Abstract
Microbiome-mediated suppression of carcinogenesis may open new avenues for identification of therapeutic targets and prevention strategies in oncology. Histidine decarboxylase (HDC) deficiency has been shown to promote inflammation-associated colorectal cancer by accumulation of CD11b+Gr-1+ immature myeloid cells, indicating a potential antitumorigenic effect of histamine. Here, we demonstrate that administration of hdc+Lactobacillus reuteri in the gut resulted in luminal hdc gene expression and histamine production in the intestines of Hdc−/− mice. This histamine-producing probiotic decreased the number and size of colon tumors and colonic uptake of [18F]-fluorodeoxyglucose by positron emission tomography in Hdc−/− mice. Administration of L. reuteri suppressed keratinocyte chemoattractant (KC), Il22, Il6, Tnf, and IL1α gene expression in the colonic mucosa and reduced the amounts of proinflammatory, cancer-associated cytokines, keratinocyte chemoattractant, IL-22, and IL-6, in plasma. Histamine-generating L. reuteri also decreased the relative numbers of splenic CD11b+Gr-1+ immature myeloid cells. Furthermore, an isogenic HDC-deficient L. reuteri mutant that was unable to generate histamine did not suppress carcinogenesis, indicating a significant role of the cometabolite, histamine, in suppression of chronic intestinal inflammation and colorectal tumorigenesis. These findings link luminal conversion of amino acids to biogenic amines by gut microbes and probiotic-mediated suppression of colorectal neoplasia.
Colorectal cancer (CRC) is the third most common cancer and the third leading cause of cancer-related mortality.1 Population-based cohort studies have shown that patients with inflammatory bowel disease have an increased lifetime risk of CRC compared with the general population.2, 3 This risk can be reduced by treatment of colitis with suppression of intestinal inflammation.4 These observations, in conjunction with studies showing that immune cells, cytokines, and other immunomodulatory agents play a role in CRC development,5 underline the association between CRC and colonic inflammation.
The role of the intestinal microbiome in colon cancer development has recently been investigated.6, 7, 8, 9 Specific gut microbes and their metabolites may contribute to the cause of CRC.10, 11, 12 Manipulation of the gut microbiome by probiotics could provide new therapeutic strategies for CRC prevention. Several probiotic strains including Bifidobacterium longum,13 Lactobacillus acidophilus NCFM,14 and Lactobacillus rhamnosus GG15 have shown beneficial effects in different murine models of colon cancer. However, the molecular mechanisms mediating suppression of colonic carcinogenesis by these microbes remain unknown.
Lactobacillus reuteri is a commensal intestinal Firmicute and probiotic that is widely prevalent in the gastrointestinal tracts of diverse avian and mammalian species.16 L. reuteri has been reported to suppress production of proinflammatory cytokines by intestinal epithelial cells17 and monocytes,18 in addition to reducing intestinal inflammation in different rodent models.17, 19, 20, 21, 22, 23 A pangenomic study showed that human-derived clade II L. reuteri strains contained a complete chromosomal hdc gene cluster (genes hdcA, hdcB, hdcP) and the genetic capacity to convert histidine to histamine.24 The ability to generate histamine by strains of this clade was correlated with suppression of human tumor necrosis factor (TNF) production.24 Studies also showed that histamine is considered to be a primary candidate immunomodulin produced by L. reuteri.18 Inactivation of histidine-to-histamine converting capacity by mutagenesis of the histidine decarboxylase gene (hdcA) or the histidine-histamine antiporter gene (hdcP) diminished the ability of hdc-positive L. reuteri strain ATCC (Manassas, VA) PTA 6475 to suppress human TNF production.18 Exploration of histidine metabolism, particularly histamine production by the gut microbes, deserves attention as a possible gateway to deepening our understanding of microbiome-mediated intestinal immunomodulation.25, 26
The lack of functional mammalian histidine decarboxylase (HDC), the enzyme converting l-histidine to histamine, yielded increased susceptibility to inflammation-associated CRC in adult mice.27 Here, we set out to address the ability of hdc+ L. reuteri to reduce the frequency and severity of inflammation-associated colon cancer in Hdc−/− mice and to investigate whether microbe-generated metabolites may suppress inflammation-associated cancer phenotypes exacerbated by mammalian enzyme deficiencies.
Materials and Methods
Association between HDC and H2R Gene Expression and Overall Survival Rates in Colon Cancer Patients
To investigate whether HDC and histamine H2 receptor (H2R; symbol: HRH2) expression is associated with changes in survival rates of colon cancer patients, the PROGgeneV2 database28 was queried by selecting gene name HDC or HRH2, cancer type colorectal, and survival measure death. Samples were divided into high and low gene expression groups, bifurcating at median expression value for mRNA expression. Data were plotted and compared by using the Coxph function to compute hazard ratio estimate and related log-rank test P value according to Goswami and Nakshatri.28 All of the CRC data sets in the database (2113 patient samples in 15 data sets) were included in this search (http://watson.compbio.iupui.edu/chirayu/proggene/database/index.php, last accessed May 12, 2017).
Bacterial Strains and Culture Conditions
Bacterial strains, including L. reuteri ATCC PTA 6475 and its isogenic hdcA mutant (generated by induction of a stop codon in hdcA gene), were described previously.18 Both strains were cultured at 37°C in deMan, Rogosa, Sharpe (MRS) media (Difco, Franklin Lakes, NJ) in an anaerobic workstation (MACS MG-500; Microbiology International, Frederick, MD) supplied with a mixture of 10% CO2, 10% H2, and 80% N2.
Mouse Care
Hdc−/− BALB/c mice in which exon 5 of the Hdc gene was replaced with a neomycin cassette were originally provided by Timothy C. Wang (Columbia University, New York, NY) and rederived at Baylor College of Medicine (Houston, TX) by embryo transfer. Four pups, including two male and two female pups, were obtained from rederivation. Mouse tail clippings were used for mouse DNA extraction. The Hdc-deficient genotype of the rederived mice was confirmed by PCR and DNA gel electrophoresis, with neomycin amplicon primers (forward, 5′-AATGGCCGCTTTTCTGGATTCA-3′; reverse, 5′-GGGAGCGGCGATACCGTAAAG-3′) and exon 5 of Hdc gene amplicon primers (forward, 5′-TTAGTCTTTGGGTGTTCCTGGTCA-3′; reverse, 5′-CCCTGTTGCTTGTCTTCCTCAATA-3′). Breeding and maintenance of Hdc−/− mice were performed under specific pathogen-free conditions at Texas Children's Hospital (Houston, TX). Mice were kept under filter top cages and had free access to distilled water and PicoLab Rodent 50IF/6F diet. All mouse experiments were performed according to an Institutional Animal Care and Use Committee–approved mouse protocol at Baylor College of Medicine.
Bacterial Preparation and Administration to Mice
L. reuteri was cultured in MRS media, and bacteria were harvested at exponential phase. Cells were centrifuged at 2500 × g for 4 minutes, and the bacterial pellets were resuspended in sterile MRS media for animal feeding. L. reuteri strains were freshly prepared before administration to mice. Each mouse received 5 × 109 colony-forming units of bacteria in 0.2 mL of MRS media or MRS media only as control by orogastric gavage. The frequency of bacteria administration was once per day for 7 days before azoxymethane (AOM; Sigma-Aldrich, St. Louis, MO) injection, followed by administration once per 3 days for 15 weeks. L. reuteri was not administered during the two cycles of dextran sulfate sodium (DSS; 36,000 to 50,000 molecular weight; MP Biomedicals, Solon, OH) challenge in drinking water (6 days per cycle).
Induction of Colon Cancer in BALB/c Mice
At 12 weeks of age, mice in the positive control group and bacteria-treated groups received a single dose of the genotoxic colonic carcinogen AOM (12.5 mg/kg body weight) by i.p. injection. These mice were challenged with two cycles of 2% (w/v) DSS in drinking water for 6 days, with one cycle immediately after AOM injection, followed by a recovery period with drinking water for 2 weeks before the second cycle. Mice in the negative control group received one dose of phosphate-buffered saline (PBS) solution instead of AOM and drinking water.
Tumor Assessment and Tissue Preparations
Fifteen weeks after AOM injection, mice were euthanized, and specimens were collected as follows. Blood was collected from sedated mice via cardiac puncture in blood sample collection tubes with K2EDTA (Becton, Dickinson and Company, Franklin Lakes, NJ) and centrifuged at 17,000 × g for 10 minutes at 4°C to isolate plasma. The gastrointestinal tract was carefully removed, and luminal contents of ileum, cecum, and colon were collected and flash-frozen in liquid nitrogen. The mouse colons were excised and opened longitudinally, and the number and size of tumors were counted and measured blindly (M.L.). Intestinal mucosa was scraped with an operating knife blade and stored in RNALater (Ambion, Austin, TX) to analyze cytokine mRNA expression. All of the samples were stored at −80°C until analyzed.
Mouse intestines were fixed in 10% formalin, embedded with paraffin, and sectioned at 3 μm by a microtome. Hematoxylin and eosin staining was performed.
Flow Cytometric Analysis
Bone marrow and spleen samples were collected immediately after euthanasia of mice. Bone marrow–derived cells from the femurs and tibia were immediately flushed with ice-cold Dulbecco's modified Eagle's medium (ATCC) containing 10% fetal bovine serum. This procedure was followed by the addition of red blood cell lysis buffer to deplete red blood cells (BD Biosciences, San Jose, CA). Spleen samples were stored in ice-cold Dulbecco's modified Eagle's medium with 10% fetal bovine serum. This step was followed by the isolation of the spleen cells using sterile glass slides and addition of red blood cell lysis buffer to the isolated cells. Single-cell suspensions were made by filtering the cells through 40-μm filter strains. For flow cytometric analysis, single-cell suspensions were stained with antibodies [1 μL of allophycocyanin-cyanine 7–conjugated anti–Gr-1 (BD Biosciences), and 5 μL of fluorescein isothiocyanate–conjugated anti-CD11b (BD Biosciences)] for 30 minutes on ice, in the dark and evaluated by multicolor flow cytometry using a BD FACSCanto cell analyzer and data collected with FACSDiva software version 6 (BD Biosciences). The accumulated data were analyzed with FlowJo software version 10 (FlowJo, Ashland, OR) gated using forward scatter and side scatter, followed by allophycocyanin-cyanine 7 (Gr-1+) on the y axis and fluorescein isothiocyanate (CD11b+) on the x axis.
PET Imaging of Living Mice
Positron emission tomography (PET)/computed tomography (CT) scanning was performed 15 weeks after AOM injection, just before euthanizing the mice as described previously.23 Briefly, mice were anesthetized with isoflurane and received 200 μCi of 18F-fluorodeoxyglucose (FDG) by i.p. injection. After 1 hour, these mice received 200 μL of MD-Gastroview as contrast agent rectally via a 3.5F catheter. CT scan was performed for 10 minutes, followed by a PET scan for 20 minutes using the Inveon PET/CT Multimodality System (Siemens, Munich, Germany). Mouse images were recorded, and FDG standardized uptake values were analyzed blindly (R.R.S.) using Inveon Research Workplace software version 1.5 (Siemens). The images of mice were generated by OsiriX Imaging software version 3.9.4 (Pixmeo, Bernex, Switzerland).
Determination of the Relative Abundances of L. reuteri, hdc Genes, and mRNA in Vivo
Twelve-week-old male Hdc−/− mice were given one dose of L. reuteri 6475 or hdcA mutant or MRS media only as described above. After 2 days, the mice were euthanized, and total DNA and RNA from luminal contents in the distal colon were isolated for bacterial DNA and mRNA extraction. Relative bacterial DNA and mRNA quantities of hdc genes were evaluated by real-time quantitative PCR and normalized to the housekeeping gene rpoB as described previously.23 Whole colons were fixed in Carnoy's solution, and fluorescence in situ hybridization was performed with a probe specific to a unique 16S rRNA sequence to L. reuteri (Reverse: 5′-GATCCATCGTCAATCAGGTGC-3′) as described previously.29 Slides were counterstained with DAPI (Sigma-Aldrich) and imaged at ×400 magnification with an Eclipse 90i fluorescence microscope (Nikon Instruments, Melville, NY).
Histamine Quantification in Hdc−/− Mouse Feces
Fecal specimens were weighed and dissolved in 200 μL of 50/50 PBS/methanol by vortexing and ultrasound disruption for 4 minutes with 30-second intervals. After spinning for 5 minutes, 14,000 × g at 4°C, the supernatant fluids were size-fractionated with Amicon Ultra-0.5 mL centrifugal filters (molecular weight cutoff, 3 kDa; Millipore, Temecula, CA). The histamine quantities in the collected supernatant fluids were determined by liquid chromatography-mass spectrometry using selected reaction monitoring (SRM).
For histamine quantification, 30 μL of each sample was dried in a SpeedVac for 2 hours. The dried samples were mixed with 30 μL of 20 ng/mL histamine-d4 (histamine-α,α,β,β-d4; CDN Isotopes, Pointe-Claire, Canada) solution by vortexing for 1 minute. The prepared samples were loaded into a Shimadzu (Kyoto, Japan) SIL-20ACxr autosampler and separation was achieved using a Shimadzu Nexera-XR HPLC system. Samples (5 μL) were loaded onto a Phenomenex (Torrance, CA) 1 mm × 50 mm phenylhexyl reversed phased column equipped with a Phenomenex phenylhexyl 4 mm × 2 mm guard column. The aqueous mobile phase (A) consisted of water/acetonitrile/formic acid/perfluoroheptanoic acid (99.3:0.5:0.1:0.1 v/v/v/v), and the organic mobile phase (B) consisted of acetonitrile/formic acid (99.9:0.1 v/v). Column flow was set at 80 μL/minute. The elution gradient was optimized as follows: started from 20% B for 0.5 minute and increased to 70% B over 5.5 minutes; ramp to 80% B after 0.1 minute and hold for 1 minute; ramp back to 20% B over 6 seconds and maintained at 20% for a total chromatographic run time of 12 minutes to re-equilibrate.
SRM was performed on a Sciex (Framingham, MA) 6500 QTRAP mass spectrometer equipped with a Turbo V ion source. The mass spectrometer was operated in the positive ion mode under the following conditions: curtain gas of 20 psi; collision gas, High; spray voltage, 4.5 kV; ion source gas 1, 20 psi; ion source gas 20, 2 psi; interface heater temperature, 175°C; Q1 and Q3 resolution, unit; scan time, 100 milliseconds; de-clustering potential, 100 V; entrance potential, 8 V; and collision exit potential, 10 V. The instrument was calibrated by using Sciex PPG calibration standard and tuned to the manufacturer's specifications. SRM transitions monitored for histamine were 112 → 95 (20 eV) and 112 → 68 (30 eV). For histamine-d4, the SRM transitions 116 → 99 (20 eV) and 116 → 72 (30 eV) were monitored. Data were acquired with Analyst Software version 1.6.2 (AB Sciex LP, Concord, ON, Canada) and quantification performed using Multiquant Software version 3.0.1 (AB Sciex LP).
Cytokine Measurement by Multiplex Immunoassay in the Mouse Plasma
The concentrations of murine interferon (Ifn)-γ, Il-1α, Il-1β, Il-4, Il-6, Il-10, Il-12, Il-13, Il-17A, keratinocyte chemoattractant (KC), Tnf, Il-21, Il-22, Il-23, and epidermal growth factor in the plasma were measured using cytokine multiplex kits (Millipore, Billerica, MA). Quantification of cytokines was performed using the Luminex system (Austin, TX) according to the manufacturer's instructions. Briefly, 25-μL plasma samples collected above from each mouse were thawed completely and diluted with the same amount of Assay Buffer provided in the kits. The assays were performed in duplicate blindly (S.V.). The reports automatically generated by MILLIPLEX Analyst software version 5.1 (Millipore) were reviewed, and only cytokines that were greater than the lower limit of detection and below the saturation value were considered.
Modulation of Mammalian Cytokine Gene Expression in the Colonic Mucosa
To quantify the relative mRNA expression levels of Ifn-γ, Tnf, Il-6, Il-12, Il-23, Il-17, Il-22, Il-1α, and KC, RNA was extracted from colonic mucosa samples using the miRNeasy mini kit (Qiagen, Hilden, Germany). One microgram of RNA was reverse-transcribed to single-stranded cDNA using the RevertAid H minus First Strand cDNA Synthesis Kit (ThermoFisher Scientific, Waltham, MA). Real-time RT-PCR was performed using Real-Time PCR system (Stratagene, La Jolla, CA). The RT-PCR reaction mix (adjusted with water to a total volume of 25 μL) contained 1 μL of template DNA, 12.5 μL of Power SYBR Green PCR master mix (ABI, Life Technologies, Carlsbad, CA), and 0.5 μL of the respective primers (10 μmol/L each). The primers used for IFN-γ, IL-12, IL-17, TNF, IL-6, and IL-23 quantification were described previously,30 and all of the primers are shown in Table 1. Relative murine mRNA target gene expression levels (Ratio = [(Etarget)dCPtarget (Control−Sample)]/[(Eref.)dCPref. (Control−Sample)]) were normalized to the housekeeping gene GAPDH and used as a reference. Subsequently, intestinal mucosal cytokine gene expression values of the control group were set to 1.0 and used as the calibrator to identify the relative mRNA fold difference between the negative control group (MRS/PBS-H2O), positive control group (MRS/AOM-DSS), L. reuteri 6475–treated group (L. reuteri 6475/AOM-DSS), and isogenic L. reuteri hdcA mutant–treated group (hdcA mutant/AOM-DSS).
Table 1.
Primers Used in qPCR-Based Gene Expression Studies
| Gene/primer name | Primer pairs |
|---|---|
| Il1α | F: 5′-CAGAGAGGGAGTCAACTCATTG-3′ |
| R: 5′-GTTTCTGGCAACTCCTTCAGC-3′ | |
| KC | F: 5′-GACTCCAGCCACACTCCAAC-3′ |
| R: 5′-TGACAGCGCAGCTCATTG-3′ | |
| Il22 | F: 5′-TGACGACCAGAACATCCAGA-3′ |
| R: 5′-AATCGCCTTGATCTCTCCAC-3′ | |
| Ifn-γ | F: 5′-GCCAAGTTTGAGGTCAACAACCC-3′ |
| R: 5′-CCGAATCAGCAGCGACTCCT-3′ | |
| Tnf | F: 5′-CCTCACACTCAGATCATCTTCTC-3′ |
| R: 5′-GTCTTTGAGATCCATGCCGT-3′ | |
| Il6 | F: 5′-CTCTGCAAGAGACTTCCATCCA-3′ |
| R: 5′-TAAGCCTCCGACTTGTGAAGTA-3′ | |
| Il12 | F: 5′-ATGTGTCAATCACGCTACCTCCTC-3′ |
| R: 5′-GGTCTTCAGCAGGTTTCGGG-3′ | |
| Il23 | F: 5′-TAGCCTGGAACGCACATGCAC-3′ |
| R: 5′-GCAAGCAGAACTGGCTGTTGTA-3′ | |
| Il17 | F: 5′-ACTCTCCACCGCAATGAAGACA-3′ |
| R: 5′-CCCTCTTCAGGACCAGGATCTC-3′ | |
| Gapdh | F: 5′-GCCAAAAGGGTCATCATCTC-3′ |
| R: 5′-CACACCCATCACAAACATGG-3′ | |
| hdcA | F: 5′-GCACTAACGATAACCGTCGTC-3′ |
| R: 5′-CACCCTTATTAGCACAAACAATGA-3′ | |
| hdcP | F: 5′-TCCCTACGGATACCAAGCAC-3′ |
| R: 5′-AGAGGAACGCTAAGACACCAAT-3′ | |
| rpoB | F: 5′-CGTGATACTTCATTACGTGTTCCT-3′ |
| R: 5′-AGTGAAGACTTTAACATCTTGGATGA-3′ |
KC, keratinocyte chemoattractant; qPCR, real-time quantitative PCR.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism software version 5 (GraphPad Inc., La Jolla, CA). For numeric variables that fit normal distribution (determined by Kolmogorov-Smirnov test), data were presented as means ± SD, and different groups were compared with the t-test (two groups) or one-way analysis of variance (more than two groups). Otherwise, data were presented as box and whiskers plots showing the median values and 10th and 90th percentiles or scatter plots showing the median values. Different groups were compared by a nonparametric U-test (two groups) or Kruskal-Wallis test.
Results
Elevated Human HDC and H2R Gene Expression Are Associated with Improved Survival Outcomes in Colon Cancer Patients
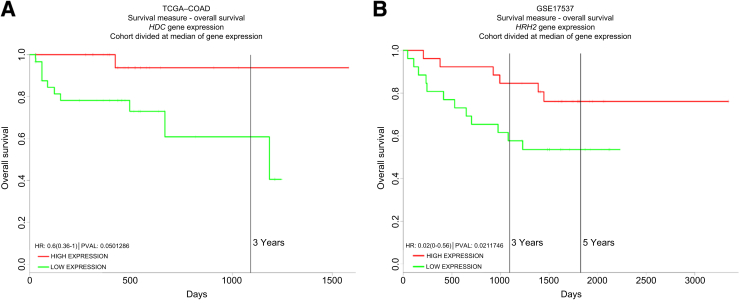
HDC is the key enzyme involved in histidine-to-histamine conversion and histamine generation in mammalian and bacterial cells. To investigate the association between HDC gene expression and survival rates in CRC patients, a large-scale human cancer gene expression database named PROGgene28 was explored. Patients with elevated patterns of HDC gene expression demonstrated improved survival in CRC (TCGA_COAD; P = 0.05) (Figure 1A).
Figure 1.
Overall survival rates increase in colon cancer patients with elevated human HDC and HRH2 gene expression. The overall survival rates in colorectal cancer patients based on HDC or HRH2 gene expression categories were plotted using the PROGgeneV2 database. Representative figures with the lowest P value are shown for HDC (A) and HRH2 (B). HR, hazard ratio; PVAL, P value.
We further investigated the association between H2R gene expression and survival rates in CRC patients because our previous study23 indicated an H2R-mediated suppression of colitis by histamine. Elevated patterns of H2R gene (HRH2) expression was correlated with enhanced survival among patients with CRC (GSE17537; P = 0.02) (Figure 1B), indicating a potential antitumorigenic effect by H2R up-regulation.
These findings based on human cancer gene expression profiling set the stage to explore whether microbial Hdc expression and histamine generation could complement histamine deficiency in Hdc−/− mice and provide mechanistic insights in murine models of CRC.
L. reuteri 6475 Administration Increases hdc Gene Expression and Histamine Production in the Intestines of Hdc−/− Mice
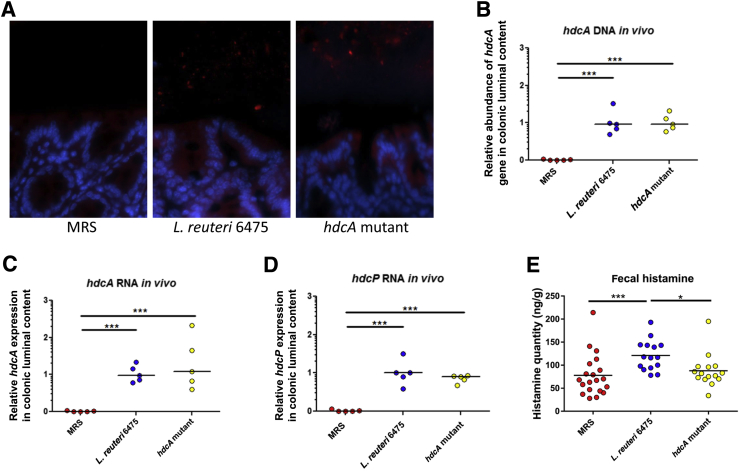
To explore whether oral administration of L. reuteri increased the relative abundance of this bacterial species in the intestines of Hdc-deficient mice, fluorescence in situ hybridization was performed in the colons of Hdc−/− mice 2 days after L. reuteri administration. Orogastric gavage with wild-type L. reuteri 6475 and HdcA-deficient L. reuteri increased the relative abundances of L. reuteri in the colonic lumen visualized by fluorescence microscopy compared with control mice that did not receive exogenous bacteria (Figure 2A). In addition, the relative abundances of hdcA DNA and hdcA and hdcP mRNA were significantly increased, demonstrating active gene expression in the intestinal lumen by viable bacteria (Figure 2, B–D). Moreover, L. reuteri 6475 administration significantly increased histamine quantities in mouse feces, in contrast to the lack of increased amounts of histamine after administration of L. reuteri defective in HDC function (hdcA mutant) (Figure 2E). These studies indicate that i) L. reuteri administration increased its relative abundance in the guts of Hdc−/− mice, and the presence of L. reuteri in the intestine was not affected by the presence or absence of an intact HCD gene in L. reuteri; ii) L. reuteri can express the HCD and histidine/histamine antiporter genes in the mammalian intestines of Hdc−/− mice; iii) hdc-positive L. reuteri is able to generate histamine in the gut, providing a mechanistic underpinning for histamine-mediated immunomodulation by the mammalian gut microbes.
Figure 2.
Increased abundance of bacterial histidine decarboxylase (Hdc) mRNA and histamine in the mouse intestine after Lactobacillus reuteri supplementation. A: Representative figures showing the microscopic visualization of L. reuteri in the colon of Hdc−/− mice. Media only, L. reuteri 6475, or HdcA-deficient L. reuteri were administered to mice orally, followed by L. reuteri species–specific fluorescence in situ hybridization after 2 days. Red indicates cyanine 3-conjugated probe to label L. reuteri; blue, DAPI-labeled host cell nuclei. B: Relative abundance of hdcA gene in mouse gut microbiome in different groups. C and D: Relative quantities of hdcA (C) and hdcP (D) mRNA in colonic luminal contents of mice. E: Histamine quantities in mouse feces determined by liquid chromatography-mass spectrometry. n = 5 for each group (A–D); n = 15 to 20 for each group (E). ∗P < 0.05, ∗∗∗P < 0.001. MRS, deMan, Rogosa, Sharpe.
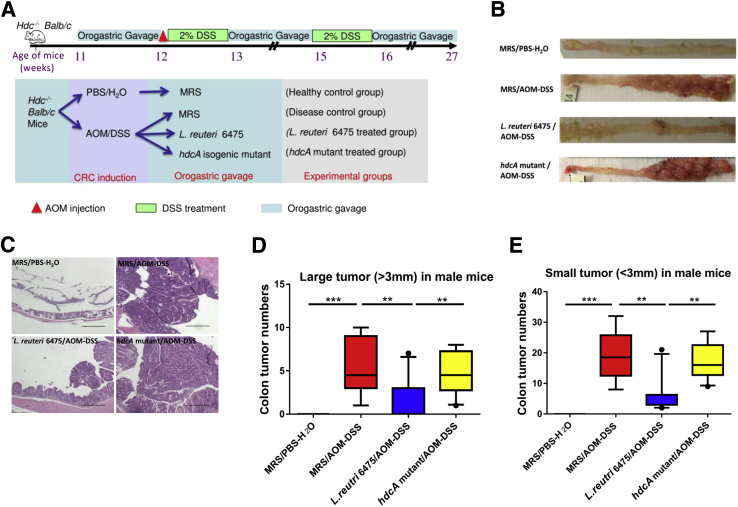
HDC Gene Is Necessary for L. reuteri 6475 to Attenuate Colonic Carcinogenesis in Vivo
To probe the ability of histamine-generating L. reuteri 6475 to suppress CRC associated with chronic intestinal inflammation, the combination of AOM and DSS was used to induce colitis-associated colon cancer in 12-week-old male Hdc−/− BALB/c mice (Figure 3A). We found that negative control mice that received PBS and drinking water, instead of AOM and 2% DSS, did not develop tumors (Figure 3, B–E). Positive control mice that were challenged with AOM/DSS and gavaged with MRS media, but did not receive exogenous bacteria, developed colonic tumors. Administration of exponential phase L. reuteri 6475 with a wild-type allele of the HDC gene significantly reduced the number and size of colonic tumors in male mice compared with the positive control group. However, oral administration of Hdc-deficient L. reuteri did not suppress colonic carcinogenesis. The relative abundances of hdcA DNA and hdcA and hdcP mRNA in the feces of the experimental mice were also determined (Supplemental Figure S1), and these bacterial genes showed a similar pattern as luminal contents (Figure 2, B–D), indicating that AOM and DSS did not affect l-histidine metabolism by L. reuteri.
Figure 3.
Lactobacillus reuteri 6475 but not the hdcA mutant attenuates colonic carcinogenesis in vivo. A: Procedures for mouse experiments. B: Representative colon images of mice in the negative control group [deMan, Rogosa, Sharpe/phosphate-buffered saline–water (MRS/PBS-H2O)], positive control group [MRS/azoxymethane-dextran sulfate sodium (AOM-DSS)], L. reuteri 6475–treated group (L. reuteri 6475/AOM-DSS), and isogenic L. reuteri hdcA mutant–treated group (hdcA mutant/AOM-DSS). C: Representative microscopic colon images (hemoxylin and eosin stained) from mice in different groups. D and E: Numbers of both large (>3 mm) and small (<3 mm) colonic tumors from mice in different groups. n = 8 to 10 for each group (D and E). ∗∗P < 0.01, ∗∗∗P < 0.001. Scale bars = 500 μm (C).
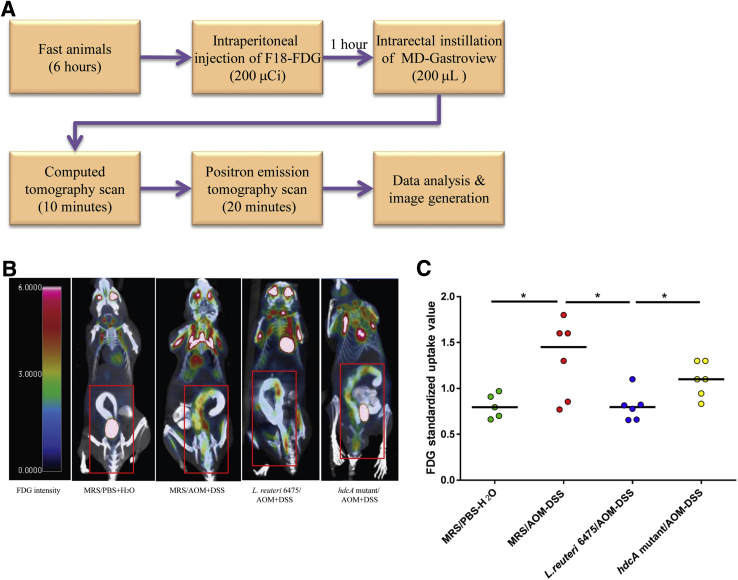
Abdominal Imaging Yields Evidence of Suppression of Colon Tumorigenesis by L. reuteri
To further confirm the antitumorigenic effects of L. reuteri in the AOM/DSS-induced Hdc-deficient mouse model of colon cancer, PET imaging was applied to evaluate the process of colonic carcinogenesis and suppression of colonic neoplasia in living mice before euthanasia (Figure 4A). Trace amounts of FDG signal were evident in the colon and surrounding abdominal sites, indicating low baseline glucose uptake in the colons of healthy mice (Figure 4, B and C, and Supplemental Video S1). In positive control mice, hot spots in the colon were observed, and the relative FDG intensity in the whole mouse colon was significantly increased compared with the negative control group (Figure 4, B and C, and Supplemental Video S2), indicating the presence of colonic tumors and increased glucose uptake in the colons of mice that received AOM plus DSS. L. reuteri 6475–treated mice showed reduced numbers of hot spots in the colon and significantly decreased abdominal FDG intensities compared with positive control mice, indicating that histamine-generating strain L. reuteri 6475 could suppress metabolic activity associated with colon carcinogenesis in live animals (Figure 4, B and C, and Supplemental Video S3). Meanwhile, the isogenic hdcA mutant strain deficient in HDC activity did not yield similar effects and showed increased numbers of hot spots and increased abdominal FDG intensities compared with the wild-type strain and consistent with unfettered colon carcinogenesis (Figure 4, B and C, and Supplemental Video S4). Together, these results showed that histamine-generating lactobacilli could suppress carcinogenesis by interkingdom complementation, whereas isogenic strains incapable of generating histamine did not suppress colonic carcinogenesis.
Figure 4.
Attenuation of colonic tumorigenesis as shown by positron emission tomography (PET) imaging. A: A schematic diagram shows the PET imaging procedures with live mice. B: Representative mouse images captured by PET-computed tomography (CT) scanning in each group. The color code bar represents fluorodeoxyglucose (FDG) signal intensity. The abdominal regions are indicated with red boxes. C: Standardized uptake values of FDG signal in the whole mouse colon in different groups. n = 5 to 6 for each group (B and C). ∗P < 0.05. AOM, azoxymethane; DSS, dextran sulfate sodium; L. reuteri, Lactobacillus reuteri; MRS, deMan, Rogosa, Sharpe; PBS, phosphate-buffered saline.
Histamine-Generating L. reuteri Suppresses Proinflammatory Cytokine Gene Expression in the Colonic Mucosa
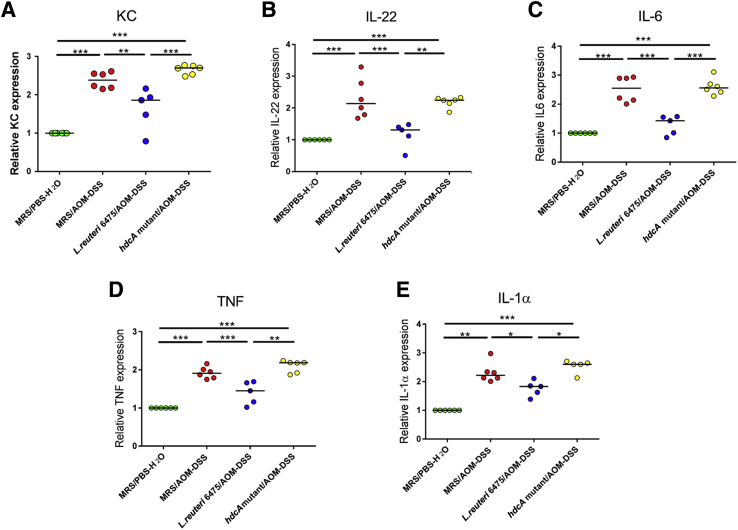
Specific proinflammatory cytokines have been reported to contribute to the development of colon tumorigenesis by promoting the formation of a tumor-supportive microenvironment.31 By analyzing cytokine gene expression of selected cytokines in the colonic mucosa (Table 1), we showed that AOM/DSS challenge induced expression of proinflammatory cytokine genes encoding KC, Il-22, Il-6, Tnf, and Il-1α compared with healthy Hdc−/− mice (Figure 5). Oral treatment of AOM/DSS-challenged mice with hdc-positive L. reuteri 6475 reduced the relative gene expression of these cytokines, whereas the isogenic L. reuteri strain lacking functional HdcA did not inhibit gene expression of these proinflammatory cytokines. Detection of other cytokine mRNAs by real-time quantitative PCR yielded either undetectable results (IL-17) or no significant differences (Il-12, Il-23, and Ifn-γ) among the groups (ie, cytokines that this probiotic intervention did not repress), indicating that histamine-generating L. reuteri suppressed a subset of proinflammatory murine cytokine genes in the colonic mucosa.
Figure 5.
Histamine-generating Lactobacillus reuteri suppresses mammalian cytokine gene expression in colonic mucosa. Wild-type L. reuteri 6475 administration significantly decreases proinflammatory cytokine keratinocyte chemoattractant (KC) (A), IL-22 (B), IL-6 (C), tumor necrosis factor (Tnf) (D), and Il-1α (E) expression in Hdc−/− mouse colonic mucosa in mRNA level determined by quantitative RT-PCR, whereas the isogenic L. reuteri hdcA mutant which lacks the histamine-producing capacity lacks such effects. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. AOM, azoxymethane; DSS, dextran sulfate sodium; MRS, deMan, Rogosa, Sharpe; PBS, phosphate-buffered saline.
Circulating Cytokine Concentrations in Plasma Are Reduced by Histamine-Generating L. reuteri
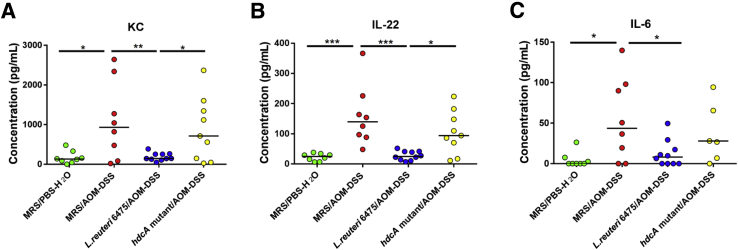
To further investigate the association between cytokines and CRC, we performed multiplex analyses of 16 cytokines (Supplemental Table S1) in the plasma by using a Luminex system (Millipore). AOM/DSS treatment in Hdc−/− mice increased the relative concentrations of KC, Il-22, and Il-6 in plasma, compared with that of control mice (Figure 6). L. reuteri 6475 administration diminished concentrations of these three cytokines (KC, Il-22, Il-6), whereas administration of isogenic L. reuteri hdcA mutant lacking histamine-producing capacity did not reduce these same cytokine concentrations in AOM/DSS-treated Hdc−/− mice. The results with KC, Il-22, and Il-6 are consistent in terms of plasma protein quantitation and mucosal gene expression. Other cytokines measured in plasma were either not detectable (Il-1β, Il-21, Il-23, epidermal growth factor) or showed no significant changes (Il-4, Il-17, Ifn, Il-1α, Il-12, Tnf, Il-10, Il-13) among groups, indicating selective reduction of circulating cytokines in plasma by histamine-generating L. reuteri. Together with cytokine gene expression in the colonic mucosa, histamine generation by lactobacilli appeared to be important for suppression of particular proinflammatory and cancer-associated cytokines.
Figure 6.
Plasma cytokine concentrations are reduced by histidine decarboxylase (Hdc)A-positive Lactobacillus reuteri. Wild-type L. reuteri 6475 administration significantly decreases the abundances of proinflammatory cytokines keratinocyte chemoattractant (KC) (A), IL-22 (B), and IL-6 (C) in Hdc−/− mouse plasma determined by Luminex assays, whereas the isogenic L. reuteri hdcA mutant that lacks the histamine-producing capacity lacks such effects. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. AOM, azoxymethane; DSS, dextran sulfate sodium; MRS, deMan, Rogosa, Sharpe; PBS, phosphate-buffered saline.
Abundance of Splenic Immature Myeloid Cells Is Suppressed by hdc+L. reuteri
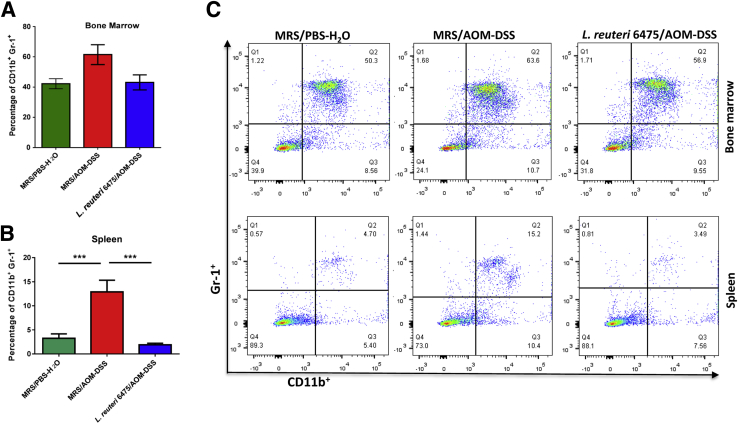
The absence of endogenous histamine leads to markedly increased numbers of immature myeloid cells (IMCs; CD11b+ Gr-1+) after AOM/DSS treatment, and this finding is associated with cancer progression in mammals.27 Here, we found that the percentage of CD11b+Gr-1+ IMCs in the spleen was significantly increased in AOM/DSS-challenged mice compared with healthy control mice (Figure 7, B and C), consistent with a previous report.27 L. reuteri 6475 administration in AOM/DSS-challenged mice significantly decreased the relative abundance of CD11b+Gr-1+ IMCs compared with mice that did not receive exogenous lactobacilli. A similar, although less dramatic pattern, was observed in the bone marrow (Figure 7, A and C). These observations are consistent with the conclusion that histamine-generating probiotic L. reuteri 6475 may attenuate AOM/DSS-induced colon carcinogenesis, at least in part, via enhanced maturation of circulating myeloid cells and concomitant reduction of proinflammatory cytokines produced by relatively mature myeloid cells.
Figure 7.
Immature myeloid cells (IMCs) in bone marrow and spleen are reduced by Lactobacillus reuteri 6475. A and B: Percentages of CD11b+Gr-1+ IMCs acquired by flow cytometric analysis of the bone marrow (A) and spleen (B) samples of Hdc−/− mice. C: Fluorescence-activated cell sorting analysis shows the relative abundance of CD11b+ Gr-1+ IMCs in the bone marrow and spleens in Hdc−/− mice gated using forward scatter and side scatter, followed by allophycocyanin-cyanine 7 (Gr-1+) on y axis and fluorescein isothiocyanate (CD11b+) on x axis. n = 3 to 4 for each group (A and B). ∗∗∗P < 0.001. AOM, azoxymethane; DSS, dextran sulfate sodium; MRS, deMan, Rogosa, Sharpe; PBS, phosphate-buffered saline.
Discussion
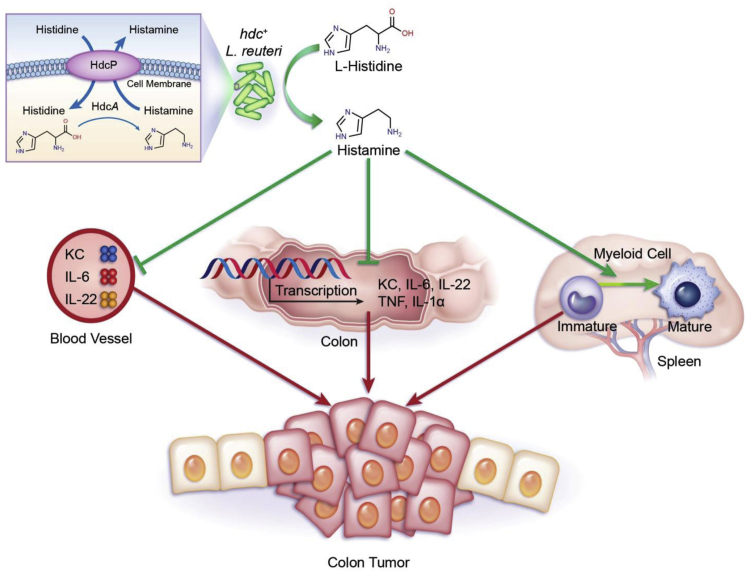
Histamine generated by intestinal microbes may supplement histamine-generating capacity by mammalian cells and may offer new possibilities for microbiome-mediated gene therapy. Our prior studies showed that microbial histamine may suppress intestinal inflammation in an acute colitis model.23 From the studies by Yang et al27 that showed an increased susceptibility of Hdc−/− mice to inflammation-associated colonic cancer induced by AOM/DSS challenge, we addressed the ability of histamine-producing gut microbes such as L. reuteri to complement the histamine deficiency in Hdc−/− mice by interkingdom complementation. Our results indicate that histamine-producing L. reuteri 6475 protected Hdc−/− mice from AOM/DSS-induced inflammation-associated colonic cancer. The bacterial enzyme, HDC, must be present in this microbe to generate histamine as the bioactive compound in the intestine, yielding anti-inflammatory and antitumorigenic effects (Figure 8).
Figure 8.
Proposed mechanism describing gut microbe–mediated suppression of inflammation-associated colon carcinogenesis by bacterial histamine production. azoxymethane (AOM)/dextran sulfate sodium (DSS) treatment induces keratinocyte chemoattractant (KC), Il6, Tnf, Il22, and Il1a gene expression in the colon, increases IL-6, IL-22, and KC protein concentrations in mouse plasma, and increases abundance of immature myeloid cells (IMCs) in the spleen. These effects may all contribute to colon carcinogenesis. When Hdc−/− mice receive hdc+ Lactobacillus reuteri, histamine is produced in the intestinal lumen via histidine decarboxylase (Hdc) protein machinery in L. reuteri: l-histidine is converted to histamine by HdcA and histamine is exported to the gut lumen by histidine-histamine antiporter (HdcP). L. reuteri–derived histamine may activate histamine H2 receptors (H2Rs) on the intestinal epithelium and trigger anticarcinogenic pathways as indicated by suppression of mammalian cytokine gene expression in the colonic mucosa and reduction of IMCs in the spleen. TNF, tumor necrosis factor.
Histamine receptors on immune cells and the intestinal epithelium are key factors driving the responsiveness of mammalian cells to histamine generated by intestinal microbes. H2R is present on the human intestinal epithelium,32 and H2R has been proposed to play a key role in responses by mammalian cells to histamine generated by luminal gut microbes.33 Our prior studies demonstrated that gut microbes such as L. reuteri suppressed intestinal inflammation via H2R.23 Gut microbes such as L. reuteri were capable of activating intracellular mammalian protein kinase A and inhibiting extracellular signal-regulated kinase signaling via histamine receptors.18 Past studies documented H2R-mediated signaling via cAMP production and protein kinase A activation, followed by inhibition of c-Raf and mitogen-activated protein kinase kinase/extracellular signal-regulated kinase signaling.18, 34, 35, 36 Besides allergic diseases, other disease phenotypes may be affected by histamine signaling. For example, H2R-mediated signaling may offer protection against nonalcoholic fatty liver disease,37 and histamine type 1 receptor–mediated signaling may contribute to irritable bowel syndrome.38 The opposing proinflammatory (histamine type 1 receptor) and anti-inflammatory (H2R) effects of histamine receptors may depend on differences in relative tissue distributions of histamine receptors. Histamine generated by microbial or mammalian cells may contribute to tissue homeostasis or pathologic processes based on relative local concentrations of histamine and its receptors. Gut microbes may represent an important source of histamine in mammals with implications for intestinal inflammation and colorectal neoplasia.
Histamine-producing L. reuteri suppressed gene expression and production of several cytokines, in contrast to the mutant strain lacking intact bacterial HDC. KC shares many functional properties with IL-839 and has been reported to promote colon cancer growth, progression, and metastasis.40, 41 IL-22 was also shown to promote gastric cancer cell invasion42, 43 and colon cancer stemness.44 IL-6 has been considered as a key regulator of CRC development,45, 46 and increased quantities of plasma IL-6 were correlated with a poor prognosis in a variety of cancers, including colon cancer.47 Cytokine signaling in CRC provides opportunities for therapeutic intervention, and some clinical trials targeting selected cytokines have been completed or are ongoing.48 Our findings on gut microbe-mediated suppression of cancer-associated cytokines may open new avenues for preventive and therapeutic strategies in oncology.
Myeloid cell populations are altered in Hdc−/− mice challenged with AOM/DSS. A marked accumulation of CD11b+ Gr-1+ IMCs in the spleen suggests that myeloid cell maturation programs may be affected by the absence of histamine. CD11b+Gr-1+ IMC populations have expanded in terms of cell numbers in the spleens of cancer-bearing mice,49 with greatly increased IMC populations observed in Hdc-deficient mice.27 Our present study shows that oral administration of histamine-producing bacteria reduced the numbers of CD11b+Gr-1+ IMCs in the bone marrow and spleen, consistent with previous studies.27 These findings support the notion that exogenous histamine from the gut microbes can stimulate maturation or proliferation of myeloid cells at distant sites. CD11b+Gr-1+ IMCs are known to produce IL-6 and other proinflammatory cytokines and may help explain how myeloid cell lineages may contribute to changes in cytokine quantities and inflammation-associated carcinogenesis.
Possible effects of histamine on cancer progression remain controversial.50, 51 Significantly increased HDC activity was found in tumor specimens in a series of 10 surgical patients with colorectal carcinoma, suggesting possible dysregulation due to genetic abnormalities (epiphenomenon) or a role for HDC enzymatic activity in oncogenesis.52 However, a deficiency of endogenous histamine was shown to promote inflammation-associated colonic cancer by promoting the accumulation of IL-6 and CD11b+Gr-1+ IMCs.27 With respect to human cancers, Lactobacillus was shown to be diminished in CRC patients compared with healthy control participants,53, 54 and some Lactobacillus species have been negatively associated with the risk of colon cancer.55, 56 Although the correlation between Lactobacillus HDC activity and CRC risk has not been reported, emerging evidence indicates that allergy and atopy are inversely associated with glioma development and CRC in patients.57, 58, 59 Histamine-generating microbes were recently demonstrated to be increased in the intestines of asthma patients.60 These findings suggest that excessive histamine and other mediators in patients with allergic disease may protect individuals from cancer. Our present study showed that patients with elevated patterns of HDC and H2R gene expression demonstrated improved survival rates in human CRC. Future investigations may clarify whether the antitumorigenic effects in the present study by L. reuteri are mediated via H2R and whether differences in H2R distribution affect cancer phenotypes.
This report highlights the potential importance of luminal conversion of l-histidine to histamine by intestinal microbes as a factor in susceptibility to colitis and colon cancer risk. Histamine represents one of the many microbial metabolites that may have profound consequences for mammalian biology, including but not limited to intestinal immunology and epithelial cell biology. Another important implication is that the mammalian microbiome may modify disease risk by complementing or supplementing deficient functions. Genetic deficiencies in specific mammalian enzymes, as demonstrated by the lack of HDC in this mouse model, may be complemented by supplying missing genes and enzymatic functions via the mammalian gut microbiome. Biogenic amines, such as histamine, may provide microbe-derived signals that augment or alter the functions of immune cells at remote sites, and these findings indicate that microbial metabolites may have an impact on mucosal biology, immune function, and cancer susceptibility. This study summarized findings with a specific microbe (L. reuteri) in the gut, microbial cometabolite (histamine), and the phenotype of inflammation-associated cancer to gain insights into probiotic-mediated modulation of oncogenesis. The importance of an amino acid, l-histidine, as the substrate for luminal conversion suggests a key link between diet and probiotic functions in vivo. Known mammalian drug targets such as histamine receptors may be reconsidered in new disease applications as a result of investigations in mammalian microbiology. These findings may point the way to a deeper understanding of colorectal neoplasia and probiotic-mediated prevention or suppression of epithelial-derived cancers.
Acknowledgments
We thank Eamonn Connolly and Stefan Roos (BioGaia AB, Stockholm) for providing the Lactobacillus reuteri strains and sharing pertinent expertise, and the staff of the Small Animal Imaging facility at Texas Children's Hospital, especially to David A Rendon and M. Waleed Gaber, for positron emission tomography (PET) imaging.
C.G. designed the study, performed experiments, analyzed and interpreted data, and wrote the manuscript; B.P.G. performed flow cytometry and gene expression studies, analyzed data, and wrote the manuscript; Z.S. performed histologic analyses; R.R.S. performed PET imaging analysis; R.F. contributed to cell isolation and flow cytometry; A.M. performed histologic examination and immunohistochemical staining; S.V. performed the Luminex assays for multiplex protein profiling; M.L. helped with mouse sample collections; K.H. and A.H. performed histamine quantification; X.C. and T.C.W. provided the Hdc−/− mice and helpful suggestions for our study and writing of the manuscript; J.V. provided guidance, helped design the experiments, co-wrote the manuscript, and provided funding.
Footnotes
Supported by the NIH (J.V.), including the National Institute of Diabetes, Digestive and Kidney Diseases grant R01 DK065075, Texas Medical Center Digestive Disease Center grant P30 DK56338, National Cancer Institute grant U01 CA170930, and unrestricted research support from BioGaia AB (J.V.).
Disclosures: J.V. receives unrestricted research support from Biogaia AB.
Current address of C.G., Immunology Research, Janssen Pharmaceutical Companies of Johnson & Johnson, Spring House, PA.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2017.06.011.
Supplemental Data
Representative three-dimensional reconstructions by positron emission tomography (PET) imaging [fluorodeoxyglucose (FDG) signal] in healthy control mice that were gavaged with deMan, Rogosa, Sharpe (MRS) media and did not receive azoxymethane/dextran sulfate sodium (AOM/DSS) challenge.
Representative three-dimensional reconstructions by positron emission tomography (PET) imaging [fluorodeoxyglucose (FDG) signal] in control mice with disease that were gavaged with deMan, Rogosa, Sharpe (MRS) media and received azoxymethane/dextran sulfate sodium (AOM/DSS) challenge.
Representative three-dimensional reconstructions by positron emission tomography (PET) imaging [fluorodeoxyglucose (FDG) signal] in Lactobacillus reuteri–treated mice that were gavaged with L. reuteri 6475 and received azoxymethane/dextran sulfate sodium (AOM/DSS) challenge.
Representative three-dimensional reconstructions by positron emission tomography (PET) imaging [fluorodeoxyglucose (FDG) signal] in hdcA mutant–treated mice that were gavaged with hdcA mutant and received azoxymethane/dextran sulfate sodium (AOM/DSS) challenge.
Lactobacillus reuteri administration yields increased abundance of hdc gene and mRNA in the feces of experimental mice. A: Relative abundance of hdcA gene in mouse gut microbiome in different groups. B and C: Relative hdcA (B) and hdcP (C) gene expression levels in mouse feces. n = 5 for each group (A–C). ∗∗P < 0.01. AOM, azoxymethane, DSS, dextran sulfate sodium; MRS, deMan, Rogosa, Sharpe; PBS, phosphate-buffered saline.
References
- 1.Siegel R., Desantis C., Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 2.Jess T., Simonsen J., Jorgensen K.T., Pedersen B.V., Nielsen N.M., Frisch M. Decreasing risk of colorectal cancer in patients with inflammatory bowel disease over 30 years. Gastroenterology. 2012;143:375–381.e1. doi: 10.1053/j.gastro.2012.04.016. quiz e13–e14. [DOI] [PubMed] [Google Scholar]
- 3.Herrinton L.J., Liu L., Levin T.R., Allison J.E., Lewis J.D., Velayos F. Incidence and mortality of colorectal adenocarcinoma in persons with inflammatory bowel disease from 1998 to 2010. Gastroenterology. 2012;143:382–389. doi: 10.1053/j.gastro.2012.04.054. [DOI] [PubMed] [Google Scholar]
- 4.Beaugerie L., Svrcek M., Seksik P., Bouvier A.M., Simon T., Allez M., Brixi H., Gornet J.M., Altwegg R., Beau P., Duclos B., Bourreille A., Faivre J., Peyrin-Biroulet L., Flejou J.F., Carrat F., CESAME Study Group Risk of colorectal high-grade dysplasia and cancer in a prospective observational cohort of patients with inflammatory bowel disease. Gastroenterology. 2013;145:166–175.e8. doi: 10.1053/j.gastro.2013.03.044. [DOI] [PubMed] [Google Scholar]
- 5.Terzic J., Grivennikov S., Karin E., Karin M. Inflammation and colon cancer. Gastroenterology. 2010;138:2101–2114.e5. doi: 10.1053/j.gastro.2010.01.058. [DOI] [PubMed] [Google Scholar]
- 6.Louis P., Hold G.L., Flint H.J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12:661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 7.Sears C.L., Garrett W.S. Microbes, microbiota, and colon cancer. Cell Host Microbe. 2014;15:317–328. doi: 10.1016/j.chom.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwabe R.F., Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tjalsma H., Boleij A., Marchesi J.R., Dutilh B.E. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. 2012;10:575–582. doi: 10.1038/nrmicro2819. [DOI] [PubMed] [Google Scholar]
- 10.Arthur J.C., Perez-Chanona E., Muhlbauer M., Tomkovich S., Uronis J.M., Fan T.J., Campbell B.J., Abujamel T., Dogan B., Rogers A.B., Rhodes J.M., Stintzi A., Simpson K.W., Hansen J.J., Keku T.O., Fodor A.A., Jobin C. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arthur J.C., Gharaibeh R.Z., Muhlbauer M., Perez-Chanona E., Uronis J.M., McCafferty J., Fodor A.A., Jobin C. Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nat Commun. 2014;5:4724. doi: 10.1038/ncomms5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodwin A.C., Destefano Shields C.E., Wu S., Huso D.L., Wu X., Murray-Stewart T.R., Hacker-Prietz A., Rabizadeh S., Woster P.M., Sears C.L., Casero R.A., Jr. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc Natl Acad Sci U S A. 2011;108:15354–15359. doi: 10.1073/pnas.1010203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rowland I.R., Rumney C.J., Coutts J.T., Lievense L.C. Effect of Bifidobacterium longum and inulin on gut bacterial metabolism and carcinogen-induced aberrant crypt foci in rats. Carcinogenesis. 1998;19:281–285. doi: 10.1093/carcin/19.2.281. [DOI] [PubMed] [Google Scholar]
- 14.Chen C.C., Lin W.C., Kong M.S., Shi H.N., Walker W.A., Lin C.Y., Huang C.T., Lin Y.C., Jung S.M., Lin T.Y. Oral inoculation of probiotics Lactobacillus acidophilus NCFM suppresses tumour growth both in segmental orthotopic colon cancer and extra-intestinal tissue. Br J Nutr. 2012;107:1623–1634. doi: 10.1017/S0007114511004934. [DOI] [PubMed] [Google Scholar]
- 15.Verma A., Shukla G. Probiotics Lactobacillus rhamnosus GG, Lactobacillus acidophilus suppresses DMH-induced procarcinogenic fecal enzymes and preneoplastic aberrant crypt foci in early colon carcinogenesis in Sprague Dawley rats. Nutr Cancer. 2013;65:84–91. doi: 10.1080/01635581.2013.741746. [DOI] [PubMed] [Google Scholar]
- 16.Casas I.A., Dobrogosz W.J. Validation of the probiotic concept: Lactobacillus reuteri confers broad-spectrum protection against disease in humans and animals. Microb Ecol Health Dis. 2000;12:247–285. [Google Scholar]
- 17.Liu Y., Fatheree N.Y., Mangalat N., Rhoads J.M. Human-derived probiotic Lactobacillus reuteri strains differentially reduce intestinal inflammation. Am J Physiol Gastrointest Liver Physiol. 2010;299:G1087–G1096. doi: 10.1152/ajpgi.00124.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas C.M., Hong T., van Pijkeren J.P., Hemarajata P., Trinh D.V., Hu W., Britton R.A., Kalkum M., Versalovic J. Histamine derived from probiotic Lactobacillus reuteri suppresses TNF via modulation of PKA and ERK signaling. PLoS One. 2012;7:e31951. doi: 10.1371/journal.pone.0031951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madsen K.L., Doyle J.S., Jewell L.D., Tavernini M.M., Fedorak R.N. Lactobacillus species prevents colitis in interleukin 10 gene-deficient mice. Gastroenterology. 1999;116:1107–1114. doi: 10.1016/s0016-5085(99)70013-2. [DOI] [PubMed] [Google Scholar]
- 20.Pena J.A., Li S.Y., Wilson P.H., Thibodeau S.A., Szary A.J., Versalovic J. Genotypic and phenotypic studies of murine intestinal lactobacilli: species differences in mice with and without colitis. Appl Environ Microbiol. 2004;70:558–568. doi: 10.1128/AEM.70.1.558-568.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schreiber O., Petersson J., Phillipson M., Perry M., Roos S., Holm L. Lactobacillus reuteri prevents colitis by reducing P-selectin-associated leukocyte- and platelet-endothelial cell interactions. Am J Physiol Gastrointest Liver Physiol. 2009;296:G534–G542. doi: 10.1152/ajpgi.90470.2008. [DOI] [PubMed] [Google Scholar]
- 22.Preidis G.A., Saulnier D.M., Blutt S.E., Mistretta T.A., Riehle K.P., Major A.M., Venable S.F., Barrish J.P., Finegold M.J., Petrosino J.F., Guerrant R.L., Conner M.E., Versalovic J. Host response to probiotics determined by nutritional status of rotavirus-infected neonatal mice. J Pediatr Gastroenterol Nutr. 2012;55:299–307. doi: 10.1097/MPG.0b013e31824d2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao C., Major A., Rendon D., Lugo M., Jackson V., Shi Z., Mori-Akiyama Y., Versalovic J. Histamine H2 receptor-mediated suppression of intestinal inflammation by probiotic Lactobacillus reuteri. MBio. 2015;6:e01358-15. doi: 10.1128/mBio.01358-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spinler J., Sontakke A., Hollister E., Venable S., Oh L.P., Balderas M., Saulnier D.M.A., Mistretta T.A., Devaraj S., Walter J., Versalovic J., Highlander S.K. From prediction to function using evolutionary genomics: human-specific ecotypes of Lactobacillus reuteri have diverse probiotic functions. Genome Biol Evol. 2014;6:1772–1789. doi: 10.1093/gbe/evu137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Preidis G.A., Versalovic J. Targeting the human microbiome with antibiotics, probiotics, and prebiotics: gastroenterology enters the metagenomics era. Gastroenterology. 2009;136:2015–2031. doi: 10.1053/j.gastro.2009.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hemarajata P., Gao C., Pflughoeft K.J., Thomas C.M., Saulnier D.M., Spinler J.K., Versalovic J. Lactobacillus reuteri-specific immunoregulatory gene rsiR modulates histamine production and immunomodulation by Lactobacillus reuteri. J Bacteriol. 2013;195:5567–5576. doi: 10.1128/JB.00261-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang X.D., Ai W., Asfaha S., Bhagat G., Friedman R.A., Jin G., Park H., Shykind B., Diacovo T.G., Falus A., Wang T.C. Histamine deficiency promotes inflammation-associated carcinogenesis through reduced myeloid maturation and accumulation of CD11b+Ly6G+ immature myeloid cells. Nat Med. 2011;17:87–95. doi: 10.1038/nm.2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goswami C.P., Nakshatri H. PROGgeneV2: enhancements on the existing database. BMC Cancer. 2014;14:970. doi: 10.1186/1471-2407-14-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Preidis G.A., Saulnier D.M., Blutt S.E., Mistretta T.A., Riehle K.P., Major A.M., Venable S.F., Finegold M.J., Petrosino J.F., Conner M.E., Versalovic J. Probiotics stimulate enterocyte migration and microbial diversity in the neonatal mouse intestine. FASEB J. 2012;26:1960–1969. doi: 10.1096/fj.10-177980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ganesh B.P., Richter J.F., Blaut M., Loh G. Enterococcus faecium NCIMB 10415 does not protect interleukin-10 knock-out mice from chronic gut inflammation. Benef Microbes. 2012;3:43–50. doi: 10.3920/BM2011.0050. [DOI] [PubMed] [Google Scholar]
- 31.Landskron G., De la Fuente M., Thuwajit P., Thuwajit C., Hermoso M.A. Chronic inflammation and cytokines in the tumor microenvironment. J Immunol Res. 2014;2014:149185. doi: 10.1155/2014/149185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sander L.E., Lorentz A., Sellge G., Coeffier M., Neipp M., Veres T., Frieling T., Meier P.N., Manns M.P., Bischoff S.C. Selective expression of histamine receptors H1R, H2R, and H4R, but not H3R, in the human intestinal tract. Gut. 2006;55:498–504. doi: 10.1136/gut.2004.061762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferstl R., Frei R., Schiavi E., Konieczna P., Barcik W., Ziegler M., Lauener R.P., Chassard C., Lacroix C., Akdis C.A., O'Mahony L. Histamine receptor 2 is a key influence in immune responses to intestinal histamine-secreting microbes. J Allergy Clin Immunol. 2014;134:744–746.e3. doi: 10.1016/j.jaci.2014.04.034. [DOI] [PubMed] [Google Scholar]
- 34.Funaki C., Hodges R.R., Dartt D.A. Identification of the Raf-1 signaling pathway used by cAMP to inhibit p42/p44 MAPK in rat lacrimal gland acini: role in potentiation of protein secretion. Invest Ophthalmol Vis Sci. 2010;51:6321–6328. doi: 10.1167/iovs.10-5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hershenson M.B., Chao T.S., Abe M.K., Gomes I., Kelleher M.D., Solway J., Rosner M.R. Histamine antagonizes serotonin and growth factor-induced mitogen-activated protein kinase activation in bovine tracheal smooth muscle cells. J Biol Chem. 1995;270:19908–19913. doi: 10.1074/jbc.270.34.19908. [DOI] [PubMed] [Google Scholar]
- 36.Waltereit R., Weller M. Signaling from cAMP/PKA to MAPK and synaptic plasticity. Mol Neurobiol. 2003;27:99–106. doi: 10.1385/MN:27:1:99. [DOI] [PubMed] [Google Scholar]
- 37.Yamada S., Guo X., Wang K.Y., Tanimoto A., Sasaguri Y. Novel function of histamine signaling via histamine receptors in cholesterol and bile acid metabolism: histamine H2 receptor protects against nonalcoholic fatty liver disease. Pathol Int. 2016;66:376–385. doi: 10.1111/pin.12423. [DOI] [PubMed] [Google Scholar]
- 38.Wouters M.M., Balemans D., Van Wanrooy S., Dooley J., Cibert-Goton V., Alpizar Y.A., Valdez-Morales E.E., Nasser Y., Van Veldhoven P.P., Vanbrabant W., Van der Merwe S., Mols R., Ghesquiere B., Cirillo C., Kortekaas I., Carmeliet P., Peetermans W.E., Vermeire S., Rutgeerts P., Augustijns P., Hellings P.W., Belmans A., Vanner S., Bulmer D.C., Talavera K., Vanden Berghe P., Liston A., Boeckxstaens G.E. Histamine receptor H1-mediated sensitization of TRPV1 mediates visceral hypersensitivity and symptoms in patients with irritable Bowel syndrome. Gastroenterology. 2016;150:875–887.e9. doi: 10.1053/j.gastro.2015.12.034. [DOI] [PubMed] [Google Scholar]
- 39.Oquendo P., Alberta J., Wen D.Z., Graycar J.L., Derynck R., Stiles C.D. The platelet-derived growth factor-inducible KC gene encodes a secretory protein related to platelet alpha-granule proteins. J Biol Chem. 1989;264:4133–4137. [PubMed] [Google Scholar]
- 40.Asfaha S., Dubeykovskiy A.N., Tomita H., Yang X., Stokes S., Shibata W., Friedman R.A., Ariyama H., Dubeykovskaya Z.A., Muthupalani S., Ericksen R., Frucht H., Fox J.G., Wang T.C. Mice that express human interleukin-8 have increased mobilization of immature myeloid cells, which exacerbates inflammation and accelerates colon carcinogenesis. Gastroenterology. 2013;144:155–166. doi: 10.1053/j.gastro.2012.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee Y.S., Choi I., Ning Y., Kim N.Y., Khatchadourian V., Yang D., Chung H.K., Choi D., LaBonte M.J., Ladner R.D., Nagulapalli Venkata K.C., Rosenberg D.O., Petasis N.A., Lenz H.J., Hong Y.K. Interleukin-8 and its receptor CXCR2 in the tumour microenvironment promote colon cancer growth, progression and metastasis. Br J Cancer. 2012;106:1833–1841. doi: 10.1038/bjc.2012.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fukui H., Zhang X., Sun C., Hara K., Kikuchi S., Yamasaki T., Kondo T., Tomita T., Oshima T., Watari J., Imura J., Fujimori T., Sasako M., Miwa H. IL-22 produced by cancer-associated fibroblasts promotes gastric cancer cell invasion via STAT3 and ERK signaling. Br J Cancer. 2014;111:763–771. doi: 10.1038/bjc.2014.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ji Y., Yang X., Li J., Lu Z., Li X., Yu J., Li N. IL-22 promotes the migration and invasion of gastric cancer cells via IL-22R1/AKT/MMP-9 signaling. Int J Clin Exp Pathol. 2014;7:3694–3703. [PMC free article] [PubMed] [Google Scholar]
- 44.Kryczek I., Lin Y., Nagarsheth N., Peng D., Zhao L., Zhao E., Vatan L., Szeliga W., Dou Y., Owens S., Zgodzinski W., Majewski M., Wallner G., Fang J., Huang E., Zou W. IL-22(+)CD4(+) T cells promote colorectal cancer stemness via STAT3 transcription factor activation and induction of the methyltransferase DOT1L. Immunity. 2014;40:772–784. doi: 10.1016/j.immuni.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rokavec M., Oner M.G., Li H., Jackstadt R., Jiang L., Lodygin D., Kaller M., Horst D., Ziegler P.K., Schwitalla S., Slotta-Huspenina J., Bader F.G., Greten F.R., Hermeking H. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J Clin Invest. 2014;124:1853–1867. doi: 10.1172/JCI73531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waldner M.J., Foersch S., Neurath M.F. Interleukin-6–a key regulator of colorectal cancer development. Int J Biol Sci. 2012;8:1248–1253. doi: 10.7150/ijbs.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagasaki T., Hara M., Nakanishi H., Takahashi H., Sato M., Takeyama H. Interleukin-6 released by colon cancer-associated fibroblasts is critical for tumour angiogenesis: anti-interleukin-6 receptor antibody suppressed angiogenesis and inhibited tumour-stroma interaction. Br J Cancer. 2014;110:469–478. doi: 10.1038/bjc.2013.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.West N.R., McCuaig S., Franchini F., Powrie F. Emerging cytokine networks in colorectal cancer. Nat Rev Immunol. 2015;15:615–629. doi: 10.1038/nri3896. [DOI] [PubMed] [Google Scholar]
- 49.Watanabe S., Deguchi K., Zheng R., Tamai H., Wang L.X., Cohen P.A., Shu S. Tumor-induced CD11b+Gr-1+ myeloid cells suppress T cell sensitization in tumor-draining lymph nodes. J Immunol. 2008;181:3291–3300. doi: 10.4049/jimmunol.181.5.3291. [DOI] [PubMed] [Google Scholar]
- 50.Kennedy L., Hodges K., Meng F., Alpini G., Francis H. Histamine and histamine receptor regulation of gastrointestinal cancers. Transl Gastrointest Cancer. 2012;1:215–227. [PMC free article] [PubMed] [Google Scholar]
- 51.Cianchi F., Vinci M.C., Masini E. Histamine in cancer: the dual faces of the coin. Cancer Biol Ther. 2008;7:36–37. doi: 10.4161/cbt.7.1.5706. [DOI] [PubMed] [Google Scholar]
- 52.Garcia-Caballero M., Neugebauer E., Campos R., Nunez de Castro I., Vara-Thorbeck C. Increased histidine decarboxylase (HDC) activity in human colorectal cancer: results of a study on ten patients. Agents Actions. 1988;23:357–360. doi: 10.1007/BF02142587. [DOI] [PubMed] [Google Scholar]
- 53.Borges-Canha M., Portela-Cidade J.P., Dinis-Ribeiro M., Leite-Moreira A.F., Pimentel-Nunes P. Role of colonic microbiota in colorectal carcinogenesis: a systematic review. Rev Esp Enferm Dig. 2015;107:659–671. doi: 10.17235/reed.2015.3830/2015. [DOI] [PubMed] [Google Scholar]
- 54.Mira-Pascual L., Cabrera-Rubio R., Ocon S., Costales P., Parra A., Suarez A., Moris F., Rodrigo L., Mira A., Collado M.C. Microbial mucosal colonic shifts associated with the development of colorectal cancer reveal the presence of different bacterial and archaeal biomarkers. J Gastroenterol. 2015;50:167–179. doi: 10.1007/s00535-014-0963-x. [DOI] [PubMed] [Google Scholar]
- 55.O'Keefe S.J., Chung D., Mahmoud N., Sepulveda A.R., Manafe M., Arch J., Adada H., van der Merwe T. Why do African Americans get more colon cancer than Native Africans? J Nutr. 2007;137:175S–182S. doi: 10.1093/jn/137.1.175S. [DOI] [PubMed] [Google Scholar]
- 56.Moore W.E., Moore L.H. Intestinal floras of populations that have a high risk of colon cancer. Appl Environ Microbiol. 1995;61:3202–3207. doi: 10.1128/aem.61.9.3202-3207.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amirian E.S., Zhou R., Wrensch M.R., Olson S.H., Scheurer M.E., Il'yasova D., Lachance D., Armstrong G.N., McCoy L.S., Lau C.C., Claus E.B., Barnholtz-Sloan J.S., Schildkraut J., Ali-Osman F., Sadetzki S., Johansen C., Houlston R.S., Jenkins R.B., Bernstein J.L., Merrell R.T., Davis F.G., Lai R., Shete S., Amos C.I., Melin B.S., Bondy M.L. Approaching a scientific consensus on the association between allergies and glioma risk: a report from the Glioma International Case-Control Study. Cancer Epidemiol Biomarkers Prev. 2016;25:282–290. doi: 10.1158/1055-9965.EPI-15-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Negri E., Bosetti C., La Vecchia C., Levi F., Tomei F., Franceschi S. Allergy and other selected diseases and risk of colorectal cancer. Eur J Cancer. 1999;35:1838–1841. doi: 10.1016/s0959-8049(99)00209-9. [DOI] [PubMed] [Google Scholar]
- 59.Prizment A.E., Folsom A.R., Cerhan J.R., Flood A., Ross J.A., Anderson K.E. History of allergy and reduced incidence of colorectal cancer, Iowa Women's Health Study. Cancer Epidemiol Biomarkers Prev. 2007;16:2357–2362. doi: 10.1158/1055-9965.EPI-07-0468. [DOI] [PubMed] [Google Scholar]
- 60.Barcik W., Pugin B., Westermann P., Perez N.R., Ferstl R., Wawrzyniak M., Smolinska S., Jutel M., Hessel E.M., Michalovich D., Akdis C.A., Frei R., O'Mahony L. Histamine-secreting microbes are increased in the gut of adult asthma patients. J Allergy Clin Immunol. 2016;138:1491–1494.e7. doi: 10.1016/j.jaci.2016.05.049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative three-dimensional reconstructions by positron emission tomography (PET) imaging [fluorodeoxyglucose (FDG) signal] in healthy control mice that were gavaged with deMan, Rogosa, Sharpe (MRS) media and did not receive azoxymethane/dextran sulfate sodium (AOM/DSS) challenge.
Representative three-dimensional reconstructions by positron emission tomography (PET) imaging [fluorodeoxyglucose (FDG) signal] in control mice with disease that were gavaged with deMan, Rogosa, Sharpe (MRS) media and received azoxymethane/dextran sulfate sodium (AOM/DSS) challenge.
Representative three-dimensional reconstructions by positron emission tomography (PET) imaging [fluorodeoxyglucose (FDG) signal] in Lactobacillus reuteri–treated mice that were gavaged with L. reuteri 6475 and received azoxymethane/dextran sulfate sodium (AOM/DSS) challenge.
Representative three-dimensional reconstructions by positron emission tomography (PET) imaging [fluorodeoxyglucose (FDG) signal] in hdcA mutant–treated mice that were gavaged with hdcA mutant and received azoxymethane/dextran sulfate sodium (AOM/DSS) challenge.
Lactobacillus reuteri administration yields increased abundance of hdc gene and mRNA in the feces of experimental mice. A: Relative abundance of hdcA gene in mouse gut microbiome in different groups. B and C: Relative hdcA (B) and hdcP (C) gene expression levels in mouse feces. n = 5 for each group (A–C). ∗∗P < 0.01. AOM, azoxymethane, DSS, dextran sulfate sodium; MRS, deMan, Rogosa, Sharpe; PBS, phosphate-buffered saline.