Abstract
Species-centric approaches to biodiversity in ecological research are limited in their ability to reflect the evolutionary history and functional diversity of community assembly. Recently, the introduction of alternative facets of biodiversity, such as phylogenetic and functional diversity, has shed light on this problem and improved our understanding of the processes underlying biodiversity patterns. Here, we investigated the phylogenetic and functional diversity patterns of α, β and γ components in woody plant assemblages along regional and local elevational gradients in South Korea. Although the patterns of phylogenetic and functional diversity varied along regional and local elevational transects, the main drivers were partitioned into two categories: regional area or climate for phylogenetic diversity, depending on whether the transect was at a regional or local scale; and habitat heterogeneity for functional diversity, which was derived in elevational bands. Moreover, environmental distance was more important than was geographic distance for phylogenetic and functional β diversity between paired elevational bands. These results support the hypothesis that niche-based deterministic processes such as environmental filtering and competitive exclusion are fundamental in structuring woody plant assemblages along temperate elevational gradients regardless of scale (regional vs. local) in our study areas.
Introduction
Biodiversity is defined as the variety of life forms at all levels of biological organization, including taxonomic, genetic, phenotypic, phylogenetic and functional diversity1. For biodiversity conservation and sustainability, understanding and investigating the patterns and drivers of biodiversity along environmental gradients are essential for determining biodiversity hotspots and management strategies in an ecosystem. In recent decades, species diversity was recognized and used as the most important component and measurement of biodiversity in several studies2. However, the definition of biodiversity contains three main components: species, functional and phylogenetic diversity. Recently, with expanded and advanced knowledge about phylogenetic relatedness and functional traits, phylogenetic and functional diversity and community structure analysis has become an important and popular new tool for differentiating the relative roles of niche-based deterministic (e.g., environmental filtering and species interaction) and neutrality-based stochastic (e.g., dispersal limitation and local extinction) processes that control plant community structure3,4. Therefore, ecologists have increasingly turned from species-centric approaches to phylogenetic and functional investigations to obtain more detailed information, such as the accumulated evolutionary and biogeographic history of communities in their study systems3,5,6. Recently, many ecological researchers have started to consider functional and phylogenetic diversity to complement the limitations and weaknesses of species diversity2 for various taxa, such as plants2,6–8, mammals9, birds10, insects11 and microorganisms7.
Mountain ecosystems provide a promising and well-organized natural laboratory for studies on biodiversity because the elevational gradients that have formed on mountains control the ecological and physiological adaptations of various organisms, such as plants, mammals, birds and invertebrates. Therefore, these gradients are recognized as the most important physical factors determining biodiversity and species distribution patterns on mountains12. Furthermore, the elevational gradient has often been found to be parallel to the latitudinal gradient13. Studies on elevational diversity patterns have been popular research subjects in ecology and biogeography for two decades, and there is extensive evidence for the patterns of species diversity and their underlying mechanisms14–17. However, to date, studies related to the other two biodiversity facets, i.e., phylogenetic and functional diversity, have been very rare. Moreover, most of the mechanisms that have been proposed to explain the relationship between diversity and elevation aim to clarify broad large-scale patterns and do not fully explain the elevational diversity patterns observed at smaller scales, such as local slope18,19. However, diversity patterns can change with spatial grain and scale15, and there is clearly a need to explore such small-scale patterns20. The lack of such analysis is partly due to constraints, such as the nature of data sets and the methodologies commonly used in macroecology, especially the dependence on secondary distribution data from the literature as well as the large number of proposed mechanisms18. Interest in studies on multifaceted approaches to biodiversity can be expanded by partitioning biodiversity into α, β and γ diversity21,22. α and γ diversity relate to the diversity patterns at a single site or in a specific habitat, sharing the same characteristics and differentiated only by scale. The complementary use of β diversity reflects the turnover among communities. Although this decomposition of biodiversity facets into α, β and γ components has been shown to be valuable in biodiversity conservation and management, it remains unclear whether these complementary components have similar trends and underlying mechanisms along the same environmental gradients.
We investigated the phylogenetic and functional diversity patterns of woody plant assemblages along one regional and two local elevational transects in temperate forests of South Korea. We also evaluated the ability of specific variables to explain these diversity patterns. Using data collected in field surveys, we investigated 1) the patterns of phylogenetic and functional α, β and γ diversity along temperate elevational gradients, 2) whether the patterns are different between regional and local transects or even between two local transects with different peaks in elevation, and 3) which environmental variables or distance matrices (environmental or geographic) play more important roles in shaping these diversity patterns.
Results
Phylogenetic signal
Regarding phylogenetic signal, Blomberg’s K and Pagel’s λ values for each of the functional traits were less than 1, with the values of Blomberg’s K being lower than those of Pagel’s λ (Table 1). However, all of the traits exhibited significant phylogenetic signal except for flowering onset according to Blomberg’s K. The results suggest that using phylogenetic distance as a proxy for differences in functional traits is appropriate for woody plant species in this study.
Table 1.
Results of tests of phylogenetic signal in the functional trait data from three elevational transects using Blomberg’s K and Pagel’s λ statistics. *P < 0.05; **P < 0.01; ***P < 0.001.
| Functional trait | Blomberg’s K | Pagel’s λ |
|---|---|---|
| Tree height (m) | 0.199*** | 0.856*** |
| Leaf length (cm) | 0.060* | 0.764*** |
| Leaf width (cm) | 0.193*** | 0.815*** |
| Flowering onset (month) | 0.029 | 0.672*** |
| Seed weight (mg) | 0.283*** | 0.984*** |
Phylogenetic and functional diversity with elevation
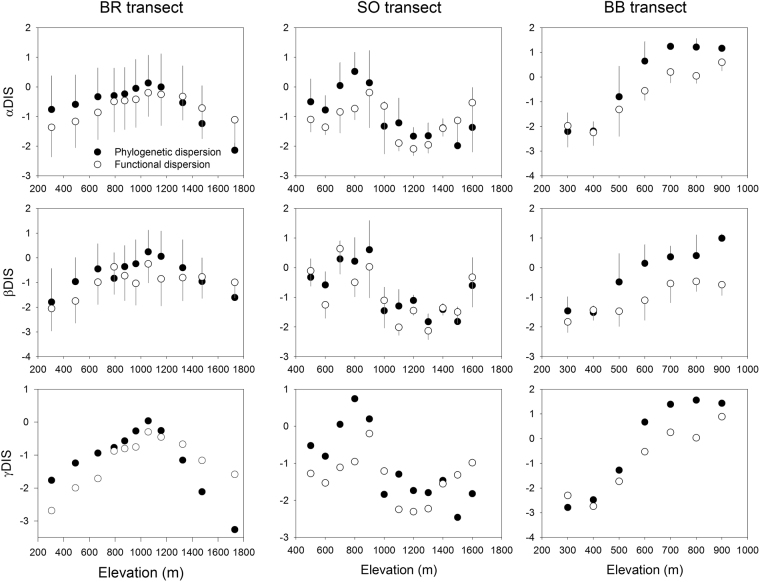
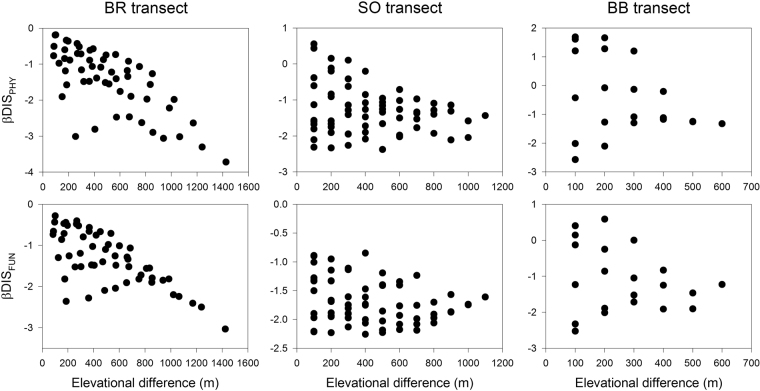
The three components of phylogenetic and functional diversity, which were derived for elevational bands (Table S1) in each study transect (Fig. S1; Table S2), exhibited significant quadratic relationships with elevation in the Baekdudaegan ridge (BR) transect (Fig. 1; Table S3). Whereas the phylogenetic diversity components were negatively correlated with elevation in the Osaek transect on Mt. Seorak (SO transect), the functional diversity components had no relationship with elevation. Furthermore, the three components of phylogenetic and functional diversity in the Bohyunsa transect on Mt. Baekhwa (BB transect) exhibited significant linear and quadratic relationships with elevation. However, the linear patterns were considered to provide better fit in the BB transect because the linear relationships had lower Akaike information criterion values than did the quadratic models (Table S3). The β components of phylogenetic and functional diversity between paired elevational bands had significant negative correlations with elevational differences except for functional β diversity in the BB transect (Fig. 2).
Figure 1.
Relationships between elevation and the three components (α, β and γ) of phylogenetic and functional diversity along the three study transects. Half lines on black and white circles in α and β components indicate standard deviations. DIS indicates phylogenetic or functional dispersion.
Figure 2.
Relationships between elevational difference and the β components of phylogenetic and functional diversity between paired elevational bands in the three study transects. The correlation coefficients and significance levels from simple Mantel tests are shown in Table 3. DISPHY and DISFUN indicate phylogenetic and functional dispersion, respectively.
Drivers of phylogenetic and functional diversity
The results of the simple ordinary least squares (OLS) regressions indicated that the α and γ components of phylogenetic diversity were related to regional area (RArea) and that the β component was correlated with habitat heterogeneity, whereas the functional diversity components were correlated only with habitat heterogeneity in the BR transect (Table 2). In the SO transect, the functional diversity components were related to habitat heterogeneity, whereas the phylogenetic diversity components were mainly correlated with climatic variables. In the BB transect, the phylogenetic diversity components were correlated with climatic variables. However, the functional diversity components were related to RArea, climatic variables and habitat heterogeneity. Analysis of the relationships of elevational difference or environmental distance matrices with β components between paired elevational bands using simple Mantel tests revealed that although the relative importance of the distance matrices differed among diversity indices and study transects, overall, environmental distances were more important than was elevational difference as a surrogate of geographic distance (Table 3). The results of the Mantel tests also indicated that the relative importance of elevational difference was higher in the regional (BR) transect than in the two local (SO and BB) transects.
Table 2.
Coefficient of determination (R2) and significance level from simple ordinary least squares regression models for environmental variables and the three components (α, β and γ) of phylogenetic and functional diversity derived in elevational bands along the three study transects. DISPHY and DISFUN indicate phylogenetic and functional dispersion, respectively. *P < 0.05; **P < 0.01; ***P < 0.001.
| Transect | Diversity index | RArea | PC1clim | PC1hetero | PC2hetero |
|---|---|---|---|---|---|
| BR | αDISPHY | 0.787*** | 0.294 | 0.195 | 0.004 |
| βDISPHY | 0.269 | 0.002 | 0.567** | 0.396* | |
| γDISPHY | 0.704*** | 0.191 | 0.291 | 0.031 | |
| αDISFUN | 0.073 | 0.079 | 0.549** | 0.567** | |
| βDISFUN | <0.001 | 0.209 | 0.508* | 0.583** | |
| γDISFUN | 0.013 | 0.192 | 0.592** | 0.694*** | |
| SO | αDISPHY | 0.398* | 0.549** | 0.195 | 0.245 |
| βDISPHY | 0.211 | 0.404* | 0.273 | 0.222 | |
| γDISPHY | 0.442* | 0.568** | 0.127 | 0.215 | |
| αDISFUN | <0.001 | 0.029 | 0.505** | 0.174 | |
| βDISFUN | 0.037 | 0.240 | 0.678*** | 0.099 | |
| γDISFUN | 0.003 | 0.052 | 0.499** | 0.195 | |
| BB | αDISPHY | 0.488 | 0.868** | 0.473 | 0.142 |
| βDISPHY | 0.687* | 0.904*** | 0.561 | 0.120 | |
| γDISPHY | 0.514 | 0.888*** | 0.515 | 0.147 | |
| αDISFUN | 0.645* | 0.832** | 0.630* | 0.229 | |
| βDISFUN | 0.516 | 0.821** | 0.670* | 0.175 | |
| γDISFUN | 0.666* | 0.818** | 0.642* | 0.244 |
Abbreviations: BR – Baekdudaegan ridge transect; SO – Osaek transect in Mt. Seorak; BB – Bohyunsa transect in Mt. Baekhwa; RArea – regional area with log-transformation; PC1clim – PC1 from climatic variables; PC1hetero – PC1 from standard deviations of topographic variables; PC2hetero – PC2 from standard deviations of topographic variables.
Table 3.
Results of simple Mantel tests to investigate the effects of elevational and environmental distances on phylogenetic and functional β diversity derived between paired elevational bands along the three study transects.
| Transect | Diversity index | Dist ele | Dist rarea | Dist clim | Dist habit |
|---|---|---|---|---|---|
| BR | βDISPHY | −0.727*** | −0.835*** | −0.747*** | 0.057 |
| βDISFUN | −0.716*** | −0.408*** | −0.635*** | −0.306*** | |
| SO | βDISPHY | −0.226** | −0.501*** | −0.166 | −0.127 |
| βDISFUN | −0.241** | −0.108 | −0.250** | −0.230** | |
| BB | βDISPHY | −0.311* | −0.121 | −0.580*** | −0.139 |
| βDISFUN | −0.226 | 0.228 | −0.449** | −0.079 |
DISPHY and DISFUN indicate phylogenetic and functional dispersion, respectively. *P < 0.05; **P < 0.01; ***P < 0.001 Abbreviations: Distele – elevational difference; Distrarea – regional area distance; Distclim – climate distance; Disthabit – habitat heterogeneity distance. The abbreviations for the study transects are defined in Table 2.
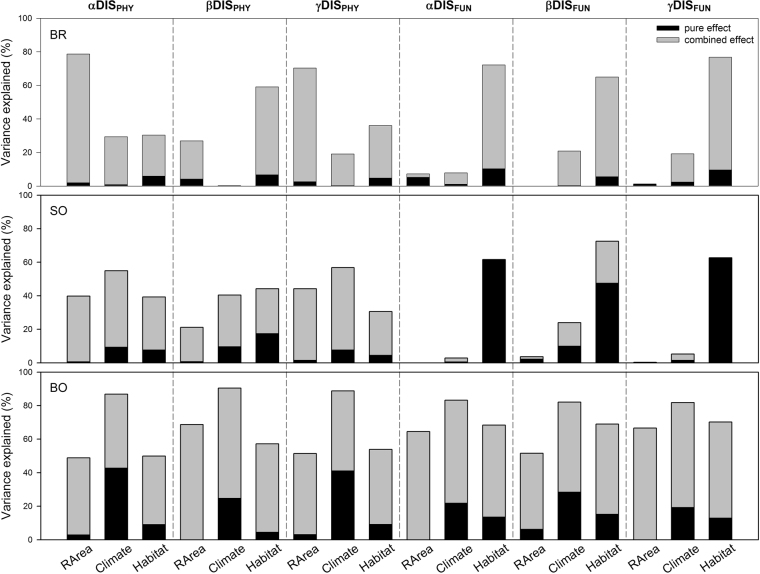
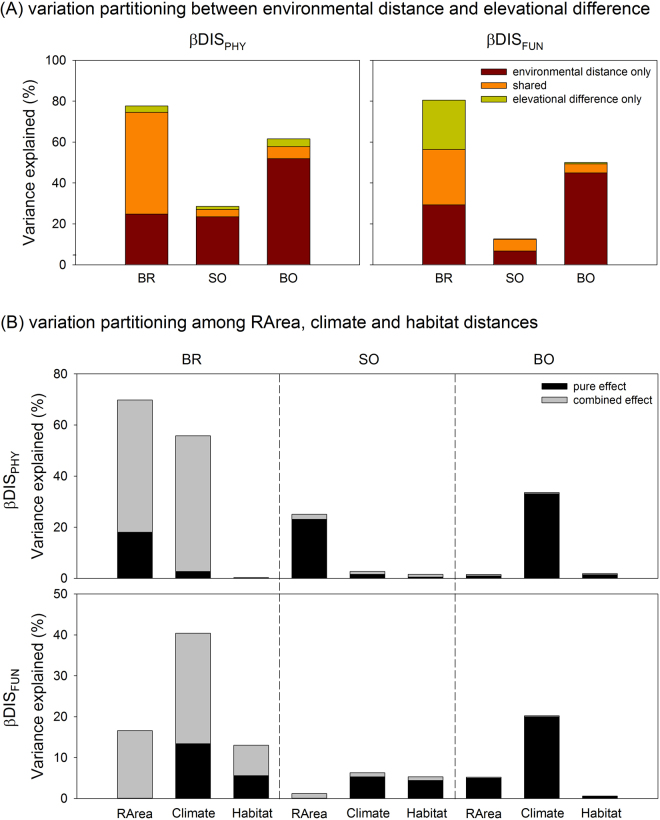
The results of stepwise multiple regression models were similar to those of the simple OLS regression and the simple Mantel tests (Table 4). In the BR transect, RArea and habitat heterogeneity were the most important factors affecting the phylogenetic diversity components derived in the elevational bands, whereas climatic variables were the most import factors in the SO and BB transects. Habitat heterogeneity was more important for the functional diversity components except the β component in the BB transect. Furthermore, the environmental distances were more important factors than was elevational difference for phylogenetic and functional β diversity between paired elevational bands in the study transects. RArea and climatic distance were important factors affecting phylogenetic and functional β diversity between paired elevational bands. The simple conditional autoregressive (CAR) models showed similar results to those of the simple OLS models (Table S4), and the results of multiple CAR models supported the results of the best models resulted from forward stepwise multiple regression models (Table S5). The results of the variation partitioning also largely reinforced the results of the stepwise multiple regression models (Figs 3 and 4).
Table 4.
Results of forward stepwise multiple regression models of the explanatory variables and the three components (α, β and γ) of phylogenetic and functional diversity along the study transects.
| Study transect | Dependent variable | Regression equation | F | R 2 | |
|---|---|---|---|---|---|
| Within elevations | BR | αDISPHY | y = −5.036 + 1.064 RArea | 33.203 | 0.787*** |
| βDISPHY | y = −1.845 + 1.009 PC1hetero | 11.781 | 0.567** | ||
| γDISPHY | y = −7.389 + 1.488 RArea | 21.398 | 0.704*** | ||
| αDISFUN | y = −1.216 − 2.111 PC2hetero | 11.798 | 0.567** | ||
| βDISFUN | y = −1.692 − 2.817 PC2hetero | 12.588 | 0.583** | ||
| γDISFUN | y = −2.285 − 4.256 PC2hetero | 20.433 | 0.694*** | ||
| SO | αDISPHY | y = −0.450 + 0.281 PC1clim | 12.167 | 0.549** | |
| βDISPHY | y = −0.348 + 0.250 PC1clim | 6.772 | 0.404* | ||
| γDISPHY | y = −0.465 + 0.350 PC1clim | 13.139 | 0.568** | ||
| αDISFUN | y = −0.985 + 0.261 PC1hetero | 10.194 | 0.505** | ||
| βDISFUN | y = −0.638 + 0.431 PC1hetero | 21.023 | 0.678*** | ||
| γDISFUN | y = −1.234 + 0.266 PC1hetero | 9.952 | 0.499** | ||
| BB | αDISPHY | y = 4.717 − 2.084 PC1clim | 32.764 | 0.868** | |
| βDISPHY | y = 2.808 − 1.302 PC1clim | 47.121 | 0.904*** | ||
| γDISPHY | y = 5.749 − 2.563 PC1clim | 39.823 | 0.888*** | ||
| αDISFUN | y = 2.542 − 1.364 PC1clim − 0.323 PC2hetero | 47.417 | 0.960* | ||
| βDISFUN | y = 0.567 − 0.699 PC1clim | 23.002 | 0.821** | ||
| γDISFUN | y = 3.186 − 1.679 PC1clim − 0.421 PC2hetero | 45.861 | 0.958* | ||
| Between elevations | BR | βDISPHY | y = − 0.381 − 1.083 Distrarea − 0.001 Distele | 80.880 | 0.757*** |
| βDISFUN | y = − 0.605 − 0.905 Distclim + 0.006 Distele | 50.982 | 0.662*** | ||
| SO | βDISPHY | y = − 0.896 − 0.705 Distrarea | 21.461 | 0.251*** | |
| βDISFUN | y = − 1.363 − 0.063 Distclim − 0.078 Disthabit | 4.506 | 0.125* | ||
| BB | βDISPHY | y = 0.473 + 1.011 Distrarea − 2.118 Distclim | 11.638 | 0.564*** | |
| βDISFUN | y = − 0.700 + 0.755 Distrarea − 1.264 Distclim | 8.270 | 0.479** |
Figure 3.
Variation partitioning of the three components (α, β and γ) of phylogenetic and functional diversity derived in elevational bands explained by regional area, climate and habitat heterogeneity along the three study transects. The black bars indicate the individual effects and the grey bars indicate the combined effects. Because the variance in αDISFUN and βDISFUN explained by regional area along the SO and BR transects, respectively, is very low (0.1%), the effects are not shown in this figure. The abbreviations for the study transects, explanatory variables and diversity indices are defined in Table 2. DISPHY and DISFUN indicate phylogenetic and functional dispersion, respectively.
Figure 4.
Variation partitioning of the β diversity components derived between paired elevational bands explained by (A) geographic and environmental distances and (B) each environmental distance, including RArea, climate and habitat distances along the three study transects. The abbreviations for the study transects, explanatory variables and diversity indices are defined in Tables 2 and 3. DISPHY and DISFUN indicate phylogenetic and functional dispersion, respectively.
Discussion
In this study, we explored regional and local elevational patterns of the α, β and γ components of phylogenetic and functional diversity in woody plant assemblages and their associated drivers using primary data along different transects on different mountains. Thus, the present study is different from many other studies that examined broad large-scale trends using secondary data from literature reviews14,15. Moreover, this study investigated whether significant phylogenetic signal was present in the functional trait data to better understand the degree to which the phylogenetic tree can estimate the functional trait similarity of species. The study of biodiversity patterns at the regional (large) scale is critical for understanding the patterns across spatial scales, whereas the study of biodiversity patterns at the local (small) scale is salient for understanding the within-domain biodiversity in biogeographic groups18,23. This study provides a valuable contribution by exploring elevational patterns and the underlying mechanisms using empirical data collected simultaneously at regional and local scales.
Phylogenetic signal
We found low levels of phylogenetic signal for the functional traits (i.e., Blomberg’s K and Pagel’s λ values < 1). Low phylogenetic signal is frequently interpreted as evolutionary trait lability or high rates of trait evolution contributing to large differences among close relatives. In this study, we quantified phylogenetic signal for five woody functional traits, and with the exception of flowering onset as based on Blomberg’s K, all of these traits exhibited significant phylogenetic signal. The general congruence of functional and phylogenetic dispersion is supported by significant phylogenetic signal in the trait data, but the congruence does not show ‘perfect correlation’ between the two dispersion patterns in our study transects (Fig. S2). This result can be understood by noting that the K and λ values of phylogenetic signal were all less than 1, suggesting that functional traits are more unstable than expected under a Brownian motion model of trait evolution. Previous studies have reported similar results in other forests, with phylogenetic and functional dispersions found to be aligned imperfectly but significantly with phylogenetic signal in trait data24–26. Thus, our results suggest that if there are significant but low values of phylogenetic signal in functional traits, phylogenetic dispersion can provide a rough approximation of functional dispersion; however, the correlations between both facets of diversity are not perfect. Moreover, if there is phylogenetic clustering in woody plant assemblages along elevational gradients in our study areas, our results might suggest the possibility of a direct connection between phylogenetic clustering and environmental filtering as one niche-based deterministic process structuring community assembly. The notion that phylogenetic clustering is mainly derived from strong environmental filtering rather than competitive exclusion relies on the assumption that the functional traits that are involved in community assembly processes have detectable phylogenetic signal27,28. Accordingly, ecologists and biogeographers increasingly recognize that testing for phylogenetic signal in functional traits is a necessary step when implementing phylogenetic community structure analysis5,27,28.
However, our results also raise the question of why these ‘not perfect correlations’ occur between phylogenetic relatedness and functional traits. First, phylogenetic relatedness is generally used as an indirect estimate of ecological similarity29. Therefore, phylogenetic relatedness serves as only an indirect proxy of the overall trait similarity of species in a community and thus may be unable to reveal similarities in individual functional traits. Second, although the estimation of general similarity for species is useful and tractable in some circumstances, this estimation is likely to overlook important information relevant to one or a few traits of species. Thus, much information can be lost when using phylogenetic relatedness as a proxy for trait similarity2,6. Moreover, an additional problem is that community assembly and species coexistence might be primarily influenced by a single resource axis, and only one functional trait might be important for understanding the processes underlying community assembly; however, phylogenetic relatedness likely cannot detect such processes29. However, these limitations similarly apply when using functional traits as a substitute for direct ecological similarity. Many studies relevant to the functional trait approach, including our study, use a few, easily measurable indirect traits and fundamental aspects of ecological strategy and functional trade-offs (e.g., morphological and structural traits, nutrient content) related to physiological processes in plants2,8. This approach is used because it is impossible to measure all traits thought to be important for physiological and defence mechanisms. Moreover, we do not know which traits are important for evolutionary processes in community assembly, and the functional trait approach also has inherent weaknesses such as the presence of intra- and inter-specific variation2,8,29. These limitations and shortcomings of both phylogenetic relatedness and functional trait approaches are likely to result in imperfect correlations between both phylogenetic and functional dispersions. Therefore, our results re-emphasize that studies on the structure of community assembly and the underlying processes should use both approaches complementarily, as emphasized in previous studies2.
Diversity patterns and drivers along elevational gradients
The α, β and γ components of phylogenetic and functional diversity derived from the elevational bands in each transect showed different patterns, including hump-shaped curves, decreasing trends, increasing trends, and no relationship, with increasing elevation across the study transects. At the regional scale, i.e., in the BR transect, the three components of phylogenetic and functional diversity showed increasing phylogenetic and functional relatedness up to intermediate elevations (1000–1100 m), and then decreased thereafter. The patterns of phylogenetic and functional diversity were different between two local transects. Along the SO transect, decreasing phylogenetic relatedness (i.e., phylogenetic overdispersion) and no relationship of functional relatedness (i.e., functional randomness) with increasing elevation were observed, whereas along the BB transection, both phylogenetic and functional relatedness increased with increasing elevation, indicating phylogenetic and functional clustering. Moreover, as drivers shaping these patterns, RArea and the climatic variables were the most important factors influencing phylogenetic diversity along the regional- (BR) and local-scale (SO and BB) transects, respectively. In addition, habitat heterogeneity, based on topographic characteristics, was the main driver shaping the patterns of functional diversity components, which were derived in elevational bands.
The hump-shaped pattern of phylogenetic diversity with increasing elevation along the BR transect differs from the findings of previous studies reporting phylogenetic clustering or overdispersion of plant communities along elevational gradients10,30–32. RArea for the α and γ components and habitat heterogeneity for the β components were the most important factors. These results indicate that mid-elevational bands of larger area or higher habitat heterogeneity along the BR transect might exhibit increased phylogenetic relatedness with stable species colonization and extinction rates, whereas lower or higher elevational bands of smaller area or lower habitat heterogeneity might show more phylogenetically overdispersed patterns, with rapid, repeated colonization and extinction33. The ecological processes governing phylogenetic community structure differ among spatial scales. It is generally recognized that small-scale dispersal and species interactions are more important at local scales and that colonization and extinctions are crucial factors at regional scales5,27,28.
There are several potential explanations for the increase in phylogenetic overdispersion with increasing elevation in the SO transect. One potential explanation involves competitive exclusion: If competitive exclusion primarily removes related species of ecological similarity that show strong niche overlap at high elevations and if the degree to which different species have traits that favour them in competition for limited resources at high elevations is positively related to phylogenetic distance, then competition will drive phylogenetic overdispersion34,35. An alternative plausible explanation involves climatic variables, such as temperature differences between the hottest and coldest months along the SO transect. The temperature difference between the hottest and coldest months is generally recognized as temperature seasonality or variability. In the SO transect, temperature difference increased with increasing elevation, whereas a declining pattern was observed in the BR and BB transects (Fig. S3). This wide temperature range at higher elevations along the SO transect might allow some phylogenetically distant species that adapted to such cold conditions and wide temperature ranges during their evolutionary history to be distributed here to a greater extent than are closely related species. Accordingly, several genera such as Lonicera, Syringa, Thuja, and Hydrangea are common at high elevations (>1100 m) along the SO transect (Table S6). These genera represent the contribution of completely novel lineages to the woody plant assemblages at higher elevations along this transect23, and many other genera are widely distributed across the entire elevation range along the SO transect (Table S6). These potential explanations are not mutually exclusive, and thus one or more might be involved in contributing to the phylogenetic overdispersion observed at higher elevations along the SO transect.
The clustering patterns of the phylogenetic diversity components along the BB transect may be explained by a stronger effect of environmental filtering than of competitive exclusion5,7 in shaping community assembly at high elevations. In particular, climate-related filtering, in which closely related species with adaptive traits to harsh and stressful environmental conditions, such as low temperature and strong wind, are filtered34,36,37, might play a strong role. The lower phylogenetic relatedness at low elevations can be interpreted as evidence that the effect of competitive exclusion among related species with ecological similarity is stronger than the environmental filtering effect such that these clades are distantly related to other temperate lineages at low elevations27,28. Accordingly, many genera were recorded at low elevations (<600 m) along the BB transect (11 genera, including Juniperus, Smilax, and Zanthoxylum), whereas a smaller number of genera (7 genera, including Carpinus, Tripterygium, and Vitis) were observed at high elevations (>600 m) (Table S6). This difference in the number of unique genera between low and high elevations represents a phylogenetic clustering pattern along the BB transect. Generally, in most previous studies on elevational patterns of phylogenetic diversity, phylogenetic clustering at higher elevations has been observed in response to abiotic filtering, such as environmental filtering10,26,30,34,36, whereas few studies have revealed phylogenetic overdispersion in response to biotic interactions, such as interspecific competition, at higher elevations31,32. Although the patterns of phylogenetic diversity and the underlying processes along elevational gradients differed among the study transects in this study and among studies, the underlying mechanisms mainly involve niche-based deterministic processes, including abiotic (e.g., environmental filtering by climate gradients) and biotic (e.g., competition) processes36. The most influential environmental factor in shaping phylogenetic diversity patterns in the two local-scale transects was the climatic factor. This finding highlights climatic variables as one of the main drivers of biodiversity36,37, which supports concerns regarding the effects of climate on species distribution and composition in many areas and on related ecosystem services and evolutionary responses38,39. Our results also indicate that the relative importance of different environmental factors on phylogenetic diversity derived in elevational bands might be scale dependent.
The functional diversity components exhibited patterns similar to those of the phylogenetic diversity components. Although the α, β and γ components of functional diversity along the SO transect did not show significant linear or quadratic relationships with elevation, suggesting random functional structure along the SO transect, we cannot infer a role of neutrality-based stochastic processes in this structure because the pattern was closely related to the gradient of habitat heterogeneity, as represented by PC1hetero. Therefore, the processes shaping the patterns of the functional diversity components were related to the gradients of climate and habitat heterogeneity along the BB transect and mainly to the gradients of habitat heterogeneity along the other transects. These results also support the hypothesis that niche-based deterministic processes are fundamental processes for functional diversity in woody plant assemblages along our study transects. Other studies have similarly documented the importance of climate40 and habitat factors26,41 in determining the functional structures of woody plant assemblages. Habitat heterogeneity is considered an important factor shaping diversity patterns, notably by driving local differences in species distribution and thereby increasing diversity via differences in environmental preferences42. Heterogeneity is also thought to significantly influence the dynamics and structure of ecological communities43. In particular, topographic heterogeneity has been recognized as creating a complex mosaic or heterogeneity of substrates and soils with varying structures, hydrology and chemistry44. In general, plant functional traits are strongly correlated with soil resources and water availability at the community scale19. Therefore, spatial heterogeneity in nutrient and water availabilities, along with other environmental factors derived from topographic heterogeneity, could influence the functional diversity of woody plants that coexist via niche partitioning45,46. In heterogeneous habitats, heterogeneous resources, including light, moisture and soil nutrients, allow plant species with different niche requirements to meet their habitat requirements, which leads to higher functional diversity47. Therefore, our results indicate that woody species distributions along topographic gradients may be partly shaped by habitat filtering through a selection of functional traits that are associated with tree resource use and growth strategies48.
Although our study emphasizes the importance of niche-based deterministic processes for structuring the patterns of phylogenetic and functional diversity in elevational bands, as evidenced in the different study transects, the implications of these processes for phylogenetic and functional diversity appear to differ. Specifically, the processes associated with phylogenetic diversity patterns varied among the study transects and the three components, whereas the processes structuring functional diversity were mainly associated with the gradients of habitat heterogeneity in the study transects. These results suggest that phylogenetic diversity, which reflects the accumulated evolutionary and biogeographic history of community assembly, is potentially associated with various abiotic and biotic processes, whereas functional diversity, which is related to ongoing ecological processes as inferred from morphological, physiological and ecological traits, is associated with a set of species with functional traits that are optimally adaptable for a given environment or set of habitat conditions regardless of the phylogenetic distance among lineages.
With the exception of functional β turnover along the BB transect, the phylogenetic and functional β turnovers between paired elevational bands along all of the study transects indicated significant linear decreases. Such patterns have been observed in many previous studies and are collectively well known as the distance-decay relationship, which describes the decrease in compositional similarity between two communities with increasing geographic distance (or equivalent elevational difference) between them49. The β components of phylogenetic and functional dispersions between paired elevational bands in the present study were mainly governed by regional area and climatic distances, although elevational difference as a proxy for geographic distance was also important in structuring the β components along the regional transect (BR). Our results indicate that environmental distance is generally a better predictor of β diversity than is geographic distance, thereby suggesting more support for deterministic processes than for stochastic processes2. Moreover, our study supports previous findings that phylogenetic or functional turnover is significantly related to environmental gradients from local to regional scales50.
In summary, we observed low but significant phylogenetic signal in functional traits, suggesting that phylogenetic dispersion can roughly approximate functional dispersion but that the two facets of diversity are not perfectly correlated. Although the patterns of phylogenetic and functional diversity differed among temperate elevational gradients at both regional and local scales in the study areas in South Korea, the main drivers were partitioned into two categories: regional area and climatic variables for phylogenetic diversity and habitat heterogeneity for functional diversity in elevational bands. Furthermore, environmental distance was a more important predictor of β components between paired elevational bands than was geographic distance. Our study generally supports the hypothesis that niche-based deterministic processes, such as biotic (e.g., competitive exclusion) and abiotic (e.g., environmental filtering by habitat and climatic factors) processes, are fundamental mechanisms structuring woody plant assemblages along temperate elevational gradients in South Korea. Moreover, the results suggest that the environmental drivers might differ between the considered facets of diversity and among scales.
Methods
Study area and plant data
To evaluate the differences in phylogenetic and functional diversity patterns and the relationships of the diversity patterns with explanatory variables along regional and local elevational transects, we selected the main ridge of the Baekdudaegan Mountains as a regional (BR) transect and two local (SO and BB) transects, one on Mt. Seorak and one on Mt. Baekhwa (Fig. S1; Table S2). A total of 256 woody plant species representing 50 families and 101 genera were recorded from 1195 400-m2 (20 m × 20 m) forest plots along the three transects during the growing season (May to August) of 2005 to 2011 (Table S2). Detailed descriptions of the study area and plant data are described in Supplementary Methods.
Phylogenetic tree and functional trait dendrogram
A phylogenetic tree of the woody plant species surveyed in this study was generated by pruning the PhytoPhylo megaphylogeny51, which is an updated version of the vascular plant phylogeny published by Zanne et al.52. This megaphylogeny is the largest phylogeny of vascular plants available and was generated based on the sequences of seven gene regions (i.e., 18S rDNA, 26S rDNA, ITS, matK, rbcL, atpB and trnL-F) from GenBank. The phylogeny includes all families of extant seed plants in the world and was time-scaled based on 39 fossil calibrations. The 101 genera surveyed in our study were present in PhytoPhylo. The S.PhyloMaker function with Scenario 3 was implemented in R software to assign those species that were not present in PhytoPhylo52.
To construct a functional trait dendrogram for quantifying functional diversity, we included five functional traits for all woody plant species: maximum height (m), leaf dimensions (cm; length and width), flowering onset (month) and seed mass (mg). Values of all traits were log transformed to improve normality and were standardized before analysis. To eliminate trait redundancy, we performed a principal component analysis (PCA) on the functional trait data (Table S7). We used the first four principal components, which explained 94.8% of the variation in the trait data, to construct a Euclidean trait distance matrix. An unweighted paired group method with arithmetic mean (UPGMA) hierarchical clustering was then applied to this matrix to produce a trait dendrogram.
The phylogenetic tree and functional trait dendrogram (Fig. S4) were separately constructed for three scenarios. Additional information regarding the importance and data sources of the five functional traits and regarding the construction of the phylogenetic tree and trait dendrogram is provided in Supplementary Methods.
Phylogenetic and functional dispersion
The abundance-weighted net relatedness index (NRI)5 was used to quantify the α and γ components of phylogenetic (or functional) dispersion. The formula is as follows:
| 1 |
where MPDobserved is the observed mean phylogenetic (or functional) distance (MPD) in each plot or elevational band, mean MPDrandom is the mean MPD of the null models, and sd MPDrandom is the standard deviation of MPD of the null models. MPD mainly reflects the deep phylogenetic (or functional) structure in a phylogeny. As such, MPD is typically thought to be more sensitive to tree-wide patterns of phylogenetic (or functional) clustering or overdispersion5,24 than to the structure near the tips.
To calculate phylogenetic (or functional) β dispersion between paired plots in each elevational band or paired elevational bands in each study transect, we quantified the standardized effect size (S.E.S.) of abundance-weighted Dpw (mean pairwise phylogenetic or functional distance) as follows:
| 2 |
| 3 |
where Dpw observed is the observed phylogenetic (or functional) dissimilarity between plots or elevational bands, mean Dpw random is the mean Dpw of the null models, and sd Dpw random is the standard deviation of Dpw of the null models. In the Dpw equation, is the mean pairwise phylogenetic (or functional) distance (MPPD) between species i in plot k1 or elevational band k1 to all species in plot k2 or elevational band k2, and is the MPPD between species j in plot k2 or elevational band k2 to all species in plot k1 or elevational band k1. fi and fj represent the relative abundances of species i and species j. This dissimilarity matrix is highly correlated with Rao’s D and is better than Rao’s D for detecting major compositional turnover between communities8,24.
To quantify phylogenetic (or functional) dispersion such as NRI and S.E.S. Dpw, we created the null model by randomly shuffling the names of the species across the tips of the phylogenetic tree (or functional trait dendrogram) 1000 times. This approach randomized the phylogenetic (or functional trait) relatedness of the species to one another while maintaining the observed community data matrix. Therefore, this null model fixes the observed levels of species occupancy rates, abundances and spatial distributions in each randomization2.
Consequently, we calculated four indices, comprising one α component, two β components and one γ component, as subsets of phylogenetic (or functional) dispersion in this study. The α component was defined as the mean value of the NRIs of all plots in each elevational band. The β components were calculated in two ways: 1) as the mean values of S.E.S. Dpw between plots in each elevational band and 2) as the S.E.S. Dpw between paired elevational bands in each study transect using species data, which were pooled and summed from multiple plots in the same elevational band. Finally, the γ component was quantified in each elevational band of each transect. The COMSTRUCT function for the α and γ components and the COMDIST function for the β components were used in Phylocom 4.253. We used the phylogenetic trees and trait dendrograms described in “Phylogenetic tree and functional trait dendrogram” to calculate the components of both dispersions.
One of the main aims of this study was to compare and contrast measurements of phylogenetic and functional diversity. Therefore, we quantified the degree of phylogenetic signal in each functional trait using Blomberg’s K54 and Pagel’s λ55 statistics to determine whether the phylogenetic tree can estimate the similarity of functional traits of species. Additional information related to the two statistics is provided in Supplementary Methods.
Environmental variables
For the environmental variables, we included the RArea, three topographic heterogeneity variables and six climatic variables for each elevational band in the three transects. Detailed information on the calculation of the environmental variables is described in Supplementary Methods. The relationships between the environmental variables and elevation are shown in Fig. S5. Before performing further statistical analyses, RArea and two precipitation-related variables were subjected to log transformation to achieve normality. Furthermore, to reduce the co-variation and possible redundancy in the data, two separate PCAs were performed on each set of standard deviations of topographic variables and climatic variables (Table S8). From the PCA, the first PCA axis for the climatic variables and the first and second PCA axes for the topographic variables were extracted, creating new PCA-derived variables as independent variables. The PCA-derived variables were labelled PC1clim, PC1hetero and PC2hetero for climate and topographic heterogeneity.
To investigate the relationships between phylogenetic or functional β dispersion and geographic or environmental distance, we calculated the elevational difference as a proxy of geographic distance between elevational bands because elevational separation is generally recognized as correlating with geographic distance49. We also calculated the climatic distance with PC1clim and the habitat distance with PC1hetero and PC2hetero using the Euclidean distance measurements, and the RArea distance was measured as the absolute value of the difference in RArea between paired elevational bands.
Data analysis
Linear and quadratic regression models were fitted to assess the relationship between elevation and each of the three components of phylogenetic and functional diversity. To test the effects of individual variables, such as RArea, PC1clim, PC1hetero and PC2hetero, on the elevation patterns of the three components of phylogenetic and functional diversity, we performed a simple OLS regression analysis. We also performed stepwise multiple regression to establish the relative importance of each environmental variable for the diversity indices. A simple Mantel test with 10000 permutations was applied to evaluate the significance of the correlation between the elevation or environmental distance matrix and each β component between the paired elevational bands. We also used simple and multiple CAR models to evaluate the influence of spatial autocorrelation, such as inflation of type I error and invalid parameter estimation, on the regression results42,56.
Studies focusing on mechanisms driving diversity patterns generally apply multiple regressions and similar statistical analyses. However, more complex strategies were applied here for the ecological data analyses because it is important to account for the lack of independence between pairs of observations across geographic space57. Therefore, in this study, we used variation partitioning57 with partial regressions for the α, β and γ components of phylogenetic and functional diversity with environmental variables, which were derived in elevational bands, and multiple regressions on distance matrices (MRM) for β components with distance matrices, which were derived between paired elevational bands.
Electronic supplementary material
Acknowledgements
We would like to thank Mr. Keun-Wook Lee, Mr. Sang-Hyouk Seo, Mr. Hyo-Hyeon Ahn and Mr. Min-Woo Park for their invaluable help during fieldwork and data analysis in this study. We also thank Dr. Hyun-Je Cho for his support and encouragement. This paper is part of the “Korean Big Tree Project” funded by the Korean Green Promotion Agency at the Korean Forest Service.
Author Contributions
C.B.L. and J.H.C. generated the main ideas and analysed the data. C.B.L. led the writing of the manuscript. C.B.L. prepared Figures 1–4 and Tables 1–4, and J.H.C. prepared Supporting Information. All of the authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-21266-4.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gaston, K. J. & Spicer, J. I. Biodiversity: an introduction. (Blackwell Publishing, 2004).
- 2.Swenson NG, et al. Phylogenetic and functional diversity alpha and beta diversity in temperate and tropical tree communities. Ecology. 2012;93:S112–S125. doi: 10.1890/11-0402.1. [DOI] [Google Scholar]
- 3.Webb CO. Exploring the phylogenetic structure of ecological communities: an example for rain forest trees. Am. Nat. 2000;156:145–155. doi: 10.1086/303378. [DOI] [PubMed] [Google Scholar]
- 4.Mo XX, Shi LL, Zhang YJ, Zhu H, Slik JWF. Chang in phylogenetic community structure during succession of traditionally managed tropical rainforest in southwest China. PLoS One. 2013;8(7):e71464. doi: 10.1371/journal.pone.0071464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Webb CO, Ackerly DD, McPeek MA, Donoghue MJ. Phylogenies and community ecology. Ann. Rev. Ecol. Syst. 2002;33:475–505. doi: 10.1146/annurev.ecolsys.33.010802.150448. [DOI] [Google Scholar]
- 6.Swenson NG. The role of evolutionary processes in producing biodiversity patterns, and the interrelationships between taxonomic, functional and phylogenetic diversity. Am. J. Bot. 2011;98:472–480. doi: 10.3732/ajb.1000289. [DOI] [PubMed] [Google Scholar]
- 7.Bryant JB, et al. Microbes on mountainsides: contrasting elevational patterns of bacterial and plant diversity. Proc. Natl. Acad. Sci. USA. 2008;105:S11505–11511. doi: 10.1073/pnas.0801920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swenson NG. Phylogenetic beta diversity metrics, trait evolution and inferring the functional beta diversity of communities. PLoS One. 2011;6:e21264. doi: 10.1371/journal.pone.0021264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Safi K, et al. Understanding global patterns of mammalian functional and phylogenetic diversity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011;366:2536–2544. doi: 10.1098/rstb.2011.0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graham CH, Parra JL, Rahbek C, McGuire JA. Phylogenetic structure in tropical hummingbird communities. Proc. Natl. Acad. Sci. USA. 2009;106:19673–19678. doi: 10.1073/pnas.0901649106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnan X, Cerdá X, Retana J. Partitioning the impact of environmental and spatial structure on alpha and beta components of taxonomic, functional and phylogenetic diversity in European ants. PeerJ. 2015;3:e1241. doi: 10.7717/peerj.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lomolino MV. Elevational gradients of species-density: historical and prospective views. Global Ecol. Biogeogr. 2001;10:3–13. doi: 10.1046/j.1466-822x.2001.00229.x. [DOI] [Google Scholar]
- 13.Begon, M., Harper J. L. & Townsend, C. R. Ecology: individuals, populations and communities. (Blackwell Publishing, 1990).
- 14.Rahbek C. The elevational gradient of species richness: a uniform pattern? Ecography. 1995;18:200–205. doi: 10.1111/j.1600-0587.1995.tb00341.x. [DOI] [Google Scholar]
- 15.Rahbek C. The role of spatial scale and the perception of large-scale species-richness patterns. Ecol. Lett. 2005;8:224–239. doi: 10.1111/j.1461-0248.2004.00701.x. [DOI] [Google Scholar]
- 16.Kluge J, Kessler M, Dunn RR. What drives elevational patterns of diversity? A test of geometric constraints, climate and species pool effects for pteridophytes on an elevational gradient in Costa Rica. Global Ecol. Biogeogr. 2006;15:358–371. doi: 10.1111/j.1466-822X.2006.00223.x. [DOI] [Google Scholar]
- 17.Wu Y, et al. Explaining the species richness of birds along a subtropical elevational gradient in the Hengduan Mountains. J. Biogeogr. 2013;40:2310–2323. doi: 10.1111/jbi.12177. [DOI] [Google Scholar]
- 18.Oommen MA, Shanker K. Elevational species richness patterns emerge from multiple local mechanisms in Himalayan woody plants. Ecology. 2005;86:3039–3047. doi: 10.1890/04-1837. [DOI] [Google Scholar]
- 19.Yang J, et al. Local-scale partitioning of functional and phylogenetic beta diversity in a tropical tree assemblage. Sci. Rep. 2015;5:12731. doi: 10.1038/srep12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Storch D, Marquet PA, Gaston MJ. Untangling an entangled bank. Science. 2005;307:684–686. doi: 10.1126/science.1106935. [DOI] [PubMed] [Google Scholar]
- 21.Whittaker RH. Vegetation of the Siskiyou Mountains, Oregon and California. Ecol. Monogr. 1960;30:280–338. doi: 10.2307/1943563. [DOI] [Google Scholar]
- 22.Jost L. Partitioning diversity into independent alpha and beta components. Ecology. 2007;88:2427–2439. doi: 10.1890/06-1736.1. [DOI] [PubMed] [Google Scholar]
- 23.Lee CB, Chun JH, Ahn HH. Elevational patterns of plant richness and their drivers on an Asian mountain. Nord. J. Bot. 2014;32:347–357. doi: 10.1111/j.1756-1051.2013.00181.x. [DOI] [Google Scholar]
- 24.Swenson NG, Enquist BJ. Opposing assembly mechanisms in a Neotrpoical dry forest: implications for phylogenetic and functional community ecology. Ecology. 2009;90:2161–2170. doi: 10.1890/08-1025.1. [DOI] [PubMed] [Google Scholar]
- 25.Kraft NJB, Ackerly DD. Functional trait and phylogenetic tests of community assembly across spatial scales in an Amazonian forest. Ecol. Monogr. 2010;80:401–422. doi: 10.1890/09-1672.1. [DOI] [Google Scholar]
- 26.Yang J, et al. Functional and phylogenetic assembly in a Chinese tropical tree community across size classes, spatial scales and habitats. Funct. Ecol. 2014;28:520–529. doi: 10.1111/1365-2435.12176. [DOI] [Google Scholar]
- 27.Cavender-Bares J, Kozak KH, Fine PVA, Kembel SW. The merging of community ecology and phylogenetic biology. Ecol. Lett. 2009;12:693–715. doi: 10.1111/j.1461-0248.2009.01314.x. [DOI] [PubMed] [Google Scholar]
- 28.Mayfield MM, Levine JM. Opposing effects of competitive exclusion on the phylogenetic structure of communities. Ecol. Lett. 2010;12:1085–1093. doi: 10.1111/j.1461-0248.2010.01509.x. [DOI] [PubMed] [Google Scholar]
- 29.Swenson NG. The assembly of tropical tree communities-the advances and shortcomings of phylogenetic and functional trait analyses. Ecolgraphy. 2013;36:264–276. doi: 10.1111/j.1600-0587.2012.00121.x. [DOI] [Google Scholar]
- 30.Wang J, Soininen J, He J, Shen J. Phylogenetic clustering increases with elevation for microbes. Environ. Microbiol. Rep. 2012;4:217–226. doi: 10.1111/j.1758-2229.2011.00324.x. [DOI] [PubMed] [Google Scholar]
- 31.Culmsee H, Leuschner C. Consistent patterns of elevational changes in tree taxonomic and phylogenetic diversity across Malesian mountain forests. J. Biogeogr. 2013;40:1997–2010. [Google Scholar]
- 32.González-Caro S, Umaña MN, Álvarez E, Stevenson PR, Swenwon NG. Phylogenetic alpha and beta diversity in tropical tree assemblages along regional scale environmental gradients in northwest South America. J. Plant Ecol. 2014;7:145–153. doi: 10.1093/jpe/rtt076. [DOI] [Google Scholar]
- 33.Helmus MR, Ives AR. Phylogenetic diversity-area curves. Ecology. 2012;93:S31–S43. doi: 10.1890/11-0435.1. [DOI] [Google Scholar]
- 34.Liu C, Dudley KL, Xu Z-H, Economo EP. Mountain metacommunities: climate and spatial connectivity shape ant diversity in a complex landscape. Ecography. 2017;40:1–11. doi: 10.1111/ecog.02974. [DOI] [Google Scholar]
- 35.Helmus MR, Savage K, Diebel MW, Maxted JT, Ives AR. Separating the determinants of phylogenetic community structure. Ecol. Lett. 2007;10:917–925. doi: 10.1111/j.1461-0248.2007.01083.x. [DOI] [PubMed] [Google Scholar]
- 36.Li XH, Zhu XX, Niu Y, Sun H. Phylogenetic clustering and overdispersion for alpine plants along elevational gradient in the Hengduan Mountains Region, southwest China. J. Syst. Evol. 2014;52:280–288. doi: 10.1111/jse.12027. [DOI] [Google Scholar]
- 37.Qian H, Hao Z, Zhang J. Phylogenetic structure and phylogenetic diversity of angiosperm assemblages in forests along an elevational gradient in Changbaishan, China. J. Plant Ecol. 2014;7:154–165. doi: 10.1093/jpe/rtt072. [DOI] [Google Scholar]
- 38.Dunn RR, et al. Climatic drivers of hemispheric asymmetry in global patterns of ant species richness. Ecol. Lett. 2009;12:324–333. doi: 10.1111/j.1461-0248.2009.01291.x. [DOI] [PubMed] [Google Scholar]
- 39.Jenkins CN, et al. Global diversity in light of climate change: the case of ants. Divers. Distrib. 2011;17:652–662. doi: 10.1111/j.1472-4642.2011.00770.x. [DOI] [Google Scholar]
- 40.Ordonez A, Svenning JC. Functional diversity of North American broad-leaved trees is codetermined by past and current environmental factors. Ecosphere. 2016;7:e01237. doi: 10.1002/ecs2.1237. [DOI] [Google Scholar]
- 41.Liu J, Yunhong T, Slik JWF. Topography related habitat associations of tree species traits, composition and diversity in a Chinese tropical forest. Forest Ecol. Manag. 2014;330:75–81. doi: 10.1016/j.foreco.2014.06.045. [DOI] [Google Scholar]
- 42.Jetz W, Rahbek C. Geographic range size and determinants of avian species richness. Science. 2002;297:1548–1551. doi: 10.1126/science.1072779. [DOI] [PubMed] [Google Scholar]
- 43.Vivian-Smith G. Microtopographic heterogeneity and floristic diversity in experimental wetland communities. J. Ecol. 1997;85:71–82. doi: 10.2307/2960628. [DOI] [Google Scholar]
- 44.Bledsoe BP, Shear TH. Vegetation along hydrologic and edaphic gradients in a North Carolina coastal plain creek bottom and implications for restoration. Wetlands. 2000;20:126–147. doi: 10.1672/0277-5212(2000)020[0126:VAHAEG]2.0.CO;2. [DOI] [Google Scholar]
- 45.Lundholm JT. Plant species diversity and environmental heterogeneity: spatial scale and competing hypotheses. J. Veg. Sci. 2009;20:377–391. doi: 10.1111/j.1654-1103.2009.05577.x. [DOI] [Google Scholar]
- 46.Laliberté E, et al. How does pedogenesis drive plant diversity? Trends Ecol. Evol. 2013;28:331–340. doi: 10.1016/j.tree.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 47.Bartels SF, Chen HYH. Is understory plant species diversity driven by resource quantity or resource heterogeneity? Ecology. 2010;91:1931–1938. doi: 10.1890/09-1376.1. [DOI] [PubMed] [Google Scholar]
- 48.Lebrija-Trejos E, Pérez-García EA, Meave JA, Bongers F, Poorter L. Functional traits and environmental filtering derive community assembly in a species-rich tropical system. Ecology. 2010;91:386–398. doi: 10.1890/08-1449.1. [DOI] [PubMed] [Google Scholar]
- 49.Soininen J, McDonald R, Hillebrand H. The distance decay of similarity in ecological communities. Ecography. 2007;30:3–12. doi: 10.1111/j.0906-7590.2007.04817.x. [DOI] [Google Scholar]
- 50.Zhang JL, et al. Phylogenetic beta diversity in tropical forests: implications for the roles of geographical and environmental distance. J. Syst. Evol. 2013;51:71–85. doi: 10.1111/j.1759-6831.2012.00220.x. [DOI] [Google Scholar]
- 51.Qian H, Jin Y. An updated megaphylogeny of plants, a tool for generating plant phylogenies and an analysis of phylogenetic community structure. J. Plant Ecol. 2016;9:233–239. doi: 10.1093/jpe/rtv047. [DOI] [Google Scholar]
- 52.Zanne AE, et al. Three keys to the radiation of angiosperms into freezing environments. Nature. 2014;506:89–92. doi: 10.1038/nature12872. [DOI] [PubMed] [Google Scholar]
- 53.Webb CO, Ackerly DD, Kembel S. Phylocom: software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics. 2008;24:2099–2101. doi: 10.1093/bioinformatics/btn358. [DOI] [PubMed] [Google Scholar]
- 54.Blomberg SP, Garland T, Jr., Ives AR. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution. 2003;57:717–745. doi: 10.1111/j.0014-3820.2003.tb00285.x. [DOI] [PubMed] [Google Scholar]
- 55.Pagel M. Inferring the historical patterns of biological evolution. Nature. 1999;401:877–884. doi: 10.1038/44766. [DOI] [PubMed] [Google Scholar]
- 56.Brown JL, Cameron A, Yoder AD, Vences M. A necessarily complex model to explain the biogeography of the amphibians and reptiles of Madagascar. Nat. Commun. 2014;5:5046. doi: 10.1038/ncomms6046. [DOI] [PubMed] [Google Scholar]
- 57.Legendre, P. & Legendre, L. Numerical ecology. (Elsevier, 1998).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.