Abstract
Background
Acute myeloid leukemia with intermediate cytogenetic risk (ICR-AML) needs to be stratified. The abnormal gene expression might be prognostic, and its cutoff value for patient grouping is pivotal.
Material/Methods
Ecotropic viral integration site 1 (EVI1) transcripts were assessed in 191 adult ICR-AML patients at diagnosis who received chemotherapy only. MLL-PTD, WT1 transcript levels, FLT3-ITD, and NPM1 mutations were simultaneously evaluated, and 27 normal bone marrow samples were tested to define normal threshold.
Results
The normal upper limit of EVI1 transcript levels was 8.0%. Receiver operating characteristic curve analysis showed that 1.0% (a 0.9-log reduction from the normal limit) was the EVI1 optimal cutoff value for significantly differentiating relapse (P=0.049). A total of 23 patients (12%) had EVI1 levels ≥1.0%. EVI1 ≥1.0% had no effect on CR achievement, whereas it was significantly associated with lower 2-year relapse-free survival (RFS), disease-free survival (DFS), and overall survival (OS) rates in the entire cohort (P=0.0003, 0.0017, and 0.0009, respectively), patients with normal karyotypes (P=0.0032, 0.0047, and 0.0007, respectively), and FLT3-ITD (−) patients (all P<0.0001). Multivariate analysis showed that EVI1 ≥1.0% was an independent adverse prognostic factor for RFS, DFS, and OS in the entire cohort. In addition, patients with EVI1 transcript levels between 1.0% and 8.0% had 2-year RFS rates similar to those with EVI1 ≥8.0%, and they both had significantly lower RFS rates than those with EVI1 <1.0% (P=0.0005 and 0.027).
Conclusions
High EVI1 expression predicts poor outcome in ICR-AML patients receiving chemotherapy. The optimal cutoff value for patient stratification is different from the normal limit.
MeSH Keywords: Gene Expression; Leukemia, Myeloid, Acute; Patient Outcome Assessment; Real-Time Polymerase Chain Reaction
Background
Acute myeloid leukemia (AML) is a heterogeneous disease, and cytogenetic analysis is the classical method and framework for stratification [1–3]. Nearly half of AML patients are defined as an intermediate cytogenetic risk group, but their outcomes greatly varied [1–3]. Therefore, further stratification is needed to guide appropriate treatment. Over the past 2 decades, dozens of gene mutations have been discovered in AML, and the characterization of their prognostic impact is ongoing [4–6].
Apart from gene mutation, abnormal gene expression might also be prognostic [7,8], and Ecotropic viral integration site 1 (EVI1), WT1, and MLL partial tandem duplications (MLL-PTD) are representatives [9–22]. The EVI1 gene is located on human chromosome 3q26 and encodes a transcription factor essential for both normal and malignant hematopoiesis [23]. It was shown that EVI1 was aberrantly highly expressed in AML patients, both with and without cytogenetic abnormalities 3q26 [24,25]. In the past decade, several studies have demonstrated an adverse prognostic role of EVI1 in adult AML and AML patients with intermediate cytogenetic risk (ICR-AML) [9–14], but its effects on complete remission (CR) achievement were contradictory [10,11,13]. To date, almost all such studies have been undertaken in European populations and its prognostic significance in other populations needs to be evaluated. In addition, the EVI1 cutoff value for patient grouping remains obscure.
The cutoff value is the key to defining abnormal expression and differentiating patients. An abnormally-expressed gene is typically expressed in normal hematopoietic cells but with different levels from those found with leukemia [26–28]. As for the determination of EVI1 high expression, some used the upper limit in normal bone marrow (NBM) as the cutoff value [13], whereas others arbitrarily selected from several values [9–12]. Therefore, the optimal threshold for clinical practice remains a challenge.
In the present study, by measuring EVI1 expression as well as additional molecular abnormalities in 191 consecutive adult ICR-AML patients receiving chemotherapy at our institute, we compared different cutoff values and evaluated their prognostic effects on outcomes.
Material and Methods
Patients and treatment
A total of 191 adult ICR-AML patients were enrolled in the present study. These patients were consecutively diagnosed from January 2009 to December 2015, had available cytogenetic results, received at least 2 cycles of chemotherapy, were followed up at our institute, and did not receive allogeneic hematopoietic stem cell transplantation (allo-HSCT). Furthermore, all patients had available RNA and DNA extracted from bone marrow samples at diagnosis. The definition of cytogenetic risk was based on NCCN guidelines [29]. Intermediate-risk cytogenetics included normal cytogenetics, +8 alone, t(9;11), and other abnormalities not classified as favorable or unfavorable. The analyzed patients included 148 (77.5%) patients with normal karyotypes and 1 (0.5%) patient with t(9;11)(p22;q23). The basic characteristics are shown in Table 1. In addition, a total of 27 NBM samples were aspirated from normal volunteers who were allo-HSCT donors and we extracted RNA.
Table 1.
Relationship between EVI1 expression and variables at diagnosis in ICR-AML.
| Variables | All | EVI1 <1.0% | EVI1 ≥1.0% | P-value** |
|---|---|---|---|---|
| N | 191 | 168 | 23 | – |
| Age (y, median, range) | 43 (17–65) | 42 (17–65) | 49 (21–59) | 0.10 |
| Males (%)* | 106 (55.5%) | 93 (55.4%) | 13 (56.5%) | 1.0 |
| WBC (×109/L; median; range) | 18.6 (0.6–321) | 19 (0.8–282.5) | 16.2 (1.8–145.1) | 0.84 |
| Hb (g/L; median; range) | 86 (40–155) | 89 (40–155) | 75 (39–135) | 0.022 |
| PLT (×109/L; median; range) | 45 (3–838) | 45 (7–838) | 67 (3–344) | 0.34 |
| Blasts in bone marrow (%, median, range) | 66% (20–95%) | 68% (20–95%) | 50% (22–94%) | 0.0090 |
| Normal karyotype (%)* | 148 (77.5%) | 131 (78.0%) | 17 (73.9%) | 0.61 |
| FLT3-ITD (+) (%)* | 40 (20.9%) | 37 (22.0%) | 3 (13.0%) | 0.42 |
| NPM1 mutation (+)(%)* | 66 (34.6%) | 61 (36.3%) | 5 (21.7%) | 0.24 |
| NPM1 mutation (+)/FLT3-ITD (−) (%)* | 49 (25.7%) | 44 (26.2%) | 5 (21.7%) | 0.80 |
| MLL-PTD transcript level ≥1.0% (%)* | 16 (8.4%) | 14 (8.3%) | 2 (8.7%) | 1.00 |
| WT1 transcript level ≥10.0% (%)* | 123 (64.4%) | 102 (60.7%) | 21 (91.3%) | 0.0043 |
| FAB type | 0.092 | |||
| M0 | 2 | 1 (0.006%) | 1 (4.3%) | |
| M1 | 9 | 8 (4.8%) | 1 (4.3%) | |
| M2 | 129 | 119 (70.8%) | 10 (43.5%) | |
| M4 | 22 | 17 (10.1%) | 5 (21.7%) | |
| M5 | 25 | 20 (11.9%) | 5 (21.7%) | |
| M6 | 4 | 3 (1.8%) | 1 (4.3%) |
Values are presented as the number of patients followed by the percentage in parentheses; other values are presented as the median followed by a range in parentheses;
The bold numbers represent P values <0.05.
All patients received idarubicin (8–10 mg/m2) for 3 days in combination with cytarabine (100 mg/m2) for 7 days as the first induction regimen. Patients who achieved partial remission repeated the first induction regimen, and those who had no response were treated with other regimens. The consolidation therapy included 4 cycles of high-dose cytarabine (2 g/m2, q12h, 3 days) followed by 2 cycles of regimens containing cytarabine and anthracyclines. The cutoff date for follow-up was September 20, 2016. This study was approved by the Ethics Committee of Peking University People’s Hospital. All patients and volunteers provided written informed consent in accordance with the Declaration of Helsinki to participate in the present study.
Polymerase chain reaction (PCR)
Nucleated cells were obtained by treating fresh bone marrow samples with 0.144 M NH4Cl, 0.01 M NH4HCO3 to lyse the red cells. Total RNA and genomic DNA were individually extracted from bone marrow nucleated cells using Trizol and DNAzol Reagent (Invitrogen, Carlsbad, CA, USA). RNA was used to test EVI1, MLL-PTD, and WT1 transcript levels by TaqMan-based real-time quantitative PCR (RQ-PCR), and DNA was used to amplify FLT3-ITD (internal tandem duplication) by qualitative PCR and NPM1 mutations (A, B, and D type) by TaqMan-based RQ-PCR [30]. RQ-PCR was performed with a ABI PRISM 7500 Sequence Detector (Applied Biosystems, Foster City, USA). The primers and probes for the EVI1 transcript were designed using Primer Express Software (Applied Biosystems, Foster City, USA) to detect all subtypes (1a, 1b, 1c, 1d, and 3L), and the sequences were as follows:
Forward primer: 5′-CCCATGTGCCAGAGGAACTT-3′ (in exon 14)
Reverse primer: 5′-CAGTGACAGCATCATAGCATATGC-3′ (in exon 15)
Probe: 5′-FAM-CAGCCGTTACACAGAAAGTCCAAATCGC-TAMRA-3′ (in exon 14)
The MLL primers and probes annealed to locations in exons 8–10 and 3 of the MLL gene to detect fusion between exons 8–10 and exon 3. The WT1 primers and probe have been previously reported [31]. ABL was used as a control gene, and the corresponding primers and probe were based on a report from the Europe Against Cancer Program [32]. The EVI1, MLL-PTD, and WT1 transcript levels were calculated as the percentage of target transcript copies/ABL copies.
Statistical analysis and definitions
Pairwise comparisons of the variables between groups were performed using the Mann–Whitney U test for continuous variables and Fisher’s exact test for categorical variables. A receiver operating characteristic (ROC) curve was used to identify optimal cutoff levels that best discriminated patients with different responses (achieving CR) and outcomes (relapse). Survival functions were estimated using the Kaplan-Meier method and compared using the log-rank test. Relapse-free survival (RFS) and disease-free survival (DFS) were measured from the date when CR was achieved. The events were relapse for RFS and death during CR1 or relapse for DFS. The event for overall survival (OS) was death (regardless of the cause), and patients were queried at the date of last follow-up to determine whether they were still alive, or were censored on the date they were last known to be alive. Variables associated with P<0.20 in the univariate analysis were entered in multivariable analysis performed by the Cox models. The level for a statistically significant difference was set at P<0.05 for all univariate tests. The SPSS 13.0 software package (SPSS Inc., Chicago, IL), and GraphPad Prism 5 (GraphPad Software Inc., La Jolla, CA) were used for data analysis.
Results
Patient outcomes
The median follow-up time for the entire cohort was 13 (2–91) months. A total of 167 (87.4%) patients achieved CR after induction during follow-up, and 117 (61.3%) patients were alive at last follow-up, with a median follow-up time of 16 (2–91) months. The 2-year RFS and DFS of the 167 patients who achieved CR were 54.2% (95% confidence interval (CI), 44.0–63.2%) and 49.4% (95% CI, 39.8–58.3%), respectively. The 2-year OS of the entire cohort was 58.4% (95% CI, 49.4–67.4%).
EVI1 expression and other molecular abnormality patterns in NBM and ICR-AML patients at diagnosis
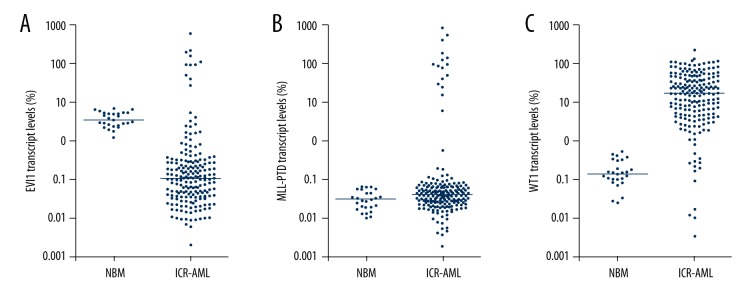
EVI1, MLL-PTD, and WT1 expression patterns in NBM and newly diagnosed ICR-AML patients are shown in Figure 1. The upper limits of EVI1, MLL-PTD, and WT1 transcript levels of 27 NBM samples were 8.0%, 0.08%, and 0.6%, respectively (Figure 1). For the entire patient cohort, the median EVI1, MLL-PTD, and WT1 transcript levels at diagnosis were 0.11% (range, 0.003–643.5%), 0.04% (range, 0.003–859.6%), and 19.4% (range, 0.004–251.2%), respectively (Figure 1). Compared with the upper limit in NBM, 5.8% (11/191), 18.3% (35/191), and 93.2% (178/191) of patients individually overexpressed EVI1, MLL-PTD, and WT1, respectively. Furthermore, the frequencies of FLT3-ITD and NPM1 mutations were 20.9% (40/191) and 34.6% (66/191), respectively.
Figure 1.
EVI1, MLL-PTD, and WT1 expression patterns in 27 normal bone marrow (NBM) samples and BM samples collected from 191 newly diagnosed ICR-AML patients. (A) EVI1; (B) MLL-PTD; (C) WT1. Y-axis indicates the percentage of target transcript copies/ABL copies.
Determination of optimal cutoff values of EVI1, MLL-PTD, and WT1
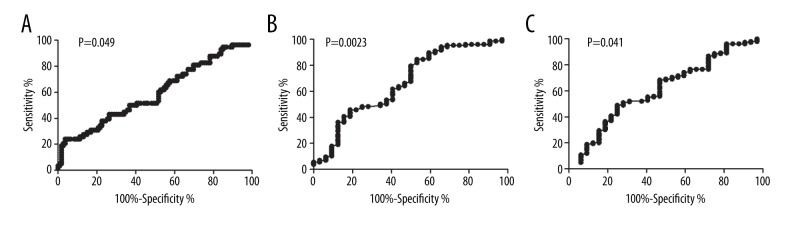
The ROC curves showed that EVI1 transcript levels significantly differentiated patients in relapse (area under curve 0.59, P=0.049, Figure 2A). A value of 1.0% (a 0.9-log reduction from the upper limit in NBM) was identified as the optimal cutoff value based on its maximal Youden index (0.20) among all values. Therefore, EVI1 ≥1.0% and <1.0% were defined as high expression and low expression, respectively. In the entire cohort, 23 (12.0%) patients had high EVI1 expression (≥1.0%). In addition, EVI1 transcript levels did not significantly differentiate patients in CR achievement after 1 and 2 courses of induction (P=0.16 and 0.42).
Figure 2.
ROC curves. (A) Relationship between EVI1 transcript levels and relapse. (B) Relationship between MLL-PTD transcript levels and 2-course induction of CR achievement. (C) Relationship between WT1 transcript levels and 2-course induction CR achievement. The optimal cutoff value was determined according to maximal Youden index (sensitivity + specificity − 1).
Similarly, both MLL-PTD and WT1 significantly differentiated patients achieving CR after 2 courses of induction (Figure 2B, 2C, P=0.023 and 0.041) but not in relapse (P=0.58 and 0.16), and the optimal cutoff value was 1.0% and 10.0%, respectively. In the entire cohort, 16 (8.4%) and 123 (64.4%) patients individually had high MLL-PTD and WT1 expression, respectively (≥1.0% and 10.0%).
Relationship between EVI1 expression and other patient characteristics and molecular abnormalities at diagnosis
As shown in Table 1, high EVI1 expression (≥1.0%) was significantly related to low hemoglobin levels, low blast percentage in bone marrow, and high WT1 expression (all P<0.05) but not age, sex, white blood cell (WBC) or platelet counts, FLT3-ITD frequency, NPM1 mutation frequency, and FAB subtype. Furthermore, EVI1 expression was not relevant to the distribution of FLT3-ITD (−)/NPM1 mutation (+) (P=0.80).
Effects of EVI1 expression and other molecular abnormalities on CR achievement
The CR rates after 1 and 2 courses of induction in the entire cohort were 65.4% (125/191) and 83.2% (159/191), respectively. As shown in Table 2, EVI1 grouped by 1.0% had no impact on CR achievement (all P>0.05). Similarly, WT1 expression did not affect CR achievement. Differing from them, both high MLL-PTD expression and NPM1 mutation (+) were significantly related to lower 1-course and 2-course induction CR rate (Table 2, EVI1: P=0.0047 and 0.0002; MLL-PTD: P=0.0022 and 0.0065), and FLT3-ITD (+) was significantly related to a lower 2-course induction CR rate (Table 2, P=0.017).
Table 2.
Impacts of molecular abnormalities on CR achievement.
| Variables | After 1 course of induction | After 2 courses of induction | ||
|---|---|---|---|---|
| CR rate | P value* | CR rate | P value* | |
| EVI1 transcript levels | ||||
| <1.0% | 67.9% | 0.16 | 83.9% | 0.55 |
| ≥1.0% | 52.2% | 78.3% | ||
| MLL-PTD transcript levels | ||||
| <1.0% | 68.6% | 0.0047 | 86.9% | 0.0002 |
| ≥1.0% | 31.3% | 43.8% | ||
| WT1 transcript levels | ||||
| <10.0% | 69.1% | 0.53 | 89.7% | 0.10 |
| ≥10.0% | 63.4% | 79.7% | ||
| NPM1 mutation | ||||
| (+) | 80.3% | 0.0022 | 93.9% | 0.0065 |
| (−) | 58.1% | 78.2% | ||
| FLT3-ITD | ||||
| (+) | 57.5% | 0.26 | 70.0% | 0.017 |
| (−) | 67.5% | 86.8% | ||
The bold numbers represent P values <0.05.
High EVI1 expression (≥1.0%) predicted poor outcomes
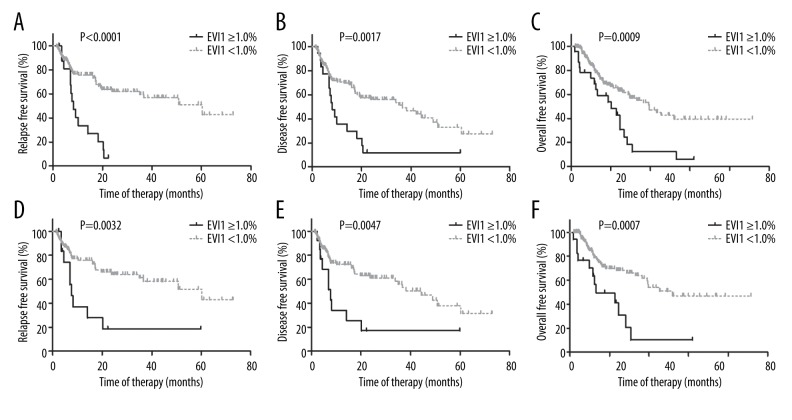
In the entire cohort, the EVI1 ≥1.0% group had significantly lower 2-year RFS, DFS, and OS rates than the EVI1 <1.0% group (RFS: 6.7% [95% CI 0.4–26.2%] vs. 62.0% [95% CI 51.2–71.1%], P<0.0001; DFS: 11.9% [95% CI 2.0–31.5%] vs. 56.0% [95% CI 45.6–65.2%], P=0.0017; OS: 43.0% [95% CI 21.6–64.4%] vs. 64.4% [95% CI 61.9–76.5%], P=0.0009; Table 3 and Figure 3A–3C).
Table 3.
Univariate analysis of relapse and survival in the entire cohort.
| Variable | RFS | DFS | OS | |||
|---|---|---|---|---|---|---|
| HR (95%CI) | P value* | HR (95%CI) | P value* | HR (95%CI) | P value* | |
| EVI1 ≥1.0% | 7.7 (3.0–19.4) | <0.0001 | 3.5 (1.6–7.6) | 0.0017 | 3.3 (1.6–6.7) | 0.0009 |
| MLL-PTD ≥1.0% | 2.0 (0.70–5.9) | 0.19 | 1.7 (0.67–4.4) | 0.26 | 1.4 (0.64–3.1) | 0.39 |
| WT1 ≥10.0% | 1.6 (0.91–2.7) | 0.10 | 1.4 (0.87–2.3) | 0.17 | 1.2 (0.73–2.0) | 0.51 |
| FLT3-ITD (+) | 5.7 (2.6–12.6) | <0.0001 | 3.4 (1.7–6.8) | 0.0005 | 2.6 (1.4–4.9) | 0.0022 |
| NPM1 mutation (+) | 1.0 (0.62–1.8) | 0.87 | 0.98 (0.61–1.6) | 0.93 | 1.0 (0.63–1.6) | 0.98 |
| Age >40 y | 1.1 (0.64–1.9) | 0.75 | 1.2 (0.73–1.9) | 0.51 | 1.1 (0.68–1.7) | 0.72 |
| Female | 1.1 (0.64–1.8) | 0.79 | 1.2 (0.78–2.0) | 0.37 | 0.96 (0.61–1.5) | 0.86 |
| WBC >10×109/L | 1.2 (0.72–2.1) | 0.47 | 1.1 (0.67–1.1) | 0.74 | 0.99 (0.61–1.6) | 0.96 |
| Hb <90 g/L | 1.2 (0.68–2.0) | 0.59 | 1.0 (0.65–1.7) | 0.89 | 1.0 (0.64–1.6) | 0.93 |
| PLT <100×109/L | 1.6 (0.91–2.9) | 0.10 | 1.7 (1.0–2.8) | 0.051 | 1.3 (0.76–2.3) | 0.33 |
| BM blast >65% | 1.7 (0.99–2.8) | 0.056 | 1.4 (0.87–2.2) | 0.17 | 1.1 (0.73–1.8) | 0.55 |
| Normal karyotype | 0.73 (0.38–1.4) | 0.35 | 0.54 (0.30–1.0) | 0.047 | 0.51 (0.29–0.88) | 0.017 |
The bold numbers represent P values <0.05.
Figure 3.
The impacts of EVI1 expression on relapse-free survival (A, D), disease-free survival (B, E), and overall survival (C, F). A–C showed the impacts in the entire cohort (n=191), and D–F showed the impacts in patients with normal karyotypes (n=148).
Patients with normal karyotypes (n=148) were further analyzed. Similarly, EVI1 ≥1.0% (n=17) was significantly associated with lower 2-year RFS, DFS, and OS rates than EVI1 <1.0% (RFS: 18.5% [95% CI 2.9–44.7%] vs. 64.0% [95% CI 52.1–73.7%], P=0.0032; DFS: 17.1% [95% CI 2.7–42.1%] vs. 61.1% [95% CI 49.5–70.8%], P=0.0047; OS: 41.3% [95% CI 16.8–64.5%] vs. 68.1% [95% CI 57.1–76.9%], P=0.0007; Figure 3D–3F).
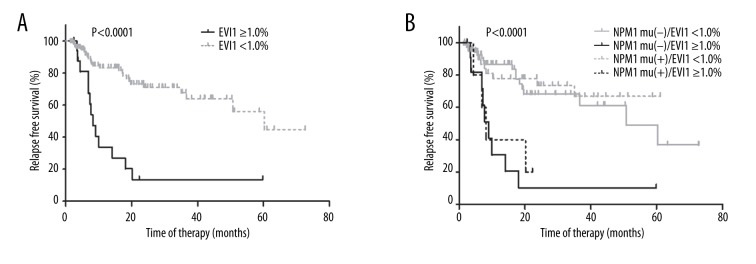
FLT3-ITD (−) patients (n=150) were analyzed. EVI1 ≥1.0% was significantly associated with lower 2-year RFS, DFS, and OS rates (RFS: 13.5% [95% CI 2.2–34.9%] vs. 71.0% [95% CI 58.9–80.1%], Figure 4A; DFS: 12.7% [95% CI 2.1–33.2%] vs. 64.2% [95% CI 52.5–73.7%]; OS: 41.1% [95% CI 19.1–62.0%] vs. 73.4% [95% CI 62.8–81.4%]. All P<0.0001). If NPM1 mutation status was simultaneously considered, 5 NPM1 mutation (+) and 15 NPM1 mutation (−) patients had EVI1 expression ≥1.0% (5/49 vs. 15/101, 10.2% vs. 14.9%), respectively. The NPM1 mutation had no impact on the RFS rate in FLT3-ITD (−) patients (P=0.53), whereas EVI1 ≥1.0% was significantly related to a higher 2-year RFS rate in both the NPM1 mutation (−) and (+) patients (P<0.0001 and P=0.0079, Figure 4B).
Figure 4.
The impact of EVI1 expression and NPM1 mutation status on RFS in FLT3-ITD (−) patients. (A) Patients were grouped according to EVI1 expression. (B) Patients were grouped according to both EVI1 expression and NPM1 mutation status.
High EVI1 expression (≥1.0%) independently predicts poor outcomes in ICR-AML patients
Univariate analysis was performed in the entire cohort and is shown in Table 3. In addition to high EVI1 expression, FLT3-ITD was significantly related to lower 2-year RFS, DFS, and OS rates (RFS: 19.8% [95% CI 5.3–40.9%] vs. 61.7% [95% CI 50.4–71.1%]; DFS: 18.9% [95% CI 5.1–39.3%] vs. 56.0% [95% CI 45.3–65.4%]; OS: 29.3% [95% CI 13.2–47.5%] vs. 65.2% [95% CI 55.0–73.7%]). However, MLL-PTD expression, WT1 expression, and NPM1 mutation all had no effects on relapse and survival.
The effects of variables associated with P<0.20 in univariate analysis were analyzed by multivariable analysis. As shown in Table 4, both high EVI1 expression (≥1.0%) and FLT3-ITD (+) were independent adverse prognostic factors for RFS, DFS, and OS in the entire cohort. Furthermore, PLT count <100×109/L was an independent adverse prognostic factor for RFS and DFS, and BM blast >65% was an independent adverse prognostic factor for RFS.
Table 4.
Independent prognostic factors for outcomes in the entire cohort.
| HR (95%CI) | P value | |
|---|---|---|
| RFS | ||
| EVI1 expression | ||
| <1.0% | 1.0 | <0.0001 |
| ≥1.0% | 4.0 (2.1–7.7) | |
| FLT3-ITD | ||
| (−) | 1.0 | <0.0001 |
| (+) | 3.4 (1.9–6.0) | |
| PLT count | ||
| ≥100×109/L | 1.0 | 0.030 |
| <100×109/L | 2.1 (1.1–4.3) | |
| Blast percentage in BM | ||
| ≤65% | 1.0 | 0.017 |
| >65% | 2.1 (1.1–3.6) | |
| DFS | ||
| EVI1 expression | ||
| <1.0% | 1.0 | 0.001 |
| ≥1.0% | 2.6 (1.5–4.7) | |
| FLT3-ITD | ||
| (−) | 1.0 | <0.0001 |
| (+) | 2.8 (1.6–4.7) | |
| PLT count | ||
| ≥100×109/L | 1.0 | 0.015 |
| <100×109/L | 2.2 (1.2–4.1) | |
| OS | ||
| EVI1 expression | ||
| <1.0% | 1.0 | 0.001 |
| ≥1.0% | 2.4 (1.4–4.1) | |
| FLT3-ITD | ||
| (−) | 1.0 | 0.002 |
| (+) | 2.2 (1.3–3.6) | |
Comparison between ROC curve and the upper limit of NBM-determined cutoff value
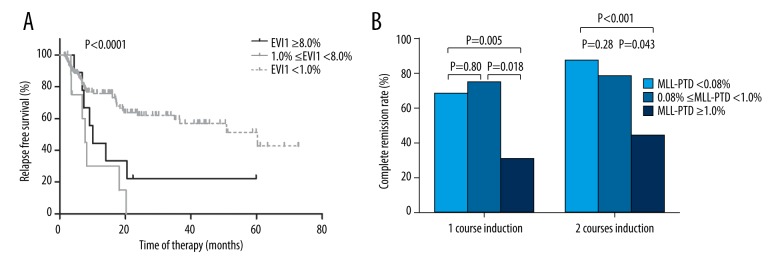
To further evaluate the impacts of EVI1 expression on relapse and MLL-PTD expression on CR achievement, the patients were individually classified into 3 groups according to the ROC curve, and the upper limit of NBM-determined cutoff values. Patients with EVI1 levels between 1.0% and 8.0% had 2-year RFS rates similar to those with EVI1 ≥8.0% (P=0.16, Figure 5A), and both patient groups had significantly lower 2-year RFS rates than those with EVI1 <1.0% (P=0.0005 and 0.027, Figure 5A). Furthermore, the 1- and 2-course induction CR rates of patients with MLL-PTD levels between 0.08% and 1.0% were similar to those with MLL-PTD <0.08% (P=0.80 and 0.28, Figure 5B), and they were all significantly higher than those of patients with MLL-PTD ≥1.0% (All P<0.05, Figure 5B).
Figure 5.
Comparisons among patients grouped according to the ROC curve and the upper limit of NBM-determined cutoff values. (A) Comparison of RFS rates among patients grouped according to EVI1 expression. (B) Comparison of CR rates among patients grouped according to MLL-PTD expression.
Discussion
AML patients with intermediate cytogenetic risk need to be differentiated [1–4]. In the present study, EVI1 expression was evaluated in combination with other molecular abnormalities in adult ICR-AML patients who received chemotherapy only. We found that the ROC curve-determined high EVI1 expression was an independent poor prognostic factor for relapse and survival in ICR-AML patients, patients with normal karyotypes, and FLT3-ITD(−) patients.
Many genes involved in leukemogenesis are expressed in both leukemic cells and normal hematopoietic stem cells/progenitors (e.g., EVI1, WT1, and MLL-PTD) [26–28]. The abnormal expression of some genes (e.g., WT1) has been widely used to monitor minimal residual disease (MRD), in which the upper limit of NBM expression was usually used to define overexpression [33]. Another role of gene overexpression is prognosis. To best differentiate patients, determining the optimal cutoff value is important. Notably, the optimal cutoff value for prognosis may not always be the same as the upper limit in NBM. For example, we recently reported that WT1 ≤5.0% (approximately 1-log increase compared with the upper limit in NBM) at diagnosis was significantly related to poor outcomes in t(8;21) AML patients [17]. In the present study, we demonstrated that a value less than the normal upper limit was the optimal cutoff value for EVI1 with the largest Youden index. Comparisons revealed that the ROC curve analysis-determined cutoff value, but not the upper limit in NBM, significantly differentiated patients with respect to relapse. It was the same for MLL-PTD levels to differentiate patients in CR achievement. Therefore, the optimal prognostic cutoff value for abnormally-expressed genes needs to be identified by patient outcome data.
Contradictory results existed for the impact of EVI1 expression on CR achievement. Lugthart et al. and Groschel et al. individually showed that EVI1 (+) AML patients had significantly lower induction CR rates than EVI1 (−) AML patients [10,11], but Haas et al. did not observe this association [13]. With respect to ICR-AML, Groschel et al. reported that the CR rate was not related to EVI1 expression [11]. Similarly, our results showed that EVI1 transcript levels had no effect on CR achievement.
Almost all relevant studies have shown an adverse impact of high EVI1 expression on outcomes in AML, despite considering different end points. High EVI1 expression was demonstrated to independently predict event-free survival (EFS), DFS, RFS, and OS [9–11,13]. As for ICR-AML, EVI1 high expression was shown to predict EFS, RFS, and OS by univariable or multivariable survival analysis [9–14]. In the present study, we confirmed the strong poor prognostic impact of EVI1 overexpression in the Chinese cohort. Therefore, high EVI1 expression strongly predicts poor outcomes for both AML and ICR-AML, which suggests its possible role in patient stratification in clinical routine.
In the AML cohort, Groschel et al. reported that EVI1 (+) was inversely correlated with both FLT3-ITD (+) and NPM1 mutation (+) [11]. However, in the present study, no significant correlations were observed. Furthermore, FLT3-ITD (−)/NPM1 mutation (+) patients were demonstrated to have better outcomes and were defined as favorable risk [29,34]. In the present study, NPM1 mutation was not found to be prognostic in FLT3-ITD (−) patients. These findings might be caused by the exclusion of patients receiving allo-HSCT. In this study, we found that high EVI1 expression was associated with a lower RFS rate in both NPM1 mutation (−) and (+) patients without FLT3-ITD. Therefore, EVI1 expression may further stratify FLT3-ITD (−) patients receiving chemotherapy, regardless of NPM1 mutation.
Although almost all relevant studies have shown an adverse impact of EVI1 high expression on outcome in AML, it was difficult to make direct comparisons among them, which hinders the direct application of EVI1 expression testing in clinical practice. In addition to the cutoff value selection method, differences also existed in the detection method and control gene for normalization. In the early studies, each subtype of EVI1 was individually tested and analyzed. Subsequently, the common site of all subtypes was amplified and quantitated. Both absolute and relative real-time quantitative PCR methods were used [9–13]. Gene expression profiling (GEP) was also used [14]. The control gene used included PBGD, cyclophilin, ubiquitin C, G6PD, and GUSB [9–13]. Thus, standardization of the EVI1 transcript testing and reporting is required. In the present study, we selected ABL as a control gene and the EVI1 transcript level was expressed as a percentage of EVI1 to ABL copies, a widely used quantitation method for other fusion genes in hematologic malignancies [32].
MLL-PTD and WT1 expression were simultaneously assessed in the present study. In contrast to previous reports [18–22,35], high MLL-PTD expression was shown to be significantly associated with a low CR achievement rate. For outcomes, the reported impact of MLL-PTD was discordant [18–22,35]. In the present study, MLL-PTD (+) was not found to be relevant to relapse and survival in ICR-AML. The therapy composition, drug dose, and race all might affect CR achievement. In addition, differences in the cutoff values for defining MLL-PTD (+) might also affect its prognostic role. The impact of WT1 expression on outcomes in AML remains controversial [15–17]. WT1 expression was not prognostic in ICR-AML in the present study. This inconsistency indicates that the prognostic value of WT1 expression is weak and is affected by other factors, such as AML subtype and treatment modality.
The included variables affected the multivariate analysis results. Great progress has been made in the discovery of prognostic gene mutations in AML in the past 2 decades [6]. The limitation of this study is that it was a retrospective study. Although we investigated the widely used overexpression markers WT1, MLL-PTD, and FLT3-ITD, we did not screen CEBPA, DNMT3A, and other newly identified mutations.
Conclusions
EVI1 expression at diagnosis could further stratify ICR-AML, and high EVI1 expression predicted poor outcomes in patients receiving chemotherapy. EVI1 transcript levels should be routinely assessed at diagnosis for stratification once a standard laboratory protocol is established and the cutoff value is determined. Furthermore, the impact of EVI1 expression should be fully investigated in the context of all newly identified gene mutations in AML.
Footnotes
Conflict of interest
None.
Source of support: This work was supported by the Natural Science Foundation of China (81370637, 81370639, and 81570130)
References
- 1.Grimwade D, Walker H, Oliver F, et al. The Medical Research Council Adult and Children’s Leukaemia Working Parties. The importance of diagnostic cytogenetics on outcome in AML: Analysis of 1,612 patients entered into the MRC AML 10 trial. Blood. 1998;92:2322–33. [PubMed] [Google Scholar]
- 2.Slovak ML, Kopecky KJ, Cassileth PA, et al. Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: A Southwest Oncology Group/Eastern Cooperative Oncology Group Study. Blood. 2000;96:4075–83. [PubMed] [Google Scholar]
- 3.Byrd JC, Mrózek K, Dodge RK, et al. Cancer and Leukemia Group B (CALGB 8461) Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: Results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100:4325–36. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- 4.Nakao M, Yokota S, Iwai T, et al. Internal tandem duplication of the flt3 gene found in acute myeloid leukemia. Leukemia. 1996;10:1911–18. [PubMed] [Google Scholar]
- 5.Patel JP, Gönen M, Figueroa ME, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366:1079–89. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grimwade D, Ivey A, Huntly BJ. Molecular landscape of acute myeloid leukemia in younger adults and its clinical relevance. Blood. 2016;127:29–41. doi: 10.1182/blood-2015-07-604496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ross ME, Mahfouz R, Onciu M, et al. Gene expression profiling of pediatric acute myelogenous leukemia. Blood. 2004;104:3679–87. doi: 10.1182/blood-2004-03-1154. [DOI] [PubMed] [Google Scholar]
- 8.Gutiérrez NC, López-Pérez R, Hernández JM, et al. Gene expression profile reveals deregulation of genes with relevant functions in the different subclasses of acute myeloid leukemia. Leukemia. 2005;19:402–9. doi: 10.1038/sj.leu.2403625. [DOI] [PubMed] [Google Scholar]
- 9.Barjesteh van Waalwijk van Doorn-Khosrovani S, Erpelinck C, van Putten WL, et al. High EVI1 expression predicts poor survival in acute myeloid leukemia: a study of 319 de novo AML patients. Blood. 2003;101:837–45. doi: 10.1182/blood-2002-05-1459. [DOI] [PubMed] [Google Scholar]
- 10.Lugthart S, van Drunen E, van Norden Y, et al. High EVI1 levels predict adverse outcome in acute myeloid leukemia: Prevalence of EVI1 overexpression and chromosome 3q26 abnormalities underestimated. Blood. 2008;111:4329–37. doi: 10.1182/blood-2007-10-119230. [DOI] [PubMed] [Google Scholar]
- 11.Gröschel S, Lugthart S, Schlenk RF, et al. High EVI1 expression predicts outcome in younger adult patients with acute myeloid leukemia and is associated with distinct cytogenetic abnormalities. J Clin Oncol. 2010;28:2101–7. doi: 10.1200/JCO.2009.26.0646. [DOI] [PubMed] [Google Scholar]
- 12.Santamaría CM, Chillón MC, García-Sanz R, et al. Molecular stratification model for prognosis in cytogenetically normal acute myeloid leukemia. Blood. 2009;114:148–52. doi: 10.1182/blood-2008-11-187724. [DOI] [PubMed] [Google Scholar]
- 13.Haas K, Kundi M, Sperr WR, et al. Expression and prognostic significance of different mRNA 5′-end variants of the oncogene EVI1 in 266 patients with de novo AML: EVI1 and MDS1/EVI1 overexpression both predict short remission duration. Genes Chromosomes Cancer. 2008;47:288–98. doi: 10.1002/gcc.20532. [DOI] [PubMed] [Google Scholar]
- 14.Rockova V, Abbas S, Wouters BJ, et al. Risk stratification of intermediate-risk acute myeloid leukemia: Integrative analysis of a multitude of gene mutation and gene expression markers. Blood. 2011;118:1069–76. doi: 10.1182/blood-2011-02-334748. [DOI] [PubMed] [Google Scholar]
- 15.Nomdedéu JF, Hoyos M, Carricondo M, et al. CETLAM Group. Bone marrow WT1 levels at diagnosis, post-induction and post-intensification in adult de novo AML. Leukemia. 2013;27:2157–64. doi: 10.1038/leu.2013.111. [DOI] [PubMed] [Google Scholar]
- 16.Miglino M, Colombo N, Pica G, et al. WT1 overexpression at diagnosis may predict favorable outcome in patients with de novo non-M3 acute myeloid leukemia. Leuk Lymphoma. 2011;52:1961–69. doi: 10.3109/10428194.2011.585673. [DOI] [PubMed] [Google Scholar]
- 17.Qin YZ, Wang Y, Zhu HH, et al. Low WT1 transcript levels at diagnosis predicted poor outcomes of acute myeloid leukemia patients with t(8;21) who received chemotherapy or allogeneic hematopoietic stem cell transplantation. Chin J Cancer. 2016;35:46. doi: 10.1186/s40880-016-0110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schnittger S, Kinkelin U, Schoch C, et al. Screening for MLL tandem duplication in 387 unselected patients with AML identify a prognostically unfavorable subset of AML. Leukemia. 2000;14:796–804. doi: 10.1038/sj.leu.2401773. [DOI] [PubMed] [Google Scholar]
- 19.Döhner K, Tobis K, Ulrich R, et al. Prognostic significance of partial tandem duplications of the MLL gene in adult patients 16 to 60 years old with acute myeloid leukemia and normal cytogenetics: A study of the Acute Myeloid Leukemia Study Group Ulm. J Clin Oncol. 2002;20:3254–61. doi: 10.1200/JCO.2002.09.088. [DOI] [PubMed] [Google Scholar]
- 20.Steudel C, Wermke M, Schaich M, et al. Comparative analysis of MLL partial tandem duplication and FLT3 internal tandem duplication mutations in 956 adult patients with acute myeloid leukemia. Genes Chromosomes Cancer. 2003;37:237–51. doi: 10.1002/gcc.10219. [DOI] [PubMed] [Google Scholar]
- 21.Schlenk RF, Döhner K, Krauter J, et al. German-Austrian Acute Myeloid Leukemia Study Group. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1909–18. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 22.Kihara R, Nagata Y, Kiyoi H, et al. Comprehensive analysis of genetic alterations and their prognostic impacts in adult acute myeloid leukemia patients. Leukemia. 2014;28:1586–95. doi: 10.1038/leu.2014.55. [DOI] [PubMed] [Google Scholar]
- 23.Hinai AA, Valk PJ. Review: Aberrant EVI1 expression in acute myeloid leukaemia. Br J Haematol. 2016;172:870–78. doi: 10.1111/bjh.13898. [DOI] [PubMed] [Google Scholar]
- 24.Morishita K, Parganas E, William CL, et al. Activation of EVI1 gene expression in human acute myelogenous leukemias by translocations spanning 300–400 kilobases on chromosome band 3q26. Proc Natl Acad Sci USA. 1992;89:3937–41. doi: 10.1073/pnas.89.9.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russell M, List A, Greenberg P, et al. Expression of EVI1 in myelodysplastic syndromes and other hematologic malignancies without 3q26 translocations. Blood. 1994;84:1243–48. [PubMed] [Google Scholar]
- 26.Matsuo H, Goyama S, Kamikubo Y, Adachi S. The subtype-specific features of EVI1 and PRDM16 in acute myeloid leukemia. Haematologica. 2015;100(3):e116–17. doi: 10.3324/haematol.2015.124396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baird PN, Simmons PJ. Expression of the Wilms’ tumor gene (WT1) in normal hemopoiesis. Exp Hematol. 1997;25(4):312–20. [PubMed] [Google Scholar]
- 28.Schnittger S, Wörmann B, Hiddemann W, Griesinger F. Partial tandem duplications of the MLL gene are detectable in peripheral blood and bone marrow of nearly all healthy donors. Blood. 1998;92(5):1728–34. [PubMed] [Google Scholar]
- 29.National Comprehensive Cancer Network Clinical Practice Guidelines in Oncology. Acute Myeloid Leukemia. Version 1.2016 (URL: https://www.nccn.org)
- 30.Ruan GR, Li JL, Qin YZ, et al. Nucleophosmin mutations in Chinese adults with acute myelogenous leukemia. Ann Hematol. 2009;88:159–66. doi: 10.1007/s00277-008-0591-8. [DOI] [PubMed] [Google Scholar]
- 31.Qin Y, Zhu H, Jiang B, et al. Expression patterns of WT1 and PRAME in acute myeloid leukemia patients and their usefulness for monitoring minimal residual disease. Leuk Res. 2009;33:384–90. doi: 10.1016/j.leukres.2008.08.026. [DOI] [PubMed] [Google Scholar]
- 32.Beillard E, Pallisgaard N, van der Velden VH, et al. Evaluation of candidate control genes for diagnosis and residual disease detection in leukemic patients using ‘real-time’ quantitative reverse-transcriptase polymerase chain reaction (RQ-PCR) – a Europe against cancer program. Leukemia. 2003;17:2474–86. doi: 10.1038/sj.leu.2403136. [DOI] [PubMed] [Google Scholar]
- 33.Cilloni D, Gottardi E, De Micheli D, et al. Quantitative assessment of WT1 expression by real time quantitative PCR may be a useful tool for monitoring minimal residual disease in acute leukemia patients. Leukemia. 2002;16:2115–21. doi: 10.1038/sj.leu.2402675. [DOI] [PubMed] [Google Scholar]
- 34.Döhner H, Estey EH, Amadori S, et al. European LeukemiaNet. Diagnosis and management of acute myeloid leukemia in adults: Recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115:453–74. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 35.Whitman SP, Ruppert AS, Marcucci G, et al. Long-term disease-free survivors with cytogenetically normal acute myeloid leukemia and MLL partial tandem duplication: A Cancer and Leukemia Group B study. Blood. 2007;109:5164–67. doi: 10.1182/blood-2007-01-069831. [DOI] [PMC free article] [PubMed] [Google Scholar]