Abstract
Metagenomics and related methods have led to significant advances in our understanding of the human microbiome. Members of the genus Lactobacillus, although best understood for essential roles in food fermentations and applications as probiotics, have also come to the fore in a number of untargeted gut microbiome studies in humans and animals. Although Lactobacillus is only a minor member of the human colonic microbiota, the proportions of those bacteria are frequently either positively or negatively correlated with human disease and chronic conditions. Recent findings on Lactobacillus species in human and animal model microbiome research, together with the increased knowledge on probiotic and other ingested lactobacilli, have resulted in new perspectives on the importance of this genus to human health.
Graphical abstract
Introduction
Members of the genus Lactobacillus were long thought to be among the most abundant microorganisms in the human gastrointestinal (GI) tract and associated with good intestinal health. Following the development of culture-independent, DNA-sequence analysis methods, the numbers of autochthonous Lactobacillus were adjusted to ≤ 1% of the total bacterial population in the distal human gut. One consequence of this change is that the relevance of this genus to human health has come under scrutiny. In contrast, there is increased acceptance of the application of allochthonous probiotic Lactobacillus in fermented foods and supplements as probiotics to maintain health and prevent and treat disease [1,2]. Although human studies frequently show a benefit with probiotic administration [3], the importance of autochthonous Lactobacillus remains under question.
Human disease is increasingly correlated with fecal microbiota composition. Similarly, intestinal bacteria are frequently correlated with numerous other host (genetics, age) and environmental (diet, medication) factors. Such associations have been useful for identifying pathobionts associated with disease as well as taxa such as Faecalibacterium prausnitzii and Akkermansia muciniphila as beneficial members of the indigenous microbiota. Similarly, a number of recent publications in which culture-independent methods were employed (e.g. 16S rRNA gene amplicon sequencing) identified Lactobacillus as being significantly enriched in the distal gut during either health or disease (Figure 1 and Table 1). Because these approaches are largely untargeted, the outcomes provide an unbiased perspective on the relative importance of this genus weighed against other bacterial inhabitants of GI tract. This review will address findings on the diversity and abundance of intestinal Lactobacillus resulting from gut microbiome studies and emerging mechanistic evidence of endogenous and ingested (probiotic) Lactobacillus species in the GI tract.
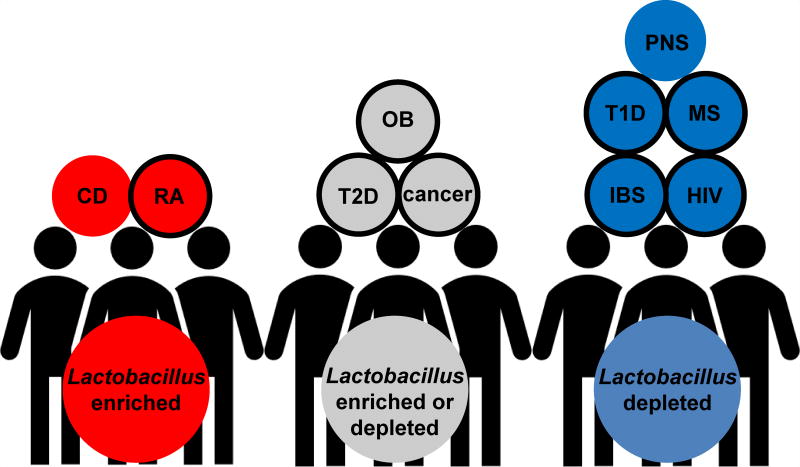
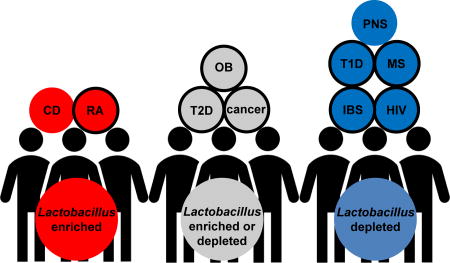
Figure 1. Alteration of intestinal Lactobacillus in health and disease.
Blue circles indicate Lactobacillus is depleted in disease compared to healthy controls. Red circles indicate Lactobacillus is increased in disease. Grey circle indicates Lactobacillus is either increased or decreased. Circles with black edges indicate a benefit for consumption of probiotics for treating disease. CD = Crohn’s disease, RA = rheumatoid arthritis, OB = obesity, T2D = type 2 diabetes, IBS = irritable bowel syndrome, T1D = type 1 diabetes, PNS = prenatal stress, HIV = human immunodeficiency virus, MS = multiple sclerosis.
Table 1.
Recent human studies analyzing microbiomes in health and disease.
| Disease/Condition | Study designa |
Lactobacillus proportionsb |
Lactobacillus speciesc |
Methodd | Reference |
|---|---|---|---|---|---|
| IBS | meta-analysis 13 studies | ↓ | qRT-PCR | [23] | |
| meta-analysis 10 Chinese studies | ↓ | fecal bacterial counts | [24] | ||
| meta-analysis 7 global studies | − | fecal bacterial counts | [24] | ||
| CD | 28 CD, 26 HC | ↑ | metagenomics | [26] | |
| 15 CD, 21 HC | ↑ | qRT-PCR | [27] | ||
| HIV | 8 infected, 8 HC | ↓ | 16S rRNA | [15] | |
| Reumatoid Arthritis | 77 RA, 80 HC | ↑ | L. salivarius, L. ruminus, L. iners | metagenomics | [30] |
| Type 1 Diabetes | 21 T1D, 32 HC | ↓ | 16S rRNA | [34] | |
| 28 T1D, 27 HC | ↓ | 16S rRNA microarray | [35] | ||
| Multiple Sclerosis | 31 MS, 36 HC | ↓ | 16S rRNA | [37] | |
| Obesity | 15 obese, 17 HC | ↓ | L. plantarum | qRT-PCR | [45] |
| 30 obese, 24 OW, 30 HC | ↑ | qRT-PCR | [42] | ||
| 42 obese, 36 HC | − | 16S rRNA | [43] | ||
| 67 obese, 67 HC | − | 16S rRNA | [44] | ||
| Type 2 Diabetes | 53 T2D, 43 HC | ↑ | L. gasseri | 16S rRNA | [46] |
| 93 Met+T2D, 106 Met−T2D, 554 HC | ↓ | metagenomics | [47] | ||
| Colon cancer | systematic review 31 studies | ↓ | 16S rRNA | [53] | |
| Breast cancer | 17 BC, 16 BD | ↑ | 16S rRNA | [55] | |
| Head and neck squamous cell cancer | 17 HNSCC, 25 HC | ↑ | 16S rRNA | [56] | |
| Prenatal stress | 56 mother-infant pairs | ↓ | 16S rRNA | [57] |
CD = Crohn's disease, HC = healthy control, OW = overweight, Met+T2D = metformin treated type 2 diabetes patients, Met−T2D = patients not treated with metformin, BC = breast cancer, BD = benign disease, HNSCC = head and neck squamous cell cancer
Proportions or numbers of Lactobacillus were lower (↓),higher (↑), or unchanged (−) in individuals with disease or chronic conditions compared to healthy individuals
Species identification was conducted by authors and not assessed by reviewers
qRT-PCR = quantitative real time-PCR of 16S rRNA, metagenomics = shotgun whole genome sequencing, 16S rRNA = 16S ribosomal RNA sequencing
Abundance and diversity of intestinal Lactobacillus
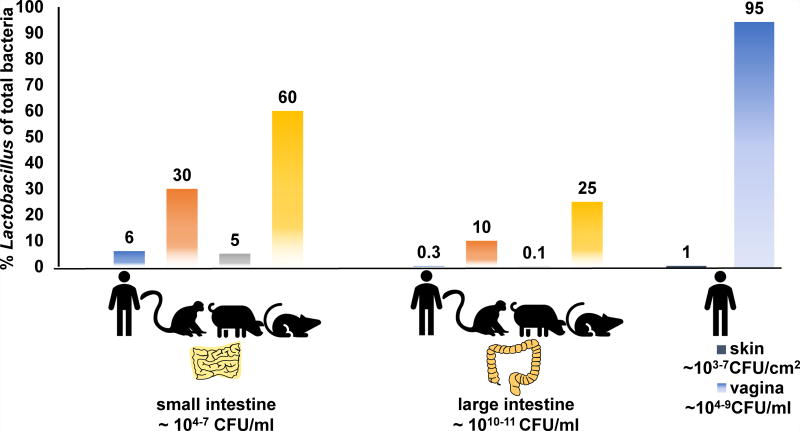
Lactobacillus species have been isolated from the entirety of the human GI tract (oral cavity to feces) as well as the skin and vagina [4,5]. This genus is estimated to constitute 6% of the total bacterial cell numbers in the human duodenum [6] and approximately 0.3% of all bacteria in the colon [4] (Figure 2). These levels are similar to the numbers of lactobacilli found in pigs, ranging from 5 to 0.1% of total bacteria in the proximal [7] and distal [8] gut, respectively. Lactobacillus was found in higher quantities in rhesus macaques (up to 30% and 10% of all bacteria in the small and large intestine, respectively) [9]. Proportions of Lactobacillus in rodent models ranged between 30 to 60% of bacterial numbers in the ileum and approximately 25% in the colon [10,11] (Figure 2). Lactobacillus can also dominate the human vaginal microbiota (90–100% of total bacteria present) and is found on the skin, but in much lower relative abundance [5] (Figure 2).
Figure 2. Relative abundance of Lactobacillus in humans and animals.
Numbers underneath anatomical locations indicate estimates for total bacterial community cell numbers.
Only a few out of the >200 known Lactobacillus species have been consistently and repeatedly associated with the human GI tract. Recently, this number was increased to over 50 Lactobacillus species that were repeatedly detected in the stools of healthy volunteers [•12]. The most abundant lactobacilli included L. casei, L. delbruckeii, L. murinus, L. plantarum, L. rhamnosus, and L. ruminus. Some of these species (e.g. L. rhamnosus and L. murinus) are rarely isolated from environments outside the intestine and are considered gut-autochthonous microorganisms. Other mucosal sites are colonized by distinct species (e.g. L. crispatus in the vagina) [•13]. There also appears to be host-specificity among some Lactobacillus species, as shown for linages of L. reuteri [14].
Infectious disease
Both human immunodeficiency virus (HIV) - infected humans and simian immunodeficiency virus (SIV) - infected rhesus macaques harbor reduced numbers of intestinal Lactobacillus [15,16] (Table 1). Lactobacillus depletion in rhesus macaques was associated with the loss of gut barrier-promoting T-helper 17 (Th17) cells and increased microbial translocation [16]. The potential of Lactobacillus to prevent or reverse intestinal damage during infection was demonstrated with the reduced interleukin-1β-mediated inflammation and improved barrier function upon inoculation of L. plantarum directly into ileal loops of SIV+ macaques shortly after SIV infection [17]. The intestinal epithelium in healthy animals responded similarly to L. plantarum, consistent with the finding that the ileal transcriptomes of L. plantarum were indistinguishable between SIV+ and SIV− animals [18]. In human populations, HIV+ patients on a multi-strain probiotic supplement exhibited higher numbers of memory Th17 cells in peripheral blood and in the intestine, and histological examination of colonic biopsies indicated increased intestinal barrier function [19].
Several recent animal studies have indicated a broader role for Lactobacillus in prevention and resolution of infectious disease. Tryptophan metabolites (indole aldehydes) produced by indigenous L. reuteri strains activate host aryl hydrocarbon receptors (AHR) to promote gut and vaginal epithelial barrier and antimicrobial responses required for limiting the expansion of Candida albicans, an opportunistic pathogen [••20]. Autochthonous Lactobacillus might also have a role in the resolution of infectious disease and recovery of immune homeostasis. Although Yerstina enterocolitica infection was cleared from toll-like receptor 1 (TLR1) knockout mice, the intestine was activated towards an inflammatory phenotype and the gut microbiota was enriched with Desulfovibrionaceae while containing lower numbers of Lactobacillus [••21]. Oral gavage with L. reuteri reduced anti-commensal antibodies, innate cytokines, and Th17 responses; thereby ameliorating immune hyper-reactivity [••21]. Conversely, post-Yersinia pseudotuberculosis infection lactobacilli were cultured from enlarged gut-associated lymphoid tissue and were associated with chronic lymphadenopathy, indicating that these bacteria might contribute to chronic, immune hyper-reactivity [22].
Irritable bowel syndrome (IBS) and inflammatory bowel disease (IBD)
A meta-analysis of reports investigating the fecal microbiomes from IBS patients and healthy subjects concluded Lactobacillus was depleted in diarrhea-dominant, IBS patients [23] (Table 1). Another meta-analysis of IBS cohort studies determined that intestinal Lactobacillus was depleted in all cases of IBS in Chinese patients, but this association was not found or was reversed in patients from other countries [24]. Consistent with these results, meta-analysis of probiotic intervention studies (43 randomized controlled trials (RCTs)) for treatment of IBS concluded that multi-species probiotics diminish symptoms (abdominal pain, bloating, and flatulence scores) [25].
Conversely, intestinal abundance of Lactobacillus and other genera including Bifidobacterium were recently positively correlated with Crohn’s disease (CD) patients [26,27] (Table 1). In both studies, Lactobacillus enrichment coincided with depletion of F. prausnitzii. Whether Lactobacillus is participating in disease or is simply adapted to survive the pro-inflammatory gut environment is not known. These findings contrast with ulcerative colitis (UC) in which probiotic Lactobacillus consumption has been associated with improved clinical symptoms [28]. Lactobacillus might be particularly supportive in CD and UC patients with caspase recruitment domain family member 9 (CARD9) risk alleles whose microbiome has a reduced production of AHR ligands [••29]. Consistent with this possibility, intestinal inflammation in CARD9 knockout (KO) mice was attenuated after inoculation of mice with Lactobacillus strains capable of metabolizing tryptophan [••29].
Rheumatoid arthritis (RA)
The intestinal microbiota of patients with severe and early onset RA were shown to have increased proportions of L. salivarius, L. ruminus, and L. iners when compared to healthy, age-matched individuals [•30] (Table 1). Enrichment of Lactobacillus spp. was also observed in collagen-induced, arthritic mice [31]. These results are in opposition to recent RCTs of probiotics in RA patients. In one study, patients consuming L. casei showed reduced disease activity scores, higher quantities of serum IL-10, and decreased levels of serum TNFα, IL- 6 and IL-12 compared to placebo [32]. The other RCT concluded that a mixed strain probiotic supplement significantly improved disease activity scores and lowered levels of serum C-reactive protein (CRP) [33]. Such findings might indicate species or strain-specific differences between autochthonous and allochthonous Lactobacillus on RA disease activity.
Type 1 Diabetes (T1D)
The proportions of Lactobacillus were lower in adults with T1D than healthy, first-degree relatives and unrelated healthy individuals according to untargeted 16S rRNA analysis [•34] (Table 1). A similar reduction in Lactobacillus was observed in children with T1D [35] (Table 1). Interestingly, children exposed to probiotic Lactobacillus early in life were found to have a significantly reduced risk of developing islet autoimmunity [36]. It is not yet understood how Lactobacillus could be regulating islet beta-cell auto-immunity, although it has been suggested that a lack of intestinal lactate-producing bacteria depletes butyrate-producing taxa leading to aberrant immune responses [35].
Multiple sclerosis (MS)
A cohort study found that the relative abundance of intestinal Lactobacillus was lower in MS patients compared to healthy adults [37] (Table 1). Similar depletions in intestinal Lactobacillus were observed in a pre-clinical, rodent model of MS [38]. Consistent with a benefit of Lactobacillus in this autoimmune disease were the findings from a recent RCT of MS patients, whereby consumption of a multi-species probiotic improved the expanded disability status score, self-reported depression, anxiety and stress, as well as decreased serum CRP compared to placebo [39]. Because circulating levels of AHR ligands are lower in MS patients compared to healthy adults [40], Lactobacillus might be useful for the maintenance or replenishment of these compounds. To this regard, Lactobacillus-produced indole aldehydes had a potent anti-inflammatory effect on brain glial cells (astrocytes) to limit central nervous system inflammation in a mouse model of human MS [•41].
Obesity and Type 2 Diabetes (T2D)
There are conflicting reports on the association of intestinal Lactobacillus with obesity in humans [42–45] (Table 1). Likewise, initial studies found increased levels of Lactobacillus in patients with T2D [46], although this trend was eliminated or reversed when controlling for metformin treatment [47] (Table 1). Also contrary to these results, meta-analysis of RCT studies found that probiotic Lactobacillus improved weight management outcomes in obese adults [48]. Consumption of yogurt and other dairy products fermented by Lactobacillus is also significantly associated with protection from T2D and obesity (recently reviewed in [2]).
Because Lactobacillus species appear to be either associated with weight gain or weight loss [49], the disparate findings among obese individuals might be due to genetic differences among the lactobacilli. Strain and species distinctions could result in variations in carbohydrate metabolism and production of fermentation end-products, such as lactate [50]. The production of bile salt hydrolases is another distinguishing feature of some Lactobacillus species, and this activity is responsible for significantly altering the activation of farnesoid x receptor (FXR) signaling and hepatic lipid metabolism [51,52].
Cancer
In a systematic review of thirty-one studies, Lactobacillus along with a limited number of butyrogenic genera were consistently diminished in colorectal cancer patients [53] (Table 1). Preventative and therapeutic roles of Lactobacillus in cancer are supported in studies with pre-clinical, rodent models, including a recently study in which a multi-strain probiotic altered Th-cell polarization away from Th17 cells in a mouse model of hepatocellular carcinoma [54]. However, Lactobacillus might not always be beneficial in certain extra-intestinal sitesas shown by the higher levels of Lactobacillus found in malignant breast cancer compared to benign-disease tissues [55]. There was also a positive association between the levels of this genus in the oral microbiome and head and neck squamous cell carcinoma [56] (Table 1).
Cognitive development and behavior
Maternal prenatal stress might influence the infant microbiome, potentially damaging cognitive development. In humans, prenatal cortisol concentrations were inversely correlated with infant levels of intestinal Lactobacillus and Lactococcus, whereas Proteobacteria were enriched [•57] (Table 1). A comparable depletion of Lactobacillus was observed in rodent models of prenatal stress, with the microbiome of the offspring remaining disrupted into adulthood [58]. Prenatal low-dose penicillin [59] or high fat diet [••60] could similarly induce long-term dysbiosis and behavioral deficits in mice. These deficits could be prevented by concurrent administration of Lactobacillus-containing probiotics to the dam [59] or by indigenous L. reuteri to offspring [••60].
In adult mouse models of microbiota-gut-brain axis deficits, administration of Lactobacillus-containing probiotics was found to beneficially impact both cognition and colonic function, while reverting intestinal dysbiosis [61,62]. Such results might also be relevant to emotional disorders and this is supported in probiotics studies which have indicated that probiotic Lactobacillus might improve symptoms of human depression [63,64]. Therefore, beneficially modulating the microbiota using Lactobacillus can impact the microbiota-gut-brain axis and should be more thoroughly studied in human mother-infant cohorts.
Conclusions
Our increased understanding of intestinal Lactobacillus from untargeted microbiome studies supports the premise that general properties conferred by this genus have far-reaching consequences on human health. Such knowledge could be further advanced via studies designed to determine the proximity of Lactobacillus to the intestinal epithelium or which focus attention on other sites on the body wherein members of this genus can constitute the majority of bacteria present (e.g. vagina). However, even without this information, strain and/or species-specific differences (e.g. tryptophan and bile metabolism) might be useful to explain variations in the involvement of this genus, either in the prevention or mitigation of disease or, alternatively, as a contributing factor to disease outcomes. Furthermore, the notable variation in intestinal abundance of this genus between healthy and diseased, or health-compromised, individuals indicates that Lactobacillus, or at least certain species or genotypes of Lactobacillus, could be useful gut biomarkers. These considerations can also inform the improved development and use of probiotics in different human populations.
Highlights.
Intestinal lactobacilli are often detected in untargeted, gut microbiome studies.
Depletion of intestinal Lactobacillus is frequently associated with disease.
Probiotics use is supported by findings on indigenous Lactobacillus populations.
Tryptophan metabolism is an emerging, beneficial trait of intestinal lactobacilli.
Acknowledgments
This manuscript is partly based upon work supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. (1148897) (DH), the American Diabetes Association ADA-GSK Award in the Microbiome and Metabolic Changes in Diabetes and Obesity grant #1-13-GSK-29 (MLM), and the NIH 1R01AT009365-01 (MGG). The contents of this review are those of the authors and do not necessarily reflect the views of the funding agencies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
None
References
- 1.Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014;11:506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 2.Marco ML, Heeney D, Binda S, Cifelli CJ, Cotter PD, Foligné B, Gänzle M, Kort R, Pasin G, Pihlanto A, et al. Health benefits of fermented foods: microbiota and beyond. Curr. Opin. Biotechnol. 2017;44:94–102. doi: 10.1016/j.copbio.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 3.Floch MH, Walker WA, Sanders E, Nieuwdorp M, Kim AS, Brenner DA, Qamar AA, Miloh TA, Guarino A, Guslandi M, et al. Recommendations for probiotic use — 2015 update proceedings and consensus opinion. J. Clin. Gastroenterol. 2015;49:S69–S73. doi: 10.1097/MCG.0000000000000420. [DOI] [PubMed] [Google Scholar]
- 4.Almonacid DE, Kraal L, Ossandon FJ, Budovskaya YV, Cardenas JP, Bik EM, Goddard AD, Richman J, Apte ZS. 16S rRNA gene sequencing and healthy reference ranges for 28 clinically relevant microbial taxa from the human gut microbiome. PLoS One. 2017;12:1–15. doi: 10.1371/journal.pone.0176555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu DM, Ma J, Prince AL, Antony KM, Seferovic MD, Aagaard KM. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat. Med. 2017;23:314–323. doi: 10.1038/nm.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nistal E, Caminero A, Herran AR, Perez-Andres J, Vivas S, Ruiz De Morales JM, Saenz de Miera LE, Casqueiro J. Study of duodenal bacterial communities by 16S rRNA gene analysis in adults with active celiac disease vs non-celiac disease controls. J. Appl. Microbiol. 2015;120:1691–1700. doi: 10.1111/jam.13111. [DOI] [PubMed] [Google Scholar]
- 7.Fan P, Liu P, Song P, Chen X, Ma X. Moderate dietary protein restriction alters the composition of gut microbiota and improves ileal barrier function in adult pig model. Sci. Rep. 2017;7:1–12. doi: 10.1038/srep43412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slifierz MJ, Friendship RM, Weese JS. Longitudinal study of the early-life fecal and nasal microbiotas of the domestic pig. BMC Microbiol. 2015;15:1–12. doi: 10.1186/s12866-015-0512-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohan M, Chow CET, Ryan CN, Chan LS, Dufour J, Aye PP, Blanchard J, Moehs CP, Sestak K. Dietary gluten-induced gut dysbiosis is accompanied by selective upregulation of microRNAs with intestinal tight junction and bacteria-binding motifs in rhesus macaque model of celiac disease. Nutrients. 2016;8:1–18. doi: 10.3390/nu8110684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li D, Chen H, Mao B, Yang Q, Zhao J, Gu Z, Zhang H, Chen YQ, Chen W. Microbial biogeography and core microbiota of the rat digestive tract. Sci. Rep. 2017;7:1–16. doi: 10.1038/srep45840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morikawa M, Tsujibe S, Kiyoshima-Shibata J, Watanabe Y, Kato-Nagaoka N, Shida K, Matsumoto S. Microbiota of the small intestine is selectively engulfed by phagocytes of the lamina propria and Peyer’s patches. PLoS One. 2016;11:1–16. doi: 10.1371/journal.pone.0163607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12•.Rossi M, Martinez-Martinez D, Amaretti A, Ulrici A, Raimondi S, Moya A. Mining metagenomic whole genome sequences revealed subdominant but constant Lactobacillus population in the human gut microbiota. Environ. Microbiol. Rep. 2016;8:399–406. doi: 10.1111/1758-2229.12405. Over 50 Lactobacillus species were repeatedly detected in quantities up to 108 cells/g in the stool of a human subject. Metagenomics enabled the identification of strain-level distinctions among the lactobacilli and individual Lactobacillus genotypes were consistently detected in the stool over a time span of nearly two years. [DOI] [PubMed] [Google Scholar]
- 13•.Gosmann C, Anahtar MN, Handley SA, Farcasanu M, Abu-Ali G, Bowman BA, Padavattan N, Desai C, Droit L, Moodley A, et al. Lactobacillus-deficient cervicovaginal bacterial communities are associated with increased HIV acquisition in young South African women. Immunity. 2017;46:29–37. doi: 10.1016/j.immuni.2016.12.013. This prospective study found that women with low-diversity vaginal bacterial communities dominated by Lactobacillus crispatus had a lower risk of acquiring HIV and had decreased numbers of activated mucosal CD4+ T cells compared to women with diverse genital bacterial communities. The results were supported by the lower numbers of activated CD4+ T cells in the genital mucosa of germ-free mice intravaginally-inoculated with L. crispatus compared with HIV-associated bacterial taxa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duar RM, Frese SA, Fernando SC, Burkey TE, Tasseva G, Peterson DA, Blom J, Wenzel CQ, Szymanski CM, Walter J. Experimental determination of host adaptation of Lactobacillus reuteri to different vertebrate species. Appl. Environ. Microbiol. 2017;83:1–44. doi: 10.1128/AEM.00132-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang L, Poles MA, Fisch GS, Ma Y, Nossa C, Phelan JA, Pei Z. HIV-induced immunosuppression is associated with colonization of the proximal gut by environmental bacteria. AIDS. 2016;30:19–29. doi: 10.1097/QAD.0000000000000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vujkovic-Cvijin I, Swainson LA, Chu SN, Ortiz AM, Santee CA, Petriello A, Dunham RM, Fadrosh DW, Lin DL, Faruqi AA, et al. Gut-resident Lactobacillus abundance associates with IDO1 inhibition and Th17 dynamics in SIV-infected macaques. Cell Rep. 2015;13:1589–1597. doi: 10.1016/j.celrep.2015.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirao LA, Grishina I, Bourry O, Hu WK, Somrit M, Sankaran-Walters S, Gaulke CA, Fenton AN, Li JA, Crawford RW, et al. Early mucosal sensing of SIV infection by paneth cells induces IL-1beta production and initiates gut epithelial disruption. PLoS Pathog. 2014;10:1–15. doi: 10.1371/journal.ppat.1004311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Golomb BL, Hirao LA, Dandekar S, Marco ML. Gene expression of Lactobacillus plantarum and the commensal microbiota in the ileum of healthy and early SIV-infected rhesus macaques. Sci. Rep. 2016;6:1–10. doi: 10.1038/srep24723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.d’Ettorre G, Rossi G, Scagnolari C, Andreotti M, Giustini N, Serafino S, Schietroma I, Scheri GC, Fard SN, Trinchieri V, et al. Probiotic supplementation promotes a reduction in T-cell activation, an increase in Th17 frequencies, and a recovery of intestinal epithelium integrity and mitochondrial morphology in ART-treated HIV-1-positive patients. Immunity, Inflamm. Dis. 2017 doi: 10.1002/iid3.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20••.Zelante T, Iannitti RG, Cunha C, DeLuca A, Giovannini G, Pieraccini G, Zecchi R, D’Angelo C, Massi-Benedetti C, Fallarino F, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. This early report singled out intestinal and vaginal lactobacilli for a potent ability to produce tryptophan metabolites that activate host-AHRs in epithelium tissue. The activation of epithelial AHRs induces the production of a myriad of antimicrobial compounds and was found to be essential for control of opportunistic pathogens. Importantly, AHR knockout mice infected with Candida albicans could not be rescued by introduction of tryptophan-metabolizing lactobacilli. [DOI] [PubMed] [Google Scholar]
- 21••.Kamdar K, Khakpour S, Chen J, Leone V, Brulc J, Mangatu T, Antonopoulos DA, Chang EB, Kahn SA, Kirschner BS, et al. Genetic and metabolic signals during acute enteric bacterial infection alter the microbiota and drive progression to chronic inflammatory disease. Cell Host Microbe. 2016;19:21–31. doi: 10.1016/j.chom.2015.12.006. In TLR1 knockout mice the acute intestinal infection of Yersinia enterocolitica destabilizes mucosal immunity allowing blooms of δ-Proteobacteria and diminished levels of other taxa, including lactobacilli. Chronic immune responses against the commensal microbiota were observed two months post-infection. Supplementation with L. reuteri was sufficient to revert the immune system towards homeostasis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.da Fonseca DM, Hand TW, Han S-J, Gerner MY, Zaretsky AG, Byrd AL, Harrison OJ, Ortiz AM, Quinones M, Trinchieri G, et al. Microbiota-dependent sequelae of acute infection compromise tissue-specific immunity. Cell. 2017;163:354–366. doi: 10.1016/j.cell.2015.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu H-N, Wu H, Chen Y-Z, Chen Y-J, Shen X-Z, Liu T-T. Altered molecular signature of intestinal microbiota in irritable bowel syndrome patients compared with healthy controls: A systematic review and meta-analysis. Dig. Liver Dis. 2017;49:331–337. doi: 10.1016/j.dld.2017.01.142. [DOI] [PubMed] [Google Scholar]
- 24.Zhuang X, Xiong L, Li L, Li M, Chen M. Alterations of gut microbiota in patients with irritable bowel syndrome: A systematic review and meta-analysis. J. Gastroenterol. Hepatol. 2017;32:28–38. doi: 10.1111/jgh.13471. [DOI] [PubMed] [Google Scholar]
- 25.Ford AC, Quigley EMM, Lacy BE, Lembo AJ, Saito YA, Schiller LR, Soffer EE, Spiegel BMR, Moayyedi P. Efficacy of prebiotics, probiotics, and synbiotics in irritable bowel syndrome and chronic idiopathic constipation: Systematic review and meta-analysis. Am. J. Gastroenterol. 2014;109:1547–1561. doi: 10.1038/ajg.2014.202. [DOI] [PubMed] [Google Scholar]
- 26.Lewis JD, Chen EZ, Baldassano RN, Otley AR, Griffiths AM, Lee D, Bittinger K, Bailey A, Friedman ES, Hoffmann C, et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host Microbe. 2015;18:489–500. doi: 10.1016/j.chom.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang W, Chen L, Zhou R, Wang X, Song L, Huang S, Wang G, Xia B. Increased proportions of Bifidobacterium and the Lactobacillus group and loss of butyrate-producing bacteria in inflammatory bowel disease. J. Clin. Microbiol. 2014;52:398–406. doi: 10.1128/JCM.01500-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ganji-Arjenaki M, Rafieian-Kopaei M. Probiotics are a good choice in remission of inflammatory bowel diseases: A meta-analysis and systematic review. J Cell Physiol. 2017 doi: 10.1002/jcp.25911. [DOI] [PubMed] [Google Scholar]
- 29••.Lamas B, Richard ML, Leducq V, Pham H-P, Michel M-L, Da Costa G, Bridonneau C, Jegou S, Hoffmann TW, Natividad JM, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 2016;22:598–605. doi: 10.1038/nm.4102. Human single nucleotide polymorphism (SNP) studies indicate caspase recruitment domain family member 9 (CARD9) alleles are associated with inflammatory bowel disease and that this risk allele may influence the development of a pro-inflammatory microbiome. By using CARD9 knockout mice, this report links CARD9 deficiencies to an altered intestinal microbiome with reduced production of AHR ligands. L. reuteri was identified as a significantly depleted taxon and replenishment of this species improved symptoms of colitis in CARD9 knockout mice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30•.Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, Wu X, Li J, Tang L, Li Y, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 2015;21:895–905. doi: 10.1038/nm.3914. This human study utilizing metagenomic sequencing on the fecal microbiota identified several species of intestinal Lactobacillus species that were elevated in the oral and fecal microbiota of RA patients compared with healthy subjects. [DOI] [PubMed] [Google Scholar]
- 31.Liu X, Zeng B, Zhang J, Li W, Mou F, Wang H, Zou Q, Zhong B, Wu L, Wei H, et al. Role of the gut microbiome in modulating arthritis progression in mice. Sci. Rep. 2016;6:1–11. doi: 10.1038/srep30594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaghef-Mehrabany E, Alipour B, Homayouni-Rad A, Sharif S-K, Asghari-Jafarabadi M, Zavvari S. Probiotic supplementation improves inflammatory status in patients with rheumatoid arthritis. Nutrition. 2014;30:430–435. doi: 10.1016/j.nut.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 33.Zamani B, Golkar HR, Farshbar S, Emadi-Baygi M, Tajabadi-Ebrahimi M, Jafari P, Akhavan R, Taghizadeh M, Memarzadeh MR, Asemi Z. Clinical and metabolic response to probiotic supplementation in patients with rheumatoid arthritis: A randomized, double-blind, placebo-controlled trial. Int. J. Rheum. Dis. 2016;19:869–879. doi: 10.1111/1756-185X.12888. [DOI] [PubMed] [Google Scholar]
- 34•.Alkanani AK, Hara N, Gottlieb PA, Ir D, Robertson CE, Wagner BD, Frank DN, Zipris D. Alterations in intestinal microbiota correlate with susceptibility to type 1 diabetes. Diabetes. 2015;64:3510–3520. doi: 10.2337/db14-1847. In this cohort study, the genetic component of T1D was partially controlled for by recruiting T1D patients and their healthy, first-degree relatives. The fecal microbiota was assessed by 16S rRNA sequencing as an environmental factor associated with disease. Lactobacillus proportions were significantly depleted in T1D patients. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Goffau MC, Fuentes S, Van Den Bogert B, Honkanen H, De Vos WM, Welling GW, Hyöty H, Harmsen HJM. Aberrant gut microbiota composition at the onset of type 1 diabetes in young children. Diabetologia. 2014;57:1569–1577. doi: 10.1007/s00125-014-3274-0. [DOI] [PubMed] [Google Scholar]
- 36.Uusitalo U, Liu X, Yang J, Hummel S, Butterworth M, Rewers M, Hagopian W, She J, Simell O, Toppari J, et al. Association of early exposure of probiotics and islet autimmunity in the TEDDY study. JAMA Pediatr. 2016;170:20–28. doi: 10.1001/jamapediatrics.2015.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen J, Chia N, Kalari KR, Yao JZ, Novotna M, Soldan MMP, Luckey DH, Marietta EV, Jeraldo PR, Chen X, et al. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci. Rep. 2016;6:1–10. doi: 10.1038/srep28484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stanisavlijević S, Lukić J, Soković S, Mihajlović S, Stojković MM, Miljković D, Golić N. Correlation of gut microbiota composition with resistance to experimental autoimmune encephalomyelitis in rats. Front. Microbiol. 2016;7:1–12. doi: 10.3389/fmicb.2016.02005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kouchaki E, Tamtaji OR, Salami M, Bahmani F, Daneshvar Kakhaki R, Akbari E, Tajabadi-Ebrahimi M, Jafari P, Asemi Z. Clinical and metabolic response to probiotic supplementation in patients with multiple sclerosis: A randomized, double-blind, placebo-controlled trial. Clin. Nutr. 2016 doi: 10.1016/j.clnu.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 40.Lim CK, Bilgin A, Lovejoy DB, Tan V, Bustamante S, Taylor BV, Bessede A, Brew BJ, Guillemin GJ. Kynurenine pathway metabolomics predicts and provides mechanistic insight into multiple sclerosis progression. Sci. Rep. 2017;7:1–9. doi: 10.1038/srep41473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41•.Rothhammer V, Mascanfroni I, Bunse L, Takenaka M, Kenison J, Mayo L, Chao C-C, Patel B, Yan R, Blain M, et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 2016;22:586–597. doi: 10.1038/nm.4106. This mouse study causally links lactobacilli to the production of AHR ligands and mitigation of experimentally induced central nervous system inflammation. Reconstitution of an antibiotic-disrupted microbiome with L. reteuri capable of metabolizing tryptophan was sufficient to improve markers of disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ignacio A, Fernandes MR, Rodrigues VAA, Groppo FC, Cardoso AL, Avila-Campos MJ, Nakano V. Correlation between body mass index and faecal microbiota from children. Clin. Microbiol. Infect. 2016;22:1–8. doi: 10.1016/j.cmi.2015.10.031. [DOI] [PubMed] [Google Scholar]
- 43.Riva A, Borgo F, Lassandro C, Verduci E, Morace G, Borghi E, Berry D. Pediatric obesity is associated with an altered gut microbiota and discordant shifts in Fiirmicutes populations. Environ. Microbiol. 2017;19:95–105. doi: 10.1111/1462-2920.13463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu HJ, Park SG, Jang HB, Choi MG, Park KH, Kang JH, Park SI, Lee HJ, Cho SH. Obesity alters the microbial community profile in Korean Adolescents. PLoS One. 2015;10:1–14. doi: 10.1371/journal.pone.0134333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teixeira TFS, Grześkowiak ŁM, Salminen S, Laitinen K, Bressan J, Gouveia Peluzio M, do C. Faecal levels of Bifidobacterium and Clostridium coccoides but not plasma lipopolysaccharide are inversely related to insulin and HOMA index in women. Clin. Nutr. 2013;32:1017–1022. doi: 10.1016/j.clnu.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 46.Karlsson FH, Tremaroli V, Nookaew I, Bergström G, Behre CJ, Fagerberg B, Nielsen J, Bäckhed F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 47.Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, Prifti E, Vieira-Silva S, Gudmundsdottir V, Krogh Pedersen H, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528:262–266. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sáez-Lara MJ, Robles-Sanchez C, Ruiz-Ojeda FJ, Plaza-Diaz J, Gil A. Effects of probiotics and synbiotics on obesity, insulin resistance syndrome, type 2 diabetes and non-alcoholic fatty liver disease: A review of human clinical trials. Int. J. Mol. Sci. 2016;17:1–15. doi: 10.3390/ijms17060928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Drissi F, Raoult D, Merhej V. Metabolic role of lactobacilli in weight modification in humans and animals. Microb. Pathog. 2016 doi: 10.1016/j.micpath.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 50.Roy CILe, Štšepetova J, Sepp E, Songisepp E, Sandrine P, Mikelsaar M. New insights into the impact of Lactobacillus population on host-bacteria metabolic interplay. Oncotarget. 2015;6:30545–30556. doi: 10.18632/oncotarget.5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gonzalez FJ, Jiang C, Patterson AD. An intestinal microbiota-farnesoid X receptor axis modulates metabolic disease. Gastroenterology. 2016;151:845–859. doi: 10.1053/j.gastro.2016.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang L, Xie C, Nichols RG, Chan SHJ, Jiang C, Hao R, Smith PB, Cai J, Simons MN, Hatzakis E, et al. Farnesoid X receptor signaling shapes the gut microbiota and controls hepatic lipid metabolism. mSystems. 2016;1:1–17. doi: 10.1128/mSystems.00070-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Borges-Canha M, Portela-Cidade JP, Dinis-Ribeiro M, Leite-Moreira AF, Pimentel-Nunes P. Role of colonic microbiota in colorectal carcinogenesis: A systematic review. Rev. Esp. Enfermedades Dig. 2015;107:659–671. doi: 10.17235/reed.2015.3830/2015. [DOI] [PubMed] [Google Scholar]
- 54.Li J, Sung CYJ, Lee N, Ni Y, Pihlajamäki J, Panagiotou G, El-Nezami H. Probiotics modulated gut microbiota suppresses hepatocellular carcinoma growth in mice. Proc. Natl. Acad. Sci. 2016;113:E1306–E1315. doi: 10.1073/pnas.1518189113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hieken TJ, Chen J, Hoskin TL, Walther-Antonio M, Johnson S, Ramaker S, Xiao J, Radisky DC, Knutson KL, Kalari KR, et al. The microbiome of aseptically collected human breast tissue in benign and malignant disease. Sci. Rep. 2016;6:1–10. doi: 10.1038/srep30751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guerrero-Preston R, Godoy-Vitorino F, Jedlicka A, Rodríguez-Hilario A, González H, Bondy J, Lawson F, Folawiyo O, Michailidi C, Dziedzic A, et al. 16S rRNA amplicon sequencing identifies microbiota associated with oral cancer, human papillomavirus infection and surgical treatment. Oncotarget. 2016;7:51320–51334. doi: 10.18632/oncotarget.9710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57•.Zijlmans MAC, Korpela K, Riksen-Walraven JM, de Vos WM, de Weerth C. Maternal prenatal stress is associated with the infant intestinal microbiota. Psychoneuroendocrinology. 2015;53:233–245. doi: 10.1016/j.psyneuen.2015.01.006. 16S rRNA sequencing of fecal microbiomes from a mother-infant cohort before and after birth allowed Zijlmans and colleagues to link maternal prenatal stress to alterations in the infant microbiome. Pathobionts were enriched in infants whose mothers were more stressed. Lactobacillus cell numbers were depleted. [DOI] [PubMed] [Google Scholar]
- 58.Golubeva AV, Crampton S, Desbonnet L, Edge D, O’Sullivan O, Lomasney KW, Zhdanov AV, Crispie F, Moloney RD, Borre YE, et al. Prenatal stress-induced alterations in major physiological systems correlate with gut microbiota composition in adulthood. Psychoneuroendocrinology. 2015;60:58–74. doi: 10.1016/j.psyneuen.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 59.Leclercq S, Mian FM, Stanisz AM, Bindels LB, Cambier E, Ben-Amram H, Koren O, Forsythe P, Bienenstock J. Low-dose penicillin in early life induces long-term changes in murine gut microbiota, brain cytokines and behavior. Nat. Commun. 2017;8:1–12. doi: 10.1038/ncomms15062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60••.Buffington SA, Di Prisco GV, Auchtung TA, Ajami NJ, Petrosino JF, Costa-Mattioli M. Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring. Cell. 2016;165:1762–1775. doi: 10.1016/j.cell.2016.06.001. Maternal high-fat diets have been linked to increased risk of disease in offspring; however, this is the first report of such diets conferring deficits to mental health. Interestingly, offspring of HF-fed dams exhibited significant reductions in cortical oxytocin receptor levels and colonization of these animals with L. reuteri was sufficient to correct this deficit. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith CJ, Emge JR, Berzins K, Lung L, Khamishon R, Shah P, Rodrigues DM, Sousa AJ, Reardon C, Sherman PM, et al. Probiotics normalize the gut-brain-microbiota axis in immunodeficient mice. Am. J. Physiol. - Gastrointest. Liver Physiol. 2014;307:G793–G802. doi: 10.1152/ajpgi.00238.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Emge JR, Huynh K, Miller EN, Kaur M, Reardon C, Barrett KE, Gareau MG. Modulation of the microbiota-gut-brain axis by probiotics in a murine model of inflammatory bowel disease. Am. J. Physiol. - Gastrointest. Liver Physiol. 2016;310:G989–G998. doi: 10.1152/ajpgi.00086.2016. [DOI] [PubMed] [Google Scholar]
- 63.Huang R, Wang K, Hu J. Effect of probiotics on depression: A systematic review and meta-analysis of randomized controlled trials. Nutrients. 2016;8:1–12. doi: 10.3390/nu8080483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Steenbergen L, Sellaro R, van Hemert S, Bosch JA, Colzato LS. A randomized controlled trial to test the effect of multispecies probiotics on cognitive reactivity to sad mood. Brain. Behav. Immun. 2015;48:258–264. doi: 10.1016/j.bbi.2015.04.003. [DOI] [PubMed] [Google Scholar]