ABSTRACT
The fungal genus Heterobasidion includes some of the most devastating conifer pathogens in the boreal forest region. In this study, we showed that the alphapartitivirus Heterobasidion partitivirus 13 from Heterobasidion annosum (HetPV13-an1) is the main causal agent of severe phenotypic debilitation in the host fungus. Based on RNA sequencing using isogenic virus-infected and cured fungal strains, HetPV13-an1 affected the transcription of 683 genes, of which 60% were downregulated and 40% upregulated. Alterations observed in carbohydrate and amino acid metabolism suggest that the virus causes a state of starvation, which is compensated for by alternative synthesis routes. We used dual cultures to transmit HetPV13-an1 into new strains of H. annosum and Heterobasidion parviporum. The three strains of H. parviporum that acquired the virus showed noticeable growth reduction on rich culturing medium, while only two of six H. annosum isolates tested showed significant debilitation. Based on reverse transcription-quantitative PCR (RT-qPCR) analysis, the response toward HetPV13-an1 infection was somewhat different in H. annosum and H. parviporum. We assessed the effects of HetPV13-an1 on the wood colonization efficacy of H. parviporum in a field experiment where 46 Norway spruce trees were inoculated with isogenic strains with or without the virus. The virus-infected H. parviporum strain showed considerably less growth within living trees than the isolate without HetPV13-an1, indicating that the virus also causes growth debilitation in natural substrates.
IMPORTANCE A biocontrol method restricting the spread of Heterobasidion species would be highly beneficial to forestry, as these fungi are difficult to eradicate from diseased forest stands and cause approximate annual losses of €800 million in Europe. We used virus curing and reintroduction experiments and RNA sequencing to show that the alphapartitivirus HetPV13-an1 affects many basic cellular functions of the white rot wood decay fungus Heterobasidion annosum, which results in aberrant hyphal morphology and a low growth rate. Dual fungal cultures were used to introduce HetPV13-an1 into a new host species, Heterobasidion parviporum, and field experiments confirmed the capability of the virus to reduce the growth of H. parviporum in living spruce wood. Taken together, our results suggest that HetPV13-an1 shows potential for the development of a future biocontrol agent against Heterobasidion fungi.
KEYWORDS: Heterobasidion, RNA-Seq, biocontrol, forest pathology, mycoviruses, wood decay
INTRODUCTION
Virus infections occur commonly in fungi. Many of them are cryptic (latent), but mutualistic or deleterious associations also are known (1–4). Fungal viruses (mycoviruses) that reduce the virulence of plant-pathogenic fungi may be used as biocontrol agents, as demonstrated by the use of hypoviruses in controlling the chestnut blight fungus, Cryphonectria parasitica, in Europe (5). Other examples of viruses associated with reduced host virulence (i.e., hypovirulence) include members of the families Narnaviridae (6), Endornaviridae (7), Megabirnaviridae (8), and Genomoviridae (9) and of the genus Botybirnavirus (10).
The Heterobasidion annosum complex (Bondarzewiaceae) consists of five species of conifer pathogenic fungi (11). Heterobasidion annosum sensu stricto is a root rot pathogenic fungus infecting mainly pines, but it also attacks spruce trees and a wide range of broadleaf trees (12). In pine trees, infection is limited to the roots and lower parts of stems. Advanced infection causes tree mortality and leads to the formation of disease centers with dead and dying trees. The pathogen spreads efficiently via airborne basidiospores that infect fresh stump surfaces or butt wounds of living trees as well as by vegetative spread of the mycelium via root contacts. Heterobasidion parviporum uses a similar dispersal strategy but infects mainly Norway spruce and causes extensive stem decay in living trees. Clonal individuals of H. parviporum are capable of spreading several meters inside spruce heartwood and may infect dozens of trees and survive for decades (13).
Heterobasidion species host members of Partitiviridae (14–20), Narnaviridae (21), and the unassigned taxon Heterobasidion RNA virus 6 (HetRV6) (22). Members of the virus family Partitiviridae have bipartite double-stranded RNA (dsRNA) genomes that infect plants, fungi, or protozoa (23). Partitiviruses were previously considered to be cryptic, but some members of this virus family mediate phenotypic alterations or hypovirulence in their host fungi (24–27). Vainio et al. (15) showed that the Heterobasidion alphapartitivirus HetPV3-ec1 causes growth reductions in Heterobasidion abietinum, and Hyder et al. (4) described both negative and positive associations for this virus species, depending on host strain and environmental conditions. Partitiviruses of Heterobasidion are readily transmitted between incompatible host strains and even distantly related species of Heterobasidion in the laboratory and in nature (14–17, 19, 28). This is rather uncommon among fungi, as somatic incompatibility often restricts virus transmission even between conspecific fungal strains (1, 5).
Here we show that the alphapartitivirus Heterobasidion partitivirus 13 from H. annosum (HetPV13-an1) causes severe growth debilitation and major alterations in the gene expression of its natural host and other sensitive hosts and reduces the growth and wood colonization efficacy of H. parviporum.
RESULTS
Phenotype of H. annosum 94233 with and without HetPV13-an1.
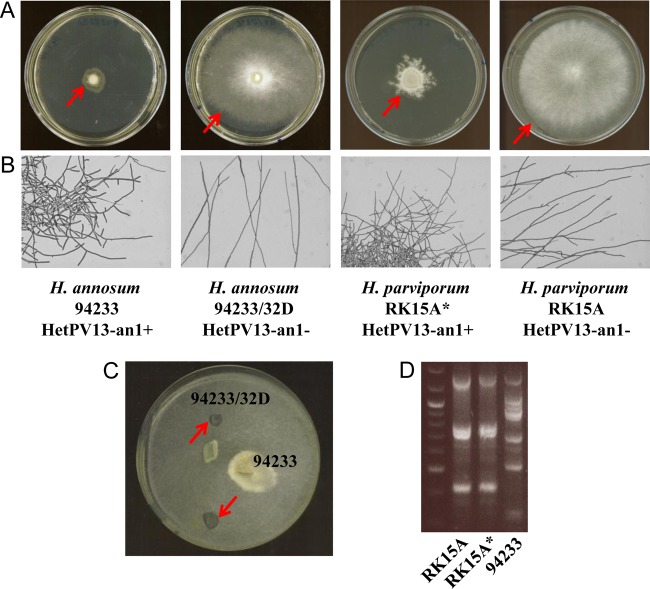
H. annosum 94233, the natural host of HetPV13-an1 (20), has a low growth rate and displays abnormal mycelial morphology characterized by increased hyphal branching (Fig. 1). Thermal treatment was used to cure H. annosum 94233 of HetPV13-an1 infection, which produced a fungal strain with normal hyphal morphology and growth, designated 94233/32D (Fig. 1). The absence of HetPV13-an1 in the cured isolate was verified by reverse transcription-PCR (RT-PCR) with specific primers PV13An1midFor2 and PV13midR (20) using four independent cDNA samples. Moreover, the introduction of HetPV13-an1 back to this cured strain regenerated the debilitated phenotype (Fig. 2). The virus reintroduction was conducted by incubating a dual culture of the cured and original strains for 3 months, after which two independent subcultures were taken from the recipient side (Fig. 1C). The subcultures were verified to be HetPV13-an1 positive based on RT-PCR with specific primers (20). The isogenic strains could be differentiated based on their original growth pattern on the agar plate (Fig. 1C). Both the original and cured strains of H. annosum 94233 are heterokaryons as determined by the presence of clamp connections.
FIG 1.
Hyphal morphology of Heterobasidion strains with and without HetPV13-an1 infection and genotype analysis. (A) Mycelial growth on MOS agar plates. The cultures were incubated for 11 days at 20°C. Arrows mark the edge of the fungal colony. (B) Light microscopy images of mycelial colonies grown on water agar plates. (C) Virus reintroduction via dual culture of the cured and original strains. Sampling sites are indicated by arrows. (D) Genotype analysis with M13 fingerprinting. The genotypes of H. parviporum RK15A free of HetPV13-an1 and virus-infected RK15A* compared to that of the virus donor strain H. annosum 94233 are shown.
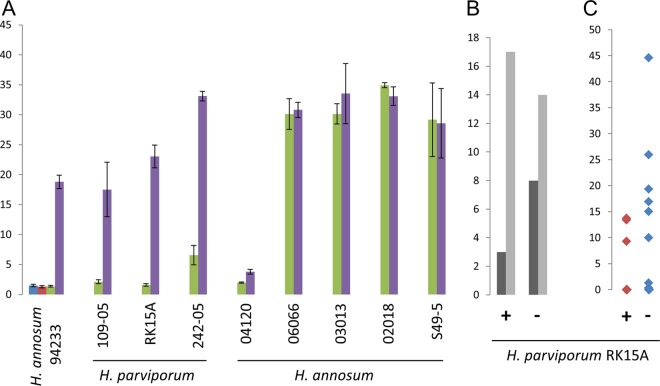
FIG 2.
Effects of HetPV13-an1 on the growth of H. annosum and H. parviporum. (A) Growth (in cm2) of Heterobasidion strains in 6 days on 2% MEA plates. Green bars show strains infected with HetPV13-an1, and purple bars indicate isogenic strains without the virus. The red and blue bars show two cultures of 94233/32D, which were reinfected with HetPV13-an1 (sampling is shown in Fig. 1C). The error bars represent standard deviation. (B) Growth of H. parviporum in Norway spruce trees in the forest, showing the number of trees with (dark gray) or without (light gray) H. parviporum growth 10 cm above the inoculation point. (C) Growth (in cm2) of H. parviporum in inoculated spruce trees. +, RK15A* (HetPV13-an1 infected); −, RK15A without HetPV13-an1.
HetPV13-an1 infection alters host transcription.
The effects of HetPV13-an1 infection on host transcription were analyzed by RNA sequencing (RNA-Seq), comparing isogenic strains of H. annosum 94233 with and without the virus. RNA-Seq was conducted with three biological replicates for each fungal strain and yielded ∼18 to 22 million reads for each replicate (see Table S1 in the supplemental material). The Illumina sequence reads were assembled to the reference genome, H. annosum v.2.0 (29) deposited at the Joint Genome Institute (JGI) (http://genome.jgi.doe.gov/Hetan2/Hetan2.home.html), which represents the North American species Heterobasidion irregulare that is closely related to H. annosum used for RNA-Seq in this study.
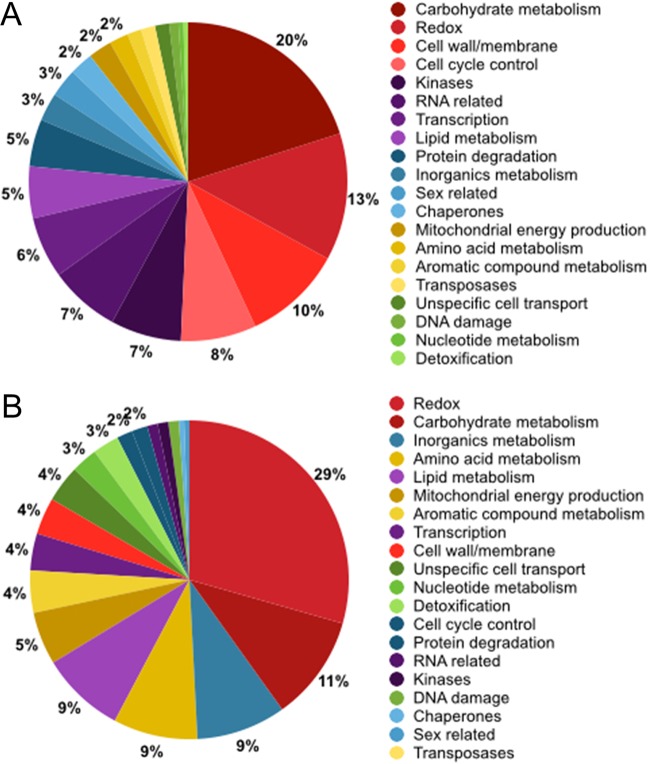
Based on the number of reads associated with each gene, a total of 683 transcripts were deregulated, with 60% (409) downregulated and 40% (274) upregulated, due to the presence of HetPV13-an1. The identified transcripts were annotated using the genome portal of the JGI webpage default analysis track option for annotated Heterobasidion annosum transcripts and NCBI's Delta-BLAST. Final annotation was based on corroborating evidence from protein domain identification, protein sequence similarity, and JGI portal user annotations. We were able to assign a probable function to 227 downregulated and 192 upregulated transcripts (see Table S2 in the supplemental material). The most prominent downregulated functional groups were carbohydrate metabolism, redox processes, cell cycle control, and cell wall- and membrane-related processes; these four groups comprised half of the downregulated annotated transcripts (Fig. 3). The most upregulated functional groups were redox processes, carbohydrate metabolism, inorganic metabolism, and amino acid metabolism.
FIG 3.
Differentially expressed genes due to the presence of HetPV13-an1 as revealed by RNA sequencing. (A) Annotated downregulated transcripts (n = 227) grouped according to transcript function. (B) Annotated upregulated transcripts (n = 192). The percentage of annotated transcripts in a functional category is indicated by the chart, and for clarity only percentages above 1 are shown.
(i) Carbohydrate metabolism.
The presence of HetPV13-an1 seems to interfere with host carbohydrate metabolism by inhibiting carbohydrate degradation and intake and to induce alternative carbohydrate pathways, such as the Leloir pathway, gluconeogenesis, and fermentation (Table S2). Carbohydrate metabolism was the largest functional group among the annotated downregulated genes (20% of the transcripts). Among the carbohydrate metabolic processes, 17 of the downregulated transcripts were related to plant carbohydrate polymer degradation (cellulose, hemicelluloses, and glycoside hydrolases), while only six of the upregulated transcripts were related to these processes. Seven carbohydrate transporter transcripts were downregulated, and two were upregulated. Two d-mannitol dehydrogenase transcripts were downregulated, possibly leading to mannitol accumulation in cells. Mannitol accumulation is required for stress tolerance in fungi, as mannitol can function as a carbohydrate storage and reactive oxygen scavenger (30).
(ii) Redox reactions and detoxification.
Several genes, such as those for P450 proteins, oxidoreductases, hydrolases, aldo/keto reductases, peroxidases, and monooxygenases, associated with redox reactions were affected by the HetPV13-an1 infection (Table S2). Redox transcripts constituted the largest functional group among the annotated upregulated genes (29% of transcripts) and the second largest group among the downregulated transcripts (13%). Redox proteins, related to various metabolic reactions, are essential, for example, for wood degradation and detoxification.
(iii) Cell wall- and membrane-related processes.
Ten percent of the annotated transcripts downregulated in the presence of HetPV13-an1 coded for proteins involved in cell wall- and membrane-related processes, such as murein transglycosylase, expansin-like protein, flotillin, several membrane-anchored proteins, hydrophobin 2, and serine-threonine-rich glycosylphosphatidyl-inositol-anchored membrane proteins (Table S2). Hydrophobins have been shown to be involved in conidium formation, while serine-threonine-rich glycosylphosphatidyl-inositol-anchored membrane proteins have a role in fruiting-body formation (31, 32). The large amount of deregulated cell membrane and cell wall transcripts is probably reflected by the striking appearance of HetPV13-an1-infected hyphae, which have a restricted and branched appearance (Fig. 1B).
(iv) Cell cycle.
Cell cycle regulator transcripts constituted 8% of all the annotated genes downregulated in the presence of HetPV13-an1. Among the affected transcripts were the cell cycle regulators Chk1 and cdc13, mitotic kinase NEK, and checkpoint proteins Mad2, Mob2, and Mrc1 (Table S2). Transcripts for separin, negative regulator of septation EzrA-like, and metacaspase were upregulated due to the presence of HetPV13-an1. Metacaspases are caspase-like mediators of apoptosis (33).
(v) Inorganic, amino acid, and lipid metabolism.
Genes affecting inorganic, amino acid, lipid, and aromatic compound metabolism were found to be uniquely upregulated due to the presence of HetPV13-an1. Transcripts related to inorganic metabolism comprised about 9% or all annotated upregulated sequences and included transcripts for polyamine transporters, ferric reductases, iron transporters, and sulfate transporters. The affected amino acid metabolic transcripts encoded, for example, amino acid transporters and proteins involved in the shikimate pathway (Table S2). The shikimate acid pathway is activated in yeast by the unavailability of amino acids, purines, or glucose (34). The lipid metabolism transcripts found to be upregulated were involved mainly in sterol synthesis (sterol o-acyltransferase, δ7-sterol-C5-desatyrase, squalene epoxidase, and terpenoid synthase).
(vi) Mitochondrial energy production.
The HetPV13-an1 infection increased the transcription of 10 genes predicted to be involved in the electron transport chain and the citric acid cycle (Table S2). Two transcripts involved in the citric acid cycle and one mitochondrial carrier protein were downregulated.
(vii) RNA-related processes.
Cells infected with HetPV13-an1 downregulated three transcripts affiliated with RNA silencing (one RNA-directed RNA polymerase QDE-1 and two Piwi domain-containing protein transcripts) (Table S2). Moreover, most of the seven annotated argonaute genes with typical domain arrangements (35) and three annotated dicer genes were somewhat (1.22- to 5.95 -fold) downregulated, and only one argonaute gene showed slight (1.88-fold) upregulation in the presence of HetPV13-an1 (Table 1).
TABLE 1.
Transcription level changes and primer sequences for the target genes used in RT-qPCR validation and for annotated genes related to the RNA silencing pathway
| TG | Predicted protein | Gene ID in H. annosum genome v.2.0 at JGI | RNA-Seq for H. annosum |
RT-qPCR validation |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Primer sequencesd | Length, bp | Annealing temp (°C) |
H. annosum |
H. parviporum |
||||||||||
| Mean expression, RPKM (94233/32D/94233)a | Fold changeb | FDR P valuec | CT (94233/32D/94233)e | Efficiencyf | Fold changeg | CT (RK15A/RK15A*)h | Efficiency | Fold change | ||||||
| 1 | Carbohydrate-binding module family 19 | fgenesh1_pm.02_#_834 | 69.59/5,528.19 | 78.99 | 1.96e−04 | GCTCCAGTGCTTCGCCTTGC, CTTCTTGGTGAACGCAGCACG | 166 | 58 | 18.9/13.3 | 1.71 | 30.74 | 20.9/16.0 | 1.72 | 30.91 |
| 2 | RTA1 domain-containing protein | Hetan1.estExt_Genewise1Plus.C_40642 | 11.74/158.68 | 13.51 | 2.30e−05 | CTATCATCCGACGTCACGACG, GAATGACAGCAGCTGGATGGC | 135 | 56 | 22.9/19.9 | 1.73 | 7.35 | 21.3/20.5 | 1.73 | 3.34 |
| 3 | Tetraspanin family protein | gw1.13.1244.1 | 55.52/765.35 | 13.80 | 4.52e−04 | CAATTGGCTGGTATCAGACTGG, CCTTCTTCTTGATGATAACG | 147 | 52 | 18.9/15.0 | 1.71 | 11.87 | 19.3/19.6 | 1.72 | 1.90 |
| 4 | Aryl-alcohol oxidase 9 | Hetan1.estExt_Genewise1Plus.C_130459 | 11.55/236.43 | 20.14 | 6.23e−05 | GCAAGAGCTATGATTACATCG, GGAATGGCGATGTTCTCGTTGC | 142 | 57 | 27.3/22.7 | 1.72 | 16.99 | 20.7/19.1 | 1.71 | 5.08 |
| 5 | Cytochrome P450 | estExt_Genemark.C_120425 | 5.17/787.86 | 158.37 | 1.03e−06 | CTTTCCTTCCCCTCTTCCAC, GCCGAGAAGGTCGATAACTG | 122 | 53 | 23.9/20.5 | 1.73 | 9.37 | NDi | ND | ND |
| 6 | MFS amino acid permease | Hetan1.fgenesh2_pm.C_scaffold_2000190 | 11.36/148.01 | 13.00 | 5.14e−06 | CGTCAACGCGCCGATCGTCG, GGATCGTGGTGCGTTTCAGCG | 80 | 60 | 22.4/18.9 | 1.72 | 9.41 | 21.2/19.9 | 1.73 | 3.48 |
| 7 | Pentafunctional AroM protein | e_gw1.13.1032.1 | 5.25/47.59 | 8.47 | 9.94e−04 | CGTCGTCGGTGATCTCGTGG, GGATTTGCACGAAAGAGACG | 68 | 53 | 18.9/16.5 | 1.69 | 4.99 | 20.3/18.3 | 1.68 | 6.30 |
| 8 | UDP-glucose 4-epimerase | estExt_Genewise1Plus.C_010240 | 82.59/1,311.17 | 14.85 | 1.64e−05 | GTGTTTGAGAAATATGGCAAGG, GTATGTCACAGGTATCTCTGTCG | 96 | 51 | 20.3/16.5 | 1.69 | 10.53 | 18.8/21.0 | 1.70 | −1.47 |
| 9 | Citrate synthase | Genemark.3471_g | 87.69/691.32 | 7.85 | 1.92e−05 | CAGTTCTCCCTCGCCGTCACC, CGGCGATGTTGGGCAGC | 142 | 57 | 19.5/15.0 | 1.73 | 17.68 | 18.6/17.0 | 1.70 | 5.21 |
| 10 | IucA/IucC (siderophore biosynthesis) | estExt_fgenesh1_pg.C_040344 | 2.49/42.20 | 16.53 | 6.53e−05 | AATATCGAGCTCCTATGTCTGG, GTGTGCGGCGCGCGCGATGC | 143 | 53 | 26.6/20.6 | 1.72 | 21.90 | ND | ND | ND |
| 11 | Catalase | Hetan1.estExt_Genewise1Plus.C_61257 | 16.92/613.30 | 35.32 | 2.81e−04 | GTTTGATAGACTTGGTACGACC, CGCATGAACTGTTCTCTCTGG | 109 | 52 | 21.6/16.5 | 1.72 | 22.78 | 21.2/19.9 | 1.71 | 4.50 |
| 12 | Fruit-body-specific gene C | Hetan1.estExt_fgenesh2_pm.C_70066 | 9.57/126.31 | 12.90 | 4.27e−04 | AGCTGTTGAAGCTCGGAGTC, AACACGTAGGTGAACCACAGC | 94 | 55 | 24.1/20.5 | 1.72 | 10.75 | 27.1/28.3 | 1.73 | 1.14 |
| 13 | MFS glucose transporter 1 | Hetan1.estExt_Genewise1Plus.C_100682 | 30.11/1.49 | −19.35 | 2.60e−05 | TTGGACATCTCGTCCATGTCTGG, GCATAGCAGCAACGATTGC | 103 | 55 | 28.2/33.2 | 1.81 | −13.51 | 26.8/26.4 | 1.75 | 2.81 |
| 14 | Lignin-expressed protein Lep1 | Hetan1.Genemark.6963_g | 8.62/1.29 | −7.02 | 1.79e−04 | CTTGGTCCGCCGACCTCTTCGG, AAGTAAGTTGTTTTGTTGAGC | 76 | 55 | 26.5/29.2 | 1.75 | −3.08 | 21.4/21.4 | 1.71 | 2.16 |
| 15 | Glycoside hydrolase family 3 protein | Hetan1.Genemark.8846_g | 27.56/2.48 | −11.01 | 9.36e−05 | CAACACAGTCAATATGTCTCC, GTCGCAAAGCTGAACTCTTCC | 138 | 54 | 19.5/23.4 | 1.72 | −5.62 | 25.8/26.0 | 1.70 | 1.98 |
| 16 | Serine/threonine kinase | gw1.08.1665.1 | 3.02/0.34 | −9.52 | 3.46e−04 | CATCGTCAAGTACGAGGGAATGG, CCTTGAGCGTCTGTCCGAGG | 95 | 57 | 20.4/20.3 | 1.71 | +1.00 | 25.0/22.2 | 1.73 | 9.99 |
| 17 | STE20-like serine/threonine kinase | fgenesh1_pm.04_#_184 | 14.91/0.01 | −593.95 | 2.22e−06 | GGTCATGTCAGATGCTATCG, GCACAAGACATAAATTGC | 204 | 54 | 25.5 | 1.71 | 23.9/25.5 | 1.68 | −1.08 | |
| 18 | RNA-directed RNA polymerase QDE-1 | estExt_fgenesh1_pg.C_020714 | 56.89/4.43 | −12.79 | 8.26e−05 | GACGATATTTGTGATTTCGTGG, CCATGCAACGCTTGTCATACG | 121 | 55 | 18.9/23.4 | 1.73 | −8.55 | 22.1/23.5 | 1.72 | −1.05 |
| 19 | Formin homology protein | Hetan1.fgenesh2_pg.C_scaffold_13000145 | 55.48/4.85 | −11.56 | 4.51e−05 | CAAGACGCAAGACGCAAGACTCG, GTTCTTGGAGTGCTGGGTGGTCG | 106 | 59 | 26.7/32.5 | 1.77 | −18.87 | ND | ND | ND |
| 20 | Piwi domain-containing protein | Hetan1.EuGene9000385 | 58.88/3.29 | −17.96 | 2.12e−04 | ATACAGTCGAAGTAGTCTTCGG, CTATCCTTTCGTCAGACTGC | 90 | 57 | 22.3/32.5 | 1.82 | −333 | 20.6/23.8 | 1.76 | −2.81 |
| 21 | HD1 homeodomain transcription factor, mating type protein | Hetan1.EuGene1000636 | 26.14/0 | −1,388.3 | 4.71e−07 | GTCAAAGGATCTTGAGGACACC, GGTATGGATAGTGAAGATTTGC | 113 | 55 | 19.5 | 1.69 | ND | ND | ND | |
| 22 | Transcription factor of the Forkhead/HNF3 family | gw1.04.2473.1 | 13.63/0.91 | −15.07 | 2.20e−04 | GATCGAGGACCGCTTCGAGTGG, GAAGCACTTGCGGAGTGACAGG | 67 | 58 | 22.7/26.9 | 1.72 | −6.85 | 24.0/23.4 | 1.78 | 3.17 |
| 23 | Glycoside hydrolase family 7 protein | Hetan1.estExt_Genewise1.C_110899 | 3,841.42/6.34 | −612.85 | 3.10e−04 | GGTTACTGCGACACGCAGTGC, GCTGTCGGAGGACGAAGG | 90 | 58 | 14.9/25.4 | 1.71 | −200 | 23.4/15.0 | 1.75 | 231 |
| 24 | Mitotic spindle checkpoint protein | e_gw1.01.3559.1 | 469.88/20.03 | −23.82 | 2.49e−05 | ATATCTCTGGGGCCATCAAA, TTGATGACGACGACCACTTT | 76 | 52 | 20.5/22 | 1.67 | −3.00 | 24.5/26.4 | 1.78 | −1.35 |
| 25 | Hydrophobin 2 HAH2 | Hetan1.estExt_fgenesh3_kg.C_90130 | 7,313.37/162.18 | −53.45 | 3.15e−05 | CGTCGCTCGTGATACCATC, CGAGCAGGCCAAGAAGAG | 113 | 57 | 13.4/19 | 1.70 | −20.83 | 14.5/14.0 | 1.67 | 2.74 |
| 26 | Cytochrome P450 monooxygenase 8 | Hetan1.estExt_Genewise1Plus.C_70955 | 18.18/0.02 | −538.92 | 3.33e−05 | CTATGTGGACTTGGCGGAGA, TAGAGCACCTGGGAACCAAC | 121 | 54 | 20.4/24.2 | 1.78 | −5.13 | 21.5/25.4 | 1.70 | −3.80 |
| 27 | Piwi domain-containing protein | Hetan1.estExt_fgenesh2_pm.C_90173 | 59.18/3.08 | −19.2 | 1.23e−04 | GGTTGCAGAAGTCCAAACAGG, CTGAGCACTTTGTCGAATTGG | 153 | 53 | 18.5/23.7 | 1.71 | −10.87 | 25.4/29.4 | 1.72 | −4.02 |
| 28 | Zfp5 (zinc finger domain containing protein) | Hetan1.EuGene4000639 | 43.76/3.57 | −13.26 | 1.63e−04 | CCGTCATGAAATCATTCATG, CACTGATCTTGCATACGTGG | 130 | 52 | 19.7/23.5 | 1.67 | −4.95 | 27.6/30.4 | 1.65 | −1.85 |
| Argonaute | e_gw1.02.2254.1 | 6.75/12.73 | 1.878 | 7.41e−05 | ACACTCCAGGGCTCACAATC, GTTCTGCACGTTGGTGAATG | 110 | 55 | 20.4/20.9 | 1.72 | 1.12 | 21.5/22.9 | 1.71 | 1.01 | |
| e_gw1.03.2120.1 | 0.62/0.16 | −4.407 | 7.87e−02 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ||
| Hetan1.estExt_Genewise1.C_80495 | 97.19/43.94 | −2.439 | 0.1390 | CCAGCATGATCACGGAACT, AGACACCGTCGCGGTAGA | 111 | 55 | 20.7/20.8 | 1.71 | 1.34 | 28.1/28.0 | 1.71 | 2.28 | ||
| estExt_Genewise1.C_033087 | 245.74/42.08 | −5.952 | 3.57e−03 | AACGAGAGTTGCGTTGCATT, GAAGAAGCGAACGTGGTGAC | 101 | 55 | 17.2/17.0 | 1.70 | −1.08 | 21.8/22.7 | 1.67 | 1.34 | ||
| fgenesh1_pg.03_#_809 | 5.55/1.35 | −4.072 | 0.1360 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ||
| estExt_fgenesh1_pg.C_030285 | 41.14/25.47 | −1.64 | 3.78e−03 | TCTTCTATCGAGATGGTGTCAGTG, GCGCAAGCCTTCTTTATAAGC | 82 | 55 | 19.3/18.0 | 1.73 | 2.832 | 24.3/26.2 | 1.73 | −1.27 | ||
| estExt_Genewise1Plus.C_110340 | 2.68/3.96 | 1.481 | 0.1467 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ||
| Dicer | Hetan1.Genemark.5915_g | 16.93/8.89 | −1.901 | 9.26e−05 | CCAGTGCCTTGCTCACACT, CGGGTCAATGTCGTATATGG | 110 | 55 | 19.3/19.8 | 1.67 | 1.07 | 19.5/20.8 | 1.71 | 1.12 | |
| Hetan1.EuGene2000966 | 11.85/11.97 | 1.007 | 0.9647 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ||
| e_gw1.03.2068.1 | 41.26/33.95 | −1.218 | 9.79e−02 | CAAAGCCACCTGCTAGATACC, TCGCATGATCGATGGAAGTA | 101 | 55 | 18.9/19.6 | 1.67 | 1.01 | 20.1/21.5 | 1.71 | 1.04 | ||
Mean expression levels in H. annosum 94233/32D and H. annosum 94233 based on three biological replicates used for RNA-Seq. RPKM, reads per kilobase of transcript per million mapped reads.
Expression ratio (fold change) between H. annosum 94233 and 94233/32D. In this analysis, the fold change results from a linear modeling process performed with the R package Limma (Bioconductor). Positive values indicate upregulated genes and negative values downregulated genes. Values in bold indicate the largest inconsistencies between the RNA-Seq and RT-qPCR data.
False-discovery rate-adjusted P value for the fold change in gene expression between H. annosum 94233 and H. annosum 94233/32D.
Primer sequences designed based on the H. irregulare genome sequence version 2.0 available at JGI.
CT, cycle threshold used for calculating fold changes (take-off point) for H. annosum 94233/32D and H. annosum 94233.
Average reaction efficiency from all replicates based on comparative quantitation analysis in RT-qPCR as determined with the Relative Expression Software Tool (REST) version 2009 (Qiagen). The integrated randomization and bootstrapping methods used in REST 2009 software test the statistical significance of calculated expression ratios. Efficiency is determined using the best fit for the standard curve and is used in the randomization process.
Fold change based on relative expression ratios at take-off points normalized with three reference genes. Two highly downregulated target genes (TGs 17 and 21) did not yield any amplification products from H. annosum 94233, and therefore the fold change could not be determined.
CT, cycle threshold used for calculating fold changes (take-off point) for H. parviporum RK15A and H. parviporum RK15A*.
ND, not detected as none of the tested primers were suitable for H. parviporum.
Validation of the RNA-Seq results using RT-qPCR and transcript levels of selected TGs in H. parviporum.
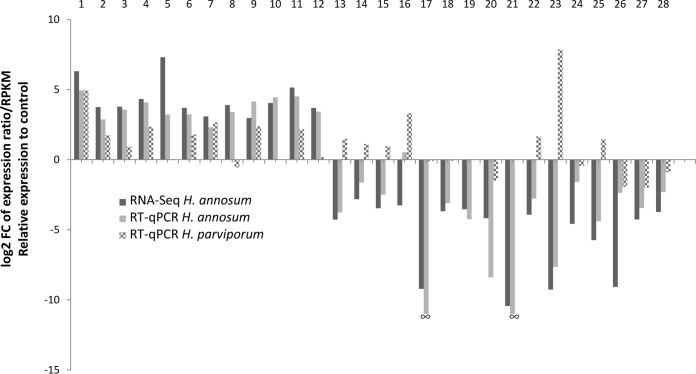
Twenty-eight target genes (TGs) that were either up- or downregulated due to the presence of HetPV13-an1 and representing diverse cellular functions were selected for reverse transcription-quantitative PCR (RT-qPCR) validation (Table 1). In H. annosum, the relative expression of genes was normalized using three internal reference genes, i.e., those for RNA polymerase III transcription factor, α-tubulin, and actin, and their expression was found to be relatively stable. The vast majority (96.4%) of genes inspected confirmed results obtained using RNA-Seq, and only one of the 28 genes tested (TG 16, encoding STE20-like serine/threonine kinase) showed contrasting results (and an ∼11-fold difference in its expression) between the two analysis methods (Table 1; Fig. 4). There were also minor differences in the magnitude of changes in relative gene expression as revealed by the two methods. Two of the highly downregulated genes tested (TG 17 and TG 21, with 594- to 1,388 -fold downregulation, respectively [Table 1]) did not show any amplification in H. annosum 94233 using RT-qPCR, which confirmed the extremely low expression levels found by RNA-Seq (amplification was successful in the cured control strain 94233/32D).
FIG 4.
Fold changes in gene expression between H. annosum 94233 and 94233/32D and H. parviporum RK15A* and RK15A. Detailed information is reported in Table 1 and in Table S3 in the supplemental material. RNA-Seq relative expression relates to log2 fold change of reads per kilobase of transcript per million mapped reads (RPKM). RT-qPCR relative expression corresponds to log2 fold change of expression ratios based on the take-off values normalized with three reference genes. Target genes 17 and 21 did not yield any amplification products from H. annosum 94233, and therefore the fold change could not be determined by RT-qPCR. Genes 1, 8, 13 to 15, and 23 were related to carbohydrate metabolism; genes 2, 4, 5, 11, and 26 were related to redox reactions and detoxification; genes 3 and 25 were related o cell wall and membrane related processes; genes 16, 17, and 24 were related to cell cycle control; gene 10 was related to inorganic metabolism; genes 6 and 7 were related to amino acid metabolism; gene 9 was related to mitochondrial energy production; genes 18, 19, and 27 were related to RNA-related processes; genes 22 and 28 were related to transcription, and genes 12 and 21 were related to sex.
There is currently no genome sequence available for H. parviporum, which is relatively distantly related to H. annosum and H. irregulare, which represent the “pine clade” of Heterobasidion annosum sensu lato (36). Therefore, the analysis of gene expression by RNA-Seq based on read mapping against the H. irregulare genome is not feasible. However, the 28 qPCR primers designed for H. annosum (Table 1) were tested for H. parviporum, and most of them (86%) showed adequate amplification by RT-qPCR. Three internal reference genes were used for the normalization of relative expression of H. parviporum genes: those for RNA polymerase III transcription factor, RNA polymerase II transcription factor, and actin. Most (90%) of the genes found to be upregulated due to the presence of HetPV13-an1 in H. annosum were also upregulated in H. parviporum. However, 43% of the genes which were downregulated in H. annosum due to the presence of HetPV13-an1 were found to be upregulated in H. parviporum (Fig. 4), suggesting that the two species of Heterobasidion differ somewhat in their response to the presence of HetPV13-an1. The most striking difference was observed in target gene 23 (encoding a glycoside hydrolase family 7 protein), which was 200-fold downregulated in H. annosum and 230-fold upregulated in H. parviporum due to the presence of HetPV13-an1 (Table 1). However, it should be noted that the Heterobasidion strains investigated differ also in their ploidy (the H. annosum isolate being heterokaryotic and the H. parviporum isolate homokaryotic), as well as in the presence of cryptic mitoviruses in H. annosum 94233/32D (see below).
In addition to the 28 target genes described above, we also designed primers for the annotated argonaute and dicer genes. Four argonaute and two dicer genes (35) showed adequate amplification by RT-qPCR in H. annosum and also in H. parviporum (Table 1), which confirmed the view obtained by RNA-Seq, revealing only less-than-3-fold changes in their transcript levels in both host species due to the presence of HetPV13-an1.
Virus transmission experiments.
We used dual cultures in an attempt to transmit HetPV13-an1 into strains of H. annosum and H. parviporum. Virus transmission into conspecific strains was efficient, and eight heterokaryotic strains (H. annosum 93160, 93173, 93177, 00002, 02018, 03013, 06066, and S49-5) and one homokaryotic strain (H. annosum 04120) successfully received HetPV13-an1 (Table 2). Strains of H. parviporum were infected less frequently, and only two homokaryotic strains (H. parviporum RK15A and 109-05) acquired the virus (Table 2). Furthermore, we could successfully transmit HetPV13-an1 from H. parviporum RK15A into the heterokaryotic H. parviporum 242-05. Based on somatic incompatibility testing and/or genetic fingerprinting, genotypes of the recipient strains remained unaltered during mycelial contact (Table 2; Fig. 1D), which demonstrated that the presence of the virus was not due to intermixture of donor and recipient hyphae.
TABLE 2.
Virus transmission experimentsa
| Strain | Species | Collection site in Finland | Host tree | Collector(s) | Yr | HetPV13-an1 transmission | Genotypeb | Growth debilitationc |
|---|---|---|---|---|---|---|---|---|
| 04120 | H. annosum sensu stricto | Myrskylä | Picea abies | K. Korhonen | 2004 | + | n/RAMS | Yes |
| 07001 | H. annosum sensu stricto | Leppävirta | Pinus sylvestris | T. Hynönen | 2007 | − | 2n | NA |
| 93177 | H. annosum sensu stricto | Toivakka, Huikko | Pinus sylvestris | K. Korhonen | 1993 | + | 2n/si | ND |
| 05008 | H. annosum sensu stricto | Dragsfjärd, Rosendal | Pinus sylvestris | J.-O. Granvik | 2005 | − | 2n | NA |
| S26-5 | H. annosum sensu stricto | Läyliäinen | Pinus sylvestris | T. Piri, H. Nuorteva | 2006 | − | 2n | NA |
| S49-5 | H. annosum sensu stricto | Läyliäinen | Pinus sylvestris | T. Piri, H. Nuorteva | 2006 | + | 2n/M13, RAMS | No |
| 00002 | H. annosum sensu stricto | Kirkkonummi, Solbacka | Picea abies | K. Korhonen | 2000 | − | 2n | NA |
| 03019 | H. annosum sensu stricto | Västanfjärd, Nivelax | Pinus sylvestris | K. Korhonen | 2003 | + | 2n/si | ND |
| 93173 | H. annosum sensu stricto | Liljendal | Pinus sylvestris | K. Korhonen | 1993 | + | 2n/si | ND |
| 06066 | H. annosum sensu stricto | Alajärvi | Pinus sylvestris | A. Pajula | 2006 | + | 2n/si | No |
| 03013 | H. annosum sensu stricto | Jämsänkoski | Picea abies | K. Korhonen | 2003 | + | 2n/si | No |
| 02018 | H. annosum sensu stricto | Kisko, Toija | Pinus sylvestris | K. Lipponen, J. Aarnio | 2002 | + | 2n/si | No |
| 93160 | H. annosum sensu stricto | Ruotsinpyhtää, Tesjoki | Picea abies | K. Korhonen | 1993 | + | 2n/si | No |
| RK15A | H. parviporum | Tuusula | Picea abies | E. Vainio, T. Piri | 2010 | + | n/M, M13 | Yes |
| RK5A | H. parviporum | Tuusula | Picea abies | E. Vainio, T. Piri | 2010 | − | 2n | NA |
| RK13A | H. parviporum | Tuusula | Picea abies | E. Vainio, T. Piri | 2010 | − | 2n | NA |
| 5R182 | H. parviporum | Tuusula | Picea abies | T. Piri | 2005 | − | 2n | NA |
| RKU3.1.26 | H. parviporum | Tuusula | Picea abies | T. Piri | 1994 | − | 2n | NA |
| RKU3.2.34 | H. parviporum | Tuusula | Picea abies | T. Piri | 1994 | − | 2n | NA |
| LAP3.11 | H. parviporum | Lapinjärvi | Picea abies | T. Piri | 1991 | − | 2n | NA |
| 15-05 | H. parviporum | Tuusula | Picea abies | T. Piri | 2005 | − | 2n | NA |
| 73-05 | H. parviporum | Tuusula | Picea abies | T. Piri | 2005 | − | n | NA |
| 184-05 | H. parviporum | Tuusula | Picea abies | T. Piri | 2005 | − | n | NA |
| 109-05 | H. parviporum | Tuusula | Picea abies | T. Piri | 2005 | + | n/M, M13 | Yes |
| 242-05d | H. parviporum | Tuusula | Picea abies | T. Piri | 2005 | + | 2n/M13, RAMS | Yes |
The natural host of HetPV13-an1 is H. annosum 94233, isolated from Poland (20).
n, homokaryotic (haploid) recipient strain; 2n, = heterokaryotic (effectively diploid) recipient strain. Genome screening method after virus transmission: si, somatic incompatibility; M, mitochondrial markers; M13, M13 minisatellite fingerprinting; RAMS, = RAMS-PCR with primers CT, CCA, and CGA. In all cases, the genotype corresponded to the recipient strain.
NA, not applicable (no virus transmission detected); ND, = not determined.
This strain was cured of indigenous viruses by thermal treatment (consecutive incubations at 32 to 35°C for ∼10 weeks), and HetPV13-an1 was transmitted from H. parviporum RK15A.
It should be noted that based on small RNA deep sequencing (21) and RNA-Seq analysis (this study), H. annosum 94233 was found to harbor cryptic mitovirus infections that were not detected by dsRNA extraction. Based on RT-PCR with specific primers, two mitovirus strains (including the previously described HetMV1-an1 and a second, previously unknown mitovirus) remained in the normally growing 94233/32D strain cured of HetPV13-an1, but the mitoviruses were not transmitted to any of the H. parviporum or H. annosum recipient strains that successfully received HetPV13-an1 via hyphal contact. We found no indication of the presence of other virus taxa (including gemycircularviruses) in H. annosum 94233 based on de novo contig assembly of RNA-Seq reads not aligned to the reference genome.
Variation of host tolerance toward HetPV13-an1.
The presence of HetPV13-an1 is associated with severe growth debilitation in its natural host, H. annosum 94233 (Fig. 1 and 2) (t test, P < 0.001). Based on the growth rate on rich culturing medium (2% malt extract agar [MEA]), one heterokaryotic and two homokaryotic strains of H. parviporum, 242-05, RK15A, and 109-05, also showed significant growth reductions due to the presence of HetPV13-an1 (Fig. 1 and 2) (P < 0.001). The slow-growing homokaryotic H. annosum 04120 was also affected by the virus (P < 0.001). However, several of the tested H. annosum strains tolerated the presence of the virus and showed no apparent growth reduction due to the presence of HetPV13-an1 (Fig. 2).
Effects of HetPV13-an1 on the growth of H. parviporum in spruce trees.
We assessed the wood colonization efficacy of H. parviporum strain RK15A using 46 large living spruce trees that were inoculated with isogenic strains with or without HetPV13 infection (Fig. 5). The spruce trees were cut two growing seasons after inoculation, and the occurrence of Heterobasidion infection in each tree was assessed by measuring the area occupied by the fungus based on the presence of conidiophores in the wood. Cultures were established from all detected Heterobasidion colonies, their identity was examined by genotyping with microsatellite markers, and any contaminants were omitted from the analysis. The numbers of acceptable trees (without preexisting Heterobasidion infections) for the HetPV13-an1-infected and uninfected strains were 20 and 22, respectively. The numbers of trees with Heterobasidion infection 10 cm above the inoculation points were three and eight (15.0% and 36.4%) (Fig. 2B), and the corresponding additive areas of wood colonization were 36.5 and 133.5 cm2, respectively (P = 0.067) (Fig. 2C). Interestingly, different tree clones differed in their susceptibility to the fungus: no Heterobasidion growth was observed 10 cm above the inoculation site in two of the clones (KU45 and KU208), while the inoculated H. parviporum strains grew efficiently in two other tree clones (KU42 and KU70).
FIG 5.
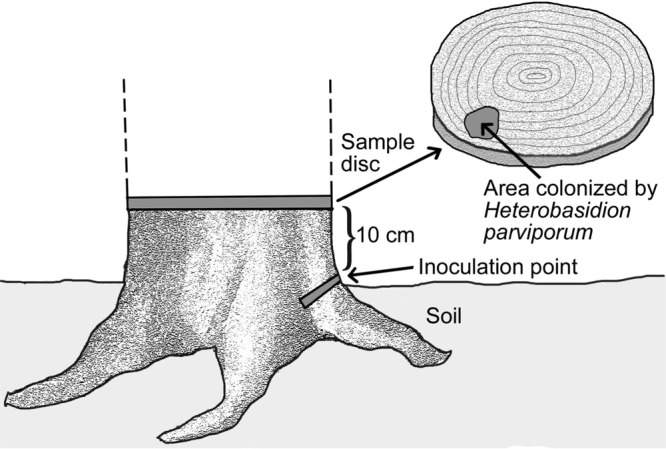
Sampling scenario for the spruce trees. The ca. 40-year-old Norway spruce trees were inoculated by inserting a spruce wood plug colonized by mycelia of H. parviporum RK15A or RK15A*. The trees were felled after two growing seasons, and sample disks were taken ∼10 cm above the inoculation point and examined for the spread of H. parviporum based on the microscopic inspection of conidiophores.
DISCUSSION
In this study, we provide evidence that the alphapartitivirus HetPV13-an1 is the main causative agent of a viral disease of an economically important root rot fungus. Detailed analysis of gene expression showed that many basic cellular functions of the original host of HetPV13-an1 were affected by the presence of the virus. The alterations observed in carbohydrate and amino acid metabolism suggest that the virus causes a state of starvation that the fungus attempts to compensate for by alternative synthesis routes.
In earlier investigations, viruses associated with hypovirulence have been shown to alter the gene expression of their host in many different ways. Cryphonectria parasitica hypovirus 1 affects host signal transduction pathways and induces the expression of dicer gene dcl2 and argonaute gene agl2. The virus also encodes a papain-like protease p29 acting as an RNA silencing suppressor (37, 38). In this study, we saw no strong response in annotated genes of the RNA silencing pathway. The polypeptide encoded by open reading frame (ORF) A of the Thanatephorus cucumeris mitovirus might interfere with specific steps of the shikimate pathway in the basidiomycete Rhizoctonia solani (6). In Fusarium graminearum, four phylogenetically different mycoviruses (FgV1 to -4) caused distinct changes in host transcriptomes, but the observed gene expression changes did not always result in phenotypic effects (39). In the case of HetPV13-an1, gene expression alterations related to many basic cellular functions from carbohydrate metabolism and chaperone functions to fungal self-defense and cell cycle control were associated with a seriously debilitated host phenotype. The mechanism behind these effects still requires further investigation, and it should be noted that both HetPV13-an1 and another Heterobasidion alphapartitivirus, HetPV3-ec1, produce a considerably larger amount of polymerase transcripts than capsid transcripts, which has been suggested to interfere with cellular processes and thereby contribute to adverse viral effects (4, 21, 40).
Partitiviruses were long considered to form cryptic and persistent associations with their host fungi. However, many recent reports challenge this view, and there are several studies describing partitiviruses associated with phenotypic changes. Magae and Sunagawa (24) found that the presence of an alphapartitivirus, Flammulina velutipes browning virus, correlates with brown discoloration of the host fungus. Bhatti et al. (25) found that the presence of a gammapartitivirus in Aspergillus fumigatus caused significant reductions in radial growth and biomass. Zheng et al. (26) described an alphapartitivirus causing hypovirulence in Rhizoctonia solani, a soilborne basidiomycete fungus infecting a wide range of hosts, including vegetables, ornamentals, and trees. Xiao et al. (27) found that the betapartitivirus Sclerotinia sclerotiorum partitivirus 1 reduces the virulence of its plant-pathogenic host fungus, a soilborne fungus infecting economically important crops, including rapeseed and soybean. Interestingly, Sasaki et al. (41) showed that a megabirnavirus of the root rot pathogen Rosellinia necatrix confers hypovirulence with the aid of a coinfecting partitivirus.
In this study, HetPV13-an1 caused phenotypic debilitation in both homokaryotic and heterokaryotic strains of H. parviporum and H. annosum, and therefore the effects were not determined by host species or nuclear condition. However, in H. annosum we found a relatively high level of natural tolerance toward HetPV13-an1. Thus, the presence of HetPV13-an1 did not alter the growth of four of the six H. annosum strains, whereas all three strains of H. parviporum were significantly debilitated by the presence of HetPV13-an1. This is consistent with results reported by Jurvansuu et al. (40) that showed more pronounced viral effects after the introduction to new host species. However, a considerably larger collection of Heterobasidion strains should be investigated for viral effects to determine the level of tolerance in both host species. Moreover, even seriously debilitated fungal strains differed somewhat in their response toward HetPV13-an1 infection at the gene expression level, although most of the genes investigated were similarly regulated in the two host species. Our results also showed that the growth debilitation observed under laboratory conditions correlates with poor capability of colonization of living trees. However, the variability of viral effects due to host strain and environmental conditions seems to be a common phenomenon for partitiviruses infecting Heterobasidion species (4). It should also be noted that the presence of coinfecting viruses may affect the host phenotype in a complex way, and multiple virus strains typically accumulate in clonally spreading Heterobasidion mycelia (19). In this study, the original host strain of HetPV13-an1 was coinfected by apparently cryptic mitoviruses, whereas HetPV13-an1 was the only virus transmitted to the recipient strains showing growth debilitation after virus introduction, indicating that HetPV13-an1 was the causative agent of the observed phenotypical changes.
HetPV13 strains closely similar to HetPV13-an1 occur naturally in H. annosum and H. parviporum (20, 42), but these strains do not cause significant levels of growth debilitation on artificial culturing media. Future studies are needed to examine which specific sequence polymorphisms between the virus strains have a contribution to host effects and to investigate these effects in natural conditions. Regarding potential biocontrol applications, the common occurrence of similar virus strains in nature suggests a low risk level of artificial introduction of HetPV13-an1.
Due to the high level of host tolerance against HetPV13-an1 observed in Finnish strains of H. annosum, we selected a homokaryotic H. parviporum strain for field experiments to test the biocontrol potential of HetPV13-an1. The virus-infected strain showed considerably less growth within the trees than the isogenic isolate without HetPV13-an1, indicating that the virus also caused growth debilitation in natural substrates. In practice, the wood colonization efficacy mainly determines the economic damage caused by this fungus, which is capable of spreading several meters inside spruce heartwood without killing the tree. Regarding putative biocontrol applications, the use of homokaryotic Heterobasidion strains as virus donors may be advantageous because Heterobasidion homokaryons are considered only weakly pathogenic as they rarely spread between living trees (43). Furthermore, the use of a homokaryon may allow more efficient virus transmission than for heterokaryotic strains, as homokaryons transmit viruses via mating in addition to anastomosis contacts without nuclear migration (44). Further studies are needed to assess the effectiveness of virus transmission from the donor strains into native Heterobasidion strains and practical inoculation methods. As Heterobasidion species cause estimated annual losses of €800 million in Europe (36), a biocontrol method restricting their spread in infected forest stands would be highly beneficial to forestry and deserves further investigation.
MATERIALS AND METHODS
Sample preparation for RNA sequencing.
The natural host of HetPV13-an1 is H. annosum 94233 from Poland (20). To examine the effects of this virus, the host strain was cured of HetPV13-an1 infection using thermal treatment, i.e., 4 weeks of incubation at 32°C on 2% MEA. The fungal strains were incubated on cellophane-covered modified orange serum (MOS) agar plates for 20 days for virus-infected 94233 and 5 days for the isogenic strain devoid of HetPV13-an1, 94233/32D. This ensured that both isogenic strains were in a state of active growth and allowed for collecting enough mycelia from the slow-growing virus-infected strain. Mycelia were collected and homogenized in TRI reagent (Molecular Research Center Inc., USA) using the Fast-Prep FP120 homogenizer (JT Baker, Holland) and quartz sand grains (1 to 2 mm in diameter) as recommended by the manufacturers. After isopropanol precipitation, total RNA was further cleaned using the E.Z.N.A. fungal RNA kit (Omega Bio-Tek), and RNA was resuspended into diethyl pyrocarbonate (DEPC)-treated water (G. Biosciences, USA). RNA quality and quantity were tested with NanoVue (GE Healthcare, USA) and a Bioanalyzer 2100 (Agilent Technologies, USA). Each analysis was conducted using three biological replicates. The samples were prepared using the TruSeq RNA Library Prep kit v2 (Illumina Inc.). The process included poly(A) selection with oligo(dT) beads without the depletion or rRNA. The samples were sequenced with an Illumina HiSeq2500 instrument using paired-end sequencing chemistry with 50-bp read length (Bioinformatics Core of Turku Centre for Biotechnology, University of Turku and Åbo Akademi University).
Bioinformatics.
Gene expression analysis was carried out in the Bioinformatics Core of the Turku Centre for Biotechnology, University of Turku and Åbo Akademi University. The Illumina sequence reads were assembled to the reference genome, H. annosum v.2.0 deposited at the JGI. Read alignment was performed in two stages. First, the reads were aligned against the H. annosum v.2.0 reference genome using TopHat version 2.0.10 (45). A summary of the mapping statistics is provided in Table S1 in the supplemental material. Next, the reads were associated with known genes based on RefSeq annotations derived from the H. annosum v.2.0 reference genome, and the number of reads associated with each gene was counted using the HTSeq tool v.0.5.4p3 (Python Software Foundation). The data were normalized to remove variation between samples caused by nonbiological reasons and to make values comparable across the sample set. Here the counts were normalized using the TMM normalization method of the edgeR R/Bioconductor package. All analyses were performed using R version 3.1.0 (R Core Team, 2014) and the bioinformatics-related Bioconductor module version 2.14.
Differentially expressed genes (DEGs) were filtered based on statistical significance and the size of the difference in the mean expression levels between sample groups. The expression level of each gene was measured by the number of sequenced reads that mapped to the sequence of the gene. The read counts were normalized as reads per kilobase of transcript per million mapped reads (RPKM). DEGs were identified based on more-than-4-fold changes and a modified t test P value (false-discovery rate P value) of 0.001.
The presence of viruses in H. annosum 94233 was reexamined based on the RNA-Seq data. Sequencing reads that remained unaligned against the H. irregulare genome sequence at JGI (H. annosum v.2.0) were de novo assembled with Geneious R10 Tadpole assembler (k-mer = 69; minimum contig length = 201). The resulting contigs (n = 15,147) were mapped against the host genome sequence (Geneious for RNA-Seq assembler), resulting in 9,383 unaligned contigs, which were examined for sequence homology against the GenBank virus database (taxid 10239) using NCBI BLASTX and BLASTN with an expect threshold of 0.1.
Validation of gene expression data with RT-qPCR.
Twenty-eight genes that were differentially expressed due to the presence of HetPV13-an1 and encoded diverse cellular functions were selected for validation by RT-qPCR (Table 1; see Table S2 in the supplemental material). Primers were designed using Primer3 (version 0.4.0) (Whitehead Institute for Biomedical Research) (Table 1). H. annosum 2.0 at JGI was used as a reference genome in primer design. Additional primers were designed for the argonaute and dicer genes described previously (35). All primers were also tested for H. parviporum.
Total RNA was extracted as described above for RNA-Seq using three biological replicates for each strain. The mycelia were grown for 2 to 3 weeks at 20°C. Two micrograms of total RNA was treated with DNase (Thermo Scientific, USA) according to the manufacturer's instructions and reverse transcribed using the RevertAid first-strand cDNA synthesis kit (Thermo Scientific, USA) and random hexamer primers (Thermo Scientific, USA), according to the manufacturer's recommendations. The cDNAs were diluted 1:1 with water before using them in RT-qPCR.
RT-qPCRs were set up using EvaGreen qPCR Mix Plus (Solis BioDyne, Estonia) with 2 μl of cDNA and 1 μl (10 μM) of each primer in a total volume of 20 μl in the 36-well rotor of Rotor-GeneQ (Qiagen, USA), according to manufacturer's recommendations. Cycling conditions were as follows: preincubation at 95°C for 15 min, denaturation at 95°C for 15 s, annealing at a primer-specific temperature (Table 1) for 20 s, and extension at 72°C for 20 s. Two technical replicates were prepared for each biological replicate. The quality and specificity of the reactions were examined using melting curve profiles and agarose gel electrophoresis.
The results were normalized using three internal reference genes and primer pairs described by Raffaello and Asiegbu (46) for RNA polymerase III transcription factor, actin, and either α-tubulin (H. annosum) or RNA polymerase II transcription factor (H. parviporum), along with the 28 target genes. Relative gene expression ratios were determined based on comparative quantitation analysis data using the Relative Expression Software Tool (REST) version 2009 (Qiagen), which uses multiple reference genes for normalization and integrated randomization and bootstrapping methods to test the statistical significance of calculated expression.
Virus transmission experiments.
HetPV13-an1 was transmitted to dsRNA-free Heterobasidion strains using dual cultures as described previously (28). Briefly, the virus-infected heterokaryotic donor strain, H. annosum 94233, and a dsRNA-free recipient strain were inoculated on a single MEA plate and incubated for 1 to 3 months. The experiment included 13 strains of H. annosum and 12 strains of H. parviporum (Table 2). Each strain combination was inoculated on two replicate plates. The absence of preexisting dsRNA infections in the recipient strains was verified using CF11 chromatography. After the incubation, a subculture was taken from the recipient side of the plate and analyzed for the presence of HetPV13-an1 using RT-PCR with the specific primers PV13midFor2 and PV13midR (20). Similarly, specific primers were designed based on the mitoviral contigs detected by RNA-Seq and used for virus screening.
To ensure that the virus-infected recipient isolates maintained their original genotype after the dual cultures, their genotypes were examined based on somatic incompatibility reactions (47), mitochondrial large-subunit ribosomal DNA markers (ML1 and ML2) (48), M13 minisatellite PCR (49), or random amplified microsatellite (RAMS) fingerprinting with CT, CCA, and CGA primers (50) as indicated in Table 2.
Laboratory growth experiments.
The phenotypes of HetPV13-an1-infected new host strains were examined by testing their growth rates on 2% MEA plates at 20°C. Growth rate testing was conducted in the laboratory using five strains of H. annosum and three strains of H. parviporum infected with HetPV13-an1. Each fungal strain and an isogenic strain free of HetPV13-an1 were inoculated at the center of a 2% MEA plate as a mycelial plug with a diameter of 5 mm (3 to 6 replicates). The area covered by the fungal mycelium was measured using a planimeter (Planix 10S; Tamaya) after 6 days of growth. Hyphal growth patterns were examined by light microscopy (Leica DMLB2) using cultures inoculated on water agar plates.
Field experiment: inoculation of spruce trees.
A slow-growing homokaryotic H. parviporum isolate infected with HetPV13-an1 (RK15A*) was selected for a field inoculation experiment. Mature (∼40-year-old) Norway spruce (Picea abies) trees (n = 46) with a mean stump diameter of 25.8 cm were infected using either H. parviporum RK15A* or an isogenic strain free of dsRNA viruses (RK15A) by inserting an 8-mm-diameter spruce wood plug colonized by Heterobasidion mycelia into a hole drilled in the root collar (inoculation date, 20 June 2013). The plugs had been infected in the laboratory by incubation on MOS agar plates covered with fungal hyphae for 12 (RK15A) or 17 (RK15A*) days. After inserting the plug, the hole was sealed with garden wax.
The inoculated trees originated from four clonal lineages designated KU42, KU45, KU70, and KU208. The experiment included 8 to 16 trees representing each clone. The Myrtillus-type forest site was located in Läyliäinen (Loppi, southern Finland; 60°37′N, 24°26′E) and were planted with 2- to 3-year-old Norway spruce saplings in 1975. The origins of the spruce clones were as follows: KU42 (V327) originated from Russia, Pskovin region (57°48′N, 28°26′E); KU45 (V330) originated from Russia, Novgorod region (58°30′N, 31°20′E); KU70 (V354) originated from Loppi; and KU208 (V483) was a cross between H3505 and E4284, originating from Loppi and Germany, Carlsfeld (50°26′N, 12°36′E).
The trees were felled two growing seasons after infection (sampling dates, 24 to 26 September 2014), and sample disks were taken ∼10 cm above the inoculation point and examined for the spread of H. parviporum (Fig. 5). The occurrence of Heterobasidion infections in the wood disks was examined based on microscopic inspection of conidiophores. Disks were taken to the laboratory, washed with running tap water, and incubated in plastic bags for 5 to 10 days at room temperature to induce the formation of Heterobasidion conidiophores. The wood area covered by conidiophores was marked under a stereomicroscope and measured using a planimeter (Planix 10S; Tamaya). Conidial and wood samples were taken from infected wood, and the obtained Heterobasidion cultures were stored in 2% MEA at 4°C. Genotypes of the cultures were determined based on vegetative compatibility testing and/or microsatellite analysis as described previously (44) using four microsatellite markers described by Johannesson and Stenlid (51): Ha-ms1, Ha-ms2, Ha-ms6, and Ha-ms10. Only cultures with the RK15A microsatellite genotype (allele sizes of 158, 134, 262, and 0 nucleotides for Ha-ms1, Ha-ms2, Ha-ms6, and Ha-ms10, respectively) were included in the analysis. Any observed spore-derived Heterobasidion contaminants and trees with suspected preexisting Heterobasidion infections were omitted from the analysis (one tree each for KU42 and KU45 and two trees for KU70).
Student's t test was used to compare the wood area occupied by H. parviporum RK15A and RK15A* in the wood disks (Fig. 2C) and the growth of Heterobasidion strains on MEA plates (Fig. 2A).
Accession number(s).
The raw sequencing data files are available from the National Center for Biotechnology Information Sequence Read Archive (SRA) under accession number SRP097618 (Bioproject accession number PRJNA362289).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Juha Puranen, Marja-Leena Santanen, Ari Rajala, Sonja Sarsila, Joonas Karppinen, Mirsad Krasniqi, Altti Tyni, and Abdi Hamid for technical assistance and to Essi Puranen for graphical editing. We thank Marja-Leena Napola for providing clonal trees for the field experiment and Kari Korhonen and P. Lakomy for providing fungal isolates.
This work was supported by the Academy of Finland (grant numbers 251193, 258520, and 309896).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01744-17.
REFERENCES
- 1.Ghabrial SA, Suzuki N. 2009. Viruses of plant pathogenic fungi. Annu Rev Phytopathol 47:353–384. doi: 10.1146/annurev-phyto-080508-081932. [DOI] [PubMed] [Google Scholar]
- 2.Xie J, Jiang D. 2014. New insights into mycoviruses and exploration for the biological control of crop fungal diseases. Annu Rev Phytopathol 52:45–68. doi: 10.1146/annurev-phyto-102313-050222. [DOI] [PubMed] [Google Scholar]
- 3.Márquez LM, Redman RS, Rodriguez RJ, Roossinck MJ. 2007. A virus in a fungus in a plant: three-way symbiosis required for thermal tolerance. Science 315:513–515. doi: 10.1126/science.1136237. [DOI] [PubMed] [Google Scholar]
- 4.Hyder R, Pennanen T, Hamberg L, Vainio EJ, Piri T, Hantula J. 2013. Two viruses of Heterobasidion confer beneficial, cryptic or detrimental effects to their hosts in different situations. Fungal Ecol 6:387–396. doi: 10.1016/j.funeco.2013.05.005. [DOI] [Google Scholar]
- 5.Milgroom MG, Cortesi P. 2004. Biological control of chestnut blight with hypovirulence: a critical analysis. Annu Rev Phytopathol 42:311–338. doi: 10.1146/annurev.phyto.42.040803.140325. [DOI] [PubMed] [Google Scholar]
- 6.Lakshman DK, Jian J, Tavantzis SM. 1998. A double-stranded RNA element from a hypovirulent strain of Rhizoctonia solani occurs in DNA form and is genetically related to the pentafunctional AROM protein of the shikimate pathway. Proc Natl Acad Sci U S A 95:6425–6429. doi: 10.1073/pnas.95.11.6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Osaki H, Nakamura H, Sasaki A, Matsumoto N, Yoshida K. 2006. An endornavirus from a hypovirulent strain of the violet root rot fungus, Helicobasidium mompa. Virus Res 118:143–149. doi: 10.1016/j.virusres.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 8.Chiba S, Salaipeth L, Lin YH, Sasaki A, Kanematsu S, Suzuki N. 2009. A novel bipartite double-stranded RNA mycovirus from the white root rot fungus Rosellinia necatrix: molecular and biological characterization, taxonomic considerations, and potential for biological control. J Virol 83:12801–12812. doi: 10.1128/JVI.01830-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu X, Li B, Fu Y, Jiang D, Ghabrial SA, Li G, Peng Y, Xie J, Cheng J, Huang J, Yi X. 2010. A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc Natl Acad Sci U S A 107:8387–8392. doi: 10.1073/pnas.0913535107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu M, Jin F, Zhang J, Yang L, Jiang D, Li G. 2012. Characterization of a novel bipartite double-stranded RNA mycovirus conferring hypovirulence in the phytopathogenic fungus Botrytis porri. J Virol 86:6605–6619. doi: 10.1128/JVI.00292-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen JJ, Cui BK, Zhou LW, Korhonen K, Dai YC. 2015. Phylogeny, divergence time estimation, and biogeography of the genus Heterobasidion (Basidiomycota, Russulales). Fungal Divers 71:185–200. doi: 10.1007/s13225-014-0317-2. [DOI] [Google Scholar]
- 12.Niemelä T, Korhonen K. 1998. Taxonomy of the genus Heterobasidion, p 27–33. In Woodward S, Stenlid J, Karjalainen R, Hütterman A (ed), Heterobasidion annosum: biology, ecology, impact and control. CAB International, Wallingford, UK. [Google Scholar]
- 13.Stenlid J, Redfern DB. 1998. Spread within the tree and stand, p 125–141. In Woodward S, Stenlid J, Karjalainen R, Hütterman A (ed), Heterobasidion annosum: biology, ecology, impact and control. CAB International, Wallingford, UK. [Google Scholar]
- 14.Ihrmark K. 2001. Double-stranded RNA elements in the root rot fungus Heterobasidion annosum. Acta Universitatis Agriculturae Sueciae 210. Ph.D. thesis Swedish University of Agricultural Sciences, Uppsala, Sweden. [Google Scholar]
- 15.Vainio EJ, Korhonen K, Tuomivirta TT, Hantula J. 2010. A novel putative partitivirus of the saprotrophic fungus Heterobasidion ecrustosum infects pathogenic species of the Heterobasidion annosum complex. Fungal Biol 114:955–965. doi: 10.1016/j.funbio.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 16.Vainio EJ, Hakanpää J, Dai YC, Hansen E, Korhonen K, Hantula J. 2011. Species of Heterobasidion host a diverse pool of partitiviruses with global distribution and interspecies transmission. Fungal Biol 115:1234–1243. doi: 10.1016/j.funbio.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 17.Vainio EJ, Keriö S, Hantula J. 2011. Description of a new putative virus infecting the conifer pathogenic fungus Heterobasidion parviporum with resemblance to Heterobasidion annosum P-type partitivirus. Arch Virol 156:79–86. doi: 10.1007/s00705-010-0823-9. [DOI] [PubMed] [Google Scholar]
- 18.Vainio EJ, Capretti P, Motta E, Hantula J. 2013. Molecular characterization of HetRV8-ir1, a partitivirus of the invasive conifer pathogenic fungus Heterobasidion irregulare. Arch Virol 158:1613–1615. doi: 10.1007/s00705-013-1643-5. [DOI] [PubMed] [Google Scholar]
- 19.Vainio EJ, Müller MM, Korhonen K, Piri T, Hantula J. 2015. Viruses accumulate in aging infection centers of a fungal forest pathogen. ISME J 9:497–507. doi: 10.1038/ismej.2014.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kashif M, Hyder R, De Vega Perez D, Hantula J, Vainio EJ. 2015. Heterobasidion wood decay fungi host diverse and globally distributed viruses related to Helicobasidium mompa partitivirus V70. Virus Res 195:119–123. doi: 10.1016/j.virusres.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 21.Vainio EJ, Jurvansuu J, Streng J, Rajamäki ML, Hantula J, Valkonen JPT. 2015. Diagnosis and discovery of fungal viruses using deep sequencing of small RNAs. J Gen Virol 96:714–725. doi: 10.1099/jgv.0.000003. [DOI] [PubMed] [Google Scholar]
- 22.Vainio EJ, Hyder R, Aday G, Hansen E, Piri T, Doğmuş-Lehtijärvi T, Lehtijärvi A, Korhonen K, Hantula J. 2012. Population structure of a novel putative mycovirus infecting the conifer root-rot fungus Heterobasidion annosum sensu lato. Virology 422:366–376. doi: 10.1016/j.virol.2011.10.032. [DOI] [PubMed] [Google Scholar]
- 23.Nibert ML, Ghabrial SA, Maiss E, Lesker T, Vainio EJ, Jiang D, Suzuki N. 2014. Taxonomic reorganization of family Partitiviridae and other recent progress in partitivirus research. Virus Res 188:128–141. doi: 10.1016/j.virusres.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 24.Magae Y, Sunagawa M. 2010. Characterization of a mycovirus associated with the brown discoloration of edible mushroom, Flammulina velutipes. Virol J 7:342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhatti MF, Jamal A, Petrou MA, Cairns TC, Bignell EM, Coutts RH. 2011. The effects of dsRNA mycoviruses on growth and murine virulence of Aspergillus fumigatus. Fungal Genet Biol 48:1071–1075. doi: 10.1016/j.fgb.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 26.Zheng L, Zhang M, Chen Q, Zhu M, Zhou E. 2014. A novel mycovirus closely related to viruses in the genus Alphapartitivirus confers hypovirulence in the phytopathogenic fungus Rhizoctonia solani. Virology 456-457:220–226. doi: 10.1016/j.virol.2014.03.029. [DOI] [PubMed] [Google Scholar]
- 27.Xiao X, Cheng J, Tang J, Fu Y, Jiang D, Baker TS, Ghabrial SA, Xie J. 2014. A novel partitivirus that confers hypovirulence on plant pathogenic fungi. J Virol 88:10120–10133. doi: 10.1128/JVI.01036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vainio EJ, Piri T, Hantula J. 2013. Virus community dynamics in the conifer pathogenic fungus Heterobasidion parviporum following an artificial introduction of a partitivirus. Microb Ecol 65:28–38. doi: 10.1007/s00248-012-0118-7. [DOI] [PubMed] [Google Scholar]
- 29.Olson Å, Aerts A, Asiegbu F, Belbahri L, Bouzid O, Broberg A, Canbäck B, Coutinho PM, Cullen D, Dalman K, Deflorio G, van Diepen LTA, Dunand C, Duplessis S, Durling M, Gonthier P, Grimwood J, Fossdal CG, Hansson D, Henrissat B, Hietala A, Himmelstrand K, Hoffmeister D, Högberg N, James TY, Karlsson M, Kohler A, Kües U, Lee Y-H, Lin Y-C, Lind M, Lindquist E, Lombard V, Lucas S, Lundén K, Morin E, Murat C, Park J, Raffaello T, Rouzé P, Salamov A, Schmutz J, Solheim H, Ståhlberg J, Vélëz H, de Vries RP, Wiebenga A, Woodward S, Yakovlev I, Garbelotto M, Martin F, Grigoriev IV, Stenlid J. 2012. Insight into trade-off between wood decay and parasitism from the genome of a fungal forest pathogen. New Phytol 194:1001–1013. doi: 10.1111/j.1469-8137.2012.04128.x. [DOI] [PubMed] [Google Scholar]
- 30.Meena M, Prasad V, Zehra A, Gupta VK, Upadhyay RS. 2015. Mannitol metabolism during pathogenic fungal-host interactions under stressed conditions. Front Microbiol 6:e1019. doi: 10.3389/fmicb.2015.01019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bayry J, Aimanianda V, Guijarro JI, Sunde M, Latgé JP. 2012. Hydrophobins—unique fungal proteins. PLoS Pathog 8:e1002700. doi: 10.1371/journal.ppat.1002700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szeto CYY., Leung GS., Kwan HS. 2007. Le. MAPK and its interacting partner, Le.DRMIP, in fruiting body development in Lentinula edodes. Gene 393:87–93. doi: 10.1016/j.gene.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 33.Carmona-Gutierrez D, Fröhlich KU, Kroemer G, Madeo F. 2010. Metacaspases are caspases. Doubt no more. Cell Death Differ 17:377–378. doi: 10.1038/cdd.2009.198. [DOI] [PubMed] [Google Scholar]
- 34.Gientka I, Duszkiewicz-Reinhard W. 2009. Shikimate pathway in yeast cells: enzymes, functioning, regulation—a review. Polish J Food Nutr Sci 59:113–118. [Google Scholar]
- 35.Hu Y, Stenlid J, Elfstrand M, Olson Å. 2013. Evolution of RNA interference proteins dicer and argonaute in Basidiomycota. Mycologia 105:1489–1498. doi: 10.3852/13-171. [DOI] [PubMed] [Google Scholar]
- 36.Woodward S, Stenlid J, Karjalainen R, Hütterman A (ed). 1998. Heterobasidion annosum: biology, ecology, impact and control. CAB International, Wallingford, UK. [Google Scholar]
- 37.Chen B, Gao S, Choi GH, Nuss DL. 1996. Extensive alteration of fungal gene transcript accumulation and elevation of G-protein-regulated cAMP levels by a virulence-attenuating hypovirus. Proc Natl Acad Sci U S A 93:7996–8000. doi: 10.1073/pnas.93.15.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Segers GC, Zhang X, Deng F, Sun Q, Nuss DL. 2007. Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc Natl Acad Sci U S A 104:12902–12906. doi: 10.1073/pnas.0702500104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee K-M, Cho WK, Yu J, Son M, Choi H, Min K, Lee Y-W, Kim K-H. 2014. A comparison of transcriptional patterns and mycological phenotypes following infection of Fusarium graminearum by four mycoviruses. PLoS One 9:e0100989. doi: 10.1371/journal.pone.0100989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jurvansuu J, Kashif M, Vaario L, Vainio E, Hantula J. 2014. Partitiviruses of a fungal forest pathogen have species-specific quantities of genome segments and transcripts. Virology 462-463:25–33. doi: 10.1016/j.virol.2014.05.021. [DOI] [PubMed] [Google Scholar]
- 41.Sasaki A, Nakamura H, Suzuki N, Kanematsu S. 2016. Characterization of a new megabirnavirus that confers hypovirulence with the aid of a co-infecting partitivirus to the host fungus, Rosellinia necatrix. Virus Res 219:73–82. doi: 10.1016/j.virusres.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 42.Hyder R, Piri T, Hantula J, Nuorteva H, Vainio EJ. 5 August 2017. Distribution of viruses inhabiting Heterobasidion annosum in a pine-dominated forest plot in southern Finland. Microb Ecol doi: 10.1007/s00248-017-1027-6. [DOI] [PubMed] [Google Scholar]
- 43.Korhonen K, Piri T. 1993. The main hosts and distribution of the S and P groups of Heterobasidion annosum in Finland, p 260–267. In Johansson M, Stenlid J (ed), Proceedings of the 8th International Conference on Root and Butt Rots. IUFRO, Vienna, Austria. [Google Scholar]
- 44.Vainio EJ, Hantula J. 2016. Taxonomy, biogeography and importance of Heterobasidion viruses. Virus Res 219:2–10. doi: 10.1016/j.virusres.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 45.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raffaello T, Asiegbu FO. 2013. Evaluation of potential reference genes for use in gene expression studies in the conifer pathogen (Heterobasidion annosum). Mol Biol Rep 40:4605–4611. doi: 10.1007/s11033-013-2553-z. [DOI] [PubMed] [Google Scholar]
- 47.Stenlid J. 1985. Population structure of Heterobasidion annosum as determined by somatic incompatibility, sexual incompatibility, and isoenzyme patterns. Can J Bot 63:2268–2273. doi: 10.1139/b85-322. [DOI] [Google Scholar]
- 48.White TJ, Bruns S, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322. In Innis MA, Gelfand DH, Sninsky JJ, White TJ (ed), PCR protocols: a guide to methods and applications. Academic Press, San Diego, CA. [Google Scholar]
- 49.Stenlid J, Karlsson J-O, Högberg N. 1994. Intraspecific genetic variation in Heterobasidion annosum revealed by amplification of minisatellite DNA. Mycol Res 98:57–63. doi: 10.1016/S0953-7562(09)80337-7. [DOI] [Google Scholar]
- 50.Hantula J, Dusabenyagasani M, Hamelin RC. 1996. Random amplified microsatellites (RAMS)—a novel method for characterizing genetic variation within fungi. Eur J For Pathol 26:159–166. doi: 10.1111/j.1439-0329.1996.tb00720.x. [DOI] [Google Scholar]
- 51.Johannesson H, Stenlid J. 2004. Nuclear reassortment between vegetative mycelia in natural populations of the basidiomycete Heterobasidion annosum. Fungal Genet Biol 41:563–570. doi: 10.1016/j.fgb.2004.01.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.