ABSTRACT
One of the first defenses against infecting pathogens is the innate immune system activated by cellular recognition of pathogen-associated molecular patterns (PAMPs). Although virus-derived RNA species, especially copyback (cb)-type defective interfering (DI) genomes, have been shown to serve as real PAMPs, which strongly induce interferon-beta (IFN-β) during mononegavirus infection, the mechanisms underlying DI generation remain unclear. Here, for the first time, we identified a single amino acid substitution causing production of cbDI genomes by successful isolation of two distinct types of viral clones with cbDI-producing and cbDI-nonproducing phenotypes from the stock Sendai virus (SeV) strain Cantell, which has been widely used in a number of studies on antiviral innate immunity as a representative IFN-β-inducing virus. IFN-β induction was totally dependent on the presence of a significant amount of cbDI genome-containing viral particles (DI particles) in the viral stock, but not on deficiency of the IFN-antagonistic viral accessory proteins C and V. Comparison of the isolates indicated that a single amino acid substitution found within the N protein of the cbDI-producing clone was enough to cause the emergence of DI genomes. The mutated N protein of the cbDI-producing clone resulted in a lower density of nucleocapsids than that of the DI-nonproducing clone, probably causing both production of the DI genomes and their formation of a stem-loop structure, which serves as an ideal ligand for RIG-I. These results suggested that the integrity of mononegaviral nucleocapsids might be a critical factor in avoiding the undesirable recognition of infection by host cells.
IMPORTANCE The type I interferon (IFN) system is a pivotal defense against infecting RNA viruses that is activated by sensing viral RNA species. RIG-I is a major sensor for infection with most mononegaviruses, and copyback (cb)-type defective interfering (DI) genomes have been shown to serve as strong RIG-I ligands in real infections. However, the mechanism underlying production of cbDI genomes remains unclear, although DI genomes emerge as the result of an error during viral replication with high doses of viruses. Sendai virus has been extensively studied and is unique in that its interaction with innate immunity reveals opposing characteristics, such as high-level IFN-β induction and strong inhibition of type I IFN pathways. Our findings provide novel insights into the mechanism of production of mononegaviral cbDI genomes, as well as virus-host interactions during innate immunity.
KEYWORDS: Sendai virus, defective interfering genome, innate immunity, interferons, nucleocapsid, paramyxovirus
INTRODUCTION
Host innate immunity is a critical defense mechanism that detects and responds rapidly to incoming pathogens. Host cells sense invading pathogens by recognizing their pathogen-associated molecular patterns (1). As for RNA viruses, structural characteristics of virus-derived RNA that differentiate it from cellular RNA are detected as pathogen-associated molecular patterns by nonself RNA sensors, such as Toll-like receptors and a family of cytosolic RNA helicases termed RIG-I-like receptors, including retinoic-acid-inducible gene I (RIG-I), melanoma differentiation-associated gene 5 (MDA5), and laboratory of genetics and physiology gene 2 (LGP2), followed by the subsequent induction of interferon-beta (IFN-β) (2–4). Autocrine or paracrine IFNs bind to IFN receptors on the cell surface, leading to the expression of hundreds of IFN-stimulated genes through the Jak/STAT signaling pathway, which ultimately exerts various antiviral effects (1). Among the RIG-I-like receptors, RIG-I has been shown to be a critical sensor to detect infection of many RNA viruses (5–7). Biochemical studies have demonstrated that a structure with 5′-triphosphate (5′-ppp), blunt-ended, double-stranded RNA (dsRNA) serves as an ideal ligand for RIG-I (8–12). Recently, the real ligands, such as mRNA, dsRNA, and 5′-ppp RNA, including leader, trailer, genome, and antigenome RNA, as well as defective interfering (DI) genomes, produced during the course of RNA viral replication, have been revealed by a combination of cross-linking between RIG-I and RNA derived from the infected cells and deep-sequencing analysis of the cross-linked RNA (13–18). Among these, copyback (cb)-type and internal-deletion (id)-type DI genomes from mononegaviruses and orthomyxoviruses, respectively, have been shown to be strong ligands of RIG-I and represent the ideal ligand structure (14, 17–19).
DI genomes are spontaneously produced owing to errors during mononegaviral replication and are released from cells packaged within an infectious form of virus-like particles (DI particles) (20, 21). Experimentally, the cbDI genomes can be easily generated by infection at high multiplicity (20, 22). The two types of DI genomes reported, id- and cb-type DI genomes, have distinct promoter orientations (21, 23). Specifically, idDI genomes retain the authentic genomic and antigenomic promoters at the ends, like intact viral genomes, whereas cbDI genomes are unique in that the 3′-genomic promoter is replaced by a complementary sequence of the 5′-antigenomic promoter. As a result of this unique structure, SDS-treated naked cbDI genomes easily form a 5′-ppp, blunt-ended, dsRNA stem-loop structure, which constitutes an ideal RIG-I ligand (21). Although template switching from the antigenome to the nascent genome strand is required for the generation of cbDI genomes during the synthesis of genomic RNAs and the switching points seem to be limited, the precise mechanisms for synthesis of the cbDI genomes during viral RNA replication, including how the switching occurs between the tightly encapsidated genomes, remain to be determined. Once generated, the cbDI genomes accumulate rapidly in the viral stocks through consecutive passages owing to their replicative advantage over intact genomes, which is caused by their short length (major species are less than 1,500 nucleotides [nt] in length) and dual antigenomic promoters that are stronger than the genomic promoter.
The role of DI genomes in natural viral infections has also not been addressed. Classically, DI genomes have been associated with the establishment of persistent mononegavirus infections (24–34). Under experimental conditions, intercalating waves of authentic and DI genomes were observed during a long-term persistent infection. The accumulation of too many DI genomes interferes with authentic genome replication, resulting in a decrease in viral replication. This, in turn, leads to a reduction in DI genomes, because replication of the DI genomes requires intact viral replication and replication of intact viral genomes dominates over replication of DI genomes. Repetition of such waves is thought to allow long-term persistent viral infection (35).
Sendai virus (SeV), a representative member of the family Paramyxoviridae, has been extensively studied in terms of virus-host interactions during innate immunity and has been shown to display opposing functions, both stimulating and inhibiting the host type I IFN system (36). One SeV strain, Cantell (CNT), is recognized for its strong stimulation of RIG-I-mediated IFN-β induction, whereas most of the other strains, such as Z, Fushimi (FSM), Nagoya (NGY), and Hamamatsu (HMT), have contributed to elucidating the counteraction function. Recently, the cbDI genome was identified as a major IFN-β-inducing factor in SeV strain CNT stocks (18).
In this study, we investigated factors affecting IFN-β inducibility and demonstrated that the presence of cbDI genomes in viral stocks strongly induced IFN-β while retaining functions antagonistic to the IFN system. To address why cbDI genomes accumulate in viral stocks of strain CNT but not in other viral stocks to such a high level, we successfully isolated for the first time a viral clone from the CNT stock that constitutively generates cbDI genomes. Further functional analysis of this clone demonstrated that a single spontaneous amino acid alteration within the N protein was responsible for cbDI production. A possible mechanism of cbDI production and the effect of generation of cbDI genomes on viral pathogenesis in mice are discussed.
RESULTS
The presence of DI particles is an exclusive determinant of IFN-β induction by the SeV stock.
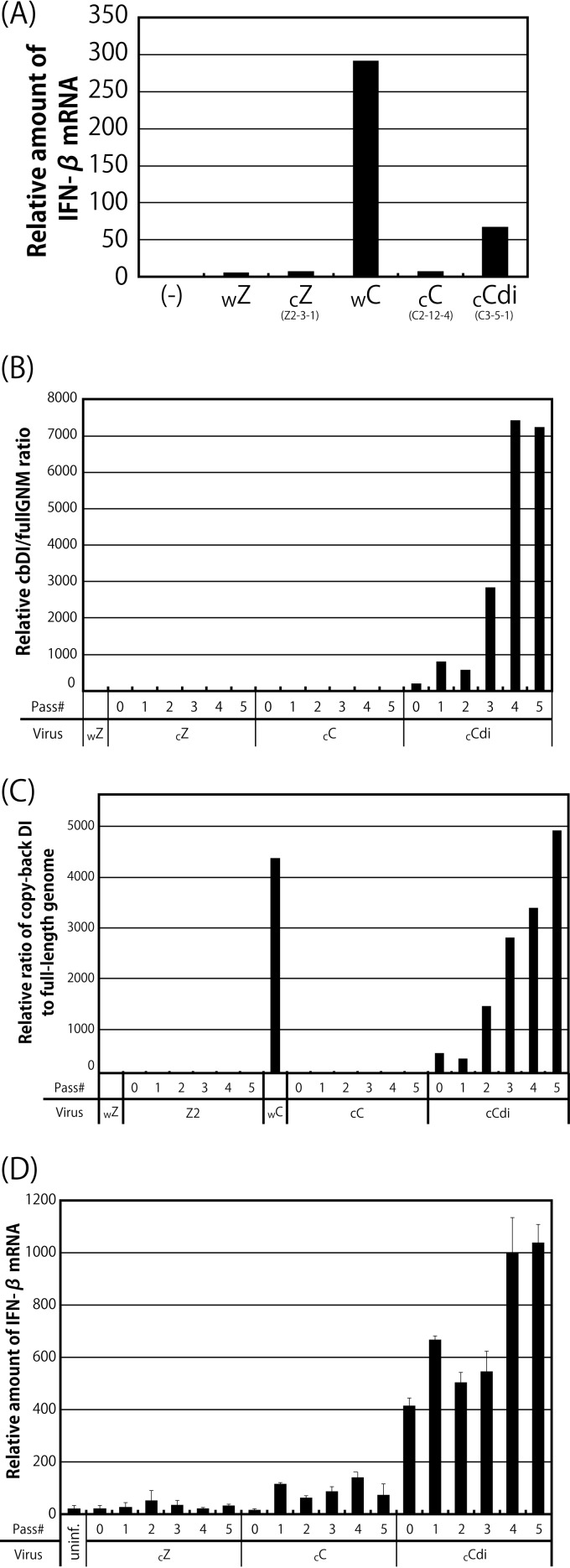
It has been shown that cbDI genomes of paramyxoviruses, such as SeV and parainfluenza virus type 5 (PIV5), serve as strong ligands for RIG-I and that viral stocks rich in DI particles, including cbDI genomes, are able to readily induce a high level of IFN-β production (18, 22, 37). Indeed, comparisons of our working stocks of SeV strains Z (wZ), CNT (wC), FSM, HMT, and NGY confirmed that their inducibility of IFN-β mRNA correlated well with the presence of cbDI particles (Fig. 1A). Ratios of the number of cbDI genomes to that of genome length viral RNA (fullGNM), including both the genome and antigenome, in the wZ and wC stocks were remarkably different, indicating that the wC stock contains cbDI genome-containing particles (DI particles) at an approximately 25,000-fold higher concentration than the wZ stock. Such a large content of cbDI genomes was also detected in the commercial stock of CNT (oriC), which had a concentration approximately 27,000-fold higher than that in the wZ stock (see Fig. 3B). The cbDI/fullGNM ratios in later experiments are presented based on that in the wZ stock.
FIG 1.
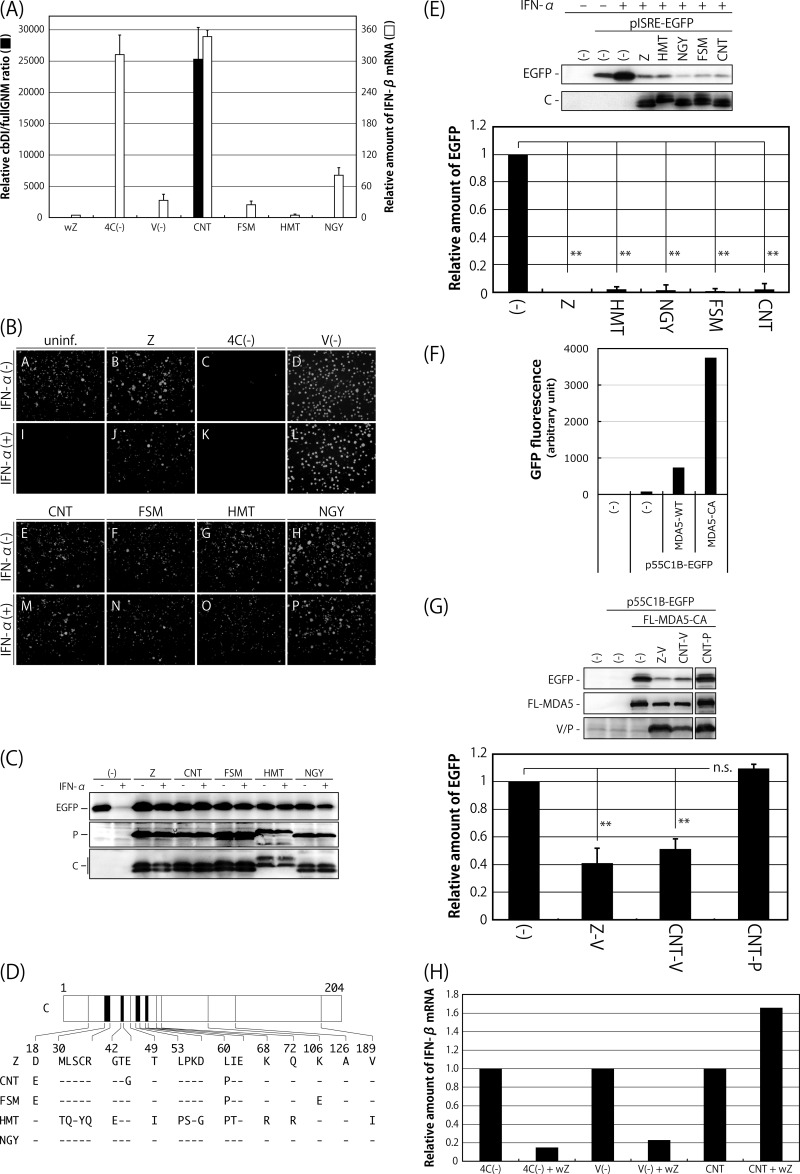
Comparison of SeV strains in terms of the content of cbDI particles in their working stocks, induction of IFN-β and the antiviral state, and counteraction against the type I IFN pathway. (A) The amounts of cbDI genomes and fullGNM in the working stocks of the indicated viruses were analyzed by one-step qRT-PCR. The cbDI/fullGNM ratios are shown. The ratio of the wZ sample was set to 1. The amounts of IFN-β and beta-actin mRNAs in the infected HeLa cells were also analyzed by one-step qRT-PCR in cells infected with the indicated viruses. The ratios of IFN-β to beta-actin mRNAs are shown. The ratio in the uninfected sample was set to 1. (B) HeLa cells were infected with the indicated viruses. At 6 h p.i., the media were replaced with fresh media containing IFN-α (1,000 IU/ml) or no IFN-α. After an additional 6-h incubation, the cells were superinfected with rVSV-GFP. After further incubation for 6 h, GFP expression derived from rVSV-GFP replication was observed under a fluorescence microscope. (C) Western blotting of the cell lysate samples in panel B using anti-GFP, -SeV P, and -SeV C polyclonal antibodies. (D) Schematic representation of the C proteins of the indicated SeV strains. Dashes indicate that the amino acids are identical to those of Z strain. (E) 293T cells were cotransfected with the C proteins derived from the indicated SeV strains, together with a reporter plasmid, pISRE-EGFP. At 18 h p.t., the cells were treated with IFN-α (1,000 IU/ml) for 8 h, and then the expression level of EGFP was analyzed by Western blotting using an anti-GFP antibody. The amount of EGFP in the cell lysates was quantitated and graphed. The value of mock-transfected and non-IFN-α-treated samples was set to 1. (F) 293T cells were cotransfected with MDA5-WT or -CA, together with a reporter plasmid, p55C1B-EGFP. At 24 h p.t., GFP fluorescence was measured using a fluorometer. (G) 293T cells were cotransfected with the V or P proteins derived from the indicated SeV strains, together with a reporter plasmid, p55C1B-EGFP, and pCAG-FL-MDA5-CA. At 24 h p.t., the expression level of EGFP was analyzed by Western blotting using an anti-GFP antibody. The amounts of EGFP in the cell lysates were quantitated and graphed. The value of the sample receiving FL-MDA5-CA but no V protein was set to 1. (H) HeLa cells were infected with SeV strain Z at an MOI of 5. At 6 h p.i., the cells were superinfected with strain Cantell, Z-4C(−), or Z-V(−). After an additional 24 h of incubation, the amounts of IFN-β and beta-actin mRNAs were analyzed by one-step qRT-PCR. The ratios of IFN-β to beta-actin mRNAs are shown. The ratio in the cells without superinfection was set to 1. All of the bar graphs represent the averages of three independent experiments, and the error bars represent the standard deviations. n.s., nonsignificant (P > 0.05); **, P < 0.01 by one-way analysis of variance (ANOVA) with Bonferroni post hoc test.
FIG 3.
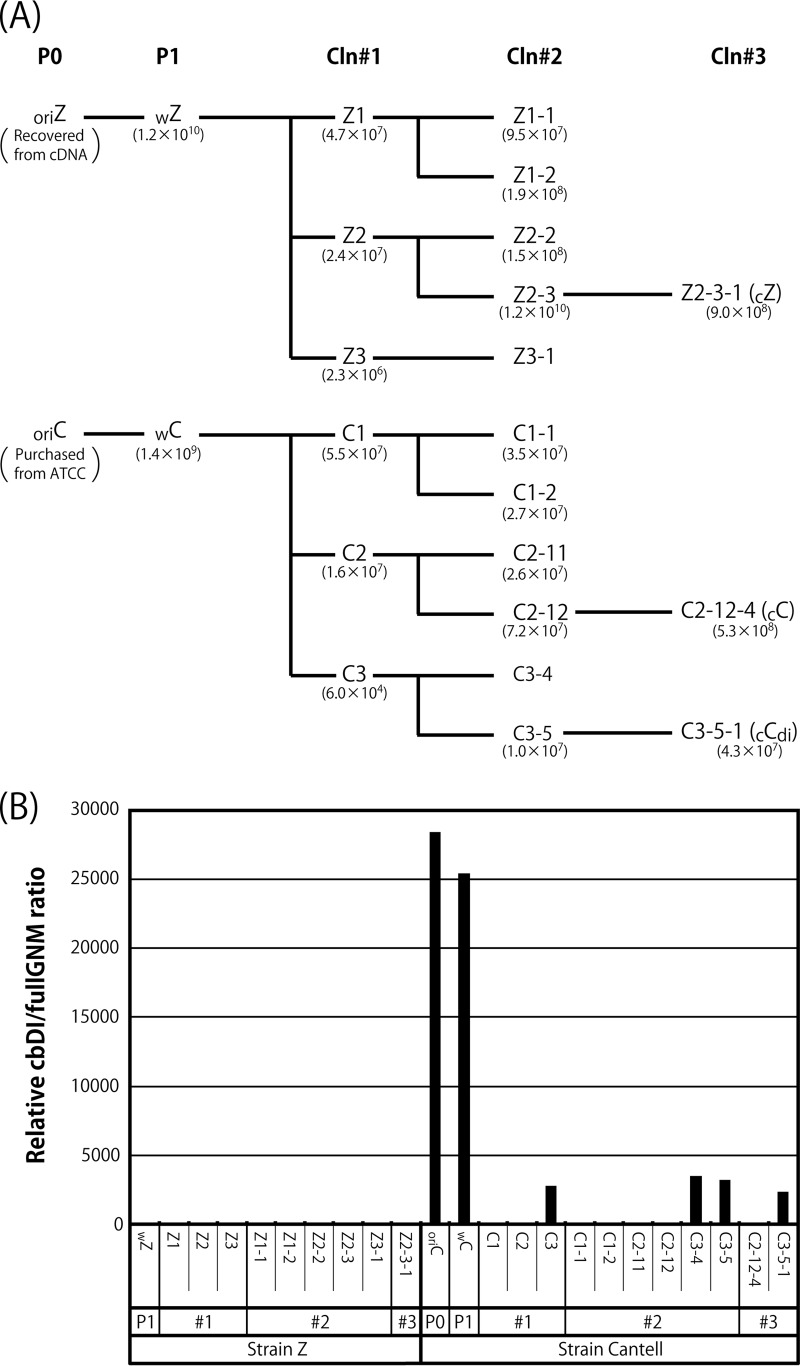
Characterization of viral clones (Cln) isolated from the wZ and wC stocks. (A) The stocks of wZ and wC were applied to three consecutive limiting dilutions as described in Materials and Methods. Representative viral samples are shown with the viral titers. (B) The relative cbDI/fullGNM ratios of the clones, as well as the parental viral stocks, are shown as in Fig. 1A. The ratio of the wZ sample was set to 1.
It has been demonstrated that SeV can effectively avoid the establishment of host antiviral states by antagonizing the host IFN system in a variety of ways (38–47). The considerable difference in IFN-β inducibility between SeV strain CNT and the other strains might be caused by their different antagonistic abilities. To elucidate this possibility, we first examined whether infection with the SeV strains themselves would lead to establishment of the antiviral state that happens after induction of IFN-β and would inhibit establishment of the antiviral state triggered by IFN-α treatment (Fig. 1B and C). In addition to the SeV strains, Z-derived IFN-β-inducing SeV recombinants Z-4C(−) and Z-V(−), lacking expression of four C and V proteins, respectively, were also tested as reference viruses. HeLa cells infected with the indicated viruses were treated or not with IFN-α and then superinfected with a recombinant vesicular stomatitis virus expressing green fluorescent protein (rVSV-GFP). Expression of GFP by replication of rVSV-GFP in the cells was examined by fluorescence microscopy (Fig. 1B) and Western blotting (Fig. 1C). GFP fluorescence was detected in uninfected cells without IFN-α treatment (Fig. 1B, A) but not in treated cells (Fig. 1B, I), because replication of rVSV-GFP was not permitted under the antiviral state induced by the treatment. GFP fluorescence was not detected in cells infected with Z-4C(−), which had lost the ability to inhibit the IFN-responding pathway because they lacked all four C proteins, regardless of IFN-α treatment (Fig. 1B, C and K), indicating that infection with Z-4C(−) itself is able to induce the antiviral state in the cells, as reported previously (43). However, preinfection of all of the other viruses tested failed to prevent replication of rVSV-GFP even in the case of IFN-α treatment, indicating that all of these viruses are able to inhibit induction of the antiviral state regardless of their varied potentials to induce IFN-β (Fig. 1B, B, D to H, J, and L to P).
The antagonism of SeV to the host IFN system has been well characterized as being exerted by the viral accessory proteins C and V (38–47). We next examined the antagonizing abilities of the C and V proteins derived from the SeV strains. The SeV C protein has been shown to inhibit induction of the antiviral state through a physical interaction with STAT1, a component of the type I IFN-responding host Jak/STAT pathway (39–41, 48). The C genes of the SeV strains were cloned, and their ability to inhibit IFN signaling was examined using an ISRE (interferon sequence response element)-driven reporter assay, as reported previously (44) (Fig. 1D and E). The amino acid sequence of the C protein of strain NGY was the same as that of strain Z, but strains CNT, FSM, and HMT differed by 3, 3, and 14 amino acids, respectively (Fig. 1D). 293T cells were cotransfected with the C and reporter pISRE-EGFP plasmids and were then treated with IFN-α. IFN-α treatment induced expression of enhanced green fluorescent protein (EGFP) in the samples that did not receive the C plasmid. However, expression of EGFP was significantly inhibited by the presence of C protein in strain Z, as well as all of the other strains (Fig. 1E). Taken together with the results shown in Fig. 1B and C, this indicated that the C proteins of all of the SeV strains were fully functional in their ability to antagonize IFN signaling triggered by IFN-α treatment.
The abilities of the V proteins of strains Z and CNT to inhibit IFN-β induction were also examined using an IRF3-driven reporter assay, as reported previously (45) (Fig. 1F and G). The SeV V protein has been reported to inhibit the induction through physical interactions with MDA5 and IRF3 (45–47). 293T cells were cotransfected with V proteins, together with FLAG-tagged wild-type MDA5 (MDA5-WT) or constitutively active MDA5 (MDA5-CA) and reporter p55C1B-EGFP plasmids. Overexpression of MDA5-CA induced production of IFN-β in the samples that did not receive V plasmid at a remarkably higher level than that of MDA5-WT (Fig. 1F). Expression of GFP induced by MDA5-CA was specifically inhibited by the V proteins from both strains Z and CNT, but not by the P protein from strain CNT (Fig. 1G), indicating that the V proteins of both strains retained the ability to inhibit IRF3-driven gene expression mediated by MDA5.
We further examined whether preinfection with strain Z would reduce IFN-β induction by strain CNT, as well as the Z-4C(−) and Z-V(−) recombinants (Fig. 1H). The levels of IFN-β mRNA induced by Z-4C(−) and Z-V(−) were reduced approximately 5-fold by preinfection with strain Z, whereas that induced by strain CNT was slightly enhanced. This result confirmed that the high levels of IFN-β induction by the Z-4C(−) and Z-V(−) viruses were caused by loss of function of the C and V proteins but that this was not the case for infection with strain CNT.
Taken together, the considerably higher level of IFN-β induction exhibited by strain CNT compared with strain Z appears to be caused almost exclusively by the extensive containment of cbDI particles but not by the loss of function of the C and V proteins in strain CNT antagonizing the host IFN system.
A major type of cbDI genome in the CNT stock displays a typical H4-like structure.
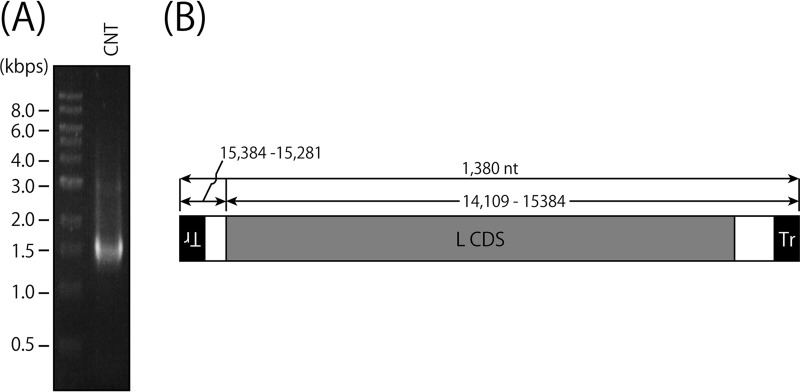
Different types of cbDI genomes have been reported to be produced during SeV replication, and two of the major types are 1,410 (H4) and 546 nucleotides in length (18, 49). To identify the nucleotide sequences of the cbDI genomes contained in the CNT stock, we tried to amplify the cbDI genomes by reverse transcription (RT)-PCR using a single primer complementary to the 3′ end of the CNT antigenome, as described in Materials and Methods (Fig. 2). Agarose gel electrophoresis of the RT-PCR product showed a major band of around 1.4 kb and some other minor bands (Fig. 2A). Nucleotide sequencing revealed that a major type of cbDI genome in the stock had a typical cb-type structure and was 1,380 nucleotides in length, which was similar to a previously reported SeV cbDI genome, H4 (Fig. 2B).
FIG 2.
Identification of cbDI genomes in the wC stock. (A) RT-PCR was performed using a virion RNA sample from the wC stock as a template and Tr80CNT as a primer. The RT-PCR product was analyzed by agarose gel electrophoresis. (B) Schematic representation of the major species of the H4-type cbDI genome.
The CNT stock contains cbDI-producing, as well as cbDI-nonproducing, viral species.
It has been reported that the content of cbDI particles in the CNT stock can be dramatically reduced by propagation at a higher dilution (37, 50). In this study, to remove the DI genomes from the viral stocks, the working stocks of strains CNT (wC) and Z (wZ) were subjected to three consecutive limiting dilutions to obtain cloned viruses, as described in Materials and Methods. The representative cloning history is shown in Fig. 3A. The cbDI/fullGNM ratios for all of the viral samples listed in Fig. 3A were examined in comparison with that of wZ (Fig. 3B). For strain Z, the cbDI genomes were reduced to an undetectable level even after the initial cloning step. Meanwhile, for strain CNT, two types of viruses were obtained after the initial cloning step. The cbDI genomes were not detected in the C1 and C2 clones, like the Z clones, but were still detected in the C3 clone, where the cbDI/fullGNM ratio was reduced approximately 10-fold compared with that in the wC stock. Surprisingly, a further two consecutive limiting dilutions were not able to eliminate or even reduce further the presence of cbDI genomes, as observed for the C3-4, C3-5, and C3-5-1 samples. These results indicated that the wC stock contains at least two distinct types of viruses, namely, cbDI-producing and cbDI-nonproducing viruses. We decided to use clones Z2-3-1 (cZ), C-2-12-4 (cC), and C3-5-1 (cCdi) as representatives of the cbDI-nonproducing Z clone and the cbDI-nonproducing and -producing CNT clones, respectively, in subsequent experiments.
The presence of cbDI-inducing virus is a potential cause of the extensive accumulation of IFN-β-inducing cbDI particles in CNT stocks.
The IFN-β inducibility of viral clones was examined using HeLa cells (Fig. 4A), as shown in Fig. 1A. The cbDI-nonproducing clones, such as cZ and cC, induced IFN-β mRNA at a low level, similar to wZ. In contrast, the cbDI-producing clone cCdi induced IFN-β mRNA at a level approximately 70-fold higher than that induced by wZ, although the level was approximately 4.5-fold lower than that observed for wC, reflecting the 10-fold-reduced content of cbDI genomes in cCdi compared with that in wC.
FIG 4.
Effects of serial passage of the viral clones on the cbDI genome content and IFN-β inducibility. (A) Ratios of IFN-β to beta-actin mRNA in cells infected with the indicated viruses. The ratio in the uninfected sample was set to 1. (B and C) The indicated clones were passaged through Vero-TMPRSS2 cells at an MOI of 10 (B) and a volume of 200 μl (C) five times. The cbDI/fullGNM ratio of each sample is shown as in Fig. 1A. The ratio in the wZ stock was set to 1. (D) Ratios of IFN-β to beta-actin mRNAs shown as in Fig. 1A. The ratio in the uninfected sample was set to 1.
It has been reported that DI genomes in viral samples are readily accumulated by serial passage of mononegaviruses at high multiplicity (20, 22). We examined the effect of serial passage of the clones at a constant multiplicity of infection (MOI) of 10 (Fig. 4B) and a constant volume of 200 μl (Fig. 4C) on accumulation of the cbDI particles in the viral samples and their IFN-β inducibility (Fig. 4D). Virtually no increase in the content of cbDI genomes or the inducibility of IFN-β was observed even after five consecutive passages of the cbDI-nonproducing clones, cZ and cC. In contrast, serial passage of the cbDI-inducing clone, cCdi, resulted in rapid accumulation of cbDI genomes and a rapid increase in IFN-β inducibility coupled with this accumulation.
These results strongly supported the idea that one of the CNT virus clones, cCdi, is more likely to produce cbDI genomes during viral replication than the other CNT clone, cC, as well as the Z clone, cZ, confirming that the presence of cbDI particles is an exclusive determinant of high IFN-β inducibility in the CNT stocks. The presence of such cbDI-producing viruses seems to cause rapid and extensive accumulation of cbDI particles in the CNT stocks.
A single amino acid difference within the N protein is responsible for the differences between CNT clones in the ability to generate cbDI genomes.
To identify amino acids and/or nucleotides responsible for the different levels of generation of cbDI genomes between the CNT clones cC and cCdi, the nucleotide sequences of the complete viral genomes of the CNT clones (cC and cCdi), as well as that of Z (cZ), were determined and compared (Table 1). Compared with the nucleotide sequence of the cDNA clone of strain Z (Z-cDNA) used for the preparation of wZ, cZ showed 6 nucleotide differences (2, 1, and 3 in the M, F, and L genes, respectively) and two amino acid differences (one each in the F and L proteins). There were a total of 121 and 118 nucleotide and 48 and 47 amino acid differences within the genomes of cC and cCdi, respectively, compared with that of Z-cDNA. Among them, T122A, A329E, and E1036K in N, HN, and L of cC and D153Y and I142N in N and HN of cCdi, respectively, were unique amino acid differences found between cC and cCdi.
TABLE 1.
Nucleotide substitutions between the genomes of the clones and fullGNM
| Geneb | Nucleotide positiona | Nucleotide differencec |
Deduced amino acid differenced | ||
|---|---|---|---|---|---|
| cZ | cC | cCdi | |||
| Leader (1–55) | 33 | G→A | G→A | ||
| 34 | T→C | T→C | |||
| 45 | T→G | T→G | |||
| N (56–1,737), N CDS (120–1,694) | 335 | G→A | G→A | ||
| 483 | A→Ge | T122Ae | |||
| 576 | G→Te | D153Ye | |||
| 598 | T→C | T→C | I160T | ||
| 953 | A→G | A→G | |||
| 1257 | T→C | T→C | |||
| 1294 | G→A | G→A | G392E | ||
| 1394 | C→T | C→T | |||
| 1410 | A→G | A→G | K431E | ||
| 1415 | C→T | C→T | |||
| 1451 | T→A | T→A | D444E | ||
| 1646 | T→C | T→C | |||
| 1708 | T→C | ||||
| P (1,741–3,633), P CDS (1,844–3,550), C CDS (1,854–2,468) | 1760 | C→T | C→T | ||
| 1907 | C→A | C→A | D18Ef | ||
| 1984 | A→G | A→G | E44Gf | ||
| 2032 | T→C | T→C | L60Pf | ||
| 2381 | A→G | A→G | S180Gg | ||
| 2500 | T→C | T→C | |||
| 2878 | A→G | A→G | |||
| 3370 | T→C | T→C | |||
| 3393 | C→A | C→A | P517H | ||
| 3453 | T→C | T→C | V537A | ||
| 3601 | C→T | C→T | |||
| 3602 | C→T | C→T | |||
| M (3,636–4,809), M CDS (3,669–4,715) | 3717 | A→G | A→G | T17A | |
| 3749 | G→A | G→A | |||
| 3768 | C→T | C→T | H34Y | ||
| 3780 | G→A | G→A | V38I | ||
| 4167 | C→T | C→T | L167F | ||
| 4220 | G→A | ||||
| 4265 | T→C | T→C | |||
| 4271 | T→C | T→C | |||
| 4272 | T→C | T→C | |||
| 4498 | A→C | A→C | N277T | ||
| 4499 | T→C | ||||
| 4587 | A→C | A→C | M307L | ||
| F (4,813–6,633), F CDS (4,866–6,563) | 4886 | A→G | A→G | ||
| 5061 | T→C | T→C | F66L | ||
| 5144 | A→G | ||||
| 5200 | C→T | C→T | A112V | ||
| 5204 | C→A | C→A | |||
| 5267 | A→G | A→G | |||
| 5512 | A→G | A→G | E216G | ||
| 5647 | C→A | C→A | C→A | S261Y | |
| 5663 | A→T | A→T | |||
| 5714 | C→T | C→T | |||
| 5756 | T→A | T→A | |||
| 5777 | A→G | A→G | |||
| 6332 | G→T | G→T | |||
| 6420 | A→G | A→G | I519V | ||
| 6442 | G→A | G→A | R526K | ||
| 6494 | C→T | C→T | |||
| 6545 | A→G | A→G | |||
| 6600 | A→G | A→G | |||
| HN (6,637–8,524), HN CDS (6,693–8,420) | 6676 | T→C | T→C | ||
| 6734 | T→C | T→C | |||
| 6752 | C→T | C→T | |||
| 6759 | C→T | C→T | P23L | ||
| 6760 | C→T | C→T | P23L | ||
| 6790 | C→T | C→T | A33V | ||
| 6791 | C→T | C→T | A33V | ||
| 6980 | A→C | A→C | |||
| 7102 | A→T | A→T | H137L | ||
| 7117 | T→Ae | I142Ne | |||
| 7136 | T→G | T→G | D148E | ||
| 7209 | G→A | G→A | E173K | ||
| 7553 | T→C | T→C | |||
| 7678 | C→Ae | A329Ee | |||
| 7691 | A→T | A→T | |||
| 7715 | A→G | A→G | |||
| 7730 | G→A | G→A | |||
| 7756 | A→G | A→G | Q355R | ||
| 7771 | T→C | T→C | V360A | ||
| 7834 | G→A | G→A | S381N | ||
| 8095 | A→C | A→C | K468T | ||
| L (8,528–15,327), L CDS (8,555–15,242) | 8747 | C→T | C→T | ||
| 8753 | A→G | A→G | |||
| 9018 | G→A | G→A | G155S | ||
| 9128 | G→A | G→A | |||
| 9140 | T→C | T→C | T→C | ||
| 9287 | A→G | A→G | |||
| 9328 | A→G | A→G | |||
| 9533 | T→C | T→C | |||
| 9595 | A→T | A→T | H347L | ||
| 9787 | G→Te | ||||
| 10049 | A→G | A→G | |||
| 10158 | G→A | G→A | E535K | ||
| 10292 | A→G | A→G | |||
| 10295 | C→T | C→T | |||
| 10298 | G→C | G→C | G→C | R581S | |
| 10316 | G→A | G→A | G→A | ||
| 10428 | G→A | G→A | E625K | ||
| 10433 | T→G | T→G | |||
| 10475 | A→G | A→G | |||
| 10952 | G→A | G→A | I800T | ||
| 10954 | T→C | T→C | |||
| 10969 | G→A | G→A | G805D | ||
| 11110 | A→G | A→G | K852R | ||
| 11525 | T→C | T→C | |||
| 11661 | G→Ae | E1036Ke | |||
| 11672 | A→C | A→C | |||
| 12024 | C→T | C→T | |||
| 12061 | T→C | T→C | |||
| 12174 | T→A | T→A | C1207S | ||
| 12821 | C→T | C→T | |||
| 12872 | T→C | T→C | |||
| 12923 | C→T | C→T | |||
| 13031 | T→C | T→C | |||
| 13137 | T→C | T←C | |||
| 13163 | T→C | T→C | |||
| 13298 | T→C | T→C | |||
| 13769 | T→C | T→C | |||
| 13850 | T→C | T→C | |||
| 14348 | C→T | C→T | |||
| 14486 | C→T | C→T | |||
| 14707 | C→T | C→T | P2051L | ||
| 15032 | A→G | A→G | |||
| 15122 | A→G | A→G | |||
| 15212 | A→G | A→G | |||
| 15303 | G→A | G→A | |||
| Trailer (15,328–15,384) | 15353 | A→T | A→T | ||
Nucleotide position on the 15,384- nt-long full-length SeV genome.
Genes and the leader and trailer regions are shown, with their nucleotide positions. Coding DNA sequences (CDS) and their nucleotide positions are also shown.
Nucleotide difference compared with fullGNM of Z-cDNA.
Amino acid difference deduced from the nucleotide sequences.
Nucleotide and amino acid substitutions found only between cC and cCdi.
Amino acid substitution found only in C, but not in P and V.
Amino acid substitution found in both P and V, but not in C.
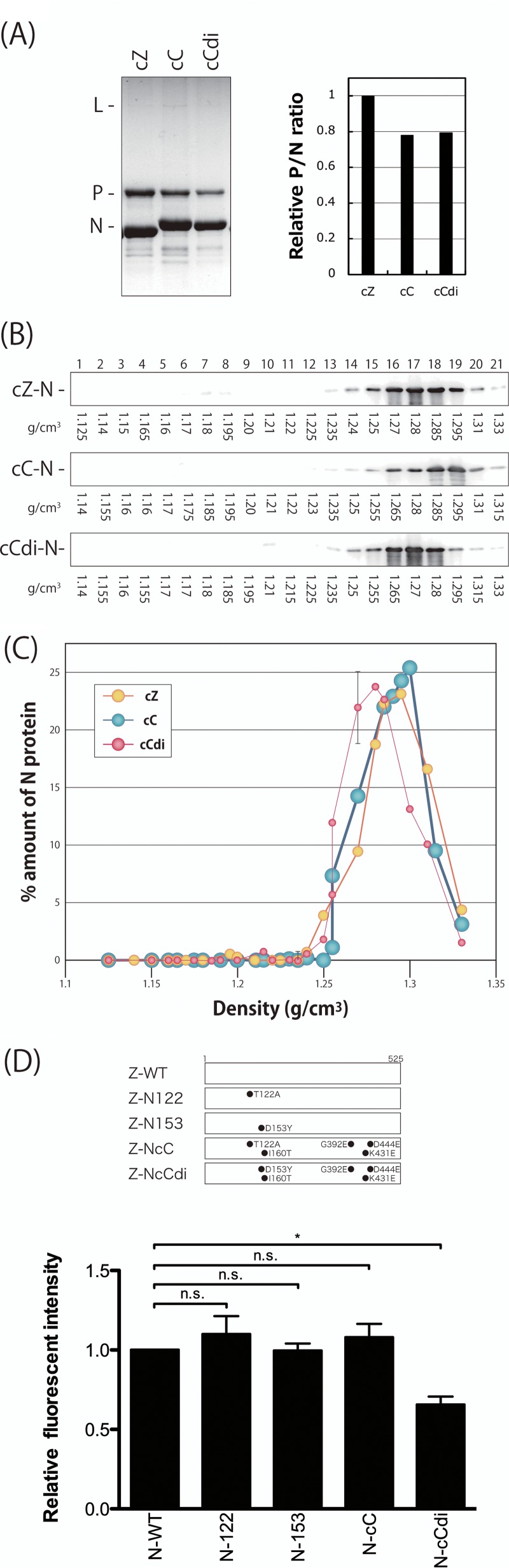
Because cZ and cC were cbDI-nonproducing clones, the D153Y and I142N substitutions in N and HN, respectively, appeared to be unique to the cbDI-producing clone, cCdi. The N protein of SeV has been shown to be a requisite protein for the formation of viral nucleocapsids (NCs), as well as being involved in the regulation of viral RNA synthesis (51), whereas the HN protein has not been assigned to viral RNA synthesis. Therefore, the D153Y substitution within N is likely to play a role in the generation of cbDI genomes by cCdi and might affect the function(s) of the N protein. To investigate this possibility, we compared the densities of NCs purified from cC and cCdi virions by CsCl density gradient fractionation (Fig. 5A and B). Although the ratios of the N and P proteins detected in the NC samples of cC and cCdi were slightly lower than that of cZ, there was no obvious difference between those of cC and cCdi, suggesting that the compositions of viral proteins in the NCs of cC and cCdi were not responsible for their different phenotypes in cbDI production.
FIG 5.
Density gradient fractionation of the nucleocapsids purified from the SeV clones. (A) Nucleocapsids of the indicated viruses were purified from virion samples as described in Materials and Methods and analyzed by SDS-10% PAGE. The densities of the N and P protein bands were measured by densitometry, and the P/N ratios relative to that of cZ are shown. (B) The purified nucleocapsid samples were subjected to 20% to 45% CsCl density gradient ultracentrifugation. Then, 21 fractions were collected from the top of the sample and analyzed by Western blotting using anti-SeV antibody to detect the nucleocapsid-associated N protein. (C) The amount of N protein in each fraction was quantitated, and the ratio of each fraction to the sum of all the fractions was plotted against the density of each fraction. (D) N mutants summarized schematically. The strength of the intermolecular interactions of each of the N mutants was examined using a bimolecular fluorescence complementation technique as described in Materials and Methods. The fluorescence intensity observed in the N-WT sample was set to 1. The graphs represent the averages of three independent experiments, and the error bars represent standard deviations. n.s., nonsignificant (P > 0.05); *, P < 0.05 by one-way ANOVA with Bonferroni post hoc test.
N protein was detected in the fractions by Western blotting using an anti-SeV polyclonal antibody (Fig. 5B), and the densities of the N protein bands were measured and plotted against the densities of the fractions (Fig. 5C). The peak amount of N protein from cZ was observed in the fraction with a density of 1.295 g/cm3, representing the peak density of NCs prepared from the cZ virions. The peak density of the NCs from cC was slightly lower (1.285 g/cm3) than that of the NCs from cZ. Interestingly, the NC of cCdi showed a density of 1.27 g/cm3, even lower than those of the cbDI-nonproducing clones cZ and cC.
SeV N protein by itself has been reported to interact intermolecularly to form viral NCs (52). Therefore, we further examined the effects of amino acid substitutions introduced into the N protein of strain Z based on the sequencing analysis performed as described above on the strength of the N-N intermolecular homologous interactions using a bimolecular fluorescence complementation assay (Fig. 5D). Fluorescence signals detected in the cells expressing N-122 and N-153, possessing T122N and D153Y substitutions found in cC and cCdi, respectively, and N-cC, which is the N protein from cC, were indistinguishable from those in the cells expressing N-WT. However, the fluorescence signal was significantly reduced in cells expressing N-cCdi, which is the N protein from cCdi, indicating that the single D153Y substitution in the N protein of strain CNT, but not that of strain Z, reduced the strength of the N-N intermolecular interaction.
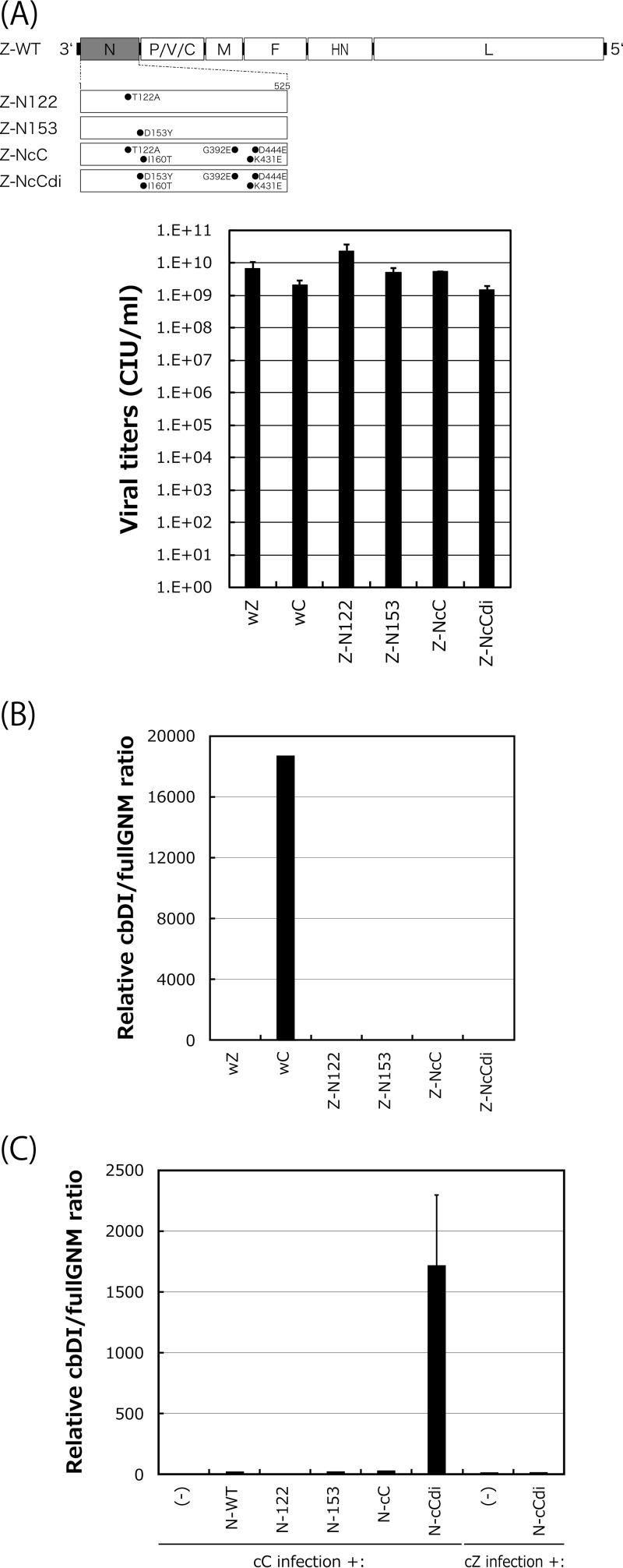
Next, we examined whether the emergence of the D153Y mutation was sufficient for the acquisition of the cbDI-producing phenotype. For this purpose, we first generated a series of strain Z-based recombinant SeVs in which the N protein was replaced with N-122, N-153, N-cC, or N-cCdi (Fig. 6A). One-step growth titers for all of these viruses did not differ significantly at 48 h postinfection (p.i.), indicating that the replacements did not exert any negative effects on viral replication. After five consecutive passages of the recombinants with high viral doses of a constant volume of 200 μl through LLC-MK2 cells, an increase in the cbDI/fullGNM ratio was not observed with any of the recombinants (Fig. 6B). The lack of an increase may have resulted from incompatibility of the mutated N proteins with the nucleotide sequence of the backbone genome of strain Z. To address this possibility, we examined whether trans-supplementation of the N mutants into cells infected with the cbDI-nonproducing clones cC and cZ would lead to the production of cbDI genomes (Fig. 6C). Unlike the Z-based recombinant viruses, an increase in cbDI genomes was observed only on supplementation with N-cCdi, but not with N-122, N-153, or N-cC, in the cC infection, while N-cCdi did not lead to an increase in cbDI genomes in the cZ infection.
FIG 6.
Effects of N mutations on production of cbDI genomes during viral replication. (A) Viral genome structures of the N-mutated recombinants summarized schematically. One-step growth titers of the N recombinants in LLC-MK2 cells at 48 h p.i. were determined as described in Materials and Methods. (B) cbDI/fullGNM ratios in cells infected with the indicated viruses shown as in Fig. 1A. The ratio in the wZ stock was set to 1. (C) Effect of transcomplementation of the N mutants on production of the cbDI genomes during cC virus replication. At 6 h p.i. of the cC clone on LLC-MK2 cells, the indicated pCAGGS-N plasmids were transfected. At 48 h after cC infection, the cbDI/fullGNM ratios in the cells were analyzed and shown as in Fig. 1A; the error bars represent standard deviations.
These results strongly suggested that the emergence of N-cCdi, in which a single D153Y mutation spontaneously occurred during cbDI-nonproducing virus replication, was sufficient to lead to production of the cbDI genomes and suggested that the generation of cbDI genomes by cCdi might be caused by its lower density of NCs than cC, probably owing to the weakness of the N-N interaction caused by the D153Y substitution within the CNT N protein.
The cbDI-producing phenotype significantly induces IFN-β and attenuates pathogenicity in mice.
It has been reported that following loss of function of the C and V proteins, antagonism of the host IFN system results in highly attenuated phenotypes of SeV recombinants (45, 53–56). However, it remained unclear whether the production of cbDI genomes during viral replication affected SeV pathogenicity in mice, owing to the lack of available viruses that differed solely in cbDI production and not in other viral phenotypes. Thus, we finally wanted to examine the pathogenicity of the viral clones cC and cCdi in mice.
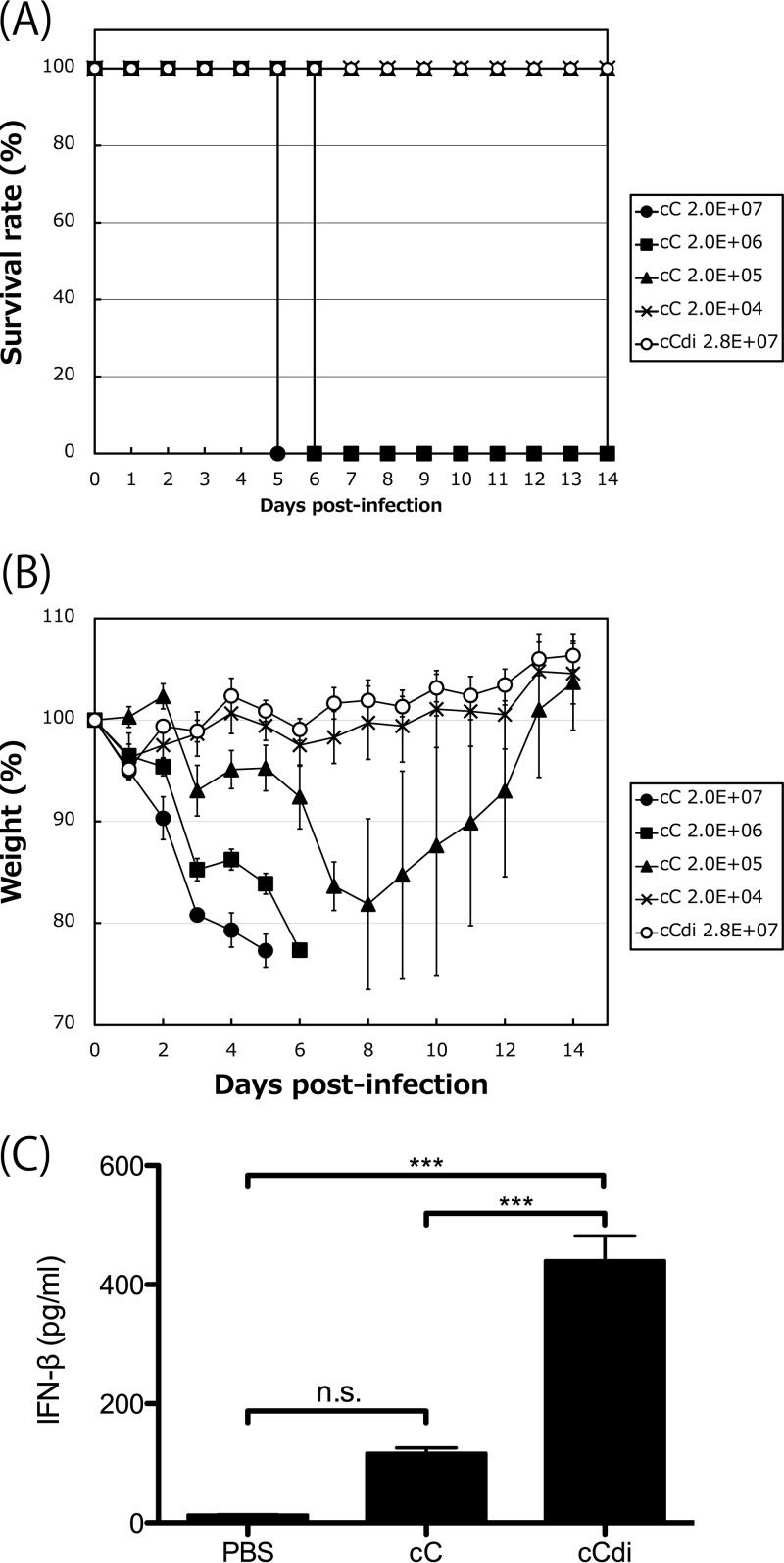
To compare the pathogenicities of these viruses in mice, mice were inoculated with 10-fold serially diluted viral samples of cC and cCdi and were assessed for weight loss and survival over a 2-week period (Fig. 7A and B). Similar to our previous observations for SeV strain Z (54, 56), all the mice infected with 2.0 × 107 and 2.0 × 106 cell-infecting units (CIU) of cC succumbed to the infection by days 6 and 7 p.i., respectively, with a mortality rate of 100%. At 2.0 × 105 CIU/mouse, mice infected with cC lost weight until day 8 p.i., but then they all recovered. At 2.0 × 104 CIU/mouse, no significant weight loss was observed. The 50% mouse lethal dose (MLD50) of cC was 6.45 × 105 CIU/mouse. Unexpectedly, cCdi dramatically lost pathogenicity in mice, with all of the infected mice surviving and no obvious weight loss observed even following infection with 2.8 × 107 CIU/mouse.
FIG 7.
(A and B) Survival curves (A) and percent weight change (B) over a 2-week period for mice infected with cC and cCdi at the indicated doses. (C) Amounts of IFN-β in BAL fluids harvested from mice infected with the indicated viruses at 1 day p.i. determined as described in Materials and Methods. n.s., nonsignificant (P > 0.05); ***, P < 0.001 by two-way ANOVA with Bonferroni post hoc test; the error bars represent standard deviations.
Finally, to examine the inducibility of IFN-β of cC and cCdi in the infected mice, the amount of IFN-β in bronchoalveolar lavage (BAL) fluid harvested at 1 day p.i. was compared by an enzyme-linked immunosorbent assay (ELISA) (Fig. 7C). We previously reported that, in a comparison of the WT and V(−) SeVs, a remarkable difference in the amounts of IFN-β induced in the infected mouse lungs was readily observed at an early time point, day 1 p.i (56). A significantly larger amount of IFN-β was detected in the BAL fluid from the cCdi-infected mice than in that from the cC-infected mice.
These results indicated that, although the growth of cC and cCdi is comparable in cell cultures, cCdi is significantly attenuated in mice compared with cC. The significantly higher IFN-β inducibility of cCdi observed in the infected cell cultures was also observed in infected mouse lungs, suggesting that the severe attenuation of cCdi might be caused by IFN-β induction by cbDI production.
DISCUSSION
DI genomes have been established as playing a role in the establishment of persistent viral infection but in recent years have received renewed interest as natural ligands for RIG-I that strongly stimulate type I IFN pathways. Accumulating evidence indicates that, in real viral infection, DI genomes serve as strong ligands for RIG-I (13, 14, 18). SeV strain CNT has been widely employed experimentally in the field of innate immunity as a representative RNA virus that strongly stimulates RIG-I-mediated IFN pathways. In contrast, most other SeV strains have been found to counteract activation of IFN pathways to varying degrees, mainly via the SeV accessory proteins C and V (36). Although cbDI genomes have recently been identified as RIG-I ligands in CNT infection, it remained unclear whether the IFN-inducing phenotype of strain CNT was determined exclusively by the presence of cbDI genomes or, additionally, by a defect in the counteraction function.
The results presented in Fig. 1 indicated that the V protein of strain CNT, as well as that of strain Z, fully inhibited the MDA5- and IRF3-mediated expression of a reporter EGFP. The SeV V protein has been shown to physically interact with MDA5 and the downstream IRF3 to suppress induction of IFN-β triggered by constitutively active mutants of RIG-I and TRIF, as well as MDA5 (45–47). Indeed, infection with Z-V(−) rSeV, harboring a negligible number of cbDI particles, increased the induction of IFN-β compared with that induced by WT SeV (Fig. 1A). However, the levels of IFN-β induction following infection with Z-4C(−) rSeV or strain CNT were dramatically higher than with Z-V(−) rSeV, despite their expression of intact V proteins (Fig. 1A). Following infection with Z-4C(−) rSeV, previously unidentified IFN-β-inducing viral dsRNA was produced (37). These IFN-β-inducing, unusual viral RNA species produced during Z-4C(−) and CNT infections seemed to trigger IFN-β production so strongly that it could not be suppressed by V protein.
The C proteins of all of the SeV strains tested retained inhibitory action against the type I IFN-triggered establishment of the antiviral state, even in the case of massive induction of IFN-β by CNT and Z-4C(−) (Fig. 1B and C). Recombinant SeV Z-4C(−) was completely attenuated in mice, owing to both the strong induction of IFN-β and the defect in counteraction against the host Jak/STAT pathway (54). Although the C and V proteins are fully functional in the ability to antagonize host innate immunity, a cbDI genome-producing, IFN-β-inducing SeV CNT clone, cCdi, had dramatically lower virulence in mice than cC (Fig. 7A and B). As observed in the cell cultures, IFN-β was significantly induced in cCdi-infected mice compared with cC-infected mice (Fig. 7C). Although both the cellular and viral factors involved in antiviral innate immunity and the viral counteraction directed against it have been extensively studied, especially using mouse infection models, it remains unclear how the emergence and/or presence of strongly IFN-β-inducing DI genomes affects viral virulence in mice. The above-described comparison between cC and cCdi suggests the potential importance of cbDI production during infection in SeV pathogenesis in mice. Although mononegaviral DI genomes have been historically associated with persistent infection, the mechanisms whereby DI genomes affect natural infections remain to be fully elucidated. DI genomes may extend the persistence of viral infection in the host, increasing the chance of systemic spread. The CNT clones cC and cCdi will be useful tools with which to address this issue because they differ only in the production of cbDI genomes.
Previous reports on the structure of cb- and id-type DI genomes of mononegaviruses and orthomyxoviruses, respectively, indicated that DI genomes are likely to form a 3′-ppp, blunt-ended, double-stranded stem-loop structure that is the ideal structure of a RIG-I ligand (18, 21, 57). DI genomes, as well as the authentic genomes of mononegaviruses, must be encapsidated by viral nucleoproteins to be replicated by viral polymerases, so cbDI genomes are unlikely to form a stem-loop structure with intramolecular base pair matching (58). However, encapsidation of the SeV cbDI genomes has been suggested to be somewhat looser than that of the authentic genomes because treatment of the purified cbDI genomes with SDS under relatively mild conditions where the encapsidated, authentic genomes are not stripped results in the spontaneous formation of a stem-loop structure (19, 21). Clones cC and cCdi differ by a single amino acid, which leads to a lower density of NCs, and the cbDI NCs of cCdi may therefore be looser, allowing base pair matching to occur more easily than with cC.
Experimentally, it has been reported that DI genomes can be generated by serial passage of mononegaviruses at high multiplicity, probably because errors occur more readily during viral genome replication when a high dose of virus is present (20, 22). However, in this study, cbDI genomes were not easily generated by such passage experiments with the non-DI-producing clones, cZ and cC, indicating that viral passage at high multiplicity itself is not a key factor for cbDI generation. Instead, the successful isolation of cCdi and the generation of cbDI by complementation of N-cCdi in the cC infection (Fig. 3 to 6) strongly suggest that the spontaneous emergence of a DI-producing variant during viral replication causes the generation and accumulation of DI particles in the viral pools.
Although template switching from the antigenome to the nascent genome strand must be required for DI generation, the precise mechanisms involved in the generation of cbDI genomes remain to be determined. Previous observations that the structures of cbDI genomes are limited suggest that the switching points may also be limited (21). As mentioned above, for switching to occur, nucleotides on the viral genome, which are tightly encapsidated within the NC structure, must be exposed before base pair matching can take place. Although the density of the NC of cCdi was lower than that of cC, no obvious difference in viral replication was found between cC and cCdi, suggesting that the structures and functions of the cC and cCdi NCs are retained without any particular abnormalities. The lower density of the cCdi NC might be due to loss of the N protein from the NC during the experimental processes, such as density centrifugation. During viral replication, looseness of the NCs may occur in specific regions that serve as the switching points, potentially allowing base pair matching between the genome and antigenome NCs. In addition to template switching, this looseness may also allow intramolecular base pair matching of cbDI nucleocapsids to form a RIG-I ligand structure.
In this study, we successfully isolated, for the first time, the cbDI-producing clone cCdi and identified a single amino acid substitution responsible for its unique phenotype. This may allow us to control cbDI production without altering any other viral functions. Further characterization of this clone may provide novel insights that may lead to a better understanding of virus-host interactions during innate immunity, viral replication, and viral pathogenesis. Recently, artificially synthesized SeV cbDI genome RNA has been shown to exert effective adjuvant activity (59). cCdi has the potential to be an effective tool for producing cbDI genomes, as well as a backbone for the generation of live recombinant vaccine vectors, thereby providing both antigen and adjuvant functions.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
LLC-MK2 cells (macaque monkey kidney-derived cells, described in reference 60), HeLa cells (human cervical cancer-derived cells, described in reference 61), and BHK-21 cells (baby hamster kidney-derived cells, described in reference 62) were originally purchased from ATCC. 293T cells (human embryonic kidney-derived cells expressing simian virus 40 [SV40] large T antigen) were purchased from Riken BRC Cell Bank, Tsukuba, Japan. These cells were maintained as reported previously (45). Vero cells stably expressing human TMPRSS2 were kindly provided by M. Takeda (National Institute of Infectious Diseases [NIID], Japan) (63). The SeV stock of strain CNT purchased from ATCC (oriC) was kindly provided by K. Takeuchi (University of Tsukuba, Tsukuba, Japan), and the working stock (wC) was prepared by propagating oriC once in 10-day-old embryonated chicken eggs. The SeV recombinants, Z-4C(−) and Z-V(−), lacking the expression of all four C and V proteins, respectively, were kindly provided by A. Kato (NIID, Japan) (55, 64). The working stocks of SeV strain Z (wZ) and the other SeV recombinants were prepared by propagating the viruses recovered from cDNA using a reverse genetics technique in LLC-MK2 cells through embryonated chicken eggs, as described previously (65). In addition to these viruses, SeV strains FSM, HMT (66), and NGY were used. Viral titers were determined as described previously (60) and represented as the number of CIU per milliliter. The growth kinetics of the SeVs was determined as described previously (67). rVSV-GFP was kindly provided by K. Shinozaki (Hiroshima Prefectural Hospital, Hiroshima City, Japan) and propagated in BHK-21 cells. The polyclonal antibodies against SeV P and C proteins were kindly provided by A. Kato. A monoclonal antibody against SeV N protein was kindly provided by E. Suzuki (NIID, Japan). The polyclonal antibody against GFP (sc-8334; Santa Cruz Biotechnology), horseradish peroxidase-conjugated anti-rabbit and anti-mouse IgG goat polyclonal antibodies (Santa Cruz Biotechnology), and a monoclonal antibody against the FLAG tag (M2; Sigma-Aldrich) were used according to the protocols of the suppliers.
Quantitative RT-PCR.
Quantitative RT-PCR (qRT-PCR) was performed as described previously (37). Briefly, HeLa, LLC-MK2, or Vero-TMPRSS2 cells were infected with the indicated viruses at an MOI of 5. At 24 h p.i., total RNA was prepared from the cells using a High Pure RNA isolation kit (Roche Diagnostics). Virion RNA was prepared from the viral stocks or culture medium harvested from infected cells using a High Pure Viral RNA kit (Roche Diagnostics). For the detection of genome length viral RNAs, cbDI RNAs, IFN-β mRNA, and beta-actin mRNA, qRT-PCR was performed using a QuantiFast SYBR green RT-PCR kit (Qiagen) with the RNA samples described above prepared as templates and the following primer sets: 5SeVZ1683 plus 3SeVZ1843, cbDIdetect15033-15014 plus cbDIdetect15312-15293 (complementary to the regions from nt 15014 to 15033 and 15193 to 15312 of the SeV antigenome), 5hIFNb1-481-500 plus 3hIFNb1-640-661, and 5hbActin249 plus 3hbActin636, respectively (Table 2), and an Eco real-time PCR system (Illumina). The amplification of specific DNA fragments during the reaction was confirmed by melting curve analysis. To examine the efficiency of qRT-PCR for the genome length viral RNAs and cbDI RNAs, amplification curves were compared by qPCR using a QuantiFast SYBR green PCR kit (Qiagen) with the primer sets described above and the RT-PCR products as templates. The RT-PCR products to be used as templates were produced in the above-mentioned qRT-PCR and were subcloned into a pUC18 cloning vector. The efficiency of qRT-PCR for the genome length RNAs relative to that of the cbDI RNAs was 1 to 0.41 ± 0.041.
TABLE 2.
Primers used in this study
| Primer | Sequence | Purpose |
|---|---|---|
| 5hIFNb1-481-500 | AAACTCATGAGCAGTCTGCA | Detection of human IFN-β1 mRNA |
| 3hIFNb1-640-661 | AGAGGCACAGGCTAGGAGATC | Detection of human IFN-β1 mRNA |
| 5hbActin249 | GAAAATCTGGCACCACACCT | Detection of human beta-actin mRNA |
| 3hbActin636 | AATGTCACGCACGATTTCCC | Detection of human beta-actin mRNA |
| cbDIdetect15033-15014 | TTTCACGGGATGATAATGAA | Detection of cbDI |
| cbDIdetect15312015293 | TCATATGGACAAGTCCAAGA | Detection of cbDI |
| 5SeVZ1 | ACCAAACAAGAGAAAAAACATGTATGGG | SeV genome sequencing |
| 5SeVZ500 | TGAGAGGACCACAGAATGGC | SeV genome sequencing |
| 5SeVZ995 | TATTAATAAGCTTAGAAGCC | SeV genome sequencing |
| fwc741ZNE2F | GTCAGGTGAGTGGTGGCCAC | SeV genome sequencing |
| 3SeVZ1560 | TCTCTATGTCTGATACATCC | SeV genome sequencing |
| 5SeVZ1683 | GGAGGAATCTAGGATCATACGAGGC | Detection of fullGNM |
| 3SeVZ1843 | GCGGTAAGTGTAGCCGAAGCCG | Detection of fullGNM |
| 5SeVZ2022 | CAGGCTCTGCTCATAGAGCC | SeV genome sequencing |
| 3SeVZ2069 | TTGAGACTTCTCCTTCGCCC | SeV genome sequencing |
| 3SeVZ2378 | TCAGGTAGGGATGTACTTCC | SeV genome sequencing |
| 5SeVZ2449 | GAAGCATGGAGCCTGGCAGC | SeV genome sequencing |
| 5SeVZ2994 | GCCGAGAAATCTTCCGCTCG | SeV genome sequencing |
| 3SeVZ3071 | ATCTCTTGTAAATATCCCGG | SeV genome sequencing |
| 5SeVZ3518 | TAAGGCAGTCATGGAACTCG | SeV genome sequencing |
| 3SeVZ3572 | GGTAGGATGCCTCACCCGGG | SeV genome sequencing |
| 5SeVZ4005 | CACCGATCTCAGAATTACGG | SeV genome sequencing |
| 3SeVZ4065 | GAGCACCAATCGAATCCACC | SeV genome sequencing |
| 3SeVM | CAGCTTTCTGATCCTTCCGATGTTCTTAGC | SeV genome sequencing |
| 5SeVFEcoRI | ATATGAATTCCACCATGACAGCATATATCC | SeV genome sequencing |
| 3SeV5318SphI | TATATGCATGCAGCGCTATGTCTCTTTTGG | SeV genome sequencing |
| 5SeVZ6315 | GCACGGAAAATCCTCTCGGAGGTAGG | SeV genome sequencing |
| 3SeVFNheI | TATATGCTAGCTCATCTTTTCTCAGCC | SeV genome sequencing |
| 5SeVZ7206 | CCTGAAATCTCATTGCTGCC | SeV genome sequencing |
| 3SeVZ7425 | CTACGGGGTTAAGATCAGGG | SeV genome sequencing |
| 5SeVZ8393 | CCCTAAATTATGCAAGGCCG | SeV genome sequencing |
| 3SeVZ8565 | GCCCATCCATGACCTATGGC | SeV genome sequencing |
| 5SeVZ9234 | ACAGGGTATATCCTAACCCC | SeV genome sequencing |
| 5SeVZ9373 | TGGATTCCCTCTTCTCAAGTCTTGGAGAGG | SeV genome sequencing |
| 3SeVZ9450 | TGAGAGCAAGTGATAGGGGC | SeV genome sequencing |
| 5SeVZ10341 | TTGACTACTCTTTCTGTCTCAGGCGTCCCC | SeV genome sequencing |
| 3SeVZ10716 | GTCGATGCATCCGGTCGGCG | SeV genome sequencing |
| 3SeVZ11482 | TGCTTGTCTAACAGATCCGC | SeV genome sequencing |
| 5SeVZ11860 | GGCTTGTCAATTATGATCTATTGCAGTACG | SeV genome sequencing |
| 3SeVZ12006 | CGTAAGTCAGGTGGATCCACATTTTCTGCC | SeV genome sequencing |
| 5SeVZ12469 | CACTAGTCCGTGCAAGTCGG | SeV genome sequencing |
| 5SeVZ-L-C901Stop | GTCAGCAGGTGTGAATATCACTAGGGATG | SeV genome sequencing |
| 3SeVZ13040 | GATTGACTAGAATACCCCCG | SeV genome sequencing |
| 5SeVZ13638 | GATTGGGATCCCGAGGCAGATAATGCACTG | SeV genome sequencing |
| 3SeVZ13886 | ACTGTTGATGCCAAAGAGCC | SeV genome sequencing |
| 5SeVZ14761 | CACACAACTGCATGATAGCTTTCAACAGGG | SeV genome sequencing |
| 3SeVZ15384 | ACCAGACAAGAGTTTAAGAGATATGTATCC | SeV genome sequencing |
| Oligo(dT) | TTTTTTTTTTTTTTTTTTTT | SeV genome sequencing |
Antiviral activity assay.
An antiviral activity assay was performed as described previously (43). Briefly, HeLa cells were infected with the indicated viruses at an MOI of 5. At 6 h p.i., the culture medium was replaced with fresh serum-free Dulbecco's minimum essential medium (DMEM) (Invitrogen) containing IFN-α (1,000 IU/ml; R&D Systems). After an additional 6-h incubation, the cells were superinfected with rVSV-GFP at an MOI of 3 and further incubated at 37°C for 6 h. GFP expression was observed using a fluorescence microscope (Nikon Eclipse TE2000-S). GFP expression in cells was also analyzed by Western blotting using polyclonal antibodies against GFP and SeV P and C proteins as primary antibodies and horseradish peroxidase-conjugated anti-rabbit IgG goat polyclonal antibody as a secondary antibody. Protein bands were visualized and analyzed using the analysis software JustTLC (Sweday, Sweden).
Construction of expression plasmids.
Plasmids encoding the C and V proteins of SeV strain Z in the pCAGGS.MCS vector were employed as described previously (44, 45). The full-length cDNA clones of C of strains CNT, FSM, HMT, and NGY were amplified from virion RNA using a OneStep RT-PCR kit (Qiagen) with specific primers and were subcloned into pCAGGS.MCS. The full-length cDNA clones of V proteins of these strains were detected by DNA sequencing from the cDNA clones of P that were amplified and cloned into pCAGGS.MCS as described above for the C proteins. Plasmid pCAGGS-FL-MDA5-CA encoding constitutively active MDA5 possessing an A946T mutation was generated by introducing a point mutation into pCAGGS-FL-MDA5-WT (47) using an AMAP site-directed mutagenesis kit (Amalgaam, Japan).
IRF3 and ISRE reporter assays.
The IRF3 reporter assay was performed as described previously (45). Briefly, 293T cells were cotransfected with a reporter plasmid, p55C1B-EGFP; an IRF3-signal-inducing plasmid, pCAGGS-FL-MDA5-CA; and an expression plasmid for the V proteins using the FuGene HD reagent (Promega). After 24 h, expression of the reporter GFP in cells was analyzed by Western blotting using polyclonal antibodies against GFP, as well as P and C proteins. Protein bands were visualized and analyzed as described above.
The ISRE reporter assay was performed as described previously (43). Briefly, 293T cells were cotransfected with a reporter plasmid, pISRE-EGFP, and the indicated expression plasmid for the C proteins. At 18 h posttransfection (p.t.), the culture medium was replaced with fresh serum-free medium containing IFN-α (1,000 IU/ml). After an additional 8-h incubation, expression of the reporter GFP in cells was analyzed as described above.
Sequencing of viral full-length and defective RNA genomes.
The complete genome sequences of the representative SeV clones of strains Z and CNT were determined by direct sequencing of the RT-PCR products amplified from the virion RNA using a OneStep RT-PCR kit with specific primers (Table 2). For determination of the 3′ and 5′ ends of the genomes, poly(A) was added to the virion RNA using an Escherichia coli poly(A) polymerase (New England BioLabs), and the polyadenylated RNAs were subjected to RT-PCR with specific primers and oligo(dT), followed by direct sequencing.
For the cbDI genomes, cDNA was amplified by one-step RT-PCR using the DI genome-containing RNA samples from the wC stock as a template and a primer, Tr80CNT, complementary to 80 nt at the 3′ end of the antigenome of strain CNT. RT-PCR products were subcloned into the pUC19 plasmid, and 20 of the plasmid clones were analyzed by DNA sequencing.
Isolation of viral clones.
Cloning of SeV strains Z and CNT was performed by three consecutive limiting dilutions. Vero-TMPRSS2 cells in 96-well plates were infected with the working stocks of Z and CNT at 0.1 CIU/well and incubated at 37°C for 2 to 3 days in the presence of 10 μg/ml trypsin (Merck) until an apparent cytopathic effect was observed. The culture medium was then harvested and titrated. This procedure was repeated three times for each strain. Representative SeV clones (cZ, cC, and cCdi) obtained after the third cloning were predominantly used in this study.
Purification of nucleocapsids and density gradient fractionation.
SeV virions were purified by ultracentrifugation through a 20% sucrose cushion using a Beckman SW28 rotor (24,000 rpm; 2 h) twice. The purified virion pellets were suspended in NP-40 lysis buffer (1% NP-40, 10 mM Tris-HCl [pH 7.8], 150 mM NaCl, 1 mM EDTA) and then subjected to 20% to 45% CsCl density gradient ultracentrifugation using a Beckman SW40 Ti rotor (35,000 rpm; 2 h). A specific band corresponding to the nucleocapsid was collected, suspended in NP-40 lysis buffer, and then again subjected to CsCl density gradient ultracentrifugation. A band corresponding to the nucleocapsid was collected, and the purity was confirmed by SDS-PAGE. The purified nucleocapsids were then subjected to CsCl density gradient ultracentrifugation (35,000 rpm; 4 h), and 21 fractions were collected from the top of the sample. The fraction samples were analyzed by SDS-10% PAGE, followed by Western blotting using an anti-SeV polyclonal antibody, and were analyzed as described above.
Bimolecular fluorescence complementation assay.
The N mutants indicated in Fig. 5 were generated by introducing point mutations using an AMAP site-directed mutagenesis kit (Amalgaam, Japan) and inserted back into the phmKGN-MC and phmKGC-MC vectors included in the CoralHue Fluo-chase kit (Amalgaam). All mutations were confirmed by DNA sequencing. 293T cells cultured in six-well plates were cotransfected with a combination of phmKGC-N plasmids encoding the indicated N proteins. At 48 h p.t., cells were suspended in cell lysis buffer (0.5% NP-40, 20 mM Tris-HCl [pH 7.4], 150 mM NaCl), and the fluorescence of the reconstructed reporter fluorescent protein Kusabira-Green was measured using a TriStar multimode microplate reader (BertholdTech, Germany).
Generation of SeV recombinants.
The pSeV(+) plasmid encoding full-length SeV cDNA of strain Z was kindly provided by A. Kato (65). Mutations were introduced by a standard PCR technique. SeV recombinants were recovered from cDNA as described previously (65). Mutations were confirmed by direct DNA sequencing of the RT-PCR products of viral genomic RNA prepared from purified virions.
Infection of mice with SeVs and ELISA.
C57BL/6N female mice were purchased from Charles River Laboratories, Japan, Ltd. (Atsugi, Japan). Six-week-old mice (n = 4 or 5) were intranasally inoculated with cC or cCdi at the indicated doses under mild anesthesia with a ketamine and xylazine mixture (20 μl/mouse), and their body weights and clinical symptoms were checked daily over a 2-week period. To examine IFN-β induction, 6-week-old mice (n = 3) were inoculated with 1.0 × 107 CIU of cC and cCdi or the same volume (20 μl) of phosphate-buffered saline (PBS). At 1 day p.i., the mice were euthanized, and BAL fluid from the lungs was prepared as described previously (68). The amount of IFN-β in the BAL fluid was determined using a Legend Max ELISA kit with mouse IFN-β-precoated plates (BioLegend) according to the manufacturer's protocol.
Ethics statement.
Infection experiments were performed under biosafety level 3 (BSL3) conditions at the animal facilities in the Natural Science Center for Basic Research and Development, Hiroshima University, and in the National Institute of Infectious Diseases and were carried out in strict accordance with the Guidelines for Animal Experimentation of the Japanese Association for Laboratory Animal Science (JALAS). The protocol was approved by the Committee on the Ethics of Animal Experiments of Hiroshima University (permit numbers A10-108 and 115116-II). The 10-day-old embryonated chicken eggs were obtained from Hiroshima Experimental Animals, Ltd., Hiroshima, Japan. All procedures regarding embryonated chicken eggs were also performed in accordance with the guidelines of the JALAS. Ethics approval was not needed for viral propagation in the eggs, because the eggs were not hatched.
Accession number(s).
The complete genome sequences for the clones cZ, cC, and cCdi have been deposited in the GenBank database under accession numbers AB855655, AB855653, and AB855654, respectively. The cbDI genome sequence has been deposited in the GenBank database under accession number AB856846.
ACKNOWLEDGMENTS
We thank all the members of our laboratory for their fruitful discussions. We also thank the staff of the Analysis Center of Life Science, Hiroshima University, for the use of their facilities. We thank Kate Fox (Edanz Group) for editing a draft of the manuscript.
This work was supported by JSPS Kakenhi grant numbers 16H05197, 25460569, and 17K19560 for T.I.; by the Japanese Agency for Medical Research and Development (AMED) for T.S.; and by a Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Science, Sports and Technology (MEXT) of Japan (grant numbers 16H06429, 16K21723, and 17H05828) for K.S. The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Akira S, Uematsu S, Takeuchi O. 2006. Pathogen recognition and innate immunity. Cell 124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M. 2006. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci U S A 103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaisho T, Akira S. 2006. Toll-like receptor function and signaling. J Allergy Clin Immunol 117:979–987. doi: 10.1016/j.jaci.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 4.Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale M Jr. 2007. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci U S A 104:582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoneyama M, Fujita T. 2007. RIG-I family RNA helicases: cytoplasmic sensor for antiviral innate immunity. Cytokine Growth Factor Rev 18:545–551. doi: 10.1016/j.cytogfr.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 6.Kawai T, Akira S. 2008. Toll-like receptor and RIG-I-like receptor signaling. Ann N Y Acad Sci 1143:1–20. doi: 10.1196/annals.1443.020. [DOI] [PubMed] [Google Scholar]
- 7.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 8.Davis WG, Bowzard JB, Sharma SD, Wiens ME, Ranjan P, Gangappa S, Stuchlik O, Pohl J, Donis RO, Katz JM, Cameron CE, Fujita T, Sambhara S. 2012. The 3′ untranslated regions of influenza genomic sequences are 5′PPP-independent ligands for RIG-I. PLoS One 7:e32661. doi: 10.1371/journal.pone.0032661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marq JB, Hausmann S, Veillard N, Kolakofsky D, Garcin D. 2011. Short double-stranded RNAs with an overhanging 5′ ppp-nucleotide, as found in arenavirus genomes, act as RIG-I decoys. J Biol Chem 286:6108–6116. doi: 10.1074/jbc.M110.186262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marq JB, Kolakofsky D, Garcin D. 2010. Unpaired 5′ ppp-nucleotides, as found in arenavirus double-stranded RNA panhandles, are not recognized by RIG-I. J Biol Chem 285:18208–18216. doi: 10.1074/jbc.M109.089425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G, Juranek S, Kato H, Kawai T, Poeck H, Fitzgerald KA, Takeuchi O, Akira S, Tuschl T, Latz E, Ludwig J, Hartmann G. 2009. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity 31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt A, Schwerd T, Hamm W, Hellmuth JC, Cui S, Wenzel M, Hoffmann FS, Michallet MC, Besch R, Hopfner KP, Endres S, Rothenfusser S. 2009. 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc Natl Acad Sci U S A 106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patel JR, Garcia-Sastre A. 2014. Activation and regulation of pathogen sensor RIG-I. Cytokine Growth Factor Rev 25:513–523. doi: 10.1016/j.cytogfr.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 14.Runge S, Sparrer KM, Lassig C, Hembach K, Baum A, Garcia-Sastre A, Soding J, Conzelmann KK, Hopfner KP. 2014. In vivo ligands of MDA5 and RIG-I in measles virus-infected cells. PLoS Pathog 10:e1004081. doi: 10.1371/journal.ppat.1004081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmolke M, Patel JR, de Castro E, Sanchez-Aparicio MT, Uccellini MB, Miller JC, Manicassamy B, Satoh T, Kawai T, Akira S, Merad M, Garcia-Sastre A. 2014. RIG-I detects mRNA of intracellular Salmonella enterica serovar Typhimurium during bacterial infection. mBio 5:e01006-. doi: 10.1128/mBio.01006-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weber M, Gawanbacht A, Habjan M, Rang A, Borner C, Schmidt AM, Veitinger S, Jacob R, Devignot S, Kochs G, Garcia-Sastre A, Weber F. 2013. Incoming RNA virus nucleocapsids containing a 5′-triphosphorylated genome activate RIG-I and antiviral signaling. Cell Host Microbe 13:336–346. doi: 10.1016/j.chom.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baum A, Garcia-Sastre A. 2011. Differential recognition of viral RNA by RIG-I. Virulence 2:166–169. doi: 10.4161/viru.2.2.15481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baum A, Sachidanandam R, Garcia-Sastre A. 2010. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc Natl Acad Sci U S A 107:16303–16308. doi: 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strahle L, Marq JB, Brini A, Hausmann S, Kolakofsky D, Garcin D. 2007. Activation of the beta interferon promoter by unnatural Sendai virus infection requires RIG-I and is inhibited by viral C proteins. J Virol 81:12227–12237. doi: 10.1128/JVI.01300-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolakofsky D. 1979. Studies on the generation and amplification of Sendai virus defective-interfering genomes. Virology 93:589–593. doi: 10.1016/0042-6822(79)90263-0. [DOI] [PubMed] [Google Scholar]
- 21.Kolakofsky D. 1976. Isolation and characterization of Sendai virus DI-RNAs. Cell 8:547–555. doi: 10.1016/0092-8674(76)90223-3. [DOI] [PubMed] [Google Scholar]
- 22.Killip MJ, Young DF, Gatherer D, Ross CS, Short JA, Davison AJ, Goodbourn S, Randall RE. 2013. Deep sequencing analysis of defective genomes of parainfluenza virus 5 and their role in interferon induction. J Virol 87:4798–4807. doi: 10.1128/JVI.03383-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leppert M, Kort L, Kolakofsky D. 1977. Further characterization of Sendai virus DI-RNAs: a model for their generation. Cell 12:539–552. doi: 10.1016/0092-8674(77)90130-1. [DOI] [PubMed] [Google Scholar]
- 24.Roux L, Waldvogel FA. 1981. Establishment of Sendai virus persistent infection: biochemical analysis of the early phase of a standard plus defective interfering virus infection of BHK cells. Virology 112:400–410. doi: 10.1016/0042-6822(81)90287-7. [DOI] [PubMed] [Google Scholar]
- 25.Roux L, Holland JJ. 1979. Role of defective interfering particles of Sendai virus in persistent infections. Virology 93:91–103. doi: 10.1016/0042-6822(79)90278-2. [DOI] [PubMed] [Google Scholar]
- 26.Rima BK, Davidson WB, Martin SJ. 1977. The role of defective interfering particles in persistent infection of Vero cells by measles virus. J Gen Virol 35:89–97. doi: 10.1099/0022-1317-35-1-89. [DOI] [PubMed] [Google Scholar]
- 27.Sekellick MJ, Marcus PI. 1978. Persistent infection. I. Interferon-inducing defective-interfering particles as mediators of cell sparing: possible role in persistent infection by vesicular stomatitis virus. Virology 85:175–186. [DOI] [PubMed] [Google Scholar]
- 28.De BK, Nayak DP. 1980. Defective interfering influenza viruses and host cells: establishment and maintenance of persistent influenza virus infection in MDBK and HeLa cells. J Virol 36:847–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grabau EA, Holland JJ. 1982. Analysis of viral and defective-interfering nucleocapsids in acute and persistent infection by rhabdoviruses. J Gen Virol 60:87–97. doi: 10.1099/0022-1317-60-1-87. [DOI] [PubMed] [Google Scholar]
- 30.Kennedy JC, Macdonald RD. 1982. Persistent infection with infectious pancreatic necrosis virus mediated by defective-interfering (DI) virus particles in a cell line showing strong interference but little DI replication. J Gen Virol 58:361–371. doi: 10.1099/0022-1317-58-2-361. [DOI] [PubMed] [Google Scholar]
- 31.Yoshida T, Hamaguchi M, Naruse H, Nagai Y. 1982. Persistent infection by a temperature-sensitive mutant isolated from a Sendai virus (HVJ) carrier culture: its initiation and maintenance without aid of defective interfering particles. Virology 120:329–339. doi: 10.1016/0042-6822(82)90034-4. [DOI] [PubMed] [Google Scholar]
- 32.DePolo NJ, Holland JJ. 1986. Very rapid generation/amplification of defective interfering particles by vesicular stomatitis virus variants isolated from persistent infection. J Gen Virol 67:1195–1198. doi: 10.1099/0022-1317-67-6-1195. [DOI] [PubMed] [Google Scholar]
- 33.Moscona A. 1991. Defective interfering particles of human parainfluenza virus type 3 are associated with persistent infection in cell culture. Virology 183:821–824. doi: 10.1016/0042-6822(91)91018-C. [DOI] [PubMed] [Google Scholar]
- 34.Calain P, Monroe MC, Nichol ST. 1999. Ebola virus defective interfering particles and persistent infection. Virology 262:114–128. doi: 10.1006/viro.1999.9915. [DOI] [PubMed] [Google Scholar]
- 35.Cave DR, Hendrickson FM, Huang AS. 1985. Defective interfering virus particles modulate virulence. J Virol 55:366–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagai Y, Takakura A, Irie T, Yonemitsu Y, Gotoh B. 2011. Sendai virus: evolution from mouse pathogen to a state-of-art tool in virus research and biotechnology, p 115–173. In Samal SK. (ed), The biology of paramyxoviruses. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 37.Yoshida A, Kawabata R, Honda T, Tomonaga K, Sakaguchi T, Irie T. 2015. IFN-beta-inducing, unusual viral RNA species produced by paramyxovirus infection accumulated into distinct cytoplasmic structures in an RNA-type-dependent manner. Front Microbiol 6:804. doi: 10.3389/fmicb.2015.00804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcin D, Latorre P, Kolakofsky D. 1999. Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J Virol 73:6559–6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcin D, Marq JB, Strahle L, le Mercier P, Kolakofsky D. 2002. All four Sendai virus C proteins bind Stat1, but only the larger forms also induce its mono-ubiquitination and degradation. Virology 295:256–265. doi: 10.1006/viro.2001.1342. [DOI] [PubMed] [Google Scholar]
- 40.Gotoh B, Takeuchi K, Komatsu T, Yokoo J. 2003. The STAT2 activation process is a crucial target of Sendai virus C protein for the blockade of alpha interferon signaling. J Virol 77:3360–3370. doi: 10.1128/JVI.77.6.3360-3370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Komatsu T, Takeuchi K, Yokoo J, Gotoh B. 2002. Sendai virus C protein impairs both phosphorylation and dephosphorylation processes of Stat1. FEBS Lett 511:139–144. doi: 10.1016/S0014-5793(01)03301-4. [DOI] [PubMed] [Google Scholar]
- 42.Komatsu T, Takeuchi K, Yokoo J, Gotoh B. 2004. C and V proteins of Sendai virus target signaling pathways leading to IRF-3 activation for the negative regulation of interferon-beta production. Virology 325:137–148. doi: 10.1016/j.virol.2004.04.025. [DOI] [PubMed] [Google Scholar]
- 43.Irie T, Nagata N, Igarashi T, Okamoto I, Sakaguchi T. 2010. Conserved charged amino acids within Sendai virus C protein play multiple roles in the evasion of innate immune responses. PLoS One 5:e10719. doi: 10.1371/journal.pone.0010719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Irie T, Yoshida A, Sakaguchi T. 2013. Clustered basic amino acids of the small Sendai virus C protein Y1 are critical to its RAN GTPase-mediated nuclear localization. PLoS One 8:e73740. doi: 10.1371/journal.pone.0073740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Irie T, Kiyotani K, Igarashi T, Yoshida A, Sakaguchi T. 2012. Inhibition of interferon regulatory factor 3 activation by paramyxovirus V protein. J Virol 86:7136–7145. doi: 10.1128/JVI.06705-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yount JS, Gitlin L, Moran TM, Lopez CB. 2008. MDA5 participates in the detection of paramyxovirus infection and is essential for the early activation of dendritic cells in response to Sendai virus defective interfering particles. J Immunol 180:4910–4918. doi: 10.4049/jimmunol.180.7.4910. [DOI] [PubMed] [Google Scholar]
- 47.Sakaguchi T, Irie T, Kuwayama M, Ueno T, Yoshida A, Kawabata R. 2011. Analysis of interaction of Sendai virus V protein and melanoma differentiation-associated gene 5. Microbiol Immunol 55:760–767. doi: 10.1111/j.1348-0421.2011.00379.x. [DOI] [PubMed] [Google Scholar]
- 48.Oda K, Matoba Y, Irie T, Kawabata R, Fukushi M, Sugiyama M, Sakaguchi T. 2015. Structural basis of the inhibition of STAT1 activity by Sendai virus C protein. J Virol 89:11487–11499. doi: 10.1128/JVI.01887-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Calain P, Curran J, Kolakofsky D, Roux L. 1992. Molecular cloning of natural paramyxovirus copy-back defective interfering RNAs and their expression from DNA. Virology 191:62–71. doi: 10.1016/0042-6822(92)90166-M. [DOI] [PubMed] [Google Scholar]
- 50.Tapia K, Kim WK, Sun Y, Mercado-Lopez X, Dunay E, Wise M, Adu M, Lopez CB. 2013. Defective viral genomes arising in vivo provide critical danger signals for the triggering of lung antiviral immunity. PLoS Pathog 9:e1003703. doi: 10.1371/journal.ppat.1003703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Myers TM, Smallwood S, Moyer SA. 1999. Identification of nucleocapsid protein residues required for Sendai virus nucleocapsid formation and genome replication. J Gen Virol 80:1383–1391. doi: 10.1099/0022-1317-80-6-1383. [DOI] [PubMed] [Google Scholar]
- 52.Myers TM, Pieters A, Moyer SA. 1997. A highly conserved region of the Sendai virus nucleocapsid protein contributes to the NP-NP binding domain. Virology 229:322–335. doi: 10.1006/viro.1996.8429. [DOI] [PubMed] [Google Scholar]
- 53.Gotoh B, Takeuchi K, Komatsu T, Yokoo J, Kimura Y, Kurotani A, Kato A, Nagai Y. 1999. Knockout of the Sendai virus C gene eliminates the viral ability to prevent the interferon-alpha/beta-mediated responses. FEBS Lett 459:205–210. doi: 10.1016/S0014-5793(99)01241-7. [DOI] [PubMed] [Google Scholar]
- 54.Kato A, Kiyotani K, Kubota T, Yoshida T, Tashiro M, Nagai Y. 2007. Importance of the anti-interferon capacity of Sendai virus C protein for pathogenicity in mice. J Virol 81:3264–3271. doi: 10.1128/JVI.02590-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kato A, Kiyotani K, Sakai Y, Yoshida T, Nagai Y. 1997. The paramyxovirus, Sendai virus, V protein encodes a luxury function required for viral pathogenesis. EMBO J 16:578–587. doi: 10.1093/emboj/16.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kiyotani K, Sakaguchi T, Kato A, Nagai Y, Yoshida T. 2007. Paramyxovirus Sendai virus V protein counteracts innate virus clearance through IRF-3 activation, but not via interferon, in mice. Virology 359:82–91. doi: 10.1016/j.virol.2006.08.053. [DOI] [PubMed] [Google Scholar]
- 57.Xu J, Mercado-Lopez X, Grier JT, Kim WK, Chun LF, Irvine EB, Del Toro Duany Y, Kell A, Hur S, Gale M Jr, Raj A, Lopez CB. 2015. Identification of a natural viral RNA motif that optimizes sensing of viral RNA by RIG-I. mBio 6:e01265-15. doi: 10.1128/mBio.01265-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lynch S, Kolakofsky D. 1978. Ends of the RNA within Sendai virus defective interfering nucleocapsids are not free. J Virol 28:584–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martinez-Gil L, Goff PH, Hai R, Garcia-Sastre A, Shaw ML, Palese P. 2013. A Sendai virus-derived RNA agonist of RIG-I as a virus vaccine adjuvant. J Virol 87:1290–1300. doi: 10.1128/JVI.02338-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kiyotani K, Takao S, Sakaguchi T, Yoshida T. 1990. Immediate protection of mice from lethal wild-type Sendai virus (HVJ) infections by a temperature-sensitive mutant, HVJpi, possessing homologous interfering capacity. Virology 177:65–74. doi: 10.1016/0042-6822(90)90460-9. [DOI] [PubMed] [Google Scholar]
- 61.Yoshida T, Hamaguchi M, Naruse H, Nishikawa K, Nagai Y. 1983. Characterization of the virus isolated from HeLa cells persistently infected with Sendai virus (HVJ). Microbiol Immunol 27:207–211. doi: 10.1111/j.1348-0421.1983.tb03575.x. [DOI] [PubMed] [Google Scholar]
- 62.Nagai Y, Yoshida T, Yoshii S, Maeno K, Matsumoto T. 1975. Modification of normal cell surface by smooth membrane preparations from BHK-21 cells infected with Newcastle disease virus. Med Microbiol Immunol 161:175–188. doi: 10.1007/BF02121008. [DOI] [PubMed] [Google Scholar]
- 63.Shirogane Y, Takeda M, Iwasaki M, Ishiguro N, Takeuchi H, Nakatsu Y, Tahara M, Kikuta H, Yanagi Y. 2008. Efficient multiplication of human metapneumovirus in Vero cells expressing the transmembrane serine protease TMPRSS2. J Virol 82:8942–8946. doi: 10.1128/JVI.00676-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kurotani A, Kiyotani K, Kato A, Shioda T, Sakai Y, Mizumoto K, Yoshida T, Nagai Y. 1998. Sendai virus C proteins are categorically nonessential gene products but silencing their expression severely impairs viral replication and pathogenesis. Genes Cells 3:111–124. doi: 10.1046/j.1365-2443.1998.00170.x. [DOI] [PubMed] [Google Scholar]
- 65.Kato A, Sakai Y, Shioda T, Kondo T, Nakanishi M, Nagai Y. 1996. Initiation of Sendai virus multiplication from transfected cDNA or RNA with negative or positive sense. Genes Cells 1:569–579. doi: 10.1046/j.1365-2443.1996.d01-261.x. [DOI] [PubMed] [Google Scholar]
- 66.Fujii Y, Sakaguchi T, Kiyotani K, Huang C, Fukuhara N, Egi Y, Yoshida T. 2002. Involvement of the leader sequence in Sendai virus pathogenesis revealed by recovery of a pathogenic field isolate from cDNA. J Virol 76:8540–8547. doi: 10.1128/JVI.76.17.8540-8547.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Irie T, Okamoto I, Yoshida A, Nagai Y, Sakaguchi T. 2014. Sendai virus C proteins regulate viral genome and antigenome synthesis to dictate the negative genome polarity. J Virol 88:690–698. doi: 10.1128/JVI.02798-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sakai K, Ami Y, Tahara M, Kubota T, Anraku M, Abe M, Nakajima N, Sekizuka T, Shirato K, Suzaki Y, Ainai A, Nakatsu Y, Kanou K, Nakamura K, Suzuki T, Komase K, Nobusawa E, Maenaka K, Kuroda M, Hasegawa H, Kawaoka Y, Tashiro M, Takeda M. 2014. The host protease TMPRSS2 plays a major role in in vivo replication of emerging H7N9 and seasonal influenza viruses. J Virol 88:5608–5616. doi: 10.1128/JVI.03677-13. [DOI] [PMC free article] [PubMed] [Google Scholar]