ABSTRACT
RNA-remodeling proteins, including RNA helicases and chaperones, act to remodel RNA structures and/or protein-RNA interactions and are required for all processes involving RNAs. Although many viruses encode RNA helicases and chaperones, their in vitro activities and their roles in infected cells largely remain elusive. Noroviruses are a diverse group of positive-strand RNA viruses in the family Caliciviridae and constitute a significant and potentially fatal threat to human health. Here, we report that the protein NS3 encoded by human norovirus has both ATP-dependent RNA helicase activity that unwinds RNA helices and ATP-independent RNA-chaperoning activity that can remodel structured RNAs and facilitate strand annealing. Moreover, NS3 can facilitate viral RNA synthesis in vitro by norovirus polymerase. NS3 may therefore play an important role in norovirus RNA replication. Lastly, we demonstrate that the RNA-remodeling activity of NS3 is inhibited by guanidine hydrochloride, an FDA-approved compound, and, more importantly, that it reduces the replication of the norovirus replicon in cultured human cells. Altogether, these findings are the first to demonstrate the presence of RNA-remodeling activities encoded by Caliciviridae and highlight the functional significance of NS3 in the noroviral life cycle.
IMPORTANCE Noroviruses are a diverse group of positive-strand RNA viruses, which annually cause hundreds of millions of human infections and over 200,000 deaths worldwide. For RNA viruses, cellular or virus-encoded RNA helicases and/or chaperones have long been considered to play pivotal roles in viral life cycles. However, neither RNA helicase nor chaperoning activity has been demonstrated to be associated with any norovirus-encoded proteins, and it is also unknown whether norovirus replication requires the participation of any viral or cellular RNA helicases/chaperones. We found that a norovirus protein, NS3, not only has ATP-dependent helicase activity, but also acts as an ATP-independent RNA chaperone. Also, NS3 can facilitate in vitro viral RNA synthesis, suggesting the important role of NS3 in norovirus replication. Moreover, NS3 activities can be inhibited by an FDA-approved compound, which also suppresses norovirus replicon replication in human cells, raising the possibility that NS3 could be a target for antinoroviral drug development.
KEYWORDS: RNA chaperone, RNA helicase, norovirus
INTRODUCTION
Noroviruses are a genetically diverse group of nonenveloped, positive-strand RNA viruses belonging to the genus Norovirus of the family Caliciviridae. They are further segregated into seven genogroups (GI to GVII), three of which (GI, GII, and GIV) cause diseases exclusively in humans (1–3). Human noroviruses (HuNoVs) are now considered the leading cause of viral gastroenteritis in humans across the globe. HuNoV infections are estimated to cause 684 million cases of diarrheal disease, 1.1 million hospitalizations, and 212,000 deaths annually worldwide, with an economic burden of approximately $60 billion per year (4, 5). They are highly contagious, infecting people of all ages, with higher incidence in young children and low-income settings (4, 5). Although HuNoV infections are usually acute and self-limiting with low mortality, they often cause chronic or life-threatening infections and symptoms in immunocompromised patients and the elderly (6). As a result of their high infectivity, stability, and resistance to common sanitizers, HuNoV has also been classified as a category B biodefense agent (6, 7). However, despite its substantial burden and potential threat, there are still no vaccines or antiviral drugs available to prevent or treat norovirus infection (4, 5, 8).
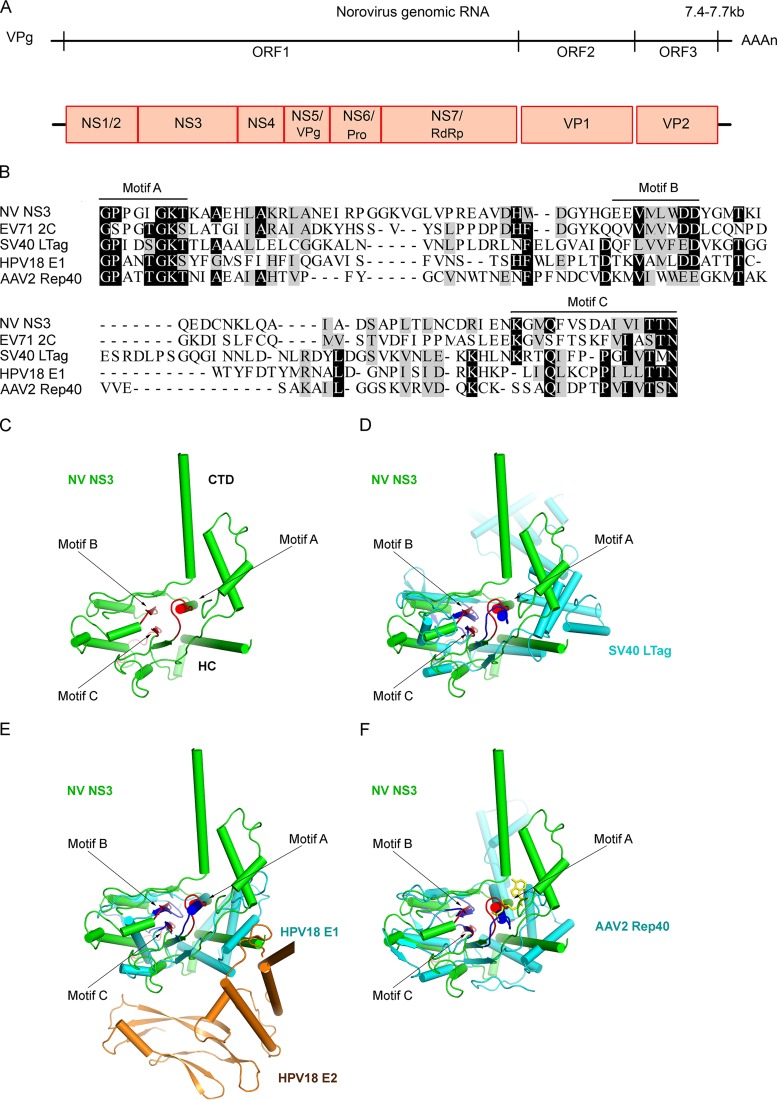
The noroviral genomic RNA is 7.4 to 7.7 kb in length and is typically organized into three open reading frames (ORF1 to -3) (8). Exceptionally, the GV murine norovirus (MNV) has an additional ORF (ORF4) within ORF2 (9, 10). The 5′-proximal ORF1 encodes a large polyprotein that is cleaved by a norovirus-encoded protease (NS6Pro) into at least six mature nonstructural proteins, while the 3′-proximal region of noroviral genomic RNA is transcribed into a subgenomic RNA that contains ORF2 and ORF3, which encode the major and minor structural proteins VP1 and VP2, respectively (Fig. 1A). Although these viruses have been extensively studied for decades, our understanding of the mechanisms of the nonstructural proteins involved in norovirus replication is very limited, particularly in comparison to other positive-strand RNA viruses, such as picornaviruses and flaviviruses.
FIG 1.
Norovirus NS3 is similar to other virus-encoded SF3 helicases in consensus motifs and protein structures. (A) The genomic RNA of norovirus contains three ORFs (ORF1, -2, and -3). The 5′-proximal ORF1 encodes a polyprotein that is subsequently self-cleaved into six nonstructural proteins (NS1/2, NS3, NS4, NS5/VPg, NS6Pro, and NS7/RdRP) by viral protease NS6Pro. ORF2 and ORF3 encode structural proteins VP1 and VP2, respectively. (B) Sequence alignment of the helicase core domains of NV NS3 and other SF3 viral helicases, EV71 2CATPase, SV40 LTag, HPV18 E1, and AAV2 Rep40. The SF3 consensus motifs A, B, and C are indicated. Completely conserved residues (100% identity) are highlighted in black, and highly conserved residues (≥50% identity) are highlighted in gray. Dashes indicate no amino acid and are included for alignment purposes. (C) The structure of NV NS3 without its N-terminal domain (NS3ΔN) was predicted by the HMMSTR/Rosetta server. NS3ΔN containing the middle HC domain and the CTD is shown in green. SF3 motifs A, B, and C are indicated and highlighted in red. (D to F) Structural alignments of NV NS3ΔN (green) and SV40 LTag, HPV18E1, or AAV2 Rep40 (cyan). The N-terminal domain of E2 binding to HPV18 E1 is shown in brown. SF3 motifs A, B, and C of these viral helicases are highlighted in blue.
For many RNA viruses, the viral RNA (vRNA) contains multiple cis-acting elements that play pivotal roles in vRNA replication, translation, and encapsidation during the viral life cycle (11–13). As with cellular RNA molecules, these elements are often highly structured in vRNAs and must be folded into the correct tertiary structures to be functional. However, this can be problematic, because RNAs can be easily trapped in stable but misfolded intermediate states (14, 15). To avoid this, cells and viruses encode a variety of RNA-remodeling proteins, such as RNA helicases and RNA chaperones, which can destabilize RNA-RNA or RNA-DNA duplexes to promote the proper folding or refolding of RNAs (16–18). In addition, viral or cellular RNA helicases are thought to participate in the unwinding of viral replicative-intermediate double-stranded RNAs (vRI-dsRNAs) formed during vRNA replication, thereby allowing recycling of vRNA templates for further rounds of vRNA synthesis. RNA helicases have nucleoside triphosphatase (NTPase) activity, using the energy of NTP binding and/or hydrolysis to unwind RNA duplexes, and are believed to participate in most ATP-dependent rearrangement of structured RNAs (19). RNA chaperones are a heterogeneous group of proteins that can destabilize RNA duplexes and promote correct RNA folding in an ATP-independent fashion (20). A wide range of RNA viruses, including flavivirus (21), picornavirus (22), alphavirus (23), coronavirus (24), reovirus (25), iflavirus (26), and alphatetravirus (27), have been found to encode their own RNA helicases and/or RNA chaperones (28).
Sequence analysis showed that the norovirus NS3 protein, also known as p41, contains signature motifs that are conserved in superfamily 3 (SF3) viral helicases, such as the human enterovirus 2CATPase (EV71 2CATPase) (22), simian virus 40 (SV40) large T antigen (LTag) (29), adeno-associated virus 2 (AAV2) Rep40 (30, 31), and the human papillomavirus 18 (HPV18) E1 protein (32) (Fig. 1). Consistent with this observation, the NS3 protein encoded by Southampton virus (SHV), a GI HuNoV, has been shown to have NTPase activity (33). In addition, norovirus NS3 has been recently reported to associate with vRI-dsRNAs in a viral replication complex (vRC) and to induce vesicular structures (34–36). However, previous work has yet to demonstrate RNA helicase or chaperoning activity associated with NS3 or any other norovirus- or calicivirus-encoded proteins or that the replication of norovirus or calicivirus requires the participation of any viral or host RNA-remodeling activities. This gap hampers our understanding of the replication mechanisms of this large group of medically important human pathogens, as well as the development of antinoroviral drugs that may specifically target viral RNA helicase/chaperoning activities.
Our previous work on enterovirus 2CATPase revealed that, in vitro, 2CATPase has RNA-remodeling activity when expressed using a baculovirus-based expression system (22). Here, we show that NS3 encoded by Norwalk virus (NV) (genotype GI.1 HuNoV), the prototype strain of the genus Norovirus, also has NTP-dependent RNA helicase activity. Moreover, we demonstrate that NS3 can also act as an RNA chaperone that is able to remodel structured RNAs and facilitate strand annealing independently of NTP. We have also found that NS3 can facilitate the in vitro synthesis of vRNA by NV NS7/RNA-dependent RNA polymerase (RdRP) on the 3′ antigenomic template, suggesting that NS3 plays an important role in norovirus RNA replication. Additionally, we have demonstrated that guanidine hydrochloride (GuHCl), which is a U.S. FDA-approved small-molecule drug and a well-known inhibitor of poliovirus 2CATPase, is able to inhibit the RNA helicase activity of NS3 in a dose-dependent manner. More importantly, GuHCl has been further determined to inhibit the replication of the NV replicon in cultured human cells, which highlights the functional significance of NS3 in the noroviral life cycle.
RESULTS
NV NS3 (NS3NV) shares similar consensus motifs and structure with other SF3 viral helicases.
A comparison of the amino acid sequence of NV NS3 with those of members of the SF3 viral helicases, including EV71 2CATPase, AAV2 Rep40, SV40 LTag, and HPV18 E1, revealed that NV NS3 contains the conserved SF3 helicase A, B, and C motifs (Fig. 1B).
Since the three-dimensional (3D) structure of norovirus NS3 has not yet been reported, we modeled the NV NS3 structure using the ROBETTA server for protein structure prediction and analysis (37). The predicted model of NV NS3 revealed that the C-terminal two-thirds, consisting of amino acids at positions 122 to 363 (i.e., NS3ΔN), is comprised of two structurally independent domains: a helicase core (HC) (amino acids [aa] 122 to 288) forming a central five-stranded β-sheet sandwiched by α-helices on both sides, and the C-terminal domain (CTD) (aa 289 to 363) comprising several α-helices. These domains are linked by flexible loops (Fig. 1C) and, interestingly, demonstrate limited similarity with the counterpart region of EV71 2CATPase (22). Moreover, the predicted SF3 motifs A, B, and C of NV NS3 nicely overlap the conserved SF3 motifs in these other SF3 viral helicases (Fig. 1D to F).
NV NS3 contains NTPase activity.
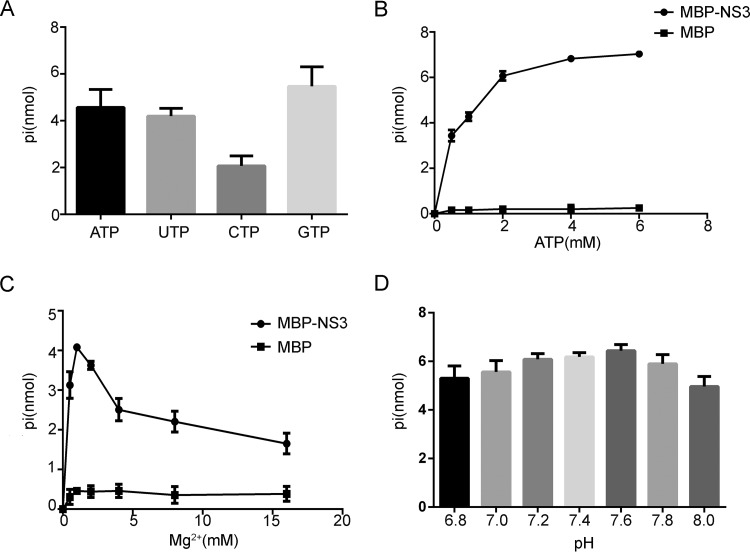
Previous studies by Pfister and Wimmer found that bacterially expressed SHV NS3 has NTPase activity (33). To confirm whether NV NS3 also has this activity, we expressed a recombinant maltose binding protein (MBP) fusion with NV NS3 (MBP-NS3) using a baculovirus expression system and then examined the NTPase activity by incubating MBP-NS3 with different NTPs. The hydrolysis of NTP was measured using a sensitive colorimetric assay that determines the total amount of released inorganic phosphate. As was found for SHV NS3, our data showed that NV NS3 hydrolyzed all four types of NTPs (Fig. 2A). However, although SHV NS3 was reported to hydrolyze UTP less well than ATP (33), NV NS3 exhibited similar efficiency in the hydrolysis of these NTPs (Fig. 2A).
FIG 2.
NV NS3 has NTPase activity. (A) MBP-NS3 was reacted with the indicated NTPs. The NTPase activity was measured as nanomoles of released inorganic phosphate. (B to D) The NTPase activity of MBP-NS3 was determined at the indicated concentrations of ATP (B), at the indicated concentrations of Mg2+ (C), or at the indicated pH (D). (B and C) MBP alone was used as the negative control. (A to D) The error bars represent standard deviation (SD) values from the results of three separate experiments.
To further characterize the NTPase activity of NV NS3, we assessed the ATPase activity of MBP-NS3 under different ATP and Mg2+ concentrations, as well as at different pHs, as ATP is the major energy source in cells. Our results showed that the amount of hydrolyzed ATP increased with increasing amounts of ATP (Fig. 2B). In addition, our data indicated that the ATPase activity of MBP-NS3 is Mg2+ dependent, since its ATP-hydrolyzing activity was undetectable in the absence of Mg2+. While the ATPase activity of MBP-NS3 was found to be optimal at a 2 mM Mg2+ concentration, higher levels of Mg2+ inhibited the ATPase activity (Fig. 2C). The optimal pH for MBP-NS3 ATPase activity was found to be ∼7.6, but broad activity was maintained between pH 6.8 and pH 8.0 (Fig. 2D).
We also expressed the MBP fusion with NV NS3 using Escherichia coli and examined its NTPase activity. We confirmed that the MBP-NS3NV expressed by bacteria can also hydrolyze ATP and other NTPs and shows biochemical properties similar to those of the MBP-NS3NV expressed by eukaryotes (data not shown). Altogether, our data demonstrate that, similar to SHV NS3, NV NS3 has NTPase activity that hydrolyzes all four NTPs and is Mg2+ dependent.
NV NS3 has nucleic acid helix-unwinding activity.
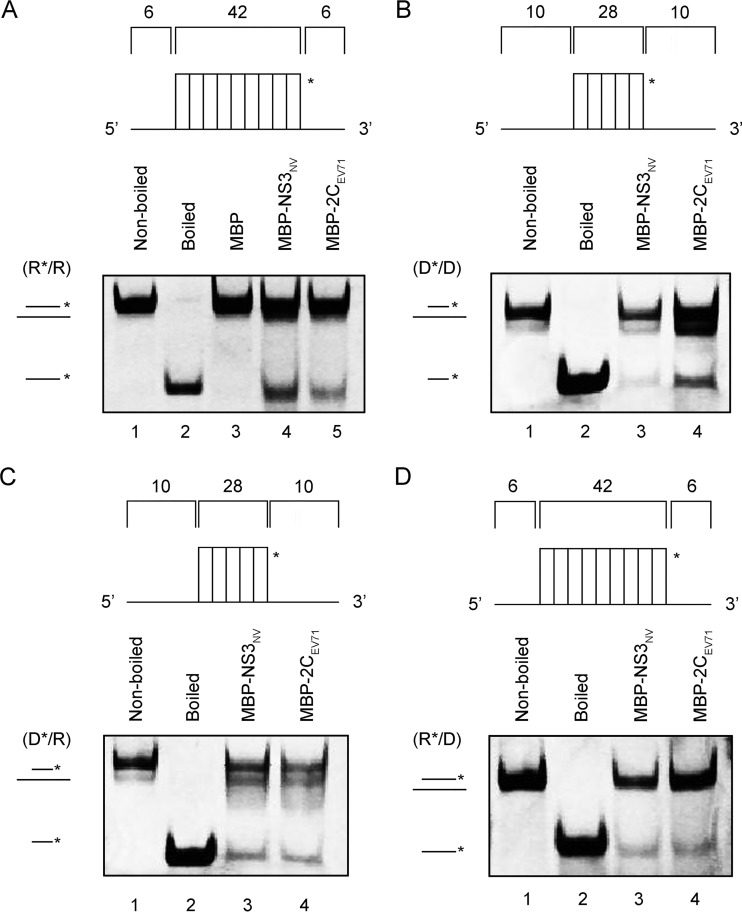
To assess the potential helix-unwinding activity of NV NS3, we annealed a short hexachlorofluorescein (HEX)-labeled RNA and a longer unlabeled RNA to generate a standard RNA helix substrate with both 5′ and 3′ single-strand protrusions (Fig. 3A and Table 1). The helix-unwinding assay was performed by incubating the standard RNA helix substrate with purified MBP-NS3 in the standard unwinding reaction and then evaluating the substrate via gel electrophoresis. As shown in Fig. 3A, the HEX-labeled RNA strand was released from the RNA helix substrate in the presence of MBP-NS3, whereas the RNA helix substrate remained annealed when MBP alone was added as a mock helicase, showing that NV NS3 can unwind the RNA helix. The reactions were performed in the presence of ATP and MgCl2. The boiled helix substrate or the addition of an MBP fusion with EV71 2CATPase was used here and in subsequent experiments as a relevant control for helix-unwinding activity.
FIG 3.
NV NS3 has nucleic acid helix-unwinding activity. (A) The standard RNA/RNA helix (R*/R substrate) is illustrated in the upper diagram. R*/R substrate (0.1 pmol) was reacted with 20 pmol of each indicated protein. Nonboiled reaction mixture (lane 1) and reaction mixture with MBP alone (lane 3) were used as negative controls, and boiled reaction mixture (lane 2) and reaction mixture with purified MBP fusion with EV71 2CATPase (lane 5) were used as positive controls. (B to D) DNA/DNA helix (D*/D substrate), DNA/RNA hybrid helix (D*/R), and RNA/DNA hybrid helix (R*/D) are illustrated in the upper diagrams; 0.1 pmol of each indicated substrate was reacted with 20 pmol of each indicted protein. Lanes 1, nonboiled reaction mixture; lanes 2, boiled reaction mixture; lanes 3, reaction mixture with purified MBP-NS3 fusion; lanes 4, reaction mixture with purified MBP-EV71 2CATPase fusion. Helix unwinding was detected via gel electrophoresis and scanning on a Typhoon 9500 imager. The asterisks indicate the HEX-labeled strands.
TABLE 1.
Oligonucleotides
| Oligonucleotide | Sequence (5′–3′)a |
|---|---|
| RNA1 | CAUUAUCGGAUAGUGGAACCUAGCUUCGACUAUCGGAUAAUC |
| RNA2 | AAUAAAGAUUAUCCGAUAGUCGAAGCUAGGUUCCACUAUCCGAUAAUGAAAUAA |
| RNA3 | UGUAGUGCUGCCAUGGUGUGGUGGUGGUGGUUGUGGUGGAGCUACGAAC |
| RNA4 | GAUUAUCCGAUAGUCGAAGCUAGGUUCCACUAUCCGAUAAUGAAAUAA |
| RNA5 | AAUAAAGAUUAUCCGAUAGUCGAAGCUAGGUUCCACUAUCCGAUAAUG |
| DNA1 | AATAAAGATTATCCGATAGTCGAAGCTAGGTTCCACTATCCGATAATGAAATAA |
| DNA2 | CACCACAACCACCACCACCACACCATGG |
| DNA3 | TGTAGTGCTGCCATGGTGTGGTGGTGGTGGTTGTGGTGGAGCTACGAAC |
| Probe | GUGAAUGAAGAUGGCGUCUAACGGCGCUUCCGCUGCCGCUGUUGCCAACAGCAACAACGACAUCGCAAAAUCUUCAAGUGACGGUGUGUUUUCUAACAUGGCUGUCACUUUUAAGCGGGCCCUCGGGGCG CGGCCUAAAC |
HEX-labeled strands are in boldface.
Because some viral RNA helicases have been reported to unwind DNA helices or RNA-DNA hybrids (22), we sought to examine if NV NS3 can unwind nucleic acid helices containing DNA. For this purpose, we constructed three helix substrates: a DNA duplex (D*/D) (Fig. 3B) and RNA-DNA hybrids with a longer unlabeled RNA (D*/R) (Fig. 3C) or DNA (R*/D) (Fig. 3D) strand. Each helix substrate was reacted with MBP-NS3 under the same condition as for Fig. 3A, and our results showed that NV NS3 could efficiently unwind either of the RNA-DNA hybrids (Fig. 3C and D) but unwound the DNA duplexes less effectively (Fig. 3B).
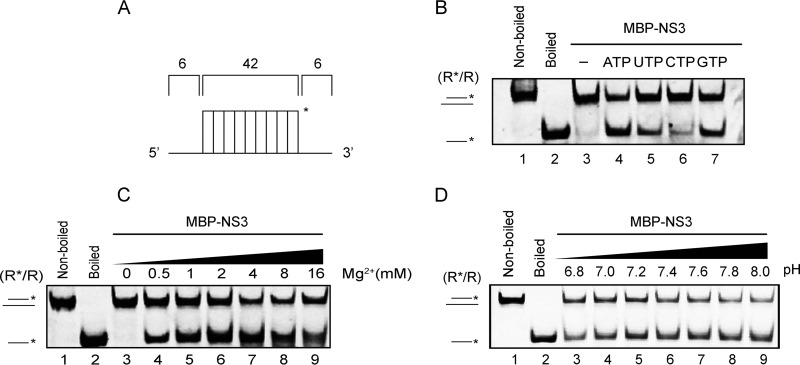
We further evaluated the RNA helix-unwinding activity of MBP-NS3 under different conditions. We observed that the helix-unwinding activity of MBP-NS3 can be promoted by the presence of NTP, particularly ATP or GTP (Fig. 4B), indicating that NV NS3 has an RNA helicase activity. Consistent with the characterization of NS3 NTPase activity (Fig. 2C and D), the optimized helix-unwinding activity of MBP-NS3 was found to be around 2 mM Mg2+ (Fig. 4C) at a pH value of around 7.6 (Fig. 4D). Collectively, these findings suggest that NV NS3 has NTP-dependent nucleic acid helix-unwinding activity.
FIG 4.
Biochemical characterization of RNA helix-unwinding activity of NV NS3. (A) Illustration of the standard RNA helix (R*/R) substrate. (B to D) R*/R substrate (0.1 pmol) was reacted with 20 pmol MBP-NS3 in the absence or presence of individual NTP (B), increasing concentrations of Mg2+ (C), or different pH values (D) as indicated. Nonboiled (lanes 1) or boiled (lanes 2) reaction mixture was used as a negative or positive control. The asterisks indicate the HEX-labeled strands. Helix-unwinding activity was detected by gel electrophoresis and scanning on a Typhoon 9500 imager.
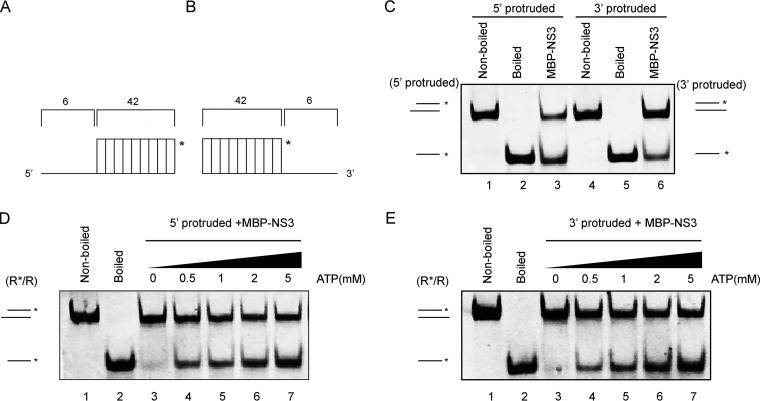
NV NS3 directs RNA helix unwinding from both 5′→3′ and 3′→5′ directions.
The directionality of helix unwinding is a fundamental characteristic of helicases (19). Previous studies of other SF3 viral helicases have revealed that the helicase activity of EV71 2CATPase has a 3′→5′ directionality, while AAV2 Rep40 and HPV18 E1 unwind helices bidirectionally (22, 31, 32). To determine the unwinding directionality of NV NS3, we generated two different RNA helix substrates, one containing a 5′ single-strand protrusion (Fig. 5A) and the other with a 3′ single-strand protrusion (Fig. 5B). Each substrate was subjected to a helix-unwinding assay in the presence of MBP-NS3. Our data showed that RNA helix substrates with either a 5′ or a 3′ protrusion could be unwound by MBP-NS3 (Fig. 5C). Moreover, the unwinding of either substrate by MBP-NS3 could be promoted by the presence of ATP in a dose-dependent manner (Fig. 5D and E). Based on these observations, we conclude that NV NS3 directs the unwinding of the RNA helix from both 5′→3′ and 3′→5′ directions in an ATP-dependent manner, similar to AAV2 Rep40, another SF3 viral helicase (22, 31, 32).
FIG 5.
NV NS3 unwinds the RNA helix in a bidirectional manner. (A and B) Schematic illustration of the RNA helix substrate with the 5′ single-strand protrusion (A) or 3′ single-strand protrusion (B). The asterisks indicate the HEX-labeled strands. (C) MBP-NS3 (20 pmol) was incubated with 5′ single-strand protruded (lane 3) or 3′ single-strand protruded (lane 6) RNA helix substrate (0.1 pmol). Nonboiled (lanes 1 and 4) or boiled (lanes 2 and 5) reaction mixtures were used as negative or positive controls. (D and E) MBP-NS3 (20 pmol) was reacted with 5′-protruded (D) or 3′-protruded (E) RNA helix substrate in the presence of the indicated concentrations of ATP. Helix unwinding was detected as described for Fig. 3.
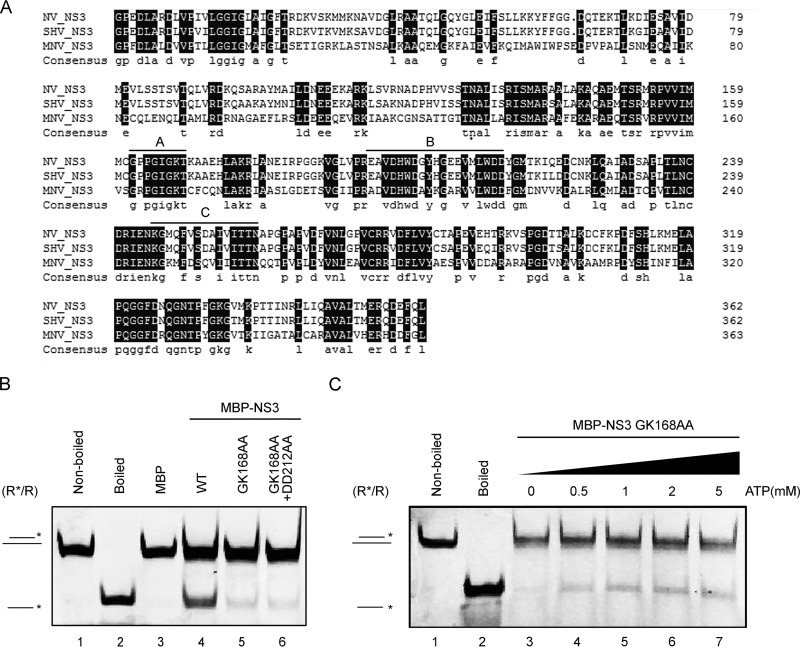
The conserved SF3 A motif is critical for the helicase activity of NS3.
The sequence analysis of NS3 indicated that NS3 contains consensus motifs that are characteristic of SF3 viral helicases (Fig. 1). Motif A has been recognized as a core element of the NTP binding and NTPase active site of SF3 helicases (19) and is conserved in different noroviruses (Fig. 6A). Previous work by Pfister and Wimmer has shown that the mutation of motif A in SHV NS3 almost completely eliminated its ATPase activity (33). Moreover, motif B of SF3 helicases has been reported to chelate Mg2+ (19). We wanted to investigate if the conserved SF3 motif A or B is important for the helicase activity associated with norovirus NS3. To this end, we generated the GK168AA and GK168AA-DD212AA mutants of the MBP-NS3 fusion (MBP-NS3GK168AA and MBP-NS3GK168AA-DD212AA) and expressed the proteins using the baculovirus expression system. In our standard helix-unwinding assays, we found that both the GK168AA and GK168AA-DD212AA mutations dramatically reduced the helix-unwinding activity of NS3 compared to the wild type (WT) (Fig. 6B). Moreover, the addition of increasing concentrations of ATP had very little effect on helix unwinding by MBP-NS3GK168AA (Fig. 6C). These findings show that the conserved SF3 motifs are critical for the helicase activity of NS3.
FIG 6.
SF3 motif A is critical for the helicase activity of NV NS3. (A) Amino acid sequence alignment of NV NS3, SHV NS3, and MNV NS3. The SF3 consensus motifs A, B, and C are indicated. Completely conserved residues (100% identity) are highlighted in black. (B) The standard RNA helix (R*/R) substrate (0.1 pmol) was reacted with 20 pmol WT (lane 4), GK168AA (lane 5), or GK168AA-DD212AA (lane 6) MBP-NS3. Nonboiled reaction mixture (lane 1) and reaction mixture with addition of MBP alone (lane 3) were used as negative controls; the boiled reaction mixture (lane 2) was used as a positive control. (C) The standard RNA helix (R*/R) substrate (0.1 pmol) was reacted with 20 pmol GK168AA mutant MBP-NS3 in the presence of the indicated concentrations of ATP. (B and C) The asterisks indicate the HEX-labeled strands.
NV NS3 also possesses NTP-independent RNA-chaperoning activity.
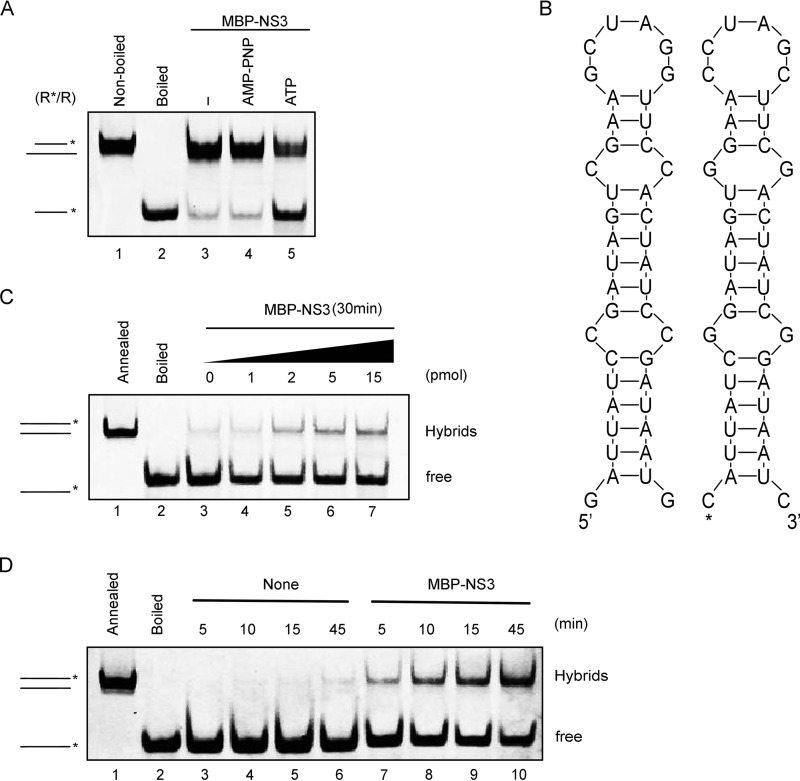
Although we had determined that NV NS3 requires NTP to reach its optimal helix-unwinding activity (Fig. 4B and 5D and E) and that the conserved SF3 A motif is critical for its helicase activity (Fig. 6), we found that a portion of the helical RNA substrate could still be unwound by MBP-NS3 in the absence of ATP (Fig. 4B and 5D and E). To confirm these observations and exclude the possible interference of trace amounts of NTP in the unwinding assays, we alternatively used AMP-PNP, an ATP analog that is nonhydrolyzable and able to block NTPase activity. The incorporation of 5 mM AMP-PNP instead of ATP or other NTPs did not completely abolish RNA helix unwinding by MBP-NS3 (Fig. 7A). While both RNA helicases and RNA chaperones possess RNA-remodeling activities, the fundamental difference between them is that the former activity requires the participation of NTP whereas the latter is NTP independent (20). These observations, therefore, suggest that NV NS3 also contains NTP-independent RNA-chaperoning activity.
FIG 7.
NV NS3 has RNA-chaperoning activity to destabilize structured RNA strands and promote annealing. (A) The standard RNA helix (R*/R) substrate (0.1 pmol) was reacted with 20 pmol MBP-NS3 in the absence or presence of 5 mM ATP or ATP analog (AMP-PNP) as indicated. Lane 1, nonboiled reaction mixture; lane 2, boiled reaction mixture; lane 3, reaction mixture with MBP-NS3 in the absence of ATP; lanes 4 and 5, reaction mixture with MBP-NS3 in the presence of AMP-PNP or ATP. (B) Schematic illustrations of the stem-loop structures of the two complementary 42-nt RNA strands. The asterisk indicates the HEX-labeled strand. (C) The two strands were mixed 1:1 (0.1 pmol each) and reacted with the indicated amounts of MBP-NS3. (D) The hybridization assay (as for panel C) was conducted in the absence (lanes 3 to 6) or presence (lanes 7 to 10) of 20 pmol MBP-NS3 for the indicated reaction times. (C and D) The preannealed (lanes 1) or boiled (lanes 2) strands were used as a positive or negative control, respectively. The hybridized and free strands are indicated.
RNA chaperones are generally thought to destabilize misfolded RNA secondary structures to facilitate the re-formation of more stable RNA structures (15). To further characterize the RNA-chaperoning activity of NV NS3, we adapted a canonical assay that evaluates the helix-destabilizing and annealing acceleration activities of RNA chaperones (38). In this assay, we designed and generated two 42-nucleotide (nt) cRNA strands, each of which forms a defined stem-loop secondary structure; in addition, one strand is 5′ HEX labeled and the other is unlabeled, as indicated in Fig. 7B. Equal amounts of the two strands were mixed and incubated in the presence or absence of MBP-NS3, and the hybridization of the two strands was then determined using a gel shift assay. In these reactions, ATP or other NTPs were not supplemented. Our data showed that the levels of the annealed hybrids gradually increased with the increasing amounts of MBP-NS3 (Fig. 7C). Moreover, although minimal spontaneous hybridization could be observed in the absence of MBP-NS3 with prolonged incubation times (Fig. 7D, lanes 3 to 6), the addition of MBP-NS3 further enhanced the hybridization at each time point (Fig. 7D, lanes 7 to 10). Altogether, our results show that NV NS3 can destabilize structured RNAs and facilitate the formation of a more stable RNA structure, confirming that NV NS3 is also an NTP-independent RNA chaperone.
NV NS3 promotes noroviral RNA synthesis in vitro.
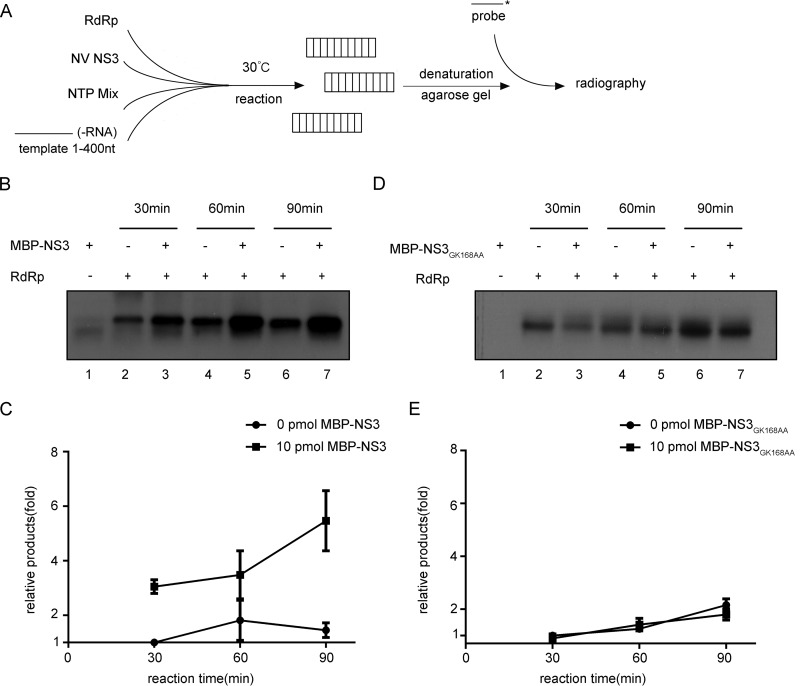
For positive-strand RNA viruses, their RdRP-mediated vRNA replication generates viral replicative-intermediate double-stranded RNAs that could be unwound by RNA helicases in order to recycle vRNA templates (39). Virus-encoded RNA chaperones may help rearrange the structured RNA elements of vRNA templates, thereby facilitating vRNA synthesis (25). After determining the RNA helicase and chaperoning activities of NV NS3, we wanted to evaluate the potential role of NS3 in norovirus RNA replication. As illustrated in Fig. 8A, we incubated a recombinantly purified MBP fusion with HuNoV NS7/RdRP protein with in vitro-transcribed RNA corresponding to the 400 nt at the 3′ end of HuNoV antigenomic (−)-vRNA, which is presumably the template for the initiation of (+)-vRNA synthesis, in the presence or absence of WT or GK168AA mutant MBP-NS3. The reaction mixture was not supplemented with an oligonucleotide RNA primer, as noroviral RNA synthesis can occur primer independently, at least in vitro (40). The newly synthesized RNA was detected by Northern blotting using a probe specific for (+)-vRNA. Here, we observed that the presence of WT MBP-NS3 promoted the NS7/RdRP-mediated production of (+)-vRNA strands from the (−)-vRNA templates and that the promoting effect of NS3 could be further enhanced by increasing the reaction time (Fig. 8B and C). On the other hand, the A motif mutation mostly eliminated the promoting effect of MBP-NS3 on in vitro RNA synthesis (Fig. 8D and E).
FIG 8.
NV NS3 facilitates noroviral RNA synthesis in vitro. (A) Schematic illustration of the experimental procedures. (B and D) The in vitro-transcribed NV 3′ end (nt 1 to 400) of (−)-vRNA template was incubated with 10 pmol recombinant NS7/RdRP and NTP mixture in the absence or presence of 10 pmol MBP-NS3 (B) or MBP-NS3GK168AA (D) at 30°C for 30, 60, or 90 min as indicated. The reaction products were detected by Northern blotting. (C and E) The synthesized (+)-RNA products in panels B and D were quantified with Bio-Rad Quantity One software. The relative RNA production was determined by comparing the level of RNA products in the presence of MBP-NS3 at each indicated time point with that in the absence of NS3 at 30 min (lanes 2). The error bars represent SD values from the results of three separate experiments.
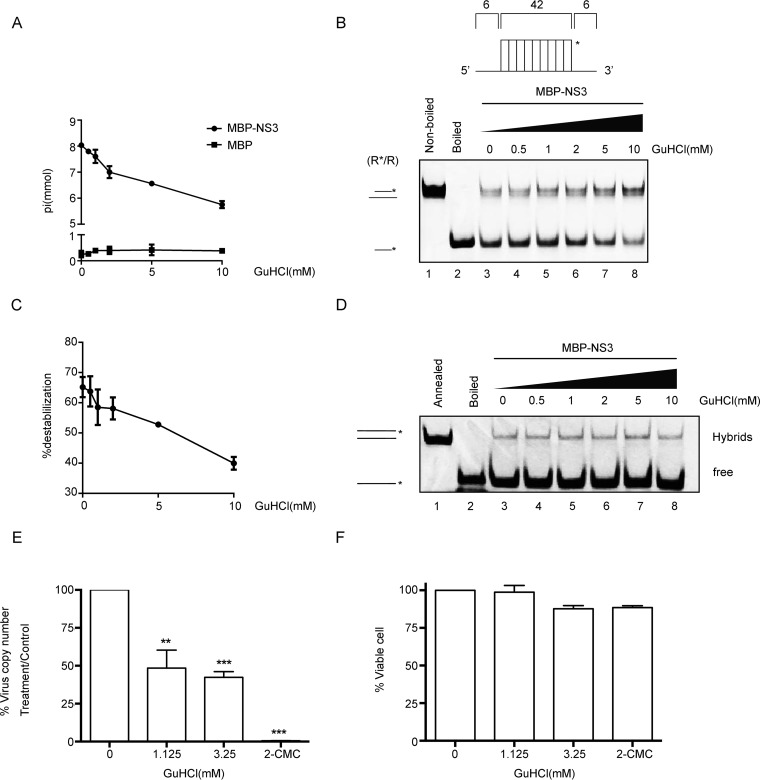
GuHCl inhibits the NTPase and RNA helix-unwinding activities of NV NS3.
Previous studies have reported that the NTPase and RNA helicase activities of enterovirus 2CATPase can be inhibited by GuHCl (22, 41–45), which also inhibits replication of the vRNAs of a number of enteroviruses (46, 47). Given the similarity between the consensus motifs in enterovirus 2CATPase and norovirus NS3, we wanted to examine whether NV NS3 could also be a target for guanidine inhibition. NTPase assays showed that GuHCl inhibited the ATPase activity of NV NS3 in a dose-dependent manner (Fig. 9A). This result is in contrast to a previous observation by Pfister and Wimmer, who demonstrated that even in the presence of up to 10 mM GuHCl, the ATPase activity of SHV NS3 was not inhibited (33). These contrasting results might be due to the different expression strategies (eukaryotic versus prokaryotic) or the inherent biochemical properties of the two NS3 proteins, which were also shown to have different hydrolysis preferences for UTP (Fig. 2A) and different responses to Mg2+ concentrations higher than 2 mM (Fig. 2C).
FIG 9.
GuHCl inhibits the biochemical activities of NV NS3. (A) The NTPase activity of MBP-NS3 was measured as nanomoles of released inorganic phosphate in the presence of the indicated concentrations (0 to 10 mM) of GuHCl. MBP alone was used as the negative control. (B) The standard RNA helix (R*/R) substrate is illustrated in the upper diagram. R*/R substrate (0.1 pmol) was reacted with 20 pmol MBP-NS3 in the presence of the indicated concentrations (0 to 10 mM) of GuHCl. Native (lane 1) or boiled (lane 2) reaction mixture was used as a negative or positive control, respectively. Helix unwinding was detected by gel electrophoresis and scanning on a Typhoon 9500 imager. The asterisk indicates the HEX-labeled strand. (C) The unwinding activities under different GuHCl concentrations were plotted as percentages of the released RNA from the total RNA helix substrate (y axis) at each indicated GuHCl concentration (x axis) with Bio-Rad Quantity One software. The error bars represent standard deviation values from the results of three separate experiments. (D) The two strands shown in Fig. 7B were mixed 1:1 (0.1 pmol each) and reacted with 10 pmol MBP-NS3 for 30 min in the presence of different concentrations of GuHCl in the absence of NTP. The hybridization of the two strands was detected by gel electrophoresis and scanning on a Typhoon 9500 imager. The preannealed (lane 1) or boiled (lane 2) reaction mixture was used as a positive or negative control, respectively. The hybridized and free strands are indicated. (E) The effect of GuHCl or 2CMC on Norwalk virus RNA levels was determined by examining the impact on NV replicon RNA levels in HGT-NV cells following 3 days of treatment. HGT-NV cells were treated with the indicated levels of either GuHCl or 2CMC, and the levels of viral RNA were calculated as a percentage of the mock-treated control. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (F) HGT-NV cells were treated as for panel D, and the number of viable cells was determined. The cell viability was calculated as a percentage of that of the mock-treated control. The error bars represent SD (n = 3). Statistical significance was determined using an unpaired t test. The experiment was repeated at least two independent times, with one representative data set presented.
To examine if GuHCl could also inhibit helix unwinding by NV NS3, the helix-unwinding reactions were performed in the presence of NS3 and increasing concentrations of GuHCl. Our results showed that the presence of GuHCl inhibited helix unwinding by NS3 in a dose-dependent manner (Fig. 9B and C). Intriguingly, we examined the effect of GuHCl on the RNA-chaperoning activity of NV NS3 using an RNA strand hybridization assay. Our data showed that the addition of GuHCl at different concentrations had no obvious effect on NS3 RNA-chaperoning activity (Fig. 9D).
It is important to note that the helicase activity and ATPase activity show similar sensitivities to GuHCl and that the inhibitory effects of GuHCl on helicase and ATPase activities of NV NS3 are moderate but similar to the observed effects on EV71 2CATPase (22). Nevertheless, these data demonstrate that GuHCl is an inhibitor of the key biochemical activities of NV NS3.
GuHCl inhibits NV replication in cultured human cells.
While we had shown that GuHCl can inhibit the NTPase and helix-unwinding activities of NV NS3, we sought to determine whether GuHCl treatment could inhibit HuNoV replication in cells. To this end, the effect of GuHCl on HuNoV replication was evaluated by using the NV replicon system in cultured human gastric tumor (HGT-1) cells. The levels of NV RNA in the presence of GuHCl at different concentrations were determined by real-time quantitative reverse transcription-PCR (qRT-PCR), and the expression levels of NV proteins were determined by Western blotting of NV NS3. 2′-C-Methylcytidine (2CMC), a nucleoside analog that has been reported to inhibit HuNoV replication (48, 49), was used as the positive control. As shown in Fig. 9E, the presence of 1.125 mM GuHCl significantly reduced the level of NV RNA to around 50% of that under mock treatment, while increasing the GuHCl concentration to 3.25 mM only moderately enhanced the inhibitory effect of GuHCl on NV RNA replication. As expected, 2CMC showed a strong inhibitory effect on NV RNA replication (Fig. 9E). We also showed that the presence of 1.125 or 3.25 mM GuHCl did not affect the viability of HGT-1 cells, as was found for 2CMC-treated cells, excluding the possibility that the inhibitory effect of GuHCl on NV RNA replication was due to inducing cell death (Fig. 9F). Hence, GuHCl can inhibit the replication of the NV replicon at 1.125 mM in cultured human cells without apparent cytotoxicity.
DISCUSSION
During the life cycle of RNA viruses, cellular or virus-encoded RNA helicases or chaperones are believed to facilitate correct folding or unfolding of RNA structures or to separate vRI-dsRNAs for more rounds of vRNA replication (14, 15, 22). Here, we report for the first time that NV-encoded NS3 has both NTP-dependent RNA helicase and NTP-independent RNA-chaperoning activities that can unwind RNA helices and destabilize and remodel structured RNA molecules. Moreover, we found that NS3 can stimulate NS7/RdRP-mediated noroviral RNA synthesis in vitro, suggesting that NS3 plays an important role in norovirus RNA replication. Our work has also shown that GuHCl, an FDA-approved small-molecule drug, can inhibit the RNA helicase activity of NS3 in vitro and, more importantly, that this small molecule has some inhibitory activity against replication of a HuNoV replicon in cultured human cells.
The RNA-remodeling activities of viral proteins are generally believed to play important roles in viral life cycles (50). For instance, during vRNA replication of RNA viruses, vRI-dsRNAs must be efficiently unwound, thereby releasing nascently synthesized progeny vRNAs from vRNA templates. Moreover, viral genomic and antigenomic RNAs contain multiple cis-acting elements, highly structured RNA regions that usually play pivotal roles in the replication, translation, and encapsidation of vRNAs (11, 51). Sometimes, a specific RNA structure required for one vRNA function, like translation or encapsidation, may need disruption and refolding to facilitate another vRNA function, like vRNA replication, and vice versa. For example, genomic RNAs or mRNAs of some RNA viruses, including norovirus (13, 52, 53), flavivirus (12), hantavirus (54), and reovirus (55), can undergo circularization, resulting in the base pairing of vRNA 5′ and 3′ ends to form panhandle-like structures that may facilitate vRNA translation, encapsidation, or RdRP recruitment to the 3′ ends of vRNAs (51, 56). However, for initiation of RNA replication, these cyclized vRNA structures need to be disrupted to make the 3′ ends of the vRNAs accessible by RdRPs; an example of this type of activity is the chaperoning effect of cypovirus VP5 on cyclized cypoviral mRNAs (25). Viral RNA-remodeling proteins may also dynamically facilitate proper folding and unfolding of different vRNA structures, thereby allowing vRNAs to switch among distinct functionalities.
For noroviruses, it has been found that their vRNAs contain multiple cis-acting elements and RNA secondary structures, which are important for norovirus replication (13, 51–53). With that in mind, it is intriguing that we have observed the ability of NS3 to enhance NS7/RdRP-mediated noroviral RNA synthesis in vitro, particularly since norovirus NS3 has recently been found to associate with vRI-dsRNAs in vRC on intracellular membranes (34–36). These findings indicate the critical roles of NS3 in norovirus RNA replication. However, owing to technical limitations, the RNA-remodeling activities of viral proteins have been mainly studied in vitro, and it is difficult to imagine how the regulation of the structures and functions of vRNAs by viral RNA helicases/chaperones can be investigated in detail in virus-infected cells. Future advances in techniques may overcome these barriers and help us understand the mechanism(s) through which the RNA helicase/chaperoning activities of NS3 remodel vRNAs at different stages of the noroviral life cycle.
Interestingly, we found that the guanidine salt GuHCl inhibits both the ATPase and RNA helix-unwinding activities of NV NS3 in a dose-dependent manner (Fig. 9), similar to the effects of the same compound on enterovirus 2CATPase (22, 41). Although previous work found that the ATPase activity of SHV NS3 was not inhibited by GuHCl (33), our data show some differences between NV NS3 and SHV NS3 in terms of ATPase activity. For example, while SHV NS3 has been found to hydrolyze UTP much less well than ATP (30), NV NS3 exhibited similar hydrolysis efficiencies for both ATP and UTP (Fig. 2A). Moreover, when the concentration of Mg2+ was higher than 2 mM, the ATPase activity of NV NS3 was inhibited in a dose-dependent manner (Fig. 2C), while that of SHV NS3 was moderately enhanced (30). It is possible that the difference between NV NS3 and SHV NS3 in ATPase activity is due to the different (eukaryotic versus prokaryotic) expression strategies.
Virus-encoded RNA-remodeling proteins, particularly helicases, have long been considered valuable targets for antiviral compounds, because they are well conserved within or even across virus families (57). In particular, enterovirus 2CATPase, another SF3 viral RNA helicase, has been found to be inhibited by ∼1 mM GuHCl, as is enterovirus replication (22, 42–45). Using an NV replicon as a model, our studies demonstrated that ∼1 mM GuHCl can significantly inhibit the replication of HuNoV in cultured human cells with negligible cell toxicity (Fig. 9). Similarly, GuHCl can inhibit the replication of poliovirus at the same concentration (22). Guanidines are ubiquitously present in nature and are categorized as organic superbases with highly basic nature and notable stability that can function as mediators of specific noncovalent binding in various catalytic processes (58, 59). Due to their physicochemical characteristics, guanidine molecules probably bind stably to the surfaces of viral RNA-remodeling proteins, such as enterovirus 2CATPase and HuNoV NS3, and may therefore alter their conformations, electrostatic states, and/or protein-protein/RNA interactions, thereby inhibiting their RNA-remodeling activities. In accordance with this idea, high concentrations of Mg2+ (>2 mM) have been found to inhibit helix unwinding by either NV NS3 (Fig. 2C) or EV71 2CATPase (22). GuHCl is a U.S. FDA-approved small compound drug that has been used to treat the symptoms of muscle weakness and fatigability associated with Eaton-Lambert syndrome (60, 61; http://www.accessdata.fda.gov/scripts/cder/daf/index.cfm?event=overview.process&ApplNo=001546). Moreover, guanidine derivatives have attracted widespread interest for developing drugs against various diseases, and some of them have been found to be potential antivirals against hepatitis C virus (HCV), human immunodeficiency virus (HIV), and flaviviruses (21, 58, 59). It is unlikely that GuHCl can be directly used as an antiviral drug against HuNoV infection due to the high concentration needed for inhibition of NS3 and NV replication. However, given the presence of multiple derivative forms of guanidine, the synthesis and screening of various guanidine derivatives and guanidine-containing compounds may help in the quest for small molecules that have better inhibitory effects on HuNoV NS3 and HuNoV replication.
In conclusion, this study provides the first demonstration of the RNA helicase and chaperoning activities associated with a norovirus- or calicivirus-encoded protein and reveals that NS3 may have a direct function (in concert with NS7/RdRP) in norovirus RNA replication. Moreover, it also provides evidence that GuHCl is an inhibitor of not only the in vitro RNA-remodeling activities of NS3, but also the replication of HuNoV in human cells. Altogether, these findings highlight the functional significance of NS3 in the noroviral life cycle and shed new light on norovirus replication.
MATERIALS AND METHODS
Construction of recombinant baculoviruses and expression and purification of recombinant proteins.
The generation of pFastBac HTB-MBP has been described previously (26). The cDNA of NV NS3 (GenBank accession no. KF039736) was synthesized by Tian Yi Hui Yuan Biotech Co. (Wuhan, China) and cloned into the vector pFastBac HTB-MBP. Site-directed mutagenesis was conducted as previously described (26). The resulting plasmids were subjected to the Bac-to-Bac baculovirus system to produce recombinant proteins. The primers used for NS3 plasmid construction were as follows: forward, GAATTCATGGGCCCCGAGGACCTTGCCAGGGATCTCGTG (EcoRI), and reverse, GTCGACCTACTGGAGTTGGAACTCATCCTGTCTCTCCAT (SalI); the primers for GK168AA and GK168AA-DD212AA of NS3 were as follows: forward, TGTGGGCCCCCTGGTATAGCTGCTACCAAG, and reverse, GTTCTGCTGCCTTGGTAGCAGCTATACCAG; forward, GCTGTGGGCTGCTTATGGAATGACAAAGAT, and reverse, TGTATCTTTGTCATTCCATAAGCAGCCCAC. Underlined letters indicate restriction endonuclease sites, and the types of the restriction endonucleases are shown in parentheses.
The expression and purification of recombinant MBP alone and MBP fusion proteins were conducted using standard procedures, as previously described (25, 26). All purified proteins were quantified with a bicinchoninic acid (BCA) protein assay kit (CWBIO, China) and stored at −80°C in aliquots. Proteins were separated on 10% SDS-PAGE and visualized with Coomassie blue.
Structural modeling and structural alignment analyses.
The 3D structure of NV NS3 was modeled by submitting its amino acid sequence to the HMMSTR/Rosetta server (ROBETTA, University of Washington [http://robetta.bakerlab.org/]). Five models were obtained, and the best one was chosen as a template based on its score and assessed by submitting it to the SWISS-MODEL server (Swiss Institute of Bioinformatics and Biozentrum, University of Basel, Basel, Switzerland [http://swissmodel.expasy.org]). The figures showing the modeled NS3ΔN and the structural alignments with SV40 LTag (Protein Data Bank [PDB] ID 4GDF), HPV18 E1 (PDB ID 1TUE), and AAV2 Rep40 (PDB ID 1U0J) were generated from coordinate files using PyMOL version 1.1 (DeLano Scientific LLC, Southern San Francisco, CA).
NTPase assay.
NTPase activities were determined by measuring the inorganic phosphate released during NTP hydrolysis using a direct colorimetric assay according to our standard procedures as previously described (26). The concentrations of released inorganic phosphate were determined by matching the A620 in a known inorganic phosphate curve. All of the results given by this quantitative assay were averages of three independently repeated experiments.
Preparation of oligonucleotide helix substrates.
The nucleic acid helix substrates were prepared according to our standard procedures as previously described (22). In brief, RNA, DNA, or RNA-DNA hybrid helix substrates were prepared by annealing two complementary nucleic acid strands, one strand of which was labeled at the 5′ end with HEX while the other was unlabeled. The two strands were mixed at a 1:1 ratio in a 10-μl reaction mixture containing 25 mM HEPES-KOH (pH 8.0) and 25 mM NaCl, and the mixture was then heated to 95°C for 5 min and gradually cooled to 25°C to produce helical duplexes. All the HEX-labeled DNA and RNA strands were purchased from TaKaRa (Dalian, China), unlabeled DNA strands were synthesized by Invitrogen, and unlabeled RNA strands were in vitro transcribed using T7 RNA polymerase (Promega, Madison, WI). The in vitro-transcribed RNA strands were purified with a Poly-Gel RNA extraction kit (Omega Bio-Tek, Guangzhou, China) according to the manufacturer's instructions. The standard RNA helix substrate with both 5′ and 3′ protrusions was annealed using RNA1 and RNA2, the DNA helix substrate was annealed using DNA2 and DNA3, the RNA*-DNA hybrid was annealed using RNA1 and DNA1, the DNA*-RNA hybrid was annealed using DNA1 and RNA3, the 3′ protruded RNA helix was annealed using RNA1 and RNA4, and the 5′ protruded RNA helix was annealed using RNA1 and RNA5. All the oligonucleotides used in this study are listed in Table 1.
Helix-unwinding and RNA strand hybridization assays.
Helix-unwinding assays were performed according to our standard procedures as previously described (25). The reaction mixtures were electrophoresed on 15% native PAGE gels and then scanned with a Typhoon 9500 imager (GE Healthcare, Piscataway, NJ).
The RNA hybridization assay was carried out according to our standard procedures as previously described (25). In brief, the indicated amount of protein, 0.2 pmol HEX-labeled stem-loop-structured RNA strand, and 0.2 pmol unlabeled stem-loop-structured RNA strand were added to reaction buffer and incubated for 1 h or the indicated time. The sequences of the stem-loop-structured RNA strands are given in Fig. 7B. The reaction products were resolved on 15% native PAGE gels, followed by scanning with a Typhoon 9500 imager (GE Healthcare, Piscataway, NJ).
In vitro RdRP assays.
The vRNA template was in vitro transcribed by T7 RNA polymerase as described above. The (−)-vRNA template contained 1 to 400 nt of 3′-end HuNoV (−)-RNA, which is complementary to the 5′-end 400 nt of HuNoV (+)-RNA. The transcribed product was purified with a Poly-Gel RNA extraction kit (Omega Bio-Tek, Guangzhou, China) according to the manufacturer's instructions. The cDNA of HuNoV NS7/RdRP (GenBank accession no. KR904238) was amplified by PCR from full-length GII.4-Sydney HuNoV cDNA, which was generously provided by Linqing Zhao (Capital Institute of Pediatrics, Beijing, China), and cloned into the vector pMAl-c2X-MBP (primers: forward, GGATCCATGGGAGGAAACACTGTCATATGTGCCACCCAG, and reverse, GTCGACTCACTCGACGCCATCTTCATTCACAAAACTGGG). The recombinant MBP fusion with NS7/RdRP was purified as described above. The RdRP assay was performed as previously described with minor modifications. In brief, the indicated amounts of MBP-NS3 were included in 10-μl reaction mixtures containing 50 mM HEPES-KOH (pH 7.5), 12.5 mM KCl, 5 mM MgCl2, 5 mM dithiothreitol (DTT), 20 U RNasin (Promega), and 5 mM NTPs. MBP-NS3 was included in the reaction mixture with or without 10 pmol NS7/RdRP at 30°C for the indicated time. After that, the reaction was terminated with 2× loading buffer (20% 10× MOPS [morpholinepropanesulfonic acid] running buffer, 10% glycerol, 50% formamide, 20% formaldehyde, and 0.05% bromophenol blue) and then denatured at 65°C for 10 min. The samples were electrophoresed on 1.5% agarose-formaldehyde denaturing gels and transferred onto N+ nylon membranes (Roche). The RdRP products were detected via Northern blots that were performed according to our standard procedures (62). The probe for the Northern blots is listed in Table 1.
Assessment of GuHCl treatment in HuNoV replicon-harboring cells.
A baby hamster kidney cell line harboring the NV replicon, provided by Kim Green (National Institutes of Health, Bethesda, MD), was used as a source of viral VPg-linked RNA (63). Total RNA was isolated from the BHK-NV replicon-containing cell line and transfected into HGT-1 cells (64). The transfected cells were placed under selection using 1 mg/ml of G418, and clonal populations were generated by limiting dilution. The levels of viral RNA were determined by qRT-PCR as described above, and a single clone, referred to as HGT-NV, was selected for future use.
To examine the effect of GuHCl on NV replication, HGT-NV cells were seeded in a 96-well plate at 1,000 cells per well without G418. The HGT-NV cells were immediately incubated with media containing various levels of GuHCl or 2CMC as a control. The cells were incubated at 37°C in 5% CO2 for 3 days. The effect of GuHCl treatment on HuNoV replicon RNA was then evaluated by qRT-PCR as described previously (65) and by Western blotting using rabbit polyclonal antisera to norovirus NS3. In addition, the effect of GuHCl on cell viability was determined using CellTiter-Blue (Promega) according to the manufacturer's instructions.
ACKNOWLEDGMENTS
We thank Linqing Zhao (Beijing, China) for generously providing reagents and Linqing Zhao and Huimin Yan (Wuhan, China) for helpful discussions.
This work was supported by the National Natural Science Foundation of China (NSFC) Excellent Young Scientists Fund (grant no. 31522004 to X.Z.), a Newton Advanced Fellowship from the United Kingdom Academy of Medical Sciences and NSFC (no. 3161101464 to X.Z. and S.C.), NSFC grant no. 31670161 (to X.Z.), the Strategic Priority Research Program of the Chinese Academy of Sciences (no. XDPB0301 to X.Z.), the National High-Tech R&D Program of China (863 Program) (grant no. 2015AA020939 to X.Z.), the National Basic Research Program of China (973 Program) (grant no. 2014CB542603 to X.Z.), and the Distinguished Young Scientists Fund of Hubei Province (grant no. 2016CFA045 to X.Z.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Zheng DP, Ando T, Fankhauser RL, Beard RS, Glass RI, Monroe SS. 2006. Norovirus classification and proposed strain nomenclature. Virology 346:312–323. doi: 10.1016/j.virol.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 2.Fankhauser RL, Noel JS, Monroe SS, Ando T, Glass RI. 1998. Molecular epidemiology of “Norwalk-like viruses” in outbreaks of gastroenteritis in the United States. J Infect Dis 178:1571–1578. doi: 10.1086/314525. [DOI] [PubMed] [Google Scholar]
- 3.Vinje J. 2015. Advances in laboratory methods for detection and typing of norovirus. J Clin Microbiol 53:373–381. doi: 10.1128/JCM.01535-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaufman SS, Green KY, Korba BE. 2014. Treatment of norovirus infections: moving antivirals from the bench to the bedside. Antiviral Res 105:80–91. doi: 10.1016/j.antiviral.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arias A, Emmott E, Vashist S, Goodfellow I. 2013. Progress towards the prevention and treatment of norovirus infections. Future Microbiol 8:1475–1487. doi: 10.2217/fmb.13.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karst SM. 2010. Pathogenesis of noroviruses, emerging RNA viruses. Viruses 2:748–781. doi: 10.3390/v2030748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leen EN, Sorgeloos F, Correia S, Chaudhry Y, Cannac F, Pastore C, Xu Y, Graham SC, Matthews SJ, Goodfellow IG, Curry S. 2016. A conserved interaction between a C-terminal motif in norovirus VPg and the HEAT-1 domain of eIF4G is essential for translation initiation. PLoS Pathog 12:e1005379. doi: 10.1371/journal.ppat.1005379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karst SM, Wobus CE, Goodfellow IG, Green KY, Virgin HW. 2014. Advances in norovirus biology. Cell Host Microbe 15:668–680. doi: 10.1016/j.chom.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McFadden N, Bailey D, Carrara G, Benson A, Chaudhry Y, Shortland A, Heeney J, Yarovinsky F, Simmonds P, Macdonald A, Goodfellow I. 2011. Norovirus regulation of the innate immune response and apoptosis occurs via the product of the alternative open reading frame 4. PLoS Pathog 7:e1002413. doi: 10.1371/journal.ppat.1002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thorne L, Bailey D, Goodfellow I. 2012. High-resolution functional profiling of the norovirus genome. J Virol 86:11441–11456. doi: 10.1128/JVI.00439-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alhatlani B, Vashist S, Goodfellow I. 2015. Functions of the 5′ and 3′ ends of calicivirus genomes. Virus Res 206:134–143. doi: 10.1016/j.virusres.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hahn CS, Hahn YS, Rice CM, Lee E, Dalgarno L, Strauss EG, Strauss JH. 1987. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J Mol Biol 198:33–41. doi: 10.1016/0022-2836(87)90455-4. [DOI] [PubMed] [Google Scholar]
- 13.McFadden N, Arias A, Dry I, Bailey D, Witteveldt J, Evans DJ, Goodfellow I, Simmonds P. 2013. Influence of genome-scale RNA structure disruption on the replication of murine norovirus: similar replication kinetics in cell culture but attenuation of viral fitness in vivo. Nucleic Acids Res 41:6316–6331. doi: 10.1093/nar/gkt334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bleichert F, Baserga SJ. 2007. The long unwinding road of RNA helicases. Mol Cell 27:339–352. doi: 10.1016/j.molcel.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 15.Jarmoskaite I, Russell R. 2014. RNA helicase proteins as chaperones and remodelers. Annu Rev Biochem 83:697–725. doi: 10.1146/annurev-biochem-060713-035546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grohman JK, Gorelick RJ, Lickwar CR, Lieb JD, Bower BD, Znosko BM, Weeks KM. 2013. A guanosine-centric mechanism for RNA chaperone function. Science 340:190–195. doi: 10.1126/science.1230715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, Jantsch MF, Konrat R, Blasi U, Schroeder R. 2007. RNA chaperones, RNA annealers and RNA helicases. RNA Biol 4:118–130. doi: 10.4161/rna.4.3.5445. [DOI] [PubMed] [Google Scholar]
- 18.Lorsch JR. 2002. RNA chaperones exist and DEAD box proteins get a life. Cell 109:797–800. doi: 10.1016/S0092-8674(02)00804-8. [DOI] [PubMed] [Google Scholar]
- 19.Kadare G, Haenni AL. 1997. Virus-encoded RNA helicases. J Virol 71:2583–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Musier-Forsyth K. 2010. RNA remodeling by chaperones and helicases. RNA Biol 7:632–633. doi: 10.4161/rna.7.6.14467. [DOI] [PubMed] [Google Scholar]
- 21.Frick DN. 2007. The hepatitis C virus NS3 protein: a model RNA helicase and potential drug target. Curr Issues Mol Biol 9:1–20. [PMC free article] [PubMed] [Google Scholar]
- 22.Xia H, Wang P, Wang GC, Yang J, Sun X, Wu W, Qiu Y, Shu T, Zhao X, Yin L, Qin CF, Hu Y, Zhou X. 2015. Human enterovirus nonstructural protein 2CATPase functions as both an RNA helicase and ATP-independent RNA chaperone. PLoS Pathog 11:e1005067. doi: 10.1371/journal.ppat.1005067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karpe YA, Aher PP, Lole KS. 2011. NTPase and 5′-RNA triphosphatase activities of Chikungunya virus nsP2 protein. PLoS One 6:e22336. doi: 10.1371/journal.pone.0022336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee NR, Kwon HM, Park K, Oh S, Jeong YJ, Kim DE. 2010. Cooperative translocation enhances the unwinding of duplex DNA by SARS coronavirus helicase nsP13. Nucleic Acids Res 38:7626–7636. doi: 10.1093/nar/gkq647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J, Cheng Z, Zhang S, Xiong W, Xia H, Qiu Y, Wang Z, Wu F, Qin CF, Yin L, Hu Y, Zhou X. 2014. A cypovirus VP5 displays the RNA chaperone-like activity that destabilizes RNA helices and accelerates strand annealing. Nucleic Acids Res 42:2538–2554. doi: 10.1093/nar/gkt1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng Z, Yang J, Xia H, Qiu Y, Wang Z, Han Y, Xia X, Qin CF, Hu Y, Zhou X. 2013. The nonstructural protein 2C of a picorna-like virus displays nucleic acid helix destabilizing activity that can be functionally separated from its ATPase activity. J Virol 87:5205–5218. doi: 10.1128/JVI.00245-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Q, Han Y, Qiu Y, Zhang S, Tang F, Wang Y, Zhang J, Hu Y, Zhou X. 2012. Identification and characterization of RNA duplex unwinding and ATPase activities of an alphatetravirus superfamily 1 helicase. Virology 433:440–448. doi: 10.1016/j.virol.2012.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J, Xia H, Qian Q, Zhou X. 2015. RNA chaperones encoded by RNA viruses. Virol Sin 30:401–409. doi: 10.1007/s12250-015-3676-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li D, Zhao R, Lilyestrom W, Gai D, Zhang R, DeCaprio JA, Fanning E, Jochimiak A, Szakonyi G, Chen XS. 2003. Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen. Nature 423:512–518. doi: 10.1038/nature01691. [DOI] [PubMed] [Google Scholar]
- 30.Zarate-Perez F, Bardelli M, Burgner JW II, Villamil-Jarauta M, Das K, Kekilli D, Mansilla-Soto J, Linden RM, Escalante CR. 2012. The interdomain linker of AAV-2 Rep68 is an integral part of its oligomerization domain: role of a conserved SF3 helicase residue in oligomerization. PLoS Pathog 8:e1002764. doi: 10.1371/journal.ppat.1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collaco RF, Kalman-Maltese V, Smith AD, Dignam JD, Trempe JP. 2003. A biochemical characterization of the adeno-associated virus Rep40 helicase. J Biol Chem 278:34011–34017. doi: 10.1074/jbc.M301537200. [DOI] [PubMed] [Google Scholar]
- 32.Lin BY, Makhov AM, Griffith JD, Broker TR, Chow LT. 2002. Chaperone proteins abrogate inhibition of the human papillomavirus (HPV) E1 replicative helicase by the HPV E2 protein. Mol Cell Biol 22:6592–6604. doi: 10.1128/MCB.22.18.6592-6604.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfister T, Wimmer E. 2001. Polypeptide p41 of a Norwalk-like virus is a nucleic acid-independent nucleoside triphosphatase. J Virol 75:1611–1619. doi: 10.1128/JVI.75.4.1611-1619.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eden JS, Tanaka MM, Boni MF, Rawlinson WD, White PA. 2013. Recombination within the pandemic norovirus GII.4 lineage. J Virol 87:6270–6282. doi: 10.1128/JVI.03464-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bull RA, Tanaka MM, White PA. 2007. Norovirus recombination. J Gen Virol 88:3347–3359. doi: 10.1099/vir.0.83321-0. [DOI] [PubMed] [Google Scholar]
- 36.Cotton BT, Hyde JL, Sarvestani ST, Sosnovtsev SV, Green KY, White PA, Mackenzie JM. 2017. The norovirus NS3 protein is a dynamic lipid- and microtubule-associated protein involved in viral RNA replication. J Virol 91:e02138-. doi: 10.1128/JVI.02138-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bystroff C, Shao Y. 2002. Fully automated ab initio protein structure prediction using I-SITES, HMMSTR and ROSETTA. Bioinformatics 18(Suppl 1):S54–S61. doi: 10.1093/bioinformatics/18.suppl_1.S54. [DOI] [PubMed] [Google Scholar]
- 38.DeStefano JJ, Titilope O. 2006. Poliovirus protein 3AB displays nucleic acid chaperone and helix-destabilizing activities. J Virol 80:1662–1671. doi: 10.1128/JVI.80.4.1662-1671.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lam AM, Frick DN. 2006. Hepatitis C virus subgenomic replicon requires an active NS3 RNA helicase. J Virol 80:404–411. doi: 10.1128/JVI.80.1.404-411.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fukushi S, Kojima S, Takai R, Hoshino FB, Oka T, Takeda N, Katayama K, Kageyama T. 2004. Poly(A)- and primer-independent RNA polymerase of norovirus. J Virol 78:3889–3896. doi: 10.1128/JVI.78.8.3889-3896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfister T, Wimmer E. 1999. Characterization of the nucleoside triphosphatase activity of poliovirus protein 2C reveals a mechanism by which guanidine inhibits poliovirus replication. J Biol Chem 274:6992–7001. doi: 10.1074/jbc.274.11.6992. [DOI] [PubMed] [Google Scholar]
- 42.Pincus SE, Diamond DC, Emini EA, Wimmer E. 1986. Guanidine-selected mutants of poliovirus: mapping of point mutations to polypeptide 2C. J Virol 57:638–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loddo B, Ferrari W, Brotzu G, Spanedda A. 1962. In vitro inhibition of infectivity of polio viruses by guanidine. Nature 193:97–98. doi: 10.1038/193097a0. [DOI] [PubMed] [Google Scholar]
- 44.Caliguiri LA, Tamm I. 1968. Action of guanidine on the replication of poliovirus RNA. Virology 35:408–417. doi: 10.1016/0042-6822(68)90219-5. [DOI] [PubMed] [Google Scholar]
- 45.Loddo B, Muntoni S, Spanedda A, Brotzu G, Ferrari W. 1963. Guanidine conditioned infectivity of ribonucleic acid extracted from a strain of guanidine-dependent polio-1-virus. Nature 197:315. [DOI] [PubMed] [Google Scholar]
- 46.Lyons T, Murray KE, Roberts AW, Barton DJ. 2001. Poliovirus 5′-terminal cloverleaf RNA is required in cis for VPg uridylylation and the initiation of negative-strand RNA synthesis. J Virol 75:10696–10708. doi: 10.1128/JVI.75.22.10696-10708.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steil BP, Barton DJ. 2009. Conversion of VPg into VPgpUpUOH before and during poliovirus negative-strand RNA synthesis. J Virol 83:12660–12670. doi: 10.1128/JVI.01676-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rocha-Pereira J, Jochmans D, Debing Y, Verbeken E, Nascimento MS, Neyts J. 2013. The viral polymerase inhibitor 2′-C-methylcytidine inhibits Norwalk virus replication and protects against norovirus-induced diarrhea and mortality in a mouse model. J Virol 87:11798–11805. doi: 10.1128/JVI.02064-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rocha-Pereira J, Jochmans D, Dallmeier K, Leyssen P, Cunha R, Costa I, Nascimento MS, Neyts J. 2012. Inhibition of norovirus replication by the nucleoside analogue 2′-C-methylcytidine. Biochem Biophys Res Commun 427:796–800. doi: 10.1016/j.bbrc.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 50.Zuniga S, Sola I, Cruz JL, Enjuanes L. 2009. Role of RNA chaperones in virus replication. Virus Res 139:253–266. doi: 10.1016/j.virusres.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lopez-Manriquez E, Vashist S, Urena L, Goodfellow I, Chavez P, Mora-Heredia JE, Cancio-Lonches C, Garrido E, Gutierrez-Escolano AL. 2013. Norovirus genome circularization and efficient replication are facilitated by binding of PCBP2 and hnRNP A1. J Virol 87:11371–11387. doi: 10.1128/JVI.03433-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yunus MA, Lin X, Bailey D, Karakasiliotis I, Chaudhry Y, Vashist S, Zhang G, Thorne L, Kao CC, Goodfellow I. 2015. The murine norovirus core subgenomic RNA promoter consists of a stable stem-loop that can direct accurate initiation of RNA synthesis. J Virol 89:1218–1229. doi: 10.1128/JVI.02432-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin X, Thorne L, Jin Z, Hammad LA, Li S, Deval J, Goodfellow IG, Kao CC. 2015. Subgenomic promoter recognition by the norovirus RNA-dependent RNA polymerases. Nucleic Acids Res 43:446–460. doi: 10.1093/nar/gku1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raju R, Kolakofsky D. 1989. The ends of La Crosse virus genome and antigenome RNAs within nucleocapsids are base paired. J Virol 63:122–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tortorici MA, Shapiro BA, Patton JT. 2006. A base-specific recognition signal in the 5′ consensus sequence of rotavirus plus-strand RNAs promotes replication of the double-stranded RNA genome segments. RNA 12:133–146. doi: 10.1261/rna.2122606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen D, Patton JT. 1998. Rotavirus RNA replication requires a single-stranded 3′ end for efficient minus-strand synthesis. J Virol 72:7387–7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kwong AD, Rao BG, Jeang KT. 2005. Viral and cellular RNA helicases as antiviral targets. Nat Rev Drug Discov 4:845–853. doi: 10.1038/nrd1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saczewski F, Balewski L. 2009. Biological activities of guanidine compounds. Expert Opin Ther Pat 19:1417–1448. doi: 10.1517/13543770903216675. [DOI] [PubMed] [Google Scholar]
- 59.Saczewski F, Balewski L. 2013. Biological activities of guanidine compounds, 2008-2012 update. Expert Opin Ther Pat 23:965–995. doi: 10.1517/13543776.2013.788645. [DOI] [PubMed] [Google Scholar]
- 60.Oh SJ, Lee YW, Rutsky E. 1977. Eaton-Lambert syndrome: reflex improvement with guanidine. Arch Phys Med Rehabil 58:457–459. [PubMed] [Google Scholar]
- 61.Oh SJ, Kim KW. 1973. Guanidine hydrochloride in the Eaton-Lambert syndrome. Electrophysiologic improvement. Neurology 23:1084–1090. [DOI] [PubMed] [Google Scholar]
- 62.Wang Z, Wu D, Liu Y, Xia X, Gong W, Qiu Y, Yang J, Zheng Y, Li J, Wang YF, Xiang Y, Hu Y, Zhou X. 2015. Drosophila Dicer-2 has an RNA interference-independent function that modulates Toll immune signaling. Sci Adv 1:e1500228. doi: 10.1126/sciadv.1500228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chang KO, Sosnovtsev SV, Belliot G, King AD, Green KY. 2006. Stable expression of a Norwalk virus RNA replicon in a human hepatoma cell line. Virology 353:463–473. doi: 10.1016/j.virol.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 64.Laboisse CL, Augeron C, Couturier-Turpin MH, Gespach C, Cheret AM, Potet F. 1982. Characterization of a newly established human gastric cancer cell line HGT-1 bearing histamine H2-receptors. Cancer Res 42:1541–1548. [PubMed] [Google Scholar]
- 65.Kageyama T, Kojima S, Shinohara M, Uchida K, Fukushi S, Hoshino FB, Takeda N, Katayama K. 2003. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J Clin Microbiol 41:1548–1557. doi: 10.1128/JCM.41.4.1548-1557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]