Abstract
Sterol regulatory element binding protein-2 (SREBP-2) is an important transcription factor in lipid homeostasis. A previous study showed that SREBP-2 also activated autophagic genes during cell-sterol depletion. Alterations in autophagy are reported to be involved in the pathogenesis of nonalcoholic fatty liver disease (NAFLD). However, whether the regulation of SREBP-2 restores dysfunctional autophagy in hepatocytes during NAFLD remains to be elucidated. In the present study, a steatosis model was established with palmitic acid (PA) treatment at the indicated times and concentrations. Autophagosomes in hepatocytes were visualized by confocal microscopy after transfection with a tandem GFP-mCherry-LC3 construct. Autophagy-associated protein levels were analyzed by western blot analysis. Loss- and gain-of-function studies were performed to examine the role of SREBP-2 in the regulation of hepatocyte autophagy. It was demonstrated that PA induced autophagy and enhanced autophagic flux at the early stage, whereas prolonged treatment with PA resulted in dysfunction of autophagy in the PA-induced steatotic hepatocytes. In addition, different cellular models presented with differing dysfunctional autophagy in response to fatty acid overload. It was also confirmed that SREBP-2 regulated autophagy-related gene expression in hepatocytes, and it was shown that the overexpression of SREBP-2 increased the expression of autophagy-related genes, but did not affect the inhibition of the autophagic flux in lipid-overloaded HL-7702 cells. By contrast, increased SREBP-2 partly restored the inhibited autophagic activity in lipid-overloaded hepatoma HepG2 cells. Taken together, the present study demonstrated that autophagic function was impaired in lipid-overloaded human hepatocytes, and the differential effect of PA on autophagy was associated with the duration of PA and the cell type. Under these conditions, the overexpression of SREBP-2 alleviated the inhibited autophagic activity rather than the inhibition of autophagic flux. Consequently, the results indicated that restoration of autophagy dysfunction via the regulation of SREBP-2 may be a potential therapeutic target for the treatment of NAFLD.
Keywords: sterol regulatory element binding protein-2, palmitic acids, hepatic steatosis, autophagy, autophagic flux
Introduction
Nonalcoholic fatty liver disease (NAFLD) has gradually emerged as the most common chronic liver disease in the majority of the developed world, which is characterized by excess accumulation of fat in hepatocytes (1). The increasing global prevalence of NAFLD may be the result of changes in dietary habits and an increase in sedentary lifestyles (2). It is reported that the prevalence of NAFLD among adults in China is ~15% (2). Although saturated fatty acid (SFA)-induced lipotoxicity and endoplasmic reticulum (ER) stress have relevant roles in the pathogenesis of NAFLD, the exact molecular mechanisms involved in liver cell injury contributing to progression from NAS to NASH remains to be elucidated (3,4).
Palmitic acid (PA) is the most abundant SFA in the circulation and a major lipotoxic inducer. Accumulating evidence supports that autophagy confers protection against the lipotoxicity induced by PA (5,6). Autophagy is a catabolic process, which is crucial in maintaining cellular homeostasis through a lysosomal degradative pathway, which is involved in various physiological and pathological processes (7). Established functions for autophagy in hepatic lipid metabolism, insulin sensitivity and cellular injury suggest multiple possible mechanisms by which autophagy may affect the lipid accumulation and hepatocellular injury which underlie NAFLD (8–10).
In NAFLD, an abnormal increase in intracellular lipid impairs autophagic degradation (8), in turn, autophagic dysfunction exacerbates NAFLD by causing ER stress and increasing insulin resistance (11). Hyperinsulinemia contributes further to the hepatic defect in autophagy (12). Although the exact mechanism remains to be fully elucidated, several mechanisms underlying NAFLD-associated autophagy dysfunction have been suggested, including decreased expression of autophagic genes and reduced levels of degradative lysosomal enzymes (13). However, the most important contributor is impaired autophagosome maturation, including vesicular fusion and acidification (14–16). The mechanism is likely to be multifactorial, however, the important implication is that a harmful cycle may exist in which hepatic lipid accumulation promotes insulin resistance, and these events impair autophagy which worsens steatosis and insulin insensitivity (13). This cycle leads to self-perpetuating worsening of cellular autophagic function and steatosis. The numerous potential mechanisms involved in autophagy in NAFLD suggest that autophagy offers novel approaches for the treatment of NAFLD.
Previous studies have shown that SREBP-2, a transcription factor that regulates cholesterol metabolism, can regulate autophagy under certain circumstances (17). However, whether the regulation of SREBP-2 enhances autophagic function in hepatocytes remains to be elucidated, particularly in lipid-overloaded hepatic cells. In the present study, it was identified that autophagic activity was impaired in lipid-overloaded human hepatocytes. It was confirmed that SREBP-2 activated autophagy-associated genes in different hepatocytes. It was also shown that the overexpression of SREBP-2 partially alleviated the inhibited autophagic activity in lipid-overloaded hepatocytes. Based on these results, the present study provided novel insights for potential NAFLD therapies.
Materials and methods
Cell culture and treatment
The HepG2 human cell line was obtained from the Department of Infectious Diseases of the Union Hospital of Wuhan (Wuhan, China). The HepG2 cells were cultured in DMEM supplemented with 10% fetal bovine serum (both from Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 37°C in a humidified atmosphere containing 5% CO2. HL-7702 cells (Cell Bank of the Chinese Academy of Sciences, Shanghai, China) were cultured in RPMI-1640 medium supplemented as above. In vitro models of hepatic steatosis were induced by treating the cells with PA at the indicated times (0, 6, 12 and 24 h) and concentrations (0, 100, 200 and 400 µM). Briefly, a 100-mM stock solution of PA (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was prepared in 0.1 N NaOH, and adjusted to 10 mM PA with fatty acid-free BSA solution (Sigma-Aldrich; Merck KGaA). The PA-BSA complex was then filtered and added to complete culture medium to achieve a final concentration of 400 µM. Cells treated with autolysosome inhibitor CQ (50 µM) were used as a positive control of autophagosome accumulation. All experimental procedures were approved by the Ethics Committee of Tongji Medical College, Huazhong University of Science and Technology (Wuhan, China).
Overexpression and silencing of SREBP-2
Firstly, a pcDNA3.1+ expression vector containing the human cDNA of SREBP-2 (pcDNA3-1-14GS0029MU-8) was designed and synthesized by Invitrogen; Thermo Fisher Scientific, Inc. (Waltham, MA, USA). For RNA silencing, the recombinant plasmid pcDNA6.2-GW/EmGFP-miR, expressing a cytomegalovirus promoter-driven micro155 short hairpin RNA (shRNA) targeting SREBP-2 (SREBP-2 shRNA), was constructed by Invitrogen (Thermo Fisher Scientific, Inc.). The sequences are shown in Table I.
Table I.
Recombinant plasmid pcDNA6.2-GW/EmGFP-microRNA sequences.
| Gene | Sequence (5′-3′) |
|---|---|
| MR0003-1F | TGCTGAGGACATTCTGATTAAAGTCCGTTTTGGCCACTGACTGACGGACTTTACAGAATGTCCT |
| MR0003-1R | CCTGAGGACATTCTGTAAAGTCCGTCAGTCAGTGGCCAAAACGGACTTTAATCAGAATGTCCTC |
| MR0003-2F | TGCTGTTCCGGTGCCTCCAGAAGGTGGTTTTGGCCACTGACTGACCACCTTCTAGGCACCGGAA |
| MR0003-2R | CCTGTTCCGGTGCCTAGAAGGTGGTCAGTCAGTGGCCAAAACCACCTTCTGGAGGCACCGGAAC |
| MR0003-3F | TGCTGTTCAAAGCCTGCCTCAGTGGCGTTTTGGCCACTGACTGACGCCACTGACAGGCTTTGAA |
| MR0003-3R | CCTGTTCAAAGCCTGTCAGTGGCGTCAGTCAGTGGCCAAAACGCCACTGAGGCAGGCTTTGAAC |
| MR0003-4F | TGCTGTCACCAAGGACTCTATAGCTCGTTTTGGCCACTGACTGACGAGCTATAGTCCTTGGTGA |
| MR0003-4R | CCTGTCACCAAGGACTATAGCTCGTCAGTCAGTGGCCAAAACGAGCTATAGAGTCCTTGGTGAC |
F, forward; R, reverse.
Cell transfection
The plasmids were transfected into HepG2 and HL-7702 cells according with Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Briefly, prior to transfection, the cells were seeded at 1×106 per well in a 6-well plate of complete medium with serum and antibiotics (100 U/ml penicillin G and 100 µg/mg streptocycin; Gibco; Thermo Fisher Scientific, Inc.). After 24 h, the cells were transfected with 4 µg of plasmids and 10 µl of Lipofectamine. Following incubation for 4–6 h, the 6-well plates were placed in 2 ml medium containing 10% FBS in a 5% CO2 incubator overnight. GFP fusion proteins were observed under a laser scanning microscope system (Olympus Corporation, Tokyo, Japan).
Western blot analysis
Cells were harvested and lysed in ice-cold RIPA lysis buffer (Applygen Technologies, Inc., Beijing, China) for 30 min before centrifugation at 12,000 × g for 15 min at 4°C. The protein concentration of the supernatants was quantified using the BCA Protein Assay Kit (Applygen Technologies, Inc., Beijing, China). Whole-cell lysates (30–50 µg) were subjected to SDS-PAGE (10 or 12%), followed by transfer onto polyvinylidene difluoride membranes. The membranes were incubated with anti-microtubule-associated protein 1 light chain 3β (LC3B) antiserum for human (cat. no. 2775; 1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA), anti-SREBP-2 (cat. no. sc-5603; 1:100; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), anti-P62 (cat. no. 7758; 1:1,000; ABclonal Biotech Co., Ltd., Cambridge, MA, USA) and anti-GAPDH (cat. no. 012; 1:1,000; AntGene Biotech Co., Ltd., Hubei, China) as primary antibodies at 4°C overnight. Following incubation with horseradish peroxidase-conjugated secondary antibodies (cat. no. 020; 1:5,000; AntGene Biotech Co., Ltd.) for 1 h at room temperature. The immunoreactive bands were visualized by enhanced chemiluminescence using an ECL western blotting starter kit (Thermo Fisher Scientific) and imaged on a chemiluminescence gel imaging system (FluorChem FC3; ProteinSimple, San Jose, CA, USA). Gray-scale values of various bands were analyzed using the AlphaView SA software version 3.4.0 (ProteinSimple).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was isolated from human hepatic cells with TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) and was reverse transcribed using PrimeScript™ RT Master mix (Takara Bio, Inc., Otsu, Japan). The mRNA for autophagic genes was estimated via RT-qPCR analysis using a Quantifast SYBR-Green PCR kit (Qiagen, Inc., Valencia, CA, USA). Briefly, RT-PCR was performed using SYBR-Green PCR Master mix in a 10 µl volume containing 5 µl SYBR Green, 1 µl cDNA, 0.5 µl forward primer, 0.5 µl reverse primer and 3 µl ddH2O. The RT-PCR cycling parameters were 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. The RT-qPCR analysis was performed on an ABI StepOne™ system. Gene expression was analyzed in triplicate and normalized to GAPDH. The relative expression levels were analyzed by the 2−ΔΔCq method as previously described (18). The primer sequences used for RT-qPCR analysis are listed in Table II.
Table II.
Primers used for autophagic genes.
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| SREBP-2 | CCCTTCAGTGCAACGGTCATTCAC | TGCCATTGGCCGTTTGTGTC |
| LC3B | AAGGCGCTTACAGCTCAATG | CTGGGAGGCATAGACCATGT |
| ATG5 | AAAGATGTGCTTCGAGATGTGT C | ACTTTGTCAGTTACCAACGTCA |
| ATG7 | CAGTTTGCCCCTTTTAGTAGTGC CC | AGCCGATACTCGTTCAGC |
| Beclin1 | GGTGTCTCTCGCAGATTCATC | TCAGTCTTCGGCTGAGGTTCT |
| GAPDH C | TGGGCTACACTGAGCACC | AAGTGGTCGTTGAGGGCAATG |
SREBP-2, sterol regulatory element binding protein-2; LC3B, microtubule-associated protein 1 light chain 3β; ATG, autophagy-related.
Oil Red O staining and triglyceride assay
Oil Red O staining was performed according to a previously described procedure (19). Representative photomicrographs were captured at 400× magnification using a system incorporated into the microscope. Intracellular triglycerides were assayed using a triglyceride assay kit (GPO-POD; Applygen Technologies, Inc., Beijing, China), according to the manufacturer's recommended protocol.
Autophagic flux assay using tandem fluorescent-tagged LC3
Human hepatic cells were transfected with GFP-mCherry-LC3 plasmid DNA (Changsha Yingrun Biotechnology Co., Ltd., Changsha, China) according to the manufacturer's protocol. After 24 h, the cells were then treated with the different treatments. The cells were then washed three times with PBS and fixed with 4% paraformaldehyde for 30 min. Images of the cells were captured and observed under a confocal laser scanning microscope (Nikon Corporation, Tokyo, Japan).
Statistical analysis
The results were analyzed using one-way analysis of variance with the SPSS 19.0 statistical software package (IBM SPSS, Armonk, NY, USA). All values are expressed as the mean ± standard deviation. P<0.05 was considered to indicate a statistically significant difference.
Results
Establishment of a cellular steatosis by treatment with PA
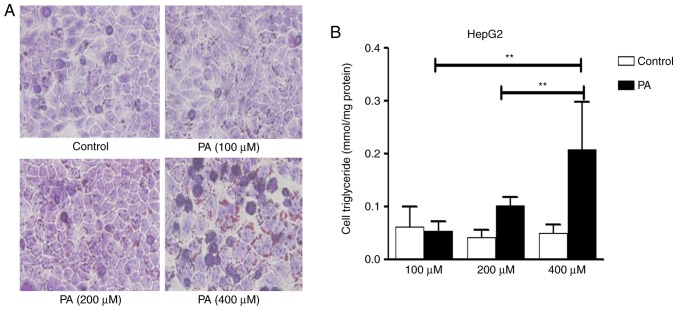
In order to determine the dysfunction of autophagy in the pathologic process of NAFLD, a steatosis model was established with PA treatment at the indicated times and concentrations. Following PA treatment for 24 h, an increased number of intracellular lipid droplets were observed in hepacytes, compared with the control (Fig. 1A). The quantitative analysis of hepatic triglycerides showed a marked increase in cellular triglyceride levels with PA (400 µM), compared with the control (Fig. 1B). With the increase of PA concentrations, cell viability gradually decreased. PA (400 µM) led to significant lipid accumulation in hepatocytes with no marked cell death under the microscope. Therefore, this concentration PA (400 µM) was used for further experiments.
Figure 1.
PA induces intracellular lipid accumulation in HepG2 cells. (A) HepG2 cells were treated with various concentrations of PA for 24 h. Cells were stained with Oil Red O and viewed under a microscope at 400× magnification. (B) Detection of triglyceride levels. HepG2 cells were treated with various concentrations of PA for 24 h for measurement of triglyceride levels with an enzymatic assay kit. Representative results from three independent experiments are shown. Data are presented as the mean standard deviation. **P<0.01, vs. control. PA, palmitic acid.
Effects of PA on autophagy in hepatocytes
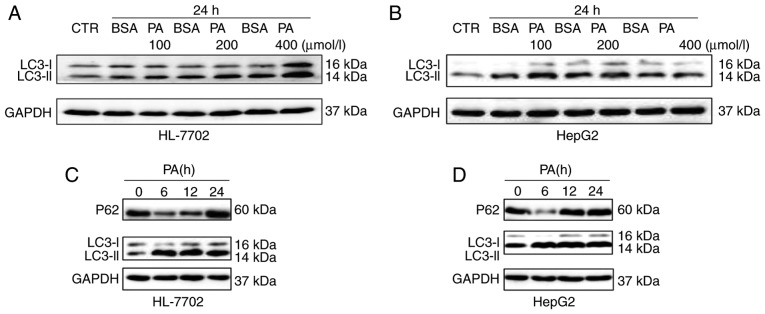
To investigate the association between autophagy and NAFLD at the cellular level, NAFLD cell models were first established by incubating HL-7702 and human HepG2 hepatoma cells with PA. In a previous study, PA was shown to induce autophagy in pancreatic β-cells (20). By contrast, it is suggested that only oleic acid, not PA, triggers autophagic responses in hepatocytes (21). However, the effects of PA on autophagy remain controversial and autophagic dysfunction in different models of cellular steatosis remains to be elucidated. In the present study, to elucidate the effects of PA on autophagic function in different cells (HL-7702 and HepG2), hepatocytes were treated with PA at different concentrations and different times. As shown in Fig. 2A, the induction of PA caused a significant increase in the levels of LC3-II in a dose-dependent manner. Similar results were observed in HepG2 cells; however, the expression levels of LC3-II were decreased at a fixed 24 h time point with 400 µM dose (Fig. 2B). The data showed that PA treatment caused a marked increase in the levels of LC3-II for up to 24 h in the HL-7702 cells (Fig. 2C). The protein levels of p62, an autophagy substrate known to be degraded as autophagic flux progresses, were also detected. As shown in Fig. 2C, a marked decrease in p62 was detected at 6 h, however, an increase in p62 was detected at 24 h. The expression of LC3-II and P62 were also examined in HepG2 cells at different time points. These results showed that PA treatment increased the levels of LC3-II at 6–12 h, but decreased levels of LC3-II at 24 h. In addition, a marked decrease in p62 was detected at 6 h, whereas the levels of P62 were increased at 12–24 h in HepG2 cells (Fig. 2D). These results indicated that PA may enhance autophagic function at the early stage in HL-7702 and HepG2, whereas long-term treatment with PA may suppress autophagic function in the above hepatocytes.
Figure 2.
Effect of PA on autophagy in different hepatocytes. (A) HL-7702 and (B) HepG2 cells were treated with PA, either for 24 h with various concentrations. (C) HL-7702 and (D) HepG2 cells were treated with a fixed PA concentration of 400 µM over a time-course. Western blot analysis was used to determine the proteins levels of p62 and LC3 following treatment with 400 µM PA. PA, palmitic acid; LC3, microtubule-associated protein 1 light chain 3; CTR, control.
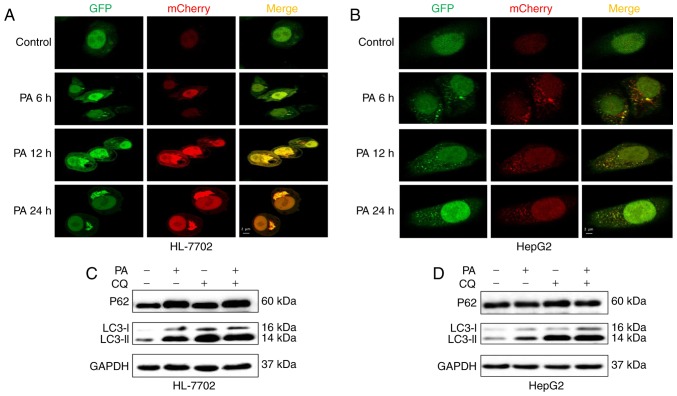
To further monitor autophagic flux induced by PA in HepG2 and HL-7702 cells, a tandem fluorescent-tagged LC3 (GFP-mCherry-LC3) expression plasmid was used. A low level of basal autophagy was observed in the hepatocytes (HL-7702 and HepG2 cells) without PA, as indicated by the low number of yellow and red puncta (Fig. 3A and B). However, when the hepatocytes were exposed to PA at an early time-point (6–12 h), yellow and red punctae were increased, suggesting that autophagic flux was increased and the accumulation of LC3-II induced by PA resulted from the induction of autophagosome formation (Fig. 3A and B). However, following treatment with PA for 24 h, the HL-7702 cells exhibited only autophagosomal LC3-II (yellow punctae without a concomitant increase in red punctae), whereas HepG2 cells showed decreased autophagosomes (yellow punctae) and autolysosomes (red puncta; Fig. 3A and B), indicating that the decrease in the numbers of autophagosomes may be due to decreased autophagosome formation, but not accelerated turnover.
Figure 3.
PA activates autophagy but impairs the autophagic pathway. Representative confocal images of (A) HL-7702 and (B) HepG2 cells stably expressing GFP-mCherry-LC3 following treatment with PA at various time points, scale bar=2 µm. LC3 turnover assay in (C) HL-7702 and (D) HepG2 cells. Protein levels of LC3B II and p62 were determined using western blot analysis following pretreatment with autophagic inhibitor CQ (50 µM) for 6 h followed by PA (400 µm) for 24 h. PA, palmitic acid; LC3, microtubule-associated protein 1 light chain 3; CQ, chlorinated quinine.
Autophagosome accumulation may also be due to lysosomal dysfunction (22). To determine whether PA impaired autophagosome turnover, the present study measured autophagic flux via analyzing LC3 turnover using CQ, which inhibits autophagosome flux at lysosomal degradation, thereby triggering the accumulation of LC3-II and P62 (23). The combination of PA and CQ induced an increase in the levels of LC3-II relative to the control, but no increased expression of p62 or LC3-II, compared with the PA-treated HL-7702 cells (Fig. 3C), indicating that PA impaired autophagosome turnover, but did not induce autophagic flux. Consistent with previous studies, PA inhibited autophagic flux by impairing lysosomal enzyme activity and reducing autophagosome clearance (14,24). However, co-treatment with PA and CQ increased the expression of p62 and induced a higher accumulation of LC3-II, compared with the PA-treated HepG2 cells (Fig. 3D), suggesting that PA-induced autophagic dysfunction was attributed to decreased autophagosome formation, but not the inhibition of autophagolysosomal maturation. These effects were coincident with decreased autophagy in hepatocytes, as shown in fluorescent microscopy images (Fig. 3A and B).
Taken together, the above in vitro experiments suggested that PA induced autophagy in the early stage of saturated fatty acid loading, whereas the long-term stimulus of PA impaired autophagic flux or inhibited autophagic activity. The differential effects of PA on autophagy may be associated with the duration of PA treatment and the cell type. Of note, it was shown that different cellular models of NAFLD exhibited different autophagic dysfunction.
SREBP-2 regulates autophagy-related gene expression in human liver cells
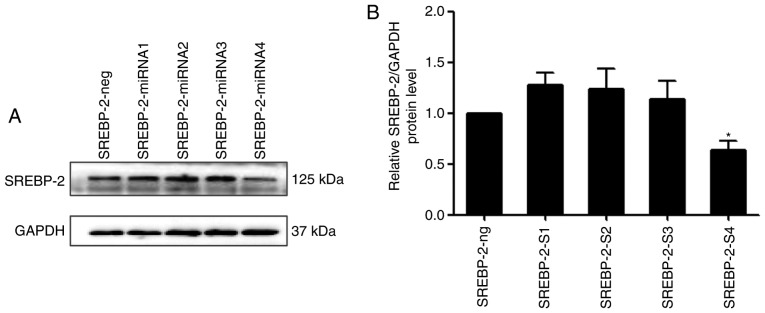
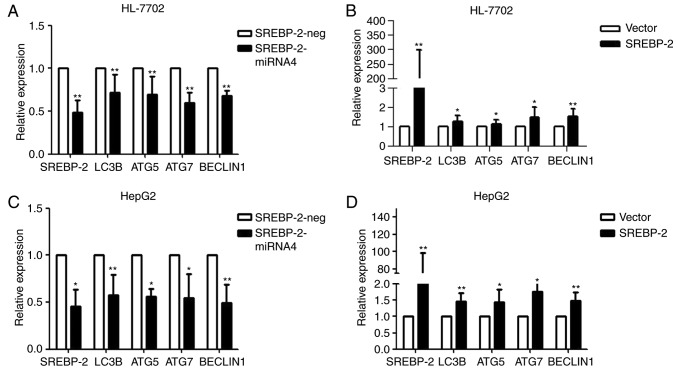
A previous study demonstrated that SREBP-2 activates the expression of key autophagic genes during cellular lipid depletion in 293 cells and HeLa cells (17). However, whether SREBP-2 also regulates autophagy in hepatocytes remains to be elucidated. To investigate the role of SREBP-2 in autophagy pcDNA6.2-GW/EmGFP-miR-SREBP-2 and pcDNA3.1-SREBP-2 expression vectors were constructed, with four miRNAs targeting the SREBP-2 gene. Immunoblot analysis showed that the inhibitory efficiency of the fourth pair of miRNAs was more marked, compared with that of the other three pairs of miRNAs, therefore SREBP-2-4miRNA was used in the subsequent experiments (Fig. 4A and B).
Figure 4.
SREBP-2 miRNA markedly reduces the protein level of SREBP-2 in HL-7702 cells. (A) SREBP-2-miRNA4 had the most marked inhibitory efficiency. (B) Whole-cell lysates were analyzed for protein levels of SREBP-2 by immunoblotting. *P<0.05 vs. control. SREBP-2, sterol regulatory element binding protein-2; miR, microRNA.
Loss- and gain-of-function experiments were performed to ascertain whether SREBP-2 regulated autophagy in hepatocytes under normal conditions. As shown in Fig. 5A, the knockdown of SREBP-2 efficiently reduced the gene expression of SREBP-2. SREBP-2-miR reduced the expression of autophagic genes. By contrast, the overexpression of SREBP-2 increased the expression of these genes (Fig. 5B). These results indicated that SREBP-2 activated the expression of autophagic genes in HL-7702 cells. Similar results were obtained via the transient overexpression or silencing of SREBP-2 in HepG2 human hepatic cells (Fig. 5C and D). These gain- and loss-of function data suggested that the biogenesis of autophagosomes may be regulated by SREBP-2.
Figure 5.
SREBP-2 regulates autophagy-related gene expression in hepatocytes. HL-7702 cells were transfected with the (A) modified SREBP-2-miR vector or negative control vector and (B) SREBP-2-overexpression plasmid vector or empty vector. After 24 h, the cells were harvested for RNA. The mRNA levels of LC3B, ATG5, ATG7 and Beclin1 were analyzed using RT-qPCR analysis. GAPDH mRNA was used as an internal control. Relative expression is plotted. HepG2 cells were harvested for RNA following transfection for 24 h with (C) modified SREBP-2-miR vector or negative control vector and (D) SREBP-2-overexpression plasmid vector or empty vector. mRNA levels of LC3B, ATG5, ATG7 and Beclin1 were analyzed using RT-qPCR analysis. GAPDH mRNA was used as an internal control. *P<0.05 and **P<0.01, vs.control. SREBP-2, sterol regulatory element binding protein-2; miR, microRNA; ATG, autophagy-related; LC3B, microtubule-associated protein 1 light chain 3β; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
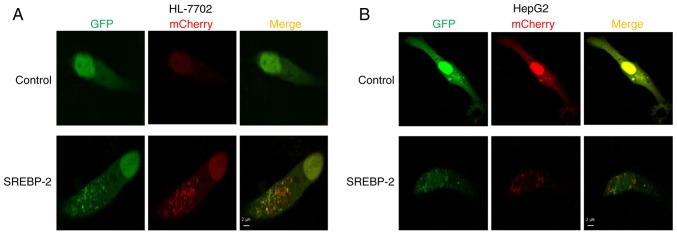
To further investigate the effect of the overexpression of SREBP-2 on autophagic influx in hepatocytes, the cells were co-transfected by GFP-mCherry-LC3 and SREBP-2 expression plasmids or mock plasmids, separately. Confocal fluorescent microscopy showed that the number of autophagosomal LC3B-II (yellow puncta on co-localization) and autolysosomal LC3B-II (red) were increased in hepatocytes transfected with the expressed vector of SREBP-2, compared with the control (Fig. 6A and B). Collectively, these results demonstrated that SREBP-2 regulated autophagy in hepatocytes.
Figure 6.
Overexpression of SREBP-2 increases the number of autophagosomes and autophagolysosomes. (A) HL-7702 cells were co-transfected with GFP-mCherry-LC3 and SREBP-2 expression plasmids or mock plasmids, separately. After 48 h, the cells were then fixed and subjected to confocal microscopy. Scale bar=2 µm. (B) HepG2 cells were co-transfected with GFP-mCherry-LC3 and SREBP-2 expression plasmids or mock plasmids, separately. After 48 h, the cells were fixed and subjected to confocal microscopy. Scale bar=2 µm. SREBP-2, sterol regulatory element binding protein-2; LC3, microtubule-associated protein 1 light chain 3.
Overexpression of SREBP-2 partly restores autophagic function in PA-induced steatotic HepG2 cells, but not in HL-7702 cells
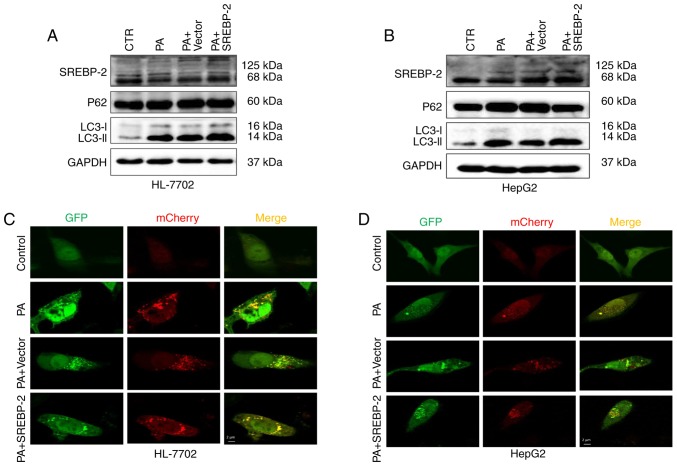
As dysfunctional autophagy in hepatocytes was observed, the present study the intracellular expression of SREBP-2 was increased via transfection with an SREBP-2-bearing plasmid to ascertain whether the overexpression of SREBP-2 restored or alleviated impaired autophagic function in fat-overloaded hepatocytes. The results showed that the expression of SREBP-2 in cells transfected with the SREBP-2-bearing vector was higher, compared with the cells transfected with the control vector (Fig. 7A). The overexpression of SREBP-2 marginally increased the levels of LC3-II in HL-7702 cells treated with PA, compared with the control, whereas no significant change in the protein expression levels of P62 were observed (Fig. 7A). Furthermore, using confocal fluorescence microscopy, it was observed that the HL-7702 cells exhibited only autophagosomal LC3B-II, but no autolysosomal LC3B-II (Fig. 7C). These data indicated that the overexpression of SREBP-2 did not restore the inhibition of autophagic flux, but increased the number of autophagosomes detected by immunoblotting of the LC3-II protein. By contrast, the overexpression of SREBP-2 marginally increased the levels of LC3-II in HepG2 cells with a concomitant decline in P62 (Fig. 7B). Confocal imaging was also performed to determine the total number of autophagosomes and autolysosomes. The results revealed that the overexpression of SREBP-2 induced an increase in the number of autophagosomes and autolysosomes (Fig. 7D). Therefore, the overexpression of SREBP-2 partly alleviated the inhibited autophagic function of HepG2 cells rather than the inhibition of autophagic flux in HL-7702 cells.
Figure 7.
Overexpression of SREBP-2 partly alleviates PA-induced autophagic dysfunction in HepG2 cells but not HL-7702 cells. Hepatocytes were transfected with SREBP-2-bearing plasmid and control vector plasmid and then treated with 400 µM PA for 24 h. The protein expression levels of LC3-II and p62 in (A) HepG2 and (B) HL-7702 cells were detected using western blot analysis. (C) HL-7702 and (D) HepG2 cells were co-transfected with GFP-mCherry-LC3 and SREBP-2 expression plasmids or mock plasmids, separately. After 24 h, the cells were treated with PA for 24 h. Confocal images show GFP (green) and mCherry (red) fluorescence of GFP-mCherry-LC3. Scale bar=2 µm. SREBP-2, sterol regulatory element binding protein-2; PA, palmitic acid; CTR, control.
Discussion
SREBPs are basic helix-loop-helix-leucine zipper transcription factors, which are key in maintaining lipid homeostasis or lipid metabolism (25). There are three mammalian SREBP isoforms: SREBP-1a, SREBP-1c and SREBP-2 (25,26). SREBP-1a and SREBP-1c are involved in fatty acid metabolism, whereas SREBP-2 primarily regulates cholesterol biosynthesis (27). A previous study showed that SREBP-2 regulates autophagy in HeLa cells (17). However, whether SREBP-2 regulates autophagic function in hepatocytes remains to be elucidated.
Autophagy, a self-catabolic process, is a conserved lysosomal degradative pathway, which degrades intracellular organelles to maintain cellular homeostasis under stress conditions (28,29). Autophagy is involved in development, differentiation, survival and homeostasis (30,31). Increasing evidence has indicated that autophagy regulates lipid metabolism (8). Studies have further demonstrated that autophagic dysfunction may lead to insulin resistance and ER stress (11,12,30). In addition, mice with chronic obesity or insulin resistance, which are prone to NAFLD, have notably decreased hepatic autophagic activity (12,32). These findings suggest that alterations in autophagy may have a relevant pathogenic role in NAFLD. Consequently, therapeutic strategies aimed at the restoration of autophagic function may provide a novel approach to preventing NAFLD.
To confirm the derangement of autophagy in the pathology of NAFLD, the present study used PA to establish an in vitro model of steatosis. Reports on the effect of PA on autophagy in different cell models of NAFLD have been controversial. In a previous study, Mei et al reported that PA does not induce autophagy in hepatocytes, whereas PA significantly induced apoptosis (21). By contrast, another study showed that the levels of autophagy were elevated in hepatoma cells upon exposure to PA, and PA-induced autophagy was identified to be independent of the regulation of mammalian target of rapamycin (mTOR) (33). In the present study, it was identified that PA induced the activation of autophagy, but also impaired autophagic function. The data showed that PA induced autophagy in the early stage of SFA loading, whereas long-term PA exposure impaired autophagic flux or inhibited autophagic activity. In the HL-7702 cellular steatosis model, a longer period of PA treatment inhibited autophagic flux and autophagosome degradation. By contrast, in the HepG2 cellular model, a short period of PA treatment activated autophagy, whereas a prolonged duration of PA inhibited autophagic activity. These results showed that two cellular steatosis models exhibited different autophagic dysfunctions: Inhibition of autophagic flux or subdued autophagic activity.
Although the exact mechanisms affecting this different outcome remain to be elucidated, several potential explanations may account for such conflicting results. One potential mechanistic explanation for the difference is that elevated ER stress triggers the disruption of autophagic flux. Accumulating data indicates that ER stress is a potent trigger of autophagy (10,34,35). A study by González-Rodríguez et al (10) identified that short-term treatment with PA triggered the activation of the unfolding protein response and autophagic flux. By contrast, prolonged treatment with PA induced ER stress and cell death, in addition to the inhibition of autophagic flux (10). These results indicated that the induction of autophagy by PA is dependent on ER stress and the mTOR signaling pathway, however, sustained or marked ER stress due to lipid overload may increase the accumulation of unfolded proteins, triggering liver injury through the disruption of the autophagic flux and leading to apoptotic cell death (10). Similar results were observed in another study. Park et al (36) identified that PA promoted mTOR complex 1-dependent autophagy at an early stage, but prolonged treatment with PA led to damage of autophagic flux and an aggravation of apoptosis in H9c2 cells. However, certain studies have shown that PA induces the activation of autophagy via the protein kinase C-mediated signaling pathway independent of the ER stress and mTOR signaling pathways (33,37). The effects of PA on autophagy remains controversial and may be cell-type dependent (21,33,36); however, accumulating evidence supports that the activation of autophagy at an early stage confers a pro-survival effect against lipotoxicity-induced cell death (38), whereas a longer period of exposure to PA leads to impaired autophagic function and apoptotic cell death. It has been reported that the autophagic process is a cell pro-survival mechanism (29) and that the evolution of NAFLD may be associated with the dynamic regulation of autophagy (30). Understanding the mechanisms of the impairment of hepatic autophagy has provided key insights into the pathogenesis of NAFLD. The modulation of autophagy may offer therapeutic potential to prevent the progression of NAFLD. Previous findings have demonstrated that SREBP-2 binds to and activates the expression of several autophagic genes, whereas the knockdown of SREBP-2 suppresses autophagosome formation and triglyceride mobilization in 293 and HeLa cells (17). In the present study, gain- and loss-of-function data demonstrated that SREBP-2 regulated autophagy-related gene expression in HepG2 cells and HL-7702 cells. In addition, the overexpression of SREBP-2 increased the number of autophagosomes. These results indicated that the initiation of autophagy may be regulated by SREBP-2. However, the mechanism of SREBP-2 in the regulation of autophagy remains to be elucidated. In a previous study, a genome-wide binding/ChIP-Seq analysis of SREBP-2 revealed that SREBP-2 occupied the promoters of several genes involved in autophagy (17). The effect of SREBP-2 on autophagy has received less attention, and its underlying mechanisms remain to be fully elucidated. There has been previous novel insight into the mechanisms; Seo et al (39) identified that SREBP-2 directly activated the gene expression of patatin-like phospholipase domain-containing enzyme 8 (PNPLA8), and PNPLA8 increased autophagy through inhibiting the phosphorylation of unc-51-like kinase 1 downstream of target of rapamycin complex 1 (TORC1). Therefore, the SREBP-2/PNPLA8 axis may regulate the initiation of autophagy through the inhibition of TORC1. These findings suggested that SREBP-2 may regulate autophagy by involving multiple signaling pathways. However, whether the modulation of SREBP-2 restored impaired autophagic activity in the steatotic hepatocytes remains to be fully elucidated. Subsequently, the present study examined the effect of the overexpression of SREBP-2 on autophagic function in fat-overloaded hepatic cells. In the HL-7702 cellular steatosis model, it was identified that the overexpression of SREBP-2 increased the levels of LC3 II, but not the expression of P62, suggesting that the modulation of SREBP-2 did not restore the inhibited autophagic flux. However, the overexpression of SREBP-2 resulted in an increase in the levels of LC3-II and a decrease in the levels of P62 in PA-treated HepG2 cells, indicating that the overexpression of SREBP-2 restored the inhibited autophagic activity. Of note, a previous study reported SREBP-2 did not affect global autophagic flux (17). Therefore, it was hypothesized that the overexpression of SREBP-2 may only be involved in the initial stage of autophagy, with no effect on the late stage of autophagy, including autophagosomes for fusion with endosomes/lysosomes and metabolite degradation.
The present study provided evidence that, in the PA-induced cellular steatosis model, PA induced autophagy and enhanced autophagic flux at the early stage, however, a longer period of exposure to PA led to impaired autophagic flux. Under the experimental conditions, two hepatic cell lines presented with different autophagic dysfunction in response to lipid overload. The results confirmed that SREBP-2 regulated the expression of hepatic autophagic genes in normal conditions. Additionally, it was identified that the overexpression of SREBP-2 partially restored the inhibited autophagic activity in lipid-overloaded hepatocytes. Although the role of SREBP-2 in the regulation of hepatocyte lipogenesis and autophagy requires further investigation, the present study demonstrated the importance of the role of SREBP-2 in autophagy in hepatocytes.
In conclusion, the present study demonstrated that SREBP-2 regulated the expression of autophagy-related genes in hepatocytes. In addition, the overexpression of SREBP-2 partly enhanced the inhibited autophagic activity in lipid-overloaded hepatocytes. Overall, the restoration of aberrant autophagic function by the modulation of SREBP-2 may be a potential therapeutic target for the prevention and treatment of NAFLD.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (grant nos. 81370525 and 81170401).
Glossary
Abbreviations
- NAFLD
nonalcoholic fatty liver disease
- SREBP-2
sterol regulatory element binding protein-2
- PA
palmitic acid
- CQ
chloroquine
Footnotes
Competing interests
The authors declare that they have no competing interests.
References
- 1.Brunt EM, Wong VW, Nobili V, Day CP, Sookoian S, Maher JJ, Bugianesi E, Sirlin CB, Neuschwander-Tetri BA, Rinella ME. Nonalcoholic fatty liver disease. Nat Rev Dis Primers. 2015;1:15080. doi: 10.1038/nrdp.2015.80. [DOI] [PubMed] [Google Scholar]
- 2.Wang FS, Fan JG, Zhang Z, Gao B, Wang HY. The global burden of liver disease: The major impact of China. Hepatology. 2014;60:2099–2108. doi: 10.1002/hep.27406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheung O, Sanyal AJ. Recent advances in nonalcoholic fatty liver disease. Curr Opin Gastroenterol. 2010;26:202–208. doi: 10.1097/MOG.0b013e328337b0c4. [DOI] [PubMed] [Google Scholar]
- 4.Neuschwander-Tetri BA. Hepatic lipotoxicity and the pathogenesis of nonalcoholic steatohepatitis: The central role of nontriglyceride fatty acid metabolites. Hepatology. 2010;52:774–788. doi: 10.1002/hep.23719. [DOI] [PubMed] [Google Scholar]
- 5.Li S, Li J, Shen C, Zhang X, Sun S, Cho M, Sun C, Song Z. Tert-Butylhydroquinone (tBHQ) protects hepatocytes against lipotoxicity via inducing autophagy independently of Nrf2 activation. Biochim Biophys Acta. 2014;1841:22–33. doi: 10.1016/j.bbalip.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu J, Wu JJ, Yang LJ, Wei LX, Zou DJ. Rosiglitazone protects against palmitate-induced pancreatic beta-cell death by activation of autophagy via 5′-AMP-activated protein kinase modulation. Endocrine. 2013;44:87–98. doi: 10.1007/s12020-012-9826-5. [DOI] [PubMed] [Google Scholar]
- 7.Ryter SW, Cloonan SM, Choi AM. Autophagy: A critical regulator of cellular metabolism and homeostasis. Mol Cells. 2013;36:7–16. doi: 10.1007/s10059-013-0140-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukuo Y, Yamashina S, Sonoue H, Arakawa A, Nakadera E, Aoyama T, Uchiyama A, Kon K, Ikejima K, Watanabe S. Abnormality of autophagic function and cathepsin expression in the liver from patients with non-alcoholic fatty liver disease. Hepatol Res. 2014;44:1026–1036. doi: 10.1111/hepr.12282. [DOI] [PubMed] [Google Scholar]
- 10.González-Rodríguez Á, Mayoral R, Agra N, Valdecantos MP, Pardo V, Miquilena-Colina ME, Vargas-Castrillón J, Lo Iacono O, Corazzari M, Fimia GM, et al. Impaired autophagic flux is associated with increased endoplasmic reticulum stress during the development of NAFLD. Cell Death Dis. 2014;5:e1179. doi: 10.1038/cddis.2014.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu HY, Han J, Cao SY, Hong T, Zhuo D, Shi J, Liu Z, Cao W. Hepatic autophagy is suppressed in the presence of insulin resistance and hyperinsulinemia: Inhibition of FoxO1-dependent expression of key autophagy genes by insulin. J Biol Chem. 2009;284:31484–31492. doi: 10.1074/jbc.M109.033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Czaja MJ. Functions of autophagy in hepatic and pancreatic physiology and disease. Gastroenterology. 2011;140:1895–1908. doi: 10.1053/j.gastro.2011.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koga H, Kaushik S, Cuervo AM. Altered lipid content inhibits autophagic vesicular fusion. FASEB J. 2010;24:3052–3065. doi: 10.1096/fj.09-144519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inami Y, Yamashina S, Izumi K, Ueno T, Tanida I, Ikejima K, Watanabe S. Hepatic steatosis inhibits autophagic proteolysis via impairment of autophagosomal acidification and cathepsin expression. Biochem Biophys Res Commun. 2011;412:618–625. doi: 10.1016/j.bbrc.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 16.Park HW, Park H, Semple IA, Jang I, Ro SH, Kim M, Cazares VA, Stuenkel EL, Kim JJ, Kim JS, Lee JH. Pharmacological correction of obesity-induced autophagy arrest using calcium channel blockers. Nat Commun. 2014;5:4834. doi: 10.1038/ncomms5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seo YK, Jeon TI, Chong HK, Biesinger J, Xie X, Osborne TF. Genome-wide localization of SREBP-2 in hepatic chromatin predicts a role in autophagy. Cell Metab. 2011;13:367–375. doi: 10.1016/j.cmet.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔC T method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Pan X, Wang P, Luo J, Wang Z, Song Y, Ye J, Hou X. Adipogenic changes of hepatocytes in a high-fat diet-induced fatty liver mice model and non-alcoholic fatty liver disease patients. Endocrine. 2015;48:834–847. doi: 10.1007/s12020-014-0384-x. [DOI] [PubMed] [Google Scholar]
- 20.Choi SE, Lee SM, Lee YJ, Li LJ, Lee SJ, Lee JH, Kim Y, Jun HS, Lee KW, Kang Y. Protective role of autophagy in palmitate-induced INS-1 beta-cell death. Endocrinology. 2009;150:126–134. doi: 10.1210/en.2008-0483. [DOI] [PubMed] [Google Scholar]
- 21.Mei S, Ni HM, Manley S, Bockus A, Kassel KM, Luyendyk JP, Copple BL, Ding WX. Differential roles of unsaturated and saturated fatty acids on autophagy and apoptosis in hepatocytes. J Pharmacol Exp Ther. 2011;339:487–498. doi: 10.1124/jpet.111.184341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaishy B, Zhang Q, Chung HS, Riehle C, Soto J, Jenkins S, Abel P, Cowart LA, Van Eyk JE, Abel ED. Lipid-induced NOX2 activation inhibits autophagic flux by impairing lysosomal enzyme activity. J Lipid Res. 2015;56:546–561. doi: 10.1194/jlr.M055152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osborne TF, Espenshade PJ. Evolutionary conservation and adaptation in the mechanism that regulates SREBP action: What a long, strange tRIP it's been. Genes Dev. 2009;23:2578–2591. doi: 10.1101/gad.1854309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shao W, Espenshade PJ. Expanding roles for SREBP in metabolism. Cell Metab. 2012;16:414–419. doi: 10.1016/j.cmet.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong TY, Lin SM, Leung LK. The flavone luteolin suppresses SREBP-2 expression and Post-translational activation in hepatic cells. PLoS One. 2015;10:e0135637. doi: 10.1371/journal.pone.0135637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang K. Autophagy and apoptosis in liver injury. Cell Cycle. 2015;14:1631–1642. doi: 10.1080/15384101.2015.1038685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lavallard VJ, Gual P. Autophagy and non-alcoholic fatty liver disease. Biomed Res Int. 2014;2014:120179. doi: 10.1155/2014/120179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Codogno P, Meijer AJ. Autophagy: A potential link between obesity and insulin resistance. Cell Metab. 2010;11:449–451. doi: 10.1016/j.cmet.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 33.Tan SH, Shui G, Zhou J, Li JJ, Bay BH, Wenk MR, Shen HM. Induction of autophagy by palmitic acid via protein kinase C-mediated signaling pathway independent of mTOR (mammalian target of rapamycin) J Biol Chem. 2012;287:14364–14376. doi: 10.1074/jbc.M111.294157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yorimitsu T, Klionsky DJ. Endoplasmic reticulum stress: A new pathway to induce autophagy. Autophagy. 2007;3:160–162. doi: 10.4161/auto.3653. [DOI] [PubMed] [Google Scholar]
- 36.Park M, Sabetski A, Kwan Chan Y, Turdi S, Sweeney G. Palmitate induces ER stress and autophagy in H9c2 cells: Implications for apoptosis and adiponectin resistance. J Cell Physiol. 2015;230:630–639. doi: 10.1002/jcp.24781. [DOI] [PubMed] [Google Scholar]
- 37.Cai N, Zhao X, Jing Y, Sun K, Jiao S, Chen X, Yang H, Zhou Y, Wei L. Autophagy protects against palmitate-induced apoptosis in hepatocytes. Cell Biosci. 2014;4:28. doi: 10.1186/2045-3701-4-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, Chang F, Li F, Fu H, Wang J, Zhang S, Zhao J, Yin D. Palmitate promotes autophagy and apoptosis through ROS-dependent JNK and p38 MAPK. Biochem Biophys Res Commun. 2015;463:262–267. doi: 10.1016/j.bbrc.2015.05.042. [DOI] [PubMed] [Google Scholar]
- 39.Kim KY, Jang HJ, Yang YR, Park KI, Seo J, Shin IW, Jeon TI, Ahn SC, Suh PG, Osborne TF, Seo YK. Corrigendum: SREBP-2/PNPLA8 axis improves non-alcoholic fatty liver disease through activation of autophagy. Sci Rep. 2016;6:35732. doi: 10.1038/srep35732. [DOI] [PMC free article] [PubMed] [Google Scholar]