Abstract
Microarray technology permits the interrogation of nearly all expressed genes under a wide range of conditions. Patterns of gene expression in response to obesity and diabetes have yielded important insights into the pathogenesis of diabetes and its relationship to obesity. In muscle, microarray studies have motivated research into mitochondrial function. In adipose tissue, clues have pointed to the importance of inflammation in obesity. New adipocyte-derived hormones involved in insulin resistance have been found; a notable example is retinol binding protein 4. In liver, genes responsive to master regulators of lipid metabolism have been identified. In β-cells, genes involved in cell survival, cell proliferation, and insulin secretion have been identified. These studies have greatly expanded our understanding of mechanisms underlying the pathogenesis of obesity-induced diabetes. When combined with genetic information, microarray data can be used to construct causal network models linking gene expression with disease.
Keywords: diabetes, obesity, insulin resistance, insulin signaling, β-cells, muscle
Introductory Comments and Words of Caution
The advent of microarray technology, ten years ago, has had a profound impact on all of biology and on the way in which biology experiments are conducted. Within a short period of time, research organizations worldwide installed the necessary equipment and acquired the expertise to conduct microarray experiments. When considered relative to the amount of data obtained, the experiments, if properly conducted, can be very informative and cost-effective (see sidebar, Guidelines for Planning a Microarray Experiment).
Numerous spectacular discoveries have been made through microarray experiments. In many cases, the discoveries were entirely serendipitous; i.e., they were not anticipated as part of an a priori hypothesis. Since many genes whose expression is assessed in microarray experiments are unannotated, the context in which differential expression (DE) is detected can help to define their biological functions.
One undesirable consequence of these success stories is that it has motivated many investigators to undertake microarray studies when they feel like they have exhausted all of their ideas. Essentially, they are employing the experiment to produce the hypothesis. Although important discoveries can still be made, this often results in a large amount of data with many hundreds of DE genes and a failure to produce the desired epiphany.
Highlights of Microarray-Derived Results in the Context of Obesity and Diabetes
This review provides a selective sampling of important discoveries made through microarray analysis of muscle, adipose tissue, liver, and pancreatic islets in the context of obesity and diabetes. We apologize to the many outstanding investigators whose work is not described here. Our goal is to highlight a small number of key observations and wherever relevant, point out some of the strengths of the experimental design that were of special importance.
Muscle
Muscle is responsible for >80% of insulin-stimulated glucose clearance from the bloodstream (161). Consequently, many gene expression studies aimed at identifying key genes that might be responsible for insulin resistance and type 2 diabetes focused on muscle.
Early microarray studies on muscle from individuals with insulin resistance and/or type 2 diabetes were disappointing. When corrected for the large multiple-testing penalty, essentially no genes emerged as differentially expressed between the experimental groups (139, 170). Mootha et al. approached this problem with a dimension-reduction approach to the data analysis (98). They organized the genes of the microarray into 146 lists according to pathways. They profiled skeletal muscle from three groups of subjects; those with normal glucose tolerance, those with impaired glucose tolerance, and those with frank type 2 diabetes. They ranked the DE lists according to the extent of DE. They tested the hypothesis that the ordering of the genes in this fashion would coincide with a particular category of genes; i.e., that there would be enrichment of the DE genes according to function, a method termed gene set enrichment analysis (GSEA) (145). This increased their statistical power to detect differential expression by about 100-fold and also allowed them to place modestly DE genes into a broader context.
Application of the GSEA method identified just one enriched pathway; genes whose products play a role in oxidative phosphorylation were modestly downregulated, but the key finding was that the expression of these genes is coregulated. This coregulated expression correlated with a modest (∼20%) reduction in the expression of a transcriptional coactivator involved in mitochondrial biogenesis, peroxisome proliferator-activated receptor γ-1α (PGC1α). A parallel study in Mexican Americans also demonstrated reduced expression of NRF-regulated oxidative phosphorylation genes, accompanied by a ≈50% reduction in expression of PGC1α and β in both insulin-resistant and diabetic subjects (111). It might be important that in both of these studies, the normal glucose tolerance groups had lower body mass index (BMI) than the other two groups. Another potentially important difference between the two studies was that the former study surveyed muscle under hyperinsulinemic clamp conditions, whereas the Mexican American subjects in the latter study were under fasting conditions.
The mitochondrial content of skeletal muscle is responsive to environmental factors such as exercise, aging, and caloric restriction (51). Nuclear magnetic resonance studies in human subjects showed a substantial decline in mitochondrial respiration associated with aging (112). Muscle from patients with type 2 diabetes has a reduction in mitochondrial density (59, 73). Offspring of type 2 diabetics also show a decline in mitochondrial function, along with insulin resistance and an increase in the accumulation of myocellular lipid (12, 99, 113). Shulman and coworkers (100) have suggested that reduced mitochondrial respiratory capacity results in diminished fatty acid β-oxidation, resulting in an accumulation of lipid signaling molecules, specifically diacylglycerols, which activate several atypical protein kinase C isoforms that blunt insulin signaling through the serine phosphorylation of IRS1.
Holloszy (60) has pointed out that skeletal muscle has an oxidative capacity far in excess of what is normally required even during strenuous exercise, enabling a 150-fold increase in oxygen consumption. He goes on to argue that insulin-resistant individuals have fewer mitochondria, but their reduction in oxidative capacity is not sufficient to present a bottleneck for substrate oxidation. However, this does not address the potential link between mitochondrial function/capacity and insulin sensitivity.
Rats and mice fed high-fat diets have long been used as a model of human insulin resistance. However, recent studies show that along with the insulin resistance, these animals experience an increase in number of muscle mitochondria and oxidative function (57, 150), especially fatty acid oxidation (165) (Figure 1a). This occurs with an increase in PGC1α protein, but not mRNA (57), emphasizing poor correlation between the transcriptional and post-transcriptional regulation of PGC1α. This result has not been uniformly reproduced; Sparks et al. (138) observed a decrease in several mitochondrial oxidative phosphorylation (ox/phos) genes and PGC1α protein in humans and mice on a high-fat diet. Even within the same laboratory, the effects of high-fat diets on muscle mitochondrial gene expression and PGC1α protein can be highly variable (D. Muoio, personal communication).
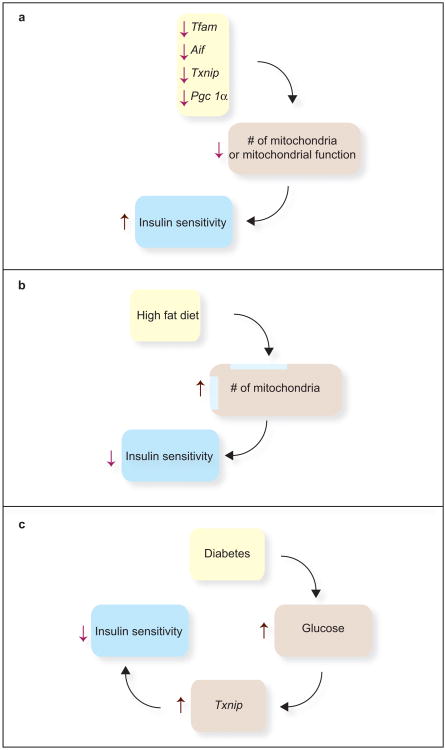
Figure 1.
Experimental observations where mitochondrial function or number of mitochondria is negatively correlated with insulin sensitivity. (a) In four different knockout mouse models that affect mitochondrial function or number of mitochondria in skeletal muscle, there is a marked improvement in insulin sensitivity. (b) High-fat diets increase the number of skeletal muscle mitochondria. The effect on PGC1α is controversial. But, these diets are associated with reduced insulin sensitivity. (c) Hyperglycemia itself is associated with reduced muscle insulin sensitivity. A highly glucose-responsive gene, Txnip, when highly expressed, reduces insulin sensitivity and conversely, as illustrated in panel a, when knocked out in skeletal muscle, is associated with improved insulin sensitivity.
To directly address the role of free fatty acids in the expression of genes for mitochondrial proteins, Holloszy's team (44) performed daily heparin injections of rats maintained on a high-fat diet to increase serum free fatty acid levels eightfold (heparin releases lipoprotein lipase from the endothelial lining of blood vessels). This resulted in an induction in the expression of skeletal muscle genes encoding TCA cycle enzymes and oxidative phosphorylation proteins and increased recruitment of the transcription factor PPARδ to the gene encoding carnitine palmitoyl transferase 1 (CPT1) (44). Overexpression of a VP16-PPARδ fusion gene leads to increased mitochondrial gene expression (85, 158), suggesting that PPARS is likely to mediate the fatty acid induction of mitochondrial biogenesis. However, Kleiner et al. (78) recently showed that although treatment of mice with the PPARS agonist GW501516 induced CPT1, PDK4, and UCP3, some other mitochondrial genes, as well as PGC1α and PGC1β, were not induced, suggesting that PPARS induces some mitochondrial genes but does not induce mitochondrial biogenesis. The response of CPT1 and PDK4 to the agonist was reduced in skeletal muscle from PGC1α-null mice. It therefore appears that PGC1α plays a crucial role in maintaining overall mitochondrial content, whereas PPARS acts specifically on several mitochondrial genes, and in the presence of PGC1α, PPARS induces expression of genes that promote fatty acid oxidation.
More recent studies by Patti and coworkers show that in obese diabetic, but not in nondiabetic insulin-resistant subjects, there is a down-regulation of mitochondrial oxidative phosphorylation genes in skeletal muscle (M.E. Patti, personal communication). These results suggest that the response tends to be reactive to diabetes and hyperglycemia rather than causal for insulin resistance. Ritov et al. (121) showed that in moderately obese (both diabetic and nondiabetic) individuals (BMI >30), there is an imbalance between TCA cycle activity and electron transport chain activity in skeletal muscle. This was inferred from the observation that citrate synthase activity was normal while NADH oxidase activity was reduced. These changes were not responsive to caloric restriction or exercise, interventions that normally improve insulin sensitivity and suggesting that obesity might affect mitochondrial function independent of its effects on insulin sensitivity.
The microarray data in human skeletal muscle (98, 111) predicted that overexpression of PGC1α in muscle should increase, whereas a knockdown or deletion of PGC1α should decrease, insulin sensitivity. Paradoxically, these predictions have not been borne out. Muscle-specific deletion of Pgc1α in mice, although it resulted in decreased mitochondrial respiratory capacity, resulted in increased glucose uptake into muscle under hyperinsulinemic conditions (58). Muscle-specific overexpression led to increased mitochondrial density and respiratory capacity and greater susceptibility to high-fat diet-induced insulin resistance (25). The high-fat diet effects on mitochondrial fatty acid oxidation capacity are not linked to any changes in insulin signaling, suggesting that these two processes may not be mechanistically linked (165).
Three other transgenic mouse models in which mitochondrial function is disrupted in muscle have increased insulin sensitivity (Figure 1b). Mice with a muscle-specific deletion of Transcription factor A (Tfam) show improved glucose tolerance and enhanced insulin-stimulated glucose uptake into extensor digitorum longus muscle (164).
Apoptosis-inducing factor (AIF), in addition to its role in apoptosis, is essential for mitochondrial electron transport, especially at Complex 1 (67). Mice deficient in the Aif gene have a reduction in mitochondrial respiratory capacity, as expected. They have a reduction in fat oxidation but increased anaerobic glucose catabolism. The mice also have increased insulin sensitivity (117).
Mice with a muscle-specific deletion of the Thioredoxin interacting protein (Txnip) gene, like the AIF-knockout mice, have a defect in aerobic glucose and fat oxidation, suggesting that their mitochondria are dysfunctional (64). The mice are extraordinarily glucose tolerant, and this correlates with an increase in phosphorylated Akt in muscle in response to insulin. A proposed mechanism is that Txnip normally prevents the thioredoxin-mediated activation of phosphatase and tensin homolog (PTEN) through reduction of its redox-sensitive disulfide. PTEN is a phophatidylinositol 3-phosphatase that blunts insulin signaling. Parikh et al. (110) identified Txnip as a regulator of glucose metabolism in humans, initially through a microarray survey. They identified TXNIP as one of just two genes repressed by insulin. The gene is induced by glucose in β-cells (18, 132) and Parikh et al. discovered that it is upregulated in the skeletal muscle of people with impaired glucose tolerance or type 2 diabetes (Figure 1c). These studies provide clues that link “redox circuitry” (102) to insulin action and offer a plausible link between mitochondrial function and the regulation of insulin signaling.
To determine whether the reduction in expression of genes encoding mitochondrial respiratory enzymes is a consequence of reduced insulin signaling or of diabetes itself, Yechoor et al. (173) surveyed gene expression in muscle from mice that were insulin-deficient and hyperglycemic due to streptozotocin treatment versus mice with a muscle-specific knockout of the insulin receptor. Diabetes was correlated with a decreased expression of these genes whereas loss of receptor signaling was not, suggesting that hyperglycemia might suppress expression of mitochondrial respiratory genes; i.e., diabetes might cause the changes in mitochondrial respiratory gene expression rather than vice versa.
It should be emphasized that although the role of mitochondrial function in insulin signaling in muscle is not fully understood, mitochondrial dysfunction in β-cells can lead to diabetes (89). Mutations in mitochondrial genes have long been known to cause insulin-dependent diabetes (87). Stimulation of insulin secretion by glucose depends upon glucose catabolism, and in β-cells, this is primarily aerobic. This was originally thought to be due to a paucity of lactate dehydrogenase in β-cells (129), but a reappraisal has shown that this is not the case (130). Thus, anything that compromises β-cell mitochondrial function will interfere with glucose oxidation and thus insulin secretion via an effect on glucose sensing.
Adipose
Two early microarray studies of adipose tissue from Leptinob/ob mice showed a coordinated decrease in the expression of genes encoding lipogenic enzymes as well as the master regulator of lipogenesis, the transcription factor Srebf1c (104, 137). In addition, both studies detected an increase in the expression of genes relevant to inflammatory and acute-phase processes and genes involved in adipocyte differentiation. These studies provided clues that obesity represents an inflammatory condition and that adipocytes in obese animals have an alteration in their differentiation state. The decrease in lipogenic gene expression has also been observed in adipose tissue of obese human subjects (34, 35, 131). It has been suggested that a limited lipogenic and/or adipogenic capacity in adipocytes might lead to spillover of excess lipids to other tissues, where lipotoxicity can contribute to the pathogenesis of diabetes (29, 103, 155).
The lipid spillover concept was tested by overexpression of the leptin receptor in adipose tissue of db/db (leptin receptor–deficient) mice under the control of the adipocyte fatty acid–binding protein (aP2) promoter (155). Although the mice were just as hyperphagic as db/db mice, they were much leaner. The remarkable result was that they were still insulin resistant, and this was correlated with steatosis in nonadipose tissues: heart, liver, and islets. Unger and coworkers (156) have suggested that for adipose tissue to play its role as a high-capacity lipid storage depot, leptin must, in an autocrine fashion, suppress functions normally associated with brown adipose tissue; i.e., reduce uncoupled lipid oxidation and thermogenesis and promote lipogenesis (Figure 2).
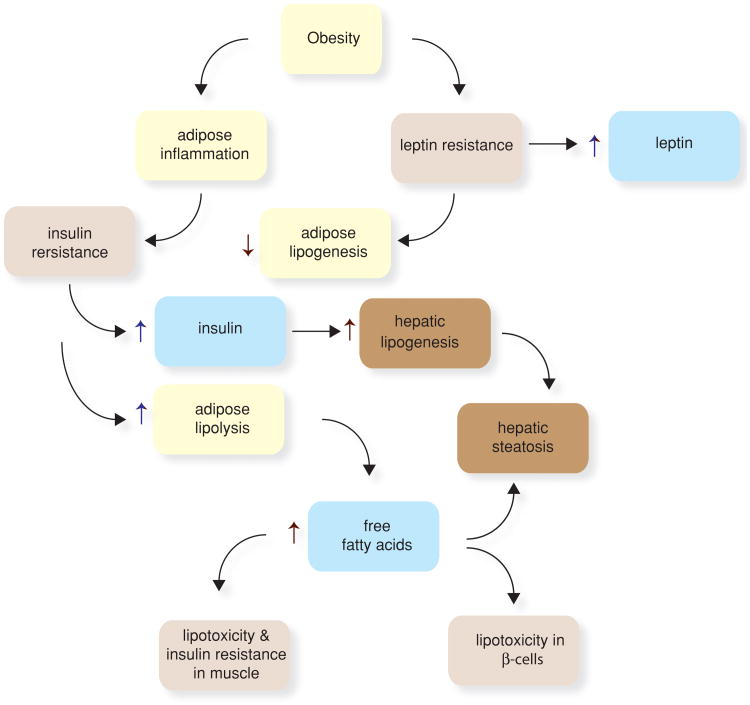
Figure 2.
Obesity causes metabolic and endocrine changes in adipose tissue. Leptin acts in an autocrine fashion on adipocytes to promote lipogenesis and reduce uncoupled oxidative phosphorylation. Obesity promotes leptin resistance, thus reducing lipogenesis in adipose tissue and promoting it in other tissues, leading to accumulation of triglycerides in the liver (hepatic steatosis). In muscle, increased fatty acid flux leads to insulin resistance. In β-cells, fatty acids can promote lipotoxicity and impair β-cell function and survival. Adipocytes in obese individuals produced adipokines that promote insulin resistance (e.g., RBP4 and resistin). Obesity promotes an inflammatory response in adipose tissue wherein macrophages and lymphocytes are recruited to adipose tissue and promote inflammation and insulin resistance. The insulin resistance in adipocytes increases the rate of triglyceride lipolysis, increasing the flux of free fatty acids to other tissues.
It is now well established that obesity is associated with the infiltration of proinflammatory (M1) macrophages in adipose tissue (82, 83, 127, 162, 166). Many of the genes whose expression are upregulated in response to obesity and are associated with inflammation are expressed in macrophages rather than adipocytes. This underscores an important caveat in the interpretation of microarray data from tissue samples. Differences in mRNA abundance between experimental groups may reflect changes in the cell composition of the tissue rather than changes in the expression of the genes in one particular cell type. Indeed, up to 40% of the cells in adipose tissue from obese animals are macrophages and lymphocytes. In addition to the altered expression profile derived from macrophage infiltration, the macrophages are able to elicit changes in the expression of adipocyte genes, as demonstrated in coculture experiments (84).
Adipocytes secrete hormones, termed adipokines, that modulate insulin signaling in an autocrine fashion in adipose tissue or in a endocrine fashion toward other tissues, principally muscle and liver (4). Leptin, adiponectin, and vaspin improve insulin sensitivity, whereas resistin, tumor necrosis factor-α, interleukin-6, plasminogen activator inhibitor-1, fasting-induced adipose factor (aka angiopoietin-like 4), and retinol binding protein-4 (RBP4) blunt insulin signaling.
The discovery of RBP4 and its role in insulin resistance illustrates a judicious application of microarray technology (168). Although adipose tissue does not contribute to insulin-stimulated glucose clearance nearly as much as muscle, overexpression or deletion of adipose tissue Glut4 results in altered insulin signaling and action in muscle and liver (2, 133). This does not occur in isolated muscle tissue, implying that a circulating factor originating in adipose tissue might be responsible for the changes in insulin responsiveness in muscle and liver. Based on this premise, Yang et al. (168) performed microarray analyses of adipose tissue from mice overexpressing Glut4 or with a deletion of the Glut4 gene specifically in adipocytes. They filtered their data, looking for genes that were modulated in opposite directions under the two conditions and encoded secreted proteins. The filter yielded just five mRNAs, including Rbp4. Follow-up studies showed that the Rbp4 protein, when introduced into the circulation of mice, is sufficient to blunt insulin signaling in muscle and liver. RBP4-overexpressing mice are insulin resistant, and RBP4 knockout mice have enhanced insulin sensitivity (168). These findings have been extended to human subjects, where serum RBP4 protein levels correlate with various measures of insulin resistance and metabolic syndrome even in large epidemi-ological studies (3, 5, 9, 10, 26, 45, 48, 49, 54, 119, 172). Furthermore, genetic studies show that a single nucleotide polymorphism (SNP) in the RBP4 promoter is associated with increased serum RBP4 levels and enhanced risk of type 2 diabetes (101, 152).
In a follow-up of the original study of the adipocyte Glut4 knockout mice, gene set enrichment analysis was applied to identify any pathway that would be enriched among transcripts that are differentially regulated when Glut4 is selectively overexpressed or knocked out in adipose tissue. The results showed that pathways regulating metabolism of other major fuels are enriched, suggesting that the differences in glucose uptake in adipose tissue lead to alterations in genetic programs in fat that regulate systemic fuel homeostasis (B.B. Kahn, personal communication).
The distribution of the increased adiposity associated with obesity is highly variable across individuals and populations. Excess adiposity can accumulate predominantly in visceral fat or in subcutaneous depots distributed around the body. Deposition in visceral fat is associated with type2diabetes, whereas subcutaneous fat deposition appears to be much more benign (66). Microarray analysis of mouse subcutaneous versus visceral adipose tissue identified 197 genes that are differentially expressed between the two depots (46). Twelve of these genes are involved in adipocyte development. These differences persisted after the adipocytes were removed from the animals and incubated in culture for six days, indicating that they are not dependent upon extrinsic factors in the intact animal. Tbxl5, Shox2, En1, Sfrp2, and Hoxc9 were more highly expressed in subcutaneous adipose tissue, whereas Nr2f1, Gpc4, Thbd, Hoxa5, and HoxC8 were more highly expressed in visceral fat. Many of the results from the mouse study were confirmed in a follow-up study in 53 lean human subjects, although in some instances the direction of the differences was not the same as seen in the mouse. The role of these genes in development led Gesta et al. (46) to suggest that adipocytes residing in different fat depots might be derived from distinct adipocyte precursor cells.
Liver
Microarray analysis helped to reliably delineate the genes regulated by two transcription factors that are master regulators of lipid metabolism gene expression. Sterol regulatory element–binding protein (SREBP) is expressed as three isoforms, SREBP-1a, SREBP-1c, and SREBP2. The latter two are the predominant isoforms in intact liver. They are translated as a protein that resides in the ER, but when ER membrane cholesterol drops below a critical threshold, the SREBP proteins are transported to the Golgi, where proteolytic cleavage releases a cytoplasmic domain, which translocates to the nucleus, where it regulates gene expression. The SREBP cleavage-activating protein (SCAP) helps to sense cholesterol and escort SREBPs to the Golgi (17).
The microarray study examined three sets of mouse livers; those from SCAP-/- mice and those from transgenic mice overexpress-ing either SREBP-1a or SREBP-2 (61). The study confirmed that SREBP-1a primarily targets genes involved in de novo fatty acid synthesis, fatty acid desaturation, and the generation of the necessary reducing equivalents to support de novo lipogenesis. SREBP-2 primarily targets enzymes that take acetate all the way through the biosynthetic pathway leading to cholesterol, as well as the LDL receptor. There were 33 genes that were upregulated by both SREBP isoforms and downregulated in the SCAP–/– mice. All but three of these genes encode enzymes in the sterol or fatty acid biosynthetic pathway or proteins related to sterol transport or regulation of sterol metabolism (LDL receptor, Pcsk9, Insig-1).
The Leptinob/ob mouse is the most widely used animal model for the study of morbid obesity. These mice have a global upregulation of expression of lipogenic enzymes and their master regulator, Srebf-1c, in the liver (39, 70, 72, 81). The upregulation of hepatic Srebf-1c also occurs in lipodystrophic mice, which, like Leptinob/ob mice, are leptin deficient (135). Like leptin deficiency, leptin resistance also induces the expression of Srebf-1c (147). These consequences of leptin deficiency and leptin resistance are opposite to what occurs in the adipose tissue, suggesting from the transcript profile that the lipogenic burden appears to be shifted from adipose tissue to the liver (103). The upregulation of lipogenic enzymes associated with leptin deficiency in the Leptinob/ob mouse, lipodystrophic mice (135), and in leptin-resistant rodent models (11, 70) is accompanied by a failure of the increased insulin levels to suppress hepatic gluconeogenesis. Thus, there is insulin insensitivity of the pathway by which insulin normally suppresses the expression of gluconeogenic genes, together with a normal lipogenic response of lipogenic genes, chiefly through the induction of Srebf-1c.
Studies to evaluate the reversal of the lipid and diabetic phenotypes of leptin deficiency showed that intracerebroventricular (ICV) administration of leptin is much more effective at regulating hepatic gene expression than is peripheral leptin administration (6). At lower doses, ICV leptin reversed hyperglycemia and hyperinsulinemia in lipodystrophic mice without ameliorating the hepatic lipid abnormalities, suggesting that the hepatic lipid abnormalities are not causal for diabetes.
In an effort to define the downstream effectors of leptin that mediate the lipogenic effects of leptin deficiency, Cohen et al. (28) identified genes that were upregulatedby leptin deficiency but not by high-fat-diet-induced obesity and are specifically repressed by leptin. Stearoyl-CoA desaturase-1 (Scd1) emerged as the gene that best fulfilled these criteria. The inhibitory effect of leptin on Scd1 expression occurs in animals with a targeted deletion of the hepatic insulin receptor, indicating that it does not depend on insulin signaling (13). These results predicted that deletion of the Scd1 gene might rescue some of the phenotypes associated with leptin deficiency. This prediction was tested in a naturally occurring animal model of Scd1 deficiency, the asebia mouse (28, 174), and in a mouse with a targeted disruption of the Scd1 gene (108). When the Leptinob/ob mutation was combined with either model of Scd1 deficiency, the mice had a reversal of their hepatic steatosis, reduced body weight, and a striking increase in O2 consumption (28, 108). Interestingly, despite these reversals in phenotypes normally associated with increased susceptibility to diabetes, the Leptinob/ob Scd-/mice actually have an increased susceptibility to diabetes (41). However, loss of Scd1 improves measures of glucose tolerance and insulin sensitivity in animals that are leptin resistant rather than leptin deficient (97). On the basis of these studies, several pharmaceutical companies have chosen Scd1 as a target for the treatment of obesity and diabetes (31).
A sharp reduction in the ability to synthesize monounsaturated fatty acids de novo appears to make animals more dependent on a critical supply of polyunsaturated fatty acids. Flowers et al. (42) showed that Scd1-/- mice on a very-low-fat diet developed cholestasis, hypercholesterolemia, and severe liver disease. Microarray analysis revealed that the liver had an activation of the unfolded protein response and inflammatory pathways (43). These studies identify potential markers that should be monitored in the development of drugs targeted to Scd.
The foregoing studies all employed mice with a loss of Scd1 in all tissues. Several of the studies strongly implied that the tissue sites responsible for the Scd1-mediated effects on hepatic steatosis, metabolic rate, body weight, and insulin sensitivity would be the liver and/or adipose tissue. Yet, tissue-specific knockout of Scd1 in the liver (96) or adipose tissue (J.M. Ntambi, personal communication) did not reproduce the phenotype of the whole-body Scd1 knockout mouse. Rather, a knockout of the gene in the skin reproduces most of the whole-body knockout phenotype (123). This result points to one of the trickiest aspects of microarray and gene expression studies: inferring causality from correlative data. We address this issue later in this article.
Glucose has a direct effect on the expression of several genes involved in converting carbohydrate into fatty acids (149). Early studies focused on pyruvate kinase and identified a site in its promoter that is responsive to glucose (91, 151). The glucose effect is not dependent on insulin. Rather, it is mediated by a pentose shunt metabolite of glucose, xylulose-5-phosphate (69, 106). This metabolite activates a phosphatase that dephosphorylates and thus activates the transcription factor carbohydrate-responsive element-binding protein (ChREBP), which has two isoforms, known as Mondo1 and Mondo 2. Both ChREBP isoforms form a heterodimer with another protein, Max-like factor X (Mlx) (140). Using a dominant negative form of Mlx, Ma et al. (86) identified genes whose expression was suppressed by the inactivation of ChREBP. They found that about half of those genes were induced by glucose. Many of the genes overlap with those induced by Srebp-1c—Acc1, Fasn, G6pdh, and Me—but also include Pyruvate kinase, Malate dehydrogenase, Transketolase, Fruc-tokinase, Glycerol phosphate dehydrogenase, and Microsomal triglyceride transfer protein, genes not responsive to SREBP-1c. It is interesting that fibroblast growth factor-21, a recently identified hepatic hormone that affects whole-body glucose homeostasis, is also induced by glucose and repressed by dominant-negative Mlx.
In contrast to muscle, liver expression of oxphos genes seems to be responsive to insulin sensitivity. Patti's group (114) showed that in morbidly obese individuals (BMI >50), the expression of several mitochondrial oxphos genes was downregulated; these were genes that are normally induced by thyroid hormone in muscle.
A recent study by Cheng et al. (24) supports a causal relationship between insulin resistance and mitochondrial function in the liver. In mice lacking both Irs1 and Irs2, there was a reduction in oxygen consumption in the presence versus the absence of adenosine diphosphate (respiratory control ratio). These physiological outcomes were associated with increased expression of genes regulated by the transcription factor Foxo1, a protein normally negatively regulated by insulin-induced phosphorylation. One of the Foxo1 target genes is Hmox1, which encodes a protein that decreases the concentration of mitochondrial heme, a key component of electron transport complexes II and IV.
β-Cells
The high content of ribonucleases in adult pancreatic acinar cells poses an extra challenge in the study of gene expression in pancreatic tissue. It makes it almost impossible to carry out in situ hybridization studies. Special care has to be exercised in obtaining islets for microarray experiments (92, 163).
During the early stages of the onset of type 2 diabetes, β-cells secrete a higher-than-normal level of insulin, but this is still not sufficient to compensate for insulin resistance and titrate blood glucose. Over time, β-cell function and/or β-cell mass diminishes, leading to more severe hyperglycemia; a feed-forward cycle of disease pathogenesis (116). Proposed mechanisms to explain the link between hyperglycemia and β-cell dysfunction fall into two categories: (a) consequences of glucose metabolism and (b) glucose effects on gene expression. The deleterious effects of excess glucose metabolism have been proposed to involve the glucosamine pathway, formation of sorbitol, and formation of reactive oxygen species (122, 146). It is interesting that reactive oxygen species have also been implicated in insulin resistance (63). In addition to glucose, fatty acids may play an important role in β-cell dysfunction and death, a process termed lipotoxicity (79, 118).
The earliest human islet microarray experiment identified genes that were upregulated in response to 16.7 versus 1.67 mM glucose. The most responsive gene was Thioredoxin interacting protein (Txnip; aka Vitamin D3 upregulated gene 1) (132), a gene discussed above in the context of muscle insulin signaling. Txnip was originally identified as a gene whose abundance is increased by 1,25-dihydroxy vitamin D3 in HL-60 leukemia cells (22), and consequently Txnip was found to act as a proapoptotic tumor suppressor gene (56). Txnip is ubiquitously expressed and plays a role in controlling the redox state in a variety of cell systems (68, 107).
Consistent with the initial findings in human islets, incubation of primary mouse islets or INS-1 rat insulinoma cells with 25 mM glucose for 24 hours led to an induction of Txnip as well as of cleaved caspase 3 protein, resulting in an increase in apoptosis as judged by terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end labeling staining (94). In fact, Txnip overexpression was sufficient to render INS-1 cells more susceptible to apoptosis (94), and microarray analysis revealed gene expression changes consistent with increased apoptosis in the Txnip-overexpressing β-cells (95). Isolated primary islets from the HcB-19 mouse, a naturally occurring model of Txnip deficiency (14, 65, 134), were completely protected from glucose toxicity–induced β-cell apoptosis (21). Interestingly, the β-cell protective effects of the antidiabetic drug Exenatide seem to be mediated in part by a reduction of Txnip expression (19).
The HcB-19 mice have also been found to have mild hypoglycemia (20, 65, 134), and their β-cell mass is increased twofold, as assessed by morphometry and measurement of whole-pancreas insulin content (20). In addition, their Txnip deficiency also protected them from β-cell death induced by streptozotocin, a model of type 1 diabetes (20). Crossing the BTBR Leptinob/ob mouse as a model of obesity-induced diabetes with the Txnip-deficient HcB-19 mouse (double-mutant congenic BTBR lepob/obtxniphcb/hcb) further revealed that lack of Txnip was also sufficient to protect against this severe model of type 2 diabetes, again primarily by reducing β-cell apoptosis (>98%) and preserving β-cell mass (20). Finally, a β-cell-specific Txnip knockout mouse generated by the Cre-LoxP system was also completely protected against diabetes (20).
Glucose-induced Txnip expression occurs at the transcriptional level, but surprisingly does not necessarily require glucose metabolism (93). Promoter analysis of the human Txnip promoter revealed a unique carbohydrate response element consisting of two nonpalindromic E-boxes (94). More recent work, including chromatin immunoprecipitation studies, demonstrated that glucose enhances binding of ChREBP to the Txnip promoter in INS-1 cells as well as in human islets (93). ChREBP, forming a heterodimer with Mlx, in turn recruits the coactivator p300, which also acts as a histone acetyltransferase and specifically leads to increased acetylation of histone H4 and recruitment of RNA polymerase II (93). In β-cells, ChREBP is necessary and sufficient for glucose-induced Txnip expression (93), whereas in skeletal muscle and renal epithelial cells, the ChREBP paralog MondoA is responsible for this function (142).
The pathogenesis of type 2 diabetes follows a feed-forward pattern whereby hyperglycemia exerts deleterious effects on β-cells and exacerbates the syndrome. A microarray study of human islets from nondiabetic versus diabetic human subjects identified the genes most altered in response to diabetes (52). A key observation was an 82% decline in the expression of ARNT, a transcription factor that plays a role in embryonic development. In addition to ARNT, one of the MODY genes, HNF4α was also sharply reduced. Genes participating in the insulin-signaling pathway, the insulin receptor, IRS2, AKT2, showed reduced expression, while a phosphatase that blunts insulin signaling, SHIP2, was upregulated, consistent with a picture of insulin resistance in the β-cells. Knockout of the Arnt genein β-cellsinmice resulted in animals with glucose intolerance, with greater severity in females than in males. The glucose intolerance was due to reduced glucose-stimulated insulin secretion. This was associated with reductions in enzymes responsible for glucose oxidation, suggesting that glucose sensing was impaired. Hnf4α expression was also reduced in the knockout islets, as were the insulin receptor and Akt2, drawing a clear parallel with the observations in islets from human diabetic subjects. Together, the effects of hyperglycemia on TXNIP and ARNT expression offer a model to explain increased β-cell apoptosis and decreased insulin secretion as a consequence of diabetes (Figure 3).
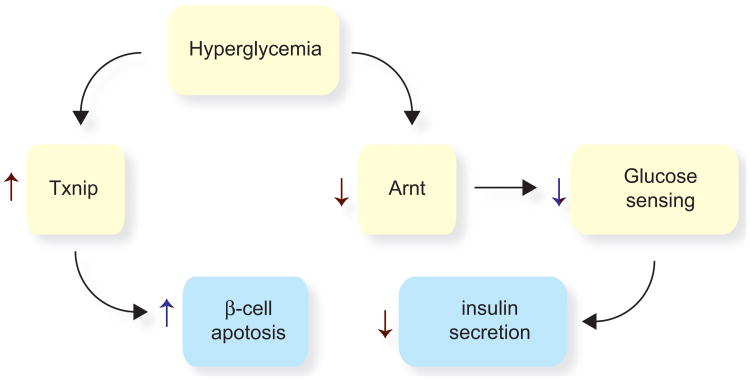
Figure 3.
Diabetes has detrimental effects on β-cells. In the early stages of type 2 diabetes, β-cells produce insulin at an elevated level in an attempt to compensate for insulin resistance. However, hyperglycemia compromises two key mechanisms by which β-cells mount a compensatory response to insulin resistance. First, glucose induces the expression of Txnip, a proapoptotic gene. Second, hyperglycemia downregulates the expression of the transcription factor Arnt. This leads to reduced glucose oxidation and reduced glucose sensing, resulting in reduced insulin secretion.
In rodent animal models (15) and perhaps also in humans, pregnancy evokes an expansion of β-cell mass through an induction of β-cell proliferation, primarily through the action of placental lactogen and prolactin (136). Rieck et al. (120) carried out microarray analyses of islet gene expression in pregnant mice at the time when β-cell proliferation is at its peak, at 14.5 days of gestation. They found that cyclins A2, B1, B2, D3, E1, F, and Cdk4 were all upregulated. In addition, genes involved in serotonin and tryptophan biosynthesis were induced. Of special interest was the gene Birc5 (aka Survivin), an antiapoptotic gene. This gene is upregulated in the pregnant mice, but also in two other models of β-cell proliferation: obesity induced by leptin deficiency in B6 mice (72) and recovery from apoptosis in a transgenic model in which caspase 8 is induced (159).
Mouse strains differ widely in their susceptibility to obesity-induced diabetes (27). One factor that affects susceptibility to diabetes is the proliferative response of β-cells to obesity. One model of obesity-induced diabetes is the BTBR strain into which the Leptinob mutation was been introgressed to produce a congenic strain, BTBR.ob. This strain develops severe diabetes by six weeks of age (72, 141). Keller et al. carried out a microarray study in islets and five other tissues and explored three variables; age (four weeks and ten weeks of age), leptin-deficiency induced obesity, and mouse strain (B6 versus BTBR). Using a method to identify transcripts whose expression is highly correlated (47), 105 gene expression “modules” were identified in the six tissues, where there was a distinct pattern of differential expression as a function of one or more of the three variables. One particular module was enriched in cells encoding genes involved in cell replication (Figure 4). In the B6 strain, the expression of these genes increased with obesity but decreased with age. In contrast, in the BTBR strain, the response to obesity was lost; the genes were only responsive to age. To establish that the behavior of this module was indeed predictive of cell proliferation, the mice were given 2H20 two weeks before sacrifice, and the enrichment of 2H in DNA was determined as a measure of cell proliferation as described by Neese et al. (105). The direct measure of cell proliferation agreed with the prediction from the behavior of the cell cycle gene-expression module (72).
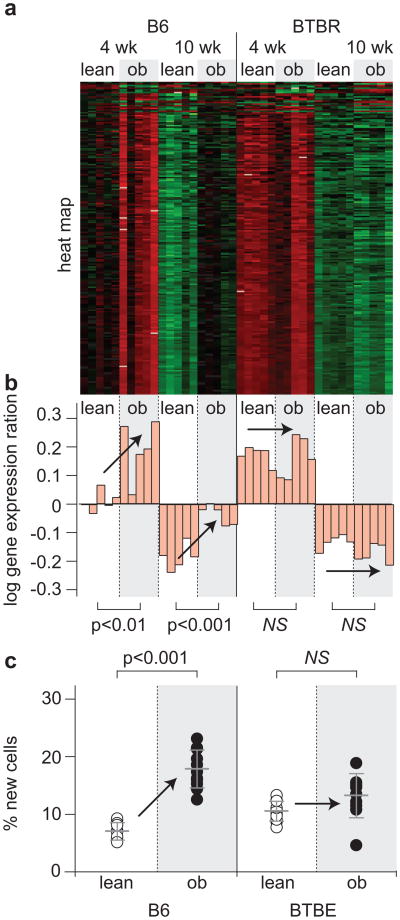
Figure 4.
Coexpression modules enriched with cell cycle regulation accurately predict diabetes and obesity. (a) Expression heat maps and (b) the PC1 on log10 scale of the cell cycle regulatory modules in islets (217 transcripts) and adipose (96 transcripts) are shown. For the heat maps, red shows increased expression, green shows decreased expression, and black is neutral. Bar plots in (b) show the PC1 for individual mice and correspond to an decreased expression for negative values and increased expression for positive values. (c) The percentage of new cells, derived from an in vivo measure of 2H incorporation into newly synthesized DNA, is shown for islets and adipose tissue. Where significant obesity-dependent differences were observed, P-values are shown. Arrows are used to show influence of obesity. NS, not significant. Figure and legend reprinted from (71).
A therapeutic objective in both type 1 and type 2 diabetes is to try to boost β-cell mass with agents that prolong β-cell survival or β-cell proliferation. An active area of research and controversy is whether or not adult β-cells proliferate in vivo or if new β-cells arise from differentiation of progenitors or transdifferentiation of other cell types (32, 167). Microarray analysis has been helpful to define some of the genes that participate in pathways leading to β-cell proliferation. Transcription factors involved in the development of the endocrine pancreas have been tested for their ability to promote β-cell proliferation. One of these transcription factors, Nkx6.1, when overexpressed in isolated islets, promotes β-cell proliferation (128). Microarray analysis identified a pattern of gene expression whereby the cell cycle genes cyclins B1, A2, and E1 are induced by this gene. A follow-up study showed that overexpression of cyclin E1 in islets is sufficient to stimulate β-cell proliferation (128).
It is difficult to obtain reliable data at the extremes of gene expression from microarrays. This is because the probes can become saturated when mRNAs are abundant, and at the low end, there is a great amount of variability in the data. An alternative albeit expensive approach is massive parallel signature sequencing (MPSS). This approach involves sequencing the transcripts and counting the number of times each transcript is sequenced. The method is extremely sensitive and has a very broad dynamic range. Using this approach, Kutlu et al. (80) identified the most enriched genes from human islets and estimated that 13% of all the mRNA in the samples was composed of insulin mRNA. Based on an estimate of 50–55% of the islet sample containing insulin-producing cells, this suggests that approximately 30% of β-cell mRNA is composed of insulin mRNA. The massive parallel signature sequencing data are available online (http://t1dbase.org).
Correlation Versus Causation
Microarray analysis delivers large datasets from which one can identify genes or sets of genes whose expression varies or covaries across a given set of conditions. For example, as described above, genes controlling lipogenesis, Srebf-1c, and its targets are upregulated in the livers of obese animals. The expression of these genes is coordinately upregulated, and this can be easily discerned in a microarray experiment. But the microarray experiment by itself does not allow one to infer the line of causality beginning with Srebf-1c and leading to genes encoding lipogenic enzymes. The microarray data simply provide correlation information for which there are multiple potential interpretations. Below, we describe the use of genetic information together with microarray data to overcome this limitation and enable the development of causal models. The method involves mapping the loci that control gene expression and then using these loci as anchor points to develop causal models.
Genetical Genomics: Expression Quantitative Trait Loci
When employed in the context of an intercross in model organisms or association mapping in an outbred population, genomic loci can be mapped that regulate the abundance of specific mRNA molecules. When the genotype at a particular locus (usually an SNP) correlates with the level of mRNA abundance, the locus is termed an expression quantitative trait locus (eQTL). The coverage provided by a genome-wide microarray, representing all known genes, means that thousands of eQTLs can be identified in these studies, which have been termed genetical genomics. The development of statistical tools required to analyze these voluminous data sets has been the subject of intense research by several groups (16, 75, 88, 175). Furthermore, integrating eQTL analyses with the more traditional disease-related QTL (cQTL) studies (e.g., loci governing body weight) has led to the development of causal gene-gene regulatory networks as part of a systems biology approach to better understand complex disease states (33, 47, 126).
There are two classes of eQTLs: cis and trans. Cis-eQTLs occur when the genomic position of the gene associated for a particular mRNA is coincident with its eQTL (Figure 5). Conceptually, a cis-eQTL implies genetic variation at or near the transcription start site, whereby the variation results in a change in the expression of the gene (153). In contrast to cis-mapping eQTLs, trans-eQTLs show linkage to genomic regions that are distant to their associated gene. A simple example would be variation in a gene encoding a transcription factor, leading to variation in the abundance of the mRNAs of its target genes. The mRNA abundance traits of the targets would map to the location of the gene encoding the transcription factor.
Figure 5.
Cis- and trans-acting expression quantitative trait loci (eQTLs). (a) When mRNA abundance maps as a phenotype near the physical location of the gene encoding the mRNA, it is termed a cis-eQTL or a proximal eQTL. (b) When the mRNA abundance maps to a genomic location that is distinctly separate from the physical location of the gene encoding the mRNA (e.g., a different chromosome, as shown), it is described as a trans-eQTL or distal eQTL. (c) When multiple mRNA abundance phenotypes map to the same genomic location in trans, then one can hypothesize that there might be one or more regulators encoded at that location that coordinately regulate this set of mRNAs. This result is one step in building network models of gene regulation.
A key premise that permits directionally causal networks to be constructed from QTL-driven expression data sets is that variation in DNA is the origin, or starting point, of the causative relationship.
This premise derives from the idea that DNA variation can cause changes in gene expression, but not vice-versa. This one-way relationship between DNA variation and changes in gene expression greatly simplifies the possible combinations that relate genomic loci, eQTLs, and cQTLs. In simplified form, three of the many possible models that interrelate these network components are (124):
Locus → eQTL → clinical trait (causal)
Locus → clinical trait → eQTL (reactive)
eQTL ← locus → clinical trait (independent)
Note that in all models, the locus is the anchor point of the causal model in a unidirectional fashion. Furthermore, if we assume that the locus is directly affecting the eQTL, we can infer it to be cis-mapping in nature. Importantly, the cQTL illustrated in causal model (a) will have an overlapping QTL with the eQTL directly upstream. Assuming the cis-eQTL depicted in this causal model is controlling the cQTL that is downstream (model a above), we would expect the expression and clinical traits associated with each QTL to be correlated (positively or negatively). Novel disease-causing genes have been identified in a variety of systems by using this integrative approach.
Identification and Validation of Causative Genes for Fat Mass in Mice
Schadt and colleagues (124) applied genetical genomics to identify and validate novel genes that regulate diet-induced changes in fat mass. An F2 intercross was generated between C57BL/6J(B6) and DBA/2J (DBA) mice, which were maintained on chow diet up to 12 months of age, followed by a high-fat diet for an additional 4 months, after which mice were sacrificed, and genome-wide expression profiling was done in liver.
Whereas the expression of thousands of genes was found to significantly vary across the panel of F2 mice, only ∼90 were identified as being key regulators of changes in fat mass in response to the dietary challenge. Such a reduction in number of candidate regulators resulted from applying several key filters: (a) determining which eQTLs favored a causal versus reactive or independent model; (b) requiring causal eQTLs to be correlated with changes in fat mass; and (c) selecting those causal and correlated eQTLs to have overlapping QTLs for fat mass. Finally, the candidate regulators were ranked according to what proportion of fat mass variance could be explained by the variation in transcript level.
The generation of genetic knockout or transgenic overexpression animal models is considered the gold standard for testing the predictive power of this integrative approach for the identification of disease-causing genes. Three genes (Zfp90, C3ar1, and Tgfbr2) were tested with genetic manipulation and found to significantly affect fat mass accumulation in response to a high-fat diet (124). Furthermore, in a series of follow-up studies, additional genes were validated as playing a key role in regulating fat mass, including Lpl and LactB (23, 169) and Gas7, Gpx3, and Me1 (169). Importantly, the mouse models utilized for these validation studies were whole-body deletions or ubiquitous promoters for the knockout and transgenic constructs, respectively.
Although the expression profiling studies used to identify the causal nature of these genes were conducted in liver, it remains unclear if their role as regulators of fat mass actually occurs in the liver. It is possible that the QTL-based causal associations established between cis-eQTLs and overlapping clinical phenotype QTLs (e.g., fat mass) translate across tissues and, thus, profiling liver can be informative for these relationships in other tissues. Identification of the tissue or cell type responsible for the effect on fat mass would require tissue-specific knockout or transgenic mouse models. It is also possible that while whole liver was used for gene expression profiling, nonhepatocyte liver cells (e.g., Kupffer cells) actually contributed to the eQTLs. Remarkably, five of the top ten candidate genes (Tgfbr2, C3ar1, Zfp90, Lpl, and LactB) originally profiled in liver (124) were recently identified in a network highly enriched for macrophage gene signatures and were showntoclosely associate with obesity and other metabolic syndrome phenotypes (23).
Several recent studies have clearly demonstrated the important role that adipose tissue macrophage infiltration plays in obesity-induced insulin resistance (30, 38, 62, 127). Very recently, it was shown that C3ar1, a leading candidate for regulating fat mass in mouse (124, 169), was found to be highly expressed in adipocytes as well as macrophages (90). Furthermore, not only were C3ar1-/- mice resistant to diet-induced obesity, but they also were protected from insulin resistance, hepatic steatosis, adipose macrophage infiltration, and increased circulating levels of proinflammatory cytokines, all of which are associated with obesity. Taken together, these results suggest that the hepatic QTL study that originally identified C3ar1 as an obesity-causing gene (124) may have actually been assessing the role of C3ar1 in macrophages that had infiltrated into adipose and potentially other tissues (90), supporting the notion that QTL-driven gene discovery in one organ or tissue type can be diagnostic about a gene's role in disease causation in multiple tissues.
Genetic Architecture of Hepatic Gene Expression in Humans and Its Use for Identification of Disease-Causing Genes
To what extent does QTL-driven discovery for disease-causing genes translate from inbred mouse strains to human biology? Schadt and colleagues (125) addressed this question by performing genome-wide expression profiling in liver samples from 427 human subjects that were genotyped at >780,000 SNPs. Approximately 3,000 cis-eQTLs were identified, classified as having linkage to SNPs within 1 Mb of the structural gene, including many genes already identified as being associated with human diseases (BRCA1, CFH) and drug responsiveness (VKORC1) or previously implicated in genome-wide association studies to the development of type I diabetes (e.g., CTLA4, HLA-DRB1, IL2RA, LONRF2, and CHST10). By leveraging the human expression and genotype data sets with eQTL-driven expression networks constructed from three mouse F2 intercrosses, some genes thought to be associated with human disease were not supported, while novel disease-causing genes were identified.
A large genome-wide association study of ∼17,000 white European subjects and seven common diseases identified a locus on chromosome 12q13, containing ERBB3 as a candidate gene that is associated with type 1 diabetes (1, 148). An additional, more directed association study in a Japanese population has confirmed the ERBB3 association, suggesting the locus on 12q13 contributes to diabetes susceptibility across ethnic subgroups (7). However, there are key differences between studies that associate disease to a particular genomic region and those that relate changes in the expression of candidate genes to disease susceptibility.
Association studies do not offer a measure of gene expression changes, nor do they identify the tissue(s) involved in the disease relationship. For example, the expression of ERBB3 was not associated with DNA variation at the 12q13 locus, whereas the expression of a flanking gene, RPS26, was found to be highly correlated to the SNP at this locus, giving rise to an eQTL in the human liver cohort (125). However, once again, we must ask whether an eQTL in liver is informative for how a gene may function in another tissue. Importantly, only ∼30% of the cis-eQTLs identified in the human liver samples were preserved across human blood and adipose tissue samples (37, 125), showing that cis-eQTLs are sometimes, but not always, tissue specific.
More than 1,000 mice were generated as part of three separate F2 intercrosses between the B6, C3H, and CAST mouse strains (125, 171). Gene expression was profiled in a number of tissues of each cross, including liver, adipose, muscle, and brain, and combined with genotype data to reconstruct causal gene-gene expression networks. Interestingly, Rps26, but not Erbb3, was identified in a subnetwork that was highly enriched with other genes (H2-Eb1, H2-Ab1, H2-T22,H2-M3, H2-DMa,H2-Aa, H2-DMb2, and H2-T23) annotated as type 1 diabetes– related genes (125). It should be noted, however, that a recent statistical analysis has challenged the conclusion that genetic variation in the expression of RPS26 is responsible for the association of type 1 diabetes at the 12q13 locus (115).
Applying the lessons learned from the type 1 diabetes locus on 12q13 and the differences observed between Erbb3 and Rps26 as likely candidate genes, additional genes (Psrc1, Sort1, and Celsr2) were identified as candidates for a locus on chromosome 1p13.3 shown to associate with coronary artery disease and LDL-cholesterol levels (125). Although uncertainties remain regarding the reproducibility of expression networks from one tissue to another as well as the degree to which cis-eQTLs are preserved across multiple tissues, the opportunity for gene discovery is enhanced by the approach of integrating the eQTL data from the human liver cohort with multiple human genome-wide association studies and with the genetical-genomic analysis of gene expression and clinical phenotypes in segregating mouse populations.
Variation in Genomic Copy Number Contributes to eQTLs
Copy number variants (CNVs) are genomic segments that have been duplicated or deleted between individual mouse strains (50). They can range in size between a few kilobases to several megabases (with an average ∼0.3 Mb) and may contain known genes (50). CNVs have been shown to affect gene expression in human lymphoblastoid cells collected from unrelated individuals from four geographically distinct populations (Utah, China, Japan, and Nigeria) (144). It is interesting to note that CFH, a gene associated with age-related macular degeneration in humans (36, 55, 77), is contained within a CNV that is well conserved across species (50) and was identified as a cis-eQTL in a study of human liver samples (125).
To determine the extent to which CNVs can influence eQTL analysis in a segregating mouse cross, the effect on gene expression of 19 CNVs, identified between the B6 and C3H mouse strains (50), was determined in an F2 intercross between the same two strains (109). A large number of eQTLs occurred within orvery near CNV regions in adipose, brain, liver, and muscle tissues. Remarkably, ∼84% of the genes known to reside within the 19 CNVs were differentially expressed as a function of genotype of the nearest marker to each CNV.
In addition to cis-eQTLs, trans-eQTLs were found to overlap several CNVs, suggesting that regulatory elements contained within the CNV may have been mediating their linkage. Both eQTLs and metabolic QTLs for body weight, plasma cholesterol, triglyceride, glucose, and insulin overlapped CNVs on chromosomes 1, 4, and 17. Furthermore, cis-eQTLs within these CNVs tested causal for the colocalized metabolic QTLs and await validation with conditional knockout and/or transgenic animal models, as discussed above. Taken together, the results indicate that CNVs represent another class of genomic polymorphism that can influence gene expression.
Concluding Remarks
The ability to interrogate the entire transcriptome in an unbiased fashion, through microarray technology, has had a profound impact in virtually every field of biology. This review highlights examples where clear expression patterns emerged that pointed to a regulatory pathway or where changes in expression pointed to an important role of a novel gene in diabetes pathogenesis. Technology enabled these dramatic developments. Now, new technology in mass spectrometry is enabling similarly dramatic developments in proteomics and metabolomics. The ability to accurately quantitate protein abundance and to assess post-translational modifications (e.g., phosphorylation, acetylation, methylation, ubiquitination, glycosylation, prenylation, and fatty acylation) promises to reveal important new regulatory mechanisms and their relation to cell function and disease. Metabolomics studies will likely identify novel metabolites. Targeted metabolomics studies are beginning to show clear patterns of metabolites associated with metabolic diseases and cancer (8). These studies might also lead to reliable biomarkers for designing and monitoring disease treatments. Finally, the integration of these various high-volume data sources can yield testable hypotheses connecting metabolites, protein, and/or transcript levels (40).
Guidelines for Planning a Microarray Experiment.
If you're planning to do a microarray study, be clear about why you're doing it, what you expect to obtain, what will you do with the data, and ask yourself if this is really the best next experiment to do.
Learn as much as you can about the physiology and biochemistry of your system before doing a microarray study. This is critical in order to decide what tissue will be profiled and under what conditions. For obesity phenotypes, the decision is not easy because many tissues can influence body weight (brain, intestine, liver, endocrine organs, and of course, adipose tissue). For example, a food intake measurement would help to understand if an obesity phenotype might be due to a change in satiety control; this would motivate the sampling of gene expression in the brain, perhaps the hypothalamus.
The ability to discern distinct DE patterns is related to the effect size of the observed phenomena and the biological experimental variance. Manipulation of the experimental conditions to maximize the effect size can greatly increase the power to detect meaningful DE patterns. Pilot experiments to test various experimental conditions can help to determine the optimal design for the microarray experiment.
Try to eliminate or minimize extraneous sources of experimental variance. If this is not possible, systematically record factors that might introduce variation; they can be used as covariates in the analysis of the data. An important factor that affects the expression of perhaps as many as 40% of all genes is sex (157). Many physiological phenomena show sexual dimorphism. For example, in most rodent models of diabetes, males are more severely affected than females. A variable that is often overlooked is time of day. A large number of genes show circadian rhythms (154). Thus, whenever possible, all animals in an experiment should be sampled at the same time of day (unless, of course, time of day is an experimental variable).
The greatest difficulty in the statistical analysis of microarray experiments is that they suffer from a large multiple testing problem (143). This issue and the many sources of experimental variation demand that there be a sufficient number of biological replicates or a well-designed pooling scheme (74, 76). Ideally, one should do a pilot study and measure the expression of several genes, covering a wide range in mRNA abundance, in order to be able to estimate the extent of variance within the experimental groups. This enables a power calculation to determine the number of replicates needed to detect a particular level of DE at a particular frequency. In our experience, fewer than five replicates is underpowered.
The dynamic range of microarrays (60 to 100-fold) is far less than that of real-time PCR or RNA-seq. The data are less reliable for transcripts of very low or very high abundance. Results obtained by microarray analysis should always be verified by real-time PCR, and whenever possible and relevant, protein abundance should also be evaluated. The overall correlation between mRNA and protein abundance is only about 0.4 (53). Discrepancies between mRNA and protein abundance can provide useful clues to investigate post-transcriptional control mechanisms.
Tissues have a mixture of cell types. It is important to be mindful of the mix of vascular tissue, blood-borne cells, tissue macrophages, and “minor” cell types when interpreting tissue-derived gene expression data. Differential expression changes might reflect changes in cell number (e.g., increased vascularization) rather than true changes in gene expression. It might be desirable to perform microdissection or cell type enrichment prior to extracting mRNA. This is especially important in the endocrine pancreas, where islet cell mass comprises only approximately 1% of total organ mass.
Of special relevance to those studying nutritional factors, it is critical to take special care in choosing the right control diet. Often overlooked is the fact that high-fat diets have more synthetic ingredients than ordinary chow and thus might not be matched for fiber content, phytoestrogens, and simple carbohydrates (160).
Summary Points.
Coexpressed genes are often coregulated.
Genes encoding mitochondrial respiratory function in muscle are dysregulated in diabetes, likely a consequence of diabetes.
Mutations that disrupt mitochondrial function in muscle are associated with improved insulin sensitivity.
Obesity is associated with an inflammatory response in adipose tissue. Adipocytes, macrophages, and lymphocytes produce hormones that affect whole-body insulin signaling. Inadequate lipid storage in adipose tissue leads to spillover and a pathological accumulation of lipids in other tissues.
Genes involved in hepatic lipogenesis are coordinately regulated through transcription factors SREBP and ChREBP. These pathways are dysregulated in obesity and diabetes.
β-cell gene expressionis responsivetohyperglycemia. Two genes that are dysregulated in diabetes are Txnip and Arnt, leading to β-cell apoptosis and impaired glucose-stimulated insulin secretion, respectively.
When combined with genetics, gene expression data can be used to construct causal network models linking gene expression with disease phenotypes.
Acknowledgments
We are grateful to Dale Abel, Klaus Kaestner, Vamsi Mootha, Debbie Muoio, Mary Elizabeth Patti, and Anath Shalev for their comments and insights. Work of the Attie laboratory is funded by NIDDK grants DK66369 and DK58037, an American Diabetes Association Mentor-Based Fellowship, and a grant from the JDRF.
Abbreviations
- DE
differential expression
- GSEA
gene set enrichment analysis
- eQTL
expression quantitative trait locus
Footnotes
Disclosure Statement: The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Literature Cited
- 1.Genome-wide association study of 14000 cases of seven common diseases and 3000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abel ED, Peroni O, Kim JK, Kim YB, Boss O, et al. Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature. 2001;409:729–33. doi: 10.1038/35055575. [DOI] [PubMed] [Google Scholar]
- 3.Aeberli I, Biebinger R, Lehmann R, L'Allemand D, Spinas GA, Zimmermann MB. Serum retinolbinding protein 4 concentration and its ratio to serum retinol are associated with obesity and metabolic syndrome components in children. J Clin Endocrinol Metab. 2007;92:4359–65. doi: 10.1210/jc.2007-0468. [DOI] [PubMed] [Google Scholar]
- 4.Ahima RS, Lazar MA. Adipokines and the peripheral and neural control of energy balance. Mol Endocrinol. 2008;22:1023–31. doi: 10.1210/me.2007-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aigner E, Bachofner N, Klein K, De Geyter C, Hohla F, et al. Retinol-binding protein 4 in polycystic ovary syndrome—association with steroid hormones and response to pioglitazone treatment. J Clin Endocrinol Metab. 2009;94:1229–35. doi: 10.1210/jc.2008-2156. [DOI] [PubMed] [Google Scholar]
- 6.Asilmaz E, Cohen P, Miyazaki M, Dobrzyn P, Ueki K, et al. Site and mechanism of leptin action in a rodent form of congenital lipodystrophy. J Clin Invest. 2004;113:414–24. doi: 10.1172/JCI19511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Awata T, Kawasaki E, Tanaka S, Ikegami H, Maruyama T, et al. Association of type 1 diabetes with two loci on 12q13 and 16p13 and the influence coexisting thyroid autoimmunity in Japanese. J Clin Endocrinol Metab. 2009;94:231–35. doi: 10.1210/jc.2008-0718. [DOI] [PubMed] [Google Scholar]
- 8.Bain JR, Stevens RD, Wenner BR, Ilkayeva O, Muoio DM, Newgard CB. Metabolomics applied to diabetes research: moving from information to knowledge. Diabetes. 2009;58:2429–43. doi: 10.2337/db09-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balagopal P, Graham TE, Kahn BB, Altomare A, Funanage V, George D. Reduction of elevated serum retinol binding protein in obese children by lifestyle intervention: association with subclinical inflammation. J Clin Endocrinol Metab. 2007;92:1971–74. doi: 10.1210/jc.2006-2712. [DOI] [PubMed] [Google Scholar]
- 10.Barber TM, Hazell M, Christodoulides C, Golding SJ, Alvey C, et al. Serum levels of retinol-binding protein 4 and adiponectin in women with polycystic ovary syndrome: associations with visceral fat but no evidence for fat mass–independent effects on pathogenesis in this condition. J Clin Endocrinol Metab. 2008;93:2859–65. doi: 10.1210/jc.2007-2759. [DOI] [PubMed] [Google Scholar]
- 11.Becker W, Kluge R, Kantner T, Linnartz K, Korn M, et al. Differential hepatic gene expression in a polygenic mouse model with insulin resistance and hyperglycemia: evidence for a combined transcriptional dysregulation of gluconeogenesis and fatty acid synthesis. J Mol Endocrinol. 2004;32:195–208. doi: 10.1677/jme.0.0320195. [DOI] [PubMed] [Google Scholar]
- 12.Befroy DE, Petersen KF, Dufour S, Mason GF, de Graaf RA, et al. Impaired mitochondrial substrate oxidation in muscle of insulin-resistant offspring of type 2 diabetic patients. Diabetes. 2007;56:1376–81. doi: 10.2337/db06-0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biddinger SB, Miyazaki M, Boucher J, Ntambi JM, Kahn CR. Leptin suppresses stearoyl-CoA desaturase 1 by mechanisms independent of insulin and sterol regulatory element-binding protein-1c. Diabetes. 2006;55:2032–41. doi: 10.2337/db05-0742. [DOI] [PubMed] [Google Scholar]
- 14.Bodnar JS, Chatterjee A, Castellani LW, Ross DA, Ohmen J, et al. Positional cloning of the combined hyperlipidemia gene Hyplip1. Nat Genet. 2002;30:110–16. doi: 10.1038/ng811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brelje TC, Parsons JA, Sorenson RL. Regulation of islet beta-cell proliferation by prolactin in rat islets. Diabetes. 1994;43:263–73. doi: 10.2337/diab.43.2.263. [DOI] [PubMed] [Google Scholar]
- 16.Broman KW. Mapping expression in randomized rodent genomes. Nat Genet. 2005;37:209–10. doi: 10.1038/ng0305-209. [DOI] [PubMed] [Google Scholar]
- 17.Brown MS, Goldstein JL. Cholesterol feedback: from Schoenheimer's bottle to Scap's MELADL. J Lipid Res. 2009;50(Suppl):S15–27. doi: 10.1194/jlr.R800054-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cha-Molstad H, Saxena G, Chen J, Shalev A. Glucose-stimulated expression of Txnip is mediated by carbohydrate response element-binding protein, p300, and histone H4 acetylation in pancreatic beta cells. J Biol Chem. 2009;284:16898–905. doi: 10.1074/jbc.M109.010504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J, Couto FM, Minn AH, Shalev A. Exenatide inhibits beta-cell apoptosis by decreasing thioredoxin-interacting protein. Biochem Biophys Res Commun. 2006;346:1067–74. doi: 10.1016/j.bbrc.2006.06.027. [DOI] [PubMed] [Google Scholar]
- 20.Chen J, Hui ST, Couto FM, Mungrue IN, Davis DB, et al. Thioredoxin-interacting protein deficiency induces Akt/Bcl-xL signaling and pancreatic beta cell mass and protects against diabetes. FASEB J. 2008;22:3581–94. doi: 10.1096/fj.08-111690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J, Saxena G, Mungrue IN, Lusis AJ, Shalev A. Thioredoxin-interacting protein: a critical link between glucose toxicity and beta cell apoptosis. Diabetes. 2008;57:938–44. doi: 10.2337/db07-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen KS, DeLuca HF. Isolation and characterization of a novel cDNA from HL-60 cells treated with 1,25-dihydroxyvitamin D-3. Biochim Biophys Acta. 1994;1219:26–32. doi: 10.1016/0167-4781(94)90242-9. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y, Zhu J, Lum PY, Yang X, Pinto S, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–35. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng Z, Guo S, Copps K, Dong X, Kollipara R, et al. Foxo1 integrates insulin signaling with mitochondrial function in the liver. Nat Med. 2009;15:1307–11. doi: 10.1038/nm.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi CS, Befroy DE, Codella R, Kim S, Reznick RM, et al. Paradoxical effects of increased expression of PGC-1alpha on muscle mitochondrial function and insulin-stimulated muscle glucose metabolism. Proc Natl Acad Sci USA. 2008;105:19926–31. doi: 10.1073/pnas.0810339105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi SH, Kwak SH, Youn BS, Lim S, Park YJ, et al. High plasma retinol binding protein-4 and low plasma adiponectin concentrations are associated with severity of glucose intolerance in women with previous gestational diabetes mellitus. J Clin Endocrinol Metab. 2008;93:3142–48. doi: 10.1210/jc.2007-1755. [DOI] [PubMed] [Google Scholar]
- 27.Clee SM, Attie AD. The genetic landscape of type 2 diabetes in mice. Endocrine Rev. 2006;28:48–83. doi: 10.1210/er.2006-0035. [DOI] [PubMed] [Google Scholar]
- 28.Cohen P, Miyazaki M, Socci ND, Hagge-Greenberg A, Liedtke W, et al. Role for stearoyl-CoA desaturase-1 in leptin-mediated weight loss. Science. 2002;297:240–43. doi: 10.1126/science.1071527. [DOI] [PubMed] [Google Scholar]
- 29.Danforth E., Jr Failure of adipocyte differentiation causes type II diabetes mellitus? Nat Genet. 2000;26:13. doi: 10.1038/79111. [DOI] [PubMed] [Google Scholar]
- 30.de Luca C, Olefsky JM. Inflammation and insulin resistance. FEBS Lett. 2008;582:97–105. doi: 10.1016/j.febslet.2007.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dobrzyn A, Ntambi JM. Stearoyl-CoA desaturase as a new drug target for obesity treatment. Obes Rev. 2005;6:169–74. doi: 10.1111/j.1467-789X.2005.00177.x. [DOI] [PubMed] [Google Scholar]
- 32.Dor Y, Brown J, Martinez OI, Melton DA. Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature. 2004;429:41–46. doi: 10.1038/nature02520. [DOI] [PubMed] [Google Scholar]
- 33.Drake TA, Schadt EE, Lusis AJ. Integrating genetic and gene expression data: application to cardiovascular and metabolic traits in mice. Mamm Genome. 2006;17:466–79. doi: 10.1007/s00335-005-0175-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dubois SG, Heilbronn LK, Smith SR, Albu JB, Kelley DE, Ravussin E. Decreased expression of adipogenic genes in obese subjects with type 2 diabetes. Obesity (Silver Spring) 2006;14:1543–52. doi: 10.1038/oby.2006.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ducluzeau PH, Perretti N, Laville M, Andreelli F, Vega N, et al. Regulation by insulin of gene expression in human skeletal muscle and adipose tissue. Evidence for specific defects in type 2 diabetes. Diabetes. 2001;50:1134–42. doi: 10.2337/diabetes.50.5.1134. [DOI] [PubMed] [Google Scholar]
- 36.Edwards AO, Ritter R, 3rd, Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–24. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 37.Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–28. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 38.Ferrante AW., Jr Obesity-induced inflammation: a metabolic dialogue in the language of inflammation. J Intern Med. 2007;262:408–14. doi: 10.1111/j.1365-2796.2007.01852.x. [DOI] [PubMed] [Google Scholar]
- 39.Ferrante AW, Jr, Thearle M, Liao T, Leibel RL. Effects of leptin deficiency and short-term repletion on hepatic gene expression in genetically obese mice. Diabetes. 2001;50:2268–78. doi: 10.2337/diabetes.50.10.2268. [DOI] [PubMed] [Google Scholar]
- 40.Ferrara CT, Wang P, Neto EC, Stevens RD, Bain JR, et al. Genetic networks of liver metabolism revealed by integration of metabolic and transcriptional profiling. PLoS Genet. 2008;4:e1000034. doi: 10.1371/journal.pgen.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flowers JB, Rabaglia ME, Schueler KL, Flowers MT, Lan H, et al. Loss of stearoyl-CoA desaturase-1 improves insulin sensitivity in lean mice but worsens diabetes in leptin-deficient obese mice. Diabetes. 2007;56:1228–39. doi: 10.2337/db06-1142. [DOI] [PubMed] [Google Scholar]
- 42.Flowers MT, Groen AK, Oler AT, Keller MP, Choi Y, et al. Cholestasis and hypercholesterolemia in SCD1-deficient mice fed a low-fat, high-carbohydrate diet. J Lipid Res. 2006;47:2668–80. doi: 10.1194/jlr.M600203-JLR200. [DOI] [PubMed] [Google Scholar]
- 43.Flowers MT, Keller MP, Choi Y, Lan H, Kendziorski C, et al. Liver gene expression analysis reveals endoplasmic reticulum stress and metabolic dysfunction in SCD1-deficient mice fed a very low-fat diet. Physiol Genomics. 2008;33:361–72. doi: 10.1152/physiolgenomics.00139.2007. [DOI] [PubMed] [Google Scholar]
- 44.Garcia-Roves P, Huss JM, Han DH, Hancock CR, Iglesias-Gutierrez E, et al. Raising plasma fatty acid concentration induces increased biogenesis of mitochondria in skeletal muscle. Proc Natl Acad Sci USA. 2007;104:10709–13. doi: 10.1073/pnas.0704024104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gavi S, Stuart LM, Kelly P, Melendez MM, Mynarcik DC, et al. Retinol-binding protein 4 is associated with insulin resistance and body fat distribution in nonobese subjects without type 2 diabetes. J Clin Endocrinol Metab. 2007;92:1886–90. doi: 10.1210/jc.2006-1815. [DOI] [PubMed] [Google Scholar]
- 46.Gesta S, Bluher M, Yamamoto Y, Norris AW, Berndt J, et al. Evidence for a role of developmental genes in the origin of obesity and body fat distribution. Proc Natl Acad Sci USA. 2006;103:6676–81. doi: 10.1073/pnas.0601752103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghazalpour A, Doss S, Zhang B, Wang S, Plaisier C, et al. Integrating genetic and network analysis to characterize genes related to mouse weight. PLoS Genet. 2006;2:e130. doi: 10.1371/journal.pgen.0020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Graham TE, Yang Q, Blüher M, Hammarstedt A, Ciaraldi TP, et al. Retinol-binding protein 4 and insulin resistance in lean, obese, and diabetic subjects. N Engl J Med. 2006;354:2552–63. doi: 10.1056/NEJMoa054862. [DOI] [PubMed] [Google Scholar]
- 49.Deleted in proof.
- 50.Graubert TA, Cahan P, Edwin D, Selzer RR, Richmond TA, et al. A high-resolution map of segmental DNA copy number variation in the mouse genome. PLoS Genet. 2007;3:e3. doi: 10.1371/journal.pgen.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guarente L. Mitochondria—a nexus for aging, calorie restriction, and sirtuins? Cell. 2008;132:171–76. doi: 10.1016/j.cell.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gunton JE, Kulkarni RN, Yim S, Okada T, Hawthorne WJ, et al. Loss of ARNT/HIF1beta mediates altered gene expression and pancreatic-islet dysfunction in human type 2 diabetes. Cell. 2005;122:337–49. doi: 10.1016/j.cell.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 53.Gygi SP, Rist B, Griffin TJ, Eng J, Aebersold R. Proteome analysis of low-abundance proteins using multidimensional chromatography and isotope-coded affinity tags. J Proteome Res. 2002;1:47–54. doi: 10.1021/pr015509n. [DOI] [PubMed] [Google Scholar]
- 54.Haider DG, Schindler K, Prager G, Bohdjalian A, Luger A, et al. Serum retinol-binding protein 4 is reduced after weight loss in morbidly obese subjects. J Clin Endocrinol Metab. 2007;92:1168–71. doi: 10.1210/jc.2006-1839. [DOI] [PubMed] [Google Scholar]
- 55.Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–21. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 56.Han SH, Jeon JH, Ju HR, Jung U, Kim KY, et al. VDUP1 upregulated by TGF-beta1 and 1,25-dihydroxyvitaminD3 inhibits tumor cell growth by blocking cell-cycle progression. Oncogene. 2003;22:4035–46. doi: 10.1038/sj.onc.1206610. [DOI] [PubMed] [Google Scholar]
- 57.Hancock CR, Han DH, Chen M, Terada S, Yasuda T, et al. High-fat diets cause insulin resistance despite an increase in muscle mitochondria. Proc Natl Acad Sci USA. 2008;105:7815–20. doi: 10.1073/pnas.0802057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Handschin C, Choi CS, Chin S, Kim S, Kawamori D, et al. Abnormal glucose homeostasis in skeletal muscle-specific PGC-1alpha knockout mice reveals skeletal muscle-pancreatic beta cell crosstalk. J Clin Invest. 2007;117:3463–74. doi: 10.1172/JCI31785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He J, Watkins S, Kelley DE. Skeletal muscle lipid content and oxidative enzyme activity in relation to muscle fiber type in type 2 diabetes and obesity. Diabetes. 2001;50:817–23. doi: 10.2337/diabetes.50.4.817. [DOI] [PubMed] [Google Scholar]
- 60.Holloszy JO. Skeletal muscle “mitochondrial deficiency” does not mediate insulin resistance. Am J Clin Nutr. 2009;89:463–66S. doi: 10.3945/ajcn.2008.26717C. [DOI] [PubMed] [Google Scholar]
- 61.Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, et al. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci USA. 2003;100:12027–32. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–67. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 63.Houstis N, Rosen ED, Lander ES. Reactive oxygen species have a causal role in multiple forms of insulin resistance. Nature. 2006;440:944–48. doi: 10.1038/nature04634. [DOI] [PubMed] [Google Scholar]
- 64.Hui ST, Andres AM, Miller AK, Spann NJ, Potter DW, et al. Txnip balances metabolic and growth signaling via PTEN disulfide reduction. Proc Natl Acad Sci USA. 2008;105:3921–26. doi: 10.1073/pnas.0800293105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hui TY, Sheth SS, Diffley JM, Potter DW, Lusis AJ, et al. Mice lacking thioredoxin-interacting protein provide evidence linking cellular redox state to appropriate response to nutritional signals. J Biol Chem. 2004;279:24387–93. doi: 10.1074/jbc.M401280200. [DOI] [PubMed] [Google Scholar]
- 66.Jensen MD. Health consequences of fat distribution. Horm Res. 1997;48(Suppl. 5):88–92. doi: 10.1159/000191335. [DOI] [PubMed] [Google Scholar]
- 67.Joza N, Oudit GY, Brown D, Benit P, Kassiri Z, et al. Muscle-specific loss of apoptosis-inducing factor leads to mitochondrial dysfunction, skeletal muscle atrophy, and dilated cardiomyopathy. Mol Cell Biol. 2005;25:10261–72. doi: 10.1128/MCB.25.23.10261-10272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Junn E, Han SH, Im JY, Yang Y, Cho EW, et al. Vitamin D3 up-regulated protein 1 mediates oxidative stress via suppressing the thioredoxin function. J Immunol. 2000;164:6287–95. doi: 10.4049/jimmunol.164.12.6287. [DOI] [PubMed] [Google Scholar]
- 69.Kabashima T, Kawaguchi T, Wadzinski BE, Uyeda K. Xylulose 5-phosphate mediates glucose-induced lipogenesis by xylulose 5-phosphate-activated protein phosphatase in rat liver. Proc Natl Acad Sci USA. 2003;100:5107–12. doi: 10.1073/pnas.0730817100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kakuma T, Lee Y, Higa M, Wang Z, Pan W, et al. Leptin, troglitazone, and the expression of sterol regulatory element binding proteins in liver and pancreatic islets. Proc Natl Acad Sci USA. 2000;97:8536–41. doi: 10.1073/pnas.97.15.8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Deleted in proof.
- 72.Keller MP, Choi Y, Wang P, Davis DB, Rabaglia ME, et al. A gene expression network model of type 2 diabetes links cell cycle regulation in islets with diabetes susceptibility. Genome Res. 2008;18:706–16. doi: 10.1101/gr.074914.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kelley DE, He J, Menshikova EV, Ritov VB. Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes. 2002;51:2944–50. doi: 10.2337/diabetes.51.10.2944. [DOI] [PubMed] [Google Scholar]
- 74.Kendziorski C, Irizarry RA, Chen KS, Haag JD, Gould MN. On the utility of pooling biological samples in microarray experiments. Proc Natl Acad Sci USA. 2005;102:4252–57. doi: 10.1073/pnas.0500607102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kendziorski C, Wang P. A review of statistical methods for expression quantitative trait loci mapping. Mamm Genome. 2006;17:509–17. doi: 10.1007/s00335-005-0189-6. [DOI] [PubMed] [Google Scholar]
- 76.Kendziorski CM, Zhang Y, Lan H, Attie AD. The efficiency of pooling mRNA in microarray experiments. Biostatistics. 2003;4:465–77. doi: 10.1093/biostatistics/4.3.465. [DOI] [PubMed] [Google Scholar]
- 77.Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–89. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kleiner S, Nguyen-Tran V, Bare O, Huang X, Spiegelman BM, Wu Z. PPARdelta agonism activates fatty acid oxidation via PGC-1alpha but does not increase mitochondrial gene expression and function. J Biol Chem. 2009;284:18624–33. doi: 10.1074/jbc.M109.008797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kusminski CM, Shetty S, Orci L, Unger RH, Scherer PE. Diabetes and apoptosis: lipotoxicity. Apoptosis. 2009;14:1484–95. doi: 10.1007/s10495-009-0352-8. [DOI] [PubMed] [Google Scholar]
- 80.Kutlu B, Burdick D, Baxter D, Rasschaert J, Flamez D, et al. Detailed transcriptome atlas of the pancreatic beta cell. BMC Med Genomics. 2009;2:3. doi: 10.1186/1755-8794-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lan H, Rabaglia ME, Stoehr JP, Nadler ST, Schueler KL, et al. Gene expression profiles of nondiabetic and diabetic obese mice suggest a role of hepatic lipogenic capacity in diabetes susceptibility. Diabetes. 2003;52:688–700. doi: 10.2337/diabetes.52.3.688. [DOI] [PubMed] [Google Scholar]
- 82.Lumeng CN, Bodzin JL, Saltiel AR. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest. 2007;117:175–84. doi: 10.1172/JCI29881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lumeng CN, Deyoung SM, Bodzin JL, Saltiel AR. Increased inflammatory properties of adipose tissue macrophages recruited during diet-induced obesity. Diabetes. 2007;56:16–23. doi: 10.2337/db06-1076. [DOI] [PubMed] [Google Scholar]
- 84.Lumeng CN, Deyoung SM, Saltiel AR. Macrophages block insulin action in adipocytes by altering expression of signaling and glucose transport proteins. Am J Physiol Endocrinol Metab. 2007;292:E166–74. doi: 10.1152/ajpendo.00284.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Luquet S, Lopez-Soriano J, Holst D, Fredenrich A, Melki J, et al. Peroxisome proliferator-activated receptor delta controls muscle development and oxidative capability. FASEB J. 2003;17:2299–301. doi: 10.1096/fj.03-0269fje. [DOI] [PubMed] [Google Scholar]
- 86.Ma L, Robinson LN, Towle HC. ChREBP*Mlx is the principal mediator of glucose-induced gene expression in the liver. J Biol Chem. 2006;281:28721–30. doi: 10.1074/jbc.M601576200. [DOI] [PubMed] [Google Scholar]
- 87.Maassen JA, 't Hart LM, Van Essen E, Heine RJ, Nijpels G, et al. Mitochondrial diabetes: molecular mechanisms and clinical presentation. Diabetes. 2004;53(Suppl. 1):S103–9. doi: 10.2337/diabetes.53.2007.s103. [DOI] [PubMed] [Google Scholar]
- 88.Mackay TF, Stone EA, Ayroles JF. The genetics of quantitative traits: challenges and prospects. Nat Rev Genet. 2009;10:565–77. doi: 10.1038/nrg2612. [DOI] [PubMed] [Google Scholar]
- 89.Maechler P, Wollheim CB. Mitochondrial function in normal and diabetic beta-cells. Nature. 2001;414:807–12. doi: 10.1038/414807a. [DOI] [PubMed] [Google Scholar]
- 90.Mamane Y, Chung Chan C, Lavallee G, Morin N, Xu LJ, et al. The C3a anaphylatoxin receptor is a key mediator of insulin resistance and functions by modulating adipose tissue macrophage infiltration and activation. Diabetes. 2009;58:2006–17. doi: 10.2337/db09-0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Marie S, Diaz-Guerra MJ, Miquerol L, Kahn A, Iynedjian PB. The pyruvate kinase gene as a model for studies of glucose-dependent regulation of gene expression in the endocrine pancreatic beta-cell type. J Biol Chem. 1993;268:23881–90. [PubMed] [Google Scholar]
- 92.Marselli L, Sgroi DC, Bonner-Weir S, Weir GC. Laser capture microdissection of human pancreatic beta-cells and RNA preparation for gene expression profiling. Methods Mol Biol. 2009;560:87–98. doi: 10.1007/978-1-59745-448-3_8. [DOI] [PubMed] [Google Scholar]
- 93.Minn AH, Couto FM, Shalev A. Metabolism-independent sugar effects on gene transcription: the role of 3-O-methylglucose. Biochemistry. 2006;45:11047–51. doi: 10.1021/bi0603625. [DOI] [PubMed] [Google Scholar]
- 94.Minn AH, Hafele C, Shalev A. Thioredoxin-interacting protein is stimulated by glucose through a carbohydrate response element and induces beta-cell apoptosis. Endocrinology. 2005;146:2397–405. doi: 10.1210/en.2004-1378. [DOI] [PubMed] [Google Scholar]
- 95.Minn AH, Pise-Masison CA, Radonovich M, Brady JN, Wang P, et al. Gene expression profiling in INS-1 cells overexpressing thioredoxin-interacting protein. Biochem Biophys Res Commun. 2005;336:770–78. doi: 10.1016/j.bbrc.2005.08.161. [DOI] [PubMed] [Google Scholar]
- 96.Miyazaki M, Flowers MT, Sampath H, Chu K, Otzelberger C, et al. Hepatic stearoyl-CoA desaturase-1 deficiency protects mice from carbohydrate-induced adiposity and hepatic steatosis. Cell Metab. 2007;6:484–96. doi: 10.1016/j.cmet.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 97.Miyazaki M, Sampath H, Liu X, Flowers MT, Chu K, et al. Stearoyl-CoA desaturase-1 deficiency attenuates obesity and insulin resistance in leptin-resistant obese mice. Biochem Biophys Res Commun. 2009;380:818–22. doi: 10.1016/j.bbrc.2009.01.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–73. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 99.Morino K, Petersen KF, Dufour S, Befroy D, Frattini J, et al. Reduced mitochondrial density and increased IRS-1 serine phosphorylation in muscle of insulin-resistant offspring of type 2 diabetic parents. J Clin Invest. 2005;115:3587–93. doi: 10.1172/JCI25151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Morino K, Petersen KF, Shulman GI. Molecular mechanisms of insulin resistance in humans and their potential links with mitochondrial dysfunction. Diabetes. 2006;55(Suppl. 2):S9–15. doi: 10.2337/db06-S002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Munkhtulga L, Nakayama K, Utsumi N, Yanagisawa Y, Gotoh T, et al. Identification of a regulatory SNP in the retinol binding protein 4 gene associated with type 2 diabetes in Mongolia. Hum Genet. 2007;120:879–88. doi: 10.1007/s00439-006-0264-4. [DOI] [PubMed] [Google Scholar]
- 102.Muoio DM. TXNIP links redox circuitry to glucose control. Cell Metab. 2007;5:412–14. doi: 10.1016/j.cmet.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 103.Nadler ST, Attie AD. Please pass the chips: genomic insights into obesity and diabetes. J Nutr. 2001;131:2078–81. doi: 10.1093/jn/131.8.2078. [DOI] [PubMed] [Google Scholar]
- 104.Nadler ST, Stoehr JP, Schueler KL, Tanimoto G, Yandell BS, Attie AD. The expression of adipogenic genes is decreased in obesity and diabetes mellitus. Proc Natl Acad Sci USA. 2000;97:11371–76. doi: 10.1073/pnas.97.21.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Neese RA, Misell LM, Turner S, Chu A, Kim J, et al. Measurement in vivo of proliferation rates of slow turnover cells by 2H2O labeling of the deoxyribose moiety of DNA. Proc Natl Acad Sci USA. 2002;99:15345–50. doi: 10.1073/pnas.232551499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nishimura M, Uyeda K. Purification and characterization of a novel xylulose 5-phosphate-activated protein phosphatase catalyzing dephosphorylation of fructose-6-phosphate,2-kinase:fructose-2,6-bisphosphatase. J Biol Chem. 1995;270:26341–46. doi: 10.1074/jbc.270.44.26341. [DOI] [PubMed] [Google Scholar]
- 107.Nishiyama A, Masutani H, Nakamura H, Nishinaka Y, Yodoi J. Redox regulation by thioredoxin and thioredoxin-binding proteins. IUBMB Life. 2001;52:29–33. doi: 10.1080/15216540252774739. [DOI] [PubMed] [Google Scholar]
- 108.Ntambi JM, Miyazaki M, Stoehr JP, Lan H, Kendziorski CM, et al. Loss of stearoyl-CoA desaturase-1 function protects mice against adiposity. Proc Natl Acad Sci USA. 2002;99:11482–86. doi: 10.1073/pnas.132384699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Orozco LD, Cokus SJ, Ghazalpour A, Ingram-Drake L, Wang S, et al. Copy number variation influences gene expression and metabolic traits in mice. Hum Mol Genet. 2009;18:4118–29. doi: 10.1093/hmg/ddp360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Parikh H, Carlsson E, Chutkow WA, Johansson LE, Storgaard H, et al. TXNIP regulates peripheral glucose metabolism in humans. PLoS Med. 2007;4:e158. doi: 10.1371/journal.pmed.0040158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: potential role of PGC1 and NRF1. Proc Natl Acad Sci USA. 2003;100:8466–71. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Petersen KF, Befroy D, Dufour S, Dziura J, Ariyan C, et al. Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science. 2003;300:1140–42. doi: 10.1126/science.1082889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Petersen KF, Dufour S, Befroy D, Garcia R, Shulman GI. Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N Engl J Med. 2004;350:664–71. doi: 10.1056/NEJMoa031314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pihlajamaki J, Boes T, Kim EY, Dearie F, Kim BW, et al. Thyroid hormone-related regulation of gene expression in human fatty liver. J Clin Endocrinol Metab. 2009;94:3521–29. doi: 10.1210/jc.2009-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Plagnol V, Smyth DJ, Todd JA, Clayton DG. Statistical independence of the colocalized association signals for type 1 diabetes and RPS26 gene expression on chromosome 12q13. Biostatistics. 2009;10:327–34. doi: 10.1093/biostatistics/kxn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Poitout V, Robertson RP. Glucolipotoxicity: fuel excess and beta-cell dysfunction. Endocr Rev. 2008;29:351–66. doi: 10.1210/er.2007-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pospisilik JA, Knauf C, Joza N, Benit P, Orthofer M, et al. Targeted deletion of AIF decreases mitochondrial oxidative phosphorylation and protects from obesity and diabetes. Cell. 2007;131:476–91. doi: 10.1016/j.cell.2007.08.047. [DOI] [PubMed] [Google Scholar]
- 118.Prentki M, Nolan CJ. Islet beta cell failure in type 2 diabetes. J Clin Invest. 2006;116:1802–12. doi: 10.1172/JCI29103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Reinehr T, Stoffel-Wagner B, Roth CL. Retinol-binding protein 4 and its relation to insulin resistance in obese children before and after weight loss. J Clin Endocrinol Metab. 2008;93:2287–93. doi: 10.1210/jc.2007-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rieck S, White P, Schug J, Fox AJ, Smirnova O, et al. The transcriptional response of the islet to pregnancy in mice. Mol Endocrinol. 2009;23:1702–12. doi: 10.1210/me.2009-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ritov VB, Menshikova EV, Azuma K, Wood RJ, Toledo FG, et al. Deficiency of electron transport chain in human skeletal muscle mitochondria in type 2 diabetes mellitus and obesity. Am J Physiol Endocrinol Metab. 2009 doi: 10.1152/ajpendo.00317.2009. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Robertson RP. Chronic oxidative stress as a central mechanism for glucose toxicity in pancreatic islet beta cells in diabetes. J Biol Chem. 2004;279:42351–54. doi: 10.1074/jbc.R400019200. [DOI] [PubMed] [Google Scholar]
- 123.Sampath H, Flowers MT, Liu X, Paton CM, Sullivan R, et al. Skin-specific deletion of stearoyl-coa desaturase-1 alters skin lipid composition and protects mice from high-fat diet-induced obesity. J Biol Chem. 2009;284:19961–73. doi: 10.1074/jbc.M109.014225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37:710–17. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Schadt EE, Molony C, Chudin E, Hao K, Yang X, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schadt EE, Zhang B, Zhu J. Advances in systems biology are enhancing our understanding of disease and moving us closer to novel disease treatments. Genetica. 2009;136:259–69. doi: 10.1007/s10709-009-9359-x. [DOI] [PubMed] [Google Scholar]
- 127.Schenk S, Saberi M, Olefsky JM. Insulin sensitivity: modulation by nutrients and inflammation. J Clin Invest. 2008;118:2992–3002. doi: 10.1172/JCI34260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schisler JC, Fueger PT, Babu DA, Hohmeier HE, Tessem JS, et al. Stimulation of human and rat islet beta-cell proliferation with retention of function by the homeodomain transcription factor Nkx6.1. Mol Cell Biol. 2008;28:3465–76. doi: 10.1128/MCB.01791-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sekine N, Cirulli V, Regazzi R, Brown LJ, Gine E, et al. Low lactate dehydrogenase and high mitochondrial glycerol phosphate dehydrogenase in pancreatic beta-cells. Potential role in nutrient sensing. J Biol Chem. 1994;269:4895–902. [PubMed] [Google Scholar]
- 130.Sener A, Mercan D, Malaisse WJ. Enzymic activities in two populations of purified rat islet beta-cells. Int J Mol Med. 2001;8:285–89. [PubMed] [Google Scholar]
- 131.Sewter C, Berger D, Considine RV, Medina G, Rochford J, et al. Human obesity and type 2 diabetes are associated with alterations in SREBP1 isoform expression that are reproduced ex vivo by tumor necrosis factor-alpha. Diabetes. 2002;51:1035–41. doi: 10.2337/diabetes.51.4.1035. [DOI] [PubMed] [Google Scholar]
- 132.Shalev A, Pise-Masison CA, Radonovich M, Hoffmann SC, Hirshberg B, et al. Oligonucleotide microarray analysis of intact human pancreatic islets: identification of glucose-responsive genes and a highly regulated TGFbeta signaling pathway. Endocrinology. 2002;143:3695–98. doi: 10.1210/en.2002-220564. [DOI] [PubMed] [Google Scholar]
- 133.Shepherd PR, Gnudi L, Tozzo E, Yang H, Leach F, Kahn BB. Adipose cell hyperplasia and enhanced glucose disposal in transgenic mice overexpressing GLUT4 selectively in adipose tissue. J Biol Chem. 1993;268:22243–46. [PubMed] [Google Scholar]
- 134.Sheth SS, Castellani LW, Chari S, Wagg C, Thipphavong CK, et al. Thioredoxin-interacting protein deficiency disrupts the fasting-feeding metabolic transition. J Lipid Res. 2005;46:123–34. doi: 10.1194/jlr.M400341-JLR200. [DOI] [PubMed] [Google Scholar]
- 135.Shimomura I, Matsuda M, Hammer RE, Bashmakov Y, Brown MS, Goldstein JL. Decreased IRS-2 and increased SREBP-1c lead to mixed insulin resistance and sensitivity in livers of lipodystrophic and ob/ob mice. Mol Cell. 2000;6:77–86. [PubMed] [Google Scholar]
- 136.Sorenson RL, Brelje TC. Adaptation of islets of Langerhans to pregnancy: beta-cell growth, enhanced insulin secretion and the role of lactogenic hormones. Horm Metab Res. 1997;29:301–7. doi: 10.1055/s-2007-979040. [DOI] [PubMed] [Google Scholar]
- 137.Soukas A, Cohen P, Socci ND, Friedman JM. Leptin-specific patterns of gene expression in white adipose tissue. Genes Dev. 2000;14:963–80. [PMC free article] [PubMed] [Google Scholar]
- 138.Sparks LM, Xie H, Koza RA, Mynatt R, Hulver MW, et al. A high-fat diet coordinately downregu-lates genes required for mitochondrial oxidative phosphorylation in skeletal muscle. Diabetes. 2005;54:1926–33. doi: 10.2337/diabetes.54.7.1926. [DOI] [PubMed] [Google Scholar]
- 139.Sreekumar R, Halvatsiotis P, Schimke JC, Nair KS. Gene expression profile in skeletal muscle of type 2 diabetes and the effect of insulin treatment. Diabetes. 2002;51:1913–20. doi: 10.2337/diabetes.51.6.1913. [DOI] [PubMed] [Google Scholar]
- 140.Stoeckman AK, Ma L, Towle HC. Mlx is the functional heteromeric partner of the carbohydrate response element-binding protein in glucose regulation of lipogenic enzyme genes. J Biol Chem. 2004;279:15662–69. doi: 10.1074/jbc.M311301200. [DOI] [PubMed] [Google Scholar]
- 141.Stoehr JP, Nadler ST, Schueler KL, Rabaglia ME, Yandell BS, et al. Genetic obesity unmasks nonlinear interactions between murine type 2 diabetes susceptibility loci. Diabetes. 2000;49:1946–54. doi: 10.2337/diabetes.49.11.1946. [DOI] [PubMed] [Google Scholar]
- 142.Stoltzman CA, Peterson CW, Breen KT, Muoio DM, Billin AN, Ayer DE. Glucose sensing by MondoA:Mlx complexes: a role for hexokinases and direct regulation of thioredoxin-interacting protein expression. Proc Natl Acad Sci USA. 2008;105:6912–17. doi: 10.1073/pnas.0712199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–45. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–53. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tanaka Y, Tran PO, Harmon J, Robertson RP. A role for glutathione peroxidase in protecting pancreatic beta cells against oxidative stress in a model of glucose toxicity. Proc Natl Acad Sci USA. 2002;99:12363–68. doi: 10.1073/pnas.192445199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tobe K, Suzuki R, Aoyama M, Yamauchi T, Kamon J, et al. Increased expression of the sterol regulatory element-binding protein-1 gene in insulin receptor substrate-2(-/-) mouse liver. J Biol Chem. 2001;276:38337–40. doi: 10.1074/jbc.C100160200. [DOI] [PubMed] [Google Scholar]
- 148.Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–64. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Towle HC. Glucose as a regulator of eukaryotic gene transcription. Trends Endocrinol Metab. 2005;16:489–94. doi: 10.1016/j.tem.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 150.Turner N, Bruce CR, Beale SM, Hoehn KL, So T, et al. Excess lipid availability increases mitochondrial fatty acid oxidative capacity in muscle: evidence against a role for reduced fatty acid oxidation in lipid-induced insulin resistance in rodents. Diabetes. 2007;56:2085–92. doi: 10.2337/db07-0093. [DOI] [PubMed] [Google Scholar]
- 151.Uyeda K, Yamashita H, Kawaguchi T. Carbohydrate responsive element-binding protein (ChREBP): a key regulator of glucose metabolism and fat storage. Biochem Pharmacol. 2002;63:2075–80. doi: 10.1016/s0006-2952(02)01012-2. [DOI] [PubMed] [Google Scholar]
- 152.van Hoek M, Dehghan A, Zillikens MC, Hofman A, Witteman JC, Sijbrands EJ. An RBP4 promoter polymorphism increases risk of type 2 diabetes. Diabetologia. 2008;51:1423–28. doi: 10.1007/s00125-008-1042-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Veyrieras JB, Kudaravalli S, Kim SY, Dermitzakis ET, Gilad Y, et al. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4:e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Walker JR, Hogenesch JB. RNA profiling in circadian biology. Methods Enzymol. 2005;393:366–76. doi: 10.1016/S0076-6879(05)93016-4. [DOI] [PubMed] [Google Scholar]
- 155.Wang MY, Grayburn P, Chen S, Ravazzola M, Orci L, Unger RH. Adipogenic capacity and the susceptibility to type 2 diabetes and metabolic syndrome. Proc Natl Acad Sci USA. 2008;105:6139–44. doi: 10.1073/pnas.0801981105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wang MY, Orci L, Ravazzola M, Unger RH. Fat storage in adipocytes requires inactivation of leptin's paracrine activity: implications for treatment of human obesity. Proc Natl Acad Sci USA. 2005;102:18011–16. doi: 10.1073/pnas.0509001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wang S, Yehya N, Schadt EE, Wang H, Drake TA, Lusis AJ. Genetic and genomic analysis of a fat mass trait with complex inheritance reveals marked sex specificity. PLoS Genet. 2006;2:e15. doi: 10.1371/journal.pgen.0020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wang YX, Zhang CL, Yu RT, Cho HK, Nelson MC, et al. Regulation of muscle fiber type and running endurance by PPARdelta. PLoS Biol. 2004;2:e294. doi: 10.1371/journal.pbio.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang ZV, Mu J, Schraw TD, Gautron L, Elmquist JK, et al. PANIC-ATTAC: a mouse model for inducible and reversible beta-cell ablation. Diabetes. 2008;57:2137–48. doi: 10.2337/db07-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Warden CH, Fisler JS. Comparisons of diets used in animal models of high-fat feeding. Cell Metab. 2008;7:277. doi: 10.1016/j.cmet.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wasserman DH. Four grams of glucose. Am J Physiol Endocrinol Metab. 2009;296:E11–21. doi: 10.1152/ajpendo.90563.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Weisberg SPD, McCann M, Desai M, Rosenbaum R, Leibel RL, Ferrante AW., Jr Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;116:1796–808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.White P, Kaestner KH. Gene expression analysis in diabetes research. Methods Mol Biol. 2009;560:239–61. doi: 10.1007/978-1-59745-448-3_16. [DOI] [PubMed] [Google Scholar]
- 164.Wredenberg A, Freyer C, Sandstrom ME, Katz A, Wibom R, et al. Respiratory chain dysfunction in skeletal muscle does not cause insulin resistance. Biochem Biophys Res Commun. 2006;350:202–7. doi: 10.1016/j.bbrc.2006.09.029. [DOI] [PubMed] [Google Scholar]
- 165.Wright JJ, Kim J, Buchanan J, Boudina S, Sena S, et al. Mechanisms for increased myocardial fatty acid utilization following short-term high-fat feeding. Cardiovasc Res. 2009;82:351–60. doi: 10.1093/cvr/cvp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Xu H, Barnes GT, Yang Q, Tan G, Yang D, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest. 2003;112:1821–30. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Xu X, D'Hoker J, Stange G, Bonne S, De Leu N, et al. Beta cells can be generated from endogenous progenitors in injured adult mouse pancreas. Cell. 2008;132:197–207. doi: 10.1016/j.cell.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 168.Yang Q, Graham TE, Mody N, Preitner F, Peroni OD, et al. Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature. 2005;436:356–62. doi: 10.1038/nature03711. [DOI] [PubMed] [Google Scholar]
- 169.Yang X, Deignan JL, Qi H, Zhu J, Qian S, et al. Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nat Genet. 2009;41:415–23. doi: 10.1038/ng.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yang X, Pratley RE, Tokraks S, Bogardus C, Permana PA. Microarray profiling of skeletal muscle tissues from equally obese, nondiabetic insulin-sensitive and insulin-resistant Pima Indians. Diabetologia. 2002;45:1584–93. doi: 10.1007/s00125-002-0905-7. [DOI] [PubMed] [Google Scholar]
- 171.Yang X, Schadt EE, Wang S, Wang H, Arnold AP, et al. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16:995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yao-Borengasser A, Varma V, Bodles AM, Rasouli N, Phanavanh B, et al. Retinol binding protein 4 expression in humans: relationship to insulin resistance, inflammation, and response to pioglitazone. J Clin Endocrinol Metab. 2007;92:2590–97. doi: 10.1210/jc.2006-0816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Yechoor VK, Patti ME, Ueki K, Laustsen PG, Saccone R, et al. Distinct pathways of insulin-regulated versus diabetes-regulated gene expression: an in vivo analysis in MIRKO mice. Proc Natl Acad Sci USA. 2004;101:16525–30. doi: 10.1073/pnas.0407574101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zheng Y, Eilertsen KJ, Ge L, Zhang L, Sundberg JP, et al. Scd1 is expressed in sebaceous glands and is disrupted in the asebia mouse. Nat Genet. 1999;23:268–70. doi: 10.1038/15446. [DOI] [PubMed] [Google Scholar]
- 175.Zhu J, Zhang B, Smith EN, Drees B, Brem RB, et al. Integrating large-scale functional genomic data to dissect the complexity of yeast regulatory networks. Nat Genet. 2008;40:854–61. doi: 10.1038/ng.167. [DOI] [PMC free article] [PubMed] [Google Scholar]