Summary
Following cessation of continuous Ebola virus (EBOV) transmission within Western Africa, sporadic EBOV disease (EVD) cases continued to re-emerge beyond the viral incubation period. Epidemiological and genomic evidence strongly suggests that this represented transmission from EVD survivors. To investigate whether persistent infections are characterized by ongoing viral replication, we sequenced EBOV from the semen of nine EVD survivors and a subset of corresponding acute specimens. EBOV evolutionary rates during persistence were either similar to or reduced relative to acute infection rates. Active EBOV replication/transcription continued during convalescence, but decreased over time, consistent with viral persistence rather than viral latency. Patterns of genetic divergence suggest a moderate relaxation of selective constraints within the sGP carboxy-terminal tail during persistent infections, but do not support widespread diversifying selection. Altogether, our data illustrate that EBOV persistence in semen, urine, and aqueous humor is not a quiescent or latent infection.
Keywords: Ebola virus, EVD survivors, persistent viral infection, evolutionary pressure, evolutionary rates, RNA hyper-editing
Graphical Abstract
Highlights
-
•
During persistence, EBOV exhibits heterogeneous evolutionary rates
-
•
Active EBOV transcription and replication occurs during persistence
-
•
RNA hyper-editing observed during viral persistence
-
•
No evidence for significant selective pressure during persistence
Whitmer et al. find that Ebola virus continues replication/transcription within the eye and male genital tract of Ebola virus disease survivors. They describe viral replication, evolutionary rates, and selective pressures experienced during acute and persistent infection.
Introduction
From December 2013 to June 2016, Sierra Leone, Guinea and Liberia experienced an Ebola virus (EBOV) outbreak causing 28,646 confirmed, probable, and suspected Ebola virus disease (EVD) cases—including 11,323 deaths and over 10,000 EVD survivors (WHO, 2016a). Despite the World Health Organization (WHO) declaring these countries disease-free 42 days (twice the 21-day viral incubation period) after the last active case, sporadic EVD cases continued to appear outside of this window and several reports strongly suggest that these unexpected re-emergences occurred due to viral transmission from persistently infected EVD survivors (Arias et al., 2016, Blackley et al., 2016, Christie et al., 2015, Diallo et al., 2016, Mate et al., 2015, Sissoko et al., 2017b). Other possible explanations, later discarded, included that sporadic cases could represent a missed transmission chain, reintroduction from an animal reservoir, or from another geographical location. Genetic data and phylogenetic analysis have been critical toward a resolution among these possibilities.
Filovirus persistence was initially observed with a single Marburg virus sexual transmission case in 1967 (Martini and Schmidt, 1968). Very scarce data from previous outbreaks suggested a prolonged presence of EBOV nucleic acids in semen and other bodily fluids collected from convalescent patients (Bausch et al., 2007, Rodriguez et al., 1999, Rowe et al., 1999). Recent EVD persistence studies in Sierra Leone, Liberia, Guinea, and the United States extended these observations and definitively demonstrated that EBOV RNA can be detected within the semen of EVD survivors months to ∼2 years after recovery, and live virus can be isolated from a subset of these specimens (Barnes et al., 2017, Deen et al., 2017, Sissoko et al., 2017a, Soka et al., 2016, Uyeki et al., 2016). Initially, the WHO and Médecins Sans Frontières (MSF) advised male survivors to abstain from sexual intercourse or use barrier protection for 3 months after recovery (Sterk, 2008, WHO, 2014), however, based on results from the current outbreak (Christie et al., 2015, Deen et al., 2017, Mate et al., 2015), the WHO revised their recommendations to include periodic EBOV RT-PCR semen testing and for survivors that cannot access EBOV RT-PCR semen testing, they should continue to practice safe sex for at least 12 months after the onset of symptoms (WHO, 2016b). Viral recrudescence outside of the male genital tract (MGT) can also develop after filovirus infection, as initially observed in 1977 for a single case of Marburg virus uveitis (Kuming and Kokoris, 1977). During the West African outbreak, recrudescent cases were again observed within the eye, and also the CNS several months after initial infection (Jacobs et al., 2016, Varkey et al., 2015). Altogether, these data suggest that after recovery from EVD, EBOV can still persist within immune-privileged sites in EVD survivors.
While much work has been done to explore the molecular evolution of EBOV during acute infection (Dudas et al., 2017, Gire et al., 2014, Ladner et al., 2015, Park et al., 2015, Simon-Loriere et al., 2015, Tong et al., 2015), little is known about the dynamics of persistent EBOV infections within immune-privileged niches. Genomes from EVD flare-ups linked to transmission from persistent infections exhibited reduced genetic divergence (Blackley et al., 2016, Diallo et al., 2016). These low levels of divergence could help to define and predict whether new outbreaks are the result of transmission from individuals with acute or persistent infections—such data could influence and guide future epidemiological investigations. Furthermore, the extraordinary discovery that EBOV can persistently infect immune-privileged sites for several months opens significant questions regarding viral replication mechanisms and the selective pressures experienced during acute and persistent infection.
To address these questions, we directly sequenced EBOV RNA from clinical specimens collected during acute EVD and during EVD convalescence (“persistence”) (Figure S1A). Using these EBOV sequences, we directly estimated viral evolutionary rates during persistent infection. We observed significant reductions in the rate of viral evolution within a subset of persistent infections, while others exhibited acute-like rates, and we present potential mechanisms to explain these results. We also examined patterns of selection during persistent infection and demonstrate that active viral replication/transcription continues during viral persistence.
Results
EBOV in Semen Specimens from Sierra Leonean EVD Survivors Exhibits Reduced Evolutionary Rates
Using a random subset of acutely acquired viral sequences (AAVS) from specimens collected from May 2014–September 2015 and sequenced directly from blood, plasma, or oral swab specimens from EVD patients with acute symptoms in Sierra Leone, we inferred a mean evolutionary rate of 0.96 × 10−3 substitutions/site/year ([0.86–1.06 × 10−3] 95% credible interval) under the uncorrelated lognormal (UCLN) model of rate variation among branches. These acute rate estimates are similar to previously reported rate estimates (Gire et al., 2014, Park et al., 2015, Simon-Loriere et al., 2015, Tong et al., 2015). Using Bayes factor values calculated from path and stepping-stone sampling, the UCLN relaxed clock models were the best fit to the data, however, evolutionary rate estimates were also similar under the fixed local clock model (Figure S1B; Table S1).
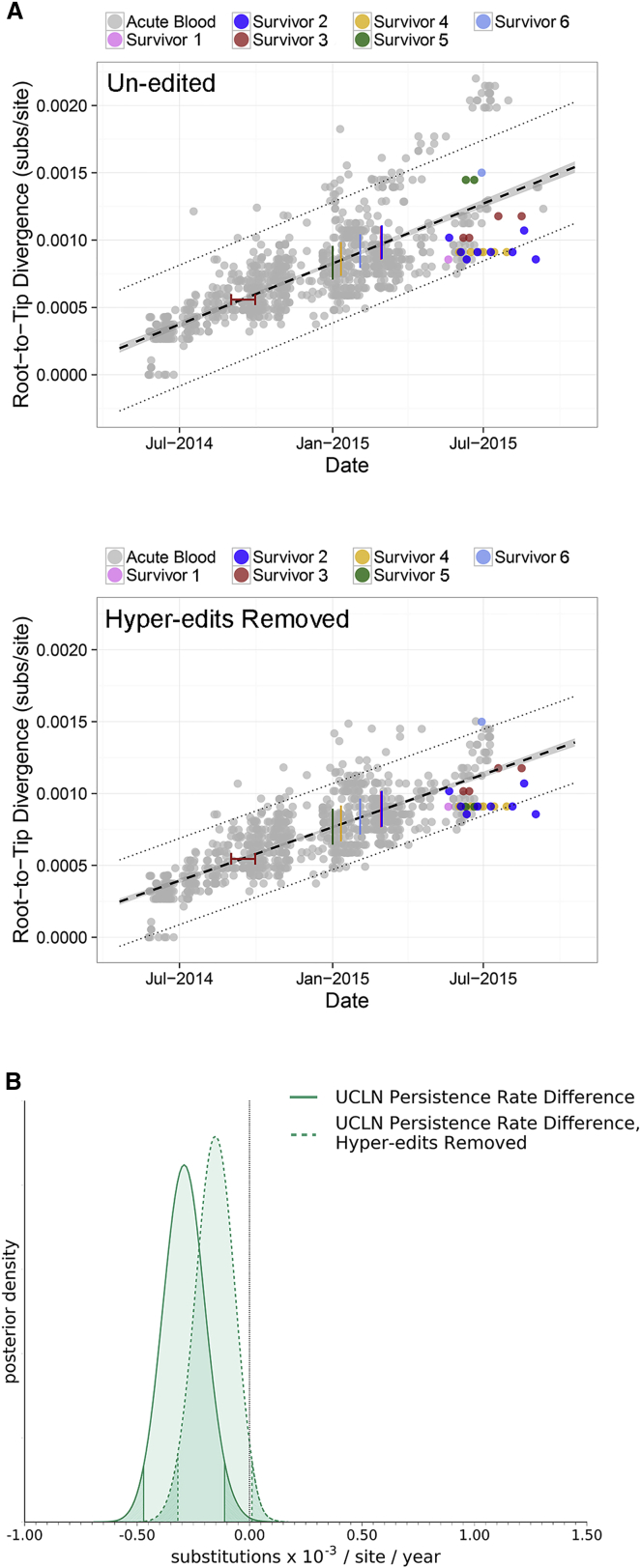
Most semen-acquired viral sequences (SAVS) exhibited lower genetic divergence, given their sampling time, than the mean AAVS divergence, although in all cases, this divergence was inside the prediction interval calculated for AAVS (Figure 1A). The average collection period for SAVS was 170 days post disease onset with a range of 82–322 days. During these collection periods, SAVS exhibited a significantly reduced evolutionary rate compared to AAVS (Figure 1B). Reversion of potential U-to-C hyper-edited sites, which may be the result of host ADAR enzymes, similar to Dudas et al. (2017), slightly decreased the acute evolutionary rate (0.89 × 10−3 subs/site/year, [0.80–0.99 × 10−3] 95% credible interval), as expected (Figure 1A). After removal of hyper-edited sites, SAVS exhibited a marginally significant reduced evolutionary rate compared to AAVS (Figure 1B).
Figure 1.
EBOV in Semen Specimens from Sierra Leonean EVD Survivors Exhibit Reduced Evolutionary Rates
(A) Genetic divergence versus specimen collection date for nearly all SLE viral sequences (n = 1,058) acquired from blood, plasma, or oral swab during acute infection (gray) and from semen during persistent infection (color). Colored bars represent survivor-reported symptom onset dates, and red whiskers represent onset date ambiguity for survivor 3. Top: includes sequences without editing. Bottom: includes sequences with reversion of potential U-to-C hyper-edited sites. Acute specimen average divergence from root is black dashed line and corresponding 95% confidence interval is gray (along black dashed line). Dotted lines represent 95% prediction intervals. EVD survivors 1, 2, 3, and 4 exhibited a reduced number of substitutions relative to the mean AAVS divergence, whereas survivors 5 and 6 exhibited an increased number of substitutions relative to the mean AAVS divergence (upper panel). Removal of hyper-edited sites reduced the number of substitutions for patient 5 (bottom).
(B) SAVS exhibit significantly reduced evolutionary rates compared to AAVS. Posterior rate distribution differences of SAVS compared to AAVS using un-edited sequences (solid line) and reversion of potential hyper-edited sites (dashed line). Shaded density tails indicate 95% highest posterior density interval (HPD) and black dotted line indicates the expectation that rate estimates are identical during acute and persistent infection.
EBOV Evolutionary Rates from Paired Acute and Convalescent Clinical Specimens
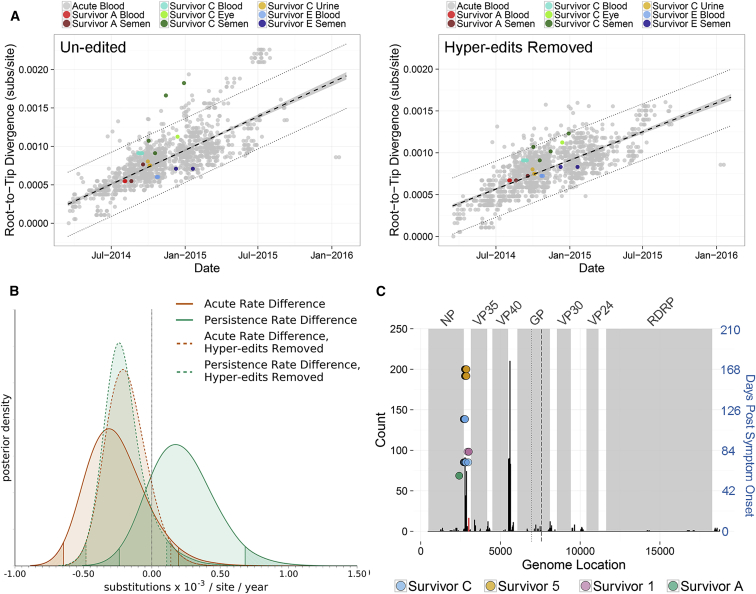
Serial specimens acquired from US EVD survivors permitted a comparison of viral sequences acquired during acute and persistent infection within a single individual. For all US survivors, AAVS were nearly identical and exhibited genetic divergence consistent with other AAVS collected during the outbreak (Figures 2A and S1C). For survivor C, concurrent viral compartmentalization was observed in the eye and MGT, and we did not observe evidence of viral exchange between these sites (Figure S1C). Using the UCLN relaxed clock model, mean posterior rate estimates from US AAVS (estimated over an average of 5 days) were slightly decreased, but not significantly different to rate estimates from other AAVS collected during the outbreak (estimated over 542 days) (Figure 2B; Table S1). In contrast to SAVS collected from EVD survivors in Sierra Leone, SAVS collected from US EVD survivors exhibited a mean evolutionary rate estimate that was ∼1.45-fold greater than acute rate estimates (Figure 2B; Table S1). We attribute this rate increase to U-to-C hyper-editing that occurred during viral persistence in survivors A and C (Figures 2A and 2C). Reversion of U-to-C hyper-edits from all sequences reduced US survivor AAVS and SAVS evolutionary rates to a level that was similar to acute-infection rate estimates (Figures 2A and 2B). While US EVD survivors received multiple therapeutic EVD treatments during early disease, we did not observe any mutations within viral regions (GP, VP35, L) targeted by these compounds (Supplemental Experimental Procedures). Thus, we hypothesize that these de novo U-to-C hyper-edits are not the result of therapeutic EVD treatments. U-to-C hyper-editing was also observed in Sierra Leone survivors 1 and 5, but from the available specimens, we cannot determine whether these changes occurred de novo during viral persistence, or during acute infection, because other AAVS from Sierra Leone (SLE) share the same set of mutations (Figure 2C).
Figure 2.
EBOV Sequenced from Acute and Persistent Clinical Specimens Acquired from US EVD Survivors Exhibits Acute-like Evolutionary Rates
(A) Genetic divergence versus specimen collection date for viral sequences from US EVD survivors and 1,498 sequences from SLE, Guinea (GIN), and Liberia (LBR). Left: includes sequences without editing. Right: includes sequences with reversion of potential hyper-edited sites. Viral sequences were acquired from blood, plasma, or oral swab specimens during acute infection (gray), or from blood, plasma, semen, urine, or eye during acute and persistent infection in EVD survivors (color). Mean divergence, 95% confidence interval, and 95% prediction intervals as in Figure 1.
(B) Prior to removal of hyper-edited U-to-C sites, SAVS (green solid line) exhibit ∼1.45-fold increased evolutionary rate compared to AAVS (orange solid line). After reversion of U-to-C hyper-edits, SAVS (green dashed line) exhibit a similar divergence as AAVS (orange dashed line). Overall, AAVS and SAVS evolutionary rates were not significantly different from the overall acute evolutionary rate (black dotted line, estimated from AAVS collected in SLE, GIN, and LBR). HPD intervals and rate distribution difference as in Figure 1.
(C) Distribution of U-to-C hyper editing sites using 1,498 sequences from SLE, GIN, and LBR. Occurrence of hyper-editing across the viral genome (black bars) and within coding regions (gray shading). GP transcriptional editing is dotted line, and GP1 and GP2 cleavage is dashed line. Hyper-edited sites from EVD survivors versus days post symptom onset is right y axis (blue). These sites only occurred within a distinct region near the untranslated 3′ nucleoprotein (NP) transcript, which was also observed with high frequency within acute specimens and is near a U-to-C editing site described in Ni et al. (2016) (red bar).
U-to-C hyper-editing is not unique to SAVS, similar patterns have also been observed within AAVS (Dudas et al., 2017, Ni et al., 2016, Park et al., 2015, Smits et al., 2015, Tong et al., 2015), however, it is currently unknown whether acute- and persistence-specific hyper-edited genomic regions exist. Here, we observed that most acute editing occurred within non-coding regions and the highest rates of hyper-editing were on the 3′ untranslated NP and VP40 transcripts. (Figure 2C). The distribution of hyper-edited sites in Figure 2C represents a combination of both de novo and ancestrally acquired hyper-edits. In contrast to AAVS, edited sites in SAVS are only within a distinct region on the 3′ untranslated NP transcript (Figure 2C). Hypermutation within this region was also observed with high frequency within AAVS and is near a U-to-C editing site (3008-11) that can upregulate NP transcription (Figure 2C) (Ni et al., 2016). Because ADAR editing deaminates adenosine to inosine, which base pairs with cytidine, canonical ADAR editing typically results in A → G mutations on the affected strand (Bass, 2002). Therefore, these U-to-C hyper-edits likely reflect ADAR editing of the viral (RNA−) genome.
Selective Pressures within Immune-Privileged Sites
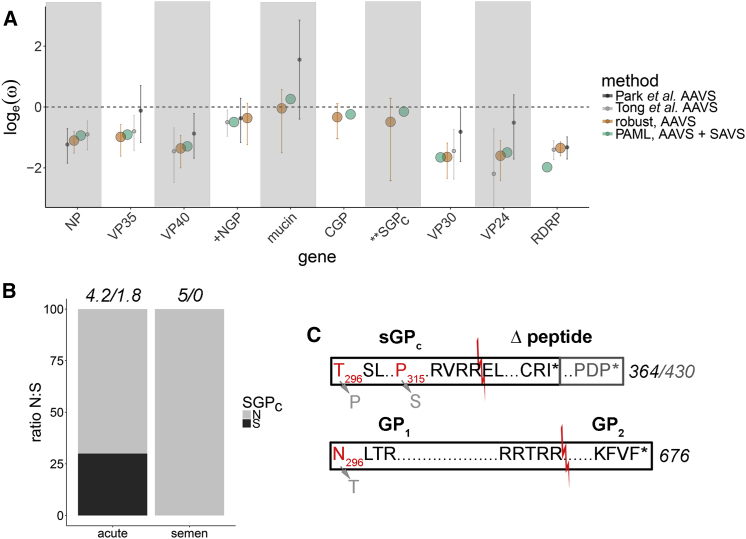
Because immune-privileged sites represent a unique niche, EBOV may experience selective pressure differences during acute and persistent infection. Selective pressures during acute infection were first estimated using Bayesian robust counting and compared to phylogenetic analysis by maximum likelihood (PAML) branch- and branch-site-specific models. To prevent rate overestimation by double-counting shared amino acids, the glycoprotein was split at the transcriptional editing site into N-terminal (NGP), C-terminal full-length (GP1 carboxy-terminus and GP2, CGP), and secreted GP (SGPc), and rates were estimated independently for each protein fragment (Figure 4). Inferred selective pressures were similar when estimated using Bayesian robust counting (AAVS only) and paml modeling (AAVS and SAVS) (Figure 3). In general, ω estimates were similar to or reduced compared to previous robust counting estimates (Park et al., 2015, Tong et al., 2015), consistent with purifying selection acting over a longer time period (Figure 3A). A comparison of the changes accumulated in the cohort, including its analysis in the context of the larger outbreak, did not reveal significant differences among groups (Figure S2; Table S2).
Figure 4.
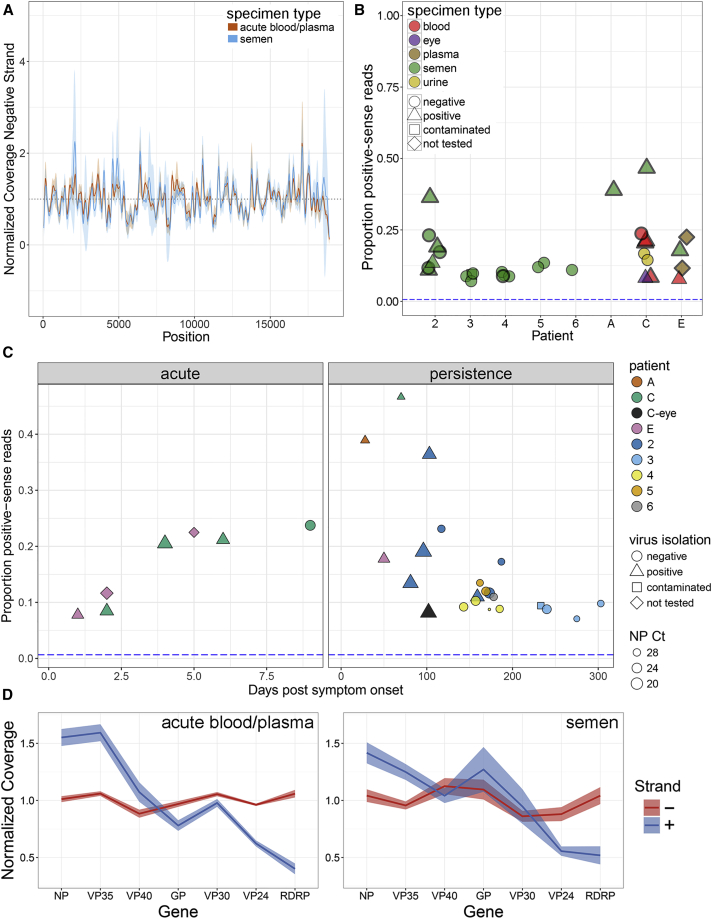
Active Viral Replication during Persistent Infection
(A) Average normalized negative-sense (viral genome) coverage for AAVS and SAVS (coverage mean [line] and standard deviations [shading]).
(B) Proportion of EBOV genome-wide positive-sense reads out of total reads from EVD survivor specimens. Specimen types indicated by color, point shape indicates virus isolation results and specimens in (D) contain thick borders. Blue dashed horizontal line indicates the proportion of positive-sense reads observed from a negative-sense viral RNA in vitro transcript (Figure S3A).
(C) Proportion of positive-sense reads versus day post symptom onset for acute specimens (left) and persistent specimens (right). Patients highlighted by color, virus isolation results highlighted by shape and nucleoprotein cycle threshold values highlighted by size.
(D) Proportion of normalized strand-specific reads per EBOV gene from AAVS (left) or SAVS (right). Negative-sense (viral genome) reads in red, and positive-sense (mRNA and viral complementary genome) reads in blue (shading is SE of the normalized coverage means).
Figure 3.
Selective Pressures within the MGT
(A) Comparison of loge(ω) estimates for viral genes calculated using PAML branch model (green) and coalescent robust counting (orange, error bars indicate 95% HPD) or from Park et al. (2015) (dark gray, error bars indicate 95% HPD) and from Tong et al. (2015) (light gray, error bars indicate 95% HPD). Rate estimates in PAML/codeml used SAVS and a subset of AAVS from SLE, GIN, and LBR (collected between 03/2014–09/2015). Robust counting estimates used a subset of AAVS from SLE, GIN and LBR collected between 03/2014–07/2015. Rate estimates from Park et al. (2015) and Tong et al. (2015) were calculated using robust counting with specimens collected between 03/2014–03/2015 and 03/2014–11/2014. In most cases, ω estimates closely agree and were reduced compared to previous estimates, consistent with purifying selection acting over a longer time period. Branch and branch-site PAML models support elevated ω in the secreted GP carboxy-tail from SAVS (“SGPc”) (stars). GP rate estimates from Park et al. (2015) and Tong et al. (2015) include full-length GP, rather than partitioned GP, as analyzed here (+ sign).
(B) Comparison of the proportion of total nonsynonymous (N, gray) and synonymous (S, black) counts across AAVS (from SLE, GIN, and LBR) and SAVS tree branches for the SGPc tail. Numbers above bars are the total count of N/S substitutions summed across AAVS and SAVS branches. Only nonsynonymous substitutions were observed in SAVS within the SGPc tail.
(C) Comparison of the glycoprotein (GP) C-terminal variants produced following transcriptional RNA editing. Sites identified with the PAML branch-site model to experience potential positive selection in SAVS are in gray and wild-type alleles are in red. Intervening amino acids (not to scale) are summarized with “……” Protease cleavage in the sGPc tail produces canonical sGPc and Δ peptide (red line) and cleavage of the full-length GP produces GP1 and GP2. Loss of the sGP stop codon is predicted to produce an extended Δ peptide for survivor 3 (gray).
Using the branch model, a moderate increase in ω was observed for the carboxy-terminal secreted glycoprotein tail (sGPc) (p = 6.13 × 10−5) of SAVS (Figure 3B; Table S2). This data were supported by the branch-site model, which provided evidence of site-based positive selection in SAVS occurring at glycoprotein amino acid residues 296N (CGP) (posterior probability 99.9%), 296T (sGPc) (posterior probability 99.9%), and 315P (sGPc) (posterior probability 78.2%) (Figure 3C). However, these mutations were each detected in only one EVD survivor (survivor 2: 296N/T and survivor 4: 315P), and thus likely represents an overestimation of ω in SAVS. Therefore, we hypothesize that nonsynonymous changes in sGPc from SAVS (Figure 3B) are suggestive of the relaxation of selection constraints, rather than evidence of positive selection at specific sites.
Additional unique glycoprotein mutations were observed during viral persistence that were not accurately captured by the PAML analysis. SAVS from survivor 2 contained an insertion in the GP transcriptional editing site (A → AC, 296N/T above) that shifts the reading frame and results in a viral genome encoding for the full-length GP tail, rather than the canonical sGP tail (Figures 3C and S2A site 6924; Table S2). This insertion was present in all 7 semen specimens from this patient, but its frequency in the SAVS population varied (34%–65%, Figures S2A–S2C). This insertion was also maintained in viral isolates (EBOV grown in tissue culture cells inoculated with survivor 2′s semen specimens) (data not shown), suggesting that it represents a true genomic mutation and not an overrepresentation of edited mRNA in consensus genomes. Interestingly, the end result of this change resembles the 7U/8U mutation that is induced by passage of some strains of EBOV (Zaire, Sudan) in Vero cell lines (Alfson et al., 2015, Volchkova et al., 2011). Additionally, survivor 3 contained a SNP that resulted in the loss of the sGP stop codon, which extends the sGP tail by an additional 66 amino acids (Figure 3C).
Evidence of Active Viral Replication within Semen Specimens from EVD Survivors
Currently there is limited data as to the extent of active viral replication during EBOV persistence and whether this replication occurs with intact or defective viral genomes. Through the use of stranded sequencing and qRT-PCR approaches, we were able to further define the strandedness of viral nucleic acids produced during acute and persistent infection (Figures 4, S3, and S4). Several studies provide support for chronic viral infection occurring due to the production of defective viral genomes (DVGs) containing internal/copy-back deletions (Calain et al., 1999, Li et al., 2011, Tapia et al., 2013) or terminal deletions (Meyer and Schmaljohn, 2000, Meyer and Southern, 1997). Overall, we observed similar depths of negative-sense (i.e., genomic) genome coverage between AAVS and SAVS (Figure 4A). Therefore, we do not see evidence for a preponderance of truncated genomes. However, we did observe a small proportion of chimeric reads containing deletions, duplications or copy back mutations (Table S3). Altogether, we did not observe any consistent trends in the proportion of chimeric reads per patient over time or during acute and persistent infection.
During acute infection, the proportion of positive-sense viral reads varied between 7%–23% (average ± SD: 16.5% ± 6.9%) and during persistent infection between 7%–46% (average 16.0% ± 10.9%) (Figure 4B). As a control, during in vitro infection with EBOV-ZsGreen we observed 78%–91% positive-sense viral reads in the monolayer (compatible with the detection of primarily mRNA) and 2%–5% positive-sense viral reads in the supernatant (compatible with the detection of primarily genomic RNA) at 18-48 hr post-infection (Figure S3A). Because SAVS contained proportions of positive-sense reads similar to or greater than that observed during acute infection, these data demonstrate the presence of active transcription/replication in all persistent survivor specimens studied herein.
During acute and persistent infections, the proportion of positive sense reads changed over time. As expected for acute infection, there was an increase in the proportion of positive sense reads over time, consistent with an increase in active viral replication/transcription during EVD (Figure 4C). After recovery from EVD, the ratio of positive-sense reads generally decreased logarithmically with time post onset (Figure 4C), however, in some instances, the proportion of positive-sense reads was higher during persistence than during acute infection (survivors A, C, and 2)—consistent with NP expression from a single survivor (Barnes et al., 2017). For a subset of clinical specimens, we isolated live virus (Spengler et al., 2015, Uyeki et al., 2016; U.S., unpublished data) and observed that the likelihood of positive virus isolation decreased over time (Figures 4B and 4C) and was significantly associated (p < 0.1) with the proportion of positive-sense reads (Figures S3B and S3C) and NP Ct value (p < 0.03) (Figures S3D and S3E).
We also observed that the relative depth of positive- and negative-sense RNA coverage was consistent with the accepted model of replication for viruses of the order Mononegavirales. For AAVS, SAVS, and during an in vitro infection, we observed a decrease in positive-sense coverage along the viral genome, consistent with mRNA expression decreasing in a roughly linear manner from the 5′ to 3′ end (Figures 4D, S3F, and S3G). In contrast, there was a steady depth of negative-sense reads across the genome for all specimens, consistent with this strand being synthesized as a continuous RNA molecule (Figures 4D, S3F, and S3G). Similar positive- and negative-sense RNA expression patterns were observed for in vitro infected cells (Figures S3F–S3G). However, a slight increase in 5′ negative-sense read coverage was observed during in vitro infection (Figures S3F–S3G), and we hypothesize that is due to interrupted/partial negative strand synthesis during active replication.
Discussion
Genomic analysis of EBOV sequences collected from acutely infected and convalescent survivors has yielded important insights into viral replication and selective pressures experienced during acute and persistent infections. During convalescence, EBOV evolutionary rates in the semen, aqueous humor, and urine were either similar to or reduced relative to the rates observed during acute infection in blood and plasma. During persistence, active EBOV replication/transcription continued, but decreased with time, consistent with viral persistence (i.e., long-term viral genome maintenance with active transcription/replication) rather than viral latency (i.e., long-term viral genome maintenance without active replication and low/no transcription). Furthermore, viral persistence did not appear to be linked to defective interfering particles with consistently truncated genomes attenuating wild-type infection (Calain et al., 1999, Li et al., 2011, Meyer and Schmaljohn, 2000, Meyer and Southern, 1997, Tapia et al., 2013). We did observe evidence for a minor population of chimeric reads in both acute and persistent specimens, however, from these short read data, we were not able to estimate the proportion of DVGs in the population, and it is currently unclear what role, if any, these DVGs may play during viral persistence. Finally, EBOV does not appear to have experienced substantially different selective pressures during persistence within immune-privileged niches (testes, eye) as compared to those experienced during acute infections. However, we did observe a moderate relaxation of selective constraints within the sGP carboxy-terminal tail during persistence.
The dichotomy of evolutionary rates observed between the Sierra Leone and US clinical specimens is of particular interest. After reversion of U-to-C hyper-edited sites, Sierra Leonean specimens, on average, exhibited a reduced evolutionary rate, while US specimens exhibited an “acute-like” rate. Our observation that SAVS can exhibit a slowed evolutionary rate is in line with a previous rate estimate from a single SAVS (Diallo et al., 2016) and supports rate estimates obtained from sexual transmission cases (Blackley et al., 2016). However, the observation of an “acute-like” evolutionary rate during EBOV persistence is a novel finding.
In general, substitution rates represent a complex product of effective population size, mutation rate, generation time, and viral fitness (Duffy et al., 2008). The US and Sierra Leone rate differences are likely due to differences in semen collection times post disease onset; US semen specimens were collected an average of 61 (minimum [min]: 28, maximum [max]: 116) days post onset, whereas Sierra Leonean semen specimens were collected an average of 188 (min: 80, max: 321) days post onset. An acute-like evolutionary rate reflects active ongoing viral replication during early convalescence, whereas the reduced rate may indicate increased pruning of deleterious alleles by purifying selection over time. However, additional factors such as a lower population size, reduced mutation rate, increased generation time, or reduced viral fitness could also contribute to a reduced substitution rate. Because the proportion of positive-sense reads decreases during convalescence, these rate differences also reflect a corresponding decrease in active viral replication over time. Within the MGT, active viral replication could be reduced by the lowered temperatures of the testes, a replication restriction, sequestration of viral nucleic acids into a cellular compartment, and/or immune/apoptotic-mediated clearance. Together, these factors will decrease the viral population size and increase generation time. While immune-privileged sites represent a novel niche, viral fitness differences likely do not contribute to the observed evolutionary rate differences, because we did not observe evidence for significant selective pressure differences in coding regions between SAVS and AAVS.
Viral nucleic acids during acute infection have been detected within the MGT (Dejucq and Jégou, 2001) and many viruses can establish persistent infections within a range of host sites (Randall and Griffin, 2017), however, despite this prevalence, relatively little is known regarding viral evolution during the acute to persistent transition. Previous studies comparing HIV sequences collected from paired blood/PBMCs or semen contained evidence of either compartmentalization or exchange between these two compartments in individual donors (Delwart et al., 1998, Gupta et al., 2000) and those patients that exhibited HIV compartmentalization also exhibited reduced genetic diversity (Pillai et al., 2005). However, abnormally low evolutionary rates for HIV and other viruses (HTLV-I, HTLV-II, SFV, GBV-C, and some plant viruses) are commonly due to a latent viral infection, or slow clonal expansion following viral integration (Duffy et al., 2008)—viral replication strategies that are distinct from models of EBOV replication.
Here, we found that two US survivors (C and A) exhibited evidence of de novo U-to-C hyper-editing in specimens acquired during viral persistence, which inflated the apparent viral evolutionary rate and likely occurred due to host-mediated ADAR1 cytoplasmic editing. Similar excessive ADAR-mediated edits within short regions were also observed within noncoding regions of AAVS (Dudas et al., 2017, Ni et al., 2016, Park et al., 2015, Tong et al., 2015), however, additional molecular studies are needed to confirm that these hyper-edits are due to host enzymes, and/or occur at sites containing secondary structure, and to evaluate the significance of these edits. Preliminary evidence suggests that a U-to-C editing site (3008-11) near those observed within SAVS can upregulate NP transcription (Ni et al., 2016). In other models, loss of ADAR1 editing activity can upregulate interferon-stimulated genes (Rice et al., 2012), thus ADAR-editing of viral transcripts may represent a proviral method to control protein production (hepatitis delta virus), or enhance viral replication (HIV), or may act through an anti-viral method to introduce excessive mutations (LCMV) (Gélinas et al., 2011, Zahn et al., 2007). Similar hyper-editing has also been observed during in vitro replication for other viruses (influenza, measles, respiratory syncytial, Epstein-Barr, and polyomavirus) (Iizasa et al., 2010, Kumar and Carmichael, 1997, Martínez and Melero, 2002, Suspène et al., 2011). Most strikingly, U-to-C and G-to-A hyper-editing has been observed following persistent measles infections in the brain 4 and 6 months after initial disease (Baczko et al., 1993, Cattaneo et al., 1988), and a similar pattern of U-to-C edits were observed on the NP 3′ untranslated region during in vitro Marburg infection (Shabman et al., 2014). Viral genomes with hyperedits in the VP40 5′ (viral genome orientation) tail were observed in the Magazine Wharf area of SLE after a disease-free 2-week period, potentially representing re-emergence from an EVD survivor, although both of these cases were also associated with “multiple high-risk contacts” (Smits et al., 2015, WHO, 2015). While there are some established links between ADAR and interferon signaling (George and Samuel, 1999, Pfaller et al., 2011, Rice et al., 2012), teasing apart the pro- and anti-viral interactions, along with their relationship to viral persistence, will be an important area for future research.
Besides on-the-ground contact tracing, there are currently no molecular signatures that would allow one to confirm whether EBOV was transmitted through contact with an acute case or from contact with an EVD survivor. Here, we observed that a delayed evolutionary rate (as suggested previously by Blackley et al. [2016] and Diallo et al. [2016]) or U-to-C hyper-editing in serial specimens could suggest transmission from persistently infected EVD survivors. However, the absence of these molecular markers does not eliminate persistently infected EVD survivors as potential sources of viral transmission.
Altogether, our data illustrate that EBOV persistence in semen and aqueous humor does not imply a quiescent or latent infection, but instead is an ongoing balance between natural selection and genetic drift within a novel intra-host niche. EBOV persistence within EVD survivors may act as a viral reservoir. Fortunately, sexual transmission of EBOV from EVD survivors is a rare mechanism for viral transmission. Ultimately, understanding the mechanisms of viral persistence in immune-privileged sites will lead to additional treatment options, clarify public health recommendations, and is critical to document whether future or past outbreaks might be due to transmission from persistently infected EVD survivors.
Experimental Procedures
Experimental Model and Subject Details
Human Subjects
Through the joint Sierra Leone Ebola Virus Persistence study (SLEVPS) with the Ministry of Health and Sanitation (MoHS) in Sierra Leone, WHO, China-CDC, and CDC, we had access to semen specimens collected from EVD survivors. The SLEVPS was reviewed and approved by the Sierra Leone Institutional Review Board and the World Health Organization Ethical Review Committee. Acute and persistent specimens from US EVD survivors were collected by their treating physicians and transported to the CDC for detection of viral RNA. This sequencing project was determined by the CDC institutional human subject advisor to be a non-research public health response activity, and institutional review board review was not required.
Method Details
Whole Genome Sequencing and Bioinformatics
RNA was extracted from blood and semen specimens and sequenced using a modified version of the Illumina TruSeq RNA Access Library Prep kit. EBOV genomes were assembled using custom scripts. Additional details are available in the Supplemental Experimental Procedures.
Sequence Analysis
Viral evolutionary rate estimates were conducted using both linear regression modeling and time-structured phylogenies. Additional sequence analysis was conducted using custom-made scripts. Evolutionary selective pressures were estimated using the renaissance counting method in beast/v1.8.2 and hypothesis testing was performed using the codeml model in paml/v4.5. Additional details are available in the Supplemental Experimental Procedures.
Consortia
Members of the Sierra Leone Ebola Virus Persistence Study Group are: Gibrilla Fadlu Deen, Nathalie Broutet, Barbara Knust, and Wenbo Xu (principal investigators), and James Bangura, Amara Jambai, Faustine James, Alie H. Wurie, Francis Yamba, Foday Sahr, Foday R. Sesay, Thomas A. Massaquoi, Tina Davies, Pierre Formenty, Anna E. Thorsen, Archchun Ariyarajah, Marylin Carino, Antoine Coursier, Kara N. Durski, Ndema Habib, Philippe Gaillard, Sihem Landoulski, Margaret O. Lamunu, Jacyln E. Marrinan, Suzanna L.R. McDonald, Dhamari Naidoo, Neetu Abad, Kyle T. Bernstein, Elizabeth Ervin, John D. Klena, Tasneem Malik, Oliver Morgan, Stuart T. Nichol, Christine Ross, Ute Ströher, Hongtu Liu, William Jun Liu, Yong Xhang, Guizhen Wu, Mifang Liang, and Patricia Ongpin (contributors).
Acknowledgments
The authors acknowledge William Davis (CDC/NCEZID/VSPB), Amy Schuh (CDC/NCEZID/VSPB), Serena Carroll (CDC/NCEZID), and Mark Whitmer (The NerdWerks, LLC) for their thoughtful review and discussion. The authors acknowledge Mike Purdy (CDC/NCHHSTP), William Fischer (LANL), and Mark Whitmer (The NerdWerks, LLC) for their software and script assistance. Work at US Army Medical Research Institute of Infectious Diseases was funded by the Defense Threat Reduction Agency, project CB10246. Financial support provided by the WHO, the CDC, the China CDC, the Paul G. Allen Family Foundation, the Sierra Leone Ministry of Health and Sanitation, and the Joint United Nations Program on HIV/AIDS. The WHO acknowledges the financial contribution of the WHO Ebola Response Program, the Paul G. Allen Family Foundation, the UNDP (United Nations Development Program), UNFPA (United Nations Population Fund), UNICEF WHO, World Bank Special Program of Research, Development and Research Training in Human Reproduction (HRP), a cosponsored program executed by the WHO in support of the Sierra Leone Ebola Virus Persistence Study. The authors acknowledge the Emory University Serious Communicable Diseases Program; the Military Hospital 34 and Lungi General Hospital staff; the Sierra Leone Association of Ebola Survivors: Yusuf Kabba; WHO: Ian Askew, Bruce Aylward, Rachel Baggaley, Anshu Banerjee, George Bindi, Rick Brennan, Mauricio Calderon, Ian Crozier, Christopher Dye, Peter Graaff, James N. Kiarie, Marie-Paule Kieny, Anders Nordström, Collins Owili, Lee Sharkey, and Lisa Thomas; and the US-CDC: Dianna Blau (IDP), Aaron Brault (ADB), Jorn Winter (VSDB), Tara Sealy (VSPB), John D. Klena (VSPB), and Lance Presser (VSPB). The content of this publication does not necessarily reflect the views or policies of the US Army. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of CDC.
Author Contributions
Conceptualization, S.L.M.W., J.T.L., M.R.W., G.P., and U.S.; Methodology, S.L.M.W., J.T.L., S.S.S., G.P., and U.S.; Software, S.L.W.M., S.S.S., J.T.L., G. Dudas, and A.R.; Validation, S.L.M.W. and J.T.L.; Formal Analysis, S.L.M.W., J.T.L., G. Dudas, A.R., G.P., and U.S.; Investigation, S.L.M.W., J.T.L., M.R.W., K. Patel, K. Prieto, and S.S.S.; Resources, F.S., E.C., B.K., D.N., G. Deen, P.F., S.T.N., G.P., and U.S.; Data Curation, S.L.M.W. and J.T.L.; Writing – Original Draft, S.L.M.W., U.S., and G.P.; Writing – Review & Editing, S.L.M.W., J.T.L., M.R.W., K. Patel, G. Dudas, A.R., B.K., P.F., S.T.N., G.P., and U.S.; Visualization, S.L.M.W., J.T.L., and G. Dudas; Supervision, F.S., B.K., D.N., G. Deen, P.F., S.T.N., G.P., and U.S.; Project Administration, S.L.M.W., B.K., P.F., G.P., and U.S.; and Funding Administration, B.K., P.F., S.T.N., G.P., and U.S.
Declaration of Interests
The authors declare no competing interests.
Published: January 30, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, and three tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.01.008.
Contributor Information
Shannon L.M. Whitmer, Email: evk3@cdc.gov.
Ute Ströher, Email: ute.stroeher@gmail.com.
Data and Software Availability
The accession numbers for the genomes acquired from clinical specimens reported in this paper are GenBank: KY401638–KY401675 and KY805810-2.
Supplemental Information
References
- Alfson K.J., Avena L.E., Beadles M.W., Menzie H., Patterson J.L., Carrion R., Jr., Griffiths A. Genetic changes at the glycoprotein editing site associated with serial passage of Sudan virus. J. Infect. Dis. 2015;212(Suppl 2):S295–S304. doi: 10.1093/infdis/jiv216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias A., Watson S.J., Asogun D., Tobin E.A., Lu J., Phan M.V.T., Jah U., Wadoum R.E.G., Meredith L., Thorne L. Rapid outbreak sequencing of Ebola virus in Sierra Leone identifies transmission chains linked to sporadic cases. Virus Evol. 2016;2:vew016. doi: 10.1093/ve/vew016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baczko K., Lampe J., Liebert U.G., Brinckmann U., ter Meulen V., Pardowitz I., Budka H., Cosby S.L., Isserte S., Rima B.K. Clonal expansion of hypermutated measles virus in a SSPE brain. Virology. 1993;197:188–195. doi: 10.1006/viro.1993.1579. [DOI] [PubMed] [Google Scholar]
- Barnes K.G., Kindrachuk J., Lin A.E., Wohl S., Qu J., Tostenson S.D., Dorman W.R., Busby M., Siddle K.J., Luo C.Y. Evidence of Ebola virus replication and high concentration in semen of a patient during recovery. Clin. Infect. Dis. 2017;65:1400–1403. doi: 10.1093/cid/cix518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass B.L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bausch D.G., Towner J.S., Dowell S.F., Kaducu F., Lukwiya M., Sanchez A., Nichol S.T., Ksiazek T.G., Rollin P.E. Assessment of the risk of Ebola virus transmission from bodily fluids and fomites. J. Infect. Dis. 2007;196(Suppl 2):S142–S147. doi: 10.1086/520545. [DOI] [PubMed] [Google Scholar]
- Blackley D.J., Wiley M.R., Ladner J.T., Fallah M., Lo T., Gilbert M.L., Gregory C., D’ambrozio J., Coulter S., Mate S. Reduced evolutionary rate in reemerged Ebola virus transmission chains. Sci. Adv. 2016;2:e1600378. doi: 10.1126/sciadv.1600378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calain P., Monroe M.C., Nichol S.T. Ebola virus defective interfering particles and persistent infection. Virology. 1999;262:114–128. doi: 10.1006/viro.1999.9915. [DOI] [PubMed] [Google Scholar]
- Cattaneo R., Schmid A., Eschle D., Baczko K., ter Meulen V., Billeter M.A. Biased hypermutation and other genetic changes in defective measles viruses in human brain infections. Cell. 1988;55:255–265. doi: 10.1016/0092-8674(88)90048-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie A., Davies-Wayne G.J., Cordier-Lassalle T., Blackley D.J., Laney A.S., Williams D.E., Shinde S.A., Badio M., Lo T., Mate S.E., Centers for Disease Control and Prevention (CDC) Possible sexual transmission of Ebola virus - Liberia, 2015. MMWR Morb. Mortal. Wkly. Rep. 2015;64:479–481. [PMC free article] [PubMed] [Google Scholar]
- Deen G.F., Knust B., Broutet N., Sesay F.R., Formenty P., Ross C., Thorson A.E., Massaquoi T.A., Marrinan J.E., Ervin E. Ebola RNA persistence in semen of Ebola virus disease survivors - preliminary report. N. Engl. J. Med. 2017;377:1428–1437. doi: 10.1056/NEJMoa1511410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejucq N., Jégou B. Viruses in the mammalian male genital tract and their effects on the reproductive system. Microbiol. Mol. Biol. Rev. 2001;65:208–231. doi: 10.1128/MMBR.65.2.208-231.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delwart E.L., Mullins J.I., Gupta P., Learn G.H., Jr., Holodniy M., Katzenstein D., Walker B.D., Singh M.K. Human immunodeficiency virus type 1 populations in blood and semen. J. Virol. 1998;72:617–623. doi: 10.1128/jvi.72.1.617-623.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diallo B., Sissoko D., Loman N.J., Bah H.A., Bah H., Worrell M.C., Conde L.S., Sacko R., Mesfin S., Loua A. Resurgence of Ebola virus disease in Guinea linked to a survivor with virus persistence in seminal fluid for more than 500 days. Clin. Infect. Dis. 2016;63:1353–1356. doi: 10.1093/cid/ciw601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudas G., Carvalho L.M., Bedford T., Tatem A.J., Baele G., Faria N.R., Park D.J., Ladner J.T., Arias A., Asogun D. Virus genomes reveal factors that spread and sustained the Ebola epidemic. Nature. 2017;544:309–315. doi: 10.1038/nature22040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy S., Shackelton L.A., Holmes E.C. Rates of evolutionary change in viruses: patterns and determinants. Nat. Rev. Genet. 2008;9:267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- Gélinas J.F., Clerzius G., Shaw E., Gatignol A. Enhancement of replication of RNA viruses by ADAR1 via RNA editing and inhibition of RNA-activated protein kinase. J. Virol. 2011;85:8460–8466. doi: 10.1128/JVI.00240-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George C.X., Samuel C.E. Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible. Proc. Natl. Acad. Sci. USA. 1999;96:4621–4626. doi: 10.1073/pnas.96.8.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gire S.K., Goba A., Andersen K.G., Sealfon R.S., Park D.J., Kanneh L., Jalloh S., Momoh M., Fullah M., Dudas G. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science. 2014;345:1369–1372. doi: 10.1126/science.1259657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P., Leroux C., Patterson B.K., Kingsley L., Rinaldo C., Ding M., Chen Y., Kulka K., Buchanan W., McKeon B., Montelaro R. Human immunodeficiency virus type 1 shedding pattern in semen correlates with the compartmentalization of viral Quasi species between blood and semen. J. Infect. Dis. 2000;182:79–87. doi: 10.1086/315644. [DOI] [PubMed] [Google Scholar]
- Iizasa H., Wulff B.E., Alla N.R., Maragkakis M., Megraw M., Hatzigeorgiou A., Iwakiri D., Takada K., Wiedmer A., Showe L. Editing of Epstein-Barr virus-encoded BART6 microRNAs controls their dicer targeting and consequently affects viral latency. J. Biol. Chem. 2010;285:33358–33370. doi: 10.1074/jbc.M110.138362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs M., Rodger A., Bell D.J., Bhagani S., Cropley I., Filipe A., Gifford R.J., Hopkins S., Hughes J., Jabeen F. Late Ebola virus relapse causing meningoencephalitis: a case report. Lancet. 2016;388:498–503. doi: 10.1016/S0140-6736(16)30386-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Carmichael G.G. Nuclear antisense RNA induces extensive adenosine modifications and nuclear retention of target transcripts. Proc. Natl. Acad. Sci. USA. 1997;94:3542–3547. doi: 10.1073/pnas.94.8.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuming B.S., Kokoris N. Uveal involvement in Marburg virus disease. Br. J. Ophthalmol. 1977;61:265–266. doi: 10.1136/bjo.61.4.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner J.T., Wiley M.R., Mate S., Dudas G., Prieto K., Lovett S., Nagle E.R., Beitzel B., Gilbert M.L., Fakoli L. Evolution and spread of Ebola virus in Liberia, 2014-2015. Cell Host Microbe. 2015;18:659–669. doi: 10.1016/j.chom.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Lott W.B., Lowry K., Jones A., Thu H.M., Aaskov J. Defective interfering viral particles in acute dengue infections. PLoS ONE. 2011;6:e19447. doi: 10.1371/journal.pone.0019447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez I., Melero J.A. A model for the generation of multiple A to G transitions in the human respiratory syncytial virus genome: predicted RNA secondary structures as substrates for adenosine deaminases that act on RNA. J. Gen. Virol. 2002;83:1445–1455. doi: 10.1099/0022-1317-83-6-1445. [DOI] [PubMed] [Google Scholar]
- Martini G.A., Schmidt H.A. [Spermatogenic transmission of the “Marburg virus”. (Causes of “Marburg simian disease”)] Klin. Wochenschr. 1968;46:398–400. doi: 10.1007/BF01734141. [DOI] [PubMed] [Google Scholar]
- Mate S.E., Kugelman J.R., Nyenswah T.G., Ladner J.T., Wiley M.R., Cordier-Lassalle T., Christie A., Schroth G.P., Gross S.M., Davies-Wayne G.J. Molecular evidence of sexual transmission of Ebola virus. N. Engl. J. Med. 2015;373:2448–2454. doi: 10.1056/NEJMoa1509773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B.J., Schmaljohn C.S. Persistent hantavirus infections: characteristics and mechanisms. Trends Microbiol. 2000;8:61–67. doi: 10.1016/s0966-842x(99)01658-3. [DOI] [PubMed] [Google Scholar]
- Meyer B.J., Southern P.J. A novel type of defective viral genome suggests a unique strategy to establish and maintain persistent lymphocytic choriomeningitis virus infections. J. Virol. 1997;71:6757–6764. doi: 10.1128/jvi.71.9.6757-6764.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M., Chen C., Qian J., Xiao H.-X., Shi W.-F., Luo Y., Wang H.-Y., Li Z., Wu J., Xu P.-S. Intra-host dynamics of Ebola virus during 2014. Nat. Microbiol. 2016;1:16151. doi: 10.1038/nmicrobiol.2016.151. [DOI] [PubMed] [Google Scholar]
- Park D.J., Dudas G., Wohl S., Goba A., Whitmer S.L., Andersen K.G., Sealfon R.S., Ladner J.T., Kugelman J.R., Matranga C.B. Ebola virus epidemiology, transmission, and evolution during seven months in Sierra Leone. Cell. 2015;161:1516–1526. doi: 10.1016/j.cell.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller C.K., Li Z., George C.X., Samuel C.E. Protein kinase PKR and RNA adenosine deaminase ADAR1: new roles for old players as modulators of the interferon response. Curr. Opin. Immunol. 2011;23:573–582. doi: 10.1016/j.coi.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai S.K., Good B., Pond S.K., Wong J.K., Strain M.C., Richman D.D., Smith D.M. Semen-specific genetic characteristics of human immunodeficiency virus type 1 env. J. Virol. 2005;79:1734–1742. doi: 10.1128/JVI.79.3.1734-1742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall R.E., Griffin D.E. Within host RNA virus persistence: mechanisms and consequences. Curr. Opin. Virol. 2017;23:35–42. doi: 10.1016/j.coviro.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice G.I., Kasher P.R., Forte G.M., Mannion N.M., Greenwood S.M., Szynkiewicz M., Dickerson J.E., Bhaskar S.S., Zampini M., Briggs T.A. Mutations in ADAR1 cause Aicardi-Goutières syndrome associated with a type I interferon signature. Nat. Genet. 2012;44:1243–1248. doi: 10.1038/ng.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez L.L., De Roo A., Guimard Y., Trappier S.G., Sanchez A., Bressler D., Williams A.J., Rowe A.K., Bertolli J., Khan A.S. Persistence and genetic stability of Ebola virus during the outbreak in Kikwit, Democratic Republic of the Congo, 1995. J. Infect. Dis. 1999;179(Suppl 1):S170–S176. doi: 10.1086/514291. [DOI] [PubMed] [Google Scholar]
- Rowe A.K., Bertolli J., Khan A.S., Mukunu R., Muyembe-Tamfum J.J., Bressler D., Williams A.J., Peters C.J., Rodriguez L., Feldmann H. Clinical, virologic, and immunologic follow-up of convalescent Ebola hemorrhagic fever patients and their household contacts, Kikwit, Democratic Republic of the Congo. Commission de Lutte contre les Epidémies à Kikwit. J. Infect. Dis. 1999;179(Suppl 1):S28–S35. doi: 10.1086/514318. [DOI] [PubMed] [Google Scholar]
- Shabman R.S., Jabado O.J., Mire C.E., Stockwell T.B., Edwards M., Mahajan M., Geisbert T.W., Basler C.F. Deep sequencing identifies noncanonical editing of Ebola and Marburg virus RNAs in infected cells. MBio. 2014;5:e02011. doi: 10.1128/mBio.02011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Loriere E., Faye O., Faye O., Koivogui L., Magassouba N., Keita S., Thiberge J.M., Diancourt L., Bouchier C., Vandenbogaert M. Distinct lineages of Ebola virus in Guinea during the 2014 West African epidemic. Nature. 2015;524:102–104. doi: 10.1038/nature14612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sissoko D., Duraffour S., Kerber R., Kolie J.S., Beavogui A.H., Camara A.M., Colin G., Rieger T., Oestereich L., Pályi B. Persistence and clearance of Ebola virus RNA from seminal fluid of Ebola virus disease survivors: a longitudinal analysis and modelling study. Lancet Glob. Health. 2017;5:e80–e88. doi: 10.1016/S2214-109X(16)30243-1. [DOI] [PubMed] [Google Scholar]
- Sissoko D., Keita M., Diallo B., Aliabadi N., Fitter D.L., Dahl B.A., Akoi Bore J., Raymond Koundouno F., Singethan K., Meisel S. Ebola virus persistence in breast milk after no reported illness: a likely source of virus transmission from mother to child. Clin. Infect. Dis. 2017;64:513–516. doi: 10.1093/cid/ciw793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits S.L., Pas S.D., Reusken C.B., Haagmans B.L., Pertile P., Cancedda C., Dierberg K., Wurie I., Kamara A., Kargbo D. Genotypic anomaly in Ebola virus strains circulating in Magazine Wharf area, Freetown, Sierra Leone, 2015. Euro Surveill. 2015;20 doi: 10.2807/1560-7917.ES.2015.20.40.30035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soka M.J., Choi M.J., Baller A., White S., Rogers E., Purpura L.J., Mahmoud N., Wasunna C., Massaquoi M., Abad N. Prevention of sexual transmission of Ebola in Liberia through a national semen testing and counselling programme for survivors: an analysis of Ebola virus RNA results and behavioural data. Lancet Glob. Health. 2016;4:e736–e743. doi: 10.1016/S2214-109X(16)30175-9. [DOI] [PubMed] [Google Scholar]
- Spengler J.R., McElroy A.K., Harmon J.R., Ströher U., Nichol S.T., Spiropoulou C.F. Relationship Between Ebola Virus Real-Time Quantitative Polymerase Chain Reaction-Based Threshold Cycle Value and Virus Isolation From Human Plasma. J. Infect. Dis. 2015;212(Suppl 2):S346–S349. doi: 10.1093/infdis/jiv187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterk, E. (2008). Filovirus Hemorrhagic Fever Guideline (http://www.slamviweb.org/es/ebola/fhffinal.pdf: Medecins Sans Frontieres).
- Suspène R., Petit V., Puyraimond-Zemmour D., Aynaud M.M., Henry M., Guétard D., Rusniok C., Wain-Hobson S., Vartanian J.P. Double-stranded RNA adenosine deaminase ADAR-1-induced hypermutated genomes among inactivated seasonal influenza and live attenuated measles virus vaccines. J. Virol. 2011;85:2458–2462. doi: 10.1128/JVI.02138-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia K., Kim W.K., Sun Y., Mercado-López X., Dunay E., Wise M., Adu M., López C.B. Defective viral genomes arising in vivo provide critical danger signals for the triggering of lung antiviral immunity. PLoS Pathog. 2013;9:e1003703. doi: 10.1371/journal.ppat.1003703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y.G., Shi W.F., Di L., Qian J., Liang L., Bo X.C., Liu J., Ren H.G., Fan H., Ni M. Genetic diversity and evolutionary dynamics of Ebola virus in Sierra Leone. Nature. 2015;524:93–96. doi: 10.1038/nature14490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeki T.M., Erickson B.R., Brown S., McElroy A.K., Cannon D., Gibbons A., Sealy T., Kainulainen M.H., Schuh A.J., Kraft C.S. Ebola virus persistence in semen of male survivors. Clin. Infect. Dis. 2016;62:1552–1555. doi: 10.1093/cid/ciw202. [DOI] [PubMed] [Google Scholar]
- Varkey J.B., Shantha J.G., Crozier I., Kraft C.S., Lyon G.M., Mehta A.K., Kumar G., Smith J.R., Kainulainen M.H., Whitmer S. Persistence of Ebola virus in ocular fluid during convalescence. N. Engl. J. Med. 2015;372:2423–2427. doi: 10.1056/NEJMoa1500306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volchkova V.A., Dolnik O., Martinez M.J., Reynard O., Volchkov V.E. Genomic RNA editing and its impact on Ebola virus adaptation during serial passages in cell culture and infection of guinea pigs. J. Infect. Dis. 2011;204(Suppl 3):S941–S946. doi: 10.1093/infdis/jir321. [DOI] [PubMed] [Google Scholar]
- WHO (2014). Clinical Management of Patients with Viral Hemorrhagic Fever: A Pocket Guide for the Front-line Health Worker. https://www.unicef.org/cbsc/files/VHF_pocket_book_Guinea-2014.pdf.
- WHO (2015). Ebola Situation Report - 24 June 2015. http://apps.who.int/ebola/current-situation/ebola-situation-report-24-june-2015.
- WHO (2016a). Ebola Situation Report - March 30, 2016. http://apps.who.int/ebola/current-situation/ebola-situation-report-30-march-2016.
- WHO . World Health Organization; Geneva, Switzerland: 2016. Clinical Care for Survivors of Ebola Virus Disease: Interim Guidance. [Google Scholar]
- Zahn R.C., Schelp I., Utermöhlen O., von Laer D. A-to-G hypermutation in the genome of lymphocytic choriomeningitis virus. J. Virol. 2007;81:457–464. doi: 10.1128/JVI.00067-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.