ABSTRACT
The β-barrel assembly machinery (BAM) complex is the core machinery for the assembly of β-barrel membrane proteins, and inhibition of BAM complex activity is lethal to bacteria. Discovery of integral membrane proteins that are key to pathogenesis and yet do not require assistance from the BAM complex raises the question of how these proteins assemble into bacterial outer membranes. Here, we address this question through a structural analysis of the type 2 secretion system (T2SS) secretin from enteropathogenic Escherichia coli O127:H6 strain E2348/69. Long β-strands assemble into a barrel extending 17 Å through and beyond the outer membrane, adding insight to how these extensive β-strands are assembled into the E. coli outer membrane. The substrate docking chamber of this secretin is shown to be sufficient to accommodate the substrate mucinase SteC.
IMPORTANCE In order to cause disease, bacterial pathogens inhibit immune responses and induce pathology that will favor their replication and dissemination. In Gram-negative bacteria, these key attributes of pathogenesis depend on structures assembled into or onto the outer membrane. One of these is the T2SS. The Vibrio-type T2SS mediates cholera toxin secretion in Vibrio cholerae, and in Escherichia coli O127:H6 strain E2348/69, the same machinery mediates secretion of the mucinases that enable the pathogen to penetrate intestinal mucus and thereby establish deadly infections.
KEYWORDS: protein secretion, bacterial pathogenesis, BAM complex, transertion, outer membrane, lipoproteins, secretin
INTRODUCTION
Enteropathogenic Escherichia coli (EPEC) and enterohemorrhagic E. coli (EHEC) are pathogens causing diseases that range from diarrhea to hemorrhagic colitis and hemolytic-uremic syndrome in humans. In order to access the epithelial layer of the human gut, these pathogens secrete mucinases that cause a thinning of the mucus layer to accelerate pathogenesis (1–3). These large mucinases are folded into functionally active forms in the bacterial periplasm before being secreted across the outer membrane (1, 4–8). Secretion of these mucinases is mediated by the type 2 secretion system (T2SS), with a defining component of the T2SS being the secretin: an approximately 70-kDa protein that homo-oligomerizes to form an ∼1-MDa secretin complex (9). The secretin complex is embedded in the outer membrane and spans the periplasm (10). Diversity in the sequence features of secretins led to the classification of two major forms of the T2SS: (i) the Klebsiella type, represented in a broad and diverse set of bacterial species and which has been functionally characterized best in Klebsiella oxytoca (11), and (ii) the Vibrio type, which is found only in a few species of Vibrio and pathovars of Escherichia coli, including EPEC and EHEC (7). Other secretins, including those from the T2SS expressed by species of Pseudomonas, Legionella, and Acinetobacter, are so diverse in sequence that their relationships have remained complex and obscure (7, 12). As the number of genome sequences available for analysis grows, these relationships too are becoming more clear (13).
The targeting pathway that delivers secretin monomers to the outer membrane has been studied intensively, particularly in Klebsiella. In K. oxytoca, the secretin (PulD) is synthesized with a signal sequence to allow its passage as an unfolded monomer into the periplasm (14). Once in the periplasm, the C-terminal S-domain of the PulD monomer is bound by a pilotin lipoprotein (PulS) and the PulS-PulD complex is trafficked across the periplasm to the inside surface of the outer membrane, utilizing the LOL pathway (15–17). The same assembly pathway is evident in bacteria such as Erwinia chrysanthemi (the secretin is OutD, and the pilotin is OutS) and many strains and pathovars of E. coli (the secretin/pilotin pair is either GspD/GspS or EtpD/EtpO [18, 19]). The targeting of the Vibrio-type secretin to the outer membrane follows equivalent steps but, through a striking example of convergent evolution, depends instead on a structurally distinct pilotin referred to as AspS that evolved independently to perform the same function (7). Thus, the S-domains in the Vibrio-type secretins and Klebsiella-type secretins have strikingly different amino acid compositions that provide recognition signals (7). These distinctive signals in the respective S-domains have been shown to dictate pilotin specificity, with chimeric secretin constructs incorporating the S-domain sequences from PulD dictating reliance on PulS (20).
How the secretin monomers then assemble to form the outer membrane channel of the T2SS has remained unclear. Typically, integral proteins are assembled into the bacterial outer membrane by the β-barrel assembly machinery (BAM complex [21, 22]). However, depletion of BamA, the core component of the BAM complex from E. coli, does not decrease the rate of assembly for secretin monomers from Klebsiella or from EPEC (23, 24). In order to address the question of how the T2SS secretin is assembled into the bacterial outer membrane, we analyzed the structure of the secretin complex from EPEC O127:H6 strain E2348/69.
The secretin complex was purified and analyzed by cryo-electron microscopy (cryo-EM), revealing structural details down to a resolution of 3.3 Å. Comparisons to monomeric and homo-oligomeric β-barrel proteins that are assembled by the BAM complex revealed that all contain a highly conserved belt of hydrophobic residues stretching around a β-barrel domain, of sufficient height to sit within the lipid phase of the outer membrane. However, in the GspD oligomer, 15 secretin subunits oligomerize into a 60-stranded pentadecamer β-barrel with each subunit contributing two full-length β-strands, one segmented β-strand, and two incomplete β-strands which are keyed into β-strands of the preceding and succeeding subunits to give a 60-stranded barrel. The length of the β-strands thereby formed in the barrel domain of the secretin complex is approximately 78 Å with an average of 22 residues in each strand, far more extensive in length than any known substrate of the BAM complex. Sitting astride this belt is an aromatic girdle, a feature known to designate the line at which the lipid phase of the bacterial outer membrane engages integral membrane proteins. The secretin complex, however, extends 50 Å below the membrane boundary with a pair of short α-helices sitting against or in the bottom face of the outer membrane. Taken together with structural information on the mucinase substrates, the size of the barrel domain so formed is sufficient to enable the passage of these very large substrates across the outer membrane in a prefolded state.
RESULTS
Architecture of the EPEC secretin complex.
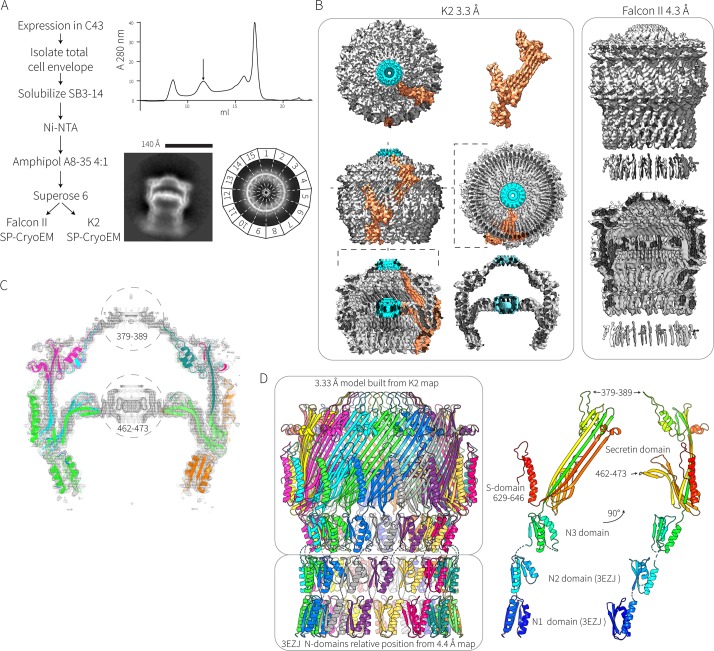
Particles of GspD from EPEC O127:H6 strain E2348/69 form a pentadecameric secretin complex (Fig. 1A and B) with no other symmetries observed in the two-dimensional (2D) class averages. Figure 1B shows how the domains from each GspD monomer are situated within the architecture of the secretin complex. The upper chamber, inner gate, and cap could be clearly resolved, whereas the N-domains were less well resolved (Fig. 1A). The N-domains contribute to a large periplasmic chamber which is known to dock onto the pseudopilus structure emanating from the inner membrane (10, 25). This N-domain chamber was less well resolved than the upper secretin domain, suggesting that there is relatively high flexibility/movement among the N-domains. Approximately 44,000 particles were used to construct a 4.4-Å map from the Falcon II data, and 8,800 particles were used to generate a 3.3-Å map from the K2 data (Fig. 1B).
FIG 1.
Structure of the EPEC secretin. (A) Flow chart summarizing the expression and purification of the secretin complex is shown alongside a final size exclusion chromatography trace. The arrow indicates the elution position of the complex at ∼1 MDa corresponding to the sample for electron microscopy: 2D class averages of “side” and “top” views from this sample show the pentadecamer arrangement of the multimer. (B) The 3.3-Å electron density map of the EPEC secretin derived from the K2 summit camera and the 4.3-Å electron density map derived from the Falcon II camera. The orange density corresponds to density that could be assigned to one GspD monomer. The blue density corresponds to density around the center of the symmetry axis that could not be assigned to any protein chains. (C) A slice through the K2 map showing how the derived model fits into the map. Regions 379 to 389 and 462 to 473, which could not be modeled, are circled. (D) The secretin gates and N3-domain were modeled from the K2 data at 3.3 Å, and the N1- and N2-domains were derived from the crystal structure (PDB accession number 3EZJ) with the N1- and N2-domains split and then individually assigned to density from the Falcon II data. The N0-domain could not be assigned. Two rotational views of a single GspD protomer extracted from the complex structure reveal the relative positioning of the characteristic secretin domains.
Individual β-strands could be easily identified in the 3.3-Å map, and much of the sequence from the N3-domain onward could be built into the map (Fig. 1C). Residues around the center of the inner gate (corresponding to residues 462 to 473) and cap (residues 379 to 389) could not be assigned (Fig. 1B and C). High-resolution density/noise was apparent in these regions, but this is probably due to amplified artifacts at the axis of the C15 symmetry imposed during map refinement. Map generation without symmetry imposed yielded poor results.
The 4.4-Å map generated from the Falcon II data was not sufficiently detailed to build an atomic model into, but the crystal structure of the N2-domain (PDB accession number 3EZJ) was readily placed in the density map below the N3-domain. Likewise, since GspD from ETEC and that from EPEC have identical primary structures, the crystal structure of the N1-domain of ETEC GspD (PDB accession number 3EZJ) was placed into the density, immediately below the N2-domain based on the position of two visible α-helices. No density for the N0-domain could be assigned. The relative position of the N1- and N2-domains was combined with the model of the N3-domain and secretin domain from the 3.3-Å map to produce a model of the complex (Fig. 1D and 2A). The domain architecture of secretin monomers (Fig. 1D) consists of four N-terminal domains (N0 to N3) followed by the highly conserved secretin domain (Pfam PF00263) and a C-terminal S-domain that is necessary and sufficient for pilotin binding (16, 17, 24). The disposition of the S-domain in the complex structure is detailed in Fig. S2 in the supplemental material.
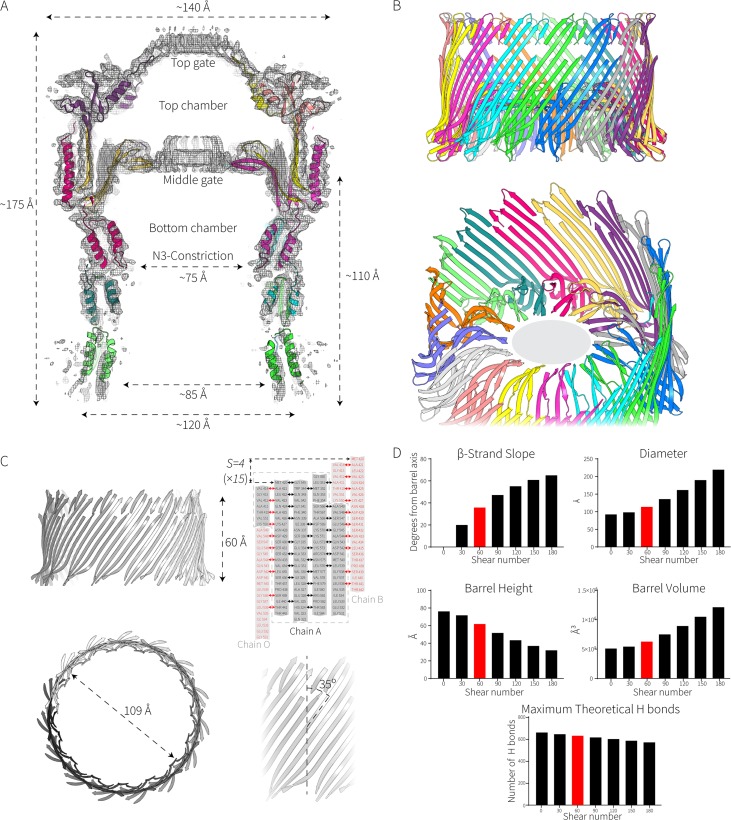
FIG 2.
Elongated β-strands in the EPEC secretin determine its cavity diameter. (A) Slice through the Falcon II map showing the fitted model of the complex. The dimensions of the complex are shown. (B) Zoom views of the double β-barrel. The gray ellipse (in the lower panel) represents the unassigned density in the middle gate corresponding to amino acids 462 to 473. (C) Properties of the giant β-barrel, including calculations of its shear number (S) and diameter. (D) How shear affects the properties of a β-barrel. The values presented here are derived from the formulas tan α = Sα/nb and R = b/[2 sin (π/n)cos α] described in the work of Murzin et al. (72), where α is the average slope of the strands in the barrel, R is the radius of the barrel, a is the Cα-to-Cα distance along the peptide, b is the interstrand distance, n is the number of strands, and S is the shear number. Shear numbers of S = 0 to S = 3n are shown in the graphs. The maximum theoretical H bonds are calculated assuming that every other residue is bonded to the adjacent residue on the next strand.
Based on sequence analysis, we previously proposed that the secretin from EPEC O127:H6 strain E2348/69 was of the same subtype as that from Vibrio cholerae and coined the term Vibrio-type secretins to describe this protein subfamily (7). Recently, Yan et al. solved the structure of the secretin from V. cholerae (26), providing a basis to assess the similarities of the two secretins.
Among the known secretin structures, the EPEC GspD T2SS secretin (covering the N3-domain to the S-domain) shares 54.3% identity to that of EspD from V. cholerae and 47.7% identity to GspD from E. coli K-12. Overall, a similar architecture can be seen in all structures, though the overall architecture of the EPEC secretin domains appears to best resemble that of V. cholerae EspD with loops extending from the extracellular side to form an exterior gate/cap (Fig. S3A). More detailed root mean square deviation (RMSD) structural analysis reveals that as expected the EPEC GspD structure is very similar to Vibrio EspD, with the only minor differences apparent at the cap and poorly resolved S-domain. Compared to the E. coli K-12 (Klebsiella-type) T2SS structure, obvious differences are apparent in the cap structure and transmembrane region (Fig. S3A and B). EPEC GspD also shares 17.5% identity to the T3SS secretin InvG from Salmonella enterica serovar Typhimurium. The architecture of the T3SS secretin resembles that of the Klebsiella-type T2SS secretins with an open cap at the surface of the cell, though the T3SS lacks the N2-domain. Also, InvG has an additional β-hairpin protruding from the N3-domain pointing up into the lumen of the lower chamber, and the RMSD of the inner barrel shows slight differences between the structures (Fig. S3B). Overall, the structures of the N3 domain and the outer barrel are strikingly well conserved among the T2SS and T3SS secretins (Fig. S3B).
The GspD β-barrel chamber.
The EPEC secretin complex is resolved into four important structural features: the cap gate, an extensive β-barrel chamber, a periplasmic constriction, and a central gate. A longitudinal cross section of the secretin complex highlights these features (Fig. 2A). The β-barrel chamber is a unique protein structure, consisting of two concentric barrels: a short inner barrel composed of 60 β-strands (4 antiparallel β-strands from each protomer) that are aligned with the long axis of the secretin complex and the large outer barrel with a diameter of approximately 110 Å (Fig. 2B). This “inner barrel” contributes to the central gate structure. The outer barrel has a complex β-strand arrangement comprised of 30 long “strands” running the full height of the barrel and 60 nonsequential β-strands interleaved among the long strands. To form this complicated outer barrel, each subunit contributes a total of 6 β-strands to the barrel. The walls of the barrel are hydrogen bonded together with the topographically first (short) strand of one subunit stacking onto the last β-strand of the preceding subunit (Fig. 2C).
The multimeric barrel is stabilized by a C-terminal α-helix, folded down over a negatively charged patch on the outer wall of the preceding subunit. This can be further stabilized by at least one intrasubunit salt bridge between the bottom of the helix and wall of the barrel (Lys598-Glu326) and one intersubunit salt bridge between Arg609 on the helix of one subunit and Glu334 on the outer wall of the preceding subunit which locks the subunits together (Fig. S2). Analysis of other T2SS secretin structures (26) revealed that these inter- and intrasubunit salt bridges that tie the subunits together are conserved, thereby providing an explanation for the well-documented thermal and chemical quaternary stability of secretin complexes (9).
In transmembrane β-barrel proteins, two parameters dictate the luminal diameter of the barrel: the number of β-strands (n) and their shear number (S) (27). The shear number indicates the stagger of each β-strand relative to its neighboring strands. In transmembrane β-barrel proteins, n is almost always an even number and must be greater than a value of 8 (28). S must be greater than or equal to 0, with S = 0 describing β-strands that sit perpendicular to the membrane. The outer barrel consists of four very long and discontiguous β-strands with a shear number of 60 (n = 4 for each of the 15 subunits), resulting in an angle of ∼35° to the long axis of the secretin complex. These strands run the length of the complex, connecting the N-terminal domains to the cap and inner gate (Fig. 2C). For a given number of strands, a greater shear number will produce a greater diameter and volume of the resultant barrel at the expense of the height and theoretical stability of the β-barrel (Fig. 2D).
Transmembrane topography of secretins.
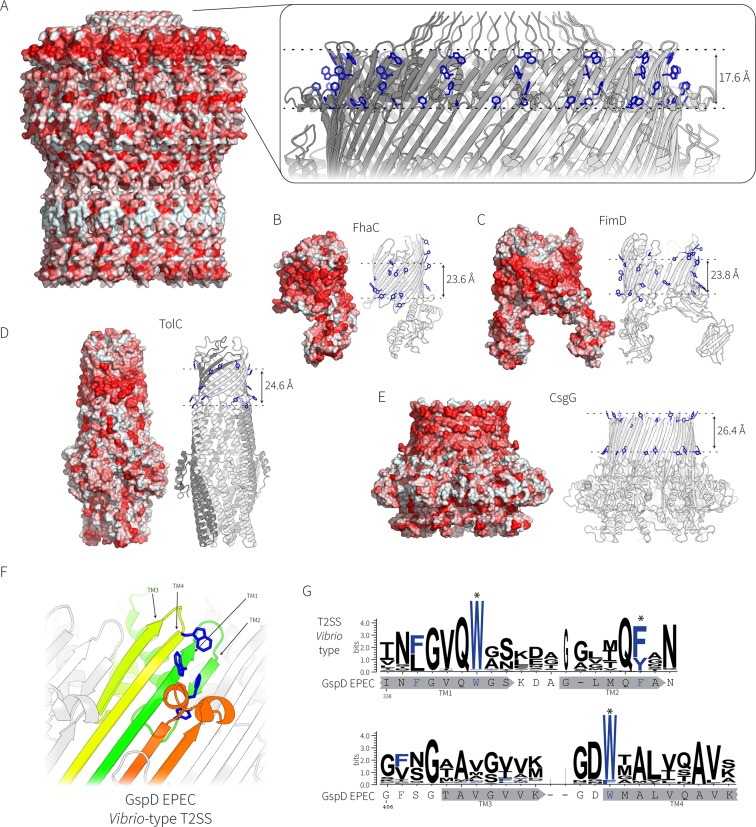
In a β-barrel protein, the side chain of every second residue points outward. In a transmembrane β-barrel protein, these residues have side chains with high hydrophobicity scores in order to be accommodated in the lipid phase of the membrane. Calculations of hydrophobicity were graphically represented on the structure of the secretin complex (Fig. 3A) and show a hydrophobic belt which is 17.6 Å wide, forming a layer across the top of the long β-strands; this is slightly narrower than the width of other well-characterized outer membrane β-barrels (Fig. 3A to E). Included in this hydrophobic belt is an aromatic residue (Trp419) whose placement forms the upper boundary of the hydrophobic belt and Phe340 and Phe354, which together form the lower boundary. An equivalent, though larger, hydrophobic belt and aromatic girdle are seen in the monomeric β-barrel proteins FhaC (Fig. 3B) and FimD (Fig. 3C) and also in the homo-oligomeric β-barrels TolC (Fig. 3D) and CsgG (Fig. 3E). The aromatic girdle present in EPEC GspD is a highly conserved feature among the sequences of 581 Vibrio-type GspD homologues (Fig. 3F and G). The Klebsiella-type T2SS secretins as shown for the structure of GspD from E. coli K-12 substrain DH5α have similar conserved aromatic residues (Fig. 4A and B). However, in addition to the residues that form the girdle, the Klebsiella-type secretins also have two additional and conserved aromatic residues (one outward facing and one inward facing) near the top of the second hairpin (Fig. 4A and B).
FIG 3.
Topological comparison of the secretin complex and other β-barrel proteins in the bacterial outer membrane. (A) Surface rendering of the secretin complex from EPEC, where residues with hydrophobic side chains are colored red according to the Eisenberg hydrophobicity scale (73). (Inset) Zoomed view of the aromatic girdle, with the side chains of aromatic residues (Phe and Tyr) represented as sticks (in blue). (B to E) Hydrophobic belts and aromatic girdles of other bacterial outer membrane proteins, shown to scale. The measurements of the transmembrane sections for all proteins are from the Orientations of Proteins in Membranes (OPM) database and the PPM server (69): FhaC (PDB accession number 2QDZ) (B), FimD (PDB accession number 4J3O) (C), TolC (PDB accession number 1EK9) (D), and CsgG (PDB accession number 4UV3) (E). (F) The transmembrane regions of the EPEC GspD secretin (PDB accession number 5W68) representing the Klebsiella-type T2SS secretin; the conserved aromatic residues are shown in blue. (G) The sequence logo showing the conservation of the aromatic residues in the transmembrane β-strands was generated from 581 Vibrio-type T2SS secretin sequences with the GspD sequence from EPEC O127:H6 strain E2348/69 shown for reference. The locations of the putative transmembrane β-strands are shown in gray below the sequence logos.
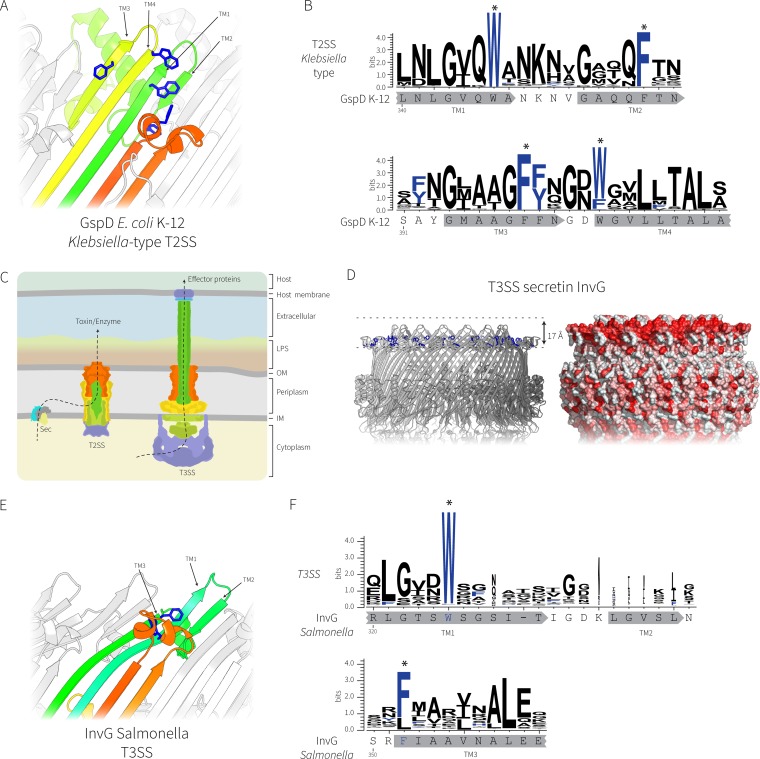
FIG 4.
Comparative analysis of the secretin complexes from T2SS and T3SS. (A) The transmembrane regions of the E. coli K-12 GspD secretin (PDB accession number 5WQ7) representing the Klebsiella-type T2SS secretin; the conserved aromatic residues are shown in blue. (B) Sequence logo showing the conservation of the aromatic residues (designated with asterisks) in the transmembrane β-strands was generated from 363 Klebsiella-type T2SS secretin sequences with the sequence from E. coli K-12 substrain DH5α shown for reference. The locations of the putative transmembrane β-strands are shown in gray below the sequence logos. (C) Cartoon depiction of the relationship between T2SS and T3SS, where the colors represent subunits with similar functions. LPS, lipopolysaccharide; OM, outer membrane; IM, inner membrane. (D) Similar transmembrane β-strands from the T3SS secretin InvG from Salmonella enterica (PDB accession number 5TCQ) with the hydrophobic belt shown. (E) The transmembrane region of InvG showing the conserved aromatic residues (blue). Note that the leftmost short β-strand is missing in the T3SS secretin, causing the upper lip of the barrel to curve inward to accommodate the missing strand. (F) Sequence logo of alignments of 1,543 T3SS secretins showing the conservation of the aromatic residues in the transmembrane β-strands. The InvG sequence and β-strands from Salmonella enterica are shown below for reference. The locations of the putative transmembrane β-strands are shown in gray below the sequence logos.
Members of the secretin protein family are also found in the needle complex of the type 3 secretion system (T3SS) (29–31). Despite showing some overall principles in common, the other components and the overall architectures of the T2SS and T3SS are quite distinct, as is the mechanism of substrate entry into the complex (Fig. 4C). In EPEC, the T3SS secretin is EscC, and the structure of the homologous protein InvG from Salmonella was recently solved (31). Comparative hydrophobicity analysis of the T3SS secretin complex revealed a major distinction from the T2SS. The T3SS secretin InvG/EscC has a hydrophobic band that could serve for membrane integration, but it is tilted inward toward the lumen of the barrel. This distinguishing transmembrane region is due to the 4 β-strands of the barrel transitioning to 3 strands around the hydrophobic band at the membrane interface (Fig. 4D and E). The transmembrane region of InvG/EscC has fewer conserved aromatic residues, with only two aromatic residues that are highly conserved in the transmembrane region (Fig. 4F).
Structural distinctions in the T2SS subtypes found in E. coli.
As a result of lateral gene transfer, two different types of the T2SS secretin protein family are found in the various pathotypes and environmental isolates of E. coli: the Vibrio-type and Klebsiella-type secretins (7). The secretin from EPEC O127:H6 strain E2348/69 belongs to the Vibrio-type secretins, and we sought to rationalize the structural features that define these two different secretin types. In their recent work, Yan et al. (26) presented the structures of the Klebsiella-type secretin from E. coli K-12 substrain DH5α (PDB accession number 5WQ7).
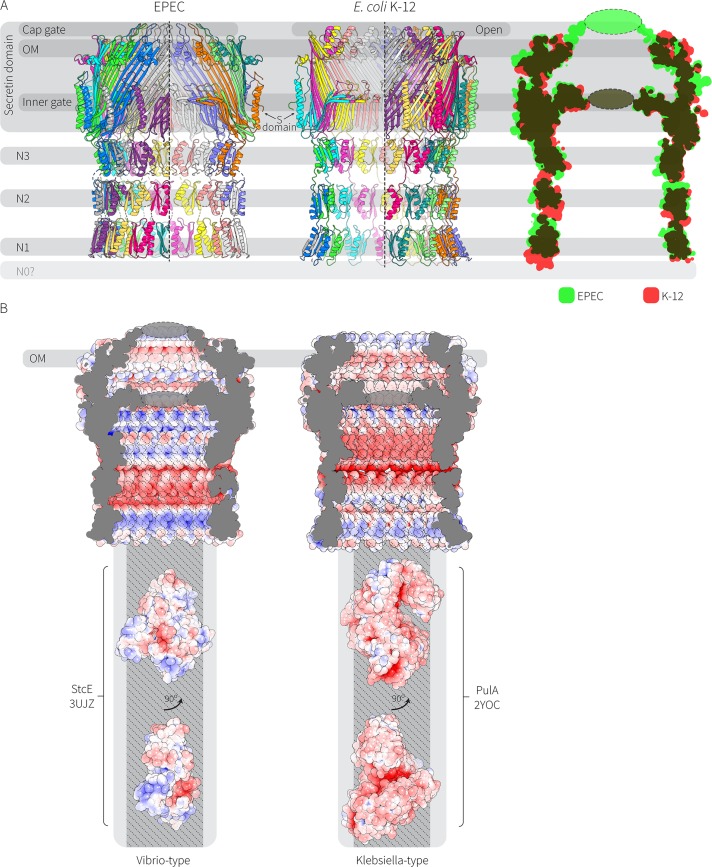
The secretin from EPEC O127:H6 strain E2348/69 has features that distinguish it from the Klebsiella-type secretin (Fig. 5A). Notably, the cap gate is longer: it protrudes above the line of the outer membrane, and it appears to be closed (based on the cryo-EM density). In the Klebsiella-type secretin from E. coli K-12 substrain DH5α, the cap gate is shorter and open to the extracellular environment. The electrostatic charge of the interior of the cavity is also a distinguishing feature, with the Vibrio-type secretins containing alternating positively and negatively charged bands, whereas the Klebsiella-type secretin is largely negatively charged with a periplasm-facing positively charged rim (Fig. 5B).
FIG 5.
Cryo-EM structures reconcile the sequence differences seen in the Vibrio-type and Klebsiella-type secretin protein families. (A) Comparison of the Vibrio-type (EPEC O127:H6 strain E2348/69) and the Klebsiella-type (E. coli K-12 substrain DH5α) T2SS secretin complexes. The structures of the aligned models are shown as an overlay with green representing EPEC O127:H6 strain E2348/69 and red representing the E. coli K-12 substrain DH5α structure. (B) Rendering of the cavity space for the Vibrio-type secretin complex, alongside rendering of the cavity space for Klebsiella-type secretin complex. The electrostatic potentials of the surfaces are shown, with red being negatively charged and blue being positively charged. Structures for the T2SS substrates protease StcE (PDB accession number 3UJZ) and pullulanase PulA (PDB accession number 2YOC) are represented to scale below the structure of the Vibrio-type and Klebsiella-type secretin complexes. The widest opening of the chamber is shown in gray, and the narrowest point (the N3 constriction) is shown in darker gray.
The Vibrio-type secretin complex found in EPEC and EHEC secretes substrates such as the mucinases StcE and SslE. The crystal structure of StcE (PDB accession number 3UJZ) is shown here represented to scale with the secretin complex structure (Fig. 5B). The Klebsiella-type secretin complex is well documented as secreting pullulanase, a starch-degrading enzyme that enables the bacterium to access sugars. The pullulanase structure from K. oxytoca (PDB accession number 2YOC) is represented to scale with the secretin complex structure (Fig. 5B). In both cases, the substrates can fit into the widest point of the barrel (the opening), a passage made even more penetrable given the apparent flexibility of the N-domain within the complex (observed in Fig. 1A).
DISCUSSION
The secretin protein superfamily has members which function in the T2SS, T3SS, type IV pili, and other secretion systems, in all cases forming the pore in the outer membrane that provides for gated transport (29, 32). We confirm here, with the structure of the T2SS secretin GspD from EPEC O127:H6 strain E2348/69, that there are defining three-dimensional (3D) features common to the Vibrio type of secretins previously described from sequence-based similarities. These features include the charge characteristics in the lumen of the complex and the specific shape and arrangements in the cap gate. Together with recently published high-resolution structures of the secretins from Vibrio cholerae (26) and the Klebsiella-type secretin found in E. coli K-12 substrain DH5α (26), these structural features suggest how these huge complexes might be assembled in the outer membrane and enable a refinement of our knowledge of how substrates are loaded into the T2SS.
Protein secretion mechanism for T2SS.
Substrate proteins enter the T2SS from the periplasm, after being translocated across the cytoplasmic membrane by either the Sec translocon or the Tat transporter (12, 33, 34). Despite the highly specific nature of protein secretion via the T2SS, how the substrate proteins are selected for engagement with the secretion machinery remains unclear. The one prerequisite for secretion is that the substrate protein must be in a folded state. Identifying secretion “signals” within T2SS substrates has been complicated since these proteins show a large variation in sequence, structure, posttranslational modifications, and oligomeric states (35). Several studies have shown the importance of surface motifs for the secretion of target substrates, and yet no general features have been identified among all T2SS substrates bioinformatically or biochemically (reviewed by Thomassin et al. [36]). That being said, several structures of secretin complexes do provide further insight into the constraints on the passage of those proteins across the outer membrane.
In the T2SS, the process of protein secretion is an active process that depends on ATP hydrolysis in the cytoplasm promoting the assembly and driving upward movement of a pseudopilus into the N-domain chamber of the secretin complex (37–39). The nature of the interactions between the N-domains of each secretin protomer allows these domains to deform considerably with respect to each other (13), and this “mobility” was observed in the 2D class averages of the EPEC secretin particles when imaged by electron microscopy. After being “actively pushed” into this accommodating chamber, the major impediment to substrate passage remains the central gate.
The structuring of the central gate from short segments of β-strand allows for at least two scenarios by which it could open. Breaking the few, interstrand hydrogen bonds that structure this gate would allow for a lateral shift, thereby opening in a mechanism analogous to that of an iris diaphragm. Alternatively, breaking the interstrand hydrogen bonds that structure the gate could simply allow the upward displacement of the sectors of the gate to align against the wall of the β-barrel, just below the region embedded in the outer membrane. A G453A mutation in the secretin from Vibrio cholerae captured the central gate open (26) but did not distinguish between the two opening mechanisms. Considering that the size of the characteristic mucinase (this study) or cholera toxin (32, 40) would be barely accommodated in the upper chamber of the secretin complex, the opening mechanism might provide elastic energy to promote the final push of the substrate through the cap gate. By comparison to the central gate, the cap gate structure would be relatively easy to deform given that it is formed of interstrand loops in the EPEC structure (this study) and other Vibrio-type secretins (26). The cap gate structure is absent from the Klebsiella-type secretins (26).
Assembly of secretin complexes into bacterial outer membranes.
GspD is not assembled into the bacterial outer membrane by the BAM complex (23, 24). Instead, secretin monomers are transferred from the SecYEG complex in the cytoplasmic membrane to a dedicated chaperone called the pilotin. For Klebsiella-type secretins, the pilotin is a homologue of the Klebsiella protein PulS, whereas Vibrio-type secretins use a structurally unrelated protein called AspS (7). While we know most about these two forms of secretin, it is becoming clear that others, such as the Hxc secretin from a subset of Pseudomonas species, might not need pilotins (41) and might represent a new subfamily of secretins (13). Pilotins selectively bind the C-terminal S-domain of their cognate secretin, through an induced fit mechanism that has been captured in biochemical assays and a crystal structure (17, 42, 43). Most of the known pilotins are lipoproteins and thus engage with the LOL pathway for transport across the periplasm to the outer membrane receptor LolB. A long standing question concerns how such a complicated protein structure as a secretin oligomer is assembled into the outer membrane. Two features common to all of the published secretin structures lead us to consider a transertion-based model for assembly of the secretin oligomer into the outer membrane.
The transertion model for protein translocation posits that the signal sequences of proteins will dictate their engagement with the closest Sec translocons in the cytoplasmic membrane. In E. coli, the RNA transcribed from the gsp operon has been mapped (44), and multiple translation initiation events (45) ensure that many molecules of GspD (and the other subunits of the T2SS) would be synthesized in close proximity, translocated into the periplasm in close proximity, and translocated to arrive at the same or a neighboring LolB receptor site within a short window of time (46–49). Computer simulations and protein localization studies confirm that transertion creates local regions of high concentration for a given membrane protein (reviewed by Matsumoto et al. [48]). In this scenario, the local concentration of GspD monomers in the outer membrane would be relatively high and would favor an allosteric activation of the monomers to polymerize. The extensive β-strand structure of the secretin complex reported here throws structure-informed support behind this model for secretin assembly. The extensive hydrogen-bond donor and acceptor contacts at the edge of an individual GspD subunit would serve as the template for β-augmentation reactions, providing favorable energetics for the assembly reaction. Each incoming monomer has to form at least 9 new hydrogen bonds between monomers in the main β-sheet region of GspD (Fig. 2C), which equates to a favorable energy change of ΔG ≈ 45 kJ/mol (50). As formation of hydrogen bonds is an enthalpically downhill process, taken together, it would appear that oligomerization of GspD monomers is energetically driven by extensive β-sheet formation.
Each protomer incorporated into the growing secretin oligomer would also add to the hydrophobic belt, creating a feature that would be increasingly costly to maintain out of a membrane environment. While not homologous, this mechanism for β-strand engagement is somewhat reminiscent of the means by which pore-forming toxins are thought to assemble into host cell membranes (51), wherein an initial engagement of monomers sets up allosteric events and conformational changes to penetrate a membrane bilayer. Past work with T2SS secretins has shown that intrinsic features in secretin protomers do allow for oligomerization without assistance from cellular machinery (52–54), and the LOL machinery may simply provide the means by which secretins can find each other for an otherwise unassisted integration into the outer membrane.
MATERIALS AND METHODS
Protein expression and purification.
The sequence of GspD was amplified from E. coli O127:H6 strain E2348/69 with the addition of a C-terminal hexahistidine tag. The gene was cloned into the plasmid pET-20b(+). This plasmid was transformed into E. coli C43 cells (55). Transformants were grown overnight in LB (Lennox) supplemented with 150 μg ml−1 ampicillin and 0.5% (wt/vol) glucose. Cells were diluted 1:50 in Terrific broth (100 mM potassium phosphate buffer, pH 7.5, 12 g liter−1 tryptone, 24 g liter−1 yeast extract, 5% [wt/vol] glycerol) and incubated at 37°C with shaking until they reached an optical density at 600 nm of approximately 0.8, at which point 0.2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added, and cells were incubated at 25°C for a further 16 h.
Cells were harvested by centrifugation at 7,000 × g, washed once in buffer A (20 mM HEPES, 300 mM NaCl), and resuspended approximately 1:5 (vol/vol) in buffer A. DNase I (10 μg ml−1) and then white lysozyme (100 μg ml−1) were added, and cells were incubated for 30 min before they were disrupted with an Avestin EmulsiFlex C3 homogenizer. Cell lysates were clarified by centrifugation at 7,000 × g for 15 min. Cell envelopes were isolated from the clarified lysates by centrifugation at 100,000 × g for 1 h. Cell envelopes were washed once with a glass homogenizer in buffer A and again centrifuged at 100,000 × g for 1 h. The washed envelope fraction was resuspended 1:10 (vol/vol) in buffer A with a glass homogenizer.
The isolated envelopes were mixed 1:1 with buffer A containing 5% Sb3-14 and stirred for 30 min. This solution was loaded onto a HiTrap nickel-nitrilotriacetic acid (Ni-NTA) column (GE Life Sciences). The column was washed with 4 column volumes of buffer A containing 50 mM imidazole and then 4 column volumes of buffer A containing 100 mM imidazole. The protein was eluted from the column with buffer A containing 400 mM imidazole. Amphipol A8-35 (Anatrace) was added to the elution fraction (4:1 protein/mass ratio) and incubated for 30 min. The high-molecular-weight fraction eluting at approximately 12 ml from the column was shown to be a pure GspD multimer when analyzed by SDS-PAGE or by electron microscopy (see Fig. S1 in the supplemental material). The GspD multimer was purified on a Superose 6 increase 10/300 GL size exclusion column running in buffer A (Fig. 1A). Fractions eluting around 12 ml were concentrated to approximately 0.5 mg ml−1 with Amicon Ultra 0.5-ml 100K centrifuge filtration units, flash frozen, and stored at −80°C until further use.
Protein complexes were flash frozen, and vitreous ice was imaged on a Titan Krios transmission electron microscope with an FEI Falcon II or Gatan K2 Summit direct electron detector (Fig. 1A).
Electron microscopy.
Quantifoil R1.2/1.3 holey carbon grids were washed and coated in a layer of lauryldimethylamine oxide (LDAO) to prevent the protein complex preferentially adsorbing to the carbon support as previously described (56). Sample (4 μl) was applied to detergent-treated grids, and frozen hydrated samples were prepared with a Vitrobot Mark IV (FEI, OR) in a 100% humidity atmosphere with 2-s blot time and no drain time before plunge-freezing into liquid ethane. Grids were imaged on a Titan Krios transmission electron microscope (FEI) at 300 kV. Falcon II data were collected as follows. Images were collected at ×127,000 magnification (1.1 Å/pixel) with 1-s exposures in 7-frame movie mode using a dose rate of 45 electrons per second. The corresponding 17 subframes were fractionated in 7 frames as follows. Subframe 1 was discarded. Subframes 2 to 7 were recorded as frames 1 to 6, respectively. Subframes 8 to 16 were pooled and integrated as frame 7. Subframe 17 was discarded. The defocus was set to a range of 0.6 μm to 3.5 μm in intervals of 0.2 μm. K2 data were collected as follows. Images were collected at ×130,000 magnification (0.53 Å/pixel in superresolution mode) with 18 frames at 0.4 s per frame with a total dose of 50 electrons/Å2. A random defocus between 0.8 and 3.5 μm was applied for each micrograph.
Data processing.
RELION 2 (57) was used as a wrapper to motion correct the movies with dose weighting using MotionCorr 2.1 (58) and estimate CTF with CTFFIND4 (59). Micrographs with acceptable contrast transfer function (CTF) estimations were used to manually pick ∼1,000 particles which were used to generate 2D class averages used for RELION autopicking. Particles were extracted with a 288- by 288-pixel box size and subjected to multiple rounds of 2D classification in RELION—only C15 symmetry could be observed in particles. Selected particles were imported into CryoSPARC (60) for ab initio three-dimensional (3D) classification and final refinement. A total of 43,526 particles were used to generate a 4.26-Å-resolution map from the Falcon II data set, and 8,896 particles were used in the final 3.33-Å-resolution map from the K2 data set. Resolution of the final reconstructions was determined using the gold standard Fourier shell correlation (FSC) = 0.143 criterion (see Fig. S4 in the supplemental material). Electron microscopy maps derived in this study are available on the Electron Microscopy Data Bank (EMDB) (EMD-8778 and EMD-8779).
Atomic model refinement.
A model of the GspD monomer was created using Rosetta (61) and the Vibrio cholerae GspD model as a template (PDB accession number 5WQ8). The monomer was symmetrized around the electron microscopy (EM) electron density map using Situs pdbsymm (62). The resulting model was then subjected to energy minimization to remove any steric clashes. Fitting the model to the EM electron density map was achieved using the MDFF routine in namd (63). The fitted model was further refined by rounds of manual model building in Coot (64) and real space refinement as implemented in the Phenix software package (65).
Bioinformatic analysis.
Protein sequences from the InterPro family “T2SS_GspD” (IPR013356) were downloaded and classified by an all-against-all BLAST search. Sequences were clustered based on pairwise similarities and visualized with CLANS (66). Sequences falling into the Vibrio and Klebsiella types (581 and 363 sequences, respectively) were extracted. Multiple sequence alignments were conducted with Clustal Omega (67) and visualized with WebLogo3 (68). For T3SS analysis, the InterPro “T3SS_OM_pore_YscC” (IPR003522) was downloaded and all proteins with a similar domain architecture (1,543 sequences, IPR005644) were used for further analysis in Clustal Omega. The predicted transmembrane regions of the proteins presented in Fig. 3 and 4 were calculated with the Orientations of Proteins in Membranes PPM server (69).
Image processing.
All images were generated with PyMOL or UCSF Chimera (70). Electrostatics were calculated with APBS (71).
Accession number(s).
The 3.3-Å model of the EPEC GspD secretin domain derived from this study is available at the PDB accession number 5W68.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the support staff and facilities at the Monash Ramaciotti Centre for Cryo-Electron Microscopy. We thank Christopher Stubenrauch for critical comments on the manuscript.
I.D.H. was the recipient of an NVIDIA Hardware grant, and we gratefully acknowledge the support of NVIDIA Corporation for the donation of the TitanX GPU used for this research. Our work in this area is supported by NHMRC program grant 1092262 and the Australian Research Council (ARC; FL130100038). I.D.H. is an ARC Laureate Postdoctoral Fellow, and T.L. is an ARC Australian Laureate Fellow.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00521-17.
For a commentary on this article, see https://doi.org/10.1128/JB.00702-17.
REFERENCES
- 1.Lathem WW, Grys TE, Witowski SE, Torres AG, Kaper JB, Tarr PI, Welch RA. 2002. StcE, a metalloprotease secreted by Escherichia coli O157:H7, specifically cleaves C1 esterase inhibitor. Mol Microbiol 45:277–288. doi: 10.1046/j.1365-2958.2002.02997.x. [DOI] [PubMed] [Google Scholar]
- 2.Nesta B, Valeri M, Spagnuolo A, Rosini R, Mora M, Donato P, Alteri CJ, Del Vecchio M, Buccato S, Pezzicoli A, Bertoldi I, Buzzigoli L, Tuscano G, Falduto M, Rippa V, Ashhab Y, Bensi G, Fontana MR, Seib KL, Mobley HL, Pizza M, Soriani M, Serino L. 2014. SslE elicits functional antibodies that impair in vitro mucinase activity and in vivo colonization by both intestinal and extraintestinal Escherichia coli strains. PLoS Pathog 10:e1004124. doi: 10.1371/journal.ppat.1004124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valeri M, Rossi Paccani S, Kasendra M, Nesta B, Serino L, Pizza M, Soriani M. 2015. Pathogenic E. coli exploits SslE mucinase activity to translocate through the mucosal barrier and get access to host cells. PLoS One 10:e0117486. doi: 10.1371/journal.pone.0117486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldi DL, Higginson EE, Hocking DM, Praszkier J, Cavaliere R, James CE, Bennett-Wood V, Azzopardi KI, Turnbull L, Lithgow T, Robins-Browne RM, Whitchurch CB, Tauschek M. 2012. The type II secretion system and its ubiquitous lipoprotein substrate, SslE, are required for biofilm formation and virulence of enteropathogenic Escherichia coli. Infect Immun 80:2042–2052. doi: 10.1128/IAI.06160-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu AC, Worrall LJ, Strynadka NC. 2012. Structural insight into the bacterial mucinase StcE essential to adhesion and immune evasion during enterohemorrhagic E. coli infection. Structure 20:707–717. doi: 10.1016/j.str.2012.02.015. [DOI] [PubMed] [Google Scholar]
- 6.Decanio MS, Landick R, Haft RJ. 2013. The non-pathogenic Escherichia coli strain W secretes SslE via the virulence-associated type II secretion system beta. BMC Microbiol 13:130. doi: 10.1186/1471-2180-13-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunstan RA, Heinz E, Wijeyewickrema LC, Pike RN, Purcell AW, Evans TJ, Praszkier J, Robins-Browne RM, Strugnell RA, Korotkov KV, Lithgow T. 2013. Assembly of the type II secretion system such as found in Vibrio cholerae depends on the novel pilotin AspS. PLoS Pathog 9:e1003117. doi: 10.1371/journal.ppat.1003117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan L, Moriel DG, Totsika M, Beatson SA, Schembri MA. 2016. Differential regulation of the surface-exposed and secreted SslE lipoprotein in extraintestinal pathogenic Escherichia coli. PLoS One 11:e0162391. doi: 10.1371/journal.pone.0162391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hardie KR, Lory S, Pugsley AP. 1996. Insertion of an outer membrane protein in Escherichia coli requires a chaperone-like protein. EMBO J 15:978–988. [PMC free article] [PubMed] [Google Scholar]
- 10.Korotkov KV, Johnson TL, Jobling MG, Pruneda J, Pardon E, Heroux A, Turley S, Steyaert J, Holmes RK, Sandkvist M, Hol WG. 2011. Structural and functional studies on the interaction of GspC and GspD in the type II secretion system. PLoS Pathog 7:e1002228. doi: 10.1371/journal.ppat.1002228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peabody CR, Chung YJ, Yen MR, Vidal-Ingigliardi D, Pugsley AP, Saier MH Jr. 2003. Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology 149:3051–3072. doi: 10.1099/mic.0.26364-0. [DOI] [PubMed] [Google Scholar]
- 12.Douzi B, Filloux A, Voulhoux R. 2012. On the path to uncover the bacterial type II secretion system. Philos Trans R Soc Lond B Biol Sci 367:1059–1072. doi: 10.1098/rstb.2011.0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hay ID, Belousoff MJ, Lithgow T. 2017. Structural basis of type 2 secretion system engagement between the inner and outer bacterial membranes. mBio 8:e01344-. doi: 10.1128/mBio.01344-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.d'Enfert C, Reyss I, Wandersman C, Pugsley AP. 1989. Protein secretion by gram-negative bacteria. Characterization of two membrane proteins required for pullulanase secretion by Escherichia coli K-12. J Biol Chem 264:17462–17468. [PubMed] [Google Scholar]
- 15.Collin S, Krehenbrink M, Guilvout I, Pugsley AP. 2013. The targeting, docking and anti-proteolysis functions of the secretin chaperone PulS. Res Microbiol 164:390–396. doi: 10.1016/j.resmic.2013.03.023. [DOI] [PubMed] [Google Scholar]
- 16.Nickerson NN, Tosi T, Dessen A, Baron B, Raynal B, England P, Pugsley AP. 2011. Outer membrane targeting of secretin PulD protein relies on disordered domain recognition by a dedicated chaperone. J Biol Chem 286:38833–38843. doi: 10.1074/jbc.M111.279851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tosi T, Nickerson NN, Mollica L, Jensen MR, Blackledge M, Baron B, England P, Pugsley AP, Dessen A. 2011. Pilotin-secretin recognition in the type II secretion system of Klebsiella oxytoca. Mol Microbiol 82:1422–1432. doi: 10.1111/j.1365-2958.2011.07896.x. [DOI] [PubMed] [Google Scholar]
- 18.Shevchik VE, Condemine G. 1998. Functional characterization of the Erwinia chrysanthemi OutS protein, an element of a type II secretion system. Microbiology 144:3219–3228. doi: 10.1099/00221287-144-11-3219. [DOI] [PubMed] [Google Scholar]
- 19.Schmidt H, Henkel B, Karch H. 1997. A gene cluster closely related to type II secretion pathway operons of gram-negative bacteria is located on the large plasmid of enterohemorrhagic Escherichia coli O157 strains. FEMS Microbiol Lett 148:265–272. doi: 10.1111/j.1574-6968.1997.tb10299.x. [DOI] [PubMed] [Google Scholar]
- 20.Daefler S, Guilvout I, Hardie KR, Pugsley AP, Russel M. 1997. The C-terminal domain of the secretin PulD contains the binding site for its cognate chaperone, PulS, and confers PulS dependence on pIVf1 function. Mol Microbiol 24:465–475. doi: 10.1046/j.1365-2958.1997.3531727.x. [DOI] [PubMed] [Google Scholar]
- 21.Knowles TJ, Scott-Tucker A, Overduin M, Henderson IR. 2009. Membrane protein architects: the role of the BAM complex in outer membrane protein assembly. Nat Rev Microbiol 7:206–214. doi: 10.1038/nrmicro2069. [DOI] [PubMed] [Google Scholar]
- 22.Hagan CL, Silhavy TJ, Kahne D. 2011. Beta-barrel membrane protein assembly by the Bam complex. Annu Rev Biochem 80:189–210. doi: 10.1146/annurev-biochem-061408-144611. [DOI] [PubMed] [Google Scholar]
- 23.Collin S, Guilvout I, Chami M, Pugsley AP. 2007. YaeT-independent multimerization and outer membrane association of secretin PulD. Mol Microbiol 64:1350–1357. doi: 10.1111/j.1365-2958.2007.05743.x. [DOI] [PubMed] [Google Scholar]
- 24.Dunstan RA, Hay ID, Wilksch JJ, Schittenhelm RB, Purcell AW, Clark J, Costin A, Ramm G, Strugnell RA, Lithgow T. 2015. Assembly of the secretion pores GspD, Wza and CsgG into bacterial outer membranes does not require the Omp85 proteins BamA or TamA. Mol Microbiol 97:616–629. doi: 10.1111/mmi.13055. [DOI] [PubMed] [Google Scholar]
- 25.Login FH, Fries M, Wang X, Pickersgill RW, Shevchik VE. 2010. A 20-residue peptide of the inner membrane protein OutC mediates interaction with two distinct sites of the outer membrane secretin OutD and is essential for the functional type II secretion system in Erwinia chrysanthemi. Mol Microbiol 76:944–955. doi: 10.1111/j.1365-2958.2010.07149.x. [DOI] [PubMed] [Google Scholar]
- 26.Yan Z, Yin M, Xu D, Zhu Y, Li X. 2017. Structural insights into the secretin translocation channel in the type II secretion system. Nat Struct Mol Biol 24:177–183. doi: 10.1038/nsmb.3350. [DOI] [PubMed] [Google Scholar]
- 27.Wimley WC. 2002. Toward genomic identification of beta-barrel membrane proteins: composition and architecture of known structures. Protein Sci 11:301–312. doi: 10.1110/ps.29402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fairman JW, Noinaj N, Buchanan SK. 2011. The structural biology of beta-barrel membrane proteins: a summary of recent reports. Curr Opin Struct Biol 21:523–531. doi: 10.1016/j.sbi.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korotkov KV, Gonen T, Hol WG. 2011. Secretins: dynamic channels for protein transport across membranes. Trends Biochem Sci 36:433–443. doi: 10.1016/j.tibs.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tosi T, Estrozi LF, Job V, Guilvout I, Pugsley AP, Schoehn G, Dessen A. 2014. Structural similarity of secretins from type II and type III secretion systems. Structure 22:1348–1355. doi: 10.1016/j.str.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 31.Worrall LJ, Hong C, Vuckovic M, Deng W, Bergeron JR, Majewski DD, Huang RK, Spreter T, Finlay BB, Yu Z, Strynadka NC. 2016. Near-atomic-resolution cryo-EM analysis of the Salmonella T3S injectisome basal body. Nature doi: 10.1038/nature20576. [DOI] [PubMed] [Google Scholar]
- 32.Korotkov KV, Sandkvist M, Hol WG. 2012. The type II secretion system: biogenesis, molecular architecture and mechanism. Nat Rev Microbiol 10:336–351. doi: 10.1038/nrmicro2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Costa TR, Felisberto-Rodrigues C, Meir A, Prevost MS, Redzej A, Trokter M, Waksman G. 2015. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat Rev Microbiol 13:343–359. doi: 10.1038/nrmicro3456. [DOI] [PubMed] [Google Scholar]
- 34.Nivaskumar M, Francetic O. 2014. Type II secretion system: a magic beanstalk or a protein escalator. Biochim Biophys Acta 1843:1568–1577. doi: 10.1016/j.bbamcr.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 35.Cianciotto NP, White RC. 2017. Expanding role of type II secretion in bacterial pathogenesis and beyond. Infect Immun 85:e00014-. doi: 10.1128/IAI.00014-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomassin JL, Santos Moreno J, Guilvout I, Tran Van Nhieu G, Francetic O. 2017. The trans-envelope architecture and function of the type 2 secretion system: new insights raising new questions. Mol Microbiol 105:211–226. doi: 10.1111/mmi.13704. [DOI] [PubMed] [Google Scholar]
- 37.Camberg JL, Sandkvist M. 2005. Molecular analysis of the Vibrio cholerae type II secretion ATPase EpsE. J Bacteriol 187:249–256. doi: 10.1128/JB.187.1.249-256.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patrick M, Korotkov KV, Hol WG, Sandkvist M. 2011. Oligomerization of EpsE coordinates residues from multiple subunits to facilitate ATPase activity. J Biol Chem 286:10378–10386. doi: 10.1074/jbc.M110.167031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Possot OM, Letellier L, Pugsley AP. 1997. Energy requirement for pullulanase secretion by the main terminal branch of the general secretory pathway. Mol Microbiol 24:457–464. doi: 10.1046/j.1365-2958.1997.3451726.x. [DOI] [PubMed] [Google Scholar]
- 40.Reichow SL, Korotkov KV, Gonen M, Sun J, Delarosa JR, Hol WG, Gonen T. 2011. The binding of cholera toxin to the periplasmic vestibule of the type II secretion channel. Channels (Austin) 5:215–218. doi: 10.4161/chan.5.3.15268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Viarre V, Cascales E, Ball G, Michel GP, Filloux A, Voulhoux R. 2009. HxcQ liposecretin is self-piloted to the outer membrane by its N-terminal lipid anchor. J Biol Chem 284:33815–33823. doi: 10.1074/jbc.M109.065938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gu S, Kelly G, Wang X, Frenkiel T, Shevchik VE, Pickersgill RW. 2012. Solution structure of homology region (HR) domain of type II secretion system. J Biol Chem 287:9072–9080. doi: 10.1074/jbc.M111.300624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rehman S, Gu S, Shevchik VE, Pickersgill RW. 2013. Anatomy of secretin binding to the Dickeya dadantii type II secretion system pilotin. Acta Crystallogr D Biol Crystallogr 69:1381–1386. doi: 10.1107/S0907444913007658. [DOI] [PubMed] [Google Scholar]
- 44.Yang J, Baldi DL, Tauschek M, Strugnell RA, Robins-Browne RM. 2007. Transcriptional regulation of the yghJ-pppA-yghG-gspCDEFGHIJKLM cluster, encoding the type II secretion pathway in enterotoxigenic Escherichia coli. J Bacteriol 189:142–150. doi: 10.1128/JB.01115-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brandt F, Etchells SA, Ortiz JO, Elcock AH, Hartl FU, Baumeister W. 2009. The native 3D organization of bacterial polysomes. Cell 136:261–271. doi: 10.1016/j.cell.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 46.Binenbaum Z, Parola AH, Zaritsky A, Fishov I. 1999. Transcription- and translation-dependent changes in membrane dynamics in bacteria: testing the transertion model for domain formation. Mol Microbiol 32:1173–1182. doi: 10.1046/j.1365-2958.1999.01426.x. [DOI] [PubMed] [Google Scholar]
- 47.Bakshi S, Choi H, Mondal J, Weisshaar JC. 2014. Time-dependent effects of transcription- and translation-halting drugs on the spatial distributions of the Escherichia coli chromosome and ribosomes. Mol Microbiol 94:871–887. doi: 10.1111/mmi.12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsumoto K, Hara H, Fishov I, Mileykovskaya E, Norris V. 2015. The membrane: transertion as an organizing principle in membrane heterogeneity. Front Microbiol 6:572. doi: 10.3389/fmicb.2015.00572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roggiani M, Goulian M. 2015. Chromosome-membrane interactions in bacteria. Annu Rev Genet 49:115–129. doi: 10.1146/annurev-genet-112414-054958. [DOI] [PubMed] [Google Scholar]
- 50.van der Spoel D, van Maaren PJ, Larsson P, Timneanu N. 2006. Thermodynamics of hydrogen bonding in hydrophilic and hydrophobic media. J Phys Chem B 110:4393–4398. doi: 10.1021/jp0572535. [DOI] [PubMed] [Google Scholar]
- 51.Reboul CF, Whisstock JC, Dunstone MA. 2016. Giant MACPF/CDC pore forming toxins: a class of their own. Biochim Biophys Acta 1858:475–486. doi: 10.1016/j.bbamem.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 52.Guilvout I, Brier S, Chami M, Hourdel V, Francetic O, Pugsley AP, Chamot-Rooke J, Huysmans GH. 2017. Prepore stability controls productive folding of the BAM-independent multimeric outer membrane secretin PulD. J Biol Chem 292:328–338. doi: 10.1074/jbc.M116.759498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huysmans GH, Guilvout I, Chami M, Nickerson NN, Pugsley AP. 2015. Lipids assist the membrane insertion of a BAM-independent outer membrane protein. Sci Rep 5:15068. doi: 10.1038/srep15068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huysmans GH, Guilvout I, Pugsley AP. 2013. Sequential steps in the assembly of the multimeric outer membrane secretin PulD. J Biol Chem 288:30700–30707. doi: 10.1074/jbc.M113.489112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miroux B, Walker JE. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol 260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 56.Cheung M, Kajimura N, Makino F, Ashihara M, Miyata T, Kato T, Namba K, Blocker AJ. 2013. A method to achieve homogeneous dispersion of large transmembrane complexes within the holes of carbon films for electron cryomicroscopy. J Struct Biol 182:51–56. doi: 10.1016/j.jsb.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kimanius D, Forsberg BO, Scheres SH, Lindahl E. 2016. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife 5:e18722. doi: 10.7554/eLife.18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. 2013. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods 10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rohou A, Grigorieff N. 2015. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J Struct Biol 192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. 2017. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 61.Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, Davis IW, Cooper S, Treuille A, Mandell DJ, Richter F, Ban YE, Fleishman SJ, Corn JE, Kim DE, Lyskov S, Berrondo M, Mentzer S, Popovic Z, Havranek JJ, Karanicolas J, Das R, Meiler J, Kortemme T, Gray JJ, Kuhlman B, Baker D, Bradley P. 2011. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol 487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wriggers W. 2012. Conventions and workflows for using Situs. Acta Crystallogr D Biol Crystallogr 68:344–351. doi: 10.1107/S0907444911049791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chan KY, Trabuco LG, Schreiner E, Schulten K. 2012. Cryo-electron microscopy modeling by the molecular dynamics flexible fitting method. Biopolymers 97:678–686. doi: 10.1002/bip.22042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Frickey T, Lupas A. 2004. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics 20:3702–3704. doi: 10.1093/bioinformatics/bth444. [DOI] [PubMed] [Google Scholar]
- 67.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL. 2012. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res 40:D370–D376. doi: 10.1093/nar/gkr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 71.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A 98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Murzin AG, Lesk AM, Chothia C. 1994. Principles determining the structure of beta-sheet barrels in proteins. I. A theoretical analysis. J Mol Biol 236:1369–1381. [DOI] [PubMed] [Google Scholar]
- 73.Eisenberg D, Schwarz E, Komaromy M, Wall R. 1984. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol 179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.