ABSTRACT
Exchange of gene segments through reassortment is a major feature of influenza A virus evolution and frequently contributes to the emergence of novel epidemic, pandemic, and zoonotic strains. It has long been evident that viral diversification through reassortment is constrained by genetic incompatibility between divergent parental viruses. In contrast, the role of virus-extrinsic factors in determining the likelihood of reassortment has remained unclear. To evaluate the impact of such factors in the absence of confounding effects of segment mismatch, we previously reported an approach in which reassortment between wild-type (wt) and genetically tagged variant (var) viruses of the same strain is measured. Here, using wt/var systems in the A/Netherlands/602/2009 (pH1N1) and A/Panama/2007/99 (H3N2) strain backgrounds, we tested whether inoculation of parental viruses into distinct sites within the respiratory tract limits their reassortment. Using a ferret (Mustella putorius furo) model, either matched parental viruses were coinoculated intranasally or one virus was instilled intranasally whereas the second was instilled intratracheally. Dual intranasal inoculation resulted in robust reassortment for wt/var viruses of both strain backgrounds. In contrast, when infections were initiated simultaneously at distinct sites, strong compartmentalization of viral replication was observed and minimal reassortment was detected. The observed lack of viral spread between upper and lower respiratory tract tissues may be attributable to localized exclusion of superinfection within the host, mediated by innate immune responses. Our findings indicate that dual infections in nature are more likely to result in reassortment if viruses are seeded into similar anatomical locations and have matched tissue tropisms.
IMPORTANCE Genetic exchange between influenza A viruses (IAVs) through reassortment can facilitate the emergence of antigenically drifted seasonal strains and plays a prominent role in the development of pandemics. Typical human influenza infections are concentrated in the upper respiratory tract; however, lower respiratory tract (LRT) infection is an important feature of severe cases, which are more common in the very young, the elderly, and individuals with underlying conditions. In addition to host factors, viral characteristics and mode of transmission can also increase the likelihood of LRT infection: certain zoonotic IAVs are thought to favor the LRT, and transmission via small droplets allows direct seeding into lower respiratory tract tissues. To gauge the likelihood of reassortment in coinfected hosts, we assessed the extent to which initiation of infection at distinct respiratory tract sites impacts reassortment frequency. Our results reveal that spatially distinct inoculations result in anatomical compartmentalization of infection, which in turn strongly limits reassortment.
KEYWORDS: compartmentalization, diversity, ferret, influenza A virus, reassortment
INTRODUCTION
Reassortment, or the exchange of intact gene segments between coinfecting viruses, plays a prominent role in the evolution and emergence of influenza A viruses (IAVs) (1, 2). While reassortment among related, cocirculating human IAVs is known to contribute to the diversification and evolution of seasonal strains (3–8), reassortment involving IAVs adapted to distinct host species poses a risk for the emergence of pandemics (9–11). For two IAVs to reassort, they must infect the same host and infect the same cell(s) within that host. In gauging the potential for reassortment in nature, it is therefore important to understand the likelihood of cellular coinfection taking place within a coinfected host. To allow rigorous examination of this issue, we previously developed a system in which reassortment between a wild-type (wt) IAV and a genetically tagged variant (var) of the same strain is measured (12). This system avoids genetic incompatibility between coinfecting viruses and thereby facilitates detection of virus-extrinsic effects on reassortment efficiency. Our prior work using wt/var coinfection in a guinea pig model revealed that reassortment can be highly efficient in a coinfected host (up to ∼65% of viruses present in nasal lavage fluid were reassortants). This efficiency was strongly dependent on the timing of superinfection, however. When viruses were introduced simultaneously, or within a 12-h time window, reassortment was prevalent. However, with a delay of greater than 18 h, neither infection with the second virus nor reassortment could be detected.
In addition to timing of infection, another important variable that may limit reassortment potential in nature is the spatial distribution of infection within the host. Examination of viral growth in ex vivo organ cultures and primary cells derived from respiratory tract tissues suggests that human seasonal IAVs can replicate in epithelial cells derived from multiple sites throughout the human respiratory tract (13–15). Within the infected human host, however, IAV is thought to replicate mainly in the upper respiratory tract (URT) in typical cases, while spread to the lower respiratory tract (LRT) is associated with severe influenza. Lower respiratory tract involvement can lead to viral pneumonia, a complication which is more common in the very young, the elderly, and individuals with underlying conditions (16–20).
In addition to host factors, viral characteristics can increase the likelihood of LRT infection. In both animal models and natural human infection, the 2009 pandemic strain has exhibited a greater propensity for growth in the LRT than seasonal IAVs (17, 21–24). In addition, zoonotic IAVs derived from avian hosts may favor the LRT over upper respiratory tract sites (25–29). This feature of infection likely contributes to severe disease in human cases of subtype H5N1 and H7N9 IAV infection. Notably, however, avian virus-like H5N1 and H7N9 subtype viruses isolated from humans have been found to attach to or replicate in cells of the upper respiratory tract ex vivo (30, 31).
Importantly, different modes of IAV transmission lead to seeding of virus into different locations within the respiratory tract. Transmission via fomites or large respiratory droplets is expected to deposit virus near the site of entry into the host, in the upper respiratory tract. In contrast, small-droplet aerosols are efficiently carried into lower regions of the respiratory tract, with a significant proportion of those that are 5 μm in diameter penetrating all the way to the alveolar region (32).
In sum, viral characteristics, host factors, and mode of viral transmission impact the site of viral replication in the human respiratory tract. It follows that, when a given host is coinfected with two distinct IAV populations, their replication may initiate at distinct anatomical locations. We therefore sought to determine the likelihood of reassortment occurring when two IAVs initiate infection separately, in the upper and lower respiratory tracts. To this end, we examined reassortment efficiency in a ferret (Mustella putorius furo) model using wt/var virus systems of two distinct strain backgrounds. The wt and var viruses either were instilled together into the nasal tract of the ferrets or were inoculated at the same time but into the URT via intranasal (IN) inoculation or the LRT via intratracheal (IT) inoculation. Our results reveal that reassortment is highly efficient in intranasally coinoculated ferrets but is severely restricted when viruses are introduced at distinct anatomical sites. These findings point to a virus-extrinsic constraint on IAV reassortment which is likely to play a role in determining the likelihood of reassortant viruses emerging in nature.
RESULTS
Coinoculation of matched wild-type and variant viruses into the nasal tract results in robust reassortment in ferrets.
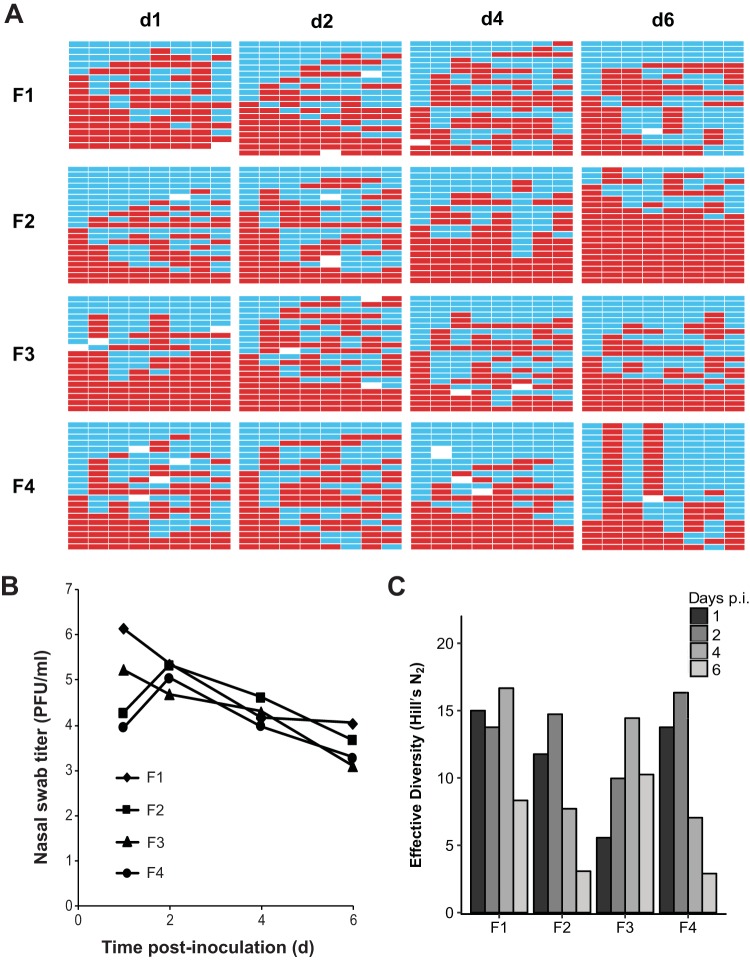
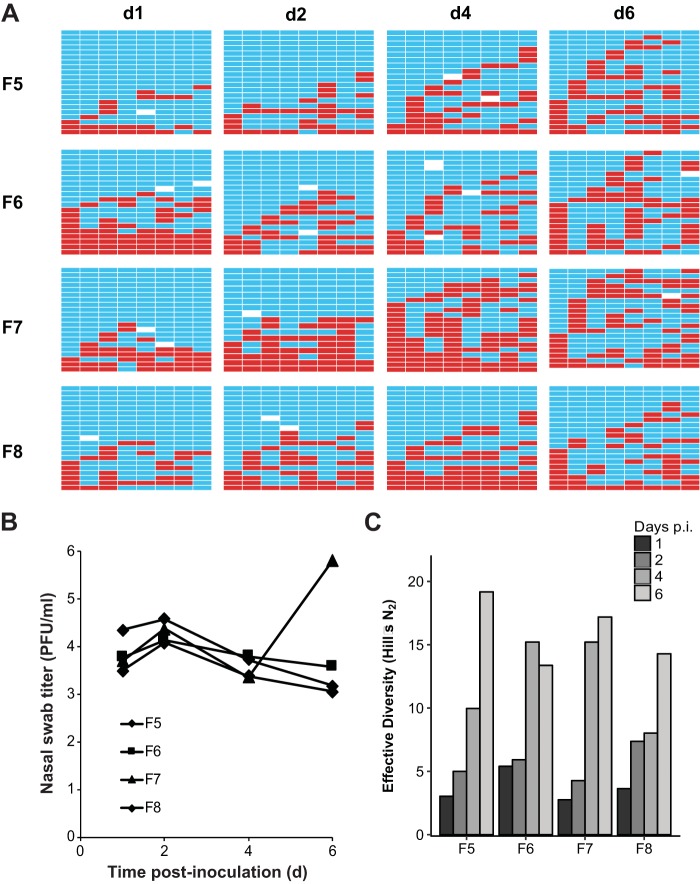
To determine baseline frequencies of reassortment in ferrets, we performed intranasal coinoculations with either NL/09wt plus NL/09var (pH1N1) viruses or with Pan/99wt plus Pan/99var (H3N2) viruses in groups of four animals. In each case, wt and var viruses were mixed in a 1:1 ratio prior to inoculation and a total dose of 2 × 105 50% tissue culture infective doses (TCID50) per ferret was used. Nasal swabs were collected daily. Samples from day 1 (d1), d2, d4, and d6 postinoculation were analyzed to evaluate reassortment levels. First, the titer of virus present in these nasal swabs was determined by plaque assay (Fig. 1B and 2B). Next, approximately 20 plaque isolates derived from each time point were genotyped by reverse transcription-quantitative PCR (RT-qPCR) followed by high-resolution melt (HRM) analysis. The results of this genotyping assay are displayed in Fig. 1 for NL/09wt plus NL/09var virus coinfections and in Fig. 2 for Pan/99wt plus Pan/99var virus coinfections. The checkerboard appearance of the genotype tables shown in both figures indicates that reassortant viruses were detected readily. It is also apparent from this pictorial view of the data that, in the case of Pan/99wt-Pan/99var virus coinfections, the prevalence of reassortant genotypes tended to increase over the course of the infection (Fig. 2).
FIG 1.
Dual intranasal inoculation with NL/09wt and NL/09var viruses resulted in high levels of reassortment. Ferrets were inoculated intranasally with a 1:1 mixture of NL/09wt and NL/09var viruses in a 40-μl volume. The dose used comprised 1 × 105 TCID50 of each virus. Nasal swabs were collected and virus therein analyzed to generate the data shown. (A) Viral genotypes of 17 to 21 plaque isolates per sample are shown in table format, where each row represents a viral isolate and each of the eight columns represents a viral gene segment in the order PB2, PB1, PA, HA, NP, NA, M, and NS. Red bars indicate segments derived from the NL/09wt parental virus, and turquoise bars show segments derived from the NL/09var virus. White bars indicate segments where the genotyping result was not clear. Day postinoculation (p.i.) is shown at the top, and the ferret from which swabs were collected is indicated at the left. (B) Infectious titers of nasal swab samples are plotted as a function of time postinoculation. (C) The diversity of viral gene constellations detected is represented for each ferret, with days p.i. indicated in grayscale. Each sample's Simpson index value was converted to a corresponding Hill number (Hill's N2) to derive its effective diversity.
FIG 2.
Dual intranasal inoculation with Pan/99wt and Pan/99var viruses resulted in high levels of reassortment. Ferrets were inoculated intranasally with a 1:1 mixture of Pan/99wt and Pan/99var viruses in a 40-μl volume. The dose used comprised 1 × 105 TCID50 of each virus. Nasal swabs were collected and virus therein analyzed to generate the data shown. (A) Viral genotypes of 17 to 21 plaque isolates per sample are shown in table format, as described in the legend to Fig. 1. Red bars indicate segments derived from the Pan/99wt virus, and turquoise bars show segments derived from the Pan/99var virus. White bars indicate segments where the genotyping result was not clear. (B) Infectious titers of nasal swab samples are plotted as a function of time postinoculation. (C) The diversity of viral gene constellations detected is represented for each ferret, with days p.i. in grayscale. Each sample's Simpson index value was converted to a corresponding Hill number (Hill's N2) to derive its effective diversity.
To give a quantitative measure of the viral genotypic diversity generated through reassortment, the Simpson's diversity index was calculated (Fig. 1C and 2C). This index takes into account both the number of different genotypes detected and their evenness or the variation in the abundance of each. Each sample's Simpson index value was then converted to a corresponding Hill number to derive its effective diversity and allow statistical analysis by basic linear regression methods. The results indicate that, even early after inoculation, an appreciable level of diversity was generated through reassortment in ferrets. Analyzing the effect of day on effective diversity (N2) using mixed effects models to account for individual ferret variation revealed a significant decrease in diversity over time in the NL/09-infected ferrets [p(day) = 0.039, estimate = −1.19/day] and a significant increase in the Pan/99-infected ferrets [p(day) = 7.4 × 10−7, estimate = +2.56/day]. The differing results observed for the NL/09 and Pan/99 strain backgrounds with respect to the kinetics of viral genotypic diversity were likely a result of differences in the kinetics of viral spread. Namely, swab titers of NL/09 wt/var-coinfected ferrets tended to be higher than the swab titers of Pan/99 wt/var-coinfected animals on d1 postinoculation (P = 0.088, t test), suggesting a more rapid expansion of the NL/09-based inoculum (see panel B in Fig. 1 and 2).
Simultaneous inoculation of ferrets via the intranasal and intratracheal routes results in little reassortment.
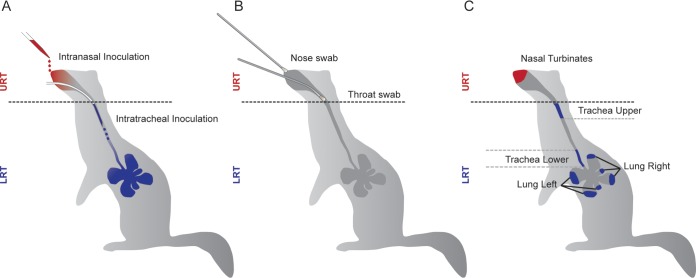
To test the extent to which reassortment is restricted when infection is initiated at distinct anatomical sites, ferrets were inoculated with one virus intranasally and with a second virus intratracheally. The sites targeted by these two routes of inoculation, as well as the sampling methods employed after infection, are shown schematically in Fig. 3. As with the dual intranasal inoculations described above, 1 × 105 TCID50 of each virus was used and the intranasal (IN) and intratracheal (IT) inoculations were performed at the same time with the aim of initiating infections at the two sites simultaneously. Again, these experiments were performed with wt/var virus pairings in the NL/09 background and, separately, in the Pan/99 background. Nasal and throat swabs were collected daily.
FIG 3.
Locations of inoculation and sampling within the ferret respiratory tract. Anatomical sites accessed for (A) inoculation, (B) swab collection, and (C) tissue collection are shown schematically.
We noted during the course of the IN/IT experiment performed with the NL/09-based viruses that the virus introduced intratracheally was not readily detected by swabbing the nose or throat. For the experiment performed with the Pan/99-based viruses, we therefore also collected tissues on day 3, 4, or 5 postinoculation. The timing of tissue collection was chosen to balance the opportunity to track reassortment over time with the need to demonstrate virus growth in the lower respiratory tract. Tissues were homogenized, and viral load was evaluated by TCID50 assay. To determine whether the viruses detected were derived from wt virus or var virus or both parental viruses, we performed deep sequencing on the PB2 segment. The results are shown in Fig. 4. In each animal, the virus instilled intranasally strongly predominated in the nasal turbinates (NT). The virus instilled intratracheally was predominant either throughout the regions of the lower respiratory tract tested (in ferrets F13, F14, and F15) or in a subset of the lower respiratory tract tissues (in ferret F16). Importantly, productive infection with both wt and var viruses was evident in all four ferrets.
FIG 4.
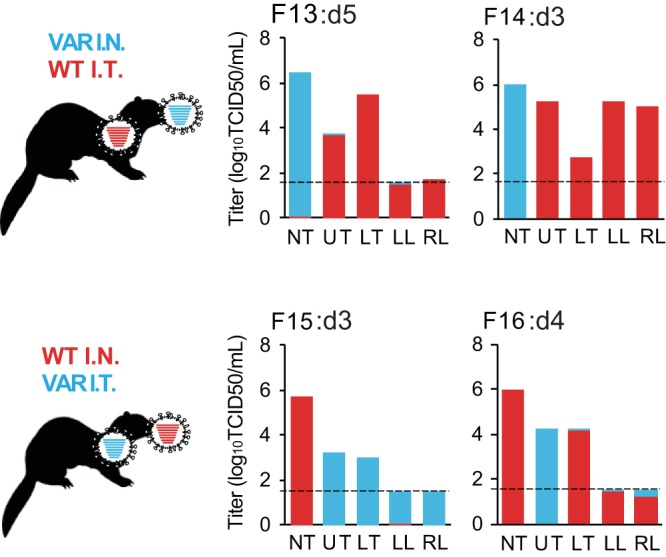
Both intranasal (I.N.) and intratracheal (I.T.) inoculations led to productive viral infection in ferrets coinfected with Pan/99wt plus Pan/99var virus. Infectious titers in tissue homogenates were determined by TCID50 assay, and the genotypes of PB2 RNAs present in each sample were determined by deep sequencing. The limit of detection for infectious virus was 1.5 log10TCID50/ml (shown with a dashed line), and samples in which infectious virus was not detected are plotted at that limit. Note that viral RNA was detected by deep sequencing in samples that were below the detection limit of the TCID50 assay. Coloring of the bars indicates the proportion of each PB2 genotype detected, with a limit of detection of 1%. wt data are shown in red and var data in turquoise. The tissues analyzed were nasal turbinate (NT), upper trachea (UT), lower trachea (LT), left lung (LL), and right lung (RL). Ferret identification numbers and day postinfection at which tissues were collected are given above each graph.
To measure reassortment in the ferrets inoculated via both the IN and IT routes, both nasal and throat swabs were analyzed. Again, plaque assays were performed to determine viral titers and approximately 20 plaques per swab sample were genotyped. In addition, deep sequencing targeting the PB2 segment was performed on swab samples collected from each animal at an early time point and a late time point.
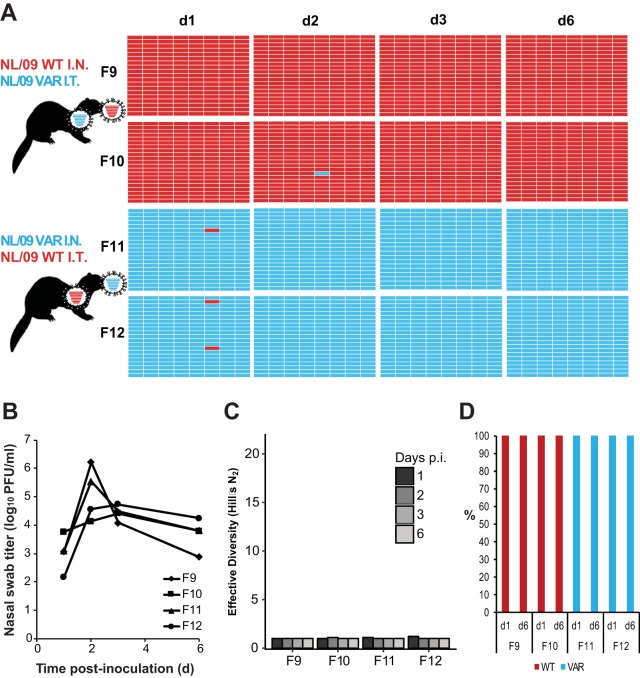
Results obtained from nasal swabs are shown in Fig. 5 for NL/09-based viruses and in Fig. 6 for Pan/99-based viruses. In both experiments, the majority of plaque isolates retained the parental genotype of the virus instilled intranasally. Very little reassortment, and therefore very low diversity, was detected. Analyzing 20 plaques per sample, the limit of detection for a given viral genotype is relatively high, at 5%. Low levels of reassortment (<5%), which were not reliably detected in our assay, could be important in a natural context if the reassortant viruses produced were to have a fitness advantage over parental strains. To determine whether we might be overlooking lower levels of reassortment, we performed deep sequencing on viral cDNA derived from the bulk swab samples with the aim of evaluating the relative prevalences of wt and var virus gene segments with an improved limit of detection. Since it is not possible to define viral gene constellations by this approach, we typed only the PB2 segment. Results were consistent with those obtained by genotyping plaques: in nasal swabs, the PB2 segment of the virus instilled intratracheally represented <5% of reads (the limit of detection for this assay was 1%).
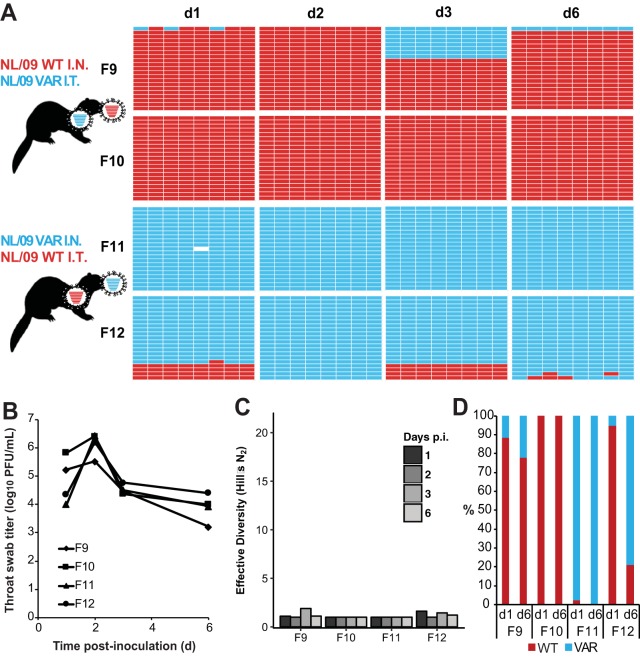
FIG 5.
Separate inoculations of NL/09wt and NL/09var viruses by the intranasal and intratracheal routes yielded little mixing and low levels of reassortment in the nasal tract. Ferrets were inoculated with 1 × 105 TCID50 of NL/09wt and 1 × 105 TCID50 of NL/09var viruses. Ferrets F9 and F10 received NL/09wt virus intranasally and NL/09var virus intratracheally. Ferrets F11 and F12 received NL/09var virus intranasally and NL/09wt virus intratracheally. Nasal swabs were collected and virus therein analyzed to generate the data shown. (A) Viral genotypes of 17 to 21 plaque isolates per sample are shown in table format, as described in the legend to Fig. 1. Red bars indicate segments derived from the NL/09wt virus, and turquoise bars show segments derived from the NL/09var virus. (B) Infectious titers of nasal swab samples are plotted as a function of time postinoculation. (C) The diversity of viral gene constellations detected is represented for each ferret, with days p.i. in grayscale. Each sample's Simpson index value was converted to a corresponding Hill number (Hill's N2) to derive its effective diversity. (D) Results of next-generation sequencing of the IAV PB2 segments present in swab samples are shown. The percentage of reads carrying wt or var sequence is indicated by the coloring of the bars. Limit of detection = 1%.
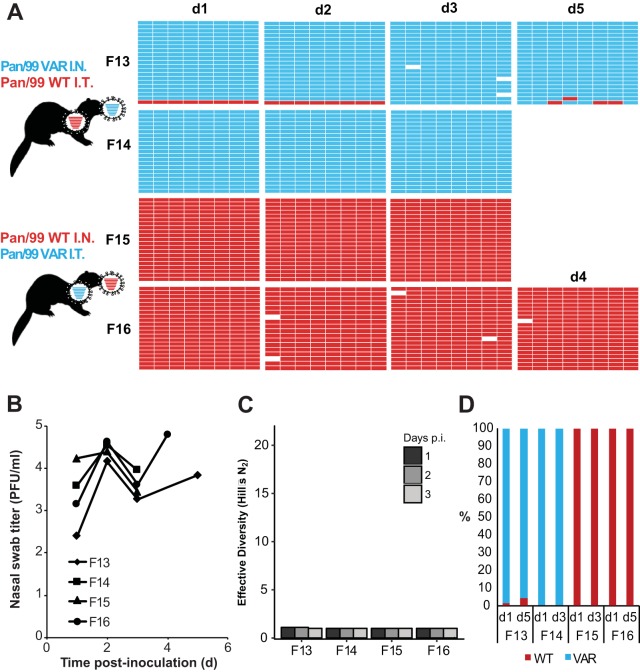
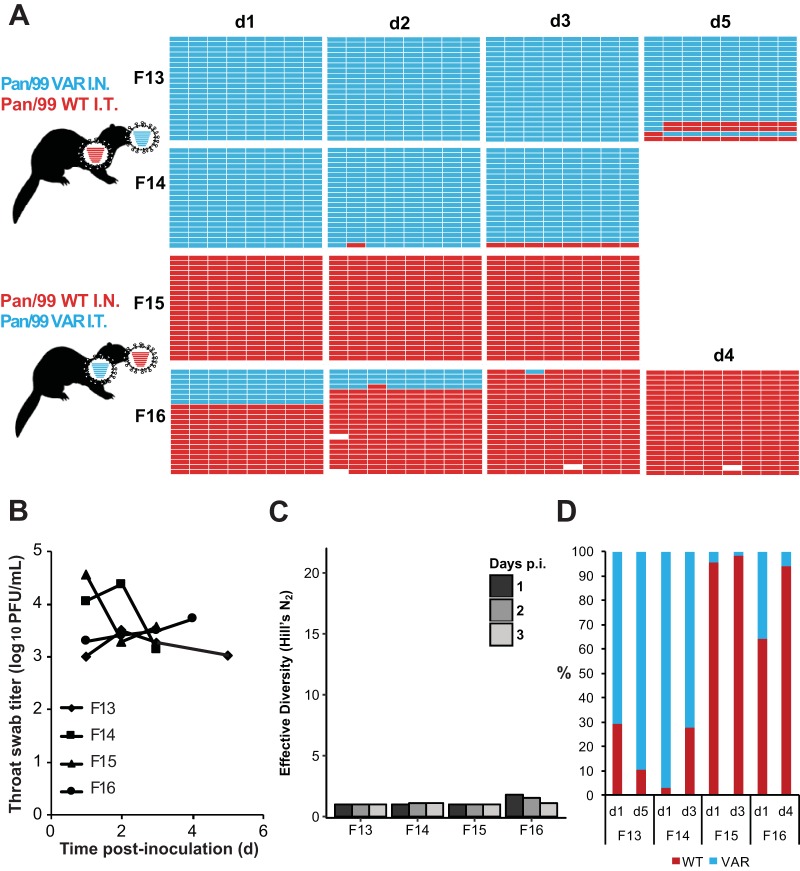
FIG 6.
Separate inoculations of Pan/99wt and Pan/99var viruses by the intranasal and intratracheal routes yielded little mixing and low levels of reassortment in the nasal tract. Ferrets were inoculated with 1 × 105 TCID50 of Pan/99wt and 1 × 105 TCID50 of Pan/99var viruses. Ferrets F13 and F14 received Pan/99var virus intranasally and Pan/99wt virus intratracheally. Ferrets F15 and F16 received Pan/99wt virus intranasally and Pan/99var virus intratracheally. Nasal swabs were collected and virus therein analyzed to generate the data shown. (A) Viral genotypes of 17 to 21 plaque isolates per sample are shown in table format, as described in the legend to Fig. 1. Red bars indicate segments derived from the Pan/99wt virus, and turquoise bars show segments derived from the Pan/99var virus. White bars indicate segments where the genotyping result was not clear. (B) Infectious titers of nasal swab samples are plotted as a function of time postinoculation. (C) The diversity of viral gene constellations detected is represented for each ferret, with days p.i. in grayscale. Each sample's Simpson index value was converted to a corresponding Hill number (Hill's N2) to derive its effective diversity. (D) Results of next-generation sequencing of the IAV PB2 segments present in swab samples are shown. The percentage of reads carrying wt or var sequence is indicated by the coloring of the bars. Limit of detection = 1%.
Results obtained from throat swabs are shown in Fig. 7 for NL/09-based viruses and in Fig. 8 for Pan/99-based viruses. These swabs were collected from a lower region of the upper respiratory tract than the nasal swabs but do not represent a true lower respiratory tract sample (Fig. 3). In throat swabs, most viruses detected were again of the same genotype as the virus instilled intranasally. In a subset of throat samples, both parental genotypes were represented by an appreciable number of isolates and an appreciable number of sequencing reads for PB2. Nevertheless, little or no reassortment was detected even where both parental viruses were present. Thus, in sharp contrast to the results seen with dual intranasal inoculation, reassortment in upper respiratory tract samples was rare and diversity was limited when viruses were introduced into distinct regions of the respiratory tract.
FIG 7.
Separate inoculations of NL/09wt and NL/09var viruses by the intranasal and intratracheal routes yielded moderate mixing but minimal reassortment in the throat. Ferrets were inoculated with 1 × 105 TCID50 of NL/09wt and 1 × 105 TCID50 of NL/09var viruses, as indicated in the legend to Fig. 4. Throat swabs were collected and virus therein analyzed to generate the data shown. (A) Viral genotypes of 17 to 21 plaque isolates per sample are shown in table format, as described in the legend to Fig. 1. Red bars indicate segments derived from the NL/09wt virus, and turquoise bars show segments derived from the NL/09var virus. White bars indicate segments where the genotyping result was not clear. (B) Infectious titers of throat swab samples are plotted as a function of time postinoculation. (C) The diversity of viral gene constellations detected is represented for each ferret, with days p.i. in grayscale. Each sample's Simpson index value was converted to a corresponding Hill number (Hill's N2) to derive its effective diversity. (D) Results of next-generation sequencing of the IAV PB2 segments present in swab samples are shown. The percentage of reads carrying wt or var sequence is indicated by the coloring of the bars. Limit of detection = 1%.
FIG 8.
Separate inoculations of Pan/99wt and Pan/99 viruses by the intranasal and intratracheal routes yielded moderate mixing but minimal reassortment in the throat. Ferrets were inoculated with 1 × 105 TCID50 of Pan/99wt and 1 × 105 TCID50 of Pan/99var viruses, as indicated in the legend to Fig. 5. Throat swabs were collected and virus therein analyzed to generate the data shown. (A) Viral genotypes of 17 to 21 plaque isolates per sample are shown in table format, as described in the legend to Fig. 1. Red bars indicate segments derived from the Pan/99wt virus, and turquoise bars show segments derived from the Pan/99var virus. White bars indicate segments where the genotyping result was not clear. (B) Infectious titers of throat swab samples are plotted as a function of time postinoculation. (C) The diversity of viral gene constellations detected is represented for each ferret, with days p.i. in grayscale. Each sample's Simpson index value was converted to a corresponding Hill number (Hill's N2) to derive its effective diversity. (D) Results of next-generation sequencing of the IAV PB2 segments present in swab samples are shown. The percentage of reads carrying wt or var sequence is indicated by the coloring of the bars. Limit of detection = 1%.
DISCUSSION
Many factors relevant to IAV infections in nature are likely to impact the frequency of coinfection within a host. Toward unraveling this complexity and achieving a greater understanding of the conditions that support IAV reassortment, we have taken a systematic approach of modulating a single variable experimentally and then measuring the impact on reassortment levels. We previously evaluated the effects of introducing a range of time intervals between infections and of adjusting the dose of coinfecting viruses (12, 33, 34). Here, we tested whether two viruses must initiate infection at the same site within the respiratory tract to give rise to robust reassortment. Our results showed that, when two viruses infected simultaneously but at distinct locations, spatial separation within the respiratory tract persisted over multiple days. Little virus transfer from lower to upper respiratory tract tissues was detected, and, as a result, minimal genetic exchange between wt and var viruses was detectable in upper respiratory tract samples.
We chose a ferret model for the present studies because it is well established that intratracheal inoculation of ferrets results in robust viral replication in the lower respiratory tract (21, 35, 36). Our previous analyses of reassortment efficiency in vivo were performed in a guinea pig model using Pan/99-based wt and var viruses. The earlier work revealed high levels of reassortment under “baseline” conditions of simultaneous, intranasal coinoculation (12, 34). Thus, the observation of extensive reassortment in ferrets coinfected intranasally with either Pan/99-based or NL/09-based wt and var viruses reported here extends our earlier findings to an additional model host and to a distinct IAV strain background. This work strengthens the conclusion that the multiplicity of infection achieved in vivo is sufficient to support efficient coinfection and reassortment (12, 33, 34, 37).
In contrast to the highly efficient reassortment seen when wt and var viruses were introduced together into the nasal tract, simultaneous inoculations into the upper and lower respiratory tract sites resulted in little reassortment detectable in upper respiratory tract swab samples. Instead, the parental virus that was introduced intranasally was found to predominate in all nasal and throat swab samples analyzed and was furthermore the only genotype detected in many of the nasal samples. Analysis of tissue samples extracted from ferrets inoculated with Pan/99-based viruses indicated that the lack of genetic exchange between wt and var viruses was due to a strong compartmentalization of infection: the virus that was instilled intratracheally replicated within lower respiratory tract tissues but was largely confined to the LRT. The virus that was instilled intranasally also typically remained in the URT but was detected in multiple LRT sites in one of the four ferrets from which tissues were collected. Similar compartmentalization was seen in a separate experiment using IN and IT inoculation of NL/09-based viruses, followed by collection of tissues from the URT and LRT (data not shown). Clearly, this lack of mixing would prevent reassortment by limiting the potential for coinfection of cells with both wt and var viruses.
The lack of viral spread from the LRT to the URT may be the result of an active exclusion of superinfection at the level of the tissue, brought about by local innate immune responses and/or activity of the viral neuraminidase (38, 39). Alternatively, a simpler mechanism in which virus fails to travel up the trachea to the URT may be responsible. In previous studies in which ferrets received a single IAV inoculation intratracheally at a dose of 106 TCID50, virus was detected in nasal and pharyngeal swabs as early as 2 days postinoculation (40, 41). The lack of spread from the lower respiratory tract to the upper respiratory tract observed here may therefore indicate that cells of the URT are refractory to infection or superinfection as a result of ongoing infection in the URT. This mechanism is consistent with our previous observations regarding the time window within which Pan/99 virus superinfection can be achieved in the upper respiratory tract of guinea pigs. When intranasal inoculation was performed first with Pan/99var virus and then at 6, 12, 18, 24, 48, or 72 h with Pan/99wt virus, we found that replication of Pan/99wt virus genes could not be detected in guinea pigs inoculated ≥24 h after the initial infection (12). Assuming similar timing of superinfection exclusion in ferrets, the spread of virus from the LRT to the URT may have been blocked beginning around 24 h postinoculation—an earlier time point than that at which virus was first detected in the URT in ferrets inoculated only via the IT route (40, 41).
Our data suggest that the site of inoculation strongly influences the site of subsequent viral replication and is therefore likely to impact the probability of reassortment in nature. In particular, within-host compartmentalization of infections may occur when coinfecting viruses have differing tissue tropisms or when two viruses infect via differing modes of transmission. A pertinent example of the former is coinfection of a human host with seasonal IAV and a typical avian IAV. While seasonal strains replicate mainly in the URT, avian IAV infections of humans are often characterized by viral growth in the lungs, likely owing to the greater prevalence of alpha-2,3-linked sialic acid receptors and the fact that the LRT is warmer than the URT (31, 42–46). The experiments reported here may therefore inform estimations of the risk of avian/human IAV reassortment in a coinfected human host. It should be noted in this context, however, that zoonotic IAVs of the H5N1 and H7N9 subtypes have been shown to bind to human olfactory mucosa and respiratory epithelia, respectively (31, 47), and to replicate in human nasopharynx ex vivo (27, 30). In addition, human viruses of the pH1N1 and seasonal H3N2 lineages spread to the lower respiratory tract in atypical cases (reviewed in references 22 and 48). Thus, while avian and human IAVs may be less likely to meet within the same region of the respiratory tract than two human IAVs, the possibility of colocalized infection should not be excluded.
The work described here also models a situation in which viruses with similar levels of tropism initiate infection separately in the URT and the LRT owing to their transit within large and small infectious droplets, respectively. Large respiratory droplets, at >20 μm in diameter, are likely to be deposited within the upper respiratory tract, with essentially no penetration beyond the trachea. In contrast, a significant proportion of droplets that are <5 μm in diameter penetrate all the way to the alveolar region (32, 49). Thus, viruses transmitted via differing modes may be less likely to meet within a coinfected host and undergo reassortment.
In summary, our data clearly show that, within a ferret model system, genetic exchange between influenza A viruses is markedly restricted in the URT when a coinfecting virus is introduced into the LRT rather than being coinoculated into the URT. This restriction is directly attributable to limited spread to infected upper respiratory tract tissues of the IAV introduced into the lower respiratory tract.
MATERIALS AND METHODS
Viruses.
All viruses used in this work were generated using reverse genetics techniques (50). The rescue system for A/Panama/2007/99 (H3N2) (Pan/99) virus was initially described in reference 51, and that for A/Netherlands/602/2009 (pH1N1) (NL/09) virus was described in reference 52. The Pan/99var virus represented the Pan/99var15 strain described previously (33) and was modified from the Pan/99wt sequence with the inclusion of 1 to 6 silent mutations per gene segment, as follows: PB2, C354T and C360T; PB1, A540G; PA, A342G and G333A; HA, T308C, C311A, C314T, A464T, C467G, and T470A; NP, C537T, T538A, and C539G; NA, C418G, T421A, and A424C; M, G586A; NS, C329T, C335T, and A341G. The NL/09var virus carries a single synonymous mutation per segment relative to the pH1N1wt virus, as follows: PB2, C273T; PB1, T288C; PA, C360T; HA, C305T; NP, A351G; NA, G336A; M, G295A; NS, C341T. These silent mutations act as genetic markers for determining the parental origin of gene segments following isolation of progeny viruses from coinfected ferrets.
Pan/99wt and Pan/99var viruses were cultured in 9-to-11-day-old embryonated chicken's eggs incubated at 33°C. The stocks used were passaged once in eggs following rescue. NL/09wt and NL/09var viruses were grown in Madin-Darby canine kidney (MDCK) cells (ATCC) incubated at 33°C. The NL/09-based viruses were passaged twice in MDCK cells following rescue.
Cells.
The MDCK cells were cultured in Eagle's minimal essential medium (EMEM; Lonza Benelux BV, Breda, The Netherlands) or minimal essential medium (MEM; Gibco) supplemented with 10% fetal bovine serum (FBS) (Greiner or Atlanta Biologicals), 100 U/ml penicillin (P; Lonza or Corning), 100 U/ml streptomycin (S; Lonza or Corning), 2 mM l-glutamine (Lonza or Corning), 1.5 mg/ml sodium bicarbonate (Lonza or Corning), and 10 mM HEPES (Lonza or Corning). EMEM was additionally supplemented with 1× nonessential amino acids (Lonza).
Virus titrations in MDCK cells.
Endpoint virus titrations were performed as described previously (24). Briefly, MDCK cells were inoculated with 10-fold serial dilutions of homogenized tissue samples. Cells were washed with phosphate-buffered saline (PBS) 1 h after inoculation and were cultured in infection media, consisting of serum-free supplemented EMEM containing 20 μg/ml trypsin (Lonza). Three days after inoculation, supernatants of cell cultures were tested for agglutinating activity using turkey erythrocytes as an indicator of virus replication. Infectious virus titers were calculated from four replicates each of the homogenized tissue samples by the method of Reed and Muench (a simple method of estimating 50 percent endpoints).
Plaque assays were performed according to standard methods. Briefly, confluent MDCK cells were washed with PBS and then inoculated with 10-fold serial dilutions of ferret swab samples. Cells were washed with PBS 1 h after inoculation and overlaid with serum-free MEM containing 3% bovine serum albumin (Sigma-Aldrich), 0.6% agar (Oxoid), and 20 μg/ml trypsin (Sigma-Aldrich). Cells were incubated for 48 h at 37°C and 5% CO2 and plaques visualized by holding dishes up to the ceiling lights. For titration of samples, plaque assays were performed in 6-well dishes. For isolation of plaques for genotyping, plaque assays were performed in 10-cm-diameter dishes with a limited number of dilutions.
Ferret experiments.
Animals were housed and experiments were performed in strict compliance with European guidelines (EU Directive on Animal Testing 86/609/EEC) and Dutch legislation (Experiments on Animals Act, 1997). Influenza virus- and Aleutian disease virus-seronegative 6-month-old female ferrets (Mustella putorius furo) were obtained from a commercial breeder. All experiments with ferrets were performed under animal biosafety level 3 conditions in class 3 isolator cages.
(i) Dual intranasal inoculations.
Four ferrets were inoculated intranasally with 1 × 105 TCID50 of NL/09wt virus and 1 × 105 TCID50 of NL/09var virus in a 40-μl volume, with 20 μl instilled in each nostril (ferrets F1, F2, F3, and F4). Similarly, four ferrets were inoculated intranasally with 1 × 105 TCID50 of Pan/99 virus and 1 × 105 TCID50 of Pan/99var virus in a 40-μl volume, with 20 μl instilled in each nostril (ferrets F5, F6, F7, and F8). Nose swabs were collected daily for 7 days and were stored at −80°C in transport medium (Hanks' balanced salt solution containing 0.5% lactalbumin [Sigma], 10% glycerol [Sigma], 200 U/ml P, 200 mg/ml S, 100 U/ml polymyxin B sulfate [Sigma], and 250 mg/ml gentamicin [Gibco]).
(ii) Intranasal/intratracheal inoculations.
Two ferrets were inoculated with NL/09wt virus intranasally and with NL/09var virus intratracheally (ferrets F9 and F10). An additional two ferrets were inoculated with the reverse placement of viruses: with NL/09var virus intranasally and with NL/09wt virus intratracheally (ferrets F11 and F12). Inoculations were performed in the same way for the Pan/99-based viruses. Specifically, two ferrets were inoculated intranasally with 1 × 105 TCID50 of Pan/99wt virus (20 μl in each nostril) and intratracheally with 1 ml containing 105 TCID50 of Pan/99var virus (ferrets F15 and F16). An additional two ferrets were inoculated with the reverse placement of viruses: with 1 × 105 TCID50 of Pan/99var virus intranasally (20 μl in each nostril) and with 1 ml containing 1 × 105 TCID50 of Pan/99wt virus intratracheally (ferrets F13 and F14). Throat and nose swabs were collected daily for 7 days and were stored at −80°C in transport medium.
In order to demonstrate replication in the lower respiratory tract of the virus that was inoculated intratracheally, ferrets were euthanized at day 3 (F14 and F15), at day 4 (F16), or at day 5 (F13). Nasal turbinates (NT), the upper part of the trachea (UT), the lower part of the trachea (LT), the left lung (LL), and the right lung (RL) were collected, homogenized in transport medium using a FastPrep system (MP Biomedicals) with 2 one-quarter-inch ceramic sphere balls, centrifuged at 1,500 × g for 10 min, aliquoted, and stored at −80°C for endpoint titration in MDCK cells and next-generation sequencing.
Genotyping of viral isolates.
Virus genotypes were determined by HRM analysis essentially as described previously (12, 53), with minor modifications as noted here. Plaque isolates were obtained by plaque assay of ferret swab samples. RNA was extracted from agar plugs using a Zymo Research ZR-96 viral RNA kit, with the following modification to the manufacturer's protocol: 40 μl water was used for the elution step. Twelve microliters of RNA was reverse transcribed using Maxima reverse transcriptase (Fermentas) according to the manufacturer's instructions. cDNA was used as the template in qPCRs with the appropriate primers (reference 54 for NL/09 primers; reference 55 for Pan/99 primers) and Precision Melt Supermix (Bio-Rad) in wells of a white, thin-wall, 384-well plate (Bio-Rad). qPCR and melt analyses were carried out in a CFX384 real-time PCR detection system per the instructions provided with Precision Melt Supermix. Data were analyzed using Precision Melt Analysis software (Bio-Rad). Viruses were scored as reassortant if the genome comprised a mixture of wt and var gene segments. Infrequently, unclear results were obtained for one or more gene segments. Isolates with one unclear segment were genotyped based on the remaining seven segments; isolates with two or more unclear segments were discarded from the analysis.
Next-generation sequencing.
Viral RNA was extracted from homogenized organs of ferrets using a High Pure RNA isolation kit (Roche). RNA was subjected to reverse transcription using Superscript III (Invitrogen) and the following primer: AGCRAAAGCAGG. Amplicons from the Pan/99 PB2 genes were generated by PCR from the cDNA using the following primers: CATAGTAGTGCAGAAATGGTTCCGGAGAGA (F) and CATAGTAGTGTTCGGCGTATCTTGACTTGA (R) (amplicon size of 239 nucleotides). Amplicons from the NL/09 PB2 genes were generated using CGCACTCAGAATGAAGTGGA (F) and GCCGAAGGTACCATGTTTCA (R) primers, producing a fragment of 265 nucleotides. These fragments were sequenced using a Roche 454 GS Junior sequencing platform. The fragment library was created for each sample according to the manufacturer's protocol (GS FLX Titanium Rapid Library Preparation; Roche) without DNA fragmentation. The emulsion PCR (Amplification Method Lib-L) and the GS Junior sequencing run were performed according to instructions of the manufacturer (Roche). Sequence reads from the GS Junior sequencing data were sorted by barcode and aligned to reference sequences using CLC Genomics software 4.6.1. The sequence reads were trimmed at 30 nucleotides from the 3′ and 5′ ends to remove all primer sequences. For quality control, sequence reads were trimmed for Phred scores of less than 20. The threshold for the detection of single nucleotide polymorphisms was manually set at 1%.
Analysis of genotypic diversity.
The diversity of genotypes in each sample was quantified by calculating Simpson's index, given by D = sum(pi2), where pi represents the proportional abundance of each genotype (56). Simpson's index accounts for both richness (raw number of species) and evenness (variation in abundance of each) but is also sensitive to the abundance of dominant species. That last feature of the index is relevant for the present study since dominance in the context examined here usually corresponds to overrepresentation of parental genotypes, with reassortment bringing about a decay of dominant species. Because Simpson's index does not scale linearly and represents a measurement of entropy rather than of diversity per se, each sample's Simpson index value was converted to a corresponding Hill number to derive its effective diversity, N2 = 1/D (57), which is defined as the number of equally abundant species required to generate the observed diversity in a sample community. Because it is linear, Hill's N2 allows a more intuitive comparison between communities (i.e., a community with N2 = 10 species is twice as diverse as one with N2 = 5) and is suitable for statistical analysis by basic linear regression methods (58).
ACKNOWLEDGMENTS
We thank Ron Fouchier for helpful discussions and support and Theo Bestebroer, Pascal Lexmond, and Dennis de Meulder for excellent technical assistance.
This work was funded in part by the NIAID Centers of Excellence in Influenza Research and Surveillance (CEIRS), contract numbers HHSN272201400004C (to A.C.L.) and HHSN272201400008C, and by R01 AI099000 (to A.C.L.). S.H. was funded in part by a NWO VIDI grant (contract number 91715372).
REFERENCES
- 1.Steel J, Lowen AC. 2014. Influenza A virus reassortment. Curr Top Microbiol Immunol 385:377–401. [DOI] [PubMed] [Google Scholar]
- 2.Lowen AC. 29 September 2017. Constraints, drivers, and implications of influenza A virus reassortment. Annu Rev Virol doi: 10.1146/annurev-virology-101416-041726. [DOI] [PubMed] [Google Scholar]
- 3.Holmes EC, Ghedin E, Miller N, Taylor J, Bao Y, St George K, Grenfell BT, Salzberg SL, Fraser CM, Lipman DJ, Taubenberger JK. 2005. Whole-genome analysis of human influenza A virus reveals multiple persistent lineages and reassortment among recent H3N2 viruses. PLoS Biol 3:e300. doi: 10.1371/journal.pbio.0030300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelson MI, Simonsen L, Viboud C, Miller MA, Taylor J, George KS, Griesemer SB, Ghedin E, Sengamalay NA, Spiro DJ, Volkov I, Grenfell BT, Lipman DJ, Taubenberger JK, Holmes EC. 2006. Stochastic processes are key determinants of short-term evolution in influenza a virus. PLoS Pathog 2:e125. doi: 10.1371/journal.ppat.0020125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nelson MI, Viboud C, Simonsen L, Bennett RT, Griesemer SB, St George K, Taylor J, Spiro DJ, Sengamalay NA, Ghedin E, Taubenberger JK, Holmes EC. 2008. Multiple reassortment events in the evolutionary history of H1N1 influenza A virus since 1918. PLoS Pathog 4:e1000012. doi: 10.1371/journal.ppat.1000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simonsen L, Viboud C, Grenfell BT, Dushoff J, Jennings L, Smit M, Macken C, Hata M, Gog J, Miller MA, Holmes EC. 2007. The genesis and spread of reassortment human influenza A/H3N2 viruses conferring adamantane resistance. Mol Biol Evol 24:1811–1820. doi: 10.1093/molbev/msm103. [DOI] [PubMed] [Google Scholar]
- 7.Westgeest KB, de Graaf M, Fourment M, Bestebroer TM, van Beek R, Spronken MI, de Jong JC, Rimmelzwaan GF, Russell CA, Osterhaus AD, Smith GJ, Smith DJ, Fouchier RA. 2012. Genetic evolution of the neuraminidase of influenza A (H3N2) viruses from 1968 to 2009 and its correspondence to haemagglutinin evolution. J Gen Virol 93:1996–2007. doi: 10.1099/vir.0.043059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Westgeest KB, Russell CA, Lin X, Spronken MIJ, Bestebroer TM, Bahl J, van Beek R, Skepner E, Halpin RA, de Jong JC, Rimmelzwaan GF, Osterhaus ADME, Smith DJ, Wentworth DE, Fouchier RAM, de Graaf M. 2014. Genome-wide analysis of reassortment and evolution of human influenza A(H3N2) viruses circulating between 1968 and 2011. J Virol 88:2844–2857. doi: 10.1128/JVI.02163-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scholtissek C. 1994. Source for influenza pandemics. Eur J Epidemiol 10:455–458. doi: 10.1007/BF01719674. [DOI] [PubMed] [Google Scholar]
- 10.Kilbourne ED. 2006. Influenza pandemics of the 20th century. Emerg Infect Dis 12:9–14. doi: 10.3201/eid1201.051254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sorrell EM, Schrauwen EJ, Linster M, De Graaf M, Herfst S, Fouchier RA. 2011. Predicting ‘airborne’ influenza viruses: (trans-) mission impossible? Curr Opin Virol 1:635–642. doi: 10.1016/j.coviro.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marshall N, Priyamvada L, Ende Z, Steel J, Lowen AC. 2013. Influenza virus reassortment occurs with high frequency in the absence of segment mismatch. PLoS Pathog 9:e1003421. doi: 10.1371/journal.ppat.1003421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan MC, Chan RW, Yu WC, Ho CC, Yuen KM, Fong JH, Tang LL, Lai WW, Lo AC, Chui WH, Sihoe AD, Kwong DL, Wong DS, Tsao GS, Poon LL, Guan Y, Nicholls JM, Peiris JS. 2010. Tropism and innate host responses of the 2009 pandemic H1N1 influenza virus in ex vivo and in vitro cultures of human conjunctiva and respiratory tract. Am J Pathol 176:1828–1840. doi: 10.2353/ajpath.2010.091087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forero A, Fenstermacher K, Wohlgemuth N, Nishida A, Carter V, Smith EA, Peng X, Hayes M, Francis D, Treanor J, Morrison J, Klein SL, Lane A, Katze MG, Pekosz A. 2017. Evaluation of the innate immune responses to influenza and live-attenuated influenza vaccine infection in primary differentiated human nasal epithelial cells. Vaccine 35:6112–6121. doi: 10.1016/j.vaccine.2017.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD. 2004. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc Natl Acad Sci U S A 101:4620–4624. doi: 10.1073/pnas.0308001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hers JF, Mulder J. 1961. Broad aspects of the pathology and pathogenesis of human influenza. Am Rev Respir Dis 83(Pt 2):84–97. [DOI] [PubMed] [Google Scholar]
- 17.Shieh WJ, Blau DM, Denison AM, Deleon-Carnes M, Adem P, Bhatnagar J, Sumner J, Liu L, Patel M, Batten B, Greer P, Jones T, Smith C, Bartlett J, Montague J, White E, Rollin D, Gao R, Seales C, Jost H, Metcalfe M, Goldsmith CS, Humphrey C, Schmitz A, Drew C, Paddock C, Uyeki TM, Zaki SR. 2010. 2009 pandemic influenza A (H1N1): pathology and pathogenesis of 100 fatal cases in the United States. Am J Pathol 177:166–175. doi: 10.2353/ajpath.2010.100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taubenberger JK, Morens DM. 2008. The pathology of influenza virus infections. Annu Rev Pathol 3:499–522. doi: 10.1146/annurev.pathmechdis.3.121806.154316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neuzil KM, Wright PF, Mitchel EF Jr, Griffin MR. 2000. The burden of influenza illness in children with asthma and other chronic medical conditions. J Pediatr 137:856–864. doi: 10.1067/mpd.2000.110445. [DOI] [PubMed] [Google Scholar]
- 20.Valdez R, Narayan KM, Geiss LS, Engelgau MM. 1999. Impact of diabetes mellitus on mortality associated with pneumonia and influenza among non-Hispanic black and white US adults. Am J Public Health 89:1715–1721. doi: 10.2105/AJPH.89.11.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van den Brand JM, Stittelaar KJ, van Amerongen G, Rimmelzwaan GF, Simon J, de Wit E, Munster V, Bestebroer T, Fouchier RA, Kuiken T, Osterhaus AD. 2010. Severity of pneumonia due to new H1N1 influenza virus in ferrets is intermediate between that due to seasonal H1N1 virus and highly pathogenic avian influenza H5N1 virus. J Infect Dis 201:993–999. doi: 10.1086/651132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Viasus D, Oteo Revuelta JA, Martínez-Montauti J, Carratalà J. 2012. Influenza A(H1N1)pdm09-related pneumonia and other complications. Enferm Infecc Microbiol Clin 30(Suppl 4):43–48. doi: 10.1016/S0213-005X(12)70104-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maines TR, Jayaraman A, Belser JA, Wadford DA, Pappas C, Zeng H, Gustin KM, Pearce MB, Viswanathan K, Shriver ZH, Raman R, Cox NJ, Sasisekharan R, Katz JM, Tumpey TM. 2009. Transmission and pathogenesis of swine-origin 2009 A(H1N1) influenza viruses in ferrets and mice. Science 325:484–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munster VJ, de Wit E, van den Brand JM, Herfst S, Schrauwen EJ, Bestebroer TM, van de Vijver D, Boucher CA, Koopmans M, Rimmelzwaan GF, Kuiken T, Osterhaus AD, Fouchier RA. 2009. Pathogenesis and transmission of swine-origin 2009 A(H1N1) influenza virus in ferrets. Science 325:481–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beigel JH, Farrar J, Han AM, Hayden FG, Hyer R, de Jong MD, Lochindarat S, Nguyen TK, Nguyen TH, Tran TH, Nicoll A, Touch S, Yuen KY; Writing Committee of the World Health Organization (WHO) Consultation on Human Influenza A/H5. 2005. Avian influenza A (H5N1) infection in humans. N Engl J Med 353:1374–1385. doi: 10.1056/NEJMra052211. [DOI] [PubMed] [Google Scholar]
- 26.Tran TH, Nguyen TL, Nguyen TD, Luong TS, Pham PM, Nguyen VV, Pham TS, Vo CD, Le TQ, Ngo TT, Dao BK, Le PP, Nguyen TT, Hoang TL, Cao VT, Le TG, Nguyen DT, Le HN, Nguyen KT, Le HS, Le VT, Christiane D, Tran TT, Menno de J, Schultsz C, Cheng P, Lim W, Horby P, Farrar J; World Health Organization International Avian Influenza Investigative Team. 2004. Avian influenza A (H5N1) in 10 patients in Vietnam. N Engl J Med 350:1179–1188. doi: 10.1056/NEJMoa040419. [DOI] [PubMed] [Google Scholar]
- 27.Chan MC, Chan RW, Chan LL, Mok CK, Hui KP, Fong JH, Tao KP, Poon LL, Nicholls JM, Guan Y, Peiris JM. 2013. Tropism and innate host responses of a novel avian influenza A H7N9 virus: an analysis of ex-vivo and in-vitro cultures of the human respiratory tract. Lancet Respir Med 1:534–542. doi: 10.1016/S2213-2600(13)70138-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ke C, Mok CKP, Zhu W, Zhou H, He J, Guan W, Wu J, Song W, Wang D, Liu J, Lin Q, Chu DKW, Yang L, Zhong N, Yang Z, Shu Y, Peiris JSM. 2017. Human infection with highly pathogenic avian influenza A(H7N9) virus, China. Emerg Infect Dis 23:1332–1340. doi: 10.3201/eid2308.170600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu L, Wang Z, Chen Y, Ding W, Jia H, Chan JF, To KK, Chen H, Yang Y, Liang W, Zheng S, Yao H, Yang S, Cao H, Dai X, Zhao H, Li J, Bao Q, Chen P, Hou X, Li L, Yuen KY. 2013. Clinical, virological, and histopathological manifestations of fatal human infections by avian influenza A(H7N9) virus. Clin Infect Dis 57:1449–1457. doi: 10.1093/cid/cit541. [DOI] [PubMed] [Google Scholar]
- 30.Nicholls JM, Chan MC, Chan WY, Wong HK, Cheung CY, Kwong DL, Wong MP, Chui WH, Poon LL, Tsao SW, Guan Y, Peiris JS. 2007. Tropism of avian influenza A (H5N1) in the upper and lower respiratory tract. Nat Med 13:147–149. doi: 10.1038/nm1529. [DOI] [PubMed] [Google Scholar]
- 31.van Riel D, Leijten LM, de Graaf M, Siegers JY, Short KR, Spronken MI, Schrauwen EJ, Fouchier RA, Osterhaus AD, Kuiken T. 2013. Novel avian-origin influenza A (H7N9) virus attaches to epithelium in both upper and lower respiratory tract of humans. Am J Pathol 183:1137–1143. doi: 10.1016/j.ajpath.2013.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tellier R. 2009. Aerosol transmission of influenza A virus: a review of new studies. J R Soc Interface 6(Suppl 6):S783–S790. doi: 10.1098/rsif.2009.0302.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tao H, Li L, White MC, Steel J, Lowen AC. 2015. Influenza A virus co-infection through transmission can support high levels of reassortment. J Virol 89:8453–8461. doi: 10.1128/JVI.01162-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tao H, Steel J, Lowen AC. 2014. Intrahost dynamics of influenza virus reassortment. J Virol 88:7485–7492. doi: 10.1128/JVI.00715-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bodewes R, Kreijtz JH, van Amerongen G, Fouchier RA, Osterhaus AD, Rimmelzwaan GF, Kuiken T. 2011. Pathogenesis of Influenza A/H5N1 virus infection in ferrets differs between intranasal and intratracheal routes of inoculation. Am J Pathol 179:30–36. doi: 10.1016/j.ajpath.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kreijtz JH, Kroeze EV, Stittelaar KJ, de Waal L, van Amerongen G, van Trierum S, van Run P, Bestebroer T, Kuiken T, Fouchier RA, Rimmelzwaan GF, Osterhaus AD. 28 June 2013. Low pathogenic avian influenza A(H7N9) virus causes high mortality in ferrets upon intratracheal challenge: a model to study intervention strategies. Vaccine doi: 10.1016/j.vaccine.2013.06.071. [DOI] [PubMed] [Google Scholar]
- 37.Brooke CB, Ince WL, Wei J, Bennink JR, Yewdell JW. 2014. Influenza A virus nucleoprotein selectively decreases neuraminidase gene-segment packaging while enhancing viral fitness and transmissibility. Proc Natl Acad Sci U S A 111:16854–16859. doi: 10.1073/pnas.1415396111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang IC, Li W, Sui J, Marasco W, Choe H, Farzan M. 2008. Influenza A virus neuraminidase limits viral superinfection. J Virol 82:4834–4843. doi: 10.1128/JVI.00079-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Riel D, Leijten LM, Kochs G, Osterhaus A, Kuiken T. 2013. Decrease of virus receptors during highly pathogenic H5N1 virus infection in humans and other mammals. Am J Pathol 183:1382–1389. doi: 10.1016/j.ajpath.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 40.Bodewes R, Kreijtz JH, van Amerongen G, Hillaire ML, Vogelzang-van Trierum SE, Nieuwkoop NJ, van Run P, Kuiken T, Fouchier RA, Osterhaus AD, Rimmelzwaan GF. 2013. Infection of the upper respiratory tract with seasonal influenza A(H3N2) virus induces protective immunity in ferrets against infection with A(H1N1)pdm09 virus after intranasal, but not intratracheal, inoculation. J Virol 87:4293–4301. doi: 10.1128/JVI.02536-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van den Brand JM, Kreijtz JH, Bodewes R, Stittelaar KJ, van Amerongen G, Kuiken T, Simon J, Fouchier RA, Del Giudice G, Rappuoli R, Rimmelzwaan GF, Osterhaus AD. 2011. Efficacy of vaccination with different combinations of MF59-adjuvanted and nonadjuvanted seasonal and pandemic influenza vaccines against pandemic H1N1 (2009) influenza virus infection in ferrets. J Virol 85:2851–2858. doi: 10.1128/JVI.01939-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Riel D, den Bakker MA, Leijten LM, Chutinimitkul S, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RA, Osterhaus AD, Kuiken T. 2010. Seasonal and pandemic human influenza viruses attach better to human upper respiratory tract epithelium than avian influenza viruses. Am J Pathol 176:1614–1618. doi: 10.2353/ajpath.2010.090949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Riel D, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RA, Osterhaus AD, Kuiken T. 2007. Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am J Pathol 171:1215–1223. doi: 10.2353/ajpath.2007.070248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murakami Y, Nerome K, Yoshioka Y, Mizuno S, Oya A. 1988. Difference in growth behavior of human, swine, equine, and avian influenza viruses at a high temperature. Arch Virol 100:231–244. doi: 10.1007/BF01487686. [DOI] [PubMed] [Google Scholar]
- 45.Lang V, Marjuki H, Krauss SL, Webby RJ, Webster RG. 2011. Different incubation temperatures affect viral polymerase activity and yields of low-pathogenic avian influenza viruses in embryonated chicken eggs. Arch Virol 156:987–994. doi: 10.1007/s00705-011-0933-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. 2006. Avian flu: influenza virus receptors in the human airway. Nature 440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 47.van Riel D, Leijten LM, Verdijk RM, GeurtsvanKessel C, van der Vries E, van Rossum AM, Osterhaus AD, Kuiken T. 2014. Evidence for influenza virus CNS invasion along the olfactory route in an immunocompromised infant. J Infect Dis 210:419–423. doi: 10.1093/infdis/jiu097. [DOI] [PubMed] [Google Scholar]
- 48.van den Brand JM, Haagmans BL, van Riel D, Osterhaus AD, Kuiken T. 2014. The pathology and pathogenesis of experimental severe acute respiratory syndrome and influenza in animal models. J Comp Pathol 151:83–112. doi: 10.1016/j.jcpa.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miguel AF. 2017. Penetration of inhaled aerosols in the bronchial tree. Med Eng Phys 44:25–31. doi: 10.1016/j.medengphy.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 50.Fodor E, Devenish L, Engelhardt OG, Palese P, Brownlee GG, Garcia-Sastre A. 1999. Rescue of influenza A virus from recombinant DNA. J Virol 73:9679–9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steel J, Lowen AC, Mubareka S, Palese P. 2009. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog 5:e1000252. doi: 10.1371/journal.ppat.1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chutinimitkul S, Herfst S, Steel J, Lowen AC, Ye J, van Riel D, Schrauwen EJA, Bestebroer TM, Koel B, Burke DF, Sutherland-Cash KH, Whittleston CS, Russell CA, Wales DJ, Smith DJ, Jonges M, Meijer A, Koopmans M, Rimmelzwaan GF, Kuiken T, Osterhaus ADME, García-Sastre A, Perez DR, Fouchier RAM. 2010. Virulence-associated substitution D222G in the hemagglutinin of 2009 pandemic influenza A(H1N1) virus affects receptor binding. J Virol 84:11802–11813. doi: 10.1128/JVI.01136-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tao H, Steel J, Lowen AC. 2014. Intrahost dynamics of influenza virus reassortment. J Virol 88:7485–7492. doi: 10.1128/JVI.00715-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Phipps KL, Marshall N, Tao H, Danzy S, Onuoha N, Steel J, Lowen AC. 10 August 2017. Seasonal H3N2 and 2009 pandemic H1N1 influenza A viruses reassort efficiently but produce attenuated progeny. J Virol doi: 10.1128/JVI.00830-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.White MC, Steel J, Lowen AC. 12 May 2017. Heterologous packaging signals on segment 4, but not segment 6 or segment 8, limit influenza A virus reassortment. J Virol doi: 10.1128/JVI.00195-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simpson EH. 1949. Measurement of diversity. Nature 163:688–688. doi: 10.1038/163688a0. [DOI] [Google Scholar]
- 57.Hill MO. 1973. Diversity and evenness: a unifying notation and its consequences. Ecology 54:427–432. doi: 10.2307/1934352. [DOI] [Google Scholar]
- 58.Jost L. 2006. Entropy and diversity. Oikos 113:363–375. doi: 10.1111/j.2006.0030-1299.14714.x. [DOI] [Google Scholar]