Abstract
Hepatic fibrosis is a physiological response to liver injury that includes a range of cell types. The pathogenesis of hepatic fibrosis currently focuses on hepatic stellate cell (HSC) activation into muscle fiber cells and fibroblasts. Baicalin is a flavone glycoside. It is the glucuronide of baicalein, which is extracted from the dried roots of Scutellaria baicalensis Georgi. Previous work focused on the anti-viral, -inflammatory and -tumor properties of baicalin. However, the potential anti-fibrotic effects and mechanisms of baicalin are not known. The present study demonstrated that baicalin influenced the activation, proliferation, apoptosis, invasion and migration of platelet-derived growth factor-BB-induced activated HSC-T6 cells in a dose-dependent manner. To investigate the anti-fibrotic effect of baicalin, a one-color micro (mi)RNA array and reverse transcription-quantitative polymerase chain reaction analyses were used. Results demonstrated that baicalin increased the expression of the miRNA, miR-3595. In addition, the inhibition of miR-3595 substantially reversed the anti-fibrotic effect of baicalin. The present data also suggested that miR-3595 negatively regulates the long-chain-fatty-acid-CoA ligase 4 (ACSL4). Furthermore, ACSL4 acted in a baicalin-dependent manner to exhibit anti-fibrotic effects. Taken together, it was concluded that baicalin induces miR-3595 expression that modulates the expression levels of ACSL4. To the best of our knowledge, the present study is the first to demonstrate that baicalin induces overexpression of human miR-3595, and subsequently decreases the expression of ACSL4, resulting in an anti-fibrotic effect.
Keywords: hepatic fibrosis, baicalin, miR-3595, long-chain-fatty-acid-CoA ligase 4
Introduction
Hepatic fibrosis, a spontaneous wound healing response, occurs when the liver has an acute or chronic injury caused by viral infection, drug damage or immune attack (1,2). Fibrosis impairs tissue and organ function and is caused by excessive deposition of the extracellular matrix (ECM) that occurs as a result of inflammation (3,4). Liver fibrosis markedly contributes to morbidity in patients with liver disease (5). Hepatic stellate cells (HSCs) are pivotal in the development of hepatic fibrosis (6) and have the ability to proliferate and differentiate into myofibroblast-like cells (7). Platelet-derived growth factor (PDGF) signaling is one of the best characterized pathways of HSC activation (8) and induces apoptosis, senescence or quiescence in HSCs (9). Thus, inhibiting the activation of HSCs may be a feasible therapeutic strategy for hepatic fibrosis.
Various agents, including malotilate, genistein, curcumin and silymarin, have been reported to be effective against fibrosis in vitro and in vivo (10,11); however, none of these are ideal candidates for inhibiting the activation of HSCs. Currently, an increasing number of natural products derived from herbal extracts are used in the treatment of diseases such as liver fibrosis; however, the different mechanisms through which they work are not known (12). Baicalin (Fig. 1), is the major bioactive flavonoid extracted from dried roots of the herb Scutellaria baicalensis Georgi (13). Baicalin exhibits a range of pharmacological effects that include antioxidative, anti-viral, and anti-inflammatory properties, as well as liver protection (14). Previous reports have indicated that baicalin has significant anti-fibrogenic effects that act by suppressing the signaling of canonical Wnts (15,16). Baicalin also inhibits hepatic fibrosis by reducing the level of pro-fibrotic cytokines, including transforming growth factor (TGF)-β1, tumor necrosis factor-α, interleukin (IL)-6 and IL-22. The reduction of these cytokines significantly enhances HSC senescence or apoptosis and reduce the activation of HSCs (17,18). To expand the application of baicalin, it is necessary to further elucidate the mechanisms of the anti-fibrotic effect of baicalin.
Figure 1.
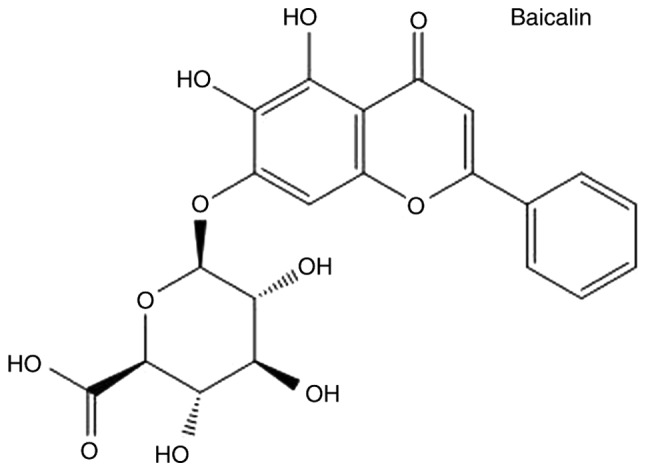
Chemical structure of baicalin.
MicroRNA (miRNA) are short RNA molecules (~22 nucleotides) (19) that typically bind to the 3′ untranslated region (3′UTR) of specific mRNA to regulate gene expression, either by repressing mRNA translation or promoting repression. They are involved in diverse physiological and developmental processes (20,21) and regulate all biological processes in all cell types, such as cells in the liver. Previous research suggests that miRNA are involved in the regulation of HSC activation (7). Thus, modulating miRNA represents a novel therapeutic approach for hepatologists in the treatment of liver disease.
Long-chain-fatty-acid-CoA ligase 4 (ACSL4) is a member of the long-chain acyl-CoA synthetase (ACSL4) family that preferentially uses arachidonate and eicosapentaenoate as substrates, and activates long-chain fatty acids for the synthesis of cellular lipids (22,23). ACSL4 is associated with various liver diseases (24) and is aberrantly regulated in liver cancer (25).
The present study demonstrated the anti-fibrogenic property of baicalin in PDGF-BB-induced HSCs. miRNA profiles of PDGF-BB-induced activated HSC-T6 cells treated with and without baicalin were compared. Results indicated that miR-3595 was upregulated in baicalin-treated cells compared with the levels in the untreated cells. It was also revealed that ACSL4 is a potential functional target of miR-3595. These findings indicate the central role of baicalin in inhibiting the activation of HSCs and suggest that baicalin has the potential to be an effective natural medicine for the treatment of hepatic fibrosis. The present results support ongoing research to identify potential therapeutic targets for liver fibrosis.
Materials and methods
Cell culture and treatment
HSC-T6 cells were purchased from American Type Culture Collection (Manassas, VA, USA) and were cultured in Dulbecco's modified Eagle medium (DMEM; cat. no. 11965-092; Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; cat. no. 10099-141; Invitrogen; Thermo Fisher Scientific, Inc.). The cells were grown in a humidified atmosphere maintained at 5% CO2 at 37°C. The cells were serum-starved in an FBS-free medium for 24 h and then induced with PDGF-BB (10 ng/ml; cat. no. P3201; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), following the protocol in a previous study (26). The cells were treated with baicalin at different concentrations (50, 100 and 150 µM; cat. no. 572667; Sigma-Aldrich; Merck KGaA) for the indicated time according to different experiments at 37°C. The same volume of diluted dimethyl sulfoxide (DMSO) was used as a negative control. While the baicalin stock solution was in DMSO, it was ensured that the concentration of DMSO did not influence cell survival. Plasmids [ACSL4 wild-type (WT) and mutant (MUT); constructed by Shanghai GenePharma Co., Ltd., Shanghai, China] and miR-3595 inhibitors (5′-AUCGUAGAGGAAAAUCCAC-3′) or negative control (NC; 5′-CAGUACUUUUGUGUAGUACAA-3′) (Shanghai GenePharma Co., Ltd.) were transfected into cells using Lipofectamine® 2000 (cat. no. 11668027; Invitrogen; Thermo Fisher Scientific, Inc.) at a concentration of 50 nM, following the manufacturer's instructions. After 48 h of transfection, cells were treated with different reagents for different experiments.
Cell proliferation assay
The induced HSC-T6 cells (1×104 cells/well) were seeded into 96-well plates and treated with baicalin for 72 h at 37°C. Cell proliferation was assessed using a Cell Counting Kit-8 (CCK-8) assay (cat. no. 96992; Sigma-Aldrich; Merck KGaA), according to the manufacturer's protocol. The optical density was measured at 450 nm.
RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA from HSC-T6 cells was extracted using TRIzol reagent (cat. no. 15596026; Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Total RNA was reverse-transcribed to cDNA by using a Bestar qPCR RT kit (cat. no. 210212; Qiagen GmbH, Hilden, Germany) following the manufacturer's protocol. Following this, cDNA was quantified by qPCR on a Mx3000p real-time PCR system with DBI Bestar® SYBRGreen qPCR master mix (cat. no. DBI-2073; DBI Bioscience, Ludwigshafen, Germany) in a 20-µl PCR reaction. qPCR was performed using the following settings: Pre-denaturation at 95°C for 10 sec, followed by 40 cycles of denaturation at 95°C for 10 sec and elongation at 60°C for 30 sec. Results were normalized to β-actin expression. The 2−ΔΔCq method was used for calculating the relative expression levels (27). The PCR primer sequences are listed in Table I.
Table I.
Primer sequences used for microRNA and mRNA expression analysis.
| Primer | Sequence (5′-3′) | Length, bp |
|---|---|---|
| β-actin F | GGAGATTACTGCCCTGGCTCCTA | 150 |
| β-actin R | GACTCATCGTACTCCTGCTTGCTG | |
| miR-3595 reverse transcription | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGATCGTAGA | |
| miR-3595 F | ACACTCCAGCTGGGGTGGATTTTCCTCTA | |
| All R | CTCAACTGGTGTCGTGGA | |
| ACSL4 F | AAGGAGAAGGGCAAAGAGA | 211 |
| ACSL4 R | GGTGGTTGTAGGAGGCTGA |
F, forward; R, reverse; ACSL4, long-chain-fatty-acid-CoA ligase 4; All R, reverse primer for miRNA qPCR.
Western blot analysis
Cells were lysed with radioimmunoprecipitation assay buffer (cat. no. R0278; Sigma-Aldrich; Merck KGaA) supplemented with protease inhibitor cocktail (cat. no. ab201111; Abcam, Cambridge, UK) for 30 min on ice. The protein concentration was determined using a Bio-Rad protein assay kit (cat no. 5000002; Bio-Rad Laboratories, Inc., Hercules, CA, USA). A total of 20 µg/lane protein was separated on 10% SDS-PAGE and transferred to a polyvinylidene difluoride membrane. The membranes were blocked with 5% skim milk for 1 h at room temperature. The membranes were incubated overnight with the following specific primary antibodies: α-smooth muscle actin (SMA; 1:1,000; cat. no. A5228; Sigma-Aldrich; Merck KGaA), collagen (Col) I (1:1,000; cat. no. NB600-408; Novus Biologicals, LLC, Littleton, CO, USA), Col III (1:1,000; cat. no. ab6310; Abcam), TGF-β1 (1:1,000; cat. no. ab92486; Abcam), desmin (1:1,000; cat. no. ab8592; Abcam), GAPDH (1:4,000; cat. no. 14C10; Cell Signaling Technology, Inc., Danvers, MA, USA) and ACSL4 (1:1,000; cat. no. sc-365230; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) at 4°C. Following this, incubation was performed with horseradish peroxidase-conjugated secondary immunoglobulin antibodies (1:10,000; cat. no. A4416 and A6154; Sigma-Aldrich; Merck KGaA) for 1 h at room temperature. Enhanced chemiluminescence chromogenic substrate (cat. no. 32109; Invitrogen; Thermo Fisher Scientific, Inc.) was used and signals were recorded on an X-ray film. β-actin was chosen as the loading control.
Cell invasion and motility assay
A cell migration assay was performed using Transwell chambers containing 8-µm pore size filters. For the invasion assay, the inserts were coated with thin Matrigel. A total of 6×104 HSC-T6 cells in 200 µl FBS-free medium were added to each upper chamber. A total of 500 µl DMEM supplemented with 10% FBS was added to the lower chamber as a chemoattractant. Following incubation for 24 and 36 h at 37°C, the cells that migrated through the filter pores were fixed with 70% ethanol at room temperature for 20 min and stained with crystal violet for 10 min at room temperature. The results were calculated from five random fields under a light microscope (magnification, ×400).
Cell cycle analysis
The cells were trypsinized, collected by centrifugation at 300 × g for 5 min at room temperature and washed with PBS. The harvested cells were subsequently fixed in pre-cooled 70% ethanol, and stored at 4°C overnight. The cells were then washed with PBS and treated with DNase-free RNase (cat. no. 11119915001; Sigma-Aldrich; Merck KGaA) and stained with propidium iodide in the dark for 15 min at room temperature. Cell samples were analyzed by using a FACSCalibur (BD Biosciences, Franklin Lakes, NJ, USA).
Analysis of apoptosis
The cells were trypsinized and collected by centrifugation at 300 × g for 5 min at room temperature. The harvested cells were washed with PBS and stained with annexin V-fluorescein isothiocyanate and propidium iodide (PI; cat. no. 556547; BD Biosciences), according to the manufacturer's protocol. Cell samples were analyzed using a flow cytometer within 1 h of staining. The results were analyzed by FlowJo version 10 (BD Biosciences).
miRNA microarray
The total RNA extracted using TRIzol was purified and RNA quality was assessed using a NanoDrop ND-1000 (Thermo Fisher Scientific, Inc., Wilmington, DE, USA). miRNA array experiments were conducted, according to the manufacturer's instructions. Total RNA was labeled using a miRCURY™ Array Power Labeling kit (cat. no. 208500; Exiqon A/S, Vedbaek, Denmark). An Axon GenePix 4000B microarray scanner (Molecular Devices, LLC, Sunnyvale, CA, USA) was utilized to obtain the scanning profiles. Following normalization, the statistical significance of differentially expressed miRNA was analyzed by a Student's t-test (GraphPad Prism 5; GraphPad Software, Inc., La Jolla, CA, USA).
Plasmid construction
To overexpress ACSL4, full-length Homo sapiens ACSL4 sequences from the National Center for Biotechnology Information database (Gene ID: 2182; ncbi.nlm.nih.gov/nuccore/NM_001318510.1) were cloned into the pcDNA3.0 vector (cat. no. V790-20; Invitrogen; Thermo Fisher Scientific, Inc.). To generate the luciferase reporter plasmid, the 3′UTR of WT ACSL4 was cloned into the pLUC vector (cat. no. 42094; Addgene, Inc., Cambridge, MA, USA). Mutations in the 3′UTR of the WT ACSL4 were designed for four nucleotides (AAAUCC to ACGUAA) of the 6-mer seed region of the putative binding site of miR-3595 (pLUC-MUT-UTR). All DNA constructs were sequenced.
Dual-luciferase reporter assay
The luciferase reporter plasmid pLUC was purchased from Addgene, Inc., (cat. no. 42094). For the dual-luciferase assay, HSC-T6 cells were transiently transfected with either the pLUC-WT-UTR plasmid or the pLUC-MUT-UTR plasmid and miR-3595 mimics or control NC mimics using Lipofectamine® 2000. After 48 h, cells were washed with PBS, lysed with lysis buffer and luciferase activity was measured by using the Dual-Luciferase Reporter Assay kit (cat. no. E1910; Promega Corporation, Madison, WI, USA). Results were normalized to Renilla luciferase activity.
miRNA targets mRNA prediction
Target mRNA of miRNA were identified using the following two websites: microRNA.org and targetscan.org.
Statistical analysis
Data were presented as the mean ± standard deviation. Statistical analyses were performed using SPSS 17.0 (SPSS, Inc., Chicago, IL, USA) in all the pertinent experiments. Differences between groups were analyzed using the Student's t-test or one-way analysis of variance followed by a Tukey's test. P<0.05 was considered to indicate a statistically significant difference.
Results
Baicalin inhibits the proliferation, activation, apoptosis and cell cycle progression of activated HSC-T6 cells induced by PDGF-BB
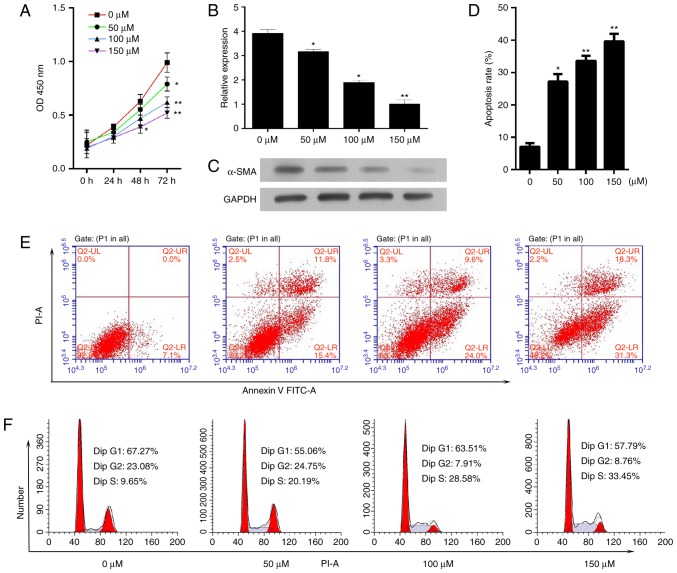
The present study aimed to determine the effect of baicalin on cell proliferation and survival. Activated HSC-T6 cells were treated with different concentrations of baicalin (50, 100 or 150 µM) for 72 h. A CCK-8 assay was utilized to measure cell proliferation and it was demonstrated that baicalin decreased the proliferative ability of the induced HSC-T6 cells (Fig. 2A). As a critical event in fibrosis is the upregulation of proteins, such as α-SMA, the mRNA and protein expression levels of α-SMA in activated HSC-T6 cells following treatment with different concentrations of baicalin for 48 h were determined. The mRNA and protein expression levels of α-SMA were significantly decreased compared with the levels in the untreated group (Fig. 2B and C). Subsequently, the effect of baicalin on apoptosis and cell cycle progression was determined. It was demonstrated that baicalin treatment significantly increased the number of apoptotic cells (Fig. 2D and E). In addition, baicalin treatment resulted in a notable increase in the number of cells in S phase and a decrease in the number of cells in the G0/G1 phase compared with the numbers in the untreated group (Fig. 2F).
Figure 2.
Baicalin inhibits the proliferation and activation and increases the apoptosis of PDGF-BB-induced HSC-T6 cells. PDGF-BB-induced HSC-T6 cells were treated with baicalin at different concentrations (0, 50, 100 or 150 µM). (A) A Cell Counting Kit-8 assay was used to determine cell proliferation every 24 h for 3 days. The expression levels of α-SMA (B) mRNA and (C) protein were determined by reverse transcription-quantitative polymerase chain reaction and western blotting, respectively. (D and E) The apoptosis rate of HSC-T6 cells was measured by flow cytometry. (F) The effect of baicalin on the cell cycle in PDGF-BB-induced HSC-T6 cells was determined. *P<0.05 and **P<0.01 vs. untreated cells. PDGF, platelet-derived growth factor; α-SMA, α-smooth muscle actin; PI, propidium iodide; FITC, fluorescein isothiocyanate; OD, optical density.
Baicalin decreases epithelial-to-mesenchymal transition (EMT) of activated HSC-T6 cells induced by PDGF-BB
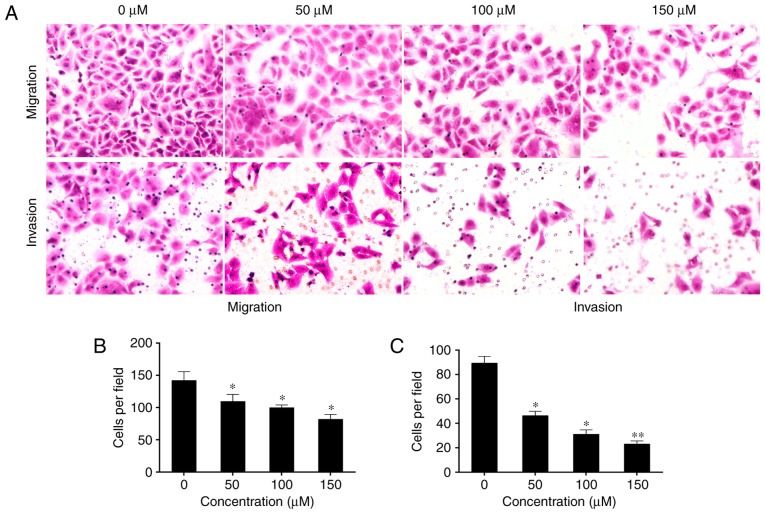
Epithelial cells lose their polarity as a result of intercellular adhesion complexes changing their phenotypes. They may then move through the ECM like mesenchymal cells, and this process is termed EMT (28). Activated HSCs exhibit a significant elevation in mesenchymal and epithelial markers, which suggests that they undergo EMT (12). Therefore, the present study aimed to determine the effect of baicalin on the invasion and migration of induced HST-T6 cells. Treatment with baicalin significantly inhibited the motility and invasive ability of the induced HST-T6 cells in a dose-dependent manner compared with that observed in untreated cells (P<0.05; Fig. 3).
Figure 3.
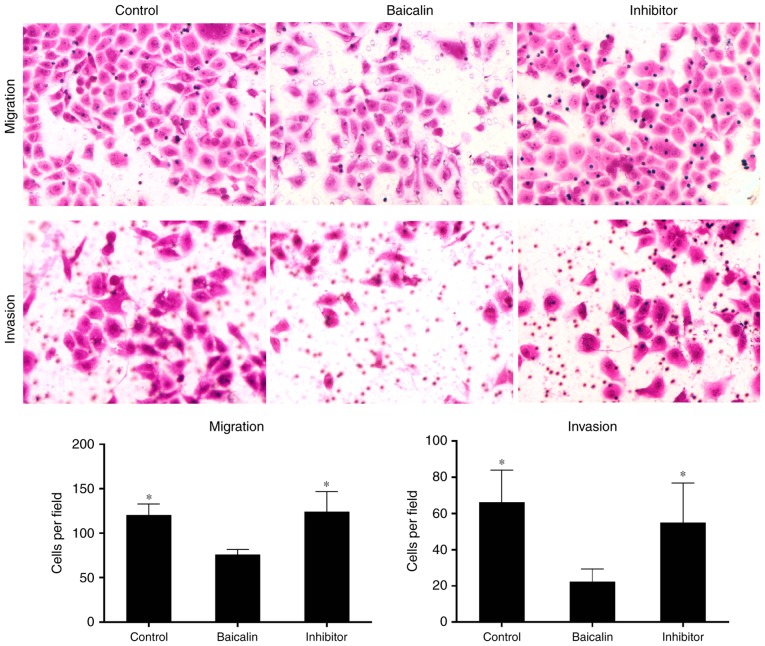
Baicalin inhibits the motility and invasive potential of PDGF-BB-induced HSC-T6 cells. (A) PDGF-BB-induced HSC-T6 cells were treated with baicalin at different concentrations (0, 50, 100 or 150 µM) and the migration and invasion rates of the cells were measured. Baicalin at 50, 100 and 150 significantly inhibited the (B) migration (magnification, ×400) and (C) invasion (magnification, ×400) of PDGF-BB-induced HSC-T6 cells compared with untreated cells. However, even though the ability of the cells to migrate and invade decreased with the increase in baicalin concentration, no significant differences were observed between the different baicalin concentrations. *P<0.05 and **P<0.01 vs. untreated cells. PDGF, platelet-derived growth factor.
miR-3595 serves an important role in the anti-fibrotic effect of baicalin
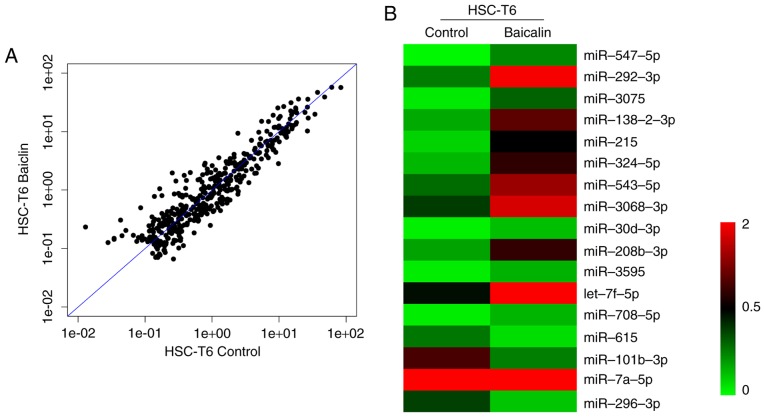
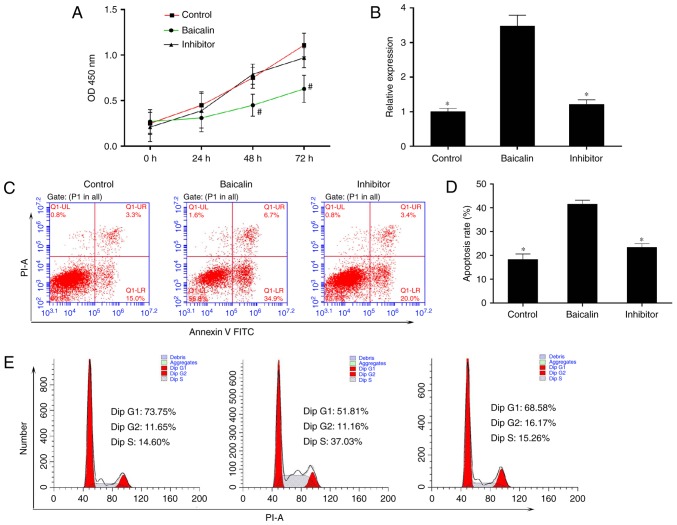
To identify the miRNA involved in the anti-fibrotic effect of baicalin, miRNA expression profiles were compared between induced HSC-T6 cells treated with 100 µM baicalin and untreated cells, using a one-color miRNA array. miR-547-5p, miR-296-3p and miR-3595 were dysregulated in the baicalin-treated group when compared with the control group (Fig. 4). To further validate the microarray data, the expression levels of miR-3595 were compared between the two groups of cells using RT-qPCR. As demonstrated in Fig. 5, miR-3595 was significantly upregulated in baicalin-treated cells compared to the level in the untreated cells (P<0.05; Fig. 5B). In addition, the cell proliferation rate was rescued by miR-3595 inhibitor compared with that in the baicalin-treated group, as measured by a CCK8 assay (Fig. 5A). Cellular apoptosis and cell cycle effects of the miR-3395 inhibitor were also detected. As demonstrated in Fig. 5C and D, baicalin significantly induced cell apoptosis compared with the level in the control group, while the miR-3595 inhibitor reversed baicalin-induced cell apoptosis. Baicalin treatment induced an increase in the number of cells in S phase and decreased the number of cells in the G0/G1 phase compared with the control group; however, miR-3595 inhibitor reversed these baicalin-induced effects (Fig. 5E). Furthermore, miR-3595 inhibitor recovered the baicalin-induced inhibitory effects on cell migration and invasion (Fig. 6). These results suggest that inhibiting miR-3595 significantly suppressed the anti-fibrotic effect of baicalin.
Figure 4.
Analysis of miRNA expression profile using a one-color miRNA array. Comparison of differentially expressed miRNA in baicalin-treated platelet-derived growth factor-BB-induced HSC-T6 cells with untreated HSC-T6 cells. (A) Scatter plot of miRNA expression profiles. (B) Clustering of differentially expressed miRNA. miRNA, microRNA.
Figure 5.
miR-3595 serves an important role in inhibiting cell proliferation and increasing apoptosis in PDGF-BB-induced HSC-T6 cells treated with baicalin. PDGF-BB-induced HSC-T6 cells were treated with baicalin at a concentration of 100 µM. The inhibitor group consisted of cells that were transfected with miR-3595 inhibitors in addition to treatment with baicalin. (A) A Cell Counting Kit-8 assay was used to determine the cell proliferation every 24 h for 3 days. (B) The expression of miR-3595 in each group was determined by quantitative reverse transcription-quantitative polymerase chain reaction. (C and D) The apoptosis rate of HSC-T6 cells in each group was measured by flow cytometry. (E) Baicalin regulated the cell cycle of PDGF-BB-induced HSC-T6 cells through miR-3595. #P<0.05 vs. control and inhibitor groups; *P<0.05 vs. baicalin only-treated group. PDGF, platelet-derived growth factor; OD, optical density; PI, propidium iodide; FITC, fluorescein isothiocyanate.
Figure 6.
miR-3595 serves an important role in inhibiting the migration and invasion of PDGF-BB-induced HSC-T6 cells treated with baicalin. PDGF-BB-induced HSC-T6 cells were treated with 100 µM baicalin. The cells in the inhibitor group were transfected with miR-3595 inhibitors in addition to treatment with baicalin. Representative numbers of cells undergoing migration or invasion was counted and captured (magnification, ×400). *P<0.05 vs. baicalin only-treated group. PDGF, platelet-derived growth factor.
To determine the role of miR-3595 on the anti-fibrotic effect of baicalin, the protein levels of α-SMA, Col I, Col III, TGF-β and desmin were measured. As demonstrated in Fig. 7, the expression levels of these proteins in activated HCS-T6 cells treated with 100 µM baicalin and miR-3595 inhibitor were similar to those in the activated HSC-T6 cells without any treatment, while the expression levels of these proteins in the baicalin-treated activated HCS-T6 cells were markedly decreased. Taken together, the present results suggest that miR-3595 regulates the anti-fibrotic effects of baicalin with respect to cell proliferation, apoptosis, invasion and migration, and fibrosis-related protein expression levels.
Figure 7.
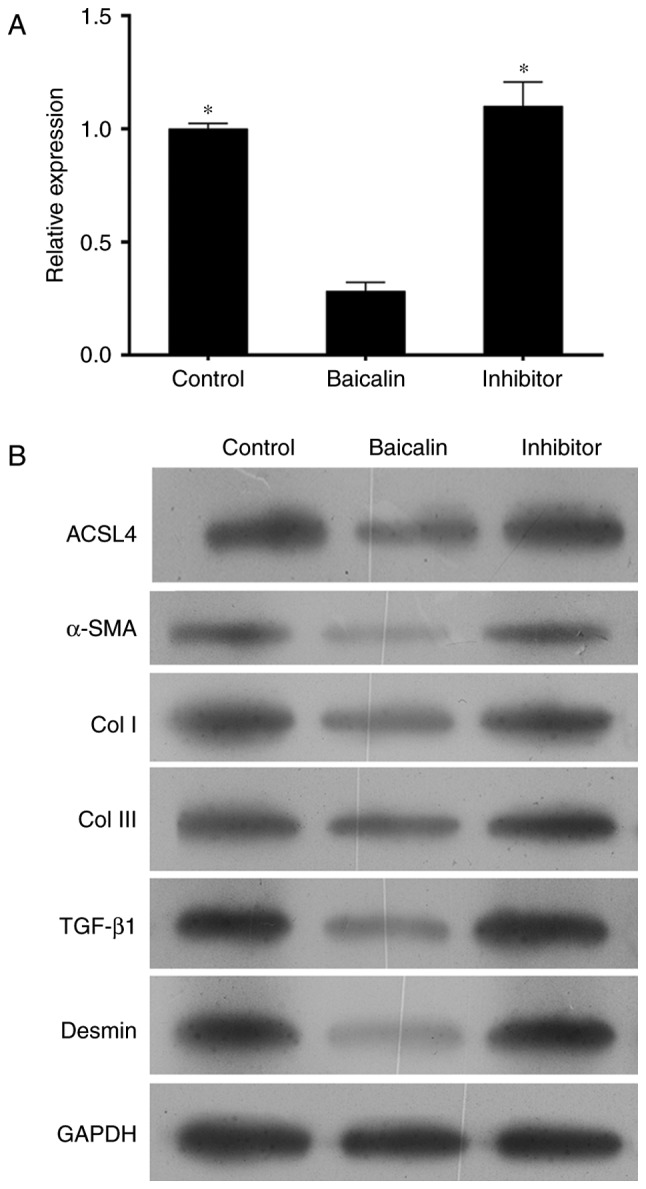
Western blot analysis of protein expression levels in activated hepatic stellate cells treated with miR-3595 inhibitors and baicalin. Platelet-derived growth factor-BB-induced HSC-T6 cells were treated with 100 µM baicalin. The inhibitor group consisted of cells that were transfected with miR-3595 inhibitors in addition to baicalin. (A) Cells were harvested to analyze the mRNA expression level of ACSL4 by reverse transcription-quantitative polymerase chain reaction and (B) the protein expression levels of ACSL4, α-SMA, Col I, Col III, TGF-β1 and desmin by western blot analysis. *P<0.05 vs. baicalin only-treated cells. ACSL4, long-chain-fatty-acid-CoA ligase 4; α-SMA, α-smooth muscle actin; TGF-β1, transforming growth factor-β1; Col, collagen.
Effect of miR-3595 on baicalin-induced anti-fibrosis is through the regulation of ACSL4 expression
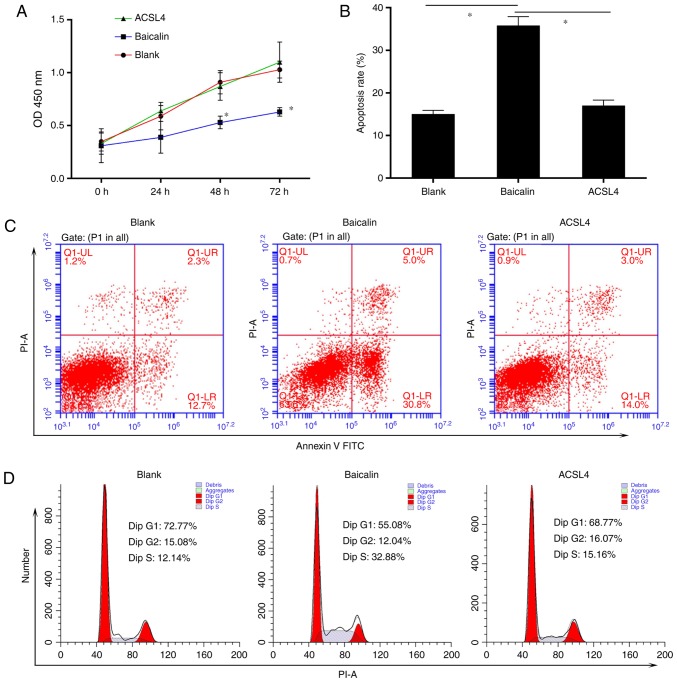
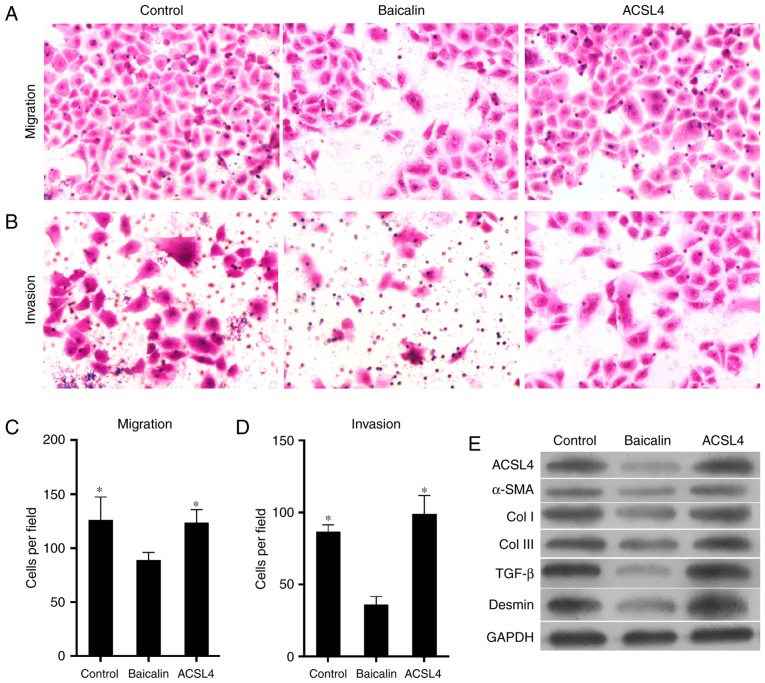
Previous studies indicate that the expression of ACSL4 is related to the synthesis of eicosanoid-CoA and the conversion of arachidonic acid into phosphatidyl ethanolamine, phosphatidyl inositol and triglyceride (29,30), and that it serves a role in liver disease. Thus, it was hypothesized that ACSL4 is a direct miR-3595 target. The perfect complementation between the miR-3595 'seed' sequence and the potential 'recognition' sequences in the 3′UTR of ACSL4 were observed (Fig. 8A), suggesting that miR-3595 downregulates ACSL4 expression by inhibiting the translation of target mRNA. To confirm this hypothesis, the induced HSC-T6 cells were co-transfected with ACSL4-3′UTR luciferase reporters and miR-3595 mimics. Results demonstrated that miR-3595 significantly decreased the luciferase activity of cells transfected with ACSL4-WT-3′UTR reporter compared with the level in the NC group (P<0.05); however, transfection of the ACSL4-MUT-3′UTR had no significant effect on luciferase activity (Fig. 8B). The mRNA and protein expression levels of ACSL4 in HSC-T6 cells decreased significantly on treatment with 100 µM baicalin compared with the level in the untreated group (P<0.05). These results indicate that miR-3595 negatively regulates ACSL4. To further validate these results, ACSL4 was overexpressed via a plasmid in HSC-T6 cells treated with 100 µM baicalin. Results demonstrated that the forced expression of ACSL4 attenuated the anti-fibrotic effect of baicalin by significantly reducing and apoptosis of drug-treated induced HST-T6 cells (P<0.05), and increasing the proliferation, invasion and migration of induced HST-T6 cells (P<0.05), and increasing expression levels of fibrosis-related proteins (Figs. 9 and 10). Therefore, miR-3595 targets ACSL4, which is involved in the regulation of the anti-fibrotic effect of baicalin.
Figure 8.
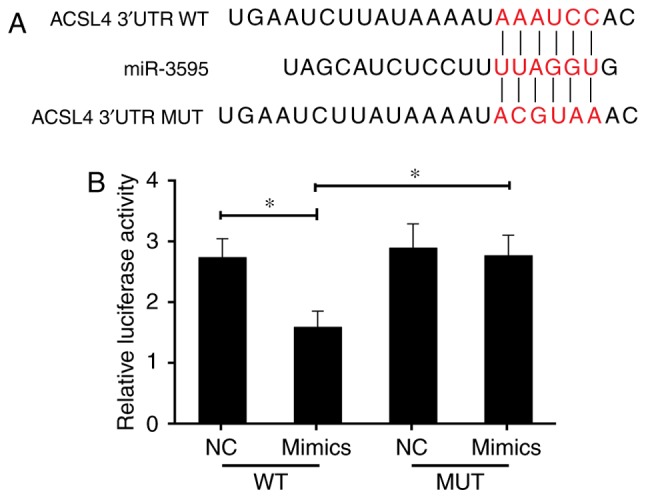
The anti-fibrosis effect of miR-3595 occurs through targeting ACSL4. (A) Sequence alignment between miR-3595 and the 3′UTR of human ACSL4 mRNA. The letters in red indicate the seed-mutated region. (B) Luciferase reporter assays demonstrated the effect of miR-3595 on the activity of the ACSL4 3′UTR and MUT ACSL4 3′UTR reporters. *P<0.05 as indicated. ACSL4, long-chain-fatty-acid-CoA ligase 4; UTR, untranslated region; WT, wild-type; MUT, mutant; NC, negative control.
Figure 9.
Exogenetic overexpression of ACSL4 attenuates the anti-fibrotic effect of baicalin on proliferation and apoptosis. Platelet-derived growth factor-BB-induced HSC-T6 cells were treated with 100 µM baicalin. One group of cells was transfected with ACSL4-overexpressing plasmid in addition to baicalin. (A) A Cell Counting Kit-8 assay was used to determine cell proliferation every 24 h for 3 days. (B and C) The apoptosis rate for each group of HSC-T6 cells was measured by flow cytometry. (D) The forced expression of ACSL4 attenuated the anti-fibrotic effect of baicalin on the cell cycle. *P<0.05 vs. ACSL4 and control group; #P<0.05 vs. baicalin-treated group. ACSL4, long-chain-fatty-acid-CoA ligase 4; OD, optical density; PI, propidium iodide; FITC, fluorescein isothiocyanate.
Figure 10.
Exogenetic overexpression of ACSL4 attenuates the anti-fibrotic effect of baicalin on cell migration and invasion, and influences the expression levels of pro-fibrotic proteins. Platelet-derived growth factor-BB-induced HSC-T6 cells were treated with 100 µM baicalin. One group of cells was transfected with ACSL4-overexpressing plasmid in addition to baicalin. Microscope images of cells undergoing (A) migration and (B) invasion (magnification, ×400). Representative number of cells undergoing (C) migration and (D) invasion was counted. (E) Protein expression levels of fibrosis-promoting proteins, including ACSL4, α-SMA, Col I, Col III, TGF-β and desmin were verified by western blotting. *P<0.05 vs. baicalin-treated group. ACSL4, long-chain-fatty-acid-CoA ligase 4; α-SMA, α-smooth muscle actin; TGF-β, transforming growth factor-β; Col, collagen.
Discussion
Liver fibrosis is a wound healing response to a variety of stimuli (31,32). The progression of liver fibrosis leads to a number of pathological and biochemical changes, such as the distortion of normal structural architecture of the liver, resulting in metabolic abnormalities (33) or hepatocellular carcinoma (34). Accumulating evidence suggests that several cell types are involved in the development of liver fibrosis (35). However, the activation of HSCs is a particularly critical process in the pathogenesis of liver fibrosis. Following liver injury, HSCs increase the expression of α-SMA, c-Myb, and myocyte enhancer factor-2, and acquire a myofibroblast-like phenotype (35,36). A complex network of intracellular events occurs during the activation of HSCs (37–43). Thus, one anti-fibrotic therapy is the modulation of the proliferation, activation and apoptosis of HSCs.
The ideal anti-fibrotic agent should be liver-specific and have low hepatotoxicity; however, currently these agents are not commercially available (33). Hence, natural products and herbal medicines are potential therapies for hepatic fibrosis. Baicalin is a bioactive flavonoid with a multitude of pharmacological activities that include inhibiting the effects of several viruses (44–46), a variety of inflammatory diseases (47–49) and various types of cancer (50). Notably, previous studies have demonstrated that baicalin serves an important role in the fibrotic liver by reducing oxidative stress and shifting the balance of cytokines from pro-fibrotic to anti-fibrotic in experimental animal models (13,51).
However, the mechanism of the anti-fibrotic effect of baicalin is not understood. To elucidate the anti-fibrotic function of baicalin and its possible mechanism, the present study treated activated HSC-T6 cells induced by PDGF-BB with baicalin at different concentrations (50, 100 or 150 µM). It was revealed that baicalin reduced the proliferation, invasion and migration of these cells. Baicalin also reduced the expression levels of α-SMA (a marker of activated HSCs), and increased the number of apoptotic cells, in a dose-dependent manner. These data suggested that baicalin inhibits the proliferation, apoptosis, invasion, migration and activation of PDGF-BB-treated HSCs.
miRNA have been implicated in various diseases, such as liver fibrosis, and several current therapeutic strategies involve the manipulation of miRNA levels (52). Thus, the present study aimed to verify whether baicalin regulated the expression of miRNA in induced HSC-T6 cells. miRNA profiling of baicalin-treated cells and untreated cells was performed, and the data indicated that some miRNA were differentially expressed between the two groups. One of these differentially expressed miRNA was miR-3595. The present study further confirmed the differential expression of miR-3595 using RT-qPCR. The present study, therefore, demonstrated that baicalin increases the expression of endogenous miR-3595. Furthermore, the present study indicated that silencing the expression of miR-3595 attenuated the anti-fibrotic effect of baicalin. These results suggested that miR-3595 upregulation serves an important role in the anti-fibrotic effect of baicalin. In humans, miRNA regulate gene expression by binding to the 3′UTR of mRNA or miRNA response elements, resulting in translational repression (53,54).
The liver is the central organ involved in fatty acid (FA) homeostasis (55). ACSL4, a member of the ACSL family, catalyzes the cellular metabolism of polyunsaturated FA in different tissues (56–58). ACSL4 was recently demonstrated to have various functions in different cell lines (59). ACSL4 also promotes cell invasion by regulating the generation of eicosanoids in breast cancer (60). Through prediction and experimental validation, the present study indicated that ACSL4 was a direct target of miR-3595. Indeed, exogenous overexpression of ACSL4 suppressed the anti-fibrotic effect of baicalin in induced HSC-T6 cells. However, the relevance of the relationship between ACSL4 and fibrosis remains unclear.
Thus, it may be speculated that the anti-fibrotic effect of baicalin is associated with the miR-3593/ACSL4 axis. Furthermore, the present study identified several differentially expressed miRNA that could be potential targets of baicalin. However, it is also possible that the anti-fibrotic effect of baicalin acts through signaling pathways other than miRNA-related ones. Baicalin may be a promising chemotherapy drug; however, further work is required to test this hypothesis.
In summary, the present findings suggest that baicalin inhibits the proliferation and activation observed in induced HSCs. One potential mechanism through which baicalin mediates its anti-fibrotic effect could be mediated via the increased expression of miR-3595, which leads to a decrease in the expression of ACSL4. Taken together, the present results demonstrate that baicalin may be a novel therapeutic in the treatment of hepatic fibrosis. To the best of our knowledge, the present study is the first to identify a role of miR-3593/ACSL4 in regulating liver fibrosis, specifically in the activation of HSCs.
Footnotes
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lee UE, Friedman SL. Mechanisms of hepatic fibrogenesis. Best Pract Res Clinical Gastroenterol. 2011;25:195–206. doi: 10.1016/j.bpg.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedman SL. Mechanisms of hepatic fibrogenesis. Gastroenterology. 2008;134:1655–1669. doi: 10.1053/j.gastro.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trappoliere M, Caligiuri A, Schmid M, Bertolani C, Failli P, Vizzutti F, Novo E, di Manzano C, Marra F, Loguercio C, Pinzani M. Silybin, a component of sylimarin, exerts anti-inflammatory and anti-fibrogenic effects on human hepatic stellate cells. J Hepatol. 2009;50:1102–1111. doi: 10.1016/j.jhep.2009.02.023. [DOI] [PubMed] [Google Scholar]
- 4.Bataller R, Brenner DA. Liver fibrosis. J Clin Invest. 2005;115:209–218. doi: 10.1172/JCI24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Popov Y, Schuppan D. Targeting liver fibrosis: Strategies for development and validation of antifibrotic therapies. Hepatology. 2009;50:1294–1306. doi: 10.1002/hep.23123. [DOI] [PubMed] [Google Scholar]
- 6.Friedman SL. Hepatic stellate cells: Protean, multifunctional, and enigmatic cells of the liver. Physiol Rev. 2008;88:125–172. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szabo G, Bala S. MicroRNAs in liver disease. Nat Rev Gastroenterol Hepatol. 2013;10:542–552. doi: 10.1038/nrgastro.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong L, Yamasaki G, Johnson RJ, Friedman SL. Induction of beta-platelet-derived growth factor receptor in rat hepatic lipocytes during cellular activation in vivo and in culture. J Clin Invest. 1994;94:1563–1569. doi: 10.1172/JCI117497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu CI, Hoffman JA, Shy BR, Ford EM, Fuchs E, Nguyen H, Merrill BJ. Function of Wnt/β-catenin in counteracting Tcf3 repression through the Tcf3-β-catenin interaction. Development. 2012;139:2118–2129. doi: 10.1242/dev.076067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takase S, Takada A, Yasuhara M, Sato H, Matsuda Y. Effects of malotilate treatment on the serum markers of hepatic fibrogenesis in liver cirrhosis. Gastroenterol Jpn. 1988;23:639–645. doi: 10.1007/BF02782949. [DOI] [PubMed] [Google Scholar]
- 11.Fu Y, Zheng S, Lin J, Ryerse J, Chen A. Curcumin protects the rat liver from CCl4-caused injury and fibrogenesis by attenuating oxidative stress and suppressing inflammation. Mol Pharmacol. 2008;73:399–409. doi: 10.1124/mol.107.039818. [DOI] [PubMed] [Google Scholar]
- 12.Chen SR, Chen XP, Lu JJ, Wang Y, Wang YT. Potent natural products and herbal medicines for treating liver fibrosis. Chin Med. 2015;10:7. doi: 10.1186/s13020-015-0036-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng XD, Dai LL, Huang CQ, He CM, Chen LJ. Correlation between anti-fibrotic effect of baicalin and serum cytokines in rat hepatic fibrosis. World J Gastroenterol. 2009;15:4720–4725. doi: 10.3748/wjg.15.4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Oliveira MR, Nabavi SF, Habtemariam S, Erdogan Orhan I, Daglia M, Nabavi SM. The effects of baicalein and baicalin on mitochondrial function and dynamics: A review. Pharmacol Res. 2015;100:296–308. doi: 10.1016/j.phrs.2015.08.021. [DOI] [PubMed] [Google Scholar]
- 15.Yang MD, Chiang YM, Higashiyama R, Asahina K, Mann DA, Mann J, Wang CC, Tsukamoto H. Rosmarinic acid and baicalin epigenetically derepress peroxisomal proliferator-activated receptor gamma in hepatic stellate cells for their antifibrotic effect. Hepatology. 2012;55:1271–1281. doi: 10.1002/hep.24792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu QJ. Research advances on baicalin and baicalein as potential therapeutic agents for fibrotic disease. Zhongguo Zhong Yao Za Zhi. 2017;42:1271–1276. doi: 10.19540/j.cnki.cjcmm.20170224.018. In Chinese. [DOI] [PubMed] [Google Scholar]
- 17.Meng F, Wang K, Aoyama T, Grivennikov SI, Paik Y, Scholten D, Cong M, Iwaisako K, Liu X, Zhang M, et al. Interleukin-17 signaling in inflammatory, Kupffer cells, and hepatic stellate cells exacerbates liver fibrosis in mice. Gastroenterology. 2012;143:765–776e3. doi: 10.1053/j.gastro.2012.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong X, Feng D, Wang H, Hong F, Bertola A, Wang FS, Gao B. Interleukin-22 induces hepatic stellate cell senescence and restricts liver fibrosis in mice. Hepatology. 2012;56:1150–1159. doi: 10.1002/hep.25744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 20.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight. Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 21.Teng KY, Ghoshal K. Role of noncoding RNAs as biomarker and therapeutic targets for liver fibrosis. Gene Expr. 2015;16:155–162. doi: 10.3727/105221615X14399878166078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewin TM, Kim JH, Granger DA, Vance JE, Coleman RA. Acyl-CoA synthetase isoforms 1,4 and 5 are present in different subcellular membranes in rat liver and can be inhibited independently. J Biol Chem. 2001;276:24674–24679. doi: 10.1074/jbc.M102036200. [DOI] [PubMed] [Google Scholar]
- 23.Lewin TM, Van Horn CG, Krisans SK, Coleman RA. Rat liver acyl-CoA synthetase 4 is a peripheral-membrane protein located in two distinct subcellular organelles, peroxisomes, and mitochondrial-associated membrane. Arch Biochem Biophys. 2002;404:263–270. doi: 10.1016/S0003-9861(02)00247-3. [DOI] [PubMed] [Google Scholar]
- 24.Yan S, Yang XF, Liu HL, Fu N, Ouyang Y, Qing K. Long-chain acyl-CoA synthetase in fatty acid metabolism involved in liver and other diseases: An update. World J Gastroenterol. 2015;21:3492–3498. doi: 10.3748/wjg.v21.i12.3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang YC, Wu CH, Chu JS, Wang CK, Hung LF, Wang YJ, Ho YS, Chang JG, Lin SY. Involvement of fatty acid-CoA ligase 4 in hepatocellular carcinoma growth: Roles of cyclic AMP and P38 mitogen-activated protein kinase. World J Gastroenterol. 2005;11:2557–2563. doi: 10.3748/wjg.v11.i17.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin X, Kong LN, Huang C, Ma TT, Meng XM, He Y, Wang QQ, Li J. Hesperetin derivative-7 inhibits PDGF-BB-induced hepatic stellate cell activation and proliferation by targeting Wnt/β-catenin pathway. Int Immunopharmacol. 2015;25:311–320. doi: 10.1016/j.intimp.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA. Epithelial-mesenchymal transitions: The importance of changing cell state in development and disease. J Clin Invest. 2009;119:1438–1449. doi: 10.1172/JCI38019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Golej DL, Askari B, Kramer F, Barnhart S, Vivekanandan-Giri A, Pennathur S, Bornfeldt KE. Long-chain acyl-CoA synthetase 4 modulates prostaglandin E(2) release from human arterial smooth muscle cells. J Lipid Res. 2011;52:782–793. doi: 10.1194/jlr.M013292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooke M, Orlando U, Maloberti P, Podestá EJ, Cornejo Maciel F. Tyrosine phosphatase SHP2 regulates the expression of acyl-CoA synthetase ACSL4. J Lipid Res. 2011;52:1936–1948. doi: 10.1194/jlr.M015552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schuppan D. Structure of the extracellular matrix in normal and fibrotic liver: Collagens and glycoproteins. Semin Liver Dis. 1990;10:1–10. doi: 10.1055/s-2008-1040452. [DOI] [PubMed] [Google Scholar]
- 32.Anthony PP, Ishak KG, Nayak NC, Poulsen HE, Scheuer PJ, Sobin LH. The morphology of cirrhosis Recommendations on definition, nomenclature, and classification by a working group sponsored by the World Health Organization. J Clin Pathol. 1978;31:395–414. doi: 10.1136/jcp.31.5.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mormone E, George J, Nieto N. Molecular pathogenesis of hepatic fibrosis and current therapeutic approaches. Chem Biol Interact. 2011;193:225–231. doi: 10.1016/j.cbi.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han YP, Zhou L, Wang J, Xiong S, Garner WL, French SW, Tsukamoto H. Essential role of matrix metalloproteinases in interleukin-1-induced myofibroblastic activation of hepatic stellate cell in collagen. J Biol Chem. 2004;279:4820–4828. doi: 10.1074/jbc.M310999200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gressner AM. Transdifferentiation of hepatic stellate cells (Ito cells) to myofibroblasts: A key event in hepatic fibrogenesis. Kidney Int. 1996;(Suppl 54):S39–S45. [PubMed] [Google Scholar]
- 36.Woodhoo A, Ir uar rizaga-Lejar reta M, Beraza N, García-Rodríguez JL, Embade N, Fernández-Ramos D, Martínez-López N, Gutiérrez-De Juan V, Arteta B, Caballeria J, et al. Human antigen R contributes to hepatic stellate cell activation and liver fibrosis. Hepatology. 2012;56:1870–1882. doi: 10.1002/hep.25828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fickert P, Fuchsbichler A, Moustafa T, Wagner M, Zollner G, Halilbasic E, Stöger U, Arrese M, Pizarro M, Solís N, et al. Farnesoid X receptor critically determines the fibrotic response in mice but is expressed to a low extent in human hepatic stellate cells and periductal myofibroblasts. Am J Pathol. 2009;175:2392–2405. doi: 10.2353/ajpath.2009.090114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharvit E, Abramovitch S, Reif S, Bruck R. Amplified inhibition of stellate cell activation pathways by PPAR-gamma, RAR and RXR agonists. PloS One. 2013;8:e76541. doi: 10.1371/journal.pone.0076541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ding N, Yu RT, Subramaniam N, Sherman MH, Wilson C, Rao R, Leblanc M, Coulter S, He M, Scott C, et al. A vitamin D receptor/SMAD genomic circuit gates hepatic fibrotic response. Cell. 2013;153:601–613. doi: 10.1016/j.cell.2013.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li T, Eheim AL, Klein S, Uschner FE, Smith AC, Brandon-Warner E, Ghosh S, Bonkovsky HL, Trebicka J, Schrum LW. Novel role of nuclear receptor Rev-erbα in hepatic stellate cell activation: Potential therapeutic target for liver injury. Hepatology. 2014;59:2383–2396. doi: 10.1002/hep.27049. [DOI] [PubMed] [Google Scholar]
- 41.Beaven SW, Wroblewski K, Wang J, Hong C, Bensinger S, Tsukamoto H, Tontonoz P. Liver X receptor signaling is a determinant of stellate cell activation and susceptibility to fibrotic liver disease. Gastroenterology. 2011;140:1052–1062. doi: 10.1053/j.gastro.2010.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu L, Asahina K, Wang J, Ueno A, Lazaro R, Miyaoka Y, Miyajima A, Tsukamoto H. Hepatic stellate cell-derived delta-like homolog 1 (DLK1) protein in liver regeneration. J Biol Chem. 2012;287:10355–10367. doi: 10.1074/jbc.M111.312751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Delgado I, Carrasco M, Cano E, Carmona R, García-Carbonero R, Marín-Gómez LM, Soria B, Martín F, Cano DA, Muñoz-Chápuli R, Rojas A. GATA4 loss in the septum transversum mesenchyme promotes liver fibrosis in mice. Hepatology. 2014;59:2358–2370. doi: 10.1002/hep.27005. [DOI] [PubMed] [Google Scholar]
- 44.Baylor NW, Fu T, Yan YD, Ruscetti FW. Inhibition of human T cell leukemia virus by the plant flavonoid baicalin (7-glucuronic acid, 5,6-dihydroxyflavone) J Infect Dis. 1992;165:433–437. doi: 10.1093/infdis/165.3.433. [DOI] [PubMed] [Google Scholar]
- 45.Li BQ, Fu T, Dongyan Y, Mikovits JA, Ruscetti FW, Wang JM. Flavonoid baicalin inhibits HIV-1 infection at the level of viral entry. Biochem Biophys Res Commun. 2000;276:534–538. doi: 10.1006/bbrc.2000.3485. [DOI] [PubMed] [Google Scholar]
- 46.Zeng Y, Song C, Ding X, Ji X, Yi L, Zhu K. Baicalin reduces the severity of experimental autoimmune encephalomyelitis. Braz J Med Biol Res. 2007;40:1003–1010. doi: 10.1590/S0100-879X2006005000115. [DOI] [PubMed] [Google Scholar]
- 47.Lin CC, Shieh DE. The anti-inflammatory activity of Scutellaria rivularis extracts and its active components, baicalin, baicalein and wogonin. Am J Chin Med. 1996;24:31–36. doi: 10.1142/S0192415X96000050. [DOI] [PubMed] [Google Scholar]
- 48.Kubo M, Matsuda H, Tanaka M, Kimura Y, Okuda H, Higashino M, Tani T, Namba K, Arichi S. Studies on Scutellariae radix. VII. Anti-arthritic and anti-inflammatory actions of methanolic extract and flavonoid components from Scutellariae radix. Chem Pharm Bull (Tokyo) 1984;32:2724–2729. doi: 10.1248/cpb.32.2724. [DOI] [PubMed] [Google Scholar]
- 49.Zhang XP, Tian H, Lai YH, Chen L, Zhang L, Cheng QH, Yan W, Li Y, Li QY, He Q, Wang F. Protective effects and mechanisms of Baicalin and octreotide on renal injury of rats with severe acute pancreatitis. World J Gastroenterol. 2007;13:5079–5089. doi: 10.3748/wjg.v13.i38.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ikezoe T, Chen SS, Heber D, Taguchi H, Koeffler HP. Baicalin is a major component of PC-SPES which inhibits the proliferation of human cancer cells via apoptosis and cell cycle arrest. Prostate. 2001;49:285–292. doi: 10.1002/pros.10024. [DOI] [PubMed] [Google Scholar]
- 51.Sun H, Che QM, Zhao X, Pu XP. Antifibrotic effects of chronic baicalein administration in a CCl4 liver fibrosis model in rats. Eur J Pharmacol. 2010;631:53–60. doi: 10.1016/j.ejphar.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 52.Hyun J, Jung Y. MicroRNAs in liver fibrosis: Focusing on the interaction with hedgehog signaling. World J Gastroenterol. 2016;22:6652–6662. doi: 10.3748/wjg.v22.i29.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Akhtar MM, Micolucci L, Islam MS, Olivieri F, Procopio AD. Bioinformatic tools for microRNA dissection. Nucleic Acids Res. 2016;44:24–44. doi: 10.1093/nar/gkv1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ambros V. microRNAs: Tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/S0092-8674(01)00616-X. [DOI] [PubMed] [Google Scholar]
- 55.Babin PJ, Gibbons GF. The evolution of plasma cholesterol: Direct utility or a 'spandrel' of hepatic lipid metabolism. Prog Lipid Res. 2009;48:73–91. doi: 10.1016/j.plipres.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 56.Soupene E, Kuypers FA. Mammalian long-chain acyl-CoA synthetases. Exp Biol Med (Maywood) 2008;233:507–521. doi: 10.3181/0710-MR-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lopes-Marques M, Cunha I, Reis-Henriques MA, Santos MM, Castro LF. Diversity and history of the long-chain acyl-CoA synthetase (Acsl) gene family in vertebrates. BMC Evol Biol. 2013;13:271. doi: 10.1186/1471-2148-13-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kang MJ, Fujino T, Sasano H, Minekura H, Yabuki N, Nagura H, Iijima H, Yamamoto TT. A novel arachidonate-preferring acyl-CoA synthetase is present in steroidogenic cells of the rat adrenal, ovary, and testis. Proc Natl Acad Sci USA. 1997;94:2880–2884. doi: 10.1073/pnas.94.7.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Doll S, Proneth B, Tyurina YY, Panzilius E, Kobayashi S, Ingold I, Irmler M, Beckers J, Aichler M, Walch A, et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat Chem Biol. 2017;13:91–98. doi: 10.1038/nchembio.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Orlando UD, Garona J, Ripoll GV, Maloberti PM, Solano ÁR, Avagnina A, Gomez DE, Alonso DF, Podestá EJ. The functional interaction between Acyl-CoA synthetase 4,5-lipooxygenase and cyclooxygenase-2 controls tumor growth: A novel therapeutic target. PloS One. 2012;7:e40794. doi: 10.1371/journal.pone.0040794. [DOI] [PMC free article] [PubMed] [Google Scholar]