ABSTRACT
HIV-1 elite controllers (EC) maintain undetectable viral loads (VL) in the absence of antiretroviral treatment. However, these subjects have heterogeneous clinical outcomes, including a proportion that loses HIV-1 control over time. In this work, we compared, in a longitudinal design, transient EC, analyzed before and after the loss of virological control, with persistent EC. The aim was to identify factors leading to the loss of natural virological control of HIV-1 infection with a longitudinal retrospective study design. Gag-specific T-cell responses were assessed by in vitro intracellular polycytokine production quantified by flow cytometry. Viral diversity determinations and sequence dating were performed in proviral DNA by PCR amplification at limiting dilution of env and gag genes. The expression profile of 70 serum cytokines and chemokines was assessed by multiplex immunoassays. We identified transient EC as subjects with low Gag-specific T-cell polyfunctionality, high viral diversity, and high proinflammatory cytokine levels before the loss of control. Gag-specific T-cell polyfunctionality was inversely associated with viral diversity in transient controllers before the loss of control (r = −0.8; P = 0.02). RANTES was a potential biomarker of transient control. This study identified virological and immunological factors, including inflammatory biomarkers associated with two different phenotypes within EC. These results may allow a more accurate definition of EC, which could help in better clinical management of these individuals and in the development of future curative approaches.
IMPORTANCE There is a rare group of HIV-infected patients who have the extraordinary capacity to maintain undetectable viral load levels in the absence of antiretroviral treatment, the so-called HIV-1 elite controllers (EC). However, there is a proportion within these subjects that eventually loses this capability. In this work, we found differences in virological and immune factors, including soluble inflammatory biomarkers, between subjects with persistent control of viral replication and EC that will lose virological control. The identification of these factors could be a key point for a right medical care of those EC who are going to lose natural control of viral replication and for the design of future immunotherapeutic strategies using as a model the natural persistent control of HIV infection.
KEYWORDS: HIV-1 elite controllers, T-cell response, viral diversity, inflammatory biomarkers, HIV-1 controllers, inflammation
INTRODUCTION
The spontaneous control of human immunodeficiency virus type 1 (HIV-1) infection is observed in a rare group of subjects known as HIV-1 elite controllers (EC) (1). As these individuals maintain undetectable viral loads (VL) in the absence of antiretroviral treatment, they have been proposed as a model of functional cure (2). The investigation of the mechanisms behind this natural control has attracted enormous interest for the identification of the host and the virological factors implicated in this phenomenon (3–6).
However, some of these individuals experience virological and immunological progression and AIDS and non-AIDS defining events (7–9). Regarding virological progression, approximately 28% of EC experience loss of viral control over time (7). The factors associated with this loss remain elusive due to the different designs of the studies. First, previous studies were limited to cross-sectional analyses in which virological progression and the heterogeneous characteristics of HIV-1 controllers were not widely taken into account (6, 10–13). Second, there have been very few longitudinal studies, mostly epidemiological (14), where the main contributors were higher ultrasensitive HIV-1 RNA VL and proviral DNA levels. Furthermore, nonconclusive results about inflammatory biomarkers have been found (15). Therefore, the specific determinants associated with virological failure in EC are not definitely established.
The objective of this work was to investigate, following a longitudinal study design, the mechanisms leading to the loss of virological control in a cohort of EC. To this end, we carried out an exhaustive analysis of virological and immunological factors, including proinflammatory cytokines, that could explain the transient or persistent nature of virological control in HIV-1 infection. The identification of biomarkers associated with the loss of viral control will allow the identification of this subgroup of EC, which should help to improve their medical care. In addition, the identification of those factors operating in the persistent control of viral replication in EC may provide new insights for the design of novel eradication and immunotherapeutic strategies.
RESULTS
Characteristics of the studied subjects.
Clinical and demographic characteristics of transient controllers (TC) and persistent controllers (PC) (see the study design in Fig. 1) are shown in Table 1. The frequency of a sexual transmission route was higher in TC (75%) than in PC (37%) (P = 0.049). The TC group presented a shorter time after diagnosis than the PC group (8 [2 to 14] years versus 18 [11 to 22] years; P = 0.002). There were no differences in the remaining variables at baseline. After the loss of control, the VL from TC were 627 (230 to 4,618) HIV RNA copies/ml at time zero (T0), 1,730 (397 to 4,420) HIV RNA copies/ml at 1 year (+T1), and 2,860 (727 to 4,920) HIV RNA copies/ml at 2 years (+T2). Therefore, half of the patients were viremic controllers because of the loss of control, and the remaining patients had low VL (<104 log HIV RNA copies/ml).
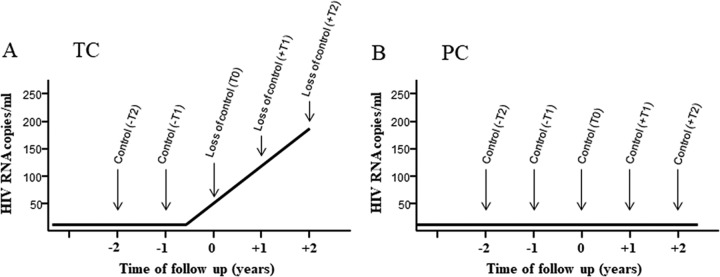
FIG 1.
Study design. Schematic representation of the longitudinal and retrospective study design in transient controllers (TC) (A) and persistent controllers (PC) (B). In TC, up to five determinations were performed: two in the “under-control period,” 2 years (−T2) and 1 year (−T1) before the loss of control, and up to three determinations in the “post-loss-of-control period,” including the closest time point to the loss of virological control (T0) and 1 year (+T1) and 2 years (+T2) after the loss of virological control. At least the −T2, −T1, and T0 samples were required for the subject to be included in the study. In total, a maximum of 54 time points were analyzed in this group. In PC, up to five determinations were performed at 1-year intervals, but at least three consecutive time points per subject were required to be included in the study. In total, a maximum of 63 time points were analyzed in this group. For Gag-specific T-cell response assays, all available follow-up time points were tested (PC, n = 14; TC, n = 14). Virological and soluble biomarkers assays were done for all available follow-up time points in the PC (n = 10 and n = 11, respectively) and only at –T2 and –T1 in the TC (n = 9 and n = 12, respectively).
TABLE 1.
Characteristics of the subjects
| Parametera | Value for groupb |
Pc | |
|---|---|---|---|
| TC (n = 14) | PC (n = 17) | ||
| Age (yr), median (IQR) | 41 (38–52) | 45 (41–48) | 0.279 |
| Male sex, no. (%) | 8 (57) | 10 (59) | 0.925 |
| Sexual transmission, no. (%) | 10 (71) | 6 (35) | 0.049 |
| Time since diagnosis (yr), median (IQR) | 8 (2–14) | 18 (11–22) | 0.002 |
| HCV RNA detected, no. (%) | 6 (43) | 10 (59) | 0.376 |
| CD4+ T cells (cells/μl), median (IQR) | 625 (392–783) | 714 (627–940) | 0.208 |
| CD8+ T cells (cells/μl), median (IQR) | 735 (548–1015) | 648 (569–970) | 0.999 |
| CD4/CD8 ratio, median (IQR) | 1 (0.58–1.28) | 1.17 (0.67–1.55) | 0.456 |
| HLA B57, no. (%)d | 3 (21) | 6 (40) | 0.280 |
| HLA B27, no. (%)d | 1 (7) | 2 (13) | 0.584 |
| HLA B35, no. (%)d | 2 (14) | 0 (0) | 0.129 |
| IL28B-CC, no. (%)e | 3 (38) | 5 (42) | 0.728 |
IQR, interquartile range.
Values from transient controllers (TC) are taken from −T2, and values from persistent controllers (PC) are taken from the first time point of follow-up.
The Mann-Whitney U and chi-square tests were used. All P values of <0.05 were considered significant and are highlighted in bold.
n = 15 for PC.
n = 8 and n = 12 for TC and PC, respectively.
Low levels of Gag-specific T-cell responses preceded the loss of virological control in TC.
The CD4+ and CD8+ T-cell counts and CD4/CD8 ratios were not different and did not change during follow-up in the two groups (Table 2). In the same way, no differences were observed throughout the follow-up in Gag-specific CD4+ and CD8+ and different T-cell subset responses in PC after multiple comparison testing (Fig. 2). Based on these data and to simplify the analyses, the variables in PC were expressed as the mean value of all longitudinal determinations.
TABLE 2.
T-cell levels and CD4/CD8 ratio of the study subjectsa
| Group | Time point | CD4 T cells (cells/μl) | CD8 T cells (cells/μl) | CD4/CD8 ratio |
|---|---|---|---|---|
| TC (n = 14)b | −T2 | 625 (391–783) | 735 (547–1,014) | 1 (0.58–1.28) |
| −T1 | 745 (491–925) | 862 (587–1,223) | 0.97 (0.66–1.16) | |
| T0 | 591 (474–761) | 686 (532–1,134) | 0.93 (0.44–1.3) | |
| +T1 | 531 (424–695) | 750 (646–1,147) | 0.73 (0.38–1.07) | |
| +T2 | 735 (547–1014) | 602 (592–1,501) | 0.56 (0.45–1.31) | |
| PC (n = 17)b | −T2 | 714 (627–940) | 648 (569–970) | 1.17 (0.67–1.55) |
| −T1 | 689 (557–940) | 720 (483–917) | 1.09 (0.81–1.72) | |
| T0 | 550 (419–958) | 629 (350–1,030) | 1.13 (0.51–1.77) | |
| +T1 | 710 (600–961) | 672 (584–1,058) | 1.43 (0.88–1.68) | |
| +T2 | 556 (489–661) | 554 (251–764) | 1.18 (0.78–2.02) | |
| PC (mean) | 651 (576–989) | 725 (591–954) | 1.09 (0.62–1.56) | |
| P | −T2 vs PC (mean) | 0.284 | 0.757 | 0.452 |
| −T1 vs PC (mean) | 0.965 | 0.292 | 0.547 | |
| T0 vs PC (mean) | 0.174 | 0.906 | 0.269 | |
| +T1 vs PC (mean) | 0.084 | 0.393 | 0.052 | |
| +T2 vs PC (mean) | 0.351 | 0.969 | 0.225 |
Values are given as medians (interquartile ranges).
The Friedman test was used to analyze differences during the follow-up in each group (TC and PC); no differences were found (P > 0.05) in all the cases. The Mann-Whitney U test was used for between-group comparisons.
FIG 2.
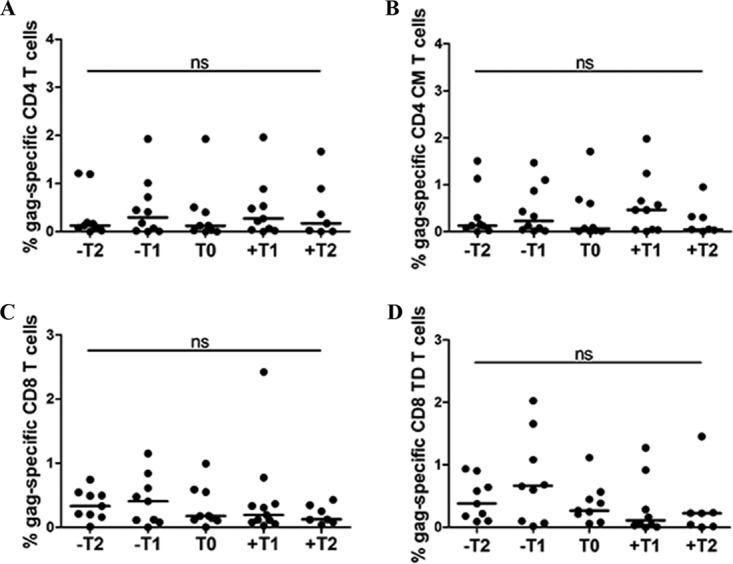
Representative longitudinal Gag-specific T-cell-associated parameters in PC. The T-cell response was defined as the frequency of cells (>0.05% after background subtraction of the unstimulated condition) with detectable IFN-γ, TNF-α, and/or IL-2 intracellular cytokine production after stimulation of cryopreserved PBMCs with Gag overlapped peptides. Gag-specific total CD4+ T-cell response (A), central memory CD4+ T-cell response (B) (CM, CD4 CD45RA CD27), total CD8+ T-cell response (C), and terminally differentiated CD8+ T-cell response (D) (TD, CD8 CD45RA CD27) levels are shown. No statistical differences were obtained throughout the follow-up. NS denotes no significant differences between multiple paired sample comparisons determined by the Wilcoxon signed-rank test (P > 0.05 in all cases). The Friedman test could not be applied due to insufficient statistical power using the five follow-up time points.
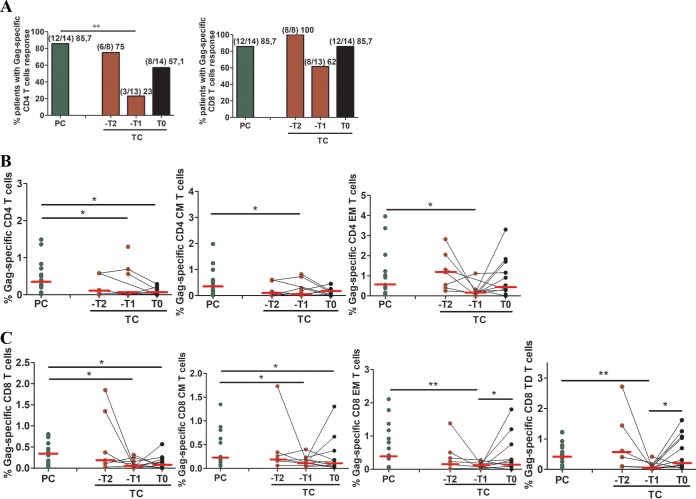
Importantly, there was a higher proportion of PC (12/14 subjects [85.7%]) who showed Gag-specific CD4+ T-cell responses than of TC (3/13 subjects [23.1%]) at 1 year before the loss of virological control (−T1), when Gag-specific CD4+ T-cell responses dramatically decreased. No statistical differences were observed between PC and TC 2 years before (−T2) the loss of virological control nor at the time point just after the loss of virological control (T0) (Fig. 3A, left panel). No differences were found in CD8+ T-cell responses, although there was a similar trend toward lower responses at –T1 in TC (Fig. 3A, right panel).
FIG 3.
CD4+ and CD8+ T-cell Gag-specific responses. The percentages of subjects with Gag-specific CD4+ and CD8+ T-cell responses are shown. (A) The T-cell response was defined as the frequency of cells (>0.05% after background subtraction of the unstimulated condition) with detectable IFN-γ, TNF-α, and/or IL-2 intracellular cytokine production after stimulation of cryopreserved PBMCs with Gag overlapped peptides. (B) Total, central memory (CM; CD4+ CD45RA− CD27+), and effector memory (EM; CD4+ CD45RA− CD27−) Gag-specific CD4+ T-cell levels; (C) total, CM, EM, and terminally differentiated (TD; CD8+ CD45RA+ CD27−) Gag-specific CD8+ T-cell levels. Differences between unpaired groups were determined by the Mann-Whitney U and chi-square tests, and differences between paired samples were determined by the Wilcoxon signed-rank test. The Friedman test was not applied due to the small number of paired samples. Only significant differences are shown. *, P < 0.05; **, P < 0.001.
By analyzing the magnitude and characteristics of the response, we observed that PC presented higher levels of Gag-specific total, central memory (CM), and effector memory (EM) CD4+ T cells than TC at –T1 (Fig. 3B). The same results were obtained for CD8 T cells (Fig. 3C). In addition, in TC, increased levels of Gag-specific EM and terminally differentiated (TD) CD8+ T cells were found at T0 compared to –T1 (Fig. 3C).
Gag-specific CD8+ T-cell polyfunctionality is decreased in TC before the loss of virological control.
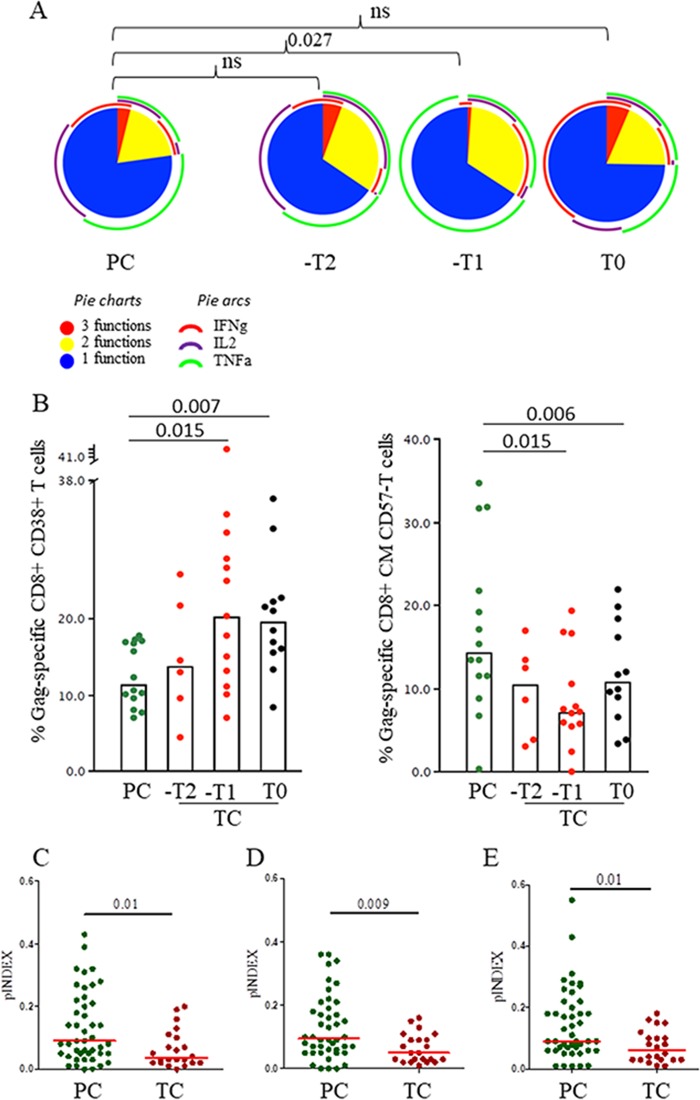
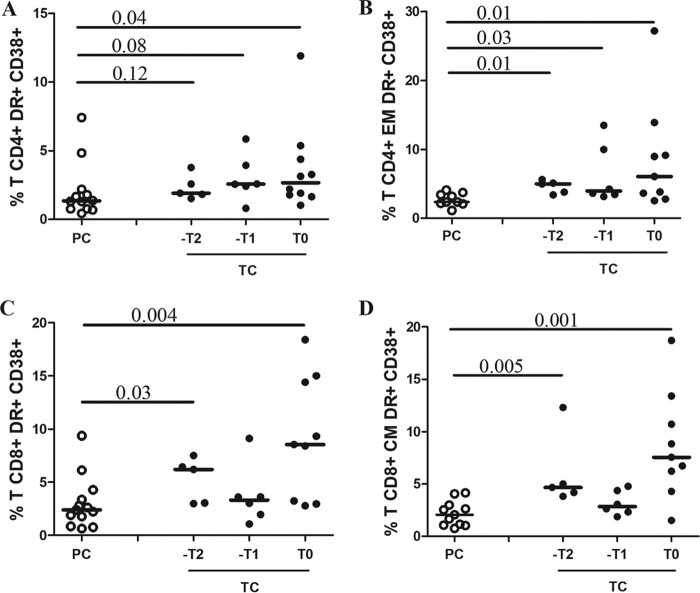
A higher frequency of polyfunctional CD8+ T cells was observed in PC than in TC at –T1, with a higher proportion of cells with three functions and a higher cytokine diversity (a representative example is shown in Fig. 4A). There were no differences between PC and TC at T0, when VL was detectable in TC. However, by analyzing the maturation and activation patterns of Gag-specific T-cell responses (Fig. 4B), we found that TC presented higher Gag-specific CD8+ CD38+ T-cell levels and lower Gag-specific CD8+ CM CD57− T-cell levels at –T1 and T0 than PC. In addition, bulk T-cell activation was higher in TC than PC at T0 (Fig. 5).
FIG 4.
HIV-1-specific CD8+ T-cell polyfunctionality. Polyfunctionality, understood as simultaneous multiple production of IFN-γ, TNF-α, and IL-2 per T cell, was studied only for subjects categorized as responders. The T-cell response was defined as the frequency of cells (>0.05% after background subtraction of the unstimulated condition) with detectable IFN-γ, TNF-α, and/or IL-2 intracellular cytokine production after stimulation. Due to the low number of Gag-specific CD4+ T-cell responders in TC, polyfunctionality analysis was not applicable. (A) Pie charts show polyfunctional distribution of HIV-1-specific CD8+ TD CD57+ T cells with up to three functional responses to Gag stimulation; IFN-γ, TNF-α, and IL-2 production in the polyfunctional distribution is shown in arcs. Pestle and Spice were used for analysis. (B) Percentages of Gag-specific CD8+ T cells expressing the activation profile, CD38+, and the maturation profile, CD45RA− CD27+ CD57−; only significant differences are shown. (C to E) Polyfunctionality index of Gag-specific total CD8+ T cells. Values from PC and preloss time points of follow-up (–T2 and –T1) in TC are based on the proportions of cells expressing combinations of IFN-γ, TNF-α, and IL-2 (three functions) (C), plus CD107a (four functions) (D), and plus perforin (five functions) (E). Single and double production of CD107a and perforin were excluded from the analyses. Differences between groups were determined by the Mann-Whitney U test.
FIG 5.
CD4+ and CD8+ T-cell activation. Percentages of total (A) and effector memory (B) CD4+ HLA-DR+ CD38+ T cells and total (C) and central memory (D) CD8+ HLA-DR+ CD38+ T cells are shown. Differences between unpaired groups were determined by the Mann-Whitney U test. The Friedman test was not applied due to the small number of paired samples. P < 0.15 are shown.
Indexes of polyfunctionality (pINDEX) in Gag-specific total CD8+ T cells were compared between PC and TC at –T2 together with –T1 (pre-loss-of-control period). The pINDEX was higher in three, four, and five functions (Fig. 4C to E) in PC than in TC.
TC displayed higher viral diversity than PC.
env and gag genes were amplified in proviral DNA by limiting-dilution PCR in all TC (9/9, 100%) but in only 5 of 10 (50%) of the PC. Afterwards, in the remaining available samples, we quantified proviral loads by an Alu real-time PCR; 33% (4/12) of TC samples showed values ranging from 5 to 108 copies/106 cells, and the remaining TC samples and 9 of 9 samples from PC (100%) showed values below the detection limit (5 copies/106 cells).
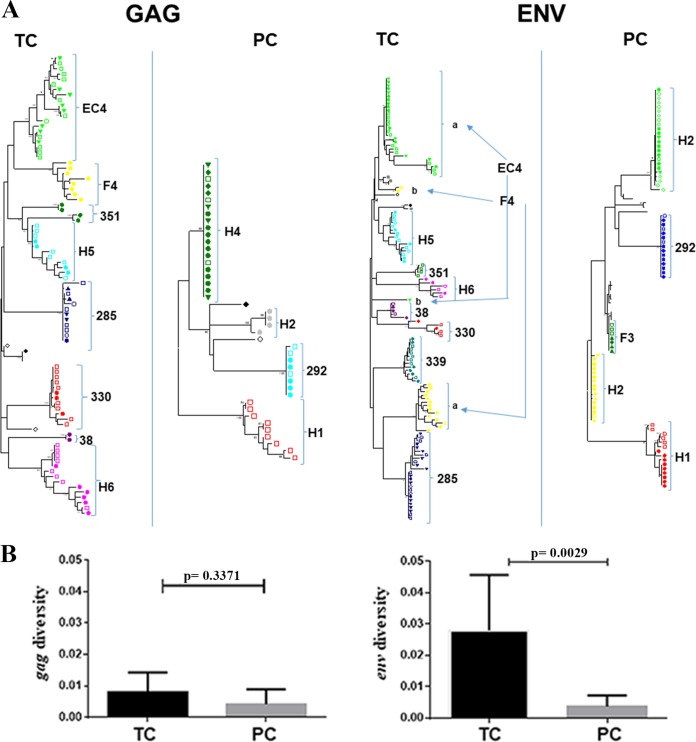
The branches in the phylogenetic tree (Fig. 6A) were longer in TC than in PC, indicating viral replication and evolution. In the sequences from PC, the branch length was minimal or zero, with many identical sequences. Although we did not analyze the integration sites, this pattern indicates a lack of viral replication and could suggest that these viral populations are the consequence of clonal expansions (16–18).
FIG 6.
Virological assays: phylogenetic analysis and virus diversity. All virological assays were performed with all time point samples in PC and with only samples from time points prior to the loss of control in TC. Phylogenetic analysis of sequences in env and gag genes from TC and PC during follow-up was done. Sequences were submitted to GenBank under accession numbers MF988754 to MF989105. (A) Phylogenetic trees were estimated by a maximum likelihood approach using the best-fit model of nucleotide substitution (GTR+G+I; jModelTest v.0.1.1) implemented in the MEGA 6 software program. Each subject is represented by a different color. Samples taken at different times are marked with different symbols. In double-infected subjects (EC4 and F4), the two viral populations are marked and labeled a and b. Bars indicate a genetic distance of 0.02. (B) Comparison between intrasample diversity in env and gag regions for TC and PC. The mean and standard errors for all pairwise nucleotide distances were determined using the MEGA 6.0 program. Differences between unpaired groups were determined by the Mann-Whitney U test. **, P < 0.05.
Regarding the evolution of gag and env gene sequences with time, TC showed intermingled sequences and no replacement of viral populations at the different time points (Fig. 6A). This evolutionary model is compatible with an atemporal mode of evolution (11). In contrast, sequences at different times from PC showed no evidence of viral evolution (Fig. 6A). These results were supported by viral dating estimation. For this analysis, available samples of PC, obtained in 2010 ± 0.9 year, a mean of 18 years after HIV-1 diagnosis, showed sequences without any viral evolution since HIV-1 diagnosis (Table 3). In these patients, the difference between viral dating and the HIV-1 diagnosis time was 5 years. In TC patients, available samples obtained a mean of 15 years after HIV-1 diagnosis showed, because of viral evolution, a viral dating (2001 ± 3.4 years) closer to the sampling time (2005 ± 3.5 years) than to the HIV diagnosis date (1989 ± 4.1 years), with a mean difference of 11.5 years (Table 3). In two TC (F4 and EC4), the nucleotide sequences were separated in two independent clades (Fig. 6A, right panel). This segregation supports that these were double-infected subjects, as previously reported for EC4 (19). All patients had an R5 tropism, and, in the case of TC, the tropism did not change after viral replication.
TABLE 3.
Year of HIV-1 diagnosis, sampling year, and viral dating in TC and PC
| Patient group and identifier | Yr of HIV-1 diagnosis | Sampling yr | Yrs since HIV-1 diagnosis | Yr estimated by viral dating | Difference in yr of viral dating and HIV-1 diagnosis (yrs) |
|---|---|---|---|---|---|
| TC | |||||
| 330 | 1991 | 2000 | 9 | 2004 | 13 |
| F4 | 1986 | 2003 | 17 | 1997 | 11.8 |
| 74 | 1996 | 2007 | 11 | 2003.2 | 7.2 |
| H5 | 1986 | 2006 | 20 | 1997 | 11 |
| H6 | 1990 | 2009 | 19 | 2003.6 | 13.6 |
| Meana | 1989 ± 4.1 | 2005 ± 3.5 | 15 ± 4.9 | 2001 ± 3.4 | 11.5 |
| PC | |||||
| H1 | 1999 | 2010 | 11 | 2004 | 5 |
| H2 | 1994 | 2011 | 17 | 1997 | 3 |
| H4 | 1986 | 2011 | 25 | 1993 | 7 |
| 292 | 1998 | 2011 | 13 | 1995 | −3 |
| F3 | 1985 | 2009 | 24 | 1994 | 9 |
| Meana | 1992 ± 6.6 | 2010 ± 0.9 | 18 ± 6.3 | 1997 ± 4.4 | 5 |
Values are expressed as means ± standard deviations. To perform the viral dating of the subjects, the genetic distance of the reconstructed most recent common ancestor (MRCA) for each patient clade was compared with that of a reconstructed MRCA for the Spanish epidemic. The viral dating time was estimated by use of a linear-correlation equation, previously developed in a large set of Spanish samples, that correlates the V3 nucleotide sequence divergence to the Spanish epidemic MRCA and the sampling year. Only four subjects in the TC group were included. The viral dating of other TC could not be estimated because the sampling time and the year of HIV-1 diagnosis were too close in time.
Viral diversity in the gag region was higher in TC than in PC, but the difference did not reach a statistical significance, probably due to the low number of sequences analyzed. In contrast, in the env C2-V5 region, diversity was statistically higher in TC than in PC (P < 0.0029), again supporting viral replication in TC (Fig. 6B), even if we exclude analysis of the sequences of the double-infected patients (P < 0.016).
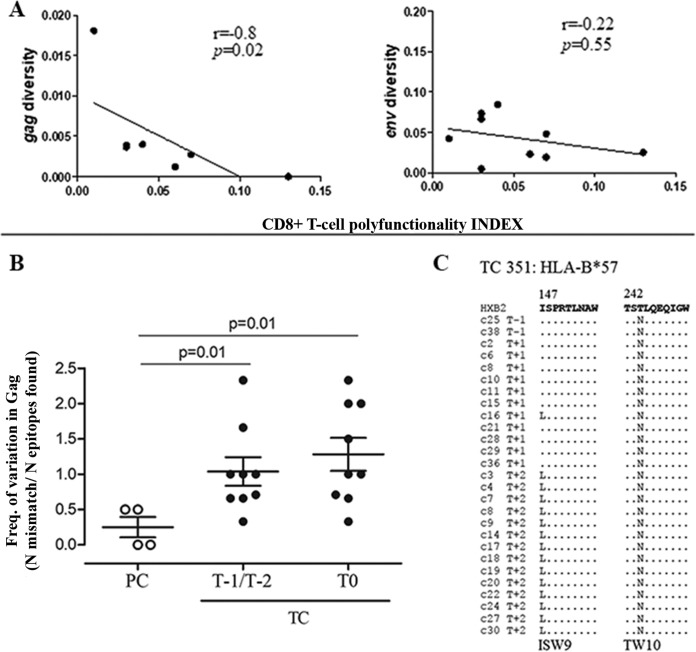
Virus diversity was associated with Gag-specific CD8+ T-cell polyfunctionality.
Three-function total CD8+ T-cell polyfunctionality was inversely associated with gag diversity (r = −0.8; P = 0.02) but not with env (Fig. 7A) in TC. Regarding four and five functions, total CD8+ T-cell polyfunctionality association with viral diversity remained in the same trend as that of three functions, although it was not statistically significant (r = −0.6, P = 0.14, and r = −0.57, P = 0.1, for four and five functions, respectively). No association was found in PC (data not shown). We evaluated the presence of HIV-1 footprints of immune escape in Gag epitopes in both pre- and post-loss-of-control samples comparing TC and PC. We found a trend toward a higher frequency of mutations in Gag epitopes in TC than in PC (Fig. 7B). In addition, for TC patient 351 expressing HLA-B*57, we tracked the temporal emergence of escape variants in the HLA-B*57 restricted epitope ISW9, which appeared after the loss of control, and the preexistence of TW10 escape mutants along with the loss of virological control over time, indicating HIV-1 evolution against CD8+ T-cell responses (Fig. 7C).
FIG 7.
Correlation of virus diversity with CD8+ T-cell polyfunctionality and analysis of Gag CD8+ T-cell epitope variation. (A) Correlations between gag and env diversity and the total CD8+ T-cell three-cytokine polyfunctionality index (pINDEX for three functions) in TC are shown. The Spearman rho correlation coefficient test was used. (B) Analysis of HIV-1 Gag CD8+ T-cell epitope variation. The frequencies (Freq.) of variation in HIV-1 Gag CD8+ T-cell epitopes in PC and TC before and after the loss of virological control are shown. Differences between groups were determined by the Mann-Whitney U test. (C) HIV-1 sequence variation in Gag ISW9 and TW10 epitopes restricted by HLA-B*57 in TC 351 during the follow-up.
High soluble biomarkers and proinflammatory cytokine levels preceded the loss of virological control.
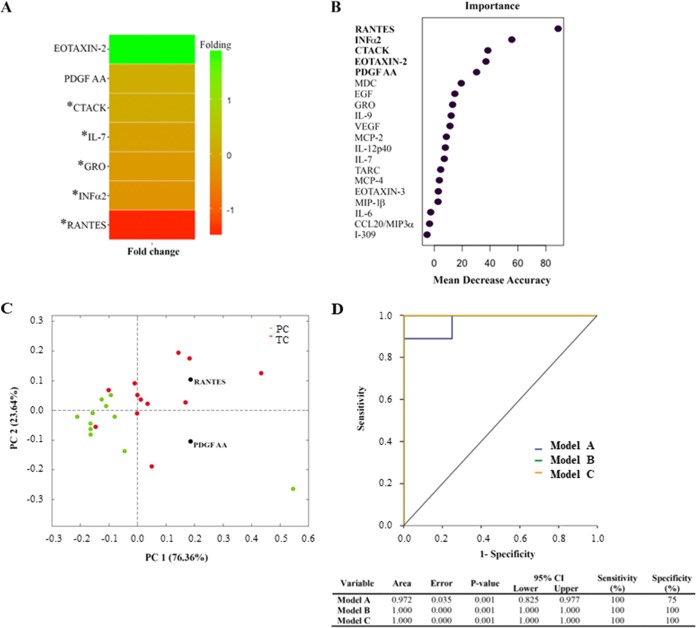
The higher levels of RANTES, interferon alpha 2 (IFN-α2), human growth-regulated oncogene (GRO), interleukin 7 (IL-7), and cutaneous T-cell-attracting chemokine (CTACK) in TC than in PC were the variables that significantly differentiated the two groups by the Mann-Whitney U test (Fig. 8A). After random forest analysis (Fig. 8B), RANTES, IFN-α2, and CTACK were similarly considered potential biomarkers together with platelet-derived growth factor AA (PDGF-AA) and eotaxin-2. The principal-component analysis (PCA) model (Fig. 8C) restricted the biomarkers to RANTES and PDFG-AA, which showed the best percentage of separation between TC and PC. Finally, analysis by receiver operating characteristic (ROC) curves (Fig. 8D) showed that the top variables of the random forest approach (model B) and the PCA (model C) displayed an area under the curve (AUC) of 1, which means that a perfect classification was achieved using only RANTES and PDGF-AA. All statistical tests defined RANTES as the most remarkable chemokine for group discrimination, with 4-fold higher levels in TC than in PC (P = 0.001) and a cutoff value of 10,464.5 pg/ml.
FIG 8.
Soluble cytokines and chemokines as potential biomarkers of the loss of virological control. Assays were performed with all time point samples in PC and with samples at preloss time points of follow-up (–T2 and –T1) in TC. (A) A fold change heat map of the relative plasma concentrations of measured inflammatory markers is shown. Positive folding (green) means higher concentrations in PC, while negative folding (red) means the opposite. Of these potential biomarkers, the five marked with an asterisk reached a concentration with a P value of <0.05 in the Mann-Whitney U test, and the remaining two were added to the list due to their classification power in the two-multivariate test. (B) Random forest analysis importance plot of the top 20 variables in importance of classification from a total of 70 cytokines and chemokines. Only the top five, highlighted in bold, were considered potential biomarkers. (C) The score plot of the PCA showed that the best percentage of separation between groups was achieved with only two variables, RANTES and PDGF AA. (D) Using logistic regression and receiver operator characteristic (ROC) curves, we assessed three different multimarker models that could accurately predict the loss of control in EC: model A (blue), which includes the statistically significant variables in the Mann-Whitney U test, model B (green), composed of the top five variables obtained from the random forest analysis, and model C (orange), compounded by the two variables obtained in the PCA analysis.
DISCUSSION
In this work, we showed that low Gag-specific T-cell polyfunctionality, high viral evolution, and high levels of proinflammatory cytokines were associated with the future loss of natural virological control. In addition, RANTES levels may be considered a potential novel biomarker predictive of the loss of virological control in EC.
The lack of HIV-1-specific T-cell responses is considered a hallmark of progressive HIV-1 infection (20). Preserved CD4+ and CD8+ T-cell multifunctional responses have been consistently associated with the control of viral replication in EC (4, 21, 22). However, this generalized assumption was derived from cross-sectional studies with heterogeneity between study subjects (23, 24). Our study demonstrates that TC show a low magnitude of Gag-specific T-cell responses together with fewer polyfunctional T cells than PC. In addition, this specific T-cell response has a particular mature and more highly activated phenotype in TC, in agreement with previous findings of T-cell activation preceding the viral breakthrough after spontaneous control in acute infection (25). These characteristics precede by 1 year the loss of spontaneous control. These results may explain the variability observed in previously studied cohorts (24, 26, 27) in which virological progression was not taken into account, and because of that, these studies could not discriminate between TC and PC. In addition, our data are in agreement with a previous work showing a decreased CD8+ T-cell breadth that was associated with loss of viral control but in viremic controllers, although we did not observe an association between functional Gag-specific CD8+ T cells and the presence of protective HLA class I alleles (28). One possible mechanism involved in the loss of control may be related to the fact that CD8+ T cells restricted by “nonprotective” HLA allele groups can be suppressed by regulatory T cells (29). Ferrando-Martinez et al. (26) showed that mature (EM) Gag-specific CD8+ T cells from EC had higher polyfunctionality than viremic controllers and non-HIV-1 controllers. In our work, we observed increased polyfunctionality in more mature (TD CD57+) Gag-specific CD8+ T cells from PC but not from TC. This result suggests that Gag-specific CD8+ T cells with a mature phenotype have increased cytotoxic abilities and points to this mechanism as one of the main causes of persistent natural control. The absence of differences in polyfunctional distribution between PC and TC in this subset at T0 may be explained by the particular mature and more highly activated phenotype of Gag-specific T-cell response and higher bulk T-cell activation at this time point. Regarding Gag-specific CD4+ T cells, the dramatic decrease in this response 1 year before the loss of control is in agreement with a previous study showing the important role of simian immunodeficiency virus (SIV)-specific CD4+ T cells in the breakthrough SIVmac239 viremia in an elite controller (30).
Among other viral properties, genetic variability and viral evolution have been associated with viral pathogenesis and disease progression (31–35). Proviral DNA from peripheral blood mononuclear cells (PBMCs) is considered to harbor a combination of recently produced and archived variants, and consequently, variability in this compartment better reflects viral evolution since primoinfection (18, 36). Analysis of env and gag proviral DNA sequences, in samples prior to the loss of control, showed evidence of viral divergence and diversity in TC, demonstrating not fully restricted viral replication despite undetectable VL. On the contrary, PC presented minimal or no proviral divergence and extremely low diversity, suggesting no viral replication since primoinfection or that replication is occurring at such a low rate that reseeding of this proviral compartment is not possible, as suggested in EC, although evolution in plasma virus was detected (37–39). Lower levels of viral diversity were observed in EC than in patients with undetectable levels of viremia due to combined antiretroviral treatment (39, 40). Moreover, the viral diversity in EC is not directed by neutralizing antibodies (40), although escape mutants to neutralizing antibodies are continuously generated and selected in these patients (41). Our results are in agreement with previous works showing a relation among viral replication and evolution, viral pathogenesis, and disease progression (33–35, 37, 42, 43). Therefore, markers of HIV-1 viral evolution (divergence and diversity) could differentiate the TC and PC phenotypes in the pool of EC. The homogeneous viral populations found in PC are also observed in viral infections with fidelity mutants of poliovirus (44, 45) or arbovirus (46), which result in less pathogenic infections (33, 34, 35).
Immunological and virological factors were intimately associated in these subjects. We found strong inverse associations between gag diversity and Gag-specific CD8+ T-cell polyfunctionality (three functions) but only at the follow-up time points with undetectable viremia in TC. The association was found in gag and not in env, probably because we analyzed Gag- but not Env-specific T-cell responses. These observations are in agreement with previous findings that demonstrate that responses targeting gag but not env are inversely associated with VL (47, 48). The results presented herein revealed that the lack of viral evolution reflects the continuous immune pressure exerted by Gag-specific T-cell responses in PC and the fine balance between viral persistence and immune control in TC. Our results are in agreement with those of Noel et al., which demonstrated the contribution of abortive viral expression in CD4+ T cells from EC for maintenance of strong HIV-1-specific CD8+ T-cell responses in the absence of HIV-1 evolution (49). Likewise, these results may be in agreement with the work of Boritz et al. showing that clonal expansion of infected cells acts to maintain the virus in HIV controllers (18). The lack of viral evolution found in this work suggests that clonal expansion may be one of the main mechanisms of virus persistence in PC. This mechanism may be enough for PC to maintain an enhanced Gag-specific T-cell response that is able to contain viral replication. On the other hand, a second mechanism proposed by Boritz et al. (18), consisting of replication of inducible proviruses of recent origin, may be the prevalent mechanism in TC before the loss of control, reflecting a less efficient Gag-specific T-cell response that is more compatible to what happened in viremic controllers.
The different patterns observed in viral diversity and DNA proviral load could be linked with different magnitudes and qualities of Gag-specific T-cell responses and with different levels of inflammatory mediators. There is increasing evidence that EC have increased levels of T-cell and myeloid activation and unique inflammatory signatures in comparison with HIV-suppressed and HIV-1-uninfected subjects (50–52). Interestingly, a lack of an association between inflammatory markers and VL in EC has been described before (13), suggesting that persistent viral replication may not be the main trigger behind the inflammation observed in EC. Our data support that despite no detectable plasma VL, the lower Gag-specific T-cell response in TC and higher viral diversity and DNA proviral load may be related to the higher levels of several inflammatory cytokines in TC.
This is the first extensive analysis of the expression profile of 70 plasma cytokines and chemokines in EC with different virological evolutions. Random forest analysis, PCA, and ROC curves which included the combination of the selected proinflammatory markers revealed that RANTES and PDGF-AA may be sufficient to clinically categorize PC and TC. Of special interest is the chemokine RANTES, the most remarkable biomarker for group discrimination, whose concentration was four times higher in TC than in PC. Despite the fact that RANTES, the natural CCR5 ligand, has been previously shown to prevent HIV infection (53), RANTES can also be considered a proinflammatory cytokine, and it has been associated with HIV disease progression (54–56). Higher levels of RANTES may reflect low-level residual viral replication in TC that could be associated with CD8+ T-cell dysregulation by exhaustion (57). Our findings not only define RANTES as a reliable biomarker for a rapid screening of potential EC with virological loss of control but also suggest that the immunomodulation of RANTES is a therapeutic target in EC.
This study has disclosed the presence of two distinct groups of EC, and we have been able to find differences between these two EC phenotypes based on immunological and virological markers. These results demonstrate that HIV-1 controllers are a heterogeneous group of subjects with different characteristics and nomenclatures (58), as we (59) and others (60) previously suggested. Accordingly, it is crucial to establish a precise definition of the EC phenotype in order to identify the correlates of persistent spontaneous control in the search for the right model of functional remission. These new insights might help in the reconsideration of the current treatment guidelines that recommend antiretroviral treatment for all HIV-1 controllers (https://aidsinfo.nih.gov/guidelines).
The main limitation of this study was sample availability, which constrained the evaluation of additional immunological and virological factors. However, these subjects are extremely rare, and even so, we were able to have a follow-up with samples before and after the loss of control.
In summary, this study has allowed the identification of several immunological, virological, and proinflammatory cytokines that will help in the accurate definition of EC and in their clinical management. In addition, the identification of important factors for the persistent natural control of HIV replication could give new clues to achieve a long-term remission status or functional cure in HIV-1-infected patients.
MATERIALS AND METHODS
Study participants.
EC were defined as subjects with three consecutive VL determinations under the detection limit (<50 HIV-1 RNA copies/ml) in the absence of antiretroviral treatment for at least 1 year of follow-up (7). Subjects were included based on frozen peripheral blood mononuclear cells (PBMCs) and plasma samples available in the Spanish HIV HGM BioBank belonging to the AIDS Research Network (61) and with data in the RIS Controllers Study Group Cohort (ECRIS) (8) (see File S1 in the supplemental material) based on the study design. Thirty-one EC were analyzed; 14 of these underwent loss of virological control (at least two consecutive measurements of VL above the detection limit in 1 year) and were named transient controllers (TC). Seventeen EC who maintained persistent virological control during the same follow-up period were designated persistent controllers (PC) (see study design in Fig. 1). All subjects participating in the study gave their informed consent, and protocols were approved by the institutional ethical committees.
Experimental procedures.
Laboratory evaluations were performed at the Laboratory of Immunovirology, Institute of Biomedicine of Seville (IBiS), Virgen del Rocío University Hospital in Seville (Spain); the Molecular Virology Unit, Laboratory of Research and Reference in Retrovirus, Centro Nacional de Microbiología, Instituto de Salud Carlos III, Madrid (Spain); the Joan XXIII University Hospital in Tarragona, IISPV, Rovira i Virgili University (Spain); and the AIDS Research Institute IrsiCaixa, Badalona (Spain).
General.
Absolute counts of CD4+ and CD8+ T cells were determined in fresh whole blood by using an Epic XL-MCL flow cytometer (Beckman-Coulter, Brea, CA) according to the manufacturer's instructions. The plasma HIV-1 RNA concentration was measured by using quantitative PCR (Cobas Ampliprep/Cobas TaqMan HIV-1 test; Roche Molecular Systems, Basel, Switzerland) according to the manufacturer's protocol. The detection limit for this assay was 50 HIV-1 RNA copies/ml. Hepatitis C virus (HCV) RNA was determined using an available PCR procedure kit (Cobas Amplicor; Roche Diagnostics, Barcelona, Spain) with a detection limit of 10 IU/ml.
Genetics.
HLA-B group alleles were genotyped using a reverse sequence-specific oligonucleotide bound to a fluorescently coded microsphere system (LABType SSO, RSSO1B; One Lambda, Canoga Park, CA) by following the manufacturer's instructions. The genotyping of the IL28B single nucleotide polymorphism (SNP) rs12979860 was performed as previously described (62), using a TaqMan 5′ allelic discrimination assay (Applied Biosystems, Foster City, CA).
Cell stimulation.
PBMCs were thawed, washed, and stimulated in vitro with 2 μg/ml of an overlapped HIV (Gag)-specific peptide pool (NIH AIDS Reagent Program [https://www.aidsreagent.org/index.cfm]) and stained with conjugated monoclonal anti-CD107a-BV786 (clone H4A3; BD Biosciences, Franklin Lakes, NJ) at the beginning of incubation as previously described (26).
Immunophenotyping and intracellular cytokine staining.
Stimulated PBMCs were washed and stained with LIVE/DEAD fixable violet dead cell stain (Life Technologies, CA, USA). The cells were then surface stained with anti-CD14-PB, anti-CD19-PB, anti-CD38-Qdot655 (Life Technologies), anti-HLA-DR-BV570 (clone L243), anti-CD56-PB (Biolegend, San Diego, CA), anti-CD8+-PerCP-Cy5.5 (clone RPA-T8), anti-CD45RA-fluorescein isothiocyanate (FITC) (clone L48), anti-CD27-BV605, and anti-CD57-phycoerythrin (PE)-CF595 (BD Biosciences). Cells were then stained intracellularly for 30 min with 100 μl of phosphate-buffered saline (PBS) with anti-CD3-allophycocyanin (APC)-H7 (clone SK7), anti-IFN-γ-PCy7, anti-tumor necrosis factor alpha (TNF-α)-Alexa 700 (clone MAb11), anti-IL-2-APC (clone MQ1-17H12), and anti-perforin-PE (clone B-D48) (BD Biosciences) and then washed twice and fixed in PBS containing 4% paraformaldehyde (PFA). Unstimulated cells and cells stimulated with staphylococcal enterotoxin B (SEB) as a positive control were included in each experiment. Lymphocytes were defined as having low forward/side scatter and expressing CD3, and/or no CD8, but not CD19, CD14, and CD56 (Fig. 9).
FIG 9.
Schematic diagram of the cytometry gating strategy. For gating strategies for Gag-specific CD4+ and CD8+ T cells, the representative plots show the functional cytokine responses to Gag peptides.
PBMCs were analyzed by using a LSR Fortessa cell analyzer (BD Biosciences, Spain). A minimum of 1,500,000 total events were recorded for each condition.
Cytokine and chemokine measurement by Milliplex bead array kits.
The compositions of Milliplex MAP human cytokine/chemokine magnetic bead panel kits were as follows. HCYTMAG-60K-PX23 included epidermal growth factor (EGF), IL-1α, fibroblast growth factor 2 (FGF-2), IL-3, eotaxin-1, IL-7, transforming growth factor alpha (TGF-α), IL-8, granulocyte colony-stimulating factor (G-CSF), IP-10, fractalkine, monocyte chemoattractant protein 1 (MCP-1), IFN-α2, MIP-1α, MIP-1β, GRO, MCP-3, vascular endothelial growth factor (VEGF), IL-12 (p40), Flt-3 ligand, MDC (CCL22), sCD40L, and IL-1ra.
HCYTMAG-60K-03 included PDGF-AA, PDGF-AB/BB, and RANTES.
HT17MG-14K-PX25 included granulocyte-macrophage colony-stimulating factor (GM-CSF), IL-21, IFN-γ, IL-4, IL-17F, IL-23, IL-10, IL-5, CCL20/MIP-3α, IL-6, IL-13, IL17E/IL-25, IL-15, IL-27, IL17A, IL-31, IL-22, TNF-α, IL-9, TNF-β, IL-1β, IL28A, IL-33, IL-2, and IL-12 (p70).
HCP2MAG-62K-19 included eotaxin-2, IL-20, MCP-2, TRAIL, BCA-1, CTACK, MCP-4, SDF-1A+b, I-309, ENA-78, IL-16, MIP-1d, TARC, 6CKINE, eotaxin-3, LIF, TPO, SCF, and TSLP6.
Median fluorescence intensities were collected on a Bio-Plex 200 instrument by using Bio-Plex Manager (Bio-Rad Laboratories, Spain) software. Cytokine concentrations were determined from the appropriate standard curves to convert fluorescence units to concentrations (pg/ml).
Nucleic acid extraction and gag and env amplification.
Proviral DNA was obtained from 2 × 106 frozen PBMCs using a Speedtools tissue DNA extraction kit (Biotools B&M Labs S.A., Spain). We used a limiting-dilution nested PCR using Phusion high-fidelity PCR master mix with HF buffer (Thermo Scientific). A first multiplex PCR for gag and env region amplification was performed using the outer primers 505-gag (5′ CGAGGGGCGGCGACTGGT 3′; HXB2 positions 728 to 745), 40-gag (5′ TTCCCTAAAAAATTAGCCTGTCT 3′; HXB2 positions 2074 to 2096), 169-env (5′ AATGTCAGCACAGTACAATGTACAC 3′; HXB2 positions 6945 to 6969), and 96-env (5′ AGACAATAATTGTCTGGCCTGTACCGT 3′; HXB2 positions 7836 to 7862). One microliter of the first PCR product was reamplified independently using primers for gag, i.e., 171-gag (5′ TTTGACTAGCGGAGGCTAG 3′; HXB2 positions 761 to 779) and 336-gag (5′ TTCCAACAGCCCTTTTTCCTAGGGG 3′; HXB2 positions 2009 to 2033), or primers for the C2-V5 region, i.e., 27-env (5′ ATAAGCTTGCAGTCTAGCAGAAGAAGA 3′; HXB2 positions 7004 to 7030) and 167-env (5′ TTCTCCAATTGTCCCTCATATCTCCTCCTCCA 3′; HXB2 positions 7634 to 7665). Nucleotide sequences were determined with the Big DyeTM terminator cycle sequencing kit (Applied Biosystems) in an ABI 3730 sequencer (Applied Biosystems) in the Genomic Unit of the CNM-ISCIII.
Proviral DNA quantification.
The DNA proviral viral load was quantified by using a nested Alu-long terminal repeat (Alu-LTR) PCR (63, 64). In brief, a first conventional PCR was performed using oligonucleotides against Alu sequence and the HIV-1 LTR, with the following conditions: 95°C for 8 min and then 12 cycles of 95°C for 1 min, 60°C for 1 min, and 72°C for 10 min, followed by 1 cycle of 72°C for 15 min. Then, a second quantitative PCR was performed using TaqMan probes with FAM/ZEN/Iowa Black and TaqMan master mix (Applied Biosystems). DNA from the 8E5 cell line was used for the standard curve. The ccr5 gene was used as a housekeeping gene for measuring the input DNA and to normalize the data.
Phylogenetic analysis of the nucleotide sequences in gag and env genes.
Nucleotides sequences were assembled using the SeqMan program (DNASTAR). Maximum likelihood trees were constructed as follows. First, nucleotide sequences were included in a global database with the sequences obtained from the patients studied in our laboratory to detect cross-contamination. All samples segregated in distinct clusters of the phylogenetic tree, excluding contamination. Phylogenies were estimated by a maximum likelihood approach using the best-fit model of nucleotide substitution (GTR+G+I; jModelTest v.0.1.1) implemented in the MEGA 6 software program (65). Internal branch support was tested with an approximate likelihood ratio test (MEGA 6). Intrasample diversity was estimated by the best-fit model of nucleotide substitution in the MEGA 6 program. An estimation of viral infection time or “viral dating” was deduced, assuming a relaxed molecular clock, from the genetic distance of the nucleotide sequence of each subject virus to the MRCA (most recent common ancestor) of the HIV-1 Spanish epidemic (6, 10, 11, 66).
Analysis of Gag CD8+ T-cell immune escape.
HIV-1 variation at the optimal Gag CD8+ T-cell epitopes was based on the best-defined CD8+ epitope summary from the Los Alamos Molecular Immunology Database in Gag proviral sequences matched to patients' HLA class I alleles with two- to four-digit resolution (35). HIV-1 variation at optimal epitopes was defined by comparison with the HIV-1 HXB2 epitope sequence at the HLA-I restriction element. The frequency of virus variation at Gag epitopes was defined as the ratio of total mismatches found at optimal epitopes to the number of total optimal epitopes found per sequence.
Statistical analysis.
Correlations between variables were assessed using the Spearman rank test. Differences between categorical values were determined by the chi-square test. Differences between unpaired groups were determined by the Mann-Whitney U test, and differences between paired samples were determined by the Wilcoxon signed rank and Friedman tests. P values of <0.05 were considered statistically significant. The Statistical Package for the Social Sciences software (SPSS) 22.0 package (IBM, Madrid, Spain) was used for the statistical analysis. Graphs were generated with Prism, version 5.0 (GraphPad Software, Inc.). Polyfunctionality was defined as the percentage of lymphocytes producing multiple cytokines. Polyfunctionality pie charts were constructed using Pestle version 1.6.2 and Spice version 5.2 (provided by M. Roederer, NIH, Bethesda, MD) and was quantified with the polyfunctionality index algorithm (67) employing the 0.1.2 beta version of the FunkyCells Boolean Dataminer software provided by Martin Larson (INSERM U1135, Paris, France).
The selection of potential biomarkers among the soluble markers of inflammation associated with the loss of spontaneous control was complemented with multivariate statistics using random forest analysis (unbiased selection of variables) and principal-component analysis (PCA) (multivariate pattern that generates the maximum degree of separation between groups), and finally, the performance of biomarkers was examined by logistic regression analysis and receiver operating characteristic (ROC) curves. The employed statistical software included the R software (http://cran.r-project.org), matrix calculation platform MATLAB (version 7.5.0; The Mathworks, Inc., Natick, MA, USA).
Accession number(s).
Nucleotide sequences were submitted to GenBank under accession numbers MF988754 to MF989105.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Instituto de Salud Carlos III (research projects PI12/02283, PI13/01912, PI13/02269, PI14/01058, PI10/2635, PI13/0796, PI16/0503, PI16CIII/0034, PI12/00506, PI12/00969, PI15/00480, and PI16/00684 and research contracts CPII014/00025 to E.R.-M., CPII15/00014 to J.G.P., and FI14/00431 to L.T.-D.), M.P. has a contract from RIS-RETIC (RD12/0017/0036), E.R.-M. was supported by Consejería de Salud y Bienestar Social of Junta de Andalucía through the Nicolás Monardes program (C-0032/17), and A.L. is a Joan Rodes consultant of the Infectious Disease Department of the Hospital Clinic of Barcelona. This work was also supported by Red Temática de Investigación Cooperativa en SIDA (RD12/0017/0002, RD12/0017/0005, RD12/0017/0029, RD12/0017/0036, RD12/0017/0037, RD 16CIII/0002/0005, and RD16/0025/0020), which is included in the Acción Estratégica en Salud, Plan Nacional de Investigación Científica, Desarrollo e Innovación Tecnológica 2008-2011, Instituto de Salud Carlos III, Fondos FEDER. F.V. received a grant from the Programa de Intensificación de Investigadores (INT15/226), Instituto de Salud Carlos III, Madrid, Spain. B.D.M. received a grant from The Spanish Ministry of Education (FPU13/02451). C.L.-G. was supported by grants SAF 2010-17226 and SAF 2016-77894-R from MINECO (Spain). F.V. was supported by Gilead Fellowship Program GLD14/293 and GLD15/00298 and Agència de Gestió d'Ajuts Universitaris i de Recerca (AGAUR 2014SGR250). We also particularly acknowledge the Spanish HIV HGM BioBank, which is supported by the Spanish Instituto de Salud Carlos III (RETIC PT13/001/0028) and is integrated in the Spanish AIDS Research Network (RD12/0017/0037).
We declare that no conflicts of interest exist.
E.R.-M. and M.L. designed the study. N.R., J.M.B., A.L., M. Planas, F.G., C.R., O.M.-M., M.D., J.A.I., J.D.R., M.M.-F., F.V., J.A., and M.L. coordinated the patient inclusion and sample management and analyzed and interpreted data from the experiments. M. Pernas, C.C., L.T.-D., C.L.-G., E.R.-M., J.G.-P., and M.C. designed the experiments. M. Pernas, I.O., L.T.-D., B.D.-M., E.R.-G., J.G., J.G.-P., and M.C. produced the experimental data in the laboratory. M. Pernas, L.T.-D., E.R.-G., C.L.-G., and E.R.-M. prepared the manuscript. All authors contributed to reviews of the manuscript.
This study would not have been possible without the collaboration of all of the patients, medical and nursery staff, and data managers who have taken part in the project.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01805-17.
REFERENCES
- 1.Lambotte O, Faroudy B, Madec Y, Nguyen A, Goujard C, Meyer L, Rouzioux C, Venet A, Delfraissy J-F, SEROCO-HEMOCO Study Group. 2005. HIV controllers: a homogeneous group of HIV-1 infected patients with a spontaneous control of viral replication. Clin Infect Dis 41:1053–1056. doi: 10.1086/433188. [DOI] [PubMed] [Google Scholar]
- 2.Shasha D, Walker BD. 2013. Lessons to be learned from natural control of HIV—future directions, therapeutic, and preventive implications. Front Immunol 4:1–8. doi: 10.3389/fimmu.2013.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pereyra F. 2010. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Migueles SA, Osborne CM, Royce C, Compton AA, Joshi RP, Weeks KA, Rood JE, Berkley AM, Sacha JB, Cogliano-Shutta NA, Lloyd M, Roby G, Kwan R, McLaughlin M, Stallings S, Rehm C, O'Shea MA, Mican J, Packard BZ, Komoriya A, Palmer S, Wiegand AP, Maldarelli F, Coffin JM, Mellors JW, Hallahan CW, Follman DA, Connors M. 2008. Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity 29:1009–1021. doi: 10.1016/j.immuni.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Machmach K, Leal M, Gras C, Viciana P, Genebat M, Franco E, Boufassa F, Lambotte O, Herbeuval JP, Ruiz-Mateos E. 2012. Plasmacytoid dendritic cells reduce HIV Production in elite controllers. J Virol 86:4245–4252. doi: 10.1128/JVI.07114-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bello G, Casado C, Sandonis V, Alonso-Nieto M, Vicario JL, García S, Hernando V, Rodríguez C, del Romero J, López-Galińdez C. 2005. A subset of human immunodeficiency virus type 1 long-term non-progressors is characterized by the unique presence of ancestral sequences in the viral population. J Gen Virol 86:355–364. doi: 10.1099/vir.0.80410-0. [DOI] [PubMed] [Google Scholar]
- 7.Leon A, Perez I, Ruiz-Mateos E, Benito JM, Leal M, Lopez-Galindez C, Rallon N, Alcami J, Lopez-Aldeguer J, Viciana P, Rodriguez C, Grau E, Iribarren J, Gatell JM, Garcia F. 2016. Rate and predictors of progression in elite and viremic HIV-1 controllers. AIDS 30:1209–1220. doi: 10.1097/QAD.0000000000001050. [DOI] [PubMed] [Google Scholar]
- 8.Dominguez-Molina B, Leon A, Rodriguez C, Benito JM, Lopez-Galindez C, Garcia F, Del Romero J, Gutierrez F, Viciana P, Alcami J, Leal M, Ruiz-Mateos E, ECRIS integrated in the Spanish AIDS Research Network 2016. Analysis of non-AIDS-defining events in HIV controllers. Clin Infect Dis 62:1304–1309. doi: 10.1093/cid/ciw120. [DOI] [PubMed] [Google Scholar]
- 9.Crowell TA, Gebo KA, Blankson JN, Korthuis PT, Yehia BR, Rutstein RM, Moore RD, Sharp V, Nijhawan AE, Mathews WC, Hanau LH, Corales RB, Beil R, Somboonwit C, Edelstein H, Allen SL, Berry SA. 2015. Hospitalization rates and reasons among HIV elite controllers and persons with medically controlled HIV infection. J Infect Dis 211:1692–1702. doi: 10.1093/infdis/jiu809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bello G, Casado C, García S, Rodríguez C, del Romero J, López-Galíndez C. 2004. Co-existence of recent and ancestral nucleotide sequences in viral quasispecies of human immunodeficiency virus type 1 patients. J Gen Virol 85:399–407. doi: 10.1099/vir.0.19365-0. [DOI] [PubMed] [Google Scholar]
- 11.Bello G, Casado C, García S, Rodríguez C, del Romero J, Carvajal-Rodriguez A, Posada D, López-Galíndez C. 2007. Lack of temporal structure in the short term HIV-1 evolution within asymptomatic naive patients. Virology 362:294–303. doi: 10.1016/j.virol.2006.11.039. [DOI] [PubMed] [Google Scholar]
- 12.Casado C, Colombo S, Rauch A, Martínez R, Günthard HF, Garcia S, Rodríguez C, del Romero J, Telenti A, López-Galíndez C. 2010. Host and viral genetic correlates of clinical definitions of HIV-1 disease progression. PLoS One 5:e11079. doi: 10.1371/journal.pone.0011079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noel N, Boufassa F, Lécuroux C, Saez-Cirion A, Bourgeois C, Dunyach-Remy C, Goujard C, Rouzioux C, Meyer L, Pancino G, Venet A, Lambotte O. 2014. Elevated IP10 levels are associated with immune activation and low CD4+ T-cell counts in HIV controller patients. AIDS 28:467–476. doi: 10.1097/QAD.0000000000000174. [DOI] [PubMed] [Google Scholar]
- 14.Chereau F, Madec Y, Sabin C, Obel N, Ruiz-Mateos E, Chrysos G, Fidler S, Lehmann C, Zangerle R, Wittkop L, Reiss P, Hamouda O, Estrada Perez V, Leal M, Mocroft A, Garcia De Olalla P, Ammassari A, D'Arminio Monforte A, Mussini C, Segura F, Castagna A, Cavassini M, Grabar S, Morlat P, De Wit S, Lambotte O, Meyer L, HIV Controllers Project Working Group for the Collaboration of Observational, HIV Epidemiological Research Europe (COHERE) in EuroCOORD. 2017. Impact of CD4 and CD8 dynamics and viral rebounds on loss of virological control in HIV controllers. PLoS One 12:e0173893. doi: 10.1371/journal.pone.0173893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noel N, Lerolle N, Lécuroux C, Goujard C, Venet A, Saez-Cirion A, Avettand-Fenoël V, Meyer L, Boufassa F, Lambotte O, Agut H, Autran B, Barin F, Costagliola D, Pancino G, Rouzioux C, Samri-Hassimi A, Taulera O, Theodorou I, Tubiana R, Viard JP, Yazdanpanah Y, Boufassa F, Lambotte O, Meyer L. 2015. Immunologic and virologic progression in HIV controllers: the role of viral “blips” and immune activation in the ANRS CO21 CODEX study. PLoS One 10:e0131922. doi: 10.1371/journal.pone.0131922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maldarelli F, Wu X, Su L, Simonetti F, Shao W, Hill S, Spindler J, Ferris A, Mellors J, Kearney M, Coffin J, Hughes S. 2014. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 345:179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wagner TA, Mclaughlin S, Garg K, Cheung CYK, Larsen B, Styrchak S, Huang HC, Edlefsen PT, Mullins JI, Frenkel LM. 2014. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science 345:570–573. doi: 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boritz EA, Darko S, Swaszek L, Wolf G, Wells D, Wu X, Henry AR, Laboune F, Hu J, Ambrozak D, Hughes MS, Hoh R, Casazza JP, Vostal A, Bunis D, Nganou-Makamdop K, Lee JS, Migueles SA, Koup RA, Connors M, Moir S, Schacker T, Maldarelli F, Hughes SH, Deeks SG, Douek DC. 2016. Multiple origins of virus persistence during natural control of HIV infection. Cell 166:1004–1015. doi: 10.1016/j.cell.2016.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pernas M, Casado C, Sandonis V, Arcones C, Rodriguez C, Ruiz-Mateos E, Ramirez de Arellano E, Rallon N, Del Val M, Grau E, Lopez-Vazquez M, Leal M, Del Romero J, Lopez Galindez C. 2013. Prevalence of HIV-1 dual infection in long-term nonprogressor-elite controllers. J Acquir Immune Defic Syndr 64:225–231. doi: 10.1097/QAI.0b013e31829bdc85. [DOI] [PubMed] [Google Scholar]
- 20.Wahren B, Morfeldt-Månsson L, Biberfeld G, Moberg L, Sönnerborg A, Ljungman P, Werner A, Kurth R, Gallo R, Bolognesi D. 1987. Characteristics of the specific cell-mediated immune response in human immunodeficiency virus infection. J Virol 61:2017–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Porichis F, Kaufmann DE. 2011. HIV-specific CD4 T cells and immune control of viral replication. Curr Opin HIV AIDS 6:174–180. doi: 10.1097/COH.0b013e3283454058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, Lederman MM, Benito JM, Goepfert PA, Connors M, Roederer M, Koup RA. 2006. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pereyra F, Addo MM, Kaufmann DE, Liu Y, Miura T, Rathod A, Baker B, Trocha A, Rosenberg R, Mackey E, Ueda P, Lu Z, Cohen D, Wrin T, Petropoulos CJ, Rosenberg ES, Walker BD. 2008. Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. J Infect Dis 197:563–571. doi: 10.1086/526786. [DOI] [PubMed] [Google Scholar]
- 24.Sáez-Cirión A, Sinet M, Shin SY, Urrutia A, Versmisse P, Lacabaratz C, Boufassa F, Avettand-Fènoël V, Rouzioux C, Delfraissy J-F, Barré-Sinoussi F, Lambotte O, Venet A, Pancino G. 2009. Heterogeneity in HIV suppression by CD8 T cells from HIV controllers: association with Gag-specific CD8 T cell responses. J Immunol 182:7828–7837. doi: 10.4049/jimmunol.0803928. [DOI] [PubMed] [Google Scholar]
- 25.Walker-Sperling VE, Pohlmeyer CW, Veenhuis RT, May M, Luna KA, Kirkpatrick AR, Laeyendecker O, Cox AL, Carrington M, Bailey JR, Arduino RC, Blankson JN. 2017. Factors associated with the control of viral replication and virologic breakthrough in a recently infected HIV-1 controller. EBioMedicine 16:141–149. doi: 10.1016/j.ebiom.2017.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferrando-Martinez S, Casazza JP, Leal M, Machmach K, Munoz-Fernandez MA, Viciana P, Koup RA, Ruiz-Mateos E. 2012. Differential Gag-specific polyfunctional T cell maturation patterns in HIV-1 elite controllers. J Virol 86:3667–3674. doi: 10.1128/JVI.07034-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hua S, Lécuroux C, Sáez-Cirión A, Pancino G, Girault I, Versmisse P, Boufassa F, Taulera O, Sinet M, Lambotte O, Venet A. 2014. Potential role for HIV-specific CD38−/HLA-DR+ CD8+ T cells in viral suppression and cytotoxicity in HIV controllers. PLoS One 9:e101920. doi: 10.1371/journal.pone.0101920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koofhethile CK, Ndhlovu ZM, Thobakgale-Tshabalala C, Prado JG, Ismail N, Mncube Z, Mkhize L, van der Stok M, Yende N, Walker BD, Goulder PJR, Ndung'u T. 2016. CD8+ T cell breadth and ex vivo virus inhibition capacity distinguish between viremic controllers with and without protective HLA class I alleles. J Virol 90:6818–6831. doi: 10.1128/JVI.00276-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elahi S, Dinges WL, Lejarcegui N, Laing KJ, Collier CA, Koelle DM, McElrath MJ, Horton H. 2011. Protective HIV-specific CD8+ T cells evade Treg cell suppression. Nat Med 17:989–995. doi: 10.1038/nm.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burwitz BJ, Giraldo-Vela J, Reed J, Newman LP, Bean AT, Nimityongskul FA, Castrovinci PA, Maness NJ, Leon EJ, Rudersdorf R, Sacha JB. 2012. CD8+ and CD4+ cytotoxic T cell escape mutations precede breakthrough SIVmac239 viremia in an elite controller. Retrovirology 9:91. doi: 10.1186/1742-4690-9-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Markham RB, Wang W-C, Weisstein AE, Wang Z, Munoz A, Templeton A, Margolick J, Vlahov D, Quinn T, Farzadegan H, Yu X-F. 1998. Patterns of HIV-1 evolution in individuals with differing rates of CD4 T cell decline. Proc Natl Acad Sci U S A 95:12568–12573. doi: 10.1073/pnas.95.21.12568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ganeshan S, Dickover R, Korber B, Bryson Y, Wolinsky S. 1997. Human immunodeficiency virus type 1 genetic evolution in children with different rates of development of disease. J Virol 71:663–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fraser C, Lythgoe K, Leventhal GE, Shirreff G, Hollingsworth TD, Alizon S, Bonhoeffer S. 2014. Virulence and pathogenesis of HIV-1 infection: an evolutionary perspective. Science 343:1243727. doi: 10.1126/science.1243727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Claiborne DT, Prince JL, Hunter E. 2014. A restriction enzyme based cloning method to assess the in vitro replication capacity of HIV-1 subtype C Gag-MJ4 chimeric viruses. J Vis Exp 90:e51506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dalmau J, Rotger M, Erkizia I, Rauch A, Reche P, Pino M, Esteve A, Palou E, Brander C, Paredes R, Phung P, Clotet B, Telenti A, Martinez-Picado J, Prado JG. 2014. Highly pathogenic adapted HIV-1 strains limit host immunity and dictate rapid disease progression. AIDS 28:1261–1272. doi: 10.1097/QAD.0000000000000293. [DOI] [PubMed] [Google Scholar]
- 36.Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, Kuo YH, Brookmeyer R, Zeiger MA, Barditch-Crovo P, Siliciano RF. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 37.O'Connell K, Brennan TP, Bailey JR, Ray SC, Siliciano RF, Blankson JN. 2010. Control of HIV-1 in elite suppressors despite ongoing replication and evolution in plasma virus. J Virol 84:7018–7028. doi: 10.1128/JVI.00548-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bailey JR, Brennan TP, O'Connell KA, Siliciano RF, Blankson JN. 2009. Evidence of CD8+ T-cell-mediated selective pressure on human immunodeficiency virus type 1 nef in HLA-B*57+ elite suppressors. J Virol 83:88–97. doi: 10.1128/JVI.01958-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bailey JR, Williams TM, Siliciano RF, Blankson JN. 2006. Maintenance of viral suppression in HIV-1-infected HLA-B*57+ elite suppressors despite CTL escape mutations. J Exp Med 203:1357–1369. doi: 10.1084/jem.20052319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bailey JR, Lassen KG, Yang H-C, Quinn TC, Ray SC, Blankson JN, Siliciano RF. 2006. Neutralizing antibodies do not mediate suppression of human immunodeficiency virus type 1 in elite suppressors or selection of plasma virus variants in patients on highly active antiretroviral therapy. J Virol 80:4758–4770. doi: 10.1128/JVI.80.10.4758-4770.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahalanabis M, Jayaraman P, Miura T, Pereyra F, Chester EM, Richardson B, Walker B, Haigwood NL. 2009. Continuous viral escape and selection by autologous neutralizing antibodies in drug-naive human immunodeficiency virus controllers. J Virol 83:662–672. doi: 10.1128/JVI.01328-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mens H, Kearney M, Wiegand A, Shao W, Schønning K, Gerstoft J, Obel N, Maldarelli F, Mellors JW, Benfield T, Coffin JM. 2010. HIV-1 continues to replicate and evolve in patients with natural control of HIV infection. J Virol 84:12971–12981. doi: 10.1128/JVI.00387-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rachinger A, Kootstra NA, Gijsbers EF, van den Kerkhof TLGM, Schuitemaker H, van ‘t Wout AB. 2012. HIV-1 envelope diversity 1 year after seroconversion predicts subsequent disease progression. AIDS 26:1517–1522. doi: 10.1097/QAD.0b013e328354f539. [DOI] [PubMed] [Google Scholar]
- 44.Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. 2006. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 439:344–348. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pfeiffer JK, Kirkegaard K. 2005. Increased fidelity reduces poliovirus fitness and virulence under selective pressure in mice. PLoS Pathog 1:e11. doi: 10.1371/journal.ppat.0010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coffey LL, Beeharry Y, Bordería AV, Blanc H, Vignuzzi M. 2011. Arbovirus high fidelity variant loses fitness in mosquitoes and mice. Proc Natl Acad Sci U S A 108:16038–16043. doi: 10.1073/pnas.1111650108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edwards BH, Bansal A, Sabbaj S, Bakari J, Mulligan MJ, Goepfert PA. 2002. Magnitude of functional CD8+ T-cell responses to the Gag protein of human immunodeficiency virus type 1 correlates inversely with viral load in plasma. J Virol 76:2298–2305. doi: 10.1128/jvi.76.5.2298-2305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, Moodley E, Reddy S, de Pierres C, Mncube Z, Mkhwanazi N, Bishop K, van der Stok M, Nair K, Khan N, Crawford H, Payne R, Leslie A, Prado J, Prendergast A, Frater J, McCarthy N, Brander C, Learn GH, Nickle D, Rousseau C, Coovadia H, Mullins JI, Heckerman D, Walker BD, Goulder P. 2007. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med 13:46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- 49.Noel N, Pena R, David A, Avettand-Fenoel V, Erkizia I, Jimenez E, Lecuroux C, Rouzioux C, Boufassa F, Pancino G, Venet A, Van Lint C, Martinez-Picado J, Lambotte O, Saez-Cirion A, Prado JG. 2016. Long-term spontaneous control of HIV-1 relates to low frequency of infected cells and inefficient viral reactivation. J Virol 90:6148–6158. doi: 10.1128/JVI.00419-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li JZ, Arnold KB, Lo J, Dugast A-S, Plants J, Ribaudo HJ, Cesa K, Heisey A, Kuritzkes DR, Lauffenburger DA, Alter G, Landay A, Grinspoon S, Pereyra F. 2015. Differential levels of soluble inflammatory markers by human immunodeficiency virus controller status and demographics. Open Forum Infect Dis 2:ofu117. doi: 10.1093/ofid/ofu117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krishnan S, Wilson EMP, Sheikh V, Rupert A, Mendoza D, Yang J, Lempicki R, Migueles SA, Sereti I. 2014. Evidence for innate immune system activation in HIV type 1-infected elite controllers. J Infect Dis 209:931–939. doi: 10.1093/infdis/jit581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pereyra F, Palmer S, Miura T, Block BL, Wiegand A, Rothchild AC, Baker B, Rosenberg R, Cutrell E, Seaman MS, Coffin JM, Walker BD. 2009. Persistent low level viremia in HIV-1 elite controllers and relationship to immunologic parameters. J Infect Dis 200:984–990. doi: 10.1086/605446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cocchi F, DeVico AL, Garzino-Demo A, Arya SK, Gallo RC, Lusso P. 1995. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science 270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 54.Zhao J, She S, Xie L, Chen X, Mo C, Huang L, Tang W, Chen X. 2016. The effects of RANTES polymorphisms on susceptibility to HIV-1 infection and disease progression: evidence from an updated meta-analysis. AIDS Res Hum Retroviruses 32:517–528. doi: 10.1089/aid.2015.0312. [DOI] [PubMed] [Google Scholar]
- 55.Vidal F, Peraire J, Domingo P, Broch M, Cairó M, Pedrol E, Montero M, Viladés C, Gutiérrez C, Sambeat MA, Fontanet A, Dalmau D, Deig E, Knobel H, Sirvent JJ, Richart C, Veloso S, Saumoy M, López-Dupla M, Olona M, Cadafalch J, Fuster M, Ochoa A, Soler A, Guelar A, González J. 2006. Polymorphism of RANTES chemokine gene promoter is not associated with long-term nonprogressive HIV-1 infection of more than 16 years. J Acquir Immune Defic Syndr 41:17–22. doi: 10.1097/01.qai.0000188335.86466.ea. [DOI] [PubMed] [Google Scholar]
- 56.Cooke GS, Tosh K, Ramaley PA, Kaleebu P, Zhuang J, Nakiyingi JS, Watera C, Gilks CF, French N, Whitworth JA, Hill AV. 2006. A polymorphism that reduces RANTES expression is associated with protection from death in HIV-seropositive Ugandans with advanced disease. J Infect Dis 194:666–669. doi: 10.1086/505875. [DOI] [PubMed] [Google Scholar]
- 57.Nakayama K, Nakamura H, Koga M, Koibuchi T, Fujii T, Miura T, Iwamoto A, Kawana-Tachikawa A. 2012. Imbalanced production of cytokines by T cells associates with the activation/exhaustion status of memory T cells in chronic HIV type 1 infection. AIDS Res Hum Retroviruses 28:702–714. doi: 10.1089/aid.2011.0073. [DOI] [PubMed] [Google Scholar]
- 58.Gurdasani D, Iles L, Dillon DG, Young EH, Olson AD, Naranbhai V, Fidler S, Gkrania-Klotsas E, Post FA, Kellam P, Porter K, Sandhu MS. 2014. A systematic review of definitions of extreme phenotypes of HIV control and progression. AIDS 28:149–162. doi: 10.1097/QAD.0000000000000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dominguez-Molina B, Tarancon-Diez L, Hua S, Abad-Molina C, Rodriguez-Gallego E, Machmach K, Vidal F, Tural C, Moreno S, Goni JM, Ramirez de Arellano E, Del Val M, Gonzalez-Escribano MF, Del Romero J, Rodriguez C, Capa L, Viciana P, Alcami J, Yu XG, Walker BD, Leal M, Lichterfeld M, Ruiz-Mateos E, ECRIS integrated in the Spanish AIDS Research Network. 2017. HLA-B*57 and IFNL4-related polymorphisms are associated with protection against HIV-1 disease progression in controllers. Clin Infect Dis 64:621–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Canouï E, Lécuroux C, Avettand-Fenoël V, Gousset M, Rouzioux C, Saez-Cirion A, Meyer L, Boufassa F, Lambotte O, Noël N, and the ANRS CO21 CODEX Study Group 2017. A subset of extreme human immunodeficiency virus (HIV) controllers is characterized by a small HIV blood reservoir and a weak T-cell activation level. Open Forum Infect Dis 4:ofx064. doi: 10.1093/ofid/ofx064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.García-Merino I, de Las Cuevas N, Jiménez JL, Gallego J, Gómez C, Prieto C, Serramía MJ, Lorente R, Muñoz-Fernández MA, Spanish HIV BioBank. 2009. The Spanish HIV BioBank: a model of cooperative HIV research. Retrovirology 6:27. doi: 10.1186/1742-4690-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Machmach K, Abad-Molina C, Romero-Sánchez MC, Abad MA, Ferrando-Martínez S, Genebat M, Pulido I, Viciana P, González-Escribano MF, Leal M, Ruiz-Mateos E, HIV Controllers Consortium of the AIDS Spanish Network. 2013. IL28B single-nucleotide polymorphism rs12979860 is associated with spontaneous HIV control in white subjects. J Infect Dis 207:651–655. doi: 10.1093/infdis/jis717. [DOI] [PubMed] [Google Scholar]
- 63.Brussel A, Sonigo P. 2003. Analysis of early human immunodeficiency virus type 1 DNA synthesis by use of a new sensitive assay for quantifying integrated provirus. J Virol 77:10119–10124. doi: 10.1128/JVI.77.18.10119-10124.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dismuke DJ, Aiken C. 2006. Evidence for a functional link between uncoating of the human immunodeficiency virus type 1 core and nuclear import of the viral preintegration complex. J Virol 80:3712–3720. doi: 10.1128/JVI.80.8.3712-3720.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Casado C, Pernas M, Sandonis V, Alvaro-Cifuentes T, Olivares I, Fuentes R, Martínez-Prats L, Grau E, Ruiz L, Delgado R, Rodríguez C, del Romero J, López-Galíndez C. 2013. Identification of a cluster of HIV-1 controllers infected with low replicating viruses. PLoS One 8:e77663. doi: 10.1371/journal.pone.0077663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Larsen M, Sauce D, Arnaud L, Fastenackels S, Appay V, Gorochov G. 2012. Evaluating cellular polyfunctionality with a novel polyfunctionality index. PLoS One 7:e42403. doi: 10.1371/journal.pone.0042403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.