Abstract
Stem cells are characterized by a number of useful properties, including their ability to migrate, differentiate, and secrete a variety of therapeutic molecules such as immunomodulatory factors. As such, numerous pre-clinical and clinical studies have utilized stem cell-based therapies and demonstrated their tremendous potential for the treatment of various human diseases and disorders. Recently, efforts have focused on engineering stem cells in order to further enhance their innate abilities as well as to confer them with new functionalities, which can then be used in various biomedical applications. These engineered stem cells can take on a number of forms. For instance, engineered stem cells encompass the genetic modification of stem cells as well as the use of stem cells for gene delivery, nanoparticle loading and delivery, and even small molecule drug delivery. The present Review gives an in-depth account of the current status of engineered stem cells, including potential cell sources, the most common methods used to engineer stem cells, and the utilization of engineered stem cells in various biomedical applications, with a particular focus on tissue regeneration, the treatment of immunodeficiency diseases, and cancer.
1. Introduction
Cellular therapies are based on the direct injection of dissociated cells or tissues into patients and have shown great potential for use in biomedical applications. [1–3] This concept is not fundamentally new, as it has been more than half a century since cellular therapies were first introduced in the form of bone marrow (BM) and organ transplants. [4] However, recent breakthroughs in genetic engineering and gene/drug delivery are now allowing for safer and more precise cellular manipulation thereby improving the feasibility and potential applicability of cellular therapies in the clinic.
Currently, various cell types are being investigated including differentiated, undifferentiated progenitor, and stem cells, wherein each presents its own unique advantages and disadvantages. However, in general, the clinical application of differentiated cells is hindered by the practical difficulties that are associated with obtaining large cell populations, their lack of self-renewal capability, and poor engraftment upon transplantation. [5] Stem cells, on the other hand, can be distinguished from all other cell types by their unique ability to continuously self-renew and differentiate into intermediate and mature cells of a variety of lineages. In addition, they are relatively easy to isolate when compared to mature cells and exhibit the ability to migrate to sites of damage and disease in vivo. [6] Finally, stem cells can often contribute directly to therapy owing to their intrinsic secretion of therapeutic and/or beneficial factors such as anti-inflammatory cytokines or angiogenic factors. [7,8]
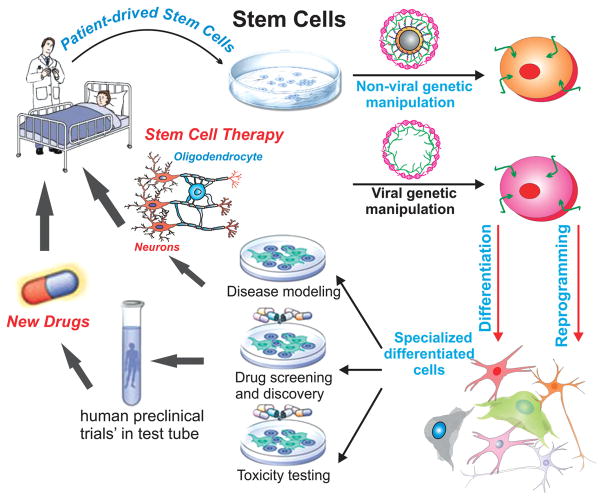
While the transplantation of unadulterated stem cells has shown great potential for the treatment of a variety of diseases and disorders, [3,9] recent efforts have increasingly focused on engineering stem cells to expand and control their innate functions. Specifically, the act of engineering stem cells can be defined as the modification of stem cells to control their behavior for a particular purpose (Figure 1). This encompasses the genetic modification of stem cells as well as the use of stem cells for gene delivery, nanoparticle delivery/loading, and even small molecule drug delivery. Currently, biomedical applications of engineered stem cells have primarily focused on regenerative medicine. In particular, studies have concentrated on engineering stem cells for the regeneration of cardiac, neural, and orthopedic tissues. [3,10] For instance, engineered neural stem cells (NSCs) can be transplanted following central nervous system (CNS) injuries such as spinal cord injury to promote neuronal cell survival and recovery or to guide NSC differentiation. Similarly, genetically modified stem cells are being developed for the treatment of more specialized genetic diseases including those related to immune deficiencies. [11] Finally, there has recently been increasing interest in engineering stem cells as potent cancer therapies, where stem cells can be used as the vehicle for gene therapy or for targeted chemotherapeutic delivery, owing to the demonstrated ability of stem cells to home to and infiltrate the tumor microenvironment. [12]
Figure 1.
Engineering stem cells for biomedical applications. Stem cells can be obtained from various sources, engineered using viral and non-viral methods, and then reintroduced back into the patients’ body. These engineered stem cells can take on a number of forms. For instance, engineered stem cells encompass the genetic modification of stem cells as well as the use of stem cells for gene delivery, nanoparticle delivery and loading, and even small molecule drug delivery. Reproduced with permission.[347] Copyright 2012, Nature.
In this Review, we will briefly discuss the strategies that have been developed to engineer stem cells, followed by a comprehensive review of their biomedical applications, with a particular focus on tissue regeneration (e.g., neural, orthopedic, and cardiac tissue regeneration), the treatment of immunodeficiency diseases (e.g., muscle dystrophy, Wiskott-Aldrich Syndrome, and leukodystrophies), and cancer. Specifically, we will highlight the astonishing progress that has been made over the last decade. While there are already a number of excellent reviews available that cover stem cell-based gene therapies, [3,10] this is a rapidly evolving area of research that is propelled by the constant expansion in our understanding of genetics and of methodologies and materials that can be used to engineer stem cells. Moreover, besides stem cell gene therapies, there have been limited reviews discussing other applications of engineered stem cell, such as their use as targeted drug and/or nanoparticle delivery vehicles. We hope that this article will inspire interest from various disciplines and highlight an exciting field wherein the use of our knowledge in genetic manipulation and nano/biotechnology to engineer stem cells can guide their behavior for use in various biomedical applications.
2. Methods for Engineering Stem Cells
Owing to the rapid advancement in our understanding of genetics and cellular behaviors, there has been an equally expeditious development of techniques with which to specifically engineer stem cells in terms of gene modification as well as for the delivery of exogenous materials such as nanoparticles, drugs, and other factors. While there are already numerous excellent and more comprehensive reviews on these topics, [13] in this section, we seek to instill the background that the reader needs in order to fully appreciate and gain a deeper understanding of the biomedical applications in which engineered stem cells are being used. To this end, we will begin by giving a broad overview of the different stem cell sources that are currently available, focusing on the intrinsic advantages and disadvantages that each source holds for engineered stem cell applications. Lastly, we will highlight the methods that have been developed to engineer these stem cells including genetic modification of stem cells via viral and non-viral methods (e.g., lipids, polymers, and nanoparticles).
2.1. Stem Cell Source
There are currently a number of stem cell sources that are being investigated for use in biomedical applications, including adult stem cells, embryonic stem cells (ESCs), and induced pluripotent stem cells (iPSCs), where each has its own advantages and disadvantages. For example, adult stem cells are a readily available source that are free from ethical concerns, are less likely to form teratomas than other stem cell sources, and can be collected from the patient, modified, and then reintroduced into the patient. On the other hand, ESCs are pluripotent cells that can be extracted from the inner cell mass of early embryos. ESCs can give rise to almost all cell lineages and, as such, are the most promising cell source for regenerative medicine. However, there are ethical issues related to their isolation. As a result, the development of iPSCs, which share many properties with ESCs but without the associated ethical concerns, also shows great promise. Unfortunately, ESCs and iPSCs have both shown the potential for teratoma formation, thereby greatly compromising their current clinical utility.
In this subsection, we will focus on these stem cell sources (Table 1 ) with a discussion of their individual advantages and disadvantages and their current unadulterated use (e.g., without any modification) in cellular transplantation applications. For a more in-depth look at stem cell sources for biomedical applications, there are also various reviews available. [1,14–16]
Table 1.
Stem Cell Sources
| Name | Sources | Advantages | Disadvantages |
|---|---|---|---|
| Neural Stem Cells | Brain and spinal cord | 1. Multipotent: can differentiate into neurons, astrocytes, and oligodendrocytes | 1. Limited differentiation potential |
| 2. Show tumor-tropic properties for various cancers | 2. Limited source | ||
| Hematopoietic Stem Cells | Bone marrow, cord blood, peripheral blood | 1. Multipotent: can form lymphoid and myeloid blood cells 2. Many sources 3. Most well-established stem cell source |
Limited differentiation potential |
| Mesenchymal Stem Cells | Bone marrow, adipose tissue, cord blood | 1. Multipotent – readily differentiates into bone, cartilage, fat, and muscle but can also be induced to differentiate into neuronal cells | 1. Limited differentiation potential but better than NSCs and HSCs |
| 2. Many sources | 2. Immunosuppressive properties | ||
| Embryonic Stem Cells | Inner cell mass of blastocyst | Pluripotent – has the highest differentiation potential | 1. Ethically controversial source (destruction of embryos) 2. Teratoma formation in vivo (requires ex vivo differentiation prior to transplantation) |
| Induced Pluripotent Stem Cells | Somatic cells | 1. Pluripotent: has similar differentiation potential as ESCs | 1. Potential tumorigenicity |
| 2. Can be derived from many cell types | 2. Low reprogramming efficiency | ||
| 3. Patient-specific | 3. Characteristics are protocol dependent |
2.1.1. Adult Stem Cells
Most of the biomedical applications that are discussed in this Review use adult stem cells. To understand the underlying reason, here, we will discuss the use of adult stem cells as a source for stem cell therapy in greater detail. Adult stem cells, also known as somatic stem cells, have been found in numerous tissues and are responsible for the maintenance and repair of the tissue in which they originate. Adult stem cell-based therapies have been successful for several decades, with the first hematopoietic stem cell (HSC) transplantation occurring over 50 years ago. [17] Adult stem cells are multi-potent and have the ability to differentiate into a number of lineages depending on their source tissue. For example, adult mesenchymal stem cells (MSCs) can readily differentiate into lineages of the mesoderm including muscle, bone, tendons, cartilage, and fat. The three main sources of stem cells that will be discussed in this subsection include: 1) NSCs, 2) HSCs, and 3) MSCs.
2.1.1.1. Neural Stem Cells
NSCs, or neural stem/precursor cells (NSPCs), are a heterogeneous population of self-renewing multipotent cells that can be found in the developing and adult CNS. [16] NSCs were first identified in the rat brain in the 1960s as proliferating neural cells. [18] Since then, NSCs have been isolated from the embryo as well as from the adult CNS. In particular, NSCs can be collected from the ganglionic eminence of embryos as well as from both the subventricular zone (SVZ) of the lateral ventricles and the subgranular zone (SGZ) of the hippocampal dentate gyrus (DG) in adults. [19] In terms of their differentiation, NSCs can differentiate into astrocytes, oligodendrocytes, as well as various types of neurons (e.g., dopaminergic). In vivo studies have demonstrated that transplanted NSCs can become incorporated into various brain regions, where they primarily differentiate into neurons and glia. [20] This lack of oligodendrocyte differentiation in vivo has been attributed to the low oligodendroglial differentiation efficiency of NSCs. [21] As such, NSCs represent a good source of stem cells for various biomedical applications, although concerns do exist owing to their limited availability and the difficult nature of their isolation.
Stem cell therapies using NSCs have primarily focused on the replacement of neurons for various nervous system disorders including Parkinson’s disease, Huntington’s disease, and spinal cord injury (SCI), which is currently being validated using numerous experimental models and a few clinical trials. [16] In terms of the experimental models, successes have been reported. However, a number of issues remain to be addressed including whether or not the transplanted NSCs can reach the target organ as well as whether, once at the target organ, the NSCs can differentiate into the appropriate lineage in sufficiently large numbers to give functional benefits. Moreover, our understanding of the in vivo differentiation process is still in its infancy. Though, it is clear that the disease microenvironment presents a complex combination of signals to the NSCs, which significantly differs from normal conditions, and, as such, may not be conducive to the survival and differentiation of NSCs into the intended lineage. [22] Furthermore, in the case of oligodendrocyte regeneration, NSC transplantation alone is unable to induce sufficient oligodendrocyte differentiation, which further confounds the use of NSCs for stem cell therapies. As such, there is significant room for investigation and improvement, which may be addressed using an engineered stem cell approach.
2.1.1.2. Hematopoietic Stem Cells
HSC transplantation is the most widely used stem cell therapy in the clinic today. It was originally developed for two purposes: 1) to treat individuals with inherited anemia or immune deficiencies by replacing the abnormal hematopoietic cells with cells from a healthy individual, and 2) to allow for the delivery of myeloablative doses of radiation and/or chemotherapy to cancer patients. [23] While effective, HSC transplantations come with a number of risks, with the most common being graft-versus-host disease (GVHD). [24]
There are three primary sources of HSCs: 1) BM, which is considered the classical source of HSCs, 2) peripheral blood, and 3) cord blood. The main differences between these sources are their reconstitutive and immunogenic potential. The first cell-surface marker that was used to enrich for human HSCs was CD34, a ligand for L-selectin.[25] In particular, in vitro assays have revealed that almost all CD34+ cells have multi-potency or oligo-potency, but also that the population is very heterogeneous. In terms of the percentage of CD34+ cells that can be collected from the different cell sources, typically, the number of circulating CD34+ cells is held at a steady state of 0.06% while 1.1% of the cells in the BM are CD34+. As such, BM is the best source of HSCs and is the primary source used clinically. [26]
Besides the applications described above, HSC transplantation is being investigated for a number of disorders including immunological and genetic blood diseases. For instance, immunosuppression followed by the transplantation of CD34+ HSCs has recently been investigated in Phase I/II clinical trials for the treatment of multiple sclerosis in order to reconstitute the immune system following the removal of active autoreactive T cells. [27] Similarly, HSC transplantation has shown promise for rheumatoid arthritis as well as Crohn’s Disease. [28] Lastly, HSC therapies are in clinical trials for sickle cell disease, where it has been demonstrated that curative levels of T cell chimerism (>50%) using HLA-matched sibling allogenic CD34+ HSC transplantations can be achieved. [29]
While HSC therapies have shown promising results in experimental models and in clinical trials, autologous HSC transplantation is not possible in every case, especially for genetic diseases. In addition, allogenic transplantation comes with significant risks of GVHD. As such, engineered HSCs may provide additional benefits such as genetically repairing autologous HSCs, which can then be transplanted to treat diseases such as Wiskott-Aldrich syndrome or muscular dystrophy as will be discussed in more detail later.
2.1.1.3. Mesenchymal Stem Cells
MSCs, which are also referred to as mesenchymal stromal cells, are a subset of non-hematopoietic adult stem cells that originate from the mesoderm. Like other adult stem cells, they possess self-renewal capabilities and can differentiate into multiple lineages. In particular, MSCs can not only differentiate into mesoderm lineages, such as chondrocytes, osteocytes and adipocytes, but also ectodermic cells (e.g., neuronal cells) and endodermic cells (e.g., pancreatic cells). [30] Importantly, MSCs exist in almost all tissues. For instance, they can be isolated from the BM, adipose tissue, the umbilical cord, liver, muscle, and lung.
To identify MSCs, there is a general consensus that human MSCs do not express the hematopoietic markers CD45, CD34, and CD14 or the co-stimulatory molecules CD80, CD86, and CD40. Instead, they express variable levels of CD105 (also known as endoglin), CD73 (ecto-5′-nucleotidase), CD44, CD90 (THY1), CD71 (transferrin receptor), the ganglioside GD2, and CD271 (low-affinity nerve growth factor receptor). Moreover, they are recognized by the monoclonal antibody STRO-1. In particular, it is thought that the observed variation in marker expression levels arise from differences in tissue source and culture conditions. [7]
As a result of the ease with which MSCs can be harvested as well as their multilineage differentiation capabilities, MSCs are currently the most widely used source for stem cell-based research and therapy. Numerous clinical trials using MSCs alone (e.g., without genetic manipulation) have been performed, with the primary applications being tissue repair and the therapy of immune disorders. In particular, MSCs have demonstrated reparative effects, where they are believed to be responsible for growth, wound healing, and the replacement of cells from everyday wear as well as from pathological conditions. [1] For instance, MSC transplantation has been shown to improve numerous musculoskeletal injuries and diseases including the regeneration of periodontal tissue defects, diabetic critical limb ischemia, bone damage caused by osteonecrosis, and burn-induced skin defects. [31] Besides musculoskeletal tissue repair, preclinical studies have also demonstrated that MSCs can effectively treat myocardial infarction as well as brain and spinal cord injuries. [32] On the other hand, MSCs also exhibit the capacity to regulate the immune response for the treatment of immune disorders. For example, MSC transplantation can reverse GVHD in patients receiving BM transplantation. [33] Similarly, the transplantation of both autologous and allogeneic MSCs was able to suppress inflammation and reduce damage to the kidneys and bowel in patients with Crohn’s disease. [34] It has also been reported that MSC transplantation can improve multiple sclerosis, amyotrophic lateral sclerosis, and stroke through their immunomodulatory effects. [35] Most importantly, MSCs for the treatment of GVHD and Crohn’s disease is currently the only stem cell-based drug approved by the FDA. [36] While already promising, similar to NSCs and HSCs, MSCs are great candidates for stem cell engineering, which can improve their survival and differentiation capacity thereby greatly enhancing the potential of MSCs for clinical applications.
Overall, adult stem cells are currently the most preferred cell type for downstream stem cell and engineered stem cell therapies as they are the most readily available and well established. Numerous studies and clinical trials have demonstrated that a large stem cell population can be obtained and expanded from patients (e.g., allogeneic source) and, following reintroduction into the patient, are less likely to form teratomas when compared to other stem cell sources upon long-term follow up. Finally, these cells are free from the ethical and moral issues associated with ESCs, which will be discussed in the following section.
2.1.2. Embryonic Stem Cells
The first successful isolation of human ESCs was achieved by Thomson and colleagues in 1998. [37] ESCs are pluripotent cells that are derived from the inner cell mass of developing blastocyst embryos and have the ability to differentiate to nearly all cell types. [38] Human ESCs are typically obtained from pre-implantation or blastocyst-stage embryos that are created during in vitro fertilization procedures and can also be generated by somatic cell nuclear transfer or parthenogenetic activation of eggs. ESCs bring great potential in terms of understanding early human development, tissue formation, and differentiation into various cell lineages. However, the derivation of ESCs from the human embryo sparked controversy in the United States and led to a presidential executive order that restricted its government funding. [39] As a result of the limited numbers of stem cell lines that were approved for research, the diversity necessary to address some of the more compelling questions, such as those related to disease modeling and treatment was unmet. [40] In addition to the moral and ethical controversy surrounding the use of ESCs, ESCs also have other significant limitations. For instance, it has been shown that transplanted ESCs will form teratomas, and thus, ES cells must first be predifferentiated ex vivo prior to grafting. [41] Lastly, as a nonautologous cell source, ESC transplantation faces the issue of immunological rejection. [22]
Despite these limitations, some ESC therapies are making their way into clinical trials. For instance, Geron conducted a Phase I clinical trial with oligodendrocyte precursor cells derived from ESCs for spinal cord injury. Advanced Cell Technology (ACT) also has Phase I/II approval for clinical trials on Stargardt’s Macular Dystrophy as well as dry macular degeneration. In these cases, they are deriving pigmented epithelial progenitor cells that can then be injected under the photoreceptor cells to redevelop and polarize the diseased retinal epithelium monolayer. As such, given the promising results that have been obtained from these preclinical and clinical studies as well as their immense differentiation potential, ESCs are also prime candidates for engineered stem cell applications. Albeit, further characterization and ESC sources, as well as a way to overcome the moral/ethical issues and teratoma formation that is associated with their use, will need to be addressed before ESCs become readily available for clinical applications.
2.1.3. Induced Pluripotent Stem Cells
While ESCs are a controversial source for pluripotent cells, iPSCs, which involve the reprogramming of adult cells towards an ESC-like state, may be able to address the downsides of ESCs. In 2006, Takahashi and Yamanaka demonstrated that the exogenous expression of at least four transcription factors (Oct4, Sox2, Klf4, and c-Myc) was able to reprogram fibroblasts into ESC-like cells, which have been dubbed iPSCs. [42] iPSCs, like ESCs, can proliferate indefinitely while maintaining their potential to give rise to virtually all cell types. These cells are therefore rapidly becoming invaluable for regenerative medicine and biomedical research.
In theory, iPSCs should be pluripotent and, as such, should have the ability to generate cell types from each of the three embryonic germ layers: the endoderm, mesoderm, and ectoderm. However, there are key differences between iPSCs and ESCs. This is corroborated by the fact that iPSCs are generally less successful in generating high percentage chimeras and even less efficient in their ability to generate live mice in tetraploid complementation experiments when compared with ESCs. [43] While high quality iPSCs and ESCs do have identical transcriptional profiles, [44] in practice, iPSCs and ESCs harbor genetic and epigenetic differences that reflect their histories and could affect the application of iPSCs to clinical situations. Lastly, just like ESCs, iPSCs are able to develop teratomas and, in fact, previous studies have shown that iPSCs develop teratomas faster and more efficiently than ESCs regardless of the site of injection. [45] As such, iPSCs cells must also first be predifferentiated ex vivo prior to grafting.
Owing to the large number of unknowns that remain to be addressed in the use of iPSCs, most studies have only utilized iPSCs in vitro for disease modeling and drug screening. iPSCs as a source for cell therapies is also being investigated, but the majority of these studies are still in a preclinical stage. For instance, Hanna and co-workers used homologous recombination to repair the genetic defect in iPSCs derived from a humanized mouse model of sickle-cell anemia. [46] However, iPSCs are slowly making their way into the clinic, where in 2014, a Japanese patient was treated with iPSCs in order to treat macular degeneration. While long-term safety and efficacy of this treatment are not yet available, no serious problems arose following surgery. As such, while limited engineered stem cell applications have utilized iPSCs, it can be argued that this stem cell source possesses the greatest potential, as they are pluripotent and can be derived from the patients’ own cells. As such, with continued optimization and investigation, we can expect to see an exponential rise in the use of iPSCs for stem cell and engineered stem cell therapies in the future. [14]
2.2. Genetically Engineering Stem Cells
The development of recombinant DNA technology in the 1970s marked the beginning of an exciting new era for biology. Molecular biologists gained the ability to manipulate DNA molecules, making it possible to study genes and harness them for the development of novel medicines and biotechnologies, which include engineering stem cells. However, to achieve the desired effects in engineered stem cells, the therapeutic genes must be carried by safe and effective vectors that can not only deliver genes specifically to the target cells but also sustain their expression thereafter. Other properties that these vectors should possess include: 1) high transfection efficiency, 2) long-term stability without integration into the host genome, 3) ability to spatiotemporally express appropriate levels of the therapeutic gene, and 4) not stimulate the host’s immune system or induce cellular transformation. [47]
For this purpose, both viral and non-viral vectors have been developed. Non-viral vectors, such as lipid-based and polymer-based vectors as well as other nanoparticles, have the advantage of being nonpathogenic and having high loading capacities but are generally associated with low transfection efficiencies. On the other hand, viral vectors such as retroviruses, lentiviruses, adenoviruses, and adenovirus-associated vectors are much more efficient, resulting in numerous preclinical and clinical gene therapy studies. Viral vectors differ in their immunogenicity, packaging capacity, ability to transduce dividing and nondividing cells, ability to insert into the host genome, and their ease of manufacturing (Table 2).[48] However, serious issues arise with their biosafety. As such, careful consideration must be taken when deciding which vectors to use for engineered stem cell applications. In this section, we will cover the techniques that have been most commonly used to genetically engineer stem cells with particular focus on viral and non-viral gene delivery methods.
Table 2.
Viral Delivery Methods.
| Feature | Retroviral Vector | Lentiviral Vector | Adenoviral Vector | Adeno-Associated Viral Vectors |
|---|---|---|---|---|
| Particle size (nm) | 100 nm | 100 | 80–120 | 20–30 |
| Genetic material | ssRNA (positive strand) | ssRNA (positive strand) | dsDNA | ssDNA |
| Cloning capacity (Kb) | 7–8 | 7–9 | Up to 36 | ≈2.4–4 |
| Chromosomal Integration | Yes | Yes | No | Yes (in about 1–10% of infected cells) |
| Immune response induction | Moderate | Low | Moderate–High (due to large size) | Low |
| Comments: | Low titers and can only primarily infect dividing cells | High efficiency and can infect both dividing and nondividing cells | High transduction efficiency in both dividing and nondividing cells | Long lasting expression and predictable chromosomal integration but small packaging ability |
2.2.1. Viral Gene Therapy
Currently, the most efficient and common method of introducing genes into stem cells is by means of viral vectors. However, the chief concerns associated with this approach involve frequent transgene silencing and the fact that integration of the transgene into the host genome can activate nearby oncogenes, leading to the selection of subclones with abnormal growth behaviors. [49] Moreover, viral vectors are severely hampered by their immunogenicity. While a number of excellent reviews covering the progress and challenges faced by viral vectors for gene therapy are available, [50–52] in this section, we will briefly highlight the various viral vectors that have been applied to engineer stem cells. Specifically, we will focus on: 1) retroviral, 2) lentiviral, 3) adenoviral, and 4) adeno-associated viral vectors.
2.2.1.1. Retroviral Vector
Retroviral vectors were the first class of viral vector to be developed and have, historically, been the most widely used in clinical trials. [51] Specifically, they are single-stranded RNA viruses that replicate in the host cell through reverse transcription, thereby producing DNA from its RNA genome. [53] Moreover, retroviruses have the ability to integrate into the host genome via an integrase enzyme. [54] However, it has been found that retroviral vectors are produced at relatively low titers, require proviral integration into the host chromosome for transduction, and can usually only infect dividing cells. As a result, these properties restrict most retroviral vector applications to ex vivo gene transfer approaches, which is not necessarily a significant limitation for the purpose of engineering stem cells.
For the purpose of engineering stem cells, retroviral vectors have traditionally been the vector of choice for the ex vivo transduction of HSCs and they offer two main advantages. First, they are non-immunogenic in nature. Second, and more importantly, they can offer constitutive transgene expression owing to their ability to integrate into the host genome. As a result, the genetically engineered stem cells can be used to treat various diseases. On the other hand, retroviral vectors are hampered by a number of significant limitations. Specifically, the use of retroviral vectors results in arbitrary integration of the inserted DNA into the host genome. This could modulate endogenous gene expression via insertional mutagenesis of a proto-oncogene or tumor suppressor resulting in carcinogenesis of the engineered stem cells. [51] As a result, in recent years, there has been a decline in the use of retroviral vectors for clinical trials (currently, only 19.7% of trials used retroviral vectors compared to 28% and 22.8% in 2004 and 2007, respectively). [52]
2.2.1.2. Lentiviral Vectors
Lentiviral vectors, such as the human immunodeficiency virus (HIV), are specialized members of the retroviral family. Like retroviral vectors, lentiviral vectors can integrate into the genome of the host cell. However, unlike other retroviruses, lentiviral vectors have the advantage of being able to transduce non-dividing cells. As such, these vectors are one of the most efficient viral methods for gene delivery.
In terms of engineering stem cells, one of the key rationales for using lentiviral vectors is their ability to transduce stem cells with a high efficiency after only a short ex vivo infection, which can favor the maintenance of stem cell properties. For example, this has been demonsrated in HSCs. [55] Moreover, lentiviruses are known to be less genotoxic than other retroviral vectors. [56] However, the potential for carcinogenesis, as induced by insertional mutation, is still a major hurdle for the clinical application of lentiviral vectors. For instance, a clinical trial using a lentiviral vector expressing β-globin to transduce hematopoietic progenitor cells was conducted for the treatment of a patient with β-thalassaemia-based anemia. [57] In this patient, following engineered stem cell transplantation, 10% of the erythroid cells contained the vector, but in 3% of cells the vector had integrated into the high mobility group AT-hook 2 (HMGA2) gene, which has previously been linked to cellular de-differentiation and metastasis of solid tumors. [58] Fortunately, at 33 months, this patient had no evidence of malignancy. Lastly, besides the potential for carcinogenesis, stem cells display low permissivity to the vector, thereby potentially requiring cytokine stimulation in order to increase transduction efficiency. [55]
2.2.1.3. Adenoviral Vectors
Adenoviral vectors are non-enveloped icosahedral viruses that are composed of a nucleocapsid and a double-stranded linear DNA genome. [59] Adenoviral vectors have a number of advantages, which make them attractive for stem cell engineering. Specifically, the 36 kb genome of the adenoviral vector provides ample space for the insertion of large sequences. [50] Moreover, adenoviral vectors have high transduction efficiency in both dividing and nondividing cells allowing for the collection of high titers with relative ease. Finally, the vector remains episomal and, as such, does not integrate into the host genome. As a result, the number of clinical trials using adenoviral vectors is growing with 23.3% of clinical trials using adenoviral vectors as of 2012. [52]
For stem cell applications, these properties may be particularly useful as the transient expression of the transduced gene can help prevent overgrowth of the transplanted stem cells (e.g., for tissue regeneration). However, there are also significant barriers that adenoviral vectors must first overcome before they can be useful in the clinic. For example, they are limited by their large size as well as their great immunogenicity. [60] Moreover, although recombinant adenoviral vectors were the first to result in high levels of systemic gene transfer in mammals, when delivered systemically they can induce severe toxicity at the dosage levels that are required for efficacy, especially in humans. To address this, second- and third-generation vectors contain additional deletions of the viral genes thereby reducing toxicities. However, even when all of the viral genes are deleted using a helper-dependent packaging system, [61] the vectors are not completely devoid of toxicity and transduction with these vectors can result in large changes in endogenous gene expression profiles. [62]
2.2.1.4. Adeno-Associated Viral Vectors
Adeno-associated viral vectors are derived from the parvovirus family and are small viruses with a single-stranded DNA genome that requires a helper virus for replication and completion of their life cycle. [63] When compared to adenoviral and other viral vectors, adeno-associated vectors are characterized by a number of advantages such as the ability to infect both dividing and non-dividing cells. In addition, the vector is largely episomal (>99%) and the <1% that is not, predictably integrates into human chromosome 19. [64] Finally, it is not currently related to any human disease and it has a lower immunogenicity.
As a result of these properties, adeno-associated viral vectors are currently the vector of choice for clinical viral transduction (4.9% in 2012, which continues to grow). [52] Previous studies have demonstrated that these vectors can mediate 10 to 100-fold higher levels of transgene expression both in vitro and in vivo compared to other vectors. However, because of their small size (2.4–4 kb), they can only accommodate small genes thereby limiting their therapeutic usefulness. [65,66] Moreover, despite their lower immunogenicity, one study reported the formation of hepatocellular carcinoma as a result of adeno-associated viral vector integration near a miRNA locus that is known to be involved in tumorigenesis. [67] On the other hand, and more significantly, a clinical trial conducted by Nathwani and colleagues demonstrated that adenovirus-associated viral vector-mediated gene transfer in Hemophilia B did not result in any acute or long-lasting toxicity but follow-up with a larger number of patients and for longer periods of time is necessary before a full evaluation of the usefulness of adeno-associated viral vectors can be made. [68]
In stem cells, studies have demonstrated that adeno-associated viruses can be used to transduce stem cells that originate from the muscle and brain. [69] However, the efficiency is significantly reduced when compared to the transduction of mature cells. For example, in muscle, Arnet et al. found that adeno-associated viral vectors were able to transduce proliferating myoblasts in culture with reduced efficiency relative to postmitotic myocytes and myotubes. [70] In addition, quiescent satellite cells were refractory to transduction in vivo in adult mice. On the other hand, for HSCs, some investigators have claimed that HSCs were impervious to adeno-associated viral transduction while others have reported that these vectors were capable of transducing HSCs but only at high vector-to-cell ratios. [69] Either way, despite their low transduction efficiency, recent efforts have focused on using directed evolution to enhance the utility of adeno-associated viruses for stem cell applications. To this end, Asuri and co-workers generated an adeno-associated virus variant with high gene delivery efficiencies (≈50%) to human pluripotent stem cells and a considerable increase in gene-targeting frequencies (up to 0.12%). [71]
2.2.2. Non-Viral Delivery Vehicles
Several limitations of viral vectors, such as safety concerns that include carcinogenesis, immunogenicity, broad tropism, as well as their relatively small capacity for therapeutic DNA, have prompted the development of synthetic non-viral vectors. [72] The ideal non-viral vector should be able to overcome the many barriers involved with systemic delivery, including: 1) targeted delivery, 2) efficient cell uptake and endosomal escape, and 3) the release of its cargo, all in a biocompatible manner while protecting the cargo from degradation. To this end, nanoparticles can provide a promising platform for gene delivery to stem cells.
Nanoparticles offer a number of advantages over viral vectors, including: 1) a lower immunogenicity, 2) the ability to deliver larger payloads, and 3) generally being easier to prepare/synthesize. [73,74] In addition, nanoparticles can be used to deliver other nucleic acids (DNA, RNA), biomolecules (e.g., peptides, proteins), small molecule drugs, and can also provide additional multifunctionalities (e.g., heating, imaging). [75] Owing to their great potential, a plethora of nanoparticle systems have been developed to overcome the physiological barriers faced by non-viral delivery methods. Specifically, these nanoparticles can be composed of various materials including metals, noble metals, semiconductors, polymers, lipids, and other inorganic materials and can have various sizes, shapes, and properties. [76] However, few of these vectors have made it through clinical trials to become FDA approved. [72] In addition, they are generally hampered by lower delivery efficiencies relative to viral vectors. [77] As such, while these vehicles possess great potential, there is still significant room for improvement before they can be widely used in the clinic. In this section, we will give a brief overview of some of the most common nanoparticle systems that have been developed for engineering stem cells with particular focus on lipid- and polymer-based vectors as well as gold and magnetic nanoparticles.
2.2.2.1. Lipid-Based Vectors
Currently, the most widely used non-viral delivery vehicle consists of lipid-based vectors. Lipid-based vectors are generally characterized of by three components: a cationic head group, a hydrophobic tail, and a linker group. [74] The liposomal delivery of DNA was first demonstrated in 1980, wherein the phospholipid phosphatidylserine was used to deliver SV40 DNA to monkey kidney cells. [78] Since then, numerous lipid-based vectors with more efficient transfection properties have been developed. Synthetic cationic lipids such as DOTMA, DOSPA, DOTAP, DMRIE and DC-cholesterol spontaneously form small, uniform liposomes that are capable of efficient encapsulation and delivery of DNA to various mammalian cells including stem cells. [72,74,79] On the other hand, neutral lipids, such as the fusogenic phospholipid DOPE or the membrane component cholesterol, have also been utilized as a component of liposomal formulations to enhance transfection activity and nanoparticle stability. [80] However, despite being the most widely used non-viral delivery vehicle, limitations do exist, including low efficacy owing to poor stability and rapid clearance, [81] as well as the generation of inflammatory or anti-inflammatory responses. [82]
2.2.2.2. Polymer-Based Vectors
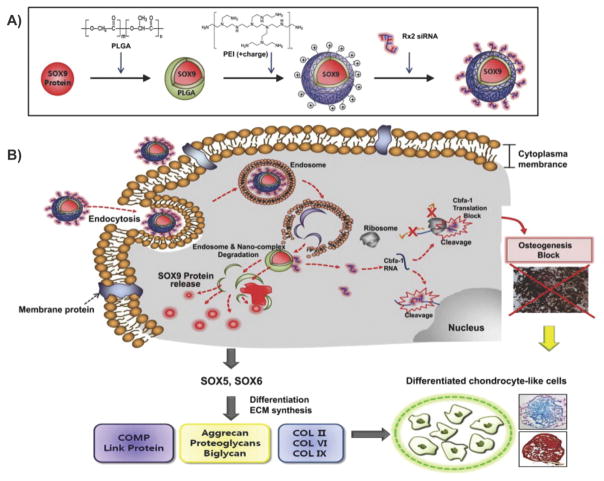
An alternative class of non-viral vectors consists of cationic polymers, which are attractive owing to their immense chemical diversity and the relative ease with which they can be functionalized. The most widely developed examples of polymeric vectors include poly(l-lysine) (PLL) and polyethylenimine (PEI), which have both been demonstrated to efficiently transfect stem cells. [72] Besides PLL and PEI, a number of other polymers, which have shown efficacy for stem cell transfection, are also available. For instance, PLGA is a popular choice and can be used to create nanoparticles via solvent evaporation. Finally, chitosan is another popular polymer with an intrinsically positive charge.
In particular, PLL is a homopolypeptide of the basic amino acid lysine although unmodified PLL shows marked in vitro cytotoxicity. [83] Moreover, in the absence of a lysosomal disruption agent such as chloroquine, PLL has fairly poor transfection ability. [74] As a result, numerous copolymer variants of PLL with enhanced gene delivery properties have been reported. [84] One example includes PLL coated with the hydrophilic polymer polyethylene glycol (PEG), which is designed to minimize nonspecific interaction with serum components and thereby increase circulation time. [85] On the other hand, PEI and its variants are among the most studied polymeric materials for gene delivery. PEI is a polymer that has a high positive charge density, especially at reduced pH values, owing to the existence of a nitrogen atom at every third position along the polymer. As a result, it has been hypothesized that this can aid in the condensation of DNA as well as enhance endosomal escape. [86] In terms of its transfection efficiency as well as its cytotoxicity, this strongly depends on the structural properties of PEI such as molecular weight and whether it is in a linear or branched form. [87] As with PLL, owing to the cytotoxicity of PEI, a range of modifications have been investigated including block co-polymers of PEG and PEI for improved stability and biocompatibility, degradable disulphide-crosslinked PEIs for reduced toxicity, and alkylated PEI to increase transfection ability. [72]
2.2.2.3. Gold Nanoparticles
Gold nanoparticles (GNPs) are one of the most widely used nanoparticles for stem cell applications. In particular, GNPs are attractive owing to their amenability to synthesis and functionalization. Moreover, they are very inert and non-toxic. Specifically, numerous studies have demonstrated that GNPs are well tolerated by stem cells depending on how they are coated and can be used to guide stem cell differentiation by delivering nucleic acids, other biomolecules, and/ or small molecule drugs. [88]
GNPs have been synthesized using an array of methods, which are mainly based on the reduction of chloroauric acid in the presence of a stabilizing agent. For example, the most commonly used method is the citrate synthesis method, which involves reduction of chloroauric acid using trisodium citrate thereby resulting into the formation of GNPs. The size of the obtained GNPs is determined mainly by the salt concentration, temperature and rate of addition of reactants resulting in a typical size range of 10–25 nm. However, a range of 1–100 nm or more can also be achieved by varying the salt concentration and temperature. [89] To utilize GNPs for drug or gene delivery, a number of functionalization have been investigated. In particular, as mentioned previously, the surface of GNPs can readily be modified using thiol-based chemistry. As such, GNPs have been stabilized via citrate as well as the more bioapplicable PEG. In addition, to allow for gene or drug delivery to stem cells, GNPs can be covalently modified with the gene or drug. Alternatively, non-covalent methods such as electrostatic interaction between PEI and nucleic acids can also be used and has been demonstrated successfully in stem cells. [90]
2.2.2.4. Magnetic Nanoparticles
Lastly, there has been considerable interest in magnetic nanoparticles (MNPs) as multifunctional nanoplatforms for stem cell applications. In particular, MNPs have many unique properties such as high biocompatibility, facile surface modification, and magnetic properties that result in an intrinsic ability to enhance MRI contrast, induce hyperthermia, [91] and be used for magnetic targeting. [92] As a result, it has been demonstrated that MNPs are biocompatible with stem cells and can actually enhance transfection efficiency via magnetically facilitated transfection (e.g., magnetofection). [93]
MNPs, such as the most common Fe3O4 MNPs, are typically synthesized through the co-precipitation of Fe 2+ and Fe 3+ ions in basic aqueous media or thermal decomposition, which results in more uniform and highly crystalline structures. [94] In addition, it has been found that doping MNPs with other metals such as Zn 2+ or Mn 2+ can greatly enhance the magnetization of the resulting MNPs, which is critical for downstream applications (4- to 14-fold increase in MRI contrast, which can be used to monitor stem cell migration, and 4-fold enhancement in hyperthermic effects for the treatment of cancer). [95] Generally, as with GNPs, these MNPs are coated with biocompatible polymers, such as dextran, dextran derivatives, or PEG, to confer stability in a biological system. In addition, nucleic acids, biomolecules, and small molecule drugs can be conjugated via covalent or non-covalent bonds (e.g., PEI via electrostatic interaction). As a result of their great potential, many MNP formulations are under clinical investigation and some formulations are already FDA approved with MRI contrast being their primary area of application. Finally, investigations have recently focused on the development of magnetic core-shell nanoparticles (MCNPs) wherein the MNP is coated with a shell that provides additional functionalities such as gold or mesoporous silica (e.g., dark-field imaging and increased drug loading, respectively). [96] As a result, MNPs and MCNPs have particularly great potential for stem cell engineering owing to their multifunctionalites and tunability.
3. Engineering Stem Cells for Tissue Regeneration
Regenerative medicine focuses on differentiating stem cells along specific lineages to effectively repair damaged or failing organs/tissues. [97] To achieve this goal, numerous strategies have been devised including the direct transplantation of various stem cells from different sources, the use of substrates, as well as engineering stem cells via genetic modification. In this Section, we will give a comprehensive review of the use of engineered stem cells for the regeneration of various tissues including the central nervous system (CNS), muscle, cartilage, and the heart.
3.1. Engineering Stem Cells for Neurological Diseases
As mentioned previously, the goal of cell therapy for neurological diseases is to replace and support neurons in diseased tissue. Stem cells, such as NSCs, have shown great potential for this purpose wherein stem cell transplantation can allay inflammation and replace tissues thereby resulting in functional benefits. However, the specific differentiation of NSCs to desired lineages (e.g., neurons and oligodendrocytes) is confounded by the microenvironment of neurological diseases and disorders. As a result, to enhance the efficacy of stem cell therapies, engineered stem cells, wherein the stem cells are modified to specifically secrete and deliver molecules that can further guide differentiation or revascularization, could greatly improve the potential of stem cell therapies for neurological diseases. In this section, we will primarily focus on the use of engineered stem cells to improve stem cell-based therapies for spinal cord injury, Alzheimer’s disease, Parkinson’s disease, and stroke.
3.1.1. Spinal Cord Injury
Spinal cord injury (SCI), which affects roughly 10 to 40 out of every million people in developed countries, [98] is a severely debilitating event that often results in permanent neurologic deficits, including partial to total sensorimotor loss, the disruption of autonomic nervous system control caudal to the injury, and chronic pain. The pathophysiology of SCI is divided into two phases: a primary and a secondary injury. The primary injury consists of the initial insult, which results in either contusion (e.g., caused by shattered vertebral bones) or compression (e.g., caused by an increased pressure) of the victim’s spinal cord. [99] Following the initial insult, a secondary injury ensues, which occurs on a cellular level. This phase begins with massive cell death due to immune response to the injury and is followed by secondary necrosis and apoptosis as well as oxidative, excitotoxicity, and axonal damage. As a result of the extensive neuronal death, axonal demyelination, as well as the limited ability of the mammalian central nervous system (CNS) to repair itself and replace lost cells, signal transduction through the spinal cord is compromised leading to the observed SCI-related functional impairments. While some treatments exist for SCI, they can only improve neurologic recovery by minimizing the secondary injury if administered promptly after SCI [100] and, as such, there are currently no cures. Therefore, cell replacement therapies represent a potential strategy that can overcome the loss of neurons and oligodendrocytes while providing neural protection thereby bridging the lesion site and creating an environment in which remyelination, axon elongation, and the formation of new circuits may occur.
For this purpose, stem cells hold great potential. Numerous studies have indicated that the grafting of NSCs into rodents following the induction of SCI can produce axonal regrowth and functional recovery. [101] As a result of these promising experimental results, a number of human clinical trials involving NSCs from various sources are now being conducted for the treatment of SCI. [102] For example, in December 2010, Stem Cells Inc. initiated a Phase I/II clinical trial utilizing human NSCs generated from the brain of an aborted human fetus to treat patients who sustained a thoracic SCI. While data has not yet been reported, a previous phase I clinical trial by Stem Cell Inc. using the same NSCs for Pelizaeus-Merzbacher disease, a rare leukodystrophy, demonstrated that the transplantation procedure was safe and resulted in modest improvements in cognitive function in three of the four patients enrolled. [103]
While the outcome of NSC transplantation for SCI appears promising, the mechanisms underlying these functional improvements have not been completely elucidated. [102] For instance, Hofstetter et al. reported that although the transplantation of naïve NSCs improved motor function, it also caused aberrant host fiber sprouting, which has been associated with allodynia-like hypersensitivity in a rodent model of SCI. [104] This suggests that the controlled differentiation of transplanted stem cells is essential in order to avoid possibly serious side effects and to achieve optimal functional improvements. In particular, engineering stem cells for the treatment of SCI can provide the benefits of stem cell transplantation while allowing for specific control over cellular behaviors such as guided differentiation or the secretion of therapeutic molecules via genetic modification. To this end, the two main strategies that have been investigated include engineering stem cells to secrete neurotrophins, which promotes neurite outgrowth and proliferation, or engineering them to secrete other factors that can specifically guide the differentiation of the transplanted stem cells.
3.1.1.1. Secretion of Neurotrophins
One commonly utilized genetic modification for the treatment of SCI is to engineer stem cells to secrete neurotrophins. Neurotrophins, such as brain-derived neurotrophic factor (BDNF), neurotrophin-3 (NT-3), neurotrophin-4 (NT-4), and ciliary-derived neurotrophic factor (CNTF), are a family of growth factors that can positively modulate the survival, development, and function of neurons. Previous studies have demonstrated that neurotrophins primary act through the Ras/MAPK and PI3K/Akt signaling pathways via the activation of Trk receptors. [105] Specifically, when used in the context of SCI, the introduction of neurotrophic factors into the site of injury has been shown to increase the extent of axonal growth thereby increasing both the length and density of projections as well as subsequent improvements in locomotor function. [106]
The earliest examples of engineering cells with a neutrophin for SCI utilized fibroblasts as a cell source. Grill and colleagues demonstrated that primary skin fibroblasts engineered to secrete NT-3, a neurotrophin that has previously been demonstrated to support the survival and differentiation of neurons as well as the formation of new synapses, [107] could enhance corticospinal tract regeneration and locomotion recovery following transplantation into SCI lesions. [108] While the transplantation of engineered fibroblasts that overexpress NT-3 could support and protect surviving neurons, thereby inducing some locomotor recovery, fibroblasts are unable to replace neurons and oligodendrocytes, which is a critical side effect of SCI that needs to be addressed. As such, Zhang and co-workers advanced the field by engineering NSCs (C17.2) [109] to express NT-3 neurotrophin. [110] Previous studies have already demonstrated that C17.2 NSCs constitutively secrete neurotrophic factors including NT-3, and in fact, can elicit a response from a wider range of host axons then engineered fibroblasts. [111] However, Zhang et al. demonstrated that engineering NSCs to secrete higher levels of NT-3, via introduction of a plasmid vector, could bring additional therapeutic benefits including enhanced cell survival and proliferation (of both transplanted NSCs and surviving neurons) over unengineered NSCs alone. Moreover, transplantation of these engineered NSCs not only promoted cellular survival and proliferation but, upon transplantation, also demonstrated an enhancement in functional recovery (via Basso, Beatie, and Bresnahan scoring) owing to the increase in axonal density.
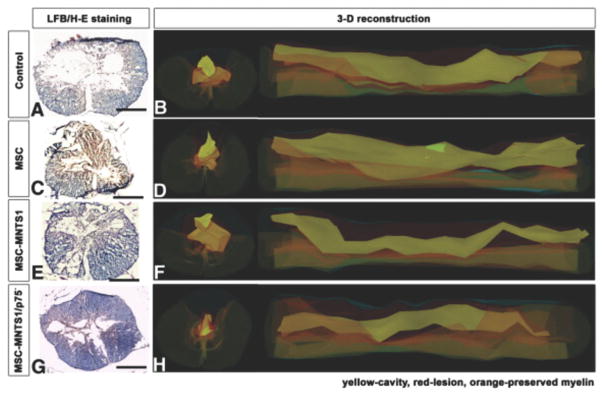
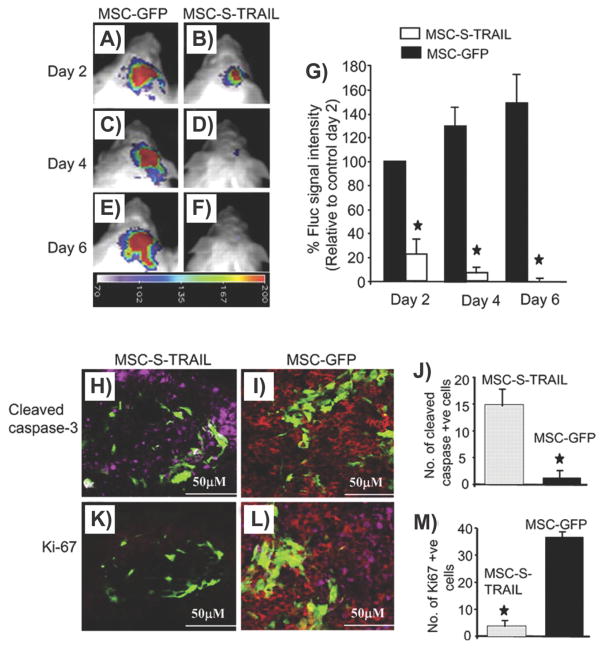
Recently, Kumagai and colleagues demonstrated that they could engineer other stem cell types for the treatment of SCI. In this case, they used a lentiviral vector to engineer MSCs to secrete the neurotrophin MNTS1, which binds to TrkA, TrkB, and TrkC, and p75 NTR.[112] In their study, engineered MSCs and control MSCs were transplanted seven days after SCI (via contusion) in rats and it was found that MSCs engineered to secrete MNTS1, but not control MSCs, were able to enhance axonal growth and significantly prevent cutaneous hypersensitivity following injury (Figure 2). In addition, the transplantation of engineered MSCs was able to promote angiogenesis and a modification of the glial scar was observed. This demonstrates the potential of MSC transplantation along with NSCs for the treatment of SCI.
Figure 2.
Engineering mesenchymal stem cells to express MNTS1 to enhance the treatment of spinal cord injury. A,C,E,G) Representative micrographs of rat spinal cord sections (axial) 1000 μm rostral to the epicenter of the insult. Sections were stained by hematoxylin and eosin and luxol fast blue. Scale bar: 500 μm. Both transplanted MSCs and engineered MSCs demonstrated reduced cavity size following SCI. However, only engineered MSCs promoted axonal growth and angiogenesis while decreasing inflammation. B,D,F,H) 3D reconstruction of injury in B) control, D) MSCs, F) engineered MSCs expressing MNTS1, and H) engineered MSCs expressing mutated MNTS1 with reduced binding to p75NTR. Reproduced with permission.[112] 2013, Elsevier.
3.1.1.2. Guided Differentiation
While neurotrophic factors can enhance neurite outgrowth and proliferation, NSCs that are transplanted into the spinal cord typically differentiate into astrocytes, which can actually hinder the effectiveness of NSC transplantation. [113] NSCs that have been engineered to express neurotrophic factors are no different. As such, another method that has been investigated to improve NSC-based SCI treatments is to engineer NSCs in order to control their differentiation following transplantation. To this end, a number of studies have sought to guide the differentiation of transplanted NSCs toward an oligodendrocyte lineage. Typically in the CNS, oligodendrocytes are responsible for the formation of the myelin sheath that surrounds axons, which, in turn, supports the fast saltatory conduction of nerve impulses in the nervous system. [114] However, the widespread apoptosis of oligodendrocytes that is typically observed following SCI has been found to be a major factor associated with the observed functional deficits, including impairment in the effective transmission of nerve impulses. [115] To address this deficit, Hwang and co-workers engineered NSCs to overexpress the Olig2 gene via retroviral transduction. [116] Olig is a family of transcription factors that are key regulators of differentiation along the oligodendrocyte lineage during development. [117] In particular, Olig2, a member of the Olig family, is more highly expressed in the spinal cord during early developmental and may play a crucial role in the differentiation of oligodendrocytes in the spinal cord. [118] As such, by overexpressing Olig2, Hwang et al. not only demonstrated that the engineered NSCs could differentiate exclusively into the oligodendrocyte lineage in vitro but also that the in vivo transplantation of these engineered NSCs improved locomotor function and increased the degree of myelination following SCI in a rodent model.
To further enhance the differentiation of NSCs to oligodendrocytes, Hu and colleagues recently demonstrated that the combination of engineered NSCs overexpressing Olig2 along with myelin basic protein-activated T (MBP-T) cells could synergistically improve the survival of transplanted NSCs thereby greatly enhancing the therapeutic outcome. [119] In this case, MBP-T cells were passively immunized for the purpose of modifying the SCI microenvironment in order to facilitate oligodendrocyte differentiation. [119] Previous work from their group had already demonstrated that T cell-based vaccination of mice with MBP, when combined with the transplantation of NSCs into the cerebrospinal fluid, synergistically promoted functional recovery following SCI. [120] The introduced MBP-T cells were then able to infiltrate the injured spinal cord thereby modulating the local T cell and microglial response. More importantly, this induced an increase in brain-derived neurotrophic factor as well as the differentiation of resident microglia and infiltrating blood monocytes into “alternatively activated” anti-inflammatory macrophages. As a result, newly formed neurons were observed from the endogenous NSC pool, substantiating the contention that immune response plays a crucial role in the recruitment of NSCs to the lesion site. As such, they hypothesized that similar immunological manipulations with MBP could also serve as a means to facilitate the differentiation of NSCs specifically towards an oligodendrocyte lineage. From the combination of MBP-T cells and lentiviral-mediated Olig2-engineered NSCs, the authors reported that five times as many transplanted NSCs cells survived and, moreover, that the number of engineered NSCs that differentiated towards an oligodendrocyte lineage was over 12-fold more than unengineered NSCs, thereby significantly increasing the number of remylinated axons. Finally, a decrease in spinal cord lesion size and an increase in myelin were observed suggesting that there was a synergistic effect in transplanting engineered stem cells and modulating the immune cells following SCI.
3.1.2. Alzheimer’s Disease
Alzheimer’s disease (AD) is the leading cause of age-related dementia, afflicting one in every eight people over the age of 65. It is characterized by a progressive loss of memory and other cognitive functions, often leading to the premature death of the patient. The hallmark pathological features of AD include the accumulation of beta-amyloid (Aβ) plaques and neurofibrillary tangles (NFTs). In addition, AD patients exhibit inflammation as well as widespread synaptic and neuronal loss. Typically, these Aβ plaques are a result of the extracellular accumulation of insoluble aggregates composed of the Aβ peptide, which induces neuronal cytotoxicity. On the other hand, NFTs consist of intraneuronal insoluble aggregates of tau, a micro-tubule binding protein. As a consequence of these Aβ plaques and NFTs, the nerve cells in the brain of AD patients progressively shrink and die, resulting in the gradual impairment of the patient’s memory and cognitive performance. [121]
Recently, a number of research groups have demonstrated that the transplantation of NSCs can markedly improve cognitive function, synaptic connectivity, and neuronal survival in experimental models of AD. [122,123] Importantly, however, it appears that the therapeutic effects of NSC transplantation are not mediated by the alteration of either Aβ or NFTs. Instead, the beneficial effects of NSC transplantation appear to be mediated by stem cell-derived neurotrophins or other neuroprotective molecules. For instance, NSC-derived cells have been observed to elevate hippocampal levels of brain-derived neurotrophic factor (BDNF), leading to an increase in synaptic density and the restoration of cognitive functions in preclinical models of AD. [122] MSCs have also been found to improve cognition in AD models by modulating cytokine levels and ameliorating brain inflammation. [5] Thus, stem cell transplantations have shown some therapeutic efficacy in preclinical models of AD by modulating complex biological systems via multiple mechanisms. Although the short-term benefits of stem cell transplantation appear promising and warrant further investigation, these studies have also shown that NSCs do not modify the underlying Aβ or tau pathology. [3,4] Moreover, given the widespread and progressive damage that is found in the brain of AD patients, it is highly unlikely that the mechanisms that are typically in place to guide the differentiation of NSCs to new neurons for neuronal cell replacement remains intact. [124] Hence, therapeutic strategies that utilize combinatorial approaches aimed at not only improving synaptic connectivity and neuronal function but also diminishing Aβ and tau accumulation as well as potentially guiding stem cell differentiation in vivo would have immense benefit.
For this purpose, Blurton-Jones et al. recently hypothesized that NSCs could provide an effective means with which to deliver disease-modifying therapeutic proteins owing to the fact that NSCs can migrate to diseased areas found in the AD brain. [125] In particular, murine NSCs were transfected with a plasmid vector encoding the Aβ-degrading enzyme, neprilysin (sNEP), using an AMAXA nucleofector. It was found that engineering the NSCs (sNEP-NSCs) with sNEP did not negatively affect their multipotency or differentiation capability. More importantly, these sNEP-NSCs were found to significantly reduce Aβ levels both in vitro and in vivo. Specifically, in the in vivo studies, these sNEP-NSCs, were transplanted (100 000 cells per animal) into the subiculum or hippocampus of AD transgenic mice (9-month or 18-month-old 3xTg-AD mice) as both of these regions have previously been shown to develop robust Aβ plaque pathology, exhibit significant synaptic degeneration, and are critical for learning and memory. One or three months following transplantation of the engineered NSCs, animals were sacrificed and their brains were examined. The sNEP-NSCs were found to engraft well and migrate into the surrounding brain tissue. Importantly, Aβ levels were assessed and significant reductions in plaque density in areas adjacent to the sNEC-NSC grafts were observed. While NFT levels were unchanged, sNEC-NSC grafts did result in a 31.8% increase in synaptic density when compared to the transplantation of control NSCs. Finally these findings were confirmed in a second AD model (Thy1-APP mice) further demonstrating the potential therapeutic benefits of engineered stem cells for AD treatment.
In support of the NEP engineered stem cell approach, Lebson and colleagues transfected CD11b+ monocytes with NEP and infused them biweekly into AD transgenic mice. [126] These engineered monocytes were able to migrate into the brain, resulting in a decrease in the rate of Aβ deposition. However, the use of monocytes as a cell source has disadvantages when compared to stem cells, as monocytes have limited half-lives (1–3 days) and thereby require repeated injections. Though, one advantage of these repeated injections is the fact that it can protect against potential adverse events that are typically associated with the transplantation of engineered stem cells such as teratoma formation or random insertion of the engineered gene into the host genome. Repeated injections can also prevent potential adverse events that could be associated with prolonged therapeutic gene expression.
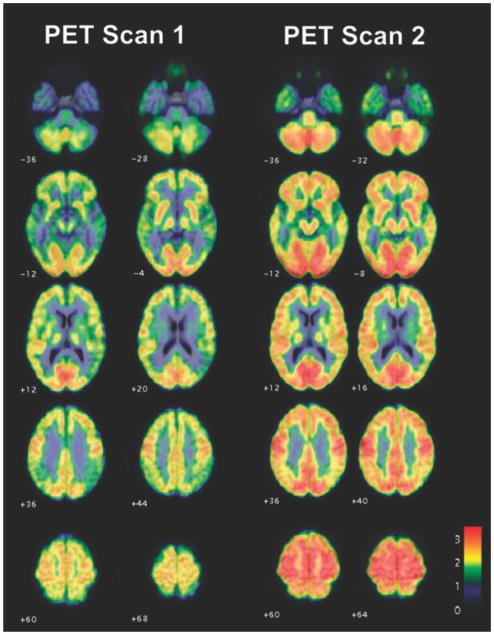
Lastly, in 2005, Tuszynski et al. conducted a phase 1 clinical trial on the suitability of nerve growth factor (NGF) gene therapy for the treatment of AD. [127] In their study, basal forebrain grafts of engineered fibroblasts that produce nerve growth factor (NGF), which counteracts cholinergic neuronal death, via modified Molony leukemia virus vectors, were injected into eight patients with mild AD. After a mean follow-up of 22 months in six subjects, no long-term adverse effects were observed. Importantly, serial PET scans showed significant increases in cortical 18-flurodeoxyglucose (Figure 3 ). Moreover, one subject demonstrated robust growth responses to NGF. These findings suggest improvement and a possible reversal of clinical outcome demonstrating the great potential of engineered cells for the treatment of AD. Although fibroblasts were engineered in this study, it is easy to imagine engineering stem cells for the same purpose, which would likely enhance the therapeutic effects seen, as the stem cells can specifically migrate to diseased areas in AD patients. In addition, besides using NSCs, other cell types may be useful for the delivery of NEP or NGF to the AD brain. For example, iPSC-derived NSCs can be used for personalized cell therapies against AD. Similarly, MSCs represent a readily available stem cell source that has seen utility in other diseases of the brain.
Figure 3.
Engineered cells expressing nerve growth factor can help reverse Alzheimer’s disease in patients. Averaged FDG PET scans in four subjects treated with NGF, overlaid on standardized MRI templates. Representative axial sections, with 6–8 months between the first and second scan, showing widespread increases in brain metabolism. Flame scale indicates FDG use/100 g tissue/min; red color indicates more FDG use than blue color. Reproduced with permission.[127] Copyright 2005, Nature.
Overall, while this field is still in the nascent stages of development, owing to the promise that stem cell transplantations have already shown for the treatment of AD via the improvement of synaptic connectivity and neuronal function, the addition of engineered stem cells that secrete factors that can reduce Aβ and tau accumulations and/or guide stem cell differentiation in vivo would have immense therapeutic benefits.
3.1.3. Parkinson’s Disease
Parkinson’s disease (PD) is a progressive neurodegenerative disorder that affects dopaminergic motor neurons of the ventral midbrain and their terminal projections thereby resulting in movement-related symptoms (e.g., shaking, rigidity, difficulty walking, and gait) and in later stages, thinking and behavioral problems (e.g., dementia and depression). [128] Strategies that focus on dopamine replacement have proven effective at remediating some motor symptoms during the course of PD. However, they ultimately fail to deliver long-term disease modification. Moreover, they lose effectiveness due to the emergence of additional side effects. [129] As such, several strategies have been investigated as alternatives for the treatment of PD, including direct cell replacement and gene transfer through viral vectors. For instance, the transplantation of human fetal ventral mesencephalic tissues, which secrete dopamine, into the striatum of Parkinson’s patients has shown promise. However, fetal tissue transplantation is problematic due to the logistics involved in acquiring large volumes of this tissue as well as the ethical questions associated with such treatments. [130] As an alternative, transplantation of stem cells that are differentiated along a dopaminergic lineage could offer a promising route of therapy with the hypothesis that these cells could act as a substitute for pharmacotherapy to directly provide long-term dopamine secretion. [131] Besides differentiation, other investigations involving gene therapy for PD have primarily focused on: 1) the restoration of dopamine synthesis and 2) neuroprotection and restoration of the surviving host dopaminergic circuitry through the introduction of trophic factors. [132]
While the majority of studies have focused on utilizing these strategies separately, recent efforts have increasingly focused on engineering stem cells to combine the benefits of stem cell and gene therapy for the treatment of PD. For instance, a number of studies have already investigated the engineering of various cell types (e.g., fibroblasts [133] and endogenous striatal cells [134]) with tyrosine hydroxylase (TH), the rate-limiting enzyme in catecholamine biosynthesis that converts tyrosine into L-DOPA. However, engineering these cell types to produce TH only resulted in a partial restoration of the behavior and biochemical deficiencies in PD animal models.
To address this, Kim and co-workers engineered NSCs to produce a combination of L-DOPA, TH, and GTP cyclohydrolase I (GTPCH1), which is a key enzyme in the synthesis of tetrahy-drobiopaterin, [135] a cofactor that supports TH activity. [136] In particular, human NSCs were transduced with retroviral vectors encoding TH and GTPCH1. Following transduction, the amount of L-DOPA produced by these engineered NSCs was significantly higher than unengineered NSCs or NSCs that were only transduced with the TH gene. HPLC results indicated that L-DOPA production in engineered NSCs (750 ng/10 6 cells/day) was 800 to 2000- fold greater than unengineered controls (0.35 ng/10 6 cells/day) or TH-transduced NSC lines (0.92 ng/10 6 cells/day). To test the ability of the engineered NSCs to produce functional improvements, they were transplanted in the striata of hemiparkinsonian rats. It was observed that the engineered NSCs survived well in the adult host brain after transplantation without any signs of rejection. More importantly, while some of the grafted cells did migrate away from the injection site into the surrounding host tissue, they maintained high levels of TH expression up to 4 weeks after transplantation. As a result, functional improvements were seen suggesting that engineered NSCs expressing both TH and GTPCH1 could have great potential for the treatment of PD.
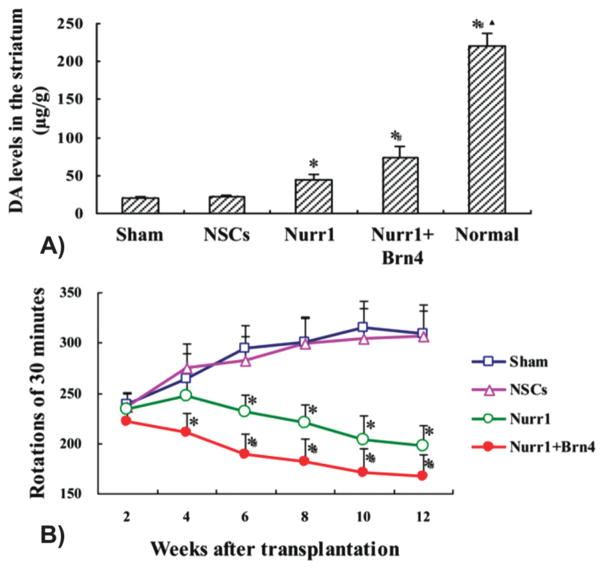
Lastly, Tan et al. recently demonstrated that engineering NSCs with both Nurr1 and Brn4 could dramatically increase the differentiation and maturity of TH-expressing dopaminergic neurons. [137,138] In particular, Nurr1 is a member of the steroid/thyroid hormone orphan nuclear receptor family and is one of the most important factors that are expressed in dopaminergic neurons. [139] On the other hand, Brn4 is a member of the POU-homeodomain family of transcription factors and plays an important role in regulating neuron migration and differentiation. [140] As such, Tan and colleagues hypothesized that the combination of Brn4 and Nurr1 could synergistically induce NSCs to differentiate into mature and functional dopaminergic neurons more effectively than either factor alone. Following the co-delivery of plasmids encoding Nurr1 and Brn4 via electroporation, as expected, engineered NSCs were found to exhibit high levels of Nurr1 and Brn4. More importantly, these cells efficiently differentiated into TH-expressing dopaminergic neurons and there were significantly more DAT positive cells when compared to controls, suggesting that the co-expression of Nurr1 and Brn4 resulted in more mature neurons. Finally, to validate this strategy in vivo, the authors investigated the effect of transplanted engineered NSCs in a rat PD model. It was observed that the overexpression of Nurr1 alone was able to promote NSC differentiation into dopaminergic neurons in vivo and increased the level of DA in the striatum (Figure 4A), resulting in behavioral improvement of PD rats. More importantly, the co-expression of both Nurr1 and Brn4 in NSCs significantly increased the maturity and viability of these dopaminergic neurons compared to all other conditions (Figure 4B).
Figure 4.
Engineered neural stem cells co-transfected with Nurr1 and Brn4 significantly increases the maturity and viability of dopaminergic neurons and reverses behavioral deficits in Parkinsonion rats. A) HPLC quantification of dopamine (DA) release in the different groups. *p < 0.01 compared with sham group, #p < 0.01 compared with Nurr1 group, p < 0.01 compared with Nurr1 + Brn4 group, n = 6. B) Rotation behavioral analysis induced by apomorphine after NSC transplantation in all groups. *p < 0.01 compared with sham group, #p < 0.01 compared with Nurr1 group, n = 12. Reproduced with permission.[138] Copyright 2013, Elsevier.
3.1.4. Stroke
Currently, the only therapies that are available for stroke are intervention to prevent inappropriate coagulation, surgical procedures to repair vascular abnormalities, and thrombolytic therapy with nothing directed at the restoration of function following stroke. As such, recent efforts have focused on the use of stem cell-based therapies to replace lost neurons and promote the survival and differentiation of both surviving and transplanted cells. [141] For instance, BM-derived MSCs have been shown to differentiate into neuronal cells, cross the blood-brain barrier (BBB), migrate to areas of damage, and secrete growth factors and cytokines. [142] Moreover, transplantation following stroke has resulted in observable improvements in functional recovery. [143] However, to further improve the efficacy of stem cell therapies for stroke, thereby enhancing their clinical potential, recent efforts have focused on engineering stem cells with neuroprotective factors as well as factors that promote neurite outgrowth.
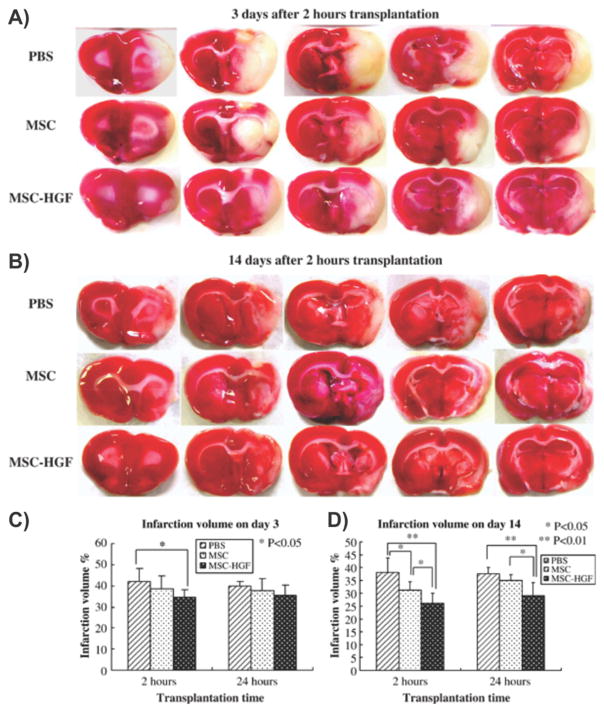
To this end, Zhao et al. engineered BM stromal cells with hepatocyte growth factor (HGF) via a multimutated herpes simplex virus type-1 (HSV-1) vector (MSC-HGF). [144] In particular, HGF has numerous functions including anti-apoptosis, angiogenesis, motogenesis, morphogenesis, tissue regeneration, and the enhancement of neurite outgrowth. Moreover, it can inhibit BBB destruction and provide neuroprotection following brain ischemia. In the present study, Zhao and co-workers found that the HSV-1 vector was able to transfer the HGF gene to the MSC population with high efficiency in vitro and, more importantly, the engineered MSCs functioned in vivo resulting in the expression and maintenance of high levels of the HGF. To evaluate the therapeutic efficacy of this engineered stem cell therapy, the authors treated brain ischemia in the superacute and acute therapeutic phases using a rat transient middle cerebral artery occlusion model. [145] The engineered stem cell therapy showed significant improvements in terms of the reversal of neurological deficits when compared to the MSC transplantation alone (Figure 5). In addition, after transplantation in the superacute therapeutic phases, Zhao and colleagues detected abundant levels of HGF protein in the ischemic brain of the MSC-HGF treated group, which was maintained for at least 2 weeks. Finally, the percentage of apoptosis-positive cells in the ischemic boundary zone was significantly decreased following treatment with MSC-HGF when compared to MSCs alone and other control conditions.
Figure 5.
Engineered bone marrow-derived mesenchymal stem cells expressing hepatocyte growth factor are more therapeutically efficient than mesenchymal stem cell therapy alone for the treatment of stroke. A,B) Reduction of infarction areas on days 3 and 14 of groups 1, 2, and 4, which received transplantation 2 h after MCAO occurred: coronal sections were stained with TTC. The red region shows the intact area while the white region shows the infarction area. C,D) Quantification of % CIV in the hemispheric lesion area on days 3 and 14. Treatment was given at 2 and 24 h after ischemia. Data are presented as means ± standard deviation. (p < 0.05; < 0.01). n = 6 for groups 1, 2, 4, and n = 5 for groups 5 to 7 at each time point. Reproduced with permission.[144] Copyright 2006, Elsevier.
Similarly, Lee and co-workers sought to provide neuroprotection by engineering human NSCs with Akt1 via a retroviral vector. [146] Akt was chosen because it is a serine/threonine kinase that plays a critical role in the modulation of cell proliferation, growth, and survival by participating in the PI3K/Akt signaling pathway. [147] Moreover, previous studies have demonstrated that Akt can promote cell survival during free radical exposure or hypoxia in hippocampal neurons. [148] As such, Lee et al. hypothesized that engineering NSCs with Akt could mediate survival and improve functional recovery following stroke. In particular, previous work has demonstrated that human NSCs can ameliorate neurological deficits in animal models of various diseases including PD [136] and stroke following their transplantation into the brain or spinal cord. [136,149] However, the survival rate of grafted NSCs in ischemia and ICH rats is very low (less than 50% of grafted NSCs survived in a mouse model of stroke 2-weeks post transplantation and 30% after 8-weeks) and, as such, is a grave concern that needs to be addressed in order to facilitate translation of NSC transplantation to the clinic. [150] Addressing this issue, NSCs expressing Akt1 were found to be highly resistant to H2O2-induced cytotoxicity in vitro. Following transplantation in the brain of a mouse model of stroke, engineered NSCs induced behavioral improvement and significantly increased cell survival (50–100% increase) at 2 and 8 weeks post- transplantation as compared to parental NSCs. Brain transplantation of NSCs overexpressing Akt1 in a mouse model of stroke provided functional recovery as demonstrated by the rotarod and neurology scores 8 weeks post-transplantation. Furthermore, it was determined that the Akt1 engineered NSCs enhanced survival as well as differentiation of the grafted NSCs into astrocytes and neurons owing to factors in the local microenvironment. Collectively, these results suggest that the Akt1 engineered NSCs could be of great value for the cellular therapy of stroke by enhancing survival of grafted NSCs.
Finally, following a slightly different strategy, Onda and colleagues demonstrated that engineering MSCs with angiopoi-etin-1 (Ang-1) could enhance functional recovery following cerebral ischemia by improving angiogenesis near the border of the ischemic lesions. [151] In particular, Ang-1 was used for this purpose as it is involved in the maturation, stabilization, remodeling of vessels, [152] and has specifically been shown to promote angiogenesis in the brain. [153] Furthermore, Ang-1 protects peripheral vasculature from leakage, [154] which may account for its anti-edematic effects if introduced following cerebral ischemia. As such, while the transplantation of unadulterated MSCs has been shown to ameliorate functional deficits via both neuroprotection and angiogenesis, Onda and coworkers hypothesized that there could be significant room to improve MSC transplantation strategies by engineering MSCs with Ang-1. Towards this objective, adenovirus-mediated gene transduction was performed to engineer MSCs with Ang-1. [143] Using a rat middle cerebral artery occlusion (MCAO) model of stroke, the authors demonstrated that intravenous infusion of either MSCs or Ang-MSCs 6 h after stroke induction resulted in a reduction in infarction volume, initiation of angiogenesis, and behavioral improvements. When comparing Ang-MSCs to MSCs, the authors reported a modest but observable improvement in Ang-MSC treated animals compared to unengineered MSC treated animals. While a significant enhancement was not seen, this suggests that MSCs alone, or with the proper genetic modification to enhance angiogenesis may provide additional functional benefits for the treatment of ischemia after stroke.
3.2. Engineering Stem Cells for Musculoskeletal Regeneration
As with other applications in regenerative medicine, stem cell-based therapies have shown great promise for the treatment of numerous musculoskeletal diseases and injuries. In particular, MSCs, ESCs, and iPSCs have all been shown to have the ability to produce tissues from various lineages that are critical for musculoskeletal regeneration, and allow for the derivation of tissues that are comprised of multiple cell types. Currently, most strategies for stem cell-based musculoskeletal regeneration have focused on direct injection of stem cells and, more recently, transplantation in combination with biomaterial scaffolds and bioreactors, which can provide an environment that better supports tissue formation. However, there is still significant room to improve stem cell-based therapies for musculoskeletal regeneration by more precisely guiding their behavior in vivo. This can be accomplished by engineering stem cells to express factors that specifically guide differentiation towards bone or cartilage lineages. As such, in this section we will highlight the progress that has been made in utilizing engineered stem cells for bone and cartilage regeneration.
3.2.1. Bone Regeneration
The primary function of bones is to provide shape, mechanical support, and protection for the body while facilitating movement. [155] On the other hand, bones also play an equally important role in mineral homeostasis and participate in the regulation of energy metabolism. [156] Typically, bones are supplied by blood and constantly undergo remodeling, allowing them to adapt to mechanical stress, maintain bone health, and repair small injuries. In particular, osteoclasts are the specialized bone cells that are responsible for the resorption of bone tissue while osteoblasts are the cells responsible for bone formation. However, clinical situations can arise where injuries, congenital malformations, or diseases cause large bone defects that cannot be repaired by natural mechanisms. Currently, autologous grafts, which contain the essential components necessary for bone regeneration (e.g., osteogenic cells, osteoinductive growth factors, and bone-supporting matrix), are the gold standard of treatment. However, they are not available in every clinical situation and autologous grafts can result in morbidity at the donor site as well as difficulties in preparing anatomically shaped grafts from the harvested bone. As such, there is a critical need for techniques that allow for specifically controlled bone regeneration.
To address these issues, a variety of engineering approaches have been investigated. Currently, the development of scaffolds is leading this area of research; wherein scaffolds using various materials, structures (e.g., pore size, roughness), cell attachment sites, and biomechanical properties have been studied extensively for musculoskeletal regeneration and have been reviewed elsewhere. [157] However, choosing which cell type with which to seed these scaffolds and achieving precise control over the differentiation of these cells are equally important issues that must be taken into consideration. In particular, stem cells, especially BM-derived MSCs, have been the most studied cells for this purpose. [158] As mentioned previously, MSCs have the ability to differentiate into bone, cartilage, adipose, muscle, tendon, ligament, and marrow stroma. BM transplantations are already used clinically in combination with osteoconductive materials to augment bone healing. [159] Moreover, MSCs have been shown to induce rapid bone regeneration and fracture repair in vivo in several models of bone loss including long bones, the calvaria, and the spine. Owing to the great promise that MSC transplantations, either with or without scaffold, hold for bone repair, one avenue of particular interest has focused on engineered MSCs as well as other cell sources to provide additional, more precise, control over their differentiation to osteoblasts and osteoclasts.
One commonly used strategy to engineer stem cells for bone regeneration has focused on genetically modifying stem cells with members from the bone morphogenetic protein (BMP) family of genes. [160] BMPs are a prime candidate as they have been found to play an important role during skeletal patterning and bone formation and are the most potent osteoinductive agents available today. For instance, recombinant BMP-2 has been approved by the FDA for the treatment of acute open tibial shaft fractures and for inducing lumbar spine fusion in patients with degenerative disk disease. [161] Following this strategy, Steinhardt and co-workers engineered maxillofacial BM-derived MSCs to overexpress MBP-2 via an adenoviral vector. [162] These engineered MSCs expressed high levels of BMP-2 protein resulting in differentiation towards an osteogenic lineage in vitro as well as significant bone formation in an ectopic site in vivo. In addition, implantation of the engineered MSCs into a mandibular defect led to regeneration of tissue at the site of the defect, which was confirmed via microcomputed tomography analysis. In particular, in vivo osteogenic differentiation as well as bone tissue regeneration was confirmed.
Further advancing BMP-based bone regeneration strategies, Virk et al. developed a “same day” methodology wherein patient-derived MSCs were engineered to overexpress BMP-2 via a lentiviral vector (Figure 6).[163] The typical viral procedure used to engineer stem cells consists of a two-step process, wherein stem cells are first harvested, expanded ex vivo, and followed by infection with the BMP-2 (or other gene) viral vector. This requires time and special culture facilities before they can be transplanted for bone regeneration or for other applications. To circumvent this step, Virk and colleages sought to determine whether MSCs could be harvested, tranduced with a lentiviral vector-expressing BMP-2, and then transplanted in the same sitting. To accomplish this, buffy coat cells were harvested from the rat BM, transduced with the lentiviral vector for 1 hour, and then implanted into a rat femoral defect (Figure 6A–E). It was found that there was no significant difference (p = 0.22) when comparing the healing rates of femoral defects that were treated with the “same day” strategy versus the traditional two-step lentiviral approach (Figure 6F,G). However, importantly, the “same day” strategy was found to induce earlier bone healing and higher bone volume (as analyzed via micro-computed tomography). As such, the “same day” strategy represents a significant advancement for the field of stem cell engineering as it offers a solution to the limitations that are typically associated with the culture expansion process that is required for the traditional ex vivo approach and can be applied to other engineered stem cell applications.
Figure 6.
“Same day” strategy wherein stem cells are engineered with BMP-2 to enhance bone repair. Steps involved in the “same day” ex vivo gene therapy. A) Harvest bone marrow from rat femur; B) Ficoll separation and preparation of “same day” rat bone marrow cells (SD-RBMCs) for viral transduction (time required = 0.5 h); C) Short-duration viral transduction of SD-RBMCs (time required = 1 h); D) Post-transduction preparation of SD-RBMCs (time required = 1 h); E) Placement of transduced SD-RBMCs on a collagen-ceramic matrix and implantation into the femoral defect. F) Representative images of healed femoral defects in animals treated with transduced SD-RBMCs. Double white arrows depict bridging bone across the femoral defect and restoration of cortex. G) Femoral defects treated with transduced cultured bone marrow cells (C-RBMCs) also exhibited healing. Defects treated with H) nontransduced SD-RBMCs, I) nontransduced C-RBMCs, and J) carrier alone demonstrated some bone formation but none exhibited complete healing. Reproduced with permission.[163] Copyright 2011, Nature.
More recently, non-viral strategies to engineer stem cells for bone regeneration have been the primary focus of investigation. To this end, Sheyn et al. engineered adult porcine adipose-derived stem cells to transiently overexpress BMP-6 via nucleofection of a BMP-6-encoding plasmid vector. [164] To test their engineered stem cells, bone void defects were created in the coccygeal vertebrate of nude rats and the engineered MSCs, which were suspended in a fibrin gel, were injected into the void. Beginning two weeks after treatment, it was found that considerable repair of the defect was observed in rats transplanted with the engineered MSCs. In addition, treatment with engineered stem cells induced bone formation at a rate that was twice as fast as the fibrin gel control group resulting in 2-fold higher bone volume when assayed at the end point. Finally, after twelve weeks, treatment with engineered MSCs resulted in complete regrowth of the void. Specifically, the engineered MSC condition reached a bone volume similar to that found in the native vertebrate and immunostaining indicated that the engineered MSCs contributed to the observed new bone formation.
To further enhance in vivo ectopic bone formation, Hosseinkhani and co-workers investigated combining engineered MSCS, which transiently express BMP-2 via a plasmid vector, with a three-dimensional (3D) cell scaffold as well as a bioreactor perfusion system thereby combining numerous beneficial factors to enhance bone formation. Specifically, the scaffold was composed of a collagen sponge reinforced with poly(glycolic acid) (PGA) fibers, which were impregnated with a cationized gelatin-DNA complex (via the introduction of spermine). [165] When cultured in vitro, it was found that BMP-2 expression was significantly enhanced in the perfusion culture condition versus static culture. Moreover, in vivo, following subcutaneous implantation into the back of rats, homogenous bone formation was observed throughout the scaffolds (from all conditions), with the extent of bone formation being highest in the engineered stem cell condition in combination with scaffold support and perfusion culturing. Finally, the level of alkaline phosphatase activity and osteocalcin content at the implanted site were significant higher in the combined engineered stem cell condition compared to the other controls. This demonstrates that engineering stem cells can act synergistically and be combined with traditional scaffold strategies to enhance bone formation.
Lastly, as bone formation typically involves multiple factors that not only include a 3D microenvironment (e.g., scaffold) to support bone growth, as well as bone-specific gene expression (e.g., BMP family), Huang et al. developed a PLGA scaffold that could be loaded with both osteogenic and angiogenic factors. [166] PLGA was chosen, as it is a widely used biomedical polymer that has been safely used as a biodegradable suture and implant for several decades. [167] In particular, loading of BMP-4 plasmid vector was achieved by precomplexing with PEI, followed by lyophilization. Freeze-dried PEI DNA condensates and a mixture of alginate and VEGF were then combined with PLGA to fabricate the scaffold, which was accomplished via a high pressure gas foaming process. [168] In particular, alginate was used because previous studies have shown that this mixture can attenuate release of VEGF from PLGA scaffolds. [169] It was found that any combination of the factors displayed increased bone formation when compared to the individual factors alone. Moreover, combining these factors led to the greatest quantity and quality of new bone tissue, suggesting that the use of a scaffold combined with angiogenic and osteogenic factors could act synergistically to regenerate bone tissue. The authors hypothesized that this could be caused by increased survival of transplanted cells owing to VEGF-mediated angiogenesis along with the direct osteogenic action of BMP-4 on this cell population.
3.2.2. Cartilage Regeneration
Defects in articular cartilage tend to heal poorly and progress to catastrophic degenerative arthritis. Typically, articular cartilage is a thin viscoelastic layer that is less than 3 mm thick, which covers the articulating surface of the bone in a joint thereby permitting smooth motion with minimal friction. At a fundamental level, cartilage is composed of a unique ECM that consists of a complex combination of specifically arranged collagen II fibrils. In addition, these fibrils have large water-retaining molecules known as aggrecan as well as its associated linked protein molecules bonded to it. This unique ECM is produced and maintained by a limited number of chondrocytes and gives articular cartilage its ability to withstand the repetitive compressive loading in daily activities without undergoing premature repair. [170] This entire structure is avascular, aneural, and alymphatic and as such, cell infiltration and repair after injury is unlikely to occur. [171] Current surgical intervention include the transplantation of autologous chondrocytes that have been expanded in vitro, which is known as autologous chondrocyte implantation (ACI). [172] Unfortunately, ACI is associated with several problems such as donor site morbidity, loss of chondrocyte phenotype upon ex vivo expansion and inferior fibrocartilage formation at the defect site. [173] Other clinical procedures are also available, including arthroscopic lavage and debridement, microfracture techniques, and osteochondral transplantation. [174] While there have been some promising results, most cartilage repair techniques lead to fibrocartilage formation and cartilage degeneration after a temporary relief of symptoms. As such, the most effective procedure utilizes surgical replacement with an autograft. However, there is a shortage of articular cartilage that can be donated for autografting.
Stem cells represent a promising cell source for cartilage repair and can be derived from two major sources: MSCs and ESCs. [175] For example, BM-derived MSCs are currently undergoing clinical trials for several orthopedic applications including articular cartilage repair. [176] However, there are still some limitations to directly transplanting stem cells for this purpose including the need for a scaffold as well as specifically guiding stem cell differentiation to cartilage. As such, there is significant room to improve stem cell-mediated cartilage repair.
3.2.2.1. Sox Family
The majority of strategies have focused on engineered MSCs with the SOX family of genes. The SOX family encodes transcription factors including SOX5, SOX6, and SOX9, which have all been shown to be the master regulators of chondrogenic differentiation. [177] In particular, SOX9 is expressed in all chondroprogenitors and chondrocytes, ensures cell survival in precartilaginous condensations, and is required to activate cartilage genes such as Col2a1, Agc1. [178] Similarly, SOX5 and SOX6 are expressed in prechondrocytes and has been shown to enhance chondrogenic specific gene transcription cooperatively with SOX9. [179] As such, this SOX trio, with a minimum of SOX9, is required and sufficient to mediate chondrogenesis via activation of cartilage-specific target enhancers such as aggrecan and Collagen-2. With this in mind, Park et al. demonstrated a non-viral method to engineer MSCs with a combination of the SOX trio to guide chondrogenesis. [180] Specifically, they utilized PEI-modified PLGA nanoparticles to complex with and deliver plasmids encoding SOX5, SOX6, and SOX9 (size distribution of 60 ± 11, 72 ± 8, and 92 ± 11 nm for the PLGA, PEI-modified PLGA, and PEI-modified PLGA/gene complexes, respectively). All three genes were efficiently delivered with a transfection efficiency of 22.21% for MSCs resulting in the synthesis of chondrocyte-related proteins well after three weeks in vitro. Moreover, engineered cells showed a high amount of staining for proteoglycans and polysaccharides, which further demonstrate their successful chondrogenic differentiation.
Similarly, Im and co-workers also engineered MSCs with the SOX trio using a PLGA-based non-viral method. [181] In this case, the plasmid (encoding the SOX trio) was complexed with a PEI-PEG polymer and then incorporated into a PLGA scaffold, [182] which allowed for slow release of the plasmid to MSCs seeded in the scaffold. The pDNA was released over 30 days and the MSCs were successfully transfected as demonstrated by a 50-fold increase in gene expression of SOX5, SOX6, and SOX9. An immunofluorescence study also demonstrated the uptake of pDNA into the MSCs and translation into protein up to 21 days after transfection. More importantly, the usefulness of their system was corroborated by in vivo implantation of the MSC/SOX trio pDNA-incorporated PLGA scaffolds into osteochondral defects created in rabbits. Increased expression of chondrogenic markers and a smooth articular surface with restoration of hyaline cartilage was observed in the engineered stem cells (MSC/pDNA-incorporated PLGA scaffolds) when compared to the PLGA scaffold alone, wherein the defect area was depressed and filled with fibrous tissue, and MSC/PLGA scaffold without plasmid, wherein regeneration was observed but the quality of repair was poorer with an irregular surface and incomplete reconstitution of subchondral bone (Figure 7). Similar to what was discussed in bone regeneration, these findings demonstrated the potential effectiveness of combining engineered stem cells (expressing SOX trio) with scaffold-based strategies for in vitro and in vivo cartilage tissue engineering.
Figure 7.
Engineered adipose stem cells expressing the SOX trio in combination with a PLGA scaffold enhances chondrogenesis for cartilage regeneration. A) Gross findings from defects implanted with PLGA scaffold only (group 1), implanted with ASCs/PLGA scaffold (group 2), implanted with ASC/SOX trio pDNA-incorporated PLGA scaffold (group 3); B) The ICRS macroscopic score; C) Histological findings; D) and O’Driscoll scores. n = 4, *p < 0.05. Reproduced with permission.[181] Copyright 2011, Elsevier.
3.2.2.2. Transforming Growth Factor Family
An alternative approach for cartilage regeneration has been the genetic modification of stem cells with members of the transforming growth factor (TGF) family. In particular, previous studies have demonstrated that the addition of TGF-β1 to MSCs can induce chondrogenesis. [183] As such, Pagnotto et al. investigated the ability of adeno-associated virus (AAV) to effectively transduce MSCs with TGF-β1 to induce chondrogenesis in vitro and in vivo. [184] To this end, adult MSCs were transduced with AAV-GFP (control) or AAV-TGF-β1 and studied in pellet cultures. For in vivo studies, AAV–GFP and AAV–TGF-β1-transduced MSCs were implanted into osteochondral defects of athymic rats. Following implantation, GFP was detected using fluorescent microscopy. GFP transgene expression was observed in 100% of the GFP implanted defects after 2 weeks, 67% after 8 weeks, and 17% after 12 weeks of implantation. More importantly, cartilage repair was assessed using gross and histological analysis at 4, 8 and 12 weeks. Improved cartilage repair was observed in osteochondral defects implanted with AAV–TGF-β1-transduced MSCs at 12 weeks (p = 0.0047). These results demonstrate that AAV, which has been proposed to be safer than other viral methods, is a suitable vector for gene delivery to improve the cartilage repair potential of MSCs.
Similarly, it has been shown that TGF-β3 can effectively induce chondrogenesis of MSCs, whereas other potential transgenes (e.g., BMP-7, SOX9) were less effective. [185] In this case, Brunger and colleagues developed a bioactive scaffold that could guide stem cell differentiation and, together with stem cells, act as a suitable replacement of musculoskeletal tissues with mechanical properties that mimic those of native tissues. [185] In particular, the motivation behind this study was to develop a scaffold that was not only able to mediate differentiation but also to guide the formation of ECM that has the biomechanical composition and mechanical features that mimic native tissue properties. Towards this objective, lentiviral vectors immobilized with PLL, which has previously been shown to facilitate efficient transduction of cells, [186] was used to functionalize poly(e-caprolactone) (PCL), one of the most commonly used polymers for tissue engineering scaffold applications. [187] The authors first demonstrated that PCL could immobilize the lentivirus to PLL films and facilitate MSC transduction (93% transfection efficiency of EGFP vs fewer than 1% in the absence of PCL). They then demonstrated that scaffold mediated gene delivery of TGF-β3, using a 3D woven PCL scaffold, induced robust cartilaginous ECM formation (Figure 8A,B). Specifically, it was found that this method resulted in the production of 17 ng/mL of TGF-β3 as well as high levels of sulfated glycosaminoglycan (sGAG) and collagen production (Figure 8C,D). Importantly, chondrogenesis induced by scaffold-mediated gene delivery was as effective as traditional differentiation protocols involving medium supplementation with TGF-β3, as assessed by gene expression (e.g., α1 chains of collagen type 1, 2, and 10 as well as aggrecan), and biochemical (e.g., total collagen and sGAG content), and biomechanical analyses.
Figure 8.
Engineered stem cells expressing TGF-β3 combined with scaffold for cartilage repair. A) Scanning electron micrograph showing the architecture of the 3D orthogonal woven PCL scaffold. (Scale bar, 500 μm). B) Fluorescence microscopy from iLVT constructs after 28 days in chondrogenic culture. C,D) Quantification of cartilaginous ECM components in the nontransduced (NT), rhTGF-β3 (rhT), and immobilized lentiviral TGF (iLVT) groups. Sulfated glycosaminoglycan content and total collagen content were normalized to DNA content. Bars represent means ± SEM (n = 6). Groups not sharing the same letter or symbol are statistically different (p < 0.05). Reproduced with permission.[185] Copyright 2014, PNAS.
3.2.2.3. Non-Viral Methods
Lastly, while the majority of approaches used to engineer stem cells for chondrogenesis have focused on viral delivery methods, Jeon et al., recently reported the development of nanoparticles for the co-delivery of Cbfa-1 siRNA and SOX9 protein to specifically guide chondrogenesis while inhibiting osteogenesis (Figure 9).[188] As mentioned previously, SOX9 is an essential chondrogenic differentiation-related protein, which triggers the expression of aggrecan and collagen type 2. [189] However, simply adding SOX9 protein to the culture medium is not a viable strategy due to rapid degradation. On the other hand, Cbfa-1 is an osteogenesis-related transcription factor that typically inhibits the chondrogenesis of MSCs. As such, to enhance chondrogenesis, it would be beneficial to silence Cbfa-1 while activating SOX9. [190] To this end, the authors fabricated PLGA nanoparticles loaded with SOX9 protein and coated with Cbfa-1 siRNA (Figure 9A). Specifically, the PLGA nanoparticles were fabricated by solvent evaporation and water-in-oil-in-water emulsion, resulting in 53 nm nanoparticles. To deliver siRNA, the PLGA nanoparticles were coated with PEI, which allowed for complexing with siRNA, resulting in 68 nm and 138 nm diameter particles, respectively with a final zeta potential of +26.3 mV. Further characterization included evaluation of the release of SOX9 protein from the PLGA nanoparticles as well as confirmation of the ability of cells to uptake/internalize the nanoparticle complexes. Importantly, upon internalization of the PLGA nanoparticles coated with Cbfa-1 siRNA and loaded with SOX9 protein into MSCs, the level of Cbfa-1 protein was reduced whereas the level of SOX9 protein was increased.
Figure 9.
Engineering stem cells via the co-delivery of Cbfa-1-targeting siRNA and SOX9 protein using PLGA nanoparticles for cartilage regeneration. Schematic showing SOX9 protein encapsulated by biodegradable PLGA nanoparticles and complexed with Cbfa-1-targeting siRNA for the induction of chondrogenesis in human mesenchymal stem cells. Reproduced with permission.[188] Copyright 2014, Elsevier.
Finally, to test the ability of these nanoparticles to induce differentiation of MSCs, both in vitro and in vivo studies were conducted and markers typically expressed in mature chondrocytes were examined (e.g., SOX9, aggrecan, COMP, and COL II). In vitro, three weeks after the initiation of differentiation via the delivery of nanoparticles, it was observed that the mature markers were highly expressed at the mRNA and protein levels in engineered MSCs compared to unengineered controls. By contrast, these cells did not express osteogenesis related markers (Cbfa-1 and COL1). In vivo, MSCs were injected into nude mice following internalization of PLGA NPs coated with Cbfa-1-targeting siRNA and loaded with SOX9 protein. When the injection site was excised, markers of chondrogenesis were found to be highly expressed at the mRNA and protein levels, corroborating the in vitro results. Moreover, the level of GAG was much higher in engineered MSCs then in control MSCs. Lastly, ECM production, as assessed by Alcian blue, Safranin-O, and Masson’s Trichrome staining, was evident in the excised samples.
3.3. Engineering Stem Cells for Cardiac Repair
Although substantial progress has been made in treating various heart conditions, the worldwide burden of heart failure is enormous and is expected to continue growing throughout this century as the aging population increases in size. [191] Specifically, heart disease is the leading cause of death in the United States, where it accounts for nearly 40% of all deaths, which is more than all cancers-related deaths combined. [192] Similarly, congenital heart defects occur in nearly 14 out of every 1000 newborn children and are the leading cause of death for children in their first years of life. [193]
Typically, the heart wall is composed of tightly packed myocytes and fibroblasts, with a dense supporting vasculature and collagen-based extracellular matrix (ECM). Owing to the high density and metabolic demand of these cells, the myocardium consumes a large amount of oxygen and is unable to tolerate hypoxia. [194] However, during myocardial infarction, a vigorous inflammatory response is elicited resulting in massive cell death. Over the weeks to months following infarction, fibroblasts and endothelial cells migrate to the site of infarction forming granulation tissue. This ultimately leads in the generation of a thick and stiff collagenous scar that reduces the contractile function of the heart thereby resulting in ventricle wall thinning and remodeling and ultimately causing heart failure. This is another clear situation where tissue engineering would be of great value, as it provides a method with which to reconstruct the heart following heart diseases or in patients with congenital heart defects. [192] However, similar to the issues presented previously with bone and cartilage regeneration, there is a need to determine which cell types to deliver and to develop methods with which to specifically control the differentiation of these cells as well as to enhance survival of remaining cells.
To this end, cell transplantations and scaffold-based tissue engineering approaches have utilized various cell types including skeletal myoblasts, [195] cardiomyocytes,[196] smooth muscle cells, [197] BM cells, [198] and HSCs [199] as promising therapeutics for cardiac regeneration following heart disease. [200] In particular, studies have demonstrated that the transplantation of these cell types into the myocardium is not only safe, but contributed to angiogenesis and improvements in cardiac function. [201] Among the various cell types investigated, MSC are an especially attractive stem cell source for cardiac regeneration as they are multipotent and are capable of differentiating into cardiomyocytes under the appropriate conditions. [202] Moreover, they express genes that encode for anti-inflammatory factors, anti-apoptotic factors, angiogenic factors, [203] and matrix-mediating factors, which may further enhance therapeutic recovery. [204]
To enhance stem cell-based therapies for cardiac repair, the development of engineered stem cells has focused on two strategies: 1) enhancing angiogenesis and the survival of remaining cardiomyocytes and 2) guiding stem cell differentiation specifically towards cardiomyocytes. In this section, we will discuss the progress that has been made with regard to both of these strategies. Moreover, we will introduce the use of optogenetics to specifically control cardiac tissue excitations and contraction and its potential in future engineered stem cell approaches for cardiac tissue regeneration.
3.3.1. Enhancing Angiogenesis and the Survival of Cardiomyocytes
Although stem cell transplantation for cardiac repair was initially aimed at inducing cardiac regeneration, our current understanding of the underlying therapeutic mechanisms suggests that stem cell therapy may limit maladaptive remodeling and improve heart function mainly through paracrine mechanisms. [205] As such, the majority of studies in this area have focused on demonstrating the use of stem cells to promote angiogenesis and promote heart function. For instance, MSC transplantation therapies can increase regional perfusion by: 1) direct effects (e.g., vasculogenesis induced by implanted MSCs), and 2) paracrine effects (e.g., angiogenic factors or ateriogenic cytokines secreted by implanted MSCs). [206] To this end, Deuse et al. demonstrated the transplantation of mouse MSCs that were engineered to secrete either hepatocyte growth factor (HGF) or vascular endothelial growth factor (VEGF) via lentiviral vectors. [207] HGF and VEGF are cytokines that have previously been shown to exert strong proproliferatory and promigratory effects on MSCs thereby suggesting that a combination of engineered MSCs expressing these cytokines could prolong the survival of transplanted MSCs and subsequent regenerative effects. [208] To test these engineered stem cells, acute myocardial infarction was induced by coronary ligation. Deuse and colleagues determined that the transplantation of engineered MSCS, expressing either HGF or VEGF, resulted in greater improvement in postinfarction myocardial function than non-engineered MSCs. [207] In particular, it was reported that HGF- and VEGF-engineered MSCs were similarly potent in initiating angiogenesis, increasing the tolerance of cardiomyocytes to ischemia, reducing cardiomyocyte apoptosis, and resulting in a decrease in scar size and improved LV function in vivo. As such, engineered MSCs strongly upregulated cytokine production and augmented both autocrine and paracrine mechanisms involved in cell survival and myocardial recovery. Similarly, Guo et al. confirmed that engineering MSCs with HGF can significantly enhance angiogenesis via the expression of VCAM-1. [209] However, engineering MSCs with both HGF as well as granulocyte colony-stimulating factor (G-CSF), which regulates BM stem cell recruitment into the peripheral circulation and has been shown to enhance angiogenesis, [210] can further enhance angiogenesis and impart a synergistic effect thereby improving myocardial endothelial density, angiogenesis, geometric preservation, and heart function in an ischemic cardiomyopathy model.
Other factors that can improve transplanted cell survival and promote angiogenesis have also been investigated. For instance, Liu et al., engineered MSCs with angiogenin, which is a heparin-binding protein that interacts with endothelial cells to promote angiogenesis via the induction of a wide range of cellular responses including migration, [211] proliferation,[212] and tube formation. [213] While angiogenin activity is relatively low when compared to VEGF and fibroblast growth factor (FGF), it has a comparable angiogenic activity and reportedly decreases fibrosis thereby making it an ideal candidate for application to myocardial infarction. [214] With this in mind, Liu and co-workers engineered primary bone-marrow derived MSCs to express angiogenin using adenoviral vectors. [215] It was observed that angiogenin modification of the MSCs greatly enhanced their survivability, allowing them to remain viable under low oxygen conditions (e.g., the number of engineered MSC deaths under hypoxic conditions was one-third that of non-engineered MSCs in vitro). In vivo, the engineered MSCs were transplanted into a myocardial infarction rat model, which was induced by ligating the left coronary artery. [216] After transplantation, they found that angiogenin modified MSCs survived over a long period of time and effectively expressed angiogenin protein for at least 6 weeks at the injected area. These results indicated that angiogenin may help the survival of transplanted MSCs, which also helps sustain the release of angiogenin. Moreover, the engineered MSCs enhanced myocardial vasculogenesis in the AMI rat model and demonstrated a significantly greater angiogenic and arteriogenic capacity than the group transplanted with untreated MSCs thereby inhibiting ventricular remodeling (Figure 10A,B). The border-zone wall was also found to be thicker and the infarction area became smaller in engineered MSC group compared to non-engineered MSCs (Figure 10C–F). Finally and most importantly, the transplantation of engineering MSCs led to significant improvement in cardiac function (e.g., improved LV systolic and diastolic functions).
Figure 10.
Engineering mesenchymal stem cells with angiogenin to enhance cardiac repair. Effects of angiogenin modified MSC transplantation on heart remodeling. A) Transverse sections of hearts treated with MSC-AdANG, with MSC-AdEGFP, and with PBS were HE stained. C) Smaller infarct sizes and D) LV chamber sizes as well as E) thicker border-zone walls were seen in the post-MI heart treated with MSC-AdANG compared with control hearts. Collagen in the infarct areas was shown by VG staining. B,F) Positively stained fibrous infarct areas were clearly observed in the PBS group, and not obviously reduced in MSC-AdEGFP group, but significantly reduced in MSC-AdANG group. Reproduced with permission.[215] Copyright 2008, Elsevier.
Similarly, several studies have reported that SDF-1α can promote the survival of cardiomyoctyes, MSCs, and other cell types via Akt activation. [217] As a result, Tang and colleagues hypothesized that MSCs engineered to secrete SDF-1α could greatly augment the survival of cardiomyoctyes and the transplanted MSCs. [218] It was found that SDF-1α engineered MSCs showed better survival ability when compared to non-engineered controls. They also demonstrated that the Ad-SDF-1-MSC transplantation enhanced VEGF expression in infarcted hearts in vivo. This suggests that the transplanted MSCs increased heart-tissue VEGF expression in the infarcted heart through paracrine mechanisms. Specifically, the authors proposed that the greater number of surviving MSCs could have produced more cytokines in the infarcted heart. [219] Overall, four weeks following transplantation, a reduced infarct size and fibrosis, greater vascular density, and thicker left ventricular wall were observed in the engineered MSC group compared to controls. Moreover, the measurement of haemodynamic parameters showed an improvement in the left ventricular performance in the Ad-SDF-MSC group as compared with other groups.
3.3.2. Controlling Stem Cell Differentiation to Myocardiocytes
As mentioned previously, the other strategy that is typically used to engineer stem cells for cardiac repair has focused on specifically guiding the differentiation of implanted stem cells to cardiomyocytes. To this end, Wang and co-workers investigated the ability of MSCs, which were engineered to secrete hypoxia-inducible factor 1α (HIF-1α), to differentiate toward cardiomyocytes. [220] Typically, HIF-1α regulates the transcription of genes that are involved in cell proliferation, survival, and differentiation. Owing to its central role in the oxygen-sensitive signaling pathway and previous findings that suggest a relationship between hypoxic microenvironments and the ability of MSCs to acquire a cardiomyocyte phenotype, [221] Wang et al. hypothesized that HIF-1α may play a key role in guiding this differentiation process. However, under normoxia, HIF-1α has a short half-life (≈5 min) and low transcriptional activity. As such, to prolong its half-life and further enhance its functionality, a HIF-1a mutant, wherein alanine (Ala) was substituted for proline (Pro) at position 564 and asparagine (Asp) at position 803, was produced to prevent HIF-1α hydroxylation resulting in a highly active form of HIF-1α. [222] Using this mutant HIF-1α, BM-derived rat MSCs were transfected via adenovirus. The authors found that when the engineered MSCs were co-cultured with cardiomyocytes, the engineered MSCs exhibited enhanced cardiac differentiation suggesting that the combination of co-culture with HIF-1α secretion was important in attaining efficient cardiac differentiation. Specifically, RT-PCR confirmed that HIF-1α activated TFG-β1 and SMAD, which are both upstream of the cardiac-specific transcription factors, NKx-2.5 and GATA-4. As a result of this high NKx-2.5 and GATA-4 expression, co-cultures with engineered MSCs exhibited 20% more differentiation than co-cultures with non-engineered MSCs.
In a subsequent study, Wang and co-workers determined that, in addition to guiding MSC differentiation towards cardiomyocytes, the use of HIF-1α could prevent apoptosis in ischemic cardiomyocytes. [223] Specifically, previous studies have demonstrated that HIF-1α can decrease apoptosis of rat cardiomyocytes following simulated ischemia-reperfusion injury by inducing multiple protective genes. [224] In agreement with these results, Wang and colleagues determined that co-culturing cardiomyocytes with engineered MSCs (wherein cobalt chloride (CoCl2) was used to mimic hypoxic/ischemic conditions including generation of reactive oxygen species (ROS)) [225] increased expression of TGF-β1 and Bcl-2, concomitant with a reduction in the expression of caspase-3, LDH release, and TUNEL-positive cardiomyocytes when compared to non-engineered MSC-cardiomyocyte co-culture and cardiomyocytes alone. Overall, this suggests that HIF-1α can not only promote the differentiation of transplanted engineered MSCs towards cardiomyocytes but also limit the apoptosis of surviving ischemic cardiomyocytes.
3.3.3. Control Cardiac Tissue with Optogenetics
Finally, optogenetics has recently been applied to control the excitation and contraction of cardiac tissue. Electrical stimulation of heart muscle is typically achieved using an external electrical field that is applied locally to induce action potentials that are then propagated to electrically coupled neighboring cells. [226] However, this approach results in irreversible Faradaic reactions that produce toxic gases such as H2, O2, or Cl2 and alters the pH. [227] As a result, electrical stimulations can only be used for short depolarizations while long-lasting depolarizations are not feasible using this method. As such, at the end of 2010, Bruegmann et al. developed a method using channelrhodopsin-2 (ChR2), which is a light-sensitive trans-membrane protein that converts photons into transmembrane voltage through proton pumping, to stimulate heart muscle both in vitro and in vivo. [228] To accomplish this, they first generated a stable transgenic mouse embryonic stem cell line [229] expressing a mutant ChR2, ChR2(H134R), [230] where they demonstrated that inward currents could be induced by illumination with 475 nm light. Next, to obtain cardiomyocytes, embryoid bodies from the transgenic ESCs were generated and cardiomyocytes were identified by staining with muscle-specific protein α-actinin antibodies. At day 7 of differentiation, application of pulsed blue light reliably induced cellular contractions. Moreover, plating of ESC-derived cardiomyocytes on multielectrode arrays demonstrated that pulsed illuminations of one region could evoke electrical activity in this area with subsequent spreading to other regions. Finally, the authors demonstrated that ChR2 engineered cells could be used for the stimulation of the adult heart in vivo. In this case, transgenic mice were generated using the ChR-2 engineered ESCs. It was found that the ChR2 protein was located in the cell membrane of the ventricular cardiomyocytes (Figure 11A). Light application induced typical ChR2 currents in ventricular cardiomyocytes and more importantly, long-term depolarizations could be achieved, which resulted in a disturbance in the regular sinus rhythm with the generation of spontaneous ventricular extra-beats (Figure 11B–D). Overall, this method enabled precise localized stimulation and constant prolonged depolarization of cardiomyocytes and cardiac tissue resulting in alterations of pacemaking, Ca 2+ homeostasis, electrical coupling, and arrhythmogenic spontaneous extrabeats.
Figure 11.
Engineering stem cells to achieve optogenetic control of heart muscle. Expression and function of ChR2 in ventricular cardiomyocytes from CAG-ChR2 mice. A) Fluorescence image of the native membrane-bound ChR2-EYFP signal (green) overlaid with α-actinin immunostaining (red) in cardiomyocytes of the ventricle and colocalization with the t-tubulus system (inset). Nuclei are shown in blue. Scale bars, 20 μm. B) Inward currents evoked at a holding potential of −40 mV by light stimulation at 0.09, 0.18, 0.45 and 1.75 mW mm−2 (from top to bottom). Monoexponential fit to measure the time constant of decay is shown in red. pA, picoampere; pF, picofarad. C) Repetitive action potential generation by 1-ms light pulses (blue bars) of 0.91 mW mm−2. D) Action potential generation by light pulses (10 ms; light blue line) of different intensities in a representative single cell. Reproduced with permission.[228] Copyright 2010, Nature.
Building upon this work, Jia and colleagues developed the first nonviral strategy involving optogenetics that does not rely on embryogenesis in order to control the excitation and contraction of cardiac muscle. [231] Specifically, they took advantage of the heart’s high coupling aspect to develop a non-viral cell delivery system using a “tandem cell unit” (TCU) strategy, wherein a unit is composed of a host cardiomyocyte and a nonexcitable donor cell that carries exogenous ion channels (e.g., ChR2). To this end, they developed, characterized, and used a stable HEK cells expressing a variant of ChR2 as the donor cell delivery system. The TCU strategy was validated in vitro in cell pairs with adult canine myocytes and in cardiac syncytium with neonatal rat cardiomyocytes. Specifically, robust response was seen and similar conduction velocities and calcium transient morphologies were observed in localized electric and optical stimulation of cardiac syncytium. Overall, using the TCU strategy, light-triggered electric waves were found to be quantitatively indistinguishable from electrically triggered waves. Moreover, the viral-free method can allow for a safer alternative for in vivo applications such as light-driven cardiac pacemakers and muscle actuations. While the authors did not use stem cells in this case, one can imagine that this non-viral method can be equally applied to stem cells with the added benefits of specifically controlling differentiation or controlling the excitation and contraction of cardiac muscle using light.
4. Engineering Stem Cells for Cancer Therapy
Cancer is one of the leading causes of death worldwide and, in the United States alone, causes one in every four deaths. [232] Currently, surgical resection of the bulk tumor is the gold standard for treatment and is typically followed by a combination of chemotherapy and radiotherapy. However, despite this aggressive therapy, late stage cancers that have already metastasized are difficult to completely eradiate, thereby ultimately resulting in recurrence. This can be attributed to a number of reasons, including the significant heterogeneity that exists between patients and within each tumor, the lack of treatment selectivity thereby resulting in considerable loss of healthy tissue, and chemoresistance. [233] Of these issues, the lack of selectivity that is seen with conventional therapies is arguable the largest drawback towards the effective treatment of cancer. Recent therapies are being developed with increased targeting in mind. For instance, hormone therapies have been developed for cancers of the sexual organs, [234] while immunotherapies have been developed to modulate the patient’s own immune cells to recognize and attack cancer cells. [235] However, these strategies are only effective against certain cancers and certain subtypes. As such, there is a great need for novel treatment strategies that can specifically target and infiltrate tumors thereby enhancing the efficacy of the delivered therapeutic while minimizing side effects.
To this end, it has been demonstrated that transplanted stem cells have the innate ability to home to tumors and metastases, enabling site-specific delivery. [236] The mechanisms that underlie stem cells tropism to tumors are far from understood; however, various chemokine–chemokine receptor pairs have been associated with tumor tropism, with stromal cell-derived factor 1 (SDF1; also known as CXCL12) and its receptor CXC-chemokine receptor 4 (CXCR4) being the most well studied. [237] As such, stem cells are attractive candidates that can act as delivery vehicles for the targeted treatment of tumors/metastases. To this end, unmodified stem cells have exhibited some intrinsic anti-tumor effects, which are attributed to the secretion of factors and physical interactions with tumor cells. [238] However, there have also been numerous conflicting reports suggesting that stem cells can actually protect cancer cells from immune recognition and treatment. [239] As such, to fully take advantage of stem cells for cancer therapy, stem cells are being engineered to stably express or deliver various anticancer agents. In this way, they can act as delivery vehicles that specifically target and infiltrate tumors/metastases while circumventing the shortcomings that plague many conventional chemotherapeutic agents such as their short half-lives. In this section, we will highlight the progress that has been made in engineering stem cells for cancer. In particular, we will focus on the genetic modifications of stem cells as well as their use as delivery vehicles for gene therapies and other therapeutic molecules.
4.1. Stem Cell-Based Gene Therapy for the Treatment of Cancer
Recently, there has been increasing interest in the development of gene therapies as a unique strategy for the treatment of cancer. Gene therapy for cancer encompasses a wide range of treatments that have the common theme of delivering genetic materials (e.g., DNA, RNA, and RNA interference molecules) in order to modify cancer cells. [240] A wide variety of gene therapies have been tested on cancers including glioma, pancreatic cancer, lung cancer, liver cancer, and many more. Examples include the creation of cancer vaccines, targeting viruses to cancer cells for the induction of lysis and death, targeting supporting cells to cutoff the blood supply, and introducing genes into cancer cells that either cause death or restore them to a normal phenotype. [240] However, as with more conventional drugs, gene therapies are hampered by our current inability to specifically target them to the cancer. As such, combining the tumor tropism/targeting ability of stem cells with gene therapy strategies is a promising way to approach gene therapy thereby using stem cells as a delivery vehicle that can improve our ability to treat cancers. In this section, we will focus on two iterations of stem cell-based gene therapy for the treatment of cancer, which includes the use of engineered stem cells as a targeted delivery vehicle for gene therapies (e.g., using stem cells to deliver viruses) and genetically engineered stem cells to secrete therapeutic molecules for cancer therapy.
4.1.1. Engineering Stem Cells as a Delivering Vehicle for Gene Therapy
Currently, one avenue of gene therapy that is being explored for the treatment of cancer is oncolytic viruses. Specifically, oncolytic viruses are viruses that are engineered to specifically replicate in and kill cancer cells while sparing healthy cells. [241] However, these viruses are quickly cleared through the bloodstream and may exhibit non-specific behaviors when directly administered. [242] Moreover, it has been demonstrated in clinical trials that engineered viruses often only affect tumor cells in close proximity to the site of injection, which significantly hampers its efficacy for metastases. [243] To address these issues, engineered stem cells that are loaded with oncolytic viruses can be used as effective targeted delivery vehicles for gene therapy. Due to their tumor-tropic properties, stem cells can carry the gene therapy vectors to tumors and sites of metastases thereby increasing the local concentration of therapeutic at the cancer site while decreasing the required dosage and subsequent side effects. [242]
To this end, multiple studies have shown that virus loaded stem cells can decrease tumor burden more effectively than direct viral injections. [244–246] In particular, MSCs have been the most frequently used stem cell source for this purpose with the most common demonstration being for gliomas. [246,247] For instance, Sonabed et al. demonstrated that MSCs can effectively deliver oncolytic conditionally replicative adenovirus (CRAd) to glioma. [243] In particular, the promoter of CRAd’s were designed to be tumor specific and, in this case, are only activated at the tumor site by C-X-C chemokine receptor 4 (CXCR4), which has been shown to be overexpressed by gliomas. [248] To infect the MSCs with CRAd, cells were simply incubated with virus-containing medium (1000 viral particles per cell) for 48 hours. It was found that CRAd-loaded MSCs effectively migrated in vitro and released CRAds that infected U87 glioma cells. More importantly, MSCs also migrated in vivo when injected away (5 mm) from the tumor site and delivered 46-fold more viral copies than CRAds injected alone.
Although the majority of studies using stem cells to deliver viruses have focused on MSCs, NSCs have also shown significant migratory ability for the treatment of gliomas. [12] As such, Ahmed and co-workers conducted a comparative study of NSC- and MSC-based carriers for oncolytic adenoviruses for GBM. [249] In this case, commercial stem cells were transduced with a variety of adenoviral vectors (AdWT, CRAd-CXCR4, etc.). Importantly, it was found that both cell sources had similar potential to function as cell carriers. However, the amount of virus released from NSCs was a log higher than from MSCs. As such, only virus loaded NSCs, which were administered intracranially to an orthotopic glioma model, significantly prolonged the survival of tumor bearing animals (68.7 days of survival for NSCs-injected animals vs 44 days for MSCs).
Besides glioma, Stoff-Khalili et al. implemented a therapy utilizing MSCs to shuttle CRAd agents to metastatic breast tumors. [250] In particular, the CRAd’s promoter was tumor specific and, as with the case in glioma, were also designed to be activated at the tumor site by CXCR4, which is overexpressed by certain breast cancer cell lines. [251] The MSCs were successfully loaded with the adenovirus via diffusion during 18 hours of incubation and were subsequently trypsinized and intravenously injected. Results indicated that mice bearing MDA-MB-231 breast cancer pulmonary metastases that were injected with the adenovirus loaded MSCs survived significantly longer than their control counterparts (approximately 3 times longer). [250] The ability of stem cells to infiltrate tumors was also hypothesized to significantly increase the tumor’s exposure time to the therapy resulting in the corresponding increase in therapeutic efficiency. [252]
Lastly, Mader and colleagues demonstrated the use of engineered patient-derived MSCs as a carrier to deliver oncolytic measles virus (MV) to ovarian tumors as optimization for a Phase I clinical trial. [253] In particular, various experimental models have previously validated the use of MV and phase I clinical trials are in progress to evaluate the safety and maximal tolerated dose of oncolytic MV for cancers such as ovarian cancer, myeloma, and glioma. [254] To further improve viral delivery to the tumor, the authors infected patient-derived MSCs with MV via centrifugation (70% infectivity with 1000 × g centrifugation for 5–10 minutes), which did not compromise cell viability. In vivo, no tumors were seen despite receiving up to 1.6 × 10 9 MSCs/kg and MSCs did not promote the growth of SKOV3 human ovarian cancer cells in mice. Using non-invasive SPECT-CT imaging, Mader et al. saw rapid co-localization of MV infected MSCs and SKOV3 tumors, within 5–8 minutes of intraperitoneal administration (Figure 12A). Importantly, MSCs could be pre-infected with MV, stored in liquid nitrogen, and thawed on the day of injection into mice without loss of activity. Finally, it was found that MV infected MSC, but not virus alone, significantly prolonged the survival of animals bearing measles immune ovarian cancer (Figure 12B).
Figure 12.
Mesenchymal stem cells as virus carriers for the treatment of ovarian cancer. A) MSCs labeled with DiR and infected with measles virus expressing RFP (MV-RFP) were injected into mice bearing SKOV3ip.1 tumors that stably expressed CFP. Representative images from mice that received MSCs from healthy donors (MSC 493B) or ovarian cancer patients (FB8) showed co-localization of MV-infected MSCs with the tumors. B) Mice with SKOV3ip.1 tumors were passively immunized with measles immune human sera and given 105 TCID50 MV-NIS or 105 MV-NIS infected MSCs at 7 days post-tumor implantation. RT = MSCs were given 20 Gy radiation immediately before MV-NIS infection. F/T = Frozen stock of MV infected MSCs were thawed, washed, and used immediately. Reproduced with permission.[253] Copyright 2013, BioMed Central.
4.1.2. Genetically Engineering Stem Cells for Cancer Therapy
Aside from delivering oncolytic viruses to cancer, stem cells can also be genetically engineered to secrete: 1) therapeutic proteins or 2) enzymes that convert a separately administered non-toxic prodrug into a cytotoxic drug. Using these approaches, engineered stem cells are capable of migrating to and continuously producing the drug or enzyme at the sites of cancer and metastases, thus bypassing restrictions such as the short half-life of drugs and the need for repeated drug dosages. [244] For this purpose, MSCs are, again, especially attractive as candidate carriers since they are relatively easy to expand and transduce. [255] Moreover, multiple studies have already shown that genetically engineered MSCs are efficient tools for delivering anticancer agents to metastatic tumors, as we will review later in this section. In particular, this section will focus on the use of genetically engineered stem cells for: 1) the secretion of therapeutic molecules and 2) the secretion of an enzyme that can then convert a separately administered prodrug.
4.1.2.1. Secretion of Therapeutic Proteins
When genetically engineering stem cells to secrete therapeutic proteins, there are a number of candidate genes including genes encoding proteins that directly act on malignant cells as well as those that affect supporting cells (e.g., blood vessel and stroma). This is typically achieved using viral methods, as although non-viral vectors have been used and offer some advantages such as lower immunogenicity, they have a much lower efficiency. [256] In particular, direct effectors include cytokines such as interferon-β (IFN-β) and tumor necrosis factor-related apoptosis inducing ligand (TRAIL). On the other hand, those that affect supporting cells typically target angiogenesis or induce an immune response via the secretion of interleukins.
In the case of IFN-β, high concentrations of IFN-β have been shown to inhibit cancer cell growth. However, the direct administration of IFN-β is limited by its short half-life and has been associated with excessive systemic toxicity. [257] Addressing these concerns, a number of studies have focused on using stem cells, especially MSCs, to deliver IFN-β specifically to tumors. [258] For instance, Studeny et al. engineered BM-derived MSCs to IFN-β via adenoviral transduction. [259] In vivo tests with mice carrying A375SM melanoma tumors demonstrated that the transplanted MSCs preferentially survive and proliferate in the presence of malignant cells and become incorporated into the tumor architecture as stromal fibroblasts. More importantly, the authors found that, on average, mice injected with engineered MSCs survived almost twice as long as control mice (60 days compared to control mice, which survived for only 37 days). On the other hand, the direct intravenous injection of recombinant IFN-β did not increase mice survival compared to control mice, which further supports the use of MSCs as a delivery vehicle for IFN-β. Similarly, Ren and colleagues reported that MSCs engineered with a recombinant adeno-associated virus encoding IFN-β could effectively treat prostate cancer lung metastasis. [260] Evaluation 30 and 75 days after transplantation indicated a significant reduction in tumor volume. In addition, a significant increase in the natural kill cell activity was observed following stem cell-based IFN-β therapy and systemic levels of IFN-β was not significantly elevated. Lastly, aside from MSCs, NSCs have also been used to deliver IFN-β but to a lesser extent. [261]
On the other hand, TRAIL has also been a cytokine of particular interest. TRAIL can induce apoptosis in a wide range of cancers while, generally, sparing normal healthy cells. [262] In particular, TRAIL has been shown to directly attach to death receptors (DR4 and DR5) that are preferentially expressed on tumor cells, activating pro-apoptotic proteases that result in cancer cell apoptosis. [263] However, translation of TRAIL into the clinic is confounded by its short half-life, inadequate delivery methods, and the fact that recent studies have found that TRAIL can cause some hepatotoxicity depending on the patient and drug combinations used. [264] As with IFN-β, MSCS have been shown to have the ability to deliver a secretable form of TRAIL, thereby enhancing the efficacy of TRAIL versus systemic administration of TRAIL alone. For example, engineered MSCs that secrete TRAIL have been utilized to treat in vivo glioma models. [265] In particular, these MSCs were transfected using a lentiviral vector and the resulting engineered MSCs secreted around 250 ng of TRAIL per every million cells over a 24-hour timespan. In addition, it was found that the engineered MSCs provided a method to facilitate the transportation of TRAIL across the BBB and continuous production of TRAIL helped mitigate the issue of TRAIL’s short half-life (Figure 13A).[266] Importantly, it was shown that MSCs were resistant to apoptosis from TRAIL making them viable targeting candidates. [265] As a result, the engineered MSCs exhibited significant anti-tumor effect over unengineered MSCs resulting in a significant reduction in glioma burden via the induction of apoptosis and a significant decrease in the number of proliferating tumor cells (Figure 13).
Figure 13.
Mesenchymal stem cells genetically engineered to secrete TRAIL to enhance the treatment of glioma. A–F) Serial in vivo bioluminescence imaging of tumor growth following intracranial implantation of Gli36-EGFRvIII-FD glioma cells mixed with MSCs expressing S-TRAIL (MSC-S-TRAIL; B,D,F) or GFP (MSC-GFP; A,C,E). G) Relative mean bioluminescent signal intensities after quantification of in vivo images. H–M) Photomicrographs show the presence of cleaved caspase-3 (H) and Ki67-positive cells (K) in brain sections from MSC-S-TRAIL-treated and control mice (I,L) 6 days after implantation. Plot shows the number of cleaved caspase-3 (J) and Ki67 (M) cells in MSC-S-TRAIL and MSC-GFP-treated tumors. (Green, MSCs; red, glioma cells; purple, Ki67 or cleaved caspase-3 expression). Reproduced with permission.[265] Copyright 2009, PNAS.
Besides direct effectors of cancer apoptosis, stem cells have also been engineered to express indirect effectors such as molecules that inhibit the formation of the tumor-associated vasculature (TSP1 [267] or PEX) or immunomodulatory molecules (IL-12 and IL-18 [269]). In addition, the delivery of growth factor inhibitors such as NK4 using MSCs has also been shown to significantly increase survival of mice in a lung metastasis model. [270] For instance, Kim et al. engineered HB1.F3 immortalized NSCs to produce PEX in order to inhibit angiogenesis for the treatment of glioma. [271] In particular, PEX is a naturally occurring fragment of human metalloproteinase-2 and acts as an inhibitor of glioma and endothelial cell proliferation, migration, and angiogenesis. [272] Following transfection of the NSCs with a plasmid for PEX via SuperFect (Qiagen) and in vivo injection, histologic analysis showed that engineered NSCs migrated to the tumor boundary and caused a 90% reduction of tumor volume. In particular, this reduction was associated with a significant decrease in angiogenesis (44.8%) and proliferation (23.6%), demonstrating the effectiveness of engineering NSCs to express PEX.
Immunomodulatory molecules such as IL-12 are also effective for the treatment of cancer. Typically, immunotherapies focus on utilizing our own immune systems or its components to attack cancer cells. In particular, the delivery of cytokines such as IL-12 has been shown to boost both the innate and adaptive immune response against tumors. However, cytokines such as IL-12 are hindered by poor in vivo distribution and are associated with serious and even life-threatening consequences as well as marginal clinical responses in most patients. [273] To improve this, MSCs were transduced with an adenovirus expressing IL-12 and the antitumor effect of these engineered MSCs, as injected via different routes, was evaluated in solid and metastatic melanoma. [274] As expected, it was reported that the engineered MSCs were more efficient than adenovirus alone as a cytokine gene delivery vehicle. Moreover, when comparing intratumoral, subcutaneous, and intravenous injection of engineered MSCs, intratumoral injection was found to be the best approach to induce a strong tumor-specific T-cell response that correlated with anti-metastatic effects as well as the inhibition of solid tumor growth. Though, interestingly, intravenous injection of engineered MSCs actually induced earlier and higher peak levels of cytokines than other routes demonstrating that this is not an indicator of subsequent antitumor effects.
4.1.2.2. Secretion of Enzymes for the Conversion of Prodrugs
Prod-rugs are another viable candidate for stem cell delivery. Prod-rugs are compounds that are normally nontoxic. Instead, they are designed to respond to tumor specific enzymes, which then convert the prodrug into its toxic form. [275] Thus, prodrugs can provide a more targeted approach towards cancer therapy as greater concentrations of the cytotoxic form of the prodrug will be located at sites of cancer rather than in healthy tissues. [276] Moreover, prodrugs exhibit the bystander effect owing to the diffusion of the activated prodrug agent further enhancing the efficacy of the prodrug. [277] As such, three major suicide gene systems are currently used. Cytosine deaminase (CD) converts 5-fluorocytosine (5-FC) to the toxic antimetabolite 5-fluorouracil. The herpes simplex virus thymidine kinase (HSV-tk) converts ganciclovir (GCV) to GCV-monophosphate, which is further phosphorylated to GCV-triphosphate thereby potently blocking DNA synthesis. Finally, carboxylesterase (CE) converts the prodrug irinotecan (CPT-11) to the potent topoisomerase inhibitor SN-38. [278]
While promising, the efficacy of prodrugs can be further improved using stem cell-based delivery thereby enhancing targeting and infiltrating. Moreover, an added benefit of stem cell-mediated prodrug delivery is that the stem cells are eliminated after conversion of the prodrug, thereby abolishing any concern over its long-term fate. Using the CD–5-FC system, engineered MSCs and NSCs have been shown to effectively treat tumors of the brain. [279–281] For instance, Aboody and colleagues engineered immortalized NB1.F3 NSCs to express CD via a retroviral vector for the treatment of glioblastoma (Figure 14A).[282] They found that these engineered NSCs retained their tumor tropism following intracerebral injection even in orthotopic glioblastoma bearing mice pretreated with radiation or dexamethasone, which mimics clinically relevant adjuvant therapies. Importantly, it was reported that the average tumor volume was one-third that of the average volume in control mice (Figure 14,C). Moreover, no toxicity associated with conversion of 5-fluorocyto-sine to 5-fluorouracil was detected and there was no evidence of tumorigenesis attributable to the NSCs. Similarly, Wang and co-workers also engineered NB1-F3 cells to express CD and demonstrated their ability to target and disseminate therapeutic agent to medulloblastoma thereby resulting in a 76% reduction of tumor volume compared to unengineered controls. [283]
Figure 14.
Engineering stem cells to secrete enzymes for the conversion of prodrugs. A) Diagram of CD-expressing NSCs localized to tumor cells, and CD conversion of 5-FC to 5-FU, which readily diffuses out of the NSCs to selectively kill the surrounding tumor cells. B,C) H&E-stained brain tumor sections from U251 glioma–bearing mice that received HB1.F3.CD NSCs only (B) or HB1.F3.CD NSCs in combination with 5-FC (C). White arrows indicate tumor region. Reproduced with permission.[282] Copyright 2013, Science. D) Brain injection of NSC1-tk cells coupled with GCV treatment prolonged the life of mice inoculated with U87 glioma cells. Statistical analysis was performed using the log rank test. E) Representative pictures of brain sections show the tumor size of different groups. NSC1-tk brain injection followed by GCV i.p. injection appears to shrink the tumor. Reproduced with permission.[280] Copyright 2012, Nature.
On the other hand, the HSV-tk system, which relies on the formation of gap junctions between the stem cell and surrounding target cells for an efficient bystander effect using the prodrug GCV, has shown efficacy in several cancer models including those of the brain, breast, and prostate. [284] For example, Yang and colleagues engineered iPSC-derived NSCs using recombinant baculovirus vectors containing the herpes HSV-tk gene expression cassette to treat metastatic breast cancer. [280,285] In particular, they demonstrated that after tail vein injection, the engineered iPSC-derived NSCs displayed robust migratory capacity even outside the CNS in both immunodeficient and immunocompetent mice and homed in on established orthotopic 4T1 mouse mammary tumors. Moreover, the engineered iPSC-derived NSCs were able to effectively inhibit the growth of orthotopic 4T1 breast tumors as well as the metastatic spread of the cancer cells, leading to prolonged survival of the tumor-bearing mice (median survival of 39 days, which was significantly greater than controls) (Figure 14D,E).
Finally, NSCs engineered using the CE–CPT-11 system have proven to be effective in the treatment of preclinical models of brain, lung, and ovarian cancers. [286] For instance, Kim et al. engineered immortalized HB1.F3 NSCs to express CE using a retroviral vector to enhance the treatment of ovarian cancer. [283,287] In this study, the authors reported that the engineered NSCs retained their ability to migrate to ovarian tumors and greatly inhibited cancer cell proliferation. Interestingly, the authors compared engineered stem cells using the CD-5-FC system to engineered NSCs expressing CE for the CE-CPT-11 system and found that the CE approach seems to be more promising than the CD approach because the CE approach decreased proliferation with a lower engineered NSC cell number and at a lower concentration of CPT-11 when compared to the concentration of cells and prodrug needed for the CD approach.
4.2. Stem Cell-Based Drug Delivery
Finally, as mentioned previously, nanoparticle delivery systems are attractive for cancer drug delivery owing to their ability to carry high concentrations of often insoluble chemotherapeutic reagents, while protecting them from degradation by the harsh biological environment. [278] Furthermore, owing to the tunability of nanoparticles, their surfaces can be specifically modified to optimize properties such as stability, solubility, and targeting for cancer applications. [288] Despite the promise that nanotechnology holds for drug delivery, the use of nanoparticles in vivo and especially in the clinic have been confounded by serious limitations such as rapid clearance by the renal system, inefficient targeting and infiltration, and an inability to target micrometastases. [278,288] To this end, stem cell-based drug delivery for cancer therapy offers a unique strategy with which to overcome these barriers. [289] In particular, stem cells can be loaded with nanoparticles carrying the particular drug of interest and injected in vivo, where they can then specifically migrate to the tumor and its metastases and deposit the loaded nanoparticles in close proximity or within the tumor. Although seemingly straight forward, as with engineered stem cell therapies, the success of this strategy depends on the ability to load stem cells with nanoparticles without negatively affecting their migration capability and then efficiently release the nanoparticles and drugs once the engineered stem cells have reached the tumor or its metastases.
This field of research is still in a nascent stage with the earliest examples of engineering stem cells by loading them with nanoparticle being solely for tracking purposes. [290] For example, Loebinger and colleagues sought to monitor the tumor homing and infiltration capability of MSCs in vivo via MRI by engineering them with iron oxide MNPs. [291] To this end, MSCs were first transfected with 200 nm MNPs in vitro with no observed effect on differentiation potential, proliferation, survival, or migration. The authors then showed that as few as 1000 MSCs carrying MNPs could be detected by MRI even one month after their coinjection with breast cancer cells that formed subcutaneous tumors. More importantly, Loebinger et al. found that i.v.-injected engineered MSCs could be tracked during their migration in vivo to lung metastases using MRI. Similarly, Gao and colleagues demonstrated that 500 nm core-shell fluorescent silica nanoparticle (C dots) could be retained in human MSCs for up to a month with minimal influence on MSC properties such as viability, proliferation, differentiation, and migration to tumors (e.g., breast tumors in zebra fish model). [292]
Building upon this, researchers have recently begun engineering stem cells with nanoparticles that are loaded with chemotherapies in order to achieve greater tumor targeting and infiltration over nanoparticle-based drug delivery methods alone. Towards this objective, one novel approach that was recently investigated involved loading the surface of MSCs with nanoparticles. [293] In particular, a 125 nm silica nanorattle–doxorubicin drug delivery system was efficiently anchored to MSCs by specific antibody–antigen recognitions at the cell membrane interface without the need for any MSC preconditioning (Figure 15). The MSCs could be loaded with up to 1500 nanoparticles while maintaining high cell viabilities as well as their tumor-tropic ability. Moreover, it was reported that the intracellular retention time of the silica nanorattle was no less than 48 h, which was a sufficient amount of time for the engineered MSCs to migrate to the U251 glioma tumors in vivo. Importantly, these in vivo studies demonstrated that the engineered MSCs could not only track U251 glioma cells more effectively than free DOX and nanoparticle-delivered DOX alone, but that the delivered DOX had a wider distribution and a longer retention time within tumor tissues. As a result, a significant enhancement in tumor-cell apoptosis and decrease in tumor burden was observed.
Figure 15.
Engineering mesenchymal stem cells with silica nanorattle-doxorubicin for glioma therapy. General scheme of silica nanorattle-doxorubicin-anchored MSCs for tumor-tropic therapy. Reproduced with permission.[293] Copyright 2011, ACS.
Lastly, another avenue of research that has engineered stem cells to carry nanoparticles is the use of the delivered nanoparticles for their other functionalities besides drug loading such as hyperthermia or photothermal therapy. For instance, Ruan et al. utilized fluorescent magnetic nanoparticle (FMNP)-labeled MSCs for the targeted imaging and hyperthermia therapy of in vivo gastric cancer. [294] In particular, the FMNPs consisted of MNPs and CdTe quantum dots embedded in an inert silica shell based on previously described methods from their lab. [295] Primary mouse marrow MSCs were then labeled with these amino-modified FMNPs and intravenously injected into a subcutaneous mouse model of gastric cancer. It was found the FMNPs remained attached to the MSCs for at least 14 days and that the engineered MSCs were able to retain their tumor targeting ability. Moreover, the engineered MSCs could be used to image in vivo gastric cancer cells even after being intravenously injected for 14 days. Once at the tumor, the engineered MSCs were exposed to an external alternating magnetic field thereby inducing magnetic hyperthermia from the embedded MNPs. In this way, the engineered MSCs significantly inhibited the growth of in vivo gastric cancer. Similarly, Schnarr and co-workers demonstrated that NSCs could also be loaded with gold nanoparticles, maintain their tumor tropism after engineering, and be used to ablate in vivo tumors via photothermal therapy. [296] As one can imagine, in future studies, these nanoparticles (e.g., MNPs and GNPs) can be loaded with a drug or other therapeutic molecule, similar to what was demonstrated by Li et al. [293] and then delivered into stem cells thereby taking advantage of the multifunctionalities offered by nanoparticles for combined imaging, hyperthermia, and chemotherapy.
5. Engineering Stem Cells for Other Diseases
The development of engineered stem cell therapies for other diseases besides those discussed previously in this Review have primarily focused on autoimmune and other inherited diseases/disorders such as muscular dystrophy, Wiskott-Aldrich Syndrome, and leukodystrophies. In particular, the treatment strategies that have been developed for these diseases fall into two general categories: 1) use of engineered stem cells to deliver genetic material that can correct the inherent genetic defects or 2) engineering the stem cells ex vivo to correct the genetic defect and then reintroducing them back into the patient.
5.1. Muscular Dystrophy
Muscular dystrophy is a group of inherited disorders that are characterized by the degeneration of muscle, which leads to variable degrees of immobility such as confinement to a wheelchair and, in the most severe cases, weakness of the heart and/ or respiratory muscles thereby leading to premature death. [297] Many muscular dystrophies arise from loss-of-function mutations in genes encoding cytoskeletal and membrane proteins, with the most common and severe being Duchenne muscular dystrophy (DMD), which is caused by mutations in the gene encoding dystrophin. In particular, dystrophin is an integral part of a complex in muscle that links the intracellular cytoskeleton with the extracellular matrix and mutations in DMD causes the disassembly of the whole multiprotein complex leading to fragility of the sarcolemma. As a result, muscular dystrophies are some of the most difficult diseases to treat, as skeletal muscle is composed of large multinucleated fibers whose nuclei cannot divide.
To this end, three main therapeutic approaches are currently being pursued: 1) the introduction of genetic material (via viral or non-viral vectors) to repair the genetic mutation, 2) transplantation of dystrophin-positive cells, or 3) modulating the synthesis of endogenous gene products to make up for the mutation. In particular, the most promising approach consists of a combination of genetic and cellular therapy, wherein the patient’s own cells can be genetically engineered and then reintroduced back into the body. However, significant challenges remain as cell therapies would have to restore proper gene expression in hundreds of millions of postmitotic nuclei. [65] In this section, we will discuss the use of genetically engineered stem cells for muscular dystrophy as well as the use of engineered stem cells as a vehicle for genetic material.
5.1.1. Engineering Stem Cells by Genetic Modification for Muscular Dystrophy
A number of studies have demonstrated that genetically engineered stem cells, which express genes that promote differentiation towards a muscle lineage or express the correct form of dystrophin, can be used to treat muscular dystrophy. As a proof-of-concept to demonstrate the utility of ESCs for skeletal muscle differentiation and eventual application to muscular dystrophy, Darabi and co-workers engineered ESCs to express Pax3 (Figure 16).[298] In particular, Pax3 is a transcription factor, whose expression results in the activation of myogenic regulatory factor (MRF) genes, Myf5, Myf6, MyoD1, and Myog. [299] Expression of Pax3 under a tetracycline transactivator in ESCs was achieved using Cre-lox. Following growth as embryoid bodies and exposure to doxycycline, cells from the engineered ESC conditions exhibited morphology resembling myogenic progenitors. This was confirmed by the upregulation of Pax3, Myf5 genes and, to a lesser extent, Myod1, and Myog as well as Myf5 and myosin heavy chain (MHC) (Figure 16B). Initial in vivo studies that sought to demonstrate the muscle regenerative potential of these engineered ESCs utilized a constitutive version of Pax3. Day 5 embryoid bodies, which demonstrated the greatest myogenic differentiation, were expanded, sorted for PDGF-α, a marker of paraxial mesoderm, and for the absence of Flk-1 (Figure 16A), and transplanted into cardiotoxin-injured tibialis anterior muscles of mice lacking Rag2 and the common γ-chain of Fc receptors 24 hours after injury. When transplanted systemically or intramuscularly into cardiotoxin-injured immunodeficient or dystrophic mice, undifferentiated Pax3-induced myogenic progenitors demonstrated considerable potential for skeletal muscle regeneration by differentiating robustly into adult myofibers without the formation of teratomas. Regardless of the route of cell delivery, dystrophin restoration in the mdx mice (11–16% of total myofibers) was accompanied by a significant increase in contractile force (Figure 16C). These data demonstrate the therapeutic potential of ESCs in muscular dystrophy.
Figure 16.
Engineering embryonic stem cells with Pax3 to induce differentiation to skeletal muscle for the treatment of muscular dystrophy. A) Doxycyline-induced cell monolayers resulting from PDGF-αR+Flk-1− sorted cells from day 5 embryoid bodies were transplanted by various routes of administration (i.m., i.v. and i.a.) into Rag2−/−γc−/− immunodeficient mice or mdx mice (with or without cardiotoxin (CTX) pre-injury). B) Analyses of Rag2−/−γc−/− mice (pre-injured with cardiotoxin) 30 d after i.m. transplantation (n = 8 mice). Shown is immunostaining for GFP (green) and MHC (red). Top and bottom rows show different magnifications. Scale bars, 100 μm. C) Performance on the rotarod was assessed in mdx mice pre-injured with cardiotoxin (both legs) and treated with PBS (control, blue) or i.v. cell transplantation (red). B/6 mice (green) also pre-injured with cardiotoxin (both legs) were analyzed as a reference. *p < 0.05. Reproduced with permission.[298] Copyright 2008, Nature.
Similarly, Goncalves et al. recently engineered human MSCs to ectopically express full-length dystrophin and demonstrated that these engineered MSCs can fuse with DMD myotubes thereby rescuing synthesis of full length dystrophin in DMD muscle cells. [300] To this end, they utilized a dual high-capacity adenovirus-adeno-associated virus hybrid vector. It was found that the engineered MSCs could participate in human myotube formation via cellular fusion when co-cultured with DMD myoblasts in an ex vivo culture model. More importantly, it was found that the engineered MSCs could rescue full length dystrophin synthesis in human dystrophin-defective myotubes.
5.1.2. Engineering Stem Cells to Deliver Genetic Materials for Muscular Dystrophy
A number of studies have also investigated engineering stem cells as delivery vehicles of genetic materials for the treatment DBD. For instance, Kazuki and colleagues reported a proof-of-concept study where they delivered the full-length dystrophin gene into iPSCs using a human artificial chromosome (HAC). [301] HACs can carry large genomic segments containing a whole genetic locus including the regulatory regions and microRNAs. Moreover, they can be engineered to express additional functional benefits and have the advantage of avoiding insertional mutagenesis as they do not become integrated into the host cell. [47] Specifically, Kazuki et al. demonstrated the complete correction of a genetic deficiency in iPSCs derived from DMD model (mdx) mice and human DBD patients using a HAC with a complete genomic dystrophic sequence (DYS-HAC) [302] via microcell-mediated chromosome transfer (MMCT) (Figure 17).[303] Using this method, when transplanted in vivo, both DMD patient- and mdx-specific iPSCs with DYS-HAC were observed to form typical teratomas that differentiated into all three germ layers. Moreover, human dystrophin expression could be detected in muscle-like tissues. Lastly, chimeric mice from mdx-iPSCs (DYS-HAC) were produced and DYS-HAC was detected in all tissues examined, with tissue-specific expression of dystrophin, which demonstrates the ability of patient-specific iPSCs and HAC to potentially treat DMD.
Figure 17.
Engineering stem cells to deliver human artificial chromosomes for the treatment of muscular dystrophy. The scheme illustrates the various steps of the process. Mesoangioblasts were first isolated from mdx dystrophic mice (1) and transduced with the DYS-HAC vector (shown in the upper part of the panel) (2); they then underwent selection. Selected clones (EGFP-positive) were transduced with two lentiviral vectors expressing MyoD and nLacZ (3). After in vitro characterization (4), the corrected mesoangioblasts were transplanted into dystrophic SCID/mdx mice (5). Afterwards, the mice were analyzed for dystrophin expression and morphological and functional recovery (6). MABs, mesoangioblasts; mdx (DYS-HAC) MABs, DYS-HAC–corrected mdx-derived mesoangioblasts; MMCT, microcell-mediated chromosome transfer; EGFP, enhanced green fluorescent protein; mDys, murine dystrophin; hDys, human dystrophin. Reproduced with permission.[304] Copyright 2011, Science.
Advancing this strategy, Tedesco and colleagues demonstrated a modified application of the typical engineered stem cell strategy wherein the stem cells were used to transfer an artificial chromosome in order to correct the dystrophin mutation in DMD cells. [304] In this case, they developed a HAC vector containing the entire human dystrophin genetic locus that can be stably maintained in recipient cells. Specifically, Tedesco and co-workers first isolated mesoangioblasts from mdx dystrophic mice and subjected them to microcell-mediated chromosome transfer, which allowed for the introduction of the DYS-HAC vector. In vitro characterization of these engineered stem cells demonstrated that following transfer of cDNA encoding MyoD, engineered mesoangioblasts were induced to differentiate resulting in multinucleated myotubes that stained positive for MyoD and myosin heavy chain. Moreover, these cells underwent terminal skeletal muscle differentiation as demonstrated by the expression of myosin heavy chain and dystrophin. More importantly, following transplantation (three intramuscular injections into the tibialis anterior, gastrocnemius, and quadriceps every three weeks) of the engineered cells into severe combined immunodeficient (SCID)/mdx mice, muscles receiving injection with engineered cells exhibited extensive engraftment as well as large areas of dystrophin-positive fibers, which produced 25% of the amount of dystorphin produced by muscles of healthy control mice. Finally, morphometric analyses revealed a marked reduction in the fibrotic and cellular infiltrates of treated dystrophic muscles along with reduced necrosis and centrally nucleated muscle fibers indicating that treated muscles underwent fewer degeneration-regeneration cycles.
On the other hand, as DMD has been determined to be a hereditary disease that is caused by mutations that disrupt the dystrophin mRNA reading frame, it has been suggested that the forced exclusion (skipping) of a single exon may restore the reading frame. In particular, the majority of DMD mutations are localized in the central rod domain of the dystrophin where in-frame removal of central spectrin-like repeats has been demonstrated to conserve functionality. [305] With this in mind, Benchaouir et al. developed a combined exon-skipping and cell-based approach. This technology uses specific anti-sense oligonucletoides (AONs) that are designed to mask the putative splicing sites of exons in the mutated region of the primary RNA transcript. [306] For this purpose, blood- and muscle-derived CD133+ myogenic progenitors obtained from DMD patients, which exhibit a frameshifting deletion of exons 49 and 50 of dystrophin, were transduced with a lentiviral vector that harbored a cassette designed to favor skipping of exon 51 of the dystrophin mRNA. It was found that 14 days after culture in myogenic conditions, both engineered CD133+ cell types (blood- and muscle-derived) generated full-length dystrophin mRNA and efficient exon-skipping was revealed. More importantly, these engineered CD133+ progenitors exhibited in vivo myogenic properties after implantation into the muscle of adult mouse recipients. For this experiment, 2 × 10 4 engineered cells were injected into the right transverse abdominis (TA) muscle of SCID/mdx mice. 21 and 45 days postinjection, the transplanted muscles were harvested and immunohistochemistry revealed that that transduced plasmid efficiently skips exon 51 of dystrophin. Finally, and most importantly, the authors demonstrated that intra-arterial delivery of the engineered CD133+ cells could result in functional recovery of dystrophic mice wherein muscle function, as evaluated by tetanic force of isolated TA and extensor digitorum longus (EDL) muscle, and in vivo treadmill exhaustion tests demonstrated that injection of engineered cells resulted in a significant increase in tetanic force and endurance capacity, respectively.
5.2. Wiskott-Aldrich Syndrome
Wiskott-Aldrich syndrome (WAS) is a rare X-linked primary immunodeficiency that is characterized by recurrent infections, microthrombocytopenia, eczema, autoimmunity, and an increased incidence of lymphoid malignancies. [307,308] It has been identified that WAS is caused by mutations in the WAS gene, which is exclusively expressed in hematopoietic cells. In particular, the WAS gene produces a WAS protein (WASP) that plays a key role in actin polymerization in hematopoietic cells, with domains involved in signaling, cell locomotion, and immunologic-synapse formation. [309,310] As such, WAS patients are characterized by an absence of WASP resulting in impairment of several immune cell functions such as leukocyte migration, [311] pathogen killing by natural killer cells and neutrophils, [312] antigen presentation by antigen-presenting cells, [313] homing of B cells, [314] and T-cell activation, which all lead to the above-mentioned effects. [315]
Owing to the wide range of hematopoietic cell types that are affected by WAS, therapeutic approaches have focused on HSCs. However, currently, the only absolute therapeutic option involves BM transplantation (BMT) from related human leukocyte antigen-identical or matched unrelated donors. [316] Moreover, owing to the lack of time, [317] patients lacking a related identical donor or a matched unrelated donor often have to undergo BMT from a mismatched related donor resulting in significant life-threatening risks such as the development of life-threatening Epstein-Barr virus lymphoproliferative syndrome, infections, autoimmunity, graft rejection, and graft-versus-host disease. [318]
To this end, an alternative therapeutic strategy involves the infusion of autologous HSCs that have been genetically engineered ex vivo to express the corrected WAS gene as these engineered HSCs would have a proliferative advantage over WASP-negative cells. [319] For instance, Marangoni et al. conducted the first long-term study wherein HSCs were engineered to express the corrected version of WAS. [320] This was accomplished using a human WAS promoter/cDNA encoding lentivirus to transduce lineage marker-depleted (lin−) cells from BM BL6-was null mice. Transduced or control lin− cells were then injected into BL6-was null mice by sublethal irradiation and donor cell engraftment resulting in high and stable engraftment (69–100%) of all hematopoietic cell types for up to 12 months. Importantly, the authors observed a selective advantage for T and B lymphocytes expressing the transgenic WASP, improvement in B lymphocyte and platelet counts, as well as functional restorations such as T-cell receptor (TCR)-driven T-cell activation and B-cell’s ability to migrate in response to CXCL13. Finally, after long-term evaluation of safety, it was found that the use of engineered stem cells did not affect the lifespan of the treated animals.
Given the efficacy that engineered stem cells have shown in preclinical studies for the treatment of WAS, [307,321] clinical trials have recently been conducted. [322–324] For instance, Aiuti and co-workers tested their lentiviral method for engineering stem cells in clinical trials. In particular, to further improve the safety of engineered stem cells for WAS treatment, Aiuti et al. developed a SIN lentiviral vector coding for human WASP under the control of a 1.6-kb reconstituted WAS gene promoter. [324] The use of this endogenous promoter combined with the SIN lentiviral vector ensured that the transgene was expressed in a physiological manner while reducing the risk of insertional mutagenesis. [309,325] In this phase I/II clinical trial, three children with WAS, as confirmed by genotyping and who did not have compatible allogenic donors, were enrolled. Autologous bone-marrow-derived CD34+ cells were collected, transduced twice with the lentiviral vector, [326] and rein-fused intravenously back into the patients 3 days after collection. All three WAS patients showed robust and multilineage engraftment (engraftment efficiency of 34%, 25%, and 48% for patient 1, 2, and 3, respectively) of the engineered HSCs in their BM and peripheral blood (PB), which was persistent for at least 30 months after therapy. In particular, WASP expression peaked in the first month after treatment and then stabilized. More importantly, all of the patients were clinically well during post-treatment follow-up of 20 to 32 months and showed substantial improvement in terms of WAS symptoms including resolution of eczema as well as decreases in the frequency and severity of infections. Moreover, platelet counts improved significantly during the first year, although they never returned to normal healthy levels. While serious adverse events did occur in patients 2 and 3 within the first 2 to 6 months of gene therapy, they were mainly due to infection. Finally, no abnormal cellular expansion was detected in the PM and PB and lentiviral gene therapy did not induce selection of integrations near oncogenes.
Clinical trials have also been performed to test retroviral methods for engineering stem cells. In a study by Boztug and co-workers, two young boys, who were at least 12 months old and had been diagnosed with severe WAS (as documented by molecular and clinical phenotype), were treated with engineered HSCs expressing the corrected WAS gene. [322] To engineer the HSCs, autologous CD34+ HSCs were collected via leukopheresis. The cells were then transduced with a WASP-expressing retroviral vector. Following gene therapy, WASP-positive cells in various leukocyte subgroups were detected by flow cytometry (increases in WASP-positive monocytes, lymphoid cells, and CD4+ and CD8+ T cells were seen 6 to 12 months after gene therapy and remained stable). Moreover, an increase in platelet count was noted starting 6 to 9 months after gene therapy and stabilized even 2.5 years after gene therapy. Owing to these functional corrections owing to sustained WASP expression, the patients’ clinical condition markedly improved with resolution of hemorrhagic diathesis, eczema, autoimmunity, and predisposition to severe infection. Lastly, comprehensive insertion-site analysis showed vector integration that targeted multiple genes controlling growth and immunologic responses and despite targeting potential oncogenes, no persistent clonal imbalance had yet been observed at the time of the study.
Lastly, to test the long-term efficacy and genotoxicity, Braun et al., from the same group as Boztug, engineered HSCs using a retroviral vector to treat 10 patients with severe WAS. [323] In particular, peripheral blood mononuclear cells were harvested by leukapheresis upon treatment with recombinant human granulocyte colony-stimulating factor alone or in combination with the CXCR4 inhibitor plerixafor. Following reintroduction of the engineered cells, patients were followed for 1.5 to 6 years. High levels of engraftment were observed with a strong increase in the proportion of WASP-corrected lymphoid cells in all patients over time. All patients demonstrated significant increases in platelet counts after gene therapy as well as improvements in lymphocyte number and function (Figure 18). However, clonality and insertion site analyses determined that retroviral insertion preferred gene loci within proto-oncogenes, where integration-driven overexpression led to the development of severe side effects such as leukemia. Specifically, seven patients developed acute leukemia (one acute myeloid leukemia (AML), four developed T cell acute lymphoblastic leukemia, and two developed primary T-ALL with secondary AML). Moreover, cytogenic analysis revealed additional genetic alterations such as chromosomal translocations. As such, while engineered HSCs show great promise for WAS treatment, retroviral transduction is associated with long-term toxicity.
Figure 18.
Stem cell gene therapy for Wiskott-Aldrich Syndrome. A) Platelet counts were assessed before GT, at 12 months after GT, and at 24 and 36 months after GT. Wilcoxon matched-pairs signed rank test was used to assess significance levels. B) Statistical analysis of NKIS formation estimated by confocal microscopy for healthy donors (HD, n = 8), for WAS patients before GT (n = 8), and for WAS patients after GT (n = 8). C) PB IgE levels are shown at different time points after GT. D) Colonoscopy pictures of patient WAS9 before and after GT (arrows indicate areas of inflammation). Reproduced with permission.[323] Copyright 2014, Science.
Overall, these findings demonstrate the great potential that engineered stem cells hold for the treatment of WAS. Owing to the selective advantage that WAS-corrected HSCs have over WAS-negative HSCs, they are able to graft and repopulate various hematopoietic cell types. As such, WAS symptoms are ameliorated, which has been confirmed in clinical trials. However, it is also clear that the method of engineered stem cells is critical to maximize long-term efficacy and genotoxicity. Studies using lentiviral vectors have demonstrated good safety but long-term studies using retroviruses resulted in the development of acute leukemia as well as additional genetic alterations such as chromosomal translocations. While more long-term follow-up using lentiviral vectors remains to be conducted, it appears that lentiviral methods are more suitable for engineering HSCs for WAS treatment.
5.3. Leukodystrophies
Lastly, leukodystrophies are a group of genetic diseases that are characterized by white matter deterioration and typically manifest during childhood or adolescence. In particular, leukodystrophies result in the degeneration of myelin sheaths in the CNS and sometimes in the peripheral nerves owing to defects in the synthesis and maintenance of the myelin membrane. Overall, most leukodystrophies fall into one of three categories: 1) lysosomsal storage diseases, 2) peroxisomal diseases, and 3) diseases caused by mitochondrial dysfunction. [327] As a result of this deterioration, clinical regression of skills are observed and in the most severe cases, neurological devastation leading to premature death. [328]
Currently, there are no available curative treatments for leukodystrophies and therapy is only supportive. [329] Various dietary regimens and pharmacological agents have not had a favorable effect on the clinical course of the disease nor its associated biochemical abnormalities. Moreover, while HSC transplantation (HSCT) can be effective in early stage leukodystrophies, it has proved to be ineffective in children with the late infantile disease and generally in all patients with evident neuropsychological and/or neurological signs. [330] On the other hand, enzyme replacement therapy has shown some efficacy but is faced by significant limitations such as the BBB and the fact that lifelong administration would be required. As such, owing to the migratory ability of stem cells as well as their ability to be engineered with various genes, efforts have focused on improving the efficiency of transduction to use these engineered stem cells to replace dysfunctional cells and to deliver the corrected enzyme.
5.3.1. Metachromatic Leukodystrophy
Metachromatic leukodystrophy (MLD) is a neurodegenerative lysosomal storage diseases caused by an arylsulfatase A (ARSA) deficiency. This enzymatic defect results in the accumulation of the ARSA substrate, galactosylceramide I 3-sulfate (sulfatide), which is a major sphingolipid of myelin, in oligodendrocytes, microglia, and certain neurons of the CNS as well as Schwann cells and macrophages of the PNS. This build up induces widespread demyelination and neurodegeneration and as a result, children affected by MLD display progressive neurologic symptoms, including ataxia, seizures, and quadriplegia, culminating in decerebration and eventual death. [329]
As mentioned previously, HSCT has seen limited effectiveness while protein replacement faces serious barriers (e.g., BBB). Previous studies have engineered HSCs to express ARSA. However, they were only used to prevent the development of major disease manifestations in mice treated at the pre-symptomatic stage. While this was promising, in most clinical cases, unless a family history is available, the diagnosis of MLD is made after the onset of symptoms. [331] To address this issue, Biffi and colleagues engineered hematopoietic stem progenitor cells (HSPCs) to overexpress ARSA via lentiviral transduction in order to treat MLD after symptoms have already been observed. [332] HSPCs from ARSA-negative donors were transduced with lentivirus vectors expressing either ARSA or GFP. These cells were then transplanted into a mouse model of MLD that was generated by targeted disruption of the murine ARSA gene. [333] The authors found that the HSC gene therapy could reverse neurological deficits and neuropathological damage in affected mice (Figure 19). However, the efficacy of engineered stem cells was dependent on ARSA overexpression in microglia progeny of transplanted HSPCs or, in other words, microglia were found to be primarily responsible for ARSA bioavailability in the CNS. On the other hand, a peripheral source of enzyme, as established by transplanting ARSA-overexpressing hepatocytes from transgenic donors, failed to effectively deliver the enzyme to the CNS further demonstrating the importance of microglia progeny.
Figure 19.
Engineering hematopoietic stem cells to express ARSA for the treatment of metachromatic leukodystrophy. A,B) GMFM score (A) and NCV index (B) of the three treated patients and of a historical cohort of LI-MLD patients (gray circles). The dotted lines indicate (inset, color code) the expected time of disease onset, according to the onset observed in their affected matched siblings; n.r., normal range of the NCV index. C) Axial T2 weighted fast spin-echo MR images (top) and FLAIR MR images (bottom) obtained from patient MLD01 at baseline (before GT) and at +2 years after treatment, and corresponding (equivalent) images of an age-matched untreated patient with LI-MLD (in parenthesis, the chronological age at imaging acquisition in months). Reproduced with permission.[334] Copyright 2013, Science.
Given their promising preclinical results, Biffi and co-workers recently performed a phase I/II clinical trial in three presymptomatic MLD patients that were biochemically characterized for ARSA deficiency. [334] To perform this study, Biffi et al. first optimized the lentiviral-mediated gene transfer of ARSA under the control of the human phosphoglycerate kinase promoter in human HSCs. To this end, transduction of human BM-derived CD34+ cells were optimized to reach ≥2 vector copy number per genome based on the ARSA overexpression levels that are required for therapeutic efficacy. [335] As a result of this optimization, a vector copy number to genome ratio of 2.5 to 4.4 was achieved and the transduction efficiency was found to be 90 to 97%. For transplantation, a myeloablative busulfan regimen was administered intravenously to the patients in 14 doses over 4 days prior to cell transplantation. The authors found a high-level of stably engrafted engineered stem cells in the BM and peripheral blood of all patients with 45 to 80% of BM-derived hematopoietic colonies harboring the vector. As a result, ARSA levels were reconstituted to above normal values. Importantly, the disease did not progress in any of the treated patients and analysis of the lentiviral integration demonstrated that there was no evidence of aberrant clonal behavior.
These results suggest that engineering HSCs with ARSA can effectively treat MLD patients when therapy begins at both the asymptomatic and symptomatic phases of their disease. In particular, engineered HSCs can correct already established neurologic disease manifestations and neuronal damage when applied to symptomatic MLD mice.
5.3.2. X-Linked Adrenoleukodystrophy
As opposed to MLD, which is a lysosomal storage disease, X-linked adrenoleukodystrophy (X-ALD) is a peroxisomal disease. X-ALD is caused by a mutation in the ABCD1 gene, which encodes a transporter (ALD protein) that is localized in the peroxisomal membrane and is involved in the metabolism of very-long-chain fatty acids (VLCFA). Deficiency in the ALD protein leads to the accumulation of VLCFA and progressive demyelination in the CNS. X-ALD typically affects boys between the ages of 5 and 12 and leads to a vegetative state or death within 2–5 years. [336]
Currently, HSCT is the only effective therapy, provided that it is performed at an early stage, as once the disease progresses, demyelination cannot be arrested. [337] However, owing to the donor-related constraints and the fact that it carries considerable risk of mortality, engineered HSCs may provide an appropriate therapeutic alterative. To this end, studies have demonstrated that lentiviral transduced human ALD CD34+ cells can be transplanted into nonobese diabetic (NOD)/SCID mice for the treatment of X-ALD. The recipient mice showed in vivo expression of ALD protein in human monocytes and macrophages derived from engrafted human stem cells. [338] More importantly, human BM–derived cells were shown to migrate into the brain of recipient mice and then differentiate into microglia expressing the human ALD protein. [339]
Owing to this promising preclinical data, Cartier et al. engineered HSCs using a lentiviral vector encoding wild-type ABCD1 in a clinical trial with X-ALD patients. [340] In particular, the study enrolled two X-ALD patients who had progressive cerebral demyelination and adrenal insufficiency and who had no human leukocyte antigen (HLA)-matched donor or cord blood for allogeneic HCT. CD34+ cells were removed from these patients, genetically corrected ex vivo, and then re-infused into the patients after they received a full myeloablative treatment to increase the engraftment of transduced HSCs as lentiviral correction of ALD HSCs does not provide a selective growth advantage. Following transplantation, 50% and 33% of CD34+ cells expressed the ALD protein 5 days after transduction in patient 1 and 2, respectively. Moreover, VLCFA levels in transduced CD34+ cells was reduced by 55% and 68% in patient 1 and 2, respectively. Hematopoietic recovery occurred at days 13 to 15 following transplantation and plateaued thereafter. Over a span of 24 to 30 months of follow-up, polyclonal reconstitution was detected, with 9 to 14% of granulocytes, monocytes, and T and B lymphocytes expressing the ALD protein. These results strongly suggest that HSCs were transduced in the patients. Beginning 14 to 16 months after infusion of the genetically corrected cells, progressive cerebral demyelination in the two patients stopped, a clinical outcome comparable to that achieved by allogeneic HCT (Figure 20). Finally, genome-wide monitoring of lentivirus-marked HSC clonality in the patients determined that no obvious clonal skewing or dominance in hematopoiesis existed, though a longer follow-up with a larger sample size is needed to verify the safety of this strategy. [341]
Figure 20.
Engineering hematopoietic stem cell with ABCD1 to enhance the treatment of X-Linked Adrenoleukodystrophy. Brain MRIs from A) P1 and B) P2 before and after gene therapy. C) Progression of cerebral demyelinating lesions in an untreated 8-year-old ALD patient. Reproduced with permission.[340] Copyright 2009, Science.
6. Conclusion
In this Review, we discussed the use of engineered stems for various biomedical applications. In particular, in addition to briefly covering the available cell sources and strategies that have been developed to engineer stem cells, we systematically reviewed the application of engineered stem cells to tissue regeneration (e.g., nervous, bone, cartilage, and cardiac tissue), the treatment of immunodeficiency diseases (e.g., muscular dystrophy, Wiskott-Aldrich Syndrome, and leukodystrophies), and cancer.
In terms of future perspectives, while engineered stem cells have shown great potential and success in preclinical and clinical studies for various biomedical applications such as tissue regeneration as well as for the treatment of genetic diseases and even for cancer, there is still a long way to go before we witness their widespread use in the clinic. In particular, there are two major limitations that must be overcome before this can become a reality. First, stem cell therapies have not yet been embraced in the clinic, as we still do not have a deep biological understanding of what these cells are. For instance, even reaching a consensus on the precise characteristics of a given type of stem cells remains a challenge. [342] The second barrier is in the use of gene therapies, which remains a great technical challenge. Specifically, it is very difficult to introduce new genetic materials (e.g., DNA, RNA, RNAi) into cells without causing detrimental side effects (e.g., tumorigenesis from integration of viral DNA into the host genome). As such, before engineered stem cell therapies can reach their full potential, stem cell and gene therapies must first, separately, prove to be safe and efficient.
To this end, there have recently been promising technological breakthroughs that could have a significant effect on our ability to engineer stem cells. The CRISPR/Cas gene-editing technology is one of these breakthroughs. In particular, CRISPR stands for clustered regularly interspaced short palindromic repeats, which are associated with RNA-guided nucleases such as Cas9 genes, hence their name (CRISPR/Cas). This system was developed for gene editing in 2013 and is generally used by delivering the Cas9 protein (either protein or plasmid) and appropriate guide RNAs (for a particular targeted site of the host genome) into the target cell resulting in the desired alteration of the genome (e.g., point mutations, insertion, etc.). [343] Current demonstrations have focused on modeling diseases such as cancer. [344] However, a few studies have moved beyond proof-of-concept studies to show the utility of CRISPR/Cas systems. For instance, Long and colleagues recently utilized CRISPR/Cas9 to edit the germline of mice thereby preventing the development of muscular dystrophy. [345]
Besides further developing our understanding of stem cells and gene therapies as well as applying new techniques to more precisely engineer stem cells, engineering stem cell approaches can also be enhanced by improving stem cell efficacy. For instance, the modification of stem cells with genes that enhance survival was briefly discussed in this review for cardiac regeneration. [146] However, in addition to this approach, other researchers have focused on enhancing stem cell migration (e.g., to tumors) via the overexpression of chemokine receptors such as CXCR. [346] Finally, strategies aimed at inducing cell lysis or apoptosis of the engineered stem cells once they have performed their role are also under investigation. An example of this was briefly discussed previously with regard to the use of prodrug systems with engineered stem cells for the treatment of cancer. [282] As such, there is significant room to develop and improve engineered stem cell approaches.
In conclusion, although it will be a long time before we see the full impact that engineered stem cells can have in the clinic, as evidenced by the preclinical and clinical studies that have been conducted up till now, engineered stem cells will ultimately offer tremendous promise for various biomedical applications. We hope that this article has inspired interest from researchers in various disciplines, whose interdisciplinary cooperation will be required to push engineered stem cells into the clinic.
Acknowledgments
K.-B.L. acknowledges financial support from the NIH Director’s Innovator Award [1DP20D006462–01], National Institute of Neurological Disorders and Stroke of the NIH [1R21NS085569–01], the N.J. Commission on Spinal Cord Research [CSR13ERG005], Burroughs Wellcome Fund Collaborative Research Travel Grant, and the Rutgers Busch Biomedical Grant Program. P.T.Y. would like to acknowledge the NIH Biotechnology Training Grant for support.
Biographies

Perry To-tien Yin received his B.S. in Biomedical Engineering from Columbia University in 2010 and is currently pursuing his Ph.D. in Biomedical Engineering at Rutgers, The State University of New Jersey, where he plans to graduate in 2015. His doctoral research, under the supervision of Prof. Ki-Bum Lee, focuses on the application of multifunctional nanoparticles for the detection and treatment of breast, ovarian, and brain cancer with particular emphasis on the application of stem cells and magnetic nanoparticles for magnetic hyperthermia-based treatments.

Edward Han is currently pursuing his B.A.Sc in Biomedical Engineering at the University of Toronto, where he plans to graduate in 2015. His research interests lie at the intersection of biomaterials and cell and tissue engineering. He spent a summer in Prof. Ki-Bum Lee’s laboratory, where he focused on developing new techniques for microparticle-based drug delivery. Other projects that he has pursued include developing a 3D bioprinter in Professor Michael Sefton’s laboratory at the University of Toronto and testing stem cell-based cancer therapies in Professor Karp’s laboratory at Harvard Medical School.

Ki-Bum Lee is an Associate Professor of Chemistry and Chemical Biology at Rutgers University, where he has been a faculty since 2008. He received his Ph.D. in Chemistry from Northwestern University (with Chad. A. Mirkin; 2004) and completed his postdoctoral training at The Scripps Research Institute (with Peter G. Schultz; 2007). The primary research interest of Prof. Lee’s group is to develop and integrate nanotechnologies and chemical functional genomics to modulate signaling pathways in cells (e.g., stem cells and cancer cells) towards specific cell lineages or behaviors.
Contributor Information
Perry T. Yin, Department of Biomedical Engineering Rutgers, The State University of New Jersey, 599 Taylor Road, Piscataway, NJ 08854, USA
Edward Han, Institute of Biomaterials and Biomedical Engineering University of Toronto, 164 College Street, Toronto, ON, M5S 3G9, Canada.
Prof. Ki-Bum Lee, Department of Biomedical Engineering Rutgers, The State University of New Jersey, 599 Taylor Road, Piscataway, NJ 08854, USA. Department of Chemistry and Chemical Biology Rutgers, The State University of New Jersey 610 Taylor Road, Piscataway, NJ 08854, USA
References
- 1.Wei X, Yang X, Han ZP, Qu FF, Shao L, Shi YF. Acta Pharmacol Sin. 2013;34:747. doi: 10.1038/aps.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim SU, de Vellis J. J Neurosci Res. 2009;87:2183. doi: 10.1002/jnr.22054. [DOI] [PubMed] [Google Scholar]
- 3.Segers VFM, Lee RT. Nature. 2008;451:937. doi: 10.1038/nature06800. [DOI] [PubMed] [Google Scholar]
- 4.a) Thomas ED, Lochte HL, Jr, Lu WC, Ferrebee JW. N Engl J Med. 1957;257:491. doi: 10.1056/NEJM195709122571102. [DOI] [PubMed] [Google Scholar]; b) Guild WR, Harrison JH, Merrill JP, Murray J. Trans Am Clin Climatol Assoc. 1955;67:167. [PMC free article] [PubMed] [Google Scholar]
- 5.a) Wang YX, Dumont NA, Rudnicki MA. J Cell Sci. 2014 doi: 10.1242/jcs.151209. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Roy NS, Cleren C, Singh SK, Yang L, Beal MF, Goldman SA. Nat Med. 2006;12:1259. doi: 10.1038/nm1495. [DOI] [PubMed] [Google Scholar]
- 6.a) Cheng ZK, Ou LL, Zhou X, Li F, Jia XH, Zhang YG, Liu XL, Li YM, Ward CA, Melo LG, Kong DL. Mol Ther. 2008;16:571. doi: 10.1038/sj.mt.6300374. [DOI] [PubMed] [Google Scholar]; b) Hoehn M, Kustermann E, Blunk J, Wiedermann D, Trapp T, Wecker S, Focking M, Arnold H, Hescheler J, Fleischmann BK, Schwindt W, Buhrle C. Proc Natl Acad Sci USA. 2002;99:16267. doi: 10.1073/pnas.242435499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uccelli A, Moretta L, Pistoia V. Nat Rev Immunol. 2008;8:726. doi: 10.1038/nri2395. [DOI] [PubMed] [Google Scholar]
- 8.Rehman J, Li JL, Orschell CM, March KL. Circulation. 2003;107:1164. doi: 10.1161/01.cir.0000058702.69484.a0. [DOI] [PubMed] [Google Scholar]
- 9.Lindvall O, Kokaia Z, Martinez-Serrano A. Nat Med. 2004;10:S42. doi: 10.1038/nm1064. [DOI] [PubMed] [Google Scholar]
- 10.a) Muller FJ, Snyder EY, Loring JF. Nat Rev Neurosci. 2006;7:75. doi: 10.1038/nrn1829. [DOI] [PubMed] [Google Scholar]; b) Gafni Y, Turgeman G, Liebergal M, Pelled G, Gazit Z, Gazit D. Gene Ther. 2004;11:417. doi: 10.1038/sj.gt.3302197. [DOI] [PubMed] [Google Scholar]
- 11.Alderuccio F, Nasa Z, Chung JY, Ko HJ, Chan J, Toh BH. Mol Pharmaceut. 2011;8:1488. doi: 10.1021/mp2001523. [DOI] [PubMed] [Google Scholar]
- 12.Ahmed AU, Alexiades NG, Lesniak MS. Curr Opin Mol Ther. 2010;12:546. [PMC free article] [PubMed] [Google Scholar]
- 13.a) Zhang Y, Satterlee A, Huang L. Mol Ther. 2012;20:1298. doi: 10.1038/mt.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Al-Dosari MS, Gao X. AAPS J. 2009;11:671. doi: 10.1208/s12248-009-9143-y. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Young LS, Searle PF, Onion D, Mautner V. J Pathol. 2006;208:299. doi: 10.1002/path.1896. [DOI] [PubMed] [Google Scholar]
- 14.Robinton DA, Daley GQ. Nature. 2012;481:295. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barker N, Bartfeld S, Clevers H. Cell Stem Cell. 2010;7:656. doi: 10.1016/j.stem.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 16.Martino G, Pluchino S. Nat Rev Neurosci. 2006;7:395. doi: 10.1038/nrn1908. [DOI] [PubMed] [Google Scholar]
- 17.Jung Y, Bauer G, Nolta JA. Stem Cells. 2012;30:42. doi: 10.1002/stem.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altman J, Das GD. J Comparative Neurol. 1965;124:319. doi: 10.1002/cne.901240303. [DOI] [PubMed] [Google Scholar]
- 19.Gage FH. Science. 2000;287:1433. doi: 10.1126/science.287.5457.1433. [DOI] [PubMed] [Google Scholar]
- 20.Zhang SC, Wernig M, Duncan ID, Brustle O, Thomson JA. Nat Biotechnol. 2001;19:1129. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- 21.Svendsen CN, Caldwell MA, Ostenfeld T. Brain Pathol. 1999;9:499. doi: 10.1111/j.1750-3639.1999.tb00538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ronaghi M, Erceg S, Moreno-Manzano V, Stojkovic M. Stem Cells. 2010;28:93. doi: 10.1002/stem.253. [DOI] [PubMed] [Google Scholar]
- 23.Jenq RR, van den Brink MRM. Nat Rev Cancer. 2010;10:213. doi: 10.1038/nrc2804. [DOI] [PubMed] [Google Scholar]
- 24.Passweg JR, Halter J, Bucher C, Gerull S, Heim D, Rovo A, Buser A, Stern M, Tichelli A. Swiss Med Weekly. 2012:142. doi: 10.4414/smw.2012.13696. [DOI] [PubMed] [Google Scholar]
- 25.a) Digiusto D, Chen S, Combs J, Webb S, Namikawa R, Tsukamoto A, Chen BP, Galy AHM. Blood. 1994;84:421. [PubMed] [Google Scholar]; b) Krause DS, Fackler MJ, Civin CI, May WS. Blood. 1996;87:1. [PubMed] [Google Scholar]
- 26.Körbling M, Anderlini P. Blood. 2001;98:2009. doi: 10.1182/blood.v98.10.2900. [DOI] [PubMed] [Google Scholar]
- 27.Capello E, Vuolo L, Gualandi F, Van Lint MT, Roccatagliata L, Bonzano L, Pardini M, Uccelli A, Mancardi G. Neurol Sci. 2009;30:175. doi: 10.1007/s10072-009-0144-5. [DOI] [PubMed] [Google Scholar]
- 28.a) Szodoray P, Varoczy L, Szegedi G, Zeher M. Scand J Rheumatol. 2010;39:1. doi: 10.3109/03009740903030324. [DOI] [PubMed] [Google Scholar]; b) Trounson A, Thakar RG, Lomax G, Gibbons D. BMC Med. 2011;9:52. doi: 10.1186/1741-7015-9-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsieh MM, Kang EM, Fitzhugh CD, Link MB, Bolan CD, Kurlander R, Childs RW, Rodgers GP, Powell JD, Tisdale JF. N Engl J Med. 2009;361:2309. doi: 10.1056/NEJMoa0904971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.a) Granero-Molto F, Weis JA, Longobardi L, Spagnoli A. Expert Opin Biol Ther. 2008;8:255. doi: 10.1517/14712598.8.3.255. [DOI] [PubMed] [Google Scholar]; b) Salem HK, Thiemermann C. Stem Cells. 2010;28:585. doi: 10.1002/stem.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.a) Lu DB, Chen B, Liang ZW, Deng WQ, Jiang YZ, Li SF, Xu J, Wu QN, Zhang ZH, Xie B, Chen SH. Diabetes Res Clin Pract. 2011;92:26. doi: 10.1016/j.diabres.2010.12.010. [DOI] [PubMed] [Google Scholar]; b) Yamada Y, Ueda M, Hibi H, Baba S. Int J Periodont Rest. 2006;26:363. [PubMed] [Google Scholar]; c) Rasulov MF, Vasil’chenkov AV, Onishchenko NA, Krasheninnikov ME, Kravchenko VI, Gorshenin TL, Pidtsan RE, Potapov IV. Bull Exp Biol Med. 2005;139:141. doi: 10.1007/s10517-005-0232-3. [DOI] [PubMed] [Google Scholar]
- 32.Lee RH, Pulin AA, Seo MJ, Kota DJ, Ylostalo J, Larson BL, Semprun-Prieto L, Delafontaine P, Prockop DJ. Cell Stem Cell. 2009;5:54. doi: 10.1016/j.stem.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prasad VK, Lucas KG, Kleiner GI, Talano JAM, Jacobsohn D, Broadwater G, Monroy R, Kurtzberg J. Biol Blood Marrow Transplant. 2011;17:534. doi: 10.1016/j.bbmt.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 34.a) Ciccocioppo R, Bernardo ME, Sgarella A, Maccario R, Avanzini MA, Ubezio C, Minelli A, Alvisi C, Vanoli A, Calliada F, Dionigi P, Perotti C, Locatelli F, Corazza GR. Gut. 2011;60:788. doi: 10.1136/gut.2010.214841. [DOI] [PubMed] [Google Scholar]; b) Duijvestein M, Vos ACW, Roelofs H, Wildenberg ME, Wendrich BB, Verspaget HW, Kooy-Winkelaar EMC, Koning F, Zwaginga JJ, Fidder HH, Verhaar AP, Fibbe WE, van den Brink GR, Hommes DW. Gut. 2010;59:1662. doi: 10.1136/gut.2010.215152. [DOI] [PubMed] [Google Scholar]
- 35.Honmou O, Houkin K, Matsunaga T, Niitsu Y, Ishiai S, Onodera R, Waxman SG, Kocsis JD. Brain. 2011;134:1790. doi: 10.1093/brain/awr063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mannon PJ. Expert Opin Biol Ther. 2011;11:1249. doi: 10.1517/14712598.2011.602967. [DOI] [PubMed] [Google Scholar]
- 37.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Science. 1998;282:1145. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 38.Evans MJ, Kaufman MH. Nature. 1981;292:154. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 39.Vogel G, Holden C. Science. 2008;321:756. doi: 10.1126/science.321.5890.756b. [DOI] [PubMed] [Google Scholar]
- 40.Mosher JT, Pemberton TJ, Harter K, Wang CL, Buzbas EO, Dvorak P, Simon C, Morrison SJ, Rosenberg NA. N Engl J Med. 2010;362:183. doi: 10.1056/NEJMc0910371. [DOI] [PubMed] [Google Scholar]
- 41.Mothe AJ, Tator CH. J Clin Invest. 2012;122:3824. doi: 10.1172/JCI64124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takahashi K, Yamanaka S. Cell. 2006;126:663. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 43.Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, Hochedlinger K. Nature. 2010;465:175. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, Ambartsumyan G, Aimiuwu O, Richter L, Zhang J, Khvorostov I, Ott V, Grunstein M, Lavon N, Benvenisty N, Croce CM, Clark AT, Baxter T, Pyle AD, Teitell MA, Pelegrini M, Plath K, Lowry WE. Cell Stem Cell. 2009;5:111. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gutierrez-Aranda I, Ramos-Mejia V, Bueno C, Munoz-Lopez M, Real PJ, Macia A, Sanchez L, Ligero G, Garcia-Parez JL, Menendez P. Stem Cells. 2010;28:1568. doi: 10.1002/stem.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hanna J, Wernig M, Markoulaki S, Sun CW, Meissner A, Cassady JP, Beard C, Brambrink T, Wu LC, Townes TM, Jaenisch R. Science. 2007;318:1920. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- 47.Kazuki Y, Oshimura M. Mol Ther. 2011;19:1591. doi: 10.1038/mt.2011.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robbins PD, Ghivizzani SC. Pharmacol Ther. 1998;80:35. [PubMed] [Google Scholar]
- 49.a) Kustikova O, Fehse B, Modlich U, Yang M, Dullmann J, Kamino K, von Neuhoff N, Schlegelberger B, Li ZX, Baum C. Science. 2005;308:1171. doi: 10.1126/science.1105063. [DOI] [PubMed] [Google Scholar]; b) Vroemen M, Weidner N, Blesch A. Exp Neurol. 2005;195:127. doi: 10.1016/j.expneurol.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 50.Kay MA. Nat Rev Genet. 2011;12:316. doi: 10.1038/nrg2971. [DOI] [PubMed] [Google Scholar]
- 51.Thomas CE, Ehrhardt A, Kay MA. Nat Rev Genet. 2003;4:346. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 52.Ginn SL, Alexander IE, Edelstein ML, Abedi MR, Wixon J. J Gene Med. 2013;15:65. doi: 10.1002/jgm.2698. [DOI] [PubMed] [Google Scholar]
- 53.Rios HF, Lin Z, Oh B, Park CH, Giannobile WV. J Periodontol. 2011;82:1223. doi: 10.1902/jop.2011.100710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gabriel R, Schmidt M, von Kalle C. Curr Opin Immunol. 2012;24:592. doi: 10.1016/j.coi.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 55.Santoni de Sio FR, Cascio P, Zingale A, Gasparini M, Naldini L. Blood. 2006;107:4257. doi: 10.1182/blood-2005-10-4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Montini E, Cesana D, Schmidt M, Sanvito F, Ponzoni M, Bartholomae C, Sergi LS, Benedicenti F, Ambrosi A, Di Serio C, Doglioni C, von Kalle C, Naldini L. Nat Biotechnol. 2006;24:687. doi: 10.1038/nbt1216. [DOI] [PubMed] [Google Scholar]
- 57.Cavazzana-Calvo M, Payen E, Negre O, Wang G, Hehir K, Fusil F, Down J, Denaro M, Brady T, Westerman K, Cavallesco R, Gillet-Legrand B, Caccavelli L, Sgarra R, Maouche-Chretien L, Bernaudin F, Girot R, Dorazio R, Mulder GJ, Polack A, Bank A, Soulier J, Larghero J, Kabbara N, Dalle B, Gourmel B, Socie G, Chretien S, Cartier N, Aubourg P, Fischer A, Cornetta K, Galacteros F, Beuzard Y, Gluckman E, Bushman F, Hacein-Bey-Abina S, Leboulch P. Nature. 2010;467:318. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Winslow MM, Dayton TL, Verhaak RG, Kim-Kiselak C, Snyder EL, Feldser DM, Hubbard DD, DuPage MJ, Whittaker CA, Hoersch S, Yoon S, Crowley D, Bronson RT, Chiang DY, Meyerson M, Jacks T. Nature. 2011;473:101. doi: 10.1038/nature09881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Partridge KA, Oreffo ROC. Tissue Eng. 2004;10:295. doi: 10.1089/107632704322791934. [DOI] [PubMed] [Google Scholar]
- 60.Douglas JT. Mol Biotechnol. 2007;36:71. doi: 10.1007/s12033-007-0021-5. [DOI] [PubMed] [Google Scholar]
- 61.Brunetti-Pierri N, Ng P. Curr Gene Ther. 2009;9:329. doi: 10.2174/156652309789753310. [DOI] [PubMed] [Google Scholar]
- 62.McCaffrey AP, Fawcett P, Nakai H, McCaffrey RL, Ehrhardt A, Pham TTT, Pandey K, Xu H, Feuss S, Storm TA, Kay MA. Mol Ther. 2008;16:931. doi: 10.1038/mt.2008.37. [DOI] [PubMed] [Google Scholar]
- 63.Ramseier CA, Abramson ZR, Jin Q, Giannobile WV. Dental Clin North Am. 2006;50:245. doi: 10.1016/j.cden.2005.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.a) Inagaki K, Piao C, Kotchey NM, Wu X, Nakai H. Journal of virology. 2008;82:9513. doi: 10.1128/JVI.01001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Surosky RT, Urabe M, Godwin SG, McQuiston SA, Kurtzman GJ, Ozawa K, Natsoulis G. J Virol. 1997;71:7951. doi: 10.1128/jvi.71.10.7951-7959.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cossu G, Sampaolesi M. Trends Mol Med. 2007;13:520. doi: 10.1016/j.molmed.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 66.McCarty DM, Monahan PE, Samulski RJ. Gene Ther. 2001;8:1248. doi: 10.1038/sj.gt.3301514. [DOI] [PubMed] [Google Scholar]
- 67.Donsante A, Miller DG, Li Y, Vogler C, Brunt EM, Russell DW, Sands MS. Science. 2007;317:477. doi: 10.1126/science.1142658. [DOI] [PubMed] [Google Scholar]
- 68.Nathwani AC, Tuddenham EG, Rangarajan S, Rosales C, McIntosh J, Linch DC, Chowdary P, Riddell A, Pie AJ, Harrington C, O’Beirne J, Smith K, Pasi J, Glader B, Rustagi P, Ng CY, Kay MA, Zhou J, Spence Y, Morton CL, Allay J, Coleman J, Sleep S, Cunningham JM, Srivastava D, Basner-Tschakarjan E, Mingozzi F, High KA, Gray JT, Reiss UM, Nienhuis AW, Davidoff AM. N Engl J Med. 2011;365:2357. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Srivastava A. Human Gene Ther. 2005;16:792. doi: 10.1089/hum.2005.16.792. [DOI] [PubMed] [Google Scholar]
- 70.Arnett ALH, Konieczny P, Ramos JN, Hall J, Odom G, Yablonka-Reuveni Z, Chamberlain JR, Chamberlain JS. Mol Ther, Methods Clin Dev. 2014:1. doi: 10.1038/mtm.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Asuri P, Bartel MA, Vazin T, Jang JH, Wong TB, Schaffer DV. Mol Ther. 2012;20:329. doi: 10.1038/mt.2011.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Nat Rev Genet. 2014;15:541. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 73.Pack DW, Hoffman AS, Pun S, Stayton PS. Nat Rev Drug Discov. 2005;4:581. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 74.Mintzer MA, Simanek EE. Chem Rev. 2009;109:259. doi: 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]
- 75.Lee DE, Koo H, Sun IC, Ryu JH, Kim K, Kwon IC. Chem Soc Rev. 2012;41:2656. doi: 10.1039/c2cs15261d. [DOI] [PubMed] [Google Scholar]
- 76.Alexis F, Pridgen E, Langer R, Farokhzad O. In: Drug Delivery. Schäfer-Korting M, editor. Vol. 197. Springer, Berlin: Heidelberg; 2010. p. 55. [Google Scholar]
- 77.Putnam D. Nat, Mater. 2006;5:439. doi: 10.1038/nmat1645. [DOI] [PubMed] [Google Scholar]
- 78.Fraley R, Subramani S, Berg P, Papahadjopoulos D. J Biol Chem. 1980;255:431. [PubMed] [Google Scholar]
- 79.Wasungu L, Hoekstra D. J Controlled Release. 2006;116:255. doi: 10.1016/j.jconrel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 80.Li WJ, Szoka FC. Pharm Res. 2007;24:438. doi: 10.1007/s11095-006-9180-5. [DOI] [PubMed] [Google Scholar]
- 81.Whitehead KA, Langer R, Anderson DG. Nat Rev Drug Discovery. 2009;8:129. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lonez C, Vandenbranden M, Ruysschaert JM. Prog Lipid Res. 2008;47:340. doi: 10.1016/j.plipres.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 83.Choi YH, Liu F, Kim JS, Choi YK, Park JS, Kim SW. J Controlled Release. 1998;54:39. doi: 10.1016/s0168-3659(97)00174-0. [DOI] [PubMed] [Google Scholar]
- 84.Kim SW. Cold Spring Harbor Protoc. 2012;2012:433. doi: 10.1101/pdb.ip068619. [DOI] [PubMed] [Google Scholar]
- 85.Alexis F, Pridgen E, Molnar LK, Farokhzad OC. Mol Pharmaceut. 2008;5:505. doi: 10.1021/mp800051m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lungwitz U, Breunig M, Blunk T, Gopferich A. Eur J Pharm Biopharm. 2005;60:247. doi: 10.1016/j.ejpb.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 87.a) Godbey WT, Wu KK, Mikos AG. J Biomed Mater Res. 1999;45:268. doi: 10.1002/(sici)1097-4636(19990605)45:3<268::aid-jbm15>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]; b) Wightman L, Kircheis R, Rossler V, Carotta S, Ruzicka R, Kursa M, Wagner E. J Gene Med. 2001;3:362. doi: 10.1002/jgm.187. [DOI] [PubMed] [Google Scholar]
- 88.a) Nativo P, Prior IA, Brust M. ACS Nano. 2008;2:1639. doi: 10.1021/nn800330a. [DOI] [PubMed] [Google Scholar]; b) Patel S, Jung D, Yin PT, Carlton P, Yamamoto M, Bando T, Sugiyama H, Lee KB. ACS Nano. 2014;8:8959. doi: 10.1021/nn501589f. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Ghosh P, Han G, De M, Kim CK, Rotello VM. Adv Drug Delivery Rev. 2008;60:1307. doi: 10.1016/j.addr.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 89.Tiwari PM, Vig K, Dennis VA, Singh SR. Nanomaterials. 2011;1:31. doi: 10.3390/nano1010031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhou JF, Ralston J, Sedev R, Beattie DA. J Colloid Interf Sci. 2009;331:251. doi: 10.1016/j.jcis.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 91.Yin PT, Shah BP, Lee KB. Small. 2014;10:4106. doi: 10.1002/smll.201400963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.a) Kim J, Piao Y, Hyeon T. Chem Soc Rev. 2009;38:372. doi: 10.1039/b709883a. [DOI] [PubMed] [Google Scholar]; b) Lu AH, Salabas EL, Schuth F. Angew Chem Int Ed. 2007;46:1222. doi: 10.1002/anie.200602866. [DOI] [PubMed] [Google Scholar]
- 93.Sapet C, Laurent N, de Chevigny A, Le Gourrierec L, Bertosio E, Zelphati O, Beclin C. Biotechniques. 2011;50:187. doi: 10.2144/000113628. [DOI] [PubMed] [Google Scholar]
- 94.Jun YW, Lee JH, Cheon J. Angew Chem Int Ed. 2008;47:5122. doi: 10.1002/anie.200701674. [DOI] [PubMed] [Google Scholar]
- 95.Jang JT, Nah H, Lee JH, Moon SH, Kim MG, Cheon J. Angew Chem Int Ed. 2009;48:1234. doi: 10.1002/anie.200805149. [DOI] [PubMed] [Google Scholar]
- 96.a) Shah B, Yin PT, Ghoshal S, Lee KB. Angew Chem Int Ed. 2013;52:6190. doi: 10.1002/anie.201302245. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Shah BP, Pasquale N, De GJ, Tan T, Ma JJ, Lee KB. ACS Nano. 2014;8:9379. doi: 10.1021/nn503431x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gimble JM, Katz AJ, Bunnell BA. Circ Res. 2007;100:1249. doi: 10.1161/01.RES.0000265074.83288.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schwartz G, Fehlings MG. J Neurosurg. 2001;94:245. doi: 10.3171/spi.2001.94.2.0245. [DOI] [PubMed] [Google Scholar]
- 99.Rowland JW, Hawryluk GW, Kwon B, Fehlings MG. Neurosurg Focus. 2008;25:E2. doi: 10.3171/FOC.2008.25.11.E2. [DOI] [PubMed] [Google Scholar]
- 100.Kwon BK, Okon E, Hillyer J, Mann C, Baptiste D, Weaver LC, Fehlings MG, Tetzlaff W. J Neurotraum. 2011;28:1545. doi: 10.1089/neu.2009.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cummings BJ, Uchida N, Tamaki SJ, Salazar DL, Hooshmand M, Summers R, Gage FH, Anderson AJ. Proc Natl Acad Sci USA. 2005;102:14069. doi: 10.1073/pnas.0507063102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mothe AJ, Tator CH. Int J Dev Neurosci. 2013;31:701. doi: 10.1016/j.ijdevneu.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 103.Gupta N, Henry RG, Strober J, Kang SM, Lim DA, Bucci M, Caverzasi E, Gaetano L, Mandelli ML, Ryan T, Perry R, Farrell J, Jeremy RJ, Ulman M, Huhn SL, Barkovich AJ, Rowitch DH. Sci Transl Med. 2012;4:155ra137. doi: 10.1126/scitranslmed.3004373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hofstetter CP, Holmstrom NA, Lilja JA, Schweinhardt P, Hao J, Spenger C, Wiesenfeld-Hallin Z, Kurpad SN, Frisen J, Olson L. Nat Neurosci. 2005;8:346. doi: 10.1038/nn1405. [DOI] [PubMed] [Google Scholar]
- 105.Huang EJ, Reichardt LF. Annu Rev Biochem. 2003;72:609. doi: 10.1146/annurev.biochem.72.121801.161629. [DOI] [PubMed] [Google Scholar]
- 106.Bregman BS, McAtee M, Dai HN, Kuhn PL. Exp Neurol. 1997;148:475. doi: 10.1006/exnr.1997.6705. [DOI] [PubMed] [Google Scholar]
- 107.Lohof AM, Ip NY, Poo MM. Nature. 1993;363:350. doi: 10.1038/363350a0. [DOI] [PubMed] [Google Scholar]
- 108.Grill R, Murai K, Blesch A, Gage FH, Tuszynski MH. J Neurosci. 1997;17:5560. doi: 10.1523/JNEUROSCI.17-14-05560.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Munoz-Elias G, Woodbury D, Black IB. Stem Cells. 2003;21:437. doi: 10.1634/stemcells.21-4-437. [DOI] [PubMed] [Google Scholar]
- 110.Zhang LY, Gu ST, Zhao CP, Wen TQ. Cell Transplant. 2007;16:475. doi: 10.3727/000000007783464902. [DOI] [PubMed] [Google Scholar]
- 111.Lu P, Jones LL, Snyder EY, Tuszynski MH. Exp Neurol. 2003;181:115. doi: 10.1016/s0014-4886(03)00037-2. [DOI] [PubMed] [Google Scholar]
- 112.Kumagai G, Tsoulfas P, Toh S, McNiece I, Bramlett HM, Dietrich WD. Exp Neurol. 2013;248:369. doi: 10.1016/j.expneurol.2013.06.028. [DOI] [PubMed] [Google Scholar]
- 113.a) Cao QL, Zhang YP, Howard RM, Walters WM, Tsoulfas P, Whittemore SR. Exp Neurol. 2001;167:48. doi: 10.1006/exnr.2000.7536. [DOI] [PubMed] [Google Scholar]; b) Blakemore WF, Gilson JM, Crang AJ. Exp Neurol. 2003;184:955. doi: 10.1016/S0014-4886(03)00347-9. [DOI] [PubMed] [Google Scholar]; c) Ricci-Vitiani L, Casalbore P, Petrucci G, Lauretti L, Montano N, Larocca LM, Falchetti ML, Lombardi DG, Gerevini VD, Cenciarelli C, D’Alessandris QG, Fernandez E, De Maria R, Maira G, Peschle C, Parati E, Pallini R. Neurol Res. 2006;28:488. doi: 10.1179/016164106X115134. [DOI] [PubMed] [Google Scholar]
- 114.Baumann N, Pham-Dinh D. Physiol Rev. 2001;81:871. doi: 10.1152/physrev.2001.81.2.871. [DOI] [PubMed] [Google Scholar]
- 115.a) Liu XZ, Xu XM, Hu R, Du C, Zhang SX, McDonald JW, Dong HX, Wu YJ, Fan GS, Jacquin MF, Hsu CY, Choi DW. J Neurosci. 1997;17:5395. doi: 10.1523/JNEUROSCI.17-14-05395.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Shuman SL, Bresnahan JC, Beattie MS. J Neurosci Res. 1997;50:798. doi: 10.1002/(SICI)1097-4547(19971201)50:5<798::AID-JNR16>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]; c) Dumont RJ, Okonkwo DO, Verma S, Hurlbert RJ, Boulos PT, Ellegala DB, Dumont AS. Clin Neuropharmacol. 2001;24:254. doi: 10.1097/00002826-200109000-00002. [DOI] [PubMed] [Google Scholar]
- 116.Hwang DH, Kim BG, Kim EJ, Lee SI, Joo IS, Suh-Kim H, Sohn S, Kim SU. BMC Neurosci. 2009:10. doi: 10.1186/1471-2202-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhou Q, Wang SL, Anderson DJ. Neuron. 2000;25:331. doi: 10.1016/s0896-6273(00)80898-3. [DOI] [PubMed] [Google Scholar]
- 118.Jakovcevski I, Zecevic N. J Neurosci. 2005;25:10064. doi: 10.1523/JNEUROSCI.2324-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hu JG, Shen L, Wang R, Wang QY, Zhang C, Xi J, Ma SF, Zhou JS, Lu HZ. Neurotherapeutics. 2012;9:422. doi: 10.1007/s13311-011-0090-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ziv Y, Avidan H, Pluchino S, Martino G, Schwartz M. Proc Natl Acad Sci USA. 2006;103:18380. doi: 10.1073/pnas.0603747103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Williams S, Chalmers K, Wilcock GK, Love S. Neuropathol Appl Neurobiol. 2005;31:414. doi: 10.1111/j.1365-2990.2005.00663.x. [DOI] [PubMed] [Google Scholar]
- 122.Blurton-Jones M, Kitazawa M, Martinez-Coria H, Castello NA, Muller FJ, Loring JF, Yamasaki TR, Poon WW, Green KN, LaFerla FM. Proc Natl Acad Sci USA. 2009;106:13594. doi: 10.1073/pnas.0901402106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang Q, Matsumoto Y, Shindo T, Miyake K, Shindo A, Kawanishi M, Kawai N, Tamiya T, Nagao S. J Med Invest. 2006;53:61. doi: 10.2152/jmi.53.61. [DOI] [PubMed] [Google Scholar]
- 124.Lindvall O, Kokaia Z. Nature. 2006;441:1094. doi: 10.1038/nature04960. [DOI] [PubMed] [Google Scholar]
- 125.Blurton-Jones M, Spencer B, Michael S, Castello NA, Agazaryan AA, Davis JL, Muller FJ, Loring JF, Masliah E, LaFerla FM. Stem Cell Res Ther. 2014;5:46. doi: 10.1186/scrt440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lebson L, Nash K, Kamath S, Herber D, Carty N, Lee DC, Li Q, Szekeres K, Jinwal U, Koren J, Dickey CA, Gottschall PE, Morgan D, Gordon MN. J Neurosci. 2010;30:9651. doi: 10.1523/JNEUROSCI.0329-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tuszynski MH, Thal L, Pay M, Salmon DP, HSU, Bakay R, Patel P, Blesch A, Vahlsing HL, Ho G, Tong G, Potkin SG, Fallon J, Hansen L, Mufson EJ, Kordower JH, Gall C, Conner J. Nat Med. 2005;11:551. doi: 10.1038/nm1239. [DOI] [PubMed] [Google Scholar]
- 128.Chaudhuri KR, Schapira AH. Lancet Neurol. 2009;8:464. doi: 10.1016/S1474-4422(09)70068-7. [DOI] [PubMed] [Google Scholar]
- 129.Lewitt PA. N Engl J Med. 2008;359:2468. doi: 10.1056/NEJMct0800326. [DOI] [PubMed] [Google Scholar]
- 130.a) Lindvall O, Brundin P, Widner H, Rehncrona S, Gustavii B, Frackowiak R, Leenders KL, Sawle G, Rothwell JC, Marsden CD, Bjorklund A. Science. 1990;247:574. doi: 10.1126/science.2105529. [DOI] [PubMed] [Google Scholar]; b) Dunnett SB, Bjorklund A. Nature. 1999;399:A32. doi: 10.1038/399a032. [DOI] [PubMed] [Google Scholar]
- 131.Lindvall O, Kokaia Z. Trends Pharmacol Sci. 2009;30:260. doi: 10.1016/j.tips.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 132.Wakeman DR, Dodiya HB, Kordower JH, Sinai Mount. J Med. 2011;78:126. doi: 10.1002/msj.20233. [DOI] [PubMed] [Google Scholar]
- 133.Wolff JA, Fisher LJ, Xu L, Jinnah HA, Langlais PJ, Iuvone PM, Omalley KL, Rosenberg MB, Shimohama S, Friedmann T, Gage FH. Proc Natl Acad Sci USA. 1989;86:9011. doi: 10.1073/pnas.86.22.9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.During MJ, Naegele JR, Omalley KL, Geller AI. Science. 1994;266:1399. doi: 10.1126/science.266.5189.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kang UJ, Fisher LJ, Joh TH, Omalley KL, Gage FH. J Neurosci. 1993;13:5203. doi: 10.1523/JNEUROSCI.13-12-05203.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kim SU, Park IH, Kim TH, Kim KS, Choi HB, Hong SH, Bang JH, Lee MA, Joo IS, Lee CS, Kim YS. Neuropathology. 2006;26:129. doi: 10.1111/j.1440-1789.2006.00688.x. [DOI] [PubMed] [Google Scholar]
- 137.Tan XF, Jin GH, Tian ML, Qin JB, Zhang L, Zhu HX, Li HM. Cell Biol Int. 2011;35:1217. doi: 10.1042/CBI20110028. [DOI] [PubMed] [Google Scholar]
- 138.Tan X, Zhang L, Qin J, Tian M, Zhu H, Dong C, Zhao H, Jin G. Int J Dev Neurosci. 2013;31:82. doi: 10.1016/j.ijdevneu.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 139.Saucedo-Cardenas O, Quintana-Hau JD, Le WD, Smidt MP, Cox JJ, DeMayo F, Burbach JPH, Conneely OM. Proc Natl Acad Sci USA. 1998;95:4013. doi: 10.1073/pnas.95.7.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Castro DS, Skowronska-Krawczyk D, Armant O, Donaldson IJ, Parras C, Hunt C, Critchley JA, Nguyen L, Gossler A, Gottgens B, Matter JM, Guillemot F. Dev Cell. 2006;11:831. doi: 10.1016/j.devcel.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 141.Bang OY, Lee JS, Lee PH, Lee G. Ann Neurol. 2005;57:874. doi: 10.1002/ana.20501. [DOI] [PubMed] [Google Scholar]
- 142.a) Chuah MKL, Brems H, Vanslembrouck V, Collen D, Vandendriessche T. Human Gene Ther. 1998;9:353. doi: 10.1089/hum.1998.9.3-353. [DOI] [PubMed] [Google Scholar]; b) Li Y, Chen J, Chen XG, Wang L, Gautam SC, Xu YX, Katakowski M, Zhang LJ, Lu M, Janakiraman N, Chopp M. Neurology. 2002;59:514. doi: 10.1212/wnl.59.4.514. [DOI] [PubMed] [Google Scholar]
- 143.Kurozumi K, Nakamura K, Tamiya T, Kawano Y, Kobune M, Hirai S, Uchida H, Sasaki K, Ito Y, Kato K, Honmou O, Houkin K, Date I, Hamada H. Mol Ther. 2004;9:766. doi: 10.1016/j.ymthe.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 144.Zhao MZ, Nonoguchi N, Ikeda N, Watanabe T, Furutama D, Miyazawa D, Funakoshi H, Kajimoto Y, Nakamura T, Dezawa M, Shibata MA, Otsuki Y, Coffin RS, Liu WD, Kuroiwa T, Miyatake S. J Cereb Blood Flow Metab. 2006;26:1176. doi: 10.1038/sj.jcbfm.9600273. [DOI] [PubMed] [Google Scholar]
- 145.Bederson JB, Pitts LH, Tsuji M, Nishimura MC, Davis RL, Bartkowski H. Stroke. 1986;17:472. doi: 10.1161/01.str.17.3.472. [DOI] [PubMed] [Google Scholar]
- 146.Lee HJ, Kim MK, Kim HJ, Kim SU. PLoS One. 2009:4. doi: 10.1371/journal.pone.0005586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.a) Dudek H, Datta SR, Franke TF, Birnbaum MJ, Yao RJ, Cooper GM, Segal RA, Kaplan DR, Greenberg ME. Science. 1997;275:661. doi: 10.1126/science.275.5300.661. [DOI] [PubMed] [Google Scholar]; b) Franke TF, Hornik CP, Segev L, Shostak GA, Sugimoto C. Oncogene. 2003;22:8983. doi: 10.1038/sj.onc.1207115. [DOI] [PubMed] [Google Scholar]
- 148.Yamaguchi A, Tamatani M, Matsuzaki H, Namikawa K, Kiyama H, Vitek MP, Mitsuda N, Tohyama M. J Biol Chem. 2001;276:5256. doi: 10.1074/jbc.M008552200. [DOI] [PubMed] [Google Scholar]
- 149.a) Chu K, Kim M, Park KI, Jeong SW, Park HK, Jung KH, Lee ST, Kang L, Lee K, Park DK, Kim SU, Roh JK. Brain Res. 2004;1016:145. doi: 10.1016/j.brainres.2004.04.038. [DOI] [PubMed] [Google Scholar]; b) Lee HJ, Kim KS, Kim EJ, Choi HB, Lee KH, Park IH, Ko Y, Jeong SW, Kim SU. Stem Cells. 2007;25:1204. doi: 10.1634/stemcells.2006-0409. [DOI] [PubMed] [Google Scholar]
- 150.Lee HJ, Kim KS, Park IH, Kim SU. PLoS One. 2007:2. doi: 10.1371/journal.pone.0000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Onda T, Honmou O, Harada K, Houkin K, Hamada H, Kocsis JD. J Cereb Blood Flow Metab. 2008;28:329. doi: 10.1038/sj.jcbfm.9600527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Suri C, McClain J, Thurston G, McDonald DM, Zhou H, Oldmixon EH, Sato TN, Yancopoulos GD. Science. 1998;282:468. doi: 10.1126/science.282.5388.468. [DOI] [PubMed] [Google Scholar]
- 153.Nourhaghighi N, Teichert-Kuliszewska K, Davis J, Stewart DJ, Nag S. Lab Invest. 2003;83:1211. doi: 10.1097/01.lab.0000082383.40635.fe. [DOI] [PubMed] [Google Scholar]
- 154.Thurston G, Rudge JS, Ioffe E, Zhou H, Ross L, Croll SD, Glazer N, Holash J, McDonald DM, Yancopoulos GD. Nat Med. 2000;6:460. doi: 10.1038/74725. [DOI] [PubMed] [Google Scholar]
- 155.Marolt D, Knezevic M, Novakovic GV. Stem Cell Res Ther. 2010;1:10. doi: 10.1186/scrt10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lee NK, Sowa H, Hinoi E, Ferron M, Ahn JD, Confavreux C, Dacquin R, Mee PJ, Mckee MD, Jung DY, Zhang Z, Kim JK, Mauvais-Jarvis F, Ducy P, Karsenty G. Cell. 2007;130:456. doi: 10.1016/j.cell.2007.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.a) Petite H, Viateau V, Bensaid W, Meunier A, de Pollak C, Bourguignon M, Oudina K, Sedel L, Guillemin G. Nat Biotechnol. 2000;18:959. doi: 10.1038/79449. [DOI] [PubMed] [Google Scholar]; b) Karageorgiou V, Kaplan D. Biomaterials. 2005;26:5474. doi: 10.1016/j.biomaterials.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 158.Friedenstein AJ, Chailakhyan RK, Gerasimov UV. Cell Tissue Kinetics. 1987;20:263. doi: 10.1111/j.1365-2184.1987.tb01309.x. [DOI] [PubMed] [Google Scholar]
- 159.Finkemeier CG. J Bone Joint Surg. 2002;84-A:454. doi: 10.2106/00004623-200203000-00020. [DOI] [PubMed] [Google Scholar]
- 160.a) Bleich NK, Kallai I, Lieberman JR, Schwarz EM, Pelled G, Gazit D. Adv Drug Delivery Rev. 2012;64:1320. doi: 10.1016/j.addr.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wilson CG, Martin-Saavedra FM, Vilaboa N, Franceschi RT. J Dent Res. 2013;92:409. doi: 10.1177/0022034513483771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Betz OB, Betz VM, Abdulazim A, Penzkofer R, Schmitt B, Schroder C, Mayer-Wagner S, Augat P, Jansson V, Muller PE. Tissue Eng Part A. 2010;16:1093. doi: 10.1089/ten.TEA.2009.0656. [DOI] [PubMed] [Google Scholar]
- 162.Steinhardt Y, Aslan H, Regev E, Zilberman Y, Kallai I, Gazit D, Gazit Z. Tissue Eng Part A. 2008;14:1763. doi: 10.1089/ten.tea.2008.0007. [DOI] [PubMed] [Google Scholar]
- 163.Virk MS, Sugiyama O, Park SH, Gambhir SS, Adams DJ, Drissi H, Lieberman JR. Mol Ther. 2011;19:960. doi: 10.1038/mt.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sheyn D, Kallai I, Tawackoli W, Yakubovich DC, Oh A, Su SS, Da XY, Lavi A, Kimelman-Beich N, Zilberman Y, Li N, Bae H, Gazit Z, Pelled G, Gazit D. Mol Pharmaceutics. 2011;8:1592. doi: 10.1021/mp200226c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hosseinkhani H, Inatsugu Y, Hiraoka Y, Inoue S, Shimokawa H, Tabata Y. Tissue Eng. 2005;11:1459. doi: 10.1089/ten.2005.11.1459. [DOI] [PubMed] [Google Scholar]
- 166.Huang YC, Kaigler D, Rice KG, Krebsbach PH, Mooney DJ. J Bone Miner Res. 2005;20:848. doi: 10.1359/JBMR.041226. [DOI] [PubMed] [Google Scholar]
- 167.Danhier F, Ansorena E, Silva JM, Coco R, Le Breton A, Preat V. J Controlled Release. 2012;161:505. doi: 10.1016/j.jconrel.2012.01.043. [DOI] [PubMed] [Google Scholar]
- 168.Huang YC, Connell M, Park Y, Mooney DJ, Rice KG. J Biomed Mater Res A. 2003;67A:1384. doi: 10.1002/jbm.a.20036. [DOI] [PubMed] [Google Scholar]
- 169.Sheridan MH, Shea LD, Peters MC, Mooney DJ. J Controlled Release. 2000;64:91. doi: 10.1016/s0168-3659(99)00138-8. [DOI] [PubMed] [Google Scholar]
- 170.Poole AR, Kojima T, Yasuda T, Mwale F, Kobayashi M, Laverty S. Clin Orthop Relat Res. 2001:S26. doi: 10.1097/00003086-200110001-00004. [DOI] [PubMed] [Google Scholar]
- 171.van der Kraan PM, Buma P, van Kuppevelt T, van den Berg WB. Osteoarthritis Cartilage. 2002;10:631. doi: 10.1053/joca.2002.0806. [DOI] [PubMed] [Google Scholar]
- 172.Brittberg M, Lindahl A, Nilsson A, Ohlsson C, Isaksson O, Peterson L. N Engl J Med. 1994;331:889. doi: 10.1056/NEJM199410063311401. [DOI] [PubMed] [Google Scholar]
- 173.Von Der Mark K, Gauss V, Von Der Mark H, Muller P. Nature. 1977;267:531. doi: 10.1038/267531a0. [DOI] [PubMed] [Google Scholar]
- 174.Clair BL, Johnson AR, Howard T. Foot Snkle Spec. 2009;2:179. doi: 10.1177/1938640009342272. [DOI] [PubMed] [Google Scholar]
- 175.Lee EH, Hui JHP. J Bone Joint Surg Br. 2006;88B:841. doi: 10.1302/0301-620X.88B7.17305. [DOI] [PubMed] [Google Scholar]
- 176.a) Wakitani S, Nawata M, Tensho K, Okabe T, Machida H, Ohgushi H. J Tissue Eng Regen Med. 2007;1:74. doi: 10.1002/term.8. [DOI] [PubMed] [Google Scholar]; b) Nejadnik H, Hui JH, Choong EPF, Tai BC, Lee EH. Am J Sport Med. 2010;38:1110. doi: 10.1177/0363546509359067. [DOI] [PubMed] [Google Scholar]
- 177.Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B. J Bone Miner Res. 2002;17:S142. [Google Scholar]
- 178.a) Amano K, Hata K, Sugita A, Takigawa Y, Ono K, Wakabayashi M, Kogo M, Nishimura R, Yoneda T. Mol Biol Cell. 2009;20:4541. doi: 10.1091/mbc.E09-03-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kupcsik L, Stoddart MJ, Li Z, Benneker LM, Alini M. Tissue Eng Part A. 2010;16:1845. doi: 10.1089/ten.TEA.2009.0531. [DOI] [PubMed] [Google Scholar]
- 179.a) Han Y, Lefebvre V. Mol Cell Biol. 2008;28:4999. doi: 10.1128/MCB.00695-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Dy P, Smits P, Silvester A, Penzo-Mendez A, Dumitriu B, Han Y, de la Motte CA, Kingsley DM, Lefebvre V. Dev Biol. 2010;341:346. doi: 10.1016/j.ydbio.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Park JS, Yang HN, Woo DG, Jeon SY, Do HJ, Lim HY, Kim JH, Park KH. Biomaterials. 2011;32:3679. doi: 10.1016/j.biomaterials.2011.01.063. [DOI] [PubMed] [Google Scholar]
- 181.Im GI, Kim HJ, Lee JH. Biomaterials. 2011;32:4385. doi: 10.1016/j.biomaterials.2011.02.054. [DOI] [PubMed] [Google Scholar]
- 182.Oh SH, Kim TH, Jang SH, Il Im G, Lee JH. J Biomed Mater Res A. 2011;97A:441. doi: 10.1002/jbm.a.33079. [DOI] [PubMed] [Google Scholar]
- 183.Kato Y, Iwamoto M, Koike T, Suzuki F, Takano Y. Proc Natl Acad Sci USA. 1988;85:9552. doi: 10.1073/pnas.85.24.9552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Pagnotto MR, Wang Z, Karpie JC, Ferretti M, Xiao X, Chu CR. Gene Ther. 2007;14:804. doi: 10.1038/sj.gt.3302938. [DOI] [PubMed] [Google Scholar]
- 185.Brunger JM, Huynh NPT, Guenther CM, Perez-Pinera P, Moutos FT, Sanchez-Adams J, Gersbach CA, Guilak F. Proc Natl Acad Sci USA. 2014;111:E798. doi: 10.1073/pnas.1321744111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Gersbach CA, Coyer SR, Le Doux JM, Garcia AJ. Biomaterials. 2007;28:5121. doi: 10.1016/j.biomaterials.2007.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sachlos E, Czernuszka JT. Eur Cells Mat. 2003;5:29. doi: 10.22203/ecm.v005a03. [DOI] [PubMed] [Google Scholar]
- 188.Jeon SY, Park JS, Yang HN, Lim HJ, Yi SW, Park H, Park KH. Biomaterials. 2014;35:8236. doi: 10.1016/j.biomaterials.2014.05.092. [DOI] [PubMed] [Google Scholar]
- 189.Sun FY, Yang QY, Weng WH, Zhang Y, Yu YC, Hong A, Ji YH, Pan QH. J Bone Miner Res. 2013;28:1950. doi: 10.1002/jbmr.1932. [DOI] [PubMed] [Google Scholar]
- 190.Jeon SY, Park JS, Yang HN, Woo DG, Park KH. Biomaterials. 2012;33:4413. doi: 10.1016/j.biomaterials.2012.02.051. [DOI] [PubMed] [Google Scholar]
- 191.Garbern JC, Lee RT. Cell Stem Cell. 2013;12:689. doi: 10.1016/j.stem.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Vunjak-Novakovic G, Tandon N, Godier A, Maidhof R, Marsano A, Martens TP, Radisic M. Tissue Eng Part B. 2010;16:169. doi: 10.1089/ten.teb.2009.0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Ren X, Ott HC. Pflug Arch Eur J Phys. 2014;466:1847. doi: 10.1007/s00424-014-1504-4. [DOI] [PubMed] [Google Scholar]
- 194.Paek S-H, Ruder WC. Encyclopedia of Molecular Cell Biology and Molecular Medicine. Wiley-VCH Verlag GmbH & Co. KGaA; Germany: 2006. [Google Scholar]
- 195.Taylor DA, Atkins BZ, Hungspreugs P, Jones TR, Reedy MC, Hutcheson KA, Glower DD, Kraus WE. Nat Med. 1998;4:929. doi: 10.1038/nm0898-929. [DOI] [PubMed] [Google Scholar]
- 196.Jia ZQ, Mickle DA, Weisel RD, Mohabeer MK, Merante F, Rao V, Li G, Li RK. Transpl Proc. 1997;29:2093. doi: 10.1016/s0041-1345(97)00247-9. [DOI] [PubMed] [Google Scholar]
- 197.Li RK, Jia ZQ, Weisel RD, Merante F, Mickle DAG. J Mol Cell Cardiol. 1999;31:513. doi: 10.1006/jmcc.1998.0882. [DOI] [PubMed] [Google Scholar]
- 198.Tomita S, Li RK, Weisel RD, Mickle DAG, Kim EJ, Sakai T, Jia ZQ. Circulation. 1999;100:247. doi: 10.1161/01.cir.100.suppl_2.ii-247. [DOI] [PubMed] [Google Scholar]
- 199.Kocher AA, Schuster MD, Szabolcs MJ, Takuma S, Burkhoff D, Wang J, Homma S, Edwards NM, Itescu S. Nat Med. 2001;7:430. doi: 10.1038/86498. [DOI] [PubMed] [Google Scholar]
- 200.Gaebel R, Furlani D, Sorg H, Polchow B, Frank J, Bieback K, Wang W, Klopsch C, Ong LL, Li W, Ma N, Steinhoff G. PLoS One. 2011;6:e15652. doi: 10.1371/journal.pone.0015652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Pittenger MF, Martin BJ. Circ Res. 2004;95:9. doi: 10.1161/01.RES.0000135902.99383.6f. [DOI] [PubMed] [Google Scholar]
- 202.Toma C, Pittenger MF, Cahill KS, Byrne BJ, Kessler PD. Circulation. 2002;105:93. doi: 10.1161/hc0102.101442. [DOI] [PubMed] [Google Scholar]
- 203.a) Nesselmann C, Li W, Ma N, Steinhoff G. Ther Adv Cardiovasc Dis. 2010;4:27. doi: 10.1177/1753944709353338. [DOI] [PubMed] [Google Scholar]; b) Wang WW, Li WZ, Ou LL, Flick E, Mark P, Nesselmann C, Lux CA, Gatzen HH, Kaminski A, Liebold A, Luetzow K, Lendlein A, Li RK, Steinhoff G, Ma N. J Cell Mol Med. 2011;15:1989. doi: 10.1111/j.1582-4934.2010.01130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Iso Y, Spees JL, Serrano C, Bakondi B, Pochampally R, Song YH, Sobel BE, Delafontaine P, Prockop DJ. Biochem Biophys, Res Commun. 2007;354:700. doi: 10.1016/j.bbrc.2007.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.a) Katritsis DG, Sotiropoulou PA, Karvouni E, Karabinos I, Korovesis S, Perez SA, Voridis EM, Papamichail M. Catheter Cardio Interv. 2005;65:321. doi: 10.1002/ccd.20406. [DOI] [PubMed] [Google Scholar]; b) Chen SL, Fang W, Ye F, Liu YH, Qian J, Shan S, Zhang J, Zhao RCH, Liao LM, Lin S, Sun JP. Am J Cardiol. 2004;94:92. doi: 10.1016/j.amjcard.2004.03.034. [DOI] [PubMed] [Google Scholar]
- 206.a) Li W, Ma N, Ong LL, Nesselmann C, Klopsch C, Ladilov Y, Furlani D, Piechaczek C, Moebius JM, Lutzow K, Lendlein A, Stamm C, Li RK, Steinhoff G. Stem Cells. 2007;25:2118. doi: 10.1634/stemcells.2006-0771. [DOI] [PubMed] [Google Scholar]; b) Jo J, Nagaya N, Miyahara Y, Kataoka M, Harada-Shiba M, Kangawa K, Tabata Y. Tissue Eng. 2007;13:313. doi: 10.1089/ten.2006.0133. [DOI] [PubMed] [Google Scholar]
- 207.Deuse T, Peter C, Fedak PWM, Doyle T, Reichenspurner H, Zimmermann WH, Eschenhagen T, Stein W, Wu JC, Robbins RC, Schrepfer S. Circulation. 2009;120:S247. doi: 10.1161/CIRCULATIONAHA.108.843680. [DOI] [PubMed] [Google Scholar]
- 208.Schrepfer S, Deuse T, Reichenspurner H, Robbins RC, Pelletier MP. J Heart Lung Transpl. 2008;27:S176. [Google Scholar]
- 209.Guo YH, He JG, Wu JL, Yang L, Zhang DS, Tan XY, Qi RD. Cytotherapy. 2008;10:857. doi: 10.1080/14653240802419278. [DOI] [PubMed] [Google Scholar]
- 210.Walter DH, Rittig K, Bahlmann FH, Kirchmair R, Silver M, Murayama T, Nishimura H, Losordo DW, Asahara T, Isner JM. Circulation. 2002;105:3017. doi: 10.1161/01.cir.0000018166.84319.55. [DOI] [PubMed] [Google Scholar]
- 211.Cho YH, Park H, Ch ES, Kim WJ, Kang BS, Park BY, Kim YJ, Lee YI, Chang SI, Park K. Exp Mol Med. 2007;39:412. doi: 10.1038/emm.2007.46. [DOI] [PubMed] [Google Scholar]
- 212.Ahn EH, Kang DK, Chang SI, Kang CS, Han MH, Kang IC. Proteomics. 2006;6:1104. doi: 10.1002/pmic.200500394. [DOI] [PubMed] [Google Scholar]
- 213.Smirnoff P, Roiz L, Angelkovitch B, Schwartz B, Shoseyov O. Cancer. 2006;107:2760. doi: 10.1002/cncr.22327. [DOI] [PubMed] [Google Scholar]
- 214.a) Xu ZP, Tsuji T, Riordan JF, Hu GF. Biochem Biophys Res Commun. 2002;294:287. doi: 10.1016/S0006-291X(02)00479-5. [DOI] [PubMed] [Google Scholar]; b) Roiz L, Smirnoff P, Bar-Eli M, Schwartz B, Shoseyov O. Cancer. 2006;106:2295. doi: 10.1002/cncr.21878. [DOI] [PubMed] [Google Scholar]
- 215.Liu XH, Bai CG, Xu ZY, Huang SD, Yuan Y, Gong DJ, Zhang JR. Microvasc Res. 2008;76:23. doi: 10.1016/j.mvr.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 216.Liu YG, Guo JX, Zhang P, Zhang SH, Chen P, Ma K, Zhou CY. Microvasc Res. 2004;68:156. doi: 10.1016/j.mvr.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 217.a) Zhang M, Mal N, Kiedrowski M, Chacko M, Askari AT, Popovic ZB, Koc ON, Penn MS. FASEB J. 2007;21:3197. doi: 10.1096/fj.06-6558com. [DOI] [PubMed] [Google Scholar]; b) Tang J, Wang J, Yang J, Kong X. Interact Cardiovasc Thorac Surg. 2008;7:767. doi: 10.1510/icvts.2007.169896. [DOI] [PubMed] [Google Scholar]
- 218.Tang JM, Wang JN, Yang JY, Kong X, Zheng F, Guo LY, Zhang L, Huang YZ. Eur J Cardio-Thorac. 2009;36:644. doi: 10.1016/j.ejcts.2009.04.052. [DOI] [PubMed] [Google Scholar]
- 219.Gnecchi M, He HM, Noiseux N, Liang OD, Zhang LM, Morello F, Mu H, Melo LG, Pratt RE, Ingwall JS, Dzau VJ. FASEB J. 2006;20:661. doi: 10.1096/fj.05-5211com. [DOI] [PubMed] [Google Scholar]
- 220.Wang Y, Feng C, Xue J, Sun A, Li J, Wu J. J Physiol Sci. 2009;59:413. doi: 10.1007/s12576-009-0050-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Xie XJ, Wang JA, Cao J, Zhang X. Acta Pharmacol Sin. 2006;27:1153. doi: 10.1111/j.1745-7254.2006.00436.x. [DOI] [PubMed] [Google Scholar]
- 222.a) Lando D, Peet DJ, Whelan DA, Gorman JJ, Whitelaw ML. Science. 2002;295:858. doi: 10.1126/science.1068592. [DOI] [PubMed] [Google Scholar]; b) Bruick RK, McKnight SL. Science. 2001;294:1337. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 223.Wang Y, Sun A, Xue J, Feng C, Li J, Wu J. Toxicol In Vitro. 2009;23:1069. doi: 10.1016/j.tiv.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 224.Date T, Mochizuki S, Belanger AJ, Yamakawa M, Luo Z, Vincent KA, Cheng SH, Gregory RJ, Jiang C. Am J Physiol Cell Physiol. 2005;288:C314. doi: 10.1152/ajpcell.00374.2004. [DOI] [PubMed] [Google Scholar]
- 225.Jung JY, Kim WJ. Neurosci Lett. 2004;371:85. doi: 10.1016/j.neulet.2004.06.069. [DOI] [PubMed] [Google Scholar]
- 226.Wikswo JP, Jr, Lin SF, Abbas RA. Biophys J. 1995;69:2195. doi: 10.1016/S0006-3495(95)80115-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Merrill DR, Bikson M, Jefferys JG. J Neurosci Methods. 2005;141:171. doi: 10.1016/j.jneumeth.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 228.Bruegmann T, Malan D, Hesse M, Beiert T, Fuegemann CJ, Fleischmann BK, Sasse P. Nat Methods. 2010;7:897. doi: 10.1038/nmeth.1512. [DOI] [PubMed] [Google Scholar]
- 229.Kolossov E, Bostani T, Roell W, Breitbach M, Pillekamp F, Nygren JM, Sasse P, Rubenchik O, Fries JWU, Wenzel D, Geisen C, Xia Y, Lu Z, Duan Y, Kettenhofen R, Jovinge S, Bloch W, Bohlen H, Welz A, Hescheler J, Jacobsen SE, Fleischmann BK. The J Exp Med. 2006;203:2315. doi: 10.1084/jem.20061469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Nagel G, Brauner M, Liewald JF, Adeishvili N, Bamberg E, Gottschalk A. Curr Biol. 2005;15:2279. doi: 10.1016/j.cub.2005.11.032. [DOI] [PubMed] [Google Scholar]
- 231.Jia Z, Valiunas V, Lu Z, Bien H, Liu H, Wang HZ, Rosati B, Brink PR, Cohen IS, Entcheva E. Circulation Arrhythmia Electrophysiol. 2011;4:753. doi: 10.1161/CIRCEP.111.964247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Siegel R, Ma J, Zou Z, Jemal A. Cancer J Clin. 2014;64:9. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 233.a) Dean M, Fojo T, Bates S. Nat Rev Cancer. 2005;5:275. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]; b) Li X, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu MF, Hilsenbeck SG, Pavlick A, Zhang X, Chamness GC, Wong H, Rosen J, Chang JC. J Natl Cancer Inst. 2008;100:672. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]; c) Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, Song E. Cell. 2007;131:1109. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 234.a) Jankowitz RC, McGuire KP, Davidson NE. Breast. 2013;22(Suppl 2):S165. doi: 10.1016/j.breast.2013.07.032. [DOI] [PubMed] [Google Scholar]; b) Potosky AL, Haque R, Cassidy-Bushrow AE, Ulcickas Yood M, Jiang M, Tsai HT, Luta G, Keating NL, Smith MR, Van Den Eeden SK. J Clin Oncol. 2014;32:1324. doi: 10.1200/JCO.2013.52.5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.a) Vassilaros S, Tsibanis A, Tsikkinis A, Pietersz GA, McKenzie IF, Apostolopoulos V. Immunotherapy. 2013;5:1177. doi: 10.2217/imt.13.126. [DOI] [PubMed] [Google Scholar]; b) Koido S, Ohkusa T, Homma S, Namiki Y, Takakura K, Saito K, Ito Z, Kobayashi H, Kajihara M, Uchiyama K, Arihiro S, Arakawa H, Okamoto M, Gong J, Tajiri H. World J Gastroenterol. 2013;19:8531. doi: 10.3748/wjg.v19.i46.8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ponte AL, Marais E, Gallay N, Langonné A, Delorme B, Hérault O, Charbord P, Domenech J. Stem Cells. 2007;25:1737. doi: 10.1634/stemcells.2007-0054. [DOI] [PubMed] [Google Scholar]
- 237.a) Suarez-Alvarez B, Lopez-Vazquez A, Lopez-Larrea C. Adv Exp Med Biol. 2012;741:152. doi: 10.1007/978-1-4614-2098-9_11. [DOI] [PubMed] [Google Scholar]; b) Guo Y, Hangoc G, Bian HM, Pelus LM, Broxmeyer HE. Stem Cells. 2005;23:1324. doi: 10.1634/stemcells.2005-0085. [DOI] [PubMed] [Google Scholar]
- 238.a) Motaln H, Gruden K, Hren M, Schichor C, Primon M, Rotter A, Lah TT. Cell Transplant. 2012;21:1529. doi: 10.3727/096368912X640547. [DOI] [PubMed] [Google Scholar]; b) Qiao L, Xu ZL, Zhao TJ, Zhao ZG, Shi MX, Zhao RC, Ye LH, Zhang XD. Cell Res. 2008;18:500. doi: 10.1038/cr.2008.40. [DOI] [PubMed] [Google Scholar]
- 239.Patel SA, Meyer JR, Greco SJ, Corcoran KE, Bryan M, Rameshwar P. J Immunol. 2010;184:5885. doi: 10.4049/jimmunol.0903143. [DOI] [PubMed] [Google Scholar]
- 240.Cross D, Burmester JK. Clin Med Res. 2006;4:218. doi: 10.3121/cmr.4.3.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Aghi M, Martuza RL. Oncogene. 2005;24:7802. doi: 10.1038/sj.onc.1209037. [DOI] [PubMed] [Google Scholar]
- 242.Nakashima H, Kaur B, Chiocca EA. Cytokine Growth Factor Reviews. 2010;21:119. doi: 10.1016/j.cytogfr.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Sonabend AM, Ulasov IV, Tyler MA, Rivera AA, Mathis JM, Lesniak MS. Stem Cells. 2008;26:831. doi: 10.1634/stemcells.2007-0758. [DOI] [PubMed] [Google Scholar]
- 244.Fritz V, Jorgensen C. Curr Stem Cell Res Ther. 2008;3:32. doi: 10.2174/157488808783489462. [DOI] [PubMed] [Google Scholar]
- 245.a) Ahmed AU, Rolle CE, Tyler MA, Han Y, Sengupta S, Wainwright DA, Balyasnikova IV, Ulasov IV, Lesniak MS. Mol Ther. 2010;18:1846. doi: 10.1038/mt.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ong HT, Federspiel MJ, Guo CM, Ooi LL, Russell SJ, Peng KW, Hui KM. J Hepatol. 2013;59:999. doi: 10.1016/j.jhep.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Yong RL, Shinojima N, Fueyo J, Gumin J, Vecil GG, Marini FC, Bogler O, Andreeff M, Lang FF. Cancer Res. 2009;69:8932. doi: 10.1158/0008-5472.CAN-08-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Duebgen M, Martinez-Quintanilla J, Tamura K, Hingtgen S, Redjal N, Wakimoto H, Shah K. J Natl Cancer Inst. 2014;106:dju090. doi: 10.1093/jnci/dju090. [DOI] [PubMed] [Google Scholar]
- 248.Ulasov IV, Rivera AA, Sonabend AM, Rivera LB, Wang M, Zhu ZB, Lesniak MS. Cancer Biol Ther. 2007;6:679. doi: 10.4161/cbt.6.5.3957. [DOI] [PubMed] [Google Scholar]
- 249.Ahmed AU, Tyler MA, Thaci B, Alexiades NG, Han Y, Ulasov IV, Lesniak MS. Mol Pharmaceutics. 2011;8:1559. doi: 10.1021/mp200161f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Stoff-Khalili M, Rivera A, Mathis JM, Banerjee NS, Moon A, Hess A, Rocconi R, Numnum TM, Everts M, Chow L, Douglas J, Siegal G, Zhu Z, Bender H, Dall P, Stoff A, Pereboeva L, Curiel D. Breast Cancer Res Treat. 2007;105:157. doi: 10.1007/s10549-006-9449-8. [DOI] [PubMed] [Google Scholar]
- 251.Furusato B, Mohamed A, Uhlen M, Rhim JS. Pathol Int. 2010;60:497. doi: 10.1111/j.1440-1827.2010.02548.x. [DOI] [PubMed] [Google Scholar]
- 252.Nakamura K, Ito Y, Kawano Y, Kurozumi K, Kobune M, Tsuda H, Bizen A, Honmou O, Niitsu Y, Hamada H. Gene Ther. 2004;11:1155. doi: 10.1038/sj.gt.3302276. [DOI] [PubMed] [Google Scholar]
- 253.Mader EK, Butler G, Dowdy SC, Mariani A, Knutson KL, Federspiel MJ, Russell SJ, Galanis E, Dietz AB, Peng KW. J Transl Med. 2013:11. doi: 10.1186/1479-5876-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Msaouel P, Dispenzieri A, Galanis E. Curr Opin Mol Ther. 2009;11:43. [PMC free article] [PubMed] [Google Scholar]
- 255.Compte M, Cuesta AM, Sanchez-Martin D, Alonso-Camino V, Vicario JL, Sanz L, Alvarez-Vallina L. Stem Cells. 2009;27:753. doi: 10.1634/stemcells.2008-0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Hu YL, Huang B, Zhang TY, Miao PH, Tang GP, Tabata Y, Gao JQ. Mol Pharmaceutics. 2012;9:2698. doi: 10.1021/mp300254s. [DOI] [PubMed] [Google Scholar]
- 257.Wakabayashi T, Natsume A, Hashizume Y, Fujii M, Mizuno M, Yoshida J. J Gene Med. 2008;10:329. doi: 10.1002/jgm.1160. [DOI] [PubMed] [Google Scholar]
- 258.Dembinski JL, Wilson SM, Spaeth EL, Studeny M, Zompetta C, Samudio I, Roby K, Andreeff M, Marini FC. Cytotherapy. 2013;15:20. doi: 10.1016/j.jcyt.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Studeny M, Marini FC, Dembinski JL, Zompetta C, Cabreira-Hansen M, Bekele BN, Champlin RE, Andreeff M. J Natl Cancer Inst. 2004;96:1593. doi: 10.1093/jnci/djh299. [DOI] [PubMed] [Google Scholar]
- 260.Ren C, Kumar S, Chanda D, Kallman L, Chen J, Mountz JD, Ponnazhagan S. Gene Ther. 2008;15:1446. doi: 10.1038/gt.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Ehtesham M, Kabos P, Kabosova A, Neuman T, Black KL, Yu JS. Cancer Res. 2002;62:5657. [PubMed] [Google Scholar]
- 262.Stuckey DW, Shah K. Trends Mol Med. 2013;19:685. doi: 10.1016/j.molmed.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Walczak H, Krammer PH. Exp Cell Res. 2000;256:58. doi: 10.1006/excr.2000.4840. [DOI] [PubMed] [Google Scholar]
- 264.Volkmann X, Fischer U, Bahr MJ, Ott M, Lehner F, MacFarlane M, Cohen GM, Manns MP, Schulze-Osthoff K, Bantel H. Hepatology. 2007;46:1498. doi: 10.1002/hep.21846. [DOI] [PubMed] [Google Scholar]
- 265.Sasportas LS, Kasmieh R, Wakimoto H, Hingtgen S, van de Water JA, Mohapatra G, Figueiredo JL, Martuza RL, Weissleder R, Shah K. Proc Natl Acad Sci USA. 2009;106:4822. doi: 10.1073/pnas.0806647106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.a) Kagawa S, He C, Gu J, Koch P, Rha SJ, Roth JA, Curley SA, Stephens LC, Fang B. Cancer Res. 2001;61:3330. [PubMed] [Google Scholar]; b) Liu L, Eckert MA, Riazifar H, Kang DK, Agalliu D, Zhao W. Stem Cells Int. 2013;2013:7. doi: 10.1155/2013/435093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.van Eekelen M, Sasportas LS, Kasmieh R, Yip S, Figueiredo JL, Louis DN, Weissleder R, Shah K. Oncogene. 2010;29:3185. doi: 10.1038/onc.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.a) Gao P, Ding Q, Wu Z, Jiang HW, Fang ZJ. Cancer Lett. 2010;290:157. doi: 10.1016/j.canlet.2009.08.031. [DOI] [PubMed] [Google Scholar]; b) Ryu CH, Park SH, Park SA, Kim SM, Lim JY, Jeong CH, Yoon WS, Oh WI, Sung YC, Jeun SS. Human Gene Ther. 2011;22:733. doi: 10.1089/hum.2010.187. [DOI] [PubMed] [Google Scholar]; c) Hong X, Miller C, Savant-Bhonsale S, Kalkanis SN. Neurosurgery. 2009;64:1139. doi: 10.1227/01.NEU.0000345646.85472.EA. [DOI] [PubMed] [Google Scholar]
- 269.Xu G, Jiang XD, Xu Y, Zhang J, Huang FH, Chen ZZ, Zhou DX, Shang JH, Zou YX, Cai YQ, Kou SB, Chen YZ, Xu RX, Zeng YJ. Cell Biol Int. 2009;33:466. doi: 10.1016/j.cellbi.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 270.Kanehira M, Xin H, Hoshino K, Maemondo M, Mizuguchi H, Hayakawa T, Matsumoto K, Nakamura T, Nukiwa T, Saijo Y. Cancer Gene Ther. 2007;14:894. doi: 10.1038/sj.cgt.7701079. [DOI] [PubMed] [Google Scholar]
- 271.Kim SK, Cargioli TG, Machluf M, Yang W, Sun Y, Al-Hashem R, Kim SU, Black PM, Carroll RS. Clin Cancer Res. 2005;11:5965. doi: 10.1158/1078-0432.CCR-05-0371. [DOI] [PubMed] [Google Scholar]
- 272.Bello L, Lucini V, Carrabba G, Giussani C, Machluf M, Pluderi M, Nikas D, Zhang J, Tomei G, Villani RM, Carroll RS, Bikfalvi A, Black PM. Cancer Res. 2001;61:8730. [PubMed] [Google Scholar]
- 273.Car BD, Eng VM, Lipman JM, Anderson TD. Toxicol Pathol. 1999;27:58. doi: 10.1177/019262339902700112. [DOI] [PubMed] [Google Scholar]
- 274.Seo SH, Kim KS, Park SH, Suh YS, Kim SJ, Jeun SS, Sung YC. Gene Ther. 2011;18:488. doi: 10.1038/gt.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Hamstra DA, Rehemtulla A. Human Gene Ther. 1999;10:235. doi: 10.1089/10430349950019020. [DOI] [PubMed] [Google Scholar]
- 276.Xu G, McLeod HL. Clin Cancer Res. 2001;7:3314. [PubMed] [Google Scholar]
- 277.Friedlos F, Court S, Ford M, Denny WA, Springer C. Gene Ther. 1998;5:105. doi: 10.1038/sj.gt.3300569. [DOI] [PubMed] [Google Scholar]
- 278.Robert H, Bahuaud J, Kerdiles N, Passuti N, Capelli M, Pujol JP, Hartman D, Locker B, Hulet C, Hardy P, Coudane H, Rochverger A, Francheschi JP, et al. Societe Francaise Arthroscopie. Rev Chir Orthop Rep Appar Mot. 2007;93:701. doi: 10.1016/s0035-1040(07)73255-5. [DOI] [PubMed] [Google Scholar]
- 279.Kosaka H, Ichikawa T, Kurozumi K, Kambara H, Inoue S, Maruo T, Nakamura K, Hamada H, Date I. Cancer Gene Therapy. 2012;19:572. doi: 10.1038/cgt.2012.35. [DOI] [PubMed] [Google Scholar]
- 280.Zhao Y, Lam DH, Yang J, Lin J, Tham CK, Ng WH, Wang S. Gene Ther. 2012;19:189. doi: 10.1038/gt.2011.82. [DOI] [PubMed] [Google Scholar]
- 281.Altaner C, Altanerova V, Cihova M, Ondicova K, Rychly B, Baciak L, Mravec B. Int J Cancer. 2014;134:1458. doi: 10.1002/ijc.28455. [DOI] [PubMed] [Google Scholar]
- 282.Aboody KS, Najbauer J, Metz MZ, D’Apuzzo M, Gutova M, Annala AJ, Synold TW, Couture LA, Blanchard S, Moats RA, Garcia E, Aramburo S, Valenzuela VV, Frank RT, Barish ME, Brown CE, Kim SU, Badie B. J Portnow, Sci Transl Med. 2013;5:184ra59. doi: 10.1126/scitranslmed.3005365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Kim SK, Kim SU, Park IH, Bang JH, Aboody KS, Wang KC, Cho BK, Kim M, Menon LG, Black PM, Carroll RS. Clin Cancer Res. 2006;12:5550. doi: 10.1158/1078-0432.CCR-05-2508. [DOI] [PubMed] [Google Scholar]
- 284.a) Martinez-Quintanilla J, Bhere D, Heidari P, He D, Mahmood U, Shah K. Stem Cells. 2013;31:1706. doi: 10.1002/stem.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ryu CH, Park KY, Kim SM, Jeong CH, Woo JS, Hou Y, Jeun SS. Biochem Biophys Res Commun. 2012;421:585. doi: 10.1016/j.bbrc.2012.04.050. [DOI] [PubMed] [Google Scholar]; c) Lee WY, Zhang T, Lau CP, Wang CC, Chan KM, Li G. Cytotherapy. 2013;15:1484. doi: 10.1016/j.jcyt.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 285.Yang J, Lam DH, Goh SS, Lee EX, Zhao Y, Tay FC, Chen C, Du SH, Balasundaram G, Shahbazi M, Tham CK, Ng WH, Toh HC, Wang S. Stem Cells. 2012;30:1021. doi: 10.1002/stem.1051. [DOI] [PubMed] [Google Scholar]
- 286.a) Hong SH, Lee HJ, An J, Lim I, Borlongan C, Aboody KS, Kim SU. Cancer Gene Ther. 2013;20:678. doi: 10.1038/cgt.2013.69. [DOI] [PubMed] [Google Scholar]; b) Gutova M, Shackleford GM, Khankaldyyan V, Herrmann KA, Shi XH, Mittelholtz K, Abramyants Y, Blanchard MS, Kim SU, Annala AJ, Najbauer J, Synold TW, D’Apuzzo M, Barish ME, Moats RA, Aboody KS. Gene Ther. 2013;20:143. doi: 10.1038/gt.2012.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Kim KY, Kim SU, Leung PCK, Jeung EB, Choi KC. Cancer Sci. 2010;101:955. doi: 10.1111/j.1349-7006.2009.01485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Bertrand N, Wu J, Xu XY, Kamaly N, Farokhzad OC. Adv Drug Delivery Rev. 2014;66:2. doi: 10.1016/j.addr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.a) Roger M, Clavreul A, Venier-Julienne MC, Passirani C, Sindji L, Schiller P, Montero-Menei C, Menei P. Biomaterials. 2010;31:8393. doi: 10.1016/j.biomaterials.2010.07.048. [DOI] [PubMed] [Google Scholar]; b) Auffinger B, Morshed R, Tobias A, Cheng Y, Ahmed AU, Lesniak MS. Oncotarget. 2013;4:378. doi: 10.18632/oncotarget.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Huang XL, Zhang F, Wang H, Niu G, Choi KY, Swierczewska M, Zhang GF, Gao HK, Wang Z, Zhu L, Choi HS, Lee S, Chen XY. Biomaterials. 2013;34:1772. doi: 10.1016/j.biomaterials.2012.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Loebinger MR, Kyrtatos PG, Turmaine M, Price AN, Pankhurst Q, Lythgoe MF, Janes SM. Cancer Res. 2009;69:8862. doi: 10.1158/0008-5472.CAN-09-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Gao Y, Wang Y, Fu A, Shi W, Yeo D, Luo KQ, Ow H, Xu C. J Mater Chem B. 2015 doi: 10.1039/c4tb01452a. [DOI] [PubMed] [Google Scholar]
- 293.Li L, Guan Y, Liu H, Hao N, Liu T, Meng X, Fu C, Li Y, Qu Q, Zhang Y, Ji S, Chen L, Chen D, Tang F. ACS Nano. 2011;5:7462. doi: 10.1021/nn202399w. [DOI] [PubMed] [Google Scholar]
- 294.Ruan J, Ji J, Song H, Qian Q, Wang K, Wang C, Cui D. Nanoscale Res Lett. 2012;7:309. doi: 10.1186/1556-276X-7-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.He R, You XG, Shao J, Gao F, Pan BF, Cui DX. Nanotechnology. 2007:18. doi: 10.1088/0957-4484/18/3/035701. [DOI] [PubMed] [Google Scholar]
- 296.Schnarr K, Mooney R, Weng YM, Zhao DH, Garcia E, Armstrong B, Annala AJ, Kim SU, Aboody KS, Berlin JM. Adv Healthcare Mater. 2013;2:976. doi: 10.1002/adhm.201300003. [DOI] [PubMed] [Google Scholar]
- 297.Emery AEH. Lancet. 2002;359:687. doi: 10.1016/S0140-6736(02)07815-7. [DOI] [PubMed] [Google Scholar]
- 298.Darabi R, Gehlbach K, Bachoo RM, Kamath S, Osawa M, Kamm KE, Kyba M, Perlingeiro RCR. Nat Med. 2008;14:134. doi: 10.1038/nm1705. [DOI] [PubMed] [Google Scholar]
- 299.Maroto M, Reshef R, Munsterberg AE, Koester S, Goulding M, Lassar AB. Dev Biol. 1997;186:B60. doi: 10.1016/s0092-8674(00)80190-7. [DOI] [PubMed] [Google Scholar]
- 300.Goncalves MAFV, de Vries AAF, Holkers M, van de Watering MJM, van der Velde I, van Nierop GP, Valerio D, Knaan-Shanzer S. Human Mol Genet. 2006;15:213. doi: 10.1093/hmg/ddi438. [DOI] [PubMed] [Google Scholar]
- 301.Kazuki Y, Hiratsuka M, Takiguchi M, Osaki M, Kajitani N, Hoshiya H, Hiramatsu K, Yoshino T, Kazuki K, Ishihara C, Takehara S, Higaki K, Nakagawa M, Takahashi K, Yamanaka S, Oshimura M. Mol Ther. 2010;18:386. doi: 10.1038/mt.2009.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Hoshiya H, Kazuki Y, Abe S, Takiguchi M, Kajitani N, Watanabe Y, Yoshino T, Shirayoshi Y, Higaki K, Messina G, Cossu G, Oshimura M. Mol Ther. 2009;17:309. doi: 10.1038/mt.2008.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Tomizuka K, Yoshida H, Uejima H, Kugoh H, Sato K, Ohguma A, Hayasaka M, Hanaoka K, Oshimura M, Ishida I. Nat Genet. 1997;16:133. doi: 10.1038/ng0697-133. [DOI] [PubMed] [Google Scholar]
- 304.Tedesco FS, Hoshiya H, D’Antona G, Gerli MFM, Messina G, Antonini S, Tonlorenzi R, Benedetti S, Berghella L, Torrente Y, Kazuki Y, Bottinelli R, Oshimura M, Cossu G. Sci Transl Med. 2011:3. doi: 10.1126/scitranslmed.3002342. [DOI] [PubMed] [Google Scholar]
- 305.Gregorevic P, Chamberlain JS. Expert Opin Biol Ther. 2003;3:803. doi: 10.1517/14712598.3.5.803. [DOI] [PubMed] [Google Scholar]
- 306.Benchaouir R, Meregalli M, Farini A, D’Antona G, Belicchi M, Goyenvalle A, Battistelli M, Bresolin N, Bottinelli R, Garcia L, Torrente Y. Cell Stem Cell. 2007;1:646. doi: 10.1016/j.stem.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 307.Catucci M, Castiello MC, Pala F, Bosticardo M, Villa A. Frontiers Immunol. 2012;3:209. doi: 10.3389/fimmu.2012.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Notarangelo LD, Miao CH, Ochs HD. Curr Opin Hematol. 2008;15:30. doi: 10.1097/MOH.0b013e3282f30448. [DOI] [PubMed] [Google Scholar]
- 309.Bosticardo M, Marangoni F, Aiuti A, Villa A, Grazia Roncarolo M. Blood. 2009;113:6288. doi: 10.1182/blood-2008-12-115253. [DOI] [PubMed] [Google Scholar]
- 310.Ochs HD, Filipovich AH, Veys P, Cowan MJ, Kapoor N. Biol Blood Marrow Transpl. 2009;15:84. doi: 10.1016/j.bbmt.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 311.Snapper SB, Meelu P, Nguyen D, Stockton BM, Bozza P, Alt FW, Rosen FS, von Andrian UH, Klein C. J Leukocyte Biol. 2005;77:993. doi: 10.1189/jlb.0804444. [DOI] [PubMed] [Google Scholar]
- 312.a) Gismondi A, Cifaldi L, Mazza C, Giliani S, Parolini S, Morrone S, Jacobelli J, Bandiera E, Notarangelo L, Santoni A. Blood. 2004;104:436. doi: 10.1182/blood-2003-07-2621. [DOI] [PubMed] [Google Scholar]; b) Zhang H, Schaff UY, Green CE, Chen H, Sarantos MR, Hu YM, Wara D, Simon SI, Lowell CA. Immunity. 2006;25:285. doi: 10.1016/j.immuni.2006.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Bouma G, Burns S, Thrasher AJ. Blood. 2007;110:4278. doi: 10.1182/blood-2007-06-096875. [DOI] [PubMed] [Google Scholar]
- 314.Westerberg L, Larsson M, Hardy SJ, Fernandez C, Thrasher AJ, Severinson E. Blood. 2005;105:1144. doi: 10.1182/blood-2004-03-1003. [DOI] [PubMed] [Google Scholar]
- 315.Massaad MJ, Ramesh N, Geha RS. Ann NY Acad Sci. 2013;1285:26. doi: 10.1111/nyas.12049. [DOI] [PubMed] [Google Scholar]
- 316.Pai SY, DeMartiis D, Forino C, Cavagnini S, Lanfranchi A, Giliani S, Moratto D, Mazza C, Porta F, Imberti L, Notarangelo LD, Mazzolari E. Bone Marrow Transpl. 2006;38:671. doi: 10.1038/sj.bmt.1705512. [DOI] [PubMed] [Google Scholar]
- 317.Ozsahin H, Cavazzana-Calvo M, Notarangelo LD, Schulz A, Thrasher AJ, Mazzolari E, Slatter MA, Le Deist F, Blanche S, Veys P, Fasth A, Bredius R, Sedlacek P, Wulffraat N, Ortega J, Heilmann C, O’Meara A, Wachowiak J, Kalwak K, Matthes-Martin S, Gungor T, Ikinciogullari A, Landais P, Cant AJ, Friedrich W, Fischer A. Blood. 2008;111:439. doi: 10.1182/blood-2007-03-076679. [DOI] [PubMed] [Google Scholar]
- 318.Kobayashi R, Ariga T, Nonoyama S, Kanegane H, Tsuchiya S, Morio T, Yabe H, Nagatoshi Y, Kawa K, Tabuchi K, Tsuchida M, Miyawaki T, Kato S. Br J Haematol. 2006;135:362. doi: 10.1111/j.1365-2141.2006.06297.x. [DOI] [PubMed] [Google Scholar]
- 319.a) Boztug K, Dewey RA, Klein C. Curr Opin Mol Ther. 2006;8:390. [PubMed] [Google Scholar]; b) Westerberg LS, de la Fuente MA, Wermeling F, Ochs HD, Karlsson MCI, Snapper SB, Notarangelo LD. Blood. 2008;112:4139. doi: 10.1182/blood-2008-02-140715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Marangoni F, Bosticardo M, Charrier S, Draghici E, Locci M, Scaramuzza S, Panaroni C, Ponzoni M, Sanvito F, Doglioni C, Liabeuf M, Gjata B, Montus M, Siminovitch K, Aiuti A, Naldini L, Dupre L, Roncarolo MG, Galy A, Villa A. Mol Ther. 2009;17:1300. doi: 10.1038/mt.2009.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Dewey RA, Diez IA, Ballmaier M, Filipovich A, Greil J, Gungor T, Happel C, Maschan A, Noyan F, Pannicke U, Schwarz K, Snapper S, Welte K, Klein C. Exp Hematol. 2006;34:1162. doi: 10.1016/j.exphem.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 322.Boztug K, Schmidt M, Schwarzer A, Banerjee PP, Diez IA, Dewey RA, Bohm M, Nowrouzi A, Ball CR, Glimm H, Naundorf S, Kuhlcke K, Blasczyk R, Kondratenko I, Marodi L, Orange JS, von Kalle C, Klein C. N Engl J Med. 2010;363:1918. doi: 10.1056/NEJMoa1003548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Braun CJ, Bozlug K, Paruzynski A, Witzel M, Schwarzer A, Rothe M, Modlich U, Beier R, Gohring G, Steinemann D, Fronza R, Ball CR, Haemmerle R, Naundorf S, Kuhlcke K, Rose M, Fraser C, Mathias L, Ferrari R, Abboud MR, Al-Herz W, Kondratenko I, Marodi L, Glimm H, Schlegelberger B, Schambach A, Albert MH, Schmidt M, von Kalle C, Klein C. Sci Transl Med. 2014:6. doi: 10.1126/scitranslmed.3007280. [DOI] [PubMed] [Google Scholar]
- 324.a) Aiuti A, Biasco L, Scaramuzza S, Ferrua F, Cicalese MP, Baricordi C, Dionisio F, Calabria A, Giannelli S, Castiello MC, Bosticardo M, Evangelio C, Assanelli A, Casiraghi M, Di Nunzio S, Callegaro L, Benati C, Rizzardi P, Pellin D, Di Serio C, Schmidt M, Von Kalle C, Gardner J, Mehta N, Neduva V, Dow DJ, Galy A, Miniero R, Finocchi A, Metin A, Banerjee PP, Orange JS, Galimberti S, Valsecchi MG, Biffi A, Montini E, Villa A, Ciceri F, Roncarolo MG, Naldini L. Science. 2013;341:865. [Google Scholar]; b) Dupre L, Trifari S, Follenzi A, Marangoni F, de Lera TL, Bernad A, Martino S, Tsuchiya S, Bordignon C, Naldini L, Aiuti A, Roncarolo MG. Mol Ther. 2004;10:903. doi: 10.1016/j.ymthe.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 325.Montini E, Cesana D, Schmidt M, Sanvito F, Bartholomae CC, Ranzani M, Benedicenti F, Sergi LS, Ambrosi A, Ponzoni M, Doglioni C, Di Serio C, von Kalle C, Naldini L. J Clin Invest. 2009;119:964. doi: 10.1172/JCI37630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Scaramuzza S, Biasco L, Ripamonti A, Castiello MC, Loperfido M, Draghici E, Hernandez RJ, Benedicenti F, Radrizzani M, Salomoni M, Ranzani M, Bartholomae CC, Vicenzi E, Finocchi A, Bredius R, Bosticardo M, Schmidt M, von Kalle C, Montini E, Biffi A, Roncarolo MG, Naldini L, Villa A, Aiuti A. Mol Ther. 2013;21:175. doi: 10.1038/mt.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Cheon JE, Kim IO, Hwang YS, Kim KJ, Wang KC, Cho BK, Chi JG, Kim CJ, Kim WS, Yeon KM. Radiographics. 2002;22:461. doi: 10.1148/radiographics.22.3.g02ma01461. [DOI] [PubMed] [Google Scholar]
- 328.Kohlschutter A, Eichler F. Exp Rev Neurother. 2011;11:1485. doi: 10.1586/ern.11.135. [DOI] [PubMed] [Google Scholar]
- 329.Biffi A, Lucchini G, Rovelli A, Sessa M. Bone Marrow Transplant. 2008;42(Suppl 2):S2. doi: 10.1038/bmt.2008.275. [DOI] [PubMed] [Google Scholar]
- 330.Peters C, Steward CG. Int Bone Marrow Transplant. 2003;31:229. doi: 10.1038/sj.bmt.1703839. [DOI] [PubMed] [Google Scholar]
- 331.Biffi A, De Palma M, Quattrini A, Del Carro U, Amadio S, Visigalli I, Sessa M, Fasano S, Brambilla R, Marchesini S, Bordignon C, Naldini L. J Clin Invest. 2004;113:1118. doi: 10.1172/JCI19205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Biffi A, Capotondo A, Fasano S, del Carro U, Marchesini S, Azuma H, Malaguti MC, Amadio S, Brambilla R, Grompe M, Bordignon C, Quattrini A, Naldini L. J Clin Invest. 2006;116:3070. doi: 10.1172/JCI28873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Hess B, Saftig P, Hartmann D, Coenen R, Lullmann-Rauch R, Goebel HH, Evers M, von Figura K, D’Hooge R, Nagels G, De Deyn P, Peters C, Gieselmann V. Proc Natl Acad Sci USA. 1996;93:14821. doi: 10.1073/pnas.93.25.14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Biffi A, Montini E, Lorioli L, Cesani M, Fumagalli F, Plati T, Baldoli C, Martino S, Calabria A, Canale S, Benedicenti F, Vallanti G, Biasco L, Leo S, Kabbara N, Zanetti G, Rizzo WB, Mehta NA, Cicalese MP, Casiraghi M, Boelens JJ, Del Carro U, Dow DJ, Schmidt M, Assanelli A, Neduva V, Di Serio C, Stupka E, Gardner J, von Kalle C, Bordignon C, Ciceri F, Rovelli A, Roncarolo MG, Aiuti A, Sessa M, Naldini L. Science. 2013;341:1233158. doi: 10.1126/science.1233158. [DOI] [PubMed] [Google Scholar]
- 335.Capotondo A, Cesani M, Pepe S, Fasano S, Gregori S, Tononi L, Venneri MA, Brambilla R, Quattrini A, Ballabio A, Cosma MP, Naldini L, Biffi A. Human Gene Ther. 2007;18:821. doi: 10.1089/hum.2007.048. [DOI] [PubMed] [Google Scholar]
- 336.Moser HW, Mahmood A, Raymond GV. Nat Clin Pract Neurol. 2007;3:140. doi: 10.1038/ncpneuro0421. [DOI] [PubMed] [Google Scholar]
- 337.Shapiro E, Krivit W, Lockman L, Jambaque I, Peters C, Cowan M, Harris R, Blanche S, Bordigoni P, Loes D, Ziegler R, Crittenden M, Ris D, Berg B, Cox C, Moser H, Fischer A, Aubourg P. Lancet. 2000;356:713. doi: 10.1016/S0140-6736(00)02629-5. [DOI] [PubMed] [Google Scholar]
- 338.Benhamida S, Pflumio F, Dubart-Kupperschmitt A, Zhao-Emonet JC, Cavazzana-Calvo M, Rocchiccioli F, Fichelson S, Aubourg P, Charneau P, Cartier N. Mol Ther. 2003;7:317. doi: 10.1016/s1525-0016(03)00002-9. [DOI] [PubMed] [Google Scholar]
- 339.Asheuer M, Pflumio FO, Benhamida S, Dubart-Kupperschmitt A, Fouquet F, Imai Y, Aubourg P, Cartier N. Proc Natl Acad Sci USA. 2004;101:3557. doi: 10.1073/pnas.0306431101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Cartier N, Hacein-Bey-Abina S, Bartholomae CC, Veres G, Schmidt M, Kutschera I, Vidaud M, Abel U, Dal-Cortivo L, Caccavelli L, Mahlaoui N, Kiermer V, Mittelstaedt D, Bellesme C, Lahlou N, Lefrere F, Blanche S, Audit M, Payen E, Leboulch P, l’Homme B, Bougneres P, Von Kalle C, Fischer A, Cavazzana-Calvo M, Aubourg P. Science. 2009;326:818. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- 341.Cartier N, Hacein-Bey-Abina S, Bartholomae CC, Bougneres P, Schmidt M, Von Kalle C, Fischer A, Cavazzana-Calvo M, Aubourg P. Method Enzymol. 2012;507:187. doi: 10.1016/B978-0-12-386509-0.00010-7. [DOI] [PubMed] [Google Scholar]
- 342.Bianco P, Cao X, Frenette PS, Mao JJ, Robey PG, Simmons PJ, Wang CY. Nat Med. 2013;19:35. doi: 10.1038/nm.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Sander JD, Joung JK. Nat Biotechnol. 2014;32:347. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Xue W, Chen S, Yin H, Tammela T, Papagiannakopoulos T, Joshi NS, Cai W, Yang G, Bronson R, Crowley DG, Zhang F, Anderson DG, Sharp PA, Jacks T. Nature. 2014;514:380. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Long C, McAnally JR, Shelton JM, Mireault AA, Bassel-Duby R, Olson EN. Science. 2014;345:1184. doi: 10.1126/science.1254445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Kim DS, Kim JH, Lee JK, Choi SJ, Kim JS, Jeun SS, Oh W, Yang YS, Chang JW. Stem Cells Dev. 2009;18:511. doi: 10.1089/scd.2008.0050. [DOI] [PubMed] [Google Scholar]
- 347.Bellin M, Marchetto MC, Gage FH, Mummery CL. Nature reviews Mol Cell Biol. 2012;13:713. doi: 10.1038/nrm3448. [DOI] [PubMed] [Google Scholar]