Abstract
U.S. Environmental Protection Agency air pollution monitoring data have been a valuable resource commonly used for investigating the associations between short-term exposures to PM2.5 chemical components and human health. However, the temporally-sparse sampling on every third or sixth day may affect health effect estimation. We examined the impact of non-daily monitoring data on health effect estimates using daily data from the Denver Aerosol Sources and Health (DASH) study. Daily concentrations of four PM2.5 chemical components (elemental and organic carbon, sulfate, and nitrate) and hospital admission counts from 2003 through 2007 were used. Three every-third-day time series were created from the daily DASH monitoring data, imitating the U.S. Speciation Trend Network (STN) monitoring schedule. A fourth, partly irregular, every-third-day time series was created by matching existing sampling days at a nearby STN monitor. Relative risks (RRs) of hospital admissions for PM2.5 components at lags 0 to 3 were estimated for each data set, adjusting for temperature, relative humidity, longer term temporal trends, and day of week using generalized additive models, and compared across different sampling schedules. The estimated RRs varied somewhat between the non-daily and daily sampling schedules and between the four non-daily schedules, and in some instances could lead to different conclusions. It was not evident which features of the data or analysis were responsible for the variation in effect estimates, although seeing similar variability in resampled data sets with relaxation of the every third day constraint suggests that limited power may have played a role. The use of non-daily monitoring data can influence interpretation of estimated effects of PM2.5 components on hospital admissions in time-series studies.
Keywords: chemical component, hospital admission, particulate matter, sampling schedule, Speciation Trends Network, time-series study
INTRODUCTION
Fine particulate matter (PM2.5) air pollution is a complex mixture of numerous chemical compounds. Investigation of health effects associated with PM2.5 chemical components may help identify the most toxic component(s) of PM2.5 (1). Because individual PM2.5 components can serve as indicators of specific pollution sources such as traffic or power plants, identification of components associated with the most toxic sources could allow for more targeted regulation to reduce health risks of PM2.5 (2).
Monitoring data from the U.S. Environmental Protection Agency (EPA) Chemical Speciation Network (CSN) have been a valuable resource for studying the association of PM2.5 components and health. The CSN has measured PM2.5 chemical components at more than 300 U.S. sites since 2000 (3). Most time-series studies on effects of short-term concentration increases of PM2.5 components have used CSN data to evaluate the association with mortality or hospital admissions (4–8). However, despite increasing use in epidemiological studies, insufficient temporal and spatial coverage of CSN data is a concern and has been cited as a limitation in investigating health effects of PM2.5 components (4,9). The temporal sparseness is mainly due to the limited sampling schedule. Fifty-three core CSN sites, termed the Speciation Trends Network (STN) sites, sample PM2.5 components on an every-third-day schedule, while the majority of remaining sites typically sample every sixth day (10). This limited sampling frequency can be expected to lead to decreasing power and poor precision of health effect estimates compared to complete daily sampling. In addition, estimated effects from the limited data may be sensitive to the sampling days included in the analysis, even for analyses with nearly equal numbers of sampling days.
In this study, we examined whether different sampling schedules affect health effect estimates in a time-series study design. Taking advantage of five years of daily PM2.5 component concentration data collected for the Denver Aerosol Sources and Health (DASH) study, we compared estimated effects of daily PM2.5 chemical component concentrations on daily hospital admissions using several related data sets: the complete daily time series, three non-daily time series based on hypothetical every-third-day schedules, and another non-daily time series based on the actual every-third-day schedule with some missing and irregular sampling days at a nearby STN site.
MATERIALS AND METHODS
In the DASH study, PM2.5 and PM2.5 components were measured at one residential monitoring station daily over five years from 2003 to 2007. This monitoring site was selected to represent residential concentrations of PM2.5 components in Denver and to not be influenced by nearby pollution emission sources, including large roadways (Figure 1). A detailed description of the DASH study is available (11). We focused on four PM2.5 components that make up the bulk of PM2.5 mass in Denver (11) and showed associations with hospital admissions in previous studies (6,7,12): elemental and organic carbon (EC and OC), sulfate and nitrate. Details on sampling and lab-analysis methods for each component have been described (13). Daily counts of hospital admissions during the corresponding study period were compiled from non-elective hospital admission discharge data for all ages collected by the Colorado Hospital Association. We aggregated daily admissions for cardiovascular and respiratory diseases based on the International Classification of Diseases, 9th revision codes within the five counties (Adams, Arapahoe, Denver, Douglas, and Jefferson counties) adjacent to the DASH monitoring site. Cardiovascular hospital admissions were identified by codes 460–519 and respiratory disease by codes 390–459, including the following subcategories: ischemic heart disease (410–414), congestive heart failure (428), cerebrovascular disease (430–438), ischemic stroke (434.01, 434.11, 434.91, 436), chronic obstructive pulmonary disease (COPD: 490–492, 496), asthma (493), pneumonia (480–486), and upper respiratory infection (460–466, 477).
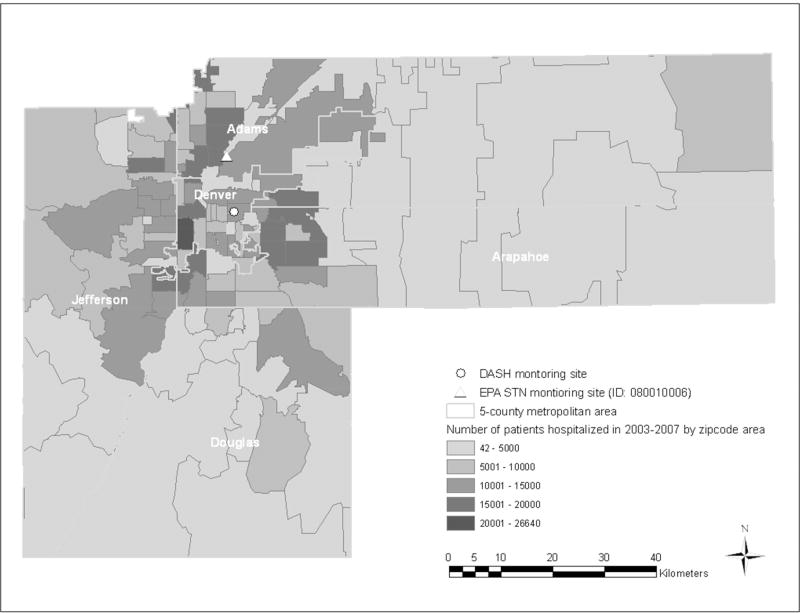
Figure 1.
Map showing the DASH and EPA STN monitoring sites within the five-county Denver metropolitan area, and the number of total hospital admissions by zip code areas for 2003–2007.
We used the complete daily DASH monitoring data to simulate data that would have been available from non-daily sampling schedules. We created four non-daily sampling schedules and populated values for the included days using the daily DASH monitoring data. Because most STN sites measure PM2.5 components every third day, three regular every-third-day schedules (E1, E2, and E3) were created, starting on January 1, 2, and 3, 2003, respectively. A fourth, more realistic scenario (E4:STN) was created by matching the days on which validated PM2.5 speciation data was actually collected from the nearby STN site shown in Figure 1. The sampling schedule at this STN site was similar to the every-third-day schedule beginning on January 3, 2003 (E3), except for six mismatched dates and 120 additional missing days throughout the five year period compared to those of E3. PM2.5 chemical component concentrations measured daily at the DASH site were then assigned to corresponding days in all four non-daily sampling schedules. There were 1826 days in the five year study period, and the DASH data were 99% complete with between 1808 and 1809 daily observations, depending on component. After pairing up the DASH data with the non-daily sampling schedules, data sets for E1, E2 and E3 contained just over 600 observations per component while the data set for E4:STN contained just under 500 observations per component (Table 1). The actual monitoring data at the STN site were not used to avoid the influence of different sampling equipment and methods on variation in health effect estimates.
Table 1.
Summary statistics of 24-hour average concentrations of PM2.5, four PM2.5 chemical components, meteorological conditions, and daily counts of hospital admissions for 2003–2007 by time-series data of complete and every third sampling days in the five-county Denver metropolitan area
| Category | Variable | N | Mean (SD) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| C1) | E12) | E23) | E34) | E4:STN5) | C | E1 | E2 | E3 | E4:STN | ||
| Pollutant (ug/m3) | PM2.5 | 1808 | 602 | 603 | 603 | 490 | 8.0 (5.1) | 7.9 (4.8) | 7.9 (5.1) | 8.1 (5.3) | 8.3 (5.7) |
| EC | 1809 | 603 | 603 | 603 | 490 | 0.5 (0.3) | 0.5 (0.4) | 0.5 (0.3) | 0.5 (0.3) | 0.5 (0.3) | |
| OC | 1809 | 603 | 603 | 603 | 490 | 3.1 (1.4) | 3.1 (1.4) | 3.1 (1.4) | 3.1 (1.3) | 3.1 (1.4) | |
| Sulfate | 1808 | 603 | 603 | 602 | 489 | 1.1 (1.0) | 1.0 (0.8) | 1.1 (1.0) | 1.1 (1.0) | 1.2 (1.2) | |
| Nitrate | 1808 | 602 | 603 | 603 | 490 | 1.0 (2.0) | 1.0 (1.9) | 1.1 (2.0) | 1.0 (2.0) | 1.1 (2.2) | |
|
| |||||||||||
| Meteorological condition | Temperature (°C) | 1813 | 605 | 605 | 603 | 491 | 10.6 (−8.1) | 10.6 (−8.0) | 10.7 (−8.2) | 10.5 (−8.0) | 10.8 (−8.0) |
| Humidity (%) | 1813 | 605 | 605 | 603 | 491 | 54.6 (20.8) | 54.8 (20.8) | 54.3 (20.8) | 54.8 (20.7) | 54.7 (20.8) | |
|
| |||||||||||
| Hospital admission (N) | Cardiovascular disease | 1826 | 609 | 609 | 608 | 494 | 44.8 (9.3) | 44.9 (9.4) | 44.9 (9.4) | 44.6 (8.9) | 44.1 (8.9) |
| Congestive heart failure | 1826 | 609 | 609 | 608 | 494 | 7.8 (3.2) | 7.8 (3.2) | 7.7 (3.2) | 7.8 (3.2) | 7.7 (3.2) | |
|
| |||||||||||
| Respiratory disease | 1826 | 609 | 609 | 608 | 494 | 37.1 (14.6) | 36.9 (14.8) | 37.2 (14.3) | 37.3 (14.6) | 36.8 (14.4) | |
| Asthma | 1826 | 609 | 609 | 608 | 494 | 5.8 (3.0) | 5.7 (3.1) | 5.8 (3.0) | 5.8 (3.0) | 5.7 (3.0) | |
|
| |||||||||||
| Weekday /Weekend | Weekday (N) | 1304 | 435 | 435 | 434 | 331 | |||||
| Weekend (N) | 522 | 174 | 174 | 174 | 163 | ||||||
| Weekday/Weekend (%) | 2.5 | 2.5 | 2.5 | 2.5 | 2.0 | ||||||
|
| |||||||||||
| Total | 1826 | 609 | 609 | 608 | 494 | ||||||
C: complete daily schedule
E1: regular every third day schedule staring on January 1, 2003
E2: regular every third day schedule staring on January 2, 2003
E3: regular every third day schedule staring on January 3, 2003
E4:STN: irregular every third day schedule equivalent to the sampling schedule (E3 except for six mismatched and 120 additional missing days) at an existing STN site 13 kilometers away from the DASH monitoring site
The time-series analysis was performed separately for the daily and four non-daily data sets. Effects of 24-hour daily average concentrations of the four PM2.5 components on daily hospital admissions were estimated using generalized additive models adjusting for time from the start of the study, day of week, and 24-hour daily averages of temperature and relative humidity. The non-linear trends of time and temperature with daily hospital admissions were accounted for using regression splines (14). Degrees of freedom (df) for the splines were given a priori as 60 df for time (12 df per year) and 3 df for temperature. We also used fewer df such as 4 and 6 df per year in a sensitivity analysis. Effects at pollution day lags 0 to 3 (i.e., pollution on the same day of to three days before the hospital admission) were estimated. Estimated effects in each data set were presented as relative risks (RRs) and 95% confidence intervals (95% CIs) for an interquartile range increase in concentration of each component. We also presented estimated effects of PM2.5 total mass for comparison with those of the PM2.5 components
To assess the influence of non-daily time-series data sets on health effect estimates, we compared the magnitude of the estimates across the daily and four non-daily data sets on the same component, hospitalization diagnosis, and lag. We also examined whether estimates across data sets lead to different conclusions based on statistical significance of the observed association defined by a lower 95% CI bound greater than 1. We focused on results for total cardiovascular and respiratory disease hospital admissions, and congestive heart failure and asthma hospital admissions, as examples from each of the general cardiovascular and respiratory diagnosis groups, to highlight some of the more discrepant health effect estimates across data sets. Results for the other specific diseases are shown in the Supplemental Information (Supplemental Figure 3).
We carried out further analyses to better understand three features that might have influenced the resulting health effect estimates across non-daily and daily data sets: data distribution, outlying measurements, and residual confounding by time. For this investigation, we chose the examples showing distinctively different health effect estimates between data sets; the examples are cases of the same component, hospitalization diagnosis, and lag that exhibited statistically significant differences between estimates in at least one pair of two data sets. Although non-overlapping confidence intervals have been commonly used to assess the statistical difference between two estimates, because this approach is overly conservative, we relied here on a standard hypothesis testing approach using a two-sample t-test (15). To assess the effects of outlying measurements, we investigated whether the difference of effect estimates across data sets lessened after removing outliers by trimming PM2.5 component, hospital admission, or meteorology data at 1, 2.5, and 5 percentiles from both ends of each distribution. Our approach to temporal smoothing in the non-daily data sets may not have adequately captured the long-term temporal trends and could therefore result in residual confounding. In an additional sensitivity analysis, we therefore adjusted for time smoothed over the complete set of days before fitting the model with every-third-day PM2.5 component data.
In addition, we relaxed the every third day structure and expanded our suite of analyses to include any temporally-limited data. Our focus on the every third day schedule was based on the sampling scheme of the EPA monitoring data which are publically available and commonly used in many time-series studies on PM2.5 components. The observed variation of health effect estimates across the every third day data sets, however, could occur with use of any limited time-series data set. To explore the impact of temporally-limited data, we sampled one third of the daily data 1000 times. Then, we performed health effect analyses for each sampled data set and compared the resulting distribution of estimates to those from the every third day data sets. The resampling employed two approaches: random selection of 603 days and random selection of a time series of 603 sequential days without replacement.
RESULTS
Summary statistics of the PM2.5 component measures, meteorology, and hospital admissions for each of the five pollution monitoring schedules are presented in Table 1. Daily PM2.5 component concentrations varied by as much as 19% across the different schedules, although the means were relatively consistent. In particular, the standard deviation of sulfate was 19% lower in the first every-third-day schedule (E1) and 19% higher in the STN schedule (E4:STN), compared to the complete schedule (C). The mean and standard deviation of meteorology in the non-daily schedules agreed within 2% compared to the complete daily schedule. The standard deviations of the non-daily hospital admission counts varied at most by 10% from the complete schedule.
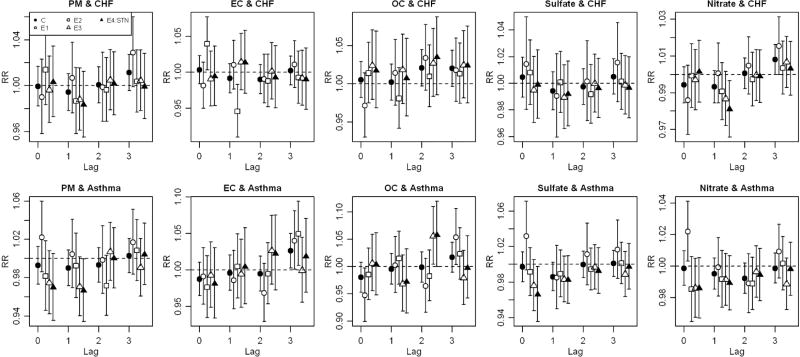
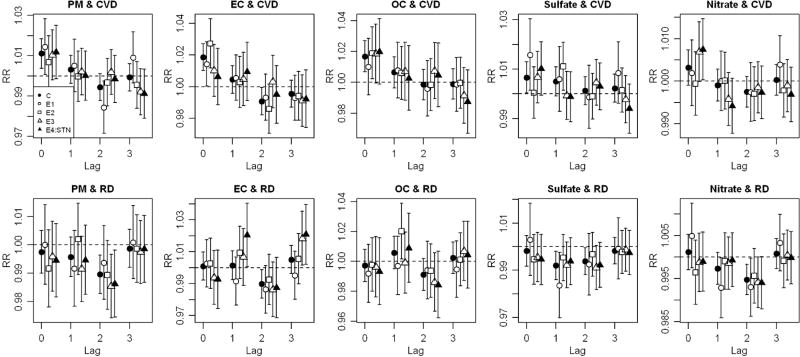
There was some variability in estimated RRs of hospital admissions across the several data sets for all PM2.5 components and hospital discharge diagnoses examined (Figures 2 and 3, Supplemental Tables 1 and 2). In most cases, this variability did not result in meaningful differences in reported findings, largely because most RRs were null. There were some notable exceptions, however, with the lower bound of a 95% CI being greater than 1 in some data sets, but not other data sets, for the same component, lag, and hospitalization diagnosis. For example, the statistically significant effect estimate of EC for total cardiovascular disease at lag 0 observed in the complete data set (C) had smaller, non-significant, point estimates in two of the incomplete data sets (E3 and E4:STN) (Figure 2, Supplemental Table 1). As another example, the effect estimate of nitrate on cardiovascular disease at lag 0 was significantly positive in an incomplete data set (E4:STN), but somewhat smaller and not statistically significant in other three (C, E1, and E2), including the complete data set. These more notable differences were also seen for the effect estimate of EC on congestive heart failure (Figure 3, Supplemental Table 2) at lag 0. For total respiratory disease (Figure 2, Supplemental Table 1), we observed null effects of EC at lags 1 and 3 in most but not all of the data sets. Similar patterns were observed for asthma (Figure 3, Supplemental Table 2) and EC at lag 3, for OC at lags 2 and 3, and for nitrate at lag 0. PM2.5 also exhibited some variation of RRs across sampling schedules. Use of fewer degrees of freedom for time did not meaningfully change these results.
Figure 2.
Relative risks (RRs) and 95% confidence intervals (CIs) of cardiovascular disease (CVD) and respiratory disease (RD) hospital admissions for an interquartile increase of PM2.5 and four PM2.5 components across lag 0 to 3 by time-series data of complete and every third sampling days in the five-county Denver metropolitan area for 2003–2007.
Figure 3.
Relative risks (RRs) and 95% confidence intervals (CIs) of congestive heart failure (CHF) and asthma hospital admissions for an interquartile increase of PM2.5 and four PM2.5 components by time-series data of complete and every third sampling days in the five-county Denver metropolitan area for 2003–2007.
We next focus attention only on differences of effect estimates using the complete data set (C) in comparison to the data set comprising days for which data were actually available from the local STN site (E4:STN). Here we observed differences for cardiovascular disease (EC lag 0 and nitrate lag 0), respiratory disease (EC lag 1 and lag 3), and asthma (EC lag 3 and OC lag 2) hospitalization diagnoses (Figures 2 and 3, Supplemental Tables 1 and 2).
To explore possible sources of discrepant estimates across data sets, we focused on the particular cases with statistically different RR estimates across data sets based on the hypothesis test for the same PM2.5 component, hospitalization diagnosis and lag. These eight cases were EC and respiratory disease at lags 1 and 3, EC and congestive heart failure at lags 0 and 1, OC and asthma at lags 2 and 3, sulfate and asthma at lag 0, and nitrate and asthma at lag 0 (Figures 2 and 3). Comparing distributions across data sets, there were no substantial differences in means or standard deviations of the PM2.5 component or health outcome data, with a few exceptions. RRs of sulfate and nitrate on asthma hospital admission at lag 0 showed differences largely between the first every-third-day data set (E1) and the other data sets; for this data set, standard deviation of daily asthma hospital admission counts was larger than for the other data sets (Table 1). However, this pattern was not found in the other selected cases. There were also no distinct differences in the distributions of meteorology, day of the week, and weekday/weekend between the data sets. A few outliers were identified in plots of PM2.5 component concentrations against fully-adjusted health effects model residuals (results not shown). Effect estimates from some every-third-day data sets were attenuated and differences between data sets were no longer statistically significant in half of the example cases when the hospitalization count data were trimmed. However, in general, differences in the health effect estimates across the data set remained after trimming (Supplemental Figure 1). Trimming the PM2.5 component data or the meteorology data had no impact on reducing differences. Removal of the temporal trend estimated from the complete data reduced the discrepancies in the effect estimates of EC on respiratory and congestive heart failure hospital admissions but not in those for other PM2.5 components and outcomes (Supplemental Figure 2).
In the resampling analysis using one third of the daily data, health effect estimates from the every third day data sets were generally within the range of the 95% coverage of the distribution of 1000 health effect estimates from 1000 resampled data sets (Supplemental Figure 4).
DISCUSSION
Our purpose in utilizing a daily time series of speciated PM2.5 data to generate hypothetical non-daily data sets was to gain insight into the possibility that conclusions reached from health effects analyses could be influenced by the availability of pollutant data. Many time-series analyses performed in the U.S. use EPA monitoring network data for PM2.5 and PM2.5 chemical components because of the widespread availability of the data (4–8). However, the PM2.5 chemical speciation data are largely collected on an every-third-day or an every-sixth-day schedule. The question, then, is whether we should be concerned that using non-daily pollution data in health analyses might lead to different conclusions than those that would have been reached had daily data or a different schedule of non-daily data been available. We found some differences in health effect estimates, and in the conclusions that in some cases would likely be drawn, when non-daily data based on every third day sampling or other temporally-limited sampling rather than daily data were used, or when one set of non-daily data was used rather than another.
Our findings suggest that some of the discrepant or inconsistent findings across time-series studies of PM2.5 components could be caused at least in part by differences in the available analysis data sets. There may be several explanations for this. A primary consideration is that using one third of the data reduces the statistical power to detect associations. We found associations between PM2.5 and four PM2.5 components with cardiovascular hospital admissions in the complete data sets but these were not present in about half of the every third day data sets, possibly due to lack of power. Several previous studies consistently found associations between four PM2.5 components and cardiovascular diseases at lag 0 using the EPA data sampled every three days (7,8). These studies, however, had longer study periods and larger populations resulting in increased power. As a reflection of this reduced power, the 95% CIs for effects estimated using non-daily data were generally wider than those based on daily data (Figures 2 and 3). The variation of estimates observed not only in the every third day data but also using any third of the complete data suggested a role of reduced power. Similarly, the use of different sets of pollution-hospitalization days will produce different estimates due to random variability. Depending upon how these are interpreted, this variability can potentially translate into bias. For example, a time-series study using the STN data in California reported an association between EC and respiratory admissions in children at lag 3 (6). In our analysis, EC at lag 3 had an estimated effect that was consistent with that finding relating to respiratory hospital admissions when the E4:STN data schedule was used (Figure 2). However, we also found much weaker and not statistically significant associations using other data subsets, including the complete and the two regular every-third-day data sets (E1 and E2). In light of many comparisons made in this paper, these diverging results are generally consistent with each other. Bias can be introduced when authors focus on reporting or emphasizing statistically significant results; this source of random bias can be particularly important in time-series studies of PM2.5 components which commonly explore the association between multiple components and outcomes.
Another possible explanation would be bias in the estimates due to features of the data used in the analysis. To investigate this, we hypothesized that aspects of the data distribution, presence of outlying values, and/or the approach to adjustment for the temporal trend could be responsible for producing differences in health effect estimates across the various data sets. However, we did not identify any of these as strongly influential. Rather, each feature contributed to some of the observed differences for particular PM2.5 components and hospitalization outcomes, but their patterns were not consistent across all examples. It is likely that our reliance on a fixed number of degrees of freedom for temporal trend resulted in less residual temporal confounding, leading to smaller differences of health effect estimates than may have been present if we relied on automated model selection criteria. A simulation study, in which the number of degrees of freedom was automatically selected using penalization, demonstrated increased bias and mean square errors as the correlation between air pollution concentrations and a confounding temporal trend increased (16). We cannot rule out the possibility that the observed differences were due to other features that we did not examine, or whether these were simply the result of random variability.
In the present study, there were differences in health effect estimates across different time-series data sets based on the lower bound of the 95% CI. For all five data set groups, for all four PM2.5 components, hospitalization diagnoses and lags, we observed that in approximately 40% of the groups (14 out of 64) the 95% CI of at least one of the effect estimates in the five data set groups did not include one. Selective reporting of statistically significant results without prior hypotheses can be misleading and is not recommended in epidemiological studies (17). One strategy for minimizing the multiple testing problem is to reduce the number of tests interpreted as key findings by relying on pre-specified choices of the components, outcomes and modeling strategies of primary interest (18). Such discipline is particularly important in studies investigating multiple different exposures as well as outcomes. Our purpose in this study, however, was to illustrate the influence that incomplete time-series data might have in drawing conclusions regarding associations between PM2.5 components and cardiorespiratory outcomes. Analysis of time-series PM2.5 component data will generally only have one of these pollutant data sets available for use in the analysis. In such analyses, findings are interpreted based on the one available data set without the benefit of knowing how well that data set reflected results that would have been obtained had a more complete data set been used. Although there are criteria other than statistical significance based on the lower bound of confidence intervals that can be used to evaluate associations and draw conclusions, we adopted this commonly-used approach in order to demonstrate our point. We evaluated statistical differences in effect estimates across data sets only as a tool to identify a subset of effect estimates for further exploration.
Sensitivity of health effect estimates to non-daily pollution data using sampling patterns that imitate the existing governmental monitoring data has been reported in other studies. Using data from the Air Pollution and Health: A Combined European and North American Approach (APHENA) study (19), investigators created every-sixth-day PM10 and ozone data from the complete daily data in four European cities and repeated their health analyses using the limited data. The results showed that using systematically missing time-series PM10 data produced smaller and less precise mortality effect estimates compared to estimates using the complete data. Similar to our approach, Klemm et al (2011) created every-third-day and every-sixth-day data sets from their own daily data from 1998 to 2007 in Atlanta, and compared mortality effect estimates from criteria air pollutants as well as EC and OC in adults over age 65 (20). They found that effect estimates fluctuated across data sets and changed signs in some cases. Effect estimates in their non-daily data sets varied not only relative to those with daily data, but also relative to those in other non-daily data sets with different starting days, as shown also in our findings. In the present study, the use of hospital admission data rather than mortality data allowed us to examine effects of non-daily data for a larger array of disease outcomes with increasing power. In addition to comparing effect estimates, we explored several possible features of the data that may produce different effect estimates for different subsets of the same pollution data set. The identification of such features is important since it may be possible to adopt an approach to analysis of time-series studies that would control these features and thus reduce the sensitivity of effect estimates in analyses based on non-daily pollution data without the need for complete daily data. The three characteristics that we hypothesized to be most likely to affect differences in effect estimates did not seem to influence the results. It is possible that future studies will be able to identify other such features.
The EPA speciation monitoring network was established to provide speciated PM2.5 data for supporting both policy decisions and health effects research (21). Although the CSN data are a useful resource for epidemiological studies, our results suggest the need for additional care in interpreting findings when analyses are based on non-daily pollution data.
Supplementary Material
Acknowledgments
This work was supported by the NIEHS research grant R01 ES010197
Footnotes
Disclaimer: The views expressed are those of the authors and do not necessarily reflect the views or policies of the U.S. Environmental Protection Agency
Conflict of interest: The authors declare no conflict of interest
Supplementary information is available at the Journal of Exposure Science and Environmental Epidemiology’s website.
References
- 1.Grahame TJ, Schlesinger RB. Health effects of airborne particulate matter: Do we know enough to consider regulating specific particle types or sources? Inhal Toxicol. 2007;19:457–481. doi: 10.1080/08958370701382220. [DOI] [PubMed] [Google Scholar]
- 2.Traffic-related air pollution: A critical review of the literature on emissions, exposure, and health effects. Boston, MA: Health Effects Institute; 2010. HEI panel on the health effects of traffic-related air pollution. (HEI special report 17) [Google Scholar]
- 3.U.S. EPA. Air quality criteria for particulate matter: Volume 1. Washington, DC: U.S. Environmental Protection Agency; 2004a. (Report No. EPA 600/P-99/002aF-bF) [Google Scholar]
- 4.Ostro BD, Feng WY, Broadwin R, Green S, Lipsett N. The effects of components of fine particulate air pollution on mortality in California: results from CALFINE. Environ Health Perspect. 2007;115:13–19. doi: 10.1289/ehp.9281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ostro BD, Feng WY, Broadwin R, Malig BJ, Green RS, Lipsett MJ. The impact of components of fine particulate matter on cardiovascular mortality in susceptible subpopulations. Occup Environ Med. 2008;65:750–706. doi: 10.1136/oem.2007.036673. [DOI] [PubMed] [Google Scholar]
- 6.Ostro BD, Roth L, Malig B, Marty M. The effects of fine particle components on respiratory hospital admissions in children. Environ Health Perspect. 2009;117:475–480. doi: 10.1289/ehp.11848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peng RD, Bell ML, Geyh AS, McDermott A, Zeger SL, Samet JM, et al. Emergency admissions of cardiovascular and respiratory diseases and the chemical composition of fine particle air pollution. Environ Health Perspect. 2009;117:957–963. doi: 10.1289/ehp.0800185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito K, Mathes R, Ross Z, Nádas A, Thurston G, Matte T. Fine particulate matter constituents associated with cardiovascular hospital admissions and mortality in New York City. Environ Health Perspect. 2011;119:467–473. doi: 10.1289/ehp.1002667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lippmann M. Semi-continuous speciation analyses for ambient air particulate matter: An urgent need for health effect studies. J Exp Sci Environ Epidemiol. 2009;19:235–247. doi: 10.1038/jes.2008.65. [DOI] [PubMed] [Google Scholar]
- 10.U.S. EPA. Integrated science assessment for particulate matter. Research Triangle Park, NC: U.S. Environmental Protection Agency, Office of Research and Development; 2009. (Report No. EPA/600/R-08/139F) [PubMed] [Google Scholar]
- 11.Vedal S, Hannigan MP, Dutton SJ, Miller SL, Milford JB, Rabinovitch N, et al. The Denver Aerosol Sources and Health (DASH) study: Overview and early findings. Atmos Environ. 2009;43:1666–1673. doi: 10.1016/j.atmosenv.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim SY, Peel JL, Hannigan MP, Dutton SJ, Sheppard L, Clark ML, et al. The Temporal Lag Structure of Short-term Associations of Fine Particulate Matter Chemical Constituents and Cardiovascular and Respiratory Hospitalizations. Environ Health Perspect. 2012;120:1094–1099. doi: 10.1289/ehp.1104721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dutton SJ, Schauer JJ, Vedal S, Hannigan MP. PM2.5 characterization for time series studies: Pointwise uncertainty estimation and bulk speciation methods applied in Denver. Atmos Environ. 2009;43:1136–1146. doi: 10.1016/j.atmosenv.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wood SN. Generalized additive models: An introduction with R. Boca Raton, FL: Chapman & Hall/CRC, Taylor & Francis Group; 2006. Some GAM theory; pp. 141–216. [Google Scholar]
- 15.Schenker N, Gentleman JF. On judging the significance of differences by examining the overlap between confidence intervals. Am Stat. 2001;55:182–186. [Google Scholar]
- 16.Peng RD, Dominici F, Louis TA. Model choice in time series studies of air pollution and mortality. J R Stat Soc Ser A. 2006;169:179–203. [Google Scholar]
- 17.Lumley T, Sheppard L. Assessing seasonal confounding and model selection bias in air pollution epidemiology using positive and negative control analyses. Environmetrics. 2000;11:705–717. [Google Scholar]
- 18.Lumley T, Sheppard L. Time series analyses of air pollution and health: straining at gnats and swallowing camels? Epidemiology. 2003;14:13–14. doi: 10.1097/00001648-200301000-00007. [DOI] [PubMed] [Google Scholar]
- 19.Katsouyanni K, Samet JM, Anderson HR, Atkinson R, Le Tertre A, Medina S, et al. HEI Health Review Committee. Air pollution and health: A European and North American approach (APHENA) Boston, MA: Health Effects Institute; 2009. (Res Rep Health Eff Inst) [PubMed] [Google Scholar]
- 20.Klemm RJ, Thomas EL, Wyzga RE. The impact of frequency and duration of air quality monitoring: Atlanta, GA, Data modeling of air pollution and mortality. J Air & Waste Manage Assoc. 2011;61:1281–1291. doi: 10.1080/10473289.2011.617648. [DOI] [PubMed] [Google Scholar]
- 21.U.S. EPA. PM2.5 Speciation Network Newsletter. Research Triangle Park, NC: U.S. Environmental Protection Agency, Office of Air Quality Planning and Standards; 2004b. ( http://www.epa.gov/ttn/amtic/files/ambient/pm25/spec/spnews1.pdf) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.