ABSTRACT
Macrophages are strategically distributed in mammalian tissues and play an essential role in priming the immune response. However, macrophages need to constantly strike a balance between activation and inhibition states to avoid a futile inflammatory reaction. Here, we identify the CBP/p300-interacting transactivator with glutamic acid/aspartic acid-rich carboxyl-terminal domain 2 (CITED2) as a potent repressor of macrophage proinflammatory activation. Gain- and loss-of-function studies revealed that CITED2 is required for optimal peroxisome proliferator-activated receptor gamma (PPARγ) activation and attendant select anti-inflammatory gene expression in macrophages. More importantly, deficiency of CITED2 resulted in significant attenuation of rosiglitazone-induced PPARγ activity, PPARγ recruitment to target gene promoters, and anti-inflammatory target gene expression in macrophages. Interestingly, deficiency of Cited2 strikingly heightened proinflammatory gene expression through stabilization of hypoxia-inducible factor 1 alpha (HIF1α) protein in macrophages. Further, overexpression of Egln3 or inhibition of HIF1α in Cited2-deficient macrophages completely reversed elevated proinflammatory cytokine/chemokine gene expression. Importantly, mice bearing a myeloid cell-specific deletion of Cited2 were highly susceptible to endotoxin-induced sepsis symptomatology and mortality. Collectively, our observations identify CITED2 as a novel negative regulator of macrophage proinflammatory activation that protects the host from inflammatory insults.
KEYWORDS: Cited2, HIF1α, inflammation, macrophage, PPARγ, innate immunity
INTRODUCTION
Inflammation is an adaptive host response that plays a critical role in maintaining tissue homeostasis by eliminating infectious or deleterious agents (1). Monocyte-derived macrophages are the major innate immune cells recruited to the site of infection or injury that orchestrate amplification for the resolution of inflammation (2). Indeed, macrophage-mediated inflammation is an explicitly robust biological response that consists of alterations in inflammatory gene expression affecting the significant part of the cellular genome (3). However, endogenous mechanisms must exist to rigorously constrain uncontrolled proinflammatory activation of macrophages to prevent unwanted tissue damage and deleterious effects on the host (4). These endogenous negative regulators of inflammation predominantly operate through cell-extrinsic and cell-intrinsic mechanisms (5). Many of the cell-extrinsic negative regulators of inflammation, such as interleukin 10 (IL-10), transforming growth factor β (TGF-β), IL-1ra, and IL-1r2, are known to diminish global proinflammatory gene expression either by inducing intracellular anti-inflammatory signaling pathways or by blocking proinflammatory cytokine activity by decoy receptor actions (6). Similarly, cell-intrinsic, signal-specific regulators, such as IRAK-M, SOCS, NOD2, A20, DUSP1, and ABIN1, attenuate global proinflammatory gene expression by dampening downstream actions of cell surface receptors (5). However, cell-intrinsic, gene-specific regulators, particularly transcriptional factors and cofactors, play a crucial role in restraining the intensity of inflammation by meticulously calibrating pro- and anti-inflammatory gene expression in myeloid cells (7).
Macrophages newly recruited to the site of infection or injury are proinflammatory in nature (8). The inflammatory milieu in the local tissue environment shapes the macrophage gene expression response by activating a specific innate immune signaling pathway (9). Many of the proinflammatory agents are known to initiate inflammation by activating a cascade of transcription factors, such as NF-κB, AP1, STAT1, hypoxia-inducible factor 1 alpha (HIF1α), interferon regulatory factor 3 (IRF3) or IRF5. Activation of these transcription factors is well known to facilitate induction of a wide range of proinflammatory gene expression in macrophages (10, 11). However, anti-inflammatory cytokines and negative regulators of inflammation mediate resolution of inflammation by activating STAT3, STAT6, peroxisome proliferator-activated receptor gamma (PPARγ), IRF4, NFIL3, ATF3, or liver X receptor (LXR) transcription factors (10, 11). Further, sumoylation of PPARγ and LXR by anti-inflammatory signaling pathways prevents proinflammatory stimulus-induced removal of the repressor complex from NF-κB and AP1 binding sites (12, 13), further advancing the repression of proinflammatory gene expression while maintaining anti-inflammatory macrophage status. In the current study, we identified CITED2 as an essential regulator that calibrates macrophage pro- and anti-inflammatory gene expression and response.
CBP/p300-interacting transactivator with glutamic acid/aspartic acid-rich carboxyl-terminal domain 2 (Cited2) is a member of the CITED family of proteins that play very critical roles in cellular development and differentiation (14). Several independent studies have revealed that mutation in the CITED2 gene is associated with human congenital heart defects (15, 16). Concordant with these observations, experimental murine studies affirmed that mutation or loss of Cited2 resulted in congenital heart defects, as well as perturbation in the left-right patterning of the body axis (17, 18). Furthermore, deficiency of Cited2 significantly impaired the development of lung, liver, placenta, and adrenal tissue at the embryonic stage (19–22). At the molecular level, CITED2 regulates the functions of several transcription factors, including LHX2, HIF1α, SMAD2/3, and HNF4a, and modulates their target gene expression (23–26). However, whether Cited2 regulates pro- or anti-inflammatory gene expression in the context of myeloid cell-mediated inflammation has not been investigated. In this study, we identify myeloid CITED2 as a critical regulator of macrophage pro- and anti-inflammatory gene expression and function. Furthermore, our studies reveal that CITED2 cooperates with PPARγ to induce anti-inflammatory gene expression while suppressing proinflammatory gene expression by diminishing HIF1α protein accumulation in macrophages.
RESULTS
CITED2 is expressed in human and murine macrophages.
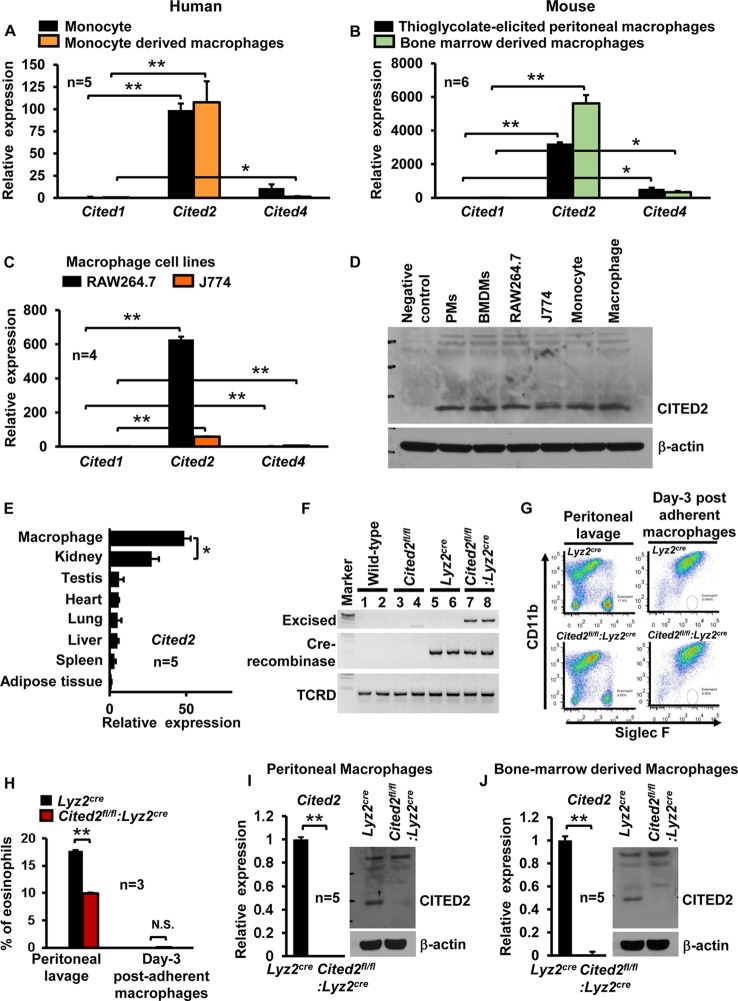
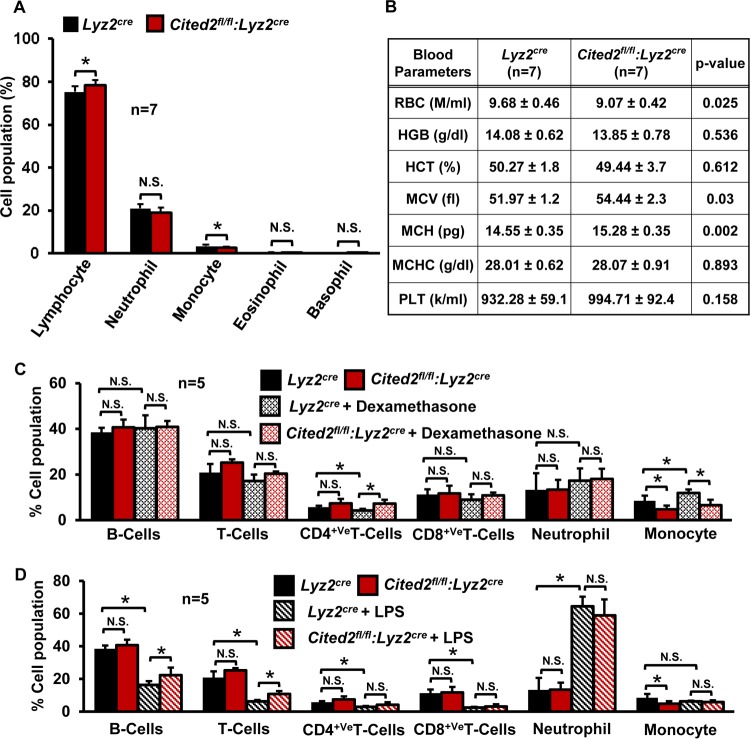
Recent studies have shown that CITED2 is required for normal hematopoiesis and plays an essential role in monocyte/macrophage cell development (20, 27). However, whether CITED2 is expressed in fully differentiated macrophages and plays any functional role has not been investigated. Therefore, we examined the expression of CITED family cofactors in human and murine primary macrophages. Analysis of human primary monocytes and monocyte-derived macrophages revealed that Cited2 mRNA expression is approximately 100-fold higher than that of Cited1 or Cited4 (Fig. 1A). Similarly, evaluation of murine thioglycolate-elicited peritoneal macrophages (PMs) and bone marrow-derived macrophages (BMDMs) revealed that Cited2 is more abundantly expressed in both cell types than Cited1 or Cited4 (Fig. 1B). Moreover, these observations are recapitulated in murine monocyte/macrophage lines (Fig. 1C). Further, we confirmed CITED2 protein expression in human/murine macrophage lines and primary macrophages (Fig. 1D). Decisively, our survey of Cited2 expression analysis across multiple murine tissues demonstrated that Cited2 is most abundantly expressed in macrophages (Fig. 1E). To investigate the functional role of CITED2 in macrophage inflammatory gene expression, we generated myeloid cell-specific Cited2-deficient mice by crossing Cited2 floxed (Cited2fl/fl) mice (28) with a mouse line expressing lysozyme M promoter-driven Cre recombinase (Lyz2cre). Our analyses confirmed the genomic excision of the Cited2 protein-coding region in Cited2fl/fl:Lyz2cre (genotype of mice with two Cited2 floxed loci and two Lyz2cre loci) mouse macrophages (Fig. 1F). Consistent with this observation, quantitative-PCR (qPCR) studies revealed greater than 99% reduction of Cited2 at the mRNA level in Cited2fl/fl:Lyz2cre mouse PMs and BMDMs compared to those of the Lyz2cre group (Fig. 1G to J). Further, Western blot analysis corroborated complete loss of CITED2 protein in Cited2fl/fl:Lyz2cre PMs and BMDMs (Fig. 1I and J). Since CITED2 is essential for normal hematopoiesis (20, 27), we examined whether myeloid deficiency of Cited2 affects the adult mouse hematopoietic cell compartment. Our results revealed a modest but significant decrease in monocyte populations in Cited2fl/fl:Lyz2cre mice (Fig. 2A). However, we did not observe any significant differences in lymphocyte, neutrophil, eosinophil, and basophil populations between the Lyz2cre and Cited2fl/fl:Lyz2cre mouse groups (Fig. 2A). Further analysis of additional hematological components revealed a modest decrease in red blood cell counts and a slight increase in mean corpuscular hemoglobin content (MCH) and volume (MCV) (Fig. 2B) in Cited2fl/fl:Lyz2cre mice. More importantly, myeloid deficiency of Cited2 did not dramatically alter anti-inflammatory (dexamethasone) or proinflammatory (lipopolysaccharide [LPS]) agent-induced shift in the lymphocyte or myeloid cell population (Fig. 2C and D). Collectively, our results revealed that CITED2 is profusely expressed in human/murine macrophages and that myeloid cell-specific deficiency of Cited2 did not dramatically alter the hematopoietic components in adult mice.
FIG 1.
CITED2 expression in macrophages and effect of myeloid Cited2 deficiency on the adult hematopoietic cell compartment. (A) Human peripheral blood monocytes and monocyte-derived macrophages were analyzed for Cited1, Cited2, and Cited4 mRNA expression by quantitative-PCR analysis. The succinate dehydrogenase complex flavoprotein subunit A (SDHA) gene was used as a housekeeping gene (n = 5). (B) Mouse thioglycolate-elicited peritoneal macrophages and bone marrow-derived macrophages were analyzed for Cited1, Cited2, and Cited4 mRNA expression by quantitative PCR. The 36B4 gene was used as a housekeeping gene (n = 6). (C) Quantitative-PCR analysis of Cited1, Cited2, and Cited4 mRNA expression in RAW 264.7 and J774 cells (n = 4). (D) CITED2 protein expression in indicated primary cells and cell lines by Western blot analysis (n = 3). (E) Cited2 mRNA expression was analyzed in mouse adipose tissue, spleen, liver, lung, heart, testis, kidney, and bone marrow-derived macrophages for relative expression of Cited2 by quantitative-PCR analysis. The 36B4 gene was used as a housekeeping gene (n = 5). (F) Wild-type, Cited2fl/fl, Lyz2cre, and Cited2fl/fl:Lyz2cre BMDM genomic DNA was subjected to reverse transcription-PCR analysis to detect the presence of Cre recombinase and excised Cited2 genomic sequence using a site-specific primer. The T-cell receptor δ chain (TCRD) gene was used as an internal control (n = 3). (G and H) Thioglycolate-elicited peritoneal lavage fluid and postadherent macrophages (day 3) derived from Lyz2cre and Cited2fl/fl:Lyz2cre mice were collected. These cell populations were examined for eosinophil and macrophage abundance by SIGLEC5 (Siglec F) and ITGAM (CD11b) staining and FACS analysis (n = 3). (I and J) Thioglycolate-elicited peritoneal macrophages and BMDMs from Lyz2cre and Cited2fl/fl:Lyz2cre mice were analyzed for CITED2 mRNA and protein expression by quantitative-PCR and Western blot analyses, respectively (n = 5). Data were analyzed by either 2-way ANOVA (A to C) or Student's t test (E to J). N.S., not significant; *, P < 0.01; **, P < 0.0001. All values are reported as means and SD.
FIG 2.
(A and B) Impact of myeloid Cited2 deficiency on the hematopoietic compartment. Age- and sex-matched Lyz2cre and Cited2fl/fl:Lyz2cre mouse total blood was analyzed with a Hemavet 950 hematology-profiling unit. The bar graph (A) and table (B) represent the mean cellular population with standard deviations (n = 7). (C and D) Ten-week-old male Lyz2cre and Cited2fl/fl:Lyz2cre mice received an intraperitoneal injection of the anti-inflammatory agent dexamethasone (20 mg/kg) or the proinflammatory agent lipopolysaccharide (5 mg/kg). Lyz2cre and Cited2fl/fl:Lyz2cre mice injected with saline served as controls. The mice were allowed to rest for 3 days, and blood samples were collected on day 4 by venipuncture. These blood samples were incubated with 1× RBC lysis buffer to remove red blood cells. The resulting white blood cells were stained with anti-CD3 (T cell marker), anti-CD4+ T cells, anti-CD8+ T cells, anti-CD11b (myeloid), anti-Ly6G (neutrophil), and anti-CD19 on ice for 30 min. The cells were washed two times with ice-cold 1× PBS and resuspended in 1× PBS containing 2% FBS and analyzed utilizing Flowsight imaging flow cytometry (EMD Millipore). The cell population was defined by utilizing two or more cell surface markers. Data were analyzed by Student's t test, and all values are reported as means and SD. N.S., not significant; *, P < 0.01.
Myeloid CITED2 deficiency attenuates anti-inflammatory gene expression.
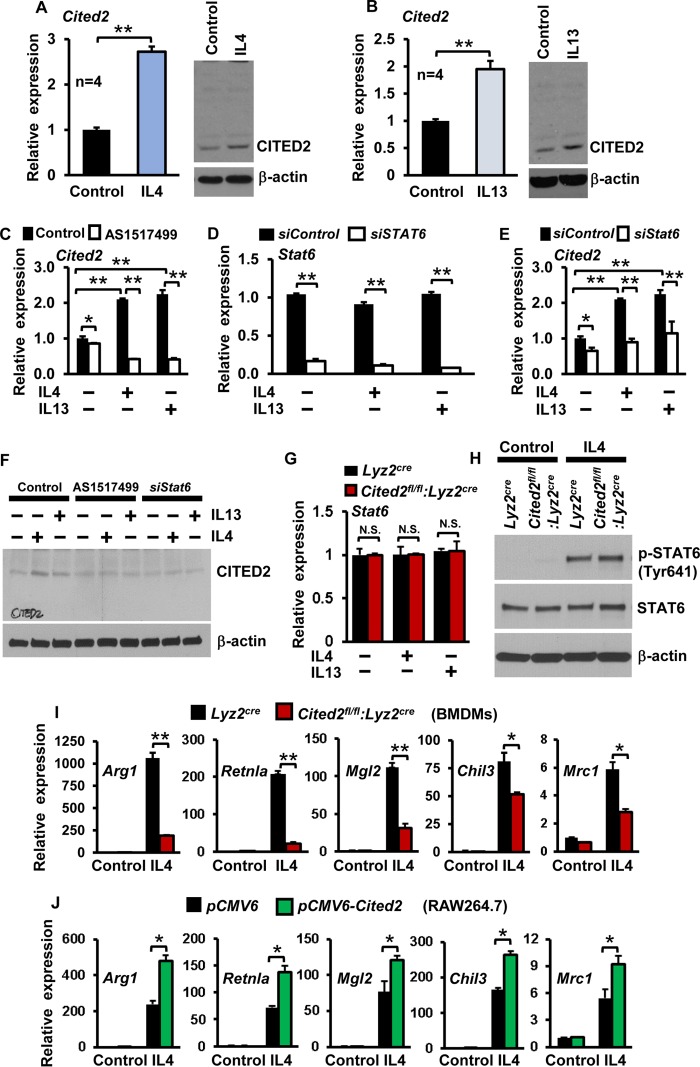
Previous studies have demonstrated that deficiency of Cited2 results in loss of quiescence in hematopoietic stem cells (29). Therefore, we examined whether anti-inflammatory cytokines, such as IL-4 and IL-13, alter CITED2 expression in macrophages. As shown in Fig. 3A and B, IL-4 or IL-13 exposure significantly induced CITED2 mRNA and protein expression in macrophages. It is well established that anti-inflammatory cytokines, such as IL-4 and IL-13, facilitate target gene expression through STAT6 (30). Therefore, we examined whether IL-4/IL-13-induced CITED2 expression is STAT6 dependent. As shown in Fig. 3C to E, pharmacological (AS1517499) or genetic (siStat6 small interfering RNA [siRNA]) blockade of STAT6 completely abrogated IL-4- or IL-13-induced Cited2 mRNA expression in macrophages. Similarly, STAT6 blockade markedly attenuated IL-4- or IL-13-induced CITED2 protein expression in macrophages (Fig. 3F). Further, deficiency of Cited2 did not alter basal or IL-4-induced STAT6 mRNA/protein expression or phosphorylation status (Fig. 3G and H). As CITED2 is induced by anti-inflammatory cytokines through a STAT6-dependent pathway, we hypothesized that CITED2 may promote Th2 cytokine-induced anti-inflammatory gene expression in macrophages. To test this hypothesis, Lyz2cre and Cited2fl/fl:Lyz2cre mouse BMDMs were stimulated with IL-4 and analyzed for anti-inflammatory gene expression. As anticipated, IL-4 stimulation robustly induced Arg1, Retnla, Mgl2, Chil3, and Mrc1 gene expression in Lyz2cre mouse BMDMs (Fig. 3I). Intriguingly, deficiency of Cited2 markedly reduced IL-4-induced Arg1, Retnla, Mgl2, Chil3, and Mrc1 gene expression (Fig. 3I). Concurrently, we evaluated the impact of Cited2 overexpression on IL-4-induced anti-inflammatory gene expression in macrophages (Fig. 3J). Consistent with our ex vivo observations, overexpression of Cited2 significantly elevated IL-4-induced Arg1, Retnla, Mgl2, Chil3, and Mrc1 gene expression (Fig. 3J). Taken together, our results reveal that CITED2 promotes IL-4-induced anti-inflammatory gene expression in macrophages.
FIG 3.
Myeloid CITED2 promotes anti-inflammatory gene expression and function. (A and B) Wild-type murine BMDMs were stimulated with 10 ng/ml of IL-4 or IL-13 separately for 6 h. CITED2 mRNA and protein expression were evaluated by quantitative-PCR and Western blot analyses, respectively (n = 4). (C to F) Wild-type murine BMDMs were pretreated with the STAT6-specific inhibitor AS1517499 (C and F) or transfected with siStat6-specific siRNA (D to F). The cells were stimulated with IL-4 or IL-13 separately and evaluated for CITED2 mRNA (C and E) and protein (F) expression (n = 3). (G) Lyz2cre and Cited2fl/fl:Lyz2cre mouse BMDMs were stimulated with IL-4 or IL-13 separately and analyzed for Stat6 mRNA expression by quantitative PCR. (H) Lyz2cre and Cited2fl/fl:Lyz2cre mouse BMDMs were stimulated with IL-4 and analyzed for STAT6 protein expression and phosphorylation (STAT6-Tyr641) by Western blotting. (I) BMDMs from Lyz2cre and Cited2fl/fl:Lyz2cre were stimulated with 10 ng/ml of IL-4 for 18 h. Total RNA was isolated, and mRNA expression of Arg1, Retnla, Mgl2, Chil3, and Mrc1 were analyzed by quantitative PCR (n = 4). (J) RAW 264.7 cells transfected with either pCMV6 or pCMV6-Cited2 were stimulated with IL-4, and expression of Arg1, Retnla, Mgl2, Chil3, and Mrc1 was analyzed by quantitative PCR (n = 4). The beta-actin and 36B4 genes were used as housekeeping genes for Western blot and quantitative-PCR analyses, respectively. Each experiment consisted of three replicates. Data were analyzed by Student's t test. N.S., not significant; *, P < 0.01; **, P < 0.0001. All values are reported as means and SD.
CITED2 cooperates with PPARγ to promote anti-inflammatory gene expression.
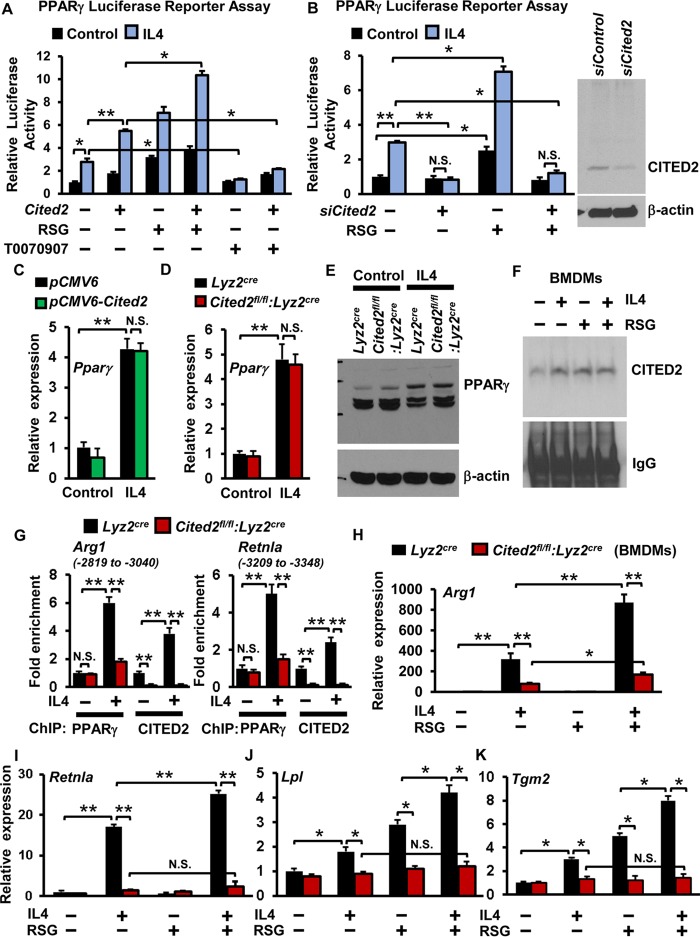
Previous studies established that IL-4-induced anti-inflammatory gene expression is broadly facilitated through PPARγ in macrophages (30). Therefore, we evaluated whether CITED2 cooperates with PPARγ to regulate IL-4-induced anti-inflammatory gene expression in macrophages. Accordingly, RAW 264.7 cells were cotransfected with PPAR luciferase reporter plasmid in the presence of pCMV6-Cited2 or siCited2 and stimulated with IL-4, and luciferase activity was documented. Our results demonstrated that overexpression of Cited2 significantly enhanced IL-4-induced PPARγ luciferase reporter activity (Fig. 4A). Interestingly, deficiency of Cited2 completely abrogated IL-4-induced PPARγ luciferase reporter activity in macrophages (Fig. 4B). Importantly, overexpression of Cited2 enhanced, and deficiency of Cited2 completely attenuated, rosiglitazone-induced PPARγ-driven luciferase reporter activity in macrophages (Fig. 4A and B). The PPARγ-specific antagonist T0070907 completely blocked IL-4-induced PPARγ-driven luciferase reporter activity and served as a negative control (Fig. 4A). Previous studies have illustrated that IL-4 induces PPARγ expression in macrophages (31). Therefore, we examined whether altering Cited2 levels affects IL-4-induced PPARγ expression in macrophages. Our analyses revealed that neither overexpression nor deficiency of Cited2 affected IL-4-induced PPARγ mRNA and protein expression in macrophages (Fig. 4C to E). Interestingly, exposure of macrophages to IL-4 or rosiglitazone markedly elevated CITED2 interaction with PPARγ (Fig. 4F). Therefore, we examined whether deficiency of Cited2 altered IL-4-induced PPARγ recruitment to its targets, such as Arg1 and Retnla gene promoters. Accordingly, Lyz2cre and Cited2fl/fl:Lyz2cre BMDMs were stimulated with IL-4, and chromatin immunoprecipitation (ChIP) was performed using anti-PPARγ and anti-CITED2 antibodies separately. These ChIP samples were analyzed for PPARγ and CITED2 enrichment in the Arg1 enhancer (−2874 to −3040) and Retnla promoter (−3209 to −3358) regions by utilizing site-specific primers. Our analysis revealed that IL-4 exposure significantly elevated PPARγ and CITED2 enrichment on the Arg1 enhancer and Retnla promoter, respectively (Fig. 4G). Interestingly, deficiency of Cited2 significantly attenuated IL-4-induced PPARγ recruitment to these genomic sites. Importantly, no significant enrichment of CITED2 was observed in Cited2fl/fl:Lyz2cre BMDMs (negative control). Taken together, these results revealed that PPARγ and CITED2 are corecruited to the Arg1 enhancer and Retnla promoter following IL-4 exposure. Next, we examined whether IL-4-induced select PPARγ target gene expression is dependent on CITED2 in macrophages. Accordingly, BMDMs from Lyz2cre and Cited2fl/fl:Lyz2cre mice were stimulated with IL-4 in the presence and absence of the PPARγ ligand rosiglitazone. Analysis of select PPARγ target gene expression revealed that deficiency of Cited2 significantly attenuated IL-4-induced expression of Arg1, Retnla, Lpl, and Tgm2 (Fig. 4H to K). More importantly, supplementation of rosiglitazone did not rescue attenuation of these PPARγ target gene expression levels in Cited2-deficient macrophages (Fig. 4H to K). Taken together, our results demonstrate that CITED2 is essential for IL-4-induced PPARγ transcriptional activity and select anti-inflammatory target gene expression in macrophages.
FIG 4.
CITED2 cooperates with PPARγ to enhance IL-4-induced anti-inflammatory gene expression. (A) RAW 264.7 cells were transfected with a PPAR luciferase reporter construct in the presence of pCMV6 or pCMV6-Cited2 plasmid. The cells were stimulated with 10 ng/ml IL-4 in the presence of rosiglitazone (RSG) or T0070907, and cell lysates were analyzed for luciferase activity (n = 3). (B) RAW 264.7 cells were transfected with a PPAR luciferase reporter construct in the presence of control or Cited2-specific siRNA. The cells were stimulated with 10 ng/ml IL-4 in the presence of 10 μM RSG, and cell lysates were analyzed for luciferase activity (n = 3). The efficiency of siCited2 siRNA was confirmed by Western blot analysis. (C) RAW 264.7 cells were transfected with either pCMV6 or pCMV6-Cited2 plasmid. The cells were stimulated with 10 ng/ml of IL-4 for 18 h, and mRNA expression of PPARγ was analyzed by quantitative PCR (n = 4). (D and E) BMDMs from Lyz2cre and Cited2fl/f:Lyz2cre mice were stimulated with 10 ng/ml of IL-4 for 18 h. PPARγ mRNA and protein expression was evaluated by quantitative-PCR and Western blot analyses, respectively (n = 4). (F) BMDMs from wild-type mice were stimulated with 10 ng/ml of IL-4 for 1 h. The cell lysates were immunoprecipitated with anti-PPARγ antibody and immunoblotted with the anti-CITED2 antibody (n = 3). (G) Lyz2cre and Cited2fl/fl:Lyz2cre BMDMs were stimulated with IL-4, and ChIP analysis was performed on the Arg1 enhancer and Retnla promoter utilizing anti-PPARγ and anti-CITED2 antibodies (n = 3). (H to K) BMDMs from Lyz2cre and Cited2fl/fl:Lyz2cre mice were stimulated with 10 ng/ml IL-4 in the presence or absence of RSG. Total RNA was isolated, and mRNA expression of Arg1, Retnla, Lpl, and Tgm2 were analyzed by quantitative PCR (n = 3). The beta-actin gene was used as a housekeeping gene for Western blotting, and the 36B4 gene was used as a housekeeping gene for quantitative-PCR analysis. Each experiment was performed at least three times with three replicates. Data were analyzed by either 2-way ANOVA (A, B, and H to K) or Student's t test (C, D, and G). N.S., not significant; *, P < 0.01; **, P < 0.0001. All values are reported as means and SD.
Myeloid CITED2 deficiency augments proinflammatory gene expression and host susceptibility to endotoxic shock.
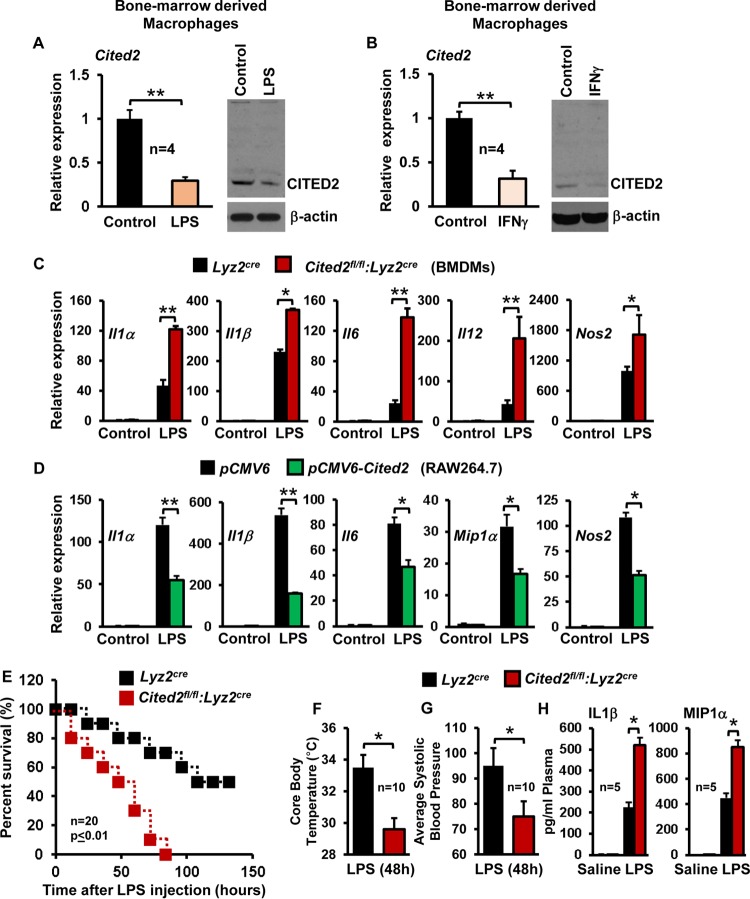
Next, we investigated whether proinflammatory agents alter CITED2 expression in macrophages. As shown in Fig. 5A and B, LPS or gamma interferon (IFN-γ) exposure significantly attenuated CITED2 mRNA and protein expression in macrophages. Based on these observations, we hypothesized that CITED2 may repress proinflammatory stimulus-induced inflammatory gene expression in macrophages. To test this hypothesis, Lyz2cre and Cited2fl/fl:Lyz2cre mouse BMDMs were stimulated with LPS and analyzed for expression of classical proinflammatory genes. As anticipated, LPS stimulation strongly induced Il1α, Il1β, Il6, Il12, and Nos2 expression in Lyz2cre mouse BMDMs (Fig. 5C). Remarkably, deficiency of Cited2 resulted in significantly elevated levels of Il1α, Il1β, Il6, Il12, and Nos2 expression following LPS treatment (Fig. 5C). Simultaneously, we examined whether overexpression of Cited2 altered LPS-induced proinflammatory gene expression in macrophages. Concordant with our ex vivo observations, overexpression of Cited2 significantly attenuated LPS-induced Il1α, Il1β, Il6, Il12, and Nos2 gene expression (Fig. 5D). It is well established that endotoxins produced from bacterial pathogens could elicit uncontrolled myeloid cell activation, which culminates in a proinflammatory cytokines storm that leads to multiorgan failure, vascular collapse, and death. Based on our observations (Fig. 5A to D), we hypothesized that myeloid Cited2 may offer host protective effects in the face of endotoxin challenge. To test this hypothesis, Lyz2cre and Cited2fl/fl:Lyz2cre mice were subjected to LPS-induced sepsis. As shown in Fig. 5E, LPS challenge of Cited2fl/fl:Lyz2cre mice produced 100% mortality by 84 h, whereas Lyz2cre mice experienced only 30% mortality. In addition, Cited2fl/fl:Lyz2cre mice exhibited significantly elevated cardinal features of sepsis, including hypothermia (Fig. 5F) and hypotension (Fig. 5G). More importantly, LPS challenge significantly elevated proinflammatory plasma cytokines in Cited2fl/fl:Lyz2cre mice compared to Lyz2cre mice (Fig. 5H). Collectively, our results reveal that myeloid CITED2 attenuates LPS-induced proinflammatory gene expression and protects the host from detrimental effects of endotoxin challenge.
FIG 5.
Myeloid CITED2 deficiency augments proinflammatory gene expression and host susceptibility to endotoxic shock. (A and B) Wild-type murine BMDMs were stimulated with 100 ng/ml of LPS or IFN-γ separately for 6 h. CITED2 mRNA and protein expression was evaluated by quantitative-PCR and Western blot analyses, respectively (n = 4). (C) BMDMs from Lyz2cre and Cited2fl/fl:Lyz2cre mice were stimulated with 100 ng/ml of LPS for 6 h. Total RNA was isolated, and mRNA expression of Il1α, Il1β, Il6, Il12, and Nos2 was analyzed by quantitative PCR (n = 4). (D) RAW 264.7 cells were transiently transfected with either pCMV6 or pCMV6-Cited2. The cells were stimulated with 100 ng/ml of LPS for 6 h, and mRNA expression of Il1α, Il1β, Il6, Il12, and Nos2 was analyzed by quantitative PCR (n = 4). (E) Age- and sex-matched Lyz2cre and Cited2fl/fl:Lyz2cre mice were subjected to LPS-induced sepsis. Host survival data were analyzed by the construction of Kaplan-Meier plots and use of the log rank test (n = 20). (F and G) Age- and sex-matched Lyz2cre and Cited2fl/fl:Lyz2cre mice were challenged with LPS, and changes in core body temperature and systolic blood pressure were recorded (n = 10). (H) Blood plasma obtained 5 h after LPS or saline administration was quantified for IL-1β and MIP1α enzyme-linked immunosorbent assay (ELISA) (n = 5). The beta-actin and 36B4 genes were used as housekeeping genes for Western blot and quantitative-PCR analyses, respectively. Data were analyzed by Student's t test (A to D and F to H). *, P < 0.01; **, P < 0.0001. All values are reported as means and SD.
CITED2 attenuates LPS-induced HIF1α transcriptional activity and protein expression.
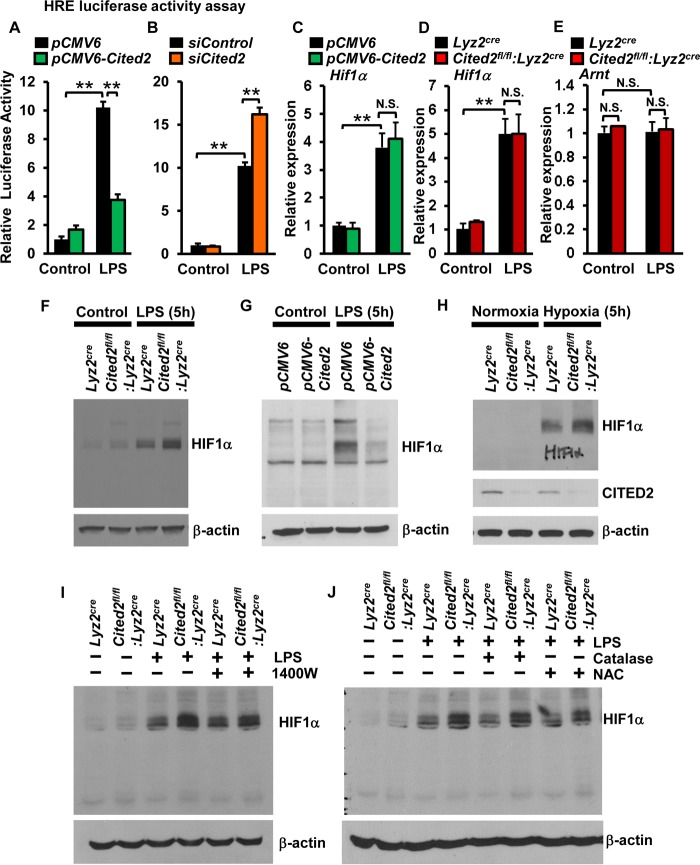
Previous studies from our group and others have demonstrated that HIF1α is essential for proinflammatory gene expression in macrophages (32, 33). Moreover, myeloid cell-specific deficiency of Hif1α is protective against LPS-induced sepsis clinical symptomatology and host mortality (34). Therefore, we examined whether altering Cited2 levels affects HIF1α transcriptional activity in macrophages. Accordingly, RAW 264.7 cells were cotransfected with an HRE (hypoxia response element)-driven luciferase reporter plasmid in the presence of pCMV6-Cited2 or siCited2. The cells were stimulated with LPS, and luciferase activity was documented. As shown in Fig. 6A and B, overexpression of Cited2 attenuated and deficiency of Cited2 enhanced LPS-induced HIF1α luciferase reporter activity in macrophages. Earlier studies showed that LPS elevates HIF1α mRNA and protein expression in macrophages (33, 35). Therefore, we examined whether altering Cited2 levels affected LPS-induced HIF1α mRNA or protein expression. As shown in Fig. 6C and D, neither overexpression nor deficiency of Cited2 altered LPS-induced Hif1α mRNA expression in macrophages. Further, deficiency of Cited2 did not modify the LPS-induced expression of the HIF1α binding partner, Arnt (Fig. 6E). Interestingly, deficiency of Cited2 markedly elevated and overexpression of Cited2 diminished LPS-induced HIF1α protein levels (Fig. 6F and G). Concordant with these observations, deficiency of Cited2 substantially elevated hypoxia-induced HIF1α protein accumulation in macrophages (Fig. 6H). Compellingly, proinflammatory conditions, such as hypoxia, considerably attenuated CITED2 protein expression in macrophages (Fig. 6H). Earlier studies had demonstrated that elevated nitric oxide (NO) or reactive oxygen species (ROS) could stabilize the HIF1α protein (36, 37). However, pharmacological inhibition of nitric oxide synthase 2 (NOS2) or ROS did not diminish elevated levels of HIF1α protein accumulation in Cited2-deficient macrophages following LPS exposure (Fig. 6I and J). Taken together, our studies revealed that CITED2 negatively regulates LPS-induced HIF1α protein accumulation in macrophages.
FIG 6.
CITED2 suppresses LPS-induced HIF1α protein accumulation. (A and B) RAW 264.7 cells were cotransfected with the HRE-luciferase reporter construct in the presence of pCMV6-Cited2 (A) or siCited2 (B). The cells were stimulated with 100 ng/ml LPS, and the cell lysates were analyzed for luciferase activity (n = 4). (C and D) RAW 264.7 cells transfected with pCMV6 and pCMV6-Cited2 (C) or BMDMs from Lyz2cre and Cited2fl/fl:Lyz2cre mice (D) were induced with 100 ng/ml of LPS for 6 h. Total RNA from the cells was analyzed for Hif1α mRNA expression by quantitative PCR (n = 4). (E) Quantitative-PCR analysis of Arnt expression in Lyz2cre and Cited2fl/fl:Lyz2cre BMDMs (n = 4). (F and G) Primary macrophages from Lyz2cre and Cited2fl/fl:Lyz2cre mice (F) or RAW 264.7 cells transfected with pCMV6 and pCMV6-Cited2 (G) were induced with 100 ng/ml of LPS for 6 h. The cell lysates were analyzed for HIF1α protein expression by immunoblotting (n = 3). (H) Primary macrophages from Lyz2cre and Cited2fl/fl:Lyz2cre mice were exposed to hypoxic condition (1% O2, 5% CO2, 94% N2). The cell lysates were analyzed for HIF1α and CITED2 protein expression by immunoblotting (n = 3). (I and J) Primary macrophages from Lyz2cre and Cited2fl/fl:Lyz2cre mice were exposed to 100 ng/ml LPS in the presence or absence of NOS2 inhibitor (1400W) and reactive oxygen species inhibitor (catalase and N-acetyl cysteine). The cell lysates were analyzed for HIF1α protein expression by immunoblotting (n = 3). The beta-actin and 36B4 genes were used as housekeeping genes for Western blot and quantitative-PCR analyses, respectively. (A to E) Data were analyzed by Student's t test. N.S., not significant; **, P < 0.0001. All values are reported as means and SD.
CITED2 curbs HIF1α protein accumulation and function by preserving EGLN3 expression.
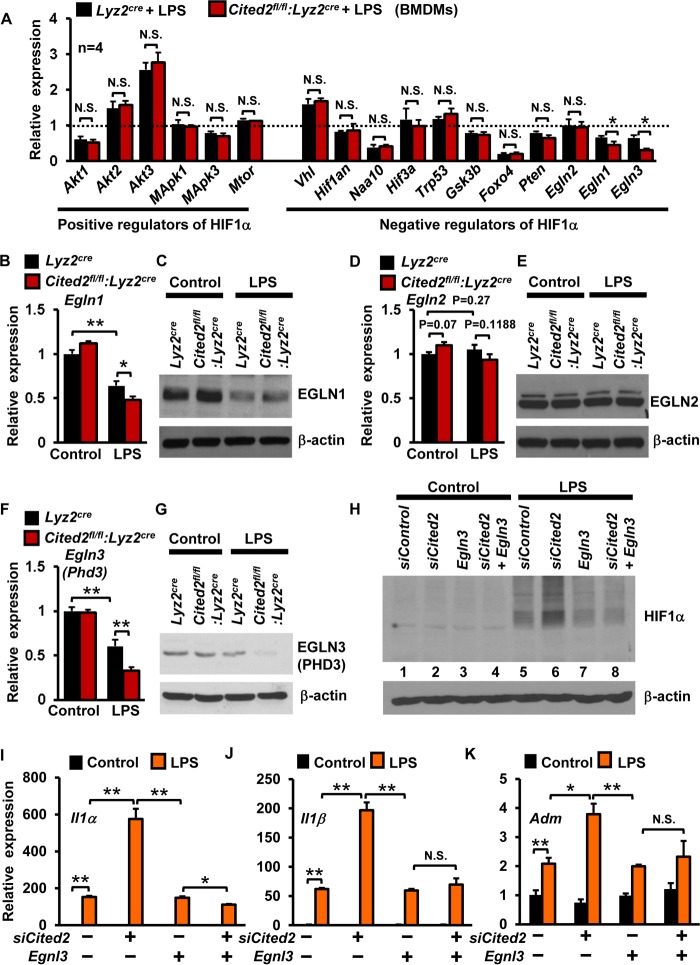
Studies from multiple laboratories have identified several positive and negative regulators of HIF1α (38). Therefore, we examined whether deficiency of Cited2 alters the expression of any critical positive and negative regulators of HIF1α following LPS stimulation. As shown in Fig. 7A, deficiency of Cited2 did not significantly alter LPS-induced expression of major positive regulators of HIF1α, including Akt1, Akt2, Akt3, Mapk1, Mapk3, and Mtor. However, examination of critical negative regulators of HIF1α revealed that deficiency of Cited2 significantly attenuated Egln1 and Egln3 expression following LPS stimulation (Fig. 7A). Therefore, we performed detailed expression analyses of EGLNs in Cited2-deficient macrophages following LPS stimulation. Our analyses revealed that LPS exposure modestly attenuated Egln1 mRNA expression in Cited2fl/fl:Lyz2cre macrophages compared to Lyz2cre cells (Fig. 7B). However, these effects were not observed at the EGLN1 protein level (Fig. 7C). Similarly, deficiency of Cited2 did not affect basal or LPS-induced EGLN2 mRNA and protein expression in macrophages (Fig. 7D and E). Interestingly, deficiency of Cited2 significantly attenuated LPS-induced EGLN3 mRNA and protein expression in macrophages (Fig. 7F and G). Based on these observations, we hypothesized that attenuation of EGLN3 in Cited2-null macrophages is responsible for heightened HIF1α protein levels following LPS exposure. To test this hypothesis, RAW 264.7 cells were overexpressed with Egln3 in the presence and absence of siRNA specifically targeting Cited2 expression (Fig. 7H). As anticipated, deprivation of Cited2 markedly enhanced LPS-induced HIF1α protein levels (Fig. 7H, lane 6). Importantly, overexpression of Egln3 attenuated LPS-induced HIF1α protein accumulation (lane 7). Compellingly, overexpression of Egln3 completely repressed LPS-induced excessive HIF1α protein accumulation in Cited2-deficient macrophages (lane 8). These results conclusively demonstrated that CITED2 attenuates HIF1α protein accumulation by preserving EGLN3 expression in macrophages. Previous studies revealed that deficiency of Egln3 dramatically enhanced myeloid cell proinflammatory response (39). Therefore, we examined whether overexpression of Egln3 could reverse elevated proinflammatory gene expression in the absence of Cited2. As expected, deficiency of Cited2 significantly elevated LPS-induced Il1α, Il1β, and Adm gene expression (Fig. 7I to K). Importantly, overexpression of Egln3 in the presence of a deficiency of Cited2 completely reversed elevated levels of Il1α, Il1β, and Adm expression following LPS stimulation (Fig. 7I to K). Taken together, our studies revealed that CITED2 attenuates LPS-induced HIF1α protein accumulation and attendant proinflammatory gene expression by preserving EGLN3 expression in macrophages.
FIG 7.
CITED2 curbs HIF1α protein level and function by preserving EGLN3 expression. (A) BMDMs from Lyz2cre and Cited2fl/fl:Lyz2cre mice were stimulated with 100 ng/ml of LPS for 6 h. Total RNA from the cells was analyzed for expression of positive and negative regulators of Hif1α at the mRNA level by quantitative PCR. Untreated Lyz2cre BMDMs were used as a control, and relative fold change over control is indicated (n = 4). (B to G) Primary macrophages from Lyz2cre and Cited2fl/fl:Lyz2cre mice were stimulated with 100 ng/ml of LPS for 6 h and analyzed for EGLN1 (B and C), EGLN2 (D and E), and EGLN3 (F and G) mRNA and protein expression by quantitative-PCR and Western blot analyses (n = 4). (H to K) RAW 264.7 cells were transfected with Cited2-specific siRNA and were further cotransfected with the Egln3 plasmid. The cells were stimulated with 100 ng/ml of LPS for 6 h. (H) Protein extracts from these experiments were evaluated for HIF1α expression by Western blotting. (I to K) Total RNA from these experiments was analyzed for Il1α, Il1β, and Adm mRNA expression by quantitative PCR (n = 4). The beta-actin and 36B4 genes were used as housekeeping genes for Western blot and quantitative-PCR analyses, respectively. Data were analyzed either by Student's t test (A, B, D, and F) or by 2-way ANOVA (I to K). N.S., not significant; *, P < 0.01; **, P < 0.0001. All values are reported as means and SD.
Inhibition of HIF1α rescues proinflammatory gene expression in Cited2-deficient macrophages.
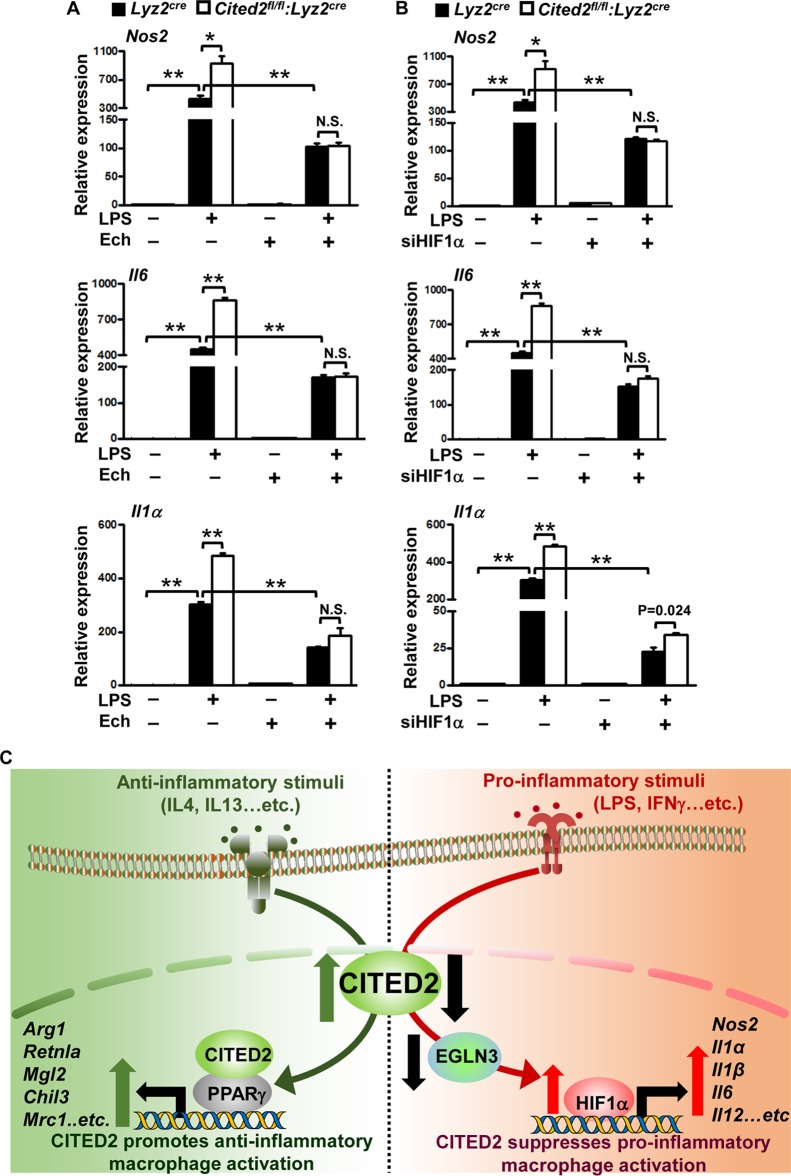
Our studies so far had revealed that deficiency of CITED2 elevated LPS-induced HIF1α protein accumulation and attendant proinflammatory gene expression in macrophages. Therefore, we examined whether elevated levels of proinflammatory gene expression in Cited2-deficient macrophages are HIF1α dependent. We employed pharmacological and genetic approaches to block HIF1α function in primary macrophages. Accordingly, BMDMs derived from Lyz2cre and Cited2fl/fl:Lyz2cre mice were stimulated with LPS in the presence and absence of a HIF1α inhibitor, echinomycin (40). As anticipated, deficiency of Cited2 significantly elevated LPS-induced HIF1α proinflammatory target gene expression, including Nos2, Il6, and Il1α (Fig. 8A). Interestingly, pharmacological inhibition of HIF1α in Cited2-null macrophages completely reversed elevated levels of these proinflammatory targets following LPS stimulation (Fig. 8A). To further corroborate these observations, we utilized a genetic approach to silence HIF1α expression in primary macrophages. Accordingly, BMDMs derived from Lyz2cre and Cited2fl/fl:Lyz2cre mice were transfected with HIF1α-specific siRNA. These cells were stimulated with LPS, and HIF1α target gene expression was analyzed. As predicted, LPS stimulation dramatically elevated HIF1α target gene expression in Cited2-null macrophages (Fig. 8B). Intriguingly, deficiency of HIF1α in Cited2-null macrophages completely reversed LPS-induced proinflammatory gene expression (Fig. 8B). Collectively, these results reveal that elevated levels of proinflammatory gene expression in Cited2-null macrophages are HIF1α dependent.
FIG 8.
Inhibition of HIF1α completely rescues proinflammatory gene expression and function in Cited2-null macrophages. (A) BMDMs from Lyz2cre and Cited2fl/fl:Lyz2cre mice were stimulated with 100 ng/ml of LPS for 6 h in the presence of the HIF1α inhibitor echinomycin. Total RNA from these experiments was analyzed for Nos2, Il6, and Il1α by quantitative PCR (n = 4). (B) BMDMs from Lyz2cre and Cited2fl/fl:Lyz2cre mice were transfected with HIF1α-specific siRNA, and the cells were stimulated with 100 ng/ml of LPS for 6 h. Total RNA from these experiments was analyzed for Nos2, Il6, and Il1α by quantitative PCR (n = 4). The 36B4 gene was used as a housekeeping gene for quantitative-PCR analyses. Each experiment was performed at least four times with three replicates. Data were analyzed by 2-way ANOVA. N.S., not significant; *, P < 0.01; **, P < 0.0001. All values are reported as means and SD. (C) (Left) CITED2 cooperates with PPARγ to elevate IL-4-induced anti-inflammatory gene expression. (Right) When macrophages are challenged with proinflammatory stimuli, such as LPS, CITED2 preserves EGLN3 expression to attenuate HIF1α-mediated proinflammatory gene expression.
DISCUSSION
Our studies identify CITED2 as a novel molecular toggle that promotes anti-inflammatory activation of macrophages while suppressing proinflammatory macrophage polarization and function. The central findings of this study are as follows: (i) CITED2 is abundantly expressed in human and murine macrophages, (ii) CITED2 promotes macrophage anti-inflammatory gene expression, (iii) IL-4/IL-13-induced CITED2 expression is STAT6 dependent, (iv) deficiency of Cited2 attenuates IL-4-induced PPARγ recruitment to Arg1 and Retnla promoters, (v) CITED2 cooperates with PPARγ to enhance IL-4-induced anti-inflammatory gene expression, (vi) myeloid deficiency of Cited2 augments proinflammatory macrophage activation and host susceptibility to endotoxic shock syndrome, (vii) CITED2 attenuates LPS-induced HIF1α protein accumulation and attendant functions, (viii) CITED2 curbs HIF1α protein accumulation and function by preserving EGLN3 expression, and (ix) abrogation of HIF1α completely rescues select proinflammatory gene expression and function in Cited2-null macrophages. Collectively, our findings establish that CITED2 cooperates with PPARγ to enhance select anti-inflammatory gene expression and antagonizes HIF1α to alleviate proinflammatory gene expression in macrophages.
Monocyte-derived macrophages are principal components of the innate immune system and are highly plastic in the inflammatory response. Recent studies have established that “classically” activated proinflammatory macrophages can self-reprogram into alternatively activated macrophages to resolve tissue inflammation (41). Here, we identify CITED2 as a novel regulator of macrophage anti-inflammatory activation and function. Previous studies have revealed that Cited2 is broadly expressed in embryonic tissue and is essential for cellular differentiation and proliferation and organ development (21, 22, 25). In particular, deficiency of Cited2 at the embryonic stage strikingly attenuated myeloid cell development and abundance (20, 27). However, as demonstrated in our studies, deletion of Cited2 in mature myeloid cells using lysozyme M Cre did not broadly affect the hematopoietic cell compartment. An earlier report also revealed that deficiency of Cited2 significantly attenuated hematopoietic stem cell quiescence (29). However, our studies indicate that deficiency of Cited2 in the mature myeloid compartment did not significantly alter the basal level of pro- or anti-inflammatory gene expression in macrophages. It is important to note that Th2 cytokines, such as IL-4, induce alternative activation of macrophages by elevating a number of transcription factors, including PPARγ (30). In this regard, our studies demonstrated that anti-inflammatory stimuli, such as IL-4 or IL-13, significantly elevate Cited2 at the mRNA and protein levels. More importantly, our results established that Cited2 is indispensable for IL-4-induced classical anti-inflammatory gene expression in macrophages. Further, our analyses demonstrated that deficiency of Cited2 completely abolished either cytokine- or agonist-induced PPARγ transcriptional activity in macrophages. In addition, anti-inflammatory stimuli enhanced CITED2 interaction with PPARγ in primary macrophages. At the functional level, it is important to note that our studies demonstrated that loss of CITED2 significantly attenuated IL-4-induced select PPARγ anti-inflammatory target gene expression in macrophages. Recent studies have established that several transcription factors, such as STAT6, IRF4, JMJD3, CEBPβ, LXRs, and retinoid X receptors, facilitate anti-inflammatory macrophage activation and functions (42). Taken together, our discoveries provide the basis for future investigations focused on the role of CITED2 in the regulation of these anti-inflammatory transcription factors in macrophages.
It is well established that many proinflammatory agents exert their effects by actively repressing negative regulators of inflammation (5). This allows derepression of many proinflammation genes that are elevated following infection or injury (7, 10). In this context, our studies demonstrate that exposure of macrophages to proinflammatory agents, such as LPS or IFN-γ, markedly attenuated CITED2 expression. These observations further establish that CITED2 might be one such critical negative regulator of inflammation in myeloid cells. Therefore, reduction of CITED2 levels is expected to boost proinflammatory gene expression in macrophages. As envisioned, our studies demonstrated that deficiency of Cited2 dramatically elevated LPS-induced classical proinflammatory gene expression in macrophages (Fig. 8C). Prior studies showed that bacterial products, such as LPS, trigger HIF1α protein accumulation in macrophages under normoxic conditions (43). However, this accumulation of HIF1α protein is primarily attributed to heightened Hif1α mRNA transcription following LPS stimulation. In this context, our studies demonstrated that neither overexpression nor deficiency of Cited2 altered LPS-induced Hif1α at the mRNA level. Interestingly, deficiency of Cited2 augmented and overexpression of Cited2 attenuated LPS-induced HIF1α protein accumulation in macrophages. These observations yield the possibility that LPS-induced HIF1α accumulation is at least in part regulated at the protein level. Earlier studies have established that EGLN family proteins regulate HIF1α protein stability by hydroxylation of proline residues (44). In this study, we established that CITED2 alleviates LPS-induced HIF1α protein accumulation by augmenting EGLN3 expression. Further, overexpression of Egln3 completely reversed elevated HIF1α protein accumulation and attendant proinflammatory gene expression in Cited2-deficient macrophages following LPS stimulation. Concordant with our observations, earlier studies demonstrated that deficiency of Egln3 significantly elevated LPS-induced proinflammatory gene expression and rendered Egln3-null mice susceptible to endotoxic shock (39). It is imperative to note that myeloid cell-specific deficiency of HIF1α is protective against LPS-induced sepsis (34). Concordant with these observations, our studies demonstrated that mice with myeloid cell-specific deficiency of Cited2 are highly susceptible to LPS-induced sepsis symptomatology and host mortality. In addition, genetic or pharmacological inhibition of HIF1α in Cited2-null macrophages completely rescued LPS-induced proinflammatory gene expression. Earlier reports suggested that CITED2 represses HIF1α transactivation by blocking CBP/p300 recruitment (26, 45, 46). Further, these studies demonstrated that the CITED2 transactivation domain (TAD) binds to the cysteine-histidine-rich 1 (CH1) domain of p300/CBP with very high affinity. This binding disrupts complex formation between the HIF1α C-terminal TAD (C-TAD) and the CH1 domain of p300/CBP, with subsequent inhibition of HIF1α functions. As we observed, proinflammatory stimuli, such as LPS or hypoxia, rapidly diminished CITED2 protein expression while elevating HIF1α protein accumulation levels in macrophages. This allowed HIF1α binding to p300/CBP and subsequent expression of HIF1α targets. With this insight, our studies uncovered a novel mechanism by which Cited2 restrains HIF1α function by preserving EGLN3 expression in macrophages. It is well established that several transcription factors, such as STAT1, IRF3, IRF5, KLF6, NF-κB, AP1, and CREB, facilitate proinflammatory macrophage activation and functions (42). It is completely conceivable that CITED2 may facilitate its function by repressing any of these proinflammatory transcription factors in macrophages. Therefore, our future investigations will focus on whether CITED2 regulates the functions of these proinflammatory transcription factors in macrophages.
MATERIALS AND METHODS
Generation of myeloid cell-specific deletion in Cited2 mice and endotoxic shock studies.
All animal procedures were approved by the Institutional Animal Care and Use Committee at Case Western Reserve University and conformed to guidelines established by the American Association for Accreditation of Laboratory Animal Care. All mice were bred and maintained under pathogen-free conditions, fed standard laboratory chow (Harlan Teklad, Indianapolis, IN), and kept on a 12-h light/dark cycle. The mouse line expressing lysozyme M promoter-driven Cre recombinase (Lyz2cre) was obtained from The Jackson Laboratory (Bar Harbor, ME). Cited2 floxed (Cited2fl/fl) mice were provided by Sally L. Dunwoodie (28). These Cited2 floxed mice were crossed with Lyz2cre mice to generate a myeloid cell-specific deletion of Cited2. The mice were further crossed to generate male and female offspring expressing two Cre and floxed Cited2 alleles. These mice with two Cited2 floxed and Cre alleles were used as the Cited2 myeloid cell-specific null group. Mice with only two Cre alleles were used as the control group. Cited2 floxed allele genotyping was performed using site-specific primers (see Table S1 in the supplemental material) (forward primer, 5′-GTCTCAGCGTCTGCTCGTTT-3′, and reverse primer, 5′-TGCTGCTGCTGGTGATGAT-3′). To examine the Cited2 genomic excision, total DNA from control and Cited2 myeloid cell-specific knockout macrophages were isolated and analyzed by PCR using the following primer pairs: forward primer, 5′-TAGGCCACTTCAGGGAACC-3′, and reverse primer, 5′-GACAGTATCGGCCTCAGGAA-3′. T-cell receptor δ chain was used as an internal control and was amplified using the forward primer 5′-CAAATGTTGCTTGTCTGGTG-3′ and the reverse primer 5′-GTCAGTCGAGTGCACAGTTT-3′. The Lyz2cre and Cited2fl/fl:Lyz2cre mice (8 to 10 weeks old) were injected intraperitoneally with 21 mg/kg of body weight of LPS or saline solution. The mice were monitored for 6 days following LPS injection. Survival data were analyzed by the construction of Kaplan-Meier plots and use of the log rank test.
Cell culture.
RAW 264.7 and J774.1 macrophages were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, 10 μg/ml streptomycin, and 2 mM glutamine in a humidified incubator (5% CO2 at 37°C). Mouse thioglycolate-elicited peritoneal macrophages were obtained by inducing peritonitis with 3% thioglycolate broth in 8- to 12-week-old mice. The peritoneal lavage fluid and adherent macrophage population were examined for contaminating cells by fluorescence-activated cell sorting (FACS) analysis using fluorescently labeled anti-CD11b and anti-SiglecF antibodies. The cells were cultured for the indicated experiments in complete DMEM as described above. Bone marrow-derived macrophages were generated by ex vivo differentiation of bone marrow cells. Briefly, bone marrow cells from 8-week-old wild-type, Lyz2cre, and Cited2fl/fl:Lyz2cre mice were harvested from the femur and tibia. These bone marrow cells were cultured in cell culture medium supplemented with recombinant mouse macrophage colony-stimulating factor (M-CSF) for 7 days. These BMDMs were collected and utilized for the indicated experiments. Human monocyte-derived macrophages were generated by culturing peripheral blood monocytes, isolated by magnetic separation, with recombinant human M-CSF for 7 days. The resulting macrophages were harvested and used for the indicated experiments. For hypoxic treatment, the cell culture plates were transferred to a modular incubator chamber that was infused with a mixture of 1% O2, 5% CO2, 94% N2 gas and placed at 37°C for the indicated period. All the studies involving human samples were approved by the Case Western Reserve University Institutional Review Board.
RNA extraction, reverse transcription (RT), and real-time quantitative PCR.
Total RNA was isolated from the indicated samples using a High Pure RNA isolation kit. One microgram of total RNA was reverse transcribed using Moloney murine leukemia virus (M-MuLV) reverse transcriptase in the presence of random hexamers and oligo(dT) primers. Real-time quantitative PCR was performed using Universal SYBR green PCR master mix or TaqMan universal master mix on an Applied Biosystems Step One Plus real-time PCR system in the presence of gene-specific primers.
Transient transfection and chromatin immunoprecipitation.
Transfection of RAW 264.7 cells was performed using Lipofectamine transfection reagent (see Table S2 in the supplemental material) according to the manufacturer's instructions. The efficiency of transfection of either pCMV6-neo (empty-vector control) or pCMV6-neo-Cited2 (overexpressing Cited2) averaged 65 to 75% in all experiments. Similarly, mouse peritoneal and bone marrow-derived macrophages were transfected with specific siRNA using TurboFect transfection reagent. These transfected cells were stimulated in vitro with either LPS (100 ng/ml), IL-4 (10 ng/ml), IL-13 (10 ng/ml), or phosphate-buffered saline (PBS) (control) and subjected to the indicated analyses. Chromatin immunoprecipitation analyses were performed using the EZ-Magna ChIP G kit according to the manufacturer's instructions. Briefly, Lyz2cre and Cited2fl/fl:Lyz2cre mouse bone marrow-derived macrophages were stimulated with 10 ng/ml IL-4. Chromatin immunoprecipitations were performed using anti-PPARγ and anti-CITED2 antibodies. Chromatin samples from these experiments were analyzed by real-time quantitative RT-PCR. Primer pairs flanking the PPARγ-binding site were targeted to amplify the mouse Arg1 (FW, 5′-CGGACACACACACAATCACA-3′; REV, 5′-TAGTCTTGCCCTGCTGACAA-3′) enhancer and Retnla (FW, 5′-TAGCAGCGAATATTAATGGGAGA-3′; REV, 5′-GTTTCCCCTCCAAAAATCC-3′) promoter regions.
Western blotting and luciferase assay.
The indicated primary cells and cell lines were lysed in ice-cold radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitor and phosphatase inhibitor. The protein concentration was measured by the bicinchoninic acid (BCA) protein assay. Equal amounts of protein samples were electrophoresed using 8% or 4 to 15% Mini-Protean TGX precast gels (Bio-Rad) and transferred to nitrocellulose membranes. The membranes were blocked with 5% nonfat dry milk or 5% bovine serum albumin (BSA) in Tris-buffered saline with Tween 20 (TBST) for 1 h at room temperature. The blots were further incubated with primary antibodies diluted in 5% BSA in TBST. After overnight incubation, the primary antibodies were removed by washing with TBST. The blots were incubated for 1 h at room temperature in horseradish peroxidase-conjugated secondary antibodies. The blots were visualized using enhanced chemiluminescence Western blotting substrate. The primary antibodies were used at the following dilutions: CITED2, 1:500; HIF1α, 1:500; NOS2, 1:1,000; PPARγ, 1:1,000; STAT6, 1:1,000; p-STAT6, 1:1,000; EGLN1, EGLN2, EGLN3, and β-actin, 1:5,000. The indicated cells were transfected with the indicated plasmids or siRNA using the Lipofectamine reagent according to the manufacturer's instructions. Luciferase reporter plasmids driven by PPAR (PPAR response element [PPRE] ×3-TK-luc) or HIF1α (hypoxia response element [HRE]-luc) were transfected alone or together with pCMV6-Cited2 or siCited2. Cells were treated with LPS (100 ng/ml for 6 h) or with IL-4 (10 ng/ml), T0070907, or rosiglitazone for 18 h. Luciferase reporter activity was measured and normalized according to the manufacturer's instructions. Results are presented as relative luciferase activity over the control group.
Flow cytometry.
Surface staining of thioglycolate-induced peritoneal lavage fluid and postadherent macrophages (day 3) was performed using antibodies against ITGAM (CD11b) and SIGLEC5 (Siglec F) in the presence of Fc blocker. The cell suspensions were then fixed, and sorting of the stained cells and multiparameter analyses were performed on an LSR II (BD). All data were analyzed with FlowJo software. For blood immunophenotyping, whole blood was collected from mice using EDTA-coated tubes, followed by red blood cell (RBC) lysing with 1× RBC lysis buffer (Biolegend, San Diego, CA). The resulting white blood cells were stained with anti-CD3, anti-CD4, anti-CD8, anti-CD11b, anti-CD19, and anti-Ly6G on ice for 30 min. After two washes with 1× PBS, the cells were suspended in 2% FBS (diluted with 1× PBS) and analyzed using Flowsight imaging flow cytometry.
Quantification and statistical analysis.
All data, unless otherwise indicated, are presented as means and standard deviations (SD). The statistical significance of differences between two groups was analyzed by paired or unpaired Student's t test. The statistical significance of differences between three or more groups was analyzed by using two-way analysis of variance (ANOVA). LPS-induced-sepsis survival data were analyzed by log rank test. A P value of <0.05 was considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant HL126626 and Crohn's and Colitis Foundation of America Senior Research Award 421904 (to G.H.M.).
The content is solely our responsibility and does not necessarily represent the official views of the National Institutes of Health.
G.H.M. conceived and designed the study. G.H.M., G.-D.K., R.D., and J.Z. performed experiments. G.H.M., G.-D.K., R.D., X.R., J.Z., J.A.D., D.L.R.-B., and S.R. analyzed and interpreted the data. G.H.M. and G.-D.K. wrote and edited the manuscript, and it was approved by all of us.
We declare that we have no conflicts of interest with the contents of this article.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00452-17.
REFERENCES
- 1.Murray PJ, Wynn TA. 2011. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol 11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi C, Pamer EG. 2011. Monocyte recruitment during infection and inflammation. Nat Rev Immunol 11:762–774. doi: 10.1038/nri3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xue J, Schmidt SV, Sander J, Draffehn A, Krebs W, Quester I, De Nardo D, Gohel TD, Emde M, Schmidleithner L, Ganesan H, Nino-Castro A, Mallmann MR, Labzin L, Theis H, Kraut M, Beyer M, Latz E, Freeman TC, Ulas T, Schultze JL. 2014. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 40:274–288. doi: 10.1016/j.immuni.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liew FY, Xu D, Brint EK, O'Neill LA. 2005. Negative regulation of Toll-like receptor-mediated immune responses. Nat Rev Immunol 5:446–458. doi: 10.1038/nri1630. [DOI] [PubMed] [Google Scholar]
- 5.Murray PJ, Smale ST. 2012. Restraint of inflammatory signaling by interdependent strata of negative regulatory pathways. Nat Immunol 13:916–924. doi: 10.1038/ni.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen HB, Mosser DM. 2013. Extrinsic and intrinsic control of macrophage inflammatory responses. J Leukoc Biol 94:913–919. doi: 10.1189/jlb.0413236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Medzhitov R, Horng T. 2009. Transcriptional control of the inflammatory response. Nat Rev Immunol 9:692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- 8.Ingersoll MA, Platt AM, Potteaux S, Randolph GJ. 2011. Monocyte trafficking in acute and chronic inflammation. Trends Immunol 32:470–477. doi: 10.1016/j.it.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gosselin D, Link VM, Romanoski CE, Fonseca GJ, Eichenfield DZ, Spann NJ, Stender JD, Chun HB, Garner H, Geissmann F, Glass CK. 2014. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 159:1327–1340. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawrence T, Natoli G. 2011. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat Rev Immunol 11:750–761. doi: 10.1038/nri3088. [DOI] [PubMed] [Google Scholar]
- 11.Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence T, Locati M, Mantovani A, Martinez FO, Mege JL, Mosser DM, Natoli G, Saeij JP, Schultze JL, Shirey KA, Sica A, Suttles J, Udalova I, van Ginderachter JA, Vogel SN, Wynn TA. 2014. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghisletti S, Huang W, Ogawa S, Pascual G, Lin ME, Willson TM, Rosenfeld MG, Glass CK. 2007. Parallel SUMOylation-dependent pathways mediate gene- and signal-specific transrepression by LXRs and PPARgamma. Mol Cell 25:57–70. doi: 10.1016/j.molcel.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jennewein C, Kuhn AM, Schmidt MV, Meilladec-Jullig V, von Knethen A, Gonzalez FJ, Brune B. 2008. Sumoylation of peroxisome proliferator-activated receptor gamma by apoptotic cells prevents lipopolysaccharide-induced NCoR removal from kappaB binding sites mediating transrepression of proinflammatory cytokines. J Immunol 181:5646–5652. doi: 10.4049/jimmunol.181.8.5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du J, Yang YC. 2013. Cited2 in hematopoietic stem cell function. Curr Opin Hematol 20:301–307. doi: 10.1097/MOH.0b013e3283606022. [DOI] [PubMed] [Google Scholar]
- 15.Sperling S, Grimm CH, Dunkel I, Mebus S, Sperling HP, Ebner A, Galli R, Lehrach H, Fusch C, Berger F, Hammer S. 2005. Identification and functional analysis of CITED2 mutations in patients with congenital heart defects. Hum Mutat 26:575–582. doi: 10.1002/humu.20262. [DOI] [PubMed] [Google Scholar]
- 16.Volcik KA, Zhu H, Finnell RH, Shaw GM, Canfield M, Lammer EJ. 2004. Evaluation of the Cited2 gene and risk for spina bifida and congenital heart defects. Am J Med Genet A 126A:324–325. doi: 10.1002/ajmg.a.20578. [DOI] [PubMed] [Google Scholar]
- 17.Bamforth SD, Braganca J, Farthing CR, Schneider JE, Broadbent C, Michell AC, Clarke K, Neubauer S, Norris D, Brown NA, Anderson RH, Bhattacharya S. 2004. Cited2 controls left-right patterning and heart development through a Nodal-Pitx2c pathway. Nat Genet 36:1189–1196. doi: 10.1038/ng1446. [DOI] [PubMed] [Google Scholar]
- 18.Weninger WJ, Lopes Floro K, Bennett MB, Withington SL, Preis JI, Barbera JP, Mohun TJ, Dunwoodie SL. 2005. Cited2 is required both for heart morphogenesis and establishment of the left-right axis in mouse development. Development 132:1337–1348. doi: 10.1242/dev.01696. [DOI] [PubMed] [Google Scholar]
- 19.Bamforth SD, Braganca J, Eloranta JJ, Murdoch JN, Marques FI, Kranc KR, Farza H, Henderson DJ, Hurst HC, Bhattacharya S. 2001. Cardiac malformations, adrenal agenesis, neural crest defects and exencephaly in mice lacking Cited2, a new Tfap2 co-activator. Nat Genet 29:469–474. doi: 10.1038/ng768. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y, Haviernik P, Bunting KD, Yang YC. 2007. Cited2 is required for normal hematopoiesis in the murine fetal liver. Blood 110:2889–2898. doi: 10.1182/blood-2007-01-066316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu B, Qu X, Gu S, Doughman YQ, Watanabe M, Dunwoodie SL, Yang YC. 2008. Cited2 is required for fetal lung maturation. Dev Biol 317:95–105. doi: 10.1016/j.ydbio.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Withington SL, Scott AN, Saunders DN, Lopes Floro K, Preis JI, Michalicek J, Maclean K, Sparrow DB, Barbera JP, Dunwoodie SL. 2006. Loss of Cited2 affects trophoblast formation and vascularization of the mouse placenta. Dev Biol 294:67–82. doi: 10.1016/j.ydbio.2006.02.025. [DOI] [PubMed] [Google Scholar]
- 23.Glenn DJ, Maurer RA. 1999. MRG1 binds to the LIM domain of Lhx2 and may function as a coactivator to stimulate glycoprotein hormone alpha-subunit gene expression. J Biol Chem 274:36159–36167. doi: 10.1074/jbc.274.51.36159. [DOI] [PubMed] [Google Scholar]
- 24.Chou YT, Yang YC. 2006. Post-transcriptional control of Cited2 by transforming growth factor beta. Regulation via Smads and Cited2 coding region. J Biol Chem 281:18451–18462. [DOI] [PubMed] [Google Scholar]
- 25.Qu X, Lam E, Doughman YQ, Chen Y, Chou YT, Lam M, Turakhia M, Dunwoodie SL, Watanabe M, Xu B, Duncan SA, Yang YC. 2007. Cited2, a coactivator of HNF4alpha, is essential for liver development. EMBO J 26:4445–4456. doi: 10.1038/sj.emboj.7601883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhattacharya S, Michels CL, Leung MK, Arany ZP, Kung AL, Livingston DM. 1999. Functional role of p35srj, a novel p300/CBP binding protein, during transactivation by HIF-1. Genes Dev 13:64–75. doi: 10.1101/gad.13.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kranc KR, Schepers H, Rodrigues NP, Bamforth S, Villadsen E, Ferry H, Bouriez-Jones T, Sigvardsson M, Bhattacharya S, Jacobsen SE, Enver T. 2009. Cited2 is an essential regulator of adult hematopoietic stem cells. Cell Stem Cell 5:659–665. doi: 10.1016/j.stem.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Preis JI, Wise N, Solloway MJ, Harvey RP, Sparrow DB, Dunwoodie SL. 2006. Generation of conditional Cited2 null alleles. Genesis 44:579–583. doi: 10.1002/dvg.20251. [DOI] [PubMed] [Google Scholar]
- 29.Du J, Chen Y, Li Q, Han X, Cheng C, Wang Z, Danielpour D, Dunwoodie SL, Bunting KD, Yang YC. 2012. HIF-1alpha deletion partially rescues defects of hematopoietic stem cell quiescence caused by Cited2 deficiency. Blood 119:2789–2798. doi: 10.1182/blood-2011-10-387902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szanto A, Balint BL, Nagy ZS, Barta E, Dezso B, Pap A, Szeles L, Poliska S, Oros M, Evans RM, Barak Y, Schwabe J, Nagy L. 2010. STAT6 transcription factor is a facilitator of the nuclear receptor PPARgamma-regulated gene expression in macrophages and dendritic cells. Immunity 33:699–712. doi: 10.1016/j.immuni.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang JT, Welch JS, Ricote M, Binder CJ, Willson TM, Kelly C, Witztum JL, Funk CD, Conrad D, Glass CK. 1999. Interleukin-4-dependent production of PPAR-gamma ligands in macrophages by 12/15-lipoxygenase. Nature 400:378–382. doi: 10.1038/22572. [DOI] [PubMed] [Google Scholar]
- 32.Cramer T, Yamanishi Y, Clausen BE, Forster I, Pawlinski R, Mackman N, Haase VH, Jaenisch R, Corr M, Nizet V, Firestein GS, Gerber HP, Ferrara N, Johnson RS. 2003. HIF-1alpha is essential for myeloid cell-mediated inflammation. Cell 112:645–657. doi: 10.1016/S0092-8674(03)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahabeleshwar GH, Kawanami D, Sharma N, Takami Y, Zhou G, Shi H, Nayak L, Jeyaraj D, Grealy R, White M, McManus R, Ryan T, Leahy P, Lin Z, Haldar SM, Atkins GB, Wong HR, Lingrel JB, Jain MK. 2011. The myeloid transcription factor KLF2 regulates the host response to polymicrobial infection and endotoxic shock. Immunity 34:715–728. doi: 10.1016/j.immuni.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peyssonnaux C, Cejudo-Martin P, Doedens A, Zinkernagel AS, Johnson RS, Nizet V. 2007. Cutting edge: essential role of hypoxia inducible factor-1alpha in development of lipopolysaccharide-induced sepsis. J Immunol 178:7516–7519. doi: 10.4049/jimmunol.178.12.7516. [DOI] [PubMed] [Google Scholar]
- 35.Peyssonnaux C, Datta V, Cramer T, Doedens A, Theodorakis EA, Gallo RL, Hurtado-Ziola N, Nizet V, Johnson RS. 2005. HIF-1alpha expression regulates the bactericidal capacity of phagocytes. J Clin Invest 115:1806–1815. doi: 10.1172/JCI23865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mateo J, Garcia-Lecea M, Cadenas S, Hernandez C, Moncada S. 2003. Regulation of hypoxia-inducible factor-1alpha by nitric oxide through mitochondria-dependent and -independent pathways. Biochem J 376:537–544. doi: 10.1042/bj20031155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chandel NS, McClintock DS, Feliciano CE, Wood TM, Melendez JA, Rodriguez AM, Schumacker PT. 2000. Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1alpha during hypoxia: a mechanism of O2 sensing. J Biol Chem 275:25130–25138. doi: 10.1074/jbc.M001914200. [DOI] [PubMed] [Google Scholar]
- 38.Bardos JI, Ashcroft M. 2005. Negative and positive regulation of HIF-1: a complex network. Biochim Biophys Acta 1755:107–120. [DOI] [PubMed] [Google Scholar]
- 39.Kiss J, Mollenhauer M, Walmsley SR, Kirchberg J, Radhakrishnan P, Niemietz T, Dudda J, Steinert G, Whyte MK, Carmeliet P, Mazzone M, Weitz J, Schneider M. 2012. Loss of the oxygen sensor PHD3 enhances the innate immune response to abdominal sepsis. J Immunol 189:1955–1965. doi: 10.4049/jimmunol.1103471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kong D, Park EJ, Stephen AG, Calvani M, Cardellina JH, Monks A, Fisher RJ, Shoemaker RH, Melillo G. 2005. Echinomycin, a small-molecule inhibitor of hypoxia-inducible factor-1 DNA-binding activity. Cancer Res 65:9047–9055. doi: 10.1158/0008-5472.CAN-05-1235. [DOI] [PubMed] [Google Scholar]
- 41.Arnold L, Henry A, Poron F, Baba-Amer Y, van Rooijen N, Plonquet A, Gherardi RK, Chazaud B. 2007. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J Exp Med 204:1057–1069. doi: 10.1084/jem.20070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murray PJ. 2017. Macrophage polarization. Annu Rev Physiol 79:541–566. doi: 10.1146/annurev-physiol-022516-034339. [DOI] [PubMed] [Google Scholar]
- 43.Blouin CC, Page EL, Soucy GM, Richard DE. 2004. Hypoxic gene activation by lipopolysaccharide in macrophages: implication of hypoxia-inducible factor 1alpha. Blood 103:1124–1130. doi: 10.1182/blood-2003-07-2427. [DOI] [PubMed] [Google Scholar]
- 44.Fong GH, Takeda K. 2008. Role and regulation of prolyl hydroxylase domain proteins. Cell Death Differ 15:635–641. doi: 10.1038/cdd.2008.10. [DOI] [PubMed] [Google Scholar]
- 45.Freedman SJ, Sun ZY, Kung AL, France DS, Wagner G, Eck MJ. 2003. Structural basis for negative regulation of hypoxia-inducible factor-1alpha by CITED2. Nat Struct Biol 10:504–512. doi: 10.1038/nsb936. [DOI] [PubMed] [Google Scholar]
- 46.Berlow RB, Dyson HJ, Wright PE. 2017. Hypersensitive termination of the hypoxic response by a disordered protein switch. Nature 543:447–451. doi: 10.1038/nature21705. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.