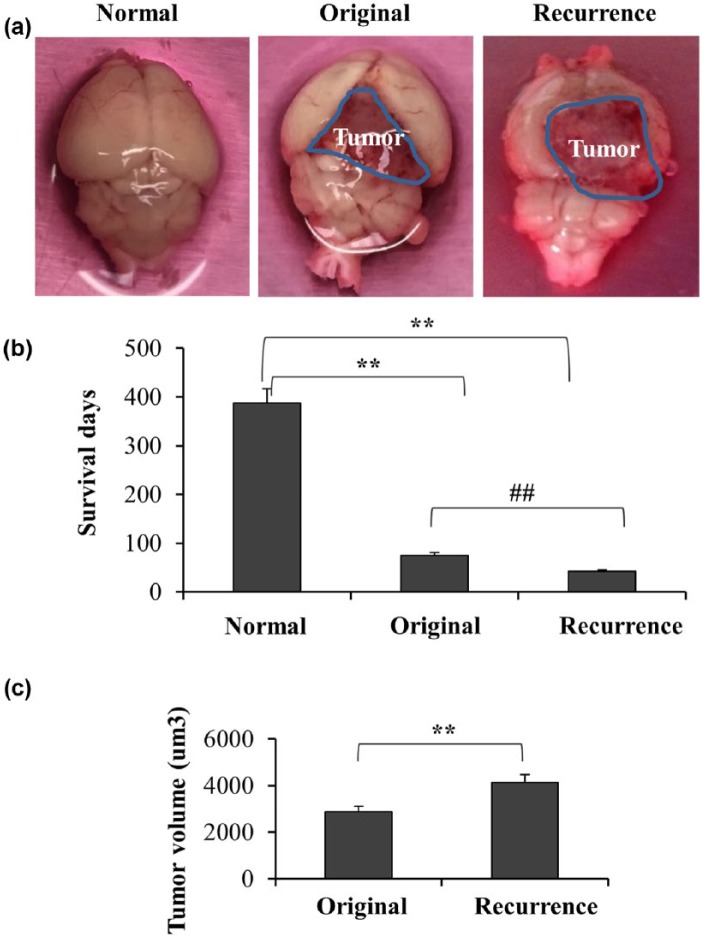
To explore the exosomes of GBM in vivo, we firstly established orthotopic xenograft model with original and recurrent human GBM cells. As shown in Figure 1(a), 1 × 104 patient-derived GBM cells were injected in the right cerebrum of mice and formed tumor eventually. We then compared the survival days among the normal mice, mice injected with original GBM cells and recurrent GBM cells, respectively. We found the survival days of the mice injected with recurrent GBM cells were significantly less than those injected with original GBM cells, suggesting the recurrent tumor cells were more aggressive than the original cells [Figure 1(b)]. Furthermore, the volume of recurrent tumors was larger than those of original tumors at 2 months after injection [Figure 1(c)].
Figure 1.
Recurrent GBM had stronger growth capacity and lethality than original GBM in mice model. (a) The xenograft orthotopic mouse model with original or recurrent GBM were established after injecting 1 × 104 patient-derived cells in the right cerebrum, respectively. A total of 4 patient-derived samples and 10 mice were used. (b) The survival times were calculated after each mouse was dead suffering sick, moribund and skinny (n = 10, **p < 0.01, ##p < 0.01). C. Tumor growth in xenografts inoculated from original or recurrent GBM after 2 months while all the mice were still survival but sick. Values are mean and SD from at least three independent experiments in duplicate (p < 0.01).
GBM, glioblastoma multiforme; SD, standard deviation.
Gene expression analysis of original tumors and blood exosomes from GBM orthotopic xenografts
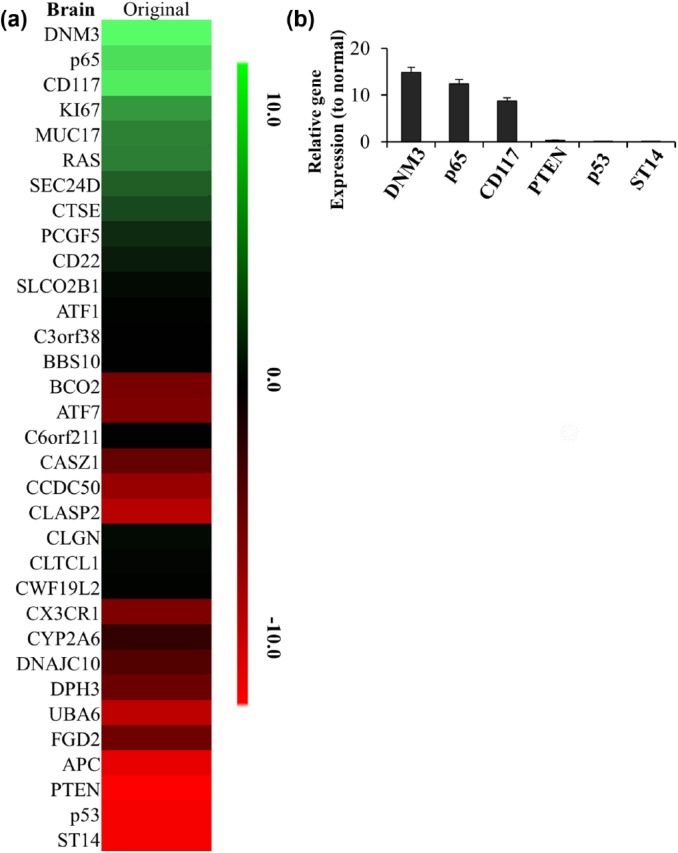
We next screened the differences in gene expression levels between original tumor and normal brain tissues. We sacrificed the orthotopic xenograft bearing original tumor 2 months after tumor cell injection. By mRNA microarray, we found that several genes showed significantly different expressions between original tumor and normal brain tissues as shown in Figure 2(a). In tumor tissues, the most upregulated genes were p65, DNM3 and CD117, which were upregulated more than 10 folds comparing with the normal brain tissues. The most downregulated genes were PTEN, p53 and ST14. Further qPCR analysis confirmed those results [Figure 2(b)].
Figure 2.
The changed gene list in brain of original GBM mouse model. (a) The heatmap of 33 differentially expressed genes identified in brain of original GBM mouse model. The decreased and increased genes are indicated by range of green and red intensities, respectively. (b) The top 3 of high-expressed or low-expressed gene ratios to normal tissue in brain of original GBM mouse model were detected by PCR. Ct values detected by real-time PCR showed p65, DNM3, CD117, PTEN, p53 and ST14 had the greatest change in brain of original GBM. Relative expression values represent mean and SD from three independent experiments.
GBM, glioblastoma multiforme; PCR, polymerase chain reaction; SD, standard deviation.
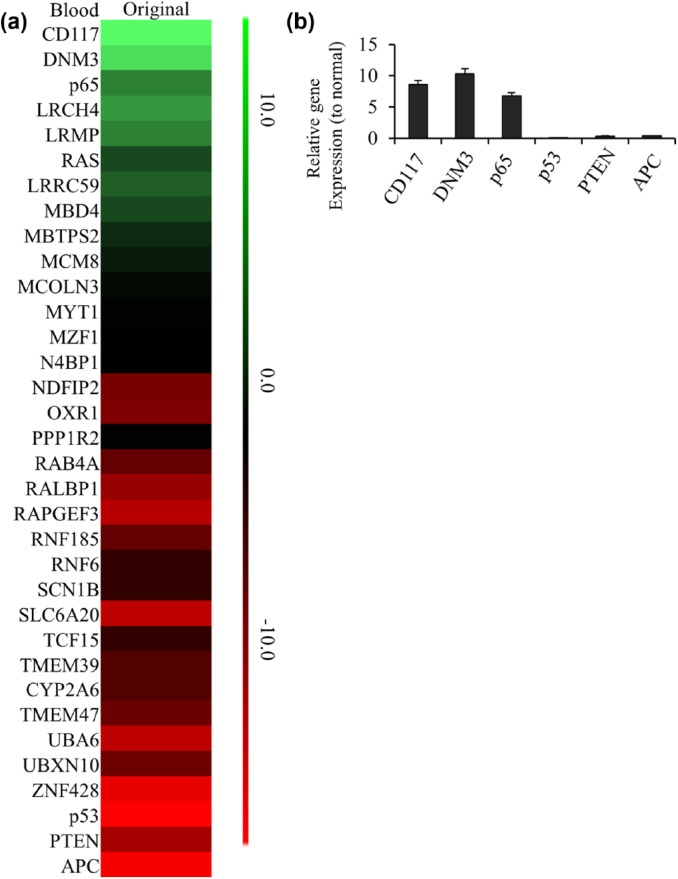
We then separated the exosomes from the blood of xenograft mice bearing original tumor and control mice. The data from mRNA array using exosomes showed several genes expressed differently between xenograft and normal mice [Figure 3(a)]. The top 3 upregulated gene in xenograft mice exosomes were p65, DNM3 and CD117; while the top 3 downregulated gene were PTEN, p53 and APC. Subsequent real-time PCR results showed those 6s genes had the greatest changes in exosomes from xenograft mice as well [Figure 3(b)].
Figure 3.
The changed gene list in blood exosome of original GBM mouse model. (a) The heatmap of 33 differentially expressed genes identified in blood exosome of original GBM mouse model. The decreased and increased genes are indicated by range of green and red intensities, respectively. (b) The top 3 of high-expressed or low-expressed gene ratios to normal tissue in blood exosome of original GBM mouse model were detected by PCR. Ct values detected by real-time PCR showed p65, DNM3, CD117, PTEN, p53 and APC had the greatest change in blood of original GBM. Relative expression values represent mean and SD from three independent experiments.
GBM, glioblastoma multiforme; PCR, polymerase chain reaction; SD, standard deviation.
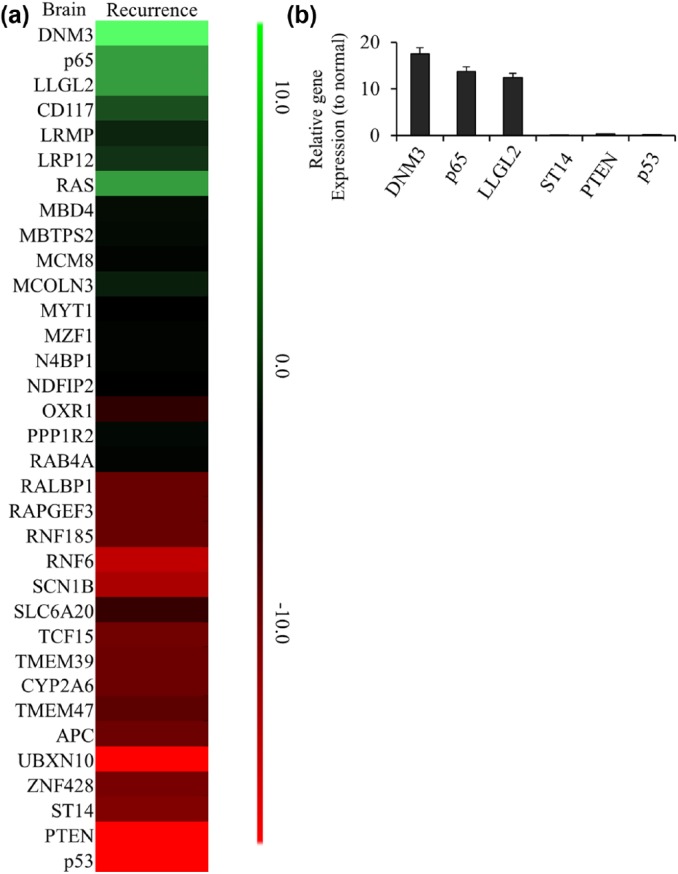
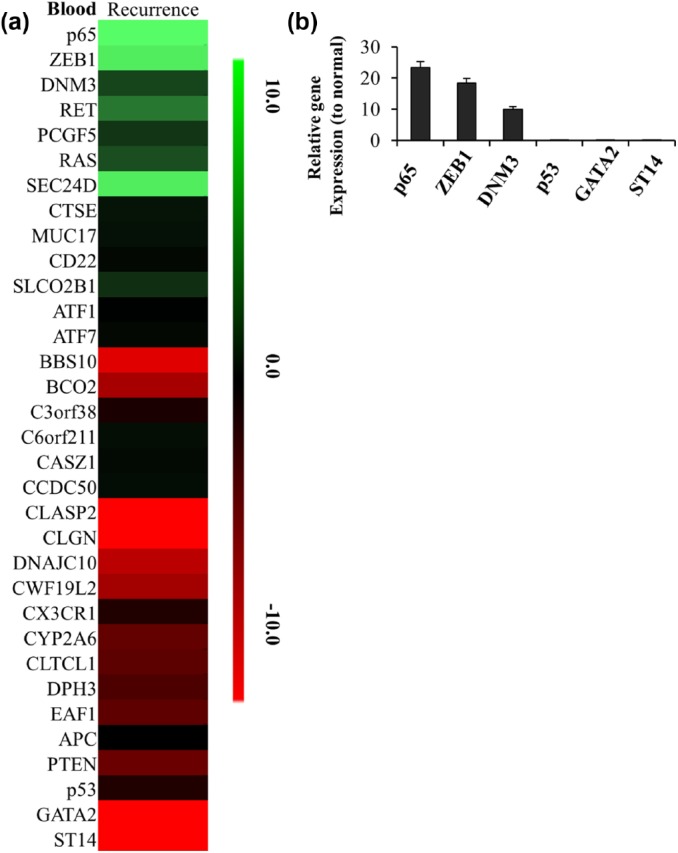
Our previous data showed the recurrent tumor cells were more aggressive than the original ones. We wonder if there were any differences in mRNA expressions in recurrent tumor or the exosomes from xenograft mice bearing recurrent tumors. We compared the mRNA levels between the recurrent tumor and normal brain tissues. Several genes were found to be dysregulated in recurrent tumor as shown in Figure 4(a). Real-time PCR results showed the top 3 upregulated genes in tumors were p65, DNM3 and LLGL2; while the top 3 downregulated gene were PTEN, p53 and ST14 [Figure 4(b)]. We separated the exosomes from the blood of xenografts bearing recurrent tumor and compared the mRNA expressions with normal exosomes as well. As shown in Figure 5(a), there were several gene dysregulated in exosomes of recurrent tumor xenografts when comparing to the healthy mice. Real-time PCR confirmed the top 3 upregulated gene were p65, DNM3 and ZEB1; while the top 3 downregulated genes were GATA2, p53 and ST14 in exosomes from xenografts bearing recurrent tumors [Figure 5(b)].
Figure 4.
The changed gene list in brain of recurrent GBM mouse model. (a) The heatmap of 33 differentially expressed genes identified in brain of recurrent GBM mouse model. The decreased and increased genes are indicated by range of green and red intensities, respectively. (b) The top 3 of high-expressed or low-expressed gene ratios to normal tissue in brain of recurrent GBM mouse model were detected by PCR. Ct values detected by real-time PCR showed p65, DNM3, LLGL2, PTEN, p53 and ST14 had the greatest change in brain of recurrent GBM. Relative expression values represent mean and SD from three independent experiments.
GBM, glioblastoma multiforme; PCR, polymerase chain reaction; SD, standard deviation.
Figure 5.
The changed gene list in blood exosome of recurrent GBM mouse model. (a) The heatmap of 33 differentially expressed genes identified in blood exosome of recurrent GBM mouse model. The decreased and increased genes are indicated by range of green and red intensities, respectively. (b) The top 3 of high-expressed or low-expressed gene ratios to normal tissue in blood exosome of recurrent GBM mouse model were detected by PCR. Ct values detected by real-time PCR showed p65, DNM3, ZEB1, GATA2, p53 and ST14 had the greatest change in blood of recurrent GBM. Relative expression values represent mean and SD from three independent experiments.
GBM, glioblastoma multiforme; PCR, polymerase chain reaction; SD, standard deviation.
We also compared the changed genes between original and recurrent tumor cells in brain tissue and blood exosomes (Figure S1). The results showed that DNM3 and p65 were highly expressed in brain and blood exosomes of original and recurrent GBM tumors, and ZEB1 were highly expressed only in blood exosomes of recurrent GBM tumor [Figure S1(a)]. PTEN, P53 and APC were downregulated in brain and blood exosomes of original and recurrent GBM tumors, and ST14 and GATA2 were downregulated only in blood exosomes of recurrent GBM tumor [Figure S1(b)].
DNM3, p65 and p53 are potential clinical diagnostic markers for GBM patients
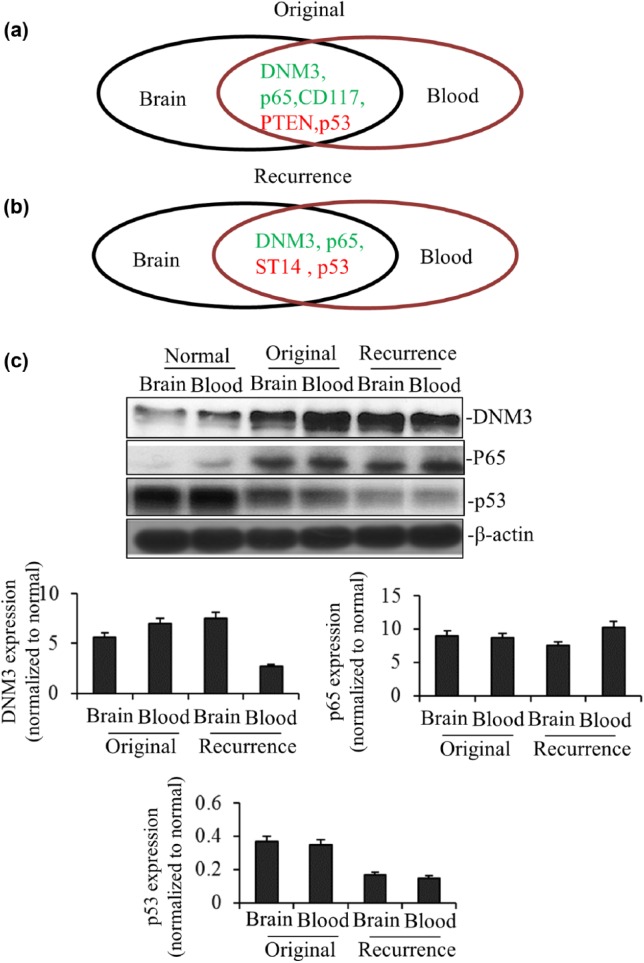
Our previous data showed DNM3, p65 and CD117 were upregulated in both tumor tissue and exosomes of xenografts bearing original GBM, while PTEN and p53 were downregulated [Figure 6(a)]. In xenografts bearing recurrent GBM, we found DNM3 and p65 were upregulated both in primary tumor site and exosomes; while ST14 and p53 were downregulated [Figure 6(b)]. Comprehensively, our results showed that DNM3 and p65 were upregulated in both primary tumor sites and exosomes in original and recurrent GBM xenografts; meanwhile, p53 was downregulated. Then we performed WB analysis to verify the above results. As shown in Figure 6(c), the protein levels of DNM3 and p65 were increased in both primary tumor sites and exosomes of xenografts bearing original and recurrent GBM, while p53 protein level was decreased, consistent with their respective changes in mRNA level.
Figure 6.
DNM3, p65 and p53 are the potential clinical diagnostic markers. In summary, DNM3, p65 and p53 had the similar trend in brain and blood exosome both for original (a) and recurrent GBM (b). (c) DNM3, p65 and p53 and internal control β-actin protein expression of brain and blood exosome both for original and recurrent GBM were analyzed by Western blotting. Relative expression values represent mean and SD from three independent experiments. Quantitation by densitometry was shown on below.
GBM, glioblastoma multiforme; PCR, polymerase chain reaction; SD, standard deviation.
Taken together, our results showed that DNM3 and p65 were upregulated in both mRNA and protein levels in xenograft exosomes, while p53 were downregulated at the same time, which suggested that the expressions of DNM3, p65 and p53 could be potential clinical markers to monitor the diagnosis and treatment of GBM patients.
Discussion
GBM is a fatal and incurable primary brain neoplasm that is characterized by necrosis, mitotic activity, nuclear atypia and microvascular proliferation. It is the most malignant brain tumor among all intracranial malignancies, as suggested by National Institutes of Health over the past decade.19 The average age of GBM at diagnosis is 64, and the annual morbidity is about 3.19/100,000.20 The survival time of most GBM patients is approximately 1 year, with only 5% of them surviving more than 5 years. GBM represents 15.4% among all the primary brain tumors and 45.6% among the malignant brain tumors.21 Treatment options of GBM are limited and there is no dramatic change of survival rate to date. Current research and technique have not led to a cure for GBM, especially when the initial treatment fails, there is no standard therapy for recurrent GBM.20 However, with improved technology, clinical trials and scientific research made a better understanding on the genomics, cellular properties, clinical behavior and disease progression of diverse subtypes of GBM.19
A number of genes are found to be related to pathogenesis of GBM, with alterations in gene expressions, epigenetics, mutations and gene copy numbers. The Cancer Genome Atlas sequenced 601 genes of 91 GBM patients to describe the genomic mutation spectrum of GBM. Except for the previously mentioned TP53 and RB1 mutations, they identified other GBM-associated mutations such as ERBB2, NF1 and PIK3R1.22 After a series of kinases had been reported to be associated with GBM, Bleeker FE and colleagues demonstrated 148 nonsynonymous somatic mutations, among which 25 had not been previously reported in GBM. These mutations include BARF, EGFR, EPHA3, FLT3,IDH1, NRAS, PIK3CA, PTEN, RPS6KC1, TGFBR2 and TP53. Furthermore, they revealed that majority of these mutated genes played essential roles in PI3K-AKT pathway, after mapping these genes into known signaling pathways.23
Exosomes are 30–100 nm vesicles derived from multiplying cells including epithelial cells, blood cells and embryonic cells.24 Exosomes are present in most eukaryotic fluids, such as blood and urine.8 The most common molecular constituents of exosomes are protein, mRNA and miRNA, and some exosomes are shown to contain double-stranded DNA.25 Valadi and colleagues first discovered that mRNA and microRNA contained in exosomes can be delivered to other cells and function in the new location.26 Exosomes play a particularly essential role in cancer progression and metastasis, acting as information transfer vehicle from tumor to local or distant sites. As exosomes transfer oncogenes between cells, they are dubbed ‘oncosomes’.27 Ilaria and colleagues revealed that the extracellular vesicles from the human glioblastoma cell line U251 had the ability to promote the formation of new blood vessels in human brain.28 In aggressive GBM, a truncated form of the epidermal growth factor receptor, known as EGFRvIII, is expressed and functions as oncogenic protein. Even though only a small portion of tumor cells express this protein, a majority of the cells exhibit a transformed phenotype. Al-Nedawi and colleagues showed that exosomes containing EGFRvIII can be released to extracellular environment, including the blood of tumor-bearing mice, which could then fuse with normal or cancer cells lacking EGFRvIII. The transferred oncoprotein functions in the recipient cells to activate downstream signaling pathways, such as AKT and MAPK, leading to changes in the expressions of EGFRvIII-regulated genes.29 One of the studies on brain tumor demonstrated that the loss of PTEN expression in brain tumor cells was associated with exosomal miRNA from brain astrocytes.30 They found that the tumor cells lost PTEN expression after metastasizing to brain microenvironment, and the PTEN expression level of these cells could be restored when leaving the brain. This reversible pattern of PTEN expression is regulated by exosomal miRNAs from brain astrocytes.
As exosomes share the properties of donor cells and can be released into body fluids, it has been investigated as an efficient biomarker for disease diagnosis or prognosis especially in certain cancers. For example, by evaluating 27 lung adenocarcinoma patients with AJCC stages I–IV and 9 controls, Rabinowits and colleagues found that the exosome and miRNA levels between lung cancer patients and controls were significantly different, while the blood exosomal miRNA and the tumor-derived miRNA patterns were consistent.31 In pancreatic studies, Ali et al., compared the exosomal miRNA expression levels in pancreatic cancer patients and healthy controls, and revealed a differential miRNA profile between these different groups. A series of miRNAs such as miR-31, miR-155 and miR-205 were upregulated in majority of tumor group, which was inversely correlated with the patient survival.32 These researches indicate that exosomal miRNA profiles are related to carcinogenesis and might be studied further as a diagnostic or prognostic biomarker.
Tissue biopsy is invasive and has limitations when reflecting current tumor dynamics or sensitivity to the treatment. Compared with tissue biopsy, blood biopsy is a promising approach that rare mutations and the heterogeneous landscape of tumor can be detected. Blood biopsy includes the circulating tumor cells (CTCs), circulating tumor DNA and exosomes that are released to the peripheral blood from the primary or metastatic tumors. Details of the genetic landscape and epigenetic profiles can be predicted from blood biopsy in a convenient and safe manner. Thus, blood biopsy can be widely used in early stage cancer screening, patient stratification and monitoring.33
In our study, the RT-PCR microarray revealed comprehensive expression profile of several mRNAs altered in GBM mouse model. Specifically, DNM3, p65 and CD117 were found upregulated in both tumor-derived tissue and exosomes of primary GBM-bearing mice, while PTEN and p53 were downregulated [Figure 6(a)]. In the mouse model bearing recurrent GBM, we found DNM3 and p65 were upregulated both in the tumor site and exosomes; while ST14 and p53 were downregulated [Figure 6(b)]. Then we chose the most differentially expressed mRNAs for individual qRT-PCR validation in each of the samples, and verified their protein levels using WB analysis. Comprehensively, we found two genes, DNM3 and p65, were upregulated and one gene, p53, was downregulated at both mRNA and protein levels in GBM tumor-bearing mouse exosomes and tumor-derived tissue.
DNM3 encodes a member of GTP-binding protein family which is associated with microtubules and involved in vesicular transport. The encoded protein functions in the development of megakaryocytes.34 There were limited studies investigating the influence of reduced DNM3 gene expression in malignant diseases. Yang and colleagues first reported that DNM3 expression level was negatively related to the glioma grades.34 They demonstrated that DNM3 was a target of miR-221 that was a pro-oncogenic factor in glioma and increasingly expressed in a variety of cancers.34–36 The result is consistent with the previous findings that DNM3 gene expression is reduced in hepatocellular cancer.37,38 However, our findings are contrary to these results, which is likely due to distinctions among different types of cancers. Moreover, this contradiction indicates that DNM3 might be a potential therapeutic target specifically for GBM.
The proto-oncogene p65, also known as nuclear factor kappa-light-chain-enhancer of activated B cells 3 (NF-κ-B3), a subunit of NF-κB, is a well-known proto-oncogene that functions as a ubiquitous transcription factor, which is held in the cytoplasm in an inactive state by specific inhibitors. The proto-oncogene p65 translocates into the nucleus and activates transcription of certain genes upon degradation of its inhibitor.39 Eukaryotic cells utilize NF-κB as a gene mediator that regulate cell proliferation and survival. Active NF-κB triggers the expression of genes that promote cell proliferation and protect cells from apoptosis. In diverse types of cancers, tumor cells show constitutively expressed NF-κB.40 Thus it is reasonable that p65 is upregulated in primary and recurrent GBM in our study.
TP53 encodes a tumor suppressor protein that contains transcriptional activation, DNA binding and oligomerization domains. The encoded protein p53 responds to cellular stresses to regulate target gene expressions that result in cell cycle arrest, DNA repair, apoptosis, senescence or metabolism changes. Most human cancers are associated with loss-of-function mutations of this TP53.41 Previous studies and our results indicate that the decreased expression of p53 is associated with the tumor progression of GBM.
Even though our study has limitations, such as the lack of sample numbers, the present study contributes to the rising understanding on the role of exosomal mRNAs in both primary and recurrent GBM. In conclusion, we found that DNM3, p65 and p53 had the similar dysregulated trends in brain and blood exosomes both for primary and recurrent GBM, which indicates that the blood exosomal mRNA levels could serve as potential clinical diagnostic markers for GBM.
Acknowledgments
The authors would like to thank Dr Li-qun Wang and Dr Zhong-qiang Lv for their technical advice.
Footnotes
Authors’ Note: Correspondence for the article can also be forwarded to Jian-kai Yang at yjk2009@hotmail.com and Guo-zhu Sun at sungzh705@163.com.
Funding: This work was funded by Provincial Clinical Medical Talents Project in 2015, and Hebei Medical Science Research Youth Project in 2018, China.
Conflict of interest statement: The authors declare that there is no conflict of interest.
Contributor Information
Jian-kai Yang, Department of Neurosurgery, The Second Hospital of Hebei Medical University, Shijiazhuang, China.
Jian Song, Department of Neurosurgery, The Second Hospital of Hebei Medical University, Shijiazhuang, China.
Hao-ran Huo, Department of Neurosurgery, The Second Hospital of Hebei Medical University, Shijiazhuang, China.
Yin-long Zhao, Department of Neurosurgery, The Second Hospital of Hebei Medical University, Shijiazhuang, China.
Guang-yu Zhang, Department of Neurosurgery, The Second Hospital of Hebei Medical University, Shijiazhuang, China.
Zong-mao Zhao, Department of Neurosurgery, The Second Hospital of Hebei Medical University, Shijiazhuang, China.
Guo-zhu Sun, Department of Neurosurgery, The Second Hospital of Hebei Medical University, Shijiazhuang, China.
Bao-hua Jiao, Department of Neurosurgery, The Second Hospital of Hebei Medical University, 215 Hepingxi Road, Shijiazhuang 050000, China.
References
- 1. McLendon RE, Halperin EC. Is the long-term survival of patients with intracranial glioblastoma multiforme overstated? Cancer 2003; 98: 1745–1748. [DOI] [PubMed] [Google Scholar]
- 2. Chou ST, Patil R, Galstyan A, et al. Simultaneous blockade of interacting CK2 and EGFR pathways by tumor-targeting nanobioconjugates increases therapeutic efficacy against glioblastoma multiforme. J Control Release 2016; 244: 14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chan DT, Hsieh SY, Kam MK, et al. Pattern of recurrence and factors associated with cerebrospinal fluid dissemination of glioblastoma in Chinese patients. Surg Neurol Int 2016; 7: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Smirniotopoulos JG, Murphy FM, Rushing EJ, et al. Patterns of contrast enhancement in the brain and meninges. Radiographics 2007; 27: 525–551. [DOI] [PubMed] [Google Scholar]
- 5. Molenaar RJ, Verbaan D, Lamba S, et al. The combination of IDH1 mutations and MGMT methylation status predicts survival in glioblastoma better than either IDH1 or MGMT alone. Neuro Oncol 2014; 16: 1263–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Keller S, Sanderson MP, Stoeck A, et al. Exosomes: from biogenesis and secretion to biological function. Immunol Lett 2006; 107: 102–108. [DOI] [PubMed] [Google Scholar]
- 7. Johnstone RM, Adam M, Hammond JR, et al. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem 1987; 262: 9412–9420. [PubMed] [Google Scholar]
- 8. van der Pol E, Boing AN, Harrison P, et al. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev 2012; 64: 676–705. [DOI] [PubMed] [Google Scholar]
- 9. Nedaeinia R, Manian M, Jazayeri MH, et al. Circulating exosomes and exosomal microRNAs as biomarkers in gastrointestinal cancer. Cancer Gene Ther. Epub ahead of print 16 December 2016. DOI: 10.1038/cgt.2016.77. [DOI] [PubMed] [Google Scholar]
- 10. Matei I, Kim HS, Lyden D. Unshielding exosomal RNA unleashes tumor growth and metastasis. Cell 2017; 170: 223–225. [DOI] [PubMed] [Google Scholar]
- 11. Kamerkar S, LeBleu VS, Sugimoto H, et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 2017; 546: 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cordonnier M, Chanteloup G, Isambert N, et al. Exosomes in cancer theranostic: diamonds in the rough. Cell Adh Migr 2017; 11: 151–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Casadei L, Calore F, Creighton CJ, et al. Exosome-derived miR-25-3p and miR-92a-3p stimulate liposarcoma progression. Cancer Res 2017; 77: 3846–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tran N. Cancer exosomes as miRNA factories. Trends Cancer 2016; 2: 329–331. [DOI] [PubMed] [Google Scholar]
- 15. Wu CY, Du SL, Zhang J, et al. Exosomes and breast cancer: a comprehensive review of novel therapeutic strategies from diagnosis to treatment. Cancer Gene Ther 2017; 24: 6–12. [DOI] [PubMed] [Google Scholar]
- 16. Pointer KB, Clark PA, Schroeder AB, et al. Association of collagen architecture with glioblastoma patient survival. J Neurosurg 2017; 126: 1812–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Giannini C, Sarkaria JN, Saito A, et al. Patient tumor EGFR and PDGFRA gene amplifications retained in an invasive intracranial xenograft model of glioblastoma multiforme. Neuro Oncol 2005; 7: 164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Baranyai T, Herczeg K, Onodi Z, et al. Isolation of exosomes from blood plasma: qualitative and quantitative comparison of ultracentrifugation and size exclusion chromatography methods. PLoS One 2015; 10: e0145686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Young RM, Jamshidi A, Davis G, et al. Current trends in the surgical management and treatment of adult glioblastoma. Ann Transl Med 2015; 3: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gallego O. Nonsurgical treatment of recurrent glioblastoma. Curr Oncol 2015; 22: e273–e281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Khosla D. Concurrent therapy to enhance radiotherapeutic outcomes in glioblastoma. Ann Transl Med 2016; 4: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010; 17: 98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bleeker FE, Lamba S, Zanon C, et al. Mutational profiling of kinases in glioblastoma. BMC Cancer 2014; 14: 718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Raposo G, Tenza D, Mecheri S, et al. Accumulation of major histocompatibility complex class II molecules in mast cell secretory granules and their release upon degranulation. Mol Biol Cell 1997; 8: 2631–2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thakur BK, Zhang H, Becker A, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res 2014; 24: 766–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Valadi H, Ekstrom K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007; 9: 654–659. [DOI] [PubMed] [Google Scholar]
- 27. Atay S, Godwin AK. Tumor-derived exosomes: a message delivery system for tumor progression. Commun Integr Biol 2014; 7: e28231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Giusti I, Delle Monache S, Di Francesco M, et al. From glioblastoma to endothelial cells through extracellular vesicles: messages for angiogenesis. Tumour Biol 2016; 37: 12743–12753. [DOI] [PubMed] [Google Scholar]
- 29. Al-Nedawi K, Meehan B, Micallef J, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol 2008; 10: 619–624. [DOI] [PubMed] [Google Scholar]
- 30. Zhang L, Zhang S, Yao J, et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature 2015; 527: 100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rabinowits G, Gercel-Taylor C, Day JM, et al. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer 2009; 10: 42–46. [DOI] [PubMed] [Google Scholar]
- 32. Ali S, Dubaybo H, Brand RE, et al. Differential expression of microRNAs in tissues and plasma co-exists as a biomarker for pancreatic cancer. J Cancer Sci Ther 2015; 7: 336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang J, Chang S, Li G, et al. Application of liquid biopsy in precision medicine: opportunities and challenges. Front Med. Epub ahead of print 25 July 2017. DOI: 10.1007/s11684-017-0526-7. [DOI] [PubMed] [Google Scholar]
- 34. Yang JK, Yang JP, Tong J, et al. Exosomal miR-221 targets DNM3 to induce tumor progression and temozolomide resistance in glioma. J Neurooncol 2017; 131: 255–265. [DOI] [PubMed] [Google Scholar]
- 35. Liu M, Liu J, Wang L, et al. Association of serum microRNA expression in hepatocellular carcinomas treated with transarterial chemoembolization and patient survival. PLoS One 2014; 9: e109347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yau TO, Wu CW, Dong Y, et al. microRNA-221 and microRNA-18a identification in stool as potential biomarkers for the non-invasive diagnosis of colorectal carcinoma. Br J Cancer 2014; 111: 1765–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang Z, Chen C, Guo W, et al. DNM3 Attenuates hepatocellular carcinoma growth by activating P53. Med Sci Monit 2016; 22: 197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Inokawa Y, Nomoto S, Hishida M, et al. Dynamin 3: a new candidate tumor suppressor gene in hepatocellular carcinoma detected by triple combination array analysis. Onco Targets Ther 2013; 6: 1417–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fuentes E, Rojas A, Palomo I. NF-kappaB signaling pathway as target for antiplatelet activity. Blood Rev 2016; 30: 309–315. [DOI] [PubMed] [Google Scholar]
- 40. Vlahopoulos SA, Cen O, Hengen N, et al. Dynamic aberrant NF-kappaB spurs tumorigenesis: a new model encompassing the microenvironment. Cytokine Growth Factor Rev 2015; 26: 389–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Basset-Seguin N, Moles JP, Mils V, et al. TP53 tumor-suppressor gene and human carcinogenesis. Exp Dermatol 1993; 2: 99–105. [DOI] [PubMed] [Google Scholar]