Abstract
Breeding for drought-tolerant crops is a pressing issue due to the increasing frequency and duration of droughts caused by climate change. Although important sources of variation for drought tolerance exist in wild relatives, the mechanisms and the key genes controlling tolerance in tomato are little known. The aim of this study is to determine the drought response of the tomato wild relative Solanum pennellii (Sp) compared with the cultivated tomato Solanum lycopersicum (Sl). The paper investigates the physiological and molecular responses in leaves of Sp and Sl plants without stress and moderate drought stress. Significant physiological differences between species were found, with Sp leaves showing greater ability to avoid water loss and oxidative damage. Leaf transcriptomic analysis carried out when leaves did not as yet show visual dehydration symptoms revealed important constitutive expression differences between Sp and Sl species. Genes linked to different physiological and metabolic processes were induced by drought in Sp, especially those involved in N assimilation, GOGAT/GS cycle and GABA-shunt. Up-regulation in Sp of genes linked to JA/ET biosynthesis and signaling pathways was also observed. In sum, genes involved in the amino acid metabolism together with genes linked to ET/JA seem to be key actors in the drought tolerance of the wild tomato species.
Introduction
Most crops are susceptible to drought stress, a main environmental factor responsible for yield losses worldwide1. The problem of drought stress will worsen in arid and semiarid regions, which are the most affected by the forecasted effects of climate change, and thus pose a serious risk to crop yields and threaten food security2,3. In spite of the great efforts invested in crop breeding for drought stress tolerance, the development of tolerant varieties has been slow and is greatly restricted by narrow genetic variations in crops, and by the complex and multigenic nature of the trait of drought tolerance4,5. In order to achieve the knowledge required to develop genotypes with enhanced tolerance to drought stress, it is essential to combine the descriptive power of the physiological analysis with the identification of key genes involved in water stress tolerance6. Transcriptomic analysis is one useful approach to identify genes playing important roles in drought tolerance and to infer the main mechanisms involved7,8. However, it is necessary to take into account that the candidate genes will be different, depending on the drought intensity and the exposure time. Severe water stress during very short periods (hours) will identify genes related to survival as there is no time for the plant to trigger the mechanisms involved in tolerance to long-term. On the other hand, plant response to moderate drought may enable a balance between growth and defence mechanisms under water-limiting conditions9, a more real scenario in agronomic conditions, albeit one that is poorly understood10,11.
Tomato is considered one of the most economically important horticultural crops grown worldwide12,13. What is more, it is an important crop in agriculture in arid and semi-arid zones. Abiotic stresses, like those promoted by water deficiency, have a negative impact on tomato production, particularly in the Mediterranean area14. Despite the economic importance of tomato, the mechanisms that govern responses to water stress in this horticultural species are not well characterized, and only a small number of genes playing a role in tomato tolerance to drought have been identified15,16. Moreover, despite the wealth of sources of variation for drought tolerance in accessions of tomato wild related species17, we still do not know the key genes controlling the tolerance. The wild species Solanum pennellii is a drought-adapted species and it constitutes an ideal experimental model to advance in our understanding of the underlying molecular mechanisms of drought adaptation and tolerance in tomato18. Moreover, the sequencing of its genome has recently been published and has assisted in the identification of candidate genes involved in stress tolerance19. Therefore, S. pennellii represents a particularly valuable genetic source for breeding programmes targeted at improving drought tolerance in tomato18.
The tolerance of plants to moderate drought stress is the sum of two processes: the plant’s ability to uptake water by the roots, which is generally associated with increased root growth, and its capacity to avoid excessive dehydration of the leaves. However, the beneficial aspects of each process are difficult to separate. The main strategy to date for increasing drought tolerance has been to increase the ability for water uptake of the root system20,21. However, the identification of drought-responsive genes expressed above and below ground may enable the elucidation of other potential drought tolerance mechanisms. Previous results from our group showed that one of the strategies used by S. pennellii against osmotic stress induced by salinity22 was the avoidance of leaf dehydration by osmotic adjustment, inducing high accumulation of solutes during the stress period23,24. Interestingly, this adaptation strategy appeared earlier in the time-course of drought response of S. pennellii than in cultivated tomato. The tomato wild species accumulated organic solutes as sugars and the diamine putrescine if inorganic solutes were not sufficient to reduce the osmotic potential24,25. Another drought tolerant strategy is to limit water loss by leaf transpiration, which is used by S. pennellii under osmotic stress, as our previous results also demonstrated26,27. Thus, it seems that S. pennellii is well adapted against osmotic stress occurring during drought and salinity, showing no visual signs of stress. This tolerance to osmotic stress may be due to either different constitutive gene expression or specific gene expression induction involved in tolerance to this stress.
The comparative study of two phylogenetically closely related tomato species, domesticated S. lycopersicum and its wild-relative S. pennellii, with a notably different degree of drought tolerance, can increase our knowledge of the genes involved in this tolerance. Furthermore, S. pennellii may serve as a model to identify key genes of drought tolerance in the shoot, as the root development of this species is poor and, consequently, its high degree of drought tolerance might be a consequence of molecular processes generated in leaves after stress. In this study we investigated the physiological and molecular responses to moderate drought stress in leaves of cultivated tomato and S. pennellii, as the analysis was carried out when the leaves did not show visual dehydration symptoms (after four days of dehydration). Here we show the important constitutive gene expression differences between both species as well as the genes specifically induced by drought stress in the tolerant species S. pennellii, which are mainly involved in amino acid metabolism and hormones.
Results
The cultivated tomato and the wild species S. pennellii show important differences at the physiological level
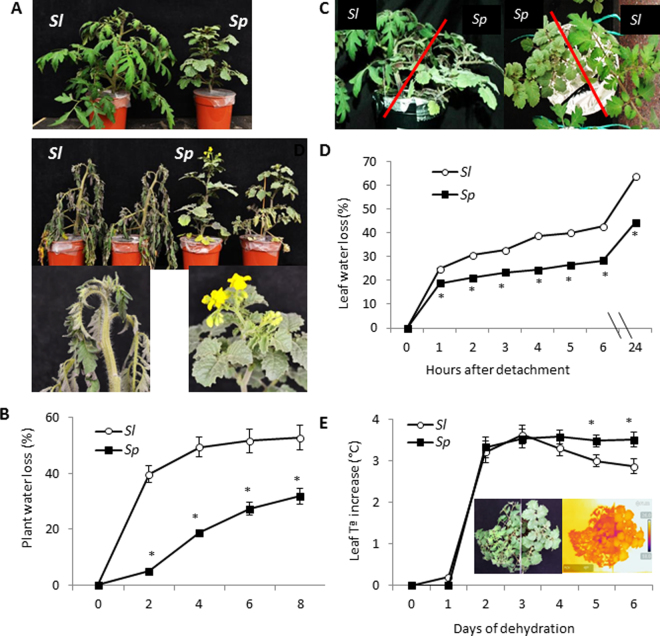
When plants of the cultivated tomato (S. lycopersicum, Sl) and the wild species S. pennellii) (Sp) were grown in separate pots, visual symptoms after 8 days of dehydration were evident in Sl leaves, including the shoot apex, while the leaves of Sp showed an aspect more similar to well irrigated plants (control) (Fig. 1A). This difference in visual symptoms was due to the lower transpired water percentages in Sp shoots compared with Sl throughout the dehydration period (Fig. 1B), which was reflected in the significantly higher shoot water content of Sp compared with Sl at the end of cycle (3.93 ± 0.06 and 6.41 ± 0.49 mL H2O g−1 DW in Sl and Sp respectively). In the following experiment, plants of Sl and Sp were grown side by side in the same pot, to ensure that both genotypes are exposed to the same severity of water stress28 (Fig. 1C). In order to corroborate that the dehydration tolerance of Sp is associated to a lower water loss occurring through the leaves, we first measured leaf water loss by using detached leaves from plants grown in control condition (Fig. 1D). Throughout the time of dehydration, leaves of Sp plants lost less water than Sl from the first hour of detachment, and differences between species held over 24 h. In addition, the leaf temperature (LT) was measured by infrared thermography during the first 6 days of dehydration (Fig. 1E). A significant increase of LT was observed between 1 and 2 days without irrigation in both tomato species. Afterwards, this parameter diverged in both species, decreasing in Sl from the 3rd to 6th days while it remained constant in Sp during this dehydration period. Representative thermal images of Sl and Sp plants at the 4th day of dehydration illustrate when Sp began to maintain higher LT than Sl (Fig. 1E).
Figure 1.
Differences in dehydration degree of cultivated tomato (Sl) and the wild species Solanum pennellii (Sp) subjected to drought stress by withholding water. (A) Representative pictures of well-watered plants in individual pots (top) and after 8 days of water withholding (bottom). Pictures of Sl and Sp apexes at the end of the dehydration cycle are shown below the images of whole plants. (B) Plant water loss (transpired water) during the dehydration cycle of plants in separate pots. (C) Picture of a pot with Sl and Sp plants, side-view and from above, after 4 days of withholding water. (D) Leaf water loss relative to day 0 from detached leaflets of Sl and Sp control plants grown in the same pot. (E) Changes in leaf temperature (LT) determined by infrared thermography in Sl and Sp plants placed in the same pot along the dehydration cycle (digital and thermal images at day 4 of water stress treatment shown within the graphic). Results are expressed as mean ± s.e.m., asterisk indicates significant differences between means values of Sl and Sp (Student t-test) for P < 0.05.
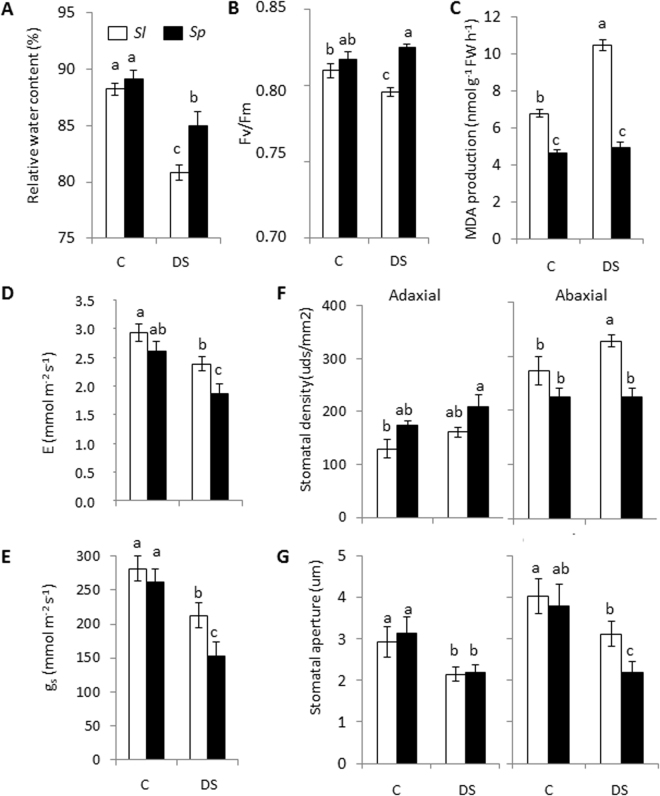
In order to study the differences found between both species at moderate drought stress more deeply, the experiment of dehydration with plants of Sl and Sp grown in the same pot was repeated again for the first 4 days of dehydration, as at this dehydration time neither the Sl nor Sp plants showed visual dehydration symptoms yet (Fig. 1C). The values of leaf relative water content (RWC) and leaf PSII photochemical efficiency, measured by Fv/Fm ratio, were similar in both species in the control, while they were significantly higher in Sp leaves compared with Sl under drought stress (Fig. 2A,B). Photosynthetic rate values were similar in both species (12.9 ± 0.8 and 13.6 ± 0.6 µmol m−2 s−1 in Sl and Sp at day 0 and 13.2 ± 0.4 and 12.4 ± 0.7 µmol m−2 s−1 in Sl and Sp after 4 days of dehydration). In addition, we determined lipid peroxidation measured as malondialdehyde (MDA) production, which is a well-recognized marker for oxidative stress and damage on plants, and we observed that MDA production increased under drought stress only in Sl leaves (Fig. 2C). Regarding gas exchange parameters stomatal conductance (gs) and transpiration rate (E), significant differences between species were observed only in drought stress, with the values being significantly lower in Sp leaves (Fig. 2D,E). In addition, stomatal density and aperture were analysed in the leaf adaxial and abaxial surfaces, since gs, a major factor determining the rate of water loss via transpiration, is modulated by both stomatal density and aperture (Fig. 2F,G). Significant changes were found only at the abaxial surface, with both stomatal density and aperture decreasing in Sp with respect to Sl under drought stress. Taken together, these results hint at the avoidance of water loss through leaves as a main response of Sp against drought stress, which is associated to its ability to reduce stomatal density and aperture in the abaxial surface.
Figure 2.
Differences induced by drought stress at the physiological and anatomical levels in leaves of Sl and Sp plants placed in the same pot. Relative water content (A), chlorophyll fluorescence (B) and lipid peroxidation degree determined by MDA production (C) in leaves of Sl and Sp plants placed in the same pot, in control (C) and drought stress (DS, 4 days of withholding water). Transpiration rate (E) (D) and stomatal conductance (gs) (E) in C and DS. Stomatal density (F) and aperture (G) determined in the abaxial and adaxial leaf side of Sl and Sp plants, in C and DS. Results are expressed as mean ± s.e.m. Values with different letters are significantly different as determined by LSD (P < 0.05).
Comparative transcriptomic analysis in leaves of cultivated and wild tomato species
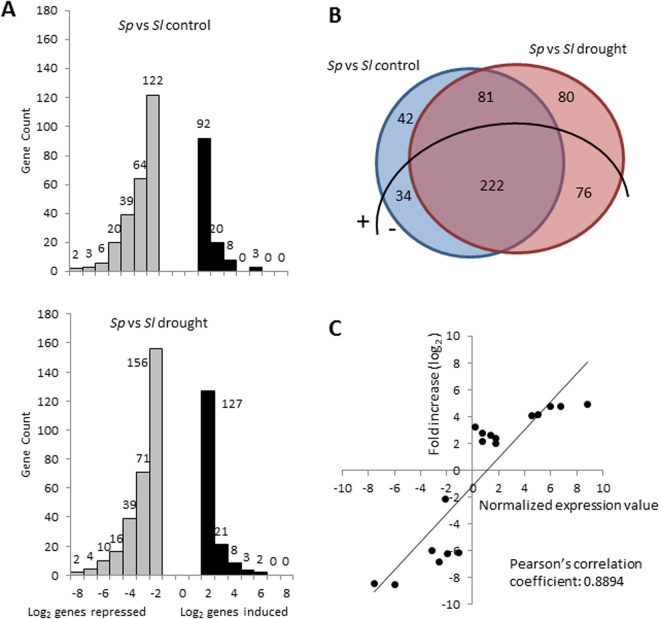
The comparative transcriptomic analysis, using the GeneChip® Tomato Genome Array (Affymetrix), was performed on leaves coming from the last water stress experiment. Samples were taken at day 0 (control) and after 4 days of dehydration, when no dehydration symptoms were observed (Fig. 1C). Four comparisons were performed: Sp vs Sl in control to identify constitutive expression differences between species; Sp vs Sl in drought stress to identify transcripts affected in Sp under stress, which may explain the drought tolerance of Sp when compared with Sl; Sp drought stress vs Sp control to identify transcripts affected specifically in Sp by drought stress; and Sl drought stress vs Sl control in order to know whether the S. pennellii transcripts are specific. The number of up- and down-regulated differentially expressed genes (DEGs) showing fold changes values between 2.0 and 3.0 outweighed the rest of the DEGs, especially in the case of up-regulated ones, and the number of down-regulated DEGs doubled that of up-regulated ones in both control and drought stress (Fig. 3A). The Venn diagram of the comparison between species showed increases of approximately two-fold in the number of genes expressed only under drought stress compared with those expressed only in control (Fig. 3B). For validation of transcriptomic data, 11 DEGs showing different expression patterns when comparing Sp with Sl (up- and down-regulated in control and drought stress) were analysed by qRT-PCR (Fig. S1). As shown in Fig. 3C, a significant correlation between the microarray data and qRT-PCR was observed, as the Pearson’s correlation coefficient was 0.8894, which upholds the transcriptomic data.
Figure 3.
Leaf transcriptome of Solanum pennellii (Sp) relative to cultivated tomato (Sl). (A) Distribution of genes induced (black) and repressed (grey) in Sp respect to Sl in control and drought (after 4 days of dehydration) by the Log2 ratio of folds expression. (B) Venn diagram showing the number of genes differentially expressed in two comparisons, as well as genes commonly expressed, discriminating up- (+) and down- (−) regulated genes, considering genes showing a FDR < 0.05 and a fold-change threshold value (Log2) of 2.0 as differentially expressed genes (DEGs). (C) Correlation analysis between microarray (y-axis) and RT-qPCR (x-axis) data. The relative expression values obtained by microarray (Log2) were compared with those obtained by RT-qPCR using the ΔΔCt method. The Pearson’s correlation coefficient between relative expression levels is shown. RNA from leaflet tissue of plants grown in control was used as calibrator sample.
Each set of DEGs distinguished in the Venn diagram was functionally classified with the MapMan software29. In Fig. S2 DEGs from the comparison Sp vs Sl are represented, classified in functional categories according to the MapMan software. The functional classifications of up- and down-regulated DEGs in Sp compared with Sl are shown separately in control, drought stress and both conditions (Tables S1–S6), as well as the up-and down-regulated DEGs in drought stress compared with control, also shown separately in Sp, Sl and both species (Tables S7–S12). The detailed information about DEGs presented in the following sections is summarized in Table S13.
Constitutive gene expression differences between cultivated and wild tomato species
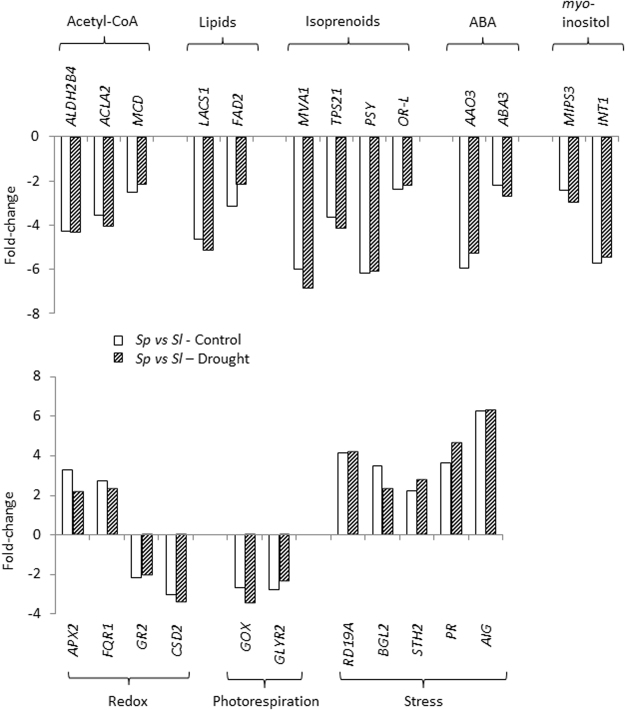
A set of constitutively down-regulated transcripts was found in Sp compared with Sl, including genes linked to acetyl-CoA production as well as to interconnected pathways of fatty acids and isoprenoid biosynthesis, and all of them were similarly reduced under drought stress (Fig. 4 and Table S5). Among them, it is interesting to point out the gene linked to the carotenoids biosynthesis, upstream of the ABA biosynthetic pathway, phytoene synthase (PSY). In addition, the abscisic aldehyde oxidase 3 (AAO3) gene involved in the last step of ABA production together with the Arabidopsis ortholog ABA3, whose activity is required for AAO functionality, were also down-regulated in Sp in both control and drought stress (Fig. 4). Two other DEGs showing similar reduced basal levels in Sp with respect to Sl in control and drought stress were involved in myo-inositol biosynthesis and transport: myo-inositol-1-phosphate synthase 3 (MIPS3) and myo-inositol transporter INT1.
Figure 4.
Constitutive expression of differentially expressed genes in Sp vs Sl. Genes are linked to different metabolic and physiological processes in leaves of Sp vs Sl. Plants were grown in control and drought (4 days of dehydration) conditions. Values of gene expression are represented as fold-changes (Log2 scale).
Different changes regarding the expression profile involved in redox homeostasis were detected (Fig. 4 and Tables S2 and S5). The genes encoding for a flavodoxin-like quinone reductase (FQR1) and a cytosolic ascorbate peroxidase (APX2) were up-regulated in Sp compared with Sl in both control and drought stress, while genes encoding for the chloroplastic forms of glutathione reductase (GR) and Cu/Zn superoxide dismutase (SOD) showed the opposite response. One source of ROS is the activity of glycolate oxidase (GOX), occurring in peroxisomes during photorespiration30. Two genes linked to photorespiration were down-regulated in Sp compared with Sl in both control and drought stress, glycolate oxidase (GOX) and glyoxylate reductase 2 (GLYR2) (Fig. 4 and Table S5).
Several stress-responsive genes were up-regulated in Sp compared with Sl, with quite similar fold-change values in control and drought stress (Fig. 4 and Table S2), as observed in the RESPONSIVE TO DEHYDRATION 19A (RD19A) gene. When expression analysis using RT-qPCR was performed for RD19A, levels were similar to the expression found in the microarray analysis (Fig. S1). There were also DEGs coding for the pathogenesis-related (PR) proteins including beta 1,3-glucanase 2 (BGL2) and SALT TOLERANCE HOMOLOG 2 (STH2), and another coding for a putative PR protein, all of them up-regulated. Another stress-responsive gene highly up-regulated in Sp compared with Sl in control and drought stress was the Arabidopsis ortholog AVIRULENCE INDUCED GENE PROTEIN (AIG), which has been described in plant responses to biotic stress.
Finally, a significant number of DEGs with unknown functions were detected in Sp in both control and drought stress (Tables S2 and S5). It is important to note the high expression levels found between species of two genes coding for elongation factors (EFs), GTP-binding elongation factor Tu (EF-Tu) and Elongation factor 1B gamma (EF-1Bg): the former down-regulated and the latter up-regulated in Sp compared with Sl (fold-change values of −8.53 and −8.41 for the former in control and drought stress respectively, and 4.97 and 4.82 for the latter). In order to corroborate their high levels of differential expression, both genes were validated by RT-qPCR, confirming the expression patterns from the microarray analysis (Fig. S1).
Genes induced by drought in the wild species S. pennellii
In relation to the C metabolism, the FRUCTOSE INSENSITIVE 1 (FINS1) gene, whose product is a cytosolic fructose-1,6-bisphosphatase, was specifically up-regulated in Sp compared with Sl under drought stress. In contrast, the gene involved in starch biosynthesis ADP-glucose pyrophosphorylase 1 (ADG1) was down-regulated (Fig. 5 and Tables S3 and S6), which suggests that Sp prevents a shift of carbon allocation towards starch synthesis.
Figure 5.
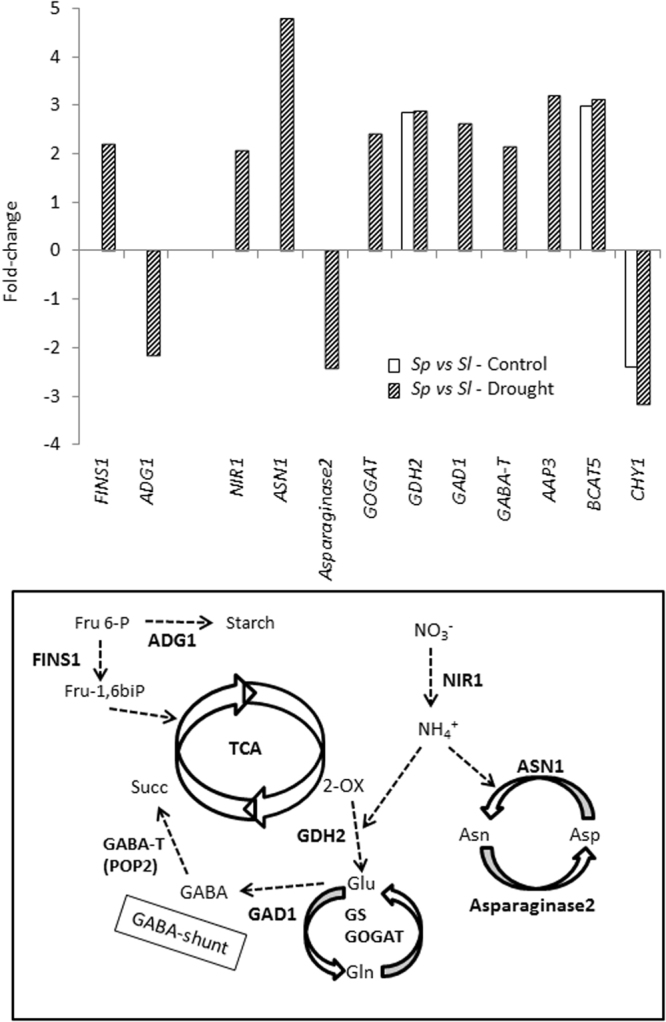
Genes linked to the C/N metabolism are upregulated by drought in leaves of Sp compared with Sl. Plants were grown in control and drought (4 days of dehydration) conditions. Values of gene expression values are represented as fold-changes (Log2 scale).
A remarkable number of genes involved in N metabolism were up-regulated in Sp respect to Sl, and most of them were only induced under drought stress (Fig. 5, and Tables S2, S3, S5 and S6). Thus, two genes involved in N assimilation, nitrite reductase 1 (NIR1) and glutamine-dependent asparagine synthase 1 (ASN1), were induced by drought stress in Sp. When expression analyses using RT-qPCR were performed for these two genes, NIR1 and ASN1, results confirmed the expression patterns of the same genes from the microarray analysis (Fig. S1). Importantly, one way of reducing the excess NH4+ caused by stress is to integrate it in the amino acid metabolism, and besides ASN1 induction, the gene asparaginase 2 involved in the reverse reaction (the production of aspartate from asparagine and release of NH4+) was specifically down-regulated by drought stress in Sp. Another gene up-regulated in Sp compared with Sl is NADH-dependent glutamate synthase 1(GLT1), whose gene product is a chloroplastic NADH-dependent glutamate synthase (also known as glutamine-oxoglutarate aminotransferase, GOGAT). Another metabolic way of assimilating most inorganic N, especially excess NH4+ occurring in stressful conditions, is through the glutamine synthetase/glutamate synthase (GS/GOGAT) cycle. A DEG coding for the Arabidopsis ortholog glutamate dehydrogenase 2 (GDH2) was already induced at basal level in Sp respect to Sl and was maintained in drought stress. In addition, genes involved in the ϒ-aminobutyric acid (GABA) metabolism, glutamate decarboxylase 1 (GAD1) and ϒ-aminobutyrate transaminase (GABA-T), were specifically induced by drought stress in Sp compared with Sl. A DEG coding for an amino acid transporter, the Arabidopsis ortholog amino acid permease 3 AAP3, has been found to be stress-responsive and it is markedly up-regulated in Sp compared with Sl, which may intervene in the amino acids transport among cytosol, mitochondrion and chloroplast, where the above mentioned gene products exert their functional activity. Moreover, a gene involved in branched-chain-amino-acid biosynthesis, the branched-chain-amino-acid transaminase 5 (BCAT5), was induced in Sp both in control and drought stress, while the opposite response was found in the 3-hydroxyisobutyryl-CoA hydrolase 1 (CHY1) gene involved in BCAA catabolism. Taken together, the strategy used by the wild species Sp to respond to drought stress is to induce the gene expression linked to N metabolism, as it is schematically shown (Fig. 5).
In relation to leaf water loss, the carbonic anhydrase 1 (CA1) gene was induced by drought stress in Sp (Fig. 6A and Table S3), which is in accordance with the water loss reduction due to reduced stomatal density and aperture (Fig. 2F,G). In addition, drought stress-responsive genes involved in cell wall metabolism were also up-regulated by drought stress in Sp (Fig. 6A and Tables S2 and S3), as XTH16 (xyloglucan endotransglucosyl/hydrolase 16), already induced at the basal level and EXPB2 (expansin B2), both linked to cell wall extensibility. Moreover, the PARVUS/GLZ1 gene involved in cell wall thickening was up-regulated by drought stress in Sp.
Figure 6.
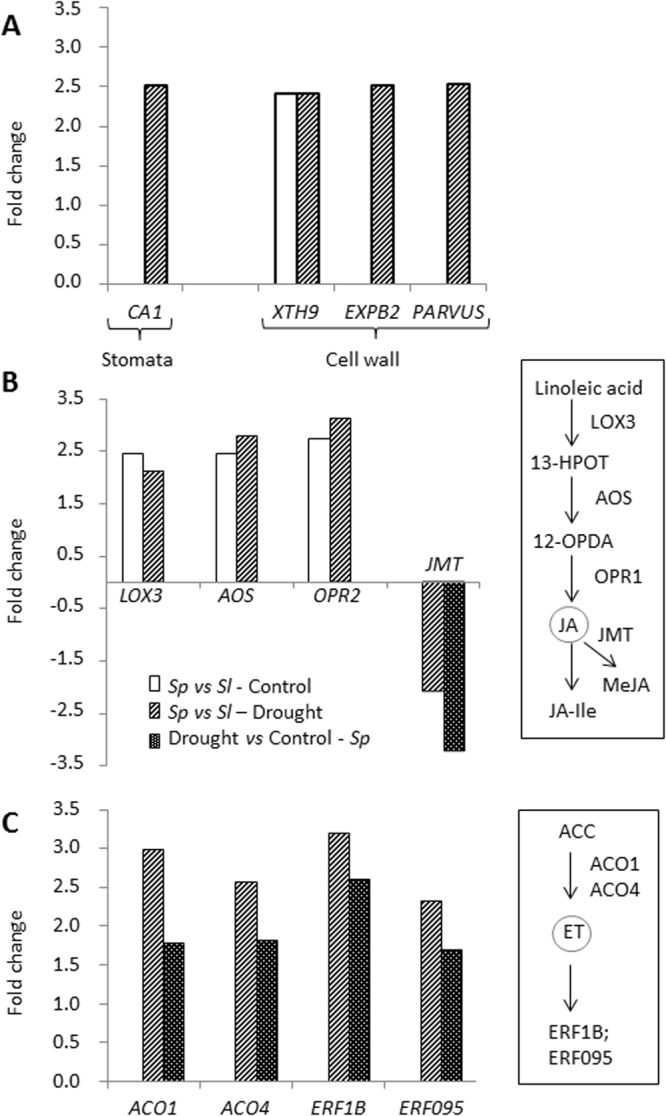
Induction of differentially expressed genes in Sp vs Sl and by drought in Sp. Genes linked to stomata movement and cell wall physiology (A), and JA (B) and ET (C) metabolism are upregulated by drought in leaves of Sp with respect to Sl. Plants were grown in control and drought (4 days of dehydration) conditions. Values of gene expression are represented as fold-changes (Log2 scale).
Regarding hormones metabolism, different genes involved in JA and ET were up-regulated in Sp compared with Sl. Thus, genes involved in JA biosynthesis lipoxygenase 3 (LOX3), allene oxide synthase (AOS) and OPDA reductase 2 (OPR2) were up-regulated in Sp compared with Sl in both control and drought stress (Fig. 6B and Table S2). However, jasmonic acid carboxyl methyltransferase (JMT) gene, whose product catalyses the conjugation of JA to form the non-bioactive compound methyl jasmonate (MeJA), was down-regulated by drought stress in Sp vs Sl, as well as in the comparison between drought stress vs control of Sp (Fig. 6B and Tables S6 and S10). Regarding ET biosynthesis and signaling, it is notable that genes induced by drought stress in Sp vs Sl were also up-regulated when comparing drought stress vs control in Sp (Fig. 6C and Tables S3 and S7). Thus, two ACC oxidases (ACO1 and ACO4), involved in the last step of ET biosynthesis, and the ethylene-responsive element binding factors ERF1B and ERF95 were all up-regulated in both comparisons, which suggests that ET biosynthesis and signaling are important issues in the drought response of the wild tomato species.
Finally, as in the case of genes with constitutive expression, we found a notable proportion of up-regulated DEGs with unknown functions, achieving percentages of around 20% with respect to the total DEGs in both comparisons, Sp vs Sl in drought stress, and drought stress compared with control in Sp.
Discussion
Differences in tolerance mechanisms between wild and cultivated tomato species may result from changes in constitutive basal levels of genes involved in different processes17. In the wild species Sp, genes involved in acetyl-CoA and the branched pathways of fatty acids and isoprenoids biosynthesis, as well as genes linked to myo-inositol and photorespiration, were constitutively down-regulated, compared with Sl, and these differences were maintained under drought (Fig. 4). The lower expression of genes in Sp could be due simply to a higher requirement of these essential components of the metabolism in domesticated tomato, since the trend of gene expressions is generally the same in control and drought. However, it could also point to a more efficient metabolism in a species where its natural habitat is already stressed. Thus, in photorespiration there is a particular reaction responsible for the highest amount of H2O2 produced in this metabolic pathway, which must be tightly regulated for the sake of redox homeostasis, which is the oxidation of glycolate to glyoxylate in peroxisome generating H2O2, making this organelle one of the main intracellular sites of ROS production30. The fact that the expression of GOX, involved in the conversion of glycolate into glyoxylate, was constitutively down-regulated in Sp compared with Sl suggests that Sp is able to reduce the H2O2 and glyoxylate accumulation. Moreover, the presumably lower amount of GOX present in Sp evidences the lack of need to activate the alternative route of glyoxylate conversion catalysed by GLYR, as the constitutive down-regulation of GLYR2 suggests (Fig. 4).
The wild species S. pennellii shows high ability to prevent leaf water loss
The changes observed at the physiological level demonstrate the ability of Sp to avoid leaf water loss under drought conditions, as Sp reduced gs and E compared with Sl, and Sp plants exhibited lower stomatal density and aperture in the leaf abaxial surface (Fig. 2D–G). Taking into account that the regulation of stomatal degree of aperture and density in the leaves must be highly controlled for the plant to be able to adapt to stressful conditions31, this response of Sp seems to be generalized to the osmotic effects induced not only by drought but also by other abiotic stress. Thus, we recently observed that Sp increased the water content of their leaves by reducing stomatal density and aperture under salinity27.
Numerous studies point out the importance of cell wall in limiting the water loss under drought32,33. In this sense, the ability of Sp to prevent leaf water loss might be associated with the up-regulation of genes involved in cell wall extensibility, like XTH16 and EXPB2, and cell wall thickening, like PARVUS/GLZ1 (Fig. 6A). In relation to water loss through stomata, carbonic anhydrases (CAs) are main regulators of stomatal movement and density, inducing stomatal closure34–36. In photosynthetic tissues, CA contributes to mesophyll conductance (gm) by maintaining the CO2-HCO3− equilibrium in the cytosol and chloroplasts, so facilitating access to CO2 for fixation by Rubisco37,38. Recently, Momayyezi and Guy39 indicated that CA activity plays a significant role in mediating the mesophyll resistance to CO2 diffusion in black cottonwood. Our study hints that the wild species is trying to reduce water loss by inducing the closure of stomata via CA, as the drought-specific upregulation of CA1 gene suggests, which is in line with the reduction of stomatal density and aperture as well as gs and E found in this wild tomato species in water stress (Fig. 2D–G).
The strategy of modifying cell wall properties or closing stomata in order to avoid water loss must be balanced since it triggers a concomitant reduction of CO2 uptake, affecting plant growth40,41. Recently, Onoda et al.42 indicated that a greater fraction of leaf mass in cell walls is associated with a lower fraction of leaf N invested in photosynthetic proteins, and lower within-leaf CO2 diffusion rates due to thicker mesophyll cell walls. In addition, stomatal closure may disturb the redox homeostasis30. However, Sp seems to be able to achieve a trade-off between water loss and growth maintenance. Thus, Sp suffers a lower degree of oxidative stress according to the MDA levels, an indicator of oxidative stress, as the values increased with drought stress in Sl but not in Sp leaves (Fig. 2C). Furthermore, the APX2 gene was induced in Sp with respect to Sl, which favours the maintenance of redox homeostasis in the wild tomato (Fig. 4). On the other hand, C depletion as consequence of stomatal closure does not seem to occur in Sp but rather the opposite, according to the upregulation of FINS1 gene, involved in the synthesis of Fru-6-P, together with the downregulation of ADG1 gene, involved in the starch biosynthesis (Fig. 5), which suggests that Sp is preventing a shift of C allocation towards starch synthesis in order to mobilize sugars. Furthermore, the down-regulation in Sp of MIPS3 gene (Fig. 4), which codifies for the rate-limiting enzyme in the synthesis of myo-inositol43, seems to contribute to the avoidance of deviating C towards others metabolites, as myo-inositol is derived from glucose-6-phosphate.
Genes involved in amino acid metabolism and JA/ET are induced by drought in the wild species S. pennellii
Although maintenance of C/N balance in plant metabolism is critical in plant growth and has to be finely regulated in stressful conditions44, to date little is known about the regulation of genes involved in N metabolism in Sp under drought. Interestingly, an important number of genes involved in N metabolism were up-regulated by drought in Sp (Fig. 5), as GDH2 and ASN1, involved in the synthesis of Glu and Asn, respectively. The production of Asn is a main way for N transport and storing in higher plants, since this amino acid has a high N/C ratio and is very stable45, with ASN playing a key role in the response to diverse stresses44,46. It is interesting to point out that the high expression level of ASN1 in drought-stressed leaves of Sp was accompanied by inhibition of Asparaginase2, whose product catalyses the opposite process. In addition, the Sp tolerance also seems to be related to induction of the GOGAT/GS cycle and the GABA-shunt, as GLT1,GAD1 and GABA-T are specifically induced by drought stress in Sp. The activation of the GABA-shunt stimulates the conversion of Glu into succinate ready to be used by the TCA cycle, so contributing to maintain the C/N balance in Sp under drought conditions. Michaeli et al.47 confirmed the importance of GABA as a precursor for multiple metabolic pathways, including the TCA cycle and polyamine metabolism. In this respect, previous studies showed that the main differences between the two tomato species lie in an earlier and greater accumulation of putrescine in Sp than in Sl under salt stress, and these changes were associated with changes in the amino acids levels related to its synthesis as Glu24. Although there are a number of studies demonstrating involvement of the GABA shunt in response to abiotic stresses48, information on the molecular mechanism underpinning the role of this pathway in the drought tolerance of Sp is practically non-existent. It is interesting to note that in a recent study carried out with tomato, the drought tolerance induced by paclobutrazol application was related to increased expression of genes involved in TCA cycle and GABA shunt49. In sum, a higher replenishment of the TCA cycle through activation of the GS/GOGAT cycle and the GABA-shunt seems to be associated to the drought tolerance of Sp. Finally, it is interesting to point out that increased N assimilation may improve water status of the plant via reductions in gs and E50,51, such as has been observed in Sp under drought (Fig. 2D,E).
Hormones are central integrators that link and control the complex stress-adaptive signaling cascades52. JA is considered a key regulator of expression of stress-responsive genes in virtually any plant species53–55. Recently, we identified the res (restored cell structure by salinity) tomato mutant that has constitutively activated the JA biosynthesis pathway, and it exhibits tolerance to abiotic stresses56,57. Interestingly, we observed that genes from the JA biosynthesis pathway, such as LOX3, AOS and OPR2, are constitutively up-regulated in Sp compared with Sl, and differences were maintained in drought stress (Fig. 6).
Another hormone involved in the plant response to abiotic stress is ET58. Note that genes involved in ET biosynthesis and signaling, specifically in the steps catalysed by ACO and regulated by ERFs, are induced by drought stress in Sp leaves (Fig. 6). Interestingly, Phukan et al.59 indicated that ERFs regulate the expression of a wide variety of down-stream target genes related to stress response and development through different mechanisms, and in our study both ERF1B and ERF095 are up-regulated in Sp. Although the role of ERFs in hormone cross-talk under drought is still scarcely known60, it has been suggested that ERF1 induction requires ET as well as JA signaling under different abiotic stress conditions61,62. In addition, it has been shown that ET/JA signaling is also required for the induction of other ERFs in response to abiotic stresses63, as observed in ERF095 (Fig. 6). Regarding the oxidative stress, overexpression of LeERF1 and LeERF2 reduced MDA levels in tomato plants under salt stress64. Our results are in concordance with those obtained in tomato, as the MDA values were lower in Sp leaves compared with Sl ones in drought stress (Fig. 2C). On the other hand, ERF1 seems to activate specific sets of stress-responsive genes by targeting their DRE elements during abiotic stress, among them the responsive to dehydration RD20 and RD29B genes62. In our study the expression of RD19A was induced in Sp compared with Sl, although this is constitutive (Fig. 4). Taken together, the induced expression of ET metabolism/signaling genes in Sp seems to be related to its drought tolerance.
It has been pointed out that ET/JA among other hormones signaling components regulates the trade-off between plant growth and stress tolerance65–67. One facet of this regulation occurs when interacting with the N metabolism, as observed by Zhang et al.68 in roots. Our results for Sp leaves also hint in this direction, as genes involved in amino acid metabolism together with genes linked to ET/JA biosynthesis-signaling are the main ones induced by drought in the drought-tolerant species (Figs 5 and 6). Based on the overall results, a model of the different processes and genes involved in the Sp drought tolerance response was proposed (Fig. 7). Besides the coordinated regulation of genes involved in leaf water loss, redox homeostasis and N metabolism, genes linked to ET/JA pathways and the downstream ERFs may participate in N metabolism, which in turn may improve the leaf water loss. Although the existence of other mechanisms involved in regulating drought tolerance in Sp is not excluded, it is evident that the different genes identified in Sp leaves may be potentially important players in the drought response of the wild tomato.
Figure 7.
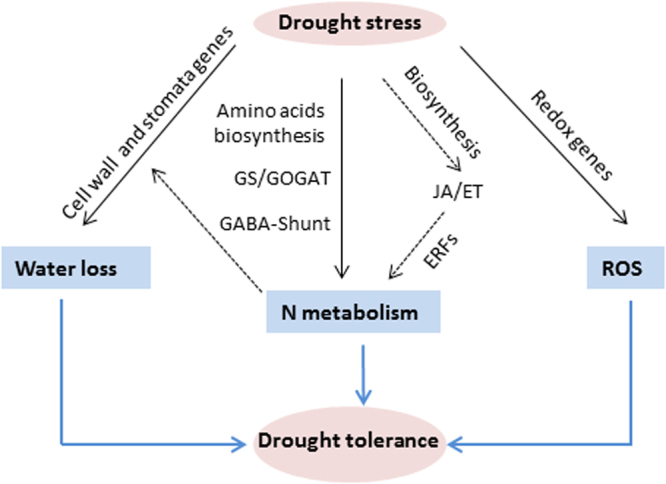
A proposed model for genes playing important roles in processes that regulate drought tolerance of Sp. Under drought, Sp reduces water loss by upregulation of genes involved in cell wall in conjunction with the stomatal regulator CA1 gene. At the same time, the low degree of oxidative stress that suffers Sp under drought stress is associated to the induction of genes involved in redox homeostasis, as APX2. In addition, genes linked to different processes of N metabolism and JA/ET pathways are induced by drought in Sp, in the manner that the downstream genes (ERFs) could participate in N metabolism, which in turn might improve the leaf water loss.
Material and Methods
Plant material and drought stress assays
Cultivated tomato (Solanum lycopersicum L.) cv. P73 and the wild species Solanum pennellii (Corell) accession PE47 (collected in Peru by Cuartero et al.69 were used in all experiments. Seeds were germinated in darkness in a 2:1 (v/v) mixture of peat:perlite, at 28 °C temperature and 90% relative humidity. The germination of S. pennellii was started 8 days earlier than cultivated tomato in order to get plants in a similar developmental stage when ready for the water stress experiments. Plant culture was carried out in a controlled conditions growth chamber, and the environmental conditions were 18–25 °C, 50–80% of relative humidity, and a photoperiod 16 h light/8 h dark. A photosynthetic photon flux density (PPFD) of 345 μmol m−2 s−1 at the plant level was provided by fluorescent tubes (Osram Lumilux daily-light 58 W and Fluora 58 W). Seedlings were transferred to plastic pots filled with peat and perlite (2:1), and were daily irrigated with half-strength Hoagland solution70.
Two different procedures were used for drought stress assays. In the first experiment plants of each species were placed in separated pots of 1.5-L capacity, while the two other experiments were carried out by placing one plant of each species in the same pot, although the volume of the pots was double (3-L). In all experiments, the control plants were irrigated daily up to pot capacity, and drought stress was applied by withholding irrigation, so water stress intensity increased with time of dehydration cycle. The drought stress was applied when the 4th true leaf was expanded. In order to avoid water evaporation from substrate, the pots were covered with parafilm, leaving room for only the plant main stem to protrude.
Three replicates per species and treatment were used in all experiments. Samples from the first two true leaves were harvested between 4–5 h after the beginning of the light period for physiological and molecular analyses. A pool was prepared from leaflets of these two leaves per sample and half of them were used for physiological analysis, and the other half were immediately frozen in liquid nitrogen and stored at −80 °C.
Physiological measurements
Plant water loss (transpired water) during dehydration cycle was determined as a percentage of the difference between the pot weight for each time and for day 0. Leaf water loss was determined in detached leaflets as described in Campos et al.71. Infrared thermography imaging and leaf surface temperatures measurements were performed following procedures described in Albaladejo et al.27. Gas exchange parameters, stomatal conductance (gs) and transpiration rate (E) were determined with an Infrared Gas Analyser (IRGA), following Campos et al.71. To assess the photochemical efficiency of photosystem II (PSII), chlorophyll fluorescent (Fv/Fm) was determined with a portable Chlorophyll Fluorometer (Opti-Sciences, Hudson, NH), following the protocol described in Garcia-Abellán et al.56. Stomatal density and aperture were measured by light microscopy, by means of the protocol described in Albaladejo et al.27.
Shoot water content was determined for the whole shoot (leaves and stems) according to the formula (FW − DW)/DW, where FW is the fresh weight and DW the dry weight obtained oven dried for 48 h at 80 °C. The leaf relative water content (RWC) was determined as (FW − DW)/(SW − DW), where SW is the saturation weight determined after 24 h re-saturation in tap water. Lipid peroxidation was determined to assess the impact of secondary oxidative stress in plants subjected to water stress. The thiobarbituric acid reactive substrates (TBARS) assay was used as previously described72.
Data were statistically analysed using ANOVA, with means separated by Student’s t-test (P < 0.05). The effects of treatment and tomato species were analysed using two-way ANOVA, with means separated by least significant difference (LSD) (P < 0.05). All data are given as mean ± standard error (s.e.m.). Significant differences between means are denoted by different lower case letters or asterisks.
Microarray hybridization and data analysis
Microarray hybridization was performed by inBIOnova Biotech S.L., a start-up company sited at the campus of the University of Murcia. RNA isolated from leaflets coming from three individual pooled plants (biological replicates) was used in hybridization to one chip, resulting in total 12 chips (three replicates per each species and treatment). RNA extraction was performed with RNeasy Mini kit and QIAshredder (Qiagen) following the manufacturer’s instructions, and the amount and quality of the RNA checked by Bioanalyzer and spectrophotometrically by Nanodrop. Biotinylated cRNA was synthetized from 200 ng of each sample using the GeneChip 3′ IVT Express kit (Affymetrix), according to the protocol supplied by the manufacturer. The amount and quality of biotinylated cRNA was checked by Nanodrop and agarose gel electrophoresis. The biotinylated cRNA targets were cleaned up and 15 µg were fragmented in order to prepare the hybridization mix, using the Hybridization, Wash and Stain kit (Affymetrix) according to recommendations of manufacturer. The resulting preparations were hybridized to GeneChip® Tomato Genome Array (Affymetrix) interrogating 10209 sequences. After applying hybridization (spike controls) and labelling (polyA controls and 3′/5′ ratios of housekeeping genes) tests, it was observed that the 12 chips have fulfilled the quality criteria.
With the aim of minimizing the discrepancies due to variables such as sample preparation, hybridization, etc. all the arrays were scaled by defining an arbitrary value of 500 as average intensity and using the Expression ConsoleTM (EC 1.1, Affymetrix®) software. The differences of scaling factor among the different samples did not exceed 3 in any case, so we conclude the results are sufficiently homogenous and feasible as required. The BioConductor affyPLM package was used to obtain Normalized Unsealed Standard Error (NUSE) plot to study the quality of the chips for each set of probes from intensity values of each probeset. All samples presented NUSE values within the required ranges. The intensity value of each probe in the array was processed and normalized according to the Robust Multichip Average (RMA) method to obtain an individual intensity value for each probeset. First filtering, normalization and second filtering of probes yielded a final list of 5050 sequences (working list). Non-supervised Principal Components Analysis (PCA) and hierarchical clustering were performed and showed that samples tend to separate according to genotype and condition. Differentially expressed genes (DEGs) were identified by the LIMMA test73, which was corrected for multiple test using the FDR74 on the working list. Genes with a FDR < 0.05 and a fold change (Log2 ratio value) of ≥2.0 when comparing genotypes in the same experimental condition were identified as DEGs. A final list of 535 genes was obtained with the above conditions. The annotation of probe sets was obtained from Affymetrix and loaded in the Partek Genomics Suite software (Partek Incorporated, St. Louis, USA). For the functional study of DEGs MapMan software was used29 since it has a more completed database for functional assignations. The Slyc_AFFY_SGN_BUILD2_070709 database was loaded and used for this functional analysis. The statistical analysis followed was of Willcoxon Rank Sum test with Benjamini-Hochberg correction.
Data deposition
The microarray data and the related analysis information from this research work were deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE97045.
Quantitative real-time PCR verification
The expression profiles obtained from microarray hybridization were further validated by RT-qPCR, using a set of 11 DEGs randomly selected (Table S14). For gene expression analysis, total RNA extracts isolated from leaflets coming from three individual pooled plants (biological replicates) from the microarray experiment were used. Total RNA was quantified in a GeneQuant II spectrophotometer (Pharmacia Biotech) and 5 μg were used for cDNA synthesis with First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). Quantitative real-time RT-qPCR was carried out as described by Garcia-Abellan et al.75 using 1 μL of undiluted cDNA mixed with iTaq™ Universal SYBR® Green Supermix (Bio-Rad) and 0.45 μL of forward and reverse primers in a CFX ConnectTM Real-Time PCR System (Bio-Rad) cycler. Serial dilutions of cDNA were used to make a standard curve to optimize amplification efficiency (Table S14). No template controls were included. All reactions were performed in triplicate with three different RNA extracts for each sample. The presence of a single band on an agarose gel electrophoresis and of a single peak in the melting temperature curve confirmed the specificity of qRT-PCR amplification. Relative expression data were calculated as described by Asins et al.76 using the tomato elongation factor 1α (LeEF1α, acc. AB061263) as housekeeping gene. Gene expression was quantified by the comparative method (2−ΔΔCt)77, using the expression level of each gene from leaflet tissue of plants grown in control condition as the calibrator sample.
Electronic supplementary material
Supplementary Figures S1 and S2, and Supplementary Tables S13 and S14
Supplementary Tables: Functional Classification of Differentially Expressed Genes
Acknowledgements
This work was supported by grants 19305/PI/14 from the SENECA Foundation (Region de Murcia, Spain) and AGL2015-64991-C3-2-R from the Spanish Ministry of Economy (MINECO, Spain). I.A. thanks the Spanish Ministry of Economy and Competitiveness for her FPI research contract (BES-2013-063526).
Author Contributions
I.E., M.C.B. and F.B.F. designed the research and conceived the project. I.A., V.M., B.M. performed the laboratory experiments. A.S. performed the bioinformatics analyses. F.B.F. and M.C.B. wrote the manuscript. I.E. and A.S. critically analysed the manuscript. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Isabel Egea and Irene Albaladejo contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-21187-2.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Singh D, Laxmi A. Transcriptional regulation of drought response: a tortuous network of transcriptional factors. Front. Plant Sci. 2015;6:895. doi: 10.3389/fpls.2015.00895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Godfray HCJ, et al. Food security: the challenge of feeding 9 billion people. Science. 2010;327:812–818. doi: 10.1126/science.1185383. [DOI] [PubMed] [Google Scholar]
- 3.Elliott J, et al. Constraints and potentials of future irrigation water availability on agricultural production under climate change. Proc. Nat. Acad. Sci. USA. 2014;111:3239–3244. doi: 10.1073/pnas.1222474110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hill CB, et al. Whole-genome mapping of agronomic and metabolic traits to identify novel quantitative trait loci in bread wheat grown in a water-limited environment. Plant Physiol. 2013;162:1266–1281. doi: 10.1104/pp.113.217851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Langridge P, Reynolds MP. Genomic tools to assist breeding for drought tolerance. Curr. Op. Biotechnol. 2015;32:130–135. doi: 10.1016/j.copbio.2014.11.027. [DOI] [PubMed] [Google Scholar]
- 6.Zhu M, et al. Molecular and systems approaches towards drought-tolerant canola crops. New Phytol. 2016;210:1169–1189. doi: 10.1111/nph.13866. [DOI] [PubMed] [Google Scholar]
- 7.Fan YY, et al. Transcriptome-wide characterization of candidate genes for improving the water use efficiency of energy crops grown on semiarid land. J. Exp. Bot. 2015;66:6415–6429. doi: 10.1093/jxb/erv353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sprenger H, et al. The drought response of potato reference cultivars with contrasting tolerance. Plant Cell Environ. 2016;39:2370–2389. doi: 10.1111/pce.12780. [DOI] [PubMed] [Google Scholar]
- 9.Claeys H, Inzé D. The agony of choice: How plants balance growth and survival under water-limiting conditions. Plant Physiol. 2013;162:1768–1779. doi: 10.1104/pp.113.220921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Des Marais DL, et al. Physiological genomics of response to soil drying in diverse Arabidopsis accessions. Plant Cell. 2012;24:893–914. doi: 10.1105/tpc.112.096180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clauw P, et al. Leaf responses to mild drought stress in natural variants of Arabidopsis. Plant Physiol. 2015;167:800–816. doi: 10.1104/pp.114.254284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwarz D, Thompson AJ, Kläring H-P. Guidelines to use tomato in experiments with a controlled environment. Front. Plant. Sci. 2014;5:625. doi: 10.3389/fpls.2014.00625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.FAOSTAT, FAO statistics. Rome, Italy. Food and Agriculture Organization of the United Nations. http://faostat3.fao.org/home/index.htlm (2014).
- 14.Saadi S, et al. Climate change and Mediterranean agriculture: Impacts on winter wheat and tomato crop evapotranspiration, irrigation requirements and yield. Agric. Water Manag. 2015;147:103–115. doi: 10.1016/j.agwat.2014.05.008. [DOI] [Google Scholar]
- 15.Iovieno P, et al. Transcriptomic changes drive physiological responses to progressive drought stress and rehydration in tomato. Front. Plant. Sci. 2016;7:371. doi: 10.3389/fpls.2016.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arms EM, Yan Z, StClair DA. Differential transcriptional regulation in roots of tomato near-isogenic lines in response to rapid-onset water stress. Front. Plant. Sci. 2017;8:166. doi: 10.3389/fpls.2017.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Escobar-Bravo R, et al. A Jasmonate-inducible defense trait transferred from wild into cultivated tomato establishes increased whitefly resistance and reduced viral disease incidence. Front. Plant Sci. 2016;7:1732. doi: 10.3389/fpls.2016.01732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Atarés A, et al. An insertional mutagenesis programme with an enhancer trap for the identification and tagging of genes involved in abiotic stress tolerance in the tomato wild-related species Solanum pennellii. Plant Cell Rep. 2011;30:1865–1879. doi: 10.1007/s00299-011-1094-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bolger A, et al. The genome of the stress-tolerant wild tomato species Solanum pennellii. Nat. Genet. 2014;46:1034–1038. doi: 10.1038/ng.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamaguchi M, Sharp RE. Complexity and coordination of root growth at low water potentials: recent advances from transcriptomic and proteomic analyses. Plant, Cell Environ. 2010;33:590–603. doi: 10.1111/j.1365-3040.2009.02064.x. [DOI] [PubMed] [Google Scholar]
- 21.Slovak R, Ogura T, Satbhai SB, Ristova D, Busch W. Genetic control of root growth: from genes to networks. Annals Bot. 2016;117:9–24. doi: 10.1093/aob/mcv160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munns R, Tester M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 23.Alarcon JJ, Sanchez-Blanco MJ, Bolarin MC, Torrecillas A. Water relations and osmotic adjustment in Lycopersicon esculentum and L. pennellii during short-term salt exposure and recovery. Physiol. Plant. 1993;89:441–447. doi: 10.1111/j.1399-3054.1993.tb05196.x. [DOI] [Google Scholar]
- 24.Santa-Cruz A, Acosta M, Rus A, Bolarin MC. Short-term salt tolerance mechanisms in differentially salt tolerant tomato species. Plant Physiol. Biochem. 1999;37:65–71. doi: 10.1016/S0981-9428(99)80068-0. [DOI] [Google Scholar]
- 25.Bolarin MC, Santa-Cruz A, Cayuela E, Perez-Alfocea F. Short-term solute changes in leaves and roots of cultivated and wild tomato seedlings under salinity. J. Plant Physiol. 1995;147:463–468. doi: 10.1016/S0176-1617(11)82184-X. [DOI] [Google Scholar]
- 26.Sanchez-Blanco MJ, Bolarin MC, Alarcón JJ, Torrecillas A. Salinity effects on water relations in Lycopersicon-esculentum and its wild salt-tolerant relative species L-pennellii. Physiol. Plant. 1991;83:269–274. doi: 10.1111/j.1399-3054.1991.tb02152.x. [DOI] [Google Scholar]
- 27.Albaladejo I, et al. Unravelling the strategies used by the wild tomato Solanum pennellii to confront salt stress: From leaf anatomical adaptations to molecular responses. Env. Exp. Bot. 2017;135:1–12. doi: 10.1016/j.envexpbot.2016.12.003. [DOI] [Google Scholar]
- 28.Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu J, Zhu J-K. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 2006;45:523–539. doi: 10.1111/j.1365-313X.2005.02593.x. [DOI] [PubMed] [Google Scholar]
- 29.Thimm O, et al. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–939. doi: 10.1111/j.1365-313X.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- 30.Miller G, Suzuki N, Cifti-Yilmaz S, Mittler R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant, Cell Environ. 2010;33:453–467. doi: 10.1111/j.1365-3040.2009.02041.x. [DOI] [PubMed] [Google Scholar]
- 31.Casson S, Gray JE. Influence of environmental factors on stomatal development. New Phytol. 2008;178:9–23. doi: 10.1111/j.1469-8137.2007.02351.x. [DOI] [PubMed] [Google Scholar]
- 32.Bray EA. Genes commonly regulated by water-deficit stress in Arabidopsis thaliana. J. Exp. Bot. 2004;55:2331–2341. doi: 10.1093/jxb/erh270. [DOI] [PubMed] [Google Scholar]
- 33.Le Gall H, et al. Cell wall metabolism in response to abiotic stress. Plants. 2015;4:112–166. doi: 10.3390/plants4010112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu H, et al. Carbonic anhydrases are upstream regulators of CO2-controlled stomatal movements in guard cells. Nat. Cell Biol. 2010;12:87–93. doi: 10.1038/ncb2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Engineer CB, et al. Carbonic anhydrases, EPF2 and a novel protease mediate CO2 control of stomatal development. Nature. 2014;513:246–250. doi: 10.1038/nature13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kollist H, Nukhat M, Roelfsema MRG. Closing gaps: linking elements that control stomatal movement. New Phytol. 2014;203:44–62. doi: 10.1111/nph.12832. [DOI] [PubMed] [Google Scholar]
- 37.Evans JR, Kaldenhoff R, Genty B, Terashima I. Resistances along the CO2 diffusion pathway inside leaves. J. Exp. Bot. 2009;60:2235–2248. doi: 10.1093/jxb/erp117. [DOI] [PubMed] [Google Scholar]
- 38.Perez-Martin A, et al. Regulation of photosynthesis and stomatal and mesophyll conductance under water stress and recovery in olive trees: correlation with gene expression of carbonic anhydrase and aquaporins. J. Exp. Bot. 2014;65:3143–3156. doi: 10.1093/jxb/eru160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Momayyezi M, Guy RD. Substantial role for carbonic anhydrase in latitudinal variation in mesophyll conductance of Populus trichocarpa Torr. & Gray. Plant Cell Environ. 2017;40:138–149. doi: 10.1111/pce.12851. [DOI] [PubMed] [Google Scholar]
- 40.Luan S. Signalling drought in guard cells. Plant Cell Environ. 2002;25:229–237. doi: 10.1046/j.1365-3040.2002.00758.x. [DOI] [PubMed] [Google Scholar]
- 41.Shabala S, Hariadi Y, Jacobsen S-E. Genotypic difference in salinity tolerance in quinoa is determined by differential control of xylem Na+ loading and stomatal density. J. Plant Physiol. 2013;170:906–914. doi: 10.1016/j.jplph.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 42.Onoda Y, et al. Physiological and structural tradeoffs underlying the leaf economics spectrum. New Phytol. 2017;214:1447–1463. doi: 10.1111/nph.14496. [DOI] [PubMed] [Google Scholar]
- 43.Luo Y, et al. D-myo-inositol-3-phosphate affects phosphatidylinositol-mediated endomembrane function in Arabidopsis and is essential for auxin-regulated embryogenesis. Plant Cell. 2011;23:1352–1372. doi: 10.1105/tpc.111.083337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rabara RC, et al. Tobacco drought stress responses reveal new targets for Solanaceae crop improvement. BMC Genomics. 2015;16:484. doi: 10.1186/s12864-015-1575-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cho C-W, et al. Molecular characterization of the soybean L-asparaginase gene induced by low temperature stress. Mol. Cells. 2007;23:280–286. [PubMed] [Google Scholar]
- 46.Gaufichon L, Reisdorf-Crena M, Rothstein SJ, Chardon F, Suzuki A. Biological functions of asparagine synthetase in plants. Plant Sci. 2010;179:141–153. doi: 10.1016/j.plantsci.2010.04.010. [DOI] [Google Scholar]
- 47.Michaeli S, et al. A mitochondrial GABA permease connects the GABA shunt and the TCA cycle, and is essential for normal carbon metabolism. Plant J. 2011;67:485–498. doi: 10.1111/j.1365-313X.2011.04612.x. [DOI] [PubMed] [Google Scholar]
- 48.Krasensky J, Jonak C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 2012;63:1593–1608. doi: 10.1093/jxb/err460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pal S, et al. Paclobutrazol induces tolerance in tomato to deficit irrigation through diversified effects on plant morphology, physiology and metabolism. Sci. Rep. 2016;6:39321. doi: 10.1038/srep39321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robredo A, et al. Elevated CO2 reduces the drought effect on nitrogen metabolism in barley plants during drought and subsequent recovery. Env. Exp. Bot. 2011;71:399–408. [Google Scholar]
- 51.Zaghdoud C, Carvajal M, Ferchichi A, Martinez-Ballesta MD. Water balance and N-metabolism in broccoli (Brassica oleracea L. var. Italica) plants depending on nitrogen source under salt stress and elevated CO2. Sci. Tot. Env. 2016;571:763–771. doi: 10.1016/j.scitotenv.2016.07.048. [DOI] [PubMed] [Google Scholar]
- 52.Golldack D, Li C, Mohan H, Probst N. Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front. Plant Sci. 2014;5:151. doi: 10.3389/fpls.2014.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wasternack C, Hause B. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Annals Bot. 2013;111:1021–1058. doi: 10.1093/aob/mct067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wasternack C. Action of jasmonates in plant stress responses and development- Applied aspects. Biotechnol. Adv. 2014;32:31–39. doi: 10.1016/j.biotechadv.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 55.Kazan K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015;20:219–229. doi: 10.1016/j.tplants.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 56.García-Abellán JO, et al. The tomato res mutant which accumulates JA in roots in non-stressed conditions restores cell structure alterations under salinity. Physiol. Plant. 2015;155:296–314. doi: 10.1111/ppl.12320. [DOI] [PubMed] [Google Scholar]
- 57.García-Abellán JO, et al. The phenotype alterations showed by the res tomato mutant disappear when the plants are grown under semi-arid conditions: Is the res mutant tolerant to multiple stresses? Plant Sig. Beh. 2017;12:e1146847. doi: 10.1080/15592324.2016.1146847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu T, et al. Ethylene and hydrogen peroxide are involved in brassinosteroid-induced salt tolerance in tomato. Sci. Rep. 2016;6:35392. doi: 10.1038/srep35392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Phukan UJ, Jeena GS, Tripathi V, Shukla RK. Regulation of Apetala 2/Ethylene Response Factors in Plants. Front. Plant Sci. 2017;8:150. doi: 10.3389/fpls.2017.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Joshi R, et al. Transcription factors and plants response to drought stress: current understanding and future directions. Front. Plant Sci. 2016;7:1029. doi: 10.3389/fpls.2016.01029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lorenzo O, Piqueras R, Sánchez-Serrano JJ, Solano R. Ethylene response factor1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell. 2003;15:165–178. doi: 10.1105/tpc.007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cheng M-C, Liao P-M, Kuo W-W, Lin T-P. The Arabidopsis ethylene response factor1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 2013;162:1566–1582. doi: 10.1104/pp.113.221911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Müller M, Munné-Bosch S. Ethylene response factors: a key regulatory hub in hormone and stress signaling. Plant Physiol. 2015;169:32–41. doi: 10.1104/pp.15.00677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu N, Tang N, Yan F, Bouzayen M, Li Z. Effect of LeERF1 and LeERF2 overexpression in the response to salinity of young tomato (Solanum lycopersicum cv. Micro-Tom) seedlings. Acta Physiol. Plant. 2014;36:1703–1712. doi: 10.1007/s11738-014-1545-5. [DOI] [Google Scholar]
- 65.Achard P, Vriezen WH, Van der Straeten D, Harberd NP. Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell. 2003;15:2816–2825. doi: 10.1105/tpc.015685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Achard P, et al. Integration of plant responses to environmentally activated phytohormonal signals. Science. 2006;311:91–94. doi: 10.1126/science.1118642. [DOI] [PubMed] [Google Scholar]
- 67.Yang DL, et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Nat. Acad. Sci. USA. 2012;109:E1192–E1200. doi: 10.1073/pnas.1201616109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang G-B, Yi H-Y, Gong J-M. The Arabidopsis ethylene/jasmonic acid-NRT signaling module coordinates nitrate reallocation and the trade-off between growth and environmental adaptation. Plant Cell. 2014;26:3984–3998. doi: 10.1105/tpc.114.129296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cuartero J, Nuez F, Diaz A. Catalog of collections of L. Lycopersicon and L. pennellii from Northwest of Peru. TGC Report. 1984;34:43–46. [Google Scholar]
- 70.Hoagland DR, Arnon DI. The water-culture method for growing plants without soil. Agric. Exp. Sta. Circ., Berkeley, CA. 1950;347:1–32. [Google Scholar]
- 71.Campos JF, et al. The tomato mutant ars1 (altered response to salt stress 1) identifies an R1-type MYB transcription factor involved in stomatal closure under salt acclimation. Plant Biotechnol. J. 2016;14:1345–1356. doi: 10.1111/pbi.12498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sanchez-Bel P, et al. Understanding the mechanisms of chilling injury in bell pepper fruits using the proteomic approach. J. Proteomics. 2012;75:5463–5478. doi: 10.1016/j.jprot.2012.06.029. [DOI] [PubMed] [Google Scholar]
- 73.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Benjamini Y, Hochberg Y. Controlling the false discovery rate—A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Series B-methodological. 1995;57:289–300. [Google Scholar]
- 75.Garcia-Abellan JO, et al. Heterologous expression of the yeast HAL5 gene in tomato enhances salt tolerance by reducing shoot Na+ accumulation in the long term. Physiol. Plant. 2014;152:700–713. doi: 10.1111/ppl.12217. [DOI] [PubMed] [Google Scholar]
- 76.Asins MJ, et al. Two closely linked tomato HKT coding genes are positional candidates for the major tomato QTL involved in Na+/K+ homeostasis. Plant Cell Environ. 2013;36:1171–1191. doi: 10.1111/pce.12051. [DOI] [PubMed] [Google Scholar]
- 77.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1 and S2, and Supplementary Tables S13 and S14
Supplementary Tables: Functional Classification of Differentially Expressed Genes