Abstract
Noncoding RNAs are pervasive in cells and contribute to diseases such as cancer. A question in biomedical research is whether noncoding RNAs are targets of medicines. Bleomycin is a natural product that cleaves DNA; however, it is known to cleave RNA in vitro. Herein, an in-depth analysis of the RNA cleavage preferences of bleomycin A5 is presented. Bleomycin A5 prefers to cleave RNAs with stretches of AU base pairs. Based on these preferences and bioinformatic analysis, the microRNA-10b hairpin precursor was identified as a potential substrate for bleomycin A5. Both in vitro and cellular experiments demonstrated cleavage. Importantly, chemical cleavage by bleomycin A5 in the microRNA-10b hairpin precursors occurred near the Drosha and Dicer enzymatic processing sites and led to destruction of the microRNA. Evidently, oncogenic noncoding RNAs can be considered targets of cancer medicines and might elicit their pharmacological effects by targeting noncoding RNA.
The Encyclopedia of DNA Elements (ENCODE) project and functional studies have shown that 80 % of the genome is transcribed into RNA, but only 1–2 % is translated into protein.[1] Thus, RNA plays major roles in both healthy cells and disease biology that go beyond protein encoding. For example, long noncoding RNAs are involved in gene silencing and activation.[2] Small RNAs such as micro (mi) RNA play critical roles in decreasing the amount of protein translated from a given messenger (m) RNA.[3] In fact, in many cancers, miRNAs have been shown to cause disease through upregulation and subsequent silencing of pro-apoptotic proteins.[4] Thus, RNA is considered an important drug target.
The main manner in which RNA is drugged is through the use of oligonucleotides that target mRNAs.[5] These compounds can have suboptimal features, such as limited tissue and cellular permeability due to their large molecular weights, and often elicit off-target effects. Small molecules could be a preferred modality to target RNA; although bacterial RNAs such as the ribosome and riboswitches are tried and true targets of small molecules,[6] human RNAs are generally not considered druggable. One challenge is to develop small molecules that target human RNAs. Previous work in this area has included screening and sequence-based rational design enabled by Inforna.[7] One salient question remains: do known drugs actually target RNA and contribute to their pharmacological responses? In support of a positive answer to this question, the aminoglycoside antibiotic streptomycin was found to target a microRNA (miR) -21 precursor, albeit streptomycin is an antibacterial, not an anticancer agent.[8]
The antitumor natural product bleomycin has been known for decades to cleave DNA (Scheme 1).[9] There is an encyclopedia of information describing structure–activity relationships (SARs) for both the cleaved DNA and the bleomycin cleaver.[10] Interestingly, RNA has also been shown to be cleaved by bleomycin in vitro,[11] with a focus on cleavage of tRNA. More detailed analysis of the preferred RNAs that are cleaved could elucidate currently unknown cellular targets. However, RNA has not been shown to be cleaved by bleomycin in cells.
Scheme 1.
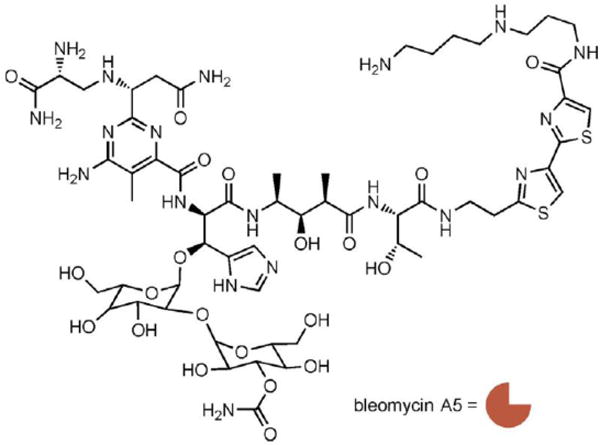
Structure of bleomycin A5, which is represented as an orange crescent.
To assess the RNAs that are cleaved by bleomycin A5, we initially tested RNAs containing stretches of base pairs that were designed to be displayed in a unimolecular hairpin structure for cleavage. These RNAs had stems that contained the canonical AU and GC base pairs in each of their 5′ and 3′ orientations, for example, 5′AU/3′UA and 5′UA/3′AU, in a unimolecular GNRA hairpin, where N indicates any nucleotide and R indicates a purine nucleotide. The initial series also included the 5′GU/3′UG wobble pair found in RNA (see Figures S1–S3 for RNA secondary structures).[12] Interestingly, bleomycin A5 cleaved the RNAs containing AU pairs most efficiently, with 5′-AUAU-3′ sequences being cleaved more than 5′-AAUU-3′ sequences (Figure 1 A). Bleomycin A5 did not significantly cleave RNAs containing only GU and GC pairs (Figures S1–S3).
Figure 1.
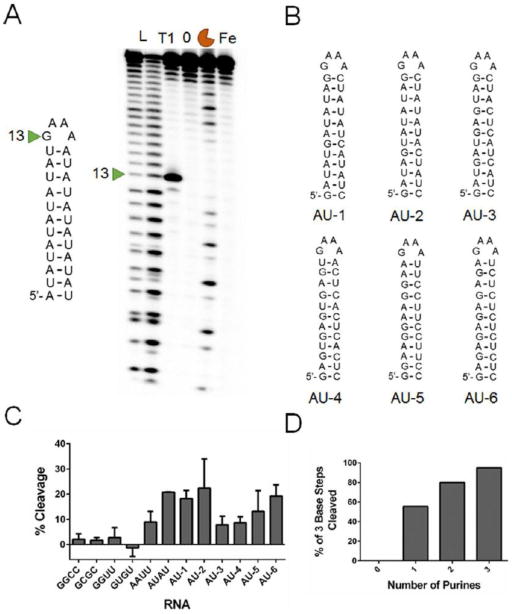
Bleomycin cleavage of hairpin RNAs. A) Representative gel image of bleomycin A5 cleavage of an RNA comprised of AU pairs. L indicates a hydrolysis ladder; T1 indicates ribonuclease T1 ladder; 0 indicates untreated RNA; the orange crescent indicates RNA treated with 10 μm bleomycin A5; and Fe indicates RNA treated with 10 μm (NH4)2Fe(SO2)2·6H2O. B) Secondary structures of the second library of RNAs. C) Quantification of cleavage for all RNAs treated with bleomycin A5. See Figures S1-S4 for full RNA secondary structures. D) Nearest-neighbor analysis of cleavage sites of each RNA in three base steps revealed a preference for cleavage of purine-rich sequences.
To further analyze the sequence selectivity of bleomycin A5 in RNA, we designed and tested six additional RNAs. These RNAs contained stretches of AU pairs of different lengths, separated by GC pairs with the GNRA hairpin loop (Figure 1 B). The RNAs with the longest stretches of AU pairs (AU-1 and AU-2) were efficiently cleaved by bleomycin A5; whereas, RNAs with only one or two AU pairs inserted between GC pairs were cleaved less efficiently (AU-3 and AU-4; Figures 1 C and S4). Interestingly, two of the RNA hairpins contain only purine bases on the 5′-side of the hairpin loop and only pyrimidine bases on the 3′-side of the hairpin loop. In both of these examples, bleomycin A5 cleaved only the purine-rich region of the RNA. Quantification of the percent cleavage of each RNA revealed that RNAs rich in AU base pairs and an RNA with a 5′-GAGA-3′ sequence (AU-6) were the most highly cleaved by bleomycin A5 (Figure 1 C).
A nearest-neighbor analysis of the cleaved sites of each RNA was completed, starting at the 5′-end of the RNA, in three base steps. This analysis revealed a strong preference for purine-rich sequences, especially those rich in AU base pairs (Figure 1 D). In the RNAs used in this study, three base sequences containing only pyrimidines were not cleaved by bleomycin A5. Three base steps containing three purines were cleaved 95 % of the time in the RNAs analyzed here, indicating that bleomycin A5 cleaves purine-rich sequences in RNA.
In studies of DNA cleavage, bleomycin prefers to cleave 5′-GC or 5′-GT sequences.[10] NMR spectroscopy and SAR studies of bleomycin and a DNA substrate have shown that this sequence selectivity in DNA is largely due to hydrogen bonding interactions between bleomycin’s metal binding pyrimidine core and the N3- and C2-amino groups of guanine.[10, 13] Bleomycin binds the minor groove of DNA and is capable of making two hydrogen bonding interactions with guanine but only one with adenine; thus, bleomycin prefers to cleave 5′GC/T sequences (Figure 2 A).[13] Although bleomycin interacts with the minor groove of DNA, the minor groove of RNA is narrower; therefore, bleomycin is more likely to interact with the major groove of RNA. Interaction with the major groove of RNA would allow hydrogen bonding to occur with the C6 amino group of adenine that is displayed in the major groove (Figure 2 B). Previous in vitro studies have demonstrated that alteration of the position of the amino group in DNA changes the cleavage pattern and cleavage intensity of bleomycin.[14] DNA substitutions of guanine to inosine and adenine to 2aminopurine altered the cleavage of DNA in vitro.[14] A 2-aminopurine substitution to an AU-paired RNA would move the amino group to the minor groove of the RNA (Figure 2 B). Indeed, 2-aminopurine substitutions to an AU-paired RNA altered the cleavage of bleomycin A5, indicating that hydrogen bonding interactions with the amino group could play a role in cleavage specificity (Figure 2 C).
Figure 2.
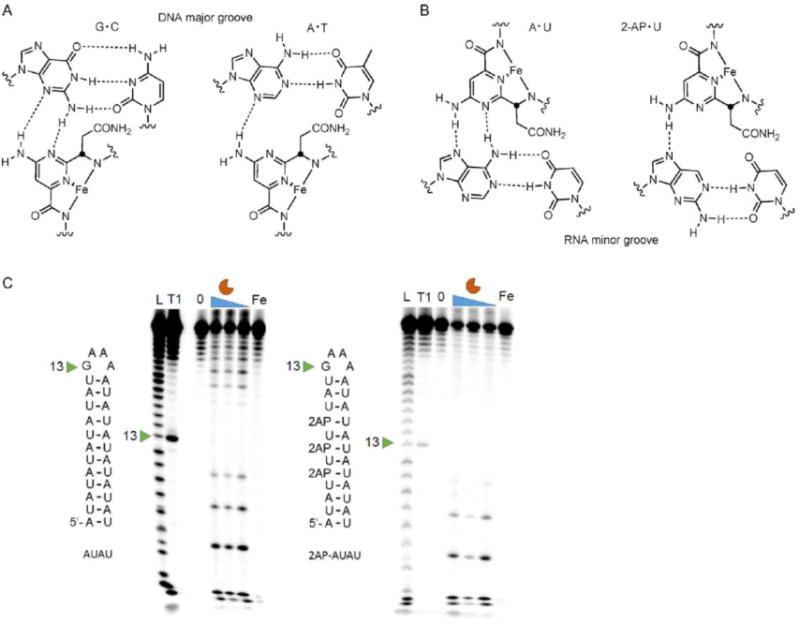
Probing the binding interaction of bleomycin with RNA. A) Bleomycin binds the minor groove of DNA and forms two hydrogen bonding interactions with guanine but only one with adenine. Figure adapted from ref. [10]. Copyright 1999, Wiley-VCH Verlag GmbH & Co. KGaA. B) Possible hydrogen binding interactions between bleomycin and adenine in the major groove of RNA. The hydrogen bond with the amino group of adenine is eliminated by 2-aminopurine substitution. C) In vitro cleavage of AU-paired RNA (left) and a 2-AP-substituted RNA (right). L indicates hydrolysis ladder; T1 indicates ribonuclease T1 ladder; 0 represents untreated pri-miR-10b; and Fe indicates RNA treated with 20 μm (NH4)3Fe(SO2)2·6H2O. Bleomycin A5 was evaluated at 20, 10, and 5 μM.
To determine whether bleomycin A5 would cleave naturally occurring, noncoding RNA containing AU-rich sequences, we searched all human miRNAs using bioinformatics to identify those with the potential to be good substrates for cleavage.[15] Among all human miRNAs, 13 miRNA precursors have a stretch of six consecutive AU base pairs in their secondary structures (Table S1). Of these 13 miRNAs, seven have been implicated in disease (Table S1). We identified the miR-10b primary hairpin precursor (pri-miR-10b) that contains a potentially reactive AU-rich region: 5′-AUAUAU/3′UAUAUA. The miR-10b is oncogenic and is overexpressed in many cancers, contributing to invasion and metastasis.[16] The putative target site for bleomycin A5 in pri-miR-10b is composed of the six consecutive AU pairs adjacent to the Drosha cleavage site (Figure 3 A). A small molecule that inhibits the biogenesis of miR-10b by binding to the Drosha cleavage site has previously been identified;[17] however, small molecule cleavage of miR-10b in particular, and miRNAs precursors in general, has not been studied.
Figure 3.
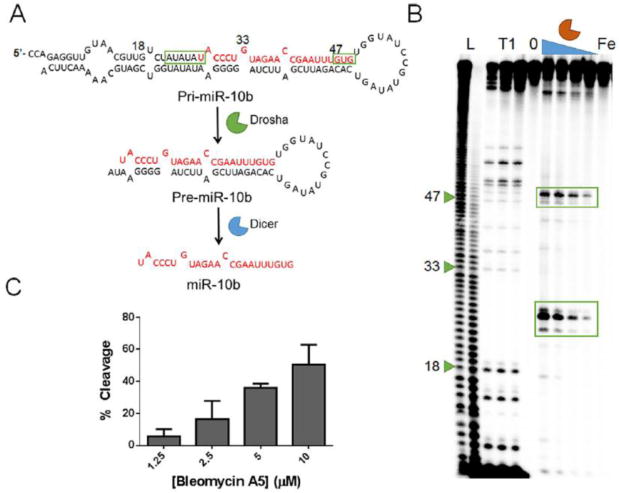
Bleomycin cleavage of pri-miR-10b. A) Pri-miR-10b is first processed by Drosha in the nucleus to produce pre-miR-10b, which is then processed by Dicer in the cytoplasm to produce mature miR-10b. B) In vitro cleavage of pri-miR-10b. L indicates hydrolysis ladder; T1 indicates ribonuclease T1 ladder; 0 represents untreated pri-miR-10b, and Fe indicates RNA treated with 10 μm (NH4)2Fe(SO2)2·6H2O. Bleomycin A5 was evaluated at 10, 5, 2.5, and 1.25 μm. C) Quantification of pri-miR-10b in vitro cleavage by bleomycin A5.
We thus analyzed in vitro cleavage of pri-miR-10b. Indeed, bleomycin A5 cleaved the target at the predicted site and also had a secondary site of cleavage near the Dicer site of 5′GUG/3′CAC. The secondary cleavage was not surprising, as the target contains two purines, and these nucleotides were cleaved at the short hairpin RNAs that were initially studied for cleavage. Other minor cleavage sites included other purine-rich sequences, such as 5′-GAA-3′ sequences. Further analysis of the cleavage sites revealed that bleomycin A5 also cleaved the 5′-AUAUAU-3′ sequence at the 3′-end of the RNA (Figure S5). To confirm the sequence-specific cleavage of pri-miR-10b by bleomycin A5, the AU pairs were mutated to GC pairs, and the 5′GUG/3′CAC sequence was mutated to a 5′GCG/3′CGC sequence. At a concentration of 10 μm, bleomycin A5 cleaved the mutated RNA 30 % less than pri-miR-10b; cleavage was completely ablated at lower concentrations (Figure S6).
With the favorable activity and selectivity data observed in vitro, we sought to assess the effect of the compound on miR-10b and its precursors in cells. Because the cleavage sites for bleomycin A5 are near the Drosha and Dicer enzyme cleavage sites in the miRNA hairpin precursor, it is possible that small molecule cleavage of the miRNA could allow it to enter the miRNA pathway, or it could cause destruction of the miRNA. Therefore, HeLa (human cervical cancer) cells overexpressing pri-miR-10b through plasmid expression were treated with bleomycin A5. Treatment with as little as 100 nm bleomycin A5 resulted in a decrease in the levels of pri-miR-10b, as determined by RT-qPCR (Figure 4 A). Cleavage of pri-miR-10b resulted in a decrease in the levels of the mature microRNA as well (Figure 4 A). Interestingly, bleomycin A5 reduced the levels of mature miRNA at concentrations as low as 10 nm. Although bleomycin A5 cleaved the RNA near the Drosha cleavage site in vitro, chemical cleavage of pri-miR-10b in cells resulted in destruction of mature miR-10b. Importantly, in this transfected system, bleomycin A5 did not affect the levels of plasmid DNA (Figure S7). In an additional cellular experiment, the effect of bleomycin A5 on the triple-negative breast cancer cell line MDA-MB-231 was assessed. This cell line was used because it endogenously expresses higher levels of miR-10b than healthy, noncancerous cells.[16a] In MDA-MB-231 cells, bleomycin A5 at concentrations of 10 and 1 μm reduced the amount of mature miR-10b, showing that bleomycin A5 can also cleave endogenous pri-miR-10b (Figure 4 B).
Figure 4.
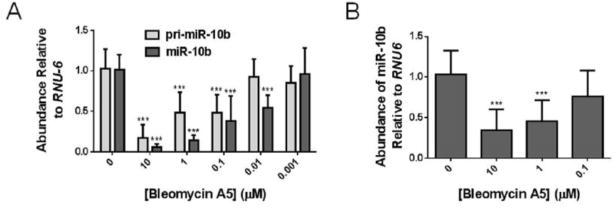
Bleomycin cleavage of miRNA-10b in cells. A) Bleomycin A5 cleaves pri-miR-10b in HeLa cells overexpressing pri-miR-10b, resulting in a decrease in mature miR-10b. B) Bleomycin A5 reduces the levels of endogenous mature miR 10b in MDA-MB-231 cells.
In summary, analysis of the preferred RNA cleavage site for bleomycin A5 revealed that bleomycin A5 cleaved 5′-AUAU-3′ sequences and other purine-rich sequences with high efficiency. The identification of a biologically relevant RNA with a stretch of six AU base pairs, pri-miR-10b, showed that bleomycin cleaved the RNA at the predicted site in vitro. In cells, treatment with bleomycin A5 also resulted in cleavage of pri-miR-10b. This study shows that noncoding RNAs such as miRNAs can be targets of known drugs. Other targets of bleomycin outside of the canonical DNA targets could include RNA, which should now be considered a druggable target from phenotypic screens, for example. Furthermore, elucidation of the preferred cleavage site of bleomycin might help in designing selective RNA cleavers by attaching an RN- binding module to bleomycin A5 to target a specific RNA, thereby reprograming its cellular targets.[18] Importantly, bleomycin A5 might also serve as a useful reagent to map RNA AU base pairs in vitro and to map RNA secondary structure in cells.
Supplementary Material
Acknowledgments
This work was funded by the US National Institutes of Health grant R01-GM097455 to M.D.D. We thank Jessica Childs-Disney and Suzanne Rzuczek for editing the manuscript and providing experimental advice.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.a. Clamp M, Fry B, Kamal M, Xie X, Cuff J, Lin MF, Kellis M, Lindblad-Toh K, Lander ES. Proc Natl Acad Sci USA. 2007;104:19428–19433. doi: 10.1073/pnas.0709013104. [DOI] [PMC free article] [PubMed] [Google Scholar]; b. The ENCODE Project Consortium. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mercer TR, Dinger ME, Mattick JS. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garzon R, Calin GA, Croce CM. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 5.a. Kole R, Krainer AR, Altman S. Nat Rev Drug Discovery. 2012;11:125–140. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]; b. Keller W, Crouch R. Proc Natl Acad Sci USA. 1972;69:3360–3364. doi: 10.1073/pnas.69.11.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]; b. Blount KF, Megyola C, Plummer M, Osterman D, O’Connell T, Aristoff P, Quinn C, Chrusciel RA, Poel TJ, Schostarez HJ, Stewart CA, Walker DP, Wuts PG, Breaker RR. Antimicrob Agents Chemother. 2015;59:5736–5746. doi: 10.1128/AAC.01282-15. [DOI] [PMC free article] [PubMed] [Google Scholar]; c. Howe JA, Wang H, Fischmann TO, Balibar CJ, Xiao L, Galgoci AM, Malinverni JC, Mayhood T, Villafania A, Nahvi A, Murgolo N, Barbieri CM, Mann PA, Carr D, Xia E, Zuck P, Riley D, Painter RE, Walker SS, Sherborne B, de Jesus R, Pan W, Plotkin MA, Wu J, Rindgen D, Cummings J, Garlisi CG, Zhang R, Sheth PR, Gill CJ, Tang H, Roemer T. Nature. 2015;526:672–677. doi: 10.1038/nature15542. [DOI] [PubMed] [Google Scholar]
- 7.a. Velagapudi SP, Gallo SM, Disney MD. Nat Chem Biol. 2014;10:291–297. doi: 10.1038/nchembio.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]; b. Disney MD, Winkelsas AM, Velagapudi SP, Southern M, Fallahi M, Childs-Disney JL. ACS Chem Biol. 2016;11:1720–1728. doi: 10.1021/acschembio.6b00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bose D, Jayaraj G, Suryawanshi H, Agarwala P, Pore SK, Banerjee R, Maiti S. Angew Chem Int Ed. 2012;51:1019–1023. doi: 10.1002/anie.201106455. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2012;124:1043–1047. [Google Scholar]
- 9.Stubbe J, Kozarich JW. Chem Rev. 1987;87:1107–1136. [Google Scholar]
- 10.Boger DL, Cai H. Angew Chem Int Ed. 1999;38:448–476. doi: 10.1002/(SICI)1521-3773(19990215)38:4<448::AID-ANIE448>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]; Angew Chem. 1999;111:470–500. [Google Scholar]
- 11.Carter BJ, de Vroom E, Long EC, van der Marel GA, van Boom JH, Hecht SM. Proc Natl Acad Sci USA. 1990;87:9373–9377. doi: 10.1073/pnas.87.23.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Varani G, McClain WH. EMBO Rep. 2000;1:18–23. doi: 10.1093/embo-reports/kvd001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu W, Vanderwall DE, Turner CJ, Kozarich JW, Stubbe J. J Am Chem Soc. 1996;118:1281–1294. [Google Scholar]
- 14.Bailly C, Waring MJ. J Am Chem Soc. 1995;117:7311–7316. [Google Scholar]
- 15.Kozomara A, Griffiths-Jones S. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.a. Ma L, Teruya-Feldstein J, Weinberg RA. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]; b. Ma L, Reinhardt F, Pan E, Soutschek J, Bhat B, Marcusson EG, Teruya-Feldstein J, Bell GW, Weinberg RA. Nat Biotechnol. 2010;28:341–347. doi: 10.1038/nbt.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Velagapudi SP, Disney MD. Chem Commun. 2014;50:3027–3029. doi: 10.1039/c3cc00173c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.S. G. Rzuczek, L. A. Colgan, Y. Nakai, M. D. Cameron, D. Furling, R. Yasuda, M. D.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


