ABSTRACT
The human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein (Env) encodes specific trafficking signals within its long cytoplasmic tail (CT) that regulate incorporation into HIV-1 particles. Rab11-family interacting protein 1C (FIP1C) and Rab14 are host trafficking factors required for Env particle incorporation, suggesting that Env undergoes sorting from the endosomal recycling compartment (ERC) to the site of particle assembly on the plasma membrane. We disrupted outward sorting from the ERC by expressing a C-terminal fragment of FIP1C (FIP1C560–649) and examined the consequences on Env trafficking and incorporation into particles. FIP1C560–649 reduced cell surface levels of Env and prevented its incorporation into HIV-1 particles. Remarkably, Env was trapped in an exaggerated perinuclear ERC in a CT-dependent manner. Mutation of either the Yxxϕ endocytic motif or the YW795 motif in the CT prevented Env trapping in the ERC and restored incorporation into particles. In contrast, simian immunodeficiency virus SIVmac239 Env was not retained in the ERC, while substitution of the HIV-1 CT for the SIV CT resulted in SIV Env retention in this compartment. These results provide the first direct evidence that Env traffics through the ERC and support a model whereby HIV-1 Env is specifically targeted to the ERC prior to FIP1C- and CT-dependent outward sorting to the particle assembly site on the plasma membrane.
IMPORTANCE The HIV envelope protein is an essential component of the viral particle. While many aspects of envelope protein structure and function have been established, the pathway it follows in the cell prior to reaching the site of particle assembly is not well understood. The envelope protein has a very long cytoplasmic tail that interacts with the host cell trafficking machinery. Here, we utilized a truncated form of the trafficking adaptor FIP1C protein to arrest the intracellular transport of the envelope protein, demonstrating that it becomes trapped inside the cell within the endosomal recycling compartment. Intracellular trapping resulted in a loss of envelope protein on released particles and a corresponding loss of infectivity. Mutations of specific trafficking motifs in the envelope protein tail prevented its trapping in the recycling compartment. These results establish that trafficking to the endosomal recycling compartment is an essential step in HIV envelope protein particle incorporation.
KEYWORDS: endosomal recycling compartment, envelope, HIV-1, Rab11-FIP1C, virus assembly
INTRODUCTION
The mechanism of human immunodeficiency virus type 1 (HIV-1) envelope (Env) incorporation into developing particles is incompletely understood. Four possible models of Env incorporation have been proposed, including passive incorporation, direct Gag-Env interaction, Gag-Env cotargeting, and indirect Gag-Env interaction models (1, 2). Lentiviral envelope glycoproteins bear unusually long cytoplasmic tails (CTs) compared to those of other retroviruses (1, 3). Truncation of the HIV-1 CT does not disrupt incorporation into the developing particle when Env is expressed in certain permissive cell types, such as 293T or HeLa cells. However, the long Env CT is required for particle incorporation, infectivity, and cell-cell spread of virus in most T cell lines, in primary T lymphocytes, and in monocyte-derived macrophages (MDMs) (4). Distinct cellular trafficking signals have been identified within the HIV-1 CT. A membrane-proximal Yxxϕ motif interacts with the AP-2 clathrin adaptor complex and mediates clathrin-dependent endocytosis of Env (5–7). This motif has been shown to be important for mediating particle incorporation, infectivity, and basolateral targeting of Env (8–10). A dileucine motif located at the C terminus of the CT mediates interactions with the AP-1 clathrin adaptor complex (5, 11). Additional conserved tyrosine-based motifs and dileucine motifs are present within the CT and have variable effects on cell surface expression and virion incorporation, as judged by mutagenesis studies (12).
We previously described a role for Rab11-family interacting protein 1C (FIP1C; also known as Rab coupling protein or RCP) and Rab14 in HIV-1 Env trafficking and particle incorporation (13). FIP1C forms a parallel homodimer capable of binding to two Rab molecules through its C-terminal Rab binding domain (RBD) (14, 15). In HeLa cells, FIP1C is largely concentrated in a perinuclear structure representing the endosomal recycling compartment (ERC) (16, 17). However, upon expression of HIV-1 Env, FIP1C redistributes to the cellular periphery along with Env (13). The redistribution of FIP1C depends upon specific sequences within the Env CT, as either a CT truncation mutant or the YW795SL mutant within the CT failed to elicit FIP1C redistribution (18). The YW795SL mutant was deficient in Env incorporation and replicated poorly in T cell lines, while a second-site revertant near the carboxyl-terminal end of the tail (L850S) restored FIP1C redistribution, Env incorporation, and replication to wild-type (WT) levels. These findings suggested to us that specific structural elements or motifs within the CT interact with FIP1C and Rab14, and that this interaction mediates Env trafficking from the ERC to the site of particle assembly on the plasma membrane (PM). However, there has been no direct evidence thus far supporting the presence of Env in the ERC, where it would need to transiently reside prior to outward sorting in complex with FIP1C and Rab14.
Expression of the carboxyl-terminal domains of FIP1C or of Rab11-FIP2 has been shown to disrupt transferrin recycling and alters the morphology of the ERC (16, 19, 20). Here, we investigated the role of the ERC in Env trafficking by disrupting outward sorting through expression of a carboxyl-terminal fragment of FIP1C. This intervention arrested HIV-1 Env trafficking, resulting in trapping of Env on perinuclear endosomal membranes that were Rab11 and Rab14 positive. Intracellular Env trapping mediated by truncated FIP1C significantly diminished Env incorporation into particles and particle infectivity. Env trapping in the aberrant ERC was specific to FIP1C, as a comparable carboxyl-terminal fragment from Rab11-FIP2 failed to trap Env. Furthermore, arrest in the ERC was specific to the CT of HIV-1, as simian immunodeficiency virus (SIV) Env was not sequestered in the ERC unless its CT was swapped with that of HIV-1.
RESULTS
A C-terminal fragment of FIP1C blocks HIV-1 Env localization at the cell surface and prevents virion incorporation.
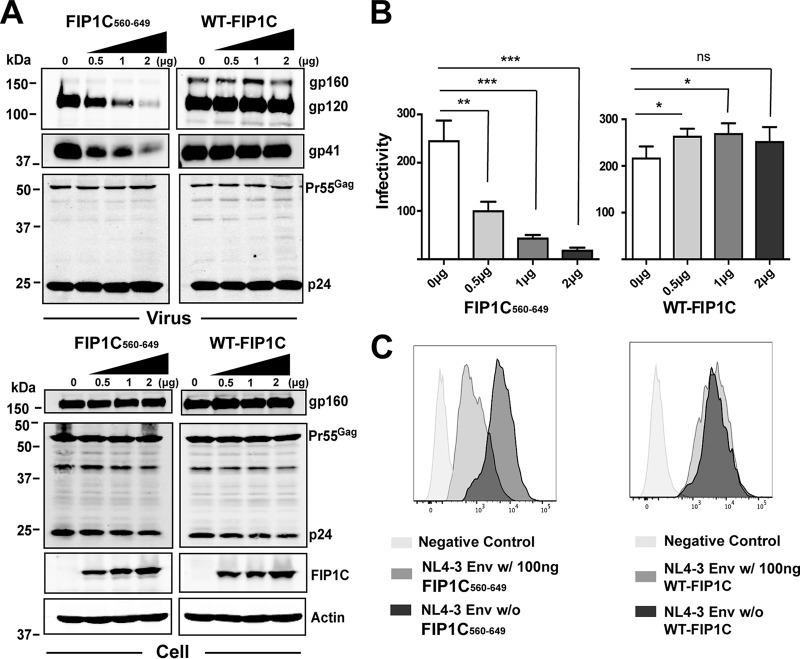
To define the role of the ERC in Env trafficking, we expressed a truncated form of FIP1C (FIP1C560–649) tagged to green fluorescent protein (GFP) in HeLa cells. Incorporation of Env into released HIV-1 particles was inhibited by FIP1C560–649 in a dose-dependent fashion (Fig. 1A, top). This effect was not due to reduced synthesis or enhanced degradation of Env, as FIP1C560–649 had little effect on cellular levels of gp160 (Fig. 1A, bottom). Titration of full-length GFP-FIP1C did not inhibit Env incorporation (Fig. 1A, right lanes). The infectivity of released particles was diminished by FIP1C560–649 in a manner proportional to the removal of Env from particles (Fig. 1B, left) and was slightly increased (rather than diminished) by expression of the full-length FIP1C (Fig. 1B, right). We next asked if truncated FIP1C altered cell surface Env levels. FIP1C560–649 expression resulted in a significant reduction of cell surface Env, as measured by flow cytometry of nonpermeabilized cells (Fig. 1C, left), while cell surface levels were not significantly altered by wild-type FIP1C expression (Fig. 1C, right). These results indicated to us that FIP1C560–649 exerts a negative effect on Env incorporation into virions through a mechanism that reduces Env cell surface levels but does not significantly alter total cellular Env, suggesting that Env is trapped in an intracellular location.
FIG 1.
Truncated FIP1C inhibits Env incorporation and particle infectivity. (A) GFP-FIP1C and truncated mutant of FIP1C (FIP1C560–649) plasmids were transfected in increasing concentrations together with a fixed amount of pNL4-3 proviral DNA into HeLa cells. Shown are results of this titration on viral protein content (upper blots) and cellular protein content (lower blots) 48 h posttransfection. (B) Infectivity of virus particles was quantified using TZM-bl cells following dose titration of FIP1C560–649 (left) or wild-type FIP1C (right). Results are shown as blue cells/nanogram of p24 and are presented as means ± standard deviations (SD). Statistical comparisons between groups were performed using the unpaired t test. Graphs shown are representative of major findings from three independent experiments. ns, ρ > 0.05; *, ρ < 0.05; **, ρ < 0.001; ***, ρ < 0.0001. (C) Cell surface staining for Env using anti-gp120 mouse monoclonal antibody BDI123 (Novus Biologicals) 48 h following transfection with the indicated constructs. Cells first were gated for GFP-FIP1C expression.
HIV-1 Env is trapped in the ERC by overexpression of FIP1C560–649.
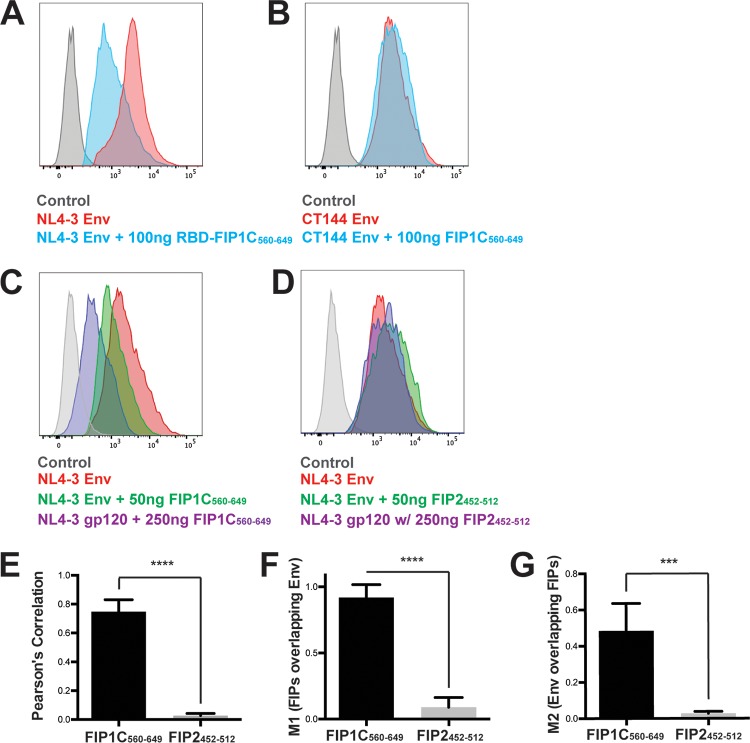
To define the mechanism underlying the disruption in Env particle incorporation, we examined the subcellular distribution of Env when coexpressed with either wild-type FIP1C or FIP1C560–649. Wild-type FIP1C demonstrated a prominent perinuclear localization but also some more diffuse and punctate signals in HeLa cells (Fig. 2A). FIP1C560–649 was also found in a pronounced perinuclear compartment with less prominent peripheral puncta (Fig. 2B). We have previously shown that WT FIP1C is redistributed from the perinuclear ERC to the cellular periphery upon expression of HIV-1 Env with a full-length CT (13, 18). Characteristic redistribution of wild-type FIP1C upon Env expression is shown in Fig. 2C. In stark contrast, FIP1C560–649 remained tightly concentrated in a perinuclear location in the presence of HIV-1 Env (Fig. 2D). Remarkably, Env was found to colocalize very strongly with FIP1C560–649, indicating trapping of Env within the ERC (Fig. 2D). Trapping of Env correlated with a loss of Env from the cell surface upon FIP1C560–649 expression (Fig. 1C and 3A). To determine if Env trapping in the ERC was dependent upon the presence of the Env CT, we examined the distribution of CT144 Env, which lacks all but the membrane-proximal six residues of the CT. CT144 Env was not trapped by FIP1C560–649 but was found on cellular membranes and largely excluded from the ERC (Fig. 2E). The level of CT144 Env on the cell surface was unaffected by FIP1C560–649 expression (Fig. 3B). We reasoned that the trapping of Env could have been due to a general disruption of recycling from the ERC by overexpression of FIP1C560–649 or could represent a specific feature of the FIP1C carboxyl-terminal segment. To examine the specificity of this finding, we expressed a similar fragment of the C terminus of Rab11-FIP2, FIP2452–512. This construct expresses the RBD of FIP2 and is similar to one (FIP2446–511) previously shown to disrupt transferrin recycling (19). The subcellular distribution of FIP2452–512 remained largely perinuclear in the presence of HIV-1 Env but did not result in Env trapping (Fig. 2F) and failed to diminish cell surface Env (Fig. 3D, compare titration to that of FIP1C560–649 in C). The colocalization of Env and FIP1C560–649 was much higher than that of Env and FIP2452–512, as represented in these images and quantified from multiple image stacks (colocalization measurements are presented in Fig. 3E, F, and G).
FIG 2.
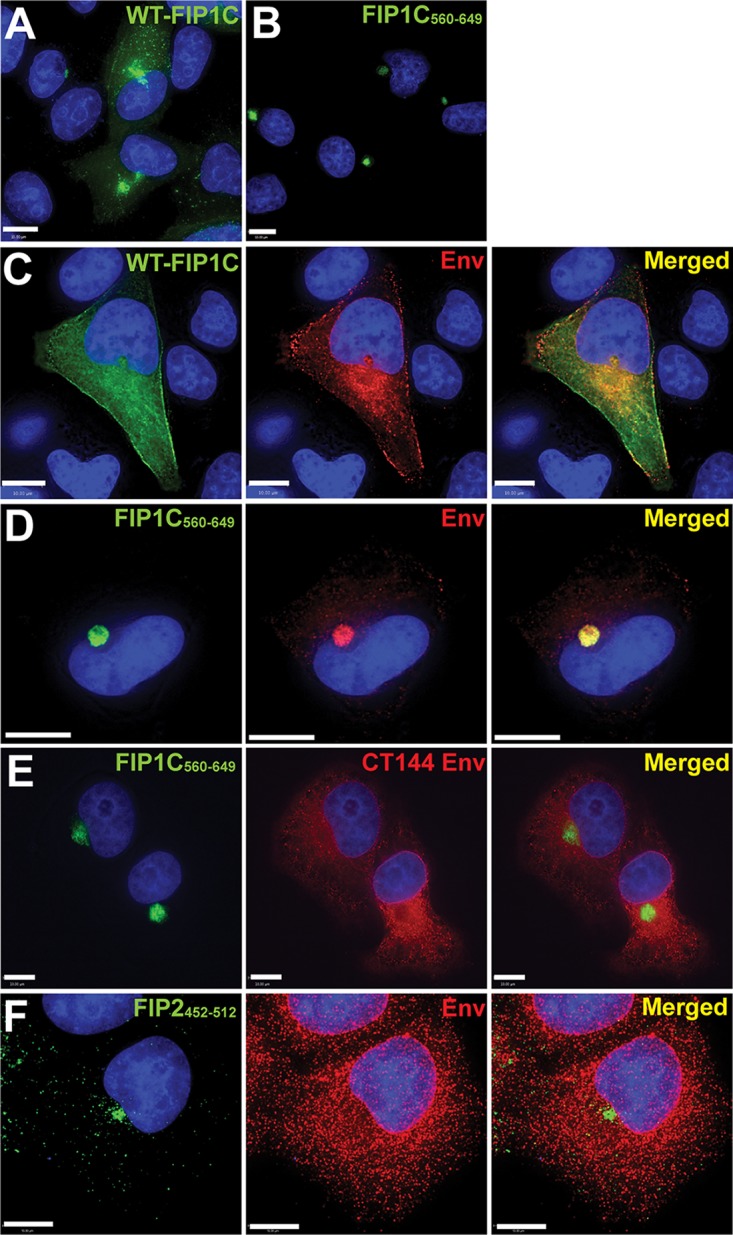
FIP1C560–649 sequesters Env in a perinuclear compartment in a CT-dependent manner. (A) Wild-type GFP-FIP1C subcellular distribution in HeLa cells when expressed alone. (B) Subcellular distribution of GFP-FIP1C560–649. (C) Distribution of wild-type GFP-FIP1C when coexpressed with NL4-3 proviral DNA. Cells were fixed and immunolabeled with human neutralizing antibody 2G12 to stain HIV-1 gp120. Green, GFP-FIP1C; red, Env; rightmost image, overlay. (D) Distribution of FIP1C560–649 when coexpressed with NL4-3 proviral DNA. Staining is as described for panel C. (E) Coexpression of FIP1C560–649 and CT144 proviral plasmids stained and imaged as described for panel C. (F) GFP-FIP2452–512 and pNL4-3 proviral expression stained and imaged as described for panel C. All panels shown are from transfected HeLa cells. Images selected are representative of the major phenotypes found for more than 100 cells examined in repeated, independent experiments. Bars represent 10 μm.
FIG 3.
Cell surface levels of CT144 Env and effect of FIP1C560–649 versus FIP2452–512. (A) HeLa cells were cotransfected with GFP-FIP1C560–649 and pNL4-3 proviral plasmid and stained for Env using anti-gp120 mouse monoclonal antibody BDI123. Histograms of cell surface staining are shown in the absence (red) or presence (other colors as indicated) of FIP1C560-649 expression. (B) CT144 proviral plasmid was substituted for wild-type Env and cell surface staining repeated with and without GFP-FIP1C560–649. (C) Dose titration of GFP-FIP1C560–649 resulted in dose-dependent reduction in cell surface Env signal. (D) Dose titration of GFP-FIP2452–512 had little effect on cell surface Env. (E) The degree of colocalization between GFP-FIPs and HIV-1 Env was measured by Pearson's correlation coefficient equation using Volocity 6.3 software after manual thresholding. Results are shown as means ± SD from a total of 10 representative images. ***, P < 0.001; ****, P < 0.0001. P values were calculated using the Student's unpaired t test using GraphPad Prism 6. (F and G) Correlation coefficients, FIP1C signal/Env (M1), or Env signal/FIP1C (M2) from the images examined for panel E.
Role of tyrosine-based motifs in Env retention in the ERC.
We previously identified a CT mutant, YW795SL, that is deficient in Env particle incorporation in a cell type-specific manner, and proposed that this mutant has a defect in FIP1C-dependent transport to the PM (18). If this is the case, then we would predict that the YW795SL Env escapes retention in the ERC elicited by FIP1C560–649. Indeed, this was the case (Fig. 4A; YW795SL is referred to here as S5). We note that the overlap of Env and FIP1C560–649 signal was partial upon expression of YW795SL Env, but clearly Env was present more abundantly on cytoplasmic membranes and the PM than was seen for WT Env (compare Fig. 4A to 2D). Cell surface Env levels reflected a slight downregulation of YW795SL Env compared to wild-type Env (Fig. 4E, compare to D). We next examined a revertant virus derived from serial passage of YW795SL in which a second-site change (L850S) near the C terminus of the CT restored Env particle incorporation and infectivity (18). The YW795SL/L850S revertant Env was retained in the ERC by FIP1C560–649 in a manner similar to that of the wild type (Fig. 4B; the revertant is referred to as S5R Env). ERC retention was associated with a significant decrease in cell surface levels (Fig. 4F). The membrane-proximal Yxxϕ motif of the CT has been shown to be involved in AP-2-dependent endocytosis and to play a role in particle incorporation (6, 7, 21). We reasoned that AP-2-dependent endocytosis may be required to deliver Env to the ERC. We therefore expressed provirus incorporating the Y712C mutant that abrogates AP-2 interaction together with FIP1C560–649. In this case, we observed no trapping of Env in the ERC (Fig. 4C), and the cell surface levels of Y712C Env were unaffected (Fig. 4G). The extent of Env retention in the ERC was quantified using Pearson's correlation coefficient (Fig. 4H) and the M2 correlation coefficient (representing Env pixels colocalizing with FIP1C pixels) from 10 representative three-dimensional (3D) image stacks for each of the trafficking mutants shown in Fig. 4A to C. The extent of trapping was greatest for YW795SL/L850S revertant Env (S5R) and least for Y712C, as can be seen from either measure (Fig. 4H and I), while differences in the degree of colocalization/trapping between S5 and S5R were more apparent when Env/FIP1C signal is measured by the M2 coefficient representing the red (Env) pixels colocalized with green pixels (Fig. 4I). The absence of Y712C trapping suggested to us that endocytosis of Env from the plasma membrane mediated by AP-2 is required for Env to reach the ERC, where FIP1C560–649 is available to sequester Env.
FIG 4.
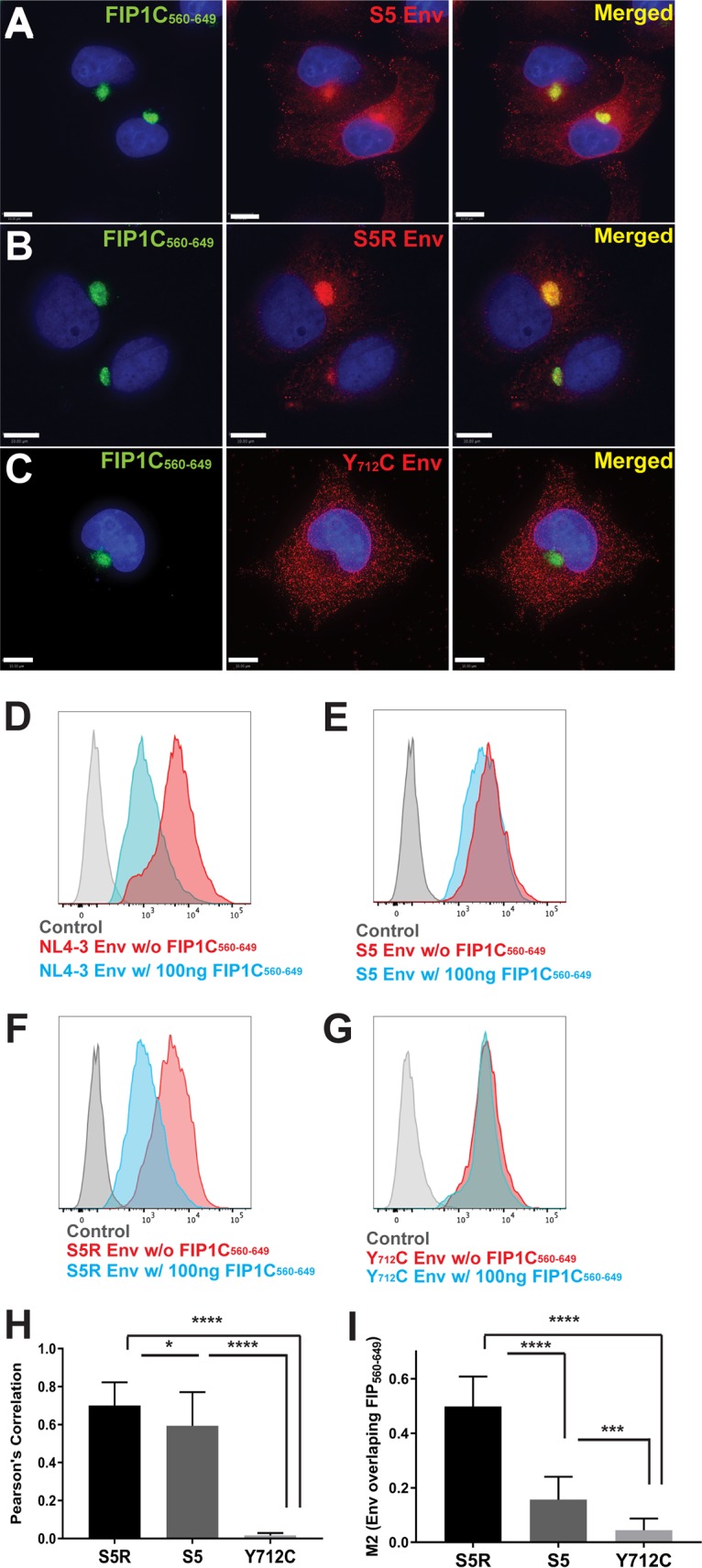
Trapping of Env by GFP-FIP1C560–649 is dependent on specific trafficking motifs in the CT. (A) HeLa cells were cotransfected with GFP-FIP1C560–649 and YW795SL (S5) proviral plasmids. Forty-eight hours posttransfection, cells were fixed and stained with anti-HIV-1 gp120 antibody 2G12 (red). (B) HeLa cells cotransfected with GFP-FIP1C560–649 and YW795SL/L850S (S5R) proviral plasmids and stained for Env as described in panel A. (C) HeLa cells were cotransfected with GFP-FIP1C560–649 and Y712C proviral plasmids and stained for Env as outlined for panel A. All size bars represent 10 μm. (D) Cell surface staining for NL4-3 Env with and without GFP-FIP1C560–649 expression. Cells were fixed and stained for Env using anti-gp120 mouse monoclonal antibody BDI123. (E) S5 designates NL4-3 Env CT mutant YW795SL. Cell surface staining performed for wild-type NL4-3 as described for panel D. (F) S5R is a second-site revertant virus in the Env CT, YW795SL/L850S. Cell surface staining was performed as described for panel D. (G) Cell surface staining of NL4-3 Y712C mutant performed with and without FIP1C560–649, as described for panel D. (H) Pearson's correlation coefficient. (I) M2 correlation coefficient, Env signal/FIP1C signal. Results are shown as means ± SD from a total of 10 representative 3D image stacks. *, P = 0.04; ***, P < 0.001; ****, P < 0.0001.
We next asked if YW795SL Env and Y712C Env incorporation into virion particles was also differentially altered upon FIP1C560–649 expression. WT NL4-3 Env incorporation was inhibited by FIP1C560–649 expression as described above (Fig. 5A, left), while YW795SL Env was resistant to this effect (Fig. 5A, middle blots). The second-site revertant YW795SL/L850S regained sensitivity to inhibition by FIP1C560–649 (Fig. 5A, right). Both CT144 and Y712C Env-bearing proviruses were resistant to the effects of FIP1C560–649 (Fig. 5B). In summary of this series of experiments, mutations disrupting FIP1C-dependent trafficking (YW795SL) or AP-2-dependent endocytosis (Y712C) allowed Env to escape trapping by FIP1C560–649 in the ERC and allowed normal levels of Env incorporation into particles budding from HeLa cells. We note that HeLa cells are permissive for Env incorporation into particles by truncated Env, such as CT144, suggesting that an alternative pathway allowing Env incorporation exists in this epithelial line that is not present in many T cell lines or in primary T cells and macrophages (4, 18). This also is likely to explain the robust incorporation of YW795SL Env into particles (Fig. 5A, middle), whereas this Env is inefficiently incorporated in most T cell lines and in primary T cells and macrophages (18).
FIG 5.
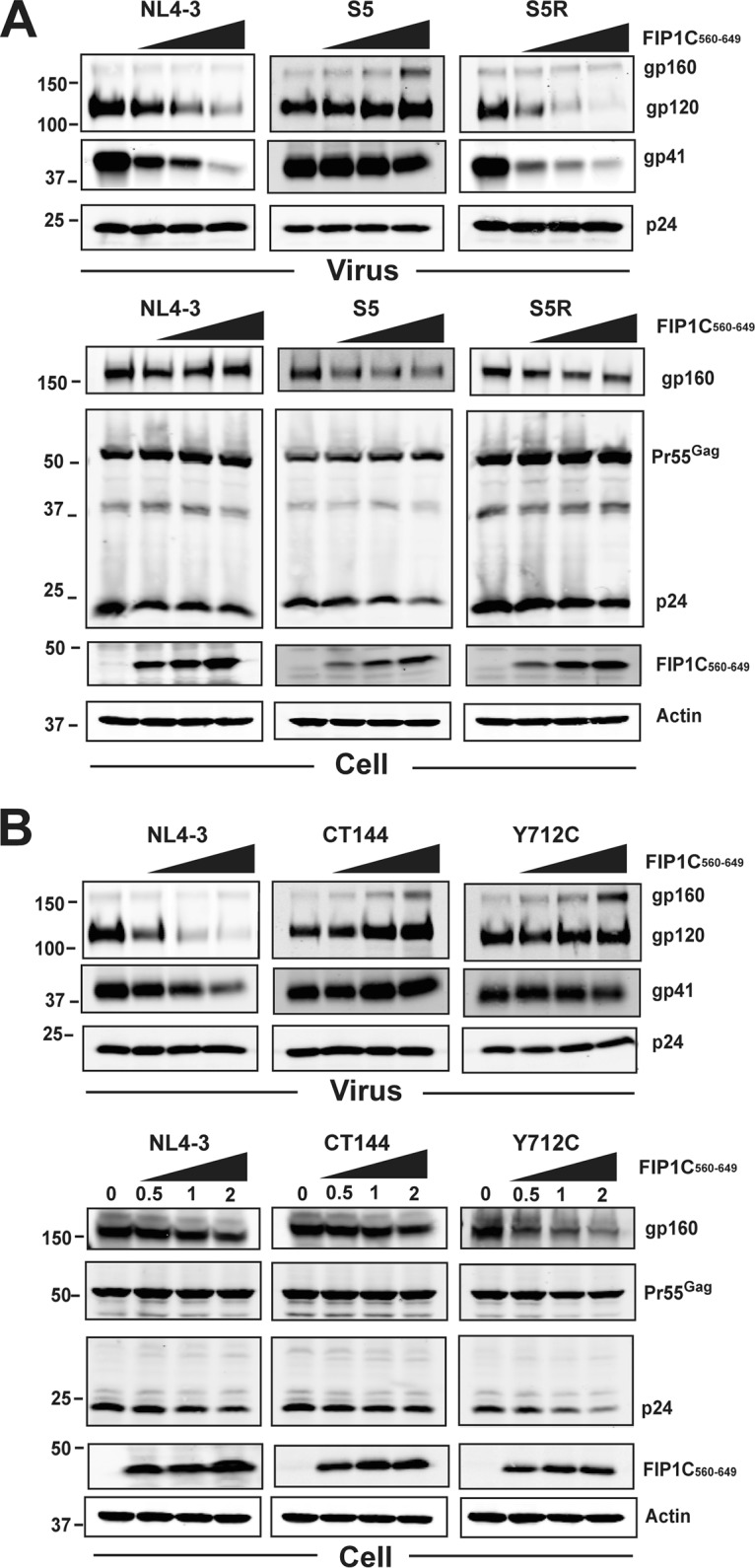
Effects of titration of FIP1C560–649 on Env particle incorporation for Env CT trafficking mutants. (A) FIP1C560–649 was cotransfected with the indicated proviral constructs at increasing amounts of DNA as indicated above the blots. Shown are Western blots for Env and Gag content in particles (top) and cell lysates (bottom). WT NL4-3 is shown as in Fig. 4 and serves as a control for inhibition of Env incorporation into particles. Note that S5 is the Env CT mutant YW795SL, while S5R (revertant) is the second-site revertant in the CT, YW795SL/L850S. (B) Dose titration of FIP1C560–649 as described for panel A, showing effects on CT144 and Y712C Env compared to wild-type NL4-3. Particle blots are shown on the top, and cell lysates are below. Results are from HeLa cells.
SIV Env avoids capture in the ERC by FIP1C560–649.
We next asked if the retention of HIV-1 Env in the ERC by FIP1C560–649 was also seen with SIVmac239 Env. Upon expression of full-length SIVmac239 provirus, we found that there was no significant trapping of Env by FIP1C560–649 (Fig. 6A), while in parallel HIV-1 Env was almost quantitatively trapped as before (Fig. 6B). Cell surface levels matched the phenotype observed by immunofluorescence, as there was no significant shift in surface fluorescence of SIVmac239 Env upon expression of FIP1C560–649 (Fig. 6E). To determine if differences in the Env CT led to this dramatically different phenotype, we employed a codon-optimized SIVmac239 Env and created a chimera bearing the ectodomain and TM of SIVmac239 fused to the CT of HIV-1 Env. SIVmac239 codon-optimized Env was not trapped in the ERC, reproducing results with expression from proviral DNA (Fig. 6C). The SIV-HIV chimera, in sharp contrast, was retained in the ERC upon FIP1C560–649 expression (Fig. 6D). Cell surface levels of SIVmac239 Env were unaffected by expression of FIP1C560–649, while the surface distribution of the SIV-HIV CT chimera was shifted significantly (compare Fig. 6G to H). Taking this further, we performed Western blots to examine SIV Env incorporation into particles (Fig. 7). FIP1C560–649 expression had no effect on SIV Env incorporation into particles, while in parallel experiments NL4-3 Env was efficiently depleted (Fig. 7A). Expression of codon-optimized HIV Env and Gag together with FIP1C560–649 led to depletion of Env from particles (Fig. 7B, left), while SIV Env incorporation onto SIV Gag particles was unaffected (Fig. 7B, middle). Strikingly, replacing the CT of SIV with that of HIV-1 resulted in inhibition of Env incorporation into particles that was FIP1C560–649 dependent (Fig. 7B, right). Thus, the lack of trapping of SIVmac239 Env by FIP1C560–649 is reflected in normal cell surface levels and particle incorporation, while the SIV-HIV chimeric Env was trapped in the ERC, unable to reach the particle assembly site on the PM and unavailable for efficient incorporation into particles.
FIG 6.
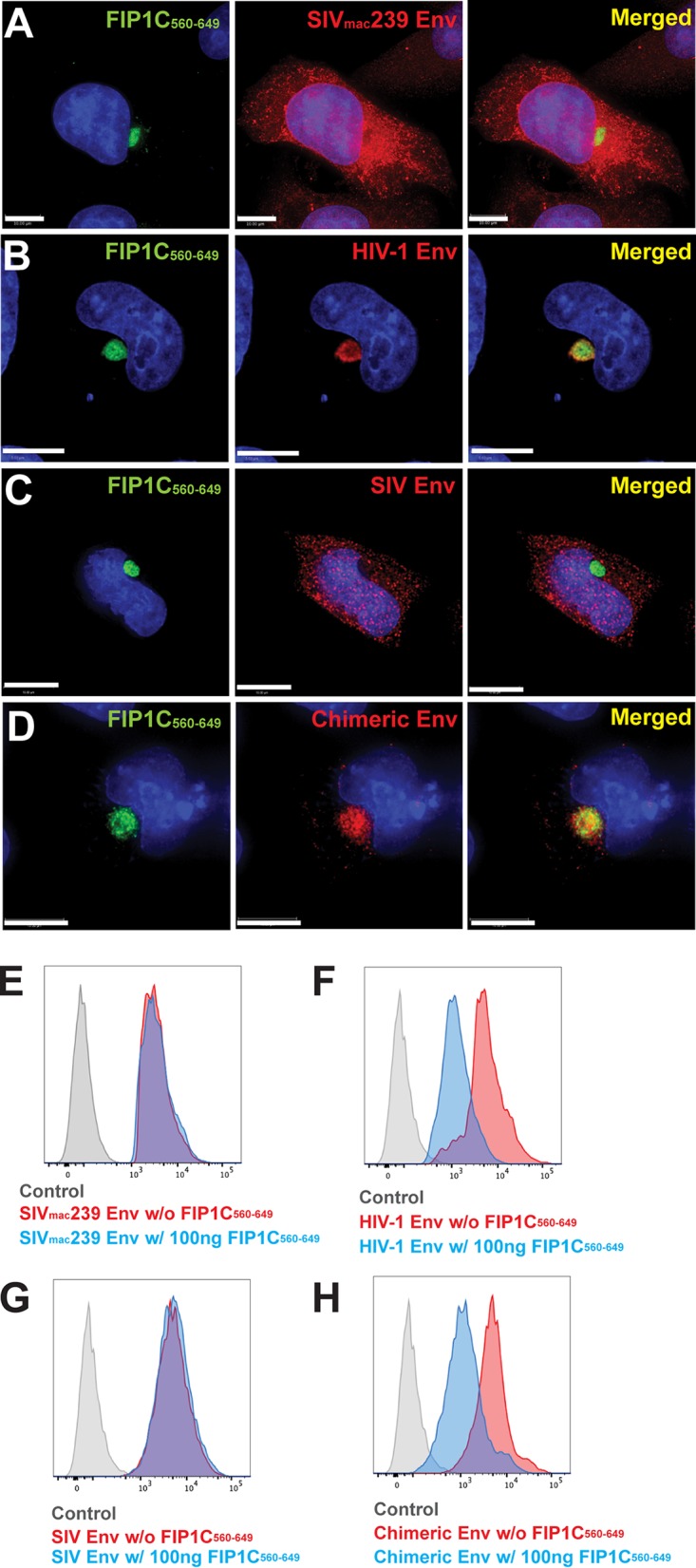
Dominant-negative FIP1C560–649 fails to trap SIV Env in the ERC. (A) HeLa cells were cotransfected with GFP-FIP1C560–649 and SIVmac239 proviral plasmids. After 48 h, cells were fixed and stained with anti-SIV gp120 rabbit serum (red). (B) Codon-optimized JR-FL Env was coexpressed with GFP-FIP1C560–649, followed by fixation and staining with 2G12 antibody (red). (C) Codon-optimized SIVmac239 Env coexpressed with GFP-FIP1C560–649 and stained with anti-SIV gp120 rabbit serum (red). (D) SIV-HIV tail chimeric Env coexpressed with GFP-FIP1C560–649 and stained with anti-SIV gp120 rabbit serum. Scale bars, 10 μm (A, C, and D) and 5 μm (B). (E) Cell surface staining of SIVmac239 Env with and without GFP-FIP1C560–649 using anti-SIV gp120 rabbit sera. (F) Surface staining of JR-FL Env expressed from codon-optimized construct with and without coexpression of GFP-FIP1C560–649. (G) Codon-optimized SIVmac239 Env cell surface staining with and without expression of GFP-FIP1C560–649. (H) Cell surface Env signal of SIV-HIV chimeric Env, stained with anti-SIV gp120 rabbit sera with and without GFP-FIP1C560–649 expression.
FIG 7.
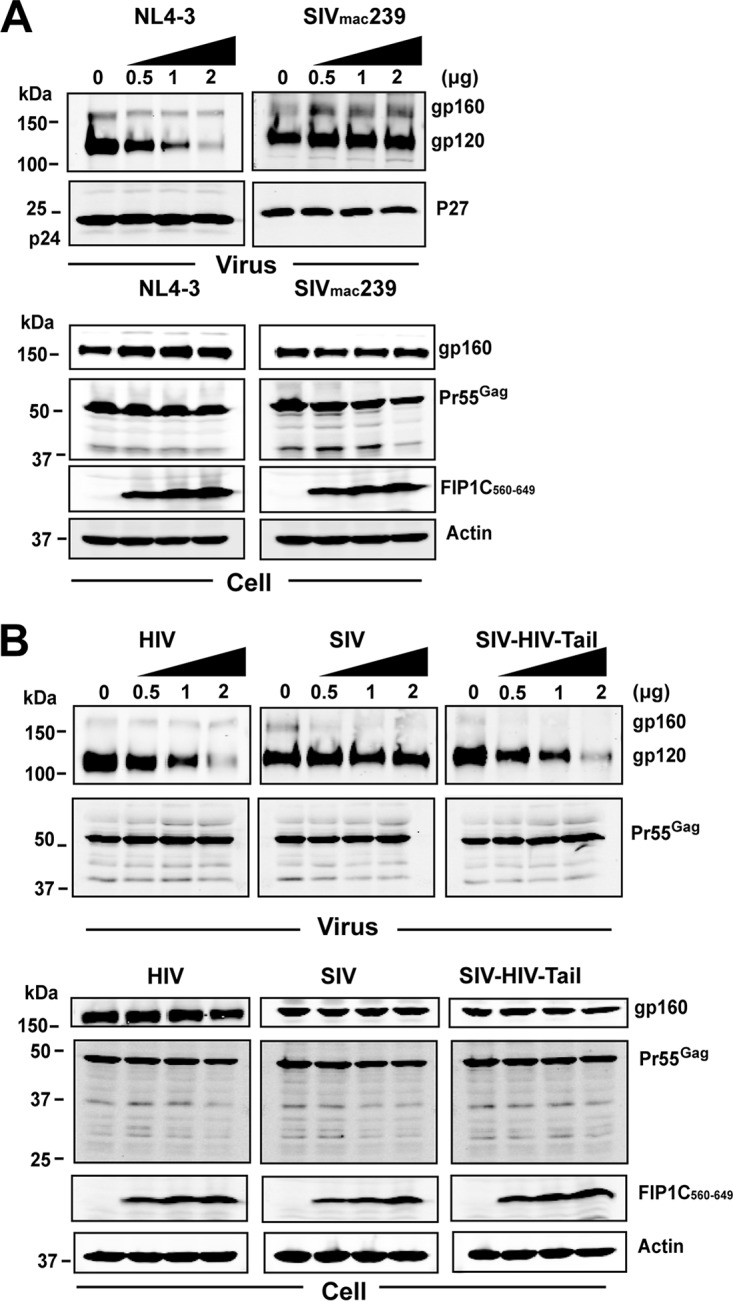
Dose titration of FIP1C560–649 and effects on SIVmac239 Env incorporation. (A) Comparison of effects of titration of FIP1C560–649 on Env incorporation by NL4-3 versus SIVmac239 in HeLa cells. In this experiment, full-length SIVmac239 proviral DNA was cotransfected with increasing amounts of FIP1C560–649. (B) CT-dependent reduction in Env incorporation illustrated by SIV-HIV Env chimera. (Left) Codon-optimized HIV-1 (JR-FL) Env was coexpressed with codon-optimized HIV Gag and increasing amounts of FIP1C560–649 DNA. (Middle) Codon-optimized SIVmac239 Env coexpressed with optimized SIVmac239 Gag and increasing amounts of FIP1C560–649. (Right) Effects of FIP1C560–649 expression on particle incorporation of a chimeric Env where the ectodomain and transmembrane domain of SIVmac239 Env was fused to the CT of HIV-1 Env. In this experiment, the Gag protein expressed was SIVmac239 Gag.
FIP1C560–649 induces an aberrant ERC and traps Env on ERC membranes.
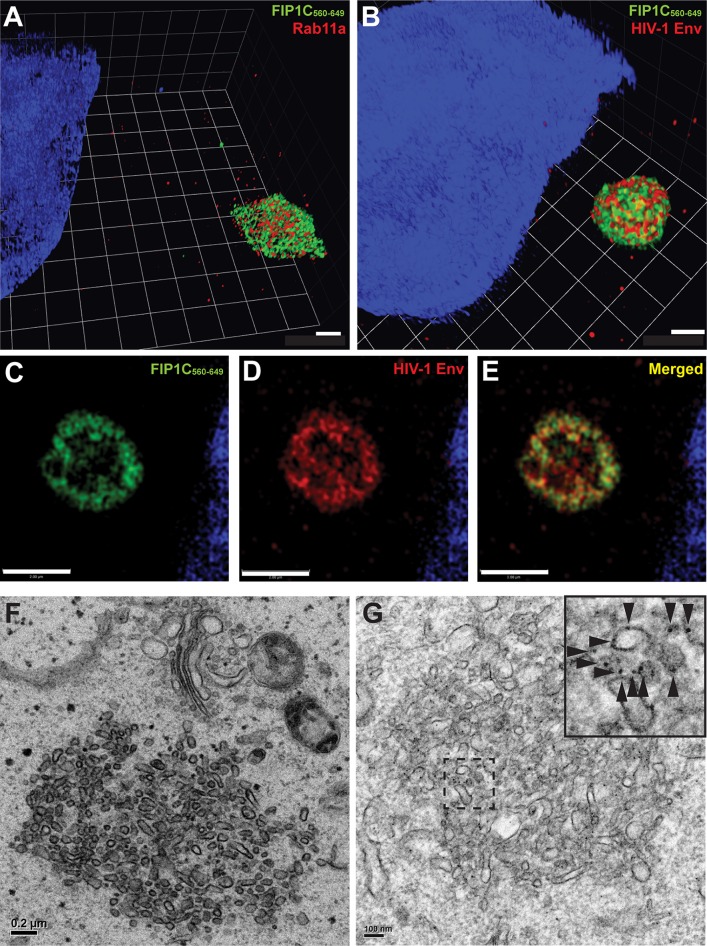
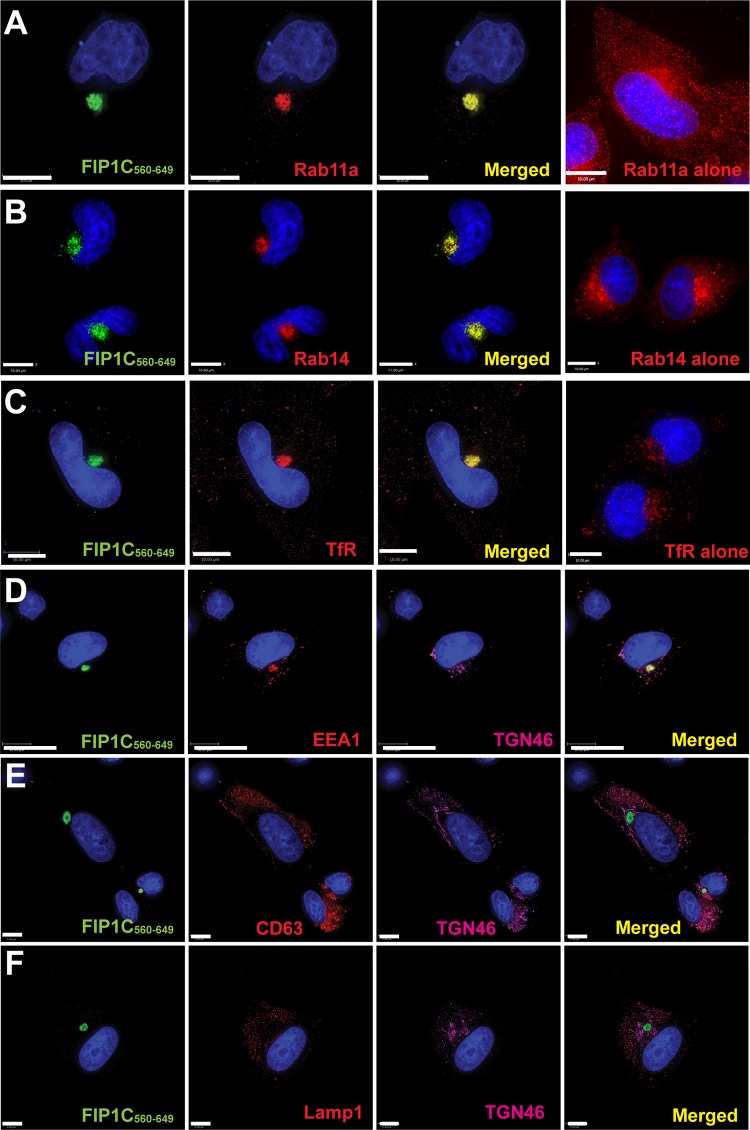
We next focused on the structure of the intracellular compartment responsible for Env trapping using superresolution optical microscopy and electron microscopy (EM). While at lower resolution the ERC appeared as a roughly spherical compartment with a diffuse distribution of FIP1C560–649, upon higher resolution it appeared to consist of intertwined and packed tubules. Rab11 and FIP1C were not uniformly distributed on all tubules but demonstrated significant colocalization (Fig. 8A). Env was similarly concentrated more strongly on a subset of tubules (Fig. 8B). The center of the ERC appeared somewhat hollow compared to the periphery (Fig. 8C to E). We extended this morphological study to the use of transmission electron microscopy (TEM). TEM images revealed compact clusters of tubules with an average diameter (or radius for those visualized on end) of 60 ± 10 nm (Fig. 8F). Control HeLa cells examined in parallel failed to reveal similar structures (not shown), indicating to us that this was an aberrant ERC similar to that described by Damiani and colleagues (20). Using immuno-EM, we found that Env was distributed along the ERC membranes (Fig. 8G). We conclude that FIP1C560–649 elicits a condensation of ERC tubules, and that Env becomes trapped on tubular membranes by this intervention and is subsequently unable to reach the particle assembly site on the plasma membrane. The identity of this structure as the ERC was further validated by colocalization of FIP1C560–649 with Rab14 and transferrin receptor and by the lack of colocalization of FIP1C560–649 with late endosomal markers (Fig. 9).
FIG 8.
GFP-FIP1C560–649 creates an aberrant ERC that traps Env on endosomal membranes. (A) Structured illumination microscopy of aberrant ERC. HeLa cells transfected with GFP-FIP1C560–649 and stained for Rab11a. Shown is a 3D projection of the aberrant ERC where both Rab11a and GFP-FIP1C concentrate. Blue color represents nuclear stain. Scale bar, 1 μm. (B) Structured illumination microscopy for morphology of ERC with HIV-1 Env. HeLa cells were cotransfected with GFP-FIP1C560–649 and pNL4-3 proviral DNA and stained for HIV-1 Env. Env is shown in red, with FIP1C in green. Scale bar, 1 μm. (C to E) A section of ERC from the experiment depicted in panel B is shown to illustrate the less densely staining inner core. (F) Transmission electron microscopy image of the abnormal ERC induced by GFP-FIP1C560–649. Scale bar, 200 nm. (G) Postembedding immunoelectron microscopy using a cocktail of anti-Env monoclonal antibodies 2G12, b12, and 447-52D, with detection using 10-nm protein A-gold beads. The hatched area in the image is shown in magnified form at the upper right corner, and arrowheads point out some of the individual gold beads. Scale bar, 100 nm.
FIG 9.
Endosomal markers present in FIP1C560–649-positive ERC. (A) To examine the enrichment of ERC markers in the FIP1C560–649 compartment, we utilized immunofluorescence microscopy for a series of endosomal markers. All images shown here are from HeLa cells. Rab11a is shown concentrated in the FIP1C560–649 compartment (left 3 images) compared with its normal distribution (rightmost image). (B) Rab14-mcherry (red) was coexpressed with GFP-FIP1C560–649. The distribution of Rab14-mcherry alone is shown in the rightmost panel. (C) The distribution of endogenous transferrin receptor is shown together with the FIP1C560–649-positive compartment compared to its normal distribution (rightmost image). (D) EEA1 distribution following expression of FIP1C560–649, together with staining for TGN46. (E) CD63 distribution in the presence of FIP1C560–649. TGN46 staining also is indicated. (F) LAMP1 and TGN46 distribution in the presence of FIP1C560–649.
Env CT-dependent internalization and capture within the aberrant ERC.
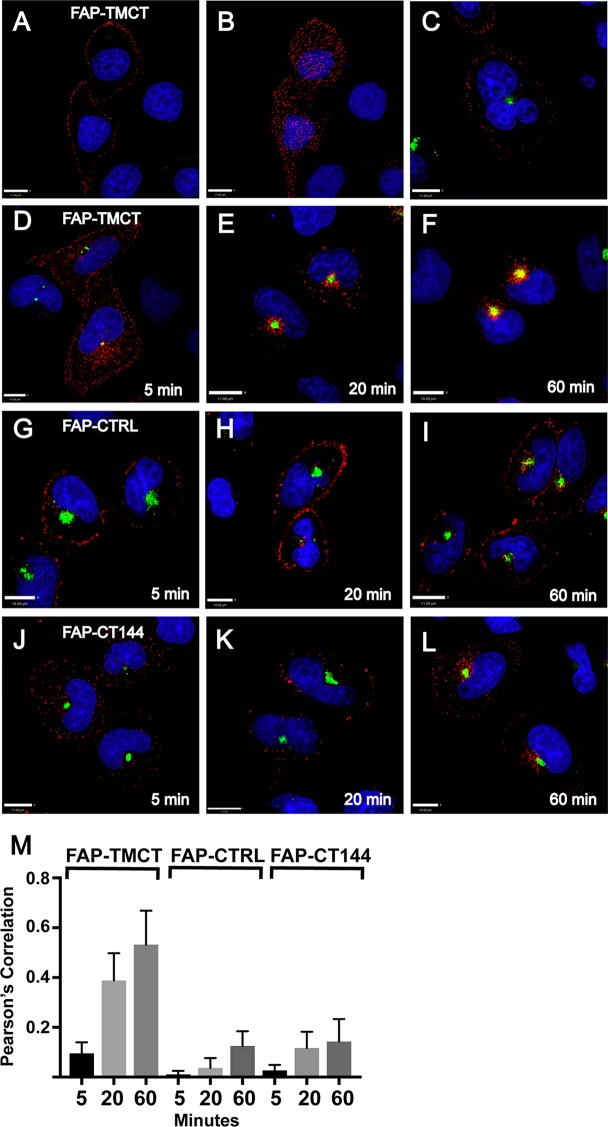
Having shown that Env is trapped in the ERC in a CT-dependent manner, we next asked if Env present on the cell surface can reach this compartment. To facilitate pulse labeling of cell surface Env, we created an artificial envelope construct composed of an N-terminal fluorogen-activating protein (FAP) domain fused to the TM and CT regions of HIV-1 Env (FAP-TMCT). This allowed fluorescent pulse labeling of FAP-TMCT on the cell surface in the absence of internal labeling by using the membrane-impermeant fluorogen MG-11p (22, 23). FAP-TMCT demonstrated PM labeling when cells were exposed to fluorogen on ice (Fig. 10A, z section in center of cell, and B, at coverslip level). Expression of FIP1C560–649 did not alter the cell surface labeling pattern of FAP-TMCT (Fig. 10C). At 37°C, however, FAP-TMCT was rapidly internalized from the cell surface. Internalized signal was apparent at the end of a 5-min pulse (Fig. 10D). FAP-TMCT signal was further internalized for 20 min and colocalized significantly with FIP1C560–649 (Fig. 10E, with quantitation in M). By 60 min, colocalization of pulsed FAP-TMCT with FIP1C560–649 was further enhanced (Fig. 10F, with quantitation in M). As controls, we employed a well-characterized membrane-anchored FAP construct that includes the platelet-derived growth factor (PDGF) receptor transmembrane domain and lacks any HIV sequence (23) and a modified version of FAP-TMCT in which the C-terminal 144 amino acids of the CT are absent (FAP-CT144). The FAP membrane control remained largely, but not entirely, on the cell surface over this time course (Fig. 10G to I). A portion of the surface-labeled FAP-CT144 was rapidly internalized (Fig. 10J) but did not appear to become significantly concentrated in the ERC over time (Fig. 10J to L). Signal from the internalized fraction of both control FAP proteins at 60 min colocalized only weakly with FIP1C560–649 compared to that with FAP-TMCT (Fig. 10M). These results support a model in which the Env CT mediates internalization from the PM to the ERC, leading to CT-dependent sequestration in the aberrant ERC formed by FIP1C560–649 expression.
FIG 10.
Endocytosis and trapping of artificial Env in ERC. FAP-TMCT is an artificial envelope bearing the FAPalpha2 module at the N terminus fused to the TM and CT of HIV-1 Env. (A) FAP-TMCT labeled following a 10-min pulse with MG-11p on ice, showing a z section at approximately the midpoint of the cell. (B) Same cells as those described for panel A, showing labeling at the PM (coverslip level). Nuclei are superimposed for reference. (C) PM labeling on ice in HeLa cells expressing GFP-FIP1C560–649. (D) HeLa cell expressing GFP-FIP1C560–649 and pulse labeled with MG-11p for 5 min, shown immediately at the end of the pulse. (E) FAP-TMCT colocalization with GFP-FIP1C560–649 after a 5-min pulse followed by a 20-min chase period. (F) FAP-TMCT colocalization with GFP-FIP1C560–649 following a 5-min pulse and 60-min chase period. (G to I) Control membrane protein produced from pMFAP-alpha2 vector using methods described for panels D and E. (J to L) FAP-TMCT144 time course with time postlabeling indicated. Scale bars, 11 μm. (M) Pearson's correlation was derived from image stacks representing 10 fields of cells from each time point. Error bars indicate standard deviations.
DISCUSSION
HIV-1 Env and Gag must coordinately reach the plasma membrane during the process of particle assembly. How they do so remains unclear. Env is translated as the precursor gp160 protein on ER-bound ribosomes and subsequently forms trimers in the endoplasmic reticulum (ER) (24–28). During transit through the trans-Golgi network (TGN), a furin-like protease cleaves gp160 into SU and TM subunits (29, 30). The form of Env that reaches the surface of the cell is therefore a trimer of SU/TM heterodimers, presumably completely modified and competent for incorporation into virions. Gag is synthesized on cytosolic ribosomes and reaches the inner face of the plasma membrane by an alternate and poorly understood route, where it interacts with phosphatitdylinositol-4,5-bisphosphate (PIP2) and cholesterol-enriched membranes (31–33). The matrix (MA) region of Gag is essential for Env incorporation, as point mutations in MA have been identified that eliminate Env incorporation without disrupting particle assembly and budding (34–36). A direct interaction between Gag and the Env CT has been reported for HIV (37) and for SIV (38), but there has been little additional evidence supporting this. Env exclusion by MA mutants may instead be regulated by a steric clash between the MA lattice and the Env CT created by MA mutants that disrupt the MA trimer interface (39). In support of this model, it was recently shown that MA trimers exist in virions, and that mutations that eliminate trimerization correlated with a loss of Env incorporation (40). This work suggests that the long Env CT must fit within a central aperture within a hexamer of trimers formed by MA at the particle budding site.
We previously reported that the Rab-related adaptor protein FIP1C directs Env incorporation in a CT-dependent manner (13). The Rab protein recruited by FIP1C relevant for Env incorporation was shown to be Rab14. This suggested to us that Env must be delivered to and sorted from the ERC prior to reaching the site of particle assembly. This model is further supported by the finding that a YW795SL mutation in the CT disrupted Env incorporation in a cell type-specific manner identical to that of the CT144 Env lacking most of the cytoplasmic tail (18). A second-site mutant near the C terminus of the CT restored FIP1C-dependent incorporation of Env in Jurkat and H9 T cell lines and in macrophages. The FIP1C-dependent trafficking model of Env incorporation proposes that Env must traffic through the ERC prior to particle incorporation, and that interactions between the CT and the FIP1C/Rab14 complex direct outward sorting of Env to the particle assembly site. This model does not conflict with the steric model of Env exclusion by MA mutants discussed above, but rather helps to incorporate the role of specific trafficking signals in the CT in directing Env to the assembly site.
Here, we employed a dominant-negative approach to further define Env trafficking and the proposed requirement for ERC trafficking. A short C-terminal fragment of FIP1C (FIP1C560–649) led to a dramatic trapping of Env in the modified ERC and inhibited incorporation of Env into particles. The YW795SL CT mutant largely escaped trapping in the ERC, while a second-site revertant in the distal CT restored the inhibitory effect of FIP1C560–649. Thus, the dominant-negative ERC trapping effect was specific to residues within the Env CT. Furthermore, a similar fragment derived from Rab11-FIP2 failed to inhibit Env incorporation. These results support the ERC trafficking model for Env incorporation and a specific requirement for FIP1C in outward sorting of Env.
The overexpression of a similar, slightly larger C-terminal fragment of FIP1C, termed H13, was reported previously to create an altered morphology of the ERC in RAW macrophages and to disrupt transferrin recycling to the PM in this cell type (20). A C2 domain deletion of FIP1C similarly caused an accumulation of phosphatidylserine-containing membranes in the pericentriolar region of HeLa cells (41). The structures we observed by fluorescence microscopy and by EM in HeLa cells are likely analogous to the altered tubulovesicular endosomes seen in these two prior studies. Both H13 and FIP1C560–649 express the FIP1C Rab binding domain, or RBD, which binds to Rab11 GTPase family members (Rab11a, Rab11b, and Rab25/Rab11c) and Rab14 (42). One potential mechanism for the dominant-negative inhibition seen upon RBD fragment overexpression is the sequestration of Rab GTPases required for outward sorting to the plasma membrane. In support of this, Rab11a and Rab14 were found in the current study to be sequestered in the aberrant ERC formed by FIP1C560–649 (Fig. 8 and 9). Because Rab14 was previously shown to be required for Env incorporation (13), it seems likely that Rab14 sequestration is involved in Env trapping by FIP1C560–649. Immuno-EM studies showed Env associated with tubular ERC membranes upon FIP1C560–649 expression. A logical interpretation of these data is that the Env CT and the Rab14-FIP1C complex normally interact on these membranes prior to outward sorting to the particle assembly site on the PM, and that sequestration of the relevant Rab GTPase-FIP1C complex leads to cosequestration of Env in this compartment. It is also possible that the complex of Env with truncated FIP1C and Rab14 is unable to recruit the motor protein required for transit out of the ERC to the plasma membrane site of assembly. Further experiments are under way to evaluate the model outlined here, including efforts to identify the relevant motor protein involved in Env transit out of the ERC and to define potential protein-protein interactions between FIP1C and the Env CT.
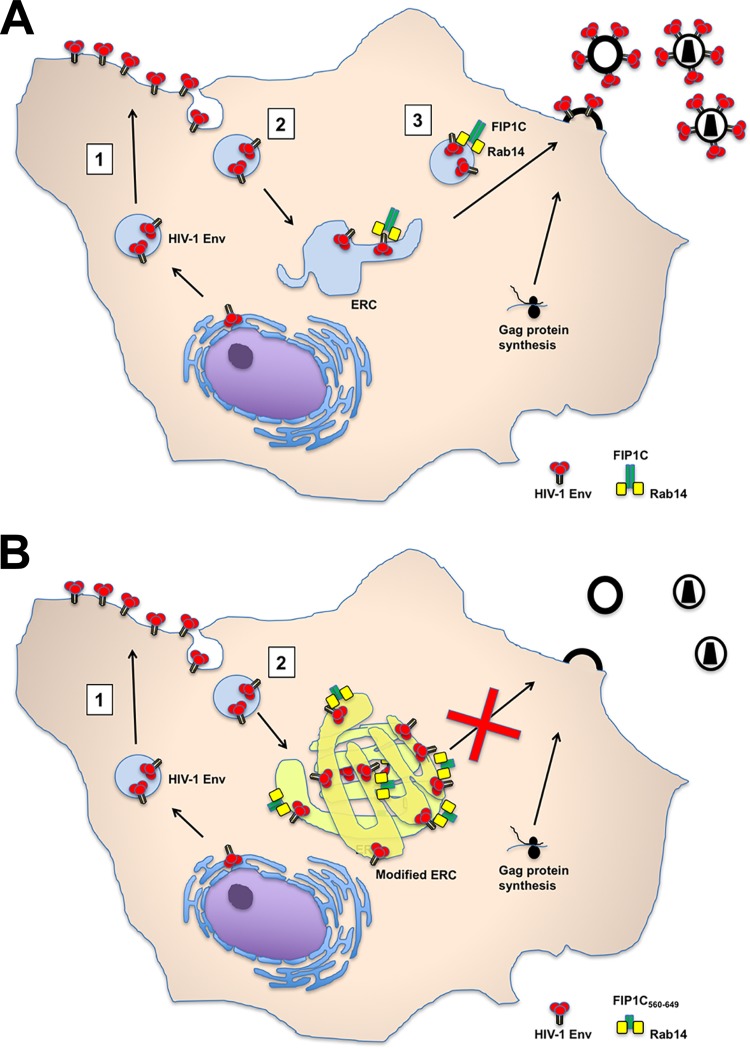
We have previously proposed that Env trimers on the PM must be endocytosed to the ERC in a step preceding outward sorting to the particle assembly site (depicted in Fig. 11). Env is known to be actively endocytosed by an AP-2/clathrin-dependent process that depends upon the Y712XXϕ motif in the proximal portion of the CT (21), and this motif plays a key role in particle infectivity (8). Remarkably, we found that a Y712C Env mutant was not trapped by FIP1C560–649, consistent with a sequential trafficking model where AP-2-dependent endocytosis to the ERC is followed by FIP1C/Rab14-directed sorting to the particle assembly site on the PM. A second plausible model, however, would not require the appearance of Env first on the PM prior to directed sorting. Notably, the Y712XXϕ motif on the Env CT also interacts with the TGN- and endosome-associated AP-1 clathrin adaptor, and AP-1 may direct post-Golgi trafficking of Env through additional CT motifs, such as the C-terminal dileucine motif (11). In this scenario, AP-1-dependent trafficking could direct Env from the TGN to the sorting endosome/ERC, followed by FIP1C/Rab14-dependent outward sorting. Either scenario would be consistent with the fact that dominant-negative FIP1C560–649 traps Env prior to outward sorting for particle incorporation. In support of the plausibility of the endocytosis model, we found that an FAP-tagged artificial Env incorporating the HIV-1 Env TM and CT domains was rapidly endocytosed and reached the aberrant ERC created by FIP1C560–649 (Fig. 10). We do not suggest that this result rules out the existence of the TGN-to-ERC pathway, but it does provide supportive evidence that Env on the PM can be endocytosed in a CT-dependent manner and subsequently reach the ERC.
FIG 11.
Model for Env trafficking and retention in ERC by truncated FIP1C. (A) Model of Env trafficking through the ERC. In this scheme, step 1 represents trafficking through the secretory pathway to the PM. Step 2 represents clathrin- and AP-2-dependent endocytosis to the ERC. Step 3 is dependent upon FIP1C and Rab14, in which Gag and Env come together at the PM assembly site. (B) Altered Env trafficking by FIP1C560–649. In this model, steps 1 and 2 remain intact but Env is retained on ERC membranes, while step 3 is absent, resulting in particles lacking Env.
We note the important limitation of performing these experiments in HeLa cells alone. As initially described by the Freed laboratory and also seen in our studies describing FIP1C's role, T cell lines such as Jurkat and H9, as well as primary T cells and macrophages, are nonpermissive for Env incorporation in the absence of the full-length CT (4, 13, 18). We expect that capture of Env with an intact CT would be retained in T cells and macrophages, while the permissive pathway allowing truncated Env to be incorporated would be absent, as in previous studies. While knockdown of FIP1C or Rab14 supports this conclusion (13, 18), unfortunately we have not been able to achieve sustained and efficient expression of FIP1C560–649 in T cell lines and primary cells in order to directly reproduce the Env incorporation defects demonstrated here for FIP1C560–649 in HeLa cells. We also recognize the potential for unintended effects upon overexpression of a protein that acts in a dominant-negative fashion. We employed overexpression of a similarly truncated FIP2 protein as a control for this but recognize that there may still be unintended effects from overexpression of truncated FIP1C. Confidence in the specificity of our findings was enhanced, however, by the lack of robust ERC trapping of the HIV-1 Env CT mutant YW795SL by FIP1C560–649 and by the HIV CT-specific sequestration of HIV/SIV chimeric Env.
How does the regulation of Env incorporation by MA fit with the sequential trafficking model proposed here? In a convergent trafficking model, the delivery of Env by FIP1C/Rab14 to the site of a developing Gag lattice would be consistent with the trimerization/steric interference model proposed by Tedbury and colleagues (40). Delivery to the common PM site would be followed in the case of MA mutants such as 29/31 KE by steric exclusion of the Env CT and lack of incorporation. Another exciting possibility is that the Gag-membrane interactions originate on tubular ERC membranes enriched in PI(4,5)P2, where Env interacts with the FIP1C/Rab14 complex, and that cotrafficking of Gag and Env on vesicles to a common PM site contributes to incorporation of Env on particles. We note that cotrafficking cannot be obligatory for Gag, however, as disruption of Env trafficking by ERC sequestration does not alter the amount of Gag particles produced. The recognition that ERC trafficking is an essential step in the Env particle incorporation pathway will facilitate future studies to define how Gag and Env reach a common assembly site on the PM.
SIVmac239 Env in this study did not appear to utilize ERC transit for incorporation into virion particles, and this was shown to be dependent on the SIV Env CT. It is possible that FIP1C-dependent outward sorting from the ERC is unique to HIV-1, or that species-specific differences in host trafficking factors exist that do not allow interactions of the SIV CT with the human FIP1C/Rab14-mediated sorting pathway. It is interesting that SIV isolates propagated in human cells frequently truncate their cytoplasmic tails (43, 44), suggesting that host cell differences limit the ability of SIV with a full-length CT to propagate in human cells. Interactions with trafficking pathways, including AP-2/clathrin-mediated endocytosis, have been implicated in determining SIV Env incorporation and infectivity (45–48). Still, the reasons that SIV CT truncations provide an advantage in human but a disadvantage in simian cells remain poorly defined. Comparisons of FIP1C-dependent trafficking pathways between human and simian cells may prove informative in understanding these differences.
The inhibition of HIV Env incorporation and particle infectivity using a dominant-negative Env trapping strategy was specific to FIP1C and the HIV-1 Env CT. There are currently no antiretroviral drugs targeting Env incorporation. Future studies establishing how the Env CT interacts with components of the FIP1C/Rab14 trafficking complex thus are likely to prove informative in understanding fundamental aspects of HIV assembly processes and should provide novel targets for antiretroviral drug development.
MATERIALS AND METHODS
Cells and plasmids.
HeLa cells were obtained from the American Type Culture Collection (ATCC; CRM-CCL-2). TZM-bl cells were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH, from John C. Kappes, Xiaoyun Wu, and Tranzyme, Inc. HeLa and TZM-bl cells were maintained in DMEM (Dulbecco's modified Eagle medium) containing 10% fetal bovine serum (FBS) and 2 mM penicillin-streptomycin. pNL4-3 proviral plasmid was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH, from Malcolm Martin. pNL4-3CTdel-144-2 Env was kindly provided by Eric Freed at NCI Frederick and is referred to as CT144 in the text. HIV-1 gp41 mutants Y712C, S5, and S5R in the pNL4-3 backbone have been described previously (12, 18). GFP-FIP1C and GFP-FIP1C560–649 have been described previously (18, 49). GFP-FIP2452–512 was generated by PCR amplification using forward primer 5′-GCAGAATTCCACCTATGAAGAGGTTCTACAGGAGCTG-3′ and reverse primer 5′-GCAGGATCCTTAACTGTTAGAGAATTTGC-3′, followed by cloning into the EcoRI and BamHI sites of the pEGFP-C1 vector (Clontech). Codon-optimized JR-FL gp160 and SIVmac239 gp160 genes were synthesized by GenScript and cloned between EcoRI and EcoRV sites of pcDNA 5/TO (Thermo Fisher Scientific). SIV-HIV chimeric Env was created by overlap extension PCR using inner primers CCATTATACTTTCAGCAGACCTCTTTTCAGACTCTGCTGCCAGCACCTAGG (forward) and TGCTGGCAGCAGATGCTGAAAAGAGGTCTGCTGAAACTAGGATGGAGGGCT (reverse) and outer primers ACGAATTCGCCACCATGGGCTGCCTGGGAAATCGA (forward) and TGGATATCTTACAGCAGTGCCCGTTCCAGCCC (reverse) and cloned between EcoRI and EcoRV sites of pcDNA 5/TO. pVRC-3900 expressing codon-optimized HIV-1 Pr55Gag polyprotein (50) was kindly provided by Gary Nabel (VRC, NIH). Fluorogen-activated protein expression constructs were obtained from Spectragenetics (Pittsburgh, PA). Plasmid pMFAP-alpha2 (Spectragenetics) was used as a control and was employed to create pMFAP-TMCT, which placed the TM and CT domains from codon-optimized JR-FL Env (synthesized by GenScript) in frame with the N-terminal alpha2 FAP domain using Sal1 and Not1 sites of pMFAP-alpha2. Oligonucleotides for amplification of the HIV-1 Env TMCT were AGAGTCGACTTCATCATGATTGTGGGAG (Sal1 site, forward oligonucleotide) and CTTCGCGGCCGCTTACAGCAGTGCCCGTT (Not1 site, reverse oligonucleotide). The same plasmid backbone and PCR cloning strategy was used to create the CT truncation construct pMFAP-CT144, using the reverse oligonucleotide CTTCGCGGCCGCTTAATAGCCCTGCCGGACTCG.
Virus release and infectivity assays.
GFP-FIP1C560–649 or GFP-FIP2452–512 and pNL4-3 were cotransfected into HeLa cells using Lipofectamine 2000 (Thermo Fisher Scientific). Cells and supernatants were harvested 48 h posttransfection for analysis by Western blotting, p24 antigen quantitation, and flow cytometry. Infectivity of cell culture supernatants was measured using TZM-bl indicator cells following p24 normalization as previously described (13). Viruses were pelleted from cell culture supernatants through a 20% sucrose cushion at 20,000 × g for 2 h at 4°C; cells were harvested and pelleted at 800 × g. Virus pellets and HeLa cell lysates were dissolved in radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitors. Virus pellets and cell lysates were separated on 8 to 12% polyacrylamide gels and subjected to Western blotting using antibodies outlined below.
Antibodies for Western blotting and immunostaining.
Goat polyclonal antibody AHP2204, from AbD Serotec (D7324), was used for detecting HIV-1 gp120 and gp160 for Western blotting. Murine monoclonal 5009 from BTI Research Reagents was used for blotting HIV-1 gp41. Mouse anti-p24 monoclonal CA-183 (provided by Bruce Chesebro and Kathy Wehrly through the NIH AIDS Research and Reference Reagent Program) was used for enzyme-linked immunosorbent assay (ELISA) to measure HIV-1 Gag in virus-containing culture supernatant and virus pellets; the capture ELISA was performed as previously described (51). Rabbit antiserum to SIV gp120 was obtained from Advanced Biosciences Laboratories. Anti-SIVmac251 p17 monoclonal KK59, from Karen Kent and Caroline Powell, was obtained through the NIH AIDS Reagent Program and was used to detect SIV Gag in Western blotting. IRDye goat anti-mouse, IRDye rabbit anti-goat, and IRDye goat anti-rabbit secondary antibodies from LiCor Biosciences were used for Western blotting. All blots were acquired and analyzed using the LiCor Odyssey infrared detection system. The following antibodies were utilized in immunofluorescence assays: human anti-gp120 antibody IgG1 2G12 was kindly provided by James Crowe (Vanderbilt University), and b12 and 447-52D were obtained from the NIH AIDS Reagent Program. Anti-Rab11a monoclonal antibody (clone 47) was from Millipore. Monoclonal antibodies detecting EEA1 (clone 14/EEA1), CD63 (clone H5C6), and Lamp1 (CD107a, clone H4A3) were obtained from BD Pharmingen. Anti-transferrin receptor monoclonal antibody (MEM-75) was from Abcam. Anti-TGN46 sheep polyclonal antiserum (AHP500) was purchased from Bio-Rad. Alexa Fluor secondary antibodies and 4′,6-diamidino-2-phenylindole (DAPI) were obtained from Molecular Probes.
Fluorescence microscopy.
HeLa cells were plated in MatTek 35-mm poly–d-lysine-treated dishes (Brooke Knapp MatTek) overnight and then were cotransfected with HIV-1 provirus and GFP-FIP constructs using Lipofectamine (ThermoFisher Scientific), followed by incubation for 48 h. Before staining, cells were rinsed with phosphate-buffered saline (PBS) three times and fixed in 4% paraformaldehyde for 10 min at room temperature. Cells were washed with PBS three times with mild agitation after fixation and then permeabilized for 10 min with 0.2% Triton X-100, followed by washing with PBS. Dako blocking solution was applied to cells for 30 min under gentle shaking conditions. 2G12 antibody against HIV-1 gp120 was diluted in Dako antibody diluent to 1:300. Anti-goat secondary antibody was diluted in Dako antibody diluent to 1:1,000. All antibodies for immunostaining endocytosis markers were used between 1:1,000 and 1:5,000 dilutions in Dako antibody diluent solution. The coverslips were mounted in Prolong Gold (Thermo Fisher) overnight and imaged on the DeltaVision imaging system (GE Healthcare) or the OMX imaging system (GE Healthcare) for structured illumination microscopy. The Volocity 6.3 software program (PerkinElmer) was employed for analysis and presentation of microscopic images obtained. Quantitation of colocalization between HIV-1 Env and GFP-FIP1C560–649 was performed using Volocity software features, including derivation of Pearson's coefficient and M1 and M2 colocalization coefficients. Statistical differences between colocalization coefficients were calculated by Student's t test as recommended by Dunn et al. (52). For labeling experiments with FAP control, FAP-TMCT, or FAP-TMCT144, HeLa cells were transfected with GFP-FIP1C560–649 and pulse labeled with the nonpermeable fluorogen MG-11p from Spectragenetics. Cells were examined the following day at 18 to 20 h after transfection. Cells were exposed to 200 nM MG-11P for 5 min, and then medium was removed and replaced with medium lacking fluorogen to prevent further labeling. Cells were incubated for the indicated times and fixed and imaged as described above, using the Cy5 channel for detection of FAP signal. Pearson's correlation coefficient was derived for 10 imaged fields of cells per construct/time point shown.
Electron microscopy.
HeLa cells were harvested 48 h following transfection, gently pelleted, and fixed in two different fixative combinations and time periods, one for morphology and one for immuno-EM. For standard TEM, as shown in Fig. 5F, fixation was 2.5% glutaraldehyde and 2.5% paraformaldehyde in cacodylate buffer (15956; EMS) for 2 h at room temperature. Cell pellets were postfixed in 1% osmium tetroxide (19100; EMS) for less than 1 h in the dark on ice. The cell samples then were dehydrated via ethanol gradient and propylene oxide. After dehydration, cell pellets were embedded in Eponate-12 resin to produce thin 70-nm sections and imaged with a Hitachi H-7500 microscope. For postembedding immuno-EM (Fig. 5G), fixation was performed with 4% paraformaldehyde freshly prepared from powder form (15959; EMS) with 0.02%, 0.05%, and 0.1% glutaraldehyde (15956; EMS) for 20 min on ice. Cell pellets were embedded in LR White (14381; EMS) without osmium postfixation to produce thick 100-nm sections on 300-mesh nickel grids supported with Formvar membrane. Sections then were etched with fresh 5% H2O2 in distilled water for 10 min, blocked with Pierce protein-free PBS (37584; Pierce) for primary antibody labeling, and then blocked with Pierce protein-free Tris-buffered saline (37585; Pierce) for secondary immunogold labeling. HIV-1 Env was visualized using a cocktail of monoclonal antibodies consisting of 2G12, b12, and 447-52D against HIV-1 gp120 at a final concentration of 1/250 to 1/500, followed by addition of protein A-gold beads of 10 nm as described previously (53).
Flow cytometry for HIV-1 Env expression on cell surface.
Two days after cotransfection with the indicated proviral or Env-expression plasmids and GFP-FIP1C560–649, cells were detached under mild conditions with Versene containing EDTA (Life Technologies) and fixed with 4% paraformaldehyde for 10 min at room temperature. Surface staining of HIV-1 Env then was performed with 0.5 μg of HIV-1 murine monoclonal antibody BDI123 from Novus Biologicals in PBS containing 1% FBS. Allophycocyanin-conjugated anti-mouse F(ab′)2 antibody from BD Pharmingen was used at 1:400 dilution for detection. Flow cytometry was performed on a FACSCanto cytometer (BD Biosciences) and analyzed using FlowJo software (Treestar, Inc.).
ACKNOWLEDGMENTS
This work was supported by R01 GM111027.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Flow cytometry was performed using the Emory-Children's Pediatric Research Center Flow Cytometry Core. Electron microscopy was performed with the help of the Robert P. Apkarian Integrated Electron Microscopy Core at Emory University.
REFERENCES
- 1.Checkley MA, Luttge BG, Freed EO. 2011. HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation. J Mol Biol 410:582–608. doi: 10.1016/j.jmb.2011.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson MC. 2011. Mechanisms for Env glycoprotein acquisition by retroviruses. AIDS Res Hum Retrovir 27:239–247. doi: 10.1089/aid.2010.0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hunter E, Swanstrom R. 1990. Retrovirus envelope glycoproteins. Curr Top Microbiol Immunol 157:187–253. [DOI] [PubMed] [Google Scholar]
- 4.Murakami T, Freed EO. 2000. The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc Natl Acad Sci U S A 97:343–348. doi: 10.1073/pnas.97.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berlioz-Torrent C, Shacklett BL, Erdtmann L, Delamarre L, Bouchaert I, Sonigo P, Dokhelar MC, Benarous R. 1999. Interactions of the cytoplasmic domains of human and simian retroviral transmembrane proteins with components of the clathrin adaptor complexes modulate intracellular and cell surface expression of envelope glycoproteins. J Virol 73:1350–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohno H, Aguilar RC, Fournier MC, Hennecke S, Cosson P, Bonifacino JS. 1997. Interaction of endocytic signals from the HIV-1 envelope glycoprotein complex with members of the adaptor medium chain family. Virology 238:305–315. doi: 10.1006/viro.1997.8839. [DOI] [PubMed] [Google Scholar]
- 7.Rowell JF, Stanhope PE, Siliciano RF. 1995. Endocytosis of endogenously synthesized HIV-1 envelope protein. Mechanism and role in processing for association with class II MHC. J Immunol 155:473–488. [PubMed] [Google Scholar]
- 8.Day JR, Munk C, Guatelli JC. 2004. The membrane-proximal tyrosine-based sorting signal of human immunodeficiency virus type 1 gp41 is required for optimal viral infectivity. J Virol 78:1069–1079. doi: 10.1128/JVI.78.3.1069-1079.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deschambeault J, Lalonde JP, Cervantes-Acosta G, Lodge R, Cohen EA, Lemay G. 1999. Polarized human immunodeficiency virus budding in lymphocytes involves a tyrosine-based signal and favors cell-to-cell viral transmission. J Virol 73:5010–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lodge R, Lalonde JP, Lemay G, Cohen EA. 1997. The membrane-proximal intracytoplasmic tyrosine residue of HIV-1 envelope glycoprotein is critical for basolateral targeting of viral budding in MDCK cells. EMBO J 16:695–705. doi: 10.1093/emboj/16.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wyss S, Berlioz-Torrent C, Boge M, Blot G, Honing S, Benarous R, Thali M. 2001. The highly conserved C-terminal dileucine motif in the cytosolic domain of the human immunodeficiency virus type 1 envelope glycoprotein is critical for its association with the AP-1 clathrin adaptor. J Virol 75:2982–2992. doi: 10.1128/JVI.75.6.2982-2992.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhakta SJ, Shang L, Prince JL, Claiborne DT, Hunter E. 2011. Mutagenesis of tyrosine and di-leucine motifs in the HIV-1 envelope cytoplasmic domain results in a loss of Env-mediated fusion and infectivity. Retrovirology 8:37. doi: 10.1186/1742-4690-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi M, Williams JA, Chu H, Chen X, Wang JJ, Ding L, Akhirome E, Wen X, Lapierre LA, Goldenring JR, Spearman P. 2013. Rab11-FIP1C and Rab14 direct plasma membrane sorting and particle incorporation of the HIV-1 envelope glycoprotein complex. PLoS Pathog 9:e1003278. doi: 10.1371/journal.ppat.1003278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallace DM, Lindsay AJ, Hendrick AG, McCaffrey MW. 2002. The novel Rab11-FIP/Rip/RCP family of proteins displays extensive homo- and hetero-interacting abilities. Biochem Biophys Res Commun 292:909–915. doi: 10.1006/bbrc.2002.6736. [DOI] [PubMed] [Google Scholar]
- 15.Lall P, Lindsay AJ, Hanscom S, Kecman T, Taglauer ES, McVeigh UM, Franklin E, McCaffrey MW, Khan AR. 2015. Structure-function analyses of the interactions between Rab11 and Rab14 small GTPases with their shared effector Rab coupling protein (RCP). J Biol Chem 290:18817–18832. doi: 10.1074/jbc.M114.612366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindsay AJ, Hendrick AG, Cantalupo G, Senic-Matuglia F, Goud B, Bucci C, McCaffrey MW. 2002. Rab coupling protein (RCP), a novel Rab4 and Rab11 effector protein. J Biol Chem 277:12190–12199. doi: 10.1074/jbc.M108665200. [DOI] [PubMed] [Google Scholar]
- 17.Hales CM, Griner R, Hobdy-Henderson KC, Dorn MC, Hardy D, Kumar R, Navarre J, Chan EK, Lapierre LA, Goldenring JR. 2001. Identification and characterization of a family of Rab11-interacting proteins. J Biol Chem 276:39067–39075. doi: 10.1074/jbc.M104831200. [DOI] [PubMed] [Google Scholar]
- 18.Qi M, Chu H, Chen X, Choi J, Wen X, Hammonds J, Ding L, Hunter E, Spearman P. 2015. A tyrosine-based motif in the HIV-1 envelope glycoprotein tail mediates cell-type- and Rab11-FIP1C-dependent incorporation into virions. Proc Natl Acad Sci U S A 112:7575–7580. doi: 10.1073/pnas.1504174112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindsay AJ, McCaffrey MW. 2002. Rab11-FIP2 functions in transferrin recycling and associates with endosomal membranes via its COOH-terminal domain. J Biol Chem 277:27193–27199. doi: 10.1074/jbc.M200757200. [DOI] [PubMed] [Google Scholar]
- 20.Damiani MT, Pavarotti M, Leiva N, Lindsay AJ, McCaffrey MW, Colombo MI. 2004. Rab coupling protein associates with phagosomes and regulates recycling from the phagosomal compartment. Traffic 5:785–797. doi: 10.1111/j.1600-0854.2004.00220.x. [DOI] [PubMed] [Google Scholar]
- 21.Boge M, Wyss S, Bonifacino JS, Thali M. 1998. A membrane-proximal tyrosine-based signal mediates internalization of the HIV-1 envelope glycoprotein via interaction with the AP-2 clathrin adaptor. J Biol Chem 273:15773–15778. doi: 10.1074/jbc.273.25.15773. [DOI] [PubMed] [Google Scholar]
- 22.Yan Q, Schmidt BF, Perkins LA, Naganbabu M, Saurabh S, Andreko SK, Bruchez MP. 2015. Near-instant surface-selective fluorogenic protein quantification using sulfonated triarylmethane dyes and fluorogen activating proteins. Org Biomol Chem 13:2078–2086. doi: 10.1039/C4OB02309A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szent-Gyorgyi C, Schmidt BF, Creeger Y, Fisher GW, Zakel KL, Adler S, Fitzpatrick JA, Woolford CA, Yan Q, Vasilev KV, Berget PB, Bruchez MP, Jarvik JW, Waggoner A. 2008. Fluorogen-activating single-chain antibodies for imaging cell surface proteins. Nat Biotechnol 26:235–240. doi: 10.1038/nbt1368. [DOI] [PubMed] [Google Scholar]
- 24.Earl PL, Doms RW, Moss B. 1990. Oligomeric structure of the human immunodeficiency virus type 1 envelope glycoprotein. Proc Natl Acad Sci U S A 87:648–652. doi: 10.1073/pnas.87.2.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Earl PL, Koenig S, Moss B. 1991. Biological and immunological properties of human immunodeficiency virus type 1 envelope glycoprotein: analysis of proteins with truncations and deletions expressed by recombinant vaccinia viruses. J Virol 65:31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otteken A, Earl PL, Moss B. 1996. Folding, assembly, and intracellular trafficking of the human immunodeficiency virus type 1 envelope glycoprotein analyzed with monoclonal antibodies recognizing maturational intermediates. J Virol 70:3407–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinter A, Honnen WJ, Tilley SA, Bona C, Zaghouani H, Gorny MK, Zolla-Pazner S. 1989. Oligomeric structure of gp41, the transmembrane protein of human immunodeficiency virus type 1. J Virol 63:2674–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schawaller M, Smith GE, Skehel JJ, Wiley DC. 1989. Studies with crosslinking reagents on the oligomeric structure of the env glycoprotein of HIV. Virology 172:367–369. doi: 10.1016/0042-6822(89)90142-6. [DOI] [PubMed] [Google Scholar]
- 29.Hallenberger S, Bosch V, Angliker H, Shaw E, Klenk HD, Garten W. 1992. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature 360:358–361. doi: 10.1038/360358a0. [DOI] [PubMed] [Google Scholar]
- 30.McCune JM, Rabin LB, Feinberg MB, Lieberman M, Kosek JC, Reyes GR, Weissman IL. 1988. Endoproteolytic cleavage of gp160 is required for the activation of human immunodeficiency virus. Cell 53:55–67. doi: 10.1016/0092-8674(88)90487-4. [DOI] [PubMed] [Google Scholar]
- 31.Ono A. 2010. HIV-1 assembly at the plasma membrane. Vaccine 28(Suppl 2):B55–B59. doi: 10.1016/j.vaccine.2009.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ono A, Freed EO. 2001. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc Natl Acad Sci U S A 98:13925–13930. doi: 10.1073/pnas.241320298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saad JS, Miller J, Tai J, Kim A, Ghanam RH, Summers MF. 2006. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc Natl Acad Sci U S A 103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Freed EO, Martin MA. 1995. Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J Virol 69:1984–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Freed EO, Martin MA. 1996. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J Virol 70:341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tedbury PR, Freed EO. 2014. The role of matrix in HIV-1 envelope glycoprotein incorporation. Trends Microbiol 22:372–378. doi: 10.1016/j.tim.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cosson P. 1996. Direct interaction between the envelope and matrix proteins of HIV-1. EMBO J 15:5783–5788. [PMC free article] [PubMed] [Google Scholar]
- 38.Vincent MJ, Melsen LR, Martin AS, Compans RW. 1999. Intracellular interaction of simian immunodeficiency virus Gag and Env proteins. J Virol 73:8138–8144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tedbury PR, Ablan SD, Freed EO. 2013. Global rescue of defects in HIV-1 envelope glycoprotein incorporation: implications for matrix structure. PLoS Pathog 9:e1003739. doi: 10.1371/journal.ppat.1003739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tedbury PR, Novikova M, Ablan SD, Freed EO. 2016. Biochemical evidence of a role for matrix trimerization in HIV-1 envelope glycoprotein incorporation. Proc Natl Acad Sci U S A 113:E182–E190. doi: 10.1073/pnas.1516618113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baetz NW, Goldenring JR. 2014. Distinct patterns of phosphatidylserine localization within the Rab11a-containing recycling system. Cell Logist 4:e28680. doi: 10.4161/cl.28680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kelly EE, Horgan CP, Adams C, Patzer TM, Ni Shuilleabhain DM, Norman JC, McCaffrey MW. 2009. Class I Rab11-family interacting proteins are binding targets for the Rab14 GTPase. Biol Cell 102:51–62. doi: 10.1042/BC20090068. [DOI] [PubMed] [Google Scholar]
- 43.Chakrabarti L, Guyader M, Alizon M, Daniel MD, Desrosiers RC, Tiollais P, Sonigo P. 1987. Sequence of simian immunodeficiency virus from macaque and its relationship to other human and simian retroviruses. Nature 328:543–547. doi: 10.1038/328543a0. [DOI] [PubMed] [Google Scholar]
- 44.Zingler K, Littman DR. 1993. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein increases env incorporation into particles and fusogenicity and infectivity. J Virol 67:2824–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.LaBranche CC, Sauter MM, Haggarty BS, Vance PJ, Romano J, Hart TK, Bugelski PJ, Marsh M, Hoxie JA. 1995. A single amino acid change in the cytoplasmic domain of the simian immunodeficiency virus transmembrane molecule increases envelope glycoprotein expression on infected cells. J Virol 69:5217–5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sauter MM, Pelchen-Matthews A, Bron R, Marsh M, LaBranche CC, Vance PJ, Romano J, Haggarty BS, Hart TK, Lee WM, Hoxie JA. 1996. An internalization signal in the simian immunodeficiency virus transmembrane protein cytoplasmic domain modulates expression of envelope glycoproteins on the cell surface. J Cell Biol 132:795–811. doi: 10.1083/jcb.132.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bowers K, Pelchen-Matthews A, Honing S, Vance PJ, Creary L, Haggarty BS, Romano J, Ballensiefen W, Hoxie JA, Marsh M. 2000. The simian immunodeficiency virus envelope glycoprotein contains multiple signals that regulate its cell surface expression and endocytosis. Traffic 1:661–674. doi: 10.1034/j.1600-0854.2000.010810.x. [DOI] [PubMed] [Google Scholar]
- 48.Byland R, Vance PJ, Hoxie JA, Marsh M. 2007. A conserved dileucine motif mediates clathrin and AP-2-dependent endocytosis of the HIV-1 envelope protein. Mol Biol Cell 18:414–425. doi: 10.1091/mbc.E06-06-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin M, Goldenring JR. 2006. The Rab11-FIP1/RCP gene codes for multiple protein transcripts related to the plasma membrane recycling system. Biochim Biophys Acta 1759:281–295. doi: 10.1016/j.bbaexp.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 50.Huang Y, Kong WP, Nabel GJ. 2001. Human immunodeficiency virus type 1-specific immunity after genetic immunization is enhanced by modification of Gag and Pol expression. J Virol 75:4947–4951. doi: 10.1128/JVI.75.10.4947-4951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hammonds JE, Beeman N, Ding L, Takushi S, Francis AC, Wang JJ, Melikyan GB, Spearman P. 2017. Siglec-1 initiates formation of the virus-containing compartment and enhances macrophage-to-T cell transmission of HIV-1. PLoS Pathog 13:e1006181. doi: 10.1371/journal.ppat.1006181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dunn KW, Kamocka MM, McDonald JH. 2011. A practical guide to evaluating colocalization in biological microscopy. Am J Physiol Cell Physiol 300:C723–C742. doi: 10.1152/ajpcell.00462.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hammonds J, Wang JJ, Yi H, Spearman P. 2010. Immunoelectron microscopic evidence for Tetherin/BST2 as the physical bridge between HIV-1 virions and the plasma membrane. PLoS Pathog 6:e1000749. doi: 10.1371/journal.ppat.1000749. [DOI] [PMC free article] [PubMed] [Google Scholar]