ABSTRACT
Porcine epidemic diarrhea virus (PEDV), an enteropathogenic Alphacoronavirus, has caused enormous economic losses in the pork industry. Nonstructural protein 1 (nsp1) is a characteristic feature of alpha- and betacoronaviruses, which exhibits both functional conservation and mechanistic diversity in inhibiting host gene expression and antiviral responses. However, the detailed structure and molecular mechanisms underlying the Alphacoronavirus nsp1 inhibition of host gene expression remain unclear. Here, we report the first full-length crystal structure of Alphacoronavirus nsp1 from PEDV. The structure displays a six-stranded β-barrel fold in the middle of two α-helices. The core structure of PEDV nsp1 shows high similarity to those of severe acute respiratory syndrome coronavirus (SARS-CoV) nsp1 and transmissible gastroenteritis virus (TGEV) nsp1, despite its low degree of sequence homology. Using ribopuromycylation and Renilla luciferase reporter assays, we showed that PEDV nsp1 can dramatically inhibit general host gene expression. Furthermore, three motifs (amino acids [aa] 67 to 71, 78 to 85, and 103 to 110) of PEDV nsp1 create a stable functional region for inhibiting protein synthesis, differing considerably from Betacoronavirus nsp1. These results elucidate the detailed structural basis through which PEDV nsp1 inhibits host gene expression, providing insight into the development of a new attenuated vaccine with nsp1 modifications.
IMPORTANCE Porcine epidemic diarrhea virus (PEDV) has led to tremendous economic losses in the global swine industry. PEDV nsp1 plays a crucial role in inhibiting host gene expression, but its functional mechanism remains unclear. Here, we report the full-length structure of PEDV nsp1, the first among coronaviruses to be reported. The 1.25-Å resolution crystal structure of PEDV nsp1 shows high similarity to severe acute respiratory syndrome coronavirus (SARS-CoV) nsp113–128 and transmissible gastroenteritis virus (TGEV) nsp11–104, despite a lack of sequence homology. Structural and biochemical characterization demonstrated that PEDV nsp1 possesses a stable functional region for inhibition of host protein synthesis, which is formed by loops at residues 67 to 71, 78 to 85, and 103 to 110. The different functional regions in PEDV nsp1 and SARS-CoV nsp1 may explain their distinct mechanisms. Importantly, our structural data are conducive to understanding the mechanism of PEDV nsp1 inhibition of the expression of host genes and may aid in the development of a new attenuated vaccine.
KEYWORDS: porcine epidemic diarrhea virus, nonstructural protein 1, structure, C terminus, host gene expression inhibition
INTRODUCTION
Coronaviruses (CoVs) belong to the order Nidovirales in the family Coronaviridae, including many important pathogens that affect humans and other animals (1). The Coronaviridae family is classified into the Alpha-, Beta-, Gamma-, and Deltacoronavirus genera (α-CoVs, β-CoVs, γ-CoVs, and δ-CoVs, respectively) (2, 3). CoVs have attracted widespread attention since the outbreaks of severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) (4, 5). Porcine epidemic diarrhea virus (PEDV), a member of the Alphacoronavirus genus, was first observed in Europe (6). Its clinical symptoms include watery diarrhea, severe enteritis, and vomiting. In 2010, massive porcine epidemic diarrhea (PED) caused by a PEDV variant occurred in Asia (7). In April 2013, PED emerged in North America, resulting in enormous economic losses (8).
CoVs carry a large, single-stranded, positive-sense RNA genome of approximately 30 kb in length (9). The CoVs replicase gene consists of two large open reading frames (ORF1a and ORF1b) which are translated into two large polyproteins (pp1a and pp1ab) (10). After these proteins are processed by papain-like and 3C-like proteases, pp1a and pp1ab yield a total of 15 or 16 mature nonstructural proteins (NSPs) (11, 12). Many of the NSPs perform essential functions in viral RNA synthesis (13–17), and nsp1, nsp3, nsp5, nsp14, nsp15, and nsp16 play important roles in the host immune system (18–39). Although the CoV genome is highly conserved within genera, nsp1, which is located at the N terminus of the replicase polyprotein pp1a, is responsible for the majority of variation (40). nsp1 exists only in α-CoVs and β-CoVs, which has prompted speculation that it may be associated with the evolution of CoVs (2, 40). In terms of both sequence homology and size, α-CoV nsp1 share low similarity with β-CoV nsp1 (41, 42). Based on a comparative sequence analysis, CoVs nsp1 can be considered a genus-specific marker (40). Betacoronavirus nsp1 inhibition of host gene expression has been extensively studied. However, study of the mechanism of Alphacoronavirus nsp1 regulation of host translation is still in its infancy. Transmissible gastroenteritis virus (TGEV) nsp1 cannot bind to the 40S subunit to affect the stability of host mRNA and suppresses translation through a putative host factor (43). The human CoV 229E and NL63 nsp1s are capable of inhibiting gene expression, possibly by binding to ribosomal subunit S6, which is part of the 40S subunit (44). PEDV nsp1 promotes the degradation of CBP and NF-κB to inhibit the interferon (IFN) response (45, 46), but the detailed mechanism of PEDV nsp1 inhibition of host protein synthesis is unclear. Here, we present the full-length crystal structure of PEDV nsp1. Detailed analysis of the structure-function relationship for PEDV nsp1 revealed the functional domain responsible for suppressing host gene expression, providing the basis for a new attenuated PEDV vaccine.
RESULTS
Crystal structure of PEDV nsp1.
The crystal structure of full-length PEDV nsp1 (residues Met1 to Gly110) was obtained, and data of sufficient quality were collected. The crystal structure was determined using the single-wavelength anomalous diffraction (SAD) method and refined to a 1.25-Å resolution in the space group P1 21 1, which was sufficient to clearly trace the whole chain (including the C-terminal His6 tag). Each asymmetric unit contained two subunits, and the solvent content value was 36%, which might explain the good crystal diffraction. Details of phasing and refinement are given in Table 1.
TABLE 1.
Data collection and refinement statistics
| Parametera | Value(s) for Se-Met-labeled nsp1b |
|---|---|
| Data collection | |
| Space group | P1 21 1 |
| Cell dimensions | |
| a, b, c (Å) | 34.71, 75.05, 37.36 |
| α, β, γ (°) | 90.00, 89.96, 90.00 |
| Wavelength | 0.97917 |
| Resolution range (Å) | 31.50–1.25 |
| Completeness (%) | 99.2 (99.2) |
| Rmerge (last shell) | 0.05 (0.509) |
| I/σ (last shell) | 29.57 (4.33) |
| Redundancy (last shell) | 3.9 (3.8) |
| Refinement | |
| Resolution (Å) | 31.50–1.25 |
| Rwork/Rfree | 17.2/19.3 |
| No. of: | |
| Reflections | 52,517 |
| Protein atoms | 1,836 |
| Solvent atoms | 348 |
| Ions/ligands | 0 |
| RMSD | |
| Bond length (Å) | 0.006 |
| Bond angle (°) | 0.991 |
| B factor (Å2) | 17.35 |
| Ramachandran plot (%) | |
| Core | 97.40 |
| Allow, disallow | 2.60, 0.00 |
Rmerge = ∑∑|Ih − 〈I〉|/∑∑Ih, where Ih is the intensity measurement of reflection h and 〈I〉 is the average intensity from multiple observations. Rwork = ∑||Fo| − |Fc||/∑|Fo|, where Fo and Fc are the observed and calculated structure factors, respectively. Rfree is equivalent to Rwork, but 5% of the measured reflections have been excluded from the refinement and set aside for cross-validation.
The highest-resolution values are indicated in parentheses.
The secondary structure of PEDV nsp1 consists of two α-helices (α1 and α2) and six β-strands (β1 to β6) (Fig. 1A). The six β-strands form a β-barrel fold flanked by α-helices (Fig. 1B), which is a common skeleton in TGEV nsp11–104 and SARS-CoV nsp113–128 (42, 47). The α1-helix, locates at the rim of the barrel, forms a separate area. The α2-helix is composed of six amino acids, making it longer than other reported CoV nsp1 helices (42, 47). Four antiparallel strands, β2, β3, β4, and β5, compose one side of the barrel. Strands β2 and β5 are loosely connected to strands β1 and β6, which form the other side of the barrel. β3 and β5 support each other to form a stable barrel, which ensures the stability of the whole structure. Several strands are irregular and have breaks in their hydrogen bonding patterns, due to a β-bulge in β3 involving the carbonyl oxygen, which creates a kink in the end of strand β5. A small cavity is located at the top of the barrel between β2 and β5, next to the α2-helix. This cavity is lined by residues Ala18, Phe19, Phe21, Ala27, Cys42, Val62, Met63, Leu90, and Phe91. Glu14, Glu58, Lys70, and Arg107 exhibit a conformation in the A chain that is different from that in the B chain, and the positions of the side chains determine the size of the cavity opening toward the solvent. The cavity is also conserved in TGEV nsp1 (42).
FIG 1.
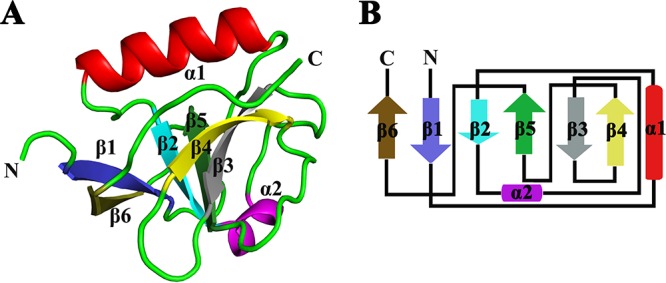
Crystal structure of PEDV nsp1. (A) Crystal structure of monomeric PEDV nsp1. The structure of nsp1 is shown as a cartoon in colors ranging from blue in the N-terminal region to brown in the C-terminal region. (B) Topology diagram. The colors correspond to those in panel A. Strands β1, β2, β3, β4, β5, and β6 make up the barrel.
PEDV nsp1 shares structural similarity with TGEV nsp11–104 and SARS-CoV nsp113–128, despite low sequence homology.
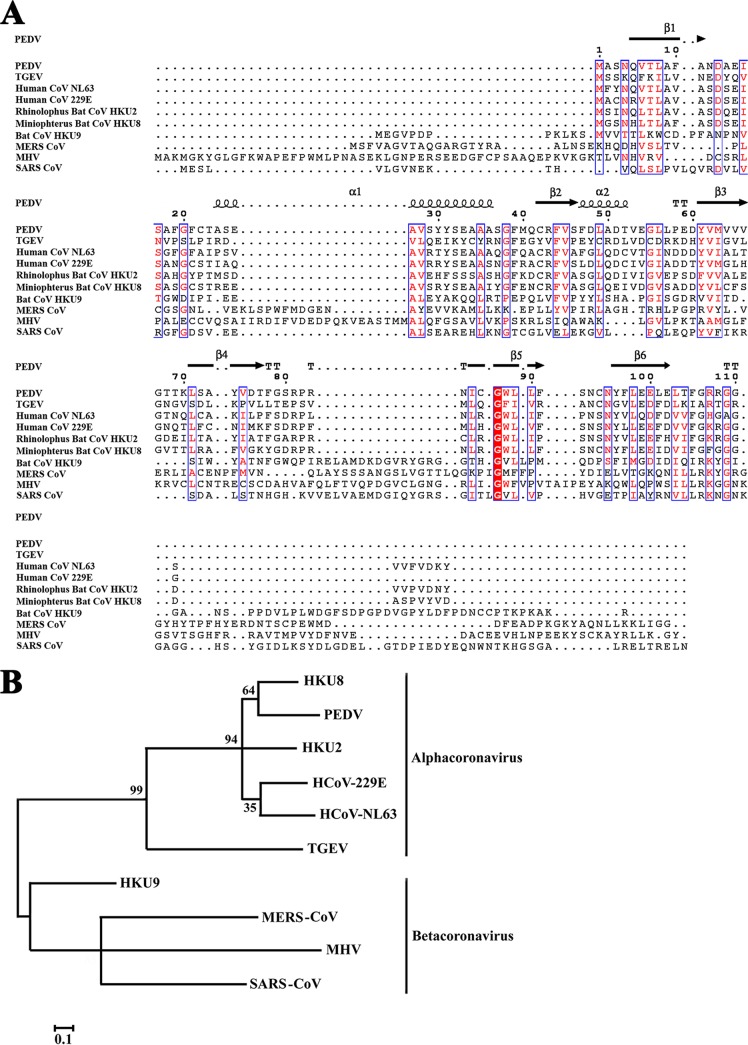
To explore the relationship between α-CoV and β-CoV nsp1 sequences, we selected representative sequences from both groups and aligned them using ClustalW2 software (Fig. 2A). Alphacoronaviruses encode nsp1 proteins of ∼9 kDa, which are substantially smaller than the 20-kDa nsp1 proteins of betacoronaviruses. However, the CoV nsp1 sequences exhibit some relatively conserved residues, including Asn4, Val6, Thr7, Leu8, Asp13, Ile16, Ser17, Gly20, Ala27, Val28, Ala35, Gly38, Phe44, Val45, Tyr61, Val62, Met63, Leu71, Val75, Ile85, Gly87, Trp88, Leu89, Leu90, Asn95, Leu98, Glu100, Leu103, Thr104, Arg107, and Gly109. We found coronavirus nsp1 structures in the Protein Data Bank, including those of TGEV nsp1 (residues 1 to 104; Protein Data Bank [PDB] accession number 3ZBD) and SARS-CoV nsp1 (residues 13 to 128; PDB accession number 2HSX). The amino acid sequence of PEDV nsp1 shared only 23% and 12% sequence identity with those of TGEV nsp1 and SARS-CoV nsp1, respectively, as demonstrated by their distance in the evolutionary tree (Fig. 2B). Thus, the sequence of PEDV nsp1 was found to share low homology with those of TGEV nsp1 and SARS-CoV nsp1 in this study.
FIG 2.
Sequence relationships among different CoV nsp1 proteins. (A) The following sequences from GenBank were used to create the sequence alignment: PEDV, accession number AJP67455.1; TGEV, ABD58989.1; human CoV NL63, AFV53147.1; human CoV 229E, CAA49377.1; rhinolophus bat CoV HKU2, YP_001552235.1; rhinolophus bat CoV HKU8, YP_001718611.1; bat CoV HKU9, YP_001039970.1; MERS CoV, YP_009047213.1; MHV, AFD97615.1; and SARS CoV, NP_828860.2. Residue numbers, referring to PEDV nsp1, are indicated above the sequences. Residues conserved in all nsp1s are shown in white on a red background. Residues conserved in most sequences are shown in red and boxed with a white background. The sequences were aligned with ClustalW2, and the figure was prepared with ESPript3.0. (B) Phylogenetic relationships were analyzed using the maximum-likelihood algorithm in the MEGA package.
Despite the lack of sequence homology, we sought to determine the level of similarity among the secondary structures of these nsp1 proteins. The X-ray crystal structure of TGEV nsp1 (PDB accession number 3ZBD) and the nuclear magnetic resonance (NMR) structure of the N-terminal domain of SARS-CoV nsp1 (PDB accession number 2HSX) were compared with the structure of PEDV nsp1. The overall structure shared the same fold with a characteristic six-stranded β-barrel, with a long α-helix on one side of the barrel (the root mean square deviations [RMSDs] with TGEV nsp11–104 and SARS-CoV nsp113–128 are1.8 Å and 2.4 Å, respectively) (42, 47). However, three-dimensional alignment also revealed several differences in the structures, including in the lengths and rotations of the loops.
The most obvious difference between the structures of PEDV nsp1 and TGEV nsp11–104 is the number of β-sheets: there are two short β-sheets unique to TGEV nsp1 that are not found in PEDV nsp1. The α2-helix is significantly longer in the PEDV nsp1 structure, consisting of six amino acids, while it is divided into the 310-helix and β4-sheet in TGEV nsp11–104. Various regions of the random coil are dissimilar, with certain loops, such as those formed by amino acids (aa) 67 to 71 and 78 to 85, being oriented in opposite directions. The C terminus of PEDV nsp1 is clearly stabilized by β4 and does not hinder crystal formation, whereas for TGEV nsp1, an appropriate crystal can be formed only after removing the C terminus (Fig. 3A and B). The structures of SARS-CoV nsp113–128 and PEDV nsp1 also exhibit many differences. The lengths and angles of the α1-helix and the α2-helix are different. There are also substantial differences in the loops, especially those located at residues 67 to 71 and 78 to 85. Additionally, the loop between β3 and β4 is significantly shorter in the PEDV nsp1 structure (Fig. 3A and C). Considering these data together (Fig. 3D and E), PEDV nsp1 shows greater similarity to TGEV nsp1 than to SARS-CoV nsp1.
FIG 3.
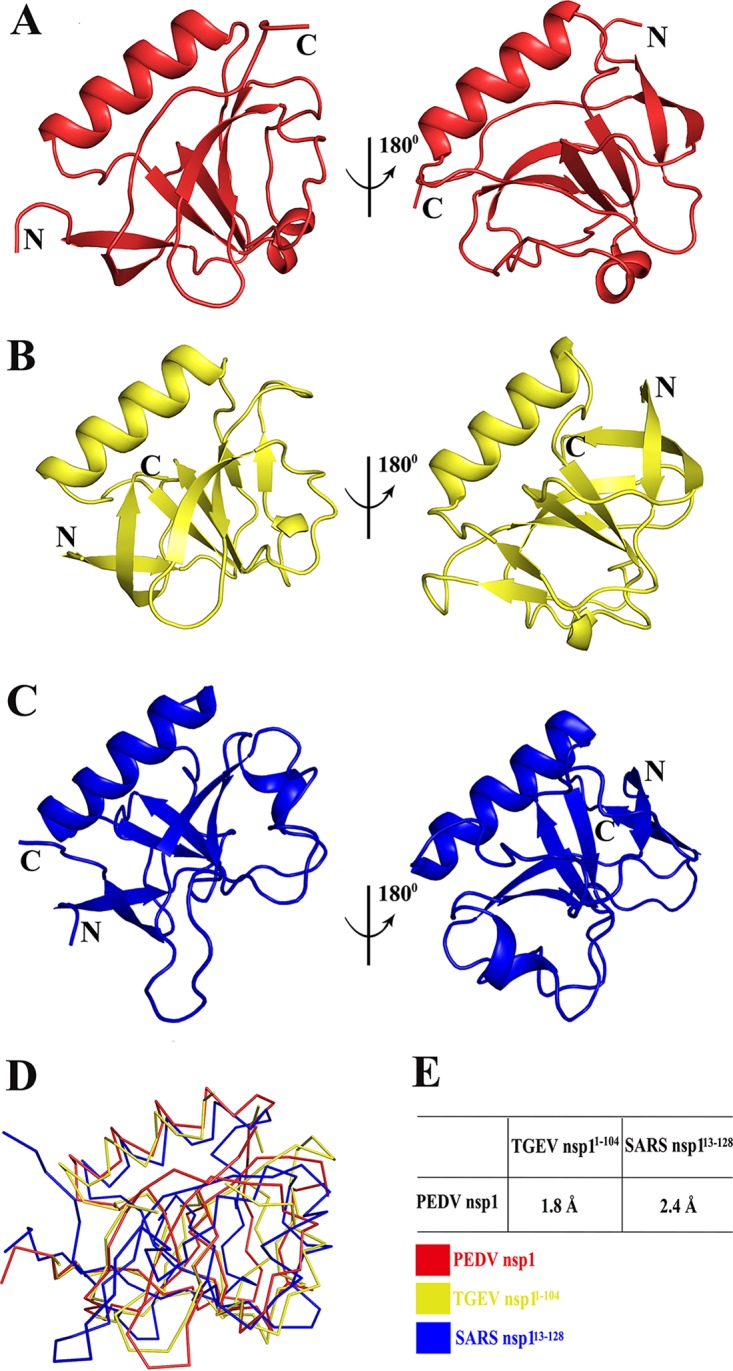
Structural comparisons of PEDV nsp1, SARS-CoV nsp113–128 (PDB accession number 2GDT) (blue), and TGEV nsp11–104 (PDB accession number 3ZBD) (red). (A) Overlay of PEDV nsp1 at 180 degrees. (B) Overlay of TGEV nsp11–104 at 180 degrees. (C) Overlay of SARS-CoV nsp113–128 at 180 degrees. PEDV nsp1 is shown in yellow, TGEV nsp11–104 is shown in red, and SARS-CoV nsp113–128 is shown in blue. (D) Ribbon diagrams for the three structures. (E) Sequence identities and RMSDs between PEDV nsp1 and TGEV nsp11–104 or between PEDV nsp1 and SARS-CoV nsp113–128. RMSDs were calculated using COOT.
Transient nsp1 expression affects cellular gene expression.
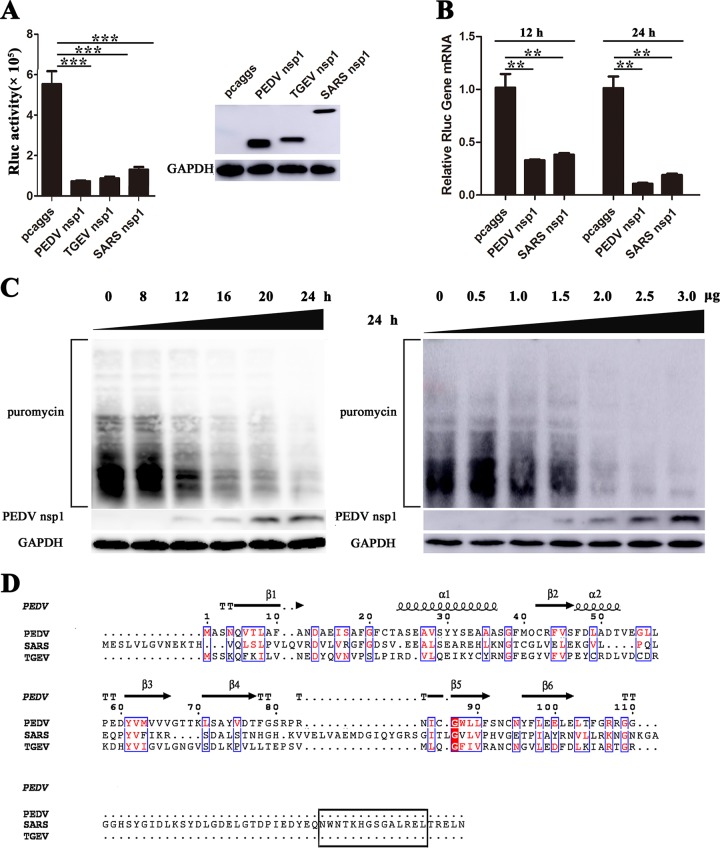
To determine whether CoV nsp1 proteins suppress host gene expression, we examined Renilla luciferase (Rluc) using luciferase assays. Porcine kidney 15 (PK-15) cells were cotransfected with the plasmids expressing the nsp1 protein and with pRL-TK, expressing thymidine kinase (TK) promoter-driven Rluc. We assessed the ability of the nsp1proteins from PEDV, TGEV, and SARS-CoV to interfere with reporter gene expression. Transient gene expression analyses showed that the PEDV, TGEV, and SARS-CoV nsp1s significantly reduced luciferase reporter gene expression, and subsequent SDS-PAGE analysis of equal amounts of intracellular proteins confirmed these findings (Fig. 4A). To examine the effects of PEDV nsp1 on host mRNA amounts, we examined Rluc mRNA in HEK-293T cells using real-time quantitative PCR and found that PEDV nsp1 could possibly degrade host mRNAs (Fig. 4B), which was consistent with data for SARS-CoV nsp1 (48). Our data validated the nsp1 antagonism of general host protein expression, rather than of the expression of only a particular protein. To further examine whether PEDV nsp1 inhibited overall gene expression in human embryonic kidney cells (HEK-293T), we examined host proteins in ribopuromycylation assays (Fig. 4C). These data showed that the inhibitory effects of nsp1 on host proteins synthesis are concentration dependent.
FIG 4.
CoVs nsp1 inhibit host protein synthesis. (A) PK-15 cells were cotransfected with pRL-TK carrying the Rluc reporter gene downstream of the TK promoter and one of the plasmids pcaggs, pcaggs-PEDV-nsp1-HA (PEDV nsp1), pcaggs-TGEV-nsp1-HA (TGEV nsp1), or pcaggs-SARS-CoV-nsp1-HA (SARS nsp1), which express PEDV nsp1, TGEV nsp1 and SARS-CoV nsp1, respectively. All the expressed proteins had N-terminal HA tags. At 24 h posttransfection, cell lysates were prepared and subjected to Rluc assays. Error bars show the standard deviations (SDs) of the results from three independent experiments. Cell extracts were also subjected to Western blot analysis using an anti-HA antibody (top) or an anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody (bottom). *, P < 0.05 (considered statistically significant); **, P < 0.01 (considered highly significant); ***, P < 0.001 (considered extremely significant). (B) HEK-293T cells were transfected with the wild-type plasmid. At 12 or 24 h posttransfection, the cells were lysed and subjected to real-time quantitative PCR analysis. Asterisks indicate statistical significance calculated by the Student t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (C) HEK-293T cells were transfected with the pcaggs-PEDV-nsp1-HA (PEDV nsp1) plasmid. Left, the cells were pulsed with 3 μM puromycin for 1 h at 0, 8, 12, 16, 20, and 24 h posttransfection and then subjected to Western blot analysis. Right, different doses of the wild-type plasmid (0 to 3.0 μg) were transfected into HEK-293T cells. After 24 h posttransfection, the cells were pulsed with 3 μM puromycin for 1 h at 37°C and then subjected to Western blot analysis. (D) The amino acid sequences of PEDV nsp1, SARS-CoV nsp1, and TGEV nsp1 were compared, and the functional region (aa 160 to 173) of SARS-CoV nsp1 is indicated with a black box.
Based on comparison of the sequences of α-CoVs nsp1 (PEDV nsp1 and TGEV nsp1) and β-CoV nsp1 (SARS-CoV nsp1), we identify a functional domain (aa 160 to 173) in SARS-CoV nsp1 that is not present in α-CoV nsp1 proteins (Fig. 4D) (49), indicating that the functional region of α-CoV nsp1 proteins is different from that of SARS-CoV nsp1.
Region of PEDV nsp1 critical for inhibiting protein synthesis.
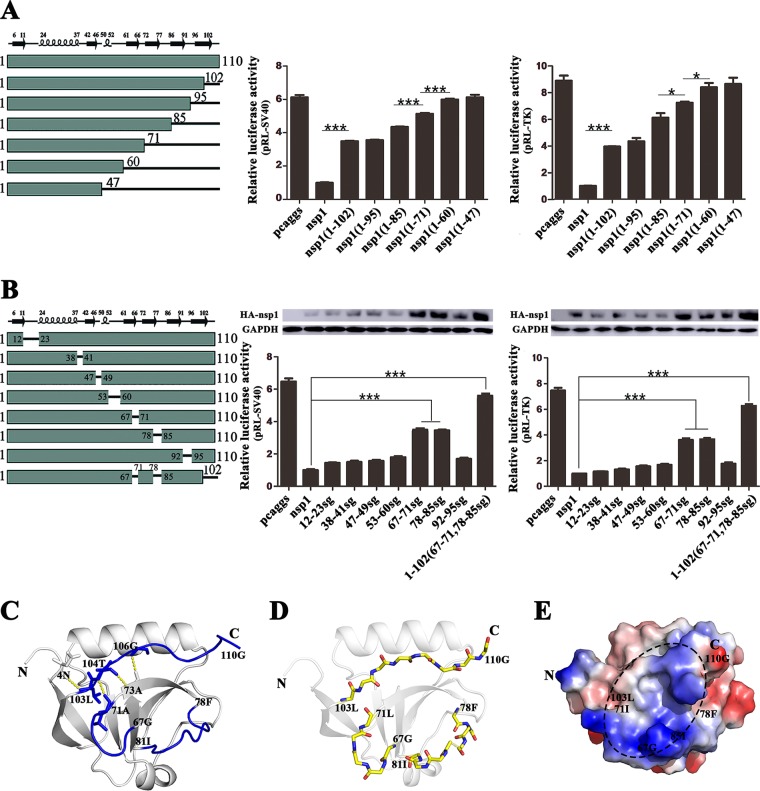
Although the highly conserved residues of PEDV and TGEV nsp1are crucial for NF-κB suppression (45), the key areas of PEDV nsp1 inhibiting the host protein synthesis are unclear. To identify which region in PEDV nsp1 was essential for its function, we designed a series of truncated and mutated nsp1 plasmids based on the crystal structure of PEDV nsp1. First, the C-terminally truncated plasmids nsp1(1–102), nsp1(1–95), nsp1(1–85), nsp1(1–71), nsp1(1–60), and nsp1(1–47) were tested, and the results showed that residues 61 to 71, 72 to 85 and 103 to 110 might be involved in inhibiting luciferase reporter gene expression under the control of the simian virus 40 (SV40) or TK promoter (Fig. 5A). To confirm the above results, we constructed the following plasmids, in which the random coils were replaced with a flexible serine-glycine (sg) linker: nsp1(12–23sg), nsp1(38–41sg), nsp1(47–49sg), nsp1(53–60sg), nsp1(67–71sg), nsp1(78–85sg), and nsp1(92–95sg). The results confirmed that nsp1 required multiple regions to function properly (Fig. 5B). The inhibitory activity of nsp1(67–71sg) and nsp1(78–85sg) was lower than that of the other truncated mutants. Furthermore, to verify that these three fragments participated in forming functional regions, we designed a plasmid with mutations in all three fragments. The results showed that the loops at aa 67 to 71, 78 to 85, and 103 to 110 formed a ring on the surface of the structure, generating a stable functional region (Fig. 5C and D). The charge of this region is positive, and it creates a small groove that may help inhibit host gene expression (Fig. 5E). The region from aa 37 to 75 in PEDV nsp1 is crucial for NF-κB suppression (45), similar to our results.
FIG 5.
Host protein synthesis is inhibited by PEDV nsp1 truncation mutants and “sg” replacement mutants. (A) Inhibition of host protein synthesis by PEDV nsp1 truncation mutants. The truncations (at the C terminus) of PEDV nsp1 are shown below the bars. PK-15 cells were cotransfected with pRL-SV40 or pRL-TK and one of the plasmids pcaggs (control), pcaggs-PEDV-nsp1, pcaggs-PEDV-nsp1(1–102), pcaggs-PEDV-nsp1(1–95), pcaggs-PEDV-nsp1(1–85), pcaggs-PEDV-nsp1(1–71), pcaggs-PEDV-nsp1(1–60), or pcaggs-PEDV-nsp1(1–47), which express nsp1, nsp1(1–102), nsp1(1–95), nsp1(1–85), nsp1(1–71), nsp1(1–60), and nsp1(1–47), respectively. At 24 h posttransfection, cell lysates were prepared and subjected to Rluc assays. Error bars show the SDs of the results from three independent experiments. Asterisks indicate statistical significance calculated by the Student t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (B) Host protein synthesis is inhibited by “sg” replacement mutants of PEDV nsp1. Each solid line between two rectangles is labeled with numbers at both termini, delineating the region replaced with the “sg” residues. The recombinant plasmids are as follows: 12–23sg, 38–41sg, 47–49sg, 53–60sg, 67–71sg, 78–85sg, 92–95sg, and 1–102(67–71, 78–85sg). PK-15 cells were transfected with the control (pcaggs) or nsp1 plasmid. At 24 h posttransfection, the cell were lysed and subjected to Rluc assays and Western blot analysis. Error bars show the SDs of the results from three independent experiments. Asterisks indicate statistical significance calculated by the Student t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (C) The loops corresponding to residues 67 to 71, 78 to 85, and 103 to 110 are indicated in blue, and the other areas are shown in gray. A detailed view of the interaction, showing residues 103 to 110 and the binding region, is indicated in yellow. (D) The main chain for the loops corresponding to residues 67 to 71, 78 to 85, and 103 to 110 is indicated. Nitrogen is shown in blue and oxygen in red. (E) The black oval area indicates the loops at residues 67 to 71, 78 to 85, and 103 to 110, which together form a small groove.
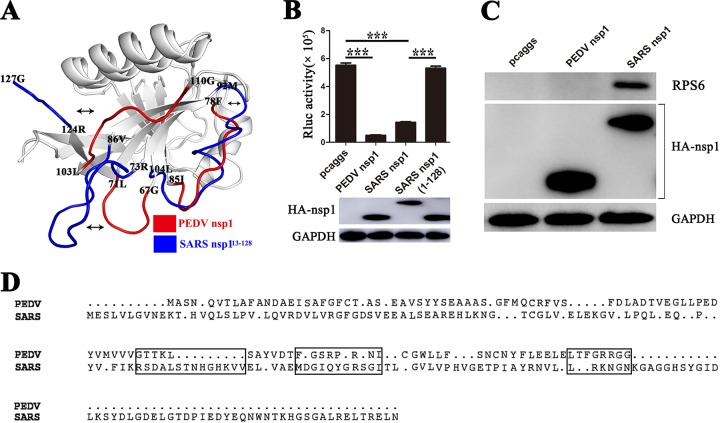
The analyses demonstrated that the region from aa 1 to 128 in SARS-CoV nsp1 played no role in inhibiting protein synthesis (Fig. 6B). The functional area of SARS-CoV nsp1 responsible for inhibiting protein synthesis is located at the C terminus (49). However, the functional area of PEDV nsp1 exhibited its own unique structure, comprising three loops. Our structural analysis indicated that the three loops (aa 67 to 71, 78 to 85, and 103 to 110) in PEDV nsp1 differ from those in SARS-CoV nsp113–128 in terms of both length and orientation (Fig. 6A and D). Moreover, immune coprecipitation confirmed that the SARS-CoV nsp1 could interact with the 40S subunit (48) but that PEDV nsp1 could not (Fig. 6C). The different functional areas indicate that the mechanisms whereby PEDV and SARS-CoV nsp1 inhibit host gene expression are diverse.
FIG 6.
Comparison of the functional domains of PEDV nsp1 and SARS-CoV nsp1. (A) The structure of PEDV nsp1 (subunit A) was superimposed on the structure of SARS-CoV nsp113–128 (PDB accession numbers 2HSX and 2GDT, respectively). The three loops of PEDV nsp1 are shown in red, and the corresponding regions in SARS-CoV nsp113–128 are shown in blue. (B) PK-15 cells were independently cotransfected with pRL-SV40 and pcaggs, pcaggs-PEDV-nsp1 (PEDV nsp1), pcaggs-SARS-CoV-nsp1 (SARS nsp1), or pcaggs-SARS-CoV-nsp1(1–128) [SARS nsp1(1–128)]. Total intracellular proteins were extracted at 24 h posttransfection, and Western blot analysis was performed using anti-HA and anti-GAPDH antibodies. At 24 h after transfection, the cells were lysed in Renilla luciferase lysis buffer, and aliquots of the lysates were used to measure Rluc activities. Asterisks indicate statistical significance calculated by the Student t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (C) Binding of nsp1 to the 40S ribosomal subunits. The control (pcaggs), PEDV nsp1, or SARS-CoV nsp1 (SARS nsp1) plasmid was transfected into 293T cells. The immunoprecipitated proteins (ribosomal protein S6 [RPS6] and CoVs nsp1 [HA-nsp1]) were examined by Western blotting using anti-S6 antibody and anti-HA antibody. The expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was detected with an anti-GAPDH MAb to confirm equal protein loading. (D) Sequence relationships of PEDV nsp1 and SARS-CoV nsp1. The corresponding loops in PEDV nsp1 and SARS-CoV nsp1 are indicated with a black box.
DISCUSSION
After invasion of host cells, some virus-encoded proteins are used to regulate the host's environment, thus helping to increase virus replication (50–53).Previous studies have revealed that the nsp1 proteins of coronaviruses can regulate the host protein synthesis and help the virus escape the host's immune interference (25, 27, 43, 45, 54). The highly variable sequence homology of the coronavirus nsp1 proteins may lead to the diverse mechanisms for inhibiting the host gene expression.
In our study, we determined the full-length structure of PEDV nsp1. The structure is characterized by an irregular six-stranded β-barrel fold in the middle of two α-helices. Despite the low sequence homology of the viruses, the core structures of PEDV nsp1, TGEV nsp11–104, and SARS-CoV nsp113–128 were highly conserved (42, 47). Furthermore, PEDV nsp1 showed higher structural similarity with TGEV nsp1 than with SARS-CoV nsp1, implying that the divergent evolution of CoV nsp1s may be explained by their structural differences (55). At the same time, their detailed structures of have some subtle differences, such as the loop length and rotation, possibly explaining why the mechanisms of CoV nsp1 inhibition of host gene expression are diverse. The structure of PEDV nsp1 provides the first clear visualization of the CoV nsp1 C terminus, which appeared to consist of a long random coil. The C terminus (aa 103 to 110) of PEDV nsp1 interacts with β4, forming a small groove, whereas the C termini of SARS-CoV nsp1 and TGEV nsp1 are unstable in the structure (42, 47). The observed structural differences suggest that the C terminus of PEDV nsp1 has a unique feature which can ensure the structure's stability. The region from aa 160 to 173 of SARS-CoV nsp1 and the region from aa 277 to 309 of murine hepatitis virus (MHV) nsp1 are necessary for inhibiting protein synthesis (49, 56), whereas the α-CoV nsp1 does not have the corresponding region. Our results indicate that two loops (aa 67 to 71 and 78 to 85) in PEDV nsp1, supported by the protein's C terminus (aa 103 to 110), form a stable functional region; these are in significantly different conformations from their counterparts in SARS-CoV nsp1, which may explain why these loops of SARS-CoV nsp1 have no function in the inhibition of host protein synthesis. The structural difference implies that the mechanisms of PEDV and SARS-CoV nsp1 inhibition of general host gene expression might be different.
At present, the various mechanisms of β-CoV nsp1 inhibition of host protein synthesis have been deeply studied. SARS-CoV nsp1 binds to the 40S ribosomal subunit to inactivate translation and promote the degradation of cellular mRNA 5′ caps, which suppresses host innate immune functions (48). MERS-CoV nsp1 inhibits host gene expression by selectively targeting transcribed mRNA instead of binding to the 40S ribosomal subunit (25). Murine hepatitis virus (MHV) nsp1 enhances the virulence of the virus by efficiently interfering with the type I IFN system (56). However, little is known about the mechanism of α-CoV nsp1 inhibition of host gene expression. This study showed that PEDV nsp1 possibly inhibits host gene expression by affecting the host RNA degradation machinery, which is consistent with the mechanisms used by SARS-CoV nsp1 and MERS-CoV nsp1 (25, 48). Furthermore, PEDV nsp1 could not interact with the S6 ribosomal protein, which indicated that PEDV nsp1 may bind with a certain host factor to cause the host gene suppression. The detailed mechanism of PEDV nsp1 inhibition of host protein synthesis requires further investigation.
In a previous study, a recombinant MHV lacking the region encoding aa 277 to 309 of the nsp1 gene was severely attenuated in infected mice, and partial nsp1 gene deletions will therefore be useful for developing attenuated vaccines (56). Similarly, the region from aa 160 to 173 of SARS-CoV nsp1 is critical for suppressing host innate immunity, providing insights into the development of attenuated vaccines (49, 57). We identified the region responsible for suppressing host gene expression based on the structure of PEDV nsp1, which is expected to contribute to the design of a new type of attenuated coronavirus vaccine based on nsp1 modifications.
MATERIALS AND METHODS
Cell lines.
HEK-293T and PK-15 cells were maintained at 37°C in a 5% CO2 incubator in Dulbecco's modified Eagle's medium (Gibco, Waltham, MA, USA) supplemented with 10% fetal bovine serum.
Plasmid construction.
For crystallization, the full-length PEDV nsp1 gene sequence (GenBank accession no. KC140102.1) was cloned via PCR amplification into pET-42b (+), with a C-terminal His6 tag, using the NdeI and XhoI restriction sites. In addition, to achieve high expression in eukaryotic cells, wild-type nsp1, flanked with an N-terminal hemagglutinin (HA) tag, was cloned into the pCAGGS vector using EcoRI and XhoI restriction sites. To confirm the essential functional regions of SARS-CoV nsp1, a fragment (encoding aa 1 to 128) was cloned into the pCAGGS vector according to the same method. Additionally, to determine the functional region of PEDV nsp1, point mutants and truncations were engineered according to the tertiary structure of PEDV nsp1. The C-terminally truncated plasmids included nsp1(1–102), nsp1(1–95), nsp1(1–85), nsp1(1–71), nsp1(1–60), and nsp1(1–47). Additionally, to avoid unexpected collapses of the structure that could potentially inactivate nsp1, plasmids in which a random coil was replaced with a flexible Ser-Gly-Ser-Gly linker were engineered, as follows: nsp1(12–23sg), nsp1(38–41sg), nsp1(47–49sg), nsp1(53–60sg), nsp1(67–71sg), nsp1(78–85sg), and nsp1(92–95sg). All constructs were validated through DNA sequencing.
Protein expression and purification.
To study the expression of PEDV nsp1 in prokaryotic cells, the recombinant plasmid was transformed into Escherichia coli Trans BL21(DE3) cells. The transformed cells were cultured at 37°C in lysogeny broth (LB) medium containing 50 μg/ml kanamycin until the optical density at 600 nm (OD600) reached 0.6 to 0.8. Protein expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and cell growth continued for an additional 5 h at 37°C. To solve the crystal phase problem, selenomethionine (Se-Met)-labeled PEDV nsp1 was expressed as previously described (58).
For protein purification, the E. coli cells were harvested, resuspended in phosphate-buffered saline (PBS), and lysed via passage through an AH-1500 homogenizer. After centrifugation at 8,500 rpm for 40 min, the supernatant was filtered and loaded onto a His Trap HP column (GE Healthcare). The target protein was eluted using a change gradient between the binding buffer (20 mM Tris [pH 7.4], 500 mM NaCl) and elution buffer A (20 mM Tris [pH 7.4], 500 mM NaCl, and 500 mM imidazole), followed by 30 ml of 100% elution buffer A. The protein was further purified using a Superdex 75 (GE Healthcare) column with elution buffer B (20 mM Tris [pH 7.4], 200 mM NaCl). All purification procedures were performed at 4°C to avoid degradation. For crystallization, the purified protein with state uniformity was concentrated to approximately 8.5 mg/ml, flash-frozen with liquid nitrogen, and stored at −80°C.
Crystallization, data collection, and structure determination.
Se-Met-labeled nsp1 was crystallized via the sitting-drop vapor diffusion method at 20°C. According to the initial conditions, which we optimized for crystallization, the best crystals of Se-Met-labeled nsp1 were obtained through vapor diffusion in sitting drops consisting of 1 μl of reservoir solution (0.1 M lithium sulfate monohydrate, 0.1 M sodium citrate tribasic dihydrate [pH 5.5], 20% [wt/vol] polyethylene glycol 1000) and 1 μl of protein solution (8.5 mg/ml in 20 mM Tris-HCl and 200 mM NaCl, pH 7.4). The crystals grew overnight and were then flash-cooled in liquid nitrogen in a cryoprotectant solution (0.1 M lithium sulfate monohydrate, 0.1 M sodium citrate tribasic dihydrate [pH 5.5], 20% [wt/vol] polyethylene glycol 1000, and 30% ethylene glycol). Native data collection was performed on a BL17U1 beamline (wavelength = 0.97917 Å, temperature = 100 K) at the Shanghai Synchrotron Radiation Facility (SSRF) using an MAR 225 CCD detector (MAR Research). All of the diffraction images were integrated, merged, and scaled using HKL-3000 software (59). The structure of nsp1 was solved using the single-wavelength anomalous-dispersion method from an Se-Met derivative (60). All three potential selenium atoms in the primary sequence of the nsp1 monomer were located, and the initial phases were calculated using the program AutoSol from the PHENIX software suite (61, 62). Manual model rebuilding was performed using COOT (63), and subsequently the structure was refined using the PHENIX software suite until the data were eligible. Structural figures were drawn using PyMOL (64). The root mean square deviation (RMSD) was analyzed using PDBeFold (http://pdbe.org/fold/). The amino acid sequences of CoV nsp1 proteins were aligned using the software program ClustalW2 and visualized with the ESPript 3 server (http://espript.ibcp.fr). Evolutionary relationships were described using the software program MEGA (65).
Reporter assay and Western blot analysis.
PK-15 cells were cotransfected at ∼80% confluence with a pRL-TK plasmid encoding the Rluc reporter gene downstream of the TK promoter, along with corresponding protein plasmids. Opti-MEM I was used to dilute Lipofectamine 2000, and the nucleic acids were mixed gently; the DNA-Lipofectamine 2000 complex was fully formed after 20 min at room temperature. At 24 h posttransfection, cell lysates were prepared and subjected to Rluc reporter activity assays (Promega).
In brief, to analyze the expression levels of nsp1 proteins, PK-15 cells were transfected with various plasmids using the same method. The cells were harvested by adding radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime), and the extracts were prepared in SDS-PAGE sample buffer. Protein expression analysis was carried out via Western blotting. The proteins were visualized using an anti-HA antibody (Ab) (ProteinTech). The expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was detected with an anti-GAPDH monoclonal Ab (MAb) (ProteinTech) to confirm equal protein loading.
Quantitative analysis of Rluc mRNA.
The quantitative analysis was performed as previously described (48). In brief, 100 ng of RNA was transferred to separate tubes for amplifying the target Rluc mRNA and the endogenous control (18S rRNA) to use with a TaqMan one-step real-time PCR master mix reagent kit. Following the manufacturer's instructions, PCR was performed on an ABI Prism 7000 real-time thermocycler (Applied Biosystems, Foster City, CA). The amount of target mRNA was normalized to an endogenous 18S rRNA.
Ribopuromycylation assay.
The ribopuromycylation assay was performed as previously described (66). Briefly, cultures of HEK-293T cells were transfected with the wild-type PEDV nsp1 plasmid. At various times posttransfection or with different doses of the wild-type plasmid, the cells were pulse-labeled with 3 μM puromycin and then incubated for an additional hour at 37°C with 5% CO2. After three washes with PBS, the cells were lysed and subjected to Western blot analysis. The analysis of total protein expression was performed using an antipuromycin Ab (Millipore).
Statistical analysis.
All data were analyzed using GraphPad Prism software (GraphPad Software Inc., CA). Significant differences were determined using Student's t test. P values of <0.05 were considered statistically significant, P values of <0.01 were considered highly significant, and P values of <0.001 were considered extremely significant.
Accession number(s).
The coordinates and structural characteristics of PEDV nsp1 were submitted to the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB) under accession number 5XBC.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grant no. 31722056 and 31372440), the National Key R&D Plan of China (program no. 2016YFD0500103), and the Huazhong Agricultural University Scientific & Technological Self-Innovation Foundation (program no. 2662015JQ003 and 2662017PY028).
We thank Yanan Li for useful discussions and the staff at the SSRF BL17U1 beamline for assistance with X-ray data collection.
REFERENCES
- 1.Weiss SR, Navas-Martin S. 2005. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol Mol Biol Rev 69:635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woo PC, Huang Y, Lau SK, Yuen KY. 2010. Coronavirus genomics and bioinformatics analysis. Viruses 2:1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belouzard S, Millet JK, Licitra BN, Whittaker GR. 2012. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan JF, Lau SK, Woo PC. 2013. The emerging novel Middle East respiratory syndrome coronavirus: the “knowns” and “unknowns”. J Formosan Med Assoc 112:372–381. doi: 10.1016/j.jfma.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stadler K, Masignani V, Eickmann M, Becker S, Abrignani S, Klenk HD, Rappuoli R. 2003. SARS—beginning to understand a new virus. Nat Rev Microbiol 1:209–218. doi: 10.1038/nrmicro775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pasick J, Berhane Y, Ojkic D, Maxie G, Embury-Hyatt C, Swekla K, Handel K, Fairles J, Alexandersen S. 2014. Investigation into the role of potentially contaminated feed as a source of the first-detected outbreaks of porcine epidemic diarrhea in Canada. Transbound Emerg Dis 61:397–410. doi: 10.1111/tbed.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun RQ, Cai RJ, Chen YQ, Liang PS, Chen DK, Song CX. 2012. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg Infect Dis 18:161–163. doi: 10.3201/eid1801.111259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L, Byrum B, Zhang Y. 2014. New variant of porcine epidemic diarrhea virus, United States, 2014. Emerg Infect Dis 20:917–919. doi: 10.3201/eid2005.140195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee HJ, Shieh CK, Gorbalenya AE, Koonin EV, La Monica N, Tuler J, Bagdzhadzhyan A, Lai MM. 1991. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology 180:567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res 117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harcourt BH, Jukneliene D, Kanjanahaluethai A, Bechill J, Severson KM, Smith CM, Rota PA, Baker SC. 2004. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J Virol 78:13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prentice E, McAuliffe J, Lu X, Subbarao K, Denison MR. 2004. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J Virol 78:9977–9986. doi: 10.1128/JVI.78.18.9977-9986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minskaia E, Hertzig T, Gorbalenya AE, Campanacci V, Cambillau C, Canard B, Ziebuhr J. 2006. Discovery of an RNA virus 3′–>5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc Natl Acad Sci U S A 103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu T, Chen C, Li H, Dou Y, Zhou M, Lu D, Zong Q, Li Y, Yang C, Zhong Z, Singh N, Hu H, Zhang R, Yang H, Su D. 2017. Structural basis for dimerization and RNA binding of avian infectious bronchitis virus nsp9. Protein Sci 26:1037–1048. doi: 10.1002/pro.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sevajol M, Subissi L, Decroly E, Canard B, Imbert I. 2014. Insights into RNA synthesis, capping, and proofreading mechanisms of SARS-coronavirus. Virus Res 194:90–99. doi: 10.1016/j.virusres.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lundin A, Dijkman R, Bergstrom T, Kann N, Adamiak B, Hannoun C, Kindler E, Jonsdottir HR, Muth D, Kint J, Forlenza M, Muller MA, Drosten C, Thiel V, Trybala E. 2014. Targeting membrane-bound viral RNA synthesis reveals potent inhibition of diverse coronaviruses including the middle East respiratory syndrome virus. PLoS Pathog 10:e1004166. doi: 10.1371/journal.ppat.1004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahn DG, Choi JK, Taylor DR, Oh JW. 2012. Biochemical characterization of a recombinant SARS coronavirus nsp12 RNA-dependent RNA polymerase capable of copying viral RNA templates. Arch Virol 157:2095–2104. doi: 10.1007/s00705-012-1404-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng X, Hackbart M, Mettelman RC, O'Brien A, Mielech AM, Yi G, Kao CC, Baker SC. 2017. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc Natl Acad Sci U S A 114:E4251–E4260. doi: 10.1073/pnas.1618310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Sun Y, Wu A, Xu S, Pan R, Zeng C, Jin X, Ge X, Shi Z, Ahola T, Chen Y, Guo D. 2015. Coronavirus nsp10/nsp16 methyltransferase can be targeted by nsp10-derived peptide in vitro and in vivo to reduce replication and pathogenesis. J Virol 89:8416–8427. doi: 10.1128/JVI.00948-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Case JB, Li Y, Elliott R, Lu X, Graepel KW, Sexton NR, Smith EC, Weiss SR, Denison MR. 18 October 2017. Murine hepatitis virus nsp14 exoribonuclease activity is required for resistance to innate immunity. J Virol doi: 10.1128/JVI.01531-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Case JB, Ashbrook AW, Dermody TS, Denison MR. 2016. Mutagenesis of S-adenosyl-l-methionine-binding residues in coronavirus nsp14 N7-methyltransferase demonstrates differing requirements for genome translation and resistance to innate immunity. J Virol 90:7248–7256. doi: 10.1128/JVI.00542-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Athmer J, Fehr AR, Grunewald M, Smith EC, Denison MR, Perlman S. 2017. In situ tagged nsp15 reveals interactions with coronavirus replication/transcription complex-associated proteins. mBio 8:e02320-. doi: 10.1128/mBio.02320-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aouadi W, Blanjoie A, Vasseur JJ, Debart F, Canard B, Decroly E. 2017. Binding of the methyl donor S-adenosyl-l-methionine to Middle East respiratory syndrome coronavirus 2′-O-methyltransferase nsp16 promotes recruitment of the allosteric activator nsp10. J Virol 91:e02217-. doi: 10.1128/JVI.02217-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Becares M, Pascual-Iglesias A, Nogales A, Sola I, Enjuanes L, Zuniga S. 2016. Mutagenesis of coronavirus nsp14 reveals its potential role in modulation of the innate immune response. J Virol 90:5399–5414. doi: 10.1128/JVI.03259-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lokugamage KG, Narayanan K, Nakagawa K, Terasaki K, Ramirez SI, Tseng CT, Makino S. 2015. Middle East respiratory syndrome coronavirus nsp1 inhibits host gene expression by selectively targeting mRNAs transcribed in the nucleus while sparing mRNAs of cytoplasmic origin. J Virol 89:10970–10981. doi: 10.1128/JVI.01352-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu X, Chen S, Hou P, Wang M, Chen Y, Guo D. 2015. VHL negatively regulates SARS coronavirus replication by modulating nsp16 ubiquitination and stability. Biochem Biophys Res Commun 459:270–276. doi: 10.1016/j.bbrc.2015.02.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narayanan K, Ramirez SI, Lokugamage KG, Makino S. 2015. Coronavirus nonstructural protein 1: common and distinct functions in the regulation of host and viral gene expression. Virus Res 202:89–100. doi: 10.1016/j.virusres.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouvet M, Lugari A, Posthuma CC, Zevenhoven JC, Bernard S, Betzi S, Imbert I, Canard B, Guillemot JC, Lecine P, Pfefferle S, Drosten C, Snijder EJ, Decroly E, Morelli X. 2014. Coronavirus Nsp10, a critical co-factor for activation of multiple replicative enzymes. J Biol Chem 289:25783–25796. doi: 10.1074/jbc.M114.577353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y, Su C, Ke M, Jin X, Xu L, Zhang Z, Wu A, Sun Y, Yang Z, Tien P, Ahola T, Liang Y, Liu X, Guo D. 2011. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2′-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog 7:e1002294. doi: 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daczkowski CM, Goodwin O, Dzimianski JV, Farhat JJ, Pegan SD. 20 September 2017. Structurally guided removal of deISGylase biochemical activity from papain-like protease originating from the Middle East respiratory syndrome virus. J Virol doi: 10.1128/JVI.01067-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alfuwaires M, Altaher A, Kandeel M. 2017. Molecular dynamic studies of interferon and innate immunity resistance in MERS CoV non-structural protein 3. Biol Pharm Bull 40:345–351. doi: 10.1248/bpb.b16-00870. [DOI] [PubMed] [Google Scholar]
- 32.Fehr AR, Channappanavar R, Jankevicius G, Fett C, Zhao JC, Athmer J, Meyerholz DK, Ahel I, Perlman S. 2016. The conserved coronavirus macrodomain promotes virulence and suppresses the innate immune response during severe acute respiratory syndrome coronavirus infection. mBio 7:e01721-. doi: 10.1128/mBio.01721-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lei J, Hilgenfeld R. 2016. Structural and mutational analysis of the interaction between the Middle-East respiratory syndrome coronavirus (MERS-CoV) papain-like protease and human ubiquitin. Virol Sin 31:288–299. doi: 10.1007/s12250-016-3742-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forni D, Cagliani R, Mozzi A, Pozzoli U, Al-Daghri N, Clerici M, Sironi M. 2016. Extensive positive selection drives the evolution of nonstructural proteins in lineage C betacoronaviruses. J Virol 90:3627–3639. doi: 10.1128/JVI.02988-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kindler E, Thiel V. 2014. To sense or not to sense viral RNA—essentials of coronavirus innate immune evasion. Curr Opin Microbiol 20:69–75. doi: 10.1016/j.mib.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bailey-Elkin BA, Knaap RC, Johnson GG, Dalebout TJ, Ninaber DK, van Kasteren PB, Bredenbeek PJ, Snijder EJ, Kikkert M, Mark BL. 2014. Crystal structure of the Middle East respiratory syndrome coronavirus (MERS-CoV) papain-like protease bound to ubiquitin facilitates targeted disruption of deubiquitinating activity to demonstrate its role in innate immune suppression. J Biol Chem 289:34667–34682. doi: 10.1074/jbc.M114.609644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Menachery VD, Debbink K, Baric RS. 2014. Coronavirus non-structural protein 16: evasion, attenuation, and possible treatments. Virus Res 194:191–199. doi: 10.1016/j.virusres.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Totura AL, Baric RS. 2012. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr Opin Virol 2:264–275. doi: 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang D, Fang LR, Shi YL, Zhang H, Gao L, Peng GQ, Chen HC, Li K, Xiao SB. 2016. Porcine epidemic diarrhea virus 3C-like protease regulates its interferon antagonism by cleaving NEMO. J Virol 90:2090–2101. doi: 10.1128/JVI.02514-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J, Poon LL, Guan Y, Rozanov M, Spaan WJ, Gorbalenya AE. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol 331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Connor RF, Roper RL. 2007. Unique SARS-CoV protein nsp1: bioinformatics, biochemistry and potential effects on virulence. Trends Microbiol 15:51–53. doi: 10.1016/j.tim.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jansson AM. 2013. Structure of alphacoronavirus transmissible gastroenteritis virus nsp1 has implications for coronavirus nsp1 function and evolution. J Virol 87:2949–2955. doi: 10.1128/JVI.03163-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang C, Lokugamage KG, Rozovics JM, Narayanan K, Semler BL, Makino S. 2011. Alphacoronavirus transmissible gastroenteritis virus nsp1 protein suppresses protein translation in mammalian cells and in cell-free HeLa cell extracts but not in rabbit reticulocyte lysate. J Virol 85:638–643. doi: 10.1128/JVI.01806-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, Shi H, Rigolet P, Wu N, Zhu L, Xi XG, Vabret A, Wang X, Wang T. 2010. Nsp1 proteins of group I and SARS coronaviruses share structural and functional similarities. Infect Genet Evol 10:919–924. doi: 10.1016/j.meegid.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Q, Ma J, Yoo D. 2017. Inhibition of NF-kappaB activity by the porcine epidemic diarrhea virus nonstructural protein 1 for innate immune evasion. Virology 510:111–126. doi: 10.1016/j.virol.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Q, Shi K, Yoo D. 2016. Suppression of type I interferon production by porcine epidemic diarrhea virus and degradation of CREB-binding protein by nsp1. Virology 489:252–268. doi: 10.1016/j.virol.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Almeida MS, Johnson MA, Wuthrich K. 2006. NMR assignment of the SARS-CoV protein nsp1. J Biomol NMR 36(Suppl 1):46. doi: 10.1007/s10858-006-9018-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kamitani W, Huang C, Narayanan K, Lokugamage KG, Makino S. 2009. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat Struct Mol Biol 16:1134–1140. doi: 10.1038/nsmb.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Narayanan K, Huang C, Lokugamage K, Kamitani W, Ikegami T, Tseng CT, Makino S. 2008. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J Virol 82:4471–4479. doi: 10.1128/JVI.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng L, Zhang C, Shi C, Yang Z, Wang Y, Zhou T, Sun F, Wang H, Zhao S, Qin Q, Qiao R, Ding Z, Wei C, Xie L, Wu J, Li Y. 2017. Rice stripe virus NS3 protein regulates primary miRNA processing through association with the miRNA biogenesis factor OsDRB1 and facilitates virus infection in rice. PLoS Pathog 13:e1006662. doi: 10.1371/journal.ppat.1006662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee N, Yario TA, Gao JS, Steitz JA. 2016. EBV noncoding RNA EBER2 interacts with host RNA-binding proteins to regulate viral gene expression. Proc Natl Acad Sci U S A 113:3221–3226. doi: 10.1073/pnas.1601773113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rawat S, Bouchard MJ. 2015. The hepatitis B virus (HBV) HBx protein activates AKT to simultaneously regulate HBV replication and hepatocyte survival. J Virol 89:999–1012. doi: 10.1128/JVI.02440-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adams S, Xing Z, Li J, Mendoza K, Perez D, Reed K, Cardona C. 2013. The effect of avian influenza virus NS1 allele on virus replication and innate gene expression in avian cells. Mol Immunol 56:358–368. doi: 10.1016/j.molimm.2013.05.236. [DOI] [PubMed] [Google Scholar]
- 54.Jauregui AR, Savalia D, Lowry VK, Farrell CM, Wathelet MG. 2013. Identification of residues of SARS-CoV nsp1 that differentially affect inhibition of gene expression and antiviral signaling. PLoS One 8:e62416. doi: 10.1371/journal.pone.0062416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peng GQ, Xu LQ, Lin YL, Chen L, Pasquarella JR, Holmes KV, Li F. 2012. Crystal structure of bovine coronavirus spike protein lectin domain. J Biol Chem 287:41931–41938. doi: 10.1074/jbc.M112.418210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zust R, Cervantes-Barragan L, Kuri T, Blakqori G, Weber F, Ludewig B, Thiel V. 2007. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog 3:e109. doi: 10.1371/journal.ppat.0030109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wathelet MG, Orr M, Frieman MB, Baric RS. 2007. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J Virol 81:11620–11633. doi: 10.1128/JVI.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi Y, Li Y, Lei Y, Ye G, Shen Z, Sun L, Luo R, Wang D, Fu ZF, Xiao S, Peng G. 2016. A dimerization-dependent mechanism drives the endoribonuclease function of porcine reproductive and respiratory syndrome virus nsp11. J Virol 90:4579–4592. doi: 10.1128/JVI.03065-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. [DOI] [PubMed] [Google Scholar]
- 60.Ogden KM, Hu L, Jha BK, Sankaran B, Weiss SR, Silverman RH, Patton JT, Prasad BV. 2015. Structural basis for 2′-5′-oligoadenylate binding and enzyme activity of a viral RNase L antagonist. J Virol 89:6633–6645. doi: 10.1128/JVI.00701-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr Sect D Biol Crystallogr 58:1948–1954. doi: 10.1107/S0907444902016657. [DOI] [PubMed] [Google Scholar]
- 63.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr Sect D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 64.Grell L, Parkin C, Slatest L, Craig PA. 2006. EZ-Viz, a tool for simplifying molecular viewing in PyMOL. Biochem Mol Biol Educ 34:402–407. doi: 10.1002/bmb.2006.494034062672. [DOI] [PubMed] [Google Scholar]
- 65.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schmidt EK, Clavarino G, Ceppi M, Pierre P. 2009. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods 6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]