Abstract
Objective:
In recent decades, the overexpression of microRNA-21 (miR-21) is found to be progressively linked with many diseases such as different types of cancers, cardiovascular diseases, and inflammation. Thereby, it has become an attractive target for pharmacological and genetic modulation in various diseases, and also for overcoming the resistance to chemotherapy in several cancers. Here, in this study, the role of molecular therapeutics of 3,3′-diindolylmethane (DIM) has been investigated for its ability to bind with the precursor miRNA as a target of miR-21 (hsa-mir-21), which may alter the catalyzation process of dicer, a RNase III enzyme, involved in miRNA transcription.
Methods:
In this context, the present study describes the potential binding and the structure alteration properties of DIM to precursor miR-21 (pre-miR-21) through Molecular Docking and Molecular Dynamics simulation techniques.
Results:
As a corollary, DIM formed both non-bonded and covalent interactions with the bases of pre-miR, while covalent interaction with guanine in the 6th position was found to be consensus in molecular dynamics simulation. Furthermore, the stability of both DIM and pre-miR-21 was found to be inversely correlated to each other in binding condition.
Conclusion:
This result indicates that DIM can be used in target-based therapy and also as a lead for further development of potent small molecule miRNA antagonist.
KEYWORDS: 3,3′-Diindolylmethane; cancer; miR-21; molecular docking; molecular dynamics simulation; pre-miR-21
INTRODUCTION
MicroRNAs (miRNAs) are the new findings of genes family consisting of 19–25 nucleotides in length which are nonprotein coding in nature but act as a major player in the regulation of gene expression by post-transcriptional interactions with mRNA.[1] Over 1000 human miRNAs have been discovered so far,[2] and they are predicted to target up to 92% of all human genes.[3] In the human genome, miR-21 (hsa-miR-21) was one of the first detected miRNAs, which was known to be upregulated in all types of human cancer including glioblastoma,[4] breast,[5] lung,[6] esophageal,[7] gastrointestinal,[8] hepatocellular,[9] cholangiocarcinoma,[10] pancreatic,[11] prostate,[12] bladder,[13] ovarian,[14] NK-cell lymphoma,[15] laryngeal carcinoma,[16] and tongue squamous cell carcinoma.[17] Until now, various experiments have been conducted to determine the physiological and pathophysiological functions of miR-21, where its major functions involved embryonic development and cell proliferation, differentiation, and death.[18,19,20,21] Recently, extensive studies are indicating that excessive expression of miR-21 is involved in all known cancer-related processes including tumor genesis, progression, and metastasis.[21,22,23,24] Furthermore, improving chemosensitivity to anticancer agents and inducing cell cycle arrest by inhibition of miR-21 concluded that miR-21 may function as an oncogene in various human cancers.[7,9,11,21,25]
In traditional theory, oligonucleotide-based gene therapy approaches such as synthetic miRNA, antagomir, and miRNA sponge/decoy are usually used for the miRNA functional studies and for therapeutic intervention.[26,27] Nevertheless, due to their limitations such as chemical instability, cellular delivery, and inappropriate biodistribution,[28] nowadays, other techniques based on small molecular chemical biology have been followed.[29,30,31,32,33] Particularly, small molecules may directly bind to the miRNA and alter its functions. In general, most of the miRNA are formed in cell by the dicer-catalyzed processing of their corresponding inactive precursor miRNA (pre-miRNA) and this step has been proposed as an appropriate target for inhibitor design.[34,35,36,37,38] Furthermore, high affinity synthetic small molecule ligands have been recently described to bind with pre-miRNA.[38,39,40] In this regard, we aim to analyze the binding properties of 3,3′-diindolylmethane (DIM) with pre-miR-21, as a potential inhibitor of miR-21. DIM is a natural compound that is abundant in cruciferous vegetables such as broccoli, cauliflower, and cabbage, and it is a major acid condensation product of indole-3-carbinol.[41] DIM has been demonstrated to reduce the expression of certain miRNAs. It inhibits cancer cell growth by controlling cell proliferation, transcription, and apoptosis through regulation of gene expression. Moreover, it is evident that it reverses the expression level of oncogenic miRNAs by inhibiting the expression of miR-21.[42] Results showed that the inhibition of endogenous miR-21 is specific, efficient in the treatment of cancer.[42]
Coupled with that, the present study is designed with some computer-aided drug discovery methods such as molecular docking and molecular dynamics simulation to investigate the bindings and the changes in the dynamics behavior of functional region of pre-miRNAs. As we know that conformational changes in the protein or macromolecules structure affect its biological function, and also it is evident that the conformational flexibility of a macromolecule influences the interactions with ligand or its biological partners at different level.[42,43]
MATERIALS AND METHODS
Data and databases
The data from databases used in this study include protein data bank (PDB)[44] and PubChem.[45] PubChem is a public repository of small molecules and their biological properties. Currently, it contains more than 25 million unique chemical structures and 90 million bioactivity outcomes associated with several thousand macromolecular targets.[46]
Ligand selection and preparation
For docking studies, compound was retrieved from Pubchem databases, i.e., DIM (CID 3071). The 3D structure for this compound was built by using Ligprep2.5 in Schrödinger Suite 2013 with OPLS_2005 force field. Their ionization states were generated at pH7.0 ± 2.0 using Epik2.2 in Schrödinger Suite. Up to 32 possible stereoisomers per ligand were retained.
MicroRNA structure preparation
The NMR solution structure of pre-miR-21 having lowest energy was obtained from (pdb id: 2MNC).[44] The solution structure of pre-miR-21 (CCGUUGAAUCUCACGG) was prepared and refined by using the Protein Preparation Wizard of Schrödinger-Maestro v9.4. Charges and bond orders were assigned, hydrogens were added to the heavy atoms, and all waters were deleted. It was optimized at neutral pH. Using force field OPLS_2005, minimization was carried out setting maximum heavy atom root mean square deviation (RMSD) to 0.30 Å.
Docking simulation study
Before docking, the structures were prepared by using dock preparation wizard integrated in UCSF Chimera,[47] by adding hydrogen and Gasteiger charges.[48] Input ligand file format was MOL2, so these were further converted into pdbqt format by Python Molecular Viewer with AutoDock tools, required by AutoDock Vina.[49] The size of grid box in AutoDock Vina was kept at 30.72, 25.00, and 25.00, respectively, for X, Y, and Z. The energy range was kept at 4 which was default setting. AutoDock Vina was implemented through shell script provided by AutoDock Vina developers. The binding affinity of ligand was observed by Kcal/Mol as a unit for a negative score.[50]
Molecular dynamics simulation
Molecular dynamics simulation was performed using the NAMD[51] engine integrated in MOE software. In this study, the AMBER12:EHT (Extended Huckel Theory Group II ion and Group VIII parameters OPLS-AA) force field was utilized, as it is widely applied to describe macromolecular system. The system was solvated and neutralized by adding Cl− and/or Na+ ions and water molecules, where the total solvent molecules was 1827; 1.023 g/cm3. The periodic boundary condition was employed to perform the simulation, where the box size was 43.3 Å × 38.0 Å × 38.1 Å. Following steepest descent energy minimization, equilibration of 100 steps done in NPT ensemble with Periodic Boundary Conditions, Particle Mesh Ewald for full-system periodic electrostatics, and constant temperature dynamics through Langevin Dynamics, constant pressure dynamics through Nose-Hoover Langevin piston, and SHAKE was used to maintain all bonds involving hydrogen atoms at their equilibrium values. Finally, the full system was subjected to molecular dynamics (MD) production run at 300 K temperature for 10 ns in NVT ensemble.
RESULTS AND DISCUSSION
Docking simulation study
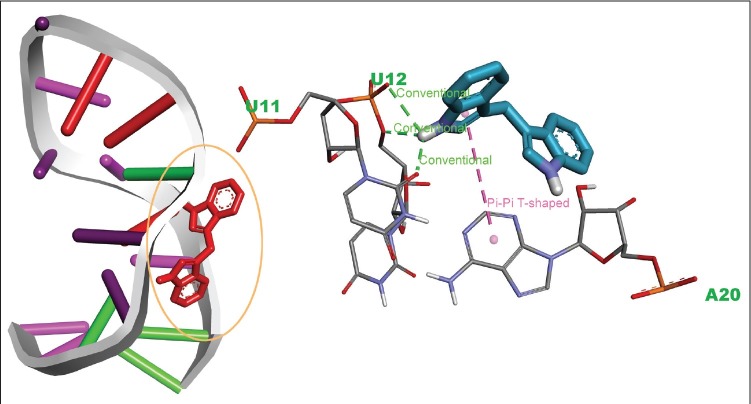
Molecular docking is one of the most computational techniques, which not only predicts the binding affinity of a ligand to a receptor but also demonstrates the binding mode in the particular active site. We therefore performed docking analysis to understand the molecular interactions of DIM in binding with miR-21. In essence, DIM formed three conventional hydrogen bonds with U11, U12 bases, where a hydrogen bond was formed with the oxygen atoms of 11th uridine and rest of two were formed with another oxygen atom of 12th uridine. Moreover, a π-π interaction was observed between the indole ring and the purine ring of 20th adenine. A detailed description of the DIM binding with miR-21 is demonstrated in Table 1 and also rendered in Figure 1. According to the docking analysis, the binding affinity of DIM was found to have −7.3 kcal/mol, which may indicate a strong binding with pre-miR-21. On this side, analysis of complex stability was carried out using long MD simulations to support the docking prediction, which is described in further sections.
Table 1.
Molecular interactions analysis of 3,3′-diindolylmethane pre-microRNA-21 complex, after docking and molecular dynamics simulation
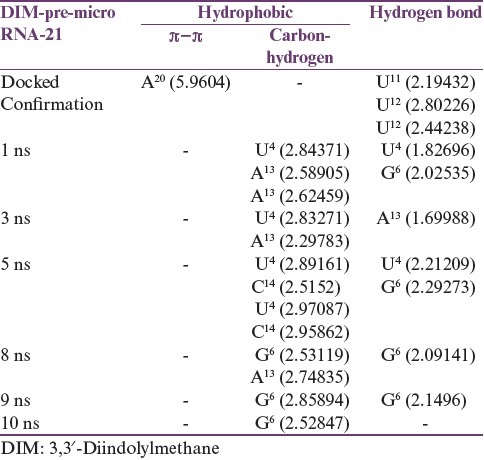
Figure 1.
Interactions and binding pose of 3,3′-diindolylmethane with precursor microRNA-21 complex were generated from AutoDock Vina. Here, green dots represent hydrogen bonding, while pink represent pi-pi T-shaped
Molecular dynamics simulation
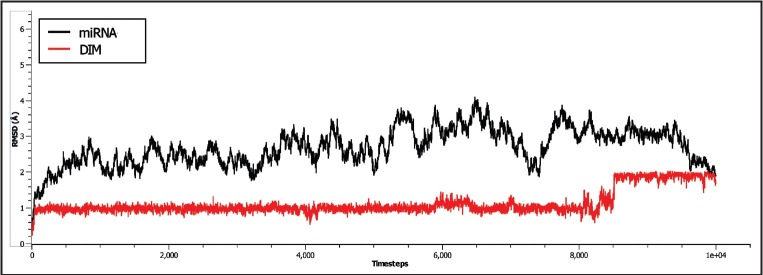
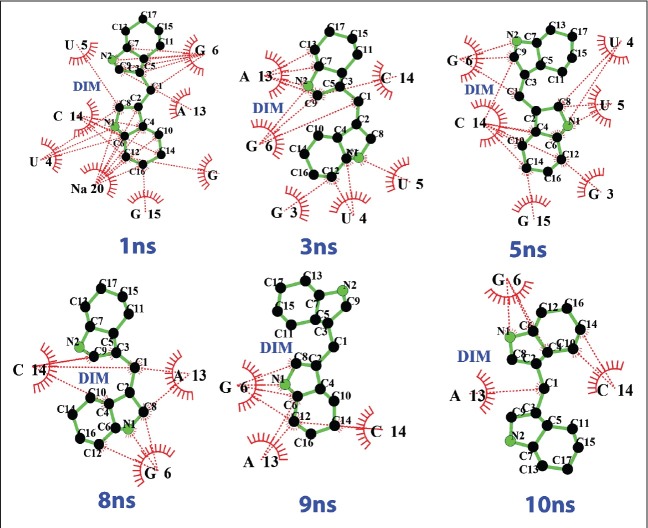
To impart stability, the pre-miR-21-DIM complex was subjected to a MD simulation for 10 ns. The energy profile of the system such as the potential energy and kinetic energy and the other factors such as temperature and pressure are rendered in Figure 2. As described in Figure 2, it can be concluded that the system had been reached at equilibration and remained stable in 10 ns simulation. Moreover, the volume of the system remained constant at the time of simulation, which is 62610.11 Å3. To describe the stability and flexibility of the ligand binding, we calculated the RMSD profile of both miR-21 and DIM along with the time steps. As can be seen in Figure 3, the RMSD of the backbone of miR-21 increased from ~1 Å at 0 ns to 3 Å at 1 ns and remained stable at 2–3 Å for 3.75 ns. After that, it then increased to 4 Å and remained stable for 8 ns. However, following 8 ns, the RMSD is tending to decrease, which means that the miRNA is somewhat stable in the simulation, afterward. In contrast, the RMSD of DIM remained stable at ~1 Å, from 0 ns to 8.5 ns. Nevertheless, it increased to ~2 Å and seems to lose its stability afterward. Interestingly, it can be assumed from the RMSD plot that in bounding with DIM, the pre-miR-21 was less stable for 8 ns, while DIM was found to be more stable at that time. In this manner, after 8 ns, the stability of DIM was decreased, while the stability of pre-miR-21 increased. To justify this hypothesis, we examined the molecular interactions of the complex in different snapshot of MD simulation. According to Table 1 and Figure 4, it is observed that the DIM formed conventional hydrogen bonds with U4 and G6 bases in 1 ns and 5 ns snapshot and also had covalent interactions by forming carbon–hydrogen bonds with U4, A13, and C4 bases. Similarly, in 8 ns and 9 ns simulation snapshot, G6 base was involved in both noncovalent and covalent interactions with DIM; however, only one covalent interaction remained in 10 ns simulation. As a corollary, the number of bonding interaction was gradually decreasing by the increasing of time step in the simulation, which means that increasing of DIM flexibility after 8 ns is due to the breaking down of its bonding interactions with pre-miR-21.
Figure 2.
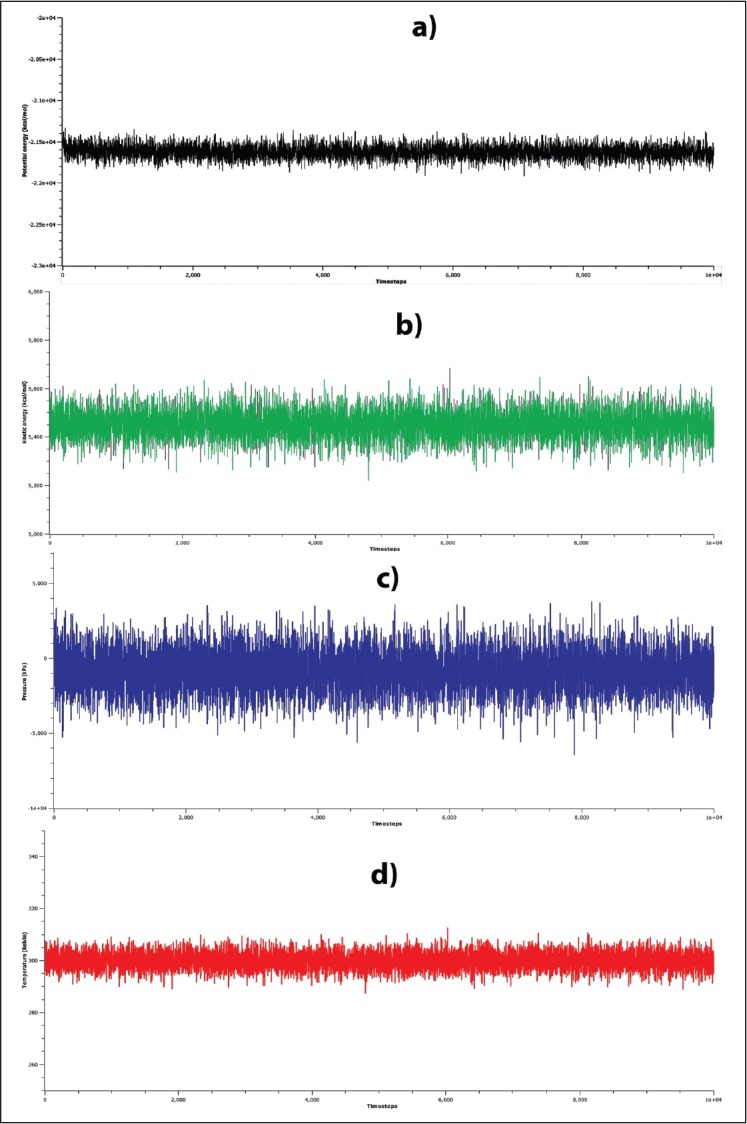
Stability of the simulation system. Stability was evaluated in terms of the (a) potential energies, (b) kinetic energies, (c) pressure, (d) temperature of the simulation as a function of simulation time
Figure 3.
Backbone root mean square deviations are shown as a function of time for precursor microRNA-21 and 3,3′-diindolylmethane at 300 K. Here, precursor microRNA-21 is shown in black and 3,3′-diindolylmethane in red
Figure 4.
Two-dimensional representation of molecular interactions between the precursor microRNA-21 and 3,3′-diindolylmethane, complexes were sampled from molecular dynamics simulation. The interactions were rendered by Ligplot
Our findings from the molecular docking and molecular dynamics simulation studies suggested that DIM has the potential chemical affinity and ability to alter the stability of pre-miRNA of miR-21. These results postulate DIM as an important ligand of potential importance for the development of potent small molecular miRNA antagonist and the remedial approaches in various cancers.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
We, all authors, are grateful to Molecular Modelling and Drug Design Laboratory, Pharmacology Research Division, Bangladesh Council of Scientific and Industrial Research (BCSIR), Chittagong-4220 for providing the all necessary facilities for this experiment. We are also grateful to Mahmuda Khatun, Director, BCSIR laboratory Chittagong, for her continuous support in this experiment. The authors are also pleased to Habibur Rahman Bhuiyan, Principal Scientific Officer, BCSIR laboratory Chittagong, for reviewing and making necessary comments during the preparation of manuscript.
REFERENCES
- 1.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–24. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 2.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 3.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, et al. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–17. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 4.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–33. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 5.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 6.Seike M, Goto A, Okano T, Bowman ED, Schetter AJ, Horikawa I, et al. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci U S A. 2009;106:12085–90. doi: 10.1073/pnas.0905234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hiyoshi Y, Kamohara H, Karashima R, Sato N, Imamura Y, Nagai Y, et al. MicroRNA-21 regulates the proliferation and invasion in esophageal squamous cell carcinoma. Clin Cancer Res. 2009;15:1915–22. doi: 10.1158/1078-0432.CCR-08-2545. [DOI] [PubMed] [Google Scholar]
- 8.Chan SH, Wu CW, Li AF, Chi CW, Lin WC. MiR-21 microRNA expression in human gastric carcinomas and its clinical association. Anticancer Res. 2008;28:907–11. [PubMed] [Google Scholar]
- 9.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T, et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–58. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, et al. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–29. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 11.Moriyama T, Ohuchida K, Mizumoto K, Yu J, Sato N, Nabae T, et al. MicroRNA-21 modulates biological functions of pancreatic cancer cells including their proliferation, invasion, and chemoresistance. Mol Cancer Ther. 2009;8:1067–74. doi: 10.1158/1535-7163.MCT-08-0592. [DOI] [PubMed] [Google Scholar]
- 12.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Catto JW, Miah S, Owen HC, Bryant H, Myers K, Dudziec E, et al. Distinct microRNA alterations characterize high- and low-grade bladder cancer. Cancer Res. 2009;69:8472–81. doi: 10.1158/0008-5472.CAN-09-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 15.Yamanaka Y, Tagawa H, Takahashi N, Watanabe A, Guo YM, Iwamoto K, et al. Aberrant overexpression of microRNAs activate AKT signaling via down-regulation of tumor suppressors in natural killer-cell lymphoma/leukemia. Blood. 2009;114:3265–75. doi: 10.1182/blood-2009-06-222794. [DOI] [PubMed] [Google Scholar]
- 16.Liu M, Wu H, Liu T, Li Y, Wang F, Wan H, et al. Regulation of the cell cycle gene, BTG2, by miR-21 in human laryngeal carcinoma. Cell Res. 2009;19:828–37. doi: 10.1038/cr.2009.72. [DOI] [PubMed] [Google Scholar]
- 17.Li J, Huang H, Sun L, Yang M, Pan C, Chen W, et al. MiR-21 indicates poor prognosis in tongue squamous cell carcinomas as an apoptosis inhibitor. Clin Cancer Res. 2009;15:3998–4008. doi: 10.1158/1078-0432.CCR-08-3053. [DOI] [PubMed] [Google Scholar]
- 18.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 19.Ramachandra RK, Salem M, Gahr S, Rexroad CE 3rd, Yao J. Cloning and characterization of microRNAs from rainbow trout (Oncorhynchus mykiss): Their expression during early embryonic development. BMC Dev Biol. 2008;8:41. doi: 10.1186/1471-213X-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carletti MZ, Fiedler SD, Christenson LK. MicroRNA 21 blocks apoptosis in mouse periovulatory granulosa cells. Biol Reprod. 2010;83:286–95. doi: 10.1095/biolreprod.109.081448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS, et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28:5369–80. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qian B, Katsaros D, Lu L, Preti M, Durando A, Arisio R, et al. High miR-21 expression in breast cancer associated with poor disease-free survival in early stage disease and high TGF-beta1. Breast Cancer Res Treat. 2009;117:131–40. doi: 10.1007/s10549-008-0219-7. [DOI] [PubMed] [Google Scholar]
- 23.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY, et al. MiR-21-mediated tumor growth. Oncogene. 2007;26:2799–803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 24.Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES, et al. Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin Chem. 2008;54:1696–704. doi: 10.1373/clinchem.2007.101741. [DOI] [PubMed] [Google Scholar]
- 25.Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68:8164–72. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- 26.Meister G, Landthaler M, Dorsett Y, Tuschl T. Sequence-specific inhibition of microRNA-and siRNA-induced RNA silencing. RNA. 2004;10:544–50. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davis S, Lollo B, Freier S, Esau C. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res. 2006;34:2294–304. doi: 10.1093/nar/gkl183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu L, Anchordoquy T. Drug delivery trends in clinical trials and translational medicine: Challenges and opportunities in the delivery of nucleic acid-based therapeutics. J Pharm Sci. 2011;100:38–52. doi: 10.1002/jps.22243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gumireddy K, Young DD, Xiong X, Hogenesch JB, Huang Q, Deiters A, et al. Small-molecule inhibitors of microrna miR-21 function. Angew Chem Int Ed Engl. 2008;47:7482–4. doi: 10.1002/anie.200801555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watashi K, Yeung ML, Starost MF, Hosmane RS, Jeang KT. Identification of small molecules that suppress microRNA function and reverse tumorigenesis. J Biol Chem. 2010;285:24707–16. doi: 10.1074/jbc.M109.062976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Georgianna WE, Young DD. Development and utilization of non-coding RNA-small molecule interactions. Org Biomol Chem. 2011;9:7969–78. doi: 10.1039/c1ob06324c. [DOI] [PubMed] [Google Scholar]
- 32.Deiters A. Small molecule modifiers of the microRNA and RNA interference pathway. AAPS J. 2010;12:51–60. doi: 10.1208/s12248-009-9159-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, He C, Jin P. Emergence of chemical biology approaches to the RNAi/miRNA pathway. Chem Biol. 2010;17:584–9. doi: 10.1016/j.chembiol.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davies BP, Arenz C. A fluorescence probe for assaying micro RNA maturation. Bioorg Med Chem. 2008;16:49–55. doi: 10.1016/j.bmc.2007.04.055. [DOI] [PubMed] [Google Scholar]
- 35.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21-and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Radmark O, et al. Ribonuclease activity and RNA binding of recombinant human dicer. EMBO J. 2002;21:5864–74. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davies BP, Arenz C. A homogenous assay for micro RNA maturation. Angew Chem Int Ed Engl. 2006;45:5550–2. doi: 10.1002/anie.200601332. [DOI] [PubMed] [Google Scholar]
- 38.Thomas JR, Hergenrother PJ. Targeting RNA with small molecules. Chem Rev. 2008;108:1171–224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 39.Chirayil S, Chirayil R, Luebke KJ. Discovering ligands for a microRNA precursor with peptoid microarrays. Nucleic Acids Res. 2009;37:5486–97. doi: 10.1093/nar/gkp549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krishnamurthy M, Simon K, Orendt AM, Beal PA. Macrocyclic helix‐threading peptides for targeting RNA. Angew Chem. 2007;119:7174–7. doi: 10.1002/anie.200702247. [DOI] [PubMed] [Google Scholar]
- 41.Verhoeven DT, Goldbohm RA, van Poppel G, Verhagen H, van den Brandt PA. Epidemiological studies on brassica vegetables and cancer risk. Cancer Epidemiol Biomarkers Prev. 1996;5:733–48. [PubMed] [Google Scholar]
- 42.Heberlé G, de Azevedo WF., Jr Bio-inspired algorithms applied to molecular docking simulations. Curr Med Chem. 2011;18:1339–52. doi: 10.2174/092986711795029573. [DOI] [PubMed] [Google Scholar]
- 43.de Azevedo WF., Jr Molecular dynamics simulations of protein targets identified in mycobacterium tuberculosis. Curr Med Chem. 2011;18:1353–66. doi: 10.2174/092986711795029519. [DOI] [PubMed] [Google Scholar]
- 44.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The protein data bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Suzek T, Zhang J, Wang J, He S, Cheng T, et al. PubChem bioAssay: 2014 update. Nucleic Acids Res. 2014;42:D1075–82. doi: 10.1093/nar/gkt978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Q, Cheng T, Wang Y, Bryant SH. PubChem as a public resource for drug discovery. Drug Discov Today. 2010;15:1052–7. doi: 10.1016/j.drudis.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF chimera – a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 48.Dunbrack RL., Jr Rotamer libraries in the 21st century. Curr Opin Struct Biol. 2002;12:431–40. doi: 10.1016/s0959-440x(02)00344-5. [DOI] [PubMed] [Google Scholar]
- 49.Sanner MF. Python: A programming language for software integration and development. J Mol Graph Model. 1999;17:57–61. [PubMed] [Google Scholar]
- 50.Trott O, Olson AJ. AutoDock vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–61. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]