Abstract
X-linked hypophosphatemic rickets (XLHR; OMIM 307800) is an X-linked dominant disorder caused by mutations in the phosphate-regulating neutral endopeptidase homolog X-linked (PHEX) gene, which is located at Xp22.11. In the present study, two novel variants of the PHEX gene were identified in two unrelated families with XLHR by directly sequencing all 22 exon regions and intron/exon boundaries of the PHEX gene. One missense variant, NM_000444.5: c.1721T>A, was identified in exon 17 of the PHEX gene in Family 1, which led to an amino acid change in the p.Ile574Lys protein. The other splicing variant identified was NM_000444.5: c.591A>G, in exon 5 in Family 2, resulting in a deletion of 77 bp in the 3′ site of exon 5 during splicing, which was verified by direct cDNA sequencing of the PHEX gene. According to the results of reverse transcription-quantitative polymerase chain reaction analysis, the affected male with the splicing variant c.591A>G showed normal gene expression of PHEX, whereas the affected female exhibited low gene expression, compared with normal females. Based on these findings, prenatal diagnoses were made for the fetuses with a family history of XLHR using the backup amniotic fluid samples. One fetus without the missense variant was confirmed to be a healthy girl in a follow-up visit 1 month following birth.
Keywords: phosphate-regulating neutral endopeptidase homolog, X-linked gene, X-linked hypophosphatemic rickets, genetic diagnosis, prenatal diagnosis, missense variant, splicing variant
Introduction
X-linked hypophosphatemic rickets (XLHR; OMIM307800) is a disorder associated with renal phosphate wasting, which results in hypophosphatemia, growth retardation, abnormal bone mineralization, rickets and osteomalacia (1,2). XLHR accounts for >80% of cases of familial hypophosphatemic rickets and occurs in ~1 per 20,000 live births (3–5). Due to loss of the physiological response to low levels of blood phosphate, patients with XLHR are reported to have normal or low serum levels of 1, 25-dihydroxyvitamin D3 (1,25(OH)2D), a normal serum level of parathyroid hormone, increased activity of serum alkaline phosphatases, and resistance to high doses of vitamin D (6,7).
XLHR is caused by variants of the phosphate-regulating neutral endopeptidase homolog, X-linked (PHEX) gene (NC_000023.11; OMIM:300550), which is located at Xp22.11 and contains 22 exons (8,9). According to the Human Gene Mutation Database (HGMD professional 2016.4 release), a total of 510 different PHEX gene variants have been identified, among which point variants, including missense/nonsense variants and splicing variants, are the most common and account for 45.09% (230 in 510) of all cases.
The PHEX gene encodes a protein, namely a phosphate-regulating neutral endopeptidase, belonging to the type II integral membrane zinc-dependent endopeptidase family. It consists of an extracellular C-terminal region, a transmembrane domain, and an N-terminal cytoplasmic tail (10). This protein is predominantly expressed in mineralized tissues, including the calvaria, long bone and teeth, and regulates the synthesis of fibroblast growth factor 23 through indistinct mechanisms (12–15). The aberrance of this protein leads to a reduction in renal phosphate reabsorption, partly via a decline in renal proximal tubular cell type II sodium-phosphate co-transporters, and results in abnormal bone mineralization by affecting fibroblast growth factor receptor signaling in osteocytes (15,16).
The present study reported on two variants of the PHEX gene in two Chinese families affected by XLHR. The primary aim was to provide a genetic diagnosis for the affected family members and to analyze the underlying genotype-phenotype correlations. Based on the results, it is likely to be possible to provide a prenatal diagnosis and medical suggestions to families carrying affected fetuses.
Materials and methods
Subjects and ethics statement
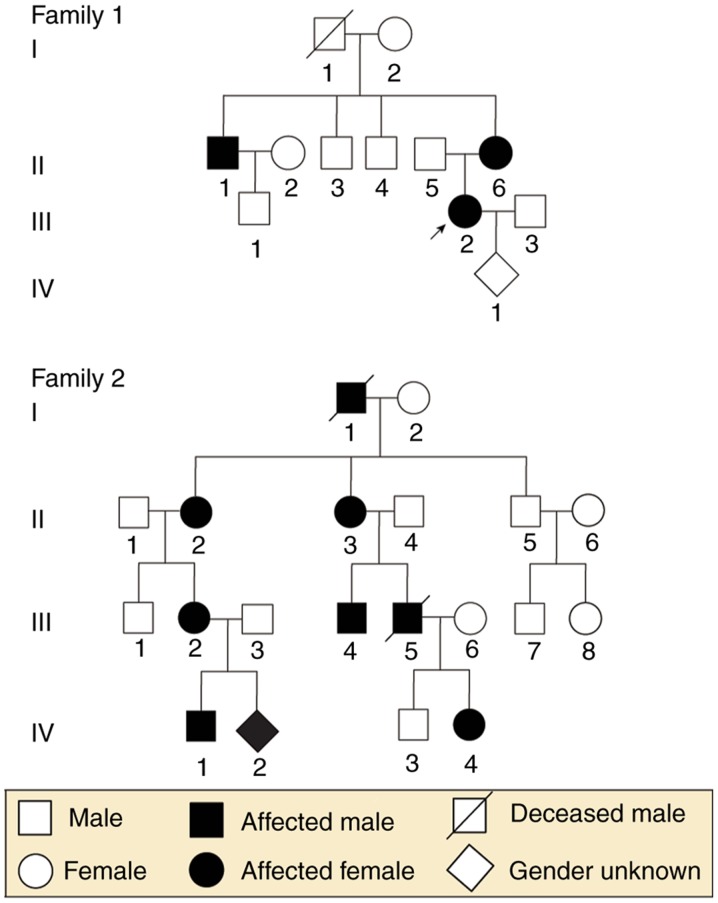
Two unrelated families, including a total of 23 individuals (nine individuals and one fetus in Family 1; 12 individuals and one fetus in Family 2) from the Department of Obstetrics and Gynecology at West China Second University Hospital, Sichuan University (Sichuan, China), were enrolled in the present study. To be specific, the nine members in Family 1 included two affected females and one affected male, and the 12 members of Family 2 included four affected females and two affected males. The present study was performed in the Prenatal Diagnosis Center of Sichuan Province (Sichuan, China) and was approved by the ethics committee of West China Second University Hospital, Sichuan University. The two families provided written informed consent prior to collection of blood samples, pedigree structures and clinical data. Fetal DNA was acquired from backup amniotic fluid samples, which were used for fetal chromosome analysis at the Prenatal Diagnosis Center of Sichuan Province. The family trees are shown in Fig. 1.
Figure 1.
Pedigrees of two unrelated families with X-linked hypophosphatemic rickets. Black symbols represent affected individuals and open symbols represent healthy individuals. Circles represent females, squares represent males, rhombi represent babies of unknown gender. Arrows indicate probands and slashes indicate deceased individuals.
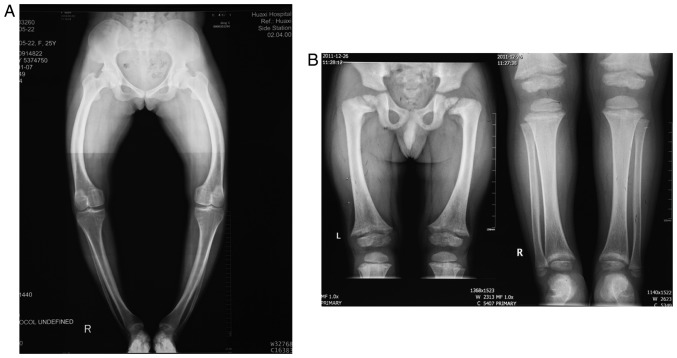
The proband from Family 1 was a 28-year-old pregnant woman with a family history of rickets, who was short in stature (146 cm) and presented with lower-extremity bowing. The T-score of her left foot was-2.0, demonstrating decreased bone mineral density (normal: T-score>-1; decreased: -1> T-score>-2.5; osteoporosis: T-score<-2.5). The X-ray of her lower extremities indicated knock knees, and a bilateral bowed femur, tibia and fibula (Fig. 2A). The patient had a low serum phosphate level (0.64 mmol/l), normal serum calcium level (2.05 mmol/l) and normal serum alkaline phosphatase (328.8 U/l). Despite the lack of laboratory tests and X-ray images of the proband's brother and sister, both were short in stature and had lower-extremity bowing, indicating the probability that they had XLHR.
Figure 2.
X-ray images of two probands. (A) X-ray of the proband from Family 1 showed knock knees, and bilateral bowed femur, tibia and fibula. (B) X-ray of the proband from Family 2 showed decreased bone mineral density, enlargement of long bones, rough metaphysis with cup-shaped change, uneven density of bilateral distal femoral ossification centers, narrowed collodiaphyseal angle and knock knees.
In Family 2, the proband was a 9-year-old boy, who also had signs and a family history of rickets. He was reported to be 110 cm and 35 kg, complained of spontaneous dental abscesses and had only three saprodontias in his mouth. An X-ray of his limbs (aged 4 years old) showed decreased bone mineral density, enlargement of the long bones, rough metaphysis with a cup-shaped change, uneven density of his bilateral distal femoral ossification centers, a narrowed collodiaphyseal angle and knock knees, suggesting active rickets (Fig. 2B). The boy had a low serum phosphate level (1.20 mmol/l), normal serum calcium level (2.20 mmol/l) and elevated serum alkaline phosphatase (412.2 U/l). He was receiving oral phosphate, calcitriol and an active form of vitamin D. His affected mother and three other family members exhibited the same distinctive clinical manifestations of a short stature with lower-extremity bowing.
DNA and RNA preparation
For the adults and children, genomic DNA was extracted from peripheral whole blood samples, and fetal DNA was isolated from amniotic fluid using the Gentra Puregene Blood kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's protocol. RNA was carefully obtained from the peripheral blood mononuclear cells using TRIzol reagent according to the manufacturer's protocol; these cells were isolated from the peripheral whole blood samples using Ficoll-Hypaque density gradient centrifugation at 800 × g for 20 min. DNA was stored at −20°C prior to use, and RNA immediately underwent reverse transcription polymerase chain reaction (RT-PCR) analysis following isolation.
Variant screening of the PHEX gene
To determine the PHEX gene variants, all 22 exons of the PHEX gene in the proband were initially amplified, followed by sequencing of the amplification products. Once a variant of the PHEX gene was identified in proband, DNA samples of the remaining healthy and affected family members were screened for this variant. Finally, the fetal PHEX gene was assessed only when the variant site was verified in all affected family members.
Information on the PHEX gene was obtained from the online NCBI database (https://www.ncbi.nlm.nih.gov/pubmed; RefSeq NM_000444.5). In total, 22 pairs of primers were designed for PCR amplification of all 22 exon regions and intron/exon boundaries of the PHEX gene using an online tool (http://www.yeastgenome.org/cgi-bin/web-primer). The sequences of the primers are listed in Table I. The PCR conditions were as follows: 95°C for 3 min, 95°C for 30 sec, 72°C for 1 min (34 cycles) and 72°C for 5 min. The PCR products were submitted to Sanger sequencing on an automated ABI PRISM 3130 sequencer (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and were then prepared with the BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems; Thermo Fisher Scientific, Inc.).
Table I.
Primers used to amplify 22 exons and cDNA of the phosphate-regulating neutral endopeptidase homolog, X-linked gene.
| Exon | Forward primer (5′–3′) | Reverse primer (5′–3′) | Annealing temperature (°C) | Product size (bp) |
|---|---|---|---|---|
| DNA | ||||
| 1 | TTCCTGACGGCAGTTTCTTA | AGGCAAACAGCCCTATACCT | 55.0 | 360 |
| 2 | TGGGTTTTGGAATACCGTGT | GGAATTCATAGCCAGCGGA | 57.0 | 530 |
| 3 | CGGCCTATCCATTCACTTTGA | TTCTGCAAACTTTCCTTTCAA | 57.2 | 513 |
| 4 | TCTGGAGGTTGGAATTGTGA | TGGCTTCTGGATTCTCTACCA | 55.5 | 486 |
| 5 | CCACCCCACCTCTTTTACCTA | ATGGCACCCCAAAAGGCTAAT | 58.9 | 436 |
| 6 | TGGGATGCAGACGATTTCAT | TCCTGGGTATCTCTTCTGACG | 56.0 | 504 |
| 7 | TTGCATAATGCCTGCACCAT | ATGGGCAATGACACAAAACG | 58.2 | 409 |
| 8 | CACCAAAGCCTTGAAAAACTT | CTGAGCCAATGCCAACAATT | 56.0 | 477 |
| 9 | TCTGTTTTGTTCTCTCTCCCC | TACACCAGACAGTGCTTTTGG | 55.0 | 343 |
| 10 | ATGGAGCTTTGCCAACTGTT | GAAAACAATGTGTGGCCTCA | 55.5 | 454 |
| 11 | TGACCTCAGGTGATCTGCTCA | AAGGCTGACATTAGCCTGTTG | 56.5 | 496 |
| 12 | GAGTCATTTCTCATGCAAGCC | CCTCCTGGTGAAACAAAAATC | 55.0 | 366 |
| 13 | CCTTCACAGTGGCTTGCATAT | TGATCTGCCTGGCATATTCA | 56.0 | 377 |
| 14 | TGTGACTGATGCAGCTTCTCT | AGAAATGGGGGACCTGTAATT | 55.5 | 423 |
| 15 | AATCTCTCCCCTACCTAACCC | CCCTGAGAAGACCCTGAAAAT | 55.0 | 394 |
| 16 | ATCTCTTAGAGGGCTCCCAGT | TAAGATGGCTTTCCTGTCCA | 55.0 | 397 |
| 17 | AGCAGTTTATCTTGGCTTTCC | AAAGTCTACCCCGATCACCAA | 56.4 | 377 |
| 18 | TGTTCCCTGCTGTTATGACTG | TGATTCAGCAGGTATGGGGTA | 56.4 | 405 |
| 19 | TTGCTGAGGATAGTTTGCCA | TGACTTCACACCCCAAAAAG | 55.0 | 452 |
| 20 | GCTAGGAATGGAAAGAACAGC | TTTCTTTGATCAAGGGAGCA | 55.0 | 451 |
| 21 | CCTGGGCACATATACGATTCT | TGGGATTTTTTTCAGATCACC | 56.0 | 462 |
| 22 | TGGCAGATAGTAATATGGGCA | ATGAAGGCTCAGTGCAGCCTC | 58.9 | 555 |
| cDNA | ||||
| 1 | ATGGAAGCAGAAACAGGGAG | TCAAGGTAGTCTTCCCTCACG | 55.5 | 710.0 |
| 2 | GCCAAAATCCTTTATTCATCC | TCTTTAAAGTACTGCGGGACG | 56.0 | 644.0 |
| 3 | TGGGCTACATCAAGAAGGTCA | TGGAAATCGGATCTGGTTGGT | 58.2 | 710.0 |
| 4 | AGAAGCCGACTACTTTGGCAA | ATAAACCAGCGTCCCAGCTA | 56.9 | 698.0 |
All sequencing results were aligned with the referenced PHEX gene sequences from the NCBI databse using the online Pairwise Sequence Alignment tool (www.ebi.ac.uk/Tools/psa/emboss_water/nucleotide.html). To predict the outcomes of variants, the SIB Bioinformatics Resource Portal database (http://www.expasy.org/), the Human Gene Variant Database (http://www.hgmd.cf.ac.uk) and the PolyPhred database (http://droog.gs.washington.edu/polyphred/) were used. Additionally, to search for the suspected aberrant splicing variants, Alamut® Alamut Interactive Biosoftware (version 2.9.0; Rouen, France) was used, which contains the four algorithms SpliceSiteFinder, MaxEntScan, NNSplice and GVGD.
RT-PCR analysis
The aberrantly spliced transcripts were verified by sequencing the entire cDNA of the PHEX gene. An initial cDNA strand was obtained from RT-PCR of total RNA using the RT reagent kit with gDNA Eraser (Takara Biotechnology Co., Kyoto, Japan) under the condition: 37°C for 15 min and 85°C for 5 sec. A total of 2 μl cDNA product was then used as a template to amplify the four overlapping fragments with the four pairs of primers. Primer sequences are listed in Table I. The PCR conditions were as follows: 95°C for 3 min, 95°C for 30 sec, 72°C for 1 min (34 cycles) and 72°C for 5 min. The final PCR products were sequenced and aligned with the coding region of the referenced PHEX gene from the NCBI database.
RT-quantitative PCR analysis
To examine the fold changes in the gene expression of PHEX affected by the suspicious abnormal splice donor site in Family 2, semi-quantitative analysis of the gene expression of PHEX in the proband and that of his affected mother was performed through qPCR analysis using the cDNA, which was derived from the peripheral blood total RNA. The samples were prepared with 10 μl of SYBR-Green Master mix (Thermo Fisher Scientific, Inc.) and 0.25 μl of primers (forward 5′-TTCGCTTGTGATGGCTGGAT-3′ and reverse 5′-CTGTATGGCTTCGGTGTCCC-3′) in a final reaction volume of 20 μl. The samples from one normal male and one normal female were used as controls, and the GAPDH gene was used as an internal reference gene. The RT-qPCR analysis was performed under the following conditions: 95°C for 5 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 30 sec, with a final extension at 72°C for 5 min. The 2−ΔΔCq method was used to assess the relative changes in the gene expression of PHEX according to the standard procedure (17).
Results
Variant screening
The PHEX gene is the only gene in which pathogenic variants known to cause XLHR have been reported. Therefore, the present study primarily focused on examining the PHEX gene in the two families. By directly sequencing all 22 exons and the intron/exon boundaries of the PHEX gene of the proband, two separate point variants were identified.
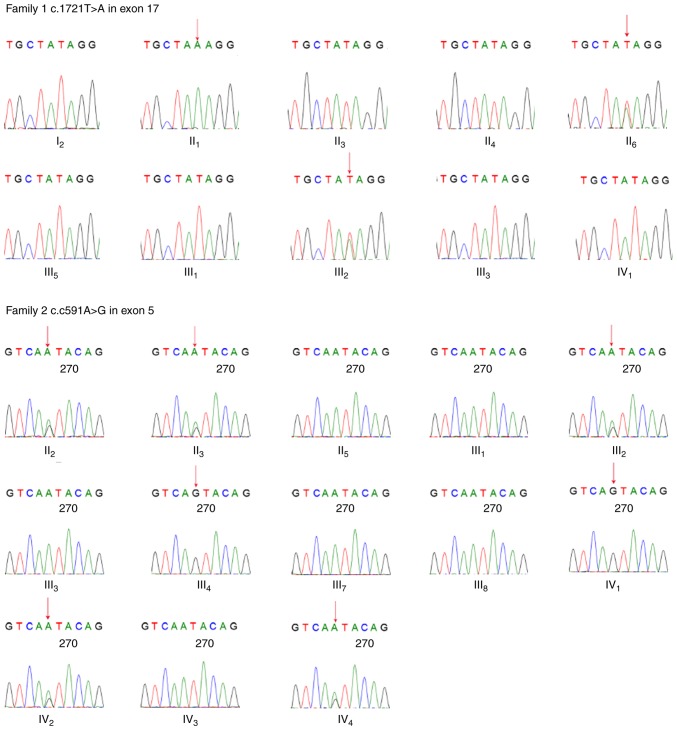
For the proband in Family 1, a novel missense heterozygous variant c.1721T>A in exon 17 was found, which resulted in a change of the 574th amino acid from isoleucine to lysine, p.574Ile>Lys. The sequencing results of exon 17 in the remaining affected family members verified the variant in this pedigree: A single peak (A) in the affected male (II1) and heterozygous peaks (A/T) in the affected female (II6) as males receive only one copy of the X chromosome. This PHEX gene variant was not found in healthy family members. Therefore, it was observed that individuals showing clinical symptoms of XLHR had this variant, whereas the healthy individuals did not. However, the proband's grandmother (I2) reported no complaints of any signs of clinical rickets, and gene analysis showed no variant in the PHEX gene. Subsequently, the present analyzed 15 short-sequence tandem repeat regions on chromosomes 21, 18,13, X and Y of the grandmother (I2) and her four children (II1, II3, II4 and II6) via RT-PCR analysis, and found the healthy grandmother (I2) was their biological mother, suggesting the possibility of the gonadal mosaicism in the grandmother.
In Family 2, the sequencing results of the PHEX gene of the proband showed a point variant c.591A>G in exon 5, which caused no change in the amino acid sequence (p.197, glutamic acid). This variant was also observed in the other affected family members, including four females (II2, II3, III2 and IV4), one male (III4) and the fetus (IV2). The affected females showed heterozygous peaks (A/G) at this site and the affected male showed only one peak (G), whereas the healthy females showed homozygous peaks (A) and the healthy male showed one peak (A). This variant was only detected in the affected male and female family members, and the inheritance patterns of the disease appearing in this pedigree strictly followed X-linked dominant inheritance. It was hypothesized that this variant likely originated in the proband's great grandfather, who had similar rickets symptoms according to his children's recall, although it is difficult to make definitive conclusions as the man had passed away. All sequencing results are shown in Fig. 3.
Figure 3.
Regions of sequencing maps of the PHEX gene in two pedigrees. Red arrows indicate variant sites, NM_000444.5: c.1721T>A in Family 1 and NM_000444.5: c.591A>G in Family 2. The two heterozygous peaks indicate the variant of the PHEX gene in females. PHEX, phosphate-regulating neutral endopeptidase homolog, X-linked.
Pathogenicity analysis of variants
In Family 1, the c.1721T>A variant resulted in the 574th amino acid of the protein (phosphate-regulating neutral endopeptidase) changing from isoleucine to lysine (p.Ile574Lys). This non-synonymous substitution is located at peptidase M13, the C-terminal domain of the protein, which is close to the mental binding (p.580, histidine) and active site (p.581, glutamic acid). According to the Align GVDV prediction (18,19), this variant scored 10.12 for Grantham variation and 93.77 for Grantham deviation, and was classified as C55, suggesting likely pathogenicity. Additionally, the SIFT prediction software sorted this variant as deleterious. In conclusion, the variant c.1721T>A of the PHEX gene was identified to cause XLHR in this pedigree, and it was possible that the variant was inherited from the grandmother, who may have had gonadal mosaicism of the PHEX gene.
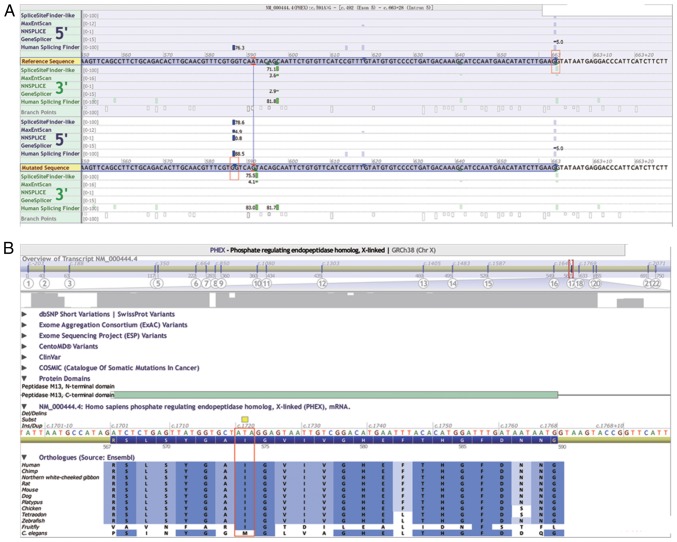
Theoretically, the c.591A>G variant in Family 2, which also encodes for the 197th glutamine residue, did not lead to a change in its amino acid sequence. However, based on the prediction of Alamut Interactive Biosoftware, this synonymous variant resulted in an abnormal splice donor site between c.586 and c.587 in the middle of exon 5, with the remainder of the sequences in the 3′ site of exon 5 being cut off during mRNA splicing activity (Fig. 4A). The Ile in this variant site is highly conserved among 12 different species, suggesting its potential importance to protein function (Fig. 4B).
Figure 4.
Prediction results of splicing variant NM_000444.5: c.591A>G in Alamut Interactive Biosoftware. (A) Normal splicing site of exon 5 is located at the end of exon 5 in the upper red square, and the aberrant splicing site was sited 5 bp before the variant site c.591A>G in the lower red square. (B) Conserved amino acid at the protein product of the phosphate-regulating neutral endopeptidase homolog, X-linked gene at the variant site NM_000444.5: c.1721T>A in different species. The isoleucinein (red square) at this variant site showed high conservation among 12 species, indicating its probable importance in protein function, and that the amino acid change may impair the protein function.
Verification of abnormal splice donor site
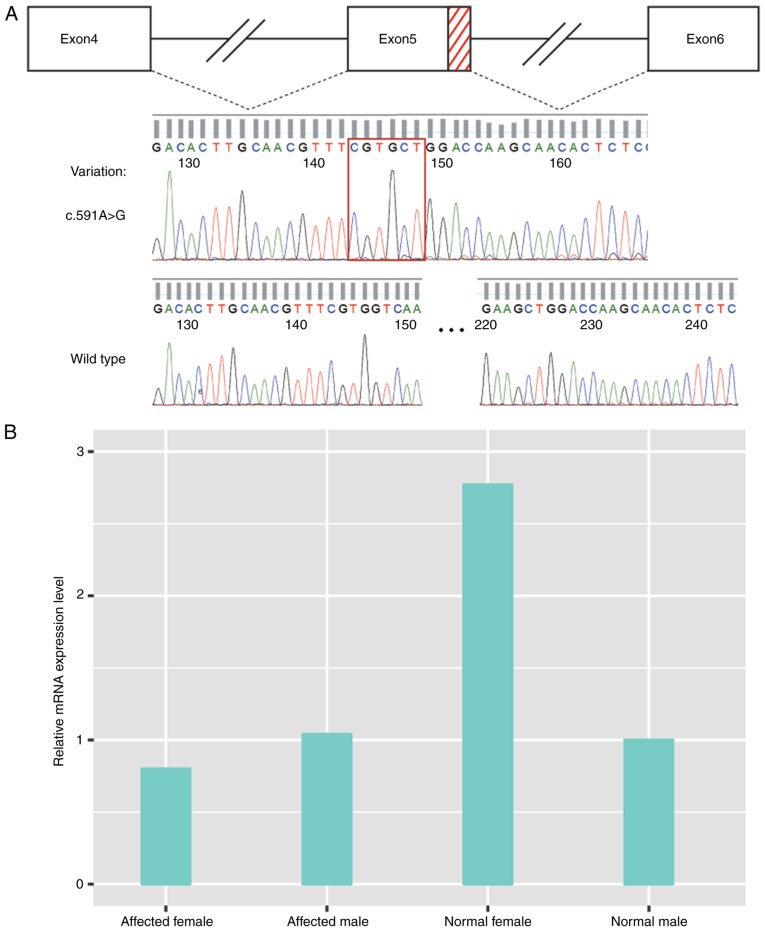
To determine the effect of the suspected splicing of variant c.591A>G in PHEX gene exon 5 in mRNA, RT-PCR was performed using peripheral blood total RNA for all affected family members from Family 2. The cDNA of the PHEX gene was amplified using the general PCR method, followed by direct sequencing. The sequencing results revealed a 77-bp deletion between c.586 and c.662 of PHEX cDNA in the patients; it occurred in the terminal region (3′ site) of exon 5, indicating an abnormal splicing donor site existing in the middle of exon 5 (Fig. 5A).
Figure 5.
Effects of splicing variant NM_000444.5: c.591A>G in mRNA splicing and gene expression. (A) Red dashes represent the deleted region of the PHEX gene exon 5 during splicing caused by the splicing variant c.591A>G. The red square indicates the ligation of exon 5 and exon 6. (B) Gene expression of PHEX was lower in the female patient, compared with that in the normal female, whereas no change was observed in male patient, compared with normal male. PHEX, phosphate-regulating neutral endopeptidase homolog, X-linked.
Analysis of relative gene expression of PHEX
To determine the change in the gene expression of PHEX in patients with the c.591A>G splicing variant in Family 2, the present study performed semi-quantitative assessment of mRNA derived from the PHEX gene of the proband and his mother through RT-qPCR. The PHEX gene was located on chromosome X, which causes the differences in expression levels in females and males; therefore, a normal female and normal male were used as controls when assessing the respective gene expression in the affected male (proband) and affected female (his mother). As shown in Fig. 5, the average 2−∆∆Cq of the proband/normal male and proband/normal female was 1.05 and 0.44, respectively, and that of the normal male/normal female was 0.36, suggesting almost the same PHEX gene expression level in the proband as in normal male. However, the average 2−∆∆Cq of the proband's mother/normal female was 0.43 and that of the proband's mother/normal male was 0.78, whereas the value for the normal female/normal male was 2.77; this indicated that the gene expression of PHEX was downregulated in the proband's mother, compared with that in the normal female. The value was even lower, compared with that of a normal male, who has only one copy of chromosome X. The results of the relative analysis of the gene expression of PHEX are shown in Fig. 5B.
Prenatal diagnosis
Two fetuses, whose mothers were genetically diagnosed with XLHR, received a prenatal diagnosis of the specific PHEX gene variant in their own family. Their DNA was acquired from the backup amniotic fluid and analyzed using direct sequencing. For the fetus (IV1) from Family 1, this sequencing map showed homozygous peaks (T) in the familial variant site (c.1721T>A), indicating that the fetus was unaffected, which was then confirmed by physical and laboratorial examinations post-delivery. For the fetus (IV2) from Family 2, heterozygous peaks (A/G) were observed in its familial variant site (c.591A>G), indicating that it was a female fetus and likely to be affected with XLHR.
Discussion
The present study reported on two unrelated families with XLHR, and identified one point variant of the PHEX gene in each family. Several other types of hypophosphatemic rickets have also been reported, including the autosomal dominant type (OMIM 193100) caused by the abnormal fibroblast growth factor 23 gene (20), and autosomal recessive HR (OMIM 241520) caused by either the ectonucleotide pyrophosphatase/phosphodiesterase-1 gene or dentin matrix protein 1 gene (21–24). However, based on the clinical symptoms, biochemical findings and modes of inheritance appearing in the family, the present study focused on the PHEX gene, which is the only causative gene for XLHR (25). In Family 1, a novel variant NM_000444.5: c.1721T>A involved an the amino acid substitution of p.Ile574Lys was identified. Due to their composition, polarity and volume, the moderate physicochemical difference between Ile and Lys is a Grantham distance 102 (26), which exceeded the average Grantham distance (92.06) for disease-associated variants (27). Considering the highly conserved amino acid Ile in this variation site among 12 species, although no current data is available concerning variant c.1721T>A, it is likely to be pathogenic. The healthy I2 member of this family had four children; two of whom were affected, including one male (II1) and one female (II6). According to the results of the PCR analysis (data not shown), I2 was the biological mother of the two affected individuals. Similarly, a father with a somatic mosaic for the PHEX variant has been reported, leaving one of his daughters affected (28); indicating the possibility of the gonadal mosaic for her PHEX gene variant.
The present study identified a splice-site variant (NM_000444.5: c.591A>G) in Family 2 with eight affected individuals of both genders. Through direct sequencing of cDNA, an abnormal splicing donor site was identified, which was 5 bp before the variant site; therefore, the aberrant mRNA failed to be translated into an intact protein due to a deletion and an advanced termination codon (TGA) at the 211th codon, which may impair the physiological function of the protein. Therefore, the variant c.591A>G is likely to be responsible for this disease in all members of Family 2. In the present study, it was found that the gene expression level of PHEX in the proband was similar to that of the normal male from the same family, almost half of that of the normal female. However, the expression level in the proband's mother, an affected female, was lower than that of the normal female, and even lower than that of the normal male. A similar expression level of PHEX was observed in another female patient (II2) from the same family. However, the general gene expression level of PHEX in female patients was not available due to the lack of RNA samples of female patients from the same family. The difference in gender may have led to the differences in expression levels of PHEX in the proband and his mother, which resulted from different splicing patterns; an alternative explanation is that it is associated with individual differences, which may explain the phenotypic diversity among patients (29,30). Further experiments concerning gene expression regulation may shed light on these results; however, it is difficult to draw clear conclusions from the present study alone.
At present, 75 splicing variants of the PHEX gene have been identified, accounting for 14.70% of the total variants (HGMD professional 2016.4 release), however, none of these occur in exons. Each mRNA is derived from pre-mRNA through a precise splicing reaction, which is one of the pivotal components of gene expression, involving a multi-component machine known as the spliceosome (30,31). The 5′ splice site (5′ss), the 3′ splice site (3′ss), and the branch point sequence constitute the three core splicing signals, which are essential to the splicing reaction (32,33). It is known that >80% of all 3′ss variants occur in the invariant AG sites of 3′ss ends (34). Therefore, the pre-mRNA of the PHEX gene in patients contained a c.586gtggtcagta sequence in the middle of exon 5, resembling a 3′ss motif, which can be recognized by the spliceosome, permitting aberrant splicing in exon 5.
XLHR is a lifelong metabolic disease, and patients begin to suffer from renal phosphate wasting at birth. To correct rickets/osteomalacia, radiographic abnormalities and skeletal deformities, treatments for children are required at the onset of diagnosis, continuing until long bone growth is complete (35). For affected parents with a family history of XLHR, prenatal diagnosis for pathogenic variants of the PHEX gene is important; doing so allows pediatricians to obtain the individual's genetic information, which is valuable for providing an accurate diagnosis and devising a treatment plan. In the present study, two pregnant women (18 weeks gestation) with XLHR were included to obtain a prenatal diagnosis for their babies. The DNA of each fetus was obtained from backup amniotic fluid and the specific PHEX gene variants were assessed. The woman from Family 2, who delivered an affected boy, conceived a baby with the same PHEX gene variant as her own; the woman from Family 1 carried a baby without the PHEX gene variant. The two babies had normal karyotypes (data not shown). Unlike the affected older brother, who received treatments until 4 years of age following diagnosis, the affected baby in Family 2 obtained sufficient medical care according to physical examinations and laboratory tests based on early genetic tests. One aim of prenatal diagnosis is to assist parents in adequately preparing for the affected baby, including financial support and psychological preparation; parents can turn to pediatricians for early treatments as soon as possible, which can be beneficial in terms of reducing disorders, including growth retardation or rickets.
The baby from Family 1 was confirmed to be a healthy girl in the follow-up visit 1 month following birth; however, the fetus from Family 2 had not yet been born, therefore, continued follow-up was planned for the baby from Family 2 following delivery. The follow-up visits for the babies planned to last for 3 years.
In conclusion, the present study reported on two unrelated pedigrees with XLHR and performed genetic analysis of the PHEX gene in the two families. Through direct sequencing of the PHEX gene, one missense variant (NM_000444.5: c.1721T>A) was identified in exon 17 in Family 1, and a splicing variant (NM_000444.5: c.591A>G) was identified in exon 5 in Family 2. The results of RT-PCR analysis verified the aberrant splicing site caused by variant c.591A>G, which resulted in a 77 bp loss in the 3′ site region of exon 5 on splicing. In the female patient, the c.591A>G variant led to downregulation of the gene expression of PHEX >2-fold, compared with that in the healthy female. However, this fold change was not observed in the male patient, who exhibited a similar gene expression level of PHEX to that of the normal male. Based on the evidence concerning the genotype-phenotype correlations between the specific variant and XLHR in each family, it was possible to provide a prenatal diagnosis for the fetuses and provide medical suggestions to the affected families.
Acknowledgments
This study was supported by a research grant from the National Natural Science Foundation of China (grant no. 81270060).
Abbreviations
- XLHR
X-linked hypophosphatemic rickets
Footnotes
Competing interests
The authors declare that they have no competing interests.
References
- 1.Pavone V, Testa G, Gioitta Iachino S, Evola FR, Avondo S, Sessa G. Hypophosphatemic rickets: Etiology, clinical features and treatment. Eur J Orthop Surg Traumatol. 2015;25:221–226. doi: 10.1007/s00590-014-1496-y. [DOI] [PubMed] [Google Scholar]
- 2.Cho HY, Lee BH, Kang JH, Ha IS, Cheong HI, Choi Y. A clinical and molecular genetic study of hypophosphatemic rickets in children. Pediatr Res. 2005;58:329–333. doi: 10.1203/01.PDR.0000169983.40758.7B. [DOI] [PubMed] [Google Scholar]
- 3.Chandran M, Chng CL, Zhao Y, Bee YM, Phua LY, Clarke BL. Novel PHEX gene mutation associated with X linked hypophosphatemic rickets. Nephron Physiol. 2010;116:17–218. doi: 10.1159/000319318. [DOI] [PubMed] [Google Scholar]
- 4.Tenenhouse HS. X-linked hypophosphataemia: A homologous disorder in humans and mice. Nephrol Dial Transplant. 1999;14:333–341. doi: 10.1093/ndt/14.2.333. [DOI] [PubMed] [Google Scholar]
- 5.Beck-Nielsen SS, Brock-Jacobsen B, Gram J, Brixen K, Jensen TK. Incidence and prevalence of nutritional and hereditary rickets in southern Denmark. Eur J Endocrinol. 2009;160:491–497. doi: 10.1530/EJE-08-0818. [DOI] [PubMed] [Google Scholar]
- 6.Albright F, Butler AM, Bloomberg E. Rickets resistant to vitamin d therapy. Am J Dis Child. 1937;54:529–547. [Google Scholar]
- 7.Stickler GB, Morgenstern BZ. Hypophosphataemic rickets: Final height and clinical symptoms in adults. Lancet. 1989;2:902–905. doi: 10.1016/S0140-6736(89)91559-6. [DOI] [PubMed] [Google Scholar]
- 8.A gene (PEX) with homologies to endopeptidases is mutated in patients with X-linked hypophosphatemic rickets. The HYP Consortium. Nat Genet. 1995;11:130–136. doi: 10.1038/ng1095-130. [DOI] [PubMed] [Google Scholar]
- 9.Filisetti D, Ostermann G, von Bredow M, Strom T, Filler G, Ehrich J, Pannetier S, Garnier JM, Rowe P, Francis F, et al. Non-random distribution of mutations in the PHEX gene, and under-detected missense mutations at non-conserved residues. Eur J Hum Genet. 1999;7:615–619. doi: 10.1038/sj.ejhg.5200341. [DOI] [PubMed] [Google Scholar]
- 10.Guo R, Quarles LD. Cloning and sequencing of human PEX from a bone cDNA library: Evidence for its developmental stage-specific regulation in osteoblasts. J Bone Miner Res. 1997;12:1009–1017. doi: 10.1359/jbmr.1997.12.7.1009. [DOI] [PubMed] [Google Scholar]
- 11.Lipman ML, Panda D, Bennett HP, Henderson JE, Shane E, Shen Y, Goltzman D, Karaplis AC. Cloning of human PEX cDNA. Expression, subcellular localization, and endopeptidase activity. J Biol Chem. 1998;273:13729–13737. doi: 10.1074/jbc.273.22.13729. [DOI] [PubMed] [Google Scholar]
- 12.Du L, Desbarats M, Viel J, Glorieux FH, Cawthorn C, Ecarot B. cDNA cloning of the murine Pex gene implicated in X-linked hypophosphatemia and evidence for expression in bone. Genomics. 1996;36:22–28. doi: 10.1006/geno.1996.0421. [DOI] [PubMed] [Google Scholar]
- 13.Liu S, Tang W, Fang J, Ren J, Li H, Xiao Z, Quarles LD. Novel regulators of Fgf23 expression and mineralization in Hyp bone. Mol Endocrinol. 2009;23:1505–1518. doi: 10.1210/me.2009-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Onishi T, Umemura S, Shintani S, Ooshima T. Phex mutation causes overexpression of FGF23 in teeth. Arch Oral Biol. 2008;53:99–104. doi: 10.1016/j.archoralbio.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 15.Farrow EG, White KE. Recent advances in renal phosphate handling. Nat Rev Nephrol. 2010;6:207–217. doi: 10.1038/nrneph.2010.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin A, Liu S, David V, Li H, Karydis A, Feng JQ, Quarles LD. Bone proteins PHEX and DMP1 regulate fibro-blastic growth factor Fgf23 expression in osteocytes through a common pathway involving FGF receptor (FGFR) signaling. FASEB J. 2011;25:2551–2562. doi: 10.1096/fj.10-177816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Mathe E, Olivier M, Kato S, Ishioka C, Hainaut P, Tavtigian SV. Computational approaches for predicting the biological effect of p53 missense mutations: A comparison of three sequence analysis based methods. Nucleic Acids Res. 2006;34:1317–1325. doi: 10.1093/nar/gkj518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tavtigian SV, Deffenbaugh AM, Yin L, Judkins T, Scholl T, Samollow PB, de Silva D, Zharkikh A, Thomas A. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J Med Genet. 2006;43:295–305. doi: 10.1136/jmg.2005.033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.ADHR Consortium Autosomal dominant hypophosphataemic rickets is associated with mutations in FGF23. Nat Genet. 2000;26:345–348. doi: 10.1038/81664. [DOI] [PubMed] [Google Scholar]
- 21.Lorenz-Depiereux B, Schnabel D, Tiosano D, Häusler G, Strom TM. Loss-of-function ENPP1 mutations cause both generalized arterial calcification of infancy and autosomal-recessive hypophosphatemic rickets. Am J Hum Genet. 2010;86:267–272. doi: 10.1016/j.ajhg.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng JQ, Ward LM, Liu S, Lu Y, Xie Y, Yuan B, Yu X, Rauch F, Davis SI, Zhang S, et al. Loss of DMP1 causes rickets and osteomalacia and identifies a role for osteocytes in mineral metabolism. Nat Genet. 2006;38:1310–1315. doi: 10.1038/ng1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorenz-Depiereux B, Bastepe M, Benet-Pagès A, Amyere M, Wagenstaller J, Müller-Barth U, Badenhoop K, Kaiser SM, Rittmaster RS, Shlossberg AH, et al. DMP1 mutations in autosomal recessive hypophosphatemia implicate a bone matrix protein in the regulation of phosphate homeostasis. Nat Genet. 2006;38:1248–1250. doi: 10.1038/ng1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beck-Nielsen SS, Brixen K, Gram J, Brusgaard K. Mutational analysis of PHEX, FGF23, DMP1, SLC34A3 and CLCN5 in patients with hypophosphatemic rickets. J Hum Genet. 2012;57:453–458. doi: 10.1038/jhg.2012.56. [DOI] [PubMed] [Google Scholar]
- 25.Econs MJ, Francis F. Positional cloning of the PEX gene: New insights into the pathophysiology of X-linked hypophosphatemic rickets. Am J Physiol. 1997;273:F489–F498. doi: 10.1152/ajprenal.1997.273.4.F489. [DOI] [PubMed] [Google Scholar]
- 26.Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 27.Subramanian S, Kumar S. Evolutionary anatomies of positions and types of disease-associated and neutral amino acid mutations in the human genome. BMC Genomics. 2006;7:306. doi: 10.1186/1471-2164-7-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goji K, Ozaki K, Sadewa AH, Nishio H, Matsuo M. Somatic and germline mosaicism for a mutation of the PHEX gene can lead to genetic transmission of X-linked hypophosphatemic rickets that mimics an autosomal dominant trait. J Clin Endocrinol Metab. 2006;91:365–370. doi: 10.1210/jc.2005-1776. [DOI] [PubMed] [Google Scholar]
- 29.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ward AJ, Cooper TA. The pathobiology of splicing. J Pathol. 2010;220:152–163. doi: 10.1002/path.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wahl MC, Will CL, Lührmann R. The spliceosome: Design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z, Burge CB. Splicing regulation: From a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 34.Krawczak M, Reiss J, Cooper DN. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: Causes and consequences. Hum Genet. 1992;90:41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- 35.Carpenter TO, Imel EA, Holm IA, Jan de Beur SM, Insogna KL. A clinician's guide to X-linked hypophosphatemia. J Bone Miner Res. 2011;26:1381–1388. doi: 10.1002/jbmr.340. [DOI] [PMC free article] [PubMed] [Google Scholar]