ABSTRACT
HIV-1-specific cytotoxic T cells (CTLs) play an important role in the control of HIV-1 subtype B or C infection. However, the role of CTLs in HIV-1 subtype A/E infection still remains unclear. Here we investigated the association of HLA class I alleles with clinical outcomes in treatment-naive Vietnamese infected with subtype A/E virus. We found that HLA-C*12:02 was significantly associated with lower plasma viral loads (pVL) and higher CD4 counts and that the HLA-A*29:01-B*07:05-C*15:05 haplotype was significantly associated with higher pVL and lower CD4 counts than those for individuals without these respective genotypes. Nine Pol and three Nef mutations were associated with at least one HLA allele in the HLA-A*29:01-B*07:05-C*15:05 haplotype, with a strong negative correlation between the number of HLA-associated Pol mutations and CD4 count as well as a positive correlation with pVL for individuals with these HLA alleles. The results suggest that the accumulation of mutations selected by CTLs restricted by these HLA alleles affects HIV control.
IMPORTANCE Most previous studies on HLA association with disease progression after HIV-1 infection have been performed on cohorts infected with HIV-1 subtypes B and C, whereas few such population-based studies have been reported for cohorts infected with the Asian subtype A/E virus. In this study, we analyzed the association of HLA class I alleles with clinical outcomes for 536 HIV-1 subtype A/E-infected Vietnamese individuals. We found that HLA-C*12:02 is protective, while the HLA haplotype HLA-A*29:01-B*07:05-C*15:05 is deleterious. The individuals with HIV-1 mutations associated with at least one of the HLA alleles in the deleterious HLA haplotype had higher plasma viral loads and lower CD4 counts than those of individuals without the mutations, suggesting that viral adaptation and escape from HLA-mediated immune control occurred. The present study identifies a protective allele and a deleterious haplotype for HIV-1 subtype A/E infection which are different from those identified for cohorts infected with HIV-1 subtypes B and C.
KEYWORDS: clinical outcome, deleterious allele, HIV-1, HLA class I, HLA-associated mutation, protective allele, subtype A/E, Vietnam
INTRODUCTION
Human leukocyte antigen (HLA) class I alleles have consistently been shown to have a significant impact on the rate of HIV-1 disease progression to AIDS. Numerous studies on HIV-1 subtype B-infected Caucasians and subtype C-infected Africans have revealed that HLA-B*57, -B*58, and -B*27 alleles are protective, whereas HLA-B*35 and -C*07:02 are detrimental (1–9). A recent study on subtype B-infected Japanese, in whom the frequencies of HLA-B*57, -B*58, and -B*27 are very low, showed that the HLA-B*52:01-C*12:02 haplotype and the HLA-B*67:01 allele had a protective effect on clinical outcomes (10).
HIV-1 subtype A/E (CRF01-AE) is the dominant subtype in Southeast Asia, especially in Myanmar, Thailand, Vietnam, Lao People's Democratic Republic, Cambodia, and Indonesia (11–14). Over 1.7 million individuals are living with HIV-1 subtype A/E, and over 100,000 individuals are still newly infected every year in these countries (UNAIDS 2016 Progress Reports [http://www.unaids.org/en/]). However, there are only two studies of HLA class I association with clinical outcomes for HIV-1 subtype A/E infection. One of these studies, in which 557 HIV-1 subtype A/E-infected individuals in Thailand were recruited in 2000 to 2002, identified HLA-B*35:05 as a protective allele in terms of plasma viral load (pVL) as the parameter of clinical outcome (15). The other study found that HLA-B*51 and HLA-C*04 were associated with mortality, with the B*51 allele being protective and the C*04 allele being deleterious, although only 99 HIV-1 subtype A/E-infected Thai individuals were analyzed (16). Thus, HLA class I associations with clinical outcomes have not been investigated thoroughly for HIV-1 subtype A/E infection.
In the present study, we analyzed the association between the presence of HLA class I alleles with frequencies of >2% and clinical outcomes for 536 treatment-naive Vietnamese individuals chronically infected with HIV-1 subtype A/E. We used both pVL and CD4 T cell count as parameters of clinical outcome. We further analyzed the role of HLA-associated mutations to test the possibility that the accumulation of cytotoxic T lymphocyte (CTL) escape mutations is involved in rapid progression to AIDS.
RESULTS
HLA class I alleles associated with clinical outcomes in a Hanoi cohort.
We recruited 536 chronically HIV-1-infected, antiretroviral therapy (ART)-naive Vietnamese individuals, and we determined their HLA genotypes at the 4-digit level and obtained the pVL and/or CD4 count at the first visit. We analyzed the association of clinical parameters with 52 HLA class I alleles (15 HLA-A, 22 HLA-B, and 15 HLA-C alleles) found in the individuals at frequencies of >2% (Table 1).
TABLE 1.
Associations of HLA alleles with clinical outcomes (n = 536)
| Allele | Population frequency (%) | Median viral load (log10 copies/ml) |
P valuea | Median CD4 count (cells/μl) |
P valuea | ||
|---|---|---|---|---|---|---|---|
| HLA+ | HLA− | HLA+ | HLA− | ||||
| HLA-A alleles | |||||||
| HLA-A*01:01 | 5.8 | 4.67 | 4.56 | 0.37 | 279 | 300 | 0.42 |
| HLA-A*02:01 | 4.1 | 4.5 | 4.56 | 0.68 | 290 | 300 | 0.88 |
| HLA-A*02:03 | 16.2 | 4.67 | 4.55 | 0.14 | 264 | 303 | 0.045 |
| HLA-A*02:06 | 8.8 | 4.46 | 4.57 | 0.31 | 293 | 300 | 0.90 |
| HLA-A*02:07 | 17.5 | 4.58 | 4.56 | 0.54 | 302 | 299 | 0.62 |
| HLA-A*03:01 | 2.4 | 4.16 | 4.57 | 0.083 | 355 | 299 | 0.32 |
| HLA-A*11:01 | 44.4 | 4.52 | 4.58 | 0.16 | 319 | 293 | 0.018 |
| HLA-A*11:02 | 5.8 | 4.58 | 4.56 | 0.89 | 280 | 300 | 0.59 |
| HLA-A*24:02 | 18.1 | 4.54 | 4.57 | 0.73 | 295 | 300 | 0.82 |
| HLA-A*24:07 | 6.7 | 4.71 | 4.56 | 0.29 | 271 | 301 | 0.36 |
| HLA-A*26:01 | 4.5 | 4.8 | 4.56 | 0.14 | 284 | 300 | 0.64 |
| HLA-A*29:01 | 13.8 | 4.82 | 4.54 | 0.012 | 248 | 304 | 0.23 |
| HLA-A*30:01 | 2.4 | 4.39 | 4.57 | 0.32 | 373 | 299 | 0.092 |
| HLA-A*31:01 | 3.2 | 4.29 | 4.57 | 0.18 | 359 | 299 | 0.23 |
| HLA-A*33:03 | 25.2 | 4.55 | 4.57 | 0.69 | 306 | 299 | 0.54 |
| HLA-B alleles | |||||||
| HLA-B*07:02 | 2.6 | 4.74 | 4.56 | 0.47 | 338 | 299 | 0.40 |
| HLA-B*07:05 | 15.3 | 4.77 | 4.54 | 0.019 | 242 | 305 | 0.009 |
| HLA-B*13:01 | 9.3 | 4.58 | 4.56 | 0.86 | 293 | 300 | 0.81 |
| HLA-B*15:01 | 2.2 | 4.57 | 4.56 | 0.93 | 388 | 298 | 0.12 |
| HLA-B*15:02 | 26.9 | 4.63 | 4.55 | 0.33 | 291 | 301 | 0.75 |
| HLA-B*15:12 | 3.9 | 4.86 | 4.56 | 0.073 | 278 | 300 | 0.51 |
| HLA-B*15:25 | 10.3 | 4.42 | 4.57 | 0.20 | 284 | 300 | 0.50 |
| HLA-B*27:04 | 2.1 | 4.35 | 4.57 | 0.33 | 248 | 300 | 0.56 |
| HLA-B*35:05 | 5.2 | 4.58 | 4.56 | 0.87 | 271 | 300 | 0.31 |
| HLA-B*37:01 | 2.2 | 4.71 | 4.56 | 0.51 | 323 | 299 | 0.60 |
| HLA-B*38:02 | 12.7 | 4.63 | 4.56 | 0.47 | 288 | 300 | 0.73 |
| HLA-B*40:01 | 9.1 | 4.42 | 4.57 | 0.25 | 317 | 299 | 0.62 |
| HLA-B*40:06 | 2.6 | 4.53 | 4.56 | 0.89 | 345 | 299 | 0.38 |
| HLA-B*44:03 | 5.8 | 4.36 | 4.57 | 0.16 | 314 | 299 | 0.80 |
| HLA-B*46:01 | 20.5 | 4.46 | 4.58 | 0.21 | 316 | 298 | 0.52 |
| HLA-B*51:01 | 4.7 | 4.51 | 4.56 | 0.60 | 292 | 300 | 0.78 |
| HLA-B*51:02 | 2.6 | 4.74 | 4.56 | 0.49 | 262 | 300 | 0.74 |
| HLA-B*52:01 | 3.7 | 4.35 | 4.57 | 0.22 | 377 | 298 | 0.059 |
| HLA-B*54:01 | 4.3 | 4.62 | 4.56 | 0.71 | 364 | 298 | 0.13 |
| HLA-B*55:02 | 6.2 | 4.73 | 4.56 | 0.21 | 298 | 300 | 0.90 |
| HLA-B*57:01 | 4.7 | 4.51 | 4.56 | 0.74 | 291 | 300 | 0.95 |
| HLA-B*58:01 | 17.0 | 4.51 | 4.57 | 0.52 | 343 | 296 | 0.087 |
| HLA-C alleles | |||||||
| HLA-C*01:02 | 27.4 | 4.54 | 4.57 | 0.85 | 316 | 297 | 0.56 |
| HLA-C*03:02 | 17.4 | 4.53 | 4.57 | 0.94 | 341 | 296 | 0.13 |
| HLA-C*03:03 | 8.6 | 4.85 | 4.55 | 0.017 | 271 | 301 | 0.38 |
| HLA-C*03:04 | 11.8 | 4.58 | 4.56 | 0.98 | 274 | 301 | 0.44 |
| HLA-C*04:01 | 6.5 | 4.49 | 4.57 | 0.56 | 290 | 300 | 0.75 |
| HLA-C*04:03 | 8.8 | 4.52 | 4.56 | 0.65 | 283 | 300 | 0.70 |
| HLA-C*06:02 | 7.3 | 4.54 | 4.56 | 0.99 | 304 | 299 | 0.96 |
| HLA-C*07:01 | 6.2 | 4.37 | 4.57 | 0.16 | 308 | 299 | 0.89 |
| HLA-C*07:02 | 30.2 | 4.52 | 4.57 | 0.33 | 312 | 297 | 0.27 |
| HLA-C*08:01 | 30.6 | 4.61 | 4.55 | 0.58 | 284 | 303 | 0.59 |
| HLA-C*12:02 | 5.6 | 4.22 | 4.57 | 0.016 | 370 | 297 | 0.040 |
| HLA-C*12:03 | 3.7 | 4.53 | 4.56 | 0.94 | 315 | 299 | 0.99 |
| HLA-C*14:02 | 4.3 | 4.52 | 4.56 | 0.93 | 258 | 300 | 0.21 |
| HLA-C*15:02 | 4.3 | 4.66 | 4.56 | 0.43 | 299 | 300 | 0.77 |
| HLA-C*15:05 | 13.6 | 4.8 | 4.54 | 0.022 | 238 | 304 | 0.014 |
Analyzed by use of a linear regression model.
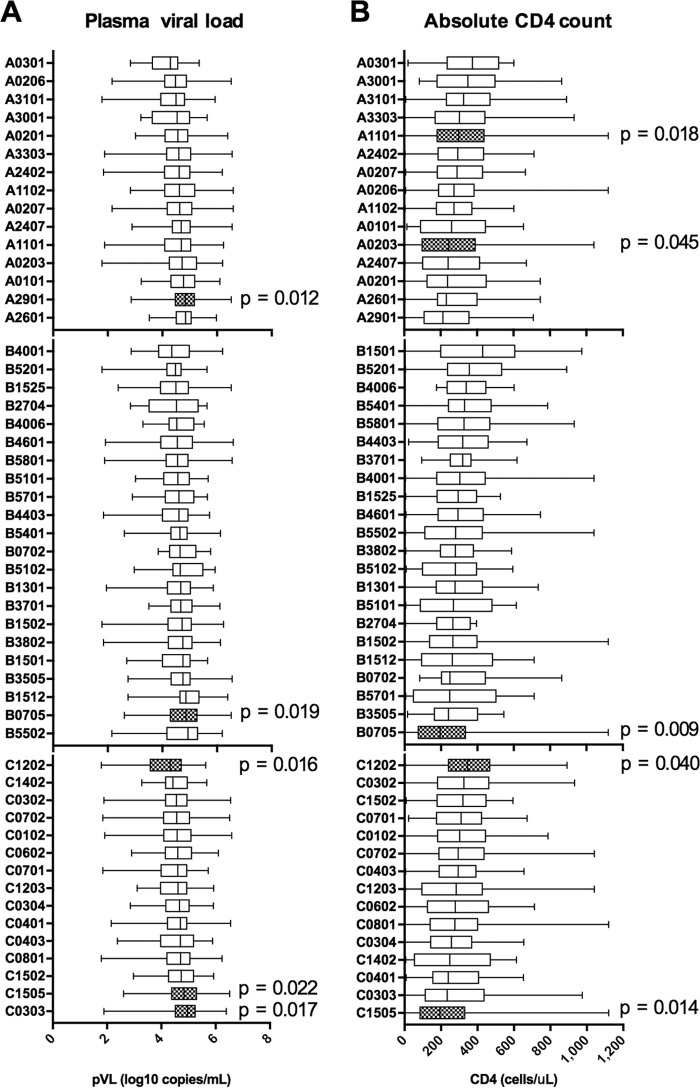
Seven HLA alleles were significantly associated with pVL and/or CD4 count (Fig. 1). Only HLA-C*12:02 was significantly associated with low pVL (Fig. 1A), while HLA-C*12:02 and HLA-A*11:01 were significantly associated with high CD4 counts (Fig. 1B). Thus, HLA-C*12:02 was significantly associated with both low VL and high CD4 counts, suggesting that it is a protective allele. Another HLA-C*12 subtype, HLA-C*12:03, showed no association with the clinical parameters tested. These two alleles differ by only a single amino acid, at position 97 in the floor of the peptide binding groove (R and W, respectively) (17), which is likely to result in differences in peptide binding repertoire between the two alleles. HLA-B*57 (B*57:01, B*57:02, and B*57:03), -B*58 (B*58:01), and -B*27 (B*27:05) are well known as protective alleles in Caucasians and Africans (1, 2, 4–9, 18). However, we did not find any association of HLA-B*57 (B*57:01), -B*58 (B*58:01), or -B*27 (B*27:04) with pVL or CD4 count in this cohort (Fig. 1 and Table 1). HLA-B*52:01, which was previously shown to be protective in Japanese (10), was not significantly associated with either of the parameters but showed a trend of lower pVL and higher CD4 counts (Fig. 1 and Table 1). This might be due to the lower frequency of this allele in Vietnamese individuals (Table 1).
FIG 1.
Association of HLA alleles with plasma viral load and absolute CD4 count in Vietnamese individuals chronically infected with HIV-1 subtype A/E. All HLA alleles occurring at phenotypic frequencies of >2% were examined for their associations with plasma viral load (A) and absolute CD4 count (B) in a cohort of 536 Vietnamese individuals chronically infected with HIV-1 subtype A/E. Associations with P values of <0.05 are highlighted with hatched boxes, and the values are shown.
HLA-A*29:01, -B*07:05, and -C*15:05 were associated with significantly higher pVL, while HLA-B*07:05 and -C*15:05 were associated with significantly lower CD4 counts (Fig. 1). These two alleles as well as HLA-A*29:01/C*15:05 and HLA-A*29:01/B*07:05 showed strong linkage disequilibrium (LD), indicating that the HLA-A*29:01-B*07:05-C*15:05 combination forms a haplotype in this cohort (F = 11.9%) (Table 2) and is likely to be detrimental in Vietnamese infected with HIV-1.
TABLE 2.
HLA linkage disequilibrium for pairs of HLA-A*29:01, -B*07:05, and -C*15:05 alleles
| First HLA allele (HLA1) | Second HLA allele (HLA2) | No. of individuals with: |
P valuea | Frequency | |||
|---|---|---|---|---|---|---|---|
| Both alleles | Only HLA1 | Only HLA2 | Neither allele | ||||
| HLA-B*07:05 | HLA-C*15:05 | 73 | 9 | 0 | 454 | 1.32 × 10−80 | 0.136 |
| HLA-A*29:01 | HLA-C*15:05 | 64 | 10 | 9 | 453 | 7.93 × 10−62 | 0.119 |
| HLA-A*29:01 | HLA-B*07:05 | 65 | 9 | 17 | 445 | 2.45 × 10−57 | 0.121 |
Analyzed by Fisher's exact test.
Effects of mutations associated with HLA-A*29:01-B*07:05-C*15:05.
It is speculated that T cells restricted by detrimental HLA alleles cannot control HIV-1. Presumably such T cells have no or weak ability to suppress HIV-1, or they may select for immune escape mutations. Therefore, we investigated the effects of mutations associated with HLA-A*29:01, -B*07:05, and -C*15:05 on clinical outcomes. Our recent study using a phylogenetically corrected logistic regression model identified several Nef and Pol mutations associated with HLA-A*29:01, -B*07:05, and/or -C*15:05 (19). However, some mutations associated with HLA alleles that are in strong LD may have been excluded in the phylogenetically corrected logistic regression model. We therefore reanalyzed mutations associated with HLA-A*29:01, -B*07:05, or -C*15:05 by Fisher's exact test, using 2 × 2 tables. We identified nine mutations at seven Pol positions and three mutations at three Nef positions that were associated with all three HLA alleles, with the exception of Nef71K, which lacked the association with HLA-A*29:01 (Table 3).
TABLE 3.
HLA-A*29:01-, -B*07:05-, and -C*15:05-associated HIV-1 mutations in HIV-1 subtype A/E-infected Vietnamese individuals
| Gene product | Position | Amino acid mutation | Consensus amino acida | Direction of adaptionb |
P valuec |
||
|---|---|---|---|---|---|---|---|
| HLA-A*29:01 | HLA-B*07:05 | HLA-C*15:05 | |||||
| Pol | 14 | R | K | Adapted | 0.03 | 0.002 | 0.002 |
| 110 | K | T | Adapted | 0.0002 | <0.0001 | <0.0001 | |
| 261 | Y | C | Adapted | 0.003 | 0.0004 | 0.0006 | |
| 272 | I | I | Nonadapted | 0.04 | 0.0006 | 0.0003 | |
| 653 | L | S | Adapted | 0.0008 | 0.002 | 0.0008 | |
| 653 | A | S | Adapted | 0.0002 | <0.0001 | <0.0001 | |
| 653 | T | S | Adapted | 0.0002 | <0.0001 | <0.0001 | |
| 655 | V | I | Adapted | <0.0001 | <0.0001 | <0.0001 | |
| 657 | R | K | Adapted | 0.0002 | 0.0004 | <0.0001 | |
| Nef | 71 | K | R | Adapted | 0.2 | <0.0001 | 0.04 |
| 156 | N | D | Nonadapted | 0.04 | 0.003 | 0.01 | |
| 173 | I | F | Adapted | 0.004 | 0.01 | 0.001 | |
Cohort consensus sequence.
Adapted, amino acids are abundant in the “presence” of a given HLA allele; nonadapted, amino acids are abundant in the “absence” of a given HLA allele.
Analyzed by Fisher's exact test.
We investigated the relationship between the presence of the HLA-associated mutations at three Nef positions (Nef71K, a non-N residue at Nef156 [Nef156nonN], or Nef173I) and pVL or CD4 count by using nef sequences from 372 Vietnamese individuals. The only mutation that showed a significant effect was Nef173I, as the presence of this mutation was associated with significantly higher pVL than those in individuals not having this mutation (Table 4). When the analysis was restricted to individuals with at least one of the HLA-A*29:01, HLA-B*07:05, and HLA-C*15:05 alleles, there was a similar effect on pVL, whereas no such effect was seen in HLA-A*29:01−B*07:05−C*15:05− individuals (see Tables S1 and S2 in the supplemental material). We further investigated the correlation of the number of HLA-associated mutations in Nef with pVL or CD4 count. No correlation was observed for these individuals (Fig. 2A and B), suggesting that only the Nef173I mutation increases pVL. Together the results suggest that the Nef173I mutation may influence the recognition of CTLs restricted by these alleles rather than the viral replication capacity.
TABLE 4.
Differences in clinical outcomes between the presence and absence of HLA-A*29:01-, -B*07:05-, and -C*15:05-associated HIV-1 mutations
| Gene product | Position | HLA-associated amino acid mutation | No. of patients with mutation | Median viral load (log10 copies/ml) |
P valuea | Median CD4 count (cells/μl) |
P valuea | ||
|---|---|---|---|---|---|---|---|---|---|
| With mutation | Without mutation | With mutation | Without mutation | ||||||
| Pol | 14 | R | 44 | 4.3 | 4.6 | 0.28 | 302 | 331 | 0.69 |
| 110 | K | 123 | 4.6 | 4.5 | 0.19 | 325 | 326 | 0.60 | |
| 261 | Y | 26 | 4.2 | 4.5 | 0.31 | 313 | 326 | 0.69 | |
| 272 | Non-I | 133 | 4.6 | 4.5 | 0.61 | 293 | 341 | 0.051 | |
| 653 | L | 18 | 4.9 | 4.5 | 0.12 | 343 | 325 | 0.74 | |
| 653 | A | 36 | 4.6 | 4.5 | 0.43 | 200 | 335 | 0.0017 | |
| 653 | T | 24 | 4.7 | 4.5 | 0.29 | 196 | 333 | 0.0083 | |
| 655 | V | 46 | 4.7 | 4.5 | 0.17 | 270 | 331 | 0.35 | |
| 657 | R | 51 | 4.8 | 4.5 | 0.11 | 213 | 337 | 0.0026 | |
| Nef | 71 | K | 53 | 4.7 | 4.5 | 0.091 | 272 | 335 | 0.075 |
| 156 | Non-N | 276 | 4.5 | 4.7 | 0.059 | 337 | 304 | 0.30 | |
| 173 | I | 38 | 4.9 | 4.5 | 0.019 | 300 | 333 | 0.99 | |
Analyzed by using the Mann-Whitney test. Values in bold indicate statistical significance (P < 0.05).
FIG 2.
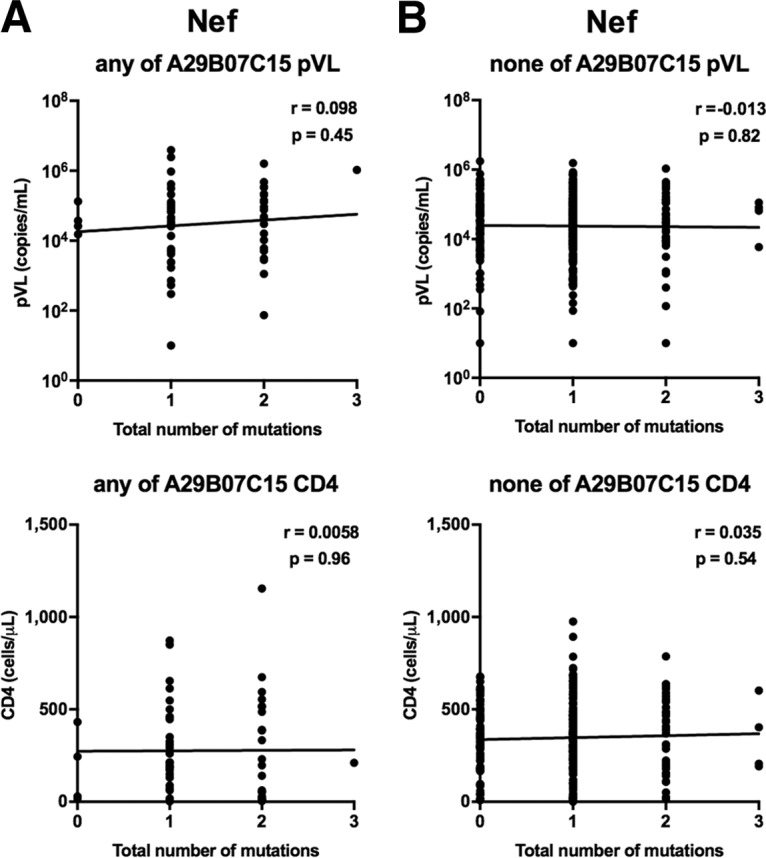
Correlations between HLA-associated mutations at three Nef positions and pVL or CD4 count for individuals having at least one of the HLA-A*29:01, -B*07:02, and -C*15:05 alleles (A) and those without any of them (B) were analyzed by using Spearman's correlation. Linear regression lines are included in the plots.
We next investigated the relationship between the presence of nine HLA-associated mutations at seven Pol positions (Pol14R, Pol110K, Pol261Y, a non-I residue at Pol272 [Pol272nonI], Pol653L/A/T, Pol655V, and Pol657R) and pVL or CD4 count by using pol sequences from 359 individuals (Table 4). The individuals with the Pol653A/T or Pol657R mutation had significantly lower CD4 counts than those of individuals without these mutations, while those having Pol272nonI showed a trend toward lower CD4 counts than those of individuals without these mutations. Similar effects of three mutations, Pol272nonI and Pol653A/T, on clinical outcome were observed in individuals with at least one of the HLA-A*29:01, HLA-B*07:05, and HLA-C*15:05 alleles (n = 61), whereas the effect of only the Pol653T mutation on CD4 count was observed in individuals with this haplotype (n = 40) (see Table S1), suggesting that the difference may result from the small number of samples from individuals with this haplotype. Further analysis of the effects of these nine mutations on pVL and CD4 count was carried out on 47 HLA-A*29:01+, 53 HLA-B*07:05+, and 47 HLA-C*15:05+ individuals stratified by the presence/absence of the HLA-associated mutations (see Table S2). The presence of Pol272nonI mutations in the HLA-B*07:05+ group was associated with significantly lower CD4 counts, but there was no effect in the other groups (HLA-A*29:01+ and HLA-C*15:05+ groups). HLA-B*07:05+ and C*15:05+ individuals having Pol653A had significantly lower CD4 counts, whereas HLA-A*29:01+ individuals having Pol653A had a trend toward lower CD4 counts. Individuals having the Pol653T mutation also had significantly lower CD4 counts and a trend toward higher pVL in all three groups. Lastly, the Pol657R mutation was associated with lower CD4 counts and higher pVL in the HLA-A*29:01+ and HLA-B*07:05+ groups, respectively. The contribution of HLA-associated Pol and Nef mutations to clinical outcomes in individuals with HLA-A*29:01, -B*07:02, or -C*15:05 is summarized in Fig. 3. Overall, the data suggest that these Pol mutations may affect the ability of allele-restricted CTLs to recognize epitopes that include these sites and thereby their ability to control HIV-1 replication. Note that there was no significant effect of these mutations in the HLA-A*29:01-B*07:05-C*15:05− group (see Table S1), suggesting that the effects of these mutations on viral fitness are minimal.
FIG 3.
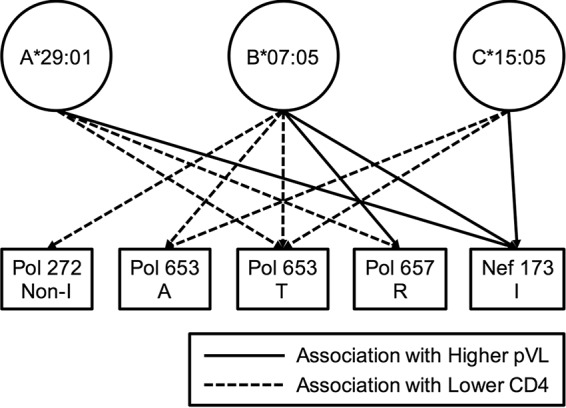
Contribution of HLA-associated mutations to clinical outcomes in individuals with HLA-A*29:01, -B*07:02, or -C*15:05. Eleven significance associations (P < 0.05) between the presence of HLA-associated mutations and the pVL or CD4 count were observed in individuals with HLA-A*29:01, -B*07:02, or -C*15:05 (see Table S2 in the supplemental material). The association was analyzed by using the Mann-Whitney test.
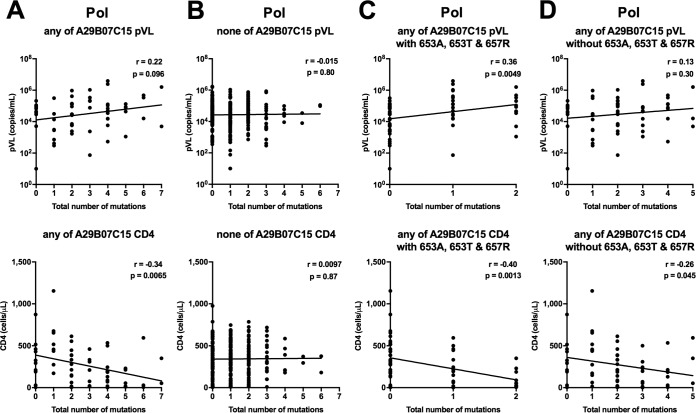
We further investigated the correlation between the number of HLA-associated mutations in Pol and pVL or CD4 count. We found a strong negative correlation between the number of HLA-associated Pol mutations and CD4 count as well as a trend toward a positive correlation with pVL for individuals having HLA-A*29:01, HLA-B*07:05, or HLA-C*15:05 but not for those without any of these alleles (Fig. 4A and B). We also found a strong negative correlation between the number of mutations and CD4 count as well as a positive correlation with pVL for the individuals with the Pol653A/T or Pol657R mutation (Fig. 4C) and a reduced correlation between the number of mutations and CD4 count or pVL for those without the Pol653A/T or Pol657R mutation (Fig. 4D). These results suggest that multiple Pol mutations, especially Pol653A/T and Pol657R, may affect the ability of specific T cells to suppress HIV-1 replication.
FIG 4.
Correlations between HLA-associated mutations at seven Pol positions and pVL or CD4 count for individuals having at least one of the HLA-A*29:01, -B*07:02, and -C*15:05 alleles (A) and those without any of them (B) were analyzed by using Spearman's correlation. Correlations between the number of mutations (0, 1, or 2) at two HLA-associated sites (Pol653A/T and Pol657R) and pVL or CD4 count (C) and between the number of mutations at six HLA-associated sites in Pol and pVL or CD4 count among individuals without the Pol653A/T and Pol657R mutations (D) were also analyzed by using Spearman's correlation. Linear regression lines are included in the plots.
DISCUSSION
We previously showed that HLA-B*52:01-C*12:02 is a protective haplotype in Japanese individuals infected with HIV-1 subtype B (10). A subsequent study demonstrated that the presence of HLA-B52:01-restricted CTLs was associated with low pVL and high CD4 T cell counts, suggesting that HLA-B*52:01 is a protective allele in Japanese infected with the subtype B virus (20). A recent study also showed that HLA-C*12:02-restricted T cells specific for two epitopes had a protective effect on HIV-1 progression to AIDS (21). In the present study of HIV-1 subtype A/E-infected Vietnamese individuals, we demonstrated that HLA-C*12:02 was significantly associated with both low pVL and high CD4 counts, whereas there was no significant association with HLA-B*52:01, although there was a trend of lower pVL and higher CD4 counts. Since the frequency of HLA-B*52:01 in Vietnamese is much lower than that in Japanese (F = 3.7% and 26.4% in Vietnamese and Japanese, respectively), this might explain the lack of a significant association of HLA-B*52:01 with pVL or CD4 count. Additional analysis with more HLA-B*52:01 samples will clarify the effect of HLA-B*52:01 in HIV-1 subtype A/E-infected Vietnamese individuals.
HLA-B*57 (B*57:01, B*57:02, and B*57:03), -B*58 (B*58:01), and -B*27 (B*27:05) are well-known protective alleles in Caucasians and Africans (1, 2, 4–9, 18). However, in this study, HLA-B*58:01 (F = 16.8%), HLA-B*57:01 (F = 4.7%), and HLA-B*27:04 (F = 2.1%) were not significantly associated with CD4 count or pVL. These findings suggest that these alleles are not protective in subtype A/E infection and imply that T cells restricted by these HLA alleles do not have a strong ability to suppress HIV-1 in subtype A/E-infected individuals. A previous Thai cohort study (n = 557) reported that individuals carrying HLA-B*35:05 (n = 17) had significantly lower pVL than those without the allele, suggesting that HLA-B*35:05 is a protective allele (15). Certain HLA-B*35 allotypes are also well known to be detrimental in HIV-1 infection (22–25). However, HLA-B*35:05 was not protective in our Hanoi cohort, even though the frequency of this allele (F = 5.2%) was higher in the Hanoi cohort than in the Thai cohort (F = 2.9%). The reason for the discrepancy between the two studies is not immediately apparent.
The present study is the first to demonstrate that HLA-A*29:01-B*07:05-C*15:05 is a detrimental haplotype in Vietnamese infected with HIV-1 subtype A/E. Since these alleles or the haplotype exist in <1% of Thai individuals, any association of these alleles with pVL or CD4 count may not have been detected in previous studies of Thai cohorts. Several HLA alleles, such as HLA-B*35, -B*53, and -B*07:02, have previously been shown to be detrimental in HIV disease progression (1, 9, 22, 23, 26), but the underlying mechanisms are not entirely known. Previous studies of HLA-B*35:01 in Africans infected with subtype C virus and Japanese, Mexican, and U.S. cohorts infected with subtype B virus demonstrated that HLA-B*35:01 has neutral and detrimental effects on the progression to AIDS in subtype C and subtype B infections, respectively (9, 10, 22, 26, 27). It was subsequently shown that the existence of Gag NY10 (Gag positions 253 to 262)-specific T cells determined the neutral effect of HLA-B*35:01 on clinical outcomes in Africans (27). This epitope is nonimmunogenic in cohorts infected with subtype B virus, as a result of the Gag-D260E mutation that is present in the majority of B clade sequences (27). Thus, the loss of T cell responses to particular epitopes might explain the lack of immune control of HIV infection in individuals with a detrimental HLA allele.
We analyzed mutations associated with HLA-A*29:01, -B*07:05 or -C*15:05 because HLA-associated mutations may affect T cell recognition. Analysis of the nef and pol genes showed that mutations at three positions in Nef and seven positions in Pol were associated with these HLA alleles. In individuals with one or more of these alleles, the presence of Nef173I, Pol272nonI, Pol653A, Pol653T, or Pol657R was associated with significantly higher pVL and/or lower CD4 counts than those of individuals without these mutations. Notably, in individuals lacking these alleles, the presence of these mutations had no significant effect on pVL or CD4 count. Together these results strongly suggest that CTLs restricted by one or more of these HLA alleles fail to control replication of these mutant viruses. This is further supported by the fact that with the increase in the number of Pol mutations present, there was a trend toward higher pVL and lower CD4 counts and that these Pol and Nef mutations are adapted mutations which are selected by HIV-1-specific CTLs (28). It is important, however, that HLA allele-restricted T cell epitopes that include the Nef173, Pol272, or Pol653/657 region have not yet been reported. Therefore, the identification of T cell epitopes in these regions that are restricted by the three HLA alleles is required to clarify the role of these mutations in the immune response to HIV.
In the present study, HLA-C*12:02 and the HLA-A*29:01-B*07:05-C*15:05 haplotype had protective and detrimental effects, respectively, on clinical outcomes for Vietnamese individuals infected with subtype A/E virus. Our previous study showed that HLA-B*52:01-C*12:02 was a protective haplotype in Japanese individuals infected with subtype B virus, whereas HLA-B*57 and HLA-B*27 were not, most likely due to the fact that these alleles are very rare in East Asian populations. Thus, protective and risk alleles are very different between Caucasians, Africans, and Asians, probably due to differences in allele frequencies across populations as well as differences in virus subtypes. These findings suggest that HIV-1 is differentially controlled by HIV-1-specific CTLs across different ethnic groups and virus subtypes, which may be important factors to consider in the development of an HIV-1 vaccine that will be globally effective.
MATERIALS AND METHODS
Subjects.
A total of 536 chronically HIV-1-infected, ART-naive Vietnamese individuals (61% and 39% of these individuals were men and women, respectively) were enrolled at their first visit to the National Hospital of Tropical Diseases (NHTD) in Hanoi from October 2012 to February 2017. All participants were adults (>18 years old) with HIV-1 infection that was confirmed by enzyme-linked immunosorbent assay (ELISA) at recruitment. Pregnant women, individuals with AIDS-related symptoms, and those with previous antiretroviral exposure were excluded. Participant CD4+ T cell counts (cells per microliter) and plasma HIV RNA loads (copies per milliliter; measured by use of the Roche Cobas TaqMan HIV monitor assay) were measured at the first visit. The median (interquartile ranges [25%/75%]) pVL and CD4+ T cell count were 48,550 (15,000/125,000) copies/ml and 284 (171/428) cells/μl, respectively.
Informed consent was obtained from all individuals according to the guidelines of the Declaration of Helsinki. This study was approved by the ethics committees of NHTD and Kumamoto University.
HLA typing.
HLA class I typing at subtype-level (four-digit) resolution was performed using a probe-based sequence-specific oligonucleotide (SSO) typing method (HLA Laboratory, Kyoto, Japan).
Identification of HLA-associated mutations and the association of these mutations with clinical markers.
We previously sequenced HIV-1 gag, pol, and nef regions in 388 Vietnamese in this cohort and analyzed the associations between HLA class I alleles and HIV-1 amino acid polymorphisms (HLA-associated HIV-1 polymorphisms [HLA-APs]) by using a phylogenetically corrected logistic regression model (19). The significance of each HLA-A*29:01-, -B*07:05-, and -C*12:02-associated mutation by 2 × 2 Fisher's exact test was determined using previously identified sequence data. The associations of these mutations with clinical markers were analyzed using nef and pol sequence data from 372 and 359 individuals, respectively, who could be analyzed for full sequences. The present study employed a P value cutoff of <0.05 to define statistical significance.
Statistical analysis.
The association of HLA alleles with the clinical outcomes pVL and CD4 count was analyzed by use of a linear regression model in the R program (P values of <0.05 were considered significant). All analyses were adjusted for HLA-A, -B, and -C alleles. Spearman's correlation was used to evaluate the relationship between the total number of HLA-associated mutations specific to each individual's HLA profiles and clinical parameters (CD4 count and pVL). The Mann-Whitney test was used to evaluate differences in the distribution of pVL or CD4 counts in the presence or absence of HLA-associated mutations.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by a JSPS KAKENHI grant-in-aid for scientific research (A) (grant 15H02658), Japan, by a grant from the Joint Usage/Research Center on Tropical Diseases, Institute of Tropical Medicine, Nagasaki University, and in part by federal funds from the National Cancer Institute, National Institutes of Health (NIH) (contract HHSN261200800001E), and the Intramural Research Program of the NIH, Frederick National Lab, Center for Cancer Research.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We have no financial conflicts of interest.
REFERENCES
- 1.Goulder PJR, Walker BD. 2012. HIV and HLA class I: an evolving relationship. Immunity 37:426–440. doi: 10.1016/j.immuni.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altfeld M, Addo MM, Rosenberg ES, Hecht FM, Lee PK, Vogel M, Yu XG, Draenert R, Johnston MN, Strick D, Allen TM, Feeney ME, Kahn JO, Sekaly RP, Levy JA, Rockstroh JK, Goulder PJ, Walker BD. 2003. Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection. AIDS 17:2581–2591. doi: 10.1097/00002030-200312050-00005. [DOI] [PubMed] [Google Scholar]
- 3.Costello C, Tang J, Rivers C, Karita E, Meizen-Derr J, Allen S, Kaslow RA. 1999. HLA-B*5703 independently associated with slower HIV-1 disease progression in Rwandan women. AIDS 13:1990. doi: 10.1097/00002030-199910010-00031. [DOI] [PubMed] [Google Scholar]
- 4.Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, Zhang K, Gumbs C, Castagna A, Cossarizza A, Cozzi-Lepri A, Luca AD, Easterbrook P, Francioli P, Mallal S, Martinez-Picado J, Miro JM, Obel N, Smith JP, Wyniger J, Descombes P, Antonarakis SE, Letvin NL, McMichael AJ, Haynes BF, Telenti A, Goldstein DB. 2007. A whole-genome association study of major determinants for host control of HIV-1. Science 317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lazaryan A, Song W, Lobashevsky E, Tang J, Shrestha S, Zhang K, Gardner LI, McNicholl JM, Wilson CM, Klein RS, Rompalo A, Mayer K, Sobel J, Kaslow RA, HIV Epidemiology Research Study, Reaching for Excellence in Adolescent Care and Health Study. 2010. Human leukocyte antigen class I supertypes and HIV-1 control in African Americans. J Virol 84:2610–2617. doi: 10.1128/JVI.01962-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leslie A, Matthews PC, Listgarten J, Carlson JM, Kadie C, Ndung'u T, Brander C, Coovadia H, Walker BD, Heckerman D, Goulder PJR. 2010. Additive contribution of HLA class I alleles in the immune control of HIV-1 infection. J Virol 84:9879–9888. doi: 10.1128/JVI.00320-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Migueles SA, Sabbaghian MS, Shupert WL, Bettinotti MP, Marincola FM, Martino L, Hallahan CW, Selig SM, Schwartz D, Sullivan J, Connors M. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci U S A 97:2709–2714. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Brien SJ, Gao X, Carrington M. 2001. HLA and AIDS: a cautionary tale. Trends Mol Med 7:379–381. doi: 10.1016/S1471-4914(01)02131-1. [DOI] [PubMed] [Google Scholar]
- 9.International HIV Controllers Study, Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PIW, Walker BD, Ripke S, Brumme CJ, Pulit SL, Carrington M, Kadie CM, Carlson JM, Heckerman D, Graham RR, Plenge RM, Deeks SG, Gianniny L, Crawford G, Sullivan J, Gonzalez E, Davies L, Camargo A, Moore JM, Beattie N, Gupta S, Crenshaw A, Burtt NP, Guiducci C, Gupta N, Gao X, Qi Y, Yuki Y, Piechocka-Trocha A, Cutrell E, Rosenberg R, Moss KL, Lemay P, O'Leary J, Schaefer T, Verma P, Toth I, Block B, Baker B, Rothchild A, Lian J, Proudfoot J, Alvino DML, Vine S, Addo MM, Allen TM, Altfeld M, Henn MR, Le Gall S, Streeck H, Haas DW, Kuritzkes DR, Robbins GK, Shafer RW, Gulick RM, Shikuma CM, Haubrich R, Riddler S, Sax PE, Daar ES, Ribaudo HJ, Agan B, Agarwal S, Ahern RL, Allen BL, Altidor S, Altschuler EL, Ambardar S, Anastos K, Anderson B, Anderson V, Andrady U, Antoniskis D, Bangsberg D, Barbaro D, Barrie W, Bartczak J, Barton S, Basden P, Basgoz N, Bazner S, Bellos NC, Benson AM, Berger J, Bernard NF, Bernard AM, Birch C, Bodner SJ, Bolan RK, Boudreaux ET, Bradley M, Braun JF, Brndjar JE, Brown SJ, Brown K, Brown ST, Burack J, Bush LM, Cafaro V, Campbell O, Campbell J, Carlson RH, Carmichael JK, Casey KK, Cavacuiti C, Celestin G, Chambers ST, Chez N, Chirch LM, Cimoch PJ, Cohen D, Cohn LE, Conway B, Cooper DA, Cornelson B, Cox DT, Cristofano MV, Cuchural G, Czartoski JL, Dahman JM, Daly JS, Davis BT, Davis K, Davod SM, DeJesus E, Dietz CA, Dunham E, Dunn ME, Ellerin TB, Eron JJ, Fangman JJW, Farel CE, Ferlazzo H, Fidler S, Fleenor-Ford A, Frankel R, Freedberg KA, French NK, Fuchs JD, Fuller JD, Gaberman J, Gallant JE, Gandhi RT, Garcia E, Garmon D, Gathe JC, Gaultier CR, Gebre W, Gilman FD, Gilson I, Goepfert PA, Gottlieb MS, Goulston C, Groger RK, Gurley TD, Haber S, Hardwicke R, Hardy WD, Harrigan PR, Hawkins TN, Heath S, Hecht FM, Henry WK, Hladek M, Hoffman RP, Horton JM, Hsu RK, Huhn GD, Hunt P, Hupert MJ, Illeman ML, Jaeger H, Jellinger RM, John M, Johnson JA, Johnson KL, Johnson H, Johnson K, Joly J, Jordan WC, Kauffman CA, Khanlou H, Killian RK, Kim AY, Kim DD, Kinder CA, Kirchner JT, Kogelman L, Kojic EM, Korthuis PT, Kurisu W, Kwon DS, LaMar M, Lampiris H, Lanzafame M, Lederman MM, Lee DM, Lee JML, Lee MJ, Lee ETY, Lemoine J, Levy JA, Llibre JM, Liguori MA, Little SJ, Liu AY, Lopez AJ, Loutfy MR, Loy D, Mohammed DY, Man A, Mansour MK, Marconi VC, Markowitz M, Marques R, Martin JN, Martin HL, Mayer KH, McElrath MJ, McGhee TA, McGovern BH, McGowan K, McIntyre D, Mcleod GX, Menezes P, Mesa G, Metroka CE, Meyer-Olson D, Miller AO, Montgomery K, Mounzer KC, Nagami EH, Nagin I, Nahass RG, Nelson MO, Nielsen C, Norene DL, O'Connor DH, Ojikutu BO, Okulicz J, Oladehin OO, Oldfield EC, Olender SA, Ostrowski M, Owen WF, Pae E, Parsonnet J, Pavlatos AM, Perlmutter AM, Pierce MN, Pincus JM, Pisani L, Price LJ, Proia L, Prokesch RC, Pujet HC, Ramgopal M, Rathod A, Rausch M, Ravishankar J, Rhame FS, Richards CS, Richman DD, Rodes B, Rodriguez M, Rose RC, Rosenberg ES, Rosenthal D, Ross PE, Rubin DS, Rumbaugh E, Saenz L, Salvaggio MR, Sanchez WC, Sanjana VM, Santiago S, Schmidt W, Schuitemaker H, Sestak PM, Shalit P, Shay W, Shirvani VN, Silebi VI, Sizemore JM, Skolnik PR, Sokol-Anderson M, Sosman JM, Stabile P, Stapleton JT, Starrett S, Stein F, Stellbrink H-J, Sterman FL, Stone VE, Stone DR, Tambussi G, Taplitz RA, Tedaldi EM, Theisen W, Torres R, Tosiello L, Tremblay C, Tribble MA, Trinh PD, Tsao A, Ueda P, Vaccaro A, Valadas E, Vanig TJ, Vecino I, Vega VM, Veikley W, Wade BH, Walworth C, Wanidworanun C, Ward DJ, Warner DA, Weber RD, Webster D, Weis S, Wheeler DA, White DJ, Wilkins E, Winston A, Wlodaver CG, van't Wout A, Wright DP, Yang OO, Yurdin DL, Zabukovic BW, Zachary KC, Zeeman B, Zhao M. 2010. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naruto T, Gatanaga H, Nelson G, Sakai K, Carrington M, Oka S, Takiguchi M. 2012. HLA class I-mediated control of HIV-1 in the Japanese population, in which the protective HLA-B*57 and HLA-B*27 alleles are absent. J Virol 86:10870–10872. doi: 10.1128/JVI.00689-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buonaguro L, Tornesello ML, Buonaguro FM. 2007. Human immunodeficiency virus type 1 subtype distribution in the worldwide epidemic: pathogenetic and therapeutic implications. J Virol 81:10209–10219. doi: 10.1128/JVI.00872-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau KA, Wang B, Saksena NK. 2007. Emerging trends of HIV epidemiology in Asia. AIDS Rev 9:218–229. [PubMed] [Google Scholar]
- 13.Nikolopoulos GK, Kostaki E-G, Paraskevis D. 2016. Overview of HIV molecular epidemiology among people who inject drugs in Europe and Asia. Infect Genet Evol 46:256–268. doi: 10.1016/j.meegid.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hemelaar J, Gouws E, Ghys PD, Osmanov S. 2006. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS 20:W13–W23. doi: 10.1097/01.aids.0000247564.73009.bc. [DOI] [PubMed] [Google Scholar]
- 15.Mori M, Wichukchinda N, Miyahara R, Rojanawiwat A, Pathipvanich P, Maekawa T, Miura T, Goulder P, Yasunami M, Ariyoshi K, Sawanpanyalert P. 2014. HLA-B*35:05 is a protective allele with a unique structure among HIV-1 CRF01_AE-infected Thais, in whom the B*57 frequency is low. AIDS 28:959–967. doi: 10.1097/QAD.0000000000000206. [DOI] [PubMed] [Google Scholar]
- 16.Gandhi RT, Bosch RJ, Rangsin R, Chuenchitra T, Sirisopana N, Kim JH, Robb ML, Vejbaesya S, Paris RM, Nelson KE. 2016. HLA class I alleles associated with mortality in Thai military recruits with HIV-1 CRF01_AE infection. AIDS Res Hum Retroviruses 32:44–49. doi: 10.1089/aid.2015.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kostyu DD, Hannick LI, Traweek JL, Ghanayem M, Heilpern D, Dawson DV. 1997. HLA class I polymorphism: structure and function and still questions. Hum Immunol 57:1–18. doi: 10.1016/S0198-8859(97)00175-4. [DOI] [PubMed] [Google Scholar]
- 18.Costello C, Tang J, Rivers C, Karita E, Meizen-Derr J, Allen S, Kaslow RA. 1999. HLA-B*5703 independently associated with slower HIV-1 disease progression in Rwandan women. AIDS 13:1990–1991. doi: 10.1097/00002030-199910010-00031. [DOI] [PubMed] [Google Scholar]
- 19.Van Tran G, Chikata T, Carlson JM, Murakoshi H, Nguyen DH, Tamura Y, Akahoshi T, Kuse N, Sakai K, Sakai S, Cobarrubias K, Oka S, Brumme ZL, Van Nguyen K, Takiguchi M, NHTD Treatment-Naive Cohort Study Group. 2016. A strong association of human leukocyte antigen-associated Pol and Gag mutations with clinical parameters in HIV-1 subtype A/E infection. AIDS 30:681–689. doi: 10.1097/QAD.0000000000000969. [DOI] [PubMed] [Google Scholar]
- 20.Murakoshi H, Akahoshi T, Koyanagi M, Chikata T, Naruto T, Maruyama R, Tamura Y, Ishizuka N, Gatanaga H, Oka S, Takiguchi M. 2015. Clinical control of HIV-1 by cytotoxic T cells specific for multiple conserved epitopes. J Virol 89:5330–5339. doi: 10.1128/JVI.00020-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chikata T, Murakoshi H, Koyanagi M, Honda K, Gatanaga H, Oka S, Takiguchi M. 2017. Control of HIV-1 by an HLA-B*52:01-C*12:02 protective haplotype. J Infect Dis 216:1415–1424. doi: 10.1093/infdis/jix483. [DOI] [PubMed] [Google Scholar]
- 22.Carrington M, Nelson GW, Martin MP, Kissner T, Vlahov D, Goedert JJ, Kaslow R, Buchbinder S, Hoots K, O'Brien SJ. 1999. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 283:1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- 23.Gao X, Nelson GW, Karacki P, Martin MP, Phair J, Kaslow R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, O'Brien SJ, Carrington M. 2001. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N Engl J Med 344:1668–1675. doi: 10.1056/NEJM200105313442203. [DOI] [PubMed] [Google Scholar]
- 24.Jin X, Gao X, Ramanathan M, Deschenes GR, Nelson GW, O'Brien SJ, Goedert JJ, Ho DD, O'Brien TR, Carrington M. 2002. Human immunodeficiency virus type 1 (HIV-1)-specific CD8+-T-cell responses for groups of HIV-1-infected individuals with different HLA-B*35 genotypes. J Virol 76:12603–12610. doi: 10.1128/JVI.76.24.12603-12610.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carrington M, O'Brien SJ. 2003. The influence of HLA genotype on AIDS. Annu Rev Med 54:535–551. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 26.Juarez-Molina CI, Valenzuela-Ponce H, Avila-Rios S, Garrido-Rodriguez D, Garcia-Tellez T, Soto-Nava M, Garcia-Morales C, Goulder P, Reyes-Teran G. 2014. Impact of HLA-B*35 subtype differences on HIV disease outcome in Mexico. AIDS 28:1687–1690. doi: 10.1097/QAD.0000000000000322. [DOI] [PubMed] [Google Scholar]
- 27.Matthews PC, Koyanagi M, Kløverpris HN, Harndahl M, Stryhn A, Akahoshi T, Gatanaga H, Oka S, Juarez Molina C, Valenzuela-Ponce H, Avila-Rios S, Cole D, Carlson J, Payne RP, Ogwu A, Bere A, Ndung'u T, Gounder K, Chen F, Riddell L, Luzzi G, Shapiro R, Brander C, Walker B, Sewell AK, Reyes-Teran G, Heckerman D, Hunter E, Buus S, Takiguchi M, Goulder PJR. 2012. Differential clade-specific HLA-B*3501 association with HIV-1 disease outcome is linked to immunogenicity of a single Gag epitope. J Virol 86:12643–12654. doi: 10.1128/JVI.01381-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carlson JM, Le AQ, Shahid A, Brumme ZL. 2015. HIV-1 adaptation to HLA: a window into virus-host immune interactions. Trends Microbiol 23:212–224. doi: 10.1016/j.tim.2014.12.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.