ABSTRACT
The recent acquisition of a novel retrovirus (KoRV) by koalas (Phascolarctos cinereus) has created new opportunities for retroviral research and new challenges for koala conservation. There are currently two major subtypes of KoRV: KoRV-A, which is believed to be endogenous only in koalas from the northern part of Australia, and KoRV-B, which appears to be exogenous. Understanding and management of these subtypes require population level studies of their prevalence and diversity, especially when coinfected in the same population, and investigations of their modes of transmission in the wild. Toward this end, we studied a wild Queensland koala population of 290 animals over a 5-year period and investigated the prevalence, diversity and mode of transmission of KoRV-A and KoRV-B. We found KoRV-A to have an infection level of 100% in the population, with all animals sharing the same dominant envelope protein sequence. In contrast, the KoRV-B infection prevalence was only 24%, with 21 different envelope protein sequence variants found in the 83 KoRV-B-positive animals. Linked to severe disease outcomes, a significant association between KoRV-B positivity and both chlamydial disease and neoplasia was found in the population. Transmission of KoRV-B was found at a rate of 3% via adult-to-adult contact per year, while there was a 100% rate of KoRV-B-positive mothers transmitting the virus to their joeys. Collectively, these findings demonstrate KoRV-B as the pathogenic subtype in this wild koala population and inform future intervention strategies with subtype variation and transmission data.
IMPORTANCE KoRV represents a unique opportunity to study a relatively young retrovirus as it goes through its molecular evolution in both an endogenous form and a more recently evolved exogenous form. The endogenous form, KoRV-A, now appears to have stably and completely established itself in Northern Australian koala populations and is progressing south. Conversely, the exogenous form, KoRV-B, is undergoing continuous mutation and spread in the north and, as yet, has not reached all southern koala populations. We can now link KoRV-B to neoplasia and chlamydial disease in both wild and captive koalas, making it an imminent threat to this already vulnerable species. This work represents the largest study of koalas in a wild population with respect to KoRV-A/KoRV-B-infected/coinfected animals and the linkage of this infection to chlamydial disease, neoplasia, viral evolution, and spread.
KEYWORDS: KoRV, koala retrovirus, Phascolarctos cinereus, chlamydia, koala, neoplasia
INTRODUCTION
All eukaryotic hosts are endogenously infected with retroviruses, with up to 10% of their genomes being retroviral elements (1). However, these retroviral infections occurred millions of years ago and the viral elements that were incorporated into the genome are now fragmented and usually nonactive. The koala (Phascolarctos cinereus) and its retrovirus (KoRV) present a unique situation, with estimates based on analysis of its long terminal repeats (LTRs) suggesting that the koala was infected with this novel retrovirus as recently as 50,000 years ago (2) and that endogenization is currently progressing through the Australian koala population (3). Studying koala retrovirus allows us to gain unique insights into how retroviruses infect their new host and evolve together in the early stages of genome invasion.
KoRV is known to exist in the koala population as both an endogenous and an exogenous virus. Northern Australian (Queensland and northern New South Wales) koala populations have been reported to have a 100% prevalence of KoRV provirus (at an estimate of 165 proviral copies per cell), while Southern Australian (southern New South Wales and Victoria) koala populations are seen to be only partially infected, with rare populations appearing to have remained KoRV free (4). The variable prevalence of endogenized KoRV in geographically distributed koala populations suggests that KoRV is in the process of endogenization, making it the only known retrovirus to be doing so (3, 5). KoRV has been putatively linked to hematopoietic neoplasia and leukemia in captive koala populations and chlamydiosis in wild koala populations (6–9), and evidence of its immunosuppressive potential has been observed in vitro (10). Taken together, KoRV presents a unique situation to study the active infection of a host by a retrovirus and to compare endogenous and exogenous strains simultaneously.
Detailed studies have revealed that at least two major subtypes of KoRV exist. KoRV-A, the first subtype described and characterized as the endogenizing form, uses a sodium-dependent phosphate transporter membrane protein (PiT1) to infect cells, similar to what is seen in its close relative, gibbon ape leukemia virus (GALV) (11). KoRV-B (originally described as KoRV-J but now added to the described KoRV-B subtype) uses the thiamine transporter protein 1 (THTR1) receptor, like feline leukemia virus (FeLV) subtype A (12), and is exogenous, with no evidence to date of it yet becoming endogenous (6, 13). Current evidence suggests that the KoRV-A variant is not overly pathogenic for its host (6, 8, 9). However, in southern Australia (where KoRV-B has not been detected [14]), an association between KoRV-A infection and “wet bottom” (the clinical presentation of urogenital chlamydial disease in koalas) has recently been reported (9). In contrast, the more recently evolved KoRV-B subtype does appear to be pathogenic, (i) potentially causing lymphoma itself (6) and (ii) contributing to the pathology caused by the other major koala infectious agent, Chlamydia (9). Additional KoRV subtypes (KoRV-C to -I) have been proposed based on differences in the envelope protein sequences detected in koala genomes; however, little to no additional experimental evidence has been collected for these subtypes at this time (6, 13, 15–20).
The effect that KoRV is having on koala health is becoming a serious threat to the koala's long-term survival. This iconic marsupial was declared “vulnerable” by the Australian Government in 2012 and was officially listed as a threatened species under the federal Environment Protection and Biodiversity Conservation Act 1999. However, with KoRV and its related complications being found in both wild Australian populations (4, 7, 9, 14, 20–22) and captive zoo populations worldwide (6, 13, 16, 23), the necessity to understand and manage KoRV impacts is now a global challenge. Three main strategies have been suggested for preventing KoRV infection and disease: (i) quarantine of uninfected koalas (for which it may already be too late); (ii) antiretroviral drug treatment (which would be extremely challenging for wild populations); and (iii) vaccination (quite promising, given the very successful vaccines available for the related FeLV [24]). However, for vaccination to have any meaningful chance of success, key parameters of KoRV subtype prevalence, virulence, diversity, and transmission need to be understood. At present, they are not.
In this project, we studied the two main KoRV subtypes (A and B) in a 290-member wild koala population over a 5-year period to obtain unique insights into diversity and transmission mechanisms. We assessed the prevalence of KoRV-A and -B subtypes in the overall population (via the presence of proviral DNA in the host genome), assessed the strain level diversity of each subtype (via partial sequencing of the envelope [p15E] protein sequence), and determined rates of adult-to-adult contact transmission and mother-to-joey contact transmission. This was an exceptional opportunity to study the active infection of a host by a retrovirus and the molecular evolution of various virus variants, to compare endogenous (KoRV-A) and exogenous (KoRV-B) subtypes, and to determine the modes of transmission of the variants in the new host.
RESULTS
Prevalence of KoRV-A and KoRV-B and association with chlamydial disease.
Our test population was a large, wild population of koalas, located in the central coastal part of Australia (Moreton Bay, Queensland, 27.25°S, 153.02°E), previously tested (n = 36) to be 100% positive for the endogenous form of KoRV-A (9). A total of 357 blood samples, collected from 290 individual koalas between 2013 and 2017, were tested for the presence of integrated KoRV envelope gene sequences that distinguish KoRV-A from KoRV-B in DNA obtained from whole blood via a subtype-specific PCR (Fig. 1A) (25, 26). All 357 samples were positive for KoRV-A, indicating a 100% prevalence of KoRV-A in this population. In contrast, only 28% of the samples (101/357) were positive for KoRV-B, representing 83 individuals.
FIG 1.
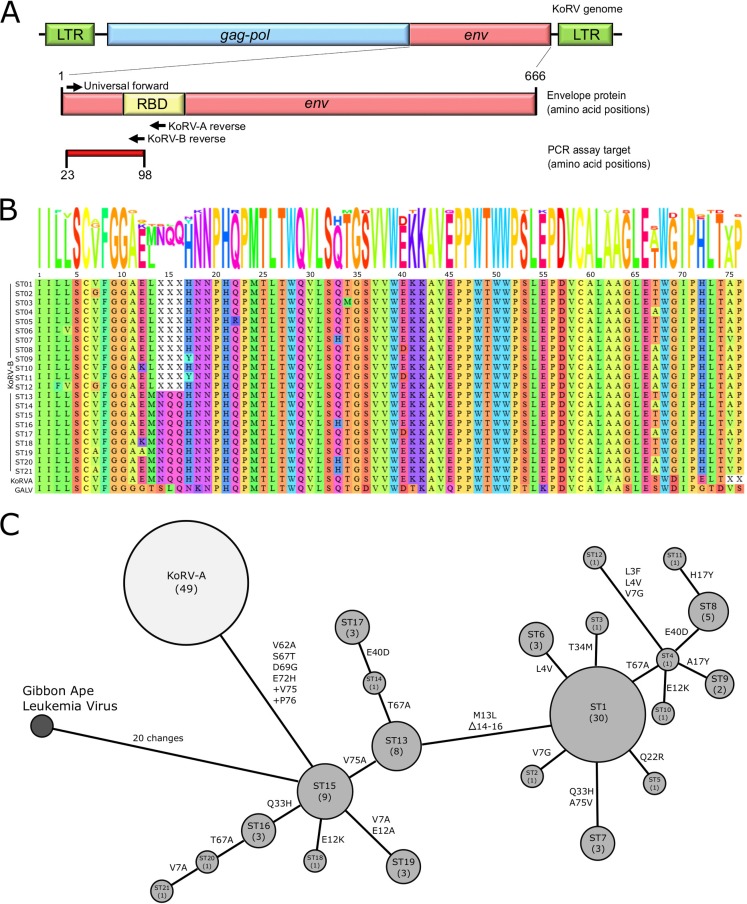
KoRV sequence variation in the MBR (Moreton Bay Rail) population. (A) Schematic of KoRV genome highlighting the targeted region of the envelope (env) gene for analysis. LTR, long terminal repeat; RBD, receptor binding domain. (B) Alignment of unique KoRV envelope fragments detected. The fragment represents amino acids 23 to 98 in the full-length sequence. One KoRV-A sequence was detected, while 21 unique KoRV-B sequence types (ST) were detected. Gibbon ape leukemia virus (GALV) is included as an outlier. Gaps are represented by “X,” and the alignment sequence logo is shown above. (C) Minimum spanning tree representing the KoRV envelope protein sequence. Medium gray nodes represent KoRV-B, with the ST indicated and the number of individual koalas with that ST stated in parentheses. Amino acid changes between node sequences are indicated on vertices.
We then narrowed down our sample collection to samples from 279 different koalas collected between 2013 and 2014 (Table 1). The age range (0.9 to 12.5 years) and sex distribution (40% males/60% females) of the koalas sampled between the 2 years were the same. All koalas in 2013 to 2014 were KoRV-A positive (100% infection rate). In 2013 to 2014, 68/279 (24%) of koalas were KoRV-B positive (Table 1). For comparison, we also examined the prevalence of chlamydial infection (as determined by PCR positivity of the samples for Chlamydia pecorum) and chlamydial disease in these koalas at the same time points. Chlamydia infection levels, as determined by C. pecorum 16S rRNA PCR on urogenital and/or ocular samples, revealed that 89/254 (35%) of PCR-tested koalas carried C. pecorum in 2013 to 2014 (Table 1). Chlamydial disease (as determined by overt signs of disease, as assessed by the field veterinarian at the time of sample collection) was 75/279 (27%) of koalas in the 2013-2014 cohort (Table 1). Interestingly, concordance of C. pecorum PCR positivity and clinical chlamydial disease was seen in only 133/254 (52%) of koalas, with 70/254 (28%) koalas carrying C. pecorum as a subclinical infection (disease negative/PCR positive) and 51/254 (20%) koalas continuing to exhibit clinical disease symptoms with C. pecorum levels below detectable limits (disease positive/PCR negative) (Table 2). This resulted in no significant association between the detection of C. pecorum by PCR and the presence of clinical chlamydial disease symptoms [χ2(1) = 2.647, P = 0.104].
TABLE 1.
Prevalence of Chlamydia pecorum infection and chlamydial disease by KoRV-B status in a population of wild koalas from 2013 to 2014a
| Chlamydia status | No. of koalas |
χ2(1); significance | ||
|---|---|---|---|---|
| KoRV-B+ | KoRV-B− | Total | ||
| By clinical symptomsb | ||||
| Disease positive | 26 | 49 | 75 | |
| Disease negative | 42 | 162 | 204 | |
| Disease total | 68 | 211 | 279 | 5.897; P = 0.015 |
| By PCR detection | ||||
| C. pecorum positive | 22 | 67 | 89 | |
| C. pecorum negative | 40 | 125 | 165 | |
| Not available | 6 | 19 | 25 | 0.007; P = 0.933 |
| PCR total | 68 | 211 | 279 | |
Koalas tested more than once in 2013–2014 had only their latest sample result included; a C. pecorum PCR result was classified as positive if there was a positive sample at either the urogenital site or the ocular site or both. Boldface indicates significance.
C. pecorum disease status was based on clinical symptoms assessed by a veterinarian at the time of sample collection.
TABLE 2.
Concordance of Chlamydia pecorum infection and chlamydial disease in a population of wild koalas from 2013 to 2014a
| Chlamydia disease statusb | No. of koalas |
χ2(1); significance | |||
|---|---|---|---|---|---|
| C. pecorum PCR positive | C. pecorum PCR negative | PCR not available | Total | ||
| Disease positive | 19 | 51 | 5 | 75 | |
| Disease negative | 70 | 114 | 20 | 204 | |
| Disease total | 89 | 165 | 25 | 279 | 2.647; P = 0.104 |
Koalas tested more than once in 2013–2014 had only their latest sample result included; a C. pecorum PCR result was classified as positive if there was a positive sample at either the urogenital site or the ocular site or both.
C. pecorum disease status was based on clinical symptoms assessed by a veterinarian at the time of sample collection.
To determine if koala age or sex was associated with KoRV-B positivity, we used an independent-samples t test and the Pearson chi-squared test, respectively, to compare these factors. There was no significant difference between the ages of KoRV-B-positive koalas (mean [M] = 3.87; standard deviation [SD] = 2.33) and KoRV-B-negative koalas [M = 4.15; SD = 2.76; t(277) = 0.765, P = 0.445]. Koalas as young as 11 months old and as old as 10 years old were found to be KoRV-B positive. Sex (male/female) was also not significantly different between KoRV-B-positive and -negative koalas [χ2(1) = 0.108; P = 0.743].
It has previously been suggested that KoRV-B infection correlates with higher levels of overt chlamydial disease (7, 9). When we examined our 2013-2014 koala population for this question, we found that indeed, the level of clinically detectable chlamydial disease was significantly different between KoRV-B-positive and -negative koalas, as determined by the Pearson chi-squared test [χ2(1) = 5.897; P = 0.015], while the underlying levels of chlamydia infection were not significantly different [χ2(1) = 0.007, P = 0.933] (Table 1).
KoRV-B infection and associations with neoplasia or AIDS-like conditions.
A link has been established between KoRV-B infection and neoplasia in captive koalas (6). To determine if a similar association could be found in our wild koala population, the veterinary records for all koalas examined in this study were searched for clinical documentation of neoplasia. Of the 290 koalas studied, 5 (1.7%) koalas developed malignant neoplasia, with 4/5 (80%) being also KoRV-B positive (Table 3). The conditions detected in the KoRV-B-positive koalas included an abdominal lymphoma, a nonspecified proliferative/bone marrow condition, osteochondroma and mesothelioma, and osteochondroma, while the KoRV-B-negative koala was affected by lymphoid leukemia and lymphoma. Despite the limited number of neoplasia cases, a significant association between neoplasia and KoRV-B positivity was detected by Fisher's exact test (P = 0.025).
TABLE 3.
Neoplasia and AIDS-like syndromea in Moreton Bay Rail koalas
| Disease status | No. of koalas |
P value by Fisher's exact test | ||
|---|---|---|---|---|
| KoRV-B+ | KoRV-B− | Total | ||
| Neoplasia | ||||
| Positive | 4 | 1 | 5 | |
| Negative | 79 | 206 | 285 | |
| Total | 83 | 207 | 290 | 0.025 |
| AIDS-like syndrome | ||||
| Positive | 3 | 4 | 7 | |
| Negative | 80 | 203 | 283 | |
| Total | 83 | 207 | 290 | 0.412 |
AIDS-like syndrome was diagnosed in cases displaying two or more of the following clinical signs: chronic ill-thrift, severe fungal infections (such as cryptococcosis), severe and/or generalized dermatitis, unexplained poor body condition, severe periodontal disease, ulcerations, gastrointestinal disease, severe chlamydiosis, and stomatitis. Boldface indicates significance.
In parallel to the neoplasia observations, the veterinary team also noted pathological conditions that they grouped into a category called “AIDS-like syndrome.” This syndrome was diagnosed in cases displaying two or more of the following clinical signs: chronic ill-thrift, severe fungal infections (such as cryptococcosis), severe and/or generalized dermatitis, unexplained poor body condition, severe periodontal disease, ulcerations, gastrointestinal disease, severe chlamydiosis, and stomatitis. Koalas affected by this condition invariably died prematurely, and veterinary treatment provided limited benefit. Of the 290 koalas in this study, 7 (2.4%) presented with “AIDS-like syndrome,” with 3/7 (42.8%) also being KoRV-B positive (Table 3). When examined by Fisher's exact test, no significant association between this syndrome and KoRV-B positivity was detected (P = 0.412). However, it should be noted that the designation of KoRV-B status for these seven koalas was based on a single time point test and it was not confirmed whether the four KoRV-B-negative “AIDS-like syndrome” koalas remained KoRV-B negative for the rest of their life.
Diversity of KoRV-A and KoRV-B strains.
We examined the molecular diversity of KoRV strains by analyzing the DNA and protein sequence of a fragment of the envelope gene from 98 of the 357 KoRV-A-positive samples and from all 101 KoRV-B-positive samples. To achieve this, we directly sequenced the products obtained from our KoRV-A and KoRV-B subtype-specific PCR, targeting the KoRV envelope protein from amino acids 23 to 98 in the full-length sequence (Fig. 1A) (9). All of the KoRV-A and 91% of the KoRV-B samples produced a single dominant envelope gene sequence, while 9% of the KoRV-B samples produced an envelope gene sequence with detectable nucleotide polymorphisms, suggesting more than one dominant envelope sequence in the genome.
For the KoRV-A-positive samples, all 98 PCR products (49 from koala samples collected in 2013 to 2014 and 49 from the same koalas sampled on average 2.8 years later) had the same DNA sequence, which was identical to the GenBank reference KoRV-A sequence LC033970.1 with one interesting feature: every sequence showed a mixed C/T polymorphism at nucleotide position 179 (position 243 in the full-length gene). This polymorphism is located at the degenerate third position for the amino acid cysteine at protein position 59 (protein position 81 in the full-length protein), resulting in a synonymous mutation (Fig. 1B).
In contrast to KoRV-A, for which all 49 koalas analyzed had identical amino acid sequences, KoRV-B sequencing identified 21 variants of the envelope amino acid sequences from just 83 koalas (Fig. 1B). At closer examination, on the DNA level, six of the KoRV-B sequences contained identifiable polymorphic sites, indicating the presence of more than one dominant proviral sequence type. Three koalas had single polymorphic sites: koala Kyarna, position 58 (C/T); koala Rhonda, position 67 (A/G); and koala Dazza, position 101 (G/C). Three koalas had two polymorphic sites: koala Sheriff, positions 79 (C/T) and 122 (A/C); koala Deedee, positions 110 (A/G) and 126 (A/C); and koala Shorty, positions 109 (T/G) and 133 (G/A). This confirmed that at least some koalas carried more than one sequence type of integrated KoRV-B envelope sequences. For further analysis with these sequences, the more-common nucleotide detected in the population at each polymorphic site was used for comparison.
A minimum spanning tree of the KoRV-B envelope sequence types was generated to determine the relatedness of the KoRV-B isolates in this population (Fig. 1C). This enabled us to predict stepwise small changes between the sequence types. Most of the differences between sequence types were created by a single amino acid substitution from their nearest neighbor, with the maximum variation between nearest neighbors composed of three amino acid substitutions or a three-amino-acid deletion (Fig. 1C). The most-dominant sequence type, ST01, was detected in 30 koalas in the population, while 11 sequence types were seen only once each, in 11 separate koalas.
Modes of KoRV-B transmission. (i) Adult-to-adult contact.
While endogenous KoRV-A is clearly transmitted in the germ line from mother to offspring, the modes of transmission for the exogenous KoRV-B are still being actively investigated. Transmission studies of FeLV have shown the retrovirus to be relatively labile in the environment, requiring very close contact between animals to share virus-containing secretions (24). For adult koalas, this type of contact could occur during mating or when males fight over territory. To evaluate adult-to-adult close-contact transmission for KoRV-B in our koala population, we selected 49 koalas that were tracked (fitted with radio collars and captured and sampled at regular intervals) and sampled over the entire 2013-2017 study period, and we compared their KoRV-B PCR results in 2013 to 2014 to their results in 2016 to 2017. The average time between samples was 34 months (2.8 years), with the minimum and maximum sampling intervals being 21 months (1.75 years) and 44 months (3.7 years), respectively. We found that 33 koalas (67.4%) were negative for KoRV-B during the entire study period, while 11 koalas (22.4%) were KoRV-B positive at both time points (Table 4). Importantly, we found four koalas (8.2%) that were KoRV-B negative at the initial time point and KoRV-B positive at the second time point. This represents an incidence rate of new infections of approximately 3% per year. Of the four koalas that acquired KoRV-B, three were females and one was male (Table 4), suggesting that contact during mating may be a more likely transmission activity than male-male fighting. There was also no indication that chlamydial infection or disease status predisposed these koalas to acquiring KoRV-B (Table 4). We also had one koala that was KoRV-B positive at the first testing and negative at the second testing. This could have been the result of a low-level infection that was within assay sensitivity at the first sampling but, given sample-to-sample variation, fell below assay sensitivity at the second sampling or could have been due to natural elimination of the virus from the circulation over time.
TABLE 4.
Transmission of KoRV-B via adult-to-adult contact in a cohort of 49 wild koalas during 2013–2014 sampling and subsequently during 2016–2017 samplinga
| KoRV-B status of koala | No. of koalas | Sex or no. F/no. M |
C. pecorum disease status, 2013–2014 |
|
|---|---|---|---|---|
| By PCR | By clinical symptoms | |||
| Negative at both time points | 33 | 23/10 | ||
| Positive at both time points | 11 | 10/1 | ||
| Negative to positive | 4 | 3/1 | ||
| Cowboy | M | Positive | Negative | |
| Mali | F | NA | Positive | |
| Princess Shrek | F | Negative | Positive | |
| Regina | F | Positive | Negative | |
| Positive to undetectable | 1 | 0/1 | ||
| Barnacles | M | NA | Negative | |
NA, not available; F, female; M, male.
(ii) Mother-to-joey transmission.
Mother-to-offspring transmission of KoRV-B has been shown from two mothers to four joeys in a captive koala setting (6). To investigate the contribution of this type of contact to KoRV-B spread in the wild, we sought out mother-joey pairs in our population. Mother-joey relationships were established when the joey was a dependent with its mother and tagged during routine veterinary assessments. The first sampling from joeys was done at or before 2 years of age. We were able to identify eight KoRV-B-positive mothers and 12 KoRV-B-negative mothers in 2013 to 2014 who went on to have 13 and 12 joeys, respectively, over a number of years. These joeys were tracked and eventually sampled during the study time period. When the first sample collected from these joeys was tested for KoRV-B, all joeys from KoRV-B-positive mothers (13/13, 100%) were KoRV-B positive. In contrast, none of the joeys from KoRV-B-negative mothers (0/12, 0%) were KoRV-B positive (Table 5). Comparison of sequence types between positive mother and joey revealed that of the 12 mother-joey pairs for which sequence data were generated, five pairs (42%) had the identical dominant envelope sequence between mother and joey, while the remaining pairs had one or two amino acid substitutions and/or a small deletion difference between mother and joey dominant envelope gene sequences (Table 6).
TABLE 5.
KoRV-B presence in mother and joey koala pairs as determined by PCR
| Name of mother | Date mother tested (day/mo/yr) | Mother KoRV-B result | Name (sex) of joey | Date joey tested (day/mo/yr) | Joey age at test (yr) | Joey KoRV-B result |
|---|---|---|---|---|---|---|
| Fu | 5/05/2014 | Positive | Bessie (F) | 26/10/2013 | 0.99 | Positive |
| Fu | 5/05/2014 | Positive | Peter (M) | 27/5/2015 | 1.42 | Positive |
| Teena | 17/10/2013 | Positive | Simon (M) | 10/12/2013 | 0.9 | Positive |
| Teena | 17/10/2013 | Positive | Wayne (M) | 21/11/2016 | 1.63 | Positive |
| Teena | 17/10/2013 | Positive | Thunder (M) | 16/3/2016 | 1.78 | Positive |
| Cindy | 27/05/2013 | Positive | Krystal (F) | 16/01/2015 | 1.16 | Positive |
| Cindy | 27/05/2013 | Positive | Lou (F) | 29/4/2014 | 1.42 | Positive |
| Rhonda | 20/05/2014 | Positive | Aerona (F) | 22/5/2015 | 1.3 | Positive |
| Rhonda | 20/05/2014 | Positive | Bicko (M) | 18/8/2016 | 1.73 | Positive |
| Red Queen | 3/11/2013 | Positive | Alice (F) | 15/7/2014 | 1.41 | Positive |
| CJ | 24/10/2013 | Positive | Carlos (M) | 19/8/2014 | 1.62 | Positive |
| Leia | 20/12/2013 | Positive | Alexander (M) | 14/7/2016 | 1.63 | Positive |
| Panda | 30/4/2014 | Positive | Ian (M) | 19/11/2014 | 1.98 | Positive |
| Adrianna | 12/8/2013 | Negative | Jordii (F) | 12/8/2013 | 0.89 | Negative |
| Bubbles | 23/4/2013 | Negative | Zoe (F) | 14/10/2013 | 0.91 | Negative |
| Summer | 18/9/2013 | Negative | Uhura (F) | 18/9/2013 | 0.95 | Negative |
| Sharon | 19/11/2013 | Negative | Miles (M) | 20/12/2013 | 1.03 | Negative |
| Paula C | 9/12/2014 | Negative | Portia (F) | 3/12/2014 | 1.23 | Negative |
| Buffy | 24/7/2013 | Negative | Spike (M) | 9/5/2014 | 1.44 | Negative |
| Raylee | 28/5/2014 | Negative | Tash (F) | 15/10/2013 | 1.44 | Negative |
| Louise M | 17/12/2013 | Negative | Satyam (M) | 30/3/2014 | 1.46 | Negative |
| Minky | 3/4/2013 | Negative | Linky (F) | 4/9/2014 | 1.5 | Negative |
| Kayla | 6/1/2014 | Negative | Morty (M) | 22/7/2014 | 1.54 | Negative |
| Maisie | 30/11/2013 | Negative | Ollie (M) | 15/7/2014 | 1.57 | Negative |
| Willow | 11/10/2013 | Negative | DSmurf (M) | 31/10/2014 | 2.05 | Negative |
TABLE 6.
KoRV-B env gene sequence type in mother and joey pairs
| Name of mother | Mother STa | Name of joey | Joey ST | Sequence difference between mother and joey | Time interval (mo) between mother and joey typing |
|---|---|---|---|---|---|
| Fu | ST13 | Bessie | ST1 | M13L, Δ14-16 | 6 |
| Fu | ST13 | Peter | ST1 | M13L, Δ14-16 | 12 |
| Teena | ST1 | Simon | ST1 | None | 1 |
| Teena | ST1 | Wayne | ST1 | None | 37 |
| Teena | ST1 | Thunder | ST1 | None | 28 |
| Cindy | ST13 | Krystal | ST1 | M13L, Δ14-16 | 19 |
| Cindy | ST13 | Lou | ST1 | M13L, Δ14-16 | 11 |
| Rhonda | ST5 | Aerona | ST1 | Q22R | 12 |
| Rhonda | ST5 | Bicko | ST13 | Q22R, M13L, Δ14-16 | 26 |
| Red Queen | NDb | Alice | ST13 | NDb | 8 |
| CJ | ST11 | Carlos | ST9 | D40E | 9 |
| Leia | ST8 | Alexander | ST8 | None | 30 |
| Panda | ST1 | Ian | ST1 | None | 6 |
ST, sequence type, as shown in Fig. 1.
ND, not determined.
DISCUSSION
The goal of this study was to characterize the prevalence, incidence, molecular diversity, and mode of transmission of KoRV-A and KoRV-B in a wild koala population. The koala retrovirus, KoRV, is unique in the fact that it currently exists in the koala population as both an endogenous (KoRV-A) and exogenous (KoRV-B) virus. This has allowed us to examine the rates and modes of spread of the two forms of the retrovirus simultaneously.
Accumulating data in the literature suggest that KoRV-A has become endogenized, is present in the koala germ line in relatively high copy numbers, and is ubiquitous in Northern and Central Australian koala populations. Our findings in a wild population located around the midcoastal region of Australia support these conclusions, with all 290 koalas in our study population being KoRV-A positive. Our sequencing of a fragment of the KoRV-A envelope protein gene revealed 100% identity between koalas, suggesting that the endogenized KoRV-A provirus is stably integrated and that the dominant sequence type in the genome is identical throughout this population (Fig. 1). This identity most likely arose from the provirus being vertically transmitted from parent to offspring via germ line DNA at a point far enough in the past for it to have become 100% distributed in this particular koala population.
Analysis of KoRV-B revealed a very different molecular epidemiology from that of KoRV-A. Our koala population had an average of only 24% KoRV-B-infected animals (Table 1). Interestingly, while there was no difference between the ages or sexes or chlamydia infection rates of koalas that were infected with KoRV-B, there was a significant difference in the clinical outcomes of chlamydial infection, with KoRV-B positivity positively correlated with expression of clinical chlamydial disease (P = 0.015) (Table 1). This reinforces previous findings, based on a much smaller group of koalas from this area, that active infection with KoRV-B is one significant contributing factor to chlamydial pathogenesis (9). In addition, although there was an overall low prevalence of neoplasia in our wild koala population, there was a significant association with KoRV-B positivity and neoplasia (P = 0.025) (Table 3). This finding adds weight to the correlation found between KoRV-B positivity and malignant neoplasia in captive koalas (6). Taken together, these results strongly suggest that intervention and prevention strategies for KoRV should focus on targeting the KoRV-B subtype.
The sequence diversity in the envelope protein further contributes to the picture of KoRV-B being an actively spreading exogenous virus, with 21 different sequence types detected in 83 KoRV-B-positive koalas in the population. Six koalas had mixed envelope protein sequence types detected, as evidenced by Sanger sequencing, indicating the presence of multiple variants of KoRV-B in the same host genome. As well, from the 11 koalas that were positive at two testing times (Table 4), only 8 had the identical dominant envelope gene sequence at both time points. Koalas Fu and TamaraO had the sequence differences of M13L and Δ14-16 between their sequenced samples (ST1 and ST13), while koala Teena had a T34M point change (ST1 and ST3). These differences most likely reflect multiple KoRV-B envelope genes being present in the host genome, which were differentially amplified in each sample.
The koalas studied in this analysis were geographically close to the animals studied by Chappell et al. (20). In their study, from the 18 koalas that they examined for KoRV envelope gene sequences using Illumina sequencing, they reported 15 KoRV-B sequence types (20). The envelope regions used in that study and ours overlap, allowing for direct comparison. Of the 21 KoRV-B sequence types in this study (ST numbers), five are identical to those listed by Chappell et al. (B numbers): ST1 is identical to B25, ST13/15 to B10/B14, ST16 to B7, and ST20 to B2. ST1, ST13, and ST15 were the sequence types shared among the most koalas in this study (30, 8, and 9 koalas, respectively) and, as evidenced by the Chappell et al. study, appear to be more widespread throughout central coastal Australian koala populations. This suggests that these sequence types are among the dominant strains of KoRV-B in the wild.
The progression of KoRV-B envelope gene mutation steps in this population was monitored by creating a minimum spanning tree (Fig. 1C). Larger nodes (representing more koalas possessing that sequence type) were seen with ST1, ST13, and ST15, suggesting that these strains may possess more-successful envelope protein variants that lead to more-successful infection between koalas. While our subtype-specific PCR targeted only a fragment of the envelope protein, comparison of the KoRV-A and KoRV-B data clearly shows that KoRV-B is undergoing more active mutation and change than KoRV-A. This result is consistent with the endogenous nature of KoRV-A and the exogenous nature of KoRV-B.
Understanding the mode of transmission of KoRV is important in managing its impacts on koala health and population viability. KoRV-A appears to be vertically transmitted from mother to offspring via germ line DNA. Alternatively, the mode of transmission of KoRV-B had previously been demonstrated in a small study of mother-joey transmission in captive zoo animals (6). Other modes to consider, based on related retroviruses and their transmission, include the transmission of FeLV infection in domestic cats through close contact to share virus-laden secretions (24).
In our koala population, we found an adult KoRV-B acquisition rate (in previously KoRV-B-negative koalas) of four animals per 49 koalas followed over a 2.8-year time frame (Table 4; Fig. 2). This translates into an approximately 3% annual incidence of new KoRV-B infection per year of previously uninfected adult koalas. Given that our study koalas were wild animals, we were unable to collect data regarding how these koalas interacted with their conspecifics to acquire their infections. However, with three of the four new infections occurring in female koalas, male-to-male fighting did not appear to be a major transmission risk in this group. Whether sexual contact or other interactions are important mechanisms of infection for adult koalas remains to be elucidated.
FIG 2.
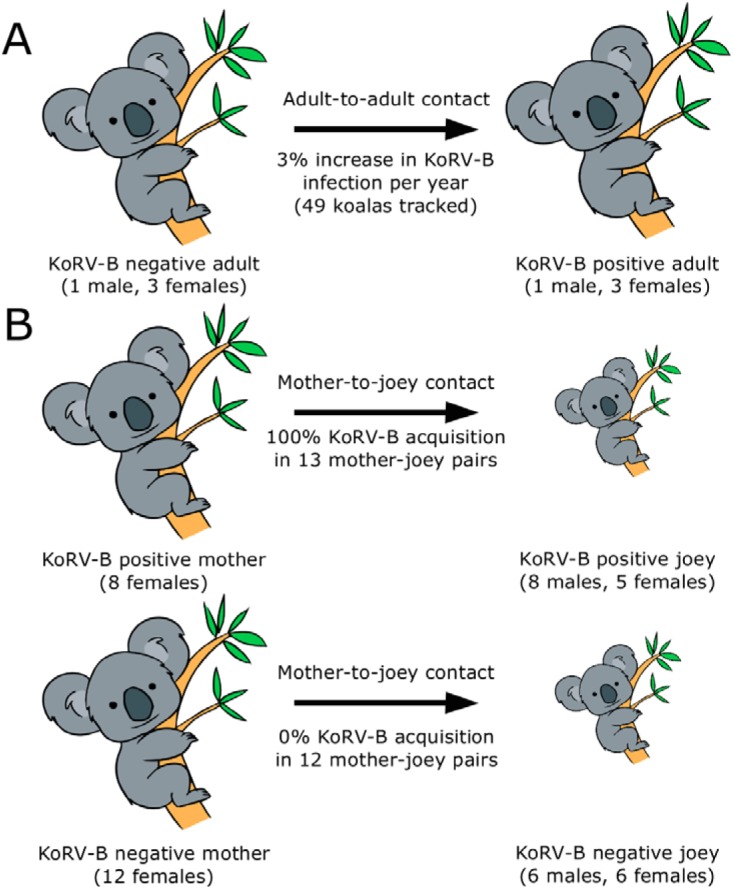
Summary of predicted KoRV-B transmission routes in koala populations via adult-to-adult (A) and mother-to-joey (B) contact. The sex of koalas tested is indicated.
Our data for mother-to-joey acquisition of KoRV-B showed a strikingly high rate of transmission, with 100% of joeys acquiring KoRV-B if their mother had KoRV-B. This is consistent with the findings of Xu et al., who noted that the joeys from two captive KoRV-B-positive mothers (dams) were also positive for KoRV-B (6). Because acquisition of KoRV-B in recently independent joeys tested for the first time could not be definitively proven to have occurred via mother-to-joey transmission (offspring were sampled close to the age of sexual maturity and independence), a second set of 12 mother-joey pairs were selected for comparison, where the mother was KoRV-B negative before and after having her joey. These recently independent offspring were sampled at approximately the same age as the KoRV-B-positive offspring, and all recently independent offspring from KoRV-B-negative mothers were also KoRV-B negative (0%) (Table 5; Fig. 2). This finding strongly validates our contention that the most important route of KoRV-B transmission is the mother-joey transmission route. In support of direct mother-to-joey acquisition of KoRV-B, 42% of joeys had envelope sequence identical to that of their mother (Table 6). Interestingly, this meant that 58% of joeys acquired a slightly mutated version of KoRV-B compared to their mother's dominant strain. We observed no pattern or relationship between the time gap of mother and joey KoRV-B typing and differences in the KoRV-B sequence (Table 6). Given that 21 different sequence variations of the envelope gene sequence have emerged in this koala population, many may have emerged as mutations from the maternal dominant provirus to the dominant provirus in the offspring. Future investigations by our group will involve determining the specific route of KoRV-B transmission from mother to joey. Exposure to secretions during birth, close contact in the pouch during joey development, and transfer through milk during the 12-month period of maternal dependence are all possible transmission routes that will require investigation to explain the high mother-to-joey transmission rate.
Unfortunately, because this study was conducted with a wild population of koalas, we have no information about the fathers (sires) of our joeys. However, the complete correlation of maternal and joey KoRV-B status in all mother-joey pairs tested suggests that sire positivity is less important than maternal positivity with respect to joey infection. This concurs with the finding from a captive koala population in which a single known KoRV-B-positive father was monitored and his two offspring tested KoRV-B negative (6). KoRV-B envelope sequence variation in mother-joey pairs can be explained by minor sequence drift during horizontal (nongenetic) transmission from mother to joey.
Given that the most important route of KoRV-B transmission is from mother to joey, with a relatively low incidence of adult infection, intervention strategies for managing KoRV-B impacts should initially target KoRV-B-positive female koalas. Reducing or eliminating breeding of KoRV-B-positive females in captive populations should result in the eventual elimination of the virus. Management of impacts in wild koala populations will be more difficult. Antiretroviral drugs have been considered for KoRV treatment in captive koalas, based on successes with anti-HIV drugs (27), but koala-specific testing is still needed. In addition, antiretroviral therapy is not a feasible approach for management of wild koalas. For KoRV-B management in a wild koala population, vaccination is currently believed to be the most-promising intervention strategy (28). The most-efficient vaccination strategy would be to target young females in the koala population, before they acquire KoRV-B as an adult. This would have a major impact on not only KoRV infection, but also the morbidity and mortality of neoplasia and chlamydiosis in the koala population. Practical considerations would probably limit the application of initial vaccine programs to local, high-profile, at-risk koala populations, for which cost-benefit analysis and appropriate scientific evaluation are feasible.
In conclusion, this study has defined prevalence and diversity metrics for the two major subtypes of KoRV in a longitudinally monitored koala population and has added valuable data to our knowledge of the modes and rates of KoRV-B transmission in koalas. These findings will not only inform future koala management strategies but also expand our understanding of this unique retrovirus and its process of infection and endogenization into a marsupial host.
MATERIALS AND METHODS
Animals.
Koalas included in the study (n = 290) were part of a 5-year population-wide management program by the Queensland Government Department of Transport and Main Roads for the Moreton Bay Rail (MBR) project, in the Moreton Bay Region, Queensland, Australia (project center point, 27.25°S, 153.02°E). A small subset of koalas (n = 36) from this population were previously analyzed by Waugh et al. (9). Koalas in this population were captured, clinically examined by experienced wildlife veterinarians, radio-collared, and released back into the wild. Koalas were monitored by remote and field telemetry and recaptured and subjected to veterinary examination and sampling at regular intervals (approximately every 6 months). Blood samples were collected from koalas under general anesthesia during veterinary examinations and stored at −20°C until transport to the laboratory, where they were stored at −80°C until processing. Determinations of koala age, sex, and chlamydial disease status at the time of sampling were documented by the examining veterinarian. Mothers with joeys were recorded so future examination of independent joeys could be linked to maternal history. All procedures were approved by the University of the Sunshine Coast (USC) Animal Ethics Committee (Animal ethics number AN/A/13/80) and by the Queensland Government (Scientific Purposes Permit, WISP11532912). All experiments were performed in accordance with relevant guidelines and regulations.
Sample processing and testing for KoRV provirus.
Total nucleic acid was extracted from 200 μl of collected whole blood using the Qiagen QIAamp DNA minikit, as per the manufacturer's instructions. Final DNA elutions (200 μl) were diluted 1:10 with sterile distilled water (dH2O) prior to testing to remove PCR inhibitors and improve sensitivity. Samples were not normalized in regard to the concentration of DNA used for KoRV PCR testing beyond using a standard volume of starting blood and a consistent protocol for processing. Given variations in blood samples and processing, small variations in the amount of KoRV proviral detection could have occurred between samples. However, consistent and overwhelming KoRV-A detection in every sample was used as an indicator of general sample quality. KoRV subtype A and B provirus was tested for using a universal KoRV envelope gene forward primer (5′-TCCTGGGAACTGGAAAAGAC-3′) and subtype-specific reverse primers (KoRV-A, 5′-GGGTTCCCCAAGTGATCTG-3′, 321-bp product; KoRV-B, 5′-GGCGCAGACTGTTGAGATTC-3′, 271-bp product), which routinely had a sensitivity of 1 to 10 copies/reaction volume (9). A diagram of the region targeted by the assay is shown in Fig. 1A, and additional diagrams of the PCR primer landing sites are shown in Fig. 6 and 7 from reference 9. This generated primer-trimmed protein fragments representing amino acids 23 to 98 of the full-length envelope protein. Conventional endpoint PCR was carried out with the HotStarTaq Plus master mix kit (Qiagen), containing 0.3 μM forward and reverse primers, under the following PCR conditions: denaturation of 95°C for 5 min and then 35 cycles of denaturing at 95°C for 30 s, annealing at 54°C for 30 s, and extension at 72°C for 30 s, with a final extension at 72°C for 1 min. Products were visualized on a 1.5% agarose gel with ethidium bromide. Only bands of the anticipated sizes were generated, and the specificity of the assay was confirmed by sequencing all KoRV-B products and a subset of KoRV-A products by Macrogen Inc. (Korea) using Sanger sequencing methods.
Sample processing and testing for Chlamydia pecorum.
Urogenital and ocular swab samples were stored at −20°C until the DNA was extracted as described by Devereaux et al. (29). The extracted samples were screened for the presence of C. pecorum using a diagnostic quantitative real-time PCR (qPCR) targeting a 204-bp fragment of the chlamydial 16S rRNA gene. Assays were as described previously (25, 26), and results in this study are reported as positive or negative. The specificity of the assay was initially confirmed by generating negative PCR results from the closely related species Chlamydia trachomatis (human origin strain) and Chlamydia pneumoniae (both human and koala origin strains), and ongoing assay specificity was monitored during sample testing by high-resolution melt analysis for the C. pecorum-specific PCR melt product. The sensitivity of the assay was determined by serial dilution of spectrophotometry-quantified purified koala C. pecorum genomic DNA to one copy/reaction volume, and a positive assay result was called if a C. pecorum-specific PCR melt product was generated within 40 amplification cycles.
Statistics.
Pearson's chi-squared tests (2-sided), Fisher's exact tests, and independent-sample t tests were used to compare KoRV-B positivity to other koala metadata, as appropriate, using SPSS software (IBM SPSS Statistics for Windows, version 22.0, released 2013; IBM Corp., Armonk, NY).
KoRV sequence analysis.
Sequences generated from the fragment of the KoRV envelope protein targets were trimmed to remove primer sequences, translated into protein, and aligned with ClustalX (default parameters) (30, 31). Aligned sequences were inputted in PHYLOViZ Online (online.phyloviz.net/index) and used with the program's default goeBURST algorithm to generate a minimum spanning tree representing the relatedness of KoRV sequences (32). The outputted tree graphic and node sequence alignment were saved, and the amino acid changes between node sequences were manually added.
Accession number(s).
Novel KoRV-B sequence types have been deposited in GenBank with the accession numbers MF741155 to MF741175.
ACKNOWLEDGMENTS
This project was significantly supported by the Queensland Government (Department of Transport and Main Roads) and specifically the Moreton Bay Rail project team. We also thank the many groups that have supported the overall koala work, including the Queensland Department of Environment and Heritage Protection, Moreton Bay Regional Council, Friends of the Koala, Lismore, Koala Action Inc., Endeavour Veterinary Ecology, Australia Zoo Wildlife Hospital, Lone Pine Koala Sanctuary, Redland City Council, and VIDO, Canada. Specifically, here we thank the Department of Transport & Main Roads—Moreton Bay Rail Project Team, for collaborating with this project, enabling access to the work undertaken as part of the Moreton Bay Rail—Koala Tagging and Monitoring Program and the dedicated staff at Endeavour Veterinary Ecology, particularly Jo Loader, for their help in capturing, radio-collaring, and tracking the koalas and undertaking the health assessments, as well as collecting samples. We also thank Jianbao Dong for assistance with chlamydia PCR testing.
Funding for this project was provided by the Australian ARC Linkage Scheme (P.T.) and the Morris Animal Foundation (P.T.).
REFERENCES
- 1.Jern P, Sperber GO, Blomberg J. 2005. Use of endogenous retroviral sequences (ERVs) and structural markers for retroviral phylogenetic inference and taxonomy. Retrovirology 2:50. doi: 10.1186/1742-4690-2-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ishida Y, Zhao K, Greenwood AD, Roca AL. 2015. Proliferation of endogenous retroviruses in the early stages of a host germ line invasion. Mol Biol Evol 32:109–120. doi: 10.1093/molbev/msu275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tarlinton RE, Meers J, Young PR. 2006. Retroviral invasion of the koala genome. Nature 442:79–81. doi: 10.1038/nature04841. [DOI] [PubMed] [Google Scholar]
- 4.Simmons G, Young P, Hanger J, Jones K, Clarke D, McKee J, Meers J. 2012. Prevalence of koala retrovirus in geographically diverse populations in Australia. Aust Vet J 90:404–409. doi: 10.1111/j.1751-0813.2012.00964.x. [DOI] [PubMed] [Google Scholar]
- 5.Stoye JP. 2006. Koala retrovirus: a genome invasion in real time. Genome Biol 7:241. doi: 10.1186/gb-2006-7-11-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu W, Stadler CK, Gorman K, Jensen N, Kim D, Zheng H, Tang S, Switzer WM, Pye GW, Eiden MV. 2013. An exogenous retrovirus isolated from koalas with malignant neoplasias in a US zoo. Proc Natl Acad Sci U S A 110:11547–11552. doi: 10.1073/pnas.1304704110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tarlinton R, Meers J, Hanger J, Young P. 2005. Real-time reverse transcriptase PCR for the endogenous koala retrovirus reveals an association between plasma viral load and neoplastic disease in koalas. J Gen Virol 86:783–787. doi: 10.1099/vir.0.80547-0. [DOI] [PubMed] [Google Scholar]
- 8.Maher IE, Higgins DP. 2016. Altered immune cytokine expression associated with KoRV B infection and season in captive koalas. PLoS One 11:e0163780. doi: 10.1371/journal.pone.0163780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waugh CA, Hanger J, Loader J, King A, Hobbs M, Johnson R, Timms P. 2017. Infection with koala retrovirus subgroup B (KoRV-B), but not KoRV-A, is associated with chlamydial disease in free-ranging koalas (Phascolarctos cinereus). Sci Rep 7:134. doi: 10.1038/s41598-017-00137-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fiebig U, Hartmann MG, Bannert N, Kurth R, Denner J. 2006. Transspecies transmission of the endogenous koala retrovirus. J Virol 80:5651–5654. doi: 10.1128/JVI.02597-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oliveira NM, Farrell KB, Eiden MV. 2006. In vitro characterization of a koala retrovirus. J Virol 80:3104–3107. doi: 10.1128/JVI.80.6.3104-3107.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendoza R, Anderson MM, Overbaugh J. 2006. A putative thiamine transport protein is a receptor for feline leukemia virus subgroup A. J Virol 80:3378–3385. doi: 10.1128/JVI.80.7.3378-3385.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shojima T, Yoshikawa R, Hoshino S, Shimode S, Nakagawa S, Ohata T, Nakaoka R, Miyazawa T. 2013. Identification of a novel subgroup of koala retrovirus from koalas in Japanese zoos. J Virol 87:9943–9948. doi: 10.1128/JVI.01385-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Legione AR, Patterson JLS, Whiteley P, Firestone SM, Curnick M, Bodley K, Lynch M, Gilkerson JR, Sansom FM, Devlin JM. 2017. Koala retrovirus genotyping analyses reveal a low prevalence of KoRV-A in Victorian koalas and an association with clinical disease. J Med Microbiol 66:236–244. doi: 10.1099/jmm.0.000416. [DOI] [PubMed] [Google Scholar]
- 15.Hanger JJ, Bromham LD, McKee JJ, O'Brien TM, Robinson WF. 2000. The nucleotide sequence of koala (Phascolarctos cinereus) retrovirus: a novel type C endogenous virus related to gibbon ape leukemia virus. J Virol 74:4264–4272. doi: 10.1128/JVI.74.9.4264-4272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyazawa T, Shojima T, Yoshikawa R, Ohata T. 2011. Isolation of koala retroviruses from koalas in Japan. J Vet Med Sci 73:65–70. doi: 10.1292/jvms.10-0250. [DOI] [PubMed] [Google Scholar]
- 17.Fiebig U, Keller M, Möller A, Timms P, Denner J. 2015. Lack of antiviral antibody response in koalas infected with koala retroviruses (KoRV). Virus Res 198:30–34. doi: 10.1016/j.virusres.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 18.Abts KC, Ivy JA, DeWoody JA. 2015. Immunomics of the koala (Phascolarctos cinereus). Immunogenetics 67:305–321. doi: 10.1007/s00251-015-0833-6. [DOI] [PubMed] [Google Scholar]
- 19.Xu W, Gorman K, Santiago JC, Kluska K, Eiden MV. 2015. Genetic diversity of koala retroviral envelopes. Viruses 7:1258–1270. doi: 10.3390/v7031258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chappell KJ, Brealey JC, Amarilla AA, Watterson D, Hulse L, Palmieri C, Johnston SD, Holmes EC, Meers J, Young PR. 2017. Phylogenetic diversity of koala retrovirus within a wild koala population. J Virol 91:e01820-16. doi: 10.1128/JVI.01820-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ávila-Arcos MC, Ho SYW, Ishida Y, Nikolaidis N, Tsangaras K, Hönig K, Medina R, Rasmussen M, Fordyce SL, Calvignac-Spencer S, Willerslev E, Gilbert MTP, Helgen KM, Roca AL, Greenwood AD. 2013. One hundred twenty years of koala retrovirus evolution determined from museum skins. Mol Biol Evol 30:299–304. doi: 10.1093/molbev/mss223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waugh C, Gillett A, Polkinghorne A, Timms P. 2016. Serum antibody response to koala retrovirus antigens varies in free-ranging koalas (Phascolarctos cinereus) in Australia: implications for vaccine design. J Wildl Dis 52:422–425. doi: 10.7589/2015-09-257. [DOI] [PubMed] [Google Scholar]
- 23.Fiebig U, Keller M, Denner J. 2016. Detection of koala retrovirus subgroup B (KoRV-B) in animals housed at European zoos. Arch Virol 161:3549–3553. doi: 10.1007/s00705-016-3064-8. [DOI] [PubMed] [Google Scholar]
- 24.Willett BJ, Hosie MJ. 2013. Feline leukaemia virus: half a century since its discovery. Vet J 195:16–23. doi: 10.1016/j.tvjl.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Marsh J, Kollipara A, Timms P, Polkinghorne A. 2011. Novel molecular markers of Chlamydia pecorum genetic diversity in the koala (Phascolarctos cinereus). BMC Microbiol 11:77. doi: 10.1186/1471-2180-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wan C, Loader J, Hanger J, Beagley K, Timms P, Polkinghorne A. 2011. Using quantitative polymerase chain reaction to correlate Chlamydia pecorum infectious load with ocular, urinary and reproductive tract disease in the koala (Phascolarctos cinereus). Aust Vet J 89:409–412. doi: 10.1111/j.1751-0813.2011.00827.x. [DOI] [PubMed] [Google Scholar]
- 27.Levy LS, Lifson JD. 2014. Anti-retroviral drugs and vaccines. Tech Rep Aust Mus Online 24:93–95. doi: 10.3853/j.1835-4211.24.2014.1625. [DOI] [Google Scholar]
- 28.Denner J, Young PR. 2013. Koala retroviruses: characterization and impact on the life of koalas. Retrovirology 10:108. doi: 10.1186/1742-4690-10-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Devereaux LN, Polkinghorne A, Meijer A, Timms P. 2003. Molecular evidence for novel chlamydial infections in the koala (Phascolarctos cinereus). Syst Appl Microbiol 26:245–253. doi: 10.1078/072320203322346092. [DOI] [PubMed] [Google Scholar]
- 30.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 32.Ribeiro-Gonçalves B, Francisco AP, Vaz C, Ramirez M, Carriço JA. 2016. PHYLOViZ Online: web-based tool for visualization, phylogenetic inference, analysis and sharing of minimum spanning trees. Nucleic Acids Res 44:W246–W251. doi: 10.1093/nar/gkw359. [DOI] [PMC free article] [PubMed] [Google Scholar]