Abstract
Glioblastoma stem-like cells (GSCs) are critical for the aggressiveness and progression of glioblastoma (GBM) and contribute to its resistance to adjuvant treatment. MicroRNAs (miRNAs) are small, non-coding RNAs controlling gene expression at the post-transcriptional level, which are known to be important regulators of the stem-like features. Moreover, miRNAs have been previously proved to be promising diagnostic biomarkers in several cancers including GBM. Using global expression analysis of miRNAs in 10 paired in-vitro as well as in-vivo characterized primary GSC and non-stem glioblastoma cultures, we identified a miRNA signature associated with the stem-like phenotype in GBM. 51 most deregulated miRNAs classified the cell cultures into GSC and non-stem cell clusters and identified a subgroup of GSC cultures with more pronounced stem-cell characteristics. The importance of the identified miRNA signature was further supported by demonstrating that a Risk Score based on the expression of seven miRNAs overexpressed in GSC predicted overall survival in GBM patients in the TCGA dataset independently of the IDH1 status. In summary, we identified miRNAs differentially expressed in GSCs and described their association with GBM patient survival. We propose that these miRNAs participate on GSC features and could represent helpful prognostic markers and potential therapeutic targets in GBM.
Introduction
Glioblastoma multiforme (GBM) is the most frequently occurring primary brain tumor of astrocytic origin in adults. Despite complex therapy consisting of maximal surgical resection, adjuvant concomitant chemoradiotherapy with temozolomide followed by temozolomide in monotherapy, the prognosis remains dismal1. The short survival of GBM patients is caused by both the impossibility of achieving “biologically” radical surgical resection and tumor resistance to adjuvant therapy. Glioblastoma stem-like cells (GSCs) are thought to be an important contributor to the poor response to the adjuvant therapy due to the higher expressions of the DNA repair enzymes, antiapoptotic factors, and multidrug transporters2,3. These rather slow proliferating cells are also capable of self -renewal and multilineage differentiation, are highly invasive, modulate immune response and promote angiogenesis. GSCs form gliomaspheres in serum-free media in vitro4,5 and have strong tumorigenic potential in immunodeficient animals recapitulating the hallmarks of the original tumors6. GSCs express, although to a variable extent, specific stemness markers such as the transcription factor Sox-2, the cytoskeletal protein nestin, and/or the cell surface glycoprotein CD1337,8, which are generally used for their identification. According to some studies, the presence of GSCs as determined by functional assays as well as the expression of GSC markers is associated with the prognosis in GBM patients9–12. Several studies have shown that microRNAs (miRNAs) are important molecular players closely related to the biological features of GSCs. MiRNAs are highly conserved, 18–25 nucleotide long non-coding RNAs that function as post-transcriptional regulators of gene expression by silencing their mRNA targets. It is estimated that miRNAs could regulate up to 60% of human genes including genes associated with the maintenance of the stem-like phenotype, differentiation, and chemo- and radioresistance13,14. Thus, miRNAs play significant roles in the functions of various types of healthy as well as cancer stem-like cells including GSCs15–17. Indeed, changes in miRNA expression were observed during the transition of GSCs to more differentiated phenotypes18 and e.g. the miR-302-367 cluster was shown to be able to abolish the stem cell characteristics of GSCs19. Our previous studies also demonstrated that miRNAs are able to predict the survival in GBM patients20,21.
In this study, we identified a set of miRNAs that is closely associated with the stem-like phenotype of GBM cells. We further corroborated the importance of the most differentially expressed miRNAs by showing their potential to predict overall survival in GBM patients independently of the IDH1 mutation status. These miRNAs may thus play an important role in the pathogenesis of brain tumors and represent potential therapeutic targets affecting GSCs and overcoming the therapeutic resistance of GBM.
Results
Characterization of the paired glioblastoma cell cultures
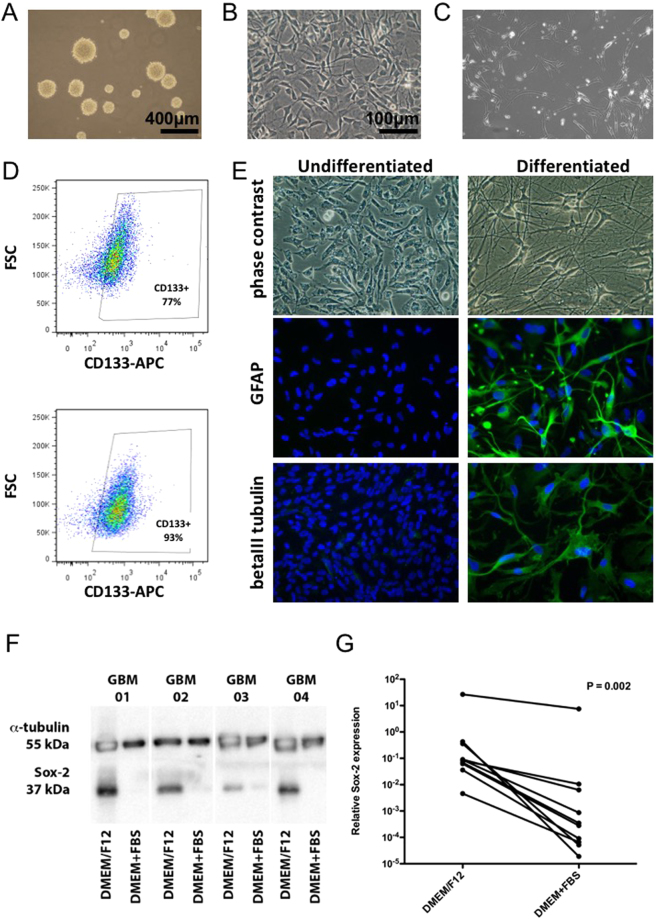
We successfully derived paired primary cell cultures from several GBMs (8 men and 2 women; median age 64 years - min 52, max 78 years), which were propagated in both defined serum-free medium favoring the expansion of GSCs and in medium supplemented with 10% FBS (non-stem cells). The cells cultured in serum-free medium initially formed gliomaspheres (Fig. 1A) and were subsequently propagated on laminin or geltrex (Fig. 1B). The matched paired primary cell cultures propagated in serum containing media grew adherently (Fig. 1C). The majority of the GSC cultures exhibited CD133 expression as determined by flow cytometry (Fig. 1D) and could undergo differentiation into GFAP and beta III tubulin expressing cells when transferred into serum containing media (Fig. 1E). All of the paired GBM cell cultures were IDH1/2 wild-type, their characteristics are summarized in Supplementary Table S1.
Figure 1.
Characterization of the primary GBM cell cultures propagated in serum-free and serum containing media. (A) Cells growing in serum-free medium as gliomaspheres, (B) cells growing in serum-free medium on laminin, (C) adherent cell growing in serum containing medium, (D) detection of CD133 in two independent serum-free medium cultures, (E) differentiation of serum-free medium cultured cells induced by 10% fetal calf serum, (F) western blot analysis of Sox-2 and α-tubulin and (G) qRT-PCR of Sox-2 expression in GBM cells propagated in serum-free (DMEM/F12) and serum containing (DMEM + FBS) media. The P value signifies the statistical significance of the difference between the paired primary cell lines as assessed by the Wilcoxon paired test.
Western blot (Fig. 1F) and qRT-PCR (P = 0.002; Wilcoxon paired test) (Fig. 1G) analyses revealed substantially higher Sox-2 expression in the cells cultured in serum-free conditions in comparison to the cells derived from the same patient sample propagated in serum containing medium. The expression of nestin, another stem cell marker, correlated with Sox-2 expression (r = 0.7955; P < 0.0001) and there was a trend for higher nestin expression in the GBM primary cells derived in serum-free conditions (P = 0.065; Wilcoxon paired test) (see Supplementary Figure S1).
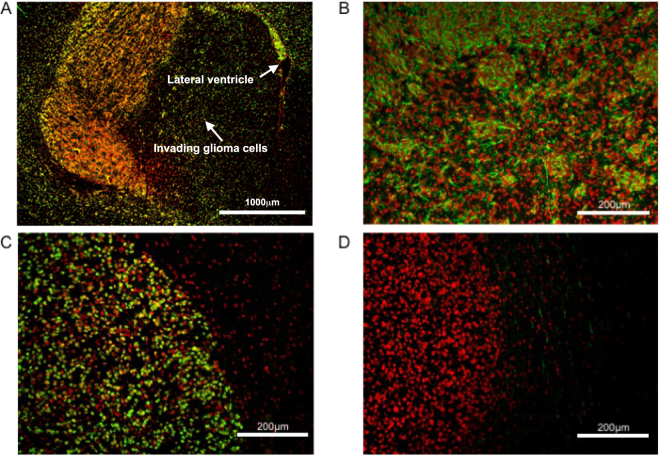
The large majority of the cultures propagated in serum-free as well as serum containing media were tumorigenic in immunodeficient mice (7/8 and 6/8 cultures, respectively). Nevertheless, the paired GBM cell cultures formed xenograft tumors with distinct features. The tumors derived from the glioma primary cell cultures propagated in serum containing media were characteristically well demarcated and GFAP negative (Fig. 2C,D). In contrast, the paired cultures derived in serum-free conditions typically produced GFAP positive tumors with single cell infiltration into the surrounding brain tissue including the contralateral hemisphere, white matter tracts and tropism towards the periventricular regions (Fig. 2A,B).
Figure 2.
Characteristics of orthotopic xenotransplants derived from primary GBM cell cultures propagated in serum-free (A,B) and serum containing (C,D) media. (A and C) Detection of human glioma cells by an antibody against human nuclei (in green). (B and D) Detection of GFAP expression (in green). ToPro3 was used for nuclear counterstaining (red).
In summary, we verified that the cell cultures derived in serum-free conditions have typical characteristics of glioma stem-like cells (GSCs).
MicroRNAs differentially expressed in the paired GSC and non-stem glioblastoma cell cultures
To identify a set of miRNAs characteristic for glioma stem-like cells we performed a genome-wide expression profiling of 2578 human miRNAs in the 10 paired GSC and non-stem GBM cell cultures derived in serum-free and serum supplemented medium, respectively. LIMMA analysis for paired samples revealed 431 significantly deregulated miRNAs in the GSCs in comparison with the non-stem GBM cells (P < 0.05) (see Supplementary Table S2). 51 miRNAs were deregulated at a significance level below 0.001 (25 miRNAs were upregulated and 26 miRNAs were downregulated). Importantly, among the 51 most deregulated miRNAs expression of 23 miRNAs correlated with Sox-2 expression at a significance level lower than 0.001, and 14 miRNAs correlated with both Sox-2 (P < 0.001) and nestin expression (P < 0.05) (Table 1). These data strongly suggest that several of the identified miRNAs are closely linked to the stemness of the glioma cell lines cultured in serum-free media.
Table 1.
MiRNAs correlating with Sox-2 and nestin expression in paired primary GBM cell cultures.
| miRNA | Sox-2 | Nestin | |||
|---|---|---|---|---|---|
| Spearman r | P value | Spearman r | P value | ||
| Negative correlation | miR-3195 | −0.85 | <10-5 | −0.60 | 0.006 |
| miR-3141 | −0.83 | <10-5 | −0.47 | 0.036 | |
| miR-4656 | −0.81 | <10-4 | −0.51 | 0.023 | |
| miR-100-5p | −0.79 | <10-4 | −0.39 | NS | |
| miR-4739 | −0.77 | <10-3 | −0.42 | NS | |
| miR-3180 | −0.75 | <10-3 | −0.55 | 0.013 | |
| miR-1260b | −0.75 | <10-3 | −0.46 | 0.043 | |
| miR-1233-5p | −0.74 | <10-3 | −0.49 | 0.029 | |
| miR-4674 | −0.73 | <10-3 | −0.54 | 0.015 | |
| miR-328-5p | −0.73 | <10-3 | −0.48 | 0.032 | |
| miR-378h | −0.72 | <10-3 | −0.48 | 0.034 | |
| miR-4505 | −0.71 | <10-3 | −0.46 | 0.045 | |
| miR-5787 | −0.71 | <10-3 | −0.47 | 0.036 | |
| miR-1207-5p | −0.70 | <10-3 | −0.37 | NS | |
| Positive correlation | miR-345-5p | 0.82 | <10-5 | 0.57 | 0.011 |
| miR-1180-3p | 0.78 | <10-4 | 0.45 | 0.048 | |
| miR-9-3p | 0.76 | <10-3 | 0.40 | NS | |
| miR-124-3p | 0.75 | <10-3 | 0.34 | NS | |
| miR-106b-3p | 0.73 | <10-3 | 0.42 | NS | |
| miR-1301-3p | 0.73 | <10-3 | 0.41 | NS | |
| miR-130b-3p | 0.71 | <10-3 | 0.49 | 0.029 | |
| miR-93-3p | 0.70 | <10-3 | 0.37 | NS | |
| miR-106b-5p | 0.70 | <10-3 | 0.32 | NS | |
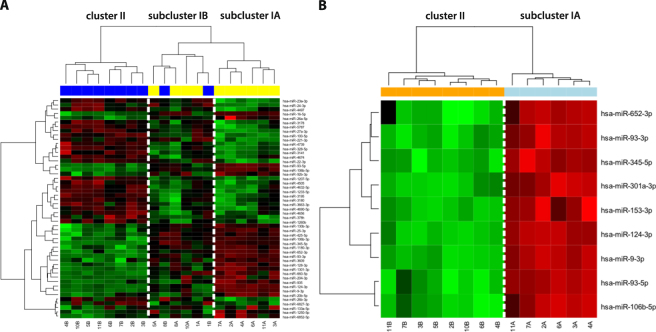
Cluster analysis based on the 51 most differentially expressed miRNAs correctly classified all GSC and 80% of the non-stem cell cultures (Fig. 3A). This analysis also revealed that the main cluster I containing all GSC samples was divided into two subclusters. Subcluster IA was exclusively composed of GSC cultures and the pattern of miRNA expression was more distinct from serum cultures contained in cluster II. All analyzed cultures in subcluster IA were tumorigenic and exhibited pronounced multilineage differentiation. Subcluster IB, which contained the remaining four serum-free derived GSC cultures, also included two serum derived non-stem cell cultures; moreover, the serum-free derived cultures in this subcluster exhibited only little differentiation when exposed to 10% serum and one of them did not form tumors in immunodeficient mice. Collectively, these data suggest somewhat less pronounced stemness characteristics of GSC cultures in subcluster IB (Table 2, Supplementary Table S1). Statistical analysis comparing only cell cultures from subcluster IA, which exhibited more pronounced stemness characteristics, and cluster II samples containing the non-stem cell cultures revealed nine miRNAs (miR-9-3p, miR-93-3p, miR-93-5p, miR-106b-5p, miR-124-3p, miR-153-3p, miR-301a-3p, miR-345-5p, and miR-652-3p), which were all upregulated in GSCs at a significance level below 0.0001 (Fig. 3B). Expression of all these miRNAs positively and statistically significantly correlated with Sox2 expression suggesting their close association with the stem cell-like phenotype of the GSCs.
Figure 3.
Hierarchical clustergram discriminating paired primary GSC (yellow) and non-stem glioblastoma cell cultures (blue) propagated in serum-free and serum containing medium, respectively. (A) Based on 51 differentially (P < 0.001) expressed miRNAs, (B)Based on 9 miRNAs differentially (P < 0.0001) expressed in GSC and non-stem cell cultures contained in subclusters IA and II, respectively. A gradient of green and red colors is used in the heatmap (green color indicates lower expression whereas red color indicates higher expression of individual miRNAs in analyzed samples).
Table 2.
Characteristics of the clusters identified based on the 51 most differentially expressed miRNAs.
| Cluster | No. of GSC cultures | No. of non-stem cell cultures | Multilineage differentiation‡ | Tumorigenicity |
|---|---|---|---|---|
| IA | 6 | 0 | 4/4** | 5/5 |
| IB | 4 | 2 | 0/3 | 2/3 (GSC) 1/2 (non-stem cells) |
| II | 0 | 8 | n.d. | 5/6 |
‡Number of cell cultures exhibiting pronounced differentiation in serum containing media/number of analyzed cell cultures, **P < 0.05, Pearson’s chi-squared test compared to cluster IB, n.d. - not determined.
MiRNAs differentially expressed in GSCs are associated with survival of GBM patients
To further support the potential importance of these miRNAs in GBM, we analyzed their relation to overall survival (OS) using the TCGA dataset comprising 485 GBM patients for whom OS and miRNA expression profiles were available. Seven out of nine of the miRNAs most differentially expressed between the GSC cluster IA and non-stem cell cluster II (Fig. 3B, miR-9-3p, miR-93-5p, miR-106b-5p, miR-153-3p, miR-301a-3p, miR-345-5p, and miR-652-3p) were represented in the TCGA dataset. First, we performed Z-score transformation on expression levels across all GBM samples for each of the aforementioned seven miRNAs; then, the seven-miRNA signature was used to calculate the Risk Score for each patient based on a linear combination of the miRNA expression level weighted by the regression coefficient derived from the multivariate Cox regression analysis22,23 as follows: risk score = 0.08270698 * hsa-miR-652 + 0.15074626 * hsa-miR-345 − 0.11310809 * hsa-miR-301 − 0.15450420 * hsa-miR-153 + 0.09238838 * hsa-miR-9* − 0.18565873 * hsa-miR-93 − 0.04894894 * hsa-miR-106b. This composite miRNA Risk Score was a statistically significant prognostic factor in the univariate Cox regression analysis (HR = 2.718; 95% CI (1.814−4.073), P < 1.26 * 10−6). Correspondingly, using the median value of the miRNA Risk Score as the threshold, GBM patients could be divided into a high-risk and a low-risk group. Kaplan–Meier analysis confirmed that OS of the high-risk patients was significantly lower in comparison with low-risk patients (P < 3.26 * 10−5, log-rank test) (Fig. 4). We further tested the prognostic power of the seven-miRNA signature with respect to the IDH1 mutation status in a subset comprising 296 GBM patients for whom exome somatic mutation data were available. The univariate Cox regression analysis revealed that the seven-miRNA signature predicted OS in these patients with higher statistical significance (P = 1.064 * 10−4; HR = 2.718; 95% CI (1.642–4.501)) in comparison with IDH1 status (P = 3.3 * 10−3; HR = 0.3147; 95% CI (0.1393–0.7109)). These results were underscored using the multivariate Cox regression analysis (P = 6.53 * 10−4; HR = 2.442; 95% CI (1.461–4.080) for the seven-miRNA signature and P = 1.67*10−2; HR = 0.367; 95% CI (0.162–0.834) for IDH1 status). The whole model based on the seven-miRNA signature and IDH1 status predicted OS in GBM patients with a P value 4.24 * 10−5 (see Supplementary Table S3). Importantly, the seven-miRNA signature was able to predict OS in both IDH1 wild-type (n = 280) and IDH1 mutated (n = 16) GBM patients (P < 1.01 * 10−3 and P < 4.62 * 10−2, respectively; log-rank test) (see Supplementary Figure S2).
Figure 4.
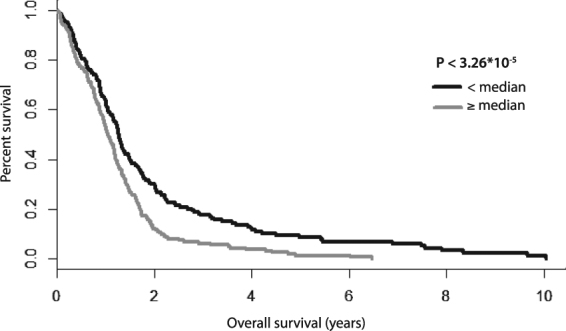
Kaplan-Meier survival curves estimating OS in GBM patients from the TCGA data set according to the 7-miRNA based Risk Score.
Discussion
Glioblastoma multiforme (GBM), the most common malignant primary brain tumor arising from glial cells, is associated with fatal prognosis caused not only by its localization in the central nervous system, but especially by the high invasiveness and resistance to conventional therapies. This biological behavior is associated with the cellular and molecular heterogeneity, which is characteristic for this disease24,25. Recent studies have suggested that GBM is driven and maintained by a subpopulation of clonogenic cells called glioblastoma stem-like cells (GSCs), which seem to play a crucial role in GBM biology26,27. These cells also contribute to GBM chemoradioresistance through the activation of DNA damage checkpoint responses and the increase in DNA repair capacity28,29. The small non-coding microRNAs (miRNAs) playing an important role in the posttranscriptional regulation of gene expression have been previously described in association with GBM initiation, progression, and resistance to therapy as well as with the maintenance of glioma stem-like cells30.
In this study, we firstly successfully derived and characterized paired GBM cell cultures from several GBMs, which were propagated either under defined serum-free conditions, or in serum containing medium. These differing cell culture conditions most likely lead to the isolation of distinct cell subpopulations from the original tumor. Consistently with the literature, the paired cell lines displayed profound biological differences31. Glioma cells cultured in serum-free conditions frequently expressed CD133, although as described by others6 the quantity was variable in individual cell lines. The serum-free medium cultured cells also showed the potential to form gliomaspheres and differentiate into GFAP and beta III tubulin positive cells. In comparison with the paired glioma cultures derived in serum containing medium, the serum-free medium derived cells expressed significantly more Sox-2, the stemness marker crucial for the tumorigenicity of GSCs32, on both mRNA and protein levels. As previously reported31, the serum-free cultured cells characteristically formed highly infiltrative tumors when implanted into immunodeficient mice.
Having established that the cell lines cultured under serum-free conditions used in this study exhibit features typical of GSC, the paired GSC and non-stem cell cultures were utilized to uncover the miRNA expression pattern specific for GSC. Using global miRNA expression analysis, we revealed 51 most differentially expressed miRNAs. These miRNAs were able to classify the cell cultures into non-stem cell cluster II and two GSC subclusters IA and IB (Fig. 3A). Analysis of these two GSC subclusters showed that the first of them (IA) consisted of cultures with more pronounced GSC features compared to the second subcluster (IB) containing among others also two non-stem cell cultures. Subsequent analysis focusing on the miRNAs most differentially expressed between the GSC cluster IA and the non-stem cell cluster II highlighted nine miRNAs (miR-9-3p, miR-93-3p, miR-93-5p, miR-106b-5p, miR-124-3p, miR-153-3p, miR-301a-3p, miR-345-5p, and miR-652-3p) that were strongly upregulated in GSCs. Several of these miRNAs were previously described to be associated with the regulation of the stemness maintenance as well as with the biological behavior of GBM and survival of patients. MiR-9-3p (referred to as miR-9*) and its hairpin counterpart miR-9-5p (referred to as miR-9), which was also upregulated in GSCs though with lower statistical significance (p < 3.3 * 10−3; log2 FC = 2.1), seem to be specifically expressed in the brain33,34, are evolutionary conserved from insects to human and are involved in vertebrate neural development35–37. Their activities are probably carried out through the effect on the Notch signaling pathway, especially by the targeting of Notch2 and the transcription factor Hes1, resulting in an enhanced differentiation and proliferation of neural stem cells (NSCs)38,39. These data seem to be somewhat contradictory to our findings as we observed higher expression of both miRNAs in GSCs. The explanation may lie in the mutual regulation of miR-9/9* and Notch signaling. It was demonstrated that expression levels of miR-9/9* depend on the activation status of Notch signaling. While Notch inhibits differentiation of NSCs, it also induces miR-9/9*38. Moreover, Hes1 expression oscillates with a period of 2–3 hours in NSCs and this oscillation is important not only for cell differentiation but also for proliferation as sustained Hes1 expression inhibits both processes39,40. Thus, it can be assumed that miR-9/9* expression levels vary in time to allow cell proliferation. It also seems that the control mechanisms in GSCs are different from those in NSCs. In line with our findings, Schraivogel et al. reported that both miRNAs were highly abundant in CD133+ GSCs and their inhibition led to the reduced neurosphere formation and stimulated cell differentiation41. Finally, the higher levels of miR-9/9* hairpin counterparts in GSCs could also contribute to the increased resistance of these cells to the conventional therapy. Munoz et al. recently showed that CD133+ GSCs expressed greater levels of miR-9 which led to the activation of the SHH/PTCH1/MDR1 axis. This axis has been shown to impart TMZ resistance. In the case of the CD133+ cells, the resistance is not acquired but seems to be inherent42.
Interestingly, miR-9 and miR-9* seem to be functionally linked with the miR-124, another most differentially expressed miRNA identified in our study. Staahl et al. published that mitotic exit in neurogenesis is inter alia partially driven by these three matured miRNAs43. Another study described the synergistic effect of miR-9 and miR-124 on the strong suppression of the GTP-binding protein Rap2a and consequent promotion of neuronal differentiation of NSCs and dendritic branching of differentiated neurons44. A very similar effect of the overexpression of miR-9/9* and miR-124 on the self-renewal and differentiation was observed by Roese-Koerner et al. in neuroepithelial-like stem cells derived from human pluripotent stem cells40.
MiR-106b-5p, miR-93-5p as well as miR-93-3p are members of the same miRNA gene cluster miR-106b~25 and it is thus not surprising that these miRNAs were jointly upregulated. Interestingly, this cluster seems to be linked to the biology of stem cells. Serum induced differentiation of GSCs was previously demonstrated to decrease the expression levels of miRNAs which belong to this cluster18 and Brett et al. showed that the expression of the entire miR-106b~25 cluster in adult mouse neuronal stem/progenitor cells increases their ability to generate new neurons45. In the CD44+ gastric cancer stem-like cells, the entire cluster was significantly upregulated and inhibition of miR-106b led to a decreased self-renewal capacity and cell invasiveness through the suppression of the TGF-β/Smad signaling pathway46.
Only few studies suggesting a direct link between the other miRNAs identified in our study and stem cell biology are available so far. Chang et al. described miR-345 to be enriched in mesenchymal stem cells found in Wharton’s jelly matrix of human umbilical cord which were able to transdifferentiate into neuronal lineage cells47. The miR-301 family has been recently shown to be the direct target of the SFRS2 splicing factor, an OCT4 regulated gene required for the pluripotency in human pluripotent stem cells48. Stappert et al. demonstrated that miR-153 contributes to the shift of long-term self-renewing neuroepithelial-like stem cells from self-renewal to neuronal differentiation49. Similarly, Tezcan et al. also demonstrated that miR-153 overexpression reduced tumorigenic capacity of GSCs by targeting the Nrf-2/GPx1/ROS pathway50. In contrast with these data, we observed higher expression of both miR-153 and the members of the miR-301 family (miR-301a-3p, miR-130b-3p, and miR-130a-3p) in GSCs. Unfortunately, there are no studies which could help to explain these discrepancies. However, it seems that the stem cell-like state is dependent on many interacting molecules in feedback loops mutually balancing one another over time.
The Risk Score utilized in this study proved that the set of the identified miRNAs is associated with GBM prognosis independently of IDH1 mutation status, further suggesting their involvement in the disease pathogenesis. Higher tissue levels of miR-652, miR-345 and miR-9* positively contributed to increased values of the risk score and thus worse prognosis; nevertheless, miR-301, miR-153, miR-93 and miR-106b which were also upregulated in GSCs were in fact negative contributors. The explanation for these results is at present speculative but may involve the following aspects. Firstly, the relation between GSCs and survival is somewhat unclear as various studies failed to show a correlation between the GSC quantity assessed by CD133, nestin or CD15 staining and survival51,52. Further, Pallini et al. observed that the percentage of CD133-positive cells somewhat paradoxically correlated with longer survival in recurrent glioblastoma, likely due to the higher presence of normal neural stem cells with possible antitumor properties, which may also apply to some newly diagnosed tumors53. Another study described that the CD133-low GBMs showed more invasive growth and gene expression profiles characteristic of mesenchymal or proliferative subtypes, whereas the CD133-high GBMs showed features of cortical and well-demarcated tumors and gene expressions typical of proneuronal subtypes. Moreover, in contrast to various reports claiming that only CD133-positive GBM cells can initiate tumor formation in vivo, Joo et al. showed that CD133-negative cells also possess tumor-initiating potential54. It should be further noted that CSCs are rather quiescent and slow-cycling, and some of the identified miRNAs may be regulators of this dormant state55. Last but not least, non-transformed stromal cells may contribute to the tissue levels of miRNAs and the function of the particular miRNAs may be different in these cells compared to glioma cells.
Taken together, we identified a set of miRNAs that are differentially expressed in GSCs as compared to non-stem glioblastoma cells, several of which correlated with the expression of the stem cell markers Sox-2 and nestin. Our findings thus suggest that a complex set of miRNAs is involved in the regulation of the stem-like characteristics in glioblastoma. Moreover, a set of the most differentially expressed miRNAs correlated with the survival of GBM patients independently of the IDH1 status indicating that they might be prognostic markers and possibly new therapeutic targets.
Material and Methods
GBM samples and primary cell cultures
The 10 paired primary GBM cell cultures were derived from fresh tumor tissues samples obtained from GBM patients who underwent surgically resection at the Departments of Neurosurgery of the Hospital Na Homolce in Prague, University Hospital Brno, and University Hospital Ostrava. Study has been approved by the local Ethical Board at Hospital Na Homolce in Prague, University Hospital Brno, and University Hospital Ostrava. Written informed consent was obtained from all patients included in the study. All experiments were performed in accordance with relevant guidelines and regulations.
The fresh tissue sample was enzymatically dissociated with TrypLE (ThermoFisher Scientific) for 20 min at 37 °C with agitation or using the Papain Dissociation System (Worthington) according to manufacturer’s instructions. Single cell suspensions were seeded into 25 cm2 tissue culture flasks (Techno Plastic Products AG) and cultured in either Dulbecco’s modified essential medium supplemented with 10% FBS, 1% Glutamax (both ThermoFisher Scientific), 100 U/mL penicillin and 100 μg/mL streptomycin, sodium pyruvate and non-essential amino acids (all GE Healthcare), or in DMEM/F12 containing bFGF 20 ng/mL, EGF 20 ng/mL (both PeproTech), B27-supplement 1:50 (ThermoFisher Scientific), 1% Glutamax, 100 U/mL penicillin and 100 μg/mL streptomycin. After 1–3 weeks, adherent cells, which covered more than 2/3 of the culture flask in DMEM, were passaged using Trypsin-EDTA solution (Sigma-Aldrich). After approximately the same time, tumor spheres formed in DMEM/F12, and these were dissociated using Accutase (Sigma-Aldrich) and up and down pipetting and then passaged. Cells that initially formed spheres were dissociated and transferred to laminin (Sigma-Aldrich) or Geltrex (ThermoFisher Scientific) coated culture flasks and propagated as monolayer cultures56,57. For the subsequent analyses, early passage cultures were used.
qRT-PCR quantification
Complementary DNA (cDNA) was synthesized from 1000 ng small RNA enriched total RNA using the High Capacity cDNA Reverse Transcription Kit (ThermoFisher Scientific) according to the manufacturer’s protocol. qRT-PCR was performed using the LightCycler 480 Instrument II (Roche) in accordance with the standard TaqMan manufacturer’s protocol using TaqMan Gene Expression Assays (GAPDH #Hs03929097_g1, SOX2 #Hs01053049_s1, NES #Hs00707120_s1; ThermoFisher Scientific). The data were evaluated using the second derivative maximum method with the arithmetic baseline adjustment. All qRT-PCR reactions were run in triplicate and average Cp and SD values were calculated. Relative expression levels were determined by the 2−ΔCp method, where ΔCp was calculated as follows: ΔCp = Cp (gene of interest) − Cp (GAPDH).
Western blot analysis
Cell pellets were lysed with RIPA buffer (Sigma-Aldrich). Protein concentration was measured using a DC Protein Assay (Bio-Rad), samples were diluted with RIPA buffer to attain the same concentration of total protein and boiled for 10 min with the Laemmli sample buffer. Proteins (15 μg per well) were separated on 10% SDS-PAGE gels, and electrophoretically transferred to the polyvinylidene difluoride (PVDF) membrane (Merck Millipore). The membranes were blocked with 5% nonfat milk in PBS with 0.1% Tween 20 (PBS-T), then incubated either with an anti-Sox2 rabbit mAb or anti-alpha/beta-tubulin rabbit mAb (No. 3579 and 2148, respectively; both Cell Signaling Technology) diluted 1:1000 in blocking solution at 4 °C overnight. Subsequently, the membranes were incubated with anti-rabbit IgG antibody HRP conjugate (No. 7074, Cell Signaling Technology) diluted 1:2500 (60 min/RT). Each step was followed by washes in PBS-T. ECL-Plus detection was performed according to the manufacturer’s instructions (Amersham).
Flow cytometry
Accutase (Sigma-Aldrich) was used to harvest adherent cells and dissociate gliomaspheres. The cell suspension was fixed with 2% paraformaldehyde for 1 hour at 4 °C, permeabilized (Intracellular Staining Permeabilization Wash Buffer, Biolegend) and stained using an anti-CD133 APC conjugated antibody (Miltenyi Biotec).
Differentiation of stem-like cell cultures, immunocytochemistry
To induce differentiation, 13 × 103 cells per cm2 were plated on geltrex coated coverslips and cultured in differentiation medium (DMEM/F12, 10% FBS, 1% Glutamax, 100 μg/mL Streptomycin and 100 U/mL Penicillin G). The medium was exchanged every 2–3 days for 10–14 days. The coverslips were subsequently fixed with 4% paraformaldehyde (10 minutes at room temperature) and stained overnight at 4 °C using the antibodies against GFAP (GF-01, Exbio, 1:200) and beta III tubulin (Exbio, 1:250).
Orthotopic xenotransplantation glioma model, immunohistochemistry
The experimental use of animals was approved by The Commission for Animal Welfare of the First Faculty of Medicine of the Charles University in Prague and The Ministry of Education, Youth and Sports of the Czech Republic according to the animal protection laws. Generation of xenotrasplants was performed as described previously58. 5 × 105 cells in 5 µL of serum free medium were injected into 6–10 week old male NOD.129S7(B6)-Rag1tm1Mom/J mice (The Jackson Laboratory) with a Hamilton syringe 1.2 mm anterior from the bregma and 2.5 mm lateral from the midline to a depth of 3 mm using a stereotactic device (Stoelting Co.). Immunohistochemistry was performed on 10 μm thick frozen sections using antibodies against human nuclei (Chemicon, 1:500) and GFAP (GF-01, Exbio, 1:200) as described59.
MiRNA microarray analysis
Small RNA enriched total RNA was isolated using the mirVana miRNA Isolation Kit (ThermoFisher Scientific). Nucleic acid concentrations and purities were controlled by UV spectrophotometry using Nanodrop ND-1000 (Thermo Scientific). To assess miRNA expression in Sox-2 high- and low-expressing GBM cells, the samples were analyzed with Affymetrix GeneChip miRNA 4.0 arrays (Affymetrix) containing 5607 probe sets for human small RNAs. Out of these probe sets the 2578 probe sets of human mature miRNAs were filtered. All steps of the procedure were performed according to the Affymetrix standardized protocol for miRNA 4.0 arrays. Intensity values for each probe cell (.cel file) were calculated using Affymetrix GeneChip Command Console (AGCC). Quality control of the microarray was performed with the Affymetrix miRNA QC Tool, version 1.1.1.0.
Microarray expression data analysis
All data were pre-processed and further analyzed by the software packages included in the R/Bioconductor60. Pre-processing was performed by the RMA method with default parameters as implemented in the Bioconductor package oligo61. All data were log2-transformed. To identify differentially expressed miRNAs, the LIMMA approach62 for paired samples was applied with additional Benjamini-Hochberg correction of P values. To determine the correlation between miRNA and Sox-2 or nestin expression, the Spearman rank correlation coefficient was used.
IDH1/2 mutation status analysis
DNA was extracted using QIAamp DNA Mini Kit (Qiagen, Germany) according to manufacturer’s instructions. Fragments of 254 bp and 293 bp lengths spanning the sequences encoding the catalytic domains of IDH1 including codon 132 and IDH2 including codon 172, respectively were amplified using 12,5 pmol each of the primers: IDH1-F ACCAAATGGCACCATACGA, IDH1-R TTCATACCTTGCTTAATGGGTGT, IDH2-F GCTGCAGTGGGACCACTATT, and IDH2-R TGTGGCCTTGTACTGCAGAG (primer sequences according to Hartmann et al., 2009). PCR was performed using standard buffer conditions, 50–250 ng of DNA input and Taq DNA Polymerase (Invitrogen, USA). PCR consisted of 35 cycles with denaturing at 95 °C for 30 s, annealing at 56 °C for 1 min and extension at 72 °C for 1 min in a total volume of 25 μl. Two microliters of the PCR amplification product were subjected to sequencing using the BigDye Terminator v3.1 Sequencing Kit (Applied Biosystems, USA). Twenty-five cycles were performed employing 0,5 μl of 10 μM primer IDH1-F ACCAAATGGCACCATACGA or IDH2-R TGTGGCCTTGTACTGCAGAG, with denaturing at 96 °C for 30 s, annealing at 50 °C for 15 s and extension at 60 °C for 4 min in a total volume of 10 μl. Sequences were determined using the sequencer (ABI 3500 Genetic Analyzer, Applied Biosystems) and the Mutation surveyor V4.0.9 software (SoftGenetics, USA).
Survival analysis
The relationship between overall survival and expression levels of the selected miRNAs was analyzed on The Cancer Genome Atlas (TCGA) data set (485 GBM patients)63. To assess the miRNAs that were identified in this study for survival prediction, a Risk Score formula for predicting survival was developed based on a linear combination of the miRNA expression level weighted by the regression coefficient derived from the multivariate Cox regression analysis22,23. Patients with high Risk Score are expected to have poor survival. Cox proportional hazards regression analysis was performed to assess the contribution of the miRNA signature to survival prediction64. Patients were further stratified into a high-risk group and a low-risk group according to the Risk Score (cutoff value median) and survival was analyzed using the Kaplan-Meier method. Subsequently, both univariate and multivariate Cox regression analyses including seven-miRNA signature and IDH1 status were performed on the data subset comprising 296 GBM patients for whom exome somatic mutation data were available.
Electronic supplementary material
Acknowledgements
This work was financially supported by the Czech Ministry of Health grants no. 15-34553A, 15-33158A, and 15-31379A; by the project MZ CR – RVO (MOU, 00209805); Grant No. LM2015064 of the EATRIS-CZ.; by the Ministry of Education, Youth and Sports of the Czech Republic under the project CEITEC 2020 (LQ1601), and by the projects UNCE 204013 and Progres Q28/1LFUK. We also acknowledge the cooperation of the staff at the Nemocnice na Homolce hospital during tissue sampling, and Kvetoslava Vlasicova and Karin Roubickova for excellent technical assistance.
Author Contributions
Conception and design: J. Sana, P. Busek, A. Sedo, O. Slaby; Development of methodology: J. Sana, P. Busek; Acquisition of data (acquired and managed patients, laboratory measurements, provided facilities, etc.): J. Sana, P. Busek, P. Fadrus, A. Besse, M. Vecera, S. Reguli, L. Sromova, M. Hilser, R. Lipina, R. Lakomy, L. Kren, M. Smrcka; Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): L. Radova, J. Sana, P. Busek, O. Slaby, A. Besse; Study supervision: O. Slaby, A. Sedo; Writing, review, and/or revision of the manuscript: J. Sana, P. Busek, O. Slaby, A. Sedo.
Competing Interests
The authors declare no competing interests.
Footnotes
Jiri Sana and Petr Busek contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-20929-6.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Aleksi Sedo, Email: aleksi@cesnet.cz.
Ondrej Slaby, Email: on.slaby@gmail.com.
References
- 1.Stupp R, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CLL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29:1203–1217. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson M, Hassiotou F, Nowak A. Glioblastoma stem-like cells: at the root of tumor recurrence and a therapeutic target. Carcinogenesis. 2015;36:177–185. doi: 10.1093/carcin/bgu243. [DOI] [PubMed] [Google Scholar]
- 4.Rahman M, et al. Neurosphere and adherent culture conditions are equivalent for malignant glioma stem cell lines. Anat. Cell Biol. 2015;48:25–35. doi: 10.5115/acb.2015.48.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pavon, L. F. et al. In vitro Analysis of Neurospheres Derived from Glioblastoma Primary Culture: A Novel Methodology Paradigm. Front. Neurol. 4 (2014). [DOI] [PMC free article] [PubMed]
- 6.Wakimoto H, et al. Maintenance of primary tumor phenotype and genotype in glioblastoma stem cells. Neuro-Oncol. 2012;14:132–144. doi: 10.1093/neuonc/nor195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iacopino F, et al. Isolation of Cancer Stem Cells from Three Human Glioblastoma Cell Lines: Characterization of Two Selected Clones. PLoS ONE. 2014;9:e105166. doi: 10.1371/journal.pone.0105166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brescia P, et al. CD133 is essential for glioblastoma stem cell maintenance. Stem Cells Dayt. Ohio. 2013;31:857–869. doi: 10.1002/stem.1317. [DOI] [PubMed] [Google Scholar]
- 9.Zhang, W., Chen, H., Lv, S. & Yang, H. High CD133 Expression Is Associated with Worse Prognosis in Patients with Glioblastoma. Mol. Neurobiol, 10.1007/s12035-015-9187-1 (2015). [DOI] [PubMed]
- 10.Dahlrot RH, et al. Clinical value of CD133 and nestin in patients with glioma: a population-based study. Int. J. Clin. Exp. Pathol. 2014;7:3739–3751. [PMC free article] [PubMed] [Google Scholar]
- 11.Miconi G, et al. Immunophenotypic characterization of human glioblastoma stem cells: correlation with clinical outcome. J. Cell. Biochem. 2015;116:864–876. doi: 10.1002/jcb.25043. [DOI] [PubMed] [Google Scholar]
- 12.Kong BH, et al. Prognostic value of glioma cancer stem cell isolation in survival of primary glioblastoma patients. Stem Cells Int. 2014;2014:838950. doi: 10.1155/2014/838950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Virant-Klun I, Ståhlberg A, Kubista M, Skutella T. MicroRNAs: From Female Fertility, Germ Cells, and Stem Cells to Cancer in Humans. Stem Cells Int. 2016;2016:3984937. doi: 10.1155/2016/3984937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Besse A, Sana J, Fadrus P, Slaby O. MicroRNAs involved in chemo- and radioresistance of high-grade gliomas. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2013;34:1969–1978. doi: 10.1007/s13277-013-0772-5. [DOI] [PubMed] [Google Scholar]
- 15.Yang HW, Xing H, Johnson MD. A major role for microRNAs in glioblastoma cancer stem-like cells. Arch. Pharm. Res. 2015;38:423–434. doi: 10.1007/s12272-015-0574-y. [DOI] [PubMed] [Google Scholar]
- 16.Kleinová R, Slabý O, Šána J. [The Relevance of MicroRNAs in GlioblastomaStem Cells] Klin. Onkol. Cas. Ceské Slov. Onkol. Spolecnosti. 2015;28:338–344. doi: 10.14735/amko2015338. [DOI] [PubMed] [Google Scholar]
- 17.Brower JV, Clark PA, Lyon W, Kuo JS. MicroRNAs in cancer: glioblastoma and glioblastoma cancer stem cells. Neurochem. Int. 2014;77:68–77. doi: 10.1016/j.neuint.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aldaz B, et al. Involvement of miRNAs in the differentiation of human glioblastoma multiforme stem-like cells. PloS One. 2013;8:e77098. doi: 10.1371/journal.pone.0077098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fareh M, et al. The miR 302-367 cluster drastically affects self-renewal and infiltration properties of glioma-initiating cells through CXCR4 repression and consequent disruption of the SHH-GLI-NANOG network. Cell Death Differ. 2012;19:232–244. doi: 10.1038/cdd.2011.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sana J, et al. Risk Score based on microRNA expression signature is independent prognostic classifier of glioblastoma patients. Carcinogenesis. 2014;35:2756–2762. doi: 10.1093/carcin/bgu212. [DOI] [PubMed] [Google Scholar]
- 21.Lakomy R, et al. MiR-195, miR-196b, miR-181c, miR-21 expression levels and O-6-methylguanine-DNA methyltransferase methylation status are associated with clinical outcome in glioblastoma patients. Cancer Sci. 2011;102:2186–2190. doi: 10.1111/j.1349-7006.2011.02092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Therneau T. Modeling Survival Data: Extending the Cox Model. Springer, New York 2000.
- 23.Therneau T. A Package for Survival Analysis in S. R Package Version 2.37-6, http://CRAN.R-project.org/package=survival (30 October 2014, date last accessed) 2014.
- 24.Aum DJ, et al. Molecular and cellular heterogeneity: the hallmark of glioblastoma. Neurosurg. Focus. 2014;37:E11. doi: 10.3171/2014.9.FOCUS14521. [DOI] [PubMed] [Google Scholar]
- 25.Eder K, Kalman B. Molecular heterogeneity of glioblastoma and its clinical relevance. Pathol. Oncol. Res. POR. 2014;20:777–787. doi: 10.1007/s12253-014-9833-3. [DOI] [PubMed] [Google Scholar]
- 26.Bayin NS, Modrek AS, Placantonakis DG. Glioblastoma stem cells: Molecular characteristics and therapeutic implications. World J. Stem Cells. 2014;6:230–238. doi: 10.4252/wjsc.v6.i2.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin C, Lv S, Chen X, Guo H. The role of glioma stem cells in glioma tumorigenesis. Front. Biosci. Landmark Ed. 2014;19:818–824. doi: 10.2741/4249. [DOI] [PubMed] [Google Scholar]
- 28.Bao S, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 29.Cheng L, et al. L1CAM regulates DNA damage checkpoint response of glioblastoma stem cells through NBS1. EMBO J. 2011;30:800–813. doi: 10.1038/emboj.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Floyd D, Purow B. Micro-masters of glioblastoma biology and therapy: increasingly recognized roles for microRNAs. Neuro-Oncol. 2014;16:622–627. doi: 10.1093/neuonc/nou049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 32.Gangemi RMR, et al. SOX2 silencing in glioblastoma tumor-initiating cells causes stop of proliferation and loss of tumorigenicity. Stem Cells Dayt. Ohio. 2009;27:40–48. doi: 10.1634/stemcells.2008-0493. [DOI] [PubMed] [Google Scholar]
- 33.Yuva-Aydemir Y, Simkin A, Gascon E, Gao F-B. MicroRNA-9: functional evolution of a conserved small regulatory RNA. RNA Biol. 2011;8:557–564. doi: 10.4161/rna.8.4.16019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lagos-Quintana M, et al. Identification of tissue-specific microRNAs from mouse. Curr. Biol. CB. 2002;12:735–739. doi: 10.1016/S0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 35.Bonev B, Pisco A, Papalopulu N. MicroRNA-9 reveals regional diversity of neural progenitors along the anterior-posterior axis. Dev. Cell. 2011;20:19–32. doi: 10.1016/j.devcel.2010.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bonev B, Stanley P, Papalopulu N. MicroRNA-9 Modulates Hes1 ultradian oscillations by forming a double-negative feedback loop. Cell Rep. 2012;2:10–18. doi: 10.1016/j.celrep.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coolen M, Thieffry D, Drivenes Ø, Becker TS, Bally-Cuif L. miR-9 controls the timing of neurogenesis through the direct inhibition of antagonistic factors. Dev. Cell. 2012;22:1052–1064. doi: 10.1016/j.devcel.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 38.Roese-Koerner B, et al. Reciprocal Regulation between Bifunctional miR-9/9(*) and its Transcriptional Modulator Notch in Human Neural Stem Cell Self-Renewal and Differentiation. Stem Cell Rep. 2016;7:207–219. doi: 10.1016/j.stemcr.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan S-L, Ohtsuka T, González A, Kageyama R. MicroRNA9 regulates neural stem cell differentiation by controlling Hes1 expression dynamics in the developing brain. Genes Cells Devoted Mol. Cell. Mech. 2012;17:952–961. doi: 10.1111/gtc.12009. [DOI] [PubMed] [Google Scholar]
- 40.Roese-Koerner B, Stappert L, Koch P, Brüstle O, Borghese L. Pluripotent stem cell-derived somatic stem cells as tool to study the role of microRNAs in early human neural development. Curr. Mol. Med. 2013;13:707–722. doi: 10.2174/1566524011313050003. [DOI] [PubMed] [Google Scholar]
- 41.Schraivogel D, et al. CAMTA1 is a novel tumour suppressor regulated by miR-9/9* in glioblastoma stem cells. EMBO J. 2011;30:4309–4322. doi: 10.1038/emboj.2011.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munoz, J. L., Rodriguez-Cruz, V. & Rameshwar, P. High expression of miR-9 in CD133(+) glioblastoma cells in chemoresistance to temozolomide. J. Cancer Stem Cell Res. 3 (2015). [DOI] [PMC free article] [PubMed]
- 43.Staahl BT, et al. Kinetic analysis of npBAF to nBAF switching reveals exchange of SS18 with CREST and integration with neural developmental pathways. J. Neurosci. Off. J. Soc. Neurosci. 2013;33:10348–10361. doi: 10.1523/JNEUROSCI.1258-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xue Q, et al. miR-9 and miR-124 synergistically affect regulation of dendritic branching via the AKT/GSK3β pathway by targeting Rap2a. Sci. Rep. 2016;6:26781. doi: 10.1038/srep26781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brett JO, Renault VM, Rafalski VA, Webb AE, Brunet A. The microRNA cluster miR-106b~25 regulates adult neural stem/progenitor cell proliferation and neuronal differentiation. Aging. 2011;3:108–124. doi: 10.18632/aging.100285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu D, Shin H-S, Lee YS, Lee YC. miR-106b modulates cancer stem cell characteristics through TGF-β/Smad signaling in CD44-positive gastric cancer cells. Lab. Investig. J. Tech. Methods Pathol. 2014;94:1370–1381. doi: 10.1038/labinvest.2014.125. [DOI] [PubMed] [Google Scholar]
- 47.Chang S-J, et al. MicroRNA-34a modulates genes involved in cellular motility and oxidative phosphorylation in neural precursors derived from human umbilical cord mesenchymal stem cells. BMC Med. Genomics. 2011;4:65. doi: 10.1186/1755-8794-4-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu Y, et al. Alternative splicing of MBD2 supports self-renewal in human pluripotent stem cells. Cell Stem Cell. 2014;15:92–101. doi: 10.1016/j.stem.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stappert L, et al. MicroRNA-based promotion of human neuronal differentiation and subtype specification. PloS One. 2013;8:e59011. doi: 10.1371/journal.pone.0059011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tezcan G, et al. microRNA expression pattern modulates temozolomide response in GBM tumors with cancer stem cells. Cell. Mol. Neurobiol. 2014;34:679–692. doi: 10.1007/s10571-014-0050-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Melguizo C, et al. MGMT promoter methylation status and MGMT and CD133 immunohistochemical expression as prognostic markers in glioblastoma patients treated with temozolomide plus radiotherapy. J. Transl. Med. 2012;10:250. doi: 10.1186/1479-5876-10-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim K-J, et al. The presence of stem cell marker-expressing cells is not prognostically significant in glioblastomas. Neuropathol. Off. J. Jpn. Soc. Neuropathol. 2011;31:494–502. doi: 10.1111/j.1440-1789.2010.01194.x. [DOI] [PubMed] [Google Scholar]
- 53.Pallini R, et al. Expression of the stem cell marker CD133 in recurrent glioblastoma and its value for prognosis. Cancer. 2011;117:162–174. doi: 10.1002/cncr.25581. [DOI] [PubMed] [Google Scholar]
- 54.Joo KM, et al. Clinical and biological implications of CD133-positive and CD133-negative cells in glioblastomas. Lab. Investig. J. Tech. Methods Pathol. 2008;88:808–815. doi: 10.1038/labinvest.2008.57. [DOI] [PubMed] [Google Scholar]
- 55.Li L, Bhatia R. Stem cell quiescence. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2011;17:4936–4941. doi: 10.1158/1078-0432.CCR-10-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fael Al-Mayhani TM, et al. An efficient method for derivation and propagation of glioblastoma cell lines that conserves the molecular profile of their original tumours. J. Neurosci. Methods. 2009;176:192–199. doi: 10.1016/j.jneumeth.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 57.Pollard SM, et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell. 2009;4:568–580. doi: 10.1016/j.stem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 58.Busek P, et al. Dipeptidyl peptidase-IV inhibits glioma cell growth independent of its enzymatic activity. Int. J. Biochem. Cell Biol. 2012;44:738–747. doi: 10.1016/j.biocel.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 59.Trylcova J, et al. Effect of cancer-associated fibroblasts on the migration of glioma cells in vitro. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2015;36:5873–5879. doi: 10.1007/s13277-015-3259-8. [DOI] [PubMed] [Google Scholar]
- 60.Team,RC. R: A Language and Environment for Statistical Computing. http://www.R-project.org/ (Foundation for Statistical Computing, 2013).
- 61.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinforma. Oxf. Engl. 2010;26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smyth G. Limma: linear models for microarray data In Gentleman R, Carey V, Dudoit S, et al. Bioinformatics and Computational Biology Solutions Using R and Bioconductor (Gentleman, R. et al.) 397–420 (Springer, 2005).
- 63.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lauss M. rocc: ROC Based Classification. R Package Version 1.2. http://CRAN.R-project.org/package=rocc (30 October 2014, date last accessed) (2010).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.