ABSTRACT
The tetraspanin protein CD63 has been recently described as a key factor in extracellular vesicle (EV) production and endosomal cargo sorting. In the context of Epstein-Barr virus (EBV) infection, CD63 is required for the efficient packaging of the major viral oncoprotein latent membrane protein 1 (LMP1) into exosomes and other EV populations and acts as a negative regulator of LMP1 intracellular signaling. Accumulating evidence has also pointed to intersections of the endosomal and autophagy pathways in maintaining cellular secretory processes and as sites for viral assembly and replication. Indeed, LMP1 can activate the mammalian target of rapamycin (mTOR) pathway to suppress host cell autophagy and facilitate cell growth and proliferation. Despite the growing recognition of cross talk between endosomes and autophagosomes and its relevance to viral infection, little is understood about the molecular mechanisms governing endosomal and autophagy convergence. Here, we demonstrate that CD63-dependent vesicle protein secretion directly opposes intracellular signaling activation downstream of LMP1, including mTOR-associated proteins. Conversely, disruption of normal autolysosomal processes increases LMP1 secretion and dampens signal transduction by the viral protein. Increases in mTOR activation following CD63 knockout are coincident with the development of serum-dependent autophagic vacuoles that are acidified in the presence of high LMP1 levels. Altogether, these findings suggest a key role of CD63 in regulating the interactions between endosomal and autophagy processes and limiting cellular signaling activity in both noninfected and virally infected cells.
IMPORTANCE The close connection between extracellular vesicles and viruses is becoming rapidly and more widely appreciated. EBV, a human gamma herpesvirus that contributes to the progression of a multitude of lymphomas and carcinomas in immunocompromised or genetically susceptible populations, packages its major oncoprotein, LMP1, into vesicles for secretion. We have recently described a role of the host cell protein CD63 in regulating intracellular signaling of the viral oncoprotein by shuttling LMP1 into exosomes. Here, we provide strong evidence of the utility of CD63-dependent EVs in regulating global intracellular signaling, including mTOR activation by LMP1. We also demonstrate a key role of CD63 in coordinating endosomal and autophagic processes to regulate LMP1 levels within the cell. Overall, this study offers new insights into the complex intersection of cellular secretory and degradative mechanisms and the implications of these processes in viral replication.
KEYWORDS: exosomes, extracellular vesicles, amphisomes, multivesicular bodies, herpesvirus, Epstein-Barr virus, herpesviruses
INTRODUCTION
Epstein-Barr virus (EBV) is a pervasive gamma human herpesvirus that contributes to the development and progression of many types of malignancies in immunocompromised or genetically predisposed humans. Although the vast majority of people are latently infected with EBV, much remains to be understood about how EBV interacts with its host to contribute to tumorigenesis. Growing evidence has implicated the exosome pathway as a major system of interaction between host cells and viruses. Indeed, many viruses, including human herpesviruses, hepatitis viruses (A, B, C, and E viruses), human papillomaviruses, and human immunodeficiency virus (HIV), can hijack and use the host cell exosome pathway to facilitate secretion of viral and altered cellular cargo (1–10).
Importantly, circulating extracellular vesicles (EVs), including exosomes, have recently emerged as players in cell-to-cell communication (11–15) and viral transmission (1–4, 16). While knowledge of the molecular mechanisms governing exosome biogenesis is similarly incomplete, recently, our work has revealed that the cluster of differentiation 63 (CD63) is a major player in small vesicle production (17–19). CD63 is a member of the tetraspanin superfamily (20, 21), a group of proteins with four transmembrane domains and a series of conserved amino acid residues (22). Assembly into tetraspanin-enriched microdomains (TEMs) allows tetraspanin proteins such as CD63 to interact with other integral membrane proteins, including integrin molecules, immunoglobulins, proteoglycans, cadherins, and signaling molecules, on the cell surface (21, 23, 24). These protein interactions likely have implications in guiding cellular functions, including motility, adhesion, proliferation, and immune response. Cell surface CD63 also may be endocytosed into clathrin-coated vesicles and subsequently delivered to early endosomes (25). As a result, CD63 is often enriched in late endosomal and lysosomal compartments and readily secreted into exosomes following enrichment in intraluminal vesicles (ILVs) that bud into multivesicular bodies (MVBs). Both endosomal sorting complex required for transport (ESCRT) machinery and ceramide accumulation have been demonstrated to guide CD63 incorporation into ILVs (26, 27).
Based on interaction of CD63 with other TEM proteins and its route of trafficking into endosomes, multiple studies have suggested a role of CD63 in intracellular transport of cell surface proteins into the endosomal system. CD63 may aid in chaperoning major histocompatibility complex (MHC) class II molecules from the plasma membrane into endosomal compartments (28–30) and is additionally recruited to phagocytic vacuoles containing foreign cargo that is internalized and subsequently trafficked to endosomes (31). Several studies have also elucidated an interaction of CD63 with viral cargo within the endosomal system (32–34). Recently, for instance, we have demonstrated that CD63 is necessary for trafficking the Epstein-Barr virus (EBV)-encoded oncoprotein LMP1 into exosomes for secretion (19). Following CD63 knockout, LMP1 packaging into EVs and intracellular localization was dramatically disrupted, supporting a role of CD63 in vesicular protein sorting. Furthermore, in the context of LMP1-mediated signal transduction, CD63 negatively regulates the activation of mitogen-activated protein kinase (MAPK)/ERK and noncanonical NF-κB pathways (19, 34). Altogether, these data suggest that CD63-dependent exosomal secretion of signaling molecules and of the viral protein limits intracellular signal transduction by EBV. Transport of signaling proteins to recipient cells may also potentiate paracrine activation of similar cellular processes. It is clear that a better understanding of the intricacies of cellular exosome machinery is necessary to understanding the interactions of viral cargo with the host endosome system.
It is also likely that intersection of the endosomal system and autophagy components will shed additional light on regulating intracellular processes during viral infection. Basal levels of autophagy serve to turn over long-lived or damaged organelles, diminish reactive oxygen species, and recycle macromolecule building blocks when cellular nutrient supplies are low (35, 36). Recent findings have described convergence of the exosome and macroautophagy pathways that appears critical for secretory function in certain cells (37). The macroautophagy pathway is initiated by many cellular signals, including nutrient deprivation, growth factor restriction, or viral infection, and results in the engulfment of cytoplasmic material following formation of a partially membrane-bound phagophore that matures into a completely membrane-enclosed autophagosome. Conversion of the cytosolic protein LC3-I into LC3-II is required for closure of the phagophore to create the autophagosome. Autophagosomes can then fuse with lysosomes where cargo is degraded and recycled back into the cytoplasm or can fuse with late endosomes to form amphisomes, an intermediate organelle whose function is unknown but can also fuse with lysosomes for degradation of contents (38). An alternative pathway of autophagy, called chaperone-mediated autophagy (CMA), relies upon a 70-kDa heat shock protein (HSC70) to traffic cytosolic proteins into lysosomes for degradation (39). Interestingly, HSC70 is also enriched in many EV populations as well as late endosomal compartments and is believed to guide endosomal microautophagy, a process that has molecular similarities to lysosomal CMA (40). Together, autophagosome fusion to MVBs through macroautophagy processes and direct endosomal delivery of cytosolic proteins by microautophagy represent two major mechanisms of endosome-mediated protein sorting and degradation.
Accumulating evidence has implicated the role of host autophagy in multiple antiviral immunity processes (41, 42). Degradation of microbes by autophagy pathways, called xenophagy (43), provides an innate immune response by directly engulfing invading pathogens, such as virions, for lysosomal degradation. In addition, transport of viral proteins through the autophagy pathway can deliver pathogen-associated molecular patterns to toll-like receptors (TLRs) that reside in the endosome, triggering the secretion of type I interferons (IFNs) and proinflammatory cytokines (44). Autophagosomes may also aid in adaptive immune responses by delivering viral antigens to MHC class II loading compartments for antigen presentation (45, 46).
However, as with many other host defense mechanisms, viruses have coevolved to evade the autophagic machinery driving antiviral immune responses (41). Particularly well adapted to suppress autophagy are the herpesviruses, likely contributing to their ability to maintain lifelong persistent infections. Viruses such as herpes simplex virus 1 (HSV-1), cytomegalovirus (CMV), Kaposi's sarcoma-associated herpesvirus (KSHV), HIV, and influenza A virus antagonize host autophagy activation that may be partially due to stimulation of the mammalian target of rapamycin (mTOR) pathway (47–52). Stimulation of mTOR reduces levels of autophagic activity (53) through interaction with lysosomal adaptor and MAPK and mTOR activator/regulator (LAMTOR) and MAPK/ERK signaling proteins that are recruited to late endosomal and lysosomal surfaces (54–57). Activation of mTOR signaling by nutrient abundance or viral infection can promote energy metabolism, enhance protein and lipid synthesis, and increase cellular proliferation, facilitating viral replication and survival (58, 59).
Many viruses additionally have evolved to take advantage of autophagic machinery. Picornaviruses, hepatitis C virus, HIV, and several herpesviruses can induce levels of early, nondegradative autophagy components, perhaps as membranous scaffolds for virion production or to facilitate transport to multivesicular bodies for release (60–65). EBV further utilizes host autophagy to regulate levels of its oncoprotein LMP1 within cells (66). High levels of LMP1 expressed in cells may trigger apoptosis or cytostasis, while lower levels can induce proliferation of host cells (66). Thus, exploitation of the autophagic degradation pathway may allow EBV to monitor and regulate levels of LMP1. We previously suggested that exosomal secretion of LMP1 similarly provides a balance of the levels of viral protein in cells. Given the close connection between the endosomal and autophagic pathways, it is likely that an understanding of the molecular mechanisms involved in the intersection of exosomal secretion and autophagy maintenance will shed further light on these intrinsic biological processes, especially in the context of viral infection.
In this study, we aimed to further investigate the endosomal cargo dependent upon CD63 for sorting into exosomes. We demonstrate the requirement of CD63-dependent exosome trafficking in negatively regulating the LAMTOR/mTOR pathways downstream of LMP1, determine the role of the LMP1 CTAR1 domain for mTOR activation and increased vesicle production, and provide a novel role of CD63 in the maintenance of autophagic activity during viral infection. Finally, we reveal new evidence of intersecting molecular mechanisms of exosome and autophagosome interaction that regulate intracellular signaling activity in the context of EBV infection.
RESULTS
CD63 is required for proper protein packaging into small extracellular vesicles.
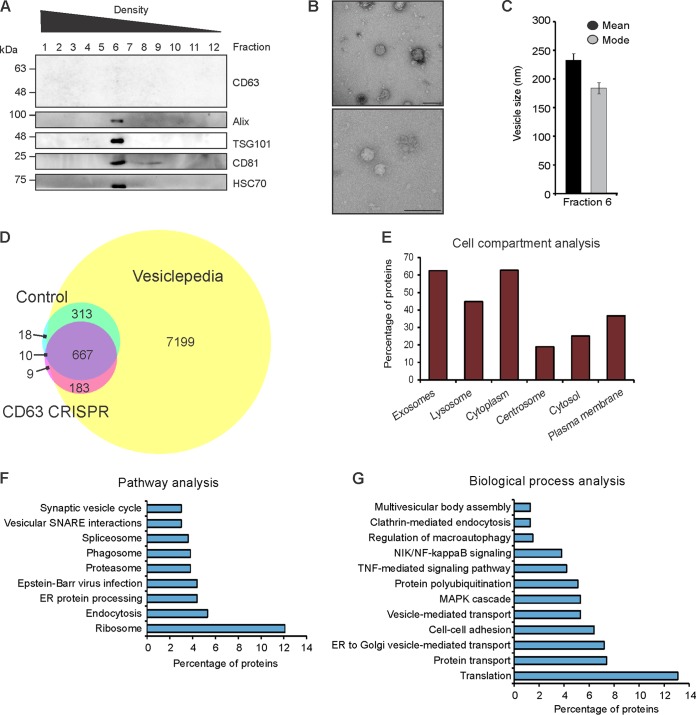
We have previously demonstrated that depletion of CD63 in HEK293 cells results in a decrease in small exosome-sized vesicle production (17), suggesting a key role of the tetraspanin protein in the endosomal pathway. In addition, CD63 is likely required for specific sorting of proteins into exosomes for secretion, including LMP1, in the context of Epstein-Barr virus infection. To assemble a global scope of CD63-dependent protein trafficking into exosomes, vesicles were harvested and purified from control or CD63 CRISPR knockout HEK293 cells by iodixanol density gradient, as described previously (17, 67). Immunoblot analysis of purified density fractions revealed the majority of EV protein from CD63 CRISPR cells to be in fraction 6, similar to what we have previously observed in control cells (Fig. 1A) (67). Electron microscopy of fraction 6 confirmed small vesicle-shaped particles ranging from 50 to 250 nm in size (Fig. 1B). Nanoparticle tracking analysis of fraction 6 showed the mode and mean sizes of EVs to be between 150 and 250 nm (Fig. 1C). Subsequent mass spectrometry analysis of three biological replicates identified a total of 1,200 unique proteins, 331 of which were specific to control EVs compared to CD63 CRISPR cell-derived EVs (Fig. 1D; see also Table S1 in the supplemental material). Comparison to the Vesiclepedia database confirmed considerable overlap of the data set with known vesicular proteins. Identified proteins were enriched in exosome, lysosome, and cytoplasmic subcellular compartments (Fig. 1E and Table S2). Further examination revealed many proteins to be decreased by greater than 2-fold in CD63 knockout EVs, including proteins involved in ribosomal, endosomal, phagosomal, and general vesicular pathways (Fig. 1F). In addition to decreased packaging of many proteins involved in protein translation and vesicle transport, key proteins involved in signal transduction pathways, including mitogen-activated protein kinase 1 (MAPK1), proteasome proteins involved in NF-κB protein activation, and regulator complex proteins LAMTOR1 and LAMTOR2, were observed to be decreased in EVs following CD63 knockout (Fig. 1G). As evident in the case of LMP1 exosomal packaging, CD63-mediated trafficking into exosomes may serve to regulate intracellular protein activity (19, 34). We previously demonstrated increases in MAPK/ERK and noncanonical NF-κB signaling activation by the EBV protein LMP1 following impaired exosomal trafficking in the absence of CD63 (19). These findings altogether support that exosomal protein packaging may be tightly linked to the negative regulation of intracellular signaling pathways.
FIG 1.
CD63 is required for proper sorting of protein cargo into small extracellular vesicles. Small EVs derived from HEK293 control or CD63 CRISPR cells were purified by iodixanol density gradient ultracentrifugation for mass spectrometry analysis. (A) The presence of EVs in the fractions was determined by immunoblot analysis for common EV markers. A representative gradient of EVs purified from CD63 CRISPR cells is shown, with equal volumes loaded. The presence of EVs in fraction six of the gradient was confirmed by electron microscopy (B) and nanoparticle tracking analysis (C). (D) Overlap of proteins found in control or CD63 CRISPR cell-derived EVs identified by mass spectrometry compared to the Vesiclepedia database of EV proteins. (E) FunRich analysis of cellular compartments enriched in all proteins identified. Analysis of pathways (F) and biological processes (G) enriched in the data set of proteins decreased by 2-fold or greater in CD63 CRISPR EVs. Size bar, 250 nm.
Moreover, many proteins were observed to be more highly secreted in EVs derived from CD63 knockout cells, representing cargo that is packaged into vesicles independent of CD63-mediated trafficking (Table S1). These components were predominately involved in cell adhesion and cytoskeletal function, including integrin subunits ITGA1, ITGA3, ITGA4, ITGAV, and ITGB5, fibronectin, actin, and collagen proteins. We previously proposed that many cytoskeletal proteins were associated with the production of vesicles not dependent on CD63 (17). These vesicles may comprise other exosome subpopulations or nonexosomal vesicles.
CD63 negatively regulates LMP1-mediated mTORC1 activation.
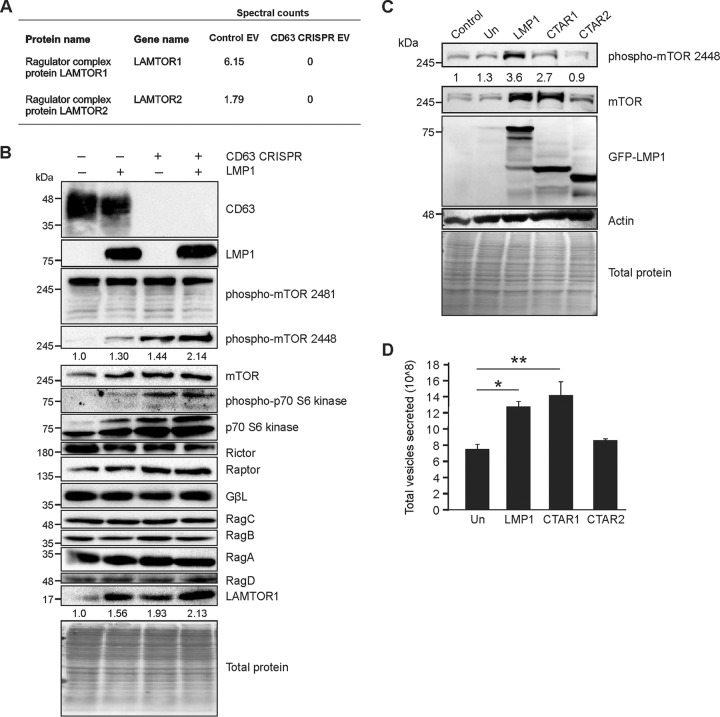
Recently, LMP1 has similarly been shown to activate the mTOR pathway (68, 69). It is likely that EBV hijacks mTOR signaling to promote host cell growth and lipid synthesis for viral replication and the establishment of latency while subduing host autophagic antiviral response. Cellular proliferation by LMP1-induced mTOR activation may also contribute to EBV-associated tumorigenesis. Here, we found key mTOR-associated proteins to be decreased in EVs following CD63 knockout (Fig. 2A), suggesting CD63 similarly provides a means to negatively regulate mTOR activation by LMP1 through vesicle sorting and secretion.
FIG 2.
CD63 negatively regulates mTORC1 signaling by Epstein-Barr virus LMP1. (A) LC-MS/MS spectral count quantification of LAMTOR proteins secreted in EVs from control and CD63 CRISPR cells. (B) HEK293 control or CD63 CRISPR cells were grown in serum-free medium and transfected with GFP or GFP-LMP1 for 24 h before cytoplasmic fractions were isolated, lysed, and analyzed by immunoblot analysis, with equal protein mass loaded. (C) Wild type (LMP1) and LMP1 C-terminal mutants containing the CTAR1 domain but lacking CTAR2 (CTAR1) or containing the CTAR2 domain but lacking CTAR1 (CTAR2) were induced by doxycycline in HK1 cells. Cell lysates were analyzed by immunoblotting for mTOR activation. All blots are representative images from 3 independent experiments. Quantitation of bands is based on averages from all experiments. (D) EVs were collected from HK1 cells following LMP1 mutant induction, and total vesicles were quantitated by nanoparticle tracking analysis. Un, uninduced. **, P < 0.01; *, P < 0.05.
Two distinct signaling complexes have been identified within the mTOR pathway. The mTORC2 complex is not well understood but is likely dependent upon upstream Akt phosphorylation for activation and consists of mTOR, GβL, rapamycin-insensitive companion of mTOR (Rictor), and other associated proteins (70–73). In contrast, the mTORC1 complex has been characterized as a major regulator of autophagy in cells. There are many upstream signals known to activate mTORC1, including the MAPK/ERK pathway (74), previously shown to be hyperactivated in the absence of CD63 (19). The mTORC1 complex consists of several protein components, including the catalytic subunit mTOR, regulatory-associated protein of mTOR (Raptor), and GβL protein (70, 72). In addition, translocation of mTORC1 protein components require Rag GTPase and LAMTOR proteins to dock on the surface of lysosomes for signaling (56, 57). Once on the lysosomal membrane, v-type H+ ATPases associate with the complex and appear to be important for relaying signals induced by the accumulation of amino acids in the lysosomal lumen (75).
Here, we observed that introduction of LMP1 into cells resulted in an increase in phosphorylation of the mTOR protein at the Ser2448 site, consistent with activation of the mTORC1 complex (76), where no change was detected in Ser2481 phosphorylation to activate mTORC2 (Fig. 2B). Noticeably, we observed increases in mTORC1 phosphorylation and subsequent increases in levels of phosphorylated and total p70 S6 kinase, a downstream target of mTOR, following CD63 knockout, augmented by the presence of the viral protein. In addition, increased accumulation of LAMTOR1, the major protein responsible for anchoring the mTORC1 complex to the lysosomal membrane, was observed in the absence of CD63 (Fig. 2B), correlating with a decrease in amount of secretion (Fig. 2A).
To determine which signaling domains of LMP1 are responsible for mTORC1 activation, inducible HK1 cell lines containing wild-type (WT) LMP1 or signaling-defective mutants CTAR1 and CTAR2 were analyzed following doxycycline induction and compared to uninduced or parental cell lysates. These data revealed that the mutant lacking CTAR2 in the C-terminal tail (called CTAR1) is sufficient to activate mTORC1, whereas the mutant lacking the CTAR1 domain (CTAR2) lost this ability (Fig. 2C). These data were not surprising, as CTAR1 is important for the activation of PI3K/AKT and MAPK/ERK, two pathways upstream of mTORC1 (77). Interestingly, LMP1-induced vesicle secretion was seen in the presence of the CTAR1-containing mutant but not with the mutant lacking CTAR1 (CTAR2) (Fig. 2D). These findings suggest that activation of mTOR and induction of vesicle secretion by LMP1 are connected and regulated through the CTAR1 domain.
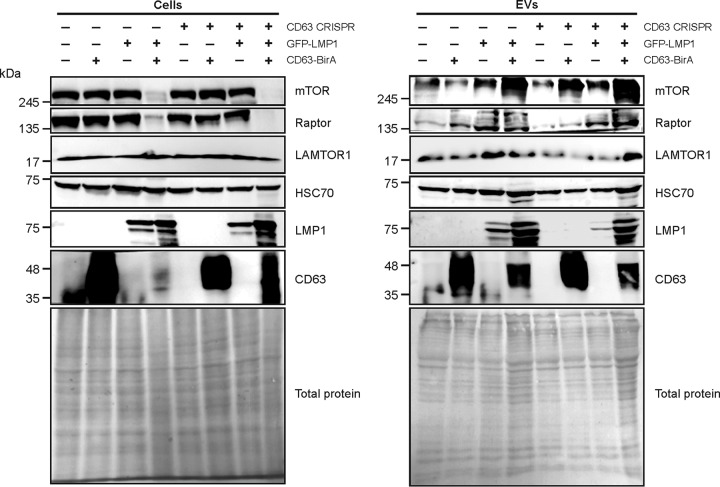
To confirm the reduced levels of mTOR-associated proteins packaged into EVs from CD63 knockout cells (Fig. 2A), immunoblot analysis of vesicles was performed (Fig. 3). Reduction of total mTOR, Raptor, and LAMTOR1 proteins secreted in EVs following CD63 knockout was seen, with little difference in HSC70 loading control levels. In attempts to rescue mTOR protein packaging, C-terminal BirA-fused CD63 protein was introduced into CD63 knockout cells to restore CD63 levels. Notably, CD63 levels were detectable in EVs secreted from cells containing CD63-BirA. Rescue or overexpression of CD63 using the fused protein increased mTOR and Raptor secretion into EVs from cells with or without LMP1. LAMTOR1 secretion was also rescued following introduction of CD63-BirA into CD63 CRISPR cells. Altogether, these findings strongly suggest that CD63 is important for trafficking components of the mTORC1 pathways, perhaps as a mechanism to negatively regulate hyperactive signaling. Interestingly, LMP1 expression also resulted in increased EV release of LAMTOR1, mTOR, and Raptor that was further increased when cotransfected with the CD63 construct. Overexpression of CD63 with LMP1 also resulted in enhanced LMP1 secretion, providing further evidence for the importance of CD63 in LMP1 exosomal trafficking and secretion.
FIG 3.
Rescue of CD63 increases vesicle secretion of LMP1 and mTOR-associated proteins. GFP-LMP1 and fused CD63-BirA proteins were transfected into HEK293 control or CD63 CRISPR cells. Cell lysates and EVs were collected for immunoblot analysis of mTOR-associated protein levels 24 h after transfection. All blots are representative images from independent experiments.
CD63 knockout disrupts cell vacuolar sorting processes.
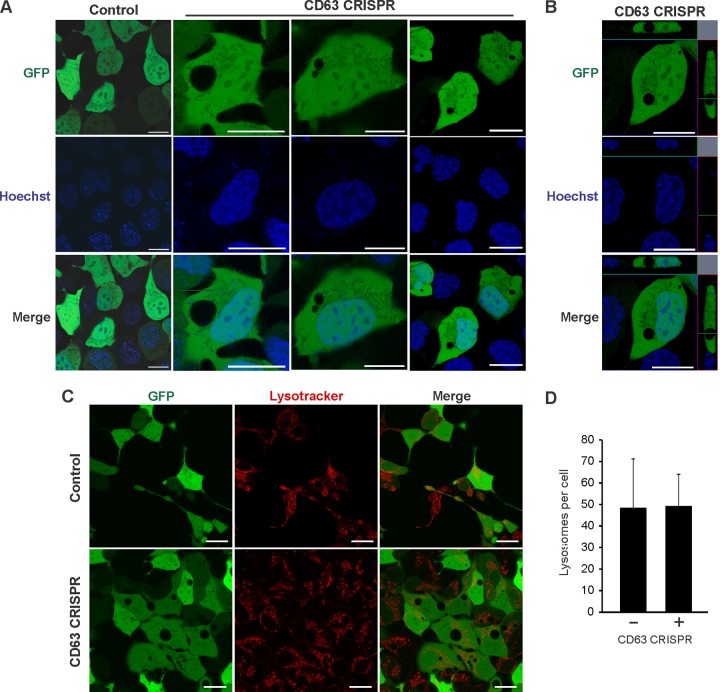
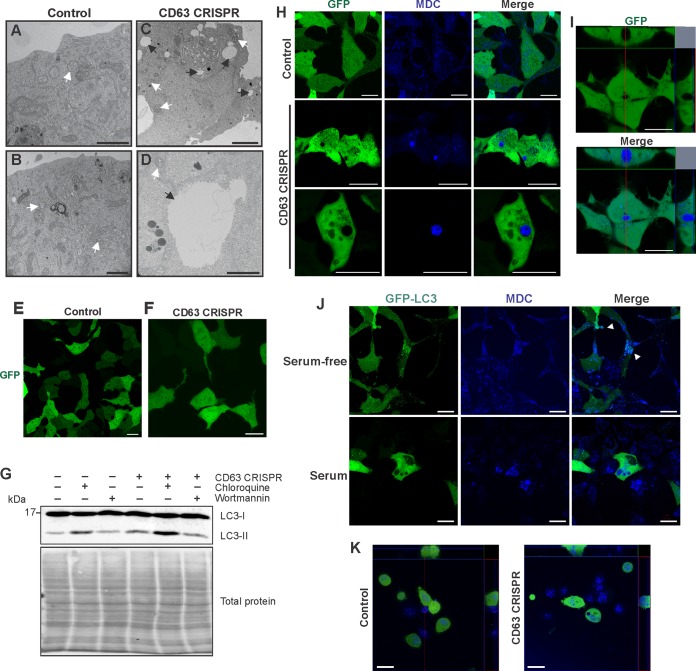
The mTOR signaling pathway is well known to regulate activity of macroautophagy. We have shown that CD63 acts as a negative regulator of mTORC1 (Fig. 2), and that many proteins involved in autophagic processes are not packaged efficiently into secreted vesicles in the absence of CD63 (Fig. 1). For instance, multiple H+ ATPase proteins involved in macroautophagy were found to be decreased in EVs secreted from CD63 knockout cells (Fig. 1 and Table S1), suggesting these proteins are more active within the cell. Strikingly, in addition to disruptions to endosomal protein trafficking and exosome secretion, morphological changes to cells were apparent following CD63 knockout. In the absence of CD63 protein, cells developed large vacuole-like compartments visible by light (data not shown) and confocal microscopy (Fig. 4A). Orthogonal projections confirmed the absence of green fluorescent protein (GFP) localized to these vacuoles (Fig. 4B). Based on our previous findings noting reductions in small vesicle secretion, and because MVBs are known to traffic either to the plasma membrane for exosome secretion or conversely to fuse to lysosomes for degradation, we hypothesized that these large cellular compartments are endocytic or lysosomal organelles. However, interrogation of the identity of the organelles with a lysosome stain (LysoTracker) suggested the vacuoles are not primarily lysosomal in origin (Fig. 4C). Notably, no significant difference was seen in the number of lysosomes following CD63 knockout (Fig. 4D).
FIG 4.
CD63 CRISPR cells develop large nonlysosomal cellular compartments. (A) HEK293 control or CD63 CRISPR cells were transfected with GFP to visualize large cellular structures by confocal microscopy. Hoechst stain was used to identify nuclear compartments. (B) Orthogonal projection of a representative CD63 CRISPR cell. (C) Cells were stained by LysoTracker to examine acidified compartments within cells. (D) Quantification of total LysoTracker-positive compartments in cells (n = 20 cells). Size bars, 20 μm.
Vacuolar compartments following CD63 knockout are linked to autophagy.
Closer inspection of these structures within CD63 knockout cells by electron microscopy revealed highly electron-lucent compartments much larger in size than observed MVBs (Fig. 5A to D) and largely resembling vacuolar organelles. Although vacuoles historically have been examined in plants and fungi, mammalian cells can develop these subcellular structures as well (78). Subcellular compartments induced by CD63 depletion contained occasional intraluminal membranes (Fig. 5D). Interestingly, following serum starvation, CD63 knockout-induced vacuolar compartments largely disappeared from cells (Fig. 5E and F), suggesting these structures are nutrient dependent. Of note, under conditions of serum starvation, LMP1 secretion was not observed to decrease (data not shown). As endosomal pathways within the cell are known to intersect the autophagy pathway, we examined levels of an autophagosomal protein marker, LC3, in CD63 knockout cells compared to those of controls (Fig. 5G). Levels of LC3-II, the activated cleaved form of LC3-I, appeared slightly but reproducibly increased following CD63 knockout. This increase in LC3-II protein was augmented in the presence of a lysosome acidification inhibitor, chloroquine, indicating a relatively higher level of autophagic flux in CD63 knockout cells, even after treatment with the phosphoinositide 3-kinase (PI3K) inhibitor wortmannin. These findings suggest that CD63 plays a direct or indirect role in regulating levels of autophagosome turnover by bridging the endosomal and autophagy pathways within the cell. To determine the origin of the vacuolar compartments, cells were stained with mono-dansylcadaverine (MDC), a marker for autophagic vacuoles (79) (Fig. 5H). Large vacuoles in CD63 knockout cells were observed to stain positive as autophagic vacuoles, confirming the role of CD63 in maintaining both endosomal and autophagic processes (Fig. 5I). Quantitation of MDC-positive and total large vacuoles in cells revealed 80% of vacuoles stained with MDC. Under serum-starved conditions, membrane-bound LC3-II was seen to localize to small MDC-positive vacuoles, as previously reported (Fig. 5J) (80). However, under full-serum conditions, LC3-GFP remained diffuse and did not localize to the large vacuoles seen in CD63 knockout cells (Fig. 5J), supporting a unique identity of these vacuoles. Finally, introduction of the CD63 CRISPR in a nasopharyngeal carcinoma line (HK1 cells) demonstrated the development of similar large MDC-positive vacuoles (Fig. 5K).
FIG 5.
CD63 knockout induces the development of large autophagic vacuoles. HEK293 control (A and B) or CD63 CRISPR (C and D) cells were fixed and stained for transmission electron microscopy to visualize large intracellular vacuoles. Black arrows denote vacuoles, and white arrows denote MVBs. Size bar, 1 μm. HEK293 control (E) or CD63 CRISPR (F) cells were transfected with GFP following serum starvation for 24 h. (G) Cells were treated with chloroquine or wortmannin under serum-deprived conditions for 2 h. LC3 levels were measured by immunoblot analysis. Blots are representative of three independent experiments. (H) Cells were transfected with GFP and stained with mono-dansylcadaverine (MDC) to examine the morphology and localization of autophagic vacuoles. (I) Orthogonal sections of CD63 CRISPR cells stained with MDC. (J) HEK293 CD63 CRISPR cells were transfected with GFP-LC3 and grown under full-serum or serum-free conditions before staining with MDC to visualize vacuoles by confocal microscopy. (K) Nasopharyngeal carcinoma cells (HK1) and an HK1 CD63 CRISPR derivative cell line were transfected with GFP and stained with MDC to visualize autophagic vacuoles. Size bars, 20 μm.
Inhibition of autophagy increases vesicular protein secretion and limits intracellular LMP1 signaling.
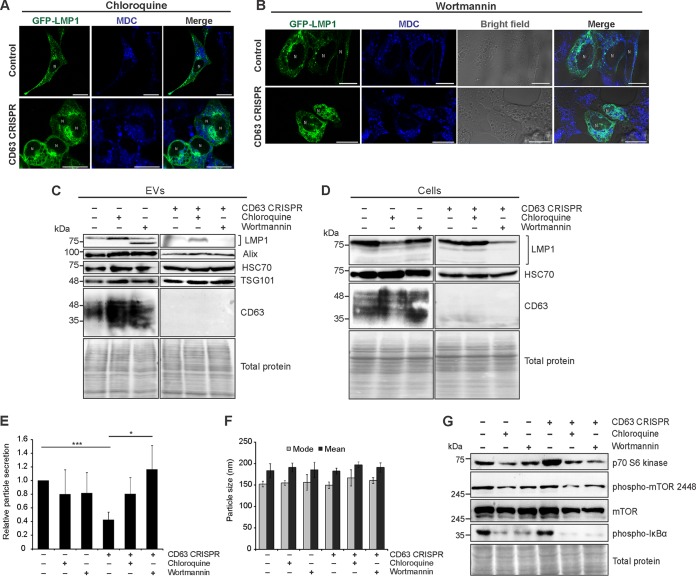
MVB secretion of ILVs into the extracellular milieu is generally believed to oppose MVB fusion to lysosomes and subsequent degradation. However, directional trafficking of endosomes to lysosomes or to the plasma membrane are not well understood. Here, we demonstrate altered cellular trafficking of autophagy-associated proteins and the appearance of autophagic vacuoles following CD63 knockout. As increasing evidence suggests exosome secretion and autophagy processes are tightly linked, we examined vesicular secretion of proteins in the presence of the autophagy inhibitors chloroquine and wortmannin. Chloroquine disrupts lysosomal acidification, preventing autophagosome fusion and degradation, while wortmannin inhibits PI3K-dependent autophagic sequestration of cytoplasmic contents into phagophores (81, 82). Inhibition of auto- and endolysosomal processes by chloroquine treatment resulted in a buildup of small MDC-positive vacuoles in control cells that were increasingly apparent in CD63 CRISPR cells (Fig. 6A). Treatment of cells with wortmannin produced large swollen compartments seen under bright field that were largely MDC negative (Fig. 6B). These compartments likely are enlarged late endosomal compartments, as previously described (82–84). Disruption of autophagy processes by treatment of control cells with chloroquine or wortmannin resulted in increases in vesicular protein present in the media of control cells, including CD63 and LMP1, as well as ESCRT-associated vesicular proteins TSG101 and Alix (Fig. 6C). Degradation of vesicular LMP1 was repeatedly noted to be increased following wortmannin treatment, perhaps due to increases in other active proteases secreted into vesicles. Cellular levels of LMP1 were decreased in the presence of autophagy inhibitors, corresponding to concurrent increases in vesicle secretion (Fig. 6D). Following CD63 knockout, chloroquine treatment rescued some secretion of LMP1, presumably into an alternative CD63-negative vesicle population (Fig. 6C). Wortmannin treatment did not increase LMP1 secretion from these cells, perhaps due in part to decreased cellular LMP1 (Fig. 6C). Nanoparticle tracking analysis of EVs released from cells treated with autophagy inhibitors demonstrated no significant change in EV secretion from control cells (Fig. 6E). However, inhibition of autophagic processes in CD63 knockout cells rescued EV secretion in CD63 knockout cells, likely by shunting endosomal vesicles toward secretion. Thus, it is conceivable that increases in vesicle protein secretion (Fig. 6C) are due to both increased cargo loading into EVs as well as increases in vesicle number. Importantly, no difference in size was seen in EVs secreted from control or CD63 knockout cells of either treatment group, suggesting that under these conditions the EV preparations did not contain increased numbers of larger microvesicles or apoptotic bodies and that cell viability was not compromised (Fig. 6F). This was further supported by no significant difference in the percentage (<5%) of cells undergoing apoptosis between control and treated cells as well as the absence of apoptotic body marker calnexin in EVs (data not shown). Finally, activated levels of phosphorylated mTOR at the Ser2448 site, the mTOR substrate p70 S6 kinase, and the canonical NF-κB protein IκBα by LMP1 were observed to decrease following treatment of cells with chloroquine or wortmannin (Fig. 6G). Altogether, these findings demonstrate that autophagic processes directly oppose vesicular secretion of proteins. In the context of EBV, blocking autophagic processes increases vesicle secretion of LMP1 and results in subsequent decreases in intracellular protein signaling.
FIG 6.
Extracellular vesicle secretion and autophagic processes regulate LMP1-mediated intracellular signaling. Confocal microscopy of GFP-LMP1-transfected control or CD63 CRISPR cells following chloroquine (A) and wortmannin (B) treatment. Vacuoles were stained with MDC before live-cell imaging. Size bars, 20 μm. N, nuclear compartments. (C) Immunoblot analysis of EVs derived from HEK293 control or CD63 CRISPR cells following transfection of GFP-LMP1 and treatment with chloroquine or wortmannin for 24 h, with equal volumes loaded. (D) Immunoblots of corresponding cell lysates, with equal protein mass loaded. Nanoparticle tracking analysis of the quantity (E) and size (F) of EVs from cells following treatment with autophagy inhibitors. (G) Immunoblot analysis of cytoplasmic cellular fractions of cells transfected with GFP-LMP1, with equal protein mass loaded. Blots are representative images from repeated independent experiments. ***, P < 0.001; *, P < 0.05.
Activation of autophagy by trehalose increases CD63-independent LMP1 secretion.
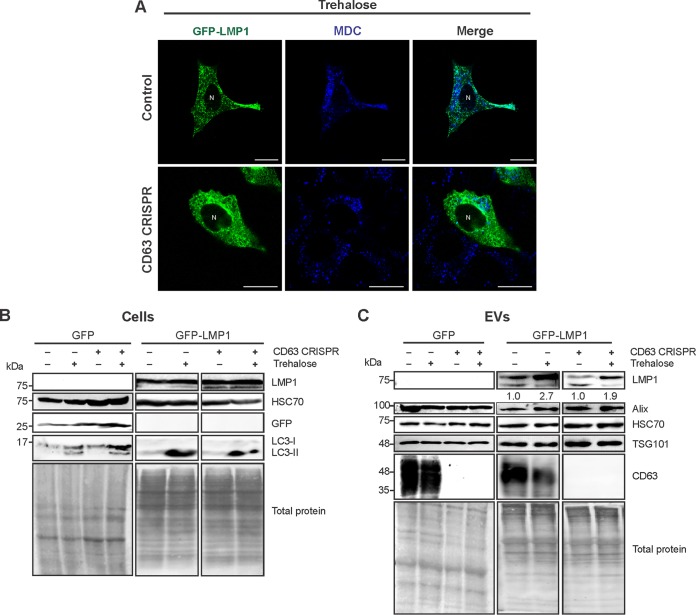
We similarly sought to examine secretion of the viral protein in the presence of an autophagy activator. Trehalose is a disaccharide that can induce autophagy through blockade of membrane glucose transporters, thereby triggering a starvation-like response. This mechanism is, at least in part, believed to be independent of mTOR inhibition (85–88). Upon trehalose treatment, the large MDC-positive vacuoles in CD63 CRISPR cells were mitigated (Fig. 7A). LC3-II levels were notably increased in trehalose-treated cells following GFP or GFP-LMP1 transfection, supporting an increase in autophagic flux (Fig. 7B). Strikingly, treatment of both control and CD63 knockout cells with trehalose resulted in reproducible increases in LMP1 secretion with no significant changes in other cases of EV protein secretion (Fig. 7C). We therefore propose that activation of autophagic flux can increase CD63-independent LMP1 release that may traffic through autophagic vacuoles.
FIG 7.
Autophagy activation by trehalose drives CD63-independent LMP1 release. (A) Confocal microscopy of GFP-LMP1-transfected control or CD63 CRISPR cells following trehalose treatment. Vacuoles were stained with MDC before live-cell imaging. Size bars, 20 μm. N, nuclear compartments. (B) Immunoblot analysis of cytoplasmic lysates from HEK293 control or CD63 CRISPR cells following transfection of GFP or GFP-LMP1 and treatment with trehalose for 24 h, with equal protein mass loaded. (C) Immunoblots of corresponding cell-derived EVs, with equal volumes loaded. Blots are representative images from repeated independent experiments. Quantitation of LMP1 secreted following trehalose treatment was normalized to parental cell secretion over multiple experiments.
Autophagic vacuoles acidify in the presence of LMP1.
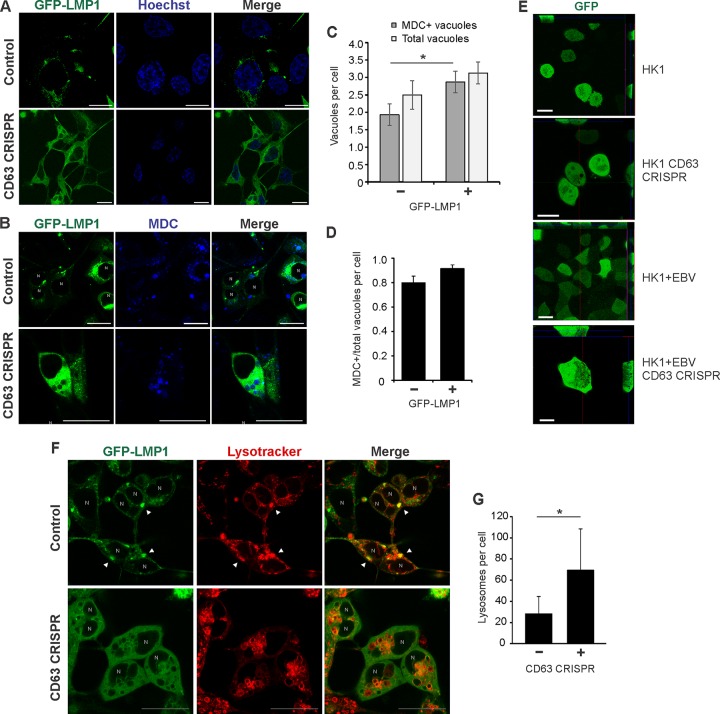
In this study, we observed LMP1 secretion to increase following disruption of autolysosomal processes, indicating an antagonism between exosome secretion and autolysosomal degradation. Furthermore, depletion of CD63 results in a decrease in vesicle production and concomitant accumulation of large intracellular vacuolar compartments. However, these vacuolar structures appeared to be nonacidic and instead are intermediate compartments representing endosome and autophagosome intersection. At high levels, LMP1 has been shown to promote late stages of autophagy, evident by increased acidic lysosomes and autolysosomes (66). Here, GFP-LMP1 similarly localized to the perinuclear area in control cells, but localization was disrupted following CD63 knockout, as previously described (Fig. 8A) (19). In the presence of high levels of GFP-LMP1, MDC-positive vacuoles again were observed in CD63 CRISPR cells (Fig. 8B). Notably, MDC-positive vacuoles appeared larger in both cell lines transfected with GFP-LMP1 than in control GFP-transfected cells (Fig. 5H). Furthermore, introduction of LMP1 into CD63 CRISPR cells resulted in a slight increase in the number of total large vacuoles and a significant increase in the number of MDC-positive vacuoles (Fig. 8C and D). An increase in the number of vacuoles in HK1 cells was similarly seen following infection with EBV (Fig. 8E). We also observed GFP-LMP1 to correspond and colocalize with increased LysoTracker signal, indicating increases in lysosomal compartments at the site of LMP1 localization (Fig. 8F). Strikingly, in the presence of LMP1, CD63 knockout-induced vacuole membranes were stained with LysoTracker, indicating acidification of these compartments (Fig. 8F). In fact, in the presence of LMP1, CD63 knockout cells contained a significantly greater number of lysosomes (Fig. 8G). These findings suggest that increased autophagic flux induced by LMP1 results in subsequent acidification of the autophagic vacuoles in CD63 knockout cells.
FIG 8.
LMP1 promotes acidification of CD63 knockout-induced autophagic vacuoles. HEK293 control and CD63 CRISPR cells were transfected with GFP-LMP1 and stained with Hoechst (A) and MDC (B). (C) MDC-positive and total vacuoles were quantified for each cell (n = 30 cells). (D) Proportion of MDC-positive over total vacuoles per CD63 CRISPR cell. (E) HK1 control and CD63 CRISPR cells were transfected with GFP to visualize vacuoles. EBV-infected HK1 (HK1 + EBV) control cells and cells containing CD63 CRISPR were treated similarly to visualize autophagic vacuoles. (F) Cells were transfected with GFP-LMP1 and stained with LysoTracker. Arrowheads represent colocalization of LMP1 with lysosomal compartments. (G) LysoTracker-positive compartments quantitated for each cell (n = 20 cells). N, nuclear compartments. *, P < 0.05.
DISCUSSION
Interactions between the host exosome pathway and viruses have recently emerged as important mechanisms of viral replication, assembly, and spread. However, many of the molecular mechanisms surrounding these interactions remain to be understood. Recently, we demonstrated that a commonly used exosome marker, tetraspanin protein CD63, plays an important role in small EV secretion and trafficking of a viral protein, LMP1, into exosomes (19). However, few studies have focused on the mechanisms of CD63 trafficking into vesicles and the role of the tetraspanin in EV production, particularly in the context of viral infection. In this study, we further illuminate the role of CD63 in global vesicle protein trafficking. Mass spectrometry analysis of small EVs derived from CD63 knockout cells revealed the packaging of many proteins to be impaired in the absence of CD63. Previously, Kowal and colleagues performed a mass spectrometry analysis of proteins in CD63-positive vesicle populations using a pulldown assay (89). A number of proteins identified in CD63-positive vesicles in the study were present in our wild-type EVs but absent from our CD63 CRISPR EVs, including autophagy-related proteins (ATG9A), vesicular integrin-membrane protein (LMAN2), multiple proteasome regulatory units, sorting nexins (SNX2), transmembrane proteins (TMED7 and TMED9), and vesicle-associated membrane proteins (VAMP2 and VAPA). This comparison further supports the dependence of many vesicular proteins upon CD63 for secretion. On the other hand, numerous proteins involved in cytoskeletal or adhesion functions were quantitatively increased in EVs following CD63 knockout. One explanation for the observation of increases in specific proteins or the presence of some proteins in vesicles only after CD63 knockout may be that CD63-independent vesicle cargo is relatively more abundant in the absence of CD63-dependent cargo sorting. As such, mass spectrometry analysis of equal vesicular protein can detect increases in abundance of these proteins. Another possibility is that increases in other vesicle populations provides a means of compensatory cargo secretion. It is worth noting that the vast majority of proteins secreted only in CD63 knockout cell-derived EVs are known vesicular proteins (Fig. 1D) and therefore likely do not represent artifactual nonvesicular cargo. Future investigation of EV subpopulations, including those dependent on CD63 trafficking, will aid in understanding global vesicle loading and secretion.
Interestingly, a number of proteins inefficiently sorted into vesicles following CD63 depletion play important roles in intracellular signal transduction, particularly in the context of EBV-associated cancers. Previous findings revealed CD63 to negatively regulate MAPK/ERK and noncanonical NF-κB pathways downstream of the viral oncoprotein LMP1. Here, we found MAPK1, tumor necrosis factor (TNF) receptors, and proteasomal proteins involved in NF-κB activation to be decreased in vesicle secretion in the absence of CD63. Similarly, we demonstrate detectable decreases in LAMTOR1/2 and v-type H+ ATPases in EVs, corresponding to increases in mTORC1 activation in CD63 knockout cells. It is likely that vesicular packaging of signal transduction molecules activated by EBV limits their activity in the cell, and vesicle secretion may be a major cellular mechanism of regulating intracellular activity. These data also support the utility of EV cargo in monitoring the biological activity occurring within progenitor cells. As accumulating evidence has implicated EVs as important mediators of cell-to-cell communication, transfer of signaling molecules may additionally serve as a means to relay the status of cellular activity or viral infection to neighboring cells and also reflect the progenitor cell status. Here, we demonstrate that vesicular secretion of signal transduction molecules is often inversely related to activation of corresponding pathways within the cell. Decreasing EV secretion by CD63 knockout results in hyperactivation of intracellular signaling pathways such as mTOR and NF-κB, while increases in EV secretion through autolysosomal inhibition conversely results in decreased signal transduction.
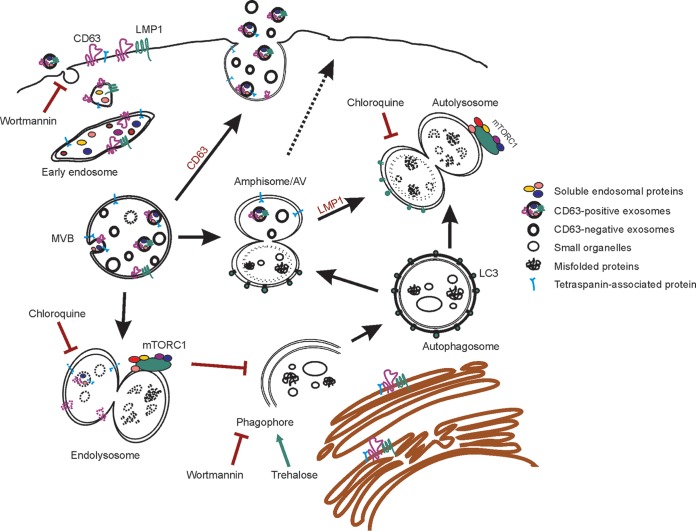
In addition to its role in endosomal protein sorting and EV production, CD63 also appears to be central in regulating endolysosomal and autophagic flux (Fig. 9). These pathways appear to be particularly important in trafficking LMP1. Altogether, we propose that CD63 provides a key link between the regulation of exosome secretion and autophagy activation, perhaps through the modulation of mTOR activity. In the absence of CD63, the autophagy-suppressing mTORC1 complex is hyperactivated, concurrent with the development of large nutrient-dependent autophagic vacuoles and increases in autophagy marker LC3-II. These findings suggest a disruption to basal autophagy levels within the cell. Recently, the convergence of endosomal and autophagy organelles has been identified (37). Fusion of late endosomes with autophagosomes can form an intermediate amphisome, also known as an autophagic vacuole, the function of which is not well understood (37, 90). Furthermore, late endosomes, autophagosomes, and amphisomes all can undergo lysosomal fusion and therefore may represent essential redundant machinery to regulate important processes, such as organelle, RNA, and protein turnover, signal transduction, cell-to-cell communication, and maintenance of global cellular homeostatic processes. Disruption of exosome production and cargo secretion by CD63 knockout may consequently result in partial compensation or accumulation of intersecting pathways, which may explain increases in LC3-II levels as well as accruing autophagic vacuoles that overburdens lysosomal fusion and degradation. Hyperactivation of the mTORC1 complex may also serve to inhibit the completion of autophagic processes, contributing to the increases in nonacidic endosomal-autophagic intermediates. In the presence of high levels of LMP1, a viral protein known to induce late stages of autophagy in host cells, these vacuoles appear to be acidified and driven toward lysosomal degradation. Induction of lysosomal degradation by the viral protein may explain why we do not observe marked increases in levels of intracellular LMP1 following decreased secretion by CD63 depletion.
FIG 9.
Proposed model of CD63-mediated intersection between endosomal and autophagic processes. Endosomal intraluminal vesicles (ILVs) are dependent in part upon CD63 for secretion as exosomes into the extracellular space. Alternatively, multivesicular bodies (MVBs) carrying ILVs can fuse to lysosomes for degradation, where mTORC1 is activated. The autophagy pathway also converges on endosomal processes. Once autophagosomes mature from phagophores, they may fuse with MVBs to form amphisomes or autophagic vacuoles (AVs). AVs may also traffic to the plasma membrane to release EVs. Treatment of cells with chloroquine or wortmannin increases Epstein-Barr virus LMP1 secretion by inhibiting lysosomal acidification or phagophore formation, respectively, and results in decreased mTORC1 activation. Wortmannin also blocks EV uptake. Activation of autophagy by trehalose drives secretion of CD63-independent vesicles containing LMP1. Depletion of CD63 results in decreased EV secretion, likely shunting MVBs toward lysosomal or autophagosomal fusion. As a result, mTORC1 activation is increased and AVs accumulate in cells. Introduction of high levels of LMP1 drives late stages of autophagy and results in the acidification of AVs.
In light of recent evidence revealing dependence of the EBV oncoprotein LMP1 on CD63 for exosome packaging, the link between CD63-mediated exosome secretion and autophagic processes also has significant implications in further understanding viral replication and transmission. In the case of EBV infection, LMP1 can specifically activate the mTORC1 complex, leading to increased cellular proliferation and therefore enhancing the ability of the virus to establish latency. CD63 acts as a negative regulator of this signaling by LMP1, likely through enhancing the packaging of many signal transduction molecules into exosomes for secretion from the cell. Moreover, EBV most likely hijacks both host cell endosomal and autophagic processes to limit intracellular levels of LMP1. It has been demonstrated that EBV-infected B cells containing intermediate levels of LMP1 comprise the majority of the clonal population of cells; cells with high levels induce later acidic stages of autophagy, while cells expressing low levels of LMP1 contain fewer autolysosomes (66). We have previously shown high and low levels of LMP1 to display the same pattern in inducing exosome secretion (19). Together, it is likely that both exosomal secretion and autophagic turnover regulate levels of LMP1 within cells to control intracellular signaling activation, and the endolysosomal tetraspanin CD63 plays a key role in coordinating these pathways.
Interestingly, disruption to several processes of autolysosomal activity led to increases in vesicle-associated LMP1 secretion. Chloroquine treatment predictably increased LMP1 release by inhibiting lysosomal-MVB degradation events. Treatment with the PI3K inhibitor wortmannin similarly increased secretion of the viral protein from control cells. However, despite increased total vesicles produced by CD63 CRISPR cells, LMP1 was not detected in these EVs. It is worth noting that PI3 kinases have been implicated in the uptake of EVs into cells, and that wortmannin treatment can inhibit vesicle uptake in a dose-dependent manner (91, 92). It is possible that the increase in secreted LMP1 seen from control cells is due in part to decreased vesicle uptake of EVs containing LMP1. Augmented LMP1 degradation observed in these vesicles may be explained by longer half-lives of EVs in the supernatant. Inhibition of vesicular LMP1 uptake may also in part explain lower levels of the viral protein in cells. Interestingly, treatment with the autophagy activator trehalose also produced increases in LMP1 secretion from both control and CD63 knockout cells. These findings suggest that autophagic activation leads to different populations of vesicles released that are not reliant upon CD63. We propose these vesicles are secreted directly from autophagic vacuoles following membrane fusion. Alternatively, LMP1 may be released directly from the cell surface in microvesicles (93). Overall, the complexities of these currently enigmatic cellular systems likely reflect the heterogeneity of EV populations and govern essential processes regulating intracellular signaling. Future investigation of the role of CD63 in endosomal and autophagic processes will help to understand the direct and indirect impacts of CD63 in mediating these intricately intersecting pathways in both noninfected and virally infected cells.
MATERIALS AND METHODS
Cell culture.
HEK293 cells were grown in Dulbecco's modified Eagle's medium (DMEM; 12-604Q; Lonza) supplemented with a 10% final concentration of fetal bovine serum (FBS; 1400-500; Seradigm), 2 mM l-glutamine (25-005-Cl; Corning), 100 IU penicillin-streptomycin (30-002-CI; Corning), and 100 μg/ml:0.25 μg/ml antibiotic-antimycotic (30-004-CI; Corning). HK1 cells were grown in RPMI 1640 cell culture medium (12-702Q; Lonza) with equivalent supplements added. The CD63 CRISPR was introduced into HK1 or EBV-infected HK1 cells as described previously (19). Serum used for extracellular vesicle enrichment experiments was depleted of EVs by ultracentrifugation at 100,000 × g for 20 h and then filtered through a 0.2-μm filter. HEK293 CD63 CRISPR and HEK293 GFP-LMP1 tetracycline-inducible cells were constructed as previously described (17, 19). To induce LMP1 expression in HEK293-inducible cells, 1 μg/μl doxycycline was added to cells for 24 h before cell lysate and EV collection. For baseline measurements of LC3-II levels in cells (Fig. 5G), medium was changed to serum-free DMEM for 24 h, and final concentrations of 50 μM chloroquine (50-63-5; Invivogen) and 5 μM wortmannin (19545-26-7; Invivogen) were added to cells for 2 h before cells were collected. For live cell imaging and EV enrichment following autophagy inhibition or activation, cells were transfected with GFP-LMP1 and incubated with 50 μM chloroquine, 5 μM wortmannin, or 100 mM trehalose dihydrate (T0167; Sigma) for 24 h before medium was harvested. HEK293 control or CD63 CRISPR cells were transfected with GFP or GFP-LMP1 plasmids using JetPrime transfection reagent (114-15; Polypus) as previously described (13). Cell viability was determined after staining cells with acridine orange (AO)-propidium iodide (PI) (CS2-0106; Nexcelom Bioscience) with an automated cell counter (Cellometer Vision, version 2.1.4.2; Nexcelom Biosciences).
DNA constructs.
GFP and GFP-LMP1 constructs were generated as described previously (19). Mutants were subcloned into the pQCXP vector (number 17386; Addgene) from pBabe-expressing plasmids previously constructed (77) (kind gifts from Nancy Raab-Traub). CD63-BirA plasmid was constructed for CD63 rescue experiments by fusing an Escherichia coli protein (BirA) to CD63 sequence obtained by PCR amplification from RFP-CD63 PQCXP vector. PCR products and the PCDNA3.1 MCS-BirA(R118G)-HA vector were cut using Nhe and EcoRI restriction enzymes and gel purified. Ligation was performed overnight using T4 DNA ligase at 16°C. Ligation products were used to transform DH5α competent cells, which were plated on LB agar plates with ampicillin and grown overnight at 37°C. DNA from transformed colonies was purified by miniprep and confirmed by diagnostic digest and sequencing. Following the generation of CD63-BirA, the target site of the guide RNA was mutated from human to mouse CD63 sequence [GCCTGTGCAGTGGGA(C to T)TGAT(T to C)GCC(G to A)TGGGTGTCGGGGCAC] using the GeneArt mutagenesis kit. Only the G-to-A mutation resulted in a change in the amino acid coding sequence (V to M).
Retrovirus production.
HEK293-T cells were transfected with expression plasmids (pQCXP GFP LMP1, pQCXP GFP-LMP1 CTAR1, pQCXP LMP1 CTAR2, and pQCXP LMP1 A5Y384G), packaging plasmid pMD2.G (number 12259; Addgene; a gift from Didier Trono), and gag/pol (plasmid 14887; Addgene; a gift from Tannishtha Reya) to produce retroviral particles for transduction. Medium was collected at 48, 72, and 96 h posttransfection, centrifuged for 10 min at 1,000 × g, filtered through a 0.45-μm filter, and frozen at −80°C until use.
Generation of LMP1 wild-type and mutant-inducible cells.
HK1 cells were initially transduced with lentivirus particles containing pLenti CMV TetR BLAST (number 17492; Addgene), and blasticidin (ant-bl-1; Invivogen) was used for selection. The generated stable cells were then transduced with retrovirus particles (pQCXP GFP LMP1, pQCXP GFP-LMP1 CTAR1, pQCXP LMP1 CTAR2, and pQCXP LMP1 A5Y384G). The subsequent stable cells were selected with medium containing blasticidin and puromycin. LMP1 expression was induced for 24 h with the addition of doxycycline (D3447; Sigma) to a final concentration of 1 μg/μl.
EV enrichment and purification.
For mass spectrometry analysis, EVs from HEK293 control or CD63 knockout cells were harvested using a modified ExtraPEG method (94) and subsequently purified on an iodixanol density gradient as previously described (17). Briefly, cell-conditioned medium was centrifuged at 500 × g for 5 min, 2,000 × g for 10 min, and then 10,000 × g for 30 min before incubating with a 1:1 volume of 2× PEG solution (16%, wt/vol, polyethylene glycol, 1 M NaCl) overnight. Solutions then were centrifuged for 1 h at 10,000 × g to obtain crude EV pellets. Pellets were resuspended in phosphate-buffered saline (PBS), ultracentrifuged at 100,000 × g for 2 h, and then layered on top of a stepwise 5 to 40% iodixanol density gradient. Vesicles were recovered from fraction 6 of the gradient, corresponding to a density of approximately 1.10 g/ml, washed in PBS, and repelleted at 100,000 × g for 2 h. Pure EV isolates were lysed in strong urea-containing lysis buffer (5% SDS, 10 mM EDTA, 120 mM Tris-HCl [pH 6.8], 8 M urea) with the addition of a protease inhibitor cocktail (Thermo). Equal volumes of fraction lysates were analyzed by immunoblotting to confirm EV protein purification. Protein concentrations were obtained by quantification using the fluorescence-based EZQ protein quantification kit (R33200; Thermo).
For immunoblot analysis of EV proteins, the post-PEG EV pellet was resuspended in 1 ml PBS and ultracentrifuged for 70 min at 100,000 × g before lysis in 2× nonreducing Laemmli sample buffer (4% SDS, 100 mM Tris-HCl [pH 6.8], 0.4 mg/ml bromophenol blue, 20% glycerol).
Nanoparticle tracking analysis.
Nanoparticle tracking was performed using a Malvern NanoSight LM10 instrument, and videos were processed using NTA 3.1 software as previously described (17).
Confocal microscopy.
HEK293 or HK1 control and CD63 knockout cells were seeded at a density of 0.2 × 106 in 35-mm glass-bottom plates (627860; Greiner Bio-One) for live cell imaging. Cells were transfected with 1 μg GFP, GFP-LMP1, or EGFP-LC3 (number 11546; Addgene) 24 h before imaging to visualize cellular structures. For serum starvation experiments, medium was changed to serum-free DMEM at the time of transfection. Hoechst 33342 nuclear staining (5 μg/ml; 62249; Thermo Scientific) was added 15 min before imaging. Lysosomal stain (LysoTracker Red DND-99; L7528; Invitrogen) was added to cells for 15 min at a final concentration of 50 nM before medium was changed. Mono-dansylcadaverine (MDC; 30432; Sigma) was added to cells at a final concentration of 50 nM for 15 min before medium was changed for visualization of autophagic vacuoles. Confocal images were taken using a Zeiss LSM 880 microscope with 488-nm and 594-nm lasers and processed using Zen 2.1 Black software. Numbers of lysosomes and vacuoles per cell were quantified using ImageJ software.
Transmission electron microscopy.
EVs for electron microscopy were prepared as described previously (19). HEK293 control and CD63 CRISPR cells were fixed in a warmed 1:1 solution of complete culture medium and fixative (2.5% glutaraldehyde–2% paraformaldehyde in 0.1 M PBS) for 10 min. Solution was removed and fresh fixative was added for another 1 h. Cells were scraped into fixative and then centrifuged at 300 × g for 5 min. Supernatant was removed and cells were resuspended in fresh fixative until postfixation. Cells were postfixed in 1% osmium tetroxide for 45 min, dehydrated in increasing concentrations of ethanol (10 min in 50%, 10 min in 70%, 10 min in 80%, 15 min in 95%, and 15 min in 100%), and cured in epoxy resin (1:1 with propylene oxide). Sections were cut between 30 and 50 nm thick, stained with uranyl acetate and lead citrate, and imaged on an FEI CM120 BioTwin transmission electron microscope.
Immunoblot analysis.
Whole-cell lysates were prepared by washing cells and then scraping them into cold PBS before pelleting at 1,000 × g for 10 min and lysis by radioimmunoprecipitation assay (RIPA) buffer as described previously (19). Insoluble material was pelleted and discarded prior to immunoblot analysis as previously detailed (19). To measure signaling activity in cells, growth medium was changed to serum-free medium at the time of GFP or GFP-LMP1 transfection. Cell pellets were harvested 24 h later, and cytoplasmic fractions were isolated as described previously (19). Total protein levels were determined by Ponceau S stain. Blots were probed using the following antibodies: Alix (Q-19; Santa Cruz Biotechnology), HSC70 (B-6; Santa Cruz), TSG101 (C-2; Santa Cruz), CD63 (TS63; Abcam), LMP1 (CS1-4; Dako), phospho-IκBα (2859; Cell Signaling), GFP (600-101-215; Rockland), beta-actin (629630; GeneTex), phospho-mTOR Ser2481 (2974; Cell Signaling), phospho-mTOR Ser2448 (5536; Cell Signaling), mTOR (2983; Cell Signaling), Raptor (2280; Cell Signaling), Rictor (2114; Cell Signaling), GβL (3274; Cell Signaling), LAMTOR1 (8975; Cell Signaling), RagA (4357; Cell Signaling), RagB (8150; Cell Signaling), RagC (5466; Cell Signaling), RagD (4470; Cell Signaling), phospho-p70 S6 kinase Thr389 (9205; Cell Signaling), p70 S6 kinase (9202; Cell Signaling), LC3B (3868; Cell Signaling), Calnexin (H-70; Santa Cruz), CD81 (SC-9158; Santa Cruz), rabbit anti-mouse IgG (26728; GeneTex), rabbit anti-goat IgG (26741; GeneTex), and goat anti-rabbit IgG (Fab fragment) (27171; GeneTex). Blots were imaged with an ImageQuant LAS4000 (General Electric) and processed using ImageQuant TL v8.1.0.0, Adobe Photoshop CS6, and CorelDraw X5.
Mass spectrometry.
Equal masses (20 μg) of iodixanol density gradient-purified EV protein were run in a 4 to 20% polyacrylamide gel (59511; Lonza) for purification and separation by SDS-PAGE. Gels were fixed and stained with a Coomassie staining protocol as described previously (95). Samples were fractionated, trypsin digested, submitted to the Florida State University Translational Science Laboratory for liquid chromatography-tandem mass spectrometry (LC-MS/MS), and analyzed as previously described in detail (94). Spectral counts were analyzed in Scaffold, version 4.7.1, using a 99.0% protein threshold, and a minimum of 2 peptides was required for identification. The protein false discovery rate was 0.7%. Spectral counts were normalized across the experiment using Scaffold protein normalization. Before enrichment analyses were performed, fold changes were calculated between control and knockout EVs after grouping biosamples.
Enrichment analysis.
Proteins identified by mass spectrometry were compared to the Vesiclepedia database of known EV proteins using FunRich, v3. Cellular compartment enrichment of all identified proteins was performed using FunRich. Proteins decreased by 2-fold in CD63 knockout cell-derived EVs were analyzed with The Database for Annotation, Visualization and Integrated Discovery (DAVID), v6.8, to identify pathways (KEGG) and biological processes (GOTERM_BP_DIRECT) enriched in control EVs.
Statistical analysis.
Significance of results was determined by Student's two-sample t test. Figures were assembled by using Microsoft Excel, Adobe Photoshop CS6, and CorelDraw X5 software programs.
Accession number(s).
The mass spectrometry data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository (96) with the data set identifiers PXD006514 and 10.6019/PXD006514.
Supplementary Material
ACKNOWLEDGMENTS
We thank Steven Carter for his help in immunoblotting experiments, Georgia Platt of the Biological Science Imaging Resource at Florida State University (FSU) for assistance with the electron microscopy work, Timothy Megraw for providing the LysoTracker stain, Xia Liu for sample preparation, the FSU Translational Science Laboratory for help in mass spectrometry data acquisition, and the FSU College of Medicine Confocal Microscopy Laboratory.
This study was supported by grants from the National Institutes of Health (RO1CA204621 and R15CA188941) awarded to D.G.M.
We have no conflicts of interest with the contents of this article.
D.G.M. and S.N.H. conceived of and designed the study. S.N.H., M.R.C., S.B.Y., D.N., and D.G.M. performed experiments and interpreted results. The manuscript was written by S.N.H. and D.G.M.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01969-17.
REFERENCES
- 1.Meckes DG, Shair KH, Marquitz AR, Kung CP, Edwards RH, Raab-Traub N. 2010. Human tumor virus utilizes exosomes for intercellular communication. Proc Natl Acad Sci U S A 107:20370–20375. doi: 10.1073/pnas.1014194107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meckes DG, Raab-Traub N. 2011. Microvesicles and viral infection. J Virol 85:12844–12854. doi: 10.1128/JVI.05853-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meckes DG. 2015. Exosomal communication goes viral. J Virol 89:5200–5203. doi: 10.1128/JVI.02470-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD, Würdinger T, Middeldorp JM. 2010. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A 107:6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gould SJ, Booth AM, Hildreth JE. 2003. The Trojan exosome hypothesis. Proc Natl Acad Sci U S A 100:10592–10597. doi: 10.1073/pnas.1831413100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mori Y, Koike M, Moriishi E, Kawabata A, Tang H, Oyaizu H, Uchiyama Y, Yamanishi K. 2008. Human herpesvirus-6 induces MVB formation, and virus egress occurs by an exosomal release pathway. Traffic 9:1728–1742. doi: 10.1111/j.1600-0854.2008.00796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen BJ, Lamb RA. 2008. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology 372:221–232. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrison MS, Sakaguchi T, Schmitt AP. 2010. Paramyxovirus assembly and budding: building particles that transmit infections. Int J Biochem Cell Biol 42:1416–1429. doi: 10.1016/j.biocel.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chahar HS, Bao X, Casola A. 2015. Exosomes and their role in the life cycle and pathogenesis of RNA viruses. Viruses 7:3204–3225. doi: 10.3390/v7062770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lenassi M, Cagney G, Liao M, Vaupotic T, Bartholomeeusen K, Cheng Y, Krogan NJ, Plemenitas A, Peterlin BM. 2010. HIV Nef is secreted in exosomes and triggers apoptosis in bystander CD4+ T cells. Traffic 11:110–122. doi: 10.1111/j.1600-0854.2009.01006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stoorvogel W, Kleijmeer MJ, Geuze HJ, Raposo G. 2002. The biogenesis and functions of exosomes. Traffic 3:321–330. doi: 10.1034/j.1600-0854.2002.30502.x. [DOI] [PubMed] [Google Scholar]
- 12.Bobrie A, Colombo M, Raposo G, Théry C. 2011. Exosome secretion: molecular mechanisms and roles in immune responses. Traffic 12:1659–1668. doi: 10.1111/j.1600-0854.2011.01225.x. [DOI] [PubMed] [Google Scholar]
- 13.Théry C, Zitvogel L, Amigorena S. 2002. Exosomes: composition, biogenesis and function. Nat Rev Immunol 2:569–579. [DOI] [PubMed] [Google Scholar]
- 14.Théry C. 2011. Exosomes: secreted vesicles and intercellular communications. F1000 Biol Rep 3:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathivanan S, Ji H, Simpson RJ. 2010. Exosomes: extracellular organelles important in intercellular communication. J Proteomics 73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 16.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. 2007. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 17.Hurwitz SN, Conlon MM, Rider MA, Brownstein NC, Meckes DG. 2016. Nanoparticle analysis sheds budding insights into genetic drivers of extracellular vesicle biogenesis. J Extracell Vesicles 5:31295. doi: 10.3402/jev.v5.31295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hurwitz SN, Rider MA, Bundy JL, Liu X, Singh RK, Meckes DG. 2016. Proteomic profiling of NCI-60 extracellular vesicles uncovers common protein cargo and cancer type-specific biomarkers. Oncotarget 7:86999–87015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hurwitz SN, Nkosi D, Conlon MM, York SB, Liu X, Tremblay DC, Meckes DG. 2017. CD63 regulates Epstein-Barr virus LMP1 exosomal packaging, enhancement of vesicle production, and noncanonical NF-κB signaling. J Virol 91:e02251-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hotta H, Ross AH, Huebner K, Isobe M, Wendeborn S, Chao MV, Ricciardi RP, Tsujimoto Y, Croce CM, Koprowski H. 1988. Molecular cloning and characterization of an antigen associated with early stages of melanoma tumor progression. Cancer Res 48:2955–2962. [PubMed] [Google Scholar]
- 21.Hemler ME. 2003. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu Rev Cell Dev Biol 19:397–422. doi: 10.1146/annurev.cellbio.19.111301.153609. [DOI] [PubMed] [Google Scholar]
- 22.Hemler ME. 2005. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol 6:801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 23.Tarrant JM, Robb L, van Spriel AB, Wright MD. 2003. Tetraspanins: molecular organisers of the leukocyte surface. Trends Immunol 24:610–617. doi: 10.1016/j.it.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 24.Boucheix C, Rubinstein E. 2001. Tetraspanins. Cell Mol Life Sci 58:1189–1205. doi: 10.1007/PL00000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunziker W, Geuze HJ. 1996. Intracellular trafficking of lysosomal membrane proteins. Bioessays 18:379–389. doi: 10.1002/bies.950180508. [DOI] [PubMed] [Google Scholar]
- 26.Saksena S, Sun J, Chu T, Emr SD. 2007. ESCRTing proteins in the endocytic pathway. Trends Biochem Sci 32:561–573. doi: 10.1016/j.tibs.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 27.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brügger B, Simons M. 2008. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 28.Vogt AB, Spindeldreher S, Kropshofer H. 2002. Clustering of MHC-peptide complexes prior to their engagement in the immunological synapse: lipid raft and tetraspan microdomains. Immunol Rev 189:136–151. doi: 10.1034/j.1600-065X.2002.18912.x. [DOI] [PubMed] [Google Scholar]
- 29.Levy S, Shoham T. 2005. The tetraspanin web modulates immune-signalling complexes. Nat Rev Immunol 5:136–148. doi: 10.1038/nri1548. [DOI] [PubMed] [Google Scholar]
- 30.Rubinstein E, Le Naour F, Lagaudrière-Gesbert C, Billard M, Conjeaud H, Boucheix C. 1996. CD9, CD63, CD81, and CD82 are components of a surface tetraspan network connected to HLA-DR and VLA integrins. Eur J Immunol 26:2657–2665. doi: 10.1002/eji.1830261117. [DOI] [PubMed] [Google Scholar]
- 31.Artavanis-Tsakonas K, Love JC, Ploegh HL, Vyas JM. 2006. Recruitment of CD63 to Cryptococcus neoformans phagosomes requires acidification. Proc Natl Acad Sci U S A 103:15945–15950. doi: 10.1073/pnas.0607528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shrivastava S, Devhare P, Sujijantarat N, Steele R, Kwon YC, Ray R, Ray RB. 2015. Knockdown of autophagy inhibits infectious hepatitis C virus release by the exosomal pathway. J Virol 90:1387–1396. doi: 10.1128/JVI.02383-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flanagan J, Middeldorp J, Sculley T. 2003. Localization of the Epstein-Barr virus protein LMP 1 to exosomes. J Gen Virol 84:1871–1879. doi: 10.1099/vir.0.18944-0. [DOI] [PubMed] [Google Scholar]
- 34.Verweij FJ, van Eijndhoven MA, Hopmans ES, Vendrig T, Wurdinger T, Cahir-McFarland E, Kieff E, Geerts D, van der Kant R, Neefjes J, Middeldorp JM, Pegtel DM. 2011. LMP1 association with CD63 in endosomes and secretion via exosomes limits constitutive NF-κB activation. EMBO J 30:2115–2129. doi: 10.1038/emboj.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vellai T. 2009. Autophagy genes and ageing. Cell Death Differ 16:94–102. doi: 10.1038/cdd.2008.126. [DOI] [PubMed] [Google Scholar]
- 36.Mizushima N. 2007. Autophagy: process and function. Genes Dev 21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 37.Patel KK, Miyoshi H, Beatty WL, Head RD, Malvin NP, Cadwell K, Guan JL, Saitoh T, Akira S, Seglen PO, Dinauer MC, Virgin HW, Stappenbeck TS. 2013. Autophagy proteins control goblet cell function by potentiating reactive oxygen species production. EMBO J 32:3130–3144. doi: 10.1038/emboj.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levine B, Mizushima N, Virgin HW. 2011. Autophagy in immunity and inflammation. Nature 469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chiang HL, Terlecky SR, Plant CP, Dice JF. 1989. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science 246:382–385. doi: 10.1126/science.2799391. [DOI] [PubMed] [Google Scholar]
- 40.Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, Follenzi A, Potolicchio I, Nieves E, Cuervo AM, Santambrogio L. 2011. Microautophagy of cytosolic proteins by late endosomes. Dev Cell 20:131–139. doi: 10.1016/j.devcel.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim HJ, Lee S, Jung JU. 2010. When autophagy meets viruses: a double-edged sword with functions in defense and offense. Semin Immunopathol 32:323–341. doi: 10.1007/s00281-010-0226-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shrivastava S, Bhanja Chowdhury J, Steele R, Ray R, Ray RB. 2012. Hepatitis C virus upregulates Beclin1 for induction of autophagy and activates mTOR signaling. J Virol 86:8705–8712. doi: 10.1128/JVI.00616-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levine B. 2005. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell 120:159–162. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 44.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. 2007. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science 315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 45.English L, Chemali M, Duron J, Rondeau C, Laplante A, Gingras D, Alexander D, Leib D, Norbury C, Lippé R, Desjardins M. 2009. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat Immunol 10:480–487. doi: 10.1038/ni.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmid D, Münz C. 2007. Innate and adaptive immunity through autophagy. Immunity 27:11–21. doi: 10.1016/j.immuni.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mulvey M, Poppers J, Sternberg D, Mohr I. 2003. Regulation of eIF2alpha phosphorylation by different functions that act during discrete phases in the herpes simplex virus type 1 life cycle. J Virol 77:10917–10928. doi: 10.1128/JVI.77.20.10917-10928.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chaumorcel M, Souquère S, Pierron G, Codogno P, Esclatine A. 2008. Human cytomegalovirus controls a new autophagy-dependent cellular antiviral defense mechanism. Autophagy 4:46–53. doi: 10.4161/auto.5184. [DOI] [PubMed] [Google Scholar]
- 49.Kudchodkar SB, Yu Y, Maguire TG, Alwine JC. 2004. Human cytomegalovirus infection induces rapamycin-insensitive phosphorylation of downstream effectors of mTOR kinase. J Virol 78:11030–11039. doi: 10.1128/JVI.78.20.11030-11039.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sinha S, Colbert CL, Becker N, Wei Y, Levine B. 2008. Molecular basis of the regulation of Beclin 1-dependent autophagy by the gamma-herpesvirus 68 Bcl-2 homolog M11. Autophagy 4:989–997. doi: 10.4161/auto.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou D, Spector SA. 2008. Human immunodeficiency virus type-1 infection inhibits autophagy. AIDS 22:695–699. doi: 10.1097/QAD.0b013e3282f4a836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gannagé M, Dormann D, Albrecht R, Dengjel J, Torossi T, Rämer PC, Lee M, Strowig T, Arrey F, Conenello G, Pypaert M, Andersen J, García-Sastre A, Münz C. 2009. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe 6:367–380. doi: 10.1016/j.chom.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. 2009. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem 284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Teis D, Wunderlich W, Huber LA. 2002. Localization of the MP1-MAPK scaffold complex to endosomes is mediated by p14 and required for signal transduction. Dev Cell 3:803–814. doi: 10.1016/S1534-5807(02)00364-7. [DOI] [PubMed] [Google Scholar]
- 55.Wunderlich W, Fialka I, Teis D, Alpi A, Pfeifer A, Parton RG, Lottspeich F, Huber LA. 2001. A novel 14-kilodalton protein interacts with the mitogen-activated protein kinase scaffold mp1 on a late endosomal/lysosomal compartment. J Cell Biol 152:765–776. doi: 10.1083/jcb.152.4.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. 2008. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. 2010. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Laplante M, Sabatini DM. 2012. mTOR signaling in growth control and disease. Cell 149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. 2010. mTOR regulation of autophagy. FEBS Lett 584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salonen A, Ahola T, Kääriäinen L. 2005. Viral RNA replication in association with cellular membranes. Curr Top Microbiol Immunol 285:139–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jackson WT, Giddings TH, Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, Kirkegaard K. 2005. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol 3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wong J, Zhang J, Si X, Gao G, Mao I, McManus BM, Luo H. 2008. Autophagosome supports coxsackievirus B3 replication in host cells. J Virol 82:9143–9153. doi: 10.1128/JVI.00641-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sir D, Chen WL, Choi J, Wakita T, Yen TS, Ou JH. 2008. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology 48:1054–1061. doi: 10.1002/hep.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kyei GB, Dinkins C, Davis AS, Roberts E, Singh SB, Dong C, Wu L, Kominami E, Ueno T, Yamamoto A, Federico M, Panganiban A, Vergne I, Deretic V. 2009. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J Cell Biol 186:255–268. doi: 10.1083/jcb.200903070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee DY, Lee J, Sugden B. 2009. The unfolded protein response and autophagy: herpesviruses rule! J Virol 83:1168–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee DY, Sugden B. 2008. The latent membrane protein 1 oncogene modifies B-cell physiology by regulating autophagy. Oncogene 27:2833–2842. doi: 10.1038/sj.onc.1210946. [DOI] [PubMed] [Google Scholar]
- 67.Hurwitz SN, Meckes DG. 2017. An adaptable polyethylene glycol-based workflow for proteomic analysis of extracellular vesicles. Methods Mol Biol 1660:303–317. doi: 10.1007/978-1-4939-7253-1_25. [DOI] [PubMed] [Google Scholar]
- 68.Chen J, Hu CF, Hou JH, Shao Q, Yan LX, Zhu XF, Zeng YX, Shao JY. 2010. Epstein-Barr virus encoded latent membrane protein 1 regulates mTOR signaling pathway genes which predict poor prognosis of nasopharyngeal carcinoma. J Transl Med 8:30. doi: 10.1186/1479-5876-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang J, Jia L, Lin W, Yip YL, Lo KW, Lau VM, Zhu D, Tsang CM, Zhou Y, Deng W, Lung HL, Lung ML, Cheung LM, Tsao SW. 2017. Epstein-Barr Virus-encoded latent membrane protein 1 upregulates glucose transporter 1 transcription via the mTORC1/NF-κB signaling pathways. J Virol 91:e02168-. doi: 10.1128/JVI.02168-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jacinto E, Loewith R, Schmidt A, Lin S, Rüegg MA, Hall A, Hall MN. 2004. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol 6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- 71.Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. 2002. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110:163–175. doi: 10.1016/S0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 72.Kim DH, Sarbassov DD, Ali SM, Latek RR, Guntur KV, Erdjument-Bromage H, Tempst P, Sabatini DM. 2003. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell 11:895–904. doi: 10.1016/S1097-2765(03)00114-X. [DOI] [PubMed] [Google Scholar]
- 73.Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. 2004. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol 14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 74.Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP. 2005. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell 121:179–193. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
- 75.Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. 2011. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Copp J, Manning G, Hunter T. 2009. TORC-specific phosphorylation of mammalian target of rapamycin (mTOR): phospho-Ser2481 is a marker for intact mTOR signaling complex 2. Cancer Res 69:1821–1827. doi: 10.1158/0008-5472.CAN-08-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mainou BA, Everly DN, Raab-Traub N. 2007. Unique signaling properties of CTAR1 in LMP1-mediated transformation. J Virol 81:9680–9692. doi: 10.1128/JVI.01001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wada Y. 2013. Vacuoles in mammals: a subcellular structure indispensable for early embryogenesis. Bioarchitecture 3:13–19. doi: 10.4161/bioa.24126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Biederbick A, Kern HF, Elsässer HP. 1995. Monodansylcadaverine (MDC) is a specific in vivo marker for autophagic vacuoles. Eur J Cell Biol 66:3–14. [PubMed] [Google Scholar]
- 80.Munafó DB, Colombo MI. 2001. A novel assay to study autophagy: regulation of autophagosome vacuole size by amino acid deprivation. J Cell Sci 114:3619–3629. [DOI] [PubMed] [Google Scholar]
- 81.Shintani T, Klionsky DJ. 2004. Autophagy in health and disease: a double-edged sword. Science 306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelárová H, Meijer AJ. 1997. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem 243:240–246. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- 83.Bright NA, Lindsay MR, Stewart A, Luzio JP. 2001. The relationship between lumenal and limiting membranes in swollen late endocytic compartments formed after wortmannin treatment or sucrose accumulation. Traffic 2:631–642. doi: 10.1034/j.1600-0854.2001.20906.x. [DOI] [PubMed] [Google Scholar]
- 84.Reaves BJ, Bright NA, Mullock BM, Luzio JP. 1996. The effect of wortmannin on the localisation of lysosomal type I integral membrane glycoproteins suggests a role for phosphoinositide 3-kinase activity in regulating membrane traffic late in the endocytic pathway. J Cell Sci 109(Part 4):749–762. [DOI] [PubMed] [Google Scholar]
- 85.Chen X, Li M, Li L, Xu S, Huang D, Ju M, Huang J, Chen K, Gu H. 2016. Trehalose, sucrose and raffinose are novel activators of autophagy in human keratinocytes through an mTOR-independent pathway. Sci Rep 6:28423. doi: 10.1038/srep28423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mardones P, Rubinsztein DC, Hetz C. 2016. Mystery solved: Trehalose kickstarts autophagy by blocking glucose transport. Sci Signal 9:fs2. doi: 10.1126/scisignal.aaf1937. [DOI] [PubMed] [Google Scholar]
- 87.DeBosch BJ, Heitmeier MR, Mayer AL, Higgins CB, Crowley JR, Kraft TE, Chi M, Newberry EP, Chen Z, Finck BN, Davidson NO, Yarasheski KE, Hruz PW, Moley KH. 2016. Trehalose inhibits solute carrier 2A (SLC2A) proteins to induce autophagy and prevent hepatic steatosis. Sci Signal 9:ra21. doi: 10.1126/scisignal.aac5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. 2007. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem 282:5641–5652. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 89.Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M, Théry C. 2016. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A 113:E968–E977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sanchez-Wandelmer J, Reggiori F. 2013. Amphisomes: out of the autophagosome shadow? EMBO J 32:3116–3118. doi: 10.1038/emboj.2013.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Feng D, Zhao WL, Ye YY, Bai XC, Liu RQ, Chang LF, Zhou Q, Sui SF. 2010. Cellular internalization of exosomes occurs through phagocytosis. Traffic 11:675–687. doi: 10.1111/j.1600-0854.2010.01041.x. [DOI] [PubMed] [Google Scholar]
- 92.Mulcahy LA, Pink RC, Carter DR. 2014. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles 3:24641. doi: 10.3402/jev.v3.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vazirabadi G, Geiger TR, Coffin WF, Martin JM. 2003. Epstein-Barr virus latent membrane protein-1 (LMP-1) and lytic LMP-1 localization in plasma membrane-derived extracellular vesicles and intracellular virions. J Gen Virol 84:1997–2008. doi: 10.1099/vir.0.19156-0. [DOI] [PubMed] [Google Scholar]
- 94.Rider MA, Hurwitz SN, Meckes DG. 2016. ExtraPEG: a polyethylene glycol-based method for enrichment of extracellular vesicles. Sci Rep 6:23978. doi: 10.1038/srep23978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Meckes DG. 2014. Affinity purification combined with mass spectrometry to identify herpes simplex virus protein-protein interactions. Methods Mol Biol 1144:209–222. doi: 10.1007/978-1-4939-0428-0_14. [DOI] [PubMed] [Google Scholar]
- 96.Vizcaíno JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Ríos D, Dianes JA, Sun Z, Farrah T, Bandeira N, Binz PA, Xenarios I, Eisenacher M, Mayer G, Gatto L, Campos A, Chalkley RJ, Kraus HJ, Albar JP, Martinez-Bartolomé S, Apweiler R, Omenn GS, Martens L, Jones AR, Hermjakob H. 2014. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol 32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.