Abstract
Mesenchymal stem cells (MSCs) can be isolated from several tissues of adults. In addition, MSCs have the potential of differentiation into several cell types. Therefore, MSCs are very useful in stem cell therapy and regenerative medicine. MSCs have also been used as gene or protein carriers. As a result, maintaining MSCs in a desirable metabolic state has been the subject of several studies. Here, we used a genome scale metabolic network model of bone marrow derived MSCs for exploring the metabolism of these cells. We analyzed metabolic fluxes of the model in order to find ways of increasing stem cell proliferation and differentiation. Consequently, the experimental results were in consistency with computational results. Therefore, analyzing metabolic models was proven to be a promising field in biomedical researches of stem cells.
Electronic supplementary material
The online version of this article (doi:10.1007/s10616-017-0148-6) contains supplementary material, which is available to authorized users.
Keywords: Metabolic networks, Mesenchymal stem cell (MSC), Proliferation, Differentiation, Modeling
Introduction
Metabolic network models have long been used for investigating metabolism of cells (Fell and Small 1986; Varma and Palsson 1993). Historically, the first metabolic models were simply collections of known reactions. Such models typically included central carbon metabolism (Poolman et al. 2004; Varma and Palsson 1993; Wiback and Palsson 2002). With the advent of next-generation sequencing and widely available genomic and gene annotation data, genome-scale metabolic networks (GEMs) have been reconstructed (King et al. 2015; Saha et al. 2014; Simeonidis and Price 2015). Such metabolic networks represent in silico models of metabolism. A standard GEM includes organism-specific reactions together with accurate gene-protein-reaction (GPR) relations.
The original models of metabolism, which were developed in the 60’s and 70’s, were generally exact ordinary differential equation models (Goldbeter and Lefever 1972; Higgins 1964). However, due to the large number of reactions in a GEM, determination of kinetic parameters for all reactions is practically impossible. Consequently, calculating the exact value of all metabolic fluxes in a cell is unfeasible.
For a GEM, an infinite number of possible flux distributions can be imagined. Constraint-based modeling (CBM) applies a limited number of constrains to the fluxes of a GEM, to narrow down the possible space of its fluxes.
Today, human cell- or tissue-specific GEMs are used to elucidate the metabolic state of that for certain cell/tissue. Such models have various biomedical applications (Fouladiha and Marashi 2017), from identifying biomarkers of diseases (Shlomi et al. 2009) to predicting drug targets (Chang et al. 2010; Hadi and Marashi 2014; Jerby and Ruppin 2012).
We have recently reconstructed a metabolic network model, iMSC1255, for bone marrow derived mesenchymal stem cells (MSCs) (Fouladiha et al. 2015). MSCs are very useful in stem cell therapy and regenerative medicine. Most of the adult tissues contain MSCs, but in a relatively low number. The time that is needed for isolation and in vitro propagation of MSCs is relatively long, usually following senescence of these cells. In the present work, we apply iMSC1255 to help us to find new ways of solving such biological problems, like low proliferation rate of MSCs. Increasing proliferation rate of MSCs can facilitate in vitro experiments of regenerative medicine and pave the way toward organ cloning. We show that this model can be used for introducing cell culture supplements which can improve cell growth rate and also, for suggesting gene up- or down-regulations for increasing stem cell differentiation. In addition, we used iMSC1255 for the analysis of internal metabolic fluxes under different oxygenation conditions. In other words, iMSC1255 was used to predict the impact of lowering concentration of oxygen on the fluxes of reactions.
Materials and methods
Materials
Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum (FBS), penicillin and streptomycin were purchased from GIBCO-BRL (Grand Island, NY, USA). Phosphatidylserine (PS) and phosphatidylethanolamine (PE) were acquired from Sigma-Aldrich (St. Louis, MO, USA). 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazoliumbromide (MTT) and dimethyl sulfoxide (DMSO) were purchased from Merck (Darmstadt, Germany) and phosphate buffer saline (PBS) was obtained from GIBCO.
Modeling the metabolic fluxes
In constraint-based modeling of metabolic networks, one applies constraints on metabolic fluxes: under stoichiometric constraints (Eq. 1, where S is the stoichiometric matrix and v is a vector of fluxes), thermodynamic constraints (Eq. 2) and capacity constraints (Eq. 3), it is possible to find a limited metabolic flux space for the network (Gutierrez and Lewis 2015; O’Brien et al. 2015). By applying additional assumptions (e.g., by assuming that the cell produces the highest possible amount of biomass), one can find a certain flux distribution for the GEM. This computational procedure is called flux balance analysis (FBA) (Orth et al. 2010). Briefly, in FBA one solves the following linear problem:
where v j is the jth element of vector v, while α j and β j are the lower bound and upper bound of the flux through reaction j.
Increasing stem cell proliferation
In a GEM, cell growth is modeled with the help of a hypothetical reaction called biomass production. This reaction consumes constituents of biomass, like lipids and amino acids, with appropriate ratios, and produces one unit of an imaginary metabolite (called biomass). Therefore, finding a strategy for increasing the flux of biomass production reaction in silico may lead to an increase in the experimental cell proliferation rate.
Increasing cell proliferation: in silico analysis
In order to simulate in vitro cell culture medium, an in silico growth medium is defined. In such definition, only the components of growth medium are allowed to be consumed, i.e., only the exchange fluxes of these components are allowed to take negative values.
In this part of our work, we searched for all those metabolites which can be added to cell culture medium to increase cell growth rate. According to the definition of in silico growth medium, by addition of a metabolite to the growth medium, we have to allow the exchange flux of that metabolite to take a negative value, which means metabolite uptake, in metabolic modeling. If uptake of certain metabolites results in an improved growth rate, we predict that supplementation of cell culture medium with the corresponding metabolite can increase experimental cell growth rate. Therefore, we perform two FBA for each metabolite, one with zero and the other with negative lower bound of exchange flux of that metabolite. In order to calculate the relative contribution of each metabolite in increasing biomass production flux, the optimum biomass production flux in the second FBA was divided to the optimum flux in the first FBA. If these relative values were more than 1, that metabolite can be chosen to supplement cell culture medium in vitro. FBA was performed with biomass production flux as objective function, using optimizeCbModel function of the COBRA Toolbox (Becker et al. 2007).
Cell culture
Human bone marrow derived MSCs were isolated and characterized according to the previously described protocols (Yazdani et al. 2013). The growth medium contained DMEM which was supplemented with 10% FBS, together with penicillin G (100 U/mL) and streptomycin (100 μg/mL). MSCs were cultured under humidified air with 5% CO2 at 37 °C in 25 cm2 culture flasks.
Cell proliferation analysis (MTT assay)
We used MTT assay to test the impact of medium supplementation on the proliferation of cells. PS and PE (the metabolites which were chosen for medium supplementation according to computational results) were used to pre-treat the 96-well cell culture plates. These metabolites were dissolved in chloroform/methanol (95/5).
We chose four concentrations of PE and PS for cell culture supplementation (20, 40, 60 and 80 μM). One μL of the solutions was dropped in each well. After evaporation of the solvents, 2 × 105 bone marrow derived MSCs of second passage were seeded into each well. Then, 100 μL of cell culture medium was added to each well. The cells were cultured in the medium supplemented with PS or PE. After three and after 6 days of incubation, the cells were used for MTT assay.
MTT assay: At the end of days 3 and 6, the culture medium was removed and 100 μL of MTT solution (0.5 mg/mL of MTT in PBS) was added to each well. After 4 h of incubation, the MTT solution was removed. Then, 100 μL of DMSO was added to each well, followed by incubation for 15 min. Finally, absorbance at 570 nm was read by ELISA microplate reader (Awareness Technology Incorporation). Increase in the absorbance of a well indicates an increase in the number of living cells. The experiments were repeated in 5 biological replicates.
Increasing stem cell differentiation: Metabolic Transition Algorithm (MTA)
Increasing the efficiency of stem cell differentiation can have a positive effect in stem cell therapy. Differentiation can be explained as the transition of cell states, e.g., cell metabolism. Recently, an algorithm named MTA has been developed for finding ways to increase transition of cells using metabolic models (Yizhak et al. 2013). In brief, MTA uses the amount of activity of reactions in the first state and a list of reaction which have to be up- or down-regulated in the second state. Then, MTA scores the impact of knock-down of each reaction at increasing cell transition.
Modeling the effect of different growth conditions on metabolic fluxes
Extracellular conditions influence and alter the metabolism of cells. In order to keep the metabolism of cells in a desired state, culture condition of the cells has to be controlled. Therefore, the exact impact of cell culture changes on intracellular state has to be elucidated. For instance, the role of different oxygenation on stem cells proliferation and differentiation has been the subject of several studies (Grayson et al. 2006; Pattappa et al. 2011; Zhao et al. 2005).
Using several gene expression profiles, Sart et al. (2014) have previously analyzed the changes in the fluxes of central carbon metabolism in different glucose and oxygen regimes. Here, we used iMSC1255 to model internal fluxes in low concentration of glucose, both in normoxia and hypoxia. In order to model each condition, the exchange fluxes of glucose and oxygen were set according to the fluxes reported in literature (Pattappa et al. 2013). The maximum uptake flux of glucose was set to 0.005 mmol/grDW/h, with a minimum of 0 mmol/grDW/h. The boundaries of oxygen exchange fluxes in different culture conditions are shown in Table 1.
Table 1.
The boundaries of exchange fluxes of oxygen in hypoxia and normoxia are shown in mmol/grDW/h
| Culture conditions | Exchange fluxes | |
|---|---|---|
| Lower bound of oxygen exchange flux (mmol/grDW/h) | Upper bound of oxygen exchange flux (mmol/grDW/h) | |
| Hypoxia | −0.2 | −0.01 |
| Normoxia | −0.5 | −0.25 |
Sampling the flux space for modeling internal fluxes
Thiele et al. (2005) had previously shown that statistical analysis of a uniform random sample of fluxes can help in understanding the physiology of cells under different conditions, e.g., diabetes, ischemia or diet. By applying a similar strategy, for each of the four above-mentioned conditions, we generated 10,000 random flux distributions in the flux space. For this purpose, we used gpsampler function of COBRA Toolbox (Becker et al. 2007). Under each of the four conditions, the median of sampled fluxes was calculated for each reaction. The computed median value for each reaction was then used as a proxy for enzyme activity, in order to compare metabolic activities under different conditions. Then, we searched the literature in order to find the effects of different oxygenation on the flux of reactions at low glucose concentration, in order to validate the sampling results.
Results and discussion
Increasing stem cell proliferation
We found that consumption of some of the metabolites can increase the maximum of biomass production flux (see “Increasing cell proliferation: in silico analysis” and Supplementary Table A.1). Then, we chose two of the most effective metabolites, phosphatidylserine (PS) and phosphatidylethanolamine (PE), to be added to cell culture medium. PE comprises about 25% of phospholipid mass in an eukaryotic cell (Bürgermeister et al. 2004). PS can be converted to PE and phosphatidylcholine, which is a precursor for several lipid second messengers and also plays a role in the structure of cell membrane (Shields et al. 2003). It is obvious that extracellular uptake of PE or PS is more efficient than their de novo synthesis. Therefore, it is expected that with extracellular availability of these metabolites, the cell stops de novo synthesis of these metabolites, until PE (or PS) is entirely consumed. This behavior of the cell is correctly predicted by the metabolic model, since PE and PS are among the biomass components of iMSC1255. In addition, such a behavior is reported for cultured cells (Nishijima et al. 1986).
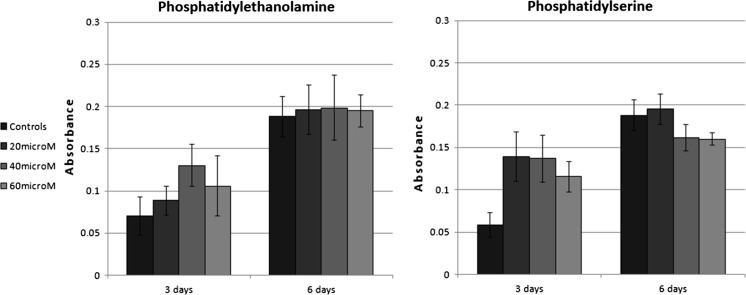
The positive effects of medium supplementation with PE and PS can be seen in Fig. 1. However, the positive trend can hardly be seen after 6 days of incubation. At this time interval, the positive effects of PE and PS on cell proliferation may have been stopped, because PE and PS have been consumed by the cells in previous days. This may approve the metabolic role of the metabolites in increasing cell proliferation, rather than a signaling influence. Overall, we can conclude that the computational results have been approved by experimental results.
Fig. 1.
MTT results of medium supplementation with phosphatidylethanolamine and phosphatidylserine. The number of living cells is directly related to its absorbance. Bone marrow derived MSCs were cultured for 3 and 6 days at different concentrations (20, 40, 60 and 80 μM) of each metabolite
It should be noted that, in the present study, we have modeled the contribution of individual metabolites to biomass production using independent FBA simulations. One may also model the metabolic consequence of simultaneous addition of a number of metabolites to the medium. However, in order to find the minimal set of metabolites that can increase growth rate, a more complicated approach, e.g., mixed integer linear programming, may be utilized.
Increasing stem cell differentiation
Here, we modeled the ways of increasing MSCs transition to chondrocytes, using MTA, iMSC1255 and transcriptome data of chondrocytes. Top score reactions which are the most appropriate candidates for knock-down are represented in Table 2. Most of these reactions (7 out of 10) are mitochondrial transport reactions, which might play related roles. According to the literature, mitochondrial metabolism is altered during cell differentiation (reviewed by Wanet et al. (2015)). Additionally, it has been shown that the transport of C4 metabolites, like malate and fumarate, is normally repressed during differentiation (Vozza et al. 2014).
Table 2.
Top ten reactions according to the scores of MTA
| Name of reactions | MTA scores of reactions |
|---|---|
| Fumarate:thiosulfate antiport, mitochondrial | 6.83512e+12 |
| Malate:thiosulfate antiport, mitochondrial | 6.83512e+12 |
| Fumarate transport, mitochondrial | 6.75313e+12 |
| Fumarate:sulfate antiport, mitochondrial | 6.41716e+12 |
| Malate:sulfate antiport, mitochondrial | 6.41716e+12 |
| Malate:sulfite antiport, mitochondrial | 1.98636e+12 |
| Fumarate:sulfite antiport, mitochondrial | 1.91809e+12 |
| Riboflavin exchange | 1.25419e+11 |
| UDP-glucose 4-epimerase | 1.25419e+11 |
| Chloride transport via bicarbonate counter transport | 5.64022e+10 |
Note that some of these reactions are involved in the transport of similar metabolites in mitochondria. Therefore, in order to investigate if knock-down of any pair of these reactions have the same effect on the flux distribution, we used flux coupling analysis (Burgard et al. 2004). Interestingly, only two of these reactions, i.e., malate:sulfate and malate:sulfite antiport are found to be coupled. Hence, except for this pair of reactions, one can conclude that changes in the fluxes of these uncoupled transport reactions have distinct (although possibly imbricated) consequences during cell differentiation. Therefore, it can be suggested for future works to knock-down top score reactions in vitro in order to see the increase in stem cell differentiation to chondrocytes.
We noted that the literature about stem cell differentiation is mainly focused on the reactions of central carbon metabolism, trying to establish a correlation between metabolic changes and stem cell differentiation. Therefore, we have also listed the MTA scores of central carbon metabolism reactions in Supplementary Table A.2. Relatively low scores in this list may suggest that alterations of certain reactions like mitochondrial transport reactions may have a more important role for differentiation, compared to the reactions of central carbon metabolism.
Modeling the effect of different growth conditions on metabolic fluxes
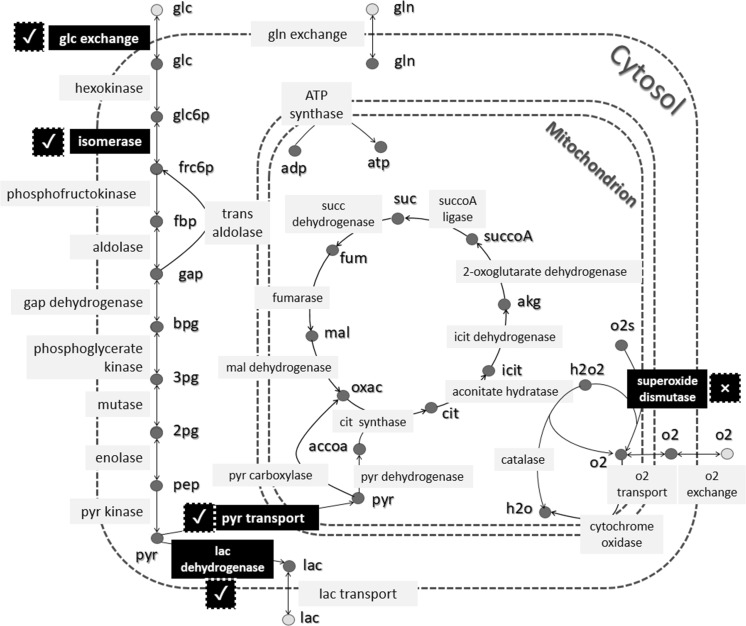
As we mentioned, GEMs can be useful for flux analysis. Here, we estimated all fluxes under two different oxygenation conditions in low glucose concentration, using sampling data of iMSC1255. Then, we tried to validate the prediction of flux changes in two conditions using literature data. We could find experimental measurements for five of the reactions. Note that in vitro measuring of fluxes, which is called Metabolic Flux Analysis (MFA), needs mass spectrometry approaches and isotope labeling techniques (Antoniewicz 2015). However, only a limited number of fluxes can be measured and available modeling approaches impose limitations on the size of the network that can be studied. According to these problems, very few studies have been performed MFA on stem cells (Sá et al. 2017). Therefore, on most of the studies, the internal fluxes and their changes in a pathway are just inferred from measurements of exchange fluxes. Here, our computational results were approved by experimental evidence for four out of the five reactions, (see Fig. 2). In other words, we could successfully predict an estimation for up- and down-regulation of fluxes with a good level of accuracy.
Fig. 2.
In our study, the effects of different oxygenation in low glucose condition on the fluxes of all reactions have been modeled using sampling data of iMSC1255. Here, we see the results for the reactions of central carbon metabolism. We have found experimental data for the changes of 5 reactions in hypoxia and normoxia (Sart et al. 2014). These five reactions are highlighted in the figure. According to the literature, the flux of glucose uptake and lactate dehydrogenase are increased, the flux of pyruvate transport to mitochondrion and superoxide dismutase are decreased and the flux of isomerase remains unchanged in hypoxia, compared to normoxia. The computational prediction for flux changes was the same as for experimental measurements for 4 out of 5 reactions, marked with tick mark in the figure. The only false prediction occurred for dismutase, which is marked with cross mark in the figure
Conclusion and future work
The concentrations of different components in cell culture medium used to be designed empirically (Castro et al. 1992). In the present study, we used a metabolic modeling approach to find novel cell culture medium supplementations to improve stem cell growth. However, it is routine to change the concentrations of amino acids in order to design cell culture medium rationally (Altamirano et al. 2004; Read et al. 2013). Therefore, we decided to propose a list of optimum concentrations of amino acids in MSCs culture medium using metabolic modeling. Yang et al. (2009) used a hepatic metabolic network model to fine-tune the concentrations of cell culture medium components. We have done the same for MSCs using iMSC1255 and literature data (See Supplementary File, Figure A.1 and Table A.3, for more information). This proposed list of optimum concentrations of amino acids should be tested in vitro as future work.
In conclusion, the present study explored some the applications of the metabolic network model of bone marrow derived MSCs, iMSC1255, and indicated that computational predictions made by metabolic modeling were in accordance with experimental findings. The metabolic models can also be integrated with regulatory or signaling networks, in order to propose a whole cell model of a stem cell (Karr et al. 2012). These models can be used in finding better approaches to optimize the induction of stem cell differentiation, which has been mostly studied from a signaling point of view.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to acknowledge the financial support of University of Tehran for this research under grant number 28791/1/2.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s10616-017-0148-6) contains supplementary material, which is available to authorized users.
References
- Altamirano C, Paredes C, Illanes A, Cairo J, Godia F. Strategies for fed-batch cultivation of t-PA producing CHO cells: substitution of glucose and glutamine and rational design of culture medium. J Biotechnol. 2004;110:171–179. doi: 10.1016/j.jbiotec.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR. Methods and advances in metabolic flux analysis: a mini-review. J Ind Microbiol Biotechnol. 2015;42:317–325. doi: 10.1007/s10295-015-1585-x. [DOI] [PubMed] [Google Scholar]
- Becker SA, Feist AM, Mo ML, Hannum G, Palsson BØ, Herrgard MJ. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA toolbox. Nat Protoc. 2007;2:727–738. doi: 10.1038/nprot.2007.99. [DOI] [PubMed] [Google Scholar]
- Burgard AP, Nikolaev EV, Schilling CH, Maranas CD. Flux coupling analysis of genome-scale metabolic network reconstructions. Genome Res. 2004;14:301–312. doi: 10.1101/gr.1926504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürgermeister M, Birner-Grünberger R, Nebauer R, Daum G. Contribution of different pathways to the supply of phosphatidylethanolamine and phosphatidylcholine to mitochondrial membranes of the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2004;1686:161–168. doi: 10.1016/j.bbalip.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Castro PM, Hayter PM, Ison AP, Bull AT. Application of a statistical design to the optimization of culture medium for recombinant interferon-gamma production by Chinese hamster ovary cells. Appl Microbiol Biotechnol. 1992;38:84–90. doi: 10.1007/BF00169424. [DOI] [PubMed] [Google Scholar]
- Chang RL, Xie L, Xie L, Bourne PE, Palsson BØ. Drug off-target effects predicted using structural analysis in the context of a metabolic network model. PLoS Comput Biol. 2010;6:e1000938. doi: 10.1371/journal.pcbi.1000938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fell DA, Small JR. Fat synthesis in adipose tissue. An examination of stoichiometric constraints. Biochem J. 1986;238:781–786. doi: 10.1042/bj2380781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouladiha H, Marashi S-A. Biomedical applications of cell-and tissue-specific metabolic network models. J Biomed Inform. 2017;68:35–49. doi: 10.1016/j.jbi.2017.02.014. [DOI] [PubMed] [Google Scholar]
- Fouladiha H, Marashi SA, Shokrgozar MA. Reconstruction and validation of a constraint-based metabolic network model for bone marrow-derived mesenchymal stem cells. Cell Prolif. 2015;48:475–485. doi: 10.1111/cpr.12197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldbeter A, Lefever R. Dissipative structures for an allosteric model. Application to glycolytic oscillations. Biophys J. 1972;12:1302–1315. doi: 10.1016/S0006-3495(72)86164-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson WL, Zhao F, Izadpanah R, Bunnell B, Ma T. Effects of hypoxia on human mesenchymal stem cell expansion and plasticity in 3D constructs. J Cell Physiol. 2006;207:331–339. doi: 10.1002/jcp.20571. [DOI] [PubMed] [Google Scholar]
- Gutierrez JM, Lewis NE. Optimizing eukaryotic cell hosts for protein production through systems biotechnology and genome-scale modeling. Biotechnol J. 2015;10:939–949. doi: 10.1002/biot.201400647. [DOI] [PubMed] [Google Scholar]
- Hadi M, Marashi SA. Reconstruction of a generic metabolic network model of cancer cells. Mol BioSyst. 2014;10:3014–3021. doi: 10.1039/C4MB00300D. [DOI] [PubMed] [Google Scholar]
- Higgins J. A chemical mechanism for oscillation of glycolytic intermediates in yeast cells. Proc Natl Acad Sci USA. 1964;51:989–994. doi: 10.1073/pnas.51.6.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerby L, Ruppin E. Predicting drug targets and biomarkers of cancer via genome-scale metabolic modeling. Clin Cancer Res. 2012;18:5572–5584. doi: 10.1158/1078-0432.CCR-12-1856. [DOI] [PubMed] [Google Scholar]
- Karr JR, et al. A whole-cell computational model predicts phenotype from genotype. Cell. 2012;150:389–401. doi: 10.1016/j.cell.2012.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King ZA, Lloyd CJ, Feist AM, Palsson BO. Next-generation genome-scale models for metabolic engineering. Curr Opin Biotechnol. 2015;35:23–29. doi: 10.1016/j.copbio.2014.12.016. [DOI] [PubMed] [Google Scholar]
- Nishijima M, Kuge O, Akamatsu Y. Phosphatidylserine biosynthesis in cultured Chinese hamster ovary cells. I. Inhibition of de novo phosphatidylserine biosynthesis by exogenous phosphatidylserine and its efficient incorporation. J Biol Chem. 1986;261:5784–5789. [PubMed] [Google Scholar]
- O’Brien EJ, Monk JM, Palsson BO. Using genome-scale models to predict biological capabilities. Cell. 2015;161:971–987. doi: 10.1016/j.cell.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth JD, Thiele I, Palsson BØ. What is flux balance analysis? Nat Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattappa G, Heywood HK, de Bruijn JD, Lee DA. The metabolism of human mesenchymal stem cells during proliferation and differentiation. J Cell Physiol. 2011;226:2562–2570. doi: 10.1002/jcp.22605. [DOI] [PubMed] [Google Scholar]
- Pattappa G, Thorpe SD, Jegard NC, Heywood HK, de Bruijn JD, Lee DA. Continuous and uninterrupted oxygen tension influences the colony formation and oxidative metabolism of human mesenchymal stem cells. Tissue Eng Part C. 2013;19:68–79. doi: 10.1089/ten.tec.2011.0734. [DOI] [PubMed] [Google Scholar]
- Poolman MG, Venkatesh KV, Pidcock MK, Fell DA. A method for the determination of flux in elementary modes, and its application to Lactobacillus rhamnosus. Biotechnol Bioeng. 2004;88:601–612. doi: 10.1002/bit.20273. [DOI] [PubMed] [Google Scholar]
- Read EK, Bradley SA, Smitka TA, Agarabi CD, Lute SC, Brorson KA. Fermentanomics informed amino acid supplementation of an antibody producing mammalian cell culture. Biotechnol Prog. 2013;29:745–753. doi: 10.1002/btpr.1728. [DOI] [PubMed] [Google Scholar]
- Sá JV, Kleiderman S, Brito C, Sonnewald U, Leist M, Teixeira AP, Alves PM. Quantification of metabolic rearrangements during neural stem cells differentiation into astrocytes by metabolic flux analysis. Neurochem Res. 2017;42:244–253. doi: 10.1007/s11064-016-1907-z. [DOI] [PubMed] [Google Scholar]
- Saha R, Chowdhury A, Maranas CD. Recent advances in the reconstruction of metabolic models and integration of omics data. Curr Opin Biotechnol. 2014;29:39–45. doi: 10.1016/j.copbio.2014.02.011. [DOI] [PubMed] [Google Scholar]
- Sart S, Agathos SN, Li Y. Process engineering of stem cell metabolism for large scale expansion and differentiation in bioreactors. Biochem Eng J. 2014;84:74–82. doi: 10.1016/j.bej.2014.01.005. [DOI] [Google Scholar]
- Shields DJ, Lehner R, Agellon LB, Vance DE. Membrane topography of human phosphatidylethanolamine N-methyltransferase. J Biol Chem. 2003;278:2956–2962. doi: 10.1074/jbc.M210904200. [DOI] [PubMed] [Google Scholar]
- Shlomi T, Cabili MN, Ruppin E. Predicting metabolic biomarkers of human inborn errors of metabolism. Mol Syst Biol. 2009;5:263. doi: 10.1038/msb.2009.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeonidis E, Price ND. Genome-scale modeling for metabolic engineering. J Ind Microbiol Biotechnol. 2015;42:327–338. doi: 10.1007/s10295-014-1576-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele I, Price ND, Vo TD, Palsson BØ. Candidate metabolic network states in human mitochondria: impact of diabetes, ischemia, and diet. J Biol Chem. 2005;280:11683–11695. doi: 10.1074/jbc.M409072200. [DOI] [PubMed] [Google Scholar]
- Varma A, Palsson BØ. Metabolic capabilities of Escherichia coli II. Optimal growth patterns. J Theor Biol. 1993;165:503–522. doi: 10.1006/jtbi.1993.1203. [DOI] [PubMed] [Google Scholar]
- Vozza A, Parisi G, De Leonardis F, Lasorsa FM, Castegna A, Amorese D, Marmo R, Calcagnile VM, Palmieri L, Ricquier D, Paradies E, Scarcia P, Palmieri F, Bouillaud F, Fiermonte G. UCP2 transports C4 metabolites out of mitochondria, regulating glucose and glutamine oxidation. Proc Natl Acad Sci USA. 2014;111:960–965. doi: 10.1073/pnas.1317400111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanet A, Arnould T, Najimi M, Renard P. Connecting mitochondria, metabolism, and stem cell fate. Stem Cells Dev. 2015;24:1957–1971. doi: 10.1089/scd.2015.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiback SJ, Palsson BØ. Extreme pathway analysis of human red blood cell metabolism. Biophys J. 2002;83:808–818. doi: 10.1016/S0006-3495(02)75210-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Roth CM, Ierapetritou MG. A rational design approach for amino acid supplementation in hepatocyte culture. Biotechnol Bioeng. 2009;103:1176–1191. doi: 10.1002/bit.22342. [DOI] [PubMed] [Google Scholar]
- Yazdani SO, Hafizi M, Zali AR, Atashi A, Ashrafi F, Seddighi AS, Soleimani M. Safety and possible outcome assessment of autologous Schwann cell and bone marrow mesenchymal stromal cell co-transplantation for treatment of patients with chronic spinal cord injury. Cytotherapy. 2013;15:782–791. doi: 10.1016/j.jcyt.2013.03.012. [DOI] [PubMed] [Google Scholar]
- Yizhak K, Gabay O, Cohen H, Ruppin E. Model-based identification of drug targets that revert disrupted metabolism and its application to ageing . Nat Commun. 2013;4:2632. doi: 10.1038/ncomms3632. [DOI] [PubMed] [Google Scholar]
- Zhao F, Pathi P, Grayson W, Xing Q, Locke BR, Ma T. Effects of oxygen transport on 3-D human mesenchymal stem cell metabolic activity in perfusion and static cultures: experiments and mathematical model. Biotechnol Prog. 2005;21:1269–1280. doi: 10.1021/bp0500664. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.