Abstract
A hypertrophic scar is the result of abnormal repair of the body after trauma. Histopathologically, it is mostly the result of the excessive proliferation of fibroblasts and the accumulation of extracellular matrix. Accumulating evidence has demonstrated that long non-coding RNAs (lncRNAs) have a critical role in the regulation of gene expression and in the pathogenesis of diseases. However, the roles of lncRNAs in hypertrophic scars have remained elusive. The present study investigated the profiles of differentially expressed lncRNAs between fibroblasts derived from a hypertrophic scar and normal skin, and explored the possible mechanisms underlying the development of hypertrophic scars. Microarray data indicated that 6,104 lncRNAs and 2,952 mRNAs were differentially expressed. A set of differentially expressed transcripts as confirmed by reverse transcription-quantitative polymerase chain reaction. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses were performed to determine the principal functions of the significantly deregulated genes. Furthermore, associated expression networks, including subgroup analysis, competing endogenous RNAs (ceRNAs) and coding-noncoding co-expression networks were constructed using bioinformatics methods. The homology between differentially expressed lncRNAs and mRNAs was assessed and two exon lncRNA were selected to explore their regulatory mechanisms. The ceRNA network inferred that NR_125715 acted as a competing endogenous RNA, bound to microRNA (miR)-141-3p, miR-200a-3p and miR-29 to regulate the expression of the miRs' targets, including transforming growth factor β2 (TGFB2). Similarly, NR_046402 acted as a competing endogenous RNA, which bound to miR-133a-3p.1 and miR-4469 to then regulate the expression of the miRs' targets, including DNA polymerase δ1, catalytic subunit (POLD1). In addition, co-expression analysis indicated that the expression of lncRNAs NR_125715 and NR_046402 was correlated with that of TGFB2 and POLD1 mRNA. The identification of these differentially expressed lncRNAs in the hypertrophic scar-derived fibroblasts in the present study, may provide novel insight into the functional interactions of lncRNA, miRNA and mRNA, and lead to novel theories for the pathogenesis and treatment of hypertrophic scars.
Keywords: hypertrophic scar, fibroblasts, long noncoding RNA, competing endogenous RNA, wound healing
Introduction
Hypertrophic scar (HS), a pathological response to skin wound healing, is characterized by the abnormal proliferation of fibroblasts and the excessive deposition of collagen (1). The formation of HS may seriously affect the bodily function and/or appearance of patients (2). A thorough understanding of the pathophysiology and clinical characteristics of HS may help determine the most appropriate treatment strategy, but at present, there is no consensus in terms of a generally accepted treatment regimen (3). Therefore, a more in-depth understanding of the pathophysiological process in the formation of HS may improve the selection process of appropriate treatments. Studies on long noncoding (lncRNA) have been emerging, based on which it may be hypothesized that lncRNAs have a key role in the development and outcome of HS (4).
lncRNAs are non-protein-coding transcripts of >200 nucleotides in length (5). According to their genomic position relative to protein-coding genes, lncRNAs may be divided into six categories: Exon sense-overlapping, intron-sense overlapping, natural antisense, bidirectional, intronic anti-sense and intergenic (6). Aggravating evidence indicates that lncRNAs regulate gene expression at the transcriptional and post-transcriptional level, and participate in various biological processes, including chromatin modification, DNA synthesis, cell proliferation, differentiation and apoptosis (7). In addition, the dysregulation of lncRNAs is closely associated with various human diseases, including cancer, as well as neurological and cardiovascular diseases (8). Wang et al (9) indicated that several lncRNAs, including HOXA distal transcript antisense RNA-005, RP11-567G11.1 and metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), are differentially expressed in pancreatic cancer tissues compared with those in healthy controls, implying their potential as diagnostic or prognostic biomarkers in pancreatic cancer. It has been reported that the perturbation of the expression of certain lncRNAs, including MALAT1 (10), HOX transcript anti-sense RNA (11) and hypoxia-inducible factor 1α-antisense RNA 2 (12), is pivotally involved in central nervous system pathologies, including glioblastoma. Li et al (13) reported that lncRNA8975-1 affected HS by inhibiting the proliferation of fibroblasts, which reduced the expression of α-smooth muscle actin (SMA) and collagens. It has been demonstrated that miRNA-21 and miRNA200b regulate the formation of HS by participating in the transforming growth factor β (TGF-β)/Smad protein signalling pathway (14). Furthermore, overexpression of miR-29b was reported to significantly reduce the expression levels of collagen type I α1 chain (COL1A1) and α-SMA, as well as to inhibit myofibroblast-like cell proliferation and induce apoptosis, suggesting that miR-29b may be involved in scarring and has a significant anti-fibrosis effect (15). An extensive network of interactions involving competing endogenous RNAs (ceRNAs) has been identified, among which lncRNAs control the repressive effect of miRNA on mRNA through competitively binding to the miRNA's binding sites (16). However, the roles of lncRNAs in HS remain largely elusive. In the present study, lncRNA and mRNA expression profiles in fibroblasts derived from HS and normal skin tissues were compared by using an Arraystar Human LncRNA Microarray v4.0 (Arraystar Inc., Rockville, MD, USA). The significant differential expression of representative mRNAs and lncRNAs was further confirmed using reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were then performed. The University of California Santa Cruz (UCSC) Genome Browser (www.genome.ucsc.edu) and the basic local alignment search tool (BLAST; https://blast.ncbi.nlm.nih.gov/Blast.cgi) were then used to identify the indirect association of lncRNAs and proteins as well as possible homologous sequences, and Targetscan (targetscan.org) was then used to identify miRNAs that may bind to these homologous sequences in order to further verify the correlation using an lncRNA-mRNA coexpression network. The present study assessed the possible roles of ceRNAs in HS and the underlying mechanism, and provided a novel way of thinking and theoretical basis to further elucidate the pathological mechanisms of the formation of HS, which may provide approaches for their prevention.
Materials and methods
Fibroblast isolation and cell culture
The present study was approved by Ethics Committee of the First Affiliated Hospital of Nanchang University (Nanchang, China). All patients provided written informed consent in advance. HS and corresponding normal skin tissues were obtained from 3 male patients (1–26 years old) who received surgery at the First Affiliated Hospital of Nanchang University (Nanchang, China) from January 2017 to June 2017. Each patient demonstrated extensive areas of hypertrophic scar, which manifested as raised, erythematous, pruritic and thickened scars restricted to the injury site. Individuals with keloids, open wounds and/or infected skin were excluded from the present study. None of the patients had received any previous treatment for HS. During the surgery, the HS was completely excised and normal skin tissue was obtained after trimming.
HS tissue and normal skin specimens were individually rinsed with PBS and the subcutaneous tissue and part of the dermis were cut off with ophthalmic scissors. Tissues were placed in a culture dish and digested in 0.25% trypsin-EDTA (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 4°C for 10–12 h. The epidermis was completely removed with tweezers and the remaining dermis was cut into 1×1 mm pieces with ophthalmic scissors. Dermal tissue pieces were attached to a Petri dish, followed by addition of DMEM with 10% fetal bovine serum (GE Healthcare Life Sciences, Logan, UT, USA) and incubation at 37°C in a humidified atmosphere containing 5% CO2. Upon confluency, cells were digested with 0.25% trypsin-EDTA and subsequently passaged. The fibroblasts at passage 3 (P3) were used for the experiments. The fibroblasts derived from HS tissue were labeled as 'BH' in the experimental group and the fibroblasts derived from normal skin were labeled as 'ZC' as the control group.
RNA extraction and quality control
When the third passage of cells had been cultured for 48 h, total RNA was extracted with TRIzol (Life Sciences; Thermo Fisher Scientific, Inc.). The absorbance of the RNA at 260, 280 and 230 nm (A260, A280 and A230, respectively) was determined using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Inc.). The A260/A280 and A260/A230 ratios were calculated to determine the concentration and evaluate the purity. The purity and integrity of total RNA in the two groups was confirmed using formaldehyde-denatured gel electrophoresis.
Microarray analysis
Following the above procedure, the total RNA samples were pooled and submitted to the Kang Cheng Bio-Tech Inc. (Shanghai, China) where they were assessed using the Arraystar human LncRNA chip (v4.0). This method is able to detect 40,173 lncRNAs, including 7,506 lncRNAs whose function has been comprehensively assessed and 32,667 frequently screened, highly reliable lncRNAs. The above lncRNAs were selected based on public transcriptome databases, including Refseq (https://www.ncbi.nlm.nih.gov/refseq/), UCSC Genome Browser (genome.ucsc.edu) and Ensembl (https://www.ensembl.org/index.html), and high impact factor papers. Also included were 20,730 protein transcripts derived from Universal Protein Resource (http://www.uniprot.org/). Each transcript is represented by a specific exon or splice probe, which is accurately detected by the probe. The chip also includes positive probes (housekeeping genes) and negative probes for hybridization quality monitoring. The ribosomal RNA was first removed from 1 µg total RNA according to the experimental procedure of the mRNA-ONLY™ Eukaryotic mRNA Isolation Kit (Epicentre, Madison, WI, USA) and then expanded using a double-stranced complementary DNA synthesis kit provided by Qiagen (Hilden, Germany) for RT. An Agilent Monochromatic DNA labeling kit (Agilent Technologies, Inc., Santa Clara, CA, USA) was used for fluorescent labelling and Agilent hybridization kit (Agilent Technologies, Inc.) was used for chip hybridization. A spectrophotometer was used to detect the efficiency of fluorescent labeling to ensure the reliability of the experimental results of the chip. The chips were hybridized and rinsed, and then scanned on an Agilent G2565BA scanner (Agilent Technologies, Inc.). The raw data were processed by Agilent Feature Extraction v11.0.1.1 software and analyzed using AgilentGeneSpring GX v12.1 software (Agilent Technologies, Inc.).
GO analysis
A list of upregulated and downregulated differentially expressed genes was prepared and imported into the GO database (geneontology.org), and calculation for human species was selected. The P-values for the enrichment of GO terms were calculated by the default statistical algorithm of the GO analysis database. The smaller the P-value, the more significant is the role of the GO term, and the terms with P≤0.05 were considered to be statistically significant. The negative base 10 logarithm of the P-value resembled the enrichment score, which signifies the probability that the differentially expressed mRNA is enriched in the term entry. The higher the enrichment score the more important is the entry. Significantly enriched features were sorted in a descending order, and the GO enrichment histogram was plotted.
Pathway analysis
The data were imported into the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (genome.jp/kegg/), and the human species was selected for calculation. The significance score of differential gene enrichment in each pathway entry was calculated by the system default statistical test to obtain significant P-values. Biological pathways with P≤0.05 were identified as being significant, and the smaller the P-value, the more significant the biological pathway. Enrichment score were calculated as above and had an equivalent meaning within the system-derived specific signaling pathways and data. Pathways were ranked in an ascending order according to their P-values, and the Log P histogram of KEGG pathways was plotted.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
RT-qPCR analysis was performed to validate selected differently expressed lncRNAs and mRNAs identified by microarray analysis. The sequences of PCR primers are listed in Table I, which was designed by Primer 5.0 (Premier Biosoft International, Palo Alto, CA, USA) Firstly, cDNA was synthesized by using a Gene Amp PCR system 9700 (Applied Biosystems; Thermo Fisher Scientific, Inc.). RT was performed in a 20 µl reaction containing 200 ng total RNA, 0.3 µl 1 µM RT primer, 2 µl 2.5 mM dNTP (HyTest Ltd., Turku, Finland), 2 µl 10× RT buffer (Epicentre; Illumina, Inc., San Diego, CA, USA), 1 µl 50 U/µl RT enzyme (Epicentre; Illumina, Inc.), 0.3 µl 40 U/µl RNase inhibitor (Epicentre; Illumina, Inc.), 20 µl nuclease free water and 0.2 µl MMLV High Performance Reverse Transcriptase (Epicentre; Illumina, Inc.). The stem-loop RT reaction was performed at 16°C for 30 min, followed by 42°C for 30 min and 85°C for 5 min. Then, RT-qPCR was performed using the SYBR Green I RT-PCR kit (Life Sciences; Thermo Fisher Scientific, Inc.) and GAPDH was used as an internal control, with the following reaction profile: Predenaturation at 95°C for 10 sec, followed by 40 cycles of 95°C for 10 sec and 60°C for 1 min. The melting curve analysis was used to determine the reaction specificity. The selected lncRNAs and mRNAs were normalized to GAPDH as the internal control using the 2−ΔΔCq method (17).
Table I.
Primers used for polymerase chain reaction.
| GenBank ID | Gene name | Sequence | Annealing temperature (°C) | Length (bp) |
|---|---|---|---|---|
| GAPDH | F: 5′-GGGAAACTGTGGCGTGAT-3′ | 60 | 299 | |
| R: 5′-GAGTGGGTGTCGCTGTTGA-3′ | ||||
| NM_198721 | COL25A1 | F: 5′-CCTACACCACCCATTGAAAC-3′ | 60 | 184 |
| R: 5′-TCTCCGCCACCTTATCTCT-3′ | ||||
| NM_020987 | ANK3 | F: 5′-TTTATCAGCCAGTCCCAGTT-3′ | 60 | 62 |
| R: 5′-TTGTTCATCGTCCACTTCC-3′ | ||||
| NR_046402 | POLD1 | F: 5′-TTCATTCCAGTCCAAGCAGA-3′ | 60 | 127 |
| R: 5′-GGCATTGAGCGTGTAGGAG-3′ | ||||
| NM_024719 | GRTP1 | F: 5′-CCAACTTTCACTGCTTTCCT-3′ | 60 | 198 |
| R: 5′-ACAAACCCACACTCACATTTT-3′ | ||||
| NR_046583 | COL4A2-AS1 | F: 5′-CCTTTTGTGACTGGCTTCTT-3′ | 60 | 104 |
| R: 5′-CCAGCCTTTATTCGGTCTT-3′ | ||||
| NM_000689 | ALDH1A1 | F: 5′-ATTTCCCGTTGGTTATGCT-3′ | 60 | 86 |
| R: 5′-AGTTTGCTCTGCTGGTTTG-3′ | ||||
| ENST00000559026 | RP11-59H7.3 | F: 5′-TCAAAAGGCTAGAAATTGTGC-3′ | 60 | 131 |
| R: 5′-CTCCAAAACTGCTGAAACTACA-3′ | ||||
| NR_125715 | TGFB2-OT1 | F: 5′-CTCATGCTGGGGTTAATAGAA-3′ | 60 | 114 |
| R: 5′-GCTGCTCTTTAGGTGAAACTG-3′ |
F, forward; R, reverse; TGFB2-OT1, transforming growth factor β2 overlapping transcript 1; POLD1, DNA polymerase δ1, catalytic subunit; COL25A1, collagen type XXV alpha 1 chain; ANK3, ankyrin 3; GRTP1, growth hormone regulated TBC protein 1; COL4A2-AS1, COL4A2 antisense RNA 1; ALDH1A1, aldehyde dehydrogenase 1 family member A1.
Analysis of lncRNA subgroups and sequences
LncRNA may be divided into different subclasses, including antisense lncRNAs, lncRNA with enhancer function, large intermediate noncoding RNA and exon overlap lncRNA. As lncRNA sequences and transcription sites may affect the mode of action, online software, including UCSC and BLAST, were used to further analyze the lncRNA sequences and their adjacent genes in order to elucidate the mode of action of these lncRNAs. Among different types of lncRNA, antisense lncRNA has been studied most extensively, with >30% of annotated human transcripts having been assigned corresponding antisense lncRNAs, the interaction of which may occur at the level of complementary base pairing mechanisms or post-transcriptional regulation of the corresponding target mRNA, and thus having a biological function.
miRNA target prediction of NR_125715 and NR_046402
The abovementioned BLAST analysis indicated that NR_125715 and TGF-β2 (TGFB2), as well as NR_046402 and DNA polymerase δ1, catalytic subunit (POLD1) had 100% similarity with a homologous gene base sequence, and certain studies have indicated that specific lncRNAs compete with certain mRNAs to bind to the same miRNA, which regulates gene expression levels. In order to further analyze the underlying mechanism of action, TargetScan online software was used to predict the possible binding of NR_125715 and NR_046402 miRNAs. The binding site of the miRNA for the target gene is located at its 5′ end encompassing nucleotides 2–8 (2–8 nt), i.e., the 'seed sequence' consisting of 7 nucleotides. The complementarity of the miRNA seed sequence and the target gene is the major mechanism for regulating gene expression, i.e., if the two are completely complementary, the target gene is degraded; if there are one or two base mismatches, the degree of complementarity is low and the translation of the target mRNA is inhibited. In TargetScan, the pairing of seed regions is divided into six types: The 8mer is fully aligned with the target gene at 2–8 nt, and the additional target gene corresponds to the first nucleotide of the miRNA at the A; 7mer-1a refers to the pair of the first 2–7 nt of the miRNA and the complementary target gene, plus the target gene corresponding to the first nucleotide of the miRNA at the A; the 7mer-m8 is the 2–8 nt of the miRNA completely matching with the target gene; the 6mer is the 2–7 nt of the miRNA exactly matching the target gene; the offset 6mer is the 3–8 nt of the miRNA fully matched with the target gene; and 'imperfect' refers to the 2–7 nt of the miRNA with the G:U pair being mismatched or deleted. The order of the binding according to the reliability score is as follows: 8mer > 7mer-m8 > 7mer-A1 > 6mer > offset 6mer > imperfect, where the 8mer is more site-specific than the 6mer, which is used to search for target genes with higher sensitivity, but which corresponds with a decline in specificity (18). The context++ model was more predictive than any published model and at least as predictive as the most informative in vivo crosslinking approaches. As the engine powering the latest version of TargetScan (v7.0; targetscan.org), this model provides a valuable resource for placing the miRNAs of human, mouse, zebrafish, and other vertebrate species into their respective gene-regulatory networks (19).
Construction of the coding-noncoding co-expression (CNC) network
In order to obtain more credible biological functions, the lncRNA-mRNA co-expression network of two important lncRNAs, including NR_125715 and NR_046402, was built. The CNC network was constructed based on the Pearson correlation analysis. LncRNAs and mRNAs with an absolute value of the Pearson correlation coefficient of >0.90 were selected to construct the network using Cytoscape software (http://www.cytoscape.org/).
Statistical analysis
Student's t-test was used for statistical analysis to compare the microarray data between the two groups. The fold changes were calculated and subjected to Student's t-tests to analyze the statistical significance of differences in expression. The false discovery rate was calculated to correct the P-values. Differential expression of lncRNAs and mRNAs was defined by setting the threshold for fold changes as >2.0 and <0.5, as well as P<0.05. P<0.05 was considered to indicate a statistically significant difference. Statistical analyzes were performed using SPSS version 13 software (SPSS, Inc., Chicago, IL, USA).
Results
Biological characteristics of the fibroblasts
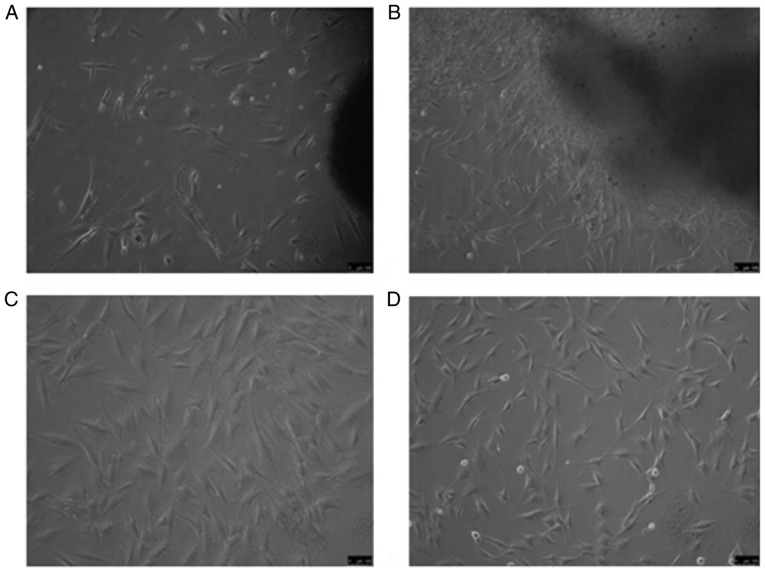
The fibroblasts were subjected to primary tissue culture in petri dishes via the tissue adherence method and the cells used in the experiment were of P3. The cells from the two groups were observed under an inverted phase contrast microscope. Fig. 1A diplays fibroblasts from HS tissue after 7 days of culture and Fig. 1B presents fibroblasts from normal skin after 7 days, which exhibited a long spindle- or star-shaped morphology. P3 fibroblasts from HS and normal skin tissues at the same time points are displayed in Fig. 1C and D, respectively.
Figure 1.
Fibroblasts derived from hypertrophic scar and normal skin under an inverted phase contrast microscope. (A) Hypertrophic scar tissue adherence after 7 days; (B) normal skin tissue adherence method for 7 days, resulting in long spindle- or star-shaped cells; (C) third generation fibroblasts from hypertro-phic scar source; (D) third generation fibroblasts of normal skin origin (magnification, ×100; scale bar, 100 µm).
Quality control of total RNA
The A260/A280 ratios of RNA extracted from the two groups were 1.86 and 1.92, respectively, which was within the required range of 1.8–2.0, indicating no degradation and protein or DNA contamination. The A260/A230 ratios were 2.35 and 2.40, respectively, indicating no organic solvent contamination. Agarose gel electrophoresis produced clear 18 and 28 S bands for the total RNA, indicating that the sample was of good quality and suitable for use in the subsequent lncRNA and mRNA analyses.
Overview of lncRNA and mRNA microarray analysis
In the scanner the high-quality cRNA probe was distributed uniformly. Subsequently, through sample labeling and array hybridization, and scatter plot (Fig. 2) were obtained. The scatter plot provided an overall indication of sample similarity between individual transcripts. For a systematic comparison of lncRNA expression between HS and normal skin, hierarchical clustering analysis was performed and samples were arranged into groups based on their expression levels. Taken together, these results suggested a significant difference in lncRNA expression between HS and normal skin.
Figure 2.
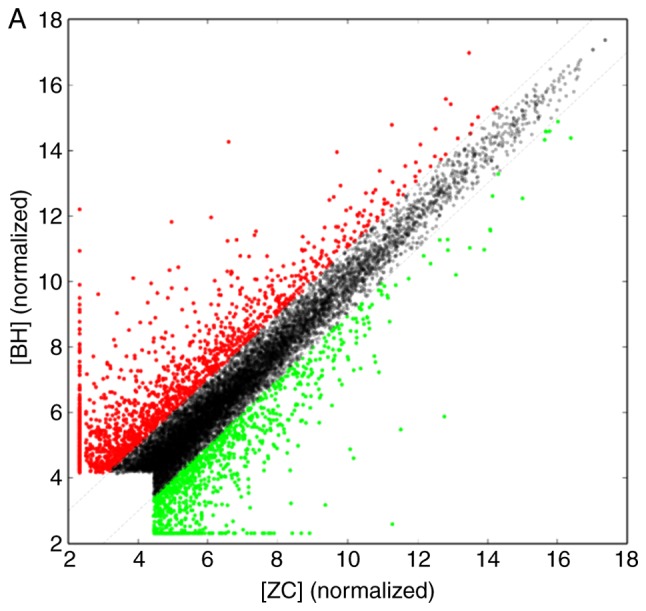
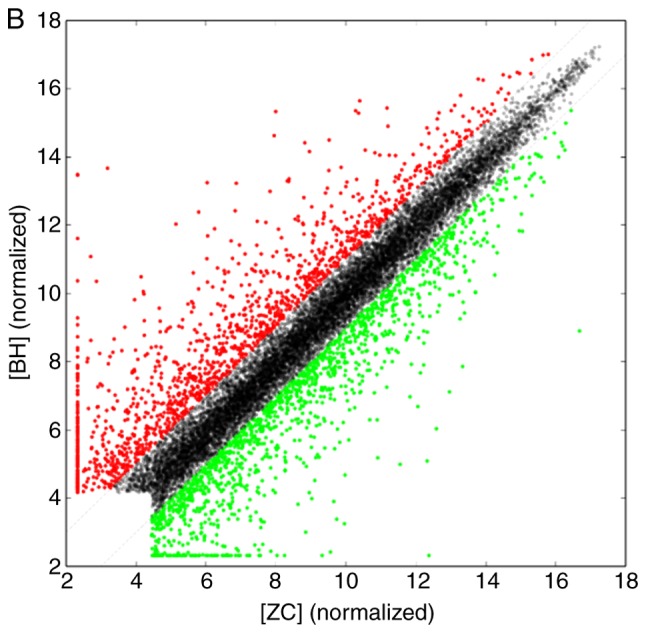
Scatter plots indicating the correlation of (A) long noncoding RNA and (B) mRNA expression between the ZC and BH groups. Red indicates that lncRNA and mRNA expression is upregulated and green indicates downregulation. ZC, fibroblasts derived from normal skin; BH, fibroblasts derived from hypertrophic scar.
Using microarray analysis, a total if 6,104 lncRNAs and 2,952 mRNAs were identified to be differentially expressed in fibroblasts derived from HS compared with those from normal skin (|fold change| ≥2.0, P<0.05 and false discocery rate <0.05). Among these, 3,709 lncRNAs and 1,424 mRNAs were upregulated in the HS group, while 2,935 lncRNAs and 1,528 mRNAs were downregulated (Table II), and various RNAs were changed >20-fold (Tables III and IV).
Table II.
Number of differentially expressed RNAs between hypertrophic scar and normal skin fibroblasts.
| A, Long noncoding RNA
| |||
|---|---|---|---|
| Aberrant RNAs (n) | Fold change
|
||
| 2–10 | 10–20 | ≥20 | |
| 6104 | |||
| 3709 upregulated | 3368 | 244 | 97 |
| 2935 downregulated | 2293 | 63 | 39 |
| B, mRNA
| |||
|---|---|---|---|
| Aberrant RNAs (n) | Fold change
|
||
| 2–10 | 10–20 | ≥20 | |
| 2952 | |||
| 1424 upregulated | 1247 | 97 | 80 |
| 1528 downregulated | 1411 | 72 | 45 |
Table III.
mRNAs with >20 fold change.
| GenBank ID | Gene name | Fold change |
|---|---|---|
| NM_014391 | ANKRD1 | +148.2 |
| NM_001165 | BIRC3 | +22.0 |
| NM_006894 | FMO3 | +57.606484 |
| NM_001147 | ANGPT2 | +154.5 |
| NM_005302 | GPR37 | −30.4 |
| NM_013381 | TRHDE | −104.6 |
| NM_003220 | TFAP2A | −41.4 |
| NM_000689 | ALDH1A1 | −219.6 |
All of these mRNAs were protein-coding. ANK3, ankyrin repeat domain 1; ALDH1A1, aldehyde dehydrogenase 1 family member A1; BIRC3, baculoviral IAP repeat containing 3; FMO3, flavin containing monooxygenase 3; ANGPT2, angiopoietin 2; GPR37, G protein-coupled receptor 37; TRHDE, thyrotropin releasing hormone degrading enzyme; TFAP2A, transcription factor AP-2α.
Table IV.
Long noncoding RNAs with >20-fold change.
| ID | Gene name | Fold change | Mode of action |
|---|---|---|---|
| T348237 | G082118 | +87.2 | Intergenic |
| NR_121621 | LINC01605 | +24.9 | Intergenic |
| NR_024376 | FAM225B | +25.6 | Intergenic |
| ENST00000522281 | RP11-44D19.1 | +107.7 | Intergenic |
| NR_040244 | LINC00547 | +21.3 | Intergenic |
| T331643 | G077712 | +20.5 | Intronic antisense |
| NR_034138 | EPHA5-AS1 | +22.8 | Natural antisense |
| T283696 | G066125 | −23.8 | Natural antisense |
| NR_122071 | LOC399886 | −410.5 | Intergenic |
| NR_002161 | FAM224A | −81.7 | Intergenic |
| ENST00000578967 | RP11-676J15.1 | −21.4 | Intergenic |
| ENST00000587099 | RP11-384O8.1 | −32.2 | Intergenic |
| ENST00000554032 | RP6-65G23.3 | −35.3 | Bidirectional |
| NR_024360 | ZFHX4-AS1 | −37.9 | Intronic antisense |
| ENST00000511165 | RP11-114H21.2 | −42.6 | Intergenic |
| NR_026836 | TRHDE-AS1 | −72.4 | Natural antisense |
| NR_131213 | XLOC-007697 | −47.5 | Natural antisense |
LINC00547, long intergenic non-protein coding RNA 1605; EPHA5, EPH receptor A5; AS1, antisense RNA1; FAM224A, family with sequence similarity 224 member A; ZFHX4, zinc finger homeobox 4; TRHDE, thyrotropin releasing hormone degrading enzyme.
RT-qPCR validation results
Based on the characteristics of the differentially expressed lncRNA, including fold changes, loci, nearby coding genes, several candidate lncRNAs (including NR_125715, NR_046402, NR_046583 and ENST00000559026) and mRNAs (including NM_19872, NM_020987, NM_024719 and NM_000689) were selected for further analysis to verify the results of the microarray analysis. As presented in Table V, the RT-qPCR results were consistent with the microarray data, which means that the results of the microarray analysis were reliable and stable.
Table V.
Hypertrophic scar vs. normal skin fibroblasts expression ratios determined by microarray analysis and RT-qPCR analysis.
| Experiment | COL25A1 | ANK3 | POLD1 | GRTP1 | COL4A2-AS1 | ALDH1A1 | RP11-59H7.3 | TGFB2-OT1 |
|---|---|---|---|---|---|---|---|---|
| Microarray | 15.23 | 118.72 | −2.76 | −28.83 | 3.91 | −219.65 | 2.22 | 8.07 |
| RT-qPCR | 53.86 | −59.87 | 0.15 | 0.10 | 6.80 | 0.00 | 9.54 | 7.72 |
RT-qPCR, reverse transcription-quantitative polymerase chain reaction analysis; TGFB2-OT1, transforming growth factor β2 overlapping transcript 1; POLD1, DNA polymerase δ1, catalytic subunit; COL25A1, collagen type XXV α1 chain; ANK3, ankyrin 3; GRTP1, growth hormone regulated TBC protein 1; COL4A2-AS1, COL4A2 antisense RNA 1; ALDH1A1, aldehyde dehydrogenase 1 family member A1.
GO and pathway analysis
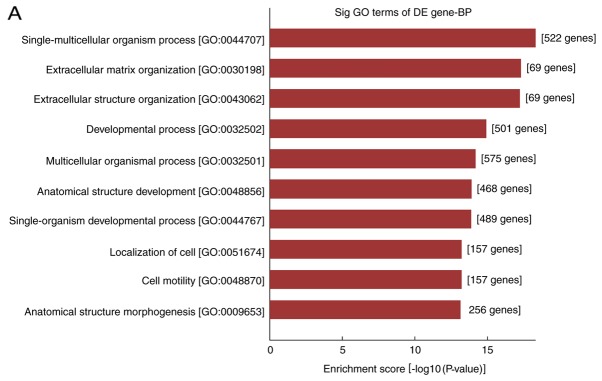
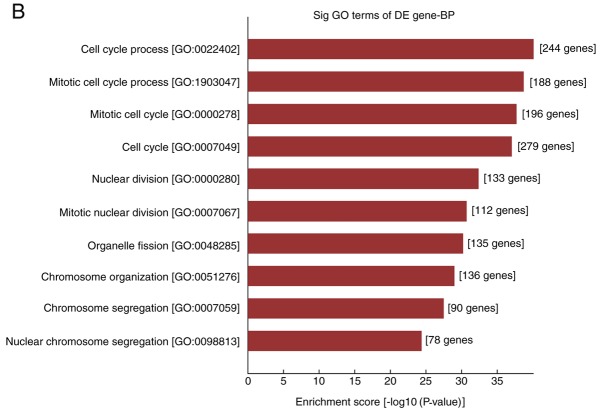
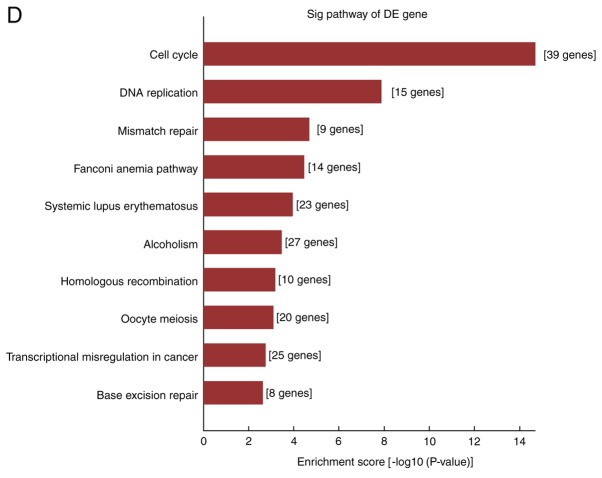
In the present study, the differentially expressed mRNAs were subjected to GO analysis and enriched genes in the categories biological processes (BP), molecular function and cellular component were identified. GO analysis indicated that the differentially expressed mRNA was involved in 2,704 GO entries, of which 2,256 were BP, including 1,042 upregulated GO terms and 1,214 downregulated GO terms accounting for 83.4%. According to this analysis, the majority of differentially expressed mRNAs in HS were implicated in BP, and the top ten GO terms in the BP category in which the upregulated and downregulated mRNAs were enriched are displayed in Fig. 3A and B, respectively. The genes that were significantly upregulated were mainly involved in cell growth and development, movement, localization, extracellular matrix (ECM) composition and tissue reconstruction, while the significantly downregulated ones were involved in cell mitotic cycle, nuclear division, organelle fission, as well as chromosome composition and segregation.
Figure 3.
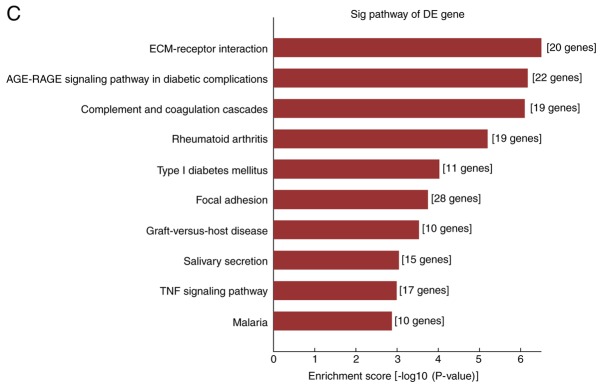
GO and pathway analysis of differentially expressed mRNAs in fibroblasts of hypertrophic scars. (A and B) The top 10 GO terms in the category BP in which (A) upregulated and (B) downregulated mRNAs are enriched; (C and D) The top 10 Kyoto Encyclopedia of Genes and Genomes pathways associated with the coding genes of (C) upregulated and (D) downregulated mRNAs. GO, gene ontology; BP, biological processes; lncRNA, long noncoding RNA; TNF, tumor necrosis factor; ECM, extracellular matrix; RAGE, receptor of advanced glycation end products.
Analysis of the differentially expressed genes using the KEGG regulatory network database indicated that 56 upregulated and 19 downregulated genes were significantly enriched in KEGG pathways. Fig. 3C and D displays the top 10 KEGG pathways in which upregulated and downregulated mRNAs, respectively, were enriched. For the upregulated mRNAs, the KEGG pathways with the highest enrichment were ECM receptor interactions, advanced glycation end products (AGE)-receptor of AGE signaling pathway in diabetic complications, tumor necrosis factor signaling pathway and cell adhesion, while downregulated mRNAs were significantly enriched in pathways including the cell cycle, DNA replication and cancer-associated signaling.
Classification and subgroup analysis of lncRNA
Antisense lncRNAs of their sense mRNAs were identified according to the previous literature in the PubMed database. (ncbi.nlm.nih.gov/geo/). A total of 166 differentially expressed antisense lncRNAs were identified by microarray analysis. Antisense lncRNA was transcribed from the antisense strand of DNA double-stranded, which may associate with the sense protein-coding strand. By pairing through a certain base complementarity and sequence overlap, these antisense lncRNAs may control the corresponding target mRNA, thus affecting their functions. In the present study, a total of 236 differentially expressed exon chain lncRNAs were detected in HS, while the exon-positive chain-type lncRNA had a similar nucleotide sequence to that of the corresponding mRNA homologous gene. These similar base sequences may regulate the characteristics of HS by competitive endogenous mechanisms, and it is therefore important to further assess whether the corresponding mRNA is differentially expressed. By sequence alignment and chromosomal localization using BLAST, lncRNAs and their associated mRNA were analysed in the present study in order to identify the function and mechanisms of action of lncRNAs. This analysis revealed significant differences in the expression of partially differentially expressed exon chain lncRNA and its homology mRNA, including NR_125715 and its homologous gene TGFB2, and their sequences were 100% similar. NR_046402 and POLD1 were also homologous genes, and their sequences were 100% similar (Table VI). Through these methods, it was identified that certain adjacent protein-coding genes of the lncRNAs may regulate processes and functions including the cell cycle and ECM composition, which may be involved in the development of HS (Table VII).
Table VI.
Similarity between parts of exon sense-overlapping lncRNAs and nearby mRNAs.
| Name of lncRNA | Chr | Strand | Nearby gene ID | Nearby gene name | Similarity (%) |
|---|---|---|---|---|---|
| NR_125715 | 1 | + | NM_003238 | TGFB2 | 100 |
| NR_046402 | 19 | + | NM_002691 | POLD1 | 100 |
lncRNA, long noncoding RNA; Chr, chromatin; TGFB2, transforming growth factor β2; POLD1, DNA polymerase δ1, catalytic subunit; +, sense strand.
Table VII.
Position between parts of lncRNAs and nearby mRNA.
| lncRNA ID | Chr | Strand | Interaction | Nearby gene ID | Nearby gene name | Strand (mRNA) |
|---|---|---|---|---|---|---|
| NR_125958 | 1 | − | Intron sense-overlapping | NM_152281 | GORAB | + |
| NR_125715 | 10 | − | Exon sense-overlapping | NM_003238 | TGFB2 | + |
| ENST00000606280 | 1 | − | Intron sense-overlapping | NM_024848 | MORN1 | − |
| NR_123723 | 12 | + | Natural antisense | NM_022468 | MMP25 | − |
| NR_037635 | 1 | − | Exon sense-overlapping | NM_012333 | MYCBP | − |
| NR_046402 | 19 | + | Exon sense-overlapping | NM_002691 | POLD1 | + |
lncRNA, long noncoding RNA; Chr, chromatin; TGFB2, transforming growth factor β2; POLD1, DNA polymerase δ1, catalytic subunit; MMP25, matrix metallopeptidase 25; MYCBP, MYC binding protein; MORN1, MORN repeat containing 1; GORAB, golgin, RAB6-interacting.
miRNA-target sites in NR_125715 and NR_046402
In order to identify the mechanisms associated with homology lncRNAs, two lncRNAs, NR-125715 and NR-046402, were selected, which were distinctly differentially expressed in HS, and were further analyzed using Targetscan to match highly complementary miRNAs forming an 8mer, 7mer-m8 or 7mer-A1. A total of 1,067 complementary miRNAs forming pairs with NR-125715 were identified, among which 16 miRNAs were highly complementary and relatively conserved. These were miR-145-5p, miR-130-3p/301-3p/454-3p, miR-141-5p, miR-141-3p/200a-3p, miR-148-3p/152-3p, miR-29-3p, miR-133a-3p.2/133b, miR-21-5p/590-5p miR-193-3p, miR-25-3p/32-5p/92-3p/363-3p/367-3p, miR-142-3p.1, miR-153-3p, miR-23-3p, miR-203a-3p.1, miR-142-5p and miR-489-3p. It was therefore confirmed that certain members of the miR family may form complementary pairs with NR-125715 (Table VIII). NR-046402 was identified to be highly complementary to 26 miRNAs, but these did not have any highly conserved sites; these were miR-3189-5p, miR-7108-3p, miR-7108-3p, miR-4258, miR-4281, miR-133a-3p, miR-7152-3p, miR-4308, miR-6791-5p, miR-7113-3p, miR-6867-3p, miR-4658-3p, miR-4287, miR-4469, miR-629-3p, miR-5571-5p, miR-4477a, miR-198, miR-6507-3p, miR-548g-5p, miR-5p, miR-548f-5p, miR-548aj-5p and miR-548p (Table IX). Furthermore, according to the abovementioned BLAST analysis, NR_125715 and TGFB2, as well as and NR_046402 and POLD1 are homologous genes, with a base sequence similarity of 100%, based on which it may be speculated that these mRNAs and their corresponding lncRNA have the same miRNA response elements (MRE).
Table VIII.
miRNA families binding to NR_125715.
| miRNA family | Conserved sites
|
Poorly conserved sites
|
6mer sites | Cumulative weighted context++ score | Total context++ scorea | Aggregate PCT | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | 8mer | 7mer-m8 | 7mer-A1 | Total | 8mer | 7mer-m8 | 7mer-A1 | |||||
| miR-145-5p | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | −0. 28 | −0.40 | 0.97 |
| miR-130-3p/301-3p/454-3p | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | −0.18 | −0.37 | 0.94 |
| miR-199-5p | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | −0.58 | −0.61 | 0.93 |
| miR-141-3p/200a-3p | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | −0.52 | −0.52 | 0.90 |
| miR-148-3p/152-3p | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | −0.24 | −0.51 | 0.89 |
| miR-29-3p | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | −0.16 | −0.32 | 0.88 |
| miR-133a-3p.2/133b | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | −0.32 | −0.34 | 0.86 |
| miR-21-5p/590-5p | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | −0.09 | −0.19 | 0.79 |
| miR-193-3p | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | −0.24 | −0.49 | 0.77 |
| miR-25-3p/32-5p/92-3p/363-3p/367-3p | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | −0.15 | −0.16 | 0.72 |
| miR-142-3p.1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | −0.09 | −0.17 | 0.54 |
| miR-153-3p | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | −0.27 | −0.28 | 0.32 |
| miR-23-3p | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | −0.06 | −0.06 | 0.27 |
| miR-203a-3p.1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | −0.16 | −0.16 | 0.24 |
| miR-142-5p | 1 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 2 | −0.01 | −0.02 | 0.22 |
| miR-489-3p | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | −0.01 | −0.02 | 0.15 |
lens.elifesciences.org/05005/index.html. miR/miRNA, microRNA.
Table IX.
miRNA families binding to NR_046402.
| miRNA family | Conserved sites
|
Poorly conserved sites
|
6mer sites | Cumulative weighted context++ score | Total context++ scorea | Aggregate PCT | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | 8mer | 7mer-m8 | 7mer-A1 | Total | 8mer | 7mer-m8 | 7mer-A1 | |||||
| miR-133a-3p.1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | −0.16 | −0.16 | <0.1 |
| miR-3189-5p | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | −0.20 | −0.20 | N/A |
| miR-7108-3p | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | −0.33 | −0.33 | N/A |
| miR-7108-3p | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | −0.78 | −0.78 | N/A |
| miR-4258 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | −0.32 | −0.32 | N/A |
| miR-4281 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | −0.39 | −0.39 | N/A |
| miR-7152-3p | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | −0.12 | −0.12 | N/A |
| miR-4308 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | −0.16 | −0.16 | N/A |
| miR-6791-5p | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | −0.17 | −0.17 | N/A |
| miR-7113-3p | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | −0.52 | −0.52 | N/A |
| miR-133a-3p.1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | −0.16 | −0.16 | <0.1 |
| miR-3189-5p | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | −0.20 | −0.20 | N/A |
| miR-7108-3p | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | −0.33 | −0.33 | N/A |
| miR-7108-3p | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | −0.78 | −0.78 | N/A |
lens.elifesciences.org/05005/index.html. miR/miRNA, microRNA; N/A not applicable.
Construction of lncRNA-mRNA co-expression network
The CNC network contained 19 pairs of co-expressed lncRNAs and mRNAs, comprising 19 mRNAs and 2 lncRNAs, in which the expression changes of lncRNAs and their associated mRNA were in the same or opposite direction. lncRNA NR_125715 and NR_046402 were positively associated with TGFB2 and POLD1, respectively (Fig. 4). Thus, it was speculated that lncRNA NR_125715 and NR_046402 may regulate the expression of TGFB2 and POLD1 mRNA by a competing endogenous RNA regulation mechanism. TGFB2 and POLD1 are involved in multiple BPs, including cell cycle, DNA replication, ECM-receptor interaction and the TGF-β signaling pathway.
Figure 4.
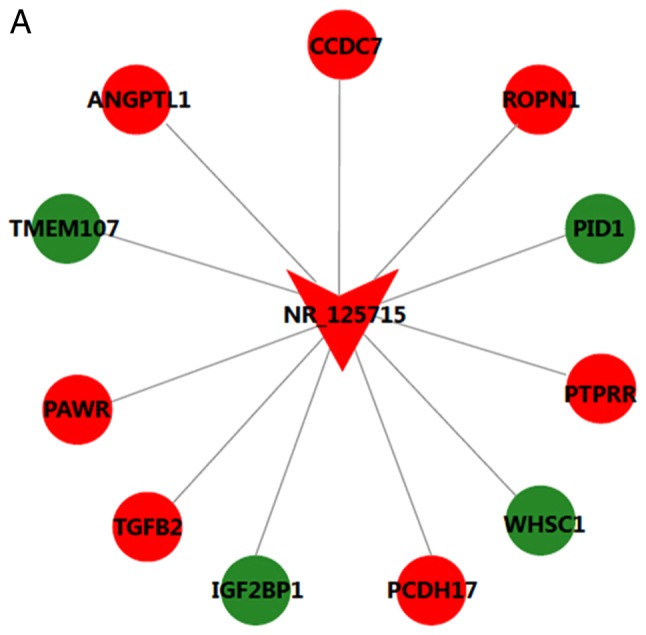
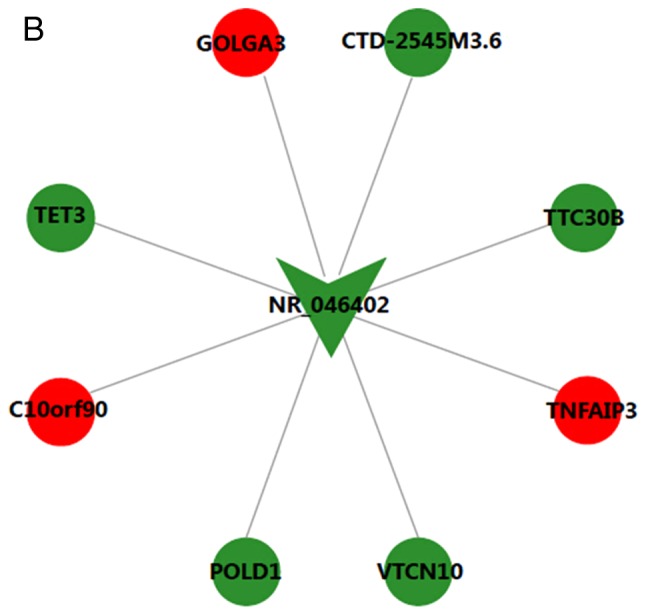
Long noncoding RNA-mRNA co-expression network for (A) NR_125715 and (B) NR_046401. Red represents upregulation and green represents downregulation. ANGPTL1, angiopoietin like 1; CCDC7, coiled-coil domain containing 7; ROPN1, rhophilin associated tail protein 1; PID1, phosphotyrosine interaction domain containing 1; PTPRR, protein tyrosine phosphatase, receptor type R; WHSC1, Wolf-Hirschhorn syndrome candidate 1; PCDH17, protocadherin 17; IGF2BP1, insulin like growth factor 2 mRNA binding protein 1; TGFB2, transforming growth factor β2; PAWR, pro-apoptotic WT1 regulator; TMEM107, transmembrane protein 107; GOLGA3, golgin A3; CTD-2545M3.6, coats disease-2545M3.6; TTC30B, tetratricopeptide repeat domain 30B; TNFAIP3, TNF-α induced protein 3; POLD1, DNA polymerase δ1; TET3, tet methylcytosine dioxygenase 3.
Discussion
HS formation is a skin condition that may cause severe functional and aesthetic defects. HS mainly arises from excessive proliferation of fibroblasts and the accumulation of ECM (20). A thorough understanding of the pathophysiology and clinical characteristics of HS may facilitate the selection of the most appropriate treatment strategy. Numerous studies have reported that ncRNAs, including miRNAs and lncRNAs, participate in a variety of basic cellular processes, including ectopic regulation of gene expression, cell proliferation and differentiation, metabolism and apoptosis. With the development of microarray techniques, it has been demonstrated that ncRNAs have a key role in HS (21).
The microarray performed in the present study indicated that 6,104 lncRNAs and 2,952 mRNAs were differentially expressed in fibroblasts derived from HS compared with those from normal skin. The RT-qPCR results validated that the expression of certain lncRNAs and mRNAs was consistent with the microarray data, which suggests that the results of the microarray are credible. GO and KEEG pathway enrichment analyses of the differentially expressed mRNAs suggested their involvement in multiple BPs and pathways, including the cell cycle, DNA replication, ECM-receptor interaction and the TGF-β signaling pathway. A previous study has indicated that lncRNAs are associated with their neighboring protein-coding genes (22). Thus, correlation analysis between lncRNAs and their neighboring mRNAs may elucidate the functions of lncRNAs in the development of HS. Indeed, thousands of differentially expressed lncRNAs were identified in the present microarray analysis. However, a review of the available literature indicated that the function of most of the differentially expressed lncRNAs remains to be elucidated. The results of the present chip microarray analysis identified that TGFB2 was upregulated, while POLD1 was downregulated. It has been reported that TGFB2 is closely associated with ECM-receptor interaction, while POLD1 is involved in DNA replication and has a key role in the development of HS. By gene sequence alignment, the present study identified that NR_125715 and NR_046402 have the same miRNA binding sequence as TGFB2 and POLD1, respectively, and their direction of change was consistent. Therefore, these genes were selected for PCR validation. The results of the present study indicated that NR_125715 and NR_046402 may participate in the development of HS and the prognosis of affected patients by controlling the endogenous mechanisms of miRNAs and regulating the expression of TGFB2 and POLD1, respectively. The present study therefore provides a theoretical basis for further clarifying the mechanisms of HS and offers a novel approach for its prevention and treatment.
The present study indicated that lncRNA NR_125715 was upregulated in HS, and the associated mRNA TGFB2, which has an identical miRNA binding sequence, exhibited the same trend of upregulation. The TGFB2 gene encodes a secreted ligand of the TGF-β superfamily of proteins. Ligands of this family bind to various TGF-β receptors, leading to the recruitment and activation of Smad family transcription factors that regulate gene expression. The encoded pre-proprotein is proteolytically processed to generate a latency-associated peptide (LAP) and a mature peptide, and occurs in either a latent form composed of a mature peptide homodimer, a LAP homodimer and a latent TGF-β binding protein, or in an active form consisting solely of the mature peptide homodimer. The mature peptide may also form heterodimers with other TGF-β family members. Disruption of the TGF-β/Smad pathway has been implicated in a variety of human cancer types. TGF-β and cell signaling pathways mediated by it are thought to have an important role in scar formation, they promote collagen synthesis and induce fibroblasts to transform into myofibroblasts (23). Studies have also demonstrated that after cataract surgery, TGF-β is released from the residual lens epithelium through the TGF-β-mediated Smad signaling pathway to promote epithelial mesenchymal transition (EMT), resulting in posterior capsule opaque (24,25). Numerous studies have confirmed the role of TGF-β in HS, such that TGF-β induces the expression of smooth muscle actin (SMA), fibronectin, as well as type I and type III collagen, and reduces the expression of vimentin, Snail1/2 and E-cadherin, which are characteristic indicators of EMT (26). Thus, TGF-β has a leading role during the formation of HS (27).
The prsent microarray analysis revealed that the lncRNA NR_046402 was downregulated in HS, and the associated mRNA POLD1 was also downregulated, which changed in the same direction. The POLD1 gene is located on chromosome 19 at q13.3-q13.4 and is ~34 kb long (28). The high fidelity of DNA replication maintains the integrity of DNA and is essential for the survival of cells and organisms. DNA polymerase in the eukaryotic cell replication process is absolutely necessary (29). POLD has a crucial role in genome maintenance by participating in replicative DNA synthesis and multiple synthetic repair processes (30). It has intrinsic 3′-5′ exon-nuclease activity, which is the basis for the function of this enzyme, and the interaction of POLD1 with proliferating cell nuclear antigen allows it to gradually replicate DNA (31). Human POLD is a complex consisting of four subunits: p125, p68, p50 and p12 (32), among which the p125 subunit has been identified as a catalytic subunit, encoded by the POLD1 gene in humans, bearing the POLD and 3′-5′ exonuclease activity sites (33). POLD1 has an important biological role in cell cycle regulation and DNA damage repair (34).
LncRNA has a variety of functions involved in the regulation of mRNA transcription and translation. ceRNA is a recently reported functional lncRNA type that has miRNA recognition sequences in common with mRNA and may compete for miRNA binding, thereby affecting the regulation and function of target mRNA (35,36). In the present study, the two exon sense-overlapping lncRNAs NR_125715 and NR_046402 were identified to have 100% sequence homology with TGFB2 and POLD1 mRNA, respectively. Given that these lncRNAs are highly homologous to these mRNAs, they may have common MREs and influence expression levels by competing for shared miRNAs. Thus, it was suggested that certain lncRNAs may act as ceRNAs by attaching to miRNAs that are part of transcriptional networks involved in the formation of HS.
In the present study, TargetScan and miRanda database analyses were performed to identify miRNAs with complementarity ro NR_125715; these included miR-145-5p, miR-130-3p/301-3p/454-3p, miR-199-5p, miR-141-3p/200a-3p, miR-148-3p/152-3p, miR-29-3p, miR-133a-3p.2/133b, miR-21-5p/590-5p, miR-193-3p, miR-25-3p/32-5p/92-3p/363-3p//367-3p, miR-142-3p.1, miR-153-3p, miR-23-3p, miR-203a-3p.1, miR-142-5p and miR-489-3p. The binding site of miRNAs to their target genes is located at its 5′ end, the 'seed sequence' consisting of 7 nucleotides (37). The complementarity of the miRNA seed sequence and the target gene is the major mechanism for regulating gene expression, i.e., if the two are completely complementary, the target gene is degraded. If the degree of complementarity is low and there are one or two base mismatches, the target gene expression is depressed (38). It has been indicated that overexpression of miR-29b significantly reduces the expression levels of COL1A1 and α-SMA, inhibits myofibroblast-like cell proliferation and induces apoptosis, suggesting that miR-29b may convey significant resistance to HS fibrosis (15). Consistently with this, Bi et al (39) reported that the miR-29 family has a role in the scar by regulating the translation of ECM mRNA. In addition, miR-200b, miR-181b, miR-145, miR-143-3p, miR-31 and miR-21 have been reported to be involved in HS formation (40). miR-200b, a member of the miR-200 family, has been associated with abnormal proliferation of fibroblasts (41). In addition, miR-200b affects HS formation by influencing collagen type I and III synthesis, fibronectin expression and TGFB1/α-SMA signaling by modulating cell proliferation and apoptosis in human HS fibroblasts (42). In previous studies, the expression of miR-143-3p in HS tissue was significantly reduced. In addition, the protein expression of Col I, Col III and α-SMA in HS fibroblasts transfected with miR-143-3p targeting connective tissue growth factor was reduced, indicating that miR-143-3p acted as an anti-fibrosis factor in HS formation (43). Another study indicated that miR-31 was highly expressed during wound healing, whereas transgenic mouse models with elevated miR-31 levels exhibited abnormal wound healing (44). Kwan et al (45) reported that miR-181b was involved in the aberrant expression of membrane proteoglycans in skin and wound healing, and transfection with miR-181b reversed TGFB1-induced downregulation of proteomic proteins and prevented the transformation of fibroblast to myofibroblasts in HS. Zhou et al (14) indicated that miR-200b and miR-21b expression may be involved in HS formation by activating the TGF-β/Smad signaling pathways.
The miRNAs identified to exhibit the strongest binding with NR_046402 included miR-3189-5p, miR-7108-3p, miR-7108-3p, miR-4258, miR-4281, miR-133a-3p.1, miR-7152-3p, miR-4308, miR-4292, miR-6791-5p, miR-7113-3p, miR-6867-3p, miR-4685-3p, miR-4287, miR-4469, miR-629-3p, miR-4713-5p, miR-5571-5p, miR-4477a, miR-198, miR-6507-3p, miR-548g-5p, miR-548x-5p, miR-548f-5p, miR-548aj-5p and miR-548p. It has been reported that miR-133a is closely associated with ECM fibrosis (46). miR-133a inhibits the mitogen-activated protein kinase kinase/extracellular signal-regulated protein kinase pathway to enhance radiosensitivity and promote cell apoptosis (47). NR_125715 and NR_046402 are likely to compete with the miRNA pool to regulate the expression of TGFB2 and POLD1 participating in HS formation. In addition, The CNC network analysis indicated that lncRNAs NR_125715 and NR_046402 were positively associated with TGFB2 and POLD1, respectively. Thus, it was spectulated that NR_125715 and NR_046402 may regulate the expression of TGFB2 and POLD1 mRNA via a ceRNA regulatory mechanism.
In conclusion, the present study identified lncRNAs that were differentially expressed in fibroblasts derived from HS compared with those in normal skin, predicted their function by constructing a network of associated mRNAs and ceRNAs. NR_125715 and NR_046402 compete with the miRNA pool to regulate the expression of TGFB2 and POLD1, which are involved in cell cycle, DNA replication, ECM-receptor interactions and the TGF-β signaling pathway, which have an important role in the development of HS. A more in-depth understanding of lncRNAs in further functional studies may lead to novel theories regarding the pathogenesis and treatment of HS.
Acknowledgments
The present study was supported by the National Natural Science Foundation of China (grant nos. 81060323 and 81460293).
Footnotes
Competing interests
The authors declare there is no competing interest.
References
- 1.Rabello FB, Souza CD, Farina Júnior JA. Update on hypertrophic scar treatment. Clinics. 2014;69:565–573. doi: 10.6061/clinics/2014(08)11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sorkin M, Cholok D, Levi B. Scar management of the burned hand. Hand Clin. 2017;33:305–315. doi: 10.1016/j.hcl.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mokos ZB, Jović A, Grgurević L, Dumić-Čule I, Kostović K, Čeović R, Marinović B. Current therapeutic approach to hypertrophic scars. Front Med. 2017;4:83. doi: 10.3389/fmed.2017.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wan DC, Wang KC. Long noncoding RNA: Significance and potential in skin biology. Cold Spring Harb Perspect Med. 2014;4:pii: a015404. doi: 10.1101/cshperspect.a015404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: Past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, McDonel PE, Guttman M, Lander ES. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539:452–455. doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 8.Gao J, Xu W, Wang J, Wang K, Li P. The role and molecular mechanism of non-coding RNAs in pathological cardiac remodeling. Int J Mol Sci. 2017;18:pii: E608. doi: 10.3390/ijms18030608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Li Z, Zheng S, Zhou Y, Zhao L, Ye H, Zhao X, Gao W, Fu Z, Zhou Q, et al. Expression profile of long non-coding RNAs in pancreatic cancer and their clinical significance as biomarkers. Oncotarget. 2015;6:35684–35698. doi: 10.18632/oncotarget.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vassallo I, Zinn P, Lai M, Rajakannu P, Hamou MF, Hegi ME. WIF1 re-expression in glioblastoma inhibits migration through attenuation of non-canonical WNT signaling by downregulating the lncRNA MALAT1. Oncogene. 2016;35:12–21. doi: 10.1038/onc.2015.61. [DOI] [PubMed] [Google Scholar]
- 11.Pastori C, Kapranov P, Penas C, Peschansky V, Volmar CH, Sarkaria JN, Bregy A, Komotar R, Laurent G, St, Ayad NG, Wahlestedt C. The bromodomain protein BRD4 controls HOTAIR, a long noncoding RNA essential for glioblastoma proliferation. Proc Natl Acad Sci USA. 2015;112:8326–8331. doi: 10.1073/pnas.1424220112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mineo M, Ricklefs F, Rooj AK, Lyons SM, Ivanov P, Ansari KI, Nakano I, Chiocca EA, Godlewski J, Bronisz A. The long non-coding RNA HIF1A-AS2 facilitates the maintenance of mesenchymal glioblastoma stem-like cells in hypoxic niches. Cell Rep. 2016;15:2500–2509. doi: 10.1016/j.celrep.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, Chen L, Cao C, Yan H, Zhou B, Gao Y, Li Q, Li J. The long non-coding RNA Lnc RNA8975-1 is upregulated in hypertrophic scar fibroblasts and controls collagen expression. Cell Physiol Biochem. 2016;40:326–334. doi: 10.1159/000452548. [DOI] [PubMed] [Google Scholar]
- 14.Zhou R, Zhang Q, Zhang Y, Fu S, Wang C. Aberrant miR-21 and miR-200b expression and its pro-fibrotic potential in hypertrophic scars. Exp Cell Res. 2015;339:360–366. doi: 10.1016/j.yexcr.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 15.Li J, Cen B, Chen S, He Y. MicroRNA-29b inhibits TGF-β1-induced fibrosis via regulation of the TGF-β1/Smad pathway in primary human endometrial stromal cells. Mol Med Rep. 2016;13:4229–4237. doi: 10.3892/mmr.2016.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomson DW, Dinger ME. Endogenous microRNA sponges: Evidence and controversy. Nat Rev Genet. 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔC T method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Ellwanger DC, Büttner FA, Mewes HW, Stümpflen V. The sufficient minimal set of miRNA seed types. Bioinformatics. 2011;27:1346–1350. doi: 10.1093/bioinformatics/btr149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J, Li Y, Bai X, Li Y, Shi J, Hu D. Recent advances in hypertrophic scar. Histol Histopathol. 2018;33:27–39. doi: 10.14670/HH-11-908. [DOI] [PubMed] [Google Scholar]
- 21.Chen L, Li J, Li Q, Yan H, Zhou B, Gao Y, Li J. Non-coding RNAs: The new insight on hypertrophic scar. J Cell Biochem. 2017;118:1965–1968. doi: 10.1002/jcb.25873. [DOI] [PubMed] [Google Scholar]
- 22.Ponjavic J, Oliver PL, Lunter G, Ponting CP. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009;5:e1000617. doi: 10.1371/journal.pgen.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu Z, Ding J, Shankowsky HA, Tredget EE. The molecular mechanism of hypertrophic scar. J Cell Commun Signal. 2013;7:239–252. doi: 10.1007/s12079-013-0195-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dawes LJ, Elliott RM, Reddan JR, Wormstone YM, Wormstone IM. Oligonucleotide microarray analysis of human lens epithelial cells: TGFbeta regulated gene expression. Mol Vis. 2007;13:1181–1197. [PubMed] [Google Scholar]
- 25.Eldred JA, Dawes LJ, Wormstone IM. The lens as a model for fibrotic disease. Philos Trans R Soc Lond B Biol Sci. 2011;366:1301–1319. doi: 10.1098/rstb.2010.0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hahn JM, McFarland KL, Combs KA, Supp DM. Partial epithelial-mesenchymal transition in keloid scars: Regulation of keloid keratinocyte gene expression by transforming growth factor-b1. Burns Trauma. 2016;4:30. doi: 10.1186/s41038-016-0055-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun Q, Guo S, Wang CC, Sun X, Wang D, Xu N, Jin SF, Li KZ. Cross-talk between TGF-β/Smad pathway and Wnt/β-catenin pathway in pathological scar formation. Int J Clin Exp Pathol. 2015;8:7631–7639. [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao H, Zhang S, Xu D, Lee MY, Zhang Z, Lee EY, Darzynkiewicz Z. Expression of the p12 subunit of human DNA polymerase δ (Pol δ), CDK inhibitor p21WAF1, Cdt1, cyclin A, PCNA and Ki-67 in relation to DNA replication in individual cells. Cell Cycle. 2014;13:3529–3540. doi: 10.4161/15384101.2014.958910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang S, Kang MS, Ryu E, Myung K. Eukaryotic DNA replication: Orchestrated action of multi-subunit protein complexes. Mutat Res. 2017 May 1; doi: 10.1016/j.mrfmmm.2017.04.002. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 30.Sekelsky J. DNA repair in drosophila: Mutagens, models, and missing genes. Genetics. 2017;205:471–490. doi: 10.1534/genetics.116.186759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Y, Meng X, Zhang S, Lee EY, Lee MY. Characterization of human DNA polymerase delta and its subassemblies reconstituted by expression in the MultiBac system. PLoS One. 2012;7:e39156. doi: 10.1371/journal.pone.0039156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee MY, Zhang S, Lin SH, Wang X, Darzynkiewicz Z, Zhang Z, Lee EY. The tail that wags the dog: p12, the smallest subunit of DNA polymerase δ, is degraded by ubiquitin ligases in response to DNA damage and during cell cycle progression. Cell Cycle. 2014;13:23–31. doi: 10.4161/cc.27407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicolas E, Golemis EA, Arora S. POLD1: Central mediator of DNA replication and repair, and implication in cancer and other pathologies. Gene. 2016;590:128–141. doi: 10.1016/j.gene.2016.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song J, Hong P, Liu C, Zhang Y, Wang J, Wang P. Human POLD1 modulates cell cycle progression and DNA damage repair. BMC Biochem. 2015;16:14. doi: 10.1186/s12858-015-0044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, Yang T, Zhang Z, Lu M, Zhao W, Zeng X, Zhang W. Long non-coding RNA TUG1 promotes migration and invasion by acting as a ceRNA of miR-335-5p in osteosarcoma cells. Cancer Sci. 2017;108:859–867. doi: 10.1111/cas.13201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang H, Ma R, Zou S, Wang Y, Li Z, Li W. Reconstruction and analysis of the lncRNA-miRNA-mRNA network based on competitive endogenous RNA reveal functional lncRNAs in rheumatoid arthritis. Mol Biosyst. 2017;13:1182–1192. doi: 10.1039/C7MB00094D. [DOI] [PubMed] [Google Scholar]
- 37.Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: A look under the hood of miRISC. Nat Struct Mol Biol. 2012;19:586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 38.Ul Hussain M. Micro-RNAs (miRNAs): Genomic organisation, biogenesis and mode of action. Cell Tissue Res. 2012;349:405–413. doi: 10.1007/s00441-012-1438-0. [DOI] [PubMed] [Google Scholar]
- 39.Bi S, Cao C, Chai LL, Li SR, Yang DY. Regulatory mechanism of miR-29 over TGF-β1 and COL1 in scar cells. Eur Rev Med Pharmacol Sci. 2017;21:2512–2517. [PubMed] [Google Scholar]
- 40.Babalola O, Mamalis A, Lev-Tov H, Jagdeo J. The role of microRNAs in skin fibrosis. Arch Dermatol Res. 2013;305:763–776. doi: 10.1007/s00403-013-1410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurashige J, Mima K, Sawada G, Takahashi Y, Eguchi H, Sugimachi K, Mori M, Yanagihara K, Yashiro M, Hirakawa K, et al. Epigenetic modulation and repression of miR-200b by cancer-associated fibroblasts contribute to cancer invasion and peritoneal dissemination in gastric cancer. Carcinogenesis. 2015;36:133–141. doi: 10.1093/carcin/bgu232. [DOI] [PubMed] [Google Scholar]
- 42.Li P, He QY, Luo CQ. Overexpression of miR-200b inhibits the cell proliferation and promotes apoptosis of human hypertro-phic scar fibroblasts in vitro. J Dermatol. 2014;41:903–911. doi: 10.1111/1346-8138.12600. [DOI] [PubMed] [Google Scholar]
- 43.Mu S, Kang B, Zeng W, Sun Y, Yang F. MicroRNA-143-3p inhibits hyperplastic scar formation by targeting connective tissue growth factor CTGF/CC N2 via the Akt/mTOR pathway. Mol Cell Biochem. 2016;416:99–108. doi: 10.1007/s11010-016-2699-9. [DOI] [PubMed] [Google Scholar]
- 44.Li D, Li X, Wang A, Meisgen F, Pivarcsi A, Sonkoly E, Ståhle M, Landén X. MicroRNA-31 promotes skin wound healing by enhancing keratinocyte proliferation and migration. J Invest Dermatol. 2015;135:1676–1685. doi: 10.1038/jid.2015.48. [DOI] [PubMed] [Google Scholar]
- 45.Kwan P, Ding J, Tredget EE. MicroRNA 181b regulates decorin production by dermal fibroblasts and may be a potential therapy for hypertrophic scar. PLoS One. 2015;10:e123054. doi: 10.1371/journal.pone.0123054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rubiś P, Totoń-Żurańska J, Wiśniowska-Śmialek S, Holcman K, Kołton-Wróż M, Wołkow P, Wypasek E, Natorska J, Rudnicka-Sosin L, Pawlak A, et al. Relations between circulating microRNAs (miR-21, miR-26, miR-29, miR-30 and miR-133a), extracellular matrix fibrosis and serum markers of fibrosis in dilated cardiomyopathy. Int J Cardiol. 2017;231:201–206. doi: 10.1016/j.ijcard.2016.11.279. [DOI] [PubMed] [Google Scholar]
- 47.Yang QS, Jiang LP, He CY, Tong YN, Liu YY. Up-regulation of microRNA-133a inhibits the MEK/ERK signaling pathway to promote cell apoptosis and enhance radio-sensitivity by targeting EGFR in esophageal cancer in vivo and in vitro. J Cell Biochem. 2017;118:2625–2634. doi: 10.1002/jcb.25829. [DOI] [PubMed] [Google Scholar]