ABSTRACT
Human parvovirus B19 (B19V) infection of human erythroid progenitor cells (EPCs) induces a DNA damage response and cell cycle arrest at late S phase, which facilitates viral DNA replication. However, it is not clear exactly which cellular factors are employed by this single-stranded DNA virus. Here, we used microarrays to systematically analyze the dynamic transcriptome of EPCs infected with B19V. We found that DNA metabolism, DNA replication, DNA repair, DNA damage response, cell cycle, and cell cycle arrest pathways were significantly regulated after B19V infection. Confocal microscopy analyses revealed that most cellular DNA replication proteins were recruited to the centers of viral DNA replication, but not the DNA repair DNA polymerases. Our results suggest that DNA replication polymerase δ and polymerase α are responsible for B19V DNA replication by knocking down its expression in EPCs. We further showed that although RPA32 is essential for B19V DNA replication and the phosphorylated forms of RPA32 colocalized with the replicating viral genomes, RPA32 phosphorylation was not necessary for B19V DNA replication. Thus, this report provides evidence that B19V uses the cellular DNA replication machinery for viral DNA replication.
IMPORTANCE Human parvovirus B19 (B19V) infection can cause transient aplastic crisis, persistent viremia, and pure red cell aplasia. In fetuses, B19V infection can result in nonimmune hydrops fetalis and fetal death. These clinical manifestations of B19V infection are a direct outcome of the death of human erythroid progenitors that host B19V replication. B19V infection induces a DNA damage response that is important for cell cycle arrest at late S phase. Here, we analyzed dynamic changes in cellular gene expression and found that DNA metabolic processes are tightly regulated during B19V infection. Although genes involved in cellular DNA replication were downregulated overall, the cellular DNA replication machinery was tightly associated with the replicating single-stranded DNA viral genome and played a critical role in viral DNA replication. In contrast, the DNA damage response-induced phosphorylated forms of RPA32 were dispensable for viral DNA replication.
KEYWORDS: DNA replication, parvovirus B19, parvovirus
INTRODUCTION
Human parvovirus B19 (B19V) is a member of the genus Erythroparvovirus within the family Parvoviridae (1). B19V is best known for causing “fifth disease” in the pediatric population. However, B19V infection can also cause hydrops fetalis in pregnant women, transient aplastic crisis in sickle cell disease patients, and chronic pure red cell aplasia in immunocompromised patients (2–5). These conditions are the direct outcomes of the death of human erythroid progenitors (EPCs) that are infected with B19V. Myocarditis, chronic fatigue syndrome, and many autoimmune diseases are also thought to be caused by B19V infection; and there are mechanisms to explain these particular manifestations of B19V; however, a direct link between these disease manifestations and the virus remains elusive (6).
B19V infection has a very narrow tropism and is restricted to EPCs from bone marrow (7–9) and fetal liver (10, 11). Erythropoietin (EPO) and EPO receptor (EPOR) signaling plays a critical role in B19V replication, which is at least partially mediated by the Janus kinase 2 (JAK2) signal transducer and the activator of transcription 5 (STAT5) pathway (12). Hypoxia significantly increases B19V infection of CD36+ EPCs and cells of human megakaryoblastoid cell line UT7/Epo-S1 through activation of STAT5 signaling and downregulation of extracellular signal-regulated kinase (ERK) signaling (13, 14). Ex vivo-expanded human primary CD36+ EPCs, which are differentiated from CD34+ hematopoietic stem cells, are highly permissive to B19V infection (15). UT7/Epo-S1 is the most B19V-permissive cell line that supports B19V replication and progeny virion production (16).
B19V infection induces a DNA damage response (DDR) with activation of all three phosphatidylinositol 3-kinase-related kinases (PI3KKs), namely, ATM (ataxia-telangiectasia mutated), ATR (ATM and Rad3 related), and DNA-PKcs (DNA-dependent protein kinase catalytic subunit) (17). B19V hijacked the induced DDR to promote viral DNA replication through ATR and the DNA-PKcs pathways (17). On the other hand, B19V infection induces cell cycle arrest at late S phase (18), supplying host factors to facilitate B19V DNA replication. Hijacking the DDR pathway for viral DNA replication was also reported in other parvoviruses (19, 20), such as minute virus of canines (MVC) (21, 22) and minute virus of mice (MVM) (23), which utilize ATM signaling for viral DNA replication, resulting in cell cycle arrest at G2/M phase (22, 24). However, the exact role of the parvovirus infection-induced DDR in viral DNA replication in dividing cells is unclear. Parvovirus can also infect nondividing cells, for example, in the form of human bocavirus 1 (HBoV1) infection of well-differentiated (nondividing) human airway epithelium (25). HBoV1 takes advantage of DDR and uses the DNA repair DNA polymerases, polymerase η (Pol η) and polymerase κ (Pol κ), to replicate viral DNA in nondividing human airway epithelial cells, which lack cellular DNA replication (S-phase) factors (26).
Here, we aimed to delineate the replication pathway of B19V. We used microarrays to systematically analyze the dynamic transcriptome of CD36+ EPCs during B19V infection for changes in cellular DNA metabolism pathways. We used confocal microscopy to localize host DNA replication factors within the replicating viral genome. Further knockdown of the key host DNA replication factors and overexpression of phosphorylated forms of replication protein A 32 subunit (RPA32) demonstrated that bona fide cellular DNA replication factors play a key role in B19V DNA replication, whereas those factors involved in DNA repair processes do not.
RESULTS
Dynamic transcriptome analysis of CD36+ EPCs during B19V infection.
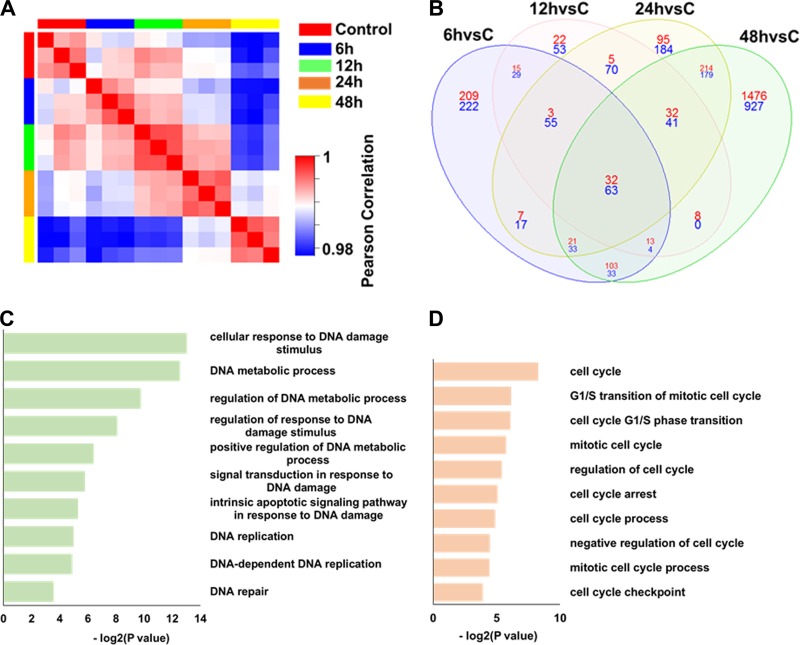
To explore the global and dynamic transcriptome profile of CD36+ EPCs infected with B19V, we performed microarray analysis of the gene expression of CD36+ EPCs infected with B19V at 6, 12, 24, and 48 h postinfection (hpi). Over 60% of the cells were infected at 48 hpi as determined by immunostaining with anti-B19V NS1 (data not shown). A Pearson correlation matrix of pairwise comparisons was used to assess the transcriptome patterns of different group of samples infected with B19V at different time points. The expression patterns in the same group had the highest correlations, and the expression patterns at 48 hpi showed the lowest correlation with those at early time points, indicating that the greatest change in gene expressions occurred at 48 hpi (Fig. 1A).
FIG 1.
Dynamic transcriptome analysis of the CD36+ EPCs infected with B19V using microarrays. (A) Pearson correlation analysis of different groups of microarray data from CD36+ EPCs infected with B19V at different time points. Red indicates high correlation; blue indicates low correlation. n = 3 for each time point. (B) Venn diagram analysis of the 4,090 significant differentially expressed gene probes. The designations 6hvsC, 12hvsC, 24hvsC, and 48hvsC indicate numbers of the differentially expressed gene probes at 6 hpi, 12 hpi, 24 hpi, and 48 hpi, respectively, versus the control group results. Numbers of upregulated gene probes are shown in red; numbers of downregulated gene probes are shown in blue. (C and D) Top 10 DNA metabolic process-associated (C) and cell cycle process-associated (D) pathways of the 4,090 differentially expressed gene probes after B19V infection.
A total of 4,090 significantly (P value < 0.05) and differentially expressed gene probes related to 2,566 genes changed more than 1.8-fold in expression in infected cells compared with their expression in the mock-infected cells (see File S1 in the supplemental material). Of these, 859 were identified at 6 hpi, 445 at 12 hpi, 1,051 at 24 hpi, and 3,179 at 48 hpi. A Venn diagram was used to visualize the distributions of the differentially expressed genes at different time points (Fig. 1B). The data show that 32 up- and 63 downregulated gene probes appeared at all four time points and that 2,403 gene probes appeared in samples only at 48 hpi (75.59%; 1,476 upregulated and 927 downregulated). This further confirms that B19V infection induced the most marked change in host gene expression at 48 hpi (Fig. 1B).
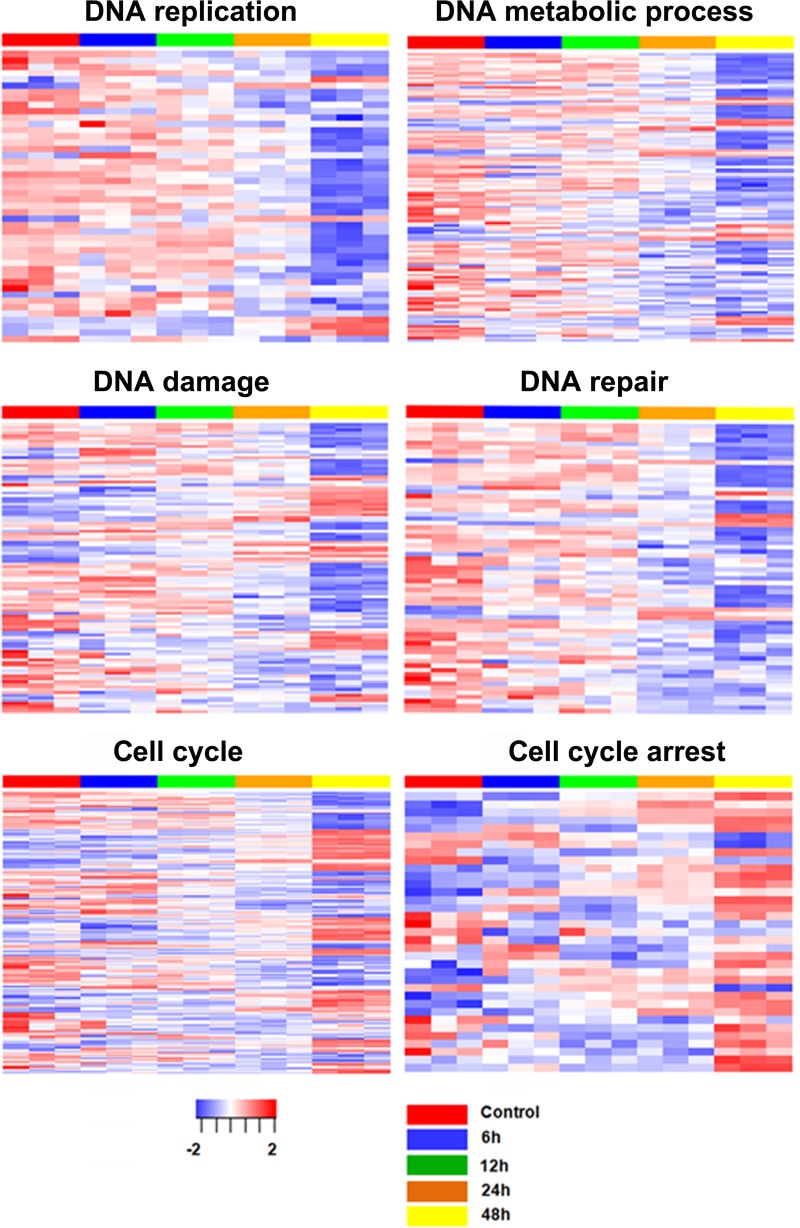
To analyze the biological function of the 2,566 differentially expressed genes, we performed biological process enrichment analysis using Database for Annotation, Visualization and Integrated Discovery (DAVID). We observed enrichment for 21 DNA metabolism-associated processes (P value = 1.18 × 10−4 to 8.56 × 10−2) and 18 cell cycle-related processes (P value = 3.24 × 10−3 to 9.34 × 10−2). The top 10 pathways for DNA metabolism-associated processes and cell cycle-related processes are shown in Fig. 1C and D. Almost all of the differentially expressed genes associated with DNA metabolic processes, DNA replication, and DNA repair pathways were downregulated, especially at 48 hpi, indicating that B19V infection shut down the cellular DNA replication and repair pathways at late infection (Fig. 2). Both up- and downregulated genes were associated with DNA damage and cell cycle pathways, indicating the changes in both DNA damage regulation and cell cycle regulation. Notably, almost all differentially expressed genes associated with cell cycle arrest were upregulated, consistent with induction of cell cycle arrest after B19V infection (Fig. 2).
FIG 2.
Heat maps of the differentially expressed genes associated with DNA replication, DNA metabolic process, DNA damage, DNA repair, cell cycle, and cell cycle arrest pathways. Each column represents gene expression data from microarray analyses of CD36+ EPCs infected with B19V at different time points (n = 3 for each). Each row represents a gene. Red indicates increased expression, and blue indicates decreased expression.
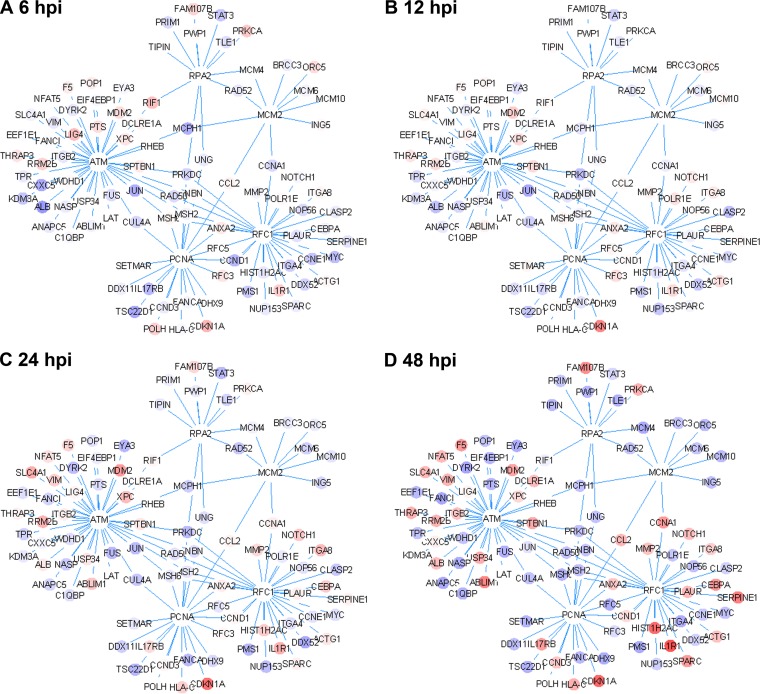
To investigate the hub genes of the differentially expressed genes, we performed PPI (protein-protein interaction) hub enrichment analysis. The RFC1, PCNA, MCM2, RPA32, and ATM genes were enriched among the identified hubs of the 2,566 differentially expressed genes, further confirming that cellular DNA replication and DNA damage pathways are altered by B19V infection (Fig. 3).
FIG 3.
Protein-protein interaction (PPI) network analysis of differentially expressed genes at different time points. The PPI networks of differential genes enriched in hub genes RPA2, PCNA, RFC1, MCM2, and ATM are shown. The node color indicates the fold change of gene expression compared with that in the mock-infected cell group. Red indicates upregulation; blue indicates downregulation.
The host DNA replication machinery is associated with the replicating B19V single-stranded DNA (ssDNA) genome.
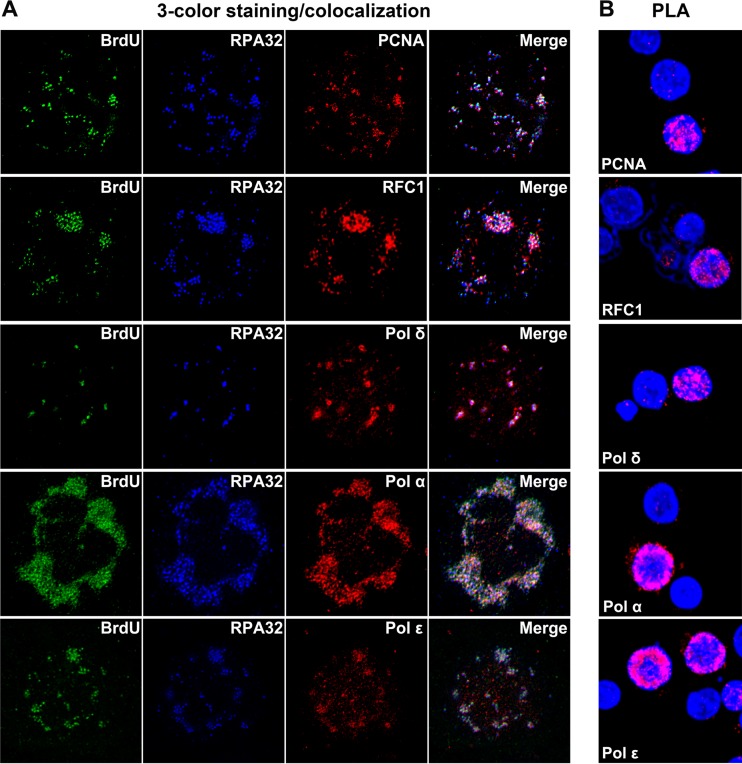
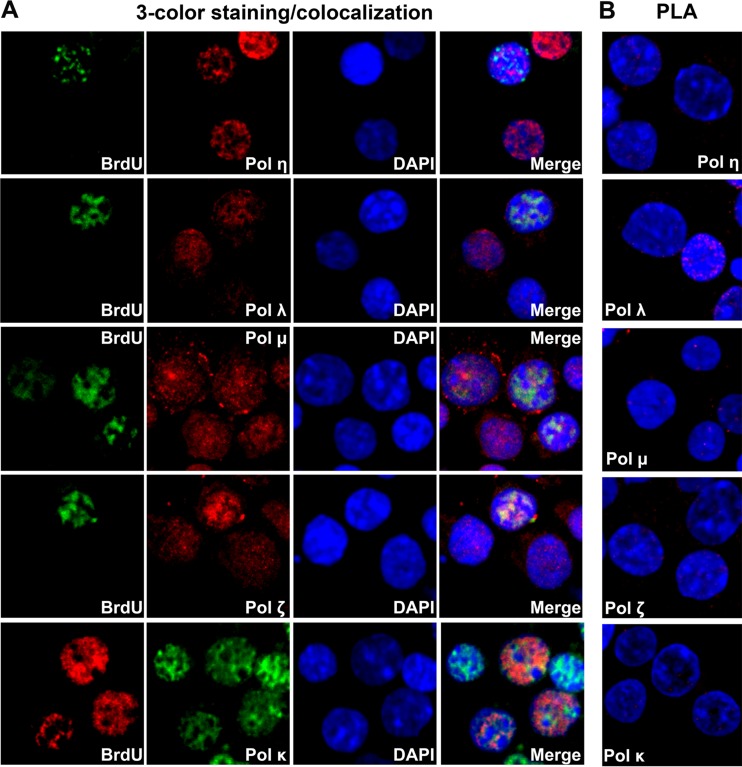
Among these identified hub genes, MCM2 plays an essential role in viral DNA replication. As a component of the minichromosome maintenance (MCM) complex, it is recruited to the viral DNA replication origin by the viral DNA replication origin (Ori)-binding protein (18, 27). In contrast, inhibition of ATM kinase phosphorylation through treatment with either an ATM-specific inhibitor or an ATM-specific short hairpin RNA (shRNA) does not affect viral replication (17). Therefore, we explored the hub genes RFC1, PCNA, and RPA2 (RPA32) in the process of B19V DNA replication. We used three-color stimulated emission depletion (STED) superresolution microscopy to examine colocalization of the proteins expressed by these genes with the replicating viral genomes, which were pulse-labeled with bromodeoxyuridine (BrdU) and with single-stranded DNA (ssDNA)-binding protein RPA32. Both PCNA and RFC1 colocalized with RPA32 and the replicating viral genome (Fig. 4A). This was confirmed by a proximity ligation assay using an anti-BrdU antibody and an antibody against one of the replication factors as indicated (Fig. 4B). The proximity ligation assay determines protein interactions within a range of proximity of 20 nm in a cell (28). As RFC1, PCNA, and RPA32 are essential cellular DNA replication genes (29), we next examined which cellular DNA replication DNA polymerases are used by B19V. Polymerase δ (Pol δ), polymerase α (Pol α), and polymerase ε (Pol ε) colocalized with RPA32 and the replicating viral genome (Fig. 4). We did not observe any colocalization of the replicating viral genome with DNA repair DNA polymerases, e.g., Pol η, Pol μ, Pol ζ, and Pol κ, as controls, and we observed only weak association of Pol λ with the replicating viral genome (Fig. 5).
FIG 4.
Colocalization of host DNA replication factors with B19V replication centers using immunofluorescence assay and proximity ligation assay (PLA). CD36+ EPCs were infected with B19V. At 24 hpi, cells were cytospun onto slides. (A) Immunofluorescence assay was performed to stain replicating viral genome (BrdU labeled), RPA32, and one of the host DNA replication proteins, Pol δ, Pol α, Pol ε, PCNA, or RFC1, as indicated. STED superresolution microscopy images were taken using a Leica TCS SP8 STED microscope under a 100× objective lens. (B) PLA was performed by costaining the cells with mouse anti-BrdU antibody and one of the rabbit antibodies against Pol δ, Pol α, Pol ε, PCNA, and RFC1, as indicated, which produces amplified signals for interacted proteins in close proximity. Confocal images were taken using an Eclipse C1 Plus confocal microscope (Nikon) that was controlled by Nikon EZ-C1 software. DAPI (4′,6-diamidino-2-phenylindole) was used to stain the nucleus.
FIG 5.
Confocal microscopy of host DNA repair DNA polymerases with replicating viral genome. CD36+ EPCs were infected with B19V. At 48 hpi, cells were cytospun onto slides. (A) An immunofluorescence assay was performed to stain the replicating viral genome (BrdU labeled) and one of the DNA repair DNA polymerases as indicated. (B) PLA was performed by costaining the cells with mouse anti-BrdU antibody and one of the rabbit antibodies against Pol η, Pol λ, Pol μ, and Pol ζ, except for testing Pol κ, for which rabbit anti-BrdU and mouse anti-Pol κ antibody was used. Confocal images were taken using an Eclipse C1 Plus confocal microscope (Nikon) that was controlled by Nikon EZ-C1 software. DAPI was used to stain the nucleus.
Taken together, these results confirmed the tight association of the cellular DNA replication machinery with the replicating B19V ssDNA genome and suggested that B19V takes advantage of the host cellular DNA replication machinery to replicate its ssDNA genome.
The host DNA replication machinery plays a key role in replicating B19V DNA.
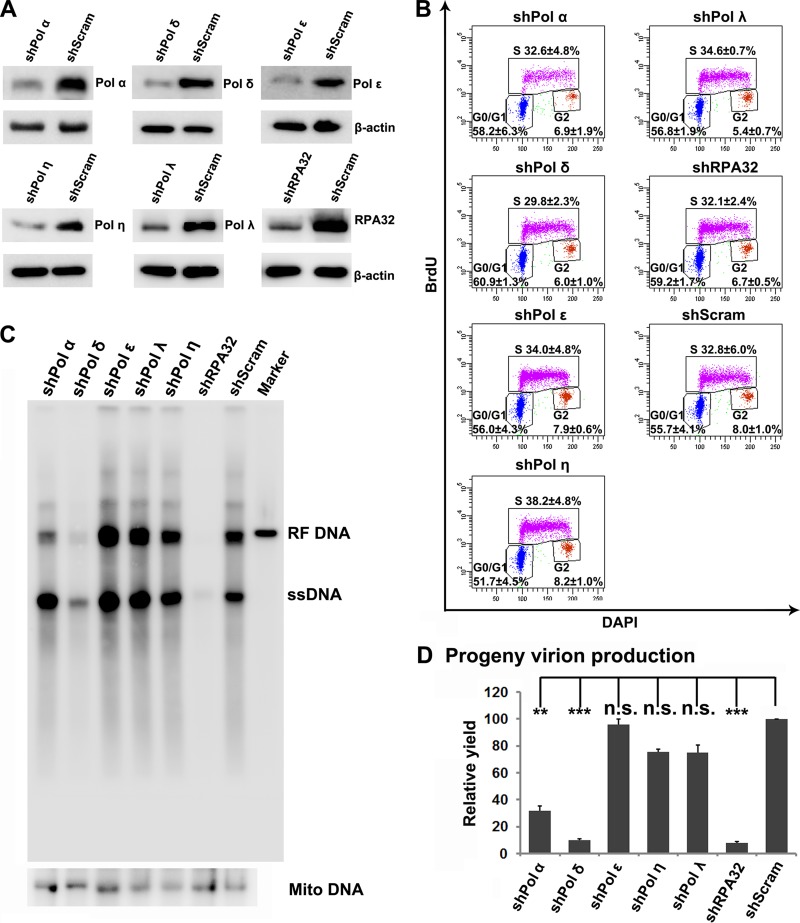
To further investigate the role of cellular DNA replication DNA polymerases in B19V DNA replication, we optimally transduced CD36+ EPCs with shRNA-expressing lentiviruses targeting cell DNA replication factors Pol α, Pol δ, and Pol ε and DNA repair factors Pol η and Pol λ and infected them with B19V for an assessment of viral DNA replication. shPol η/shPol λ and shRPA32 were used as negative and positive controls, respectively. The shRNA-expressing lentiviruses knocked down the expression of the targeted genes more than 4-fold, as shown by Western blotting (Fig. 6A). This level of knockdown did not significantly affect cellular DNA replication of these transduced cells, as shown by the absence of any significantly changed percentages of the cells in the S phase (determined using one-way analysis of variance [ANOVA]) compared to those of the scrambled control (shScram)-transduced cells (Fig. 6B). However, viral DNA replication, as shown by the replicative form (RF) DNA and ssDNA on the Southern blot, was drastically inhibited in shPol δ- and shRPA32-transduced cells (Fig. 6C). There was a moderate inhibition of RF DNA in shPol α-transduced cells but not at the level of the ssDNA (Fig. 6C). Consistent with the level of replicated viral DNA on the Southern blot, shPol δ- and shRPA32-transduced cells showed significantly lower production of progeny virions than the shScram-transduced control cells (Fig. 6D). ShPol α-transduced cells also showed inhibited progeny virion production but to a lesser degree. shPol ε-, shPol η-, and shPol λ-transduced cells produced progeny virions at a level similar to that seen with the shScram-transduced cells (Fig. 6D). These results confirmed that the key host DNA replication factor DNA polymerases, Pol δ and Pol α, and ssDNA-binding protein RPA32 play important roles in B19V DNA replication but not the cellular DNA replication-essential Pol ε (30) and these DNA repair DNA polymerases.
FIG 6.
Pol δ and RPA32 play an important role in B19V DNA replication. (A) Western blot analysis of shRNA knockdown. CD36+ EPCs transduced with shRNA-expressing lentiviruses, as indicated, were collected at 2 days postransduction. The cells were analyzed for expression of Pol α, Pol δ, Pol ε, Pol η, Pol λ, and RPA32, as indicated, by Western blotting. β-Actin was used as a loading control. (B) Cell cycle analysis. At 2 days postransduction, shRNA-transduced CD36+ EPCs were labeled with BrdU and analyzed for cell proliferation using flow cytometry with anti-BrdU and DAPI costaining. The percentages of the cells in each phase of the cell cycle are shown as means ± standard deviations (STD) of data obtained from three independent experiments. (C) Southern blot analysis of B19V DNA replication. CD36+ EPCs were transduced with the indicated shRNA for 2 days, and the cells were infected with B19V. At 48 hpi, Hirt DNA was extracted from infected cells and analyzed by Southern blotting with the M20 DNA probe (upper) and the mitochondrial DNA (Mito DNA) probe (lower). (D) B19V progeny virion production. CD36+ EPCs were transduced with the indicated shRNAs for 2 days, and the cells were infected with B19V. After 3 h of incubation, the infected cells were washed twice with phosphate-buffered saline (PBS) and cultured with fresh medium. At 48 hpi, both cells and medium were collected for quantification of B19V virions using qPCR. The value for virus (vgc) produced from shScram-transduced CD36+ EPCs is arbitrarily set at 100. **, P < 0.005; ***, P < 0.001; n.s., not significant.
Phosphorylation of RPA32 is not necessary for B19V DNA replication.
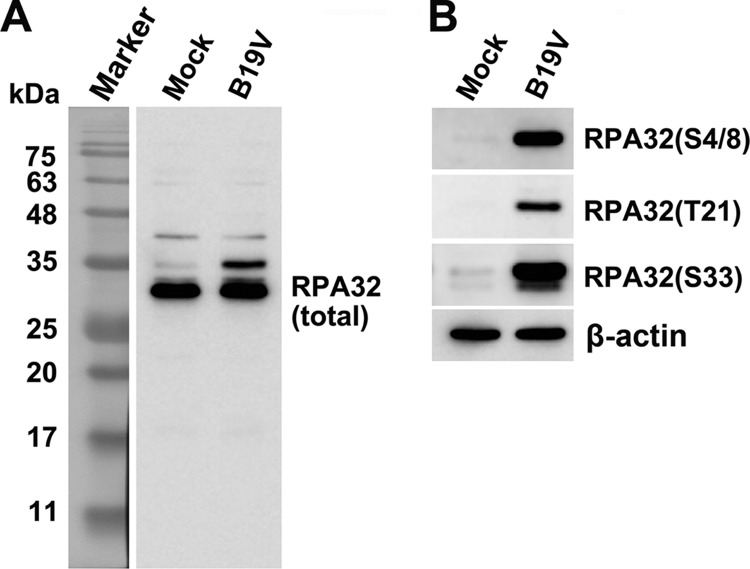
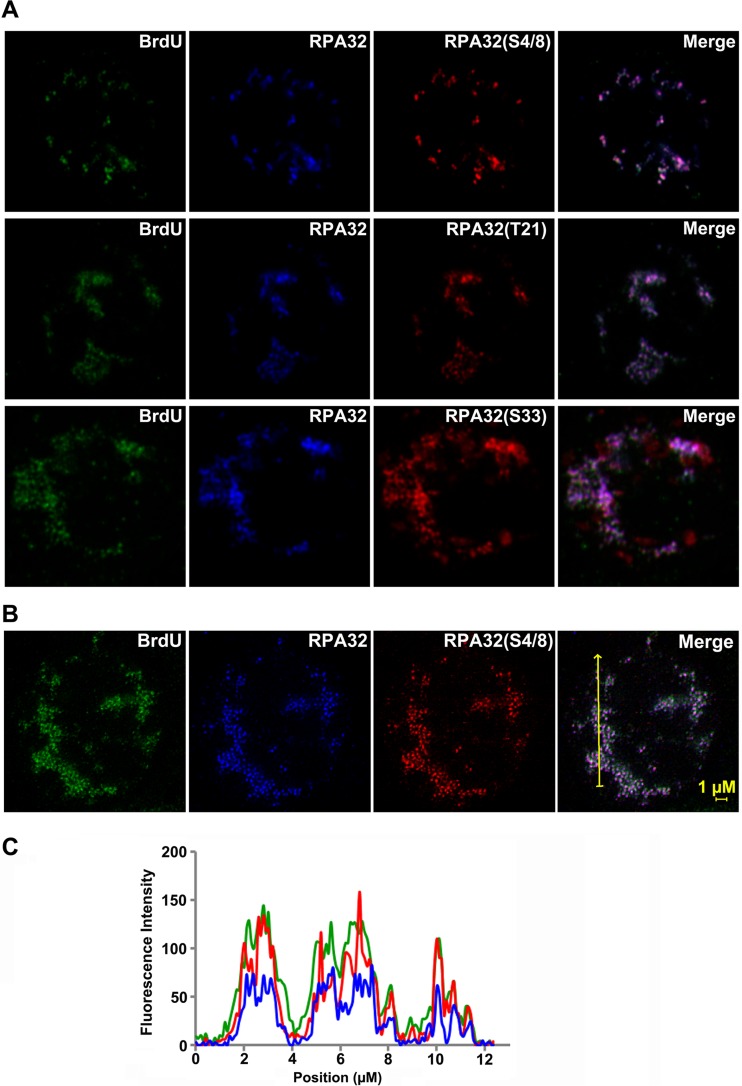
RPA32 was essential for B19V DNA replication (Fig. 6); however, it was hyperphosphorylated in B19V-infected CD36+ EPCs (Fig. 7A) (17, 31). Thus, we investigated the role of phosphorylated RPA32 in viral DNA replication. We first tested the various forms of RPA32 phosphorylation in B19V-infected cells. All four different RPA32 phosphorylation forms (S4, S8, T21, and S33) were expressed in B19V-infected cells but not in mock-infected cells (Fig. 7B). We then tested the colocalization of the major forms of phosphorylated RPA32 with the replicating viral genome. All four phosphorylated RPA32 forms were associated with the replicating viral genome (Fig. 8A). In infected cells, all of the RPA32 proteins which were stained with an antibody against unmodified RPA32 colocalized with the replicating viral genome (Fig. 4 and 8A). We further performed STED superresolution microscopy to quantify the association of RPA32(S4/8) with the BrdU-labeled viral DNA. Not only RPA32(S4/8) but also total RPA32 precisely colocalized with viral DNA foci (Fig. 8B), as evidenced by the quantification of the signals of the three colors (Fig. 8C). These results suggested that both the phosphorylated and unphosphorylated forms of RPA32 are tightly associated with the replicating viral genome.
FIG 7.
All forms of phosphorylated RPA32 are expressed in B19V-infected cells. CD36+ EPCs were infected with B19V or mock infected. At 48 hpi, the cells were collected and lysed for Western blotting of total RPA32 (A) and of various forms of phosphorylated RPA32 as indicated (B). β-Actin was used as a loading control.
FIG 8.
All forms of phosphorylated RPA32 are associated with the BrdU-labeled replicating viral genome. CD36+ EPCs were infected with B19V. At 24 hpi, the cells were labeled with BrdU and collected for immunofluorescence (IF) assay with antibodies against RPA32(S4/8), RPA32(T21), RPA32(S33), total RPA32, and anti-BrdU. (A) Images were taken using a Leica TCS SPE confocal microscope at ×63 magnification. (B and C) (B) Images were taken using a Leica TCS SP8 STED superresolution microscope under a 100× objective lens and processed with deconvolution editing using Huygens software. (C) Quantification of the intensity of each stained signaling was performed using Huygens Software.
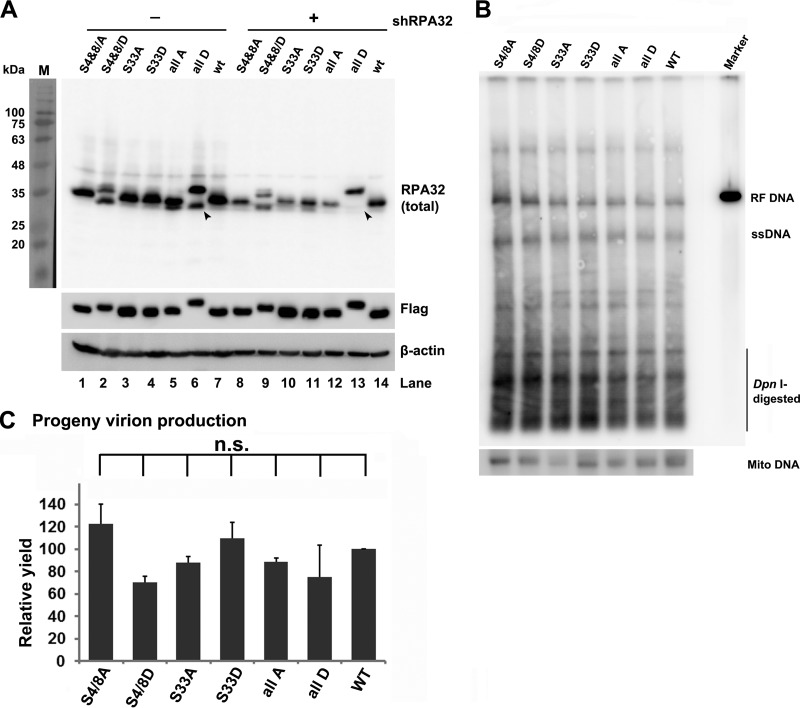
To explore the role of phosphorylated RPA32 in B19V DNA replication, we generated a series of B19V-permissive UT7/Epo-S1 cell lines that had endogenous RPA32 knocked down and ectopically expressed Flag-tagged wild-type (WT) RPA32 and different phosphorylated RPA32 mutant forms separately or together. The endogenous RPA32 knockdown was confirmed by Western blotting (Fig. 9A; compare the lower bands in lanes 6 and 13, as indicated by arrowheads). Expression of total RPA32 was detected (Fig. 9A). B19V infectious DNA (M20) replicated in the mutant RPA32-expressing cell lines at a level similar to that seen in the WT RPA32-expressing cells (Fig. 9B). In parallel, the results seen after generation of progeny virions were similar in all the cell lines (Fig. 9C). These results strongly suggest that mutant RPA32 with changes of serine (S)4/8 to alanine (A), S4/8 to aspartic acid (D), S33 to A, and S33 to D or with all of changes [S4/8, S33, and threonine (T)21 to A or D] supports progeny virion production. Both the A and D mutations, which are phosphodeficient and phosphomimetic mutations, respectively, have been used widely to study the role of RPA32 phosphorylation in response to replication stress (32).
FIG 9.
Both phosphorylated and unphosphorylated RPA32 proteins play a role in B19V replication. (A) Expression of RPA32 mutants. Flag-tagged wild-type RPA32 and phosphorylation mutants, namely, RPA32(S4/8A, S4/8D, S33A, S33D, S4/8T21S33 to A or D mutations [all A or D]), were expressed in UT7/Epo-S1 cells by lentivirus transduction. Endogenous RPA32 was knocked down in these cell lines by transducing a shRPA32-expressing lentivirus. Protein expression of RPA32 mutants was detected using anti-RPA32 or anti-Flag antibodies. β-Actin was used as a loading control. Arrowheads indicate endogenous unphosphorylated RPA32. (B) Southern blot analysis of B19V DNA replication. RPA32 mutant-expressing UT7/Epo-S1 cells were cultured under hypoxia conditions and electroporated with SalI-linearized pM20. At 48 hpi, Hirt DNA was extracted from the transfected cells, digested with DpnI enzyme, and analyzed by Southern blotting using the M20 probe (upper panel) and the mitochondrial (Mito) DNA probe (lower panel). (C) B19V virion production. RPA32 mutant-expressing UT7/Epo-S1 cells were cultured under hypoxia conditions for 2 days and electroporated with SalI-linearized M20. At 48 hpi, the cell samples were collected and quantified for B19V virions using qPCR. The value for virus (vgc) produced from M20-transfected RPA32(WT)-expressing UT7/Epo-S1 cells was arbitrarily set at 100. n.s., not significant.
Collectively, our results suggest that, although phosphorylated RPA32 proteins are recruited to the replicating viral genome, RPA32 phosphorylation at S4/8, S33, T21, or all sites is dispensable for B19V DNA replication, as both the phosphodeficient and phosphomimetic mutants of RPA32 fully supported B19V DNA replication and progeny virion production.
DISCUSSION
In this study, we systematically studied the dynamic transcriptome profile of CD36+ EPCs during B19V infection. Most differentially expressed genes appeared in infected cells at 48 hpi. B19V infection nearly shut down cellular DNA metabolic processes and DNA replication pathways during late infection, which is consistent with the G2 or late S phase arrest induced by B19V infection (18, 33). Interestingly, B19V infection also nearly shut down the cellular DNA repair pathway. However, cellular DNA damage is not observed although the DDR is induced after B19V infection (18). Nevertheless, PPI hub enrichment analysis revealed that cellular DNA replication-essential genes RFC1, PCNA, MCM2, and RPA32 were enriched. Confocal microscopy and gene knockdown analyses of the cellular DNA replication machinery demonstrated that B19V DNA replication utilizes essential cellular DNA replication factors RFC1, PCNA, RPA32, Pol δ, and Pol α. Interestingly, our analysis of RPA32 phosphorylation suggested that phosphorylated forms of RPA32 are dispensable for viral DNA replication.
This was the first study to report the cellular gene expression profile of B19V-infected primary human erythroid progenitors. The gene ontology (GO) analysis of the gene expression profile showed that at the early time point of infection (6 hpi), the most significantly enriched pathways were those associated with the response to oxygen levels (GO 0070482) and the response to hypoxia (GO 0001666) (see File S2 in the supplemental material), which is consistent with the hypoxia-increased B19V replication (13, 14). At the 12 hpi, the differentially expressed genes were significantly enriched in apoptosis regulation- and cell death-associated pathways (GO 0042981 and 0043067 and GO 0010941, respectively), a result which continued at 24 hpi and 48 hpi (File S2), confirming B19V infection-induced apoptotic cell death (34–36). At 24 hpi, the response to DNA damage stimulus was also enriched, echoing the B19V infection-induced DDR (17). At 48 hpi, except for the apoptosis and cell death regulation pathways, the most significantly enriched pathways were the immune response (GO 0006955)-related regulation pathways, including the inflammatory response (GO 0006954) (File S2). Apparently, there is limited knowledge regarding how the host cells recognize the infected B19V genome and how the virus evades the host immune clearance, which warrant further investigation.
Parvovirus DNA replication is S phase dependent (37–40), except for HBoV1 replication in nondividing human airway epithelia (26). Parvovirus infection causes DDR and cell cycle arrest of infected cells (19, 20, 41). For minute virus of mice (MVM) and B19V, the infection-induced DDR has been shown to facilitate viral DNA replication (17, 18, 22, 23). However, the role of the DDR in replication of other parvoviruses has not been closely examined so far. Therefore, understanding of how parvoviruses hijack host replication factors to replicate viral DNA is limited. HBoV1, which belongs to the Bocaparvovirus genus of the Parvoviridae family, infects mitotically quiescent human airway epithelial cells and elicits a DDR with activation of all three PI3KKs, which are essential for viral DNA replication (26). Two Y-family DNA repair polymerases, Pol η and Pol κ, are involved in HBoV1 genome amplification, providing evidence that parvovirus replication in nondividing cells is dependent on cellular DNA repair pathways. However, the case is quite different from parvovirus replication in dividing cells, in which the cellular replication machinery is available to the virus. An in vitro DNA replication study suggested that cyclin A is essential for activation of MVM DNA complementary strand synthesis and that PCNA can rescue the inhibition of cyclin-dependent kinase inhibitor p21 upon MVM DNA replication (37). Furthermore, in vitro studies confirmed that MVM uses Pol δ, PCNA, RPA, and parvovirus initiation factor (PIF) to replicate viral DNA (42, 43). Clearly, the results of the present study demonstrate that B19V DNA replication uses cellular DNA replication factors Pol δ, PCNA, RFC, and RPA, in addition to the MCM complex (18, 27).
B19V infection triggers a DDR with activation of ATM, ATR, and DNA-PKcs signaling (17). Notably, only the ATR and DNA-PKcs signaling pathways are important for B19V replication, whereas the ATM pathway is dispensable (17). ATR activation plays an important role in B19V NS1-induced G2-phase arrest through the ATR-CHK1-CDC25C-CDK1 pathway (33). Additionally, B19V replication-induced DDR induces cell cycle arrest at late S phase (18). Together with the NS1-induced G2 arrest, B19V infection induces a unique cell cycle arrest of cell cycle with 4N DNA content and cellular DNA replication activity. Combined with the results presented in the current study, we propose that the DDR-mediated late S phase is important for maintaining expression of cellular DNA replication factors, such as DNA polymerases, RPA32, RFC1, and the MCM complex, in infected cells and, more importantly, in recruiting them (in particular, RPA32) to the replicating genome. This recruitment may occur through the ATR and DNA-PK signaling pathways. It is interesting that phosphorylated forms of RPA32 are dispensable for viral DNA replication. We speculate that phosphorylation of RPA32 is the second stage of the DDR after its recruitment to the viral ssDNA genome.
Phosphorylated STAT5 specifically binds the STAT5 binding sites in viral DNA replication origins both in vivo and in vitro and is actively recruited to the viral DNA replication centers (27). More importantly, STAT5 interacts with the MCM complex and recruits the helicase complex of the cellular DNA replication machinery to viral DNA replication centers, which facilitates viral DNA replication (27), further supporting our hypothesis that B19V takes advantage of the cellular DNA replication machinery (Pol δ, PCNA, RPA, RFC, MCM complex) for viral DNA replication. These requirements for cellular factors are the same for in vitro DNA replication of adeno-associated virus 2 (AAV2) (44).
In summary, we provide evidence that B19V DNA replication uses cellular DNA replication factors. Although RPA32 is required for viral DNA replication, RPA32 phosphorylation is dispensable. We speculate that RPA32 phosphorylation is a phenotype of virus infection-induced DDR, which warrants further investigation. We believe that parvovirus DNA replication in dividing cells uses the host DNA replication mechanism for viral DNA replication. The infection-induced DDR may play an indirect role in viral DNA replication by arresting infected cells at S phase or G2 phase, during which host DNA replication factors are available.
MATERIALS AND METHODS
Primary cells and cell lines.
Primary human CD133+ hematopoietic stem cells were isolated from bone marrow of healthy donors according to a protocol (04-H-0179) approved by the National Heart, Lung, and Blood Institute institutional review board. Primary human CD36+ EPCs were expanded ex vivo as previously described (12, 15). Briefly, hematopoietic stem cells were cultured in Wong medium under normoxia conditions (5% CO2 and 21% O2) up to day 4 and frozen in liquid nitrogen. In each experiment, day 4 cells were seeded under normoxia conditions for 2 to 3 days, prior to incubation under hypoxia conditions (5% CO2 and 1% O2) for 2 days.
The human UT7/Epo-S1 megakaryoblastoid cell line was obtained from Kevin Brown with permission from Kazuo Sugamura (45) and was grown in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (Sigma, St. Louis, MO) and 2 U/ml of EPO (Amgen, Thousand Oaks, CA). UT7/Epo-S1 cell lines expressing RPA32 mutants were cultured under the same conditions but in the presence of puromycin before pM20 electroporation. All UT7/Epo-S1 cells were cultured under hypoxia conditions for 2 days before electroporation.
Virus and infection.
Plasma sample 347 containing B19V at 1 × 1012 viral genomic copies per ml (vgc/ml) was obtained from ViraCor Eurofins Laboratories (Lee's Summit, MO). After 2 days under hypoxia incubation, CD36+ EPCs were infected with B19V at a multiplicity of infection (MOI) of 1,000 vgc per cell. At various hpi, the infected cells were collected for microarray analysis, immunofluorescence assay, Southern blot analysis, or quantification of virus production.
RNA extraction and microarray hybridization.
The mock- and B19V-infected CD36+ EPCs were collected at 6, 12, 24, and 48 hpi. RNA samples were extracted using an miRNeasy minikit by following the instructions of the manufacturer (Qiagen, Valencia, CA). At each time point, three repeat samples were prepared. All RNA samples were analyzed on an Agilent 2100 Bioanalyzer with an Agilent RNA 6000 Nano kit to ensure an RNA Integrity Number (RIN) value of ≥8.0. Microarray hybridization was performed on a GeneChip Human Genome U133A 2.0 array using an Affymetrix GeneChip system (Affymetrix, Santa Clara, CA) at the Microarray Core Facility, University of Kansas Medical Center.
Microarray data preprocessing.
After microarray hybridization, the produced signal intensity fluorescent images were read using an Agilent Gene Array Scanner and were converted to GeneChip probe result files (CEL) using MAS 5.0 software (Affymetrix). Bioconductor ReadAffy was used to read CEL files into an AffyBatch, and robust multiarray average (RMA) calculations were applied to convert AffyBatch objects to an expression matrix. Quantile values were used to normalize expression intensities and to ensure that the intensities had similar distributions across different time points. Limma eBayes was applied to access statistically differentially regulated probes (46). DAVID (the Database for Annotation, Visualization and Integrated Discovery) Bioinformatics Resources (https://david.ncifcrf.gov/) were used to perform Gene Ontology (GO) biological process analysis. Enrichr was used to identify enriched pathways for protein-protein interacting proteins (PPI Hub Proteins). Cytoscape was used to visualize protein-protein interacting networks and to integrate gene expression profiles.
BrdU labeling, immunofluorescence assay, and confocal microscopy imaging.
Immunofluorescence assays were carried out as described previously (47). Briefly, at 24 or 48 hpi, 1 × 105 CD36+ cells were labeled with BrdU at a final concentration of 30 μM for 30 min and fixed. The fixed cells were then incubated with mouse or rabbit anti-BrdU, goat anti-RPA32, and another antibody targeting a host protein, followed by incubation with anti-mouse IgG conjugated with Alexa Fluor 488, anti-rabbit IgG conjugated with Alexa Fluor 594, and anti-goat IgG conjugated with Alexa Fluor 647 secondary antibodies. After washing, the slide was mounted with Mowoil or Prolong Diamond Antifade Mountant (Invitrogen) with a cover glass. Confocal images were taken using a Leica TPE confocal microscope or a Nikon Eclipse C1 Plus confocal microscope. In some instances, STED superresolution microscopy images were taken using a Leica TCS SP8 STED microscope under a 100× objective lens (HC PL APO STED White 100×/1.4 oil). Images were deconvoluted using the Classic Maximum Likelihood Estimation (CMLE) algorithm and Huygens software (Scientific Volume Imaging B.V. Netherlands).
Proximity ligation assay (PLA).
A Duolink In-Situ Red mouse/rabbit kit (catalog no. DUO92101) was purchased from Millipore Sigma (St. Louis, MO). PLA was performed following the manufacturer's instructions, as described previously (47). Briefly, B19V-infected CD36+ EPCs were labeled with BrdU as described above. The labeled cells were fixed with 3.7% paraformaldehyde for 15 min, permeabilized with 0.2% Triton X-100 for 5 min, and blocked with Duolink blocking buffer for 30 min. The cells were then incubated with primary antibodies, a mouse/rabbit anti-BrdU antibody, and a rabbit/mouse antibody against a cellular DNA replication/repair factor for 1 h. Two diluted PLA probes (specific to mouse and rabbit IgG, respectively) were applied to the cells and incubated for 60 min at 37°C. Next, the hybridized oligonucleotides were ligated in Ligation Solution at 37°C for 30 min and amplified in Amplification Solution for 100 min. Finally, the cells were washed and mounted using Mounting Medium. The slides were visualized under a Nikon Eclipse C1 Plus confocal microscope.
Establishment of RPA32 mutant-expressing UT7/Epo-S1 cell lines.
A Flag-tagged wild-type (WT) RPA32 open reading frame (ORF) or ORFs of different mutant forms of RPA32 (S4/8A, S4/8D, S33A, S33D, S4/8T21S33 to A or D mutations [all A or D]) were cloned into lentiviral vector pLenti-CMV-MCS-IRES-GFP-WPRE (12). Lentiviruses were produced and purified as previously described (12). UT7/Epo-S1 cells were transduced with lentivirus followed by selection of green fluorescent protein (GFP)-expressing cells by flow cytometry. Then, the endogenous RPA32 in these cell lines was knocked down by transduction with a shRPA32-expressing lentivirus.
shRNA-expressing lentivirus production.
shRNA sequences targeting Pol α, Pol δ, Pol ε, Pol η, Pol λ, and RPA32 were cloned into the pLKO.1 vector, and lentiviruses were produced and purified as previously described (12).
The shRNA sequences for knocking down target proteins were as follows: for shRPA32, 5′-CCU AGU UUC ACA AUC UGU UGU-3′, located at the 3′ untranscribed region (UTR) of the mRNA; for shPol α, 5′-GCC AAU UAA ACC CGG UCU AAA-3′; for shPol δ, 5′-GCU UAU CAG CAA GAA GCG CUA-3′; for shPol ε, 5′-CCU GUG CUA ACA ACU UUG CAA-3′; for shPol η, 5′-AGU UAU GAA GCU CGU GCA UUU-3′; for shPol λ, 5′-UGG UGA UGU CGA CGU GCU CAU-3′.
Cell cycle analysis by flow cytometry.
For cell cycle analysis of CD36+ EPCs, a BrdU incorporation assay was performed, and the cells were analyzed by flow cytometry analysis as described previously (18).
Electroporation.
UT7/Epo-S1 cells were electroporated with 3 μg of SalI-linearized B19V infectious clone pM20 (48) in solution V using Amaxa Nucleofector (Lonza, Basel, Switzerland), as described previously (27). After transfection, cells were cultured under hypoxia conditions (1% O2).
Southern blot analysis.
Low-molecular-weight DNA (Hirt DNA) was extracted from either B19V-infected CD36+ EPCs or UT7/Epo-S1 cells, as described previously (49). B19V RF DNA M20 excised from SalI-digested pM20 was used as a probe.
Quantification of progeny virion production.
For progeny virion production from B19V-infected CD36+ EPCs, 1 × 106 shRNA-expressing lentivirus-transduced CD36+ EPCs were infected with B19V. For progeny virion production from pM20-transfected UT7/Epo-S1 cells, various RPA32 mutant-expressing cells were electroporated with SalI-linearized M20 DNA. The cells were washed and cultured under hypoxia conditions. At 48 hpi or 48 h postelectroporation, the cultures were frozen and thawed three times and were treated with Benzonase nuclease (250 U/ml) at 37°C for 2 h. The reaction was stopped by adding EDTA to 10 mM, and the nuclease-protected viral genome was extracted using a DNeasy blood and tissue kit (Qiagen, Germantown, MD), following the manufacturer's instructions. The produced B19V genome numbers (vgc) were determined by real-time quantitative PCR (qPCR), as described previously (49).
Western blotting.
Western blotting was performed according to a previously reported method with antibodies for the proteins indicated in each figure (18, 50).
Antibodies used.
Anti-Pol α (sc-48818) and anti-RFC1 (sc-20993) were purchased from Santa Cruz (Dallas, TX); anti-Pol δ (catalog no. A304-007A-T), anti-total RPA32 (catalog no. A300-244A), anti-RPA32(S4/8) (catalog no. A300-245A-T), and anti-RPA32(S33) (catalog no. A300-246A-T) were from Bethyl Laboratories (Montgomery, TX); anti-Pol ε (catalog no. GTX116557), anti-RPA32(T21) (catalog no. GTX62664), and anti-ATR(pT1989) (catalog no. GTX128145) were from GeneTex (Irvine, CA); anti-PCNA (44434) was from One World Lab (San Diego, CA); and anti-ATM(pS1981) (ab81292) and anti-DNA-PK (ab18192) were from Abcam (Cambridge, MA).
The secondary antibodies used were horseradish peroxidase (HRP)-conjugated anti-mouse IgG and HRP-conjugated anti-rabbit IgG (Sigma) and Alexa Fluor 488-conjugated anti-mouse IgG, Alexa Fluor 594-conjugated anti-rabbit IgG, and Alexa Fluor 647conjugated anti-goat IgG (Jackson Immuno Research Inc., West Grove, PA).
Statistical analyses.
For cell cycle analysis, the percentage of cells at each phase of the cell cycle was determined from three independent experiments and expressed as the mean ± standard deviation (mean ± STD). The statistical significance of the percentage of the cells in S phase (Fig. 6B) and B19V progeny virion production (Fig. 6 and 9) was determined using one-way ANOVA (GraphPad Prism Version 7.0). Error bars show mean ± STD unless otherwise specified.
Accession number(s).
The complete raw and normalized microarray data are available in the Gene Expression Omnibus of the National Center for Biotechnology Information (GSE103460).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to members of the Qiu laboratory for technical support and valuable discussions. We are indebted to Susan Wong at the Hematology Branch, NHLBI, NIH, for providing CD133+ human hematopoietic stem cells. We thank Clark Bloomer and Yafen Niu of the Microarray Core Facility, University of Kansas School of Medicine, for performing the microarray experiments and Alexander Jurkevich at the Molecular Cytology Core, University of Missouri—Columbia, for STED superresolution microscopy.
This study was supported in full by PHS grant R01 AI070723 from the National Institute of Allergy and Infectious Diseases to J.Q. and in part by PHS grants R01 NS078214 from the National Institute of Neurological Disorders and Stroke and R01 AG051470 from the National Institute of Aging to H.N. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01881-17.
REFERENCES
- 1.Cotmore SF, Agbandje-McKenna M, Chiorini JA, Mukha DV, Pintel DJ, Qiu J, Söderlund-Venermo M, Tattersall P, Tijssen P, Gatherer D, Davison AJ. 2014. The family Parvoviridae. Arch Virol 159:1239–1247. doi: 10.1007/s00705-013-1914-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Young NS, Brown KE. 2004. Parvovirus B19. N Engl J Med 350:586–597. doi: 10.1056/NEJMra030840. [DOI] [PubMed] [Google Scholar]
- 3.Brown KE, Young NS. 1997. Parvovirus B19 in human disease. Annu Rev Med 48:59–67. doi: 10.1146/annurev.med.48.1.59. [DOI] [PubMed] [Google Scholar]
- 4.Gallinella G. 2013. Parvovirus B19 achievements and challenges. ISRN Virology 2013:898730. doi: 10.5402/2013/898730. [DOI] [Google Scholar]
- 5.Young N.S. 1995. B19 parvovirus. Baillieres Clin Haematol 8:25–56. doi: 10.1016/S0950-3536(05)80231-8. [DOI] [PubMed] [Google Scholar]
- 6.Qiu J, Söderlund-Venermo M, Young NS. 2017. Human parvoviruses. Clin Microbiol Rev 30:43–113. doi: 10.1128/CMR.00040-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ozawa K, Kurtzman G, Young N. 1986. Replication of the B19 parvovirus in human bone marrow cell cultures. Science 233:883–886. doi: 10.1126/science.3738514. [DOI] [PubMed] [Google Scholar]
- 8.Young NS, Mortimer PP, Moore JG, Humphries RK. 1984. Characterization of a virus that causes transient aplastic crisis. J Clin Invest 73:224–230. doi: 10.1172/JCI111195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young N, Harrison M, Moore J, Mortimer P, Humphries RK. 1984. Direct demonstration of the human parvovirus in erythroid progenitor cells infected in vitro. J Clin Invest 74:2024–2032. doi: 10.1172/JCI111625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yaegashi N, Shiraishi H, Takeshita T, Nakamura M, Yajima A, Sugamura K. 1989. Propagation of human parvovirus B19 in primary culture of erythroid lineage cells derived from fetal liver. J Virol 63:2422–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morey AL, Fleming KA. 1992. Immunophenotyping of fetal haemopoietic cells permissive for human parvovirus B19 replication in vitro. Br J Haematol 82:302–309. doi: 10.1111/j.1365-2141.1992.tb06422.x. [DOI] [PubMed] [Google Scholar]
- 12.Chen AY, Guan W, Lou S, Liu Z, Kleiboeker S, Qiu J. 2010. Role of erythropoietin receptor signaling in parvovirus B19 replication in human erythroid progenitor cells. J Virol 84:12385–12396. doi: 10.1128/JVI.01229-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pillet S, Le GN, Hofer T, NguyenKhac F, Koken M, Aubin JT, Fichelson S, Gassmann M, Morinet F. 2004. Hypoxia enhances human B19 erythrovirus gene expression in primary erythroid cells. Virology 327:1–7. doi: 10.1016/j.virol.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 14.Chen AY, Kleiboeker S, Qiu J. 2011. Productive parvovirus B19 infection of primary human erythroid progenitor cells at hypoxia is regulated by STAT5A and MEK signaling but not HIF alpha. PLoS Pathog 7:e1002088. doi: 10.1371/journal.ppat.1002088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong S, Zhi N, Filippone C, Keyvanfar K, Kajigaya S, Brown KE, Young NS. 2008. Ex vivo-generated CD36+ erythroid progenitors are highly permissive to human parvovirus B19 replication. J Virol 82:2470–2476. doi: 10.1128/JVI.02247-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morita E, Nakashima A, Asao H, Sato H, Sugamura K. 2003. Human parvovirus B19 nonstructural protein (NS1) induces cell cycle arrest at G(1) phase. J Virol 77:2915–2921. doi: 10.1128/JVI.77.5.2915-2921.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo Y, Lou S, Deng X, Liu Z, Li Y, Kleiboeker S, Qiu J. 2011. Parvovirus B19 infection of human primary erythroid progenitor cells triggers ATR-Chk1 signaling, which promotes B19 virus replication. J Virol 85:8046–8055. doi: 10.1128/JVI.00831-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo Y, Kleiboeker S, Deng X, Qiu J. 2013. Human parvovirus B19 infection causes cell cycle arrest of human erythroid progenitors at late S phase that favors viral DNA replication. J Virol 87:12766–12775. doi: 10.1128/JVI.02333-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cotmore S. F., Tattersall P. 2013. Parvovirus diversity and DNA damage responses. Cold Spring Harb Perspect Biol 5:a012989. doi: 10.1101/cshperspect.a012989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo Y, Qiu J. 2013. Parvovirus infection induced DNA damage response. Future Virol 8:245–257. doi: 10.2217/fvl.13.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo Y, Chen AY, Qiu J. 2011. Bocavirus infection induces a DNA damage response that facilitates viral DNA replication and mediates cell death. J Virol 85:133–145. doi: 10.1128/JVI.01534-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo Y, Deng X, Cheng F, Li Y, Qiu J. 2013. SMC1-mediated intra-S phase arrest facilitates bocavirus DNA replication. J Virol 87:4017–4032. doi: 10.1128/JVI.03396-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adeyemi RO, Landry S, Davis ME, Weitzman MD, Pintel DJ. 2010. Parvovirus minute virus of mice induces a DNA damage response that facilitates viral replication. PLoS Pathog 6:e1001141. doi: 10.1371/journal.ppat.1001141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adeyemi RO, Pintel DJ. 2012. Replication of minute virus of mice (MVM) in murine cells is facilitated by virally induced depletion of p21. J Virol 86:8328–8332. doi: 10.1128/JVI.00820-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Q, Deng X, Yan Z, Cheng F, Luo Y, Shen W, Lei-Butters DC, Chen AY, Li Y, Tang L, Söderlund-Venermo M, Engelhardt JF, Qiu J. 2012. Establishment of a reverse genetics system for studying human bocavirus in human airway epithelia. PLoS Pathog 8:e1002899. doi: 10.1371/journal.ppat.1002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng X, Yan Z, Cheng F, Engelhardt JF, Qiu J. 2016. Replication of an autonomous human parvovirus in non-dividing human airway epithelium is facilitated through the DNA damage and repair pathways. PLoS Pathog 12:e1005399. doi: 10.1371/journal.ppat.1005399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ganaie SS, Zou W, Xu P, Deng X, Kleiboeker S, Qiu J. 2017. Phosphorylated STAT5 directly facilitates parvovirus B19 DNA replication in human erythroid progenitors through interaction with the MCM complex. PLoS Pathog 13:e1006370. doi: 10.1371/journal.ppat.1006370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Söderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, Landegren U. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods 3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 29.Zhang D, O'Donnell M. 2016. The eukaryotic replication machine. Enzymes 39:191–229. doi: 10.1016/bs.enz.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 30.Johnson RE, Klassen R, Prakash L, Prakash S. 2015. A major role of DNA polymerase delta in replication of both the leading and lagging DNA strands. Mol Cell 59:163–175. doi: 10.1016/j.molcel.2015.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lou S, Luo Y, Cheng F, Huang Q, Shen W, Kleiboeker S, Tisdale JF, Liu Z, Qiu J. 2012. Human parvovirus B19 DNA replication induces a DNA damage response that is dispensable for cell cycle arrest at G2/M phase. J Virol 86:10748–10758. doi: 10.1128/JVI.01007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu S, Opiyo SO, Manthey K, Glanzer JG, Ashley AK, Amerin C, Troksa K, Shrivastav M, Nickoloff JA, Oakley GG. 2012. Distinct roles for DNA-PK, ATM and ATR in RPA phosphorylation and checkpoint activation in response to replication stress. Nucleic Acids Res 40:10780–10794. doi: 10.1093/nar/gks849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu P, Zhou Z, Xiong M, Zou W, Deng X, Ganaie SS, Kleiboeker S, Peng J, Liu K, Wang S, Ye SQ, Qiu J. 2017. Parvovirus B19 NS1 protein induces cell cycle arrest at G2-phase by activating the ATR-CDC25C-CDK1 pathway. PLoS Pathog 13:e1006266. doi: 10.1371/journal.ppat.1006266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moffatt S, Yaegashi N, Tada K, Tanaka N, Sugamura K. 1998. Human parvovirus B19 nonstructural (NS1) protein induces apoptosis in erythroid lineage cells. J Virol 72:3018–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sol N, Le JJ, Vassias I, Freyssinier JM, Thomas A, Prigent AF, Rudkin BB, Fichelson S, Morinet F. 1999. Possible interactions between the NS-1 protein and tumor necrosis factor alpha pathways in erythroid cell apoptosis induced by human parvovirus B19. J Virol 73:8762–8770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen AY, Zhang EY, Guan W, Cheng F, Kleiboeker S, Yankee TM, Qiu J. 2010. The small 11kDa non-structural protein of human parvovirus B19 plays a key role in inducing apoptosis during B19 virus infection of primary erythroid progenitor cells. Blood 115:1070–1080. doi: 10.1182/blood-2009-04-215756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bashir T, Horlein R, Rommelaere J, Willwand K. 2000. Cyclin A activates the DNA polymerase delta-dependent elongation machinery in vitro: a parvovirus DNA replication model. Proc Natl Acad Sci U S A 97:5522–5527. doi: 10.1073/pnas.090485297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bashir T, Rommelaere J, Cziepluch C. 2001. In vivo accumulation of cyclin A and cellular replication factors in autonomous parvovirus minute virus of mice-associated replication bodies. J Virol 75:4394–4398. doi: 10.1128/JVI.75.9.4394-4398.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cotmore SF, Tattersall P. 1987. The autonomously replicating parvoviruses of vertebrates. Adv Virus Res 33:91–174. doi: 10.1016/S0065-3527(08)60317-6. [DOI] [PubMed] [Google Scholar]
- 40.Berns KI, Parrish CR. 2015. Parvoviridae, p 1768–1791. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 41.Chen AY, Qiu J. 2010. Parvovirus infection-induced cell death and cell cycle arrest. Future Virol 5:731–741. doi: 10.2217/fvl.10.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Christensen J, Tattersall P. 2002. Parvovirus initiator protein NS1 and RPA coordinate replication fork progression in a reconstituted DNA replication system. J Virol 76:6518–6531. doi: 10.1128/JVI.76.13.6518-6531.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Christensen J, Cotmore SF, Tattersall P. 1997. Parvovirus initiation factor PIF: a novel human DNA-binding factor which coordinately recognizes two ACGT motifs. J Virol 71:5733–5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nash K, Chen W, Muzyczka N. 2008. Complete in vitro reconstitution of adeno-associated virus DNA replication requires the minichromosome maintenance complex proteins. J Virol 82:1458–1464. doi: 10.1128/JVI.01968-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morita E, Tada K, Chisaka H, Asao H, Sato H, Yaegashi N, Sugamura K. 2001. Human parvovirus B19 induces cell cycle arrest at G(2) phase with accumulation of mitotic cyclins. J Virol 75:7555–7563. doi: 10.1128/JVI.75.16.7555-7563.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smyth GK. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 47.Deng X, Xu P, Zou W, Shen W, Peng J, Liu K, Engelhardt JF, Yan Z, Qiu J. 2017. DNA damage signaling is required for replication of human bocavirus 1 DNA in dividing HEK293 cells. J Virol 91:e01831-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhi N, Zadori Z, Brown KE, Tijssen P. 2004. Construction and sequencing of an infectious clone of the human parvovirus B19. Virology 318:142–152. doi: 10.1016/j.virol.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 49.Guan W, Wong S, Zhi N, Qiu J. 2009. The genome of human parvovirus B19 virus can replicate in non-permissive cells with the help of adenovirus genes and produces infectious virus. J Virol 83:9541–9553. doi: 10.1128/JVI.00702-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zou W, Cheng F, Shen W, Engelhardt JF, Yan Z, Qiu J. 2016. Nonstructural protein NP1 of human bocavirus 1 plays a critical role in the expression of viral capsid proteins. J Virol 90:4658–4669. doi: 10.1128/JVI.02964-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.